Abstract
Existing traditional Chinese medicine (TCM)-related databases are still insufficient in data standardization, integrity and precision, and need to be updated urgently. Herein, an Encyclopedia of Traditional Chinese Medicine version 2.0 (ETCM v2.0, http://www.tcmip.cn/ETCM2/front/#/) was constructed as the latest curated database hosting 48,442 TCM formulas recorded by ancient Chinese medical books, 9872 Chinese patent drugs, 2079 Chinese medicinal materials and 38,298 ingredients. To facilitate the mechanistic research and new drug discovery, we improved the target identification method based on a two-dimensional ligand similarity search module, which provides the confirmed and/or potential targets of each ingredient, as well as their binding activities. Importantly, five TCM formulas/Chinese patent drugs/herbs/ingredients with the highest Jaccard similarity scores to the submitted drugs are offered in ETCM v2.0, which may be of significance to identify prescriptions/herbs/ingredients with similar clinical efficacy, to summarize the rules of prescription use, and to find alternative drugs for endangered Chinese medicinal materials. Moreover, ETCM v2.0 provides an enhanced JavaScript-based network visualization tool for creating, modifying and exploring multi-scale biological networks. ETCM v2.0 may be a major data warehouse for the quality marker identification of TCMs, the TCM-derived drug discovery and repurposing, and the pharmacological mechanism investigation of TCMs against various human diseases.
Key words: Traditional Chinese medicine, Database, Target identification, Network visualization, New drug research and development, Molecular mechanism, Quality marker, Drug repurposing
Graphical abstract
An updated ETCM v2.0 database may be a major data warehouse for quality marker identification and pharmacological mechanism investigation of TCMs against human diseases, and TCM-derived drug discovery and repurposing.
1. Introduction
Traditional Chinese medicine (TCM) has been extensively used in the prevention and treatment of various human diseases, and also offers a great resource for modern drug discovery and development. Many U.S. Food and Drug Administration (FDA)-approved drugs, such as artemisinin from Herba Artemisiae Annuae as a first-line drug for malaria1,2 and ephedrine form Herba Ephedrae as an anti-asthmatic drug3, were derived from TCM. Notably, the clinical efficacy and safety of various TCM formulas, such as Huashi Baidu Granules, Xuanfei Baidu Granules, etc., for the treatment of COVID-19 have also been verified by accumulating high-quality clinical trials.4, 5, 6, 7 Recently, big data and artificial intelligence technologies have driven the rapid development of TCM.8 To better dissect the pharmacological mechanisms and identify the active ingredients in TCM, we originally established an Encyclopaedia of Traditional Chinese Medicine (ETCM) in 20199, which has been highly recognized among pharmacologists and scholars in TCM researches worldwide. Since then, several TCM-related databases, such as HERB10, SuperTCM11 and HIT 2.012, have been constructed and provided useful data and tools for TCM-based research and drug discovery. However, they are still insufficient in data standardization, integrity and precision. None of the existing databases provide the quality control information of Chinese patent drugs and herbs which may be one of the most important factors for the effectiveness of TCM, and the similarity evaluation data among various TCM formulas/Chinese patent drugs/herbs/ingredients which may be useful in summarizing the rules of prescription use and finding alternative drugs for endangered Chinese medicinal materials. Therefore, a database with more comprehensive and accurate recording related data and information is in demand.
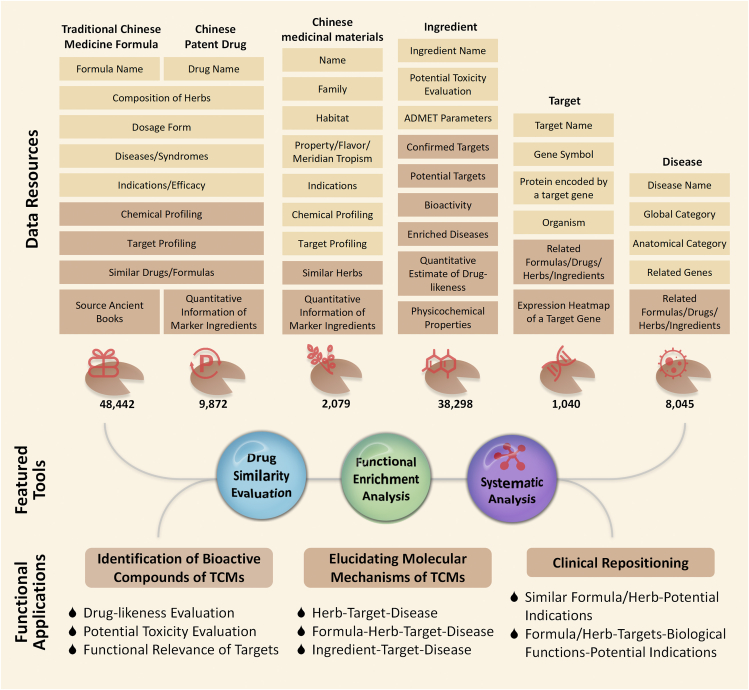
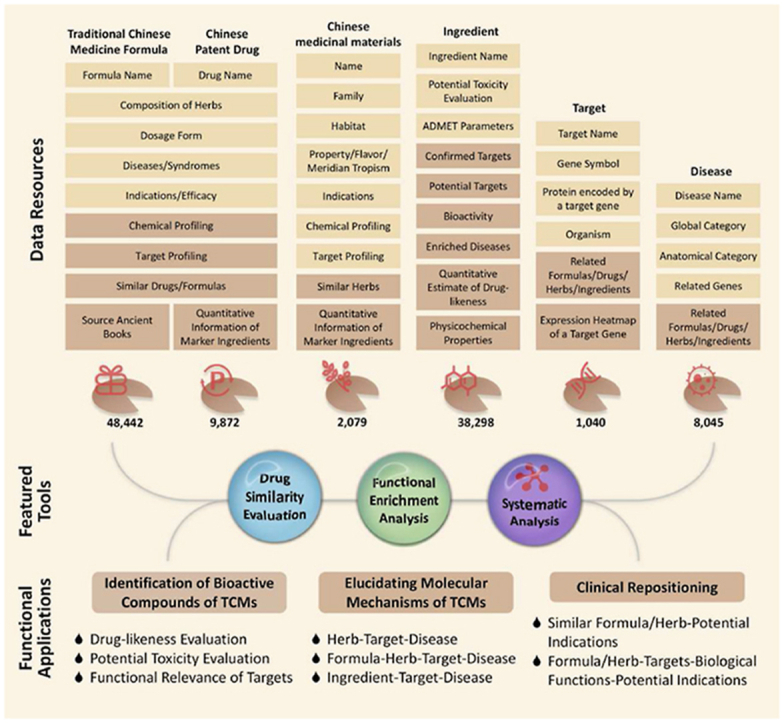
In the updated ETCM v2.0 (freely accessible at http://www.tcmip.cn/ETCM2/front/#/) as shown in Fig. 1, the original data is largely expanded while a new data field “Traditional Chinese Medicine Formulas” based on ancient Chinese medical books has been added, and a novel method for target identification and prediction has been utilized. Currently, ETCM v2.0 hosts 48,442 TCM formulas recorded by ancient Chinese medical books, 9872 Chinese patent drugs, 2079 Chinese medicinal materials and 38,298 ingredients, 1040 confirmed or potential drug targets, as well as 8045 related diseases. Moreover, ETCM v2.0 also provides various pairwise associations and cross retrievals between different data sections by combing diverse strategies including database mining, reference mining and bioinformatics mining. This new release provides more comprehensive and more efficient TCM information with downloadable and visualized knowledge graphs, which may facilitate the deep mining of implicit knowledge on TCM, and the systematic integration of TCM with modern medicine at both molecular and phenotypic levels (Fig. 1).
Figure 1.
The schema and objectives of ETCM v2.0. This database includes 48,442 TCM formulas recorded by ancient Chinese medical books, 9872 Chinese patent drugs, 2079 Chinese medicinal materials and 38,298 ingredients. Functionally, ETCM v2.0 may be a major data warehouse for the quality marker identification of TCMs, the TCM-derived drug discovery and repurposing, and the pharmacological mechanism investigation of TCMs against various human diseases.
2. Data updates and extensions
ETCM v2.0 is composed of six data sections, including TCM formulas recorded by ancient Chinese medical books, Chinese patent drugs, Chinese medicinal materials, ingredients, targets and diseases as summarized in Table 1.
Table 1.
Content statistics of ETCM v2.0.
| Data type | Number | Resources |
|---|---|---|
| TCM Formulas | 48,442 | Traditional Chinese Medicine Formulas |
| Chinese Patent Drugs | 9872 | China Food and Drug Administration (http://eng.sfda.gov.cn/WS03/CL0755/) |
| Chinese Patent Drugs with quantitative information of marker ingredients | 865 | Pharmacopoeia of the People's Republic of China (2020 version) |
| Chinese medicinal materials | 2079 | |
| Herbs | 2005 | The Fourth National Survey on Chinese Materia Medica Resources (http://www.zyzypc.com.cn/), Pharmacopoeia of the People's Republic of China (2020 version), Authoritative Chinese Medical Books and Dictionaries |
| Animal medicine | 44 | Pharmacopoeia of the People's Republic of China (2020 version) |
| Mineral medicine | 30 | Pharmacopoeia of the People's Republic of China (2020 version) |
| Chinese medicinal materials with quantitative information of marker ingredients | 504 | Pharmacopoeia of the People's Republic of China (2020 version) |
| Herbs with referenced targets | 990 | Identified by D3CARP platform based on BindingDB database (updated on 2021-11-01) |
| Ingredients | 38,298 | Pharmacopoeia of the People's Republic of China (2020 version) and other literatures |
| Ingredients with referenced targets | 25,647 | Identified by D3CARP platform based on BindingDB database (updated on 2021-11-01) |
| Ingredients with drug-likeness evaluation | 38,265 | A quantitative estimate model of drug-likeness reported by Bickerton group [Nat Chem. 2012 Jan 24; 4 (2):90–8]14 |
| Drug target genes | 1040 | Identified by D3CARP platform based on BindingDB database (updated on 2021-11-01) |
| Targets with references | 1040 | |
| Diseases | 8045 | MalaCards v5.0, Human Phenotype Ontology (HPO, Released in 2018), Online Mendelian Inheritance in Man (OMIM, Released in April 2018), Database of gene-disease associations (DisGeNET v5.0), the portal for rare diseases and orphan drugs (ORPHANET v5.49.0) |
| Diseases with relevant genes | 8038 |
2.1. The TCM formula and Chinese patent drug information
TCM formulas were originated from the clinical experience of TCM during the thousands of years. A large number of ancient TCM formulas were recorded in ancient books by different doctors in different dynasties, which may be the source of research and development of new TCM-based drugs. At present, some TCM formulas are still widely used with definite curative effects, obvious characteristics and advantages. To this end, the Chinese governments have promulgated the “first batch of ancient classic famous prescriptions catalogue” (including 100 TCM formulas) and promoted their listing as proprietary TCM formulas. Especially, some of them have been developed into Chinese patent drugs. Since 1985, more than 10,000 Chinese patent drugs have been approved for marketing, and have become the mainstream product in TCM market and one of the three pillar industries of Chinese pharmaceutical industry along with biological drugs and chemical drugs. In this context, a completely new data section was integrated to ETCM v2.0, namely “Traditional Chinese Medicine Formulas”. The most comprehensive list of 48,442 TCM formulas (including the first batch of 100 proprietary TCM formulas) and 9872 Chinese patent drugs with manually integrating and normalizing information respectively obtained from ancient Chinese medical books and package insert of drugs are provided. Notably, the TCM formula/Chinese patent drug-related clinical data, such as the corresponding symptoms and syndromes, and clinical efficacy were standardized to obtain accurate query results. Both the chemical profiling and target profiling of each TCM formula/Chinese patent drug, as well as the quantitative information of marker ingredients, the related properties and functional information are also provided.
2.2. The Chinese medicinal materials information
The number of herbs has increased from 403 (ETCM) to 2005 (ETCM v2.0), which includes 548 commonly used herbs recorded in the Chinese Pharmacopoeia (2020 version) and investigated in the Fourth National Survey on Chinese Materia Medica Resources (http://www.zyzypc.com.cn/). In addition, 44 animal medicines and 30 mineral medicines are also included. The general and drug effectiveness-related information, including the name, type, species, collection time, property, flavor, meridian tropism, indication, specification, medicinal part, classification based on efficacy, references and habitat map of Chinese medicinal materials, are provided. Of note, the quantitative information of marker ingredients of each herb is provided according to the Chinese pharmacopoeia (2020 version), which are necessary for herbal quality control. Moreover, the cross-links of herbs to HERB, HIT2.0 and SymMap databases are also established in herbal detailed information page.
2.3. The ingredient information
The ingredients of herbs, TCM formulas and Chinese patent drugs recorded in ETCM v2.0 were collected manually according to the Pharmacopoeia of the People's Republic of China (2020 version) and other literatures. The number of ingredients has increased from 7274 (ETCM) to 38,298 (ETCM v2.0). For each ingredient, its name, 18 physicochemical properties [molecular formula, molecular weight, volume, density, number of hydrogen bond acceptors (nHA), number of hydrogen bond donors (nHD), number of rotatable bonds (nRot), number of rings (nRing), number of atoms in the biggest ring (MaxRing), number of heteroatoms (nHet), formal charge (fChar), number of rigid bonds (nRig), flexibility, number of stereocenters, topological polar surface area (TPSA), the logarithm of aqueous solubility value (logS), the logarithm of the n-octanol/water distribution coefficient (logP), the logarithm of the n-octanol/water distribution coefficients at pH = 7.4 (logD7.4)], 6 medicinal chemistry properties [quantitative estimate of drug-likeness (QED), synthetic accessibility score (SAscore), natural product-likeness score (NPscore), Lipinski Rule, GSK Rule, Golden Triangle], 22 ADME (absorption, distribution, metabolism, excretion) properties [Caco-2 permeability, MDCK permeability, Pgp-inhibitor, Pgp-substrate, human intestinal absorption (HIA), 20% oral bioavailability (F20%), plasma protein binding (PPB), volume distribution (VD), penetration blood brain barrier penetration (BBB), the fraction unbound in plasms (Fu), CYP 1A2 inhibitor, CYP 2C19 inhibitor, CYP 2C9 inhibitor, CYP 2D6 inhibitor, CYP 3A4 inhibitor, CYP 1A2 substrate, CYP 2C19 substrate, CYP 2C9 substrate, CYP 2D6 substrate, CYP 3A4 substrate, the clearance of a drug (CL), T1/2], 32 toxicity endpoints [FAF-drugs 4 rule, NonBiodegradable rule, Pfizer rule, SR-MMP, SureChEMBL rule, FDA maximum (recommended) daily dose (FDAMDD), IGC50, LC50FM, LC50DM, NR-AR, NR-AR-LBD, NR-Aromatase, NR-ER, NR-ER-LBD, skin sensitization rule, skin sensitization, acute toxicity rule, rat oral acute toxicity, hERG blockers, the human hepatotoxicity (H-HT), drug-induced liver injury (DILI), eye corrosion/irritation, respiratory toxicity, AMES toxicity, genotoxic carcinogenicity rule, nongenotoxic carcinogenicity rule, carcinogenicity, antioxidant response element (SR-ARE), ATPase family AAA domain-containing protein 5 (SR-ATAD5), SR-p53] calculated by the robust and accurate models in ADMETlab 2.013 (https://admetmesh.scbdd.com/) are provided. Especially, the drug-likeness based on desirability (QED)14, 15, 16 and the maximum recommended daily dose by FDA (FDAMDD)17 of herbal ingredients are provided to evaluate the potentials and the safety as drug candidates, respectively. Among them, the mean QED is 0.67 for the attractive compounds and 0.49 for the unattractive compounds. Thus, we classified all 38,298 ingredients collected in ETCM v2.0 into three groups according to their QED scores, excellent (QED>0.67), medium (0.49 ≤ QED ≤ 0.67) and poor (QED<0.49). FDAMDD provides an estimate of the toxic dose threshold of chemicals in humans. The output value of FDAMDD is the probability of being toxic, within the range of 0–1. Our ingredients are divided into excellent (0 ≤ FDAMDD ≤ 0.3), medium (0.3<FDAMDD ≤ 0.7) and poor (FDAMDD>0.7).
2.4. The target information
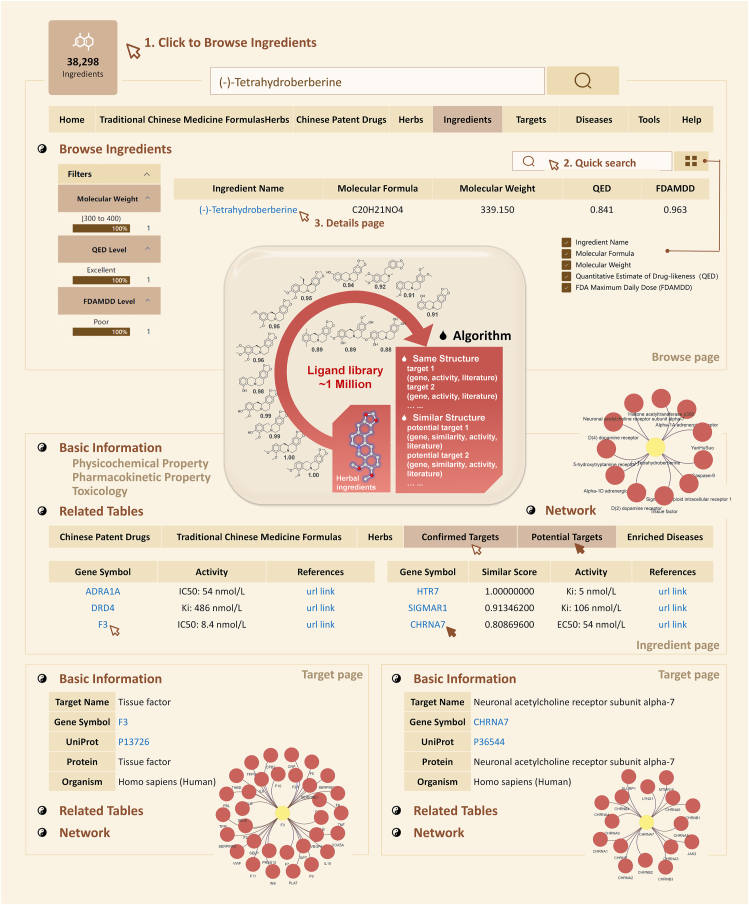
ETCM v2.0 provides the detailed information on the confirmed and/or potential targets for ingredients, herbs, Chinese patent drugs and TCM formulas to facilitate the mechanistic study of TCM and the new drug discovery. We improved the target identification method in ETCM v2.0 based on a two-dimensional ligand similarity search module in our indoor D3CARP platform (the article of D3CARP will be submitted and published in the near future) and the BindingDB database18 (updated on 2021-11-01) (Fig. 2). Specifically, the platform ligand database is derived from the BindingDB database, which is a public, web-accessible database of measured binding affinities and contains approximately one million small molecules annotated with more than 2.5 million binding data and related literature information18. If the calculated similarity value is 1.0, the targets of the reference compound are considered as the confirmed targets for the submitted ingredient. If the calculated similarity value is between 0.8 and 1.0, the targets of the reference compound are considered as potential targets of the submitted ingredient. Notably, the binding activities between each ingredient and its targets are also provided to facilitate the identification of bioactive compounds and the investigation of pharmacological mechanisms of TCMs. Comparing with the existing TCM target prediction tools mainly based on manual retrieval or intelligent retrieval10,12,19, 20, 21, 22, 23, our method may obtain more sufficient and more reliable target information under the appropriate similarity threshold. Moreover, the biological functions and participated pathways of the confirmed and/or potential targets of ingredients/herbs/Chinese patent drugs/TCM formulas are also provided according to the enrichment analysis based on the Gene Ontology24 and Reactome pathway database25 using in house python scripts with hypergeometric test.
Figure 2.
Pipeline of ETCM v2.0 target identification and prediction, and detailed information of target genes of a certain ingredient. To facilitate the mechanistic research and new drug discovery, the target identification method of ETCM v2.0 was improved based on a two-dimensional ligand similarity search module, which provides the confirmed and/or potential targets of each ingredient, as well as their binding activities.
2.5. The disease information
The number of human diseases has increased from 2266 (ETCM) to 8045 (ETCM v2.0). The detailed information of each disease, including disease name, global category, anatomical category, symptoms, disease-related genes and hallmark gene set annotations, collected from the human disease database (MalaCards v5.0)26, Human Phenotype Ontology (HPO, Released in 2018)27, Online Mendelian Inheritance in Man (OMIM, Released in April 2018)28, Database of gene-disease associations (DisGeNET v5.0)29, the portal for rare diseases and orphan drugs (ORPHANET v5.49.0)30. The inconsistent gene or protein IDs of different resources were manually inspected and converted into Official Gene Symbols and UniProt Accession Numbers. The diseases are linked to ingredients, herbs, Chinese patent drugs and TCM formulas according to the enrichment analysis based on the disease-causing genes and the confirm/potential drug target genes using in house python scripts with hypergeometric test.
3. Direct associations & indirect associations among various data fields
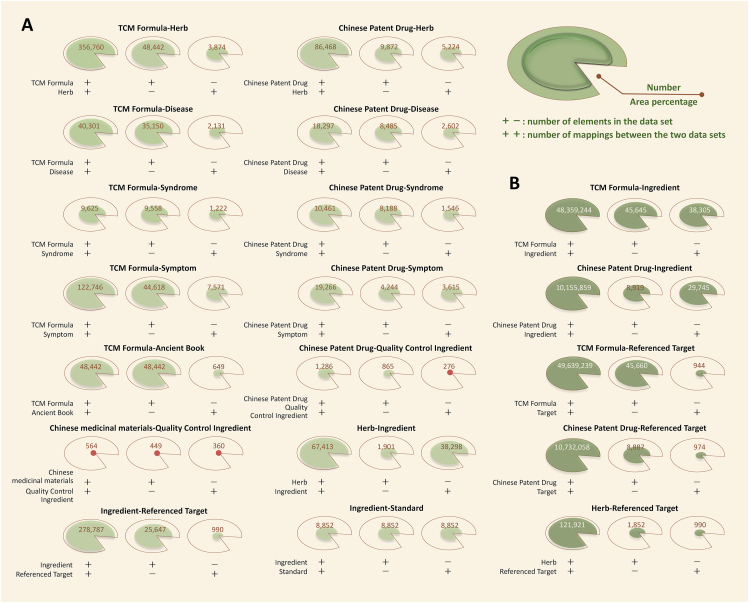
ETCM v2.0 provides comprehensive pairwise associations and cross retrievals between different data sections by combing diverse strategies including database mining, reference mining and bioinformatics mining (Fig. 3). We manually collected a total of 14 direct associations of “TCM Formula/Chinese Patent Drug-Herb”, “TCM Formula/Chinese Patent Drug-Disease”, “TCM Formula/Chinese Patent Drug-Syndrome”, “TCM Formula/Chinese Patent Drug-Symptom”, “TCM Formula-Ancient Book”, “Chinese Patent Drug/Herb-Quality Control Ingredient”, “Herb-Ingredient”, “Ingredient-Referenced Target” and “Ingredient-Standard” from ancient Chinese medical books, package insert of drugs and Chinese Pharmacopoeia (2020 version). Among them, the modern diseases, TCM syndromes and clinical symptoms associated with TCM formulas, Chinese patent drugs and herbs are rigorously standardized by clinical expert consensus and subsequent manual verification. The direct associations for “Ingredient-Referenced Target” were identified based on a two-dimensional ligand similarity search module in our indoor D3CARP platform and the BindingDB database18.
Figure 3.
The number of curated TCM formulas, Chinese patent drugs, herbs, ingredients, targets and diseases, and their associations shown as pie charts (A) Includes direct associations of “TCM Formula/Chinese Patent Drug-Herb”, “TCM Formula/Chinese Patent Drug-Disease”, “TCM Formula/Chinese Patent Drug-Syndrome”, “TCM Formula/Chinese Patent Drug-Symptom”, “TCM Formula-Ancient Book”, “Chinese Patent Drug/Herb-Quality Control Ingredient”, “Herb-Ingredient”, “Ingredient-Referenced Target” and “Ingredient-Standard” associations (B) Includes indirect associations of “TCM Formula/Chinese Patent Drug-Ingredient”, “TCM Formula/Chinese Patent Drug/Herb-Referenced Target”.
In addition to the above direct associations involving adjacent characteristic parameters, there are also five indirect associations involving non-adjacent parameters, such as “TCM Formula/Chinese Patent Drug-Ingredient” and “TCM Formula/Chinese Patent Drug/Herb-Referenced Target”. The indirect associations between TCM formula/Chinese patent drugs and ingredients were obtained using herbs as the middle parameters. Based on the Ingredient-Referenced Target relationship, the TCM formulas/Chinese patent drugs/Herbs were connected to the referenced targets.
4. Drug similarity evaluation
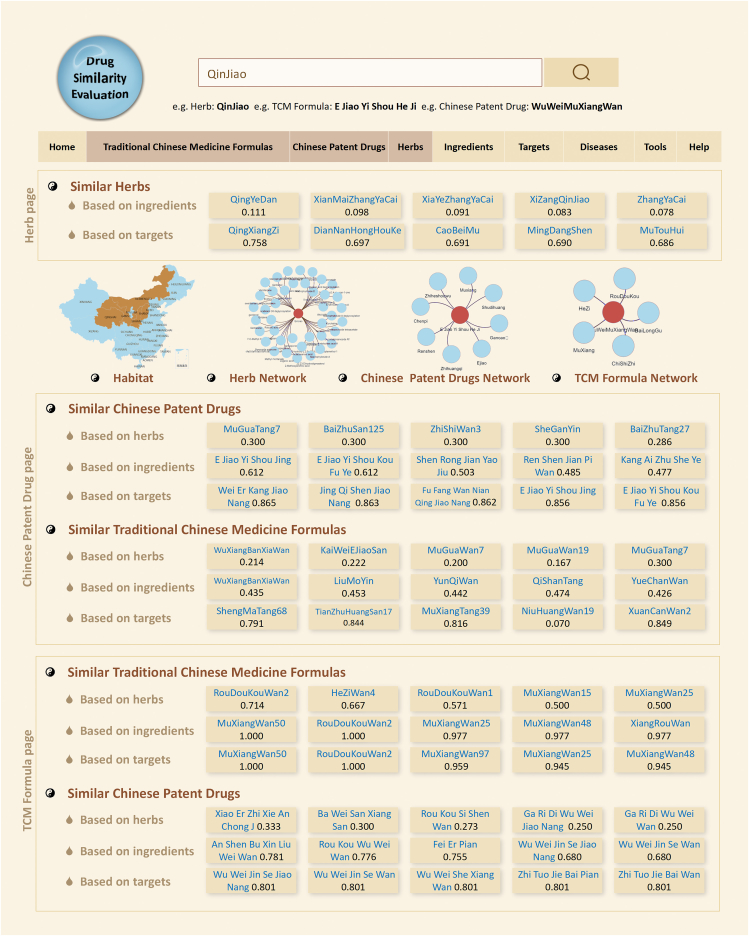
The Jaccard similarity scores among different TCM formulas, Chinese patent drugs and herbs calculated based on their herb compositions, chemical and target profilings are provided in the corresponding sections of ETCM v2.0. In each detailed information page, five TCM formulas/Chinese patent drugs/herbs with the highest similarity to the submitted TCM formula/Chinese patent drug/herb are displayed and also can be linked to their respective detailed information pages (Fig. 4). These similarity evaluation data may be of significance to identify prescriptions/herbs with similar clinical efficacy, to summarize the rules of prescription use, and to find alternative herbs for endangered Chinese medicinal materials.
Figure 4.
Drug similarity evaluation results in each detailed information page. Five TCM formulas/Chinese patent drugs/herbs with the highest similarity to the submitted TCM formula/Chinese patent drug/herb are displayed and also can be linked to their respective detailed information pages.
5. Improved browsing and inquiry functions
ETCM v2.0 not only includes detailed medicinal properties of TCM formulas, Chinese patent drugs, herbs and ingredients, but also provides potential links between TCM with target genes and modern diseases to facilitate TCM related basic researches, clinical applications and drug development. It offers the browse and search pages for users to navigate each data section, and provides the examples and the types of searchable keywords in or below the search menu.
For the novel data section “Traditional Chinese Medicine Formulas” added in ETCM v2.0, we provide additional browsing and searching pages. All 48,442 TCM formulas are classified according to their dosage forms (such as powder preparation, decoction, pill preparation, etc.) and their source of ancient Chinese medical books (such as “ShenJiZongLu”, “TaiPingShengHuiFang”, “PuJiFang”, etc.), which are presented in the browse page of the “Traditional Chinese Medicine Formulas” section. Users can view a full list of all TCM formulas belonging to each category by clicking on each above mentioned category. Detailed information of each TCM formula can be retrieved by clicking on its Chinese or Pinyin name, including dosage form, herb-chemical ingredient-target profiling, indications, major syndromes and main symptoms treated by the formula, the source ancient books of TCM, etc. Users can link to the information pages of the herbs, chemical ingredients and targets by clicking the corresponding name in the TCM formula information page. It is worth to note that the TCM formula indication information is described according to the records in the ancient books of TCM, these indications are different from modern diseases, therefore we tried to use genes related to both TCM ingredients and modern diseases to build the links between TCM formula indications and modern diseases. Similarly, TCM formulas collected in the ETCM v2.0 database are also classified based on the corresponding TCM syndromes, such as blood deficiency, heat syndrome, etc.
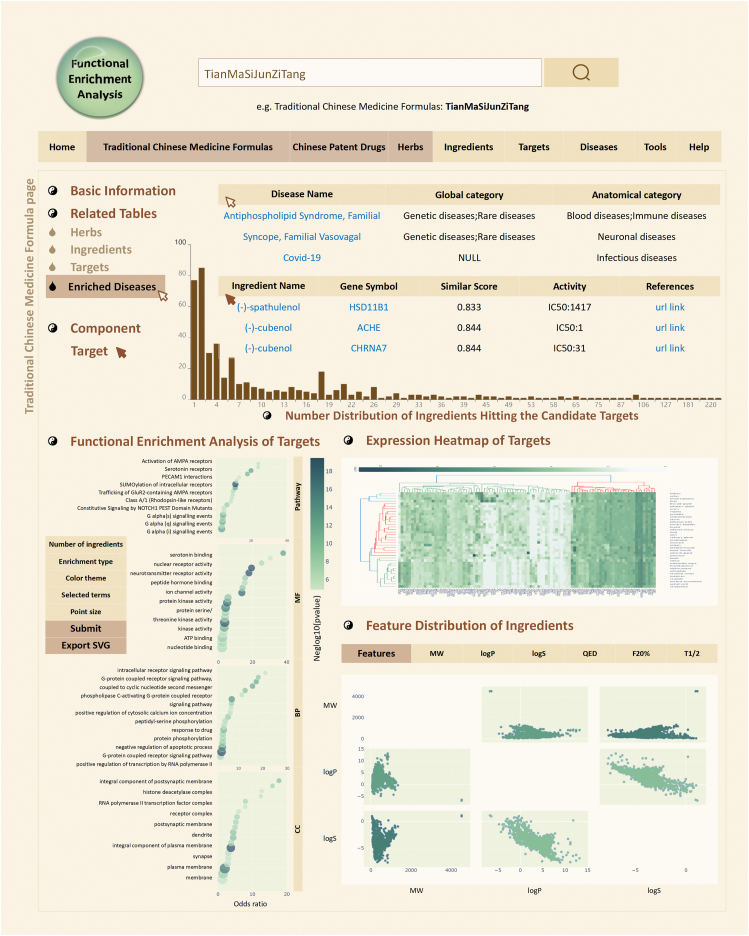
To facilitate the understanding of TCM functions from the modern science point of view, we improved our target identification model for ingredients of the collected herbs, TCM formulas and Chinese patent drugs according to the two-dimensional ligand similarity search module between TCM ingredients and ligands derived from the BindingDB database, and considered the target genes of ligands as the confirmed and/or potential targets of TCM ingredients based on the calculated similarity values. Besides the information of the target profiling, the binding activities between each herbal ingredient and its targets are also provided (Fig. 5). Moreover, the collective targets of all ingredients of a herb, a TCM formula or a Chinese patent drug are indicated as the confirmed and/or potential targets of the herb, TCM formula or Chinese patent drug. Diseases significantly associated with those target genes in the enrichment analysis are also shown as diseases that may be cured by the herbs, TCM formulas or Chinese patent drugs. Gene Ontology terms or pathways enriched by genes targeted by certain ingredients, herbs, TCM formulas, Chinese patent drugs or associated with certain diseases, are also included in ETCM v2.0 (Fig. 5).
Figure 5.
Improved biological relevance mining functions. In the detailed information page, the target profiling of each herb, Chinese patent drug and TCM formula, and the corresponding enriched diseases and biological functions are provided. In addition, the expression heatmap of targets and feature distribution of ingredients are also displayed for each herb, Chinese patent drug and TCM formula.
6. Enhanced network visualization capabilities
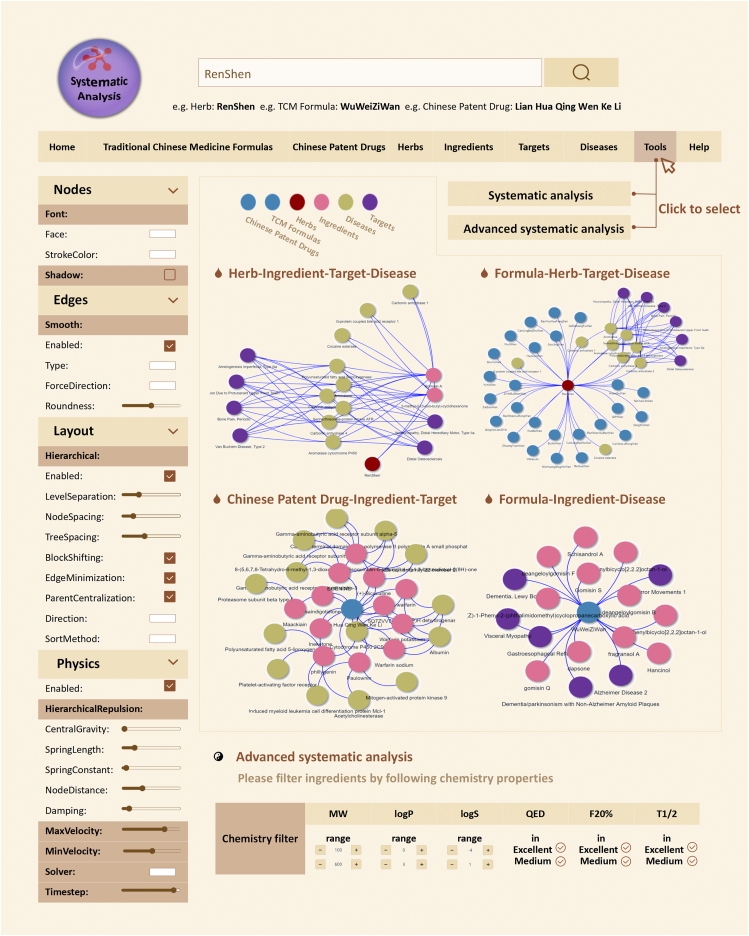
To better illustrate the relationships among TCM formulas, Chinese patent drugs, herbs, ingredients, target genes, gene-involved pathways and diseases, ETCM v2.0 provides an enhanced JavaScript-based network visualization tool for creating, modifying, and exploring multi-scale biological networks, which may be the characteristic function of our database and not available in other databases of the same type, such as HERB10, SuperTCM11 and HIT 2.012, YaTCM31, TCMID 2.019 and TCMSP32. Users are allowed to choose to display relationships among TCM formulas, Chinese patent drugs, herbs, ingredients, targets, disease-related genes, and diseases according to their research aims, as well as design, add, delete, shape, and edit the details of network nodes and edges as they like (Fig. 6). The gene-gene interaction data were collected from five existing molecular interaction databases, including STRING (https://cn.string-db.org/, version 11.5)33, Reactome (https://reactome.org/, version 81)34, Human Protein Reference Database (HPRD, http://www.hprd.org/, Release 9)35, IntAct molecular interaction database (https://www.ebi.ac.uk/intact, updated in August 2021)36 and Database of Interacting Proteins (DIP, https://dip.doe-mbi.ucla.edu/dip/, updated in Feb 13, 2017)37. Links among TCM formulas, Chinese patent drugs, herbs, pathways and diseases are established by the ingredients and the confirmed/potential targets of each ingredient. The dynamic layout of the network can be feasibly customized and the final network can be exported in SVG and JPG formats.
Figure 6.
Improved systematic analysis function. Examples of various networks constructed by the Systematic Analysis tool of ETCM v2.0. Users can also filter the components of the network to be constructed by various features in the Advanced Systematic Analysis tool.
7. Discussion
With a long history of widespread clinical applications, TCM has made important contributions to the prosperity of human beings, especially in East Asia. In recent years, TCM has also been frequently used in Western countries. On the basis of the systematic associations among TCM formulas, Chinese patent drugs, herbs, ingredients, targets and diseases, TCM gives specific prescriptions to each individual patient according to their disease conditions, which may be consistent with the goals of precision medicine. Although a growing number of TCM databases have been established with various data sources and similar functions, they may be lack of standardization and comprehensiveness. Herein, ETCM v2.0 is the latest curated database focusing on comprehensive resource and rich annotations for the TCM-derived drug discovery and repurposing, and the investigation of underlying mechanisms of TCMs against various human diseases (Table 2).
Table 2.
Comparisons of ETCM v2.0 to other existing TCM databases.
| Database | TCM Formulas |
Chinese Patent Drugs |
Chinese medicinal materials |
Ingredients |
Targets |
Diseases |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total No. | No. of sourcing ancient TCM books | Total No. | No. of drugs with mark ingredients | Total No. | No. of Chinese medicinal materials with mark ingredients | Average No. of ingredients per herb | No. of herbs with referenced targets | Total No. | No. of ingredients with standards | No. of ingredient–referenced target pairs | Total No. | No. of referenced targets | Total No. | No. of disease–gene pairs | |
| ETCM 2.0 | 48,442 | 649 | 9872 | 865 | 2079 | 504 | 35 | 990 | 38,298 | 6417 | 278,787 | 1040 | 1040 | 8045 | 22,122,972 |
| HIT 2.0 | – | – | – | – | 1250 | – | – | – | 1237 | – | 10,031 | 2208 | 2208 | – | – |
| TCMBank | – | – | – | – | 9191 | – | – | – | 61,965 | – | – | 15,179 | – | 32,529 | – |
| SuperTCM | 242 | – | – | – | 6516 | – | – | – | 55,772 | – | – | 543 | – | 8634 | – |
| HERB | – | – | – | – | 7263 | – | 16 | 291 | 49,258 | – | 4815 | 12,933 | 1241 | 28,212 | – |
| ETCM 1.0 | – | – | 3962 | 478 | 403 | 263 | – | – | 7274 | – | – | 2266 | – | 4323 | – |
| YaTCM | 1813 | 2 | – | – | 6220 | – | – | – | 47,696 | – | – | 18,697 | – | – | – |
| TCMID 2.0 | 46,929 | – | – | – | 8159 | – | 2 | – | 43,413 | – | – | 17,521 | – | 4633 | – |
| TCMSP | – | – | – | – | 499 | – | – | – | 29,384 | – | – | 3311 | – | 837 | – |
Using ETCM v2.0, researchers and drug developers can view primary data as well as the data- and algorithm-driven mapping results among TCM formulas/Chinese patent drugs, herbs, ingredients, targets and diseases, allowing easy identification of new potentially effective therapeutics and discovery of novel drug combination with synergistic effects. In addition, ETCM v2.0 offers the ingredients of Chinese herbs and prescriptions, and the evaluation results on their drug-likeness and safety, as well as the confirmed and/or potential target lists and the corresponding biological function enrichment analysis, which may be helpful in discovering the novel drug candidates.
Polypharmacological prediction of known TCM prescriptions by the similarity comparison based on herbal compositions, chemical and target profiling is very useful to find new therapeutic applications of existing TCM prescriptions and to discover multi-target drugs with improved efficacies. ETCM v2.0 provides highly similar TCM formulas/Chinese patent drugs/herbs/ingredients and the corresponding similar scores which may help screening TCMs that connect to targets of known drugs and discovering their new therapeutic effects.
Accumulating studies have revealed that TCM network pharmacology may play a crucial role in elucidating the network-based biological basis of complex diseases, TCM formula and herb therapeutics38,39, and the first international standard of network pharmacology—Network Pharmacology Evaluation Method Guidance has been issued40. On this context, ETCM v2.0 also provides the confirmed and/or potential drug targets, the disease-related genes, and the interactions among them, as well as user-friendly network visualization, which all may be of great significance to promote the exploration of the “multi-component, multi-target and multi-pathway” integrated therapeutic paradigm of TCM medicinal prescriptions.
Over the past three years since ETCM was released, our database has attracted an increasing attention of pharmacologists and scholars in the research field of TCM, and function as an important connector to link TCM with modern researches41,42. With more and diverse data becoming available, we updated our ETCM database to ETCM v2.0 to provide a more comprehensive and integrated resource of TCM big data for identifying bioactive constituents and quality markers of TCMs, promoting new drug research and development, accelerating the drug repurposing, and facilitating the mechanistic investigation and clinical application of TCMs. With a convenient web interface, users can browse, search, visualize and download key information from ETCM v2.0. In the future, we plan to continuously improve this database with new types of data fields and analysis toolkits as much as possible.
All data of ETCM is available at http://www.tcmip.cn/ETCM2/front/#/.
Acknowledgments
This work was supported by Key project at the National Natural Science Foundation of China (Grant Nos. 81830111 and 82030122, China), the Innovation Project of China Academy of Chinese Medical Sciences (Grant No. CI2021A04907, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Author contributions
Yanqiong Zhang participated in the database design, data preparation and validation, and prepared the manuscript. Xin Li and Yulong Shi performed data collection and processing, and developed the target prediction algorithm, respectively. Haiyu Xu participated in the study design and coordination, and offered material support for the obtained funding and supervised the study. The other authors performed parts of the database construction. All authors reviewed and approved the final manuscript.
Conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Ekiert H., Świątkowska J., Klin P., Rzepiela A., Szopa A. Artemisia annua—importance in traditional medicine and current state of knowledge on the chemistry, biological activity and possible applications. Planta Med. 2021;87:584–599. doi: 10.1055/a-1345-9528. [DOI] [PubMed] [Google Scholar]
- 2.Su X.Z., Miller L.H. The discovery of artemisinin and the nobel prize in physiology or medicine. Sci China Life Sci. 2015;58:1175–1179. doi: 10.1007/s11427-015-4948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo M., Wu Z., An Q., Li H., Wang L., Zheng Y., et al. Comparison of volatile oils and primary metabolites of raw and honey-processed Ephedrae Herba by GC–MS and chemometrics. J AOAC Int. 2022;105:576–586. doi: 10.1093/jaoacint/qsab139. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Yang W., Liu Y., Lu C., Ruan L., Zhao C., et al. Combination of Hua Shi Bai Du granule (Q-14) and standard care in the treatment of patients with coronavirus disease 2019 (COVID-19): a single-center, open-label, randomized controlled trial. Phytomedicine. 2021;91 doi: 10.1016/j.phymed.2021.153671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi N., Guo L., Liu B., Bian Y., Chen R., Chen S., et al. Efficacy and safety of Chinese herbal medicine versus lopinavir–ritonavir in adult patients with coronavirus disease 2019: a non-randomized controlled trial. Phytomedicine. 2021;81 doi: 10.1016/j.phymed.2020.153367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Lu C., Li H., Qi W., Ruan L., Bian Y., et al. Efficacy and safety assessment of severe COVID-19 patients with Chinese medicine: a retrospective case series study at early stage of the COVID-19 epidemic in Wuhan, China. J Ethnopharmacol. 2021;277 doi: 10.1016/j.jep.2021.113888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong W.Z., Wang G., Du J., Ai W. Efficacy of herbal medicine (Xuanfei Baidu decoction) combined with conventional drug in treating COVID-19: a pilot randomized clinical trial. Integr Med Res. 2020;9 doi: 10.1016/j.imr.2020.100489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H., Zhang Y., Wang P., Zhang J., Chen H., Zhang L., et al. A comprehensive review of integrative pharmacology-based investigation: a paradigm shift in traditional Chinese medicine. Acta Pharm Sin B. 2021;11:1379–1399. doi: 10.1016/j.apsb.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H., et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47:D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang S., Dong L., Liu L., Guo J., Zhao L., Zhang J., et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49:D1197–D1206. doi: 10.1093/nar/gkaa1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q., Springer L., Gohlke B.O., Goede A., Dunkel M., Abel R., et al. SuperTCM: a biocultural database combining biological pathways and historical linguistic data of Chinese Materia Medica for drug development. Biomed Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112315. [DOI] [PubMed] [Google Scholar]
- 12.Yan D., Zheng G., Wang C., Chen Z., Mao T., Gao J., et al. Hit 2.0: an enhanced platform for herbal ingredients' targets. Nucleic Acids Res. 2022;50:D1238–D1243. doi: 10.1093/nar/gkab1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doak B.C., Zheng J., Dobritzsch D., Kihlberg J. How beyond rule of 5 drugs and clinical candidates bind to their targets. J Med Chem. 2016;59:2312–2327. doi: 10.1021/acs.jmedchem.5b01286. [DOI] [PubMed] [Google Scholar]
- 16.Doak B.C., Over B., Giordanetto F., Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem Biol. 2014;21:1115–1142. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Contrera J.F., Matthews E.J., Kruhlak N.L., Benz R.D. Estimating the safe starting dose in phase I clinical trials and no observed effect level based on QSAR modeling of the human maximum recommended daily dose. Regul Toxicol Pharmacol. 2004;40:185–206. doi: 10.1016/j.yrtph.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., et al. Tcmid 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018;46:D1117–D1120. doi: 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y., Gao B., Liu L., Liu J., Zhu Y. AMFormulaS: an intelligent retrieval system for traditional Chinese medicine formulas. BMC Med Inf Decis Making. 2021;21:56. doi: 10.1186/s12911-021-01419-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Yang K., Liu Z., Lai X., Yang Z., Zeng J., et al. DrugAI: a multi-view deep learning model for predicting drug–target activating/inhibiting mechanisms. Briefings Bioinf. 2023;24:bbac526. doi: 10.1093/bib/bbac526. [DOI] [PubMed] [Google Scholar]
- 22.Öztürk H., Özgür A., Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I., Keum J., Nam H. DeepConv-DTI: prediction of drug–target interactions via deep learning with convolution on protein sequences. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappaport N., Twik M., Plaschkes I., Nudel R., Iny Stein T., Levitt J., et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45:D877–D887. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köhler S., Gargano M., Matentzoglu N., Carmody L.C., Lewis-Smith D., Vasilevsky N.A., et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021;49:D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:789–798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer-Mehren A., Rautschka M., Sanz F., Furlong L.I. DisGeNET: a cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics. 2010;26:2924–2926. doi: 10.1093/bioinformatics/btq538. [DOI] [PubMed] [Google Scholar]
- 30.Nguengang Wakap S., Lambert D.M., Olry A., Rodwell C., Gueydan C., Lanneau V., et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B., Ma C., Zhao X., Hu Z., Du T., Xu X., et al. YaTCM: yet another traditional Chinese medicine database for drug discovery. Comput Struct Biotechnol J. 2018;16:600–610. doi: 10.1016/j.csbj.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ru J., Li P., Wang J., Zhou W., Li B., Huang C., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Toro N., Shrivastava A., Ragueneau E., Meldal B., Combe C., Barrera E., et al. The IntAct database: efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022;50:D648–D653. doi: 10.1093/nar/gkab1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Wang Z.Y., Zheng J.H., Li S. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19:1–11. doi: 10.1016/S1875-5364(21)60001-8. [DOI] [PubMed] [Google Scholar]
- 39.Li S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11:110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 40.Li Shao. Network pharmacology evaluation method guidance—draft. World J Tradit Chin Med. 2021;7 165-6+146-54. [Google Scholar]
- 41.Zhang Z., Liu J., Liu Y., Shi D., He Y., Zhao P. Virtual screening of the multi-gene regulatory molecular mechanism of Si-Wu-tang against non-triple-negative breast cancer based on network pharmacology combined with experimental validation. J Ethnopharmacol. 2021;269 doi: 10.1016/j.jep.2020.113696. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y., Liang B., Chen W., Wu X.R., Liu-Huo W.S., Zhao L.Z. Potential mechanism of Dingji Fumai Decoction against atrial fibrillation based on network pharmacology, molecular docking, and experimental verification integration strategy. Front Cardiovasc Med. 2021;8 doi: 10.3389/fcvm.2021.712398. [DOI] [PMC free article] [PubMed] [Google Scholar]