Abstract
Induction of cancer cell ferroptosis has been proposed as a potential treatment in several cancer types. Tumor-associated macrophages (TAMs) play a key role in promoting tumor malignant progression and therapy resistance. However, the roles and mechanisms of TAMs in regulating tumor ferroptosis is still unexplored and remains enigmatic. This study shows ferroptosis inducers has shown therapeutic outcomes in cervical cancer in vitro and in vivo. TAMs have been found to suppress cervical cancer cells ferroptosis. Mechanistically, macrophage-derived miRNA-660-5p packaged into exosomes are transported into cancer cells. In cancer cells, miRNA-660-5p attenuates ALOX15 expression to inhibit ferroptosis. Moreover, the upregulation of miRNA-660-5p in macrophages depends on autocrine IL4/IL13-activated STAT6 pathway. Importantly, in clinical cervical cancer cases, ALOX15 is negatively associated with macrophages infiltration, which also raises the possibility that macrophages reduce ALOX15 levels in cervical cancer. Moreover, both univariate and multivariate Cox analyses show ALOX15 expression is independent prognostic factor and positively associated with good prognosis in cervical cancer. Altogether, this study reveals the potential utility of targeting TAMs in ferroptosis-based treatment and ALOX15 as prognosis indicators for cervical cancer.
KEY WORDS: Cervical cancer, Tumor-associated macrophage, ALOX15, Ferroptosis, Exosome, miRNA-660-5p, Macrophages infiltration, STAT6
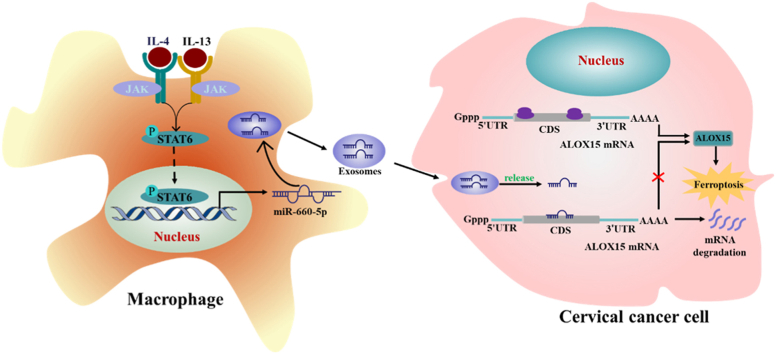
Graphical abstract
In cervical cancer, autocrine IL4/IL13-activated STAT6 induced miRNA-660-5p expression in macrophages. miRNA-660-5p was packaged into exosomes and transported into cancer cells to attenuate ALOX15 expression to inhibit cancer cells ferroptosis.
1. Introduction
Cervical cancer is one of the most common female malignant tumors and a prevalent cause of malignant tumor-related death in women worldwide1. As a major treatment modality in cervical cancer, chemotherapy exerts an important role in improving patients' survival by inducing cancer cells apoptosis2. However, under the action of continuous chemotherapy drugs, cancer cells gradually resist chemotherapy-induced apoptosis3. Resistance to cell apoptosis is a major obstacle for effective drug treatment in various types of tumors including cervical cancer4. Ferroptosis, a novel nonapoptotic form of regulated cell death, involves iron-dependent lipid peroxides accumulation and causes lethal damage of cells5. However, the function of ferroptosis in cervical cancer is largely unknown. Recent studies showed that induction of ferroptosis by some drugs can effectively prevent tumor progression and promote effects of chemotherapy, targeted therapy and immunotherapy6,7. For example, dihydroartemisinin prevents glioma progression by inducing cancer cells ferroptosis8. This raises the possibility that induction of ferroptosis may be a novel treatment for cervical cancer. Therefore, we investigated the antitumor effects of inducers of ferroptosis in cervical cancer in this study.
It is well known that in vivo tumor has unique tumor microenvironment (TME), which is consists of several immune cells including tumor-associated macrophages (TAMs) and nonimmune cell populations including cancer-associated fibroblasts (CAFs)9. Growing evidence uncovered CAFs and TAMs in TME play important roles in promoting tumor resistance to anti-cancer agents. For example, in hepatocellular carcinoma, TAMs mediate sorafenib resistance by secreting hepatocyte growth factor10. In pancreatic cancer, TAMs promote chemoresistance by secreting 14-3-3ζ11. CAF has been reported to suppress ferroptosis and enhance chemotherapy resistance in gastric cancer12. Therefore, we investigated the effects of TAMs and CAFs in anti-cancer effects of ferroptosis inducers in cervical cancer. We found that cell death induced by ferroptosis inducers was not significantly changed by CAFs, but was obviously suppressed by TAMs. Therefore, we speculated that TAMs might attenuate cervical cancer ferroptosis.
TAMs are the main type of immune cells in TME and exhibit distinct tumorigenic properties9. TAMs surrounding tumor cells release anti-inflammatory cytokines such as IL-10 and TGF-β, and growth factors such as IGF-1 to promote tumor cell growth, migration and invasion, even enhance cancer cell resistance to drugs9. However, whether TAMs prevent cancer cell ferroptosis remains unclear. In this study, we showed the cells ferroptosis induced by erastin, RSL3 and sorafenib was blocked by TAMs in cervical cancer in vivo and in vitro. Moreover, exosomes released by TAMs were involved in the TAMs-inhibited cervical cancer ferroptosis. Exosomes are membrane vesicles with 30–200 nm in diameter are secreted by most cell types including macrophages13. Exosomes exert important roles in regulating cell biological functions by delivering proteins, mRNAs and microRNAs to recipient cells14,15. Exosomes derived from macrophages have been reported to promote malignant tumors progression by delivering microRNAs16. Here, we found that TAMs-derived exosome carried miR-660-5p into cervical cancer cells to suppress arachidonate 15-lipoxygenase (ALOX15) expression and thus attenuating ferroptosis.
Taken together, these findings provided evidence for TAMs-inhibited ferroptosis in cervical cancer. Moreover, we revealed a novel and important communication mechanism between TAMs and cancer cells. Our results suggest targeted TAMs combined with ferroptosis inducers might be an attractive treatment for the cervical cancer therapy.
2. Materials and methods
2.1. Patients and samples
The study was approved by the Institutional Review Board, and informed consent was obtained from all subjects (IRB20150305). Paraffin-embedded samples from patients diagnosed with cervical cancer (n = 147) were collected between January 2010 and August 2015. Follow-up data were collected from postoperative patients to monitor and assess the overall rate of cancer and recurrence. The correlation between ALOX15 expression in cervical cancer patients and patients' clinical parameters and prognoses was analyzed.
2.2. Cell culture and the reagents
HeLa, SiHa and THP-1 cell lines used in this study were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). THP-1 was grown in PRMI Medium 1640 (Gibco). HeLa and SiHa were cultured in Dulbecco's modified Eagle's medium (Gibco). All medium was supplemented with 10% fetal bovine serum (Gibco) and antibiotics (penicillin and streptomycin) (Gibco). Erastin, RSL3, sorafenib, AS1517499 (STAT6 inhibitor) and STAT3-IN-1 (STAT3 inhibitor) were purchased from MedChemExpress.
2.3. Cell viability and cell death assays
Cell viability was assessed with Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Japan) according to the manufacturer's instructions. Briefly, cells were plated into 96-well plates at a density of 3 × 103 cells/well and cultured for 12 h. Cells were subsequently incubated with 10 μL CCK8 for 1 h. And then the cell viability was determined by measuring the absorbance of each well at a wavelength of 450 nm using a microplate reader (Infinite M200, Tecan). Cell death was determined using propidium iodide (PI) (Roche) staining on a BD FACS Canto II Flow Cytometer (Becton Dickinson). Briefly, cells treated with indicated agents were collected in the centrifuge tube at a density of 1 × 106, washed with PBS for three times. The cells collected after centrifugation were fixed by 70% alcohol for about 1 h, followed by staining with 5 μg/mL of PI in the dark for 30 min. And then the cell death was determined on a BD FACS Canto II Flow Cytometer. Cell death was observed by confocal microscope (LSM 880 Meta; Zeiss). Cells were plated in 2-cm confocal dishes (BIOFIL, Guangzhou, China) or 10-cm dish and then treated as indicated. Adherent cells were incubated with 5 μmol/L of PI for 30 min at 37 °C, then the cells were washed once with PBS. PI-positive cell was assessed by confocal microscope.
2.4. Lipid ROS analysis
Cells were plated in 2-cm confocal dishes or 10-cm dish and then treated as indicated. Adherent cells were incubated with 5 μmol/L of C11-BODIPY 581/591 (D3861, ThermoFisher Scientific, Shanghai, China) for 30 min at 37 °C, then the cells were washed once with PBS. Lipid ROS generation was assessed by confocal microscope. For flow cytometer detection, stained cells were collected by trypsinization and resuspended in PBS, and then cells were detected by BD FACS Canto II Flow Cytometer.
2.5. Malondialdehyde (MDA) and glutathione determination
To assay lipid peroxidation, the MDA lipid peroxidation microplate assay (Merck KGaA) was used according to manufacturers' instructions. Briefly, cells were homogenized in lysis solution containing butylated hydroxytoluene. The insoluble fraction was removed by centrifugation, and the supernatant was used for analysis. The supernatants were mixed with thiobarbituric acid solution reconstituted in glacial acetic acid and then incubated at 95 °C for 60 min. The supernatants containing MDA-thiobarbituric acid adduct were added into a 96-well microplate for analysis. A microplate reader was used to measure the absorbance at OD 532 nm.
For glutathione determination, intracellular reduced form glutathione (GSH) and its oxidized form (GSSG) were assessed using a GSH/GSSG Ratio Detection Assay Kit II (Abcam, Cambridge, UK) according to the manufacturer's instructions. Briefly, cells were homogenized in 5% 5-sulfosalicylic acid, and the insoluble fraction was removed by centrifugation. The resultant supernatant was added to double-deionized H2O (ddH2O) to reduce the 5-sulfosalicylic acid concentration to 0.5% for the assay. A microplate reader was used to measure absorbance at OD 415 nm. The concentration of GSH was calculated by subtracting 2 × GSSG from the total glutathione concentration.
2.6. RNA interference
siRNA transfections were performed with Lipofectamine 3000 reagent (Thermo Fisher-Invitrogen) according to the manufacturer's instructions. Control siRNA and siRNAs targeting human ALOX15 (106384, 116945, ThermoScientific, Karlsruhe, Germany) were transfected into cervical cancer cells at a final concentration of 50 nmol/L.
2.7. Dual-luciferase activity assay
To generate the pGL3 luciferase reporter vector of ALOX15, the wild type (WT) and mutated (MT) human coding sequence (CDS) of ALOX15 genes were amplified by PCR and cloned into the XbaI site of the pGL3-control vector (Promega, Madison, WI, USA), which is the downstream of the luciferase gene. For luciferase assay, HeLa cells were cultured in 24-well plates and transfected with 500 ng of pGL3 luciferase reporter vector of ALOX15 and control vector along with 50 nmol/L of miR-660-5p mimics and inhibitor, respectively. Transfection of vectors and miRNAs was carried out using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA). After 24 h, firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega) as previously described17. The above experiment was repeated at least three times.
2.8. Real-time quantitative PCR analysis
RT-qPCR was performed as described previously18. In brief, total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Following DNaseI treatment, 2 μg of total RNA was reverse transcribed using cDNA synthesis Kit (Bio-Rad, Richmond, CA, USA) to synthesize cDNA specimens. RT-PCR was performed using 2 μL of cDNA and SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) as recommended by the manufacturer, and conducted by means of the SYBR on the CFX96 system (Bio-Rad). For detection of miRNA, a Ploy-A tail was added to the miRNA, which was then transcribed into cDNA using a universal adaptor primer that included oligo-dT. The generated cDNA was then combined with the Uni-miR RT-PCR primer and a miR-660-5p primer (sequence complementary to the miR-660-5p) to complete the RT-PCR. The miRNA expression was normalized using the endogenous U6 snRNA. The primers sequences of ferroptosis-associated gene were available in Supporting Information Table S1 19.
IL-10: 5′-GGCGCTGTCATCGATTTCTT-3′ (forward) and 5′-GGCTTTGTAGATGCCTTTCTCTTG-3′ (reverse); IL-1β: 5′-CTCGCCAGTGAAATGATGGCT-3′ (forward) and 5′-GTCGGAGATTCGTAGCTGGCT-3′ (reverse); CD206: 5′-AAGGCGGTGACCTCACAAG-3′ (forward) and 5′-AAAGTCCAATTCCTCGATGGTG-3′ (reverse); CD163: 5′-ACATAGATCATGCATCTGTCATTTG-3′ (forward) and 5′-CATTCTCCTTGGAATCTCACTTCTA-3′ (reverse); U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); For β-actin primers: 5′-TTCTACAATGAGCTGCGTGTG-3′ (forward) and 5′-GGGGTGTTGAAGGTCTCAAA-3′ (reverse). The PCR was run in triplicate at 95 °C for 2 min followed by 35–38 cycles of 95 °C for 15 s, 56 °C for 20 s and 72 °C for 15–25 s. Comparative quantification was performed using the 2–ΔΔCt method. Each sample was analyzed in triplicate.
2.9. Western blot
Western blot was performed as previously described20. In brief, cells or exosomes were lysed in radioimmune precipitation assay buffer (Sigma–Aldrich, US) at 4 °C, supplementing with protease and phosphatase inhibitor cocktail tablets (Roche, UK). And protein concentration was measured using bicinchoninic acid protein assay. The protein sample was mixed with loading buffer (10% SDS, 40% Glycerol, 3% Bromophenyl Blue, and 10% β-mercaptoethanol) and boiled for 10 min. Proteins were separated on 8%–12% SDS-PAGE, transferred onto the PVDF membrane (EMS Millipore, USA), blocked with 5% bovine serum albumin, and then immunoblotted with specific antibodies. Antibodies used are listed here: CD63 (Abcam ab68418), CD81 (Abcam ab109201), ALOX15 (Abcam ab119774), β-actin (Santa Cruz sc-47778), Phospho-STAT3 (Tyr705) (CST, #9145), STAT3 (CST, #9132), Phospho-STAT6 (Tyr641) (CST, #56554), STAT6 (CST, #9362).
2.10. Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min, permeabilization with 0.2% Triton-X 100 for 10 min at 4 °C, and blocked with 3% bovine serum albumin for 1 h at room temperature, followed by incubation overnight at 4 °C with ALOX15 (Abcam ab119774, 1:100) and CD68 (CST, #23308, 1:150). Cells were washed 3 times for 5 min in PBS, and then incubated for 2 h with FITC-conjugated goat anti-mouse secondary antibody (diluted 1:200 in blocking buffer; Abcam). Acquired images with a laser scanning confocal microscope.
2.11. To visualize the shuttling of miRNAs
MiR-660-5p mimics labeled by Cy3 were obtained from GenePharma. The labeled miRNAs were transfected into macrophages by Lipo3000. After 24 h, macrophages were washed three times using PBS to remove the residual labeled miRNAs and transfection reagent. Macrophages carrying Cy3-miRNA mimics were then placed onto Transwell upper inserts, and cervical cancer cells were seeded in the lower inserts. After incubation for 48 h, cancer cells were collected and analyzed by fluorescence microscopy or flow cytometric analyses.
2.12. miRNA target prediction
The analysis of miRNA-660 predicted targets was performed using the algorithms miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
2.13. Exosome purification and labeling, microparticles purification, and treatment
Exosome-free growth medium containing 10% FBS was produced by ultra-centrifuging at 100,000 × g for 18 h to deplete exosomes. Macrophages (8 × 105 cells) with 70%–80% confluence were cultured in exosome-free medium in 2.5 cm dish for 48 h. This medium as condition media (CM) was collected. Exosomes were purified by differential centrifugation. Briefly, the CM were centrifuged at 500 × g for 30 min to eliminate cells, and then at 16,500 × g for 20 min to eliminate cellular debris. Supernatants were filtered through 0.22 μm filters. Exosomes were pelleted by ultracentrifugation at 120,000 × g for 3 h. After washing the exosomes once with PBS, exosomes were stained with specific plasma membrane fluorescent dye CMDiI (CellTracker, C7000), according to the manufacturer's instructions. Next, exosomes were diluted in 2 mL of growth medium with exosome-free. And then, in 2.5 cm dish, the growth medium with exosomes were used to treat the same number of tumor cells (8 × 105 cells) as macrophages which were used to extract exosomes for indicated time. And then, the fluorescence of tumor cells was observed under a confocal microscope and analyzed using flow cytometry. For exosomes treatment in ferroptosis functions study, the number of macrophages used to extract exosomes was consistent with the number of tumor cells.
The microparticles were purified as follows. The condition media was centrifuged to remove cell, cell debris and apoptotic bodies by 300 × g for 10 min, 2000 × g for 10 min, and 10,000 × g for 10 min in turn. And then the supernatants were centrifuged for 60 min at 14,000 × g to pellet microparticles (MP).
2.14. Xenografted cervical cancer model in mice
Athymic nu/nu nude mice were acquired from Medical Laboratory Animal Center of Guangdong (Guangzhou, China). The animals used to test the treatment were 4 to 5-week-old female mice. Briefly, nude mice were randomly allocated to each group and subcutaneously injected into the animal's right flank with 1 × 106 HeLa cells alone or HeLa cells in combination with 1 × 106 THP-1-derived macrophages (cells diluted in Matrigel for each mouse, 5 mice per group). Ten days later, mice were intraperitoneally injected with PBS, 10 mg/kg erastin every three days for 15 days, and the length and width of tumor were monitored and recorded every 2–3 days. For combination administration of erastin and STAT6 inhibitor AS1517499 (AS), AS was dissolved in 20% DMSO and 80% normal saline, the dose of 10 mg/kg AS and erastin were intraperitoneally injected once every two days. The tumor volumes were estimated using Eq. (1):
| Tumor volume = 0.5 × Length × Width2 | (1) |
All procedures were approved by the Institutional Animal Care and Use Committee.
2.15. Co-culture system with macrophages and cervical cancer cells
Human peripheral blood mononuclear cell (PBMC) from the buffy coats of healthy donors was obtained by density gradient centrifugation with Lymphocyte Isolation Solution (Qiagen) by centrifugation at 450 × g for 25 min at room temperature. Monocytes (Mo) were isolated from PBMC cells by positive magnetic separation using CD14 immunomagnetic beads (Miltenyi Biotec, Bergisch, Germany). CD14+ cells (106/mL) were cultured in 1640 media with 10% FBS in 48-well flat-bottom culture plates. Subsequently, non-adherent cells were washed with Hank's solution and discarded, and the adherent monocytes were incubated for 7 days in RPMI-1640 medium supplemented with 50 ng/mL of M-CSF (Peprotech Inc., Rocky Hill, NJ, USA) to be differentiated into macrophages which were considered to be M0 macrophages. The fresh growth media with same concentration of M-CSF was replaced every 2 days for a total of 7 days. The written informed consents were from all donors at their recruitment time. This study was approved by the Clinical Ethics Review Board at The Affiliated Cancer Hospital of Zhengzhou University (IRB20150305). For THP-1-derived macrophages, THP-1 monocytes (Mo) were incubated with 200 nmol/L phorbol 12-myristate 13-acetate acquired from Sigma for 48 h to differentiate into adhered macrophages which were considered to be M0 macrophages. M0 macrophages were incubated with IL-4 (20 ng/mL) + IL-13 (20 ng/mL) for 72 h to be differentiated into M2 macrophages. M0 macrophages were incubated with IFN-γ (10 ng/mL) and LPS (100 ng/mL) for 72 h to be differentiated into M1 macrophages. For TAM generation, in co-culture system, macrophages were co-cultured with cervical cancer cells or condition medium from cancer cells to generate TAMs. Macrophages were seeded in lower inserts of 6-well transwell plate [0.4 μm pore size polycarbonate transwell filters (Corning BV Life Sciences, Schiphol-Rijk, The Netherlands)], and cancer cells were seeded in upper inserts. The two cells were co-cultured without direct contact. After 48 h, the macrophages in the lower inserts were used for next treatment.
2.16. Transmission electron microscopy (TEM) assay
For conventional TEM, the purified exosome was placed in a droplet of 2.5% glutaraldehyde in PBS and fixed overnight at 4 °C. Samples were rinsed three times using PBS and post-fixed in 1% osmium tetroxide for 1 h at 25 °C. Then, the samples were embedded in 10% gelatin and fixed in glutaraldehyde at 4 °C and cut into several blocks (<1 mm3). The samples were successively dehydrated in 30%–100% of alcohol. Pure alcohol was then exchanged by propylene oxide, and specimens were soaked into 25%–100% of Quetol-812 epoxy resin mixed with propylene oxide for more than 3 h per step. Samples were embedded in pure, fresh Quetol-812 epoxy resin and polymerized at 35 °C for 12 h, 45 °C for 12 h, and 60 °C for 24 h. Samples were cut into 100-nm sections using a Leica UC6 ultra-microtome and stained with uranyl acetate for 10 min and with lead citrate for 5 min at 25 °C. The samples were observed in a JEM-1400 plus transmission electron microscope (JEOL, Japan), operated at 120 kV21.
2.17. Nanoparticle tracking analysis (NTA)
The particle size distribution and density of exosomes were tracked by Nanosight NS 300 system (NanoSight technology, Malvern, UK)21. Captures and analysis were achieved by using the built-in NanoSight software NTA3.3.301 (Malvern Panalytical Ltd., Malvern, UK). The camera level was set at 14 and the detection threshold was fixed at 5 min. Samples were diluted in PBS to a final volume of 1 mL, and their concentration was adjusted by observing a particles/frame rate of around 50.
2.18. Immunohistochemical (IHC) analysis
Immunohistochemical analysis was performed to assess alterations in ALOX15 protein expression in the 147 human cervical cancer tissues as previously described18. For primary antibody, the anti-ALOX15 mouse monoclonal antibody (1:200, Novus Biologicals, NBP2-01740) was used. ALOX15 staining was scored as the product of the proportion of positively stained cells and the staining intensity by two independent pathologists. The scores assigned by the two investigators were averaged (range from 0 to 12). The proportion of tumor cells was scored as follows: 1 (<10% positive tumor cells), 2 (10%–50% positive tumor cells), 3 (50%–75% positive tumor cells), and 4 (>75% positive tumor cells). Staining intensity was graded according to the following criteria: 0 (no staining); 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). Here, we used the receiver operating characteristic curve with respect to overall survival (OS) to calculate the cut-off value to divide patients into groups of high and low expression of ALOX15. The optimal cut-off values were assigned as follows: staining scores of ≥6 described tumors with high ALOX15 expression, and scores of ≤4 were assigned to those with low ALOX15 expression.
2.19. Data source and preprocessing
Clinical information and transcriptional profiles of patients' data from The Cancer Genome Atlas (TCGA) was downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). First, the samples with OS and survival status of 0 in the clinical information were filtered out. And then, the RNA-seq count data were transformed into the Transcripts Per Million to calculate the immune related metagene signature scores.
2.20. Isolation and culture of CAFs and normal fibroblasts and TAM from cancer sample
Cancer-associated fibroblasts and TAM were isolated from 3 tumor specimens of cervical cancer patients. Normal fibroblasts (NFs) were isolated from samples of nontumorigenic tissue areas. These specimens were washed three times with PBS, then minced into less than 2 mm2 sized pieces and digested using 10 mL of 0.1% trypsin solution (Gibco, CA, USA) with 0.75 μg/μL collagenase I (Sigma Chemical Co., St. Louis, MO, USA). Subsequently, the homogenate was collected and sequentially passed through 500 μm mesh, 100 μm and 70 μm strainer. Then, CAFs, TAM and NFs were purified using magnetic activated cell sorting. The cells were then centrifuged in a centrifuge (5804R, Eppendorf, MA) at 1127 g for 20 min with 1 mL cell suspension in upper, 5 mL 45% Percoll (GE Healthcare) in the middle and 5 mL 60% Percoll at the bottom in a 15 mL tube. CAFs and NF and mononuclear cells were collected from the cell layer in the interphase between 45% and 60% Percoll. Cells were incubated with anti-fibroblast and anti-CD14 microbeads (Miltenyi Biotec, Auburn, CA, USA) for 0.5 h at room temperature. After passing through a MiniMACS separator, positive cells were resuspended and plated in RPMI 1640 with 10% FBS (Gibco). After 24 h culture on the first day, the medium was replaced to eliminate floating cells, purified CAFs, TAM and NFs were obtained.
2.21. Assessment of immune cell infiltration level and signature score
We used single-sample gene set enrichment analysis (ssGSEA) and the xCell to estimate immune infiltration in TCGA RNA-seq cohorts22,23. The xCell algorithm was used to quantify the abundance of major immune cells in cervical cancer23. xCell integrated a large number of human cell type transcriptomes from various sources and employed a curve fitting approach for linear comparison of cell types and introduced a novel spillover compensation technique for separating them23. Patients with TCGA cohorts were divided into two groups based on median ALOX15 expression value. The ssGSEA was used to identify differences in 28 immune cell types in the tumor microenvironment between patients with high and low ALOX15 expression. The immune related metagenes was derived from Charoentong et al.24, and the infiltration level of each immune cell type was expressed by the ssGSEA scores, which was normalized as a z-score.
2.22. Statistical analysis
Statistical analysis was performed using SPSS version 20.0 (SPSS, Inc.). Student's t test was used to make a statistical comparison between groups. One-way analysis of variance (ANOVA) followed by Dunnett's post hoc tests were used to compare the differences among more than two groups. P < 0.05 was considered statistically significant. Results were representative of at least three experiments under identical conditions. The Chi square test and Fisher's exact test were used to analyze the relationship between ALOX15 expression and the clinicopathological characteristics. The log-rank test was used to compare Kaplan–Meier survival curves. Univariate and multivariate Cox regression analyses were used to evaluate the survival data. P < 0.05 was considered statistically significant.
3. Results
3.1. Tumor-associated macrophages suppress ferroptosis in cervical cancer
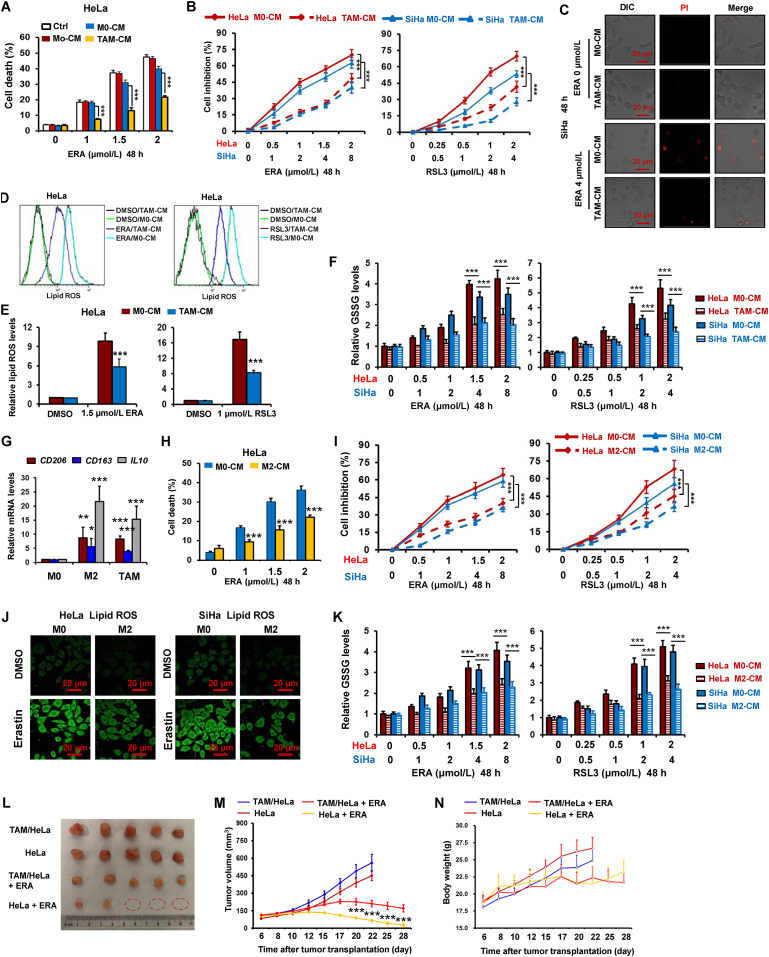
To test the function of macrophages in the regulation of ferroptosis of cervical cancer cells, we endeavored to test the sensitivity of HeLa cells treated with macrophage CM to ferroptosis inducer erastin, which inhibits cystine/glutamate antiporter system Xc activity. As shown in Fig. 1A and C, by cell death detection using PI staining, HeLa and SiHa cells treated with CM from TAMs (TAMs-CM) exerted higher resistance to erastin relative to the cells treated with monocytes CM (Mo-CM) and M0 macrophages CM (M0-CM). By using the CCK-8 assay to measure the cell viability in both cervical cancer cells HeLa and SiHa treated with TAMs-CM and three ferroptosis inducers, we found that TAMs-CM significantly inhibited the reduction of cell viability by erastin, RSL3 (GPX4 inhibitor) and sorafenib (system Xc inhibitor) in these two cells (Fig. 1B and Supporting Information Fig. S1A).
Figure 1.
Tumor-associated macrophages suppress ferroptosis in cervical cancer. (A) HeLa cells were treated with condition medium (CM) from human monocytes (Mo), PBMC-derived M0 macrophages, tumor-associated macrophage (TAM) and growth media (Control, Ctrl). And then cells were treated with indicated dose of erastin (ERA) for 48 h, and cell death was detected by PI staining using flow cytometer. (B, C) HeLa and/or SiHa cells were treated with CM from PBMC-derived M0 macrophages (Control) and TAM. And then cells were treated with indicated dose of ERA and/or RSL3 for 48 h. Inhibition ratio of cell viability was detected by CCK-8 assay (B). Cell staining PI was observed by confocal microscope (C). (D–F) HeLa cells were treated with TAM-CM. And then cells were treated with indicated dose of ERA and RSL3 for 48 h, lipid ROS was detected by staining C11-BODIPY 581/591 using flow cytometer (D, E), GSSG levels were detected by using a GSH/GSSG Ratio Detection Assay Kit (F). (G) The relative mRNA levels of CD206, CD163 and IL10 were detected in human PBMC-derived M0 macrophage, IL4-induced M2 and TAM. (H) HeLa cells were treated with CM from human monocyte, PBMC-derived IL4-induced M2 macrophages. And then cells were treated with indicated dose of ERA for 48 h, and cell death were detected. (I) HeLa and SiHa cells were treated with CM from human PBMC-derived M0 and M2 macrophages. And then inhibition ratio of cell viability was detected. (J, K) HeLa and SiHa cells were treated with M2-CM. And then cells were treated with ERA or RSL3 for 48 h, lipid ROS was detected by staining C11-BODIPY 581/591 using confocal microscope (J), GSSG levels were detected (K). (L–N) TAM decreased anti-tumor effects of ERA in mouse xenograft cervical cancer model. Representative images of xenograft tumors and tumor growth curve from different groups (L, M). Change in mice body weight was displayed (N). Data represented the mean ± SEM of at least three times biological replicates. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared to control.
Given lipid peroxidation is the essential event during ferroptosis25, we therefore investigated the effects of TAMs on the production of lipid ROS by erastin and RSL3. As shown in Fig. 1D and E, TAMs significantly inhibited the burst of lipid ROS by the two agents in HeLa cells. The same result was observed in sorafenib-treated HeLa cells (Fig. S1B). By detecting another two ferroptosis markers MDA and GSSG, we further validated TAMs inhibited ferroptosis in both cervical cancer cells HeLa and SiHa (Fig. 1F and Supporting Information Fig. S2A).
Previous study showed TAMs resembled M2-type macrophage9. We therefore determined whether M2 macrophage could also attenuated ferroptosis in cervical cancer cells. As shown in Fig. 1G, the levels of M2 macrophage markers CD206, CD163 and IL10 were obviously increased in TAMs and IL4-induced M2 macrophage (M2). Fig. 1H and I indicate that the cell death induced by erastin and RSL3 was significantly reduced by M2 CM. In addition, the induction of lipid ROS, GSSG and MDA by erastin and/or RSL3 was significantly weakened in HeLa and SiHa cells (Fig. 1J and K and Fig. S2B). In addition, we determined whether M1 macrophage also regulated ferroptosis in cervical cancer cells. As shown in Supporting Information Fig. S3A, the mRNA levels of M1 macrophage markers IL-1β and IL12 were obviously increased, while M2 macrophage marker IL10 were significantly decreased in M1 macrophage. Fig. S3B indicates that the cell death induced by erastin was not significantly changed by M1 macrophage.
To further test the effects of TAMs on cervical cancer cells ferroptosis in vivo, we evaluated the antitumor activity of erastin in HeLa cells mixed TAMs xenografts in nude mice. As shown in Fig. 1L and M and Supporting Information Fig. S4, erastin significantly inhibited the tumors growth, the antitumor effect of erastin were clearly attenuated by TAMs. In addition, no significant loss in body weight was observed in the mice treated with the erastin (Fig. 1N), suggesting that the toxic side effects of the drug were minimal in vivo.
3.2. TAMs suppress cervical cancer ferroptosis by exosomes
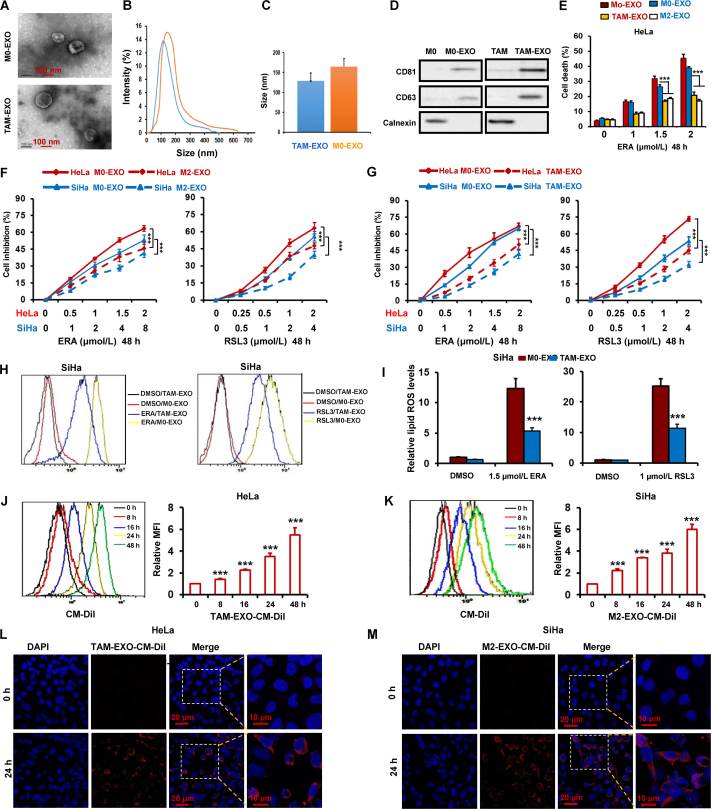
Previous studies demonstrated that microvesicles and exosomes in the tumor microenvironment promote tumor progression and metastasis26. To explore whether exosomes secreted from TAMs enhanced cervical cancer resistance to ferroptosis, the exosomes in TAMs-CM were removed by ultra-high-speed centrifugation, and then the TAMs-CM combined with erastin and RSL3 were used to treat HeLa and SiHa cells. As shown in Supporting Information Fig. S5A and S5B, both exosomes-free TAMs-CM (TAM supernatant, TAM-SP) and TAM-derived microparticles (TAM-MP) failed to abate the reduction of cell viability by the two ferroptosis inducers. Thus, we speculated that TAMs-derived exosomes could prevent ferroptosis. To test this hypothesis, exosomes in TAMs-CM were collected by centrifugation. The size distribution and morphology of exosomes were characterized by NTA and TEM respectively. As show in Fig. 2A–C, exosomes were heterogeneous in size with an intensity-weighted z-average diameter of 128 ± 25 nm in TAMs-CM and159 ± 21 nm in M0-CM. By Western blot, we further detected the positive markers CD63 and CD81 of exosomes, and negative marker Calnexin to characterize exosomes. Fig. 2D indicates that both CD63 and CD81 were high expressed in exosome of both M0 and TAM, low expressed in M0 and TAM. In addition, there was no expression of Calnexin in exosome of both M0 and TAM (Fig. 2D).
Figure 2.
TAMs suppress cervical cancer ferroptosis by exosomes. (A) Exosomes (EXO) secreted from TAM and M0 macrophages were collected and observed with an electron microscope. Scale bars are as indicated. (B) The intensity of exosomes of different sizes were quantified. (C) The diameter of exosomes derived from TAMs were quantified. Data are averages of exosomes from each group. (D) Exosomal markers were detected in the exosomes isolated from PBMC-derived M0, M2 macrophage and TAM by Western blot. (E) HeLa cells were treated with exosomes derived from human monocytes (Mo), PBMC-derived M0, M2 macrophages and TAM. And then cells were treated with ERA for 48 h, cell death were detected. (F, G) HeLa and SiHa cells were treated with exosomes from PBMC-derived M0, M2 macrophages and TAM. And then cells were treated with ERA and RSL3 for 48 h, the inhibition ratio of cell viability were detected. (H, I) SiHa cells were treated with exosomes from PBMC-derived M0 macrophages and TAM. And then cells were treated with ERA and RSL3 for 48 h, and then lipid ROS was detected. (J–M) HeLa and SiHa cells were treated with CM-Dil-labeled exosomes from PBMC-derived TAM and M2 macrophages for the indicated times. The internalization of fluorescently labeled exosomes in the two cells was detected by flow cytometer (J, K) and confocal microscopy (L, M). Scale bars are as indicated. Data represented the mean ± SEM of at least three times biological replicates. ∗∗∗P < 0.001, compared to control.
To evaluate the function of exosomes derived from macrophages in the regulation of cervical cancer cells ferroptosis, macrophages exosomes were isolated and co-cultured with HeLa cells treated with erastin. We found that TAMs- and M2-macrophages-derived exosomes significantly inhibited erastin-induced ferroptosis (Fig. 2E). Moreover, by CCK-8 assay, the cell viability was measured in HeLa and SiHa cells treated with TAMs- or M2-macrophaes-derived exosomes combined with erastin or RSL3 or sorafenib. Results indicate that both TAMs- and M2-macrophaes-derived exosomes obviously attenuated the decrease of cell viability by the three ferroptosis inducers (Fig. 2F, G and Fig. S5E and S5F). By detecting lipid ROS, MDA and GSSG, we further validated TAMs-derived exosomes inhibited ferroptosis in both cervical cancer cells HeLa and SiHa (Fig. 2H, I and Fig. S5C, S5D and S5G).
To visualize exosome uptake in cervical cancer cells, exosomes were isolated from TAMs and labeled with CM-DiI, and then HeLa and SiHa cells were incubated with CM-DiI-labeled exosomes for different time. And then, internalization of exosomes was detected by flow cytometry. As show in Fig. 2J and K, the number of cells with CM-DiI-labeled exosomes were increased in a time-dependent manner. By confocal microscopy, internalized fluorescence-labeled exosomes were observed at 0 and 24 h. As show in Fig. 2L and M, we found both HeLa and SiHa cells could absorb exosomes effectively.
3.3. Attenuation of cervical cancer ferroptosis by TAMs is involved in downregulation of ALOX15
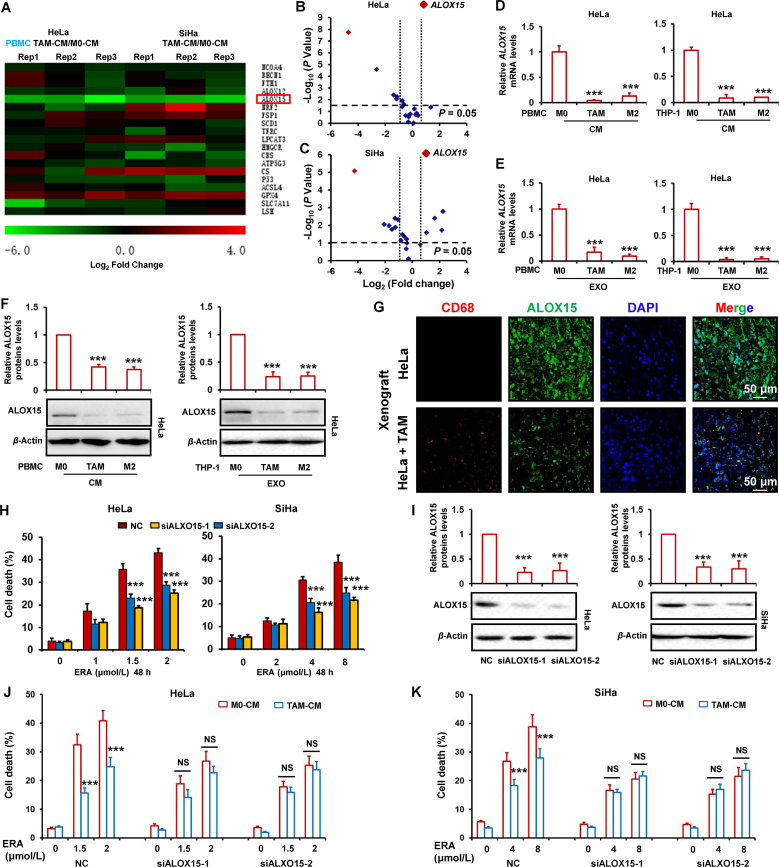
To explore the molecular mechanism of ferroptosis inhibition by TAMs, we performed RT-PCR to detect the mRNA levels of ferroptosis-associated gene including NCOA4, BECN1, FTH1, ALOX12, ALOX15, NRF2, FSP1, SCD1, TFRC, LPCAT3, HMGCR, CBS, ATP5G3, CS, P53, ACSL4, GPX4, SLC7A11 and LSH7. As shown in Fig. 3A, by comparing to M0-CM, we found TAMs-CM observably decreased the ALOX15 mRNA levels and slightly increased the GPX4 mRNA levels in both HeLa and SiHa cells. To simultaneously correlate the fold change on gene levels changed by TAMs-CM and the statistical significance at the global level, volcano plot was delineated by log2-fold change as x axis and a log10 (P value) as y axis. As shown in Fig. 3B and C, volcano plot shows the difference of gene expression in TAMs-CM treated groups relative to M0-CM group in HeLa and SiHa cells. The red dots represented the selected ALOX15 gene. These results show that ALOX15 was the most significant down-regulated gene by TAMs-CM in both cervical cancer cells. To further confirm expression of ALOX15 decreased by TAMs-CM, HeLa cells were treated with the condition media or purified exosomes of PBMC- and THP-1-derived TAMs or M2 macrophage. And then, ALOX15 mRNA and protein levels were detected by RT-PCR and Western blot respectively. As shown in Fig. 3D–F, the mRNA and protein levels of ALOX15 were markedly decreased by the condition media or exosomes form TAMs or M2 macrophages. Several vital ferroptosis regulators FSP1, ACSL4, DHODH and GPX4 protein expression were detected in HeLa cells treated with TAM-CM. Supporting Information Fig. S6A indicates that TAM failed to significantly change these regulators levels. Although TAMs-CM slightly increased the GPX4 mRNA levels in both HeLa and SiHa cells (Fig. 3A), no significant increase of GPX4 mRNA levels was found in HeLa cells treated with exosomes form TAMs (Fig. S6B).
Figure 3.
Attenuation of cervical cancer ferroptosis by TAM is involved in downregulation of ALOX15. (A–C) HeLa and SiHa cells were treated with CM from M0 macrophage and TAM for 48 h. Ferroptosis-associated gene was detected by RT-PCR. The ratio of genes expressions in cells treated with TAMs-CM relative to M0-CM were shown in the heatmap (A). Volcano map indicated the relationship between the observed fold change in gene expression and the P value significance of such changes in cells treated with CM. The horizontal dotted lines represented the P = 0.05, and vertical dotted line represented the 2-fold change cut-offs, the red dots represented the selected ALOX15 gene (B, C). (D, E) HeLa cells were treated with CM or exosome (EXO) from PBMC or THP-1-derived M0, M2 macrophage and TAM for 48 h. ALOX15 mRNA levels was detected by RT-PCR. (F) HeLa cells were treated with CM or EXO from PBMC or THP-1-derived M0, M2 macrophage and TAM for 48 h. ALOX15 protein levels was detected by Western blot. The relative quantification of protein was analyzed by scanning densitometry using Image J software and β-actin was used as the loading control. (G) TAM (THP-1) and HeLa in combination or HeLa alone were inoculated subcutaneously in nude mice. And then, the ALOX15 and macrophage marker CD68 expression was detected by immunofluorescence. (H, I) HeLa and SiHa cells were transfected with ALOX15 siRNAs for 48 h. Cell death induced by ERA was detected (H). ALOX15 protein levels were detected by Western blot. The relative quantification of protein was analyzed by scanning densitometry using Image J software and β-actin was used as the loading control (I). (J, K) HeLa and SiHa cells were transfected with ALOX15 siRNAs and treated by TAM-CM for 48 h. Cell death induced by ERA was detected. Data represented the mean ± SEM of at least three times biological replicates. NS, not significant; ∗∗∗P < 0.001, compared to control.
Moreover, we also detected the ALOX15 expression in xenograft tumor tissues. As shown in Fig. 3G, the ALOX15 expression was significantly reduced in HeLa cells mixed TAMs xenografts tumor tissues compared to only HeLa cells xenografts. To test the downregulation of ALOX15 was involved in ferroptosis inhibition, the ALOX15 expression was depleted by siRNAs in two cervical cancer cells, and then the cells were treated with erastin. As shown in Fig. 3H and I, knockdown of ALOX15 obviously reduced cell death induced by erastin. The induction of MDA and GSSG levels by erastin were significantly reduced by ALOX15 siRNAs in HeLa and SiHa cells (Supporting Information Fig. S7). To further prove that ALOX15 was involved in TAMs-inhibited cervical cancer cells ferroptosis, the ALOX15 was depleted by siRNAs in cancer cells treated with TAMs and the erastin-induced ferroptosis was investigated. As shown in Fig. 3J and K, when ALOX15 was depleted, TAMs failed to significantly attenuate cervical cancer ferroptosis.
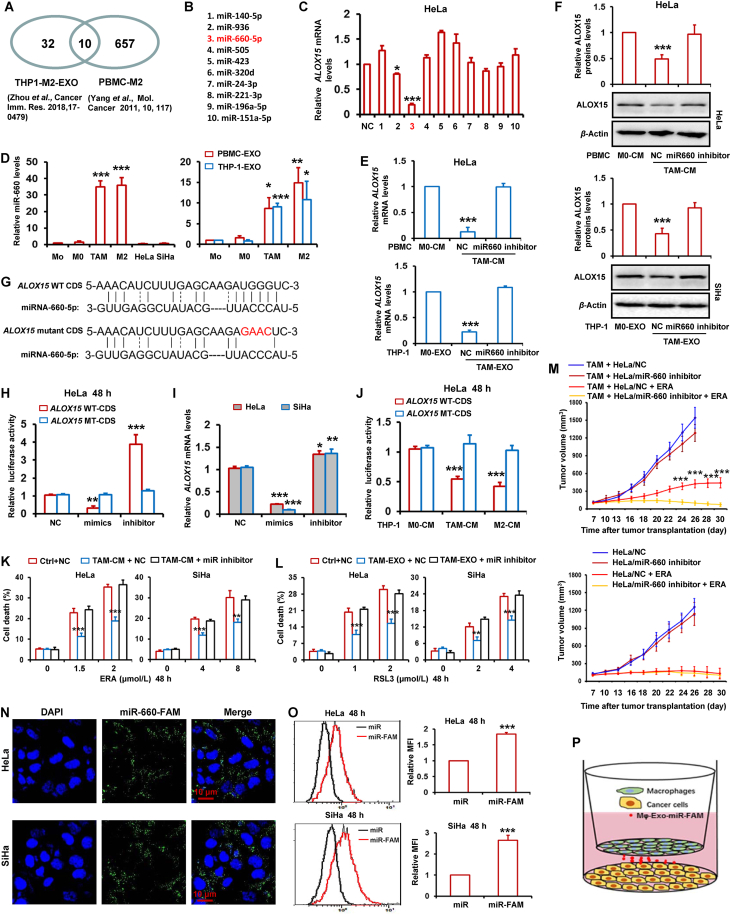
3.4. TAMs-derived exosomal miR-660-5p attenuates ALOX15 expression to inhibit ferroptosis in cervical cancer cells
Previous studies reported that macrophages release exosomes to shuttle microRNAs into adjacent cancer cells in tumor microenvironment26. Zhou et al.27 reported that 42 microRNAs were significantly upregulated in M2 macrophages-derived exosomes. Yang et al.26 reported 667 microRNAs were significantly increased in PBMC-derived M2 macrophages. We took the intersection of the microRNAs from these two studies and found 10 common microRNAs (Fig. 4A and B). Next, we transfected these 10 microRNAs mimics into HeLa cells and detected the ALOX15 expression. As shown in Fig. 4C, ALOX15 levels were attenuated by miR-936 and miR-660-5p. MiR-660-5p has the most obvious inhibitory effect on ALOX15 expression. In addition, miR-660-5p expression was strikingly higher in TAMs and M2 macrophages than Monocytes (Mo) and M0 macrophage, as well as HeLa and SiHa cells (Fig. 4D). The miR-660-5p levels was also detected in exosomes from TAMs, Mo, M0 and M2 macrophages. As shown in Fig. 4D, the miR-660-5p levels was significantly increased in TAMs and M2 macrophages-derived exosomes relative to Mo and M0 macrophages. To investigate the induction of miR-660-5p in cervical cancer cell by TAM or M2 macrophages, HeLa and SiHa cells were treated with CM from M2 macrophages and TAM. And then miRNA-660-5p levels were detected by RT-PCR in HeLa and SiHa cells. As shown in Supporting Information Fig. S8A, TAM and M2 macrophages significantly increased miRNA-660-5p levels of both cervical cancer cells.
Figure 4.
TAMs-derived exosomal miR-660-5p attenuates ALOX15 expression to inhibit ferroptosis in cervical cancer cells. (A, B) Flowchart for the selection of ten miRNAs. (C) HeLa cells were transfected with the ten miRNAs mimics, and then ALOX15 mRNA levels were detected by RT-PCR. (D) The miR-660-5p levels were detected by RT-PCR in PBMC-derived monocytes (Mo), M0, TAM, M2, HeLa, SiHa cells and exosomes (EXO) from PBMC and THP-1-derived Mo, M0, TAM, M2. (E, F) HeLa cells were transfected with miR-660-5p inhibitor, and then treated with CM and EXO from M0 and TAM. ALOX15 mRNA levels were detected by RT-PCR (E). ALOX15 protein levels were detected by Western blot. The relative quantification of protein was analyzed by scanning densitometry using Image J software and β-actin was used as the loading control (F). (G) The predicted miR-660-5p target sites in the coding sequence (CDS) of ALOX15 mRNA and the mutated version. (H) HeLa cells were co-transfected with pGL3 vector containing the wild type (WT) or mutated (MT) CDS of ALOX15, or pGL3-control vector, along with miR-660-5p mimics and inhibitor. After 48 h, luciferase activity was detected. Data were normalized to luciferase activity in the corresponding cells transfected with NC. (I) HeLa and SiHa cells were transfected with miR-660-5p mimics and inhibitor. ALOX15 mRNA levels were detected by RT-PCR. (J) HeLa cells were co-transfected with pGL3 vector containing the WT or MT CDS of ALOX15, and then cells were treated with CM from THP-1 derived M0, M2 macrophages and TAM. After 48 h, luciferase activity was detected. (K, L) HeLa and SiHa cells were transfected with miR-660-5p inhibitor, and then treated with TAM-CM or TAM-EXO and indicated dose of ERA or RSL3 for 48 h. Cell death was detected. (M) HeLa cells stably expressing miR-660-5p inhibitor alone or mixed with TAM were injected into nude mice. Nude mice were administered with ERA as indicated at each time point. Tumor volume was measured once per 2–3 days by using calipers for 30 days. (N–P) HeLa and SiHa cells were co-cultured with TAM transfected with miR-660-5p-FAM for 48 h. And then the capture of TAM-exosome delivered miR-660-5p-FAM by HeLa or SiHa cells were observed by confocal microscope (N), and quantitatively analyzed by flow cytometry (O). Schematic diagram for the cell co-culture model of TAM and HeLa or SiHa cells (P). Data are represented as the mean ± SEM of three replicates. For mice model studying, data are represented as the mean ± SEM of 5 mice. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared to control.
To investigate the role of miR-660-5p in mediating TAMs-inhibited the ALOX15 expression of cervical cancer cells, HeLa cells were transfected with miR-660-5p inhibitor, and then treated with the CM and exosomes of M0 macrophage and TAMs. By RT-PCR and Western blot, the mRNA and protein levels of ALOX15 were detected respectively. Fig. 4E and F indicate that ALOX15 expression was significantly suppressed by the CM or exosomes of TAMs relative to that of control M0 macrophages, while the reduction of ALOX15 expression by the CM or exosomes of TAMs was clearly reversed by miR-660-5p inhibitor.
Since microRNAs attenuated gene expression mainly through binding to the mRNA of gene, we thus predicted whether miR-660-5p bound to the mRNA of ALOX15 gene. By using online miRNA target bioinformatics prediction databases miRWalk, the conserved miR-660-5p target sites in the coding sequence (CDS) of ALOX15 were predicted (Fig. 4G). To further confirm targeting of ALOX15 by miR-660-5p, luciferase activity assay was performed. The wild type (WT) or mutated (MT) CDS of ALOX15 were cloned into the downstream of firefly luciferase coding region in pGL3 luciferase reporter vector. The vectors were cotransfected with miR-660-5p mimics or inhibitor into HeLa cells. As expected, we showed that miR-660-5p mimics sharply decreased luciferase activity, whereas miR-660-5p inhibitor obviously increased luciferase activity (Fig. 4H). Additionally, no obvious change of luciferase activities by miR-660-5p mimics and inhibitor was observed in HeLa cells transfected with MT reporter vectors (Fig. 4H). In HeLa and SiHa cells, we also showed mimics and inhibitor could evidently decrease and induced ALOX15 mRNA expression respectively (Fig. 4I). HeLa cells transfected with luciferase reporter vector with the WT or MT CDS of ALOX15 was further treated with TAMs- and M2 macrophages-CM, and then luciferase activity was detected. Fig. 4J shows that TAMs- and M2 macrophages-CM significantly down-regulated luciferase activity in HeLa cells transfected reporter vector with WT CDS but not MT CDS.
To investigate the role of miR-660-5p in TAMs-inhibited ferroptosis, HeLa and SiHa cells were transfected with miR-660-5p inhibitor, and incubated with the CM and exosomes of TAMs. And then these cells were treated with erastin and RSL3. By detecting cell death, we found that the reduction of cell death by CM and exosomes were completely blocked by miR-660-5p inhibitor (Fig. 4K and L). Next, TAMs-derived exosomes were treated with RNase plus Triton X-100 and incubated those exosomes and erastin with HeLa cells. Fig. S8B indicates that TAMs-derived exosomes significantly inhibited ferroptosis, whereas TAMs-derived exosomes treated with RNase plus Triton X-100 failed to attenuate erastin-induced cell death compared to the control M0 macrophages exosomes group. By RT-PCR and luciferase activity assay, we also showed that TAMs-derived exosomes treated with RNase plus Triton X-100 failed to attenuate ALOX15 mRNA expression (Fig. S8C and S8D). In addition, we treated HeLa and SiHa cells with miRNA-660-5p inhibitor plus ferroptosis inducer erastin, and then detected cell death. As shown in Supporting Information Fig. S9A and S9B, miRNA-660-5p inhibitor failed to significantly increased cell death. This result suggests that the expression of endogenous miRNA-660-5p in cervical cancer cells was too low to affect cell ferroptosis response.
Moreover, a tumor xenograft nude mouse model was used to evaluate the role of miR-660-5p in mediating the TAMs-induced ferroptosis resistance of cervical cancer in vivo. As indicated in Fig. 4M and Supporting Information Fig. S10C and S10D, after erastin treatment, xenograft tumors with miR-660-5p inhibitor high-expressed HeLa cells and TAMs in combination grew much faster and larger than the tumors with TAMs and control HeLa. However, in those tumors with HeLa alone, miR-660-5p inhibitor failed to change tumor sensitivity to erastin. Additionally, there was no significant change in body weight of mice injected with erastin (Fig. S10A and S10B).
To directly observe the TAMs-derived miRNA into tumor cells, HeLa and SiHa cells were co-cultured with TAMs transfected with FAM-labeled miR-660-5p mimics in cervical cancer cells (Fig. 4P), and observed by fluorescence microscopy. Fig. 4N displayed that the presence of FAM fluorescence in both HeLa and SiHa cells. Furthermore, by Flow cytometry analysis, we also showed that FAM fluorescence was increased in both cancer cells transfected with FAM-labeled microRNA (Fig. 4O).
Previous study has demonstrated that CAFs-secreted miR-522 suppresses cancer cell ferroptosis by reducing ALOX15 expression12. Thus, we investigated whether CAFs could regulate cervical cancer ferroptosis. Supporting Information Fig. S11A indicates that cell death induced by erastin was not significantly changed by CAFs and NFs. Moreover, the miR-660-5p was low expressed in CAFs and NF relative to TAM (Fig. S11B).
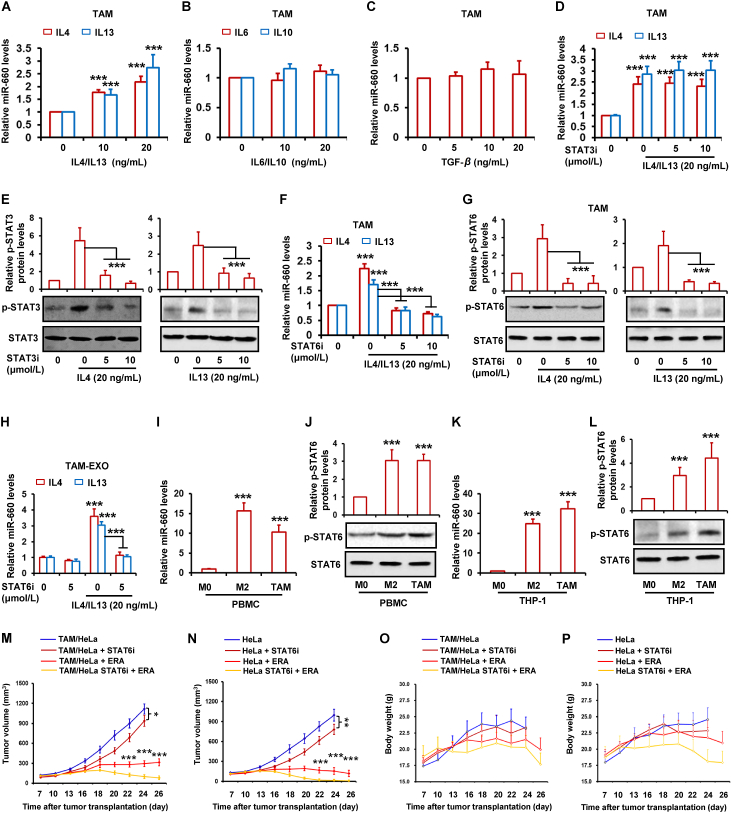
3.5. IL4/IL13 induces miR-660-5p expression by activating STAT6 pathway
Here, we found that miR-660-5p was overexpressed in both TAMs and M2 macrophages. It has been reported that many of the properties of TAMs are also the characteristics of M2 macrophages. Several cytokines are expressed in both cells, such as IL6, IL10, IL4, IL13 and TGF-β. Therefore, we wanted to know whether the expression of these factors was related to miR-660-5p. By RT-PCR assay, we showed that miR-660-5p levels were increased by IL4 and IL13 in dose-dependent manner in TAMs (Fig. 5A), whereas IL6, IL10 and TGF-β did not influence the miR-660-5p levels (Fig. 5B and C). Previous studies indicated STAT3 and STAT6 activation are responsible for IL4 and IL13. Therefore, we supposed the activated STAT3 and STAT6 by IL4 and IL13 in TAMs was relevant to miR-660-5p expression. By inhibiting the activation of STAT3 and STAT6 in TAMs, we found that the induction of miR-660-5p by IL4 and IL13 was significantly attenuated by STAT6 inhibitor not STAT3 inhibitor (Fig. 5D and F). Results of Western blot show that the p-STAT6 and p-STAT3, which is the activated STAT6 and STAT3, were obviously decreased by the two inhibitors respectively (Fig. 5E and G). Fig. 5H also indicates that both IL4 and IL13 significantly increased miR-660-5p levels in TAMs exosomes, whereas the induction of miR-660-5p levels by IL4 and IL13 was distinctly abated. By detecting miR-660-5p and p-STAT6 expression in macrophages, we found the levels of both miR-660-5p and p-STAT6 were obviously higher in TAMs and M2 macrophages relative to M0 macrophages (Fig. 5I–L). To investigate whether inhibition of miR-660-5p expression by STAT6 inhibitor attenuated TAM-induced ferroptosis resistance, the xenograft tumors with Hela mixed TAM were treated with STAT6 inhibitor and erastin in combination. As shown in Fig. 5M, STAT6 inhibitor significantly attenuated TAM-induced resistance to erastin. However, in xenograft tumors with Hela alone, we found STAT6 inhibitor also enhanced antitumor effect of erastin (Fig. 5N). In both tumor models, STAT6 inhibitor alone exerted certain antitumor effect (Fig. 5M and N). This suggests STAT6 inhibitor enhanced erastin-antitumor effect might be involved in both inhibition of miR-660-5p expression in TAM and inhibition of STAT6 activity in tumor cells. Additionally, there was no significant change in body weight of mice injected with erastin and STAT6 inhibitor (Fig. 5O and P).
Figure 5.
IL4/IL13 induces miR-660-5p expression by activating STAT6 pathway. (A–C) THP-1-derived TAM was treated with indicated dose of IL4, IL13, IL6, IL10 and TGF-β for 48 h. RT-PCR was performed to detect the expression of miR-660-5p. (D–H) THP-1-derived TAM was treated with indicated dose of STAT3 inhibitor (STAT3i) or STAT6 inhibitor (STAT6i) for 2 h, and then treated with 20 ng/mL of IL4 or IL13 for 48 h. RT-PCR was performed to detect the expression of miR-660-5p in TAM (D, F) and TAM-exosome (H). STAT3, p-STAT3, STAT6 and p-STAT6 protein levels were detected by Western blot (E, G). (I–L) miR-660-5p levels were detected in PBMC or THP-1-derived M0, M2 and TAM by RT-PCR (I, K). STAT6 and p-STAT6 protein levels were detected by Western blot (J, L). (M–P) HeLa alone or mixed with TAM cells were injected into nude mice (n = 5). Nude mice were administered with ERA and STAT6i as indicated at each time point. Tumor volume was measured once per 2–3 days by using calipers for 30 days (M, N). Change in mice body weight was displayed (O, P). The relative quantification of protein was analyzed by scanning densitometry using Image J software and β-actin was used as the loading control. Data are represented as the mean ± SEM of three replicates. For mice model studying, data are represented as the mean ± SEM of 5 mice. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared to control.
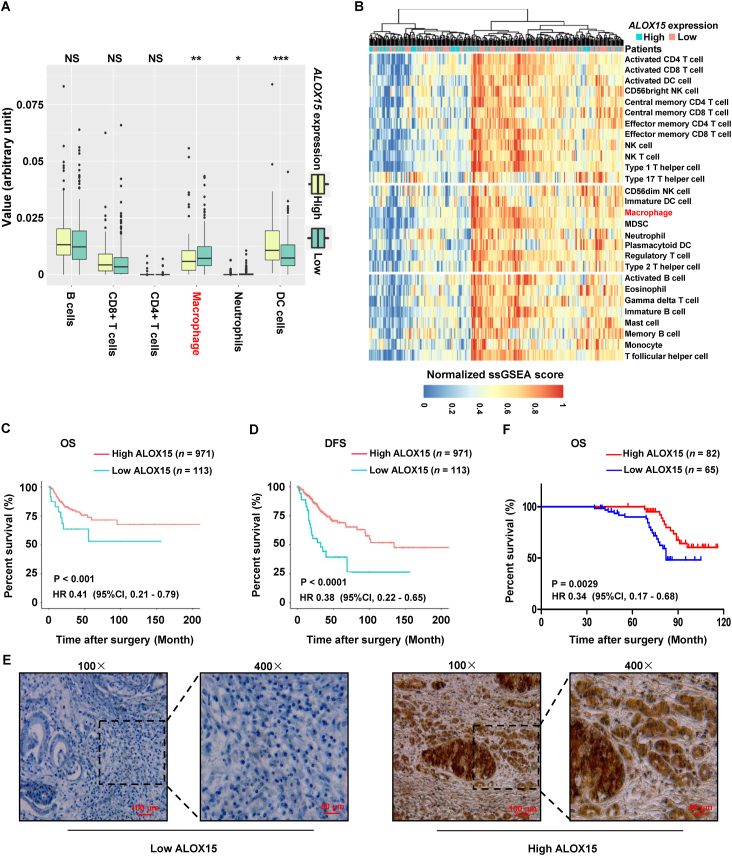
3.6. ALOX15 is negatively associated with macrophages infiltration and positively linked with good prognosis
To further confirm TAMs suppressed ALOX15 expression in cervical cancer, abundance of immune cell populations was analyzed in high-expressed versus low-expressed ALOX15 cervical cancer tissues in TCGA. Fig. 6A shows that abundance of macrophages in high-expressed ALOX15 cervical cancer tissues was significantly lower. Moreover, we composed a heatmap to visualize the relative abundance of 28 infiltrating immune cell populations (Fig. 6B). Results show that normalized ssGSEA score of macrophages was significantly higher in low-expressed ALOX15 cervical cancer tissues relative to high-expressed ALOX15 tissues. Above results raised the possibility that infiltrated TAMs suppressed ALOX15 expression of cervical cancer cell.
Figure 6.
ALOX15 is associated with macrophages infiltration and good prognosis. (A) Infiltration of immune cells including TAMs in cervical cancer tissues with high or low expression of ALOX15 by using ssGSEA and CIBERSORT. The relative numerical values corresponding to the height of the histogram indicated the different levels of abundance and the proportions. NS, not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, high expression of ALOX15 compared to low expression. (B) Single-sample GSEA analysis identifying the relative infiltration of immune cell populations for 311 cervical cancer samples with available RNA-sequencing data in TCGA. The relative infiltration of each cell type is normalized into a z-score. (C, D) Association of ALOX15 mRNA expression with the disease-free survival (DFS) and overall survival (OS) in cervical cancer patients, respectively. These data of cervical cancer samples were acquired from TCGA. (E) Representative immunohistochemical staining for high and low expression of ALOX15 in human cervical cancer sample. (F) Association of ALOX15 protein expression with the OS in 147 cervical cancer patients.
Additionally, ALOX15 expression is positively associated with OS and disease-free survival in cervical cancer in TCGA (Fig. 6C and D). Moreover, the association of ALOX15 expression and prognosis were further analyzed in cervical cancer from Zhengzhou University. By using IHC staining, ALOX15 expression was investigated in 147 paraffin-embedded archived cervical cancer tissues. The tissues included 53 stage Ib1, 28 stage Ib2, 31 stage IIa1 and 35 stage IIa2 tumors. Among the 147 samples, high expression of ALOX15 protein were detected in 82 samples, and low levels of ALOX15 were observed in the remaining 65 samples (Table 1 and Fig. 6E). Patient survival analysis showed a clear positive correlation between ALOX15 expression and OS of cervical cancer patients (Fig. 6F).
Table 1.
Correlation between ALOX15 expression and clinicopathological features of cervical cancer.
| Variables | Total | ALOX15 expression |
Pa | |
|---|---|---|---|---|
| High | Low | |||
| Age, years | ||||
| ≥42 | 120 | 71 | 49 | |
| <42 | 27 | 11 | 16 | 0.1268 |
| FIGO stage | ||||
| Ib1 | 53 | 28 | 25 | |
| Ib2 | 28 | 14 | 14 | |
| IIa1 | 31 | 15 | 16 | |
| >IIa2 | 35 | 25 | 10 | 0.098 |
| Tumor size, cm | ||||
| <4 | 79 | 56 | 23 | |
| ≥4 | 68 | 26 | 42 | 0.0001 |
| Histological type | ||||
| Squamous cell carcinoma | 122 | 69 | 53 | |
| Adeno cell carcinoma | 25 | 13 | 12 | 0.8438 |
| Histological differentiation | ||||
| Well | 42 | 24 | 18 | |
| Moderate | 53 | 26 | 27 | |
| Poor | 52 | 32 | 20 | 0.3891 |
| Deep stromal invasion | ||||
| <1/2 | 36 | 24 | 12 | |
| >1/2 | 111 | 58 | 53 | 0.1867 |
| Lymphovascular space involvement | ||||
| No | 139 | 78 | 61 | |
| Yes | 8 | 4 | 4 | 0.9781 |
| Pelvic lymph node metastasis | ||||
| No | 98 | 61 | 37 | |
| Yes | 49 | 17 | 32 | 0.0381 |
| Positive parametrium | ||||
| No | 140 | 76 | 64 | |
| Yes | 7 | 6 | 1 | 0.2135 |
| Positive surgical margin | ||||
| No | 139 | 79 | 60 | |
| Yes | 8 | 3 | 5 | 0.4810 |
| Vaginal involvement | ||||
| No | 136 | 77 | 59 | |
| Yes | 11 | 5 | 6 | 0.688 |
| T classificationb | ||||
| T1b1 | 51 | 28 | 23 | |
| T1b2 | 31 | 15 | 16 | |
| T2a | 32 | 14 | 18 | |
| >T2b | 33 | 25 | 8 | 0.0775 |
| Recurrence | ||||
| No | 101 | 67 | 34 | |
| Yes | 46 | 15 | 31 | 0.0003 |
| Vital status in 5 years | ||||
| Alive | 133 | 81 | 52 | |
| Dead | 14 | 1 | 13 | 0.0005 |
P value from Fisher's exact test; The italic number inside the table reflected P < 0.05.
FIGO 2009 was used.
According to the IHC score in cervical cancer patients, the correlation between ALOX15 levels and clinicopathological features was analyzed. As shown in Table 1, high ALOX15 expression was significantly negatively correlated with tumor size, recurrence and vital status in 5 years. However, other poor clinicopathological features, such as age and FIGO stage were not significantly correlated with ALOX15 expression. To verify the relationship between ALOX15 expression and the prognosis in cervical cancer, we performed univariate and multivariate Cox analyses. Univariate analysis showed that tumor size, lymphovascular space involvement, T classification, pelvic lymph node metastasis and ALOX15 expression were prognostic factors for OS (Table 2). Multivariate analysis showed that lymphovascular space involvement, T classification and ALOX15 expression were independent prognostic factors (Table 2).
Table 2.
Univariate multivariate Cox regression analysis of clinicopathological features associated with overall survival in cervical cancer.
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI)a | P | HR (95% CI) | P | |
| Tumor size, cm | ||||
| <4 | 1 | 1 | ||
| ≥4 | 0.384 (0.191–0.773) | 0.007 | 0.579 (0.281–1.192) | 0.138 |
| Lymphovascular space involvement | ||||
| No | 1 | 1 | ||
| Yes | 0.27 (0.117–0.623) | 0.002 | 0.203 (0.077–0.533) | 0.001 |
| T classification | ||||
| T1b1, T1b2 | 1 | 1 | ||
| T2a, >T2b | 0.35 (0.167–0.733) | 0.005 | 0.343 (0.149–0.788) | 0.012 |
| Pelvic lymph node metastasis | ||||
| No | 1 | 1 | ||
| Yes | 0.428 (0.216–0.848) | 0.016 | 1.308 (0.583–2.934) | 0.515 |
| ALOX15 | ||||
| Low | 1 | 1 | ||
| High | 0.311 (0.151–0.642) | 0.002 | 4.69 (1.939–11.343) | 0.001 |
95% CI, 95% confidence interval; HR, hazard ratio.
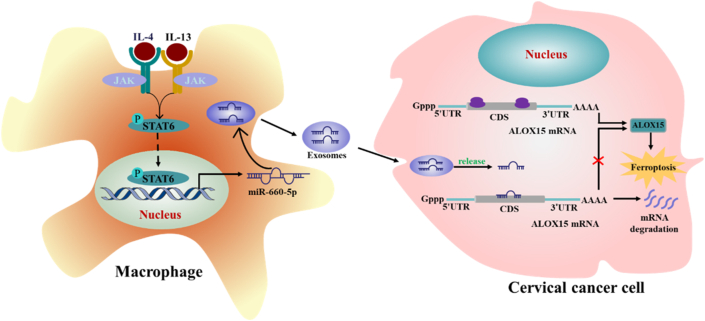
Finally, we provided a schematic diagram to show mechanism that TAM enhanced cervical cancer resistance to ferroptosis (Fig. 7).
Figure 7.
Schematic diagram that TAM enhanced cervical cancer resistance to ferroptosis. A proposed model illustrating the role of TAM-derived exosomal miR-660-5p in regulating ALOX15-induced ferroptosis in cervical cancer cells.
4. Discussion
Ferroptosis plays an essential role in mediating tumor development and drugs resistance28,29. Activation of ferroptosis related pathways enhances effects of chemotherapy, radiotherapy and targeted therapy to effectively prevent tumor progression7. Recent reports showed that targeting ferroptosis is a very promising treatment for cancer therapy in some types of cancer, such as breast cancer and cervical cancer30,31. For example, in in vitro cervical cancer cell, propofol increases cells ferroptosis induced by paclitaxel31. Oleanolic acid has been reported to induce Hela cell ferroptosis in ACSL4-dependent manner32. However, ferroptosis inducer is rarely reported in the treatment of cervical cancer in vivo. Here, we firstly showed that ferroptosis inducers exerted potent antitumor effects in vivo in cervical cancer. The tumor microenvironment has been showed to play a crucial role in cancer drugs resistance33. For example, CD8+ T cells in microenvironment are found to induce tumor ferroptosis during cancer immunotherapy by releasing IFNγ. Mechanistically, IFNγ downregulates the levels of SLC3A2 and SLC7A11, two subunits of the glutamate–cystine antiporter system Xc-in tumor cells28. As a result, the uptake of cystine is impaired, which promotes tumor cell lipid peroxidation and ferroptosis28. Recent study showed that cancer-associated fibroblasts secrete exosomal miR-522 to inhibit gastric cancer cells ferroptosis12. Several studies reported that there is crosstalk between cancer cells and macrophage in the tumor microenvironment by exosomes. For example, in pancreatic cancer, tumor cells-derived exosome delivers FGD5-AS1 to promotes macrophage M2 polarization, while M2 macrophage feedback promotes cancer cell proliferation and metastasis34. In hepatocellular carcinoma, the exosomes derived from cancer cells transport PD-L1 into macrophages and thus upregulating the expression of PD-L1 on macrophages. Macrophages further inhibit the activity of CD8+ T cell to promote tumor growth35. In cervical cancer, cancer cell-secreted exosomes deliver TIE2 protein to TAMs to induce angiogenesis36. Macrophages-derived exosomes can also affect tumor cells function26. However, the regulation of TAMs-derived exosomes on tumor cell function in cervical cancer has not been reported. Moreover, the potential functions of macrophages in regulating ferroptosis of tumor cells are still unclear. Macrophage as an important component of tumor microenvironment exerts dual functions to modulate the efficacy of various forms of anticancer drugs33. Some studies showed that M1 type macrophage increases the tumor sensitivity to drugs, while others showed that M2 type macrophage attenuates the cytotoxicity of drugs33. TAMs that resembles the alternatively activated M2 type macrophage also showed to enhance tumor resistance to drugs33. In this study, we found both M2 type macrophages and TAMs increased cervical cancer cell sensitivity to ferroptosis inducers.
The signal communication modes between macrophages and tumor cells are usually considered to be physical contact and cytokine or chemokine secretion37. There is also compelling evidence showed TAMs attenuate the efficacy of drugs by directly secreting several cytokines such as IL6 and EGF in tumor microenvironment38,39. In addition, exosomes or microvesicles secreted by TAMs has been reported to mediate the drugs resistance in several cancer types40. Exosomes or microvesicles exert its biological functions mainly through delivering proteins or microRNAs into adjacent cells41. Here, the RNA in macrophages-derived exosomes were degraded by using RNase and Triton X-100. And then cervical cancer cells were incubated with the RNA-depleted exosomes. Results showed the depletion of RNAs in exosomes significantly reversed the effects of exosomes-decreased ferroptosis. These results suggested TAMs or M2 macrophages-derived exosomes carried miRNAs to shuttle into cervical cancer cells, and thus promoting cancer cells resistance to ferroptosis inducers. MiRNAs acting as oncogenes or tumor suppressors are involved in the pathogenesis of cancer by negatively regulating gene expression. In this study, macrophages-secreted miR-660-5p was found to inhibit cervical cancer cells ferroptosis by downregulating ALOX15 expression. Previous studies reported miR-660 is downregulated in several cancer types such as lung cancer42. We also found that miR-660-5p was expressed at a relatively low levels in cervical cancer cells. However, in hepatocellular carcinoma and breast cancer, miR-660-5p was significantly high-expressed and promoted cancer progression43,44. Although we showed TAMs-derived miR-660-5p attenuated cervical cancer sensitivity to ferroptosis inducers, the others function of the miRNA in cervical cancer remained unclear and needed further evaluation. In this study, we showed that inhibition of STAT6 activity decreased miR-660-5p levels in TAM. However, we had not been elucidated the underlying molecular mechanism. Previous study found that STAT6 regulates microRNA expression at the transcriptional or mRNA maturation level in both human and mouse macrophage45. This hinted STAT6 might promote miR-660-5p expression at the transcriptional or mRNA maturation level. This needs further validation in the further study.
By detecting the mRNA levels of ferroptosis-associated gene, we indicated that attenuation of cervical cancer ferroptosis by TAMs was involved in downregulation of ALOX15. Further study certified that TAMs-derived miR-660-5p was shuttled into cancer cells to inhibit ALOX15 expression. ALOX15 is a member of lipoxygenases family, the lipid peroxidizing enzymes, and has been implicated in a number of physiological processes46. Recent studies showed that ALOX15 is capable of inducing cells ferroptosis by oxidizing membrane phospholipids in some cancers such as colon cancer47. However, the association between ALOX15 and tumor progression has not been reported in most of solid tumor except colon cancer. Here, we show ALOX15 expression was significantly negatively associated with overall patient survival and disease-free survival in cervical cancer. ALOX15 expression has been reported to be downregulated in colorectal cancer and suppress premetastatic stages of colonic tumorigenesis48. Hypoxia is a common feature in solid tumor microenvironment. ALOX15 has been proved to inhibit colon cancer progression through modulation of hypoxia response49. Moreover, ALOX15 can also downregulate the expression of the oncogenes XIAP and Bcl-XL, to trigger colon cancer cell apoptosis and inhibit cell survival in vitro and in vivo47. The above results were consistent with our finding that ALOX15 was a tumor suppressor gene. Additionally, we also found that there was a significantly negative correlation between macrophage infiltration and ALOX15 expression in cervical cancer. Infiltration of macrophages has been reported to be conferred poor prognosis in many tumor types such as breast cancer48. In cervical cancer, recent study found TAMs promoted cancer metastasis by secreting IL1049, and mediated anti-tumor immune responses50. Another study found that hypoxia-induced ZEB1 in cervical cancer cells promoted cancer metastasis and progression by recruiting TAMs to foster a prometastatic environment51. These studies proposed another possibility that the downregulation of ALOX15 was not the cause of cervical cancer progression, but was only a results of macrophages infiltration.
5. Conclusions
Altogether, we illustrated the novel function of TAMs in suppressing ferroptosis of cervical cancer cells by secreting exosomes to deliver special signaling molecule. Our study also provides new insights into the mechanisms of TME-inhibited cancer cell death. Moreover, STAT6 inhibitor enhanced antitumor effect of ferroptosis inducer, suggesting that the combined application of STAT6 inhibitor and ferroptosis inducer may be a new promising treatment for cervical cancer. ALOX15 expression was found be significantly negatively associated with overall patient survival and disease-free survival, suggesting ALOX15 might be a prognostic marker in cervical cancer.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81972479, U2004118, 82072899, 81772643 and 81772803), Natural Science Foundation of Guangdong province (2019A1515011100 and 2021A1515012576, China), Henan Natural Science Foundation (202300410359, China) and Henan Medical Program (SBGJ2020002081, China). Guangzhou High-Level Clinical Key Specialty Construction Project; Clinical Key Specialty Construction Project of Guangzhou Medical University (202005, China), the Innovation Project of Universities in Guangdong Province (NO. 2021KTSCX026, China). Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University (QNCXTD2023019, China)
Author contributions
Zhengzhi Zou, Yanlin Luo, Yibing Chen and Quentin Liu developed the concept. Yanlin Luo, Yibing Chen, Huan Jin, Yong Sang Song and Zhengzhi Zou designed and performed experiments, analyzed and interpreted data, and co-wrote the manuscript. Benxin Hou, Xiang Li, Lingfeng Liu, Yuan Zhou, Hongsheng Li and Yonghua Li contributed to experimental and helped with data interpretation. All authors edited and reviewed the final manuscript.
Conflicts of interest
The authors declare no conflicts of interests.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.03.025.
Contributor Information
Yibing Chen, Email: chenyibing@zzu.edu.cn.
Zhengzhi Zou, Email: zouzhengzhi@m.scnu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Cohen P.A., Jhingran A., Oaknin A., Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 2.Rose P.G., Bundy B.N., Watkins E.B., Thigpen J.T., Deppe G., Maiman M.A., et al. Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med. 1999;340:1144–1153. doi: 10.1056/NEJM199904153401502. [DOI] [PubMed] [Google Scholar]
- 3.Ferrandina G., Lauriola L., Distefano M.G., Zannoni G.F., Gessi M., Legge F., et al. Increased cyclooxygenase-2 expression is associated with chemotherapy resistance and poor survival in cervical cancer patients. J Clin Oncol. 2002;20:973–981. doi: 10.1200/JCO.2002.20.4.973. [DOI] [PubMed] [Google Scholar]
- 4.Leisching G., Loos B., Botha M., Engelbrecht A.M. Bcl-2 confers survival in cisplatin treated cervical cancer cells: circumventing cisplatin dose-dependent toxicity and resistance. J Transl Med. 2015;13:328. doi: 10.1186/s12967-015-0689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L.N., Chen Y.B., Mi Y.J., Qiao J.H., Jin H., Li J., et al. ATF2 inhibits ani-tumor effects of BET inhibitor in a negative feedback manner by attenuating ferroptosis. Biochem Biophys Res Commun. 2021;558:216–223. doi: 10.1016/j.bbrc.2020.08.113. [DOI] [PubMed] [Google Scholar]
- 7.Tang D.L., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y.B., Mi Y.J., Zhang X., Ma Q., Song Y.C., Zhang L., et al. Dihydroartemisinin-induced unfolded protein response feedback attenuates ferroptosis via PERK/ATF4/HSPA5 pathway in glioma cells. J Exp Clin Cancer Res. 2019;38:402. doi: 10.1186/s13046-019-1413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y.B., Jin H., Song Y.C., Huang T., Cao J., Tang Q., et al. Targeting tumor-associated macrophages: A potential treatment for solid tumors. J Cell Physiol. 2021;236:3445–3465. doi: 10.1002/jcp.30139. [DOI] [PubMed] [Google Scholar]
- 10.D’Errico G., Alonso-Nocelo M., Vallespinos M., Hermann P.C., Alcalá S., García C.P., et al. Tumor-associated macrophage-secreted 14-3-3ζ signals via AXL to promote pancreatic cancer chemoresistance. Oncogene. 2019;38:5469–5485. doi: 10.1038/s41388-019-0803-9. [DOI] [PubMed] [Google Scholar]
- 11.Dong N.Y., Shi X.Y., Wang S.H., Gao Y.J., Kuang Z.Z., Xie Q., et al. M2 macrophages mediate sorafenib resistance by secreting HGF in a feed-forward manner in hepatocellular carcinoma. Br J Cancer. 2019;121:22–33. doi: 10.1038/s41416-019-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H.Y., Deng T., Liu R., Ning T., Yang H., Liu D.Y., et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi X.F., Xu R.Y., Hong S.Z., Xu T.T., Zhang W.F., Liu M. M2 macrophage-derived exosomal LNCRNA AFAP1-AS1 and microRNA-26a affect cell migration and metastasis in esophageal cancer. Mol Ther Nucleic Acids. 2020;22:779–790. doi: 10.1016/j.omtn.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan W., Jiang S. Immune cell-derived exosomes in the cancer-immunity cycle. Trends Cancer. 2020;6:506–517. doi: 10.1016/j.trecan.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Yan Y., Du C.C., Duan X.X., Yao H.H., Wan J.J., Jiang A.M., et al. Inhibiting collagen I production and tumor cell colonization in the lung via miR-29a-3p loading of exosome-/liposome-based nanovesicles. Acta Pharm Sin B. 2022;12:939–951. doi: 10.1016/j.apsb.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z., Ma T.T., Huang B.W., Lin L.H., Zhou Y.H., Yan J.Y., et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. 2019;38:310. doi: 10.1186/s13046-019-1313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ao X., Nie P.P., Wu B.Y., Xu W., Zhang T., Wang S.M., et al. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463. doi: 10.1038/cddis.2016.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou Z.Z., Yuan Z.Y., Zhang Q.X., Long Z.J., Chen J., Tang Z., et al. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798–1810. doi: 10.4161/auto.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z.L., Chen X.W., Liu N., Shi Y., Liu Y.T., Ouyang L., et al. A nuclear long non-coding RNA LINC00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol Ther. 2021;29:263–274. doi: 10.1016/j.ymthe.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P.J., Luo X.Y., Nie P.P., Wu B.Y., Xu W., Shi X., et al. CQ synergistically sensitizes human colorectal cancer cells to SN-38/CPT-11 through lysosomal and mitochondrial apoptotic pathway via p53-ROS cross-talk. Free Radic Biol Med. 2017;104:280–297. doi: 10.1016/j.freeradbiomed.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H.Y., Deng T., Liu R., Bai M., Zhou L.K., Wang X., et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2018;2018:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M., Chen J.Q., Su F., Yu B., Su F.X., Lin L., et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J.R., Li X.D., Wu X.L., Zhang T., Zhu Q.Y., Wang X.J., et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6:1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 28.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X.M., Zou L.B., Liao H.Y., Luo J.Q., Yang T.W., Wu J., et al. Abrogation of HnRNP L enhances anti-PD-1 therapy efficacy via diminishing PD-L1 and promoting CD8+ T cell-mediated ferroptosis in castration-resistant prostate cancer. Acta Pharm Sin B. 2022;12:692–707. doi: 10.1016/j.apsb.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Y.H., Chen X.P., Liu C., Ge W.Z., Wang Q., Hao X., et al. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J Hematol Oncol. 2021;14:19. doi: 10.1186/s13045-020-01016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M.Y., Liu P., Sun C., Pei L.J., Huang Y.G. Propofol augments paclitaxel-induced cervical cancer cell ferroptosis in vitro. Front Pharmacol. 2022;13:816432. doi: 10.3389/fphar.2022.816432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X.F., Shi M.Q., Sui M., Yuan Y.Z., Zhang S., Xia Q., et al. Oleanolic acid inhibits cervical cancer Hela cell proliferation through modulation of the ACSL4 ferroptosis signaling pathway. Biochem Biophys Res Commun. 2021;545:81–88. doi: 10.1016/j.bbrc.2021.01.028. [DOI] [PubMed] [Google Scholar]
- 33.Ruffell B., Coussens L.M. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z.W., Wang J., Zhu C.H., Xu J., Chen P., Jiang X.Y., et al. Exosome-derived FGD5-AS1 promotes tumor-associated macrophage M2 polarization-mediated pancreatic cancer cell proliferation and metastasis. Cancer Lett. 2022;548:215751. doi: 10.1016/j.canlet.2022.215751. [DOI] [PubMed] [Google Scholar]
- 35.Chen J., Lin Z.H., Liu L.F., Zhang R., Geng Y., Fan M.H., et al. GOLM1 exacerbates CD8+ T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal Transduct Target Ther. 2021;6:397. doi: 10.1038/s41392-021-00784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du S., Qian J.X., Tan S.R., Li W.H., Liu P., Zhao J., et al. Tumor cell-derived exosomes deliver TIE2 protein to macrophages to promote angiogenesis in cervical cancer. Cancer Lett. 2022;529:168–179. doi: 10.1016/j.canlet.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Wei C., Yang C.G., Wang S.Y., Shi D.D., Zhang C.X., Lin X.B., et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y., Yao S.R., Hu Y.L., Feng Y.Y., Li M., Bian Z.H., et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin Cancer Res. 2017;23:7375–7387. doi: 10.1158/1078-0432.CCR-17-1283. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J.H., Fan J.J., Zeng X., Nie M.M., Luan J.Y., Wang Y.C., et al. Hedgehog signaling in gastrointestinal carcinogenesis and the gastrointestinal tumor microenvironment. Acta Pharm Sin B. 2021;11:609–620. doi: 10.1016/j.apsb.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie F., Zhou X.X., Fang M.Y., Li H.Y., Su P., Tu Y.F., et al. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv Sci (Weinh) 2019;6:1901779. doi: 10.1002/advs.201901779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Li X.J., Zeng C., Liu C.L., Hao Q., Li W.N., et al. CD63+ cancer-associated fibroblasts confer tamoxifen resistance to breast cancer cells through exosomal miR-22. Adv Sci (Weinh) 2020;7:2002518. doi: 10.1002/advs.202002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortunato O., Boeri M., Moro M., Verri C., Mensah M., Conte D., et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis. 2014;5:e1564. doi: 10.1038/cddis.2014.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y.Y., Zhang Y., Wang F., Ni Q.F., Li M. MiR-660-5p promotes the progression of hepatocellular carcinoma by interaction with YWHAH via PI3K/Akt signaling pathway. Biochem Biophys Res Commun. 2020;531:480–489. doi: 10.1016/j.bbrc.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 44.Shen Y., Ye Y.F., Ruan L.W., Bao L., Wu M.W., Zhou Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet Mol Res. 2017;16:1. doi: 10.4238/gmr16019479. [DOI] [PubMed] [Google Scholar]
- 45.Czimmerer Z., Varga T., Kiss M., Vázquez C.O., Doan-Xuan Q.M., Rückerl D., et al. The IL-4/STAT6 signaling axis establishes a conserved microRNA signature in human and mouse macrophages regulating cell survival via miR-342-3p. Genome Med. 2016;8:63. doi: 10.1186/s13073-016-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y., Mao F., Zuo X., Moussalli M.J., Elias E., Xu W., et al. 15-LOX-1 suppression of hypoxia-induced metastatic phenotype and HIF-1α expression in human colon cancer cells. Cancer Med. 2014;3:472–484. doi: 10.1002/cam4.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y., Fang B., Yang X.Q., Wang L., Chen D., Krasnykh V., et al. Therapeutic molecular targeting of 15-lipoxygenase-1 in colon cancer. Mol Ther. 2008;16:886–892. doi: 10.1038/mt.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y.B., Song Y.C., Du W., Gong L.L., Chang H.C., Zou Z.Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26:78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X.J., Wei W.F., Wang Z.C., Wang N., Guo C.H., Zhou C.F., et al. A novel lymphatic pattern promotes metastasis of cervical cancer in a hypoxic tumour-associated macrophage-dependent manner. Angiogenesis. 2021;24:549–565. doi: 10.1007/s10456-020-09766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J.L., Li L.L., Yu B.F., Xu E.W., Sun N.P., Li X.N., et al. Extracellular vesicles mediated proinflammatory macrophage phenotype induced by radiotherapy in cervical cancer. BMC Cancer. 2022;22:88. doi: 10.1186/s12885-022-09194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X.J., Deng Y.R., Wang Z.C., Wei W.F., Zhou C.F., Zhang Y.M., et al. Hypoxia-induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis. 2019;10:508. doi: 10.1038/s41419-019-1748-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.