Abstract
Small cell lung cancer (SCLC) accounts for approximately 15% of all lung cancer cases and features a strong predilection for early metastasis and extremely poor prognosis. Despite being highly sensitive to chemotherapy and/or radiotherapy initially, most SCLC patients develop therapeutic resistance within one year and die of distant metastases. Multiple studies have revealed the high heterogeneity and strong plasticity of SCLC associated with frequent metastases and early development of therapeutic resistance as well as poor clinical outcome. Importantly, different SCLC subtypes are associated with different therapeutic vulnerabilities, and the inflamed subtype tends to have the best response to immunotherapy, which highlights the importance of precision medicine in the clinic. Here, we review recent advances in SCLC heterogeneity and plasticity and their link to distant metastases and chemotherapy resistance. We hope that a better understanding of the molecular mechanisms underlying SCLC malignant progression will help to develop better intervention strategies for this deadly disease.
Keywords: small cell lung cancer, heterogeneity, plasticity, metastases, chemotherapy resistance
Introduction
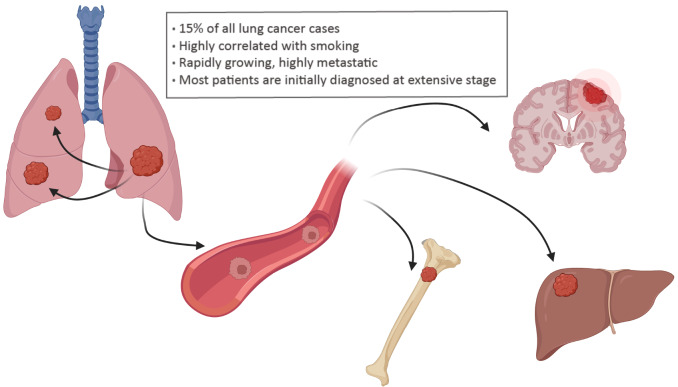
Small cell lung cancer (SCLC) accounts for approximately 15% of all lung cancer cases [1]. SCLC frequently occurs in lifetime heavy smokers (5 or more cigarettes a day), with only 2% of cases arising in never-smokers [ 2– 4] , which is possibly linked to exposure to air pollution [5] and radon [6] and histological transformation from non-small cell lung cancer (NSCLC) [7]. SCLC is rapidly growing, highly metastatic, and relatively immune-cold, with an extremely poor prognosis compared to other solid tumors [8]. SCLC can be classified into limited stage and extensive stage. Most patients are initially diagnosed at an extensive stage, characterized by nearby lung and/or distant organ metastases [9]. The most common organs for SCLC metastases include the contralateral lung, brain, bone and liver [ 10, 11] ( Figure 1). A high frequency of early metastasis and therapeutic resistance contributes to poor clinical outcomes of SCLC, with a 5-year survival rate of less than 7% [12]. Current first-line SCLC therapy remains chemotherapy and radiotherapy, which were established decades ago [ 13– 16] . Although SCLC patients initially exhibit a strong response to chemotherapy, most relapse with acquired drug resistance within one year [ 17– 20] .
Figure 1 .
Various organs for SCLC metastasis
Based on studies of human specimens and mouse models, emerging recognition of the high heterogeneity and plasticity of SCLC has implicated the complexity of this disease [ 21– 33] . In this review, we will summarize the progression of current findings in SCLC heterogeneity and plasticity, as well as their link to distant metastases and chemotherapy resistance. The improved understanding of the molecular mechanisms underlying SCLC heterogeneity and plasticity might help the development of novel therapeutic strategies for clinical management.
SCLC Subtyping and Therapeutic Vulnerabilities
Although neuroendocrine (NE) cells serve as the predominant cell of origin of SCLC, alveolar type II (AT2) cells and club cells are also endowed with this ability [ 34– 36] . Growing evidence has supported the multiple cells of origin of SCLC, indicating the potential heterogeneity of this disease. Since the 1980s, both “classic” and “variant” phenotypes have been detected in established human SCLC cell lines [ 37, 38] . Nearly 70% of these cell lines feature a “classic” phenotype, grow as tight aggregates and highly express NE-associated proteins [39]. The rest belong to “variant” cell lines, which can be further classified into morphological and chemical variant subtypes, with the former adherent to the dishes in cell culture and the latter growing as tightly aggregates with reduced NE markers [39].
The differential expression of four lineage-related transcription factors distinguishes SCLC into the achaete-scute homolog 1 (ASCL1) (SCLC-A), neuronal differentiation 1 (NEUROD1) (SCLC-N), POU class 2 homeobox 3 (POU2F3) (SCLC-P) and Yes1 associated transcriptional regulator (YAP1) (SCLC-Y) subtypes, which are associated with distinct therapeutic vulnerabilities [30]. Delta-like ligand 3 (DLL3), a direct transcriptional target of ASCL1, tends to be highly expressed in the SCLC-A subtype and minimally expressed in normal tissues [40], which enables the development of therapeutics to specifically target SCLC cells ( Table 1). The DLL3-targeted antibody drug conjugate (ADC) rovalpituzumab tesirine (Rova-T) has been evaluated in clinical trials [ 41] ( Table 1). Unfortunately, the following phase II study demonstrated associated toxicities [42], and additional DLL3 targeting approaches are currently under development [43] ( Table 1). Other important aberrations in the SCLC-A subtype include amplifications of BCL2 apoptosis regulator (BCL2) [44] and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) [ 45, 46] and a decrease in CREB binding protein (CREBBP) [47] ( Table 1).
Table 1 SCLC subtyping and therapeutic strategies
|
Subtype |
Characteristic |
Therapeutic strategies |
|
SCLC-A |
High DLL3 level [40] Amplifications of BCL2 [44] or EZH2 [ 45, 46] Decrease of CREBBP [47] |
|
|
SCLC-N |
Upregulation of C-MYC [30] |
Aurora A/B kinase, CHK1, IMPDH1/2 inhibitors, arginine deprivation [ 48‒ 56] |
|
SCLC-P |
Activation of IGF-1R or PARP signaling [ 44, 57] High MCL1 level [58] |
|
|
SCLC-Y |
Inflamed tumor microenvironment [59] |
Immunotherapy [59] |
|
SCLC-I a |
Low expression of ASCL1, NEUROD1 and POU2F3 [44] |
Combined chemotherapy and immunotherapy [44] |
|
SCLC-I b |
Immunosuppressive feature and high genomic instability, high POU2F3 level [60] |
Combined chemotherapy and immunotherapy [60] |
Upregulation of C-MYC is related to the SCLC-N subtype [30] and serves as a potential target for therapeutic agents [ 61, 62] . SCLC with high C-MYC expression is selectively vulnerable to aurora A/B kinase, checkpoint kinase 1 (CHK1), inosine monophosphate dehydrogenase 1/2 (IMPDH1/2) inhibitor treatment, and arginine deprivation [ 48– 56] ( Table 1). Although C-MYC shares major features with its paralogues N-MYC and L-MYC, the sensitivity to aurora kinase inhibitors seems unique for C-MYC-driven SCLC based on the results of a clustered regularly interspaced short palindromic repeats (CRISPR) activation model [54]. A recent double-blind clinical study confirmed that C-MYC may be a potential predictive biomarker of response to the aurora A inhibitor alisertib [63]. The SCLC-P subtype has been shown to preferentially depend on insulin-like growth factor 1 receptor (IGF-1R) and poly(ADP-ribose) polymerase 1 (PARP) signaling [ 44, 57] and is enriched with myeloid cell leukemia 1 (MCL1) expression, indicating a potential response to targeted therapy with the MCL1 inhibitor S63845 [58] ( Table 1). The SCLC-Y subtype is associated with an inflamed tumor microenvironment, suggesting a potential benefit from immune checkpoint blockade treatment [59] ( Table 1).
Studies show the close association between increased tumor mutational burden (TMB) and response to immunotherapy in multiple cancer types [ 64, 65] . Considering the high TMB of SCLC [66] and better prognosis of patients with increased tumor-infiltrating lymphocytes (TILs) [ 67, 68] , immunotherapy is approved for patients with extensive stage or relapsed SCLC [ 69– 71] . However, response to anti-PD-1/PD-L1 therapy only occurs in a small group of patients with SCLC [ 71, 72] . A phase III clinical trial of anti-PD-1 (nivolumab) in combination with anti-CTLA-4 (ipilimumab) in patients with extensive stage SCLC failed to meet its primary endpoint of overall survival [70]. Moreover, the correlation between PD-L1 expression and the effect of immunotherapy is ambiguous [ 73, 74] . These studies imply that the efficacy of immunotherapy for unselected patients with SCLC is modest; thus, it is urgently needed to identify the patients who may benefit most from immunotherapy. Using tumor expression data and nonnegative matrix factorization, a previous study identified a SCLC subtype (SCLC-I a) with low expression of ASCL1, NEUROD1 and POU2F3 and featured an inflamed gene signature [44] ( Table 1). SCLC-I a is sensitive to the addition of immunotherapy to chemotherapy [44] ( Table 1). Through integrative analysis of multi-omics data, we also uncovered the immune features of SCLC (SCLC-I b) with immunosuppressive features and high genomic instability [60]. Importantly, we found that POU2F3 is effective in predicting the SCLC-I b subtype, and patients with high POU2F3 expression exhibit better responses to immunotherapy [60] ( Table 1). SCLC-A (70%), SCLC-N (10-15%) and SCLC-P (12%) are the dominant subtypes of SCLC [ 57, 75, 76] . However, only 2% of SCLC shows YAP1 expression at quite low levels relative to ASCL1, NEUROD1 and POU2F3 [ 76, 77] . YAP1 and its transcriptional targets are higher in both POU2F3 and SCLC-I a subtypes than in the other two subtypes [44]. Therefore, it is proposed that YAP1 alone may not define a single group [ 78– 80] . In contrast to the SCLC-I a subtype with low POU2F3 expression, the SCLC-I b subtype shows high POU2F3 level [60]. However, the relationship between the SCLC-P and SCLC-I b subtypes is unclear and needs further study.
Notch signaling is positively correlated with the non-NE phenotype and significantly predicts the clinical benefit of immunotherapy [67]. Cyclin-dependent kinase 7 (CDK7) is a central regulator of the cell cycle and gene transcription [81]. Combining CDK inhibitors with anti-PD-1 offers a significant survival benefit in SCLC, providing a rationale for new combination regimens and immunotherapies [82]. More recently, certain combination chemotherapy plus immunotherapy regimens (including the anti-PD-L1 drugs atezolizumab or durvalumab) have been recommended in the National Comprehensive Cancer Network (NCCN) Guidelines for SCLC as preferred options for patients with extensive stage SCLC [ 69, 83– 88] .
Molecular and Cellular Mechanisms Involved in SCLC Metastasis
Concurrent loss of p53 and RB1 occurs frequently in SCLC [ 89, 90] . Homozygous deletion of these two alleles in mouse lung epithelia promotes SCLC development and dramatic metastasis, which closely recapitulates the clinical disease [91]. SCLC derived from the Rb1 L/L/ Trp53 L/L ( RP) mouse model typically expresses NE markers, including neural cell adhesion molecule 1 (NCAM) and ASCL1, and frequently metastasizes into distant organs. Concurrent deletion of RB family members p107 and p130 or Pten in the RP model significantly accelerates malignant progression and SCLC metastasis [ 22, 92– 94] .
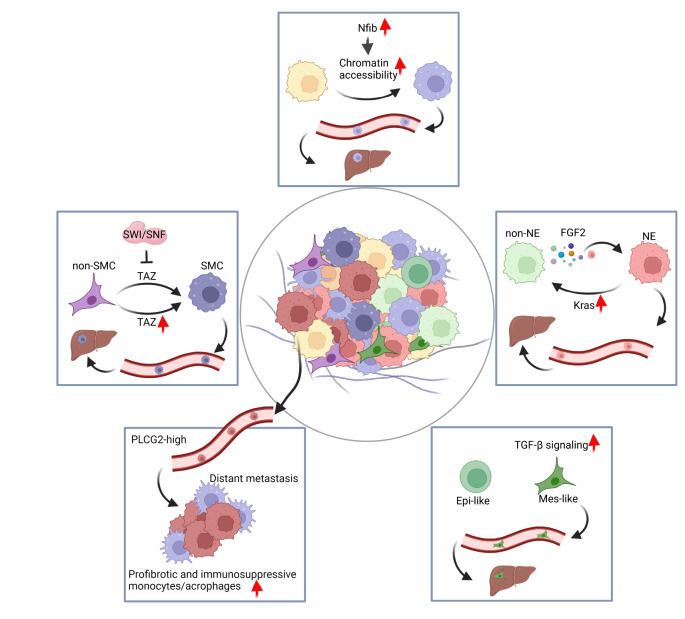
Based on the study of these autochthonous SCLC mouse models, we recently identified NCAM hiCD44 lo cells as the major subpopulation responsible for liver metastasis [32] ( Figure 2). During SCLC malignant progression, the phenotypic transition of NCAM hiCD44 lo cells (SCLC metastasizing cells, SMCs) from NCAM loCD44 hi cells (non-SCLC metastasizing cells, non-SMCs) is driven by the down-regulation of the Hippo pathway co-activator Taz/ Wwtr1. Moreover, the SWI/SNF chromatin remodelling complex plays an important role in silencing Taz during this process. Liver metastasis from SCLC patients showed decreased Taz expression and an increased NCAM hiCD44 lo phenotype. To study the heterogeneity and tumor microenvironment of clinical SCLC specimens, Chan et al. [95] used single-cell transcriptome sequencing and imaging techniques and identified a phospholipase C gamma 2 (PLCG2)-high-expressing subpopulation linked to increased brain metastasis and poor prognosis, as well as an enrichment of a monocyte/macrophage population with a profibrotic, immunosuppressive phenotype.
Figure 2 .
The link of SCLC heterogeneity and plasticity to distant metastases
SMC, SCLC metastasizing cell. non-SMC, non-SCLC metastasizing cell. NE, neuroendocrine. non-NE, nonneuroendocrine. FGF2, fibroblast growth factor 2. Epi, epithelial. Mes, mesenchymal.
A previous study showed that NE and nonneuroendocrine (non-NE) share common cell origins and play different roles during SCLC metastasis [21]. Further study revealed that non-NE cells secrete fibroblast growth factor 2 (FGF2) and enhance the expression of polyomavirus enhancer activator 3 (Pea3) in NE cells, resulting in metastatic dissemination of the NE subclone to the liver [26] ( Figure 2). Interestingly, we recently found that the non-NE subtype of SCLC is composed of mesenchymal and epithelial-like subsets, and the activation of TGF-β signaling in the mesenchymal subset promotes cancer metastasis to the liver [96] ( Figure 2). Moreover, depletion of the TGF-β signaling pathway in mice depresses the liver metastasis capability of SCLC [96]. In addition, Nfib is oncogenic in SCLC, promotes pro-metastatic neuronal gene expression programs and drives SCLC liver metastasis [ 23, 28, 29] . Mechanistically, NFIB promotes SCLC metastasis by increasing the accessibility of global chromatin during cancer progression [23] ( Figure 2).
Mechanistic Insights into SCLC Chemotherapy Resistance and Overcoming Strategy
The current first-line treatment for SCLC, combining etoposide and cisplatin (E/P), has been the clinical standard of care since the 1970s [97]. Despite the initial sensitivity to chemotherapy, resistance emerges rapidly. The paucity of tumor specimens from chemotherapy-resistant SCLC patients has greatly hindered the current mechanistic understanding of chemotherapy resistance in the clinic. Therefore, the clinical outcome has not significantly improved over the past few decades, and chemotherapy resistance remains the central problem for SCLC treatment [8].
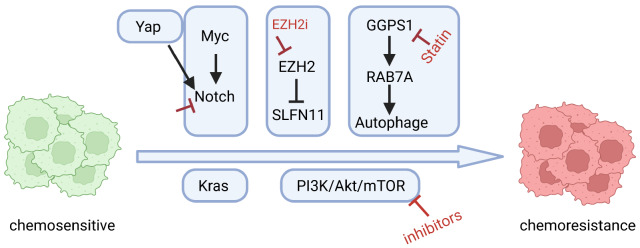
Through comprehensive bioinformatics analyses, we found that adherent or semi-adherent SCLC cells are enriched with increased PI3K/Akt/mTOR pathway activity and high chemotherapy resistance [98] ( Figure 3). Activation of this pathway promotes the transition from the suspension to adhesion growth pattern of SCLC cells and confers chemotherapy resistance [98]. Such chemotherapy resistance could be largely overcome by combining chemotherapy with PI3K/Akt/mTOR pathway inhibitors [98]. A phase I/II clinical trial (NCT03366103) targeting mammalian target of rapamycin kinase (mTOR) and BCL-2 is currently ongoing [1].
Figure 3 .
Mechanisms in regulating SCLC chemoresistance
Activation of the Kras or Notch pathway results in an NE to non-NE fate switch, which enhances SCLC chemotherapy resistance [ 21, 23, 24, 51] ( Figure 3). MYC drives the temporal evolution of SCLC subtypes by reprogramming neuroendocrine fate through activation of the Notch pathway and promotes SCLC chemotherapy resistance [ 31, 51, 99, 100] ( Figure 3). YAP can signal through Notch-dependent or Notch-independent pathways to promote the fate conversion from NE to non-NE tumor cells [101] ( Figure 3). Notch blockade in combination with chemotherapy suppresses tumor growth and delays relapse in pre-clinical models [24]. Moreover, EZH2 is upregulated in chemoresistant SCLC and promotes drug resistance through epigenetic silencing of schlafen family member 11 ( SLFN11) [102]. Combined EZH2 inhibition and chemotherapy treatment is currently being explored in a phase I/II clinical trial of recurrent SCLC (NCT038979798) [103].
Recently, we found that chemoresistant SCLC undergoes metabolic reprogramming relying on the mevalonate (MVA)–geranylgeranyl diphosphate (GGPP) pathway, which can be targeted by clinically approved statins [104]. Mechanistically, statins induce oxidative stress accumulation and apoptosis through the GGPP synthase 1 (GGPS1)–RAB7A–autophagy axis [104]. Statin treatment overcomes both intrinsic and acquired SCLC chemotherapy resistance in vivo across multiple SCLC patient-derived xenograft (PDX) models bearing high GGPS1 levels. Importantly, GGPS1 expression is negatively associated with survival in SCLC patients, and combined statin and chemotherapy treatment resulted in durable responses in three SCLC patients who relapsed from first-line chemotherapy [104].
Perspective
SCLC is the most malignant type of lung cancer with an extremely poor prognosis, and most patients are diagnosed at an extensive stage. Patients with SCLC exhibit a remarkable initial response to chemotherapy and/or radiotherapy followed by the quick development of drug resistance. Although an increasing number of targeted therapies have emerged in many other cancer types [ 105, 106] , treatments for recurrent or refractory SCLC are limited and unsatisfactory [ 12, 107– 109] . Most SCLC patients eventually die of distant metastases and chemotherapy resistance. Recent advances in SCLC have revealed the heterogeneity and plasticity of SCLC and have uncovered pivotal roles during cancer metastasis and chemotherapy resistance. The phenotypic evolution during malignant cancer progression emphasizes the importance of timely and precise diagnosis and related therapeutic interventions in the clinic.
Different molecular subtypes of SCLC have been defined by gene expression profiling and exhibit distinct vulnerabilities to targeted therapies. Precise analysis of the patients at initial diagnosis may help to improve the therapeutic outcomes. Immunotherapy has been recommended for extensive-stage and recurrent SCLC clinical treatment, and recent studies have identified the inflamed or immune subtype that may benefit from immunotherapy. The combination of molecularly targeted therapy or immunotherapy with traditional chemotherapy may improve the clinical outcomes in the future. A better understanding of SCLC biology will hopefully uncover novel vulnerabilities that might be amenable to clinical therapeutic approaches.
Acknowledgments
The figures were created with BioRender.com, and the license numbers are as follows: IG24YJF9QO, AP24YJFXAL, KC24Z9JSZU.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Key R&D Program of China (Nos. 2022YFA1103900 and 2020YFA0803300 to H.J.), the National Natural Science Foundation of China (Nos. 82341002, 81872312, 82011540007, 31621003, 32293192, and 82030083 to H.J., 81871875 and 82173340 to L.H.), the Basic Frontier Scientific Research Program of Chinese Academy of Science (No. ZDBS-LY-SM006 to H.J.), the International Cooperation Project of Chinese Academy of Sciences (No. 153D31KYSB20190035 to H.J.), the Innovative research team of high-level local universities in Shanghai (No. SSMU-ZLCX20180500 to H.J.), and the Science and Technology Commission of Shanghai Municipality (No. 21ZR1470300 to L.H.).
References
- 1.Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. . 2021;7:3. doi: 10.1038/s41572-020-00235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang R, Wei Y, Hung RJ, Liu G, Su L, Zhang R, Zong X, et al. Associated links among smoking, chronic obstructive pulmonary disease, and small cell lung cancer: a pooled analysis in the international lung cancer consortium. EBioMedicine. . 2015;2:1677–1685. doi: 10.1016/j.ebiom.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pesch B, Kendzia B, Gustavsson P, Jöckel KH, Johnen G, Pohlabeln H, Olsson A, et al. Cigarette smoking and lung cancer-relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int J Cancer. . 2012;131:1210–1219. doi: 10.1002/ijc.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varghese AM, Zakowski MF, Yu HA, Won HH, Riely GJ, Krug LM, Kris MG, et al. Small-cell lung cancers in patients who never smoked cigarettes. J Thoracic Oncol. . 2014;9:892–896. doi: 10.1097/JTO.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamichhane DK, Kim HC, Choi CM, Shin MH, Shim YM, Leem JH, Ryu JS, et al. Lung cancer risk and residential exposure to air pollution: a korean population-based case-control study. Yonsei Med J. . 2017;58:1111–1118. doi: 10.3349/ymj.2017.58.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Martinez A, Torres-Duran M, Barros-Dios JM, Ruano-Ravina A. Residential radon and small cell lung cancer. A systematic review. Cancer Lett. . 2018;426:57–62. doi: 10.1016/j.canlet.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Quintanal-Villalonga Á, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, Rudin CM. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol. . 2020;17:360–371. doi: 10.1038/s41571-020-0340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. . 2017;17:725–737. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 9.Micke P, Faldum A, Metz T, Beeh KM, Bittinger F, Hengstler JG, Buhl R. Staging small cell lung cancer: Veterans Administration Lung Study Group versus International Association for the Study of Lung Cancer—what limits limited disease? Lung Cancer. . 2002;37:271–276. doi: 10.1016/s0169-5002(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 10.Obenauf AC, Massagué J. Surviving at a distance: organ-specific metastasis. Trends Cancer. . 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. . 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 12.Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. . 2015;121:664–672. doi: 10.1002/cncr.29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demedts IK, Vermaelen KY, van Meerbeeck JP. Treatment of extensive-stage small cell lung carcinoma: current status and future prospects. Eur Respiratory J. . 2010;35:202–215. doi: 10.1183/09031936.00105009. [DOI] [PubMed] [Google Scholar]
- 14.Planchard D, Le Péchoux C. Small cell lung cancer: new clinical recommendations and current status of biomarker assessment. Eur J Cancer. . 2011;47:S272–S283. doi: 10.1016/S0959-8049(11)70173-3. [DOI] [PubMed] [Google Scholar]
- 15.Paumier A, Le Péchoux C. Radiotherapy in small-cell lung cancer: where should it go? Lung Cancer. . 2010;69:133–140. doi: 10.1016/j.lungcan.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Kalemkerian GP, Loo Jr BW, Akerley W, Attia A, Bassetti M, Boumber Y, Decker R, et al. NCCN guidelines insights: small cell lung cancer, version 2.2018. J Natl Compr Canc Netw. . 2018;16:1171–1182. doi: 10.6004/jnccn.2018.0079. [DOI] [PubMed] [Google Scholar]
- 17.Faivre-Finn C, Snee M, Ashcroft L, Appel W, Barlesi F, Bhatnagar A, Bezjak A, et al. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): an open-label, phase 3, randomised, superiority trial. Lancet Oncol. . 2017;18:1116–1125. doi: 10.1016/S1470-2045(17)30318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell HA, Tata LJ, Baldwin DR, Potter VA, Stanley RA, Khakwani A, Hubbard RB. Treatment decisions and survival for people with small-cell lung cancer. Br J Cancer. . 2014;110:908–915. doi: 10.1038/bjc.2013.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. . 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 20.Jett JR, Schild SE, Kesler KA, Kalemkerian GP. Treatment of small cell lung cancer. Chest. . 2013;143:e400S–e419S. doi: 10.1378/chest.12-2363. [DOI] [PubMed] [Google Scholar]
- 21.Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, Berns A. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. . 2011;19:244–256. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 22.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. . 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Grüner BM, Chiou SH, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. . 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O′Young G, Cristea S, Luca VC, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. . 2017;545:360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, Xu L, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep. . 2016;16:644–656. doi: 10.1016/j.celrep.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon M, Proost N, Song JY, Sutherland KD, Zevenhoven J, Berns A. Paracrine signaling between tumor subclones of mouse SCLC: a critical role of ETS transcription factor Pea3 in facilitating metastasis. Genes Dev. . 2015;29:1587–1592. doi: 10.1101/gad.262998.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu N, Jia D, Ibrahim AH, Bachurski CJ, Gronostajski RM, MacPherson D. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer . Oncotarget. . 2016;7:57514–57524. doi: 10.18632/oncotarget.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semenova EA, Kwon M, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, Kuilman T, et al. Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell Rep. . 2016;16:631–643. doi: 10.1016/j.celrep.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dooley AL, Winslow MM, Chiang DY, Banerji S, Stransky N, Dayton TL, Snyder EL, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. . 2011;25:1470–1475. doi: 10.1101/gad.2046711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. . 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, Conley CC, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. . 2020;38:60–78.e12. doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Y, Zhao Q, Zhu W, Feng Y, Xiao T, Zhang P, Jiang L, et al. Identification of TAZ as the essential molecular switch in orchestrating SCLC phenotypic transition and metastasis. Natl Sci Rev. . 2022;9:nwab232. doi: 10.1093/nsr/nwab232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu S, Fetsch P, Thomas A, Pommier Y, Schrump DS, Miettinen MM, Chen H. Molecular subtypes of primary SCLC tumors and their associations with neuroendocrine and therapeutic markers. J Thoracic Oncol. . 2022;17:141–153. doi: 10.1016/j.jtho.2021.08.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park KS, Liang MC, Raiser DM, Zamponi R, Roach RR, Curtis SJ, Walton Z, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle. . 2011;10:2806–2815. doi: 10.4161/cc.10.16.17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. . 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Guanizo A, Luong Q, Jayasekara WSN, Jayasinghe D, Inampudi C, Szczepny A, et al. Lineage-restricted neoplasia driven by Myc defaults to small cell lung cancer when combined with loss of p53 and Rb in the airway epithelium. Oncogene. . 2022;41:138–145. doi: 10.1038/s41388-021-02070-3. [DOI] [PubMed] [Google Scholar]
- 37.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, Zweig MH, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985, 45: 2913–2923 . [PubMed]
- 38.Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985, 45: 2924–2930 . [PubMed]
- 39.Bepler G, Jaques G, Koehler A, Gropp C, Havemann K. Markers and characteristics of human SCLC cell lines. J Cancer Res Clin Oncol. . 1987;113:253–259. doi: 10.1007/BF00396382. [DOI] [PubMed] [Google Scholar]
- 40.Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black KA, Desai R, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo . Sci Transl Med. . 2015;7:302ra136. doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol. . 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgensztern D, Besse B, Greillier L, Santana-Davila R, Ready N, Hann CL, Glisson BS, et al. Efficacy and safety of rovalpituzumab tesirine in third-line and beyond patients with DLL3-expressing, relapsed/refractory small-cell lung cancer: results from the phase II TRINITY study. Clin Cancer Res. . 2019;25:6958–6966. doi: 10.1158/1078-0432.CCR-19-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen DH, Giffin MJ, Bailis JM, Smit MAD, Carbone DP, He K. DLL3: an emerging target in small cell lung cancer. J Hematol Oncol. . 2019;12:61. doi: 10.1186/s13045-019-0745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, Nabet BY, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. . 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, Rudin CM. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. . 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murai F, Koinuma D, Shinozaki-Ushiku A, Fukayama M, Miyaozono K, Ehata S. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-β-Smad-ASCL1 pathway. Cell Discov. . 2015;1:15026. doi: 10.1038/celldisc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, Kim KB, et al. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition . Cancer Discovery. . 2018;8:1422–1437. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sos ML, Dietlein F, Peifer M, Schöttle J, Balke-Want H, Müller C, Koker M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci USA. . 2012;109:17034–17039. doi: 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hook KE, Garza SJ, Lira ME, Ching KA, Lee NV, Cao J, Yuan J, et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol Cancer Ther. . 2012;11:710–719. doi: 10.1158/1535-7163.MCT-11-0184. [DOI] [PubMed] [Google Scholar]
- 50.Helfrich BA, Kim J, Gao D, Chan DC, Zhang Z, Tan AC, Bunn Jr PA. Barasertib (AZD1152), a small molecule aurora B inhibitor, inhibits the growth of SCLC cell lines in vitro and in vivo . Mol Cancer Ther. . 2016;15:2314–2322. doi: 10.1158/1535-7163.MCT-16-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, Marx A, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. . 2017;31:270–285. doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, Ireland AS, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget. . 2017;8:73419–73432. doi: 10.18632/oncotarget.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalishazar MD, Wait SJ, Huang F, Ireland AS, Mukhopadhyay A, Lee Y, Schuman SS, et al. MYC-driven small-cell lung cancer is metabolically distinct and vulnerable to arginine depletion. Clin Cancer Res. . 2019;25:5107–5121. doi: 10.1158/1078-0432.CCR-18-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dammert MA, Brägelmann J, Olsen RR, Böhm S, Monhasery N, Whitney CP, Chalishazar MD, et al. MYC paralog-dependent apoptotic priming orchestrates a spectrum of vulnerabilities in small cell lung cancer. Nat Commun. . 2019;10:3485. doi: 10.1038/s41467-019-11371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, Shi X, et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab. . 2018;28:369–382.e5. doi: 10.1016/j.cmet.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, Redwood AB, et al. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res. . 2017;77:3870–3884. doi: 10.1158/0008-5472.CAN-16-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, Somerville TDD, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. . 2018;32:915–928. doi: 10.1101/gad.314815.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda Y, Ozasa H, Kim YH, Yamazoe M, Ajimizu H, Yamamoto Funazo T, Nomizo T, et al. MCL1 inhibition is effective against a subset of small-cell lung cancer with high MCL1 and low BCL-XL expression. Cell Death Dis. . 2020;11:177. doi: 10.1038/s41419-020-2379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owonikoko TK, Dwivedi B, Chen Z, Zhang C, Barwick B, Ernani V, Zhang G, et al. YAP1 expression in SCLC defines a distinct subtype with T-cell–Inflamed phenotype. J Thoracic Oncol. . 2021;16:464–476. doi: 10.1016/j.jtho.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Fang Z, Tang Y, Jin Y, Guo C, Hu L, Xu Y, et al. Integrative analysis of multi‐omics data reveals the heterogeneity and signatures of immune therapy for small cell lung cancer. Clin Transl Med. . 2021;11:e620. doi: 10.1002/ctm2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masso-Valles D, Beaulieu ME, Soucek L. MYC, MYCL, and MYCN as therapeutic targets in lung cancer. Expert Opin Therapeutic Targets. . 2020;24:101–114. doi: 10.1080/14728222.2020.1723548. [DOI] [PubMed] [Google Scholar]
- 62.Schwendenwein A, Megyesfalvi Z, Barany N, Valko Z, Bugyik E, Lang C, Ferencz B, et al. Molecular profiles of small cell lung cancer subtypes: therapeutic implications. Mol Ther Oncolytics. . 2021;20:470–483. doi: 10.1016/j.omto.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owonikoko TK, Niu H, Nackaerts K, Csoszi T, Ostoros G, Mark Z, Baik C, et al. Randomized phase II study of paclitaxel plus alisertib versus paclitaxel plus placebo as second-line therapy for SCLC: primary and correlative biomarker analyses. J Thoracic Oncol. . 2020;15:274–287. doi: 10.1016/j.jtho.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 64.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. . 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, Rizvi NA, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. . 2018;33:853–861.e4. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, et al. Signatures of mutational processes in human cancer. Nature. . 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roper N, Velez MJ, Chiappori A, Kim YS, Wei JS, Sindiri S, Takahashi N, et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat Commun. . 2021;12:3880. doi: 10.1038/s41467-021-24164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iams WT, Shiuan E, Meador CB, Roth M, Bordeaux J, Vaupel C, Boyd KL, et al. Improved prognosis and increased tumor-infiltrating lymphocytes in patients who have SCLC with neurologic paraneoplastic syndromes. J Thoracic Oncol. . 2019;14:1970–1981. doi: 10.1016/j.jtho.2019.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, Huemer F, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. . 2018;379:2220–2229. doi: 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- 70.Owonikoko TK, Park K, Govindan R, Ready N, Reck M, Peters S, Dakhil SR, et al. Nivolumab and ipilimumab as maintenance therapy in extensive-disease small-cell lung cancer: checkmate 451. J Clin Oncol. . 2021;39:1349–1359. doi: 10.1200/JCO.20.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ready N, Farago AF, de Braud F, Atmaca A, Hellmann MD, Schneider JG, Spigel DR, et al. Third-line nivolumab monotherapy in recurrent SCLC: checkmate 032. J Thoracic Oncol. . 2019;14:237–244. doi: 10.1016/j.jtho.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung HC, Piha-Paul SA, Lopez-Martin J, Schellens JHM, Kao S, Miller Jr WH, Delord JP, et al. Pembrolizumab after two or more lines of previous therapy in patients with recurrent or metastatic SCLC: results from the KEYNOTE-028 and KEYNOTE-158 studies. J Thoracic Oncol. . 2020;15:618–627. doi: 10.1016/j.jtho.2019.12.109. [DOI] [PubMed] [Google Scholar]
- 73.Antonia SJ, López-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, Jäger D, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. . 2016;17:883–895. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 74.Ott PA, Elez E, Hiret S, Kim DW, Morosky A, Saraf S, Piperdi B, et al. Pembrolizumab in patients with extensive-stage small-cell lung cancer: results from the phase Ib KEYNOTE-028 study. J Clin Oncol. . 2017;35:3823–3829. doi: 10.1200/JCO.2017.72.5069. [DOI] [PubMed] [Google Scholar]
- 75.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. . 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato Y, Okamoto I, Kameyama H, Kudoh S, Saito H, Sanada M, Kudo N, et al. Integrated immunohistochemical study on small-cell carcinoma of the lung focusing on transcription and co-transcription factors. Diagnostics. . 2020;10:949. doi: 10.3390/diagnostics10110949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ito T, Matsubara D, Tanaka I, Makiya K, Tanei Z, Kumagai Y, Shiu S, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci. . 2016;107:1527–1538. doi: 10.1111/cas.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baine MK, Hsieh MS, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, Beras A, et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thoracic Oncol. . 2020;15:1823–1835. doi: 10.1016/j.jtho.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, Humphrey S, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer. . 2020;1:437–451. doi: 10.1038/s43018-020-0046-2. [DOI] [PubMed] [Google Scholar]
- 80.Yatabe Y. Reassessing the SCLC subtypes. J Thorac Oncol. . 2020;15:1819–1822. doi: 10.1016/j.jtho.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 81.Malumbres M. Cyclin-dependent kinases. Genome Biol. . 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, Li F, et al. CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer Cell. . 2020;37:37–54.e9. doi: 10.1016/j.ccell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu SV, Reck M, Mansfield AS, Mok T, Scherpereel A, Reinmuth N, Garassino MC, et al. Updated overall survival and PD-L1 subgroup analysis of patients with extensive-stage small-cell lung cancer treated with atezolizumab, carboplatin, and etoposide (IMpower133) J Clin Oncol. . 2021;39:619–630. doi: 10.1200/JCO.20.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. . 2019;394:1929–1939. doi: 10.1016/S0140-6736(19)32222-6. [DOI] [PubMed] [Google Scholar]
- 85.Goldman JW, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, et al. Durvalumab, with or without tremelimumab, plus platinum–etoposide versus platinum–etoposide alone in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): updated results from a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. . 2021;22:51–65. doi: 10.1016/S1470-2045(20)30539-8. [DOI] [PubMed] [Google Scholar]
- 86.Paz-Ares L, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, Hochmair MJ, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open. . 2022;7:100408. doi: 10.1016/j.esmoop.2022.100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathieu L, Shah S, Pai-Scherf L, Larkins E, Vallejo J, Li X, Rodriguez L, et al. FDA approval summary: atezolizumab and durvalumab in combination with platinum-based chemotherapy in extensive stage small cell lung cancer. Oncologist. . 2021;26:433–438. doi: 10.1002/onco.13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ganti AKP, Loo BW, Bassetti M, Blakely C, Chiang A, D′Amico TA, D′Avella C, et al. Small cell lung cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J Natl Comprehensive Cancer Network. . 2021;19:1441–1464. doi: 10.6004/jnccn.2021.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. . 1988;241:353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bunn PA Jr, Minna JD, Augustyn A, Gazdar AF, Ouadah Y, Krasnow MA, Berns A, et al. Small cell lung cancer: can recent advances in biology and molecular biology be translated into improved outcomes? J Thoracic Oncol. . 2016;11:453–474. doi: 10.1016/j.jtho.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. . 2003;4:181–189. doi: 10.1016/S1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 92.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. . 2010;70:3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ng SR, Rideout Iii WM, Akama-Garren EH, Bhutkar A, Mercer KL, Schenkel JM, Bronson RT, et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci USA. . 2020;117:513–521. doi: 10.1073/pnas.1821893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, Ingolia N, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. . 2014;12:654–659. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O, Masilionis I, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. . 2021;39:1479–1496.e18. doi: 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin Y, Xiao T, Feng Y, Yang J, Guo C, Hu L, Ji H. A mesenchymal-like subpopulation in non-neuroendocrine SCLC contributes to metastasis. J Genet Genomics. . 2021;48:571–581. doi: 10.1016/j.jgg.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 97.Murray N, Turrisi Iii AT. A review of first-line treatment for small-cell lung cancer. J Thoracic Oncol. . 2006;1:270–278. doi: 10.1016/S1556-0864(15)31579-3. [DOI] [PubMed] [Google Scholar]
- 98.Li X, Li C, Guo C, Zhao Q, Cao J, Huang HY, Yue M, et al. PI3K/Akt/mTOR signaling orchestrates the phenotypic transition and chemo-resistance of small cell lung cancer. J Genet Genomics. . 2021;48:640–651. doi: 10.1016/j.jgg.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 99.Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, Phat S, et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov. . 2018;8:600–615. doi: 10.1158/2159-8290.CD-17-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng J, Sutherland KD. NOTCH your usual suspect: MYC charged with controlling neuroendocrine cell-fate in small cell lung cancer. Cancer Cell. . 2020;38:17–20. doi: 10.1016/j.ccell.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 101.Wu Q, Guo J, Liu Y, Zheng Q, Li X, Wu C, Fang D, et al. YAP drives fate conversion and chemoresistance of small cell lung cancer. Sci Adv. . 2021;7:eabg1850. doi: 10.1126/sciadv.abg1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, Ni A, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. . 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sutherland KD, Ireland AS, Oliver TG. Killing SCLC: insights into how to target a shapeshifting tumor. Genes Dev. . 2022;36:241–258. doi: 10.1101/gad.349359.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo C, Wan R, He Y, Lin SH, Cao J, Qiu Y, Zhang T, et al. Therapeutic targeting of the mevalonate–geranylgeranyl diphosphate pathway with statins overcomes chemotherapy resistance in small cell lung cancer. Nat Cancer. . 2022;3:614–628. doi: 10.1038/s43018-022-00358-1. [DOI] [PubMed] [Google Scholar]
- 105.Xie J, Zhang X. The impact of genomic profiling for novel cancer therapy – recent progress in non-small cell lung cancer. J Genet Genomics. . 2016;43:3–10. doi: 10.1016/j.jgg.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shulman M, Shi R, Zhang Q. Von Hippel-Lindau tumor suppressor pathways & corresponding therapeutics in kidney cancer. J Genet Genomics. . 2021;48:552–559. doi: 10.1016/j.jgg.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koinis F, Agelaki S, Karavassilis V, Kentepozidis N, Samantas E, Peroukidis S, Katsaounis P, et al. Second-line pazopanib in patients with relapsed and refractory small-cell lung cancer: a multicentre phase II study of the Hellenic Oncology Research Group. Br J Cancer. . 2017;117:8–14. doi: 10.1038/bjc.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gadgeel SM, Pennell NA, Fidler MJ, Halmos B, Bonomi P, Stevenson J, Schneider B, et al. Phase II study of maintenance pembrolizumab in patients with extensive-stage small cell lung cancer (SCLC) J Thoracic Oncol. . 2018;13:1393–1399. doi: 10.1016/j.jtho.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saito M, Shiraishi K, Goto A, Suzuki H, Kohno T, Kono K. Development of targeted therapy and immunotherapy for treatment of small cell lung cancer. Jpnese J Clin Oncol. . 2018;48:603–608. doi: 10.1093/jjco/hyy068. [DOI] [PubMed] [Google Scholar]