Abstract
Background
A methylation-based classification of ependymoma has recently found broad application. However, the diagnostic advantage and implications for treatment decisions remain unclear. Here, we retrospectively evaluate the impact of surgery and radiotherapy on outcome after molecular reclassification of adult intracranial ependymomas.
Methods
Tumors diagnosed as intracranial ependymomas from 170 adult patients collected from 8 diagnostic institutions were subjected to DNA methylation profiling. Molecular classes, patient characteristics, and treatment were correlated with progression-free survival (PFS).
Results
The classifier indicated an ependymal tumor in 73.5%, a different tumor entity in 10.6%, and non-classifiable tumors in 15.9% of cases, respectively. The most prevalent molecular classes were posterior fossa ependymoma group B (EPN-PFB, 32.9%), posterior fossa subependymoma (PF-SE, 25.9%), and supratentorial ZFTA fusion-positive ependymoma (EPN-ZFTA, 11.2%). With a median follow-up of 60.0 months, the 5- and 10-year-PFS rates were 64.5% and 41.8% for EPN-PFB, 67.4% and 45.2% for PF-SE, and 60.3% and 60.3% for EPN-ZFTA. In EPN-PFB, but not in other molecular classes, gross total resection (GTR) (P = .009) and postoperative radiotherapy (P = .007) were significantly associated with improved PFS in multivariable analysis. Histological tumor grading (WHO 2 vs. 3) was not a predictor of the prognosis within molecularly defined ependymoma classes.
Conclusions
DNA methylation profiling improves diagnostic accuracy and risk stratification in adult intracranial ependymoma. The molecular class of PF-SE is unexpectedly prevalent among adult tumors with ependymoma histology and relapsed as frequently as EPN-PFB, despite the supposed benign nature. GTR and radiotherapy may represent key factors in determining the outcome of EPN-PFB patients.
Keywords: adult, DNA methylation, ependymoma, intracranial, radiotherapy
Key Points.
Molecular classification improves diagnostic accuracy in tumors with ependymoma histology.
The molecular profile of subependymoma is highly prevalent in adult infratentorial ependymoma.
Radiotherapy appears to be effective in posterior fossa ependymoma group B.
Importance of the Study.
Little is known about the possible implications of a novel methylation-based classification for diagnosis and treatment of adult intracranial ependymoma. In this multi-institutional study, 170 intracranial tumors of adult patients with histological diagnosis of ependymoma were molecularly reclassified, and patient characteristics were correlated with clinical outcomes. We found molecular profiles of both ependymal and non-ependymal tumors as well as non-classifiable cases. Ependymal tumor classes most prevalent were posterior fossa ependymoma group B (EPN-PFB), posterior fossa subependymoma (PF-SE), and supratentorial ZFTA fusion-positive ependymoma. All PF-SE tumors had pure or partial ependymoma histology and relapsed as frequently as EPN-PFB. Gross total resection and radiotherapy are associated with improved progression-free survival in EPN-PFB tumors. DNA methylation profiling appears beneficial for diagnostic accuracy in tumors with ependymoma morphology and stratification into risk groups. Future studies are necessary to investigate the impact of different treatment approaches (e.g., delay of radiotherapy) on survival in subgroups of ependymoma.
Ependymomas are glial tumors arising from the ependymal lining of the ventricles and spinal canal. Intracranial ependymoma is usually diagnosed in childhood and to a lesser extent in adult patients.1 Until recently, ependymomas were diagnosed and graded by histopathology alone. On the World Health Organization (WHO) malignancy scale, subependymomas are designated WHO grade 1 whereas ependymomas are defined as grade 2 or 3 tumors. Studies on DNA methylation and gene expression profiling have revealed at least 7 molecular classes of ependymoma and 3 classes of subependymoma across the central nervous system (CNS).2–7 Each of these molecular classes represents a biological group of ependymal tumors characterized by distinct molecular features and is mostly restricted to 1 of the 3 anatomic compartments of the CNS: Supratentorial (ST), posterior fossa (PF), and spinal (SP).8 In the ST compartment, molecular classes are primarily defined by the presence of specific gene fusions, namely YAP1 fusion and ZFTA fusion (previously known as c11orf95, recurrently fused to RELA). Molecular classes in the PF compartment comprise posterior fossa ependymoma group A (EPN-PFA) and group B (EPN-PFB), while molecular classes of the SP compartment include myxopapillary ependymoma (SP-EPN-MPE), spinal ependymoma (SP-EPN), and a more aggressive variant of SP ependymoma characterized by MYCN amplification (SP-EPN-MYCN). In addition, there is a distinct molecular class of subependymoma (SE) for each anatomic site (termed ST-SE, PF-SE, and SP-SE).2 Molecular classes have been implemented in the 2021 WHO classification of CNS tumors and should be used in combination with histopathological features for diagnosis and tumor grading.9 Molecular classes of ependymoma show a characteristic distribution among age groups with PF-SE, EPN-PFB, and ZFTA fusion-positive ependymomas (EPN-ZFTAs) being the predominant intracranial classes in adult patients.10
Standard of treatment remains maximal safe surgical resection for all ependymal tumors regardless of grade, whereas postoperative radiotherapy is recommended in WHO grade 3 and incompletely resected grade 2 ependymomas.11,12 However, evaluation of ependymoma by histopathology alone has been demonstrated to be of limited clinical utility due to substantial interobserver variability regarding diagnosis and grading.13,14 For this reason, DNA methylation profiling is becoming more and more accepted for diagnostic assessment of ependymoma as it allows confirmation of diagnosis and additional assignment of an underlying biological class that is of prognostic relevance.2,8 Clinical data on outcomes within molecularly defined ependymoma classes are lacking, especially for adults. To date, there is only one molecular study of adult ependymoma with a total of 56 intracranial tumors reporting on clinical outcomes.10 In particular, the role of surgery and adjuvant radiotherapy remains unresolved in molecularly defined classes.
The present study outlines the landscape of DNA methylation profiles in a large multicenter cohort of adult patients with intracranial tumors histopathologically classified as ependymoma WHO grade 2 and 3. All tumors were reclassified utilizing DNA methylation profiling. Within these molecularly defined classes, clinical parameters, and treatment approaches were evaluated with regard to oncological outcomes.
Materials and Methods
For a detailed version of the methods section, see Supplementary Methods.
Patient Population
Formalin-fixed, paraffin-embedded (FFPE) intracranial tumor samples from 170 adult patients (age ≥18 years) histopathologically classified as “Ependymoma WHO Grade 2 or 3” between 1990 and 2020 were collected from 7 centers for neuropathology in Germany (Berlin, Hamburg, Münster, Tübingen, München, and Potsdam) and Switzerland (Bern). Additionally, we obtained previously published DNA methylation data of confirmed adult intracranial ependymoma from the National Cancer Center of Japan.15 Incoming diagnoses were based on the WHO classification for CNS tumors valid at the time of diagnosis. The study was approved by the institutional review board of Charité-Universitätsmedizin Berlin (EA2/236/19), as well as the respective institutional review boards of the participating centers.
Genome-wide DNA Methylation Analysis
Genome-wide DNA methylation analysis was performed using an Illumina HumanMethylation450k or EPIC bead chip array (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The Heidelberg Brain Tumor Classifier was used via the website www.molecularneuropathology.org for the assignment of methylation class for each tumor.16 Tumors were assessed as matching the corresponding methylation class if the classifier indicated a calibrated class score ≥0.84.17
Unsupervised t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis was performed as previously described using the Rtsne package (version 0.15).18
We derived copy-number profiles from the DNA methylation data using R, version 4.2.0 “Vigorous Calesthenics” and the conumee package, version 1.30.0 according to the developer’s recommendation.19 Chromosomal gains and losses were examined by manual inspection of each profile.
DNA methylation array data are available from the Gene Expression Omnibus (GEO) repository under accession number GSE224218 (https://www.ncbi.nlm.nih.gov/geo/).
Clinical Data
Clinical data were collected retrospectively and included demographic information, treatment, and clinical outcome data. Tumor progression was defined as local recurrence or metastatic spread in a contrast-enhanced MRI or CT scan as judged by the respective neuroradiologist. The main endpoint was progression-free survival (PFS). PFS was calculated as the time span from the date of primary surgery to the date of progression. Patients who died without signs of progression were censored. Patients who were still alive and showed no signs of progression at the time of analysis were censored on the day of the last follow-up. Overall survival (OS) was determined as the time between date of primary surgery and date of death. Living patients were censored at the time of the last follow-up. Gross total resection (GTR) was defined as the complete absence of residual tumor as assessed by the neuroradiologist and neurosurgeon.
Statistical Analyses
Statistical tests were done using IBM SPSS Statistics (Version 28.0, Armonk, NY, USA: IBM Corp.). Survival was calculated using the Kaplan–Meier estimate. Effects on survival were analyzed using either the log-rank test or univariate and multivariable Cox regression. Continuous variables were tested for normal distribution and subsequent analyses were performed using the unpaired t-test. Comparison between groups was done using the chi-square test. A P-value <.05 was considered statistically significant.
Results
Patient characteristics of the cohort are summarized in Supplementary Table 1. Median age at diagnosis was 45 years (range 18-81 years). The age distribution was relatively homogenous across 10-year age groups with 2 small peaks in patients 18-30 years (24%) and 51-60 years (21%), with only a few patients (6%) older than 70 years (Supplementary Figure 1). There was slightly more male than female patients (59% and 41%). Incoming histopathological diagnosis was WHO grade 2 ependymoma in 68% of tumors and WHO grade 3 ependymoma in the remaining cases (32%). Tumors were more frequently located in the PF (68%) than in the ST (32%) compartment. CNS metastases were found in 14/139 patients (10%). Detailed patient data are provided in Supplementary Table 2. GTR at initial surgery was achieved in 99/156 patients (64%). Sixty-eight of 163 patients (42%) received adjuvant radiotherapy and one patient received adjuvant radiochemotherapy. A median total dose of 59.4 Gy (range 35-72.0 Gy) in median single doses of 1.8 Gy (range 1.0-3.0 Gy) was delivered to the primary tumor site. Ninety-four of 163 patients (58%) did not receive further adjuvant treatment.
Molecular Subgrouping of Ependymoma
Reclassification of incoming tumors with histological diagnosis of ependymoma or anaplastic ependymoma by DNA methylation profiles are shown in Figure 1. We observed a higher diversity of overall tumor diagnoses among ST tumors (11 ST classes vs. 8 classes of the PF).
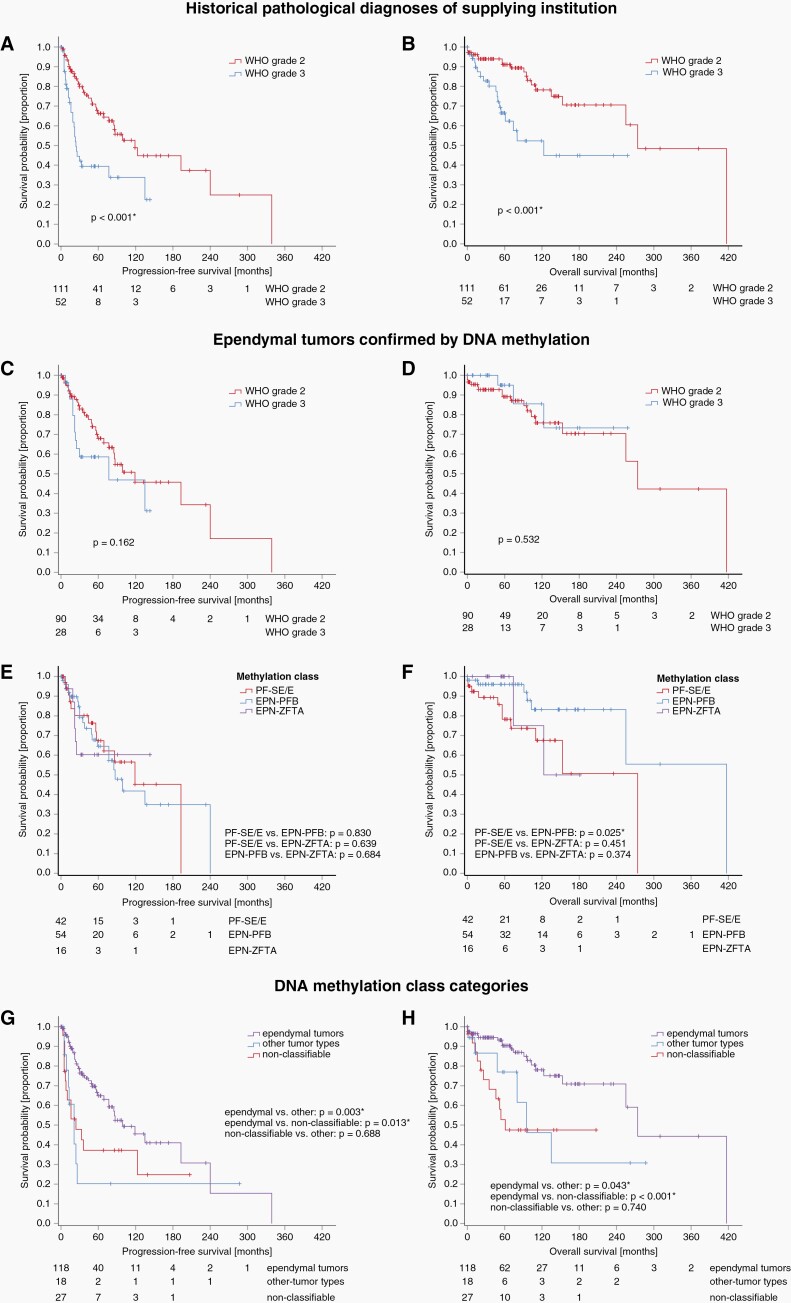
Figure 1.
Reassessment of histologically diagnosed supratentorial (A) and infratentorial (B) ependymoma in adults by DNA methylation profiling. Molecular analyses resulted in the classification of tumors as either belonging to an ependymal class (ependymoma and subependymoma, green), or cases were reclassified into other defined molecular brain tumor classes (yellow) or were not classifiable (gray/black).
Abbreviations: A IDH = astrocytoma, IDH-mutant, C-NC = central neurocytoma, CNS BCOR = neuroepithelial tumor with BCOR alteration, CPP = choroid plexus papilloma A, EPN-PFA = ependymoma posterior fossa group A, EPN-PFB = ependymoma posterior fossa group B, GBM IDHwt = glioblastoma IDH wild type, HGG NEC = high-grade glioma not elsewhere classified, HGNET MN1 = high grade neuroepithelial tumor with MN1 alteration, PF-SE = posterior fossa subependymoma, PTPR = papillary tumor of the pineal region, PXA = pleomorphic xanthoastrocytoma, RGNT = rosette-forming glioneuronal tumor, EPN-ZFTA = supratentorial ZFTA fusion-positive ependymoma, spinal EPN = spinal ependymoma, ST-SE = supratentorial subependymoma.
In 125 of 170 cases (73.5%), epigenetic profiling confirmed the diagnosis of an ependymal tumor. The ependymal tumor class most prevalent was EPN-PFB (56/170, 32.9%). The second largest group (44 of 170, 25.9%) showed a molecular profile of PF-SE. Notably, all PF-SE tumors had ependymoma histology (tumors with pure subependymoma histology were not included in this study). To highlight this aspect, we designate this group hereafter “posterior fossa subependymoma with ependymoma histology” (PF-SE/E). The remaining molecularly confirmed ependymal tumors were distributed among the methylation classes EPN-ZFTA (n = 19, 11.2%), EPN-PFA (n = 3, 1.8%), ST-SE (n = 2, 1.2%), and SP-EPN (n = 1, 0.6%). Clinical patient data according to DNA methylation class are shown in Table 1 for the three most prevalent classes (EPN-PFB, PF-SE/E, and EPN-ZFTA) and additionally for less prevalent classes in Supplementary Table 3. In 18/170 cases (10.6%), the classifier indicated a methylation class outside of the ependymal tumor spectrum with high confidence (classifier score ≥0.84, Figure 1). All these cases were pathologically reviewed and diagnoses were changed as suggested by DNA methylation profiling. The histological appearance and corresponding methylation profile of 4 representative cases are shown in Supplementary Figure 2. Twenty-seven of 170 tumors (15.9%) did not reach a calibrated score ≥0.84 in any methylation class and were therefore considered non-classifiable.
Table 1.
Patient characteristics within the most prevalent ependymal tumor classes
| EPN-PFB | PF-SE/E | EPN-ZFTA | |
|---|---|---|---|
| Number, n (%) | 56 (32.9) | 44 (25.9) | 19 (11.2) |
| Age, [y] median (range) | 39 (18-66) | 60 (29-81) | 37 (20-63) |
| Sex ratio | |||
| Male to female | ~1:1 | ~8:1 | ~2:1 |
| KPS, n (%) | |||
| <70 | 4 (11) | 9 (25) | 1 (12) |
| 70–80 | 11 (31) | 14 (39) | 2 (25) |
| 90–100 | 21 (58) | 13 (36) | 5 (63) |
| No data | 20 | 8 | 11 |
| Extent of surgery, n (%) | |||
| Subtotal | 18 (34) | 18 (45) | 2 (13) |
| Gross total | 35 (66) | 22 (55) | 13 (87) |
| No data | 3 | 4 | 4 |
| Adjuvant therapy, n (%) | |||
| None | 36 (67) | 35 (81) | 4 (25) |
| RT alone | 18 (33) | 8 (19) | 9 (56) |
| Chemotherapy alone | 0 (0) | 0 (0) | 0 (0) |
| RT + Chemotherapy | 0 (0) | 0 (0) | 3 (19) |
| No data | 2 | 1 | 3 |
| CNS WHO Grade, n (%) | |||
| 2 | 49 (88) | 41 (93) | 1 (5) |
| 3 | 7 (12) | 3 (7) | 18 (95) |
| Tumor location, n (%) | |||
| Supratentorial | 2 (4) | 2 (5) | 19 (100) |
| Infratentorial | 54 (96) | 42 (95) | 0 (0) |
| Drop metastases, n (%) | |||
| No | 45 (88) | 34 (97) | 15 (88) |
| Present at diagnosis | 2 (4) | 0 (0) | 0 (0) |
| During disease course | 4 (8) | 1 (3) | 2 (12) |
| No data | 5 | 9 | 2 |
| Disease progression (local recurrence/ metastases) | |||
| Events, n (%) | 21 (38) | 13 (31) | 6 (38) |
| No data | 1 | 2 | 3 |
| Death, n (%) | |||
| Events, n (%) | 7 (13) | 11 (28) | 2 (11) |
| Died of disease | 2 (4) | 1 (3) | 2 (11) |
| Perioperative complications | 1 (2) | 3 (8) | 0 (0) |
| Other reason | 1 (2) | 2 (5) | 0 (0) |
| No data | 3 | 5 | 0 |
| 5y-PFS-rates (%) | 64.5 | 67.4 | 60.3 |
| 10y-PFS-rates (%) | 41.8 | 45.3 | 60.3 |
| 5y-OS-rates (%) | 96.0 | 78.3 | 100.0 |
| 10y-OS-rates (%) | 83.2 | 67.5 | 75.0 |
Abbreviations: EPN-PFB = posterior fossa ependymoma group B, EPN-ZFTA = ZFTA fusion-positive ependymoma, KPS = Karnofsky performance status, OS = overall survival, PFS = progression-free survival, PF-SE/E = posterior fossa subependymoma molecular class with ependymoma histology, RT = radiotherapy.
Reclassification of ependymoma was observed in almost all participating centers with differences in frequency of reclassification to ependymal, non-ependymal, and non-classifiable tumors (Supplementary Figure 3). Cases of PF-SE/E were common in all diagnostic institutions with frequencies ranging between 29% and 67% of ependymal tumors (Supplementary Figure 4).
Major Molecular Classes of Adult Intracranial Ependymoma
Patient characteristics varied between the 3 most prevalent tumor classes of adult intracranial ependymal tumors (EPN-PFB, PF-SE/E, and EPN-ZFTA) and are summarized in Table 1. Age differed significantly between PF-SE/E vs. EPN-PFB and vs. EPN-ZFTA (each P < .001). There was a male predominance in PF-SE/E and EPN-ZFTA tumors, whereas sex was balanced in EPN-PFB tumors. Male patients were significantly older than female patients at initial diagnosis in PF-SE/E tumors (mean age 60.7 vs. 50.8 years, P = .038). WHO grade 2 was the predominant incoming tumor grade in PF-SE/E and EPN-PFB tumors, whereas all but one EPN-ZFTA tumor were designated WHO grade 3 at initial histopathologic grading. Drop metastases were found in equal proportions in EPN-PFB (6 of 51) and EPN-ZFTA tumors (2 of 15, both approximately 12%) and in 1 of the 35 cases (3%) of PF-SE/E. Within EPN-PFB tumors, 5 molecular subclasses have been defined (designated EPN-PFB1-5).3 All of these subclasses were found in this cohort, with EPN-PFB1 being the most prevalent subclass (46.4% of all EPN-PFB tumors, Supplementary Table 4).
Chromosomal Aberrations Within Molecular Classes of Adult Intracranial Ependymoma
Copy-number alterations were found with varying frequency in the different molecular classes. Cytogenetic events across the 3 major molecular classes of adult intracranial ependymoma are summarized in Supplementary Figure 5.
EPN-PFB tumors showed a high degree of genomic instability, which is in accordance with previous reports.2,3 Copy-number changes most common in this class included chromosome 6 loss (75%), 12 gain (62%), 15q gain (60%), 18 gain (58%), and 7 gain (51%).
In PF-SE/E, the most frequent copy-number alteration was the loss of whole chromosome 6 (28/44 cases, 64%), which is in line with earlier observations.2,20
In EPN-ZFTA tumors, chromosomal aberrations were less frequent than in EPN-PFB tumors with chromosome 9 loss being the most common change (seen in 74% of tumors). Deletions of CDKN2A/B (both homozygous and hemizygous) were common in these tumors (11/15, 73%).
Outcome Analysis by Histology, Molecular Subgrouping, and Time of Diagnosis
Among the entire cohort of tumors with histological diagnosis of ependymoma prior to DNA methylation profiling, WHO grade 3 was significantly associated with worse PFS and OS (each P < .001, Figure 2A and B). However, after the exclusion of non-ependymal and non-classifiable tumors by DNA methylation profiling WHO grade was no longer a predictor of survival (Figure 2C and D). Exclusion of these groups was also relevant for prognosis of the entire cohort since reclassification by DNA methylation profiling identified included tumors with very poor prognosis like glioblastoma IDH wildtype: Risk of progression was significantly lower in confirmed ependymal tumors than in the group of non-classifiable tumors and the group of other tumor types (P = .013 and P = .003, respectively, Figure 2G). Given the 30-year span of diagnoses, we also investigated the possible impact of advances in radiology, surgery, and radiation approaches on prognosis. Outcome analysis of ependymal tumors using the Kaplan–Meier method did not show any differences for PFS or OS when patients were grouped by the decade of their initial diagnosis (Supplementary Figure 6).
Figure 2.
Kaplan–Meier curves showing progression-free survival (PFS, left) and overall survival (OS, right). Above diagrams show survival by histological tumor grade for the whole cohort of adult patients with diagnosis of ependymoma prior to DNA methylation profiling (A and B) and for the group of ependymal tumors confirmed by DNA methylation profiling (C and D). Notably, histological tumor grade was no longer a predictor of survival after exclusion of tumors that were non-ependymal and non-classifiable after DNA-methylation profiling. Panels (E and F) show that PFS and OS did not differ significantly between the three major molecular classes of adult intracranial ependymoma. Panels (G and H) show that confirmed ependymal tumors generally had a better PFS and OS in comparison to the groups of other tumor types and non-classifiable tumors as indicated by the classifier demonstrating the clinical importance of DNA methylation profiling.
Abbreviations: EPN-PFB = posterior fossa ependymoma group B, EPN-ZFTA = supratentorial ZFTA fusion-positive ependymoma, PF-SE/E = posterior fossa subependymoma molecular class with ependymoma histology.
Outcome Analysis of Molecular Ependymal Tumor Classes
The number of patients was sufficient for survival analysis in the 3 most prevalent classes in this cohort (EPN-PFB, PF-SE/E, and EPN-ZFTA). However, the retrospective nature with missing data concerning the cause of death for the majority of patients limited the analysis to PFS. Despite these limitations, OS analysis was performed for the largest group (EPN-PFB) and is shown in the supplements (Supplementary Figure 7).
Progression was recorded in 21/55 EPN-PFB patients (38%), 13/42 PF-SE/E patients (31%), and 6 of 16 EPN-ZFTA patients (38%) (Table 1). The 5- and 10-year-PFS rates were 64.5% and 41.8% for EPN-PFB, 67.4% and 45.3% for PF-SE/E, and 60.3% and 60.3% for EPN-ZFTA tumors. PFS did not differ significantly between the 3 classes, notably, not even between PF-SE/E and the class of EPN-ZFTA tumors, the latter associated with poor prognosis at least in children (Figure 2E).2
Disease and non-disease related death occurred in 7 of 53 EPN-PFB patients- (13%), in 11 of 39 PF-SE/E patients- (26%), and in 2 of 19 EPN-ZFTA patients (12%) with available follow-up on OS (Table 1).
Predictors of Oncologic Outcome in Adult EPN-PFB Patients
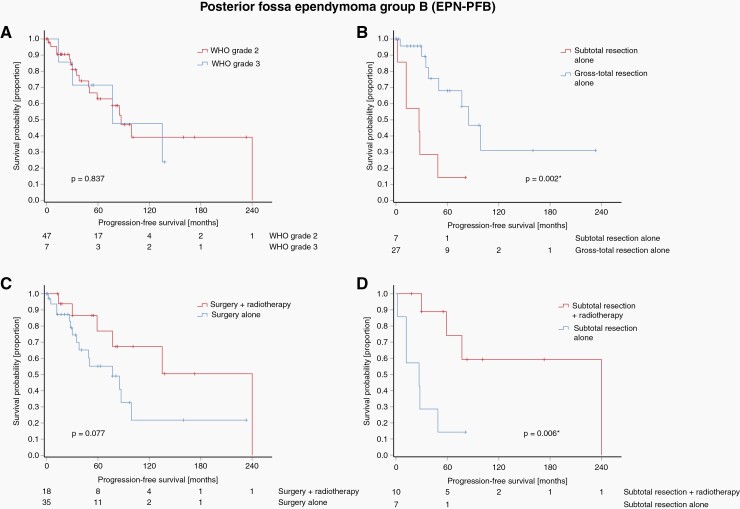
We further analyzed factors associated with PFS in adult EPN-PFB. Median follow-up was 81 months (range 0-407 months). WHO grade was no predictor of PFS in adult EPN-PFB on univariate analysis (Figure 3A). In the EPN-PFB subgroup receiving only surgery GTR was significantly associated with longer PFS compared to incompletely resected tumors (P = .002 Figure 3B). Adjuvant radiotherapy in EPN-PFB tumors regardless of the extent of resection showed a trend towards improved PFS (P = .077, Figure 3C). However, in the subset of incompletely resected EPN-PFB tumors, radiotherapy was significantly associated with improved PFS on univariate analysis (P = .006, Figure 3D). Finally, in multivariable analysis both incomplete resection and lack of adjuvant radiotherapy showed to be independent prognostic factors of worse PFS (HR = 5.92 95% CI 1.84-19.03 P = .003 and HR = 21.26 95% CI 2.29-197.25 P = .007, respectively, Table 2). To what extent these observations can be translated into an effect on the OS could not be clearly determined in this study: In general, patients with EPN-PFB tumors have a rather favorable prognosis (Figure 2F). Neither tumor grade, resection status nor postoperative radiation showed a significant association with overall survival (Supplementary Figure 7). Besides the low number of patients, the analysis was hampered by partially incomplete data regarding the causes of death and an observation period that was likely too short for these slow-growing tumors.
Figure 3.
Kaplan–Meier diagrams showing progression-free survival (PFS) for EPN-PFB tumors. No difference in PFS was seen by histological tumor grade in this molecular class (A). In EPN-PFB patients receiving only surgery gross total resection was associated with improved PFS (B). Use of postoperative radiotherapy showed a tendency to a longer PFS in EPN-PFB tumors regardless of resection status (C). In the subgroup of incompletely resected EPN-PFB tumors, adjuvant radiotherapy was significantly associated with PFS (D).
Table 2.
Multivariable analysis of progression-free survival for EPN-PFB tumors
| P | HR | 95% CI | |
|---|---|---|---|
| Age at diagnosis | .998 | 1.00 | 0.95-1.05 |
| Sex (female vs. male) | .433 | 1.61 | 0.49-5.32 |
| WHO tumor grade (grade 3 vs. grade 2) | .087 | 7.89 | 0.74-84.24 |
| 1q gain (yes vs. no) | .159 | 2.14 | 0.74-6.16 |
| 13q loss (yes vs. no) | .260 | 2.07 | 0.58-7.31 |
| Extent of resection (subtotal vs. gross total resection) | .003* | 5.92 | 1.84-19.03 |
| Postoperative radiotherapy (no vs. yes) | .007* | 21.26 | 2.29-197.25 |
Abbreviations:
* P-value ≤.05, HR = hazard ratio, CI = confidence interval.
In earlier studies on EPN-PFB tumors either gain of chromosome 1q and loss of 13q was associated with worse PFS.2,3 In this study, however, neither 1q gain nor 13q loss was a predictor for PFS in EPN-PFB (Supplementary Figure 8).
Predictors of Oncologic Outcome in PF-SE/E Patients
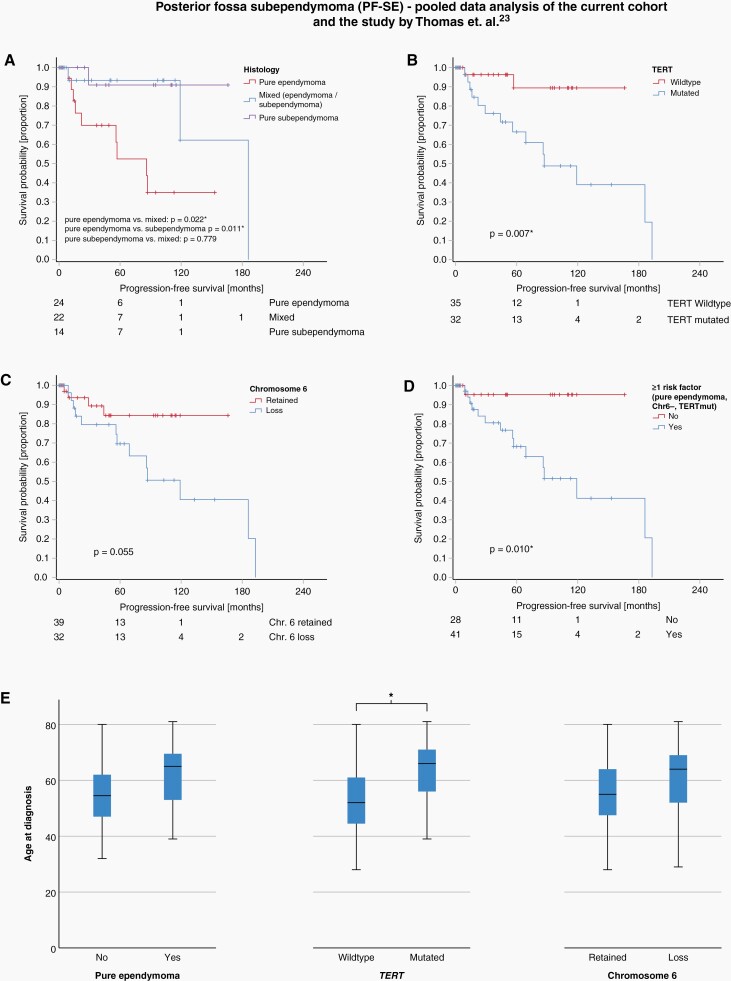
We previously showed that TERT promoter mutations and loss of chromosome 6 are characteristic of PF-SE tumors with ependymoma histology.20 Both TERT promoter mutations, loss of chromosome 6, and pure ependymoma histology were independently associated with reduced PFS in our previous study.20 In the current cohort, we studied tumors initially diagnosed as ependymomas or anaplastic ependymomas and detected a surprisingly high fraction of tumors with PF-SE molecular profile (44/170, 25.9%, subsequently termed PF-SE/E). We histologically re-assessed available PF-SE/E tumor samples (n = 33) and observed no histological indication of subependymoma in 19 cases (58%), whereas in 14 cases (42%) areas with subependymoma morphology were present (ranging from 10% to 95% of the tumor area). To align with our previous series, we performed additional TERT promoter sequencing for all PF-SE/E with available material of this series (n = 40). TERT promoter hotspot mutations were observed with a high frequency (present in 26 of 40 tumors, 59%). In the current series, both TERT promoter mutations and chromosome 6 loss were significantly more frequent in PF-SE tumors with pure ependymoma histology than in tumors with mixed histology (both P < .001).
To optimally investigate the factors influencing PFS in PF-SE tumors, we performed a combined data analysis of the present study and the cohort by Thomas et al. which also included PF-SE tumors of pure subependymoma histology. Their cohort consisted of 50 PF-SE tumors with the following histology: 14 subependymomas, 12 ependymomas, and 24 cases of mixed ependymoma-subependymoma.20 In this combined analysis, factors significantly associated with reduced PFS were pure ependymoma histology vs. pure subependymoma (P = .011) and pure ependymoma histology vs. mixed ependymoma/subependymoma (P = .022), (Figure 4A), TERT promoter mutation (P = .007, Figure 4B) and, with borderline significance, loss of chromosome 6 (P = .055, Figure 4C). The absence of any of these factors were also associated with a favorable PFS (P = .010, Figure 4D). The factors pure ependymoma histology, TERT promoter mutations, and chromosome 6 loss were associated with older age, with a significant association for TERT promoter mutations (P < .001, Figure 4E) indicating an accumulation of genetic alterations increasing with age. These findings further support the hypothesis of supependymoma evolution over time to a more aggressive variant as previously proposed.20
Figure 4.
Kaplan–Meier diagrams showing progression-free survival (PFS) of PF-SE tumors in a combined data analysis of the current series and the series by Thomas et al.20 PFS is shorter in the presence of pure ependymoma histology (A), TERT promoter mutation (B) and, with borderline significance, loss of chromosome 6 (C). The absence of all these factors is associated with a favorable prognosis (D). The changes were more common in elderly patients. This correlation was significant for TERT promoter mutations (P < .001*, E).
In the current series, deaths were observed in a relatively high fraction of PF-SE/E patients (11/39, 28%). Although data were incomplete with regard to the cause of death and the sample size was small, high rates of perioperative complications (3 of 6 with data on cause of death) and nontumor-associated causes of death (2 of 6 with data on cause of death) were observed in patients, which is most likely related to the general vulnerability of this group of patients given their advanced age.
Predictors of Oncologic Outcome in EPN-ZFTA Patients
The low number of EPN-ZFTA patients with available outcome data (n = 16) did not allow for profound statistical assessment of prognostic factors. With a median follow-up of 56 months (range 8-143 months), no differences in PFS or OS in comparison to the other classes of ependymal tumors were identified (Figure 2E). GTR was significantly more often achieved in EPN-ZFTA tumors than in PF-SE/E tumors (P = .03), with 13 of 15 (87%) cases with GTR possibly because of better surgical accessibility on ST tumors. Since all but one EPN-ZFTA tumor (18 of 19, 95%) were designated WHO grade 3, adjuvant treatment was frequent: Radiotherapy was conducted in 12 of 16 (75%) patients with available treatment data, 3 of them (25%) with additional chemotherapy. Adjuvant radiotherapy was significantly more common in EPN-ZFTA patients than in PF-SE/E patients (P < .001) and EPN-PFB patients (P = .003). Progression was observed in 6 of 16 (38%) patients. These patients had a median time to progression of 22 months and all relapses occurred within 25 months after initial treatment. Deletion of CDKN2A/B was earlier identified as a risk factor for poor outcomes in pediatric EPN-ZFTA tumors.21 In this study, we did not detect a significant impact on survival due to the low number of patients.
Assessment of Non-classifiable Tumors With Ependymoma Histology Using t-SNE and CNV Profiles
As the classifier did not indicate a sufficient score (<0.84) for the assignment of a known tumor type in 15.9% of tumors, uncertainty exists regarding the risk of recurrence and survival in these cases. To further clarify the nature of non-classifiable cases, we performed an unsupervised t-SNE analysis of the diagnostic cohort and compared it to a reference set comprising 2801 samples and 82 distinct molecular CNS tumor types (Supplementary Figure 9).16 Using this approach, we found an overlap or close relation with an established molecular tumor class suggestive for biology similar to the corresponding class. Non-classifiable cases were related to IDH wild-type glioblastoma (n = 6), EPN-PFB (n = 5), SP-SE (n = 3), EPN-ZFTA (n = 2), neuroepithelial tumors with BCOR alteration (CNS BCOR, n = 2), ST-SE (n = 1), YAP1 fusion-positive ependymoma (n = 1), high-grade astrocytoma with piloid features (n = 1), chordoid glioma (n = 1), liponeurocytoma (n = 1), low-grade glioma subtype ganglioglioma (n = 1), low-grade glioma subtype posterior fossa pilocytic astrocytoma (n = 1), and low-grade glioma subtype dysembryoplastic neuroepithelial tumor (n = 1). Concordance of related molecular class as assessed by t-SNE and the class with the highest classifier score was seen in 13 (48%) of non-classifiable tumors.
We further visually inspected the CNV profiles of the non-classifiable tumors for tumor-type specific chromosomal alterations and compared the results with the grouping from t-SNE. In 6 of 27 cases (22%) highly specific copy-number alterations were identified that confirmed the t-SNE grouping and allowed a final classification. Five of these were classified as glioblastoma, IDH wildtype because of glioblastoma defining copy-number alterations. Eight of 27 cases (30%) showed a well-compatible but not per se diagnostic chromosomal profile and in 13 of 27 cases (48%) the profile was not specific for a certain tumor class and thus non-informative (Supplementary Table 2).
Relation to molecular subclasses of glioblastoma IDH wild type or CNS BCOR was associated with dismal prognosis. In contrast, no progression occurred in non-classifiable tumors related to a class of SE (SP and STN).
Discussion
This multi-institutional study represents the largest cohort of molecularly classified adult intracranial ependymomas published so far. Reclassification of tumors with ependymoma histology using the brain tumor classifier yielded promising results: First, implementation of this technique reduced the number of misclassifications in almost all supplying centers. Second, it detected tumors with PF-SE molecular profile at a considerable frequency. Third, it identified tumors that currently cannot be allocated to a specific class and should consequently be designated as NEC (not elsewhere classified). The advantages of DNA methylation profiling are particularly evident when compared to classic assessment of ependymoma by histology alone: In our cohort, histological WHO grade is only of prognostic relevance if misdiagnoses are included. In contrast, within the group of molecularly confirmed ependymal tumor classes, no differences in survival were observed between WHO grades 2 and 3. This is of relevance when interpreting studies on adult ependymoma without additional molecular verification. In particular, most treatment recommendations established to date are based on studies that did not account for molecular classes, but included WHO histological tumor grade. However, the numbers of grade 3 tumors within the ependymal tumor classes used for this analysis in this study were relatively small and the statistical power thus remains limited. Still, our data indicate that DNA methylation profiling should be performed at an early point during the diagnostic workup to avoid misclassification and for stratification in clinical studies.
EPN-PFB tumors represent the largest group among intracranial ependymoma in adult patients. Median recurrence-free survival was 87 months, which is best compatible with grade 2 on the WHO malignancy scale. Both, 5y PFS and OS survival rates of our EPN-PFB series were at the lower end compared to previous multicenter cohorts (5y-PFS-rate 64% in this study vs. 73%, ~80% and ~65% in earlier studies; 5y-OS-rate 96% vs. 100%, 100% and 100%).2,10,22 The reason for this could not be elucidated. Additional histological grading within this class was not of prognostic significance. However, the relatively small number of grade 3 tumors in this cohort limited the statistical power of this analysis. On multivariable analysis, both GTR and postoperative radiotherapy were associated with improved PFS. Therefore, postoperative radiotherapy should be considered at least in incompletely resected tumors.
Conclusions regarding a possible OS benefit of adjuvant radiotherapy in EPN-PFB tumors could not be shown based on this study.
Unexpectedly, PF-SE tumors constitute the second largest molecular group in adult intracranial ependymoma (“posterior fossa subependymoma with ependymoma histology”, PF-SE/E). Patients with PF-SE/E tumors were significantly older than patients in other classes of this cohort. Median PFS in PF-SE/E patients was 119 months. Some of these tumors showed partial subependymoma histology (mixed ependymoma/subependymoma), although pure ependymoma was the most common histology. As proposed earlier these tumors likely represent progressed subependymomas characterized by a loss of chromosome 6 and/or TERT promoter mutations that may drive tumor progression.20 Twenty-three tumors with at least one of these changes showed a higher risk of relapse in a combined data analysis. We, therefore, propose that PF-SE tumors with either pure ependymoma histology, chromosome 6 loss, or TERT promoter mutations should be considered as WHO grade 2. Extent of resection and adjuvant radiotherapy was not associated with improved PFS, however, only eight of 44 PF-SE patients (19%) were irradiated following initial surgery. Whether postoperative radiotherapy may be required in PF-SE patients with risk factors of recurrence (ependymoma histology, chromosome 6 loss, TERT promoter mutations) has to be investigated in future studies. Furthermore, radiation may represent an alternative therapeutical approach to extensive surgery for elderly patients who may be at higher risk for surgery-associated complications.
EPN-ZFTA represented the third largest group in this cohort. All of these tumors were located in the supratentorial compartment and macroscopic GTR was achieved in almost all cases possibly due to better surgical accessibility. Adjuvant radiotherapy was performed in two-thirds of patients, only a few received additional chemotherapy. Histopathological grading of all but one EPN-ZFTA was WHO grade 3, leading to a high rate of adjuvant radio- or radiochemotherapy in this class. Median PFS was not reached at the end of follow-up. Notably, all cases of recurrence occurred within 25 months after diagnosis.
A considerable proportion (15.9%) of tumors in this cohort could not be assigned a molecular class by the classifier and was considered non-classifiable. Non-classifiable tumors seem to represent a heterogeneous group that requires additional assessment. Performing t-SNE analysis in these cases may help to further characterize tumor biology, if they fall into—or near the cluster of an established molecular class. T-SNE analysis retrospectively identified tumors mimicking ependymoma on histology with relation to a more malignant class (especially glioblastoma and CNS BCOR) with dismal prognosis and more benign classes (such as SP-SE or ST-SE) with favorable outcomes.
Supplementary Material
Acknowledgments
We thank Katja von Hoff, Andrea Mathis, Daniel Teichmann, Carola Geiler, Peggy Wolkenstein, Christin Siewert, Felix Thierfelder, and Ida Lösche for their support in this project.
Contributor Information
Malte Träger, Department of Radiation Oncology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
Leonille Schweizer, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany; German Cancer Consortium (DKTK), Partner Site Frankfurt/Mainz, German Cancer Research Center (DKFZ), Heidelberg, Germany; Edinger Institute, Institute of Neurology, University of Frankfurt am Main, Frankfurt am Main, Germany; Frankfurt Cancer Institute (FCI), Frankfurt am Main, Germany.
Eilís Pérez, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
Simone Schmid, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Elisabeth G Hain, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Carsten Dittmayer, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
Julia Onken, German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurosurgery, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
Kohei Fukuoka, Division of Brain Tumor Translational Research, National Cancer Center Research Institute, Tokyo, Japan.
Koichi Ichimura, Division of Brain Tumor Translational Research, National Cancer Center Research Institute, Tokyo, Japan; Department of Brain Disease Translational Research, Juntendo University Graduate School of Medicine, Tokyo, Japan.
Ulrich Schüller, Institute of Neuropathology, University Medical Center, Hamburg-Eppendorf, Hamburg, Germany; Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Research Institute Children’s Cancer Center Hamburg, Hamburg, Germany.
Lasse Dührsen, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Michael Müther, Department of Neurosurgery, University Hospital Münster, Münster, Germany.
Werner Paulus, Institute of Neuropathology, University Hospital Münster, Münster, Germany.
Christian Thomas, Institute of Neuropathology, University Hospital Münster, Münster, Germany.
Marielena Gutt-Will, Department of Neurosurgery, Inselspital, Bern University Hospital, Bern, Switzerland.
Philippe Schucht, Department of Neurosurgery, Inselspital, Bern University Hospital, Bern, Switzerland.
Theoni Maragkou, Institute of Tissue Medicine and Pathology, University of Bern, Bern, Switzerland.
Jens Schittenhelm, Department of Neuropathology, Institute of Pathology and Neuropathology, University Hospital Tübingen, Tübingen, Germany.
Franziska Eckert, Department of Radiation Oncology, University of Tübingen, Tübingen, Germany; Department of Radiation Oncology, Medical University Vienna, AKH, Comprehensive Cancer Center, Vienna, Austria.
Maximilian Niyazi, Department of Radiation Oncology, University Hospital, Ludwig-Maximilians-University Munich, Munich, Germany; German Cancer Consortium (DKTK) partner site Munich, Munich, Germany; Bavarian Cancer Research Center (BZKF), Munich, Germany.
Daniel F Fleischmann, Department of Radiation Oncology, University Hospital, Ludwig-Maximilians-University Munich, Munich, Germany; German Cancer Consortium (DKTK) partner site Munich, Munich, Germany.
Mario M Dorostkar, Center for Neuropathology, Ludwig-Maximilians-University Munich, Munich, Germany.
Petra Feyer, Department of Radiation Oncology, Vivantes Hospital Neukölln, Berlin, Germany.
Sven-Axel May, Department of Neurosurgery, Klinikum Chemnitz, Chemnitz, Germany.
Dag Moskopp, Department of Neurosurgery, Vivantes Klinikum Im Friedrichshain, Berlin, Germany.
Harun Badakhshi, Department of Clinical Radiation Oncology, Ernst Von Bergmann Medical Center Potsdam, Potsdam, Germany.
Cornelia Radke, Department of Pathology, Ernst Von Bergmann Medical Center Potsdam, Potsdam, Germany.
Jan Walter, Department of Neurosurgery, Jena University Hospital, Jena, Germany; Department of Neurosurgery, Medical Center Saarbrücken, Saarbrücken, Germany.
Felix Ehret, Department of Radiation Oncology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
David Capper, Department of Neuropathology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
David Kaul, Department of Radiation Oncology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany; German Cancer Consortium (DKTK), Partner Site Berlin, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Funding
This study was supported by the Berlin Cancer Society (Berliner Krebsgesellschaft e.V.) and the German Consortium for Translational Cancer Research (DKTK), partner site Berlin.
Conflict of interest statement: D.C. declares a patent on DNA methylation-based methods for molecular classification of tumor species and holds shares of Heidelberg Epignostix GmbH, Heidelberg, Germany. The other authors have no conflicts of interest to declare.
Authorship statement: Conception and design: M.T., D.C., D.K., and L.S. Collection and assembly of data: M.T., D.C., D.K., L.S., C.D., E.G.H., J.O., K.F., K.I., U.S., L.D., M.M., W.P., C.T., M.G.-W., P.S., T.M., J.S., F.E., M.N., D.F.F., M.M.D., P.F., S.-A.M., D.M., H.B., C.R., and J.W. Histological review: D.C., L.S., C.D., and E.G.H. Data analysis and interpretation: M.T., D.K., D.C., S.S., and E.P. Manuscript writing, final approval of manuscript, accountability for all aspects of the work: All authors.
References
- 1. Rodríguez D, Cheung MC, Housri N, et al. Outcomes of malignant CNS ependymomas: an examination of 2408 cases through the surveillance, epidemiology, and end results (SEER) database (1973–2005). J Surg Res. 2009;156(2):340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavalli FMG, Hubner JM, Sharma T, et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018;136(2):227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pajtler KW, Wen J, Sill M, et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018;136(2):211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer cell. 2011;20(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghasemi DR, Sill M, Okonechnikov K, et al. MYCN amplification drives an aggressive form of spinal ependymoma. Acta Neuropathol. 2019;138(6):1075–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kresbach C, Neyazi S, Schüller U.. Updates in the classification of ependymal neoplasms: the 2021 WHO classification and beyond. Brain Pathol. 2022;32(4):e13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pajtler KW, Mack SC, Ramaswamy V, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133(1):5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Witt H, Gramatzki D, Hentschel B, et al. DNA methylation-based classification of ependymomas in adulthood: implications for diagnosis and treatment. Neuro Oncol. 2018;20(12):1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert MR, Ruda R, Soffietti R.. Ependymomas in adults. Curr Neurol Neurosci Rep. 2010;10(3):240–247. [DOI] [PubMed] [Google Scholar]
- 12. Rudà R, Reifenberger G, Frappaz D, et al. EANO guidelines for the diagnosis and treatment of ependymal tumors. Neuro Oncol. 2017;20(4):445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellison DW, Kocak M, Figarella-Branger D, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Metellus P, Barrie M, Figarella-Branger D, et al. Multicentric french study on adult intracranial ependymomas: prognostic factors analysis and therapeutic considerations from a cohort of 152 patients. Brain. 2007;130(Pt 5):1338–1349. [DOI] [PubMed] [Google Scholar]
- 15. Fukuoka K, Kanemura Y, Shofuda T, et al. Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta neuropathol Commun. 2018;6(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capper D, Stichel D, Sahm F, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van der Maaten L, Hinton G.. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(11):2579–2605. [Google Scholar]
- 19. Hovestadt V, Zapatka M.. Conumee: enhanced copy-number variation analysis using Illumina DNA methylation arrays. (Version 1.9.0). Retrieved from http://bioconductor.org/packages/conumee/
- 20. Thomas C, Thierfelder F, Träger M, et al. TERT promoter mutation and chromosome 6 loss define a high-risk subtype of ependymoma evolving from posterior fossa subependymoma. Acta Neuropathol. 2021;141(6):959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jünger ST, Andreiuolo F, Mynarek M, et al. CDKN2A deletion in supratentorial ependymoma with RELA alteration indicates a dismal prognosis: a retrospective analysis of the HIT ependymoma trial cohort. Acta Neuropathol. 2020;140(3):405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramaswamy V, Hielscher T, Mack SC, et al. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol. 2016;34(21):2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.