Abstract
Background
Glioblastoma is one of the most lethal forms of cancer, with 5-year survival rates of only 6%. Glioblastoma-targeted therapeutics have been challenging to develop due to significant inter- and intra-tumoral heterogeneity. Telomerase reverse transcriptase gene (TERT) promoter mutations are the most common known clonal oncogenic mutations in glioblastoma. Telomerase is therefore considered to be a promising therapeutic target against this tumor. However, an important limitation of this strategy is that cell death does not occur immediately after telomerase ablation, but rather after several cell divisions required to reach critically short telomeres. We, therefore, hypothesize that telomerase inhibition would only be effective in glioblastomas with low tumor burden.
Methods
We used CRISPR interference to knock down TERT expression in TERT promoter-mutant glioblastoma cell lines and patient-derived models. We then measured viability using serial proliferation assays. We also assessed for features of telomere crisis by measuring telomere length and chromatin bridge formation. Finally, we used a doxycycline-inducible CRISPR interference system to knock down TERT expression in vivo early and late in tumor development.
Results
Upon TERT inactivation, glioblastoma cells lose their proliferative ability over time and exhibit telomere shortening and chromatin bridge formation. In vivo, survival is only prolonged when TERT knockdown is induced shortly after tumor implantation, but not when the tumor burden is high.
Conclusions
Our results support the idea that telomerase inhibition would be most effective at treating glioblastomas with low tumor burden, for example in the adjuvant setting after surgical debulking and chemoradiation.
Keywords: Telomerase, glioblastoma, targeted therapy, adjuvant therapy, tumor burden, target validation
Key Points.
TERT knockdown leads to a reduction in the proliferation of TERT promoter-mutant glioblastomas.
TERT loss only leads to prolonged survival in vivo if initiated in animals with low tumor burden.
Importance of the Study.
Given the high prevalence and clonal nature of TERT promoter mutations in glioblastoma, telomerase is considered a promising therapeutic target for this deadly cancer. Prior studies have validated this hypothesis, demonstrating that knockout of the transcription factor GABP, which selectively binds to the mutant TERT promoter, as well as base editing-mediated correction of TERT promoter mutations, are selectively toxic to TERT promoter-mutant glioblastomas. However, an important limitation of this strategy is that cancer cell death upon telomerase inhibition occurs only after multiple cell divisions. For this reason, it is important to define the appropriate clinical setting that would maximize the therapeutic efficacy of telomerase inhibitors. In this study, we use CRISPR interference to demonstrate that TERT promoter-mutant glioblastoma cells are sensitive to telomerase inhibition and undergo telomere crisis. Furthermore, we demonstrate that telomerase inhibition in vivo prolongs survival of xenografted mice only if initiated shortly after tumor implantation, supporting the idea that telomerase inhibition would be a suitable therapeutic strategy for glioblastoma patients with low tumor burden.
Glioblastoma is an aggressive cancer in dire need of therapeutic progress. Despite extensive research efforts, standard therapies for this tumor have not changed substantially in over 10 years1 and 5-year survival rates continue to be less than 10%.2,3 Strategies successfully employed in other cancers, such as inhibiting mutated oncogenic drivers in the RTK-Ras-Raf pathway, have shown very little efficacy.4,5 Additionally, immunotherapeutic agents such as checkpoint inhibitors have achieved some benefit in patients with germline mismatch repair deficiencies6 but have had minimal success in patients whose tumors do not harbor mismatch repair deficiencies.6,7 These challenges can partly be explained by the low mutational rate of glioblastomas compared to epithelial malignancies, such as lung, bladder, endometrial, or colorectal carcinomas.8 Finally, when oncogenic mutations are present, they often exhibit intra-tumoral heterogeneity.9 For example, single-cell sequencing analysis of glioblastomas revealed that multiple activating mutations in EGFR can be found within the same tumor as part of different subclones, which may explain the lack of response or resistance to tyrosine kinase inhibitors.10
Interestingly, while many activated oncogenes in glioblastoma are subclonal, telomerase reverse transcriptase (TERT) promoter mutations commonly occur as clonal events.11,12TERT promoter mutations were discovered in melanoma13 and later found in up to 80% of IDH-wild-type glioblastomas.14,15 These mutations are thought to be responsible for oncogenic re-activation of telomerase, a reverse transcriptase ribonucleoprotein complex that maintains telomere length in cells with high replicative potential.16,17 Without telomerase, cells have a finite number of divisions before telomere erosion and deprotection occurs, with activation of the DNA damage response pathway and induction of senescence and apoptosis.18,19TERT promoter mutations result in the transition of cytidine to thymidine and occur most frequently at 2 “hotspot” loci, named c.-124C and c.-146C, upstream of the transcriptional start site.13 Transcriptional activation was found to occur by recruiting the E26-transformation-specific family transcription factor GA-binding protein,20 which selectively binds to the mutant TERT promoter.
Given that TERT promoter mutations are frequent and among the few clonal oncogenic events in glioblastoma, we hypothesized that telomerase inhibition will be detrimental to the survival of tumor cells. Even before the TERT promoter mutations were discovered, telomerase was explored as an anticancer target because it is expressed in tumors but not most somatic cells.21 The presence of TERT promoter mutations further strengthens the idea that telomerase expression in cancer is an active process rather than simply a marker of immortality. Multiple studies have analyzed cellular responses to short telomeres in normal cells through the use of transgenic mouse models.19,22,23 In addition, there have been several studies that explored the effects of telomerase ablation in cancer cells. Early studies, using a dominant negative form of telomerase24 and anti-telomerase modified oligomers,25 have shown that telomerase loss is detrimental to cancer cells. In transgenic mice, T-cell lymphomas on a telomerase-null background display a less aggressive phenotype with lower penetrance and longer latency than control tumors from telomerase wild-type mice, however, they eventually resume growth through activation of the alternative telomere lengthening (ALT) pathway.26 In glioblastoma, loss of the β1L isoform of the GA-binding protein transcription factor that drives TERT expression leads to cell death in TERT promoter-mutant cells in a telomerase-dependent manner.27 Most recently, TERT promoter mutation correction using programmable base editing was shown to lead to decreased proliferation, telomere length reduction, and senescence in glioblastoma cells, both in vitro and in vivo.28
In this study, we used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) interference (CRISPRi) to demonstrate that telomerase ablation can lead to cell lethality in TERT promoter-mutant glioblastoma cells, both in vitro and in vivo. This occurs over several cell divisions required to cause telomere dysfunction, with telomere shortening, and formation of chromatin bridges. Additionally, we utilize an inducible CRISPRi system to demonstrate that in vivo therapeutic efficacy is only achieved when telomerase expression is turned off early in the tumorigenic process. These results highlight the importance of selecting a patient population with low tumor burden when considering potential clinical applications of telomerase inhibitors.
Materials and Methods
Plasmids
Plasmids used in this study include newly described plasmids including pRDA355 (Addgene # pending), and pLV407 (Addgene # pending), as well as previously described plasmids including pLX_311-KRAB-dCas9 (Addgene plasmid #96918), pLenti-dCas9_KRAB-MeCP2,29 pXPR_023d (in press), lentiGuide-Puro (Addgene plasmid # 52963), and px458 (Addgene plasmid # 48138).
Cell Culture
LN18, T98G, and SF295 glioblastoma cells were obtained from ATCC in December 2019 and genotyped using short tandem repeat analysis. The most recent date of Mycoplasma testing was 9/29/2021 for T98G and SF295 and 11/2/22 for LN18, and the results were negative. Cells were cultured in Dulbecco-modified eagle medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. CPDM0095 and BT112 glioblastoma neurospheres were obtained from the Dana Farber Center for Patient Derived Models. Most recent date of mycoplasma testing was 3/22/22 and the results were negative. Cells were cultured in neural stem cell media supplemented with epidermal growth factor at 20 ng/mL, fibroblast growth factor at 20 ng/mL, and 0.2% heparin.
Genotyping
Genomic DNA was extracted from glioblastoma cell lines LN18, T98G, SF295, CPDM0095, and BT112. PCR was performed using the primers annotated in Supplementary Table 1. The products were then sequenced using Sanger sequencing.
CRISPR Interference
Transcriptional silencing using CRISPRi was performed as previously described.30 Cells were first transduced with pLX_311-KRAB-dCas9 or Lenti_dCas9-KRAB-MeCP231 for in vivo studies. Cells expressing these constructs were then transduced with LentiGuide puro harboring short guide RNAs (sgRNAs) targeting TERT exon 1 (sgTERTe) or the TERT promoter (sgTERTp) (Figure 1B), or as controls, the hypoxanthine phosphoribosyltransferase 1 (HPRT1) promoter or a non-coding region of chromosome 2 (sgCh2.4). For inducible CRISPRi, cells expressing dCas9-KRAB-MeCP2 were transduced with pRDA355 harboring sgTERTe. For rescue experiments, cells were first transduced with pLV407 lentiviral vectors encoding either GFP or TERT. They were then transduced with pXPR_023d harboring sgRNA sgCh2.2 as well as sgTERTe (Supplementary Table 1).
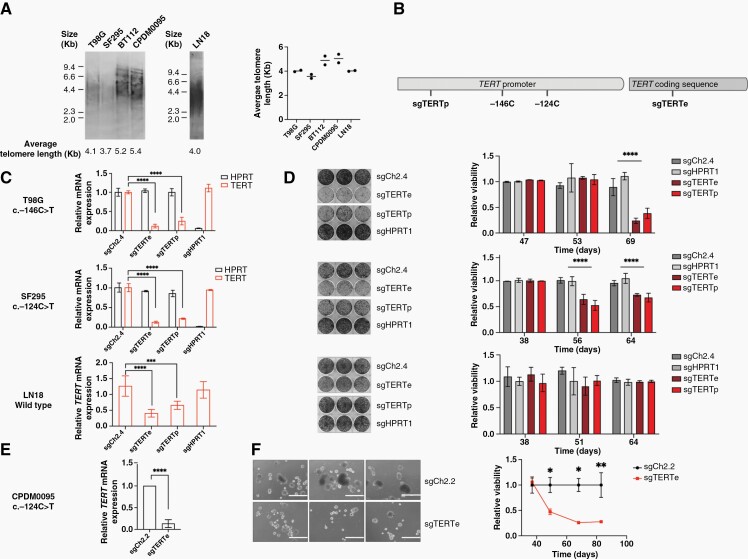
Figure 1.
(A) Average telomere length of TERT promoter-mutant glioblastoma cell lines T98G and SF295 and glioblastoma neurospheres BT112 and CPDM0095, as well as TERT promoter-wild-type cell line LN18. (B) Two sgRNAs targeting the TERT locus: sgTERTp binds to the TERT promoter and sgTERTe binds to TERT exon 1. (C) Relative TERT and HPRT mRNA expression after CRISPR interference treatment of T98G, SF295, and LN18 cells. Two biological replicates were used. (D) Crystal violet-stained plates (left panel) and proliferation curves (right panel) of CRISPR interference-treated T98G, SF295, and LN18 cells. Illustrated plates were stained 69, 65, and 64 days after transduction with sgRNAs for T98G (upper panel) SF295 (middle panel) and LN18 (lower panel). Three technical replicates were used, and the experiment was repeated for validation. (E) Relative TERT mRNA expression for CPDM0095 treated with sgTERTe versus sgCh2.2. Four technical replicates were used. (F) Representative images of CPDM0059 cells harboring sgCh2.2 and sgTERTe 69 days post-transduction (left panel) and proliferation curve (right panel). Scale bars represent 1 mm. Two biological replicates were used. * = P < .05, ** = P < .005, **** = P < .0001.
Generation of TERT-knockout Clones Using CRISPR/Cas9
T98G cells were transfected with the px458 plasmid harboring sgRNAs targeting TERT exon 2 or the AAVS1 locus (Supplementary Table 1). GFP-positive cells were isolated using fluorescence-activated cell sorting and seeded into 96-well plates. Clones were then expanded and the CRISPR target region was amplified using PCR (Supplementary Table 1); amplicons showing evidence of genomic editing based on gel electrophoresis were then sequenced using next-generation sequencing (Illumina paired-end sequencing). Analysis of next-generation sequencing results was done using the NGS Genotyper v1.4.0.
Real-Time PCR
Knockdown efficiency was validated using real-time PCR. Total RNA was extracted from cells and 1 μg of RNA was used for the reverse transcriptase reaction. Real-time PCR products were detected using SYBR green dye and primers targeting TERT, HPRT as well as actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as controls (see Supplementary Table 1 for sequence information).
Colony Formation Assays
Two-dimensional colony formation assays were performed by seeding 8000 cells/well as 3 technical replicates in a 6-well plate. After 8–10 days, the cells were fixed and stained as previously described.32 They were first washed with phosphate-buffered saline (PBS), then fixed in a solution of 4% paraformaldehyde in PBS for 15 minutes, then stained in a solution of 0.2% crystal violet and 2% ethanol for 30 minutes. Dye extraction was performed by adding 2 mL of 10% acetic acid solution to the fixed and stained cells and incubating for 20 min. Quantification was then performed by measuring absorbance at 580 nm.
Growth Curve Generation
TERT-knockout T98G clones and control clones were seeded at a density of 40 000 cells/well in a 24-well plate. The following day, they were transferred to the Incucyte chamber and images were taken every 6 hours (25 images per well). Growth curves were plotted using the Incucyte software based on percent confluency.
Cell Cycle Analysis
Cells were seeded at a density of 250 000 cells/well in 6-well plates. The next day, they were trypsinized and fixed in cold 70% ethanol for 2 hours. They were then washed with PBS and resuspended in a staining solution of 100 μg/mL RNAse A and 50 μg/mL propidium iodide in PBS; incubation was for 30 minutes at 37°C. Data were collected using a Beckman CytoFLEX flow cytometer (5000 events per sample) and analyzed using FloJo.
Chromatin Bridge Analysis
Cells were trypsinized and seeded on silicone-based coverslips in a 6-well plate at a density of 200 000 cells/well. The following day, they were fixed in a solution of 4% paraformaldeyde in PBS for 15 minutes and stained using 4ʹ,6-diamidino-2-penylindole. Images were captured on a Nikon Ti-E inverted microscope with an Andor CSU-X1 spinning disc confocal system using a 60x oil immersion objective. For each condition, 10 separate fields were photographed, and the number of chromatin bridges was counted in each field by 2 independent observers.
Protein Expression Analysis by Immunoblotting
Protein lysates were prepared using CHAPS lysis buffer supplemented with protease inhibitor (Millipore Sigma 11697498001) and 2.5 mM MgCl2. Fifty micrograms of protein were loaded for each sample and transferred to a PVDF membrane (Millipore Sigma IPVH00010). The following antibodies were used: Anti-TERT (Rockland 600-401-252S), anti-PARP (Cell Signaling Technologies #9532), anti-cleaved PARP (Cell Signaling Technologies #5625), and anti-actin (Cell Signaling Technologies #4967). Secondary antibodies included goat anti-rabbit (LI-COR Biosciences 926-32211) and goat anti-mouse (LI-COR Biosciences 926-68020).
Telomere Length Measurements
Telomere length was measured using the Telo TTAGGG telomere length assay (Millipore Sigma 12209136001), based on telomere restriction fragment analysis.33 Briefly, genomic DNA was extracted from cells and 1.5 μg of DNA was digested using HinFI and RsaI. Digestion products were separated using agarose gel electrophoresis (0.8% agarose in TAE buffer), transferred overnight onto a nylon membrane using capillary action in 20× SSC buffer, and crosslinked using ultraviolet light. Hybridization was performed for 3 hours using a digoxigenin-linked telomere probe. The membrane was then incubated in a solution containing anti-digoxigenin antibody fragments linked to alkaline phosphatase. Luminescence signal was generated using the CDP-Star chemiluminescence substrate and detected using a chemiluminescence scanner. Developed films were scanned and quantified using Fiji (ImageJ).
Intracranial Mouse Injections
Animal studies were performed in compliance with the guidelines and regulations of the Broad Institute Institutional Animal Care and Use Committee. Six-week-old female NOD-scid ILRgammanull (NSG) mice weighing between 15 and 20 g were purchased from The Jackson Laboratory. Intracranial tumor cell injections were performed as previously described.27 Mice were anesthetized using isoflurane until not responsive to pinch reflex test. After preparing the surgical field, a 1-cm skin incision was made in the scalp and the skull was penetrated using a drill with a 1.4-mm burr, 2 mm to the right of the bregma, directly posterior to the right suture. The needle was then inserted at 2 mm depth and 300 000 cells in 2 μL of PBS were injected. The injection was performed over 1 minute and the needle was kept in place for 1 minute after injection. The surgical site was closed by suturing with 4–0 monofilament sutures. Perioperative care included a subcutaneous injection of 1 mg/kg buprenorphine directly after the procedure and 3 daily subcutaneous doses of 1 mg/kg meloxicam starting on the day of surgery. Animals were euthanized once they met humane endpoints of lethargy, neurological symptoms, or weight loss of 20% from initial weight. For doxycycline-inducible experiments, T98G cells harboring Lenti_dCas9-KRAB-MeCP2 as well as inducible sgTERTe were injected intracranially in mice. Animals in the control group received regular feed, while animals in the experimental group received feed supplemented with doxycycline at 625 ppm.
Tumor Imaging
Animals were anesthetized with isoflurane and received intraperitoneal injections of 150 mg/kg luciferin. They were then placed in the imaging chamber of the Perkin Elmer in vivo imaging system and bioluminescent images were captured. Luminescence was quantified using the Living Image software.
Statistical Analysis
Statistical methods were not used to predetermine the sample size. Data in all graphs shown are presented as the mean of independent biological or technical replicates as indicated in the figure legends and error bars represent standard deviations. For Figures 1C, D, and F, 2D, 3A and B, and 4D, H, and J (bioluminescence curves), Supplementary Figures 3B, D and 5, P-values were calculated using the unpaired t-test (GraphPad Prism 9). In Figure 4E, I, and K and Supplementary Figure 7C (survival curves), survival analysis was performed using the Kaplan–Meier method and P-values were calculated using the Log-rank test (GraphPad Prism 9).
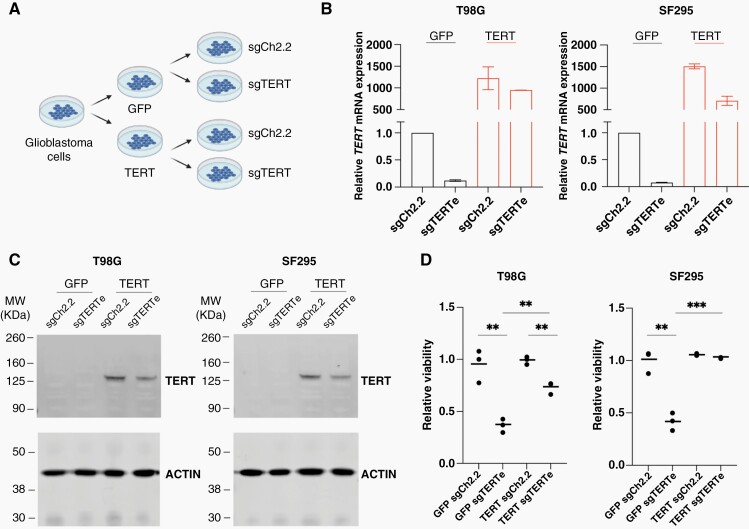
Figure 2.
(A) Design of rescue experiments. (B) Real-time qRT-PCR analysis of TERT mRNA levels in sgCh2.2-treated and sgTERTe-treated T98G and SF295 cells, with ectopic GFP expression (left) or ectopic TERT expression (right). (C) Immunoblot with anti-TERT antibody 600-491-252 (D) Normalized 580 nm absorbance of crystal violet-stained plates seeded with GFP-expressing T98G and SF295 cells harboring sgCh2.2 versus sgTERTe (left) or TERT-expressing cells harboring sgCh2.2 versus sgTERTe (right). Colony formation assays were stained 32 and 35 days after transduction with sgRNAs for T98G and SF295 cells, respectively. Three technical replicates were used, and the experiment was repeated for validation. ** = P < .005, *** = P < .001.
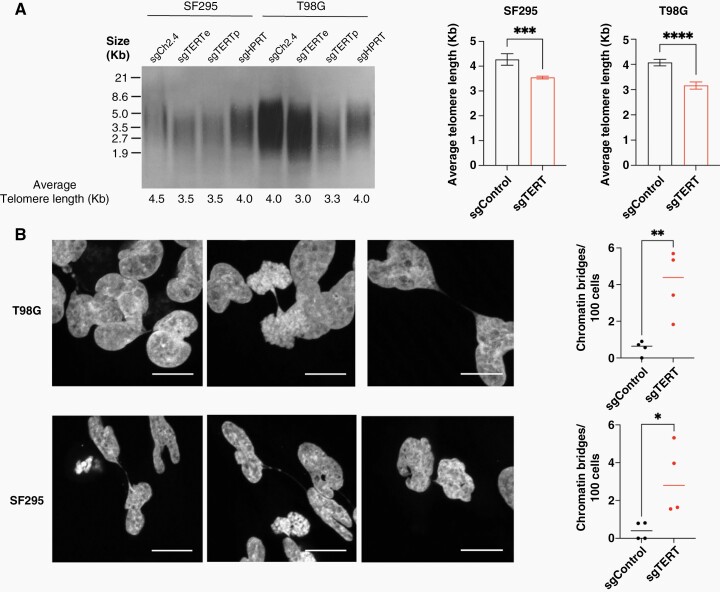
Figure 3.
(A) Telomere restriction fragment analysis of SF295 and T98G glioblastoma cells with and without TERT knockdown, performed 46 days after induction of knockdown. Two technical replicates were used for this analysis. (B) Representative images showing chromatin bridges in TERT-knockdown T98G and SF295 cells. Scale bars represent 2.17 μm. (C) Quantification of chromatin bridges in TERT-knockdown T98G and SF295 cells compared to controls. * = P < .05, ** = P < .005, *** = P < .001, **** = P < .0001.
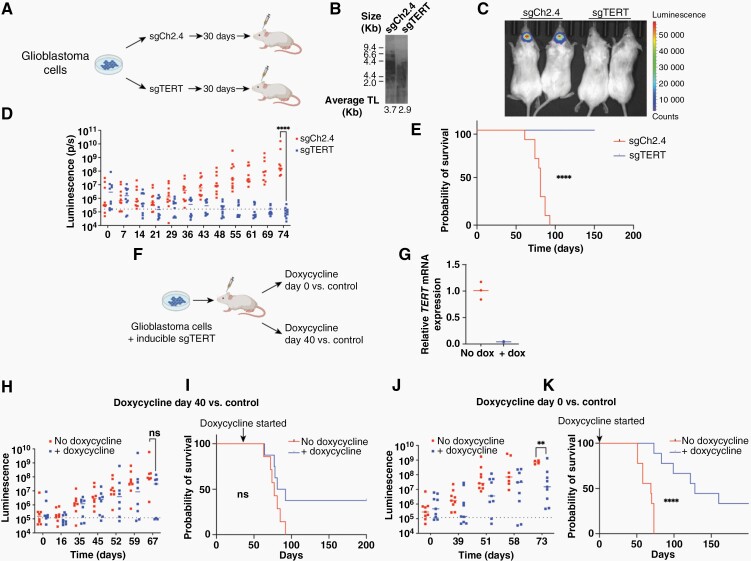
Figure 4.
(A) Schematic diagram of in vivo xenograft experiments. (B) telomere restriction fragment analysis showing average telomere lengths of T98G cells treated with sgCh2.4 and sgTERTe prior to implantation into mice. (C) Representative image of intracranial luminescence of animals harboring control and TERT-knockdown T98G cells; images were taken 55 days after tumor implantation. (D) Serial measurements of intracranial luminescence of animals harboring control and sgTERTe-treated T98G cells. (E) Kaplan–Meier curve showing overall survival of animals harboring sgTERTe-treated T98G cells compared to mice harboring control cells. (F) Schematic of in vivo experiment using a doxycycline-inducible CRISPR interference system. (G) Relative TERT mRNA expression of intracranial tumors of animals treated with doxycycline feed versus control feed. (H) Serial measurements of intracranial luminescence of animals treated with doxycycline feed at day 40 post-surgery versus control feed. (I) Overall survival of animals treated with doxycycline feed at day 40 post-surgery versus control. (J) Serial measurements of intracranial luminescence of animals treated with doxycycline feed on the day of surgery versus control feed. (K) Overall survival of animals treated with doxycycline feed on the day of surgery versus control feed. ** = P < .005, **** = P < .0001, ns = nonsignificant.
Results
Telomerase Loss Halts Proliferation of TERT Promoter-Mutant Glioblastoma Cells In Vitro
We selected TERT promoter-mutant glioblastoma cell lines T98G and SF295 and TERT promoter-wild type LN18 cells for this study. We also selected glioblastoma patient-derived neurospheres BT112 and CPDM0095. LN18 was confirmed to be TERT promoter wild type, T98G and SF295 were confirmed to be heterozygous for the c.-146C > T and c.-124C > T mutations, respectively, and BT112 and CPDM0095 were found to be heterozygous for the c.-124C > T mutation using Sanger sequencing (Supplementary Figure 1). We then measured telomere length in these cell lines using the telomere restriction fragment assay33 (Figure 1A) and found that the average telomere length is 4.1 Kb for T98G, 3.7 Kb for SF295, 5.2 Kb for BT112, 5.4 Kb for CPDM0095 and 4.0 Kb for LN18. We then applied CRISPR interference30 to inhibit expression of the telomerase protein TERT in cell lines T98G, SF295, and LN18. Two different sgRNAs were used, sgTERTe targeting TERT exon 1 and sgTERTp targeting the TERT promoter (Figure 1B), leading to reduction in TERT mRNA levels of >70% for the TERT promoter-mutant lines and >50% for LN18 (Figure 1C). Two sgRNAs were used as control, sgCh2.4, targeting a non-coding region on chromosome 2, as well as sgHPRT1, targeting the promoter of HPRT1, which is not known to be an essential gene for cell survival. TERT knockdown led to a decrease in proliferation manifesting over a period of 69 days for T98G and 65 days for SF295. We did not detect a significant reduction in proliferation for LN18 cells over a period of over 64 days (Figure 1D). T98G and SF295 cells harboring sgTERTe or sgTERTp eventually restored TERT expression (Supplementary Figure 2A) by decreasing Cas9 expression, restoring viability and proliferative capacity compared to control cells with HPRT1 knockdown (Supplementary Figure 2B), which in contrast retained Cas9 expression and HPRT1 loss (Supplementary Figure 2C). The restoration of survival and proliferation by loss of Cas9 expression supports the idea that telomerase-null cells are under negative selective pressure. We validated these results using glioblastoma patient-derived neurosphere CPDM0095. CPDM0095 cells harboring sgTERTe exhibited a reduction in TERT mRNA levels of >90% compared to cells harboring sgCh2.2 (Figure 1E). These cells also exhibit a loss of proliferation over a period of 50–80 days (Figure 1F).
To further validate the effect of TERT knockdown on proliferation in clonal rather than polyclonal populations, we generated T98G single-cell clones harboring homozygous frameshift edits in TERT exon 2 using CRISPR/Cas9. We identified 2 clones with frameshift edits in TERT exon 2 corresponding to the CRISPR sgRNA binding site (Supplementary Figure 3A). TERT-edited clones proliferated at a lower rate when compared to control clones (Supplementary Figure 3B). These results further support the conclusion that telomerase is essential for cell survival in TERT promoter-mutant glioblastoma cells.
To verify whether the viability defect caused by anti-TERT sgRNAs in T98G and SF295 cells was due to reduction of TERT expression, we asked whether ectopic expression of TERT would rescue this growth defect. We ectopically expressed GFP and TERT in T98G and SF295 cells (Figure 2A). This ectopic expression led to a significant increase in TERT mRNA levels even when the TERTe sgRNA was also expressed (Figure 2B). When we attempted to assess TERT protein levels by immunoblotting, we saw a band at approximately 125 kDa only in the cells with TERT overexpression (Figure 2C). This result indicates that TERT ectopic expression was successful and endogenous TERT protein is not detectable by immunoblot in T98G and SF295 cells under our experimental conditions. Next, we used crystal violet staining of colony formation to assess the proliferation status of T98G and SF295 cells transduced with sgTERTe and the sgCh2.2 control. Overexpression of wild-type TERT in both T98G and SF295 cells rescued the proliferation defect induced by sgTERTe (Figure 2D).
TERT-knockdown Glioblastoma Cells Exhibit Telomere Shortening and Evidence of Telomere Dysfunction
To understand the mechanism of proliferation arrest in TERT-knockdown glioblastoma cells, we measured telomere length using the telomere restriction fragment assay. We measured telomere length 46 days after TERT knockdown in control and telomerase-deficient cells. We found that the average telomere length of TERT-knockdown cells was on average ~900 base pairs shorter than the controls for T98G and ~700 base pairs shorter than the controls for SF295 (Figure 3A). Similarly, TERT-edited single-cell clones had shorter telomere lengths compared to control clones (Supplementary Figure 3C). Short telomeres are known to cause growth arrest by senescence, apoptosis, or telomere crisis; telomere crisis can occur in the absence of a functioning p53 pathway.34–37 Alterations in the p53 pathway are frequent in glioblastomas, occurring in up to 85% of cases through TP53 mutations, CDKN2A deletion, and MDM1/2/4 amplification.38 Both T98G and SF295 cells carry TP53 loss of function mutations (Supplementary Figure 4A) as well as homozygous CDKN2A deletions. Upon telomere shortening and growth arrest, we did not observe an increase in apoptosis markers by immunoblot (Supplementary Figure 4B). We found that telomerase-deficient cells undergo cell cycle arrest, with an accumulation of cells in the S or G2/M phases of the cycle, a phenotype that was pronounced in TERT-deficient T98G clones (Supplementary Figure 3D) but not in cell populations treated with CRISPR interference (Supplementary Figure 5). This difference may be due to the fact that the population of cells treated with CRISPR interference is more heterogeneous than in the clones.
Regarding the mechanism of cell death induced by telomere shortening, on the chromosomal level, we observed a significant increase in chromatin bridges in telomerase-deficient cells compared to control cells (Figure 3B). Chromatin bridges are thought to occur from fusions between dysfunctional telomeres that have become deprotected and have been described as precursors to catastrophic genomic events in telomere crisis, including chromothripsis and katagesis.37 Together, these findings suggest that upon telomerase ablation, glioblastoma cells undergo a telomere crisis.
Telomerase Inhibition In Vivo Prolongs Survival Only When Induced in the Low Tumor Burden Setting
To further validate telomerase dependency in glioblastoma, we generated luciferase-expressing TERT-knockdown and control T98G cell populations and performed intracranial xenograft injections into immunocompromised mice (Figure 4A). We allowed cells to proliferate in vitro for 30 days before injecting them into mice. Shortly before implantation, TERT mRNA levels were reduced by >99% in TERT-knockdown cells compared to controls, and their average telomere length was 3.7 Kb for control cells and 2.9 Kb for TERT-knockdown cells (Figure 4B). We observed a significant reduction in tumor-forming abilities in TERT-knockdown cells, which did not form intracranial tumors in over 60 days (Figure 4C, D). This in turn led to significantly prolonged survival for animals injected with telomerase-deficient cells versus control cells (Figure 4E). It is possible that the reduction in tumor-forming abilities of TERT-knockdown cells was due to telomere shortening that took place while the cells were proliferating in culture prior to implantation, rather than by the impact of the loss of telomerase activity after implantation. We then sought to determine the degree of tumor burden that would be required to achieve a therapeutic benefit from telomerase inhibition. For this purpose, we generated an inducible CRISPRi system using sgTERTe, which successfully suppressed TERT expression in vitro (Supplementary Figure 6). We then performed intracranial xenograft injections of T98G cells harboring the inducible CRISPRi system. We divided our animals into 2 cohorts, one where we started doxycycline feeding 40 days post-surgery and one where were started doxycycline on the day of surgery (Figure 4F). We found that in vivo TERT expression was successfully suppressed (Figure 4G). While there was no statistically significant difference in intracranial luminescence in animals treated with doxycycline at day 40 (Figure 4H), we detected a significant difference in intracranial luminescence 73 days after tumor implantation in animals treated at day 0 (Figure 4J). Similarly, we detected a survival benefit only for the group that received doxycycline at day 0 (Figures 4I and K). In a follow-up experiment, we administered doxycycline feed at additional intermediate time points (days 10 and 25) (Supplementary Figure 7A). We did not observe a significant difference in intracranial luminescence signal between the groups (Supplementary Figure 7B), and we only observed a statistically significant prolongation in survival for animals that were treated at days 0 and 10 (Supplementary Figure 7C). Longer-term follow-up suggests a survival advantage for a subset of mice with high tumor burden treated with doxycycline to induce TERT silencing. There were no long-term surviving mice in a 200-day experiment, in the group without doxycycline induction of TERT silencing. In contrast, after 200 days of doxycycline treatment, there were 3 surviving mice in the group that was treated with doxycycline at day 0 (Figure 4K) as well as 3 surviving mice in the group that was treated with doxycycline at day 40 (Figure 4I). In addition, there were 2 mice in the day 0 and day 10 induction arms, as well as 1 mouse in the day 25 induction arm, still surviving at day 100 in the follow-up experiment (Supplementary Figure 7C). Overall, these results suggest that the most appropriate clinical setting for the deployment of a telomerase inhibitor might be for glioblastoma patients with low tumor burden, but a subset of patients with high tumor burden may benefit as well if the human disease would recapitulate the observations seen in this mouse orthotopic model.
Discussion
Glioblastoma is among the deadliest of all cancers, with a median duration of survival of only 14 months.1 In the past decade, there has been significant progress in understanding the genomic landscape of glioblastoma and glioblastomas were among the first tumors to be studied in The Cancer Genome Atlas project (TCGA).38 Despite these advances, standard therapeutic options have not changed significantly since 2005, when the addition of the alkylating agent temozolomide to radiation therapy was found to confer an overall survival benefit of 2.6 months for all patients1 and 6.4 months for patients whose tumors harbor methylation at the MGMT promoter.39 Clinical trials of targeted therapeutics aimed towards mutant and amplified oncogenic drivers have shown very little benefit.4,5 These results can be explained by a unique feature of glioblastomas, which is their genomic heterogeneity as evidenced by independent amplifications of multiple oncogenic driver genes in distinct tumor cells9 or by multiple activating mutations of the same driver gene in distinct tumor cells.10
In contrast, multiple studies have reported that TERT promoter mutations are the most common clonal activating mutations in glioblastoma.11 The TERT promoter mutations are therefore thought to arise early in tumor evolution.11 For this reason, TERT promoter mutations could provide a unique therapeutic opportunity with a lower probability of exhibiting intrinsic resistance from intra-tumoral heterogeneity. Prior studies have demonstrated that silencing the TERT promoter by CRISPR-mediated ablation of the GA-binding protein transcription factor,27 or by correction of TERT promoter mutations using base editing28 is deleterious to glioblastoma cells. In this study, we silenced the TERT promoter using CRISPR interference. This method leads to the reliable and substantial reduction of TERT mRNA levels. CRISPR interference can be helpful to understand the effects of telomerase loss in a population of cells rather than individual knockout clones. Its advantage over traditional CRISPR editing is that the degree of knockdown can be readily measured and followed using real-time PCR. This is particularly useful when studying telomerase since TERT protein levels are challenging to detect due to low endogenous expression in cells.40 The limitation of CRISPR interference relative to TERT-knockout clones is that telomerase-null cells are gradually lost in the population over cells with wild-type TERT expression and low Cas9 expression. The phenotype of cells in telomere crisis is therefore more pronounced in TERT knockout clones, which are a more appropriate model to perform mechanistic evaluations of cell lethality.
We found that TERT loss in TERT promoter-mutant glioblastoma cells leads to a reduction in cell viability associated with features of telomere crisis, including the formation of chromatin bridges and cell cycle arrest. This suggests that telomerase is not only an important driver of glioma initiation, but it is also key for tumor maintenance, raising the possibility that telomerase-targeted therapeutics may be effective at treating this deadly cancer. An important limitation of telomerase inhibitors as anticancer therapeutics is that cell death upon telomerase loss does not occur immediately but requires several cell divisions. Before considering this strategy, it is, therefore, crucial to demonstrate whether telomerase inhibition can offer a therapeutic benefit in vivo, and if so in what specific clinical setting. With this study, we showed that telomerase loss does not lead to a survival benefit in animals with high tumor burden, but it provides a significant benefit in the low tumor burden setting. Here, we should mention the limitation that our current animal model data represent only the study of a single cell line, albeit under many experimental conditions. This supports the idea that telomerase inhibitors could be employed in the adjuvant setting when tumor debulking has recently occurred and the tumor burden is low. A recent study showing that telomerase loss sensitizes glioblastoma cells to DNA damage20 further supports the idea that telomerase inhibitors could be offered to glioblastoma patients in conjunction with adjuvant temozolomide.
In conclusion with this study we describe the results of TERT knockdown in a population of cells using CRISPR interference. Using this approach, we showed that TERT promoter-mutant glioblastoma cells are dependent on telomerase and exhibit classic features of telomere crisis upon telomerase loss. Using orthotopic xenograft models, we also showed that only animals with low tumor burden achieve a survival benefit from telomerase inhibition. These results support the value of preclinical and eventually clinical investigations of anti-telomerase compounds to treat glioblastoma, and they help in the identification of the patient population that would most benefit from this therapeutic strategy.
Supplementary Material
Acknowledgments
We thank Andrew Allen for technical assistance with the confocal microscope at the Broad Institute. We thank Dr. Douglas Wheeler, Dr. Gizem Uzumbas and Dr. Charlie Lee for their insightful discussion. We thank Dr. Tsukasa Shibue for plasmid pXPR_023D.
Contributor Information
Elisa Aquilanti, Division of Neuro Oncology, Department of Medical Oncology, Dana Farber Cancer Institute, Boston, Massachusetts, USA; Department of Medical Oncology, Dana Farber Cancer Institute, Boston, Massachusetts, USA; Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Lauren Kageler, Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Jacqueline Watson, Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Duncan M Baird, Division of Cancer and Genetics, School of Medicine, Cardiff University, Cardiff, UK.
Rhiannon E Jones, Division of Cancer and Genetics, School of Medicine, Cardiff University, Cardiff, UK.
Marie Hodges, Division of Cancer and Genetics, School of Medicine, Cardiff University, Cardiff, UK.
Zsofia M Szegletes, Genetic Perturbation Platform, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
John G Doench, Genetic Perturbation Platform, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Craig A Strathdee, Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Jose Ricardo Mc Faline Figueroa, Division of Neuro Oncology, Department of Medical Oncology, Dana Farber Cancer Institute, Boston, Massachusetts, USA.
Keith L Ligon, Department of Pathology, Brigham and Women’s Hospital, Boston Children’s Hospital, Dana Farber Cancer Institute, Boston, Massachusetts, USA.
Matthew Beck, Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Patrick Y Wen, Division of Neuro Oncology, Department of Medical Oncology, Dana Farber Cancer Institute, Boston, Massachusetts, USA.
Matthew Meyerson, Department of Medical Oncology, Dana Farber Cancer Institute, Boston, Massachusetts, USA; Cancer Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA; Center for Cancer Genomics, Dana Farber Cancer Institute, Boston, Massachusetts, USA; Department of Genetics and Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Funding
E.A. is the Ben and Catherine Ivy Foundation Physician-Scientist of the Damon Runyon Cancer Research Foundation (PST-28-20). The Baird laboratory was supported by Cancer Research UK (A18246/A29202) and the Wales Cancer Research Centre.
Conflict of interest statement
P.W.: Research Support: Agios, Astra Zeneca/Medimmune, Beigene, Celgene, Eli Lily, Genentech/Roche, Kazia, MediciNova, Merck, Novartis, Nuvation Bio, Oncoceutics, Vascular Biogenics, VBI Vaccines. Advisory Board: Agios, Astra Zeneca, Bayer, Boston Pharmaceuticals, CNS Pharmaceuticals, Elevate Bio Immunomic Therapeutics, Imvax, Karyopharm, Merck, Novartis, Nuvation Bio, Vascular Biogenics, VBI Vaccines, Voyager, QED, Celularity, Sapience. J.G.D.: consults for Microsoft Research, Abata Therapeutics, Servier, Maze Therapeutics, BioNTech, Sangamo, and Pfizer; consults for and has equity in Tango Therapeutics; serves as a paid scientific advisor to the Laboratory for Genomics Research, funded in part by GSK; receives funding support from the Functional Genomics Consortium: Abbvie, Bristol Myers Squibb, Janssen, Merck, and Vir Biotechnology. M.M. is the scientific advisory board chair of Isabl; consults for Bayer, DelveBio and Interline; is an inventor of patents licensed to LabCorp and Bayer; and receives research funding from Bayer and Janssen. K.L. Research Support: BMS; Consulting : BMS, Travera, Integragen Equity: Travera.
Author Contributions
E.A., P.W., and M.M. generated the idea for the study. E.A., J.W., and L.K. executed most of the experimental work. D.B., R.J., and M.H. provided advice and expertise on telomere length measurement methods. Z.M.S. and J.G.D. developed the dox-inducible system. C.A.S developed the overexpression construct. K.L. provided the glioblastoma neurospheres. M.B. helped with in vivo experiments. J.R.M.F. helped with CRISPR knockdown glioblastoma neurospheres. E.A. and L.K. wrote the manuscript. M.M supervised the work and edited the manuscript.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Weller M, Lee EQ, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raizer JJ, Giglio P, Hu J, et al. ; Brain Tumor Trials Collaborative. A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J Neurooncol. 2016;126(1):185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reardon DA, Nabors LB, Mason WP, et al. ; BI 1200 36 Trial Group and the Canadian Brain Tumour Consortium. Phase I/randomized phase II study of afatinib, an irreversible ErbB family blocker, with or without protracted temozolomide in adults with recurrent glioblastoma. Neuro Oncol. 2015;17(3):430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 7. Reardon DA, Brandes AA, Omuro A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the checkmate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020;6(7):1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 10. Francis JM, Zhang CZ, Maire CL, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4(8):956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korber V, Yang J, Barah P, et al. Evolutionary trajectories of IDH(WT) glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell. 2019;35(4):692–704.e12. [DOI] [PubMed] [Google Scholar]
- 12. Brastianos PK, Nayyar N, Rosebrock D, et al. Resolving the phylogenetic origin of glioblastoma via multifocal genomic analysis of pre-treatment and treatment-resistant autopsy specimens. NPJ Precis Oncol. 2017;1(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4(2185). doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 16. Greider CW, Blackburn EH.. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337(6205):331–337. [DOI] [PubMed] [Google Scholar]
- 17. Meyerson M, Counter CM, Eaton EN, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–795. [DOI] [PubMed] [Google Scholar]
- 18. AS IJ, Greider CW.. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(3):987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. [DOI] [PubMed] [Google Scholar]
- 20. Amen AM, Fellmann C, Soczek KM, et al. Cancer-specific loss of TERT activation sensitizes glioblastoma to DNA damage. Proc Natl Acad Sci U S A. 2021;118(13). doi: 10.1073/pnas.2008772118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn WC, Meyerson M.. Telomerase activation, cellular immortalization and cancer. Ann Med. 2001;33(2):123–129. [DOI] [PubMed] [Google Scholar]
- 22. Lee HW, Blasco MA, Gottlieb GJ, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392(6676):569–574. [DOI] [PubMed] [Google Scholar]
- 23. Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. [DOI] [PubMed] [Google Scholar]
- 24. Hahn WC, Stewart SA, Brooks MW, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5(10):1164–1170. [DOI] [PubMed] [Google Scholar]
- 25. Herbert B, Pitts AE, Baker SI, et al. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci USA. 1999;96(25):14276–14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu JH SS, Liesa M, Gan B, et al. Anti-telomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148(4):651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mancini A, Xavier-Magalhaes A, Woods WS, et al. Disruption of the beta1L isoform of GABP reverses glioblastoma replicative immortality in a TERT promoter mutation-dependent manner. Cancer Cell. 2018;34(3):513–528.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X, Qian X, Wang B, et al. Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nat Cell Biol. Mar 2020;22(3):282–288. [DOI] [PubMed] [Google Scholar]
- 29. Liu Y, Wu Z, Zhou J, et al. A predominant enhancer co-amplified with the SOX2 oncogene is necessary and sufficient for its expression in squamous cancer. Nat Commun. 2021;12(1):7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larson MH, Gilbert LA, Wang X, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeo NC, Chavez A, Lance-Byrne A, et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat Methods. 2018;15(8):611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wheeler DB, Zoncu R, Root DE, Sabatini DM, Sawyers CL.. Identification of an oncogenic RAB protein. Science. 2015;350(6257):211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mender I, Shay JW.. Telomere restriction fragment (TRF) analysis. Bio Protoc. Nov 20 2015;5(22). doi: 10.21769/bioprotoc.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chin L, Artandi SE, Shen Q, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. [DOI] [PubMed] [Google Scholar]
- 35. Maser RS, DePinho RA.. Connecting chromosomes, crisis, and cancer. Science. 2002;297(5581):565–569. [DOI] [PubMed] [Google Scholar]
- 36. Smogorzewska A, de Lange T.. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21(16):4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T.. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163(7):1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 40. Xi L, Schmidt JC, Zaug AJ, Ascarrunz DR, Cech TR.. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015;16(231). doi: 10.1186/s13059-015-0791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.