Metabolic rewiring of liver metabolism by extrahepatic tumors via innate immune cells occurs at the early disease stage, promoting tumor progression and cancer-related systemic manifestations, such as weight loss.
Abstract
Multiple studies have identified metabolic changes within the tumor and its microenvironment during carcinogenesis. Yet, the mechanisms by which tumors affect the host metabolism are unclear. We find that systemic inflammation induced by cancer leads to liver infiltration of myeloid cells at early extrahepatic carcinogenesis. The infiltrating immune cells via IL6–pSTAT3 immune–hepatocyte cross-talk cause the depletion of a master metabolic regulator, HNF4α, consequently leading to systemic metabolic changes that promote breast and pancreatic cancer proliferation and a worse outcome. Preserving HNF4α levels maintains liver metabolism and restricts carcinogenesis. Standard liver biochemical tests can identify early metabolic changes and predict patients’ outcomes and weight loss. Thus, the tumor induces early metabolic changes in its macroenvironment with diagnostic and potentially therapeutic implications for the host.
Significance:
Cancer growth requires a permanent nutrient supply starting from early disease stages. We find that the tumor extends its effect to the host's liver to obtain nutrients and rewires the systemic and tissue-specific metabolism early during carcinogenesis. Preserving liver metabolism restricts tumor growth and improves cancer outcomes.
This article is highlighted in the In This Issue feature, p. 1501
INTRODUCTION
The liver communicates with all the organs in our body and with the tumor either directly via messenger molecules or indirectly via the immune system (1, 2). Notably, the liver is also an immunogenic organ containing diverse resident immune cells that can respond to systemic or tissue-specific immune-related vulnerabilities by producing acute-phase proteins, complement components, cytokines, and chemokines (3, 4). Recent work demonstrates that the immune system, while aiming to be protective, can also exert selective pressures that promote cancerous features in normal tissue-resident cells (3).
Unrestricted cancer growth requires a permanent supply of glucose, amino, and fatty acids. These nutrient demands are obtained from cancer-mediated reprogramming of metabolism in the tumor and its microenvironment (5). Unraveling these interactions led to synergistic therapy combining chemotherapy with drugs targeting metabolic dependencies between the tumor and the microenvironment (5). In addition to the tumor microenvironment, the tumor connects with the host with networks of nerves, blood, and lymph vessels and extends its effect from the microenvironment to external organs, such as the liver. Consequently, the tumor can dysregulate tissue-specific metabolism and induces systemic metabolic rewiring, potentially contributing to cancer manifestations (6).
Because the liver is a central metabolic organ essential for maintaining body homeostasis, it senses and responds to systemic nutrient-level fluctuations by facilitating tissue-specific adaptations that preserve systemic equilibrium (4, 7–10). At the cellular level, hepatocytes play significant roles in carbohydrate, protein, amino acid, and lipid metabolism (11). Some of these metabolic reactions are mostly liver-specific such as the complete urea cycle (UC), which disposes of excess nitrogen in the form of ammonia by converting it to urea (12). We previously found evidence of decreased UC activity in the livers of 4T1 breast cancer–bearing mice and plasma of children with cancer (13), supporting a potential metabolic communication between extrahepatic tumors and the liver. These findings intrigued us to decipher how cancer regulates liver metabolism and for what benefit.
RESULTS
Nonliver Cancers Induce Early Metabolic Changes in the Liver During Carcinogenesis
To evaluate liver metabolism for changes during the carcinogenesis of nonliver cancers, we utilized the orthotopic 4T1-luciferase breast cancer model that rarely metastasizes to the liver. Using this approach enabled us to differentiate between the liver metabolic changes induced by the primary tumor to those initiated by the metastases. As a control for potential side effects resulting from the orthotopic injection of cancer cells, we injected the mice with sham saline and used the MMTV-PyMT mouse, which is a genetic model of autochthonous breast cancer (14). We analyzed the UC enzymes and intermediates as a readout of liver-specific metabolic changes in the host during the first 3 weeks of carcinogenesis. We excluded arginase 1 from our analysis to accurately differentiate the UC from the nitric oxide metabolism (ref. 15; Supplementary Fig. S1A). In addition, a certified pathologist analyzed and ruled out the existence of metastasis in the livers at all experimental time points.
In the 4T1 breast cancer model, we found a decrease in the expression of the UC enzymes in the host's liver, starting as early as day 4 after the orthotopic injection. The decrease in UC enzyme expression was dynamic along a 3-week course, increasing in both the magnitude of the reduction and the number of enzymes involved (Fig. 1A and B). Interestingly, the genetic MMTV-PyMT breast cancer model also had reduced expression levels of UC enzymes compared with controls, ruling out the possibility that the injection caused the change in UC expression levels (Fig. 1C). To verify the significance of the results in UC expression on UC function, we performed live infusion with 15N2-labeled glutamine and measured the labeled urea to glutamine and glutamate ratio. We found both these ratios to be decreased in the plasma of breast cancer–bearing mice, and furthermore in the livers, whereas total glutamate was elevated, the labeled urea to glutamate ratio was reduced in the 4T1 mice, confirming a malfunctioning UC (Fig. 1D; Supplementary Fig. S1B). In addition, we measured UC-related upstream substrates—glutamate and aspartate—and the downstream metabolite fumarate in the plasma and livers of 4T1. As expected from a dysfunctional UC, we found elevated levels of UC substrates, whereas the levels of fumarate, a UC product, were decreased (Supplementary Fig. S1C). We further found elevated levels of plasma ammonia and reduced urea levels in the urine, confirming a failing UC (Fig. 1E). Notably, although the levels of the 15N-labeled aspartate were lower in the tumor compared with its levels in the plasma and the liver, the ratio of labeled uracil to labeled aspartate was higher in the tumor, suggesting utilization of aspartate for nucleotide synthesis (Fig. 1F). Indeed, the high levels of UC substrates and the decreased levels of UC products resulting from the depletion of UC function in the host liver directly potentiated cancer cell proliferation (Fig. 1G). Interestingly, ex vivo supplementation of ammonia reduced the survival and activation of T-cell splenocytes excised from wild-type (WT) mice (Fig. 1H). Thus, the rewiring of the liver UC during carcinogenesis potentially benefits cancer growth by increasing substrate availability for tumor growth and by reducing the systemic immune response.
Figure 1.
Breast cancers induce metabolic changes in the liver during early carcinogenesis. A, RT-PCR of livers from breast cancer–bearing mice demonstrates decreased RNA expression of UC enzymes along cancer progression (n = 5, Student t test). Day 4, P = 0.003, day 14, P = 0.033, 0.01, and 0.028 (respectively), day 21, P = 0.0002, 0.019, 0.0002, 0.013, and 0.007 (respectively). B, Left, Western blots demonstrating decreased protein expression levels of ASS1 and OTC in livers of breast cancer–bearing mice vs. WT PBS-injected mice (n = 5, Student t test). Right, quantification of band intensities: ASS1, P = 0.002; OTC, P = 0.011. C, RT-PCR of liver from MMTV-PyMT of 14-week-old cancer-bearing mice demonstrate decreased RNA expression of UC enzymes compared with livers of WT mice (WT n = 5, MMTV-PyMT n = 3); P = 0.033, 3.14E−05, 0.038 (respectively). D, Measurements of UC-related metabolites following glutamine 15N2 infusion to 4T1 breast cancer–bearing or PBS-injected mice in the plasma and livers. Left, a decreased ratio of urea to glutamine and glutamate m + 1 isotopologues in the plasma of 4T1 breast cancer–bearing mice (WT n = 5, 4T1 n = 7, Student t test); P = 0.019, 0.042, respectively. Right, an increase in the levels of glutamate (AUC/internal standard/dry weight) in 4T1 breast cancer–bearing mice and a decrease in the ratio of urea to glutamate following infusion in 4T1 breast cancer–bearing mice, supporting a malfunctioning UC (n = 7, Student t test); P = 0.016, 0.048, respectively. E, Left, measurements of ammonia levels, using an ammonia assay kit, in the plasma of breast cancer–bearing mice compared with WT PBS-injected mice (WT n = 4, an outlier – interquartile range method, 4T1 n = 5, Student t test). Right, urea levels in the urine of breast cancer–bearing mice compared with WT PBS-injected mice. P = 0.033. F, Left, decreased levels of m + 1–labeled aspartate in the tumor of 4T1 breast cancer–bearing mice compared with the levels in the plasma and liver following infusion of 15N2 glutamine (n = 8, plasma n = 7, two-way ANOVA); P values: plasma vs. tumor = 0.041, liver vs. tumor = 0.007. Right, increased ratio of uracil m + 1 to aspartate (Asp) m + 1 isotopologues in the tumors of 4T1 breast cancer–bearing mice (n = 8, plasma n = 7, two-way ANOVA); P values: plasma vs. tumor = 0.042, liver vs. tumor = 0.008. G, XTT assay for 4T1 cancer cell proliferation following supplementation of the medium with UC intermediates (n = 3, two-way ANOVA). P values: 24 hours: aspartate: not significant, ammonia = 0.007, glutamate and fumarate <0.0001; 48 and 72 <0.0001 for all measurements. Metabolite concentrations were supplemented in the following concentrations: ammonia, 0.75 mmol/L as published (49); aspartate, 0.25 mmol/L; fumarate, 0.35 mmol/L; glutamate, 0.25 mmol/L. H,Ex vivo FACS analysis of CD8+ splenocyte cell survival (left) and activation (right) following supplementation with 1 mmol/L ammonia (Amn; ref. 59; measured plasma ammonia levels 1.34 mmol/L; n = 5, Student t test); P = 0.011, 0.043, respectively. I, Heat map for differential gene expression in hepatocytes demonstrates a unique pattern on day 21 in the livers of 4T1 breast cancer mice compared with day 4. J, 4T1 day 21 vs. 4T1 day 4 pathway enrichment analysis. Each bar shows the fold enrichment of a specific pathway. TCA, tricarboxylic acid.
To broadly evaluate whether liver-specific metabolic pathways other than the UC are affected by cancer in vivo, we performed RNA sequencing (RNA-seq) of hepatocytes isolated from perfused livers of WT and 4T1 cancer–bearing mice on days 4 and 21 following cancer cell injection. In corroboration of our previous findings, we found a reduction in the expression of the UC enzymes argininosuccinate synthetase (ASS1), ornithine transcarbamylase (OTC), and carbamoyl phosphate synthetase I (CPS1), and a significant and distinctive gene expression signature with disease at day 21 (Supplementary Fig. S1D and S1E; Fig. 1I). In this signature, we found reduced expression of multiple other enzymes participating in central metabolic pathways, supporting a general reprogramming of hepatocyte metabolism during carcinogenesis (Fig. 1J). Further analysis of the hepatocyte RNA-seq demonstrated that not all liver genes changed in expression, supporting the specificity of the changes in the metabolic genes (Supplementary Fig. S1F). Interestingly, the broad changes in liver metabolism were associated with a significant elevation in AST levels (Supplementary Fig. S1G).
As several of the metabolic pathways we found altered, including the UC, depend on adequately functioning mitochondria, we evaluated whether there is a decrease in mitochondrial functions or number following carcinogenesis. In isolated mitochondria, we found a reduction in the activities of respiratory chain complexes that include mitochondrial DNA (mtDNA)–encoded subunits (I, II–III, IV) relative to succinate dehydrogenase (SDH; complex II), which is entirely nuclear-encoded (Supplementary Fig. S1H). This finding was corroborated by the observed reduction in liver mtDNA levels in breast cancer–bearing mice (Supplementary Fig. S1I). To further understand the perturbation in mitochondrial metabolism, we analyzed the protein and RNA levels of Mitochondrial Transcription Factor A (TFAM), which regulates mtDNA levels and transcription, and found it significantly downregulated in breast cancer tumor–bearing mouse livers (Supplementary Fig. S1J and S1K; ref. 16). Yet, the total mitochondrial amount, estimated by citrate synthase activity in liver homogenate, was unaltered (Supplementary Fig. S1L). Thus, it is likely that transcriptional changes cause the observed metabolic changes rather than mtDNA depletion.
Collectively, our data suggest that in breast cancer mouse models, there are early transcriptional alterations in the expression of metabolic enzymes that cause global changes in liver metabolism at the pathway and organelle levels.
Innate Immune Cells Infiltrate the Liver of the Host during Early Carcinogenesis
To understand what potentially causes such an early and extensive metabolic rewiring, we first examined the livers for morphologic changes following orthotopic injections of breast cancer cells. Interestingly, we found evidence of immune cell infiltration to the livers that start as early as day 4 after breast cancer cell injections and increase along the disease course (Fig. 2A). FACS analysis further confirmed the gradual increase in the liver immune cell (CD45+) population in livers of breast cancer–bearing mice between days 0 and 21 (Supplementary Fig. S2A). Notably, liver histology sections of the MMTV-PyMT genetic breast cancer model also showed infiltration of immune cells compared with livers of WT mice, supporting that the liver-immune infiltration was not caused by a reaction to the injection (Supplementary Fig. S2B).
Figure 2.
Innate immune cells infiltrate the host's liver during early breast carcinogenesis. A, Hematoxylin and eosin staining of liver sections demonstrates increased immune cell infiltration along a time course following breast cancer cell injection. The black arrows point to the infiltrating immune cells. Magnifications: 10 and 40× as detailed in the figure. B, Cell types annotated from the scRNA-seq analysis are projected on three uniform manifold approximation and projections (UMAP), indicating which cells appear at which time point. The arrows mark the neutrophil and monocyte subsets that were absent in the liver of WT mice and gradually accumulated in the liver until day 21. DC, dendritic cell; moDC, monocyte-derived dendritic cell; NK, natural killer; pDC, plasmacytoid dendritic cell; Treg, regulatory T cell. C–E, CyTOF-t-distributed stochastic neighbor embedding (t-SNE) analysis (C and E) and (D) quantification of liver and blood CD45+ populations show increased levels of innate immune cells and reduction of lymphocytes in the liver (C) and blood (E) of breast cancer–bearing mice compared with WT PBS-injected mice (WT n = 3, 4T1 = 5, Student t test). P values for blood <0.0001. P values for liver: B cells = 0.013, T cells = 0.0014, neutrophils = 0.0001. F, Multiplex ELISA immunoassay demonstrates increasing levels of IL6 (left) and TNFα (right) in breast cancer–bearing mice from day 4 to day 21 following cancer cell injection (n = 5, Student t test). P values for IL6: day 4 = 0.024, day 14 <0.0001, and day 21 = 0.0009. P values for TNFα: day 4 = 0.05, day 14, and day 21 <0.0001. G, ELISA assay measurements of CCL2 levels in plasma, livers, spleens, and lungs of breast cancer–bearing mice (n = 5, Student t test). P values: plasma = 0.0005, liver = 0.017, spleen = 0.04. H, ELISA assay demonstrates a significant interaction effect between the time following cancer cell injection and elevation of CCL2 in the liver of breast cancer–bearing mice relative to WT PBS-injected mice (n = 5, two-way ANOVA); P = 0.0034. I, Expression of CCR2 in immune cells from the scRNA-seq experiment, projected on the UMAP, demonstrates that CCR2 is mainly expressed in monocyte subpopulations.
To identify which immune cells infiltrate the liver, we performed single-cell RNA-seq (scRNA-seq) analysis of the nonhepatic parenchyma cells in the liver. We complemented the results at the protein level by cytometry time of flight (CyTOF). Using both technologies, we found a significant decrease in lymphocyte infiltration and an increase in liver-infiltrating innate immune cells—neutrophils and monocytes (Fig. 2B–D). The reduction in liver lymphocytes may result from blood lymphopenia due to high sensitivity to elevated ammonia levels (Fig. 2E), which was potentially caused by sensitivity to elevated ammonia levels (Fig. 1H). Conversely, the increased infiltration of innate immune cells to the liver has been described during the formation of premetastatic niches (17) or could be part of inflammation-derived extramedullary hematopoiesis (18). We excluded the option of extramedullary hematopoiesis, as we did not find erythrocytes, megakaryocytes, and young granulocytes in the livers. In contrast, we found a significant elevation in the number of mature neutrophils (CXCR2+) and a reduction in CXCR2− neutrophils (Supplementary Fig. S2C). Furthermore, FACS analyses excluded bone marrow (BM) failure and demonstrated an extensive maturation of neutrophils, correlating with a significant elevation in plasma levels of granulocyte colony-stimulating factor (G-CSF; Supplementary Fig. S2D and S2E).
It may be that the liver infiltration of immune cells during carcinogenesis increases liver susceptibility to late metastasis formation. Still, our data suggest that liver involvement at this early stage is more likely to be part of a systemic inflammatory response.
Our scRNA data demonstrate that the neutrophils in the liver can be clustered into four subgroups based on substantial differences in gene expression along the time course of carcinogenesis (Supplementary Fig. S2F). Neutrophil subset 1, found in the liver at day 4, likely represents immature neutrophils based on the high expression levels of maturation and chemotaxis genes, granules genes, and elevated inflammatory markers such as IL1β (19). Following 3 weeks of carcinogenesis, we found that more mature neutrophils accumulate in the liver in high numbers (subsets 2–4; Supplementary Fig. S2F–S2H). Additionally, measurement of cytokine levels in the plasma of breast cancer–bearing mice demonstrated a significant increase in IL6 and TNFα, supporting the notion that tumorigenesis induces a systemic inflammatory response that involves the liver (Fig. 2F). Because it is usually challenging to detect direct elevation in RNA levels of secreted cytokines in scRNA of in vivo samples, we analyzed the data for elevations in the RNA of their downstream target. Encouragingly, we found in macrophages at day 21 a significant elevation in the expression of ANXA1, which is known to be driven by IL6 (ref. 20; Supplementary Fig. S2I).
The chemokine CCL2 and its primary receptor CCR2 have been linked to the pathogenesis of inflammation and cancer (21). Indeed, we found elevated levels of CCL2 in livers, plasma, and spleens of breast cancer–bearing mice in the first week following the injection of cancer cells and less so in the lungs (Fig. 2G). Notably, although the increase in plasma CCL2 levels in breast cancer–bearing mice compared with healthy mice remained constant, the relative elevation of CCL2 in the livers of breast cancer–bearing mice continued to increase along the cancer course (Fig. 2H; Supplementary Fig. S2J). In addition, we found increased CCR2+ cells in liver-infiltrating monocytes and upregulation in the expression of genes involved in migration pathways (Fig. 2I; Supplementary Fig. S2K).
Thus, following carcinogenesis, there is an early induction of systemic immune response, in which immune tissues such as the liver secrete increasing levels of CCL2, resulting in the infiltration of immune cells to different organs.
Phospho-ERK Activation in Myeloid Cells Results in the Depletion of HNF4α from Hepatocytes
In addition to the alterations we found in metabolic pathways, the bulk RNA-seq analysis we performed on isolated hepatocytes of breast cancer mice demonstrated an upregulation of signaling pathways (Fig. 3A). IL6, NFκβ, and ERK, which we found to be elevated in our scRNA pathway analysis, were described to participate together as a signaling pathway (22). In addition, IL6 was shown to downregulate TFAM (23), and we found its levels elevated in the plasma of 4T1 mice (Fig. 2F; Supplementary Fig. S1G and S1H). Thus, we further evaluated the activation state of the ERK–IL6 pathway in the livers. Immunostaining and FACS analysis of phospho-ERK (pERK) and CD45 cells in the livers of breast cancer–bearing mice revealed increased costaining of pERK and CD45+ in infiltrating liver cells (Fig. 3B and C). Notably, we did not find pERK activation in the blood cells of breast cancer–bearing mice, suggesting that the activation occurred in the liver (Fig. 3C).
Figure 3.
Activated liver-infiltrating myeloid cells perturb liver metabolism via HNF4α depletion. A, Differentially expressed gene pathway enrichment analysis of hepatocytes from breast cancer–bearing mice on day 21 compared with day 4. The bar shows the Z score of a specific pathway. B, Immunofluorescence staining demonstrates increased levels of pERK in CD45+ cells in the livers of breast cancer–bearing mice. C, Mean fluorescence intensity (MFI) of pERK in CD45+ cells demonstrates a specific increase for pERK in the livers of breast cancer–bearing mice compared with WT PBS-injected mice (left) and no significant difference in the staining for CD45+ pERK in the blood (middle; n = 4, Student t test). Quantification is shown on the right. P < 0.0001. ns, not significant. D, Normalized RNA-seq counts for three integrin binders expressed on hepatocytes on days 4 and 21. P < 0.0001, 0.001, 0.027 (respectively). E, Normalized RNA expression counts of integrins (left: Itga4, middle: Itgal, right: Itgam) in neutrophils, monocytes, macrophages, and hepatocytes. F, Western blots demonstrating increased protein expression levels of pSTAT3 and decreased expression of HNF4α in livers of breast cancer–bearing mice compared with livers from WT PBS-injected mice (n = 5, Student t test). G, RT-PCR of livers from breast cancer–bearing mice demonstrates an increase in miR-24 levels (n = 5, Student t test); P = 0.005. H, RT-PCR of livers from breast cancer–bearing mice demonstrates a significant decrease in Hnf4a levels (n = 5, Student t test); P = 0.005. I, RT-PCR of primary hepatocytes demonstrates increased RNA expression of Ccl2 following supplementation of IL6 (n = 3, Student t test, representative experiment of two independent biological replicates); P = 0.009.
Integrins can induce pERK activation upon interaction with other cells (24). Thus, we performed a ligand–receptor interaction analysis (25) of our scRNA data of liver-infiltrating immune cells together with the bulk RNA-seq of hepatocytes. We found support for intercellular cross-talk between immune cells and hepatocytes via integrins and their receptors that may stabilize their hepatic localization (Fig. 3D and E). Activated ERK has been shown to cause secretion of IL6 from myeloid cells with multiple effects on other cells (26, 27), one of which is a consequent elevation of pSTAT3 levels. Elevated pSTAT3 via miR-24 causes the downregulation of HNF4α, a master regulator of liver metabolism (28–30). Indeed, we found significant elevations in pSTAT3 and miR-24 levels in the livers of breast cancer tumor–bearing mice together with complete depletion of HNF4α, at both the RNA and protein levels (Fig. 3F–H). Notably, supplementing primary hepatocytes with IL6 increased CCL2 RNA levels (Fig. 3I). Hence, cancer-induced inflammation recruits innate immune cells to the liver at least partly via CCL2. The consequent immune activation and secretion of IL6 activate pSTAT3 in hepatocytes, leading to HNF4α depletion and perturbation of liver metabolism.
HNF4α Depletion Disrupts Liver Metabolism during Extrahepatic Carcinogenesis
Further analysis of the RNA-seq data from the livers of breast cancer mice confirmed that many of the genes we found dysregulated and responsible for the perturbed metabolic and signaling pathways are indeed regulated by HNF4α (Fig. 4A; Supplementary Table S1). Because albumin is an established downstream target of HNF4α (31), we confirmed its decrease at both the RNA and protein levels in the liver and plasma of 4T1 mice, respectively (Fig. 4B).
Figure 4.
IL6–STAT3–HNF4α signaling causes changes in liver metabolism during breast cancer carcinogenesis. A, Differentially expressed gene pathway enrichment analysis for genes regulated by HNF4α (Supplementary Table S1) in livers of breast cancer–bearing mice on day 21 compared with day 4. The bar shows the Z score of a specific pathway. B, Left, plasma albumin (Alb) measurements demonstrate decreased levels in breast cancer–bearing mice compared with WT PBS-injected mice (n = 5, Student t test); P = 0.002. Right, normalized RNA-seq analysis of albumin expression in hepatocytes on days 4 and 21. P = 0.013. C, RT-PCR of primary hepatocytes demonstrates decreased RNA expression levels of Otc following IL6 supplementation (n = 3, Student t test, representative experiment of two independent biological replicates); P = 0.0002. D, STAT3 inhibitor HJCO152 restores Otc levels even in the presence of IL6 (n = 3, one-way ANOVA); P < 0.0001 for CTRL + DMSO vs. IL6 + DMSO and STAT3 inhibitor vs. STAT3 inhibitor + IL6. inh, CTRL + DMSO vs. STAT3 inh + IL6; ns, not significant. E, Western blots (left) demonstrate increased expression levels of HNF4α in breast cancer–bearing mice treated with an ERK inhibitor compared with livers from breast cancer–bearing mice treated with vehicle (vehicle n = 4, trametinib n = 3, Student t test). Right, quantification of band intensities. P = 0.024. F, Measurement of tumor weight on days 8 and 14 following treatment with an ERK inhibitor (day 8 n = 5, day 14 n = 4, Student t test). P values: day 8 = 0.007 and day 14 = 0.014. G, RT-PCR of livers from breast cancer–bearing mice treated with AAV8-HNF4α demonstrates increased RNA expression of Hnf4a, UC enzymes, and Alb in comparison with AAV8-GFP–treated breast cancer–bearing mice (AAV8-GFP n = 4, AAV8-HNF4α n = 7, Student t test); P = 0.004, 0.004, 0.002, 0.001, respectively. H, 4T1 tumor growth is significantly higher following AAV-GFP injection compared with AAV-HNF4α (AAV8-GFP n = 12, AAV8-HNF4α n = 13, Student t test). P = 0.049. I, Left, Western blots showing decreased levels of PCNA in tumors from breast cancer–bearing mice treated with HNF4α compared with mice treated with GFP. Right, quantification of band intensity relative to actin. P = 0.003.
To validate the causality between the signaling cascade initiated by immune cells and the consequent metabolic changes in the livers of cancer-bearing mice, we measured the expression levels of the UC enzyme OTC, a known target gene of HNF4α (32), in isolated primary hepatocytes. We found that IL6 supplementation decreased the expression levels of OTC and that this effect can be rescued with a STAT3 inhibitor, HJCO152 (Fig. 4C and D; ref. 33). Although trametinib (MEK inhibitor; ref. 34) may have an oncolytic effect, treating 4T1 mice with trametinib maintained the expression levels of HNF4α and liver UC enzymes and reduced tumor size, further supporting the proposed signaling cascade (Fig. 4E and F; Supplementary Fig. S3A).
Finally, in vivo reexpression of HNF4α via viral transduction in breast cancer–bearing mice increased liver HNF4α levels, reversed the changes in the expression of UC enzymes, and restricted breast cancer tumor growth (Fig. 4G and H). Furthermore, tumors from breast cancer–bearing mice treated with HNF4α had lower levels of PCNA and lower pCAD, suggesting that preservation of liver metabolism by reexpressing HNF4α decreases tumor growth by restricting proliferation (Fig. 4I; Supplementary Fig. S3B). HNF4α injection decreased glutamate and aspartate levels in the liver, plasma, and tumor, suggesting that preserved liver UC metabolism restricted tumor growth (Supplementary Fig. S3C). Notably, HNF4α-AAV injection increased the expression of viral HNF4α in the liver but did not increase the levels of viral HNF4α in tumors, supporting the liver specificity of the AAV-HNF4 (Supplementary Fig. S3D). Thus, liver-infiltrating immune cells with activated pERK secrete IL6, activating pSTAT3 in hepatocytes, which leads to HNF4α depletion and subsequently to metabolic perturbations in breast cancer–bearing mouse livers. Importantly, reexpressing HNF4α could restrict the changes in liver metabolism.
Similar Changes in Liver Metabolism Occur via HNF4α in Pancreatic Cancer Mice
To evaluate our results in another cancer mouse model, we used the KrasG12D/Trp53R172H/Pdx-1-Cre pancreatic cancer (KPC) orthotopic mouse (35). Encouragingly, we found that the early metabolic findings we demonstrated in the 4T1 breast cancer model also occur in the pancreatic cancer model. Indeed, we found in the pancreatic cancer model a significant decrease in the levels of OTC, a direct target of HNF4α, already in the first week after injection of KPC cells, as well as an increase in UC substrates and reduced levels of UC products on day 21 (Supplementary Fig. S4A and S4B). Additionally, we found in the pancreatic cancer mice a gradual increase in immune cell infiltration to the liver during the first 3 weeks of tumorigenesis (Supplementary Fig. S4C). FACS analysis confirmed that the liver-infiltrating immune cells were predominantly of innate immunity and associated with increased CCR2+ monocytes and lymphocyte depletion (Supplementary Fig. S4D). Consistent with our findings in the breast cancer model, our results in the KPC model were associated with an increase in the plasma levels of IL6, elevation in pSTAT3 protein levels, a decrease in HNF4α RNA levels, and consequently decreased albumin levels (Supplementary Fig. S4E–S4H). In further support, treating KPC mice with anti-IL6 antibodies rescued albumin levels (Supplementary Fig. S4I). Notably, these changes in liver metabolism occurred when there were no changes in the weight of the KPC mice (Supplementary Fig. S4J) and when a certified pathologist ruled out liver metastasis. Thus, the KPC mouse model demonstrates changes in liver metabolism that are similar to the ones we describe in breast cancer mouse models.
CCR2 Knockout and HNF4α Reexpression Rescue the Early Metabolic Changes in the Liver during Carcinogenesis in Pancreatic Cancer–Bearing Mice
To further confirm that CCL2 drives the immune infiltration to the liver and is responsible for the metabolic changes we find, we orthotopically injected the KPC cells into C57/Bl6 WT and CCR2−/− knockout (KO) mice, which do not express the receptor for CCL2 and cannot recruit CCL2+ immune cells (36). In contrast to KPC CCR2+/+ mice, we found that the KPC CCR2−/− mice developed pancreatic cancer but did not demonstrate liver infiltration by immune cells (Fig. 5A). Consequently, we found decreased levels of IL6 in the plasma of CCR2−/− pancreatic cancer–bearing mice (Fig. 5B), whereas the expression levels of UC enzymes, albumin, and HNF4α were preserved (Fig. 5C). Notably, the CCR2−/− mice had reduced levels of UC substrates in their livers and did not develop systemic metabolic manifestations associating with carcinogenesis, such as weight loss and changes in body composition, correlating with the preservation of albumin level. Importantly, CCR2−/− mice had significantly decreased tumor growth (Fig. 5D–F).
Figure 5.
CCR2 KO mice and HNF4α rescue the early changes in liver metabolism in pancreatic cancer–bearing mice. A, Hematoxylin and eosin staining demonstrates decreased immune cell infiltration to the livers of CCR2−/− pancreatic cancer–bearing mice compared with livers of pancreatic cancer–bearing WT mice. Pancreatic cancer–bearing mice were mice injected with KPC. Magnifications: 10, 20, and 40× as detailed in the figure. B, ELISA demonstrates decreased IL6 levels in the plasma of pancreatic cancer CCR2−/−–bearing mice relative to pancreatic cancer–bearing WT mice (PC WT n = 7, CCR2−/−n = 6, Student t test). P = 0.0125. C, RT-PCR of livers from CCR2−/− pancreatic cancer–bearing mice demonstrates preserved RNA expression levels of Ass1, Otc, Alb, and Hnf4a compared with decreased expression levels of these genes in livers of pancreatic cancer–bearing WT mice (PC WT n = 5, CCR2−/−n = 6, Student t test). P = 0.019, 0.011, 0.002, respectively. D, Decreased levels of aspartate and glutamate in the liver of pancreatic cancer CCR2−/−–bearing mice. UC intermediate levels were measured with gas chromatography–mass spectrometry (n = 7, Student t test). P = 0.011, 0.029, respectively. E, Weight measurements and nuclear magnetic resonance (NMR) body composition analysis of CCR2−/− pancreatic cancer–bearing mice demonstrate significantly less weight loss (left), increased fat tissue (middle), and decreased free fluids (right) in comparison with WT pancreatic cancer on day 21 (relative to day 0; n = 7 in each group, Student t test). P = 0.014, 0.006, and 0.004, respectively. F, Tumor weights measured on the day of sacrifice are significantly lower in the CCR2−/− mice with pancreatic cancer compared with WT pancreatic cancer (n = 7, Student t test). P = 0.009. G, Pancreatic cancer tumor growth is significantly more prominent in mice injected with AAV-GFP compared with AAV-HNF4α (AAV-GFP n = 4, AAV-HNF4α n = 9, Student t test). P = 0.006. H, Mice with pancreatic cancer injected with AAV-HNF4α demonstrate a significant increase in survival compared with those injected with AAV-GFP [log-rank (Mantel–Cox) test]. P = 0.02. I, Mice with pancreatic cancer injected with AAV-HNF4α maintain weight compared with those injected with AAV-GFP (AAV-GFP n = 8, AAV-HNF4α n = 9, two-way ANOVA). P = 0.05. J, NMR body composition analysis of mice with pancreatic cancer injected with AAV-HNF4α demonstrate less fat tissue loss (left) and decreased free fluid accumulation (right) in comparison with mice with pancreatic cancer injected with AAV-GFP (AAV-GFP n = 5, AAV-HNF4α n = 6, Student t test). P = 0.007, 0.012, respectively.
Finally, to evaluate the potential therapeutic relevance of our findings, we injected the AAV8-HNF4α virus into KPC mice. The AAV8-HNF4α virus significantly restricted pancreatic cancer tumor growth and improved survival (Fig. 5G and H). Notably, reexpressing HNF4α reduced weight loss, decreased fat loss, and restricted the accumulation of body fluids, as demonstrated by nuclear magnetic resonance (NMR; Fig. 5I and J; Supplementary Fig. S4K and S4L). Thus, preserving the levels of HNF4α in the liver by preventing CCL2+ immune cell infiltration or by reexpressing HNF4α alleviated the systemic manifestations of tumorigenesis, such as weight loss and changes in body composition.
A Biochemical Liver Score Based on Routine Tests Can Predict Outcomes in Patients with Breast Cancer and Pancreatic Ductal Adenocarcinoma
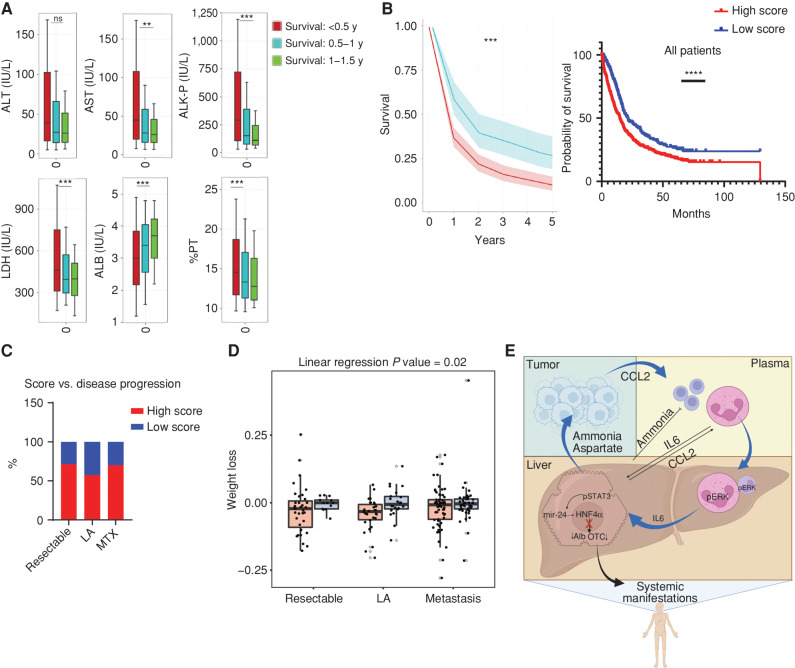
To understand the translational relevance of our findings in liver metabolism for patients with cancer, we performed analyses of the Clalit health maintenance organization dataset, which encompasses digital health data of 5 million Israeli subjects for 18 years. We found that patients with nonmetastatic breast cancer and pancreatic ductal adenocarcinoma (PDAC) with abnormal liver parameters on the day of diagnosis survive for a shorter time than patients with normal liver parameters (Fig. 6A; Supplementary Fig. S5A). Notably, some of these liver changes such as lactate dehydrogenase (LDH) and alkaline phosphatase (ALK-P) could be significantly detected in the plasma of patients with breast cancer and predict the outcomes, even a year before diagnosis (Supplementary Fig. S5B).
Figure 6.
Routine liver tests predict weight loss in patients with PDAC. A, Analysis of the Clalit HealthCare database for liver biochemical tests demonstrate that patients with nonmetastatic PDAC with shorter survival time have abnormal liver parameters at the time of diagnosis compared with those with longer survival (n = 2,037, survival of <0.5 years; n = 659, survival of 0.5–1 year; n = 342, survival of 1–1.5 years; **, P < 0.01; ***, P < 0.001). ALB, albumin; ns, not significant; PT, prothrombin time. B, Kaplan–Meier survival curves of patients with PDAC from the Clalit database (left) and from the Sheba and Souraski medical centers (right) demonstrate decreased survival in PDAC patients with high liver function scores. P = 0.0003 and P < 0.0001, respectively. C and D, Analysis of data from the Sheba and Sourasky medical centers. C, Liver score does not correlate with pancreatic cancer disease stage. LA, locally advanced; MTX, metastatic disease. D, Correlation of weight loss during PDAC for high (red) and low (blue) liver scores of patients with PDAC from the Sheba and Sourasky medical centers, as binned by stage of disease at diagnosis (linear regression P = 0.02). E, Graphical summary of the findings proposing a mechanism for the tumor-induced systemic metabolic changes in the livers of patients with cancer and cancer-bearing mice that can lead to cancer-associated cachexia (created with BioRender.com).
In addition, we analyzed datasets of patients with PDAC specifically from two independent medical centers in Israel: the Sheba Medical Center and Souraski Medical Center, two of Israel's largest oncology centers. We first confirmed that this cohort behaves as published in the literature and shows a correlation between decreased survival and weight loss (Supplementary Fig. S5C). Similar to a score we previously generated for the expression of UC enzymes (13), we here developed a “liver score” that is based on measured levels of liver biochemical and functional parameters [AST, ALT, ALK-P, albumin, international normalized ratio (INR)] and trained it on a small dataset (Supplementary Fig. S5D). We next tested the correlation potential of our score with survival in the Clalit and the hospitals’ datasets. We found that in all three cohorts, our liver score correlates with decreased survival in patients with PDAC (Fig. 6B). Interestingly, the liver score did not correlate with the disease stage but did correlate with weight loss (Fig. 6C and D; Supplementary Fig. S5E; Supplementary Table S2). Thus, the decreased survival predicted by our liver score coincides with systemic manifestations induced by cancer development.
DISCUSSION
Tumor-promoting inflammation is a recognized hallmark of cancer (37). The inflammatory response induced by cancers is orchestrated in part through the secretion of cytokines and chemokines and involves the infiltration of immune cells into different organs, such as the liver. While in the healthy liver exposure to foreign molecules results in regulated inflammation (38), we find that following carcinogenesis and immune infiltration, the liver recruits innate immune cells by secreting an increasing amount of CCL2. Activated immune cells positive for pERK and elevated IL6 levels lead to transcriptional changes in the expression of metabolic enzymes in the liver. Tumor-induced IL6 has been shown to impair the liver ketogenic response (39). Here we demonstrate a broad rewiring of liver metabolism via the IL6–pSTAT3 immune–hepatic axis, which leads to the depletion of HNF4α, a master regulator of liver metabolism. Consequently, there are changes in systemic metabolism, increasing the availability of substrates that promote cancer growth and contributing to systemic manifestations, such as weight loss and changes in body composition (Fig. 6E).
Complex clinical phenotypes can gain insights from genetic disorders in which an isolated gene is mutated (40). Indeed, in support of our findings, patients with HNF4A germline mutations have been linked with maturity-onset diabetes of the young and associated with weight loss (41, 42). In addition, mutations in the binding site of HNF4α to the OTC promoter and enhancer have been demonstrated to manifest clinically in OTC deficiency (32). Thus, our finding in extrahepatic cancers that HNF4α depletion in hepatocytes dysregulates central metabolic pathways such as the UC and contributes to a weight loss phenotype is consistent along these lines.
Our results demonstrate that HNF4α depletion induces liver alterations in protein metabolism involving the decreased activity of the UC and reduced albumin synthesis. Indeed, patients with cancer with lower albumin levels accumulate fluids and have poor outcomes (43). We propose a biochemical liver score that includes albumin and can predict survival and weight loss independent of the cancer stage. Therapeutically, our data suggest that giving clinically available drugs such as ERK inhibitors, STAT inhibitors, or IL6 antibodies to the identified patients at risk at an early cancer stage may preserve liver metabolism and restrict cancer progression. Moreover, we allude to a future option of using exogenous HNF4α to maintain liver metabolism and limit systemic manifestations.
It is well established that obesity, diabetes, and diet influence tumor development. Our findings that early changes in liver metabolism induce systemic metabolic changes that affect the tumor and the host highlight the essentiality of understanding the tumor macroenvironment for optimizing cancer diagnosis and therapy. Furthermore, they provide metabolic insights to better understand cancer complications such as metastasis, resistance, and cachexia. A comprehensive understanding of the tumor metabolic macroenvironment could identify early disease biomarkers, advance cancer patients’ management, and provide new treatment approaches to improve survival and quality of life for patients with cancer. Because cancer hijacks physiologic mechanisms, our findings may suggest an immune-mediated metabolic liver response that could be potentially relevant to other systemic conditions, such as viral infections requiring a general metabolic adaptation.
METHODS
In Vivo Animal Studies
Animal experiments were approved by the Weizmann Institute Animal Care and Use Committee following the U.S. National Institute of Health, European Commission, and Israeli guidelines. To generate syngeneic mouse cancer models, 8- to 12-week-old C57BL/6 or BALB/c male and female mice were purchased from Envigo and randomly assigned to experimental groups. For the breast cancer model, 8-week-old BALB/c female mice were injected with 1 × 106 4T1 breast cancer cells [in phosphate-buffered saline (PBS)] in the mammary fat pad. For the pancreatic cancer model, 12-week-old C57BL/6 male mice and CCR2-RFP KO mice were injected with 0.3–0.4 × 106 KPC pancreatic cancer cells (in DMEM 50% Matrigel) in the pancreas tail. After sacrifice, livers, spleens, BM, and lungs were removed from the mice, and blood was collected for further analysis by quantitative PCR, Western blot, and IHC.
Cell Lines
4T1-luciferase cells derived from mouse breast cancer cells were kindly provided by Professor Yosef Yarden, Department of Immunology and Regenerative Biology, Weizmann Institute of Science, Rehovot, Israel. KPC-luciferase cells derived from mouse solid PDAC were kindly provided by Professor Avigdor Scherz, Department of Plant and Environmental Sciences, Weizmann Institute of Science. All cells were tested routinely for Mycoplasma using a Mycoplasma EZ-PCR test kit (#20-700-20, Biological Industries, Kibbutz Beit Ha'emek).
Western Blotting
The tissues were ground and lysed in a RIPA lysis buffer (Sigma-Aldrich) containing 1% protease inhibitor cocktail (Calbiochem) and 1% phosphatase inhibitor cocktail (P5726, Sigma-Aldrich). Following centrifugation, the supernatant was collected and protein content was evaluated by the Bradford assay or a BCA Protein Assay Kit (Thermo Fisher Scientific, cat # 23225). Twenty to 50 μg from each sample under reducing conditions was loaded into each lane and separated by electrophoresis on a 10% SDS polyacrylamide gel. Following electrophoresis, proteins were transferred to Cellulose Nitrate membranes (Tamar). Nonspecific binding was blocked by incubation with TBST [10 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 0.1% Tween 20] containing 5% skim milk for 1 hour at room temperature (RT). Membranes were subsequently incubated with antibodies (Western blot antibodies list; Supplementary Table S3).
Antibody was detected using peroxidase-conjugated AffiniPure goat anti-rabbit IgG or goat anti-mouse IgG (Jackson ImmunoResearch) and enhanced chemiluminescence Western blotting detection reagents (EZ-Gel, Biological Industries). Gels were quantified by Gel Doc XR+ (Bio-Rad) and analyzed by ImageLab 5.1 software (Bio-Rad). The relative intensity of each band was calculated by dividing the specific band intensity by the value obtained from the loading control.
Liver Perfusions and Hepatocyte Dissociation
Livers of anesthetized mice were perfused as previously described (44), with specific adjustments. The vena cava was inserted with a 27-G syringe, fixed on the perfusion line. Ten milliliters of prewarmed to 42°C PPML buffer followed by 25 mL of prewarmed to 42°C PM buffer with Liberase (Roche, cat. # 05401127001) was perfused through the vena cava. Immediately at the beginning of the perfusion, the portal vein was cut. Following perfusion of 25 mL of PM buffer and Liberase solution, livers were harvested into a Petri dish with 10 mL of prewarmed PM buffer and chopped by forceps. Dissociated liver cells were collected and filtered through a slanted 100-μm cell strainer. Cells were spun down at 30 g for 3 minutes at 4°C to get hepatocyte enriched pellet. Pellet was resuspended in 25 μL cold PM buffer. To enrich live hepatocytes, 21.6 mL cold Percoll (GE HealthCare; #17-0891-01) mixed with 2.4 mL DPBSX10 + CaCl2 + MgCl2 was added to the cells. Cells were centrifuged at 600 rpm for 10 minutes at 4°C. The supernatant containing the dead cells was aspirated, and cells were resuspended in 25 mL of cold Williams E + GlutaMax-TM-1 (Gibco, cat. #32551, 1% penicillin and streptomycin, 10 FBS, 1% L-glutamine). Cells were centrifuged at 600 rpm for 5 minutes at 4°C. The supernatant was aspirated, and cells were resuspended in 3 mL cold Williams E + GlutaMax-TM-1.
Primary Hepatocytes Culture
Following perfusion, 1 × 106 isolated hepatocytes were seeded in 3 mL Williams E + GlutaMax-TM-1 in a 6-well plate. Four hours after, the medium was aspirated, and the cells were washed with PBS and incubated with 2 mL Williams E + GlutaMax-TM-1 overnight. Cells were treated with 10 μmol/L of the STAT3 inhibitor HJC0152 (Selleckchem # S8561) for 30 minutes and then treated with 10 ng/mL recombinant mouse IL6 (R&D Systems; # 406-ML-005) for 48 hours.
Histopathologic and IHC Staining Analyses
Following 4, 14, and 21 days of 4T1 breast cancer cell injection and 7, 14, and 21 days of KPC pancreatic cancer cell injection, PFA-fixed liver and lung tissues were embedded in paraffin blocks. The blocks were sectioned into 4 μm, and tissue sections were backed at 37°C overnight. Hematoxylin and eosin staining was performed according to a standard protocol including the following steps: deparaffinization, rehydration, staining with hematoxylin and eosin, followed by dehydration. The slides were cleaned with xylene and mounted.
Immunofluorescence
Following 21 days of 4T1 breast cancer cell injection, livers were collected, fixed in 4% paraformaldehyde, and embedded in paraffin blocks and 4-μm sections were made. Slides were deparaffinized, and antigen retrieval was done using citric acid pH 6. Blocking for unspecific binding was done with 20% normal horse serum (NHS) and 0.1% Triton in PBS. Rat anti-CD45 (Bio-Rad; #MCA1031G) and mouse anti-pERK 1:100 (Sigma; #M8159) were diluted in 2% NHS and 0.1% Triton and were incubated overnight. Slides were then incubated with biotinylated donkey anti-rat 1:100 (Jackson ImmunoResearch; #712-065-153) and HRP-conjugated goat anti-mouse 1:100 (PerkinElmer; #NEF822001EA) diluted 2% NHS for 1.5 hours. Slides were then incubated with 1:500 OPAL 690 (Akoya Biosciences; #FP1497001KT) and streptavidin Cy3 (016-160-084; Jackson ImmunoResearch). Slides were imaged with a Leica Mi8 microscope equipped with a motorized stage and a Leica DFC365 FX camera. Single ×20 magnification images were tiled to receive a full scan of the tumor section. The quantification in the liver sections stained with pERK was done by ImageJ.
RNA Processing and Quantitative PCR
RNA was extracted from liver tissue using QIAzol Lysis Reagent (according to a QIAzol Handbook) or the Direct-zol MiniPrep Plus Kit (Zymo Research ZR-R2070). For hepatocyte RNA-seq, RNA was extracted from dissociated hepatocytes. Following liver perfusion as previously described, hepatocytes were resuspended in QIAzol and frozen at −80°C. RNA was extracted using QIAzol Lysis Reagent. RNA from cultured primary hepatocytes was extracted by using RNeasy Mini Kit (Qiagen; #74104). To evaluate mtDNA copy number, total DNA was isolated using a DNA purification kit (DNeasy Blood and Tissue Kits, Qiagen; #69504). cDNA was synthesized from 1 μg RNA using the qScript cDNA Synthesis Kit (Quanta; #95749). Detection on cDNAs was performed using SYBR Green FastMix Perfect CT (Quantabio; #95073) with the required primers (qPCR primer list; Supplementary Table S4).
Perfusion for CyTOF and FACS Assays
For blood collection, once anesthetized, mice were either injected with 10 μL of heparin on the left ventricle and ∼700 μL blood was withdrawn by cardiac puncture on the right ventricle using a 27-G needle in a 1 mL syringe coated with heparin or via retro-orbital bleeding procedure using heparinized micro hematocrit capillary tubes for immune cell isolation. Mice were intracardially perfused with ice-cold PBS. These assays were performed 14 days following the injection of 4T1 breast cancer cells and control PBS, and 21 days following the injection of KPC pancreatic cancer cells and control PBS.
Blood Immune Cell Isolation
Following perfusion, blood was withdrawn and transferred into 15-mL tubes. Red blood cell lysis buffer (5 mL; Thermo Fisher, ACK Lysing Buffer, A104920) was added and incubated at RT for 5 to 10 minutes. Blood was then centrifuged at 300 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in residual volume. Cells were washed with 5-mL ice-cold FACS buffer [Ca/Mg2 free PBS + 2 mmol/L EDTA + 0.5% bovine serum albumin (BSA) or 5% FCS] and centrifuged at 300 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in residual volume.
Liver Immune Cell Isolation
Following perfusion, livers were extracted and transferred to a Petri dish. Livers were then minced into ∼1-mm pieces. DMEM-F12 (3 mL; Thermo Fisher, 31330038) was added to 15-mL tubes on ice. Three milliliter of collagenase (Worthington, LS004188) cocktail (1 mg/mL collagenase IV + 0.2 mg/mL DNAse I + 20% FBS in DMEM/gF12) was added, and the tissue homogenates were incubated at 37°C for 60 minutes shaking at 250 rpm, with brief vortex every 15 minutes. Cell suspension was filtered with a 40-μm strainer into a 50-mL tube and washed with 20 mL of ice-cold FACS buffer. Cells were then centrifuged at 600 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in 5-mL red blood cell lysis buffer. At the end of the incubation, 15 mL of ice-cold FACS buffer was added and samples were centrifuged at 600 × g for 5 minutes at 4°C. Leukocyte enrichment based on the Percoll gradient was performed. Isotonic Percoll (9 parts of Percoll, 1 part of sterile 10× PBS), 80% Percoll (8 parts of isotonic Percoll, 2 parts of 1× PBS), and 40% Percoll (5 parts of 80% Percoll, 5 parts of DMEM-F12) solutions were prepared. Pellet was resuspended in 8 mL of 40% Percoll and carefully transferred to 15 mL containing 5 mL of 80% Percoll. Cells were centrifuged at 1,500 × g for 30 minutes at 4°C (acceleration 5/brake 0). The middle layer containing immune cells was collected and transferred into a new 15-mL tube containing 5 mL of ice-cold PBS buffer. Volumes were evened out to 10 mL with ice-cold PBS, and cells were centrifuged at 600 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in residual volume.
Spleen Immune Cell Isolation
Following perfusion, the spleen was squashed over a 70-μm strainer and filtered with 10 mL FACS buffer. Cells were centrifuged at 400 × g for 5 minutes at 4°C. The supernatant was aspirated, and cells were resuspended in 1 mL red blood cell lysis buffer. After 5 minutes of incubation at RT, 10 mL of ice-cold PBS was added and cells were centrifuged at 300 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in residual volume.
BM Immune Cell Isolation
Following perfusion, the femur was dislocated and transferred into a Petri dish containing ice-cold PBS. Condyles, patella, and epiphysis were removed to expose the metaphysis. The BM was flushed with 2 mL of medium over a 70-μm strainer. BM was smashed with a syringe plunger, and the filter was washed with 10 mL of RPMI + 10% FBS + 2 mmol/L EDTA. Cells were centrifuged at 400 × g for 5 minutes at 4°C. The supernatant was aspirated, and the pellet was resuspended in 1 mL red blood cell lysis buffer for 5 minutes at RT. RPMI (10 mL) containing 10% FBS + 2 mmol/L EDTA was added, and the cells were centrifuged at 400 × g for 5 minutes at RT.
Flow Cytometry
Immune cells were washed with ice-cold PBS and stained with LIVE/DEAD Fixable Aqua Dead (Thermo Fisher) according to the manufacturer's instructions. After Fc blocking (BioLegend, BLG-101320), cells were stained for surface antigen (FACS antibodies list, Supplementary Table S5). Flow cytometry data were acquired on CytoFLEX (Beckman Coulter) and analyzed using FlowJo software. In each experiment, relevant negative, single-stained, and fluorescence-minus-one controls were used to identify the populations of interest (FACS gating strategy, Supplementary Extended Data S1).
Mass Cytometry
For mass cytometry analysis, 3 × 106 cells per each liver sample and all blood-derived immune cells were stained as described before (45). Cisplatin viability stain was used prior to the barcoding of samples with palladium metal isotopes. Briefly, individual samples were incubated with Human TruStain FcX (BioLegend), followed by staining with a panel of antibodies (CyTOF antibody list, Supplementary Table S6), for 30 minutes at RT, washed with 5 mL of Maxpar Cell Staining Buffer, fixed with Fix I Buffer, and permeabilized with Barcode Perm Buffer. Samples were then incubated with their respective barcodes for 45 minutes at RT, after which they were washed with Maxpar Barcode Perm Buffer and combined into a composite sample. After washing, the mixed sample was incubated with formaldehyde 4% overnight at 4°C. Before acquisition in a Helios II CyTOF system, samples were stained with iridium to detect cells and washed with cell staining buffer and mass cytometry–grade water. Multidimensional datasets were analyzed using the Cytobank cloud-based platform, FlowJo software (Tree Star, Inc), and R (R Core Team, 2017).
Algorithm-Based High-Dimensional Analysis
Mass cytometry data were normalized and debarcoded with the Fluidigm CyTOF software version 6.7. Individual samples were manually gated using Cytobank to exclude normalization beads, cell debris, and dead cells. Only CD45+ cells were used for downstream analyses. All analyses on CyTOF data were performed after arcsinh (with cofactor = 5) transformation of marker expression. Clustering, data visualization, and dimension reduction (uniform manifold approximation and projection) were performed using the CyTOF workflow package (46). All plots were drawn using ggplot2 or GraphPad Prism (version 8.0.1).
L-Glutamine-15N2 Infusion
Isotope infusion experiments were performed 3 weeks following orthotopic tumor inoculation. The mice were fasted for 4 hours, followed by 5 hours of infusion with L-Glutamine-15N2 (Sigma). Infusion solutions containing 1.725 g of L-Glutamine-15N2 per kg of body weight were prepared in saline. The mice were anesthetized on a heating pad, and a catheter connected to the infusion solution was inserted in the lateral tail vein. Each mouse was initially infused with a bolus of 150 μL/minute for 1 minute, followed by a continuous infusion of 2.5 μL/minute for 5 hours. Mice were kept awake throughout the infusion in individual infusion cages as described by Martini and colleagues (47). At the end of the infusion, mice were anesthetized and blood was collected into heparin tubes. Organs were harvested and snap-frozen in liquid nitrogen.
Gas Chromatography–Mass Spectrometry
Plasma was collected from blood samples through centrifugation at 1,000 rcf for 15 minutes at 4°C. Plasma (20 μL) was resuspended in an ice-cold MeOH/H2O mixture, 8:1 with ribitol, incubated on ice for 20 minutes, and centrifuged for 10 minutes at 15,000 rpm. The supernatants were vacuum-dried overnight, and the dried samples were incubated with 20 μL of methoxyamine hydrochloride solution (20 mg/mL in pyridine) at 37°C for 90 minutes, followed by incubation of 40 μL of N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane (Sigma) for 60 minutes at 60°C.
Tissue samples of about 5 to 25 mg were vacuum-dried and powdered using two stainless steel 3.2-mm balls on the CryoMill (Retscht). The powdered tissues were resuspended with methanol and ribitol as internal standard and sonicated for 20 minutes. Polar metabolites were extracted following the addition of 1 volume of water and 0.5 volume of chloroform. The mixture was vortexed and centrifuged at 15,000 rpm for 15 minutes at 4°C. The samples’ supernatants were dried overnight before derivatization with 40 μL methoxyamine hydrochloride solution (20 mg/mL in pyridine) at 37°C for 90 minutes while shaking, followed by incubation with 70 mL N,O-bis (trimethylsilyl) trifluoroacetamide (Sigma) at 37°C for an additional 30 minutes.
For the samples that were derivatized with BSTFA, instrument details and run were as described in Joo and colleagues (12). The samples that were derivatized with TBDMS were run on the same instrument using the same flow rate. One microliter of the sample was injected either in splitless or in 1:25 split mode, using an inlet temperature of 270°C, and the gas chromatography oven was held at 100°C for 3 minutes and then ramped to 300°C with a gradient of 3°C/minute followed by 5 minutes after run at 315°C. The MS system was operated under electron impact ionization at 70 eV, and a mass range of 100 to 650 amu was scanned. The resulting chromatograms were analyzed in MassHunter software (Agilent Technologies). Isotopologue distribution of the metabolites was corrected for naturally occurring isotopes using IsoCor software (48).
Amino Acid Analyzer
Frozen liver samples were lyophilized and ground to powder and extracted with 50% methanol homogenized in bullet blunder following 10 minutes of sonication in ice-cold water. The samples were centrifuged at 15,000 × g for 15 minutes and lyophilized again as described in the “Gas Chromatography–Mass Spectrometry” section. The samples were resuspended with lithium loading buffer (Biochrom), and proteins were precipitated by the addition of (v/v) cold 5% 5-sulphosalicylic acid (SSA) solution supplemented with 500 μmol/L of norleucine as internal standard. The mixtures were incubated for 30 minutes at 4°C following centrifugation at 15,000 × g for 15 minutes. The supernatants were filtered through a 0.22-μm filter and subsequently injected into Biochrom 30 series amino acid analyzer (Biochrom) with a Lithium Accelerated cation-exchange column (200 × 4.6). A mixture of amino acids at known concentrations (calibration standards, Biochrom) was supplemented with glutamine and used as standard. Amino acids were post-column derivatized with ninhydrin reagent and detected by absorbance at 440 nm (proline and hydroxyproline) or 570 nm (all the other amino acids).
Extraction of Polar Metabolites from Urine and Plasma
To extract polar metabolites from urine (20–100 μL) samples, 1 mL methanol (with labeled amino acids as internal standard) was added, respectively, into a biological sample-containing Eppendorf tube. Then, the resulting mixture was vortexed and sonicated for 15 minutes, vortexed again, and centrifuged at 14,000 rpm for 10 minutes. The liquid phase was transferred into a new tube and lyophilized. Then, the pellets were dissolved using 150 μL DDW-methanol (1:1), centrifuged twice to remove possible precipitants, and injected into the LC-MS system.
LC-MS Polar Metabolite Analysis
Metabolic profiling of the polar phase was done as described (45) with minor modifications as described below. Briefly, analysis was performed using the Acquity I Class UPLC System combined with a mass spectrometer (Thermo Exactive Plus Orbitrap), which was operated in a negative ionization mode. The LC separation was done using the SeQuant Zic-pHilic (150 mm × 2.1 mm) with the SeQuant guard column (20 mm × 2.1 mm; Merck). There were two mobile phases performed: mobile phase A with acetonitrile and mobile phase B with 20 mmol/L ammonium carbonate plus 0.1% ammonia hydroxide in water. The flow rate was kept at 200 μL/minute and gradient as follows: 0 to 2 minutes 75% of B, 17 minutes 12.5% of B, 17.1 minutes 25% of B, 19 minutes 25% of B, 19.1 minutes 75% of B, and 19 minutes 75% of B.
Polar Metabolite Data Analysis
The data processing was done using TraceFinder Thermo Fisher software, and detected compounds were identified by retention time and fragments and verified using an in-house mass spectra library. Urine metabolites were normalized by creatinine peak area.
Cytokine Detection
Cytokine levels were measured by either ProcartaPlex Immunoassays (Thermo Fisher ProcartaPlex Panel) or by an IL6 ELISA kit (Thermo Fisher 88-7064-22) according to the manufacturer's instructions.
CCL2 and Ammonia Levels
CCL2 and ammonia levels were measured by a CCL2 ELISA kit (R&D Systems; #MJE00B) and Ammonia Assay Kit (Abcam; ab83360), respectively, according to the manufacturer's instructions.
Activation of T Cells from Mice Spleens
Ten-week-old, WT, female Balb/c mice were sacrificed, and spleens were harvested into cold PBS on ice. Spleens were homogenized by a syringe plunger through a 70-μmol/L strainer and washed with PBS. Following centrifugation at 1,200 rpm for 5 minutes, pellets were treated with red blood cell lysis buffer according to the manufacturer's instructions. Cells were resuspended in 2 × 106 cells/mL in splenocyte medium (complete RPMI medium supplemented with 50 μmol/L β-mercaptoethanol, 10% sodium pyruvate and nonessential amino acids) supplemented with 6,000 IU/mL IL2 (Chiron, rhIL2) and seeded in 24-well plates precoated with CD3 (BLG #100302). After 72 hours, cells were collected, centrifuged at 1,200 rpm for 5 minutes, washed with splenocyte medium ×2, and analyzed using a CytoFLEX (Beckman Coulter) FACS analyzer.
Cancer Cell Proliferation
4T1 cells (20 × 104) were seeded in 100 μL of complete RPMI medium. On the following day, cells were washed with PBS and the medium was replaced with DMEM glutamine-free medium (Biological Industries; #01-057-1A), supplemented with ammonia (0.75 mmol/L) as described previously (46, 49), aspartate (0.25 mmol/L), fumarate (0.35 mmol/L), or glutamine (0.25 mmol/L). Proliferation assays (XTT cell proliferation kit; Biological Industries; #20-300-1000) were used according to the manufacturer's instructions 24, 48, and 72 hours following metabolite supplementation.
Measurements of Respiratory Chain Complex Activity
The enzymatic activities of respiratory chain complexes were measured at 37°C by standard spectrophotometric methods as described (50). Briefly, Complex I was measured as rotenone-sensitive NADH-CoQ reductase monitoring the oxidation of NADH at 340 nm in the presence of coenzyme Q1. Complex II was measured as succinate dehydrogenase based on the succinate-mediated phenazine methosulfate reduction of dichloroindophenol at 600 nm. Complex II + III was measured as succinate cytochrome c reductase after the reduction of oxidized cytochrome c at 550 nm. Complex IV (cytochrome c oxidase) was measured by following the oxidation of reduced cytochrome c at 550 nm.
Citrate synthase, a ubiquitous mitochondrial matrix enzyme, was measured in the presence of acetyl-CoA and oxaloacetate by monitoring the liberation of CoASH coupled to 5′,5′-dithiobis (2-nitrobenzoic) acid at 412 nm. Protein concentration was determined by the Lowry method and calculated according to a BSA standard curve.
In Vivo ERK Inhibition
Following 24 hours of 4T1 breast cancer cell injection, mice were injected i.p. with the 1 mg/kg of the ERK inhibitor trametinib (GSK1120212; Selleckchem; #S2673) in 4% DMSO corn oil or 4% DMSO corn oil only 6 more times a week. Mice were sacrificed 8 or 14 days following tumor injection.
In Vivo IL6 Inhibition
Four days after KPC cell injection, mice were injected i.p. with 200 μg/mice of IL6 antibody [InVivoMab anti-mouse IL6 (Bio X Cell; #BE0046)] or control IgG [InVivoMab rat IgG1 isotype control (anti-HRP; Bio X Cell; #BE0088)] every 2 days. Mice were sacrificed 21 days after tumor injection.
rAAV-HNF4α
Cells.
Low-passage HEK293T were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle medium supplemented with 10% FBS.
Production of rAAV.
To produce rAAV8, a triple cotransfection procedure was used to introduce an rAAV vector plasmid (pAAV-CMV-mHNF4α or pAAV-CMV-GFP) together with pXR8, AAV8 helper plasmid carrying AAV rep and cap genes and pXX6-80, and Ad helper plasmid at a 1:1:1 molar ratio (51).
Briefly, HEK293T cells were transfected using poly-ethylenimine (PEI; linear; molecular weight, 25,000; Polysciences, Inc.), and the medium was replaced at 18 hours after transfection. Cells were harvested 72 hours after transfection, subjected to 3 rounds of freeze–thawing, and then digested with 100 U/mL Benzonase (EMD Millipore) at 37°C for 1 hour. Viral vectors were purified by iodixanol (Serumwerk Bernburg AG) gradient ultracentrifugation (52), followed by further concentration using Amicon ultra-15 100K (100,000-molecular-weight cutoff, Merck Millipore) and washed with PBS (−/−). The final concentration of rAAV8 particles was 2.78E+10 vg per microliter (AAV-CMV-mHNF4α) and 2.35E+10 vg per microliter (pAAV- CMV-GFP). Mice were injected via tail vein with 5E11 vg 48 hours following inoculation with cancer cells.
RNA-seq
Total RNA was fragmented, followed by reverse transcription and second-strand cDNA synthesis. The double-strand cDNA was subjected to end repair, a base addition, adapter ligation, and PCR amplification to create libraries. Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of 12 samples on one lane of Illumina HiSeq 2500 V4 instrument, resulting in ∼23 million single-end 60-bp reads per sample.
Bioinformatics.
Poly-A/T stretches and Illumina adapters were trimmed from the reads using cutadapt (53); resulting reads shorter than 30 bp were discarded. Reads were mapped to the M. musculus reference genome GRCm38 using STAR (54), supplied with gene annotations downloaded from Ensembl (with the option EndToEnd and outFilterMismatchNoverLmax set to 0.04). Expression levels for each gene were quantified using htseq-count (55), using the gtf above. Differentially expressed genes were identified using DESeq2 (56) with the betaPrior, cooksCutoff, and independentFiltering parameters set to false. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. The pipeline was run using snakemake (57).
Figure Captions.
Principal component analysis was performed (using the R Stats package) on the DESeq2 variance stabilizing transformed values of the 1,000 most variable genes. The figure depicts the first versus the second principal component in a scatter plot. The first principal component explains 73%, and the second principal explains 9% of the variance of the data. Heat map analysis was performed on a total of 2,829 genes that came up significant in any of the comparisons (4T1 breast cancer bearing–mice and CTRL WT mice on days 4 and 21) are shown. A gene was considered to be significant if its absolute fold change was above 1.5, if the FDR was below 0.05, and if the gene had a count of at least 30 in one of the samples.
The log2-normalized counts were standardized to have for each gene zero mean and unit SD. Gap Statistic was used for estimating the number of clusters. K-means clustering of the standardized values was performed. The expression profile is accompanied by a colored bar indicating the standardized log2-normalized counts. For pathway enrichment analysis, we used Qiagen's Ingenuity Pathway Analysis.
Identifying Genes under HNF4α Regulation.
Differentially expressed genes between 4T1 hepatocytes on day 21 and day 4 were calculated and normalized to control mice in both time points (|logFC|> = 1.5 and FDR < 0.05). A total of 1,914 genes were upregulated in the day 21 time point compared with day 4, and 514 genes were downregulated at this time point. The list of downregulated genes was crossed with the list of target genes of the HNF4α transcription factor from the Harmonizome tool (https://maayanlab.cloud/Harmonizome/gene_set/HNF4A/ENCODE+Transcription+Factor+Targets) built using chromatin immunoprecipitation sequencing datasets from the ENCODE Transcription Factor Targets dataset. The final list contained 149 genes (Supplementary Table S1).
The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (58) and are accessible through GEO series accession number GSE212113 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212113).
scRNA-seq Using the Chromium 10x Genomics Platform
For liver NPC enrichment, the liver of mice injected with PBS or 4T1 breast cancer cells 4 and 21 days after injection was perfused as previously described above. Following 3 minutes of centrifugation at 30 × g, supernatant was collected and centrifuged at 300 × g for 5 minutes. Cell pellet was treated with red blood cell lysis buffer (Thermo Fisher, ACK Lysing Buffer #A104920) according to the manufacturer's instructions. scRNA-seq libraries were prepared using the chromium scRNA-seq platform (10x Genomics). Cells were counted and diluted to a final concentration in PBS supplemented with 0.04% BSA. Cellular suspension was loaded onto Next GEM Chip G targeting liver nonparenchymal cells and then run on a Chromium Controller instrument to generate Gel-bead-in-EMulsion (10x Genomics). Single-cell 3′ RNA-seq libraries were generated according to the manufacturer's protocol (10x Genomics Chromium Single-Cell 3′ Reagent Kit User Guide v3 Chemistry). Final libraries were quantified using the NEBNext Library Quant Kit for Illumina (NEB) and high-sensitivity D1000 TapeStation (Agilent). Libraries were pooled according to targeted cell number, aiming for ∼50,000 reads per cell. Pooled libraries were sequenced on a NovaSeq 6000 instrument using an SP 100 cycles reagent kit (Illumina). The scRNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (58) and are accessible through GEO series accession number GSE223835 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223835).
scRNA Analysis
MetaCell Pipeline.
The MetaCell pipeline36 was used to derive informative genes and to compute cell-to-cell similarity to compute k-nearest neighbor graph covers and derive distribution of RNA in cohesive groups of cells (or metacells) and to derive strongly separated clusters using bootstrap analysis and computation of graph covers on resampled data. We removed specific mitochondrial genes, immunoglobulin genes, and genes linked with poorly supported transcriptional models (annotated with the prefix “Rp-”). Gene features were selected using the threshold value for the normalized var/mean (Tvm) = 0.3 and a minimum total unique molecular identifier (UMI) count >50. We subsequently performed hierarchical clustering of the correlation matrix between metacells and grouped them into clusters representing cell types and states. We used K = 100, 500 bootstrap iterations, and otherwise standard parameters.
Neutrophil Subset Analysis.
Neutrophil subsets were identified according to the maturation score and chemotaxis score, as well as granule identification, as described (19).
Pathway Analysis.
To evaluate pathways enriched in differentially expressed genes, we used Gene Ontology 37, 38, 39 for genes differentially upregulated in macrophages on day 21 compared with day 0 and for HNF4α-regulated genes out of differentially downregulated genes in hepatocytes between day 21 and 4.Statistical Analyses. Differential gene expression analysis was performed on UMIs divided by the median UMI count using a Mann–Whitney U test with FDR correction.
Human Data from Clalit Healthcare
The analysis of the data from the Clalit Healthcare environment was under Helsinki approval no. 195-17COM2. The cohort definition for breast cancer was as follows: Female patients with a diagnosis of breast cancer (all stages, both right and left breasts, all quadrants, all histologic subtypes) between the years 2002 and 2019 were included in breast cancer cohorts. Cohort 1 included breast cancer patients with survival of less than 2 years from diagnosis, N = 4,732. Cohort 2 included breast cancer patients with survival between 2 and 5 years from diagnosis, N = 4,086. Cohort 3 included breast cancer patients with survival between 5 and 10 years from diagnosis, N = 3,984. Cohort 4 included breast cancer patients with a complete blood count (CBC) test within 1 year of diagnosis (−365 days before diagnosis to 365 days after diagnosis) indicating the percentage of neutrophils higher than 80% or percentage of lymphocytes lower than 10%, N = 10,556. Cohort 5 included breast cancer patients with a CBC test within 1 year of diagnosis (−365 days before diagnosis to 365 days after diagnosis) indicating the percentage of neutrophils lower than 80% and percentage of lymphocytes higher than 10%, N = 35,723. The cohort definition for pancreatic cancer was as follows: Male patients with the diagnosis of pancreatic cancer (all stages, all locations in the pancreas, all histologic subtypes) between the years 2002 and 2019 were included in pancreatic cancer cohorts. Cohort 1 included pancreatic cancer patients with survival of less than half a year from diagnosis, N = 2037. Cohort 2 included pancreatic cancer patients with survival between half a year and 1 year from diagnosis, N = 659. Cohort 3 included pancreatic cancer patients with survival between 1 and 1.5 years from diagnosis, N = 342. Cohort 4 included pancreatic cancer patients with a CBC test within 1 year of diagnosis (−365 days before diagnosis to 365 days after diagnosis) indicating a percentage of neutrophils higher than 80% or percentage of lymphocytes lower than 10%. N = 4,238. Cohort 5 included pancreatic cancer patients with a CBC test within one year of diagnosis (−365 days before diagnosis to 365 days after diagnosis) indicating the percentage of neutrophils lower than 80% and percentage of lymphocytes higher than 10%, N = 4,218.
Liver Function Lab Data.
For cohorts 1 to 3 of each cancer type, where available, results of lab tests for selected variables taken in two time points (1 year before diagnosis and at the time of diagnosis) were obtained for each patient between the ages of 60 and 70. The variables for liver function were AST, ALT, albumin, ALK-P, LDH, and PT-SEC.
Survival Analysis for Cohorts.
Four to five of each cancer type, percentage of patients alive at the end of every year (0 = time of diagnosis) was calculated and a survival curve was created as an X–Y plot of %live patients at each year.
Score for Liver Function in Pancreatic Patient Data
The Sheba and Souraski medical center data on participants’ demographics, past surgical procedures, blood test values, and survival were extracted from patients’ medical records under Institutional Review Board approval (4474 and 5073-18 and 0551-17-TLV). Written informed consent was obtained from all patients prior to study enrollment. The protocols were approved by the Institutional Review Board at the Sheba and Sourasky medical centers, and the studies were conducted in accordance with the Good Clinical Practice guidelines and the Declaration of Helsinki. Pancreatic cancer patients with either liver metastases or ascites were excluded to perform the analysis on nonliver cancer data: Souraski: N = 732 (n for resectable = 255, n for locally advanced and metastatic disease = 362) and Sheba: N = 252 (n for stages 1 + 2 = 82, n for stages 3 + 4 = 170). For each sample, we then determined its liver function score, which is a weighted sum of the normalized expression of 5 liver enzymes and function-based molecules, the normalization into the average and SD in patients with pancreatic cancer (as studied from this cohort), and the weights defined based on correlation of each value with the survival of a random small cohort (50 patients)—that is, 2*|ALT(IU/I)−25.5|19.5+2*|AST(IU/I)−25|15+5*|INR−1.025|0.175+|ALKP(IU/I)−79.5|35.5+7*|4.46−Albumin(g/dL)|0.8617, where the names of genes denote their lab result value. A cutoff to separate high from the low score was set at cutoff = 0.6, where approximately 66% of patients had a high score and 33% a low score. For high-score and low-score patients, a Kaplan–Meier survival curve was plotted for patients from all cancer stages (stage on diagnosis) and also stratified by stage.
Weight Loss Analysis
For weight loss analysis, we excluded patients who underwent either Whipple or distal pancreatectomy surgeries and followed patients who had at least two weight measurements during the course of the disease (N = 369). The association between liver enzyme score and body mass index (BMI) change was evaluated through a linear regression analysis in which tumor stage and patient age were controlled. BMI change, ∆BMI, was defined as the (second BMI − first BMI)/first BMI. The linear regression follows the form:
where “score” stands for liver enzyme score.
Statistical Analysis
Unless otherwise specified, all statistical analyses were performed using two-way ANOVA, Student t test, or Wilcoxon rank sum test of multiple or two groups, with Dunnett correction when required. The sample size was chosen in advance based on the common practice of the described experiment and is mentioned for each experiment. Each experiment was conducted with biological and technical replicates and repeated at least three times unless specified otherwise. The results are presented as mean values ± SD. All error bars represent SD. P < 0.05 was considered significant in all analyses (*, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001).
Data Availability
Due to privacy regulations, all data analyses were conducted on a secured deidentified dedicated server within the Clalit Healthcare environment. Requests for access to all or parts of the Clalit datasets should be addressed to Clalit Healthcare Services via the Clalit Research Institute and addressed to Professor Shay Ben-Shachar at shayb@clalit.org.il. Requests will be considered by the Clalit Data Access Committee per the Clalit data-sharing policy.
Supplementary Material
Supplementary Table S1 details clinical data of PDAC patients
Supplementary Table S2 lists the antibodies used for western blots
Supplementary Table S3 lists the primers used for qPCR
Supplementary Table S4 lists the antibodies used for FACS
Supplementary Table S5 lists the antibodies used for CyTOF
Supplementary Table S6 lists the genes regulated by HNF4a
Supplementary Figure S1 demonstrates global metabolic changes in liver metabolism during early BC carcinogenesis
Supplementary Figure S2 demonstrates the infiltration of mature innate immune cells to the liver during breast carcinogenesis
Supplementary Figure S3 shows that targeting the IL-6-pSTAT-HNF4a pathway preserves liver metabolism and restricts BC growth
Supplementary Figure S4 demonstrates similar transcriptional and metabolic changes following liver infiltration of immune cells to the liver of PDAC mice
Supplementary Figure S5 demonstrate the significance of our liver score in predicting outcomes of BC and PDAC patients
Supplementary Extended Data 1 describes the FACS gating startegy
Acknowledgments
We acknowledge and thank the Weizmann Institute for providing financial and infrastructural support. The CCR2 KO mice were kindly provided to us by Ziv Shulman (Department of System Immunology, Weizmann Institute of Science, Rehovot, Israel). We thank Eli Pikarsky, Michal Schwartz, and Yaron Carmi for their intellectual input and Tomer Meir Salame, Keren Sasson, Jarmila Sekeresova Karlova, Yael Kuperman, Reinat Nevo, and Shani Ben-Moshe for their technical guidance. We thank Eytan Ruppin and Steffen Jung for their valuable comments and suggestions. A. Erez is supported by research grants from the European Research Council (ERC818943), the Israel Science Foundation (860/18), and the Israel Cancer Research Fund (837124). A. Erez received additional support from The Moross Integrated Cancer Center, the Blumberg Family Research Fellow Chair in Honor of Talia Lynn Steckman, Manya and Adolph Zarovinsky, and the Koret Foundation. We have a paid license to use BioRender.com through the Weizmann Institute.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
O. Goldman reports a patent for reexpression of HNF4A to alleviate cancer-associated cachexia issued. E. Hajaj reports a patent for reexpression of HNF4A to alleviate cancer-associated cachexia pending. T. Golan reports grants and personal fees from AstraZeneca, grants from Merck, and personal fees from AbbVie, Teva, and Roche outside the submitted work. I. Wolf reports grants and personal fees from Roche, and personal fees from Novartis, Sanofi, AstraZeneca, Bristol Myers Squibb, and Beyond Cancer outside the submitted work. A. Erez reports a patent for reexpression of HNF4A to alleviate cancer-associated cachexia issued. No disclosures were reported by the other authors.
Authors’ Contributions
O. Goldman: Conceptualization, validation, investigation, visualization, methodology. L.N. Adler: Formal analysis, supervision, validation, investigation, methodology, writing–original draft. E. Hajaj: Data curation, formal analysis, writing–original draft. T. Croese: Formal analysis, validation, investigation, methodology, writing–original draft. N. Darzi: Validation, investigation. S. Galai: Resources. H. Tishler: Methodology. Y. Ariav: Methodology. D. Lavie: Resources, validation, investigation, methodology. L. Fellus-Alyagor: Methodology. R. Oren: Supervision, investigation, methodology. Y. Kuznetsov: Investigation, methodology. E. David: Data curation, formal analysis, methodology. R. Jaschek: Data curation, formal analysis, investigation. C. Stossel: Resources, data curation, software, formal analysis, supervision. O. Singer: Resources, supervision, investigation, methodology. S. Malitsky: Validation, investigation, methodology. R. Barak: Resources, data curation, formal analysis, validation, investigation. R. Seger: Resources, data curation, formal analysis, supervision, methodology. N. Erez: Resources, supervision. I. Amit: Software, formal analysis, supervision. A. Tanay: Resources, data curation, software, formal analysis, supervision, investigation. A. Saada: Resources, data curation, formal analysis, validation, investigation. T. Golan: Resources, data curation, supervision. T. Rubinek: Resources, data curation. J. Sang Lee: Formal analysis. S. Ben-Shachar: Resources, data curation, supervision. I. Wolf: Conceptualization, resources, data curation, software, formal analysis, supervision. A. Erez: Conceptualization, resources, supervision, funding acquisition, validation.
References
- 1. McAllister SS, Weinberg RA. Tumor-host interactions: a far-reaching relationship. J Clin Oncol 2010;28:4022–8. [DOI] [PubMed] [Google Scholar]
- 2. Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010;18:884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin TD, Patel RS, Cook DR, Choi MY, Patil A, Liang AC, et al. The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 2021;373:1327–35. [DOI] [PubMed] [Google Scholar]
- 4. Rui L. Energy metabolism in the liver. Compr Physiol 2014;4:177–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell 2020;78:1019–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al-Zoughbi W, Hoefler G. Tumor macroenvironment: an update. Pathobiology 2020;87:58–60. [DOI] [PubMed] [Google Scholar]
- 7. Dong V, Nanchal R, Karvellas CJ. Pathophysiology of acute liver failure. Nutr Clin Pract 2020;35:24–29. [DOI] [PubMed] [Google Scholar]
- 8. Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol 2009;27:147–63. [DOI] [PubMed] [Google Scholar]
- 9. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology 2006;43( Suppl 1):S54–62. [DOI] [PubMed] [Google Scholar]
- 10. Jensen-Cody SO, Potthoff MJ. Hepatokines and metabolism: deciphering communication from the liver. Mol Metab 2021;44:101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ringseis R, Gessner DK, Eder K. The gut-liver axis in the control of energy metabolism and food intake in animals. Annu Rev Anim Biosci 2020;8:295–319. [DOI] [PubMed] [Google Scholar]
- 12. Schutz Y. Protein turnover, ureagenesis and gluconeogenesis. Int J Vitam Nutr Res 2011;81:101–7. [DOI] [PubMed] [Google Scholar]
- 13. Lee JS, Adler N, Karathia H, Carmel N, Rabinovich S, Auslander N, et al. Urea cycle dysregulation generates clinically relevant genomic and biochemical signatures. Cell 2018;174:1559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiu TH, Chandramouli GVR, Hunter KW, Alkharouf NW, Green JE, Liu ET, et al. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res 2004;64:5973–81. [DOI] [PubMed] [Google Scholar]
- 15. Erez A, Nagamani SC, Lee B. Argininosuccinate lyase deficiency-argininosuccinic aciduria and beyond. Am J Med Genet C Semin Med Genet 2011;157:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor a regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 2012;1819:921–9. [DOI] [PubMed] [Google Scholar]
- 17. Zheng Z, Li Y, Jia S, Zhu M, Cao L, Tao M, et al. Lung mesenchymal stromal cells influenced by Th2 cytokines mobilize neutrophils and facilitate metastasis by producing complement C3. Nat Commun 2021;12:6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caiado F, Pietras EM, Manz MG. Inflammation as a regulator of hematopoietic stem cell function in disease, aging, and clonal selection. J Exp Med 2021;218:e20201541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol 2020;21:1119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang YH, Aeberli D, Dacumos D, Xue JR, Morand EF, et al. Annexin-1 regulates macrophage IL-6 and TNF via glucocorticoid-induced leucine zipper. J Immunol 2009;183:1435–45. [DOI] [PubMed] [Google Scholar]
- 21. Hao Q, Vadgama JV, Wang P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal 2020;18:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu SL, Huang CC, Tzeng TT, Liu SC, Tsai SC, Fong YC, et al. S1P promotes IL-6 expression in osteoblasts through the PI3K, MEK/ERK and NF-kappaB signaling pathways. Int J Med Sci 2020;17:1207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang S, He X, Zhao J, Wang D, Guo S, Gao T, et al. Mitochondrial transcription factor A plays opposite roles in the initiation and progression of colitis-associated cancer. Cancer Commun (Lond) 2021;41:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juliano RL, Reddig P, Alahari S, Edin M, Howe A, Aplin A, et al. Integrin regulation of cell signalling and motility. Biochem Soc Trans 2004;32:443–6. [DOI] [PubMed] [Google Scholar]
- 25. Wang K, Patkar S, Lee JS, Gertz EM, Robinson W, Schischlik F, et al. Deconvolving clinically relevant cellular immune cross-talk from bulk gene expression using CODEFACS and LIRICS stratifies patients with melanoma to anti–PD-1 therapy. Cancer Discov 2022;12:1088–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee MM, Chui RKS, Tam IYS, Lau AHY, Wong YH, et al. CCR1-mediated STAT3 tyrosine phosphorylation and CXCL8 expression in THP-1 macrophage-like cells involve pertussis toxin-insensitive Galpha(14/16) signaling and IL-6 release. J Immunol 2012;189:5266–76. [DOI] [PubMed] [Google Scholar]
- 27. Martinez-Perez C, Kay C, Meehan J, Gray M, Dixon JM, Turnbull AK, et al. The IL6-like cytokine family: role and biomarker potential in breast cancer. J Pers Med 2021;11:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T, et al. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 2010;285:4415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh MM, Bosch DE, Daoud SS. Role of hepatocyte nuclear factor 4-alpha in gastrointestinal and liver diseases. World J Gastroenterol 2019;25:4074–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147:1233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu X, Xie P, Li W, Li Z, Shan H, et al. Direct induction of hepatocyte-like cells from immortalized human bone marrow mesenchymal stem cells by overexpression of HNF4alpha. Biochem Biophys Res Commun 2016;478:791–7. [DOI] [PubMed] [Google Scholar]
- 32. Jang YJ, LaBella AL, Feeney TP, Braverman N, Tuchma M, Morizono H, et al. Disease-causing mutations in the promoter and enhancer of the ornithine transcarbamylase gene. Hum Mutat 2018;39:527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang X, Wu M, Xu Z, Wang H, Wang H, Yu X, et al. HJC0152, a novel STAT3 inhibitor with promising anti-tumor effect in gastric cancer. Cancer Manag Res 2018;10:6857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 2011;17:989–1000. [DOI] [PubMed] [Google Scholar]
- 35. Michaelis KA, Zhu X, Burfeind KG, Krasnow SM, Levasseur PR, Morgan TK, et al. Establishment and characterization of a novel murine model of pancreatic cancer cachexia. J Cachexia Sarcopenia Muscle 2017;8:824–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Izhak L, Wildbaum G, Jung S, Stein A, Shaked Y, Karin N, et al. Correction: dissecting the autocrine and paracrine roles of the CCR2-CCL2 axis in tumor survival and angiogenesis. PLoS One 2018;13:e0195170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 38. Robinson MW, Harmon C, O'Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol 2016;13:267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Flint TR, Janowitz T, Connell CM, Roberts EW, Denton AE, Coll AP, et al. Tumor-induced IL-6 reprograms host metabolism to suppress anti-tumor immunity. Cell Metab 2016;24:672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Erez A, DeBerardinis RJ. Metabolic dysregulation in monogenic disorders and cancer-finding method in madness. Nat Rev Cancer 2015;15:440–8. [DOI] [PubMed] [Google Scholar]
- 41. Gaal Z, Szűcs Z, Kántor I, Luczay A, Tóth-Heyn P, Benn O, et al. A comprehensive analysis of Hungarian MODY patients-part I: gene panel sequencing reveals pathogenic mutations in HNF1A, HNF1B, HNF4A, ABCC8 and INS genes. Life (Basel) 2021;11:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y, Zhou TC, Liu YY, Li X, Wang W, Irwin DM, et al. Identification of HNF4A mutation p.T130I and HNF1A mutations p.I27L and p.S487N in a Han Chinese family with early-onset maternally inherited type 2 diabetes. J Diabetes Res 2016;2016:3582616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu XY, Zhang X, Ruan G, Zhang K, Tang M, Zhang Q, et al. One-year mortality in patients with cancer cachexia: association with albumin and total protein. Cancer Manag Res 2021;13:6775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mederacke I, Dapito DH, Affò S, Uchinami H, Schwabe RF, et al. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc 2015;10:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun 2015;6:6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FH, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 2017;6:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marini JC, Lee B, Garlick PJ. Non-surgical alternatives to invasive procedures in mice. Lab Anim 2006;40:275–81. [DOI] [PubMed] [Google Scholar]
- 48. Millard P, Delépine B, Guionnet M, Heuillet M, Bellvert F, Létisse F, et al. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 2019;35:4484–7. [DOI] [PubMed] [Google Scholar]
- 49. Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC, et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 2017;358:941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rustin P, Chretien D, Bourgeron T, Gérard B, Rötig A, Saudubray JM, et al. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 1994;228:35–51. [DOI] [PubMed] [Google Scholar]
- 51. Zincarelli C, Soltys S, Rengo G, Rabinowitz JE, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16:1073–80. [DOI] [PubMed] [Google Scholar]
- 52. Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 1999;6:973–85. [DOI] [PubMed] [Google Scholar]
- 53. Kechin A, Boyarskikh U, Kel A, Filipenko M.. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 2017;24:1138–43. [DOI] [PubMed] [Google Scholar]
- 54. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koster J, Rahmann S. Snakemake: a scalable bioinformatics workflow engine. Bioinformatics 2012;28:2520–2. [DOI] [PubMed] [Google Scholar]
- 58. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bell HN, Huber AK, Singhal R, Korimerla N, Rebernick RJ, Kumar R, et al. Microenvironmental ammonia enhances T cell exhaustion in colorectal cancer. Cell Metab 2023;35:134–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 details clinical data of PDAC patients
Supplementary Table S2 lists the antibodies used for western blots
Supplementary Table S3 lists the primers used for qPCR
Supplementary Table S4 lists the antibodies used for FACS
Supplementary Table S5 lists the antibodies used for CyTOF
Supplementary Table S6 lists the genes regulated by HNF4a
Supplementary Figure S1 demonstrates global metabolic changes in liver metabolism during early BC carcinogenesis
Supplementary Figure S2 demonstrates the infiltration of mature innate immune cells to the liver during breast carcinogenesis
Supplementary Figure S3 shows that targeting the IL-6-pSTAT-HNF4a pathway preserves liver metabolism and restricts BC growth
Supplementary Figure S4 demonstrates similar transcriptional and metabolic changes following liver infiltration of immune cells to the liver of PDAC mice
Supplementary Figure S5 demonstrate the significance of our liver score in predicting outcomes of BC and PDAC patients
Supplementary Extended Data 1 describes the FACS gating startegy
Data Availability Statement
Due to privacy regulations, all data analyses were conducted on a secured deidentified dedicated server within the Clalit Healthcare environment. Requests for access to all or parts of the Clalit datasets should be addressed to Clalit Healthcare Services via the Clalit Research Institute and addressed to Professor Shay Ben-Shachar at shayb@clalit.org.il. Requests will be considered by the Clalit Data Access Committee per the Clalit data-sharing policy.



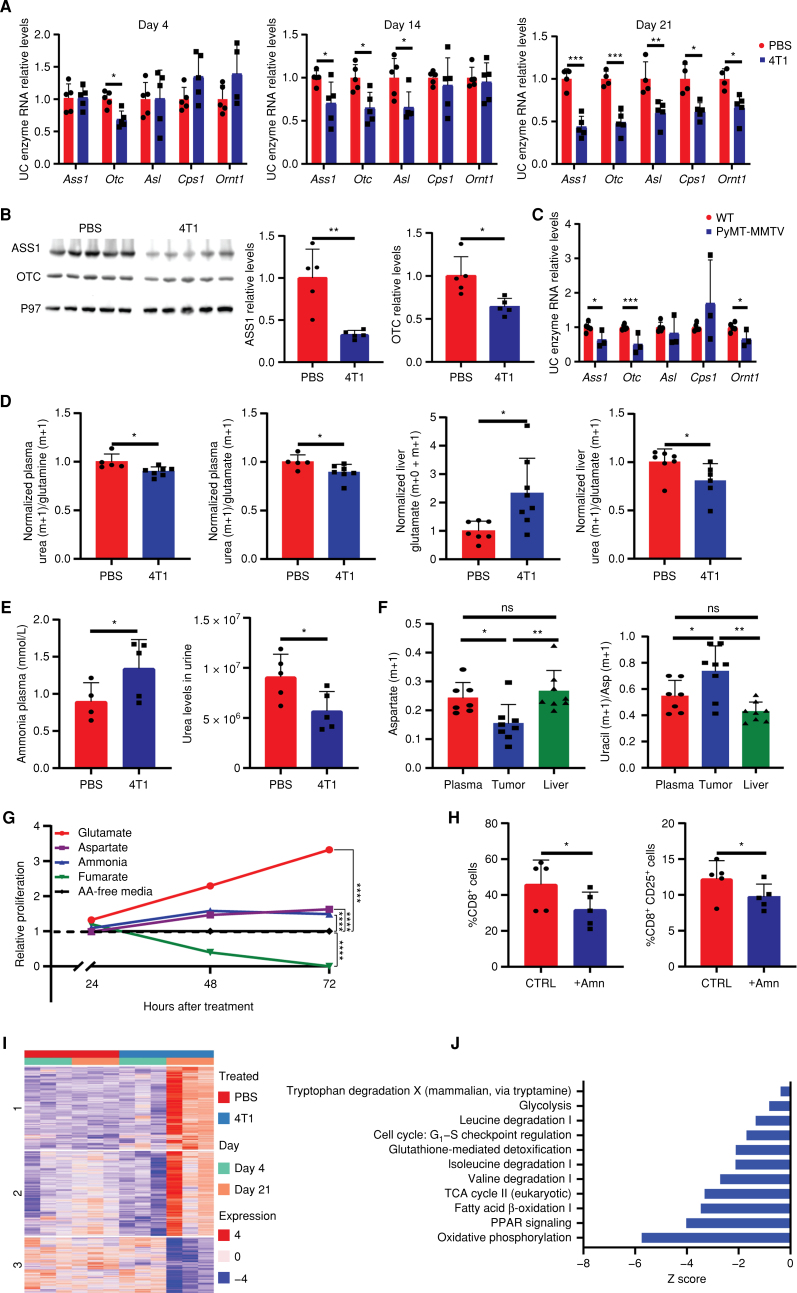
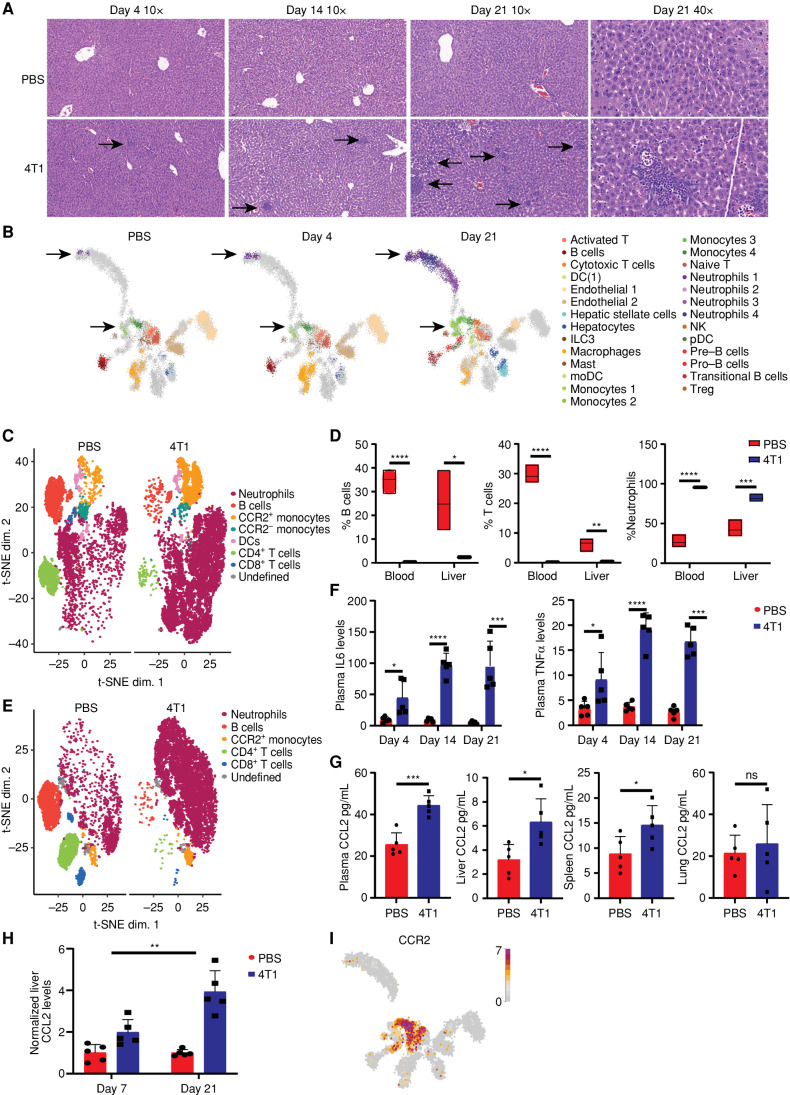
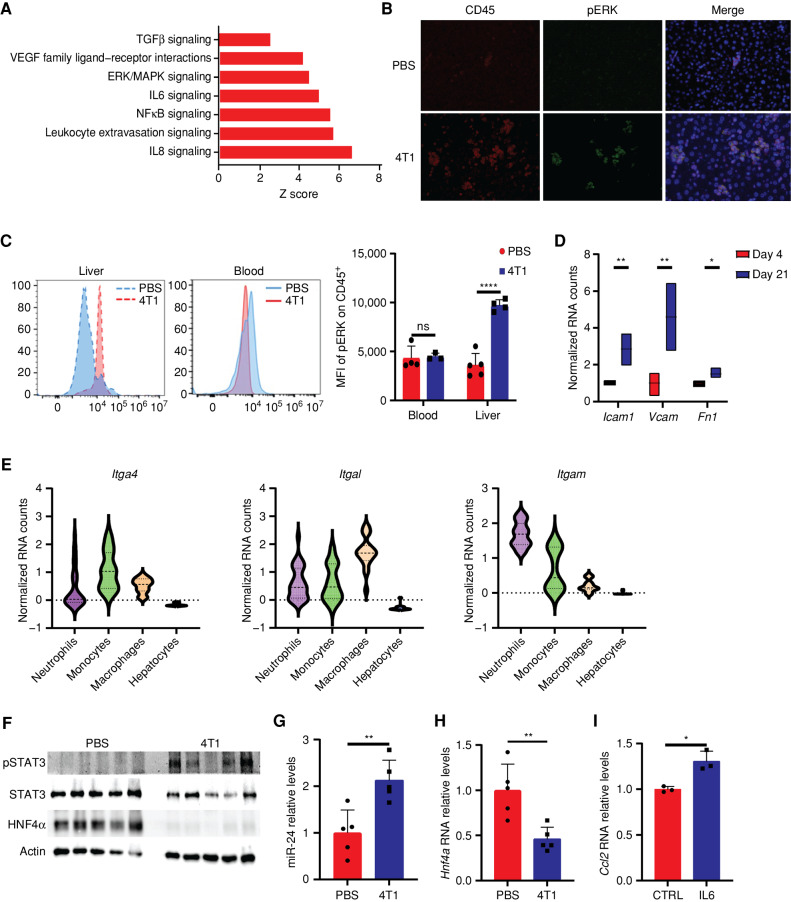
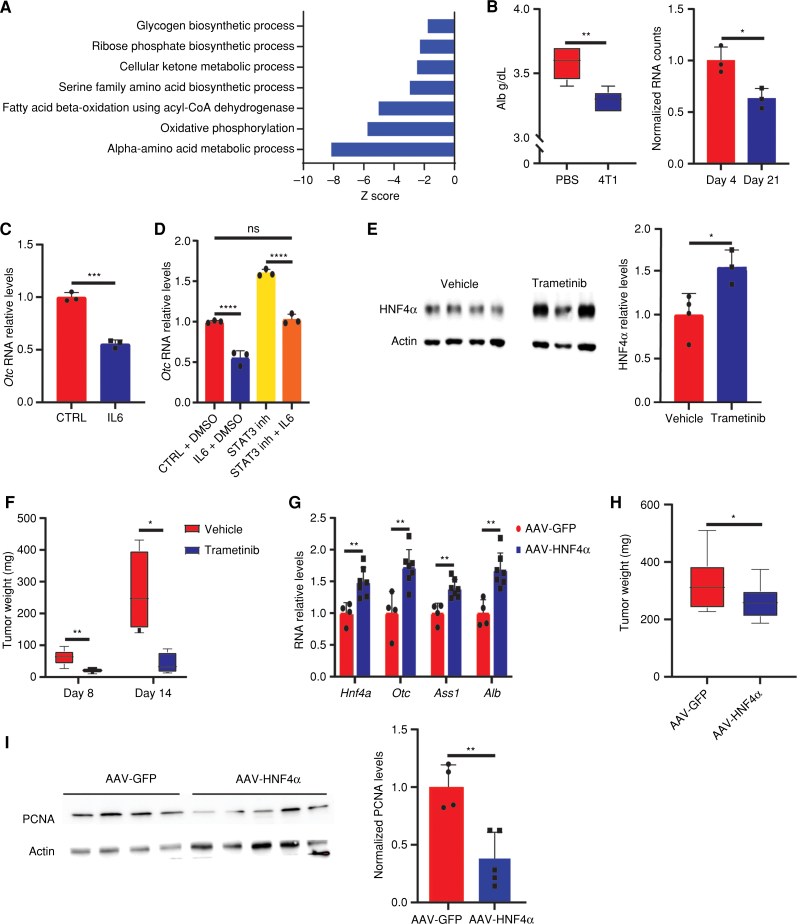
![Figure 5. CCR2 KO mice and HNF4α rescue the early changes in liver metabolism in pancreatic cancer–bearing mice. A, Hematoxylin and eosin staining demonstrates decreased immune cell infiltration to the livers of CCR2−/− pancreatic cancer–bearing mice compared with livers of pancreatic cancer–bearing WT mice. Pancreatic cancer–bearing mice were mice injected with KPC. Magnifications: 10, 20, and 40× as detailed in the figure. B, ELISA demonstrates decreased IL6 levels in the plasma of pancreatic cancer CCR2−/−–bearing mice relative to pancreatic cancer–bearing WT mice (PC WT n = 7, CCR2−/− n = 6, Student t test). P = 0.0125. C, RT-PCR of livers from CCR2−/− pancreatic cancer–bearing mice demonstrates preserved RNA expression levels of Ass1, Otc, Alb, and Hnf4a compared with decreased expression levels of these genes in livers of pancreatic cancer–bearing WT mice (PC WT n = 5, CCR2−/− n = 6, Student t test). P = 0.019, 0.011, 0.002, respectively. D, Decreased levels of aspartate and glutamate in the liver of pancreatic cancer CCR2−/−–bearing mice. UC intermediate levels were measured with gas chromatography–mass spectrometry (n = 7, Student t test). P = 0.011, 0.029, respectively. E, Weight measurements and nuclear magnetic resonance (NMR) body composition analysis of CCR2−/− pancreatic cancer–bearing mice demonstrate significantly less weight loss (left), increased fat tissue (middle), and decreased free fluids (right) in comparison with WT pancreatic cancer on day 21 (relative to day 0; n = 7 in each group, Student t test). P = 0.014, 0.006, and 0.004, respectively. F, Tumor weights measured on the day of sacrifice are significantly lower in the CCR2−/− mice with pancreatic cancer compared with WT pancreatic cancer (n = 7, Student t test). P = 0.009. G, Pancreatic cancer tumor growth is significantly more prominent in mice injected with AAV-GFP compared with AAV-HNF4α (AAV-GFP n = 4, AAV-HNF4α n = 9, Student t test). P = 0.006. H, Mice with pancreatic cancer injected with AAV-HNF4α demonstrate a significant increase in survival compared with those injected with AAV-GFP [log-rank (Mantel–Cox) test]. P = 0.02. I, Mice with pancreatic cancer injected with AAV-HNF4α maintain weight compared with those injected with AAV-GFP (AAV-GFP n = 8, AAV-HNF4α n = 9, two-way ANOVA). P = 0.05. J, NMR body composition analysis of mice with pancreatic cancer injected with AAV-HNF4α demonstrate less fat tissue loss (left) and decreased free fluid accumulation (right) in comparison with mice with pancreatic cancer injected with AAV-GFP (AAV-GFP n = 5, AAV-HNF4α n = 6, Student t test). P = 0.007, 0.012, respectively.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b437/10326600/f9b973233bef/1616fig5.jpg)