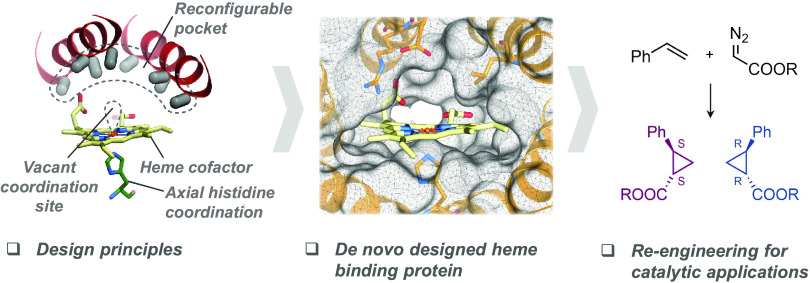
Abstract
The catalytic versatility of pentacoordinated iron is highlighted by the broad range of natural and engineered activities of heme enzymes such as cytochrome P450s, which position a porphyrin cofactor coordinating a central iron atom below an open substrate binding pocket. This catalytic prowess has inspired efforts to design de novo helical bundle scaffolds that bind porphyrin cofactors. However, such designs lack the large open substrate binding pocket of P450s, and hence, the range of chemical transformations accessible is limited. Here, with the goal of combining the advantages of the P450 catalytic site geometry with the almost unlimited customizability of de novo protein design, we design a high-affinity heme-binding protein, dnHEM1, with an axial histidine ligand, a vacant coordination site for generating reactive intermediates, and a tunable distal pocket for substrate binding. A 1.6 Å X-ray crystal structure of dnHEM1 reveals excellent agreement to the design model with key features programmed as intended. The incorporation of distal pocket substitutions converted dnHEM1 into a proficient peroxidase with a stable neutral ferryl intermediate. In parallel, dnHEM1 was redesigned to generate enantiocomplementary carbene transferases for styrene cyclopropanation (up to 93% isolated yield, 5000 turnovers, 97:3 e.r.) by reconfiguring the distal pocket to accommodate calculated transition state models. Our approach now enables the custom design of enzymes containing cofactors adjacent to binding pockets with an almost unlimited variety of shapes and functionalities.
Introduction
Heme-binding proteins carry out a wide range of chemical reactions in biology1−5 and directed evolution of cytochrome P450s, peroxidases, peroxygenases, globins, and other heme enzymes has led to a wealth of new catalytic activities.6−18 The key structural feature that gives rise to this catalytic versatility is a pentacoordinate heme iron cofactor positioned adjacent to an open substrate binding pocket. De novo design efforts have taken advantage of the simplicity and designability of helical bundle scaffolds19 to generate porphyrin-containing catalysts.20−29 However, this simplicity is also a limitation as such scaffolds cannot support large, open, and customizable active site pockets adjacent to the transition metal. These systems can also display considerable conformational flexibility/heterogeneity making them difficult to structurally characterize or rationally engineer, and covalent attachment of the heme cofactor may be needed to achieve selective catalysis. The design of porphyrin-binding proteins with large central cavities is a more difficult design challenge, as the side chain interactions that determine protein folding and stability generally reside in the protein core.
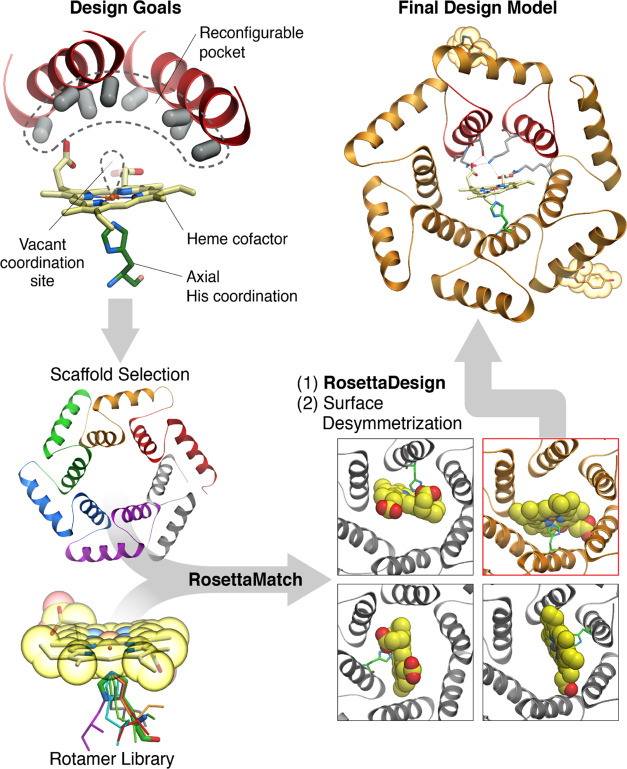
Advances in de novo protein design now enable the design of a wide variety of protein scaffold geometries extending far beyond helical bundles.30−36 With this control over structure and stability, protein functional sites can now be designed starting from a specification of an ideal site rather than being limited to particular scaffolds.37,38 We reasoned that this control could enable the design of hyperstable porphyrin-binding proteins with large reconfigurable pockets adjacent to an open transition metal coordination site. Such proteins could provide starting points for extending the catalytic potential of porphyrin-bound transition metals beyond natural scaffolds, as the shape of the catalytic pocket and arrangements of functional residues can be more extensively tuned. To achieve this goal, we aimed to design proteins with several key properties. First, a large central cavity lined by many reconfigurable side chains to enable the binding of a wide variety of substrates and tuning of active site geometry and chemistry. Second, very high stability, with the side chain interactions driving folding independent of the central cavity, to enable active site optimization free from the constraints of protein folding and stability. Third, overall backbone structure modularity to enable extensive reconfiguration of the structure lining one part of the active site without disruption of the overall fold to accommodate an even wider range of substrate sizes, shapes, and chemistries (Figure 1).
Figure 1.
Schematic of the approach to computational design. To generate heme-binding proteins containing a reconfigurable pocket near a free coordination site on the Fe atom (top left), a Heme-HIS moiety was matched into the pore of helical solenoid scaffolds of appropriate size (bottom left and right). The sequence around the heme cofactor was then optimized using Rosetta (top right).
Results and Discussion
Design of a Heme-Binding Protein
We chose closed α helical solenoid repeat architectures as optimal structural solutions to the general porphyrin-based protein catalyst design challenge (Figure 1). These hyperstable proteins have large central cavities lined by side chains emanating from repeating structural units; the stability and structure of the fold are determined by hydrophobic packing interactions within the repeat units, and hence, the cavity lining residues are almost entirely reconfigurable. The protein backbone itself is reconfigurable in repeat proteins as individual repeats can be structurally diversified.39 Within the family of closed helical solenoids, we selected a toroidal scaffold (PDB id: 4YXX)40 with an interior cavity sufficiently large to position the heme cofactor and an adjacent substrate, but compact enough that side chains at the opposite end of the cavity from the heme can modulate catalysis, substrate binding affinity, and specificity.
We sought to incorporate a heme-binding site within the central cavity of the toroid, with a proximal histidine ligand directly coordinating the iron atom of the cofactor, the distal site open for catalysis, and sufficient non-polar and hydrogen bonding side chain–cofactor interactions at the base and edges of the binding pocket to hold the heme in place without obstructing the free coordination site or interfering with potential substrate binding. We used Rosetta Match41 to place a histidine-coordinated heme into the toroid, limiting the matched positions only to those that would place the cofactor into the center of the pore (Figure 1). The heme model contained a methyl group bound to the iron atom, opposite to the histidine, acting as a steric placeholder to ensure free volume for a future substrate. In a subset of calculations, a supporting Glu or Asp side chain hydrogen bonding to the heme-ligating histidine was included to increase active site pre-organization. The remaining amino acid identities and conformations flanking the heme-binding site were optimized using Rosetta combinatorial design calculations42 for binding the hemin cofactor and supporting the coordinating histidine. The resulting design models were evaluated based on the His–Fe interaction geometry, shape complementarity, and pre-organization of the heme-binding pocket (assessed by side chain packing calculations in the absence of the heme), and pre-organized side chain hydrogen bonds with the propionate groups of the hemin to enable precise control of heme-binding orientation (Supplementary Figure 30). For designs having these features, ligand docking calculations were carried out using Rosetta GALigandDock,43 and those 22 for which the designed heme-binding orientation was lower in energy than any alternative binding modes were selected for experimental characterization (see SI for further details on the selection process). To aid with structure determination, the surface of the protein was desymmetrized by introducing two mutations K29W and K165Y.
Structural and Spectroscopic Characterization
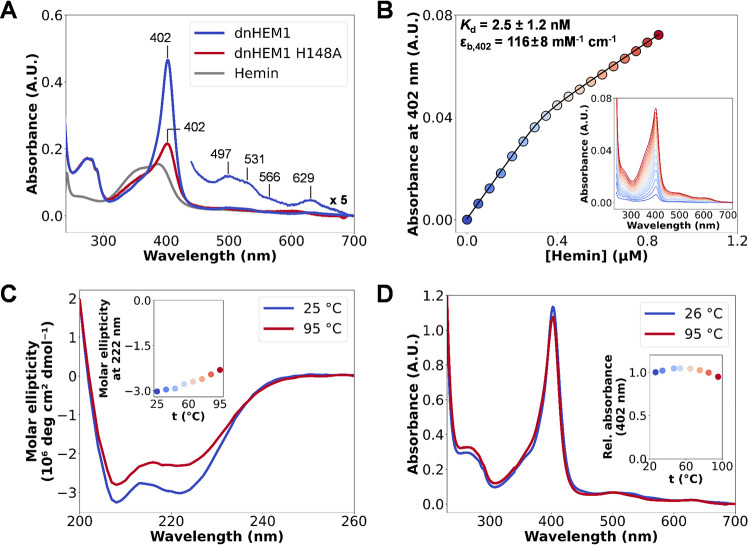
We selected twenty-two designs for expression in Escherichia coli (E. coli) and found that fifteen of them were well-expressed, soluble, and had size exclusion chromatography (SEC) elution volumes consistent with the intended monomeric state (Supplementary Figures 1 and 2). To identify potential binders, we performed a qualitative assay by incubating 30 μM of purified apo-proteins with 10 μM hemin and directly recording the UV/Vis spectra (Supplementary Figure 3). One of the designs, dnHEM1, yielded a promisingly sharp Soret band (λSoret = 402 nm), together with Q band features at 497, 531, 566, and 629 nm, reminiscent of the spectral features of natural heme proteins (Figure 2A).44 The UV/Vis spectrum of the ferrous-CO bound state of dnHEM1 has a sharp Soret feature at 417 nm, which is also consistent with a His-ligated heme iron (Supplementary Figure 12).45 Upon isolating a fully heme-loaded protein, we measured a ReinheitszahlRZ-value (ASoret/A280) of 5.62.46 The dissociation constant (Kd) of this design for hemin was determined to be <10 nM by titrating hemin into a solution of the apo-protein and monitoring absorbance changes at 402 nm (Figure 2B); three independent titrations yielded an average Kd of 2.5 ± 1.2 nM (Supplementary Figure 10). Replacement of the putative heme-coordinating His148 ligand of dnHEM1 by Phe or Ala leads to a substantial broadening of the Soret feature of proteins loaded with heme in vitro (Figure 2A and Supplementary Figure 5). dnHEM1 is a hyperthermostable protein (Tm > 95 °C), as evidenced by minimal changes in the circular dichroism (CD) spectrum in the 25–95 °C temperature range (Figure 2C). The structural integrity of the protein is not dependent on heme binding, with no differences in CD Tm measurements observed between apo and holo variants (Supplementary Figure 7). UV/Vis spectra collected at increasing temperatures showed minimal changes in the Soret band intensity and wavelength, suggesting that dnHEM1 retains most of its heme-binding ability even at 95 °C (Figure 2D). Similarly, minimal spectral changes are observed across a wide range of pH values (Supplementary Figure 13). Changing the surface charge of dnHEM1 from positive (pI 10) to slightly negative (pI 6) through 12 mutations, bringing it closer to most naturally occurring proteins, also had no detrimental effect on its expression level, solubility, or heme-binding ability (as judged by the sharpness and λmax value of the Soret band; see Supplementary Figure 8).
Figure 2.
Characterization of Heme binding. (A) UV/Vis spectra of dnHEM1 (blue) and its H148A mutant (red), after mixing 10 μM protein with 2 μM hemin, indicating the importance of H148 for heme binding. (B) Kd determination by heme titration into dnHEM1 (0.4 μM) and following absorbance changes at 402 nm, which were plotted against heme concentration and fitted to a one-site binding equation (see SI). (C) CD spectra of holo-dnHEM1 at increasing temperatures. (D) UV/Vis spectra of holo-dnHEM1 collected at increasing temperatures indicate that heme-binding ability is retained at temperatures up to 95 °C.
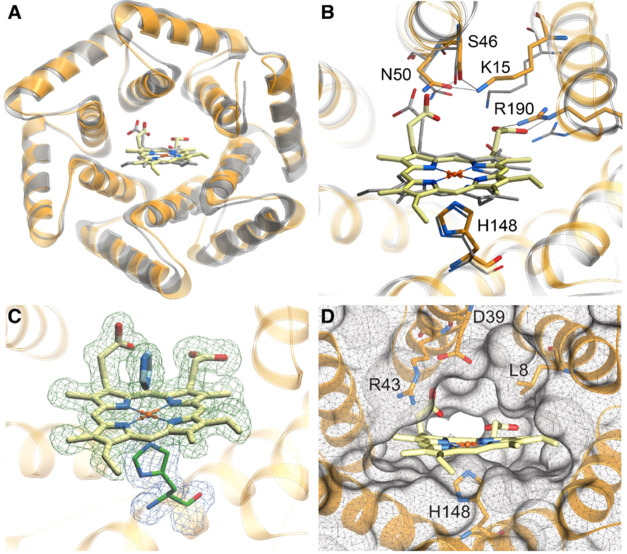
The crystal structure of heme-loaded dnHEM1 was solved to 1.6 Å resolution (Figure 3A, PDB id: 8C3W) and closely matches the computational design model (1.18 Å backbone RMSD). The heme cofactor is positioned as designed but adopts a flipped conformation, which alters the relative positioning of the heme vinyl and methyl substituents, but preserves the positioning of the heme propionates. Similar flipped heme orientations have previously been observed in natural and engineered heme proteins.44,47,48 The propionate groups interact with a network of polar residues (K15, S46, N50, R190) (Figure 3B). The heme iron is coordinated by His148 as designed, which in turn forms a hydrogen bond with the backbone carbonyl oxygen of Ile144. The open coordination site is occupied by an exogenous imidazole in the dnHEM1 crystal structure (Figure 3C). Overall, the crystal structure shows that we were successful in achieving our design goals; the heme is held in place by numerous polar and hydrophobic side chain–cofactor interactions, and the heme iron is coordinated on one side by a designed histidine and, on the other, by a small molecule from a solution that occupies a central binding cavity (Figure 3C,D).
Figure 3.
The crystal structure of dnHEM1 (PDB id: 8C3W) closely matches the design model. (A) Crystal structure of dnHEM1 (orange) overlaid with the design model (silver). (B) Superposition of the heme-binding site of the dnHEM1 design model (silver) and the crystal structure (orange). Hydrogen bond interactions with heme are indicated with dashed lines. (C) Crystal structure of dnHEM1 showing the electron density corresponding to heme and the exogenous imidazole in blue (Fo – Fc omit map contoured at 1σ) and His148 shown in green (2Fo – Fc map contoured at 1σ). (D) Open substrate binding pocket at the distal site upon omission of co-crystallized imidazole.
Engineering Peroxidase Activity
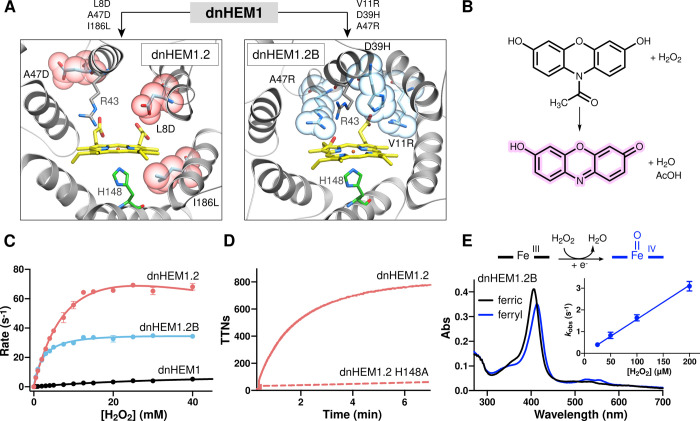
As a first step in exploring the catalytic potential of our designed heme protein, we assessed the peroxidase activity of dnHEM1 using Amplex Red as a model substrate (Figure 4B).49 Arg43 in the distal pocket of dnHEM1 sits in a similar position, relative to the heme cofactor, to a conserved catalytic arginine found in natural peroxidases. We found that under saturating concentrations of Amplex Red, dnHEM1 catalyzed the peroxidase reaction with a kcat of 9.5 ± 0.2 s–1 and KM of 36.7 ± 1.6 mM for H2O2 (Figure 4C). To increase activity, we performed two rounds of directed evolution, targeting residues lining the heme-binding cavity, which were individually randomized using NNK degenerate codons. Beneficial mutations identified during each round were subsequently combined by DNA shuffling (Supplementary Figure 14). Following evolution, we arrived at two triple mutants (dnHEM1.2: L8D-A47D-I186L and dnHEM1.2B: V11R-D39H-A47R) with significantly improved peroxidase activity (Figure 4A). There are minimal changes to the CD spectra and thermostability in either variant (Supplementary Figures 15 and 16), and in both cases, the catalytic function is strongly dependent on the proximal His148 ligand (Figure 4D and Supplementary Figure 17). These observations suggest that both protein structure and heme-binding geometry are well conserved in these evolved enzymes. The most active variant (dnHEM1.2) has two additional carboxylate groups situated in the distal heme pocket and catalyzes the oxidation of Amplex Red with a kcat of 129.5 ± 8.7 s–1 and KM[H2O2] of 11.5 ± 1.2 mM. This kcat value is ca. 20-fold and 5-fold lower than the rate of Amplex Red oxidation reported for HRP and the heavily engineered myoglobin-based peroxidase MbQ2.1 NMH, respectively, but is ca. 200-fold faster than Amplex Red oxidation by APX (see Table S8 of reference (44)).44 In contrast, improvements in catalytic efficiency observed in dnHEM1.2B (kcat of 37.0 ± 0.3 s–1 and KM[H2O2] of 2.0 ± 0.1 mM) are achieved by embedding two (positively charged) arginines and a His39 residue into the distal cavity (Supplementary Figure 18). Improvements in catalytic efficiency coincided with stabilization of a neutral ferryl [Fe(IV) = O] heme state (λSoret = 414 nm and optical features at 524/553 nm) akin to Compound II of natural peroxidases, which can be readily generated in this variant upon mixing of the ferric enzyme with hydrogen peroxide (Figure 4E, rate constants of (1.6 ± 0.05) × 104 M–1 s–1 and 0.03 ± 0.005 s–1 for ferryl intermediate formation and decay, respectively, see Supplementary Figures 19 and 21). For comparison, the Compound II state of ascorbate peroxidase has a Soret maximum at 418 nm and Q bands at 528 and 559 nm. This stabilization of the ferryl state could plausibly arise from additional hydrogen bonding interactions with the ferryl oxygen as a result of distal pocket mutations introduced during evolution.50,51 Analysis of the dnHEM1.2B evolutionary trajectory reveals a single V11R mutation installed in the first round of evolution was sufficient to allow accumulation of the neutral ferryl heme state, with the additional D39H and A47R mutations increasing the rate of ferryl formation (kobs of 1.2 and 3.0 s–1 for dnHEM1 V11R and dnHEM1.2B, respectively, using 200 μM hydrogen peroxide, Supplementary Figures 20 and 21). These combined data highlight the potential for modulating the structures and reactivities of key metal-oxo intermediates within our dnHEM scaffolds.
Figure 4.
Directed evolution converts dnHEM1 into a proficient peroxidase with a stable high valent ferryl intermediate. (A) AlphaFold252 predicted models of dnHEM1.2 and dnHEM1.2B indicating the mutations accumulated as a result of divergent directed evolution. Sites of mutation for dnHEM1.2 and dnHEM1.2B are shown as atom colored ball and sticks with purple or blue carbons, respectively. The heme cofactor is shown as gray atom colored ball and sticks and CPK spheres. (B) Chemical scheme showing the conversion of Amplex Red dye to resorufin mediated by hydrogen peroxide and dnHEM1 variants. (C) Michaelis–Menten plots under saturating concentrations of Amplex Red for dnHEM1 (black, kcat = 9.5 ± 0.2 s–1, KM[H2O2] = 36.7 ± 1.6 mM), dnHEM1.2 (red, kcat = 129.5 ± 8.7 s–1, KM[H2O2] = 11.5 ± 1.2 mM, KI = 58.9 ± 10.7 mM) and dnHEM1.2B (blue, kcat = 37.0 ± 0.3 s–1, KM[H2O2] = 2.0 ± 0.1 mM). Error bars represent SD n = 3. (D) Time course of resorufin formation by dnHEM1.2 (red solid line, TTNs = 788 ± 18) and dnHEM1.2 H148A (red dotted line). (E) Formation of a ferryl intermediate (blue) in dnHEM1.2B upon oxidation of the ferric enzyme (black line) with H2O2. The observed neutral ferryl heme state is most likely formed via rapid single electron transfer to a transient porphyrin-π cation radical species, with a redox active amino acid side chain being the most likely electron donor. Inset: A linear fit of kobs vs [H2O2] was used to derive a bimolecular rate constant of (1.6 ± 0.05) × 104 M–1 s–1 for ferryl heme formation. Error bars represent SD n = 3.
Computational Design of Enantiocomplementary Carbene Transferases
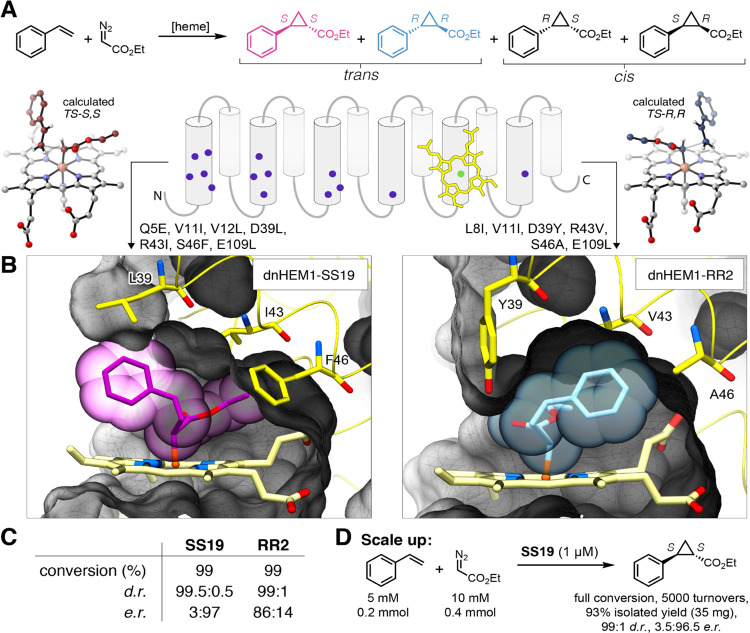
We next sought to computationally design new catalytic activities into dnHEM1, choosing as a model reaction the cyclopropanation of olefins, in particular, the reaction of ethyl diazoacetate (EDA) and styrene (Figure 5A). This reaction creates two new stereocenters, providing a stringent test of our control over reaction selectivity, and provides access to building blocks useful for the synthesis of bioactive molecules.53 The reaction pathways leading to the S,S and R,R enantiomers were modeled using density functional theory (DFT) calculations ((Supplementary Figure 32); as proof of principle, we considered only the energetically more facile trans-addition of styrene to the Fe-carbenoid intermediate). For each enantiomer, five TS conformers based on the rotation around the Fe-C(COOEt) bond were located and found to be within ΔG‡ = 3.6 kcal/mol (Supplementary Table 8). We overlaid these transition state models with the heme cofactor of dnHEM1 and then optimized the sequence in the immediate vicinity of the reaction substrates using Rosetta FastDesign54 (a total of 14 residues in the distal pocket were targeted for in silico engineering, Figure 5B, Supplementary Figure 33). There was very little overlap between the pocket sequences designed for each of the two enantiomers. We used the metrics described above to select designs for experimental testing (His–Fe constraint score, shape complementarity, pocket stability). This yielded 35 designs that expressed well in E. coli, in addition to being soluble and monomeric.
Figure 5.
Computational redesign of dnHEM1 for enantioselective olefin cyclopropanation activity. (A) Enantiocomplementary transition states for R,R-and S,S-cyclopropane formation were computed with DFT and selected positions in the distal pocket were redesigned with Rosetta. (B) Complementarity of the designed pocket with most selective enzymes for S,S (left) and R,R (right) transition state models. (C) Selectivities obtained with best variants for the synthesis of S,S and R,R cyclopropanes. Standard reaction condition: 1 μM catalyst, 1 mM styrene, 10 mM EDA, 100 μM dithionite, under N2 in aqueous potassium phosphate buffer (50 mM, NaCl 200 mM, pH 7.2) and 5% MeCN cosolvent for 2 h at 25 °C. See SI for the results with other designs. (D) Preparative scale synthesis of the S,S stereoisomer, catalyzed by dnHEM1-SS19 over 2 h at r.t.
We then evaluated the isolated heme-loaded proteins for cyclopropanation activity with styrene and EDA as the coupling partners in the presence of Na2S2O4 as a reductant. All designs had high diastereoselectivity (d.r. > 95:5) in favor of the trans-product, which contrasts the modest selectivity (d.r. 76:24) achieved with free hemin in solution (Supplementary Figure 23 and Table 4). Furthermore, all proteins designed to mediate S,S-selective catalysis did so, with 2 designs giving enantiomeric ratios (e.r.) > 95:5. Designing R,R-selective catalysts proved to be more challenging. Nevertheless, we were able to generate R,R-selective designs with up to 86:14 e.r., highlighting how with computational design it is possible to create substrate binding pockets that are able to discriminate between the two enantiomeric transition states. For comparison, neither the parent design dnHEM1 nor the evolved peroxidases (dnHEM1.2 and dnHEM1.2B) gave any appreciable levels of enantiocontrol for olefin cyclopropanation (Supplementary Figure 24). Redesign of the dnHEM1 catalytic pocket for cyclopropanation activity through 6–7 mutations, of which four were from polar to apolar amino acids, had no detrimental effect on protein stability, with the most selective S,S- and R,R-designs (dnHEM1-SS19 and dnHEM1-RR2) retaining their secondary structures and heme-binding abilities at 95 °C (Supplementary Figures 25 and 26). To demonstrate synthetic utility, we performed a preparative scale cyclopropanation using dnHEM1-SS19 as a biocatalyst. Using only 0.02 mol% enzyme, complete conversion of styrene (5 mM) was achieved within 2 h, affording optically enriched S,S-product (3.5:96.5 e.r., 35 mg) as a single diastereomer in 93% isolated yield (Figure 5D and Supplementary Figure 27). The high reaction yields, total turnover numbers, and stereocontrol achievable with dnHEM1-SS19 is particularly impressive given that this enzyme was designed in silico and has not been subjected to any evolutionary optimization. By way of comparison, the turnover numbers and selectivity achieved by dnHEM1-SS19 compare favorably to those reported for the de novo carbene transferase C45,24 which contains a covalently bound heme iron cofactor, and are only marginally lower than the values reported for the engineered myoglobin H64V V68A variant.8
Conclusions
The crystal structure of dnHEM1 demonstrates the attainment of our design goal: the creation of high-affinity heme-binding proteins with an open coordination site adjacent to a large reconfigurable substrate binding cavity. The ability to alter the surface charge, embed polar residues in the distal cavity to facilitate peroxidase catalysis, and computationally design substrate binding pockets for enantiocomplementary olefin cyclopropanations all testify to the engineerability of our designed systems. There are exciting routes moving forward from this initial study. First, we have not yet taken advantage of the reconfigurability of the individual repeat units in the designed solenoid - this should enable the more complete customization of the binding pocket to accommodate new substrates of interest. Second, using cysteine rather than histidine as an axial ligand to the heme iron will open up new catalytic possibilities by allowing the generation of more potent ferryl intermediates, as illustrated by the catalytic prowess of P450s and peroxygenases. Finally, our methods can be readily extended to design customized proteins that bind a range of biological or non-natural catalytic cofactors to extend the range of accessible chemistries;55−57 the pore size and internal geometry of α helical solenoids can readily be varied.40 We anticipate the enzymes and design principles presented here will serve as a platform for the creation of hyperstable biocatalysts for diverse applications.
Acknowledgments
The authors thank Xinting Li, Ryanne Ballard, and Mila Lamb for analytical services, Gyu Rie Lee for assistance with GALigandDock, and Yakov Kipnis for helpful discussions. The authors also thank Luki Goldschmidt and Kandise VanWormer, respectively, for maintaining the computational and wet lab resources at the Institute for Protein Design. The authors thank Florence J. Hardy, Richard Obexer, and Sarah L. Lovelock for helpful discussions throughout this project and for assistance in manuscript preparation. The authors thank Reynard Spiess (Manchester Institute of Biotechnology) for acquiring protein mass spectra and Mark Dunstan (Manchester Institute of Biotechnology) for guidance on automating directed evolution workflows.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c02742.
Data supporting the findings of this work, including methods and protocols, supplementary figures and tables, sequence, and crystallographic data are available within the Supplementary Information files (PDF)
Energies and molecular coordinates of calculated structures are provided in the Supplementary Information files (XYZ)
Computational models of designed proteins (ZIP)
Scripts and files associated with computational design are available for download on GitHub58
Accession Codes
PDB id 8C3W contains the coordinates and associated structure factors of dnHEM1 that have been deposited in the Protein Data Bank (PDB) database.
Author Contributions
⊥ I.K., M.O., and J.Z. contributed equally.
This work was supported by the Open Philanthropy Project Improving Protein Design Fund (I.K., D.B.), The Washington Research Foundation (A.R.), the Department of Energy ARPA-E Grant # 2459-1671 (N.M.E., D.B.), a Human Frontier Science Program Cross-Disciplinary Fellowship (LT000838/2018-C, I.K.), the European Research Council (ERC Starting Grant no. 757991, A.P.G.), and the Biotechnology and Biological Sciences Research Council (David Phillips Fellowship BB/M027023/1, A.P.G.)
The authors declare no competing financial interest.
Supplementary Material
References
- Rittle J.; Green M. T. Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science 2010, 330, 933–937. 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- Poulos T. L. Heme Enzyme Structure and Function. Chem. Rev. 2014, 114, 3919–3962. 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadei C. M.; Gumiero A.; Metcalfe C. L.; Murphy E. J.; Basran J.; Concilio M. G.; Teixeira S. C. M.; Schrader T. E.; Fielding A. J.; Ostermann A.; Blakeley M. P.; Raven E. L.; Moody P. C. E. Heme Enzymes. Neutron Cryo-Crystallography Captures the Protonation State of Ferryl Heme in a Peroxidase. Science 2014, 345, 193–197. 10.1126/science.1254398. [DOI] [PubMed] [Google Scholar]
- Moody P. C. E.; Raven E. L. The Nature and Reactivity of Ferryl Heme in Compounds I and II. Acc. Chem. Res. 2018, 51, 427–435. 10.1021/acs.accounts.7b00463. [DOI] [PubMed] [Google Scholar]
- Huang X.; Groves J. T. Oxygen Activation and Radical Transformations in Heme Proteins and Metalloporphyrins. Chem. Rev. 2018, 118, 2491–2553. 10.1021/acs.chemrev.7b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan R. Tuning P450 Enzymes as Oxidation Catalysts. ACS Catal. 2012, 2, 647–666. 10.1021/cs300001x. [DOI] [Google Scholar]
- Coelho P. S.; Brustad E. M.; Kannan A.; Arnold F. H. Olefin Cyclopropanation via Carbene Transfer Catalyzed by Engineered Cytochrome P450 Enzymes. Science 2013, 339, 307–310. 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- Bordeaux M.; Tyagi V.; Fasan R. Highly Diastereoselective and Enantioselective Olefin Cyclopropanation Using Engineered Myoglobin-Based Catalysts. Angew. Chem. Int. Ed. 2015, 54, 1744–1748. 10.1002/anie.201409928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan S. B. J.; Lewis R. D.; Chen K.; Arnold F. H. Directed Evolution of Cytochrome c for Carbon-Silicon Bond Formation: Bringing Silicon to Life. Science 2016, 354, 1048–1051. 10.1126/science.aah6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan S. B. J.; Huang X.; Gumulya Y.; Chen K.; Arnold F. H. Genetically Programmed Chiral Organoborane Synthesis. Nature 2017, 552, 132–136. 10.1038/nature24996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.; Wang B.; Ilie A.; Dubey K. D.; Bange G.; Korendovych I. V.; Shaik S.; Reetz M. T. A Redox-Mediated Kemp Eliminase. Nat. Commun. 2017, 8, 14876 10.1038/ncomms14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberg O. F.; Fasan R.; Arnold F. H. Exploiting and Engineering Hemoproteins for Abiological Carbene and Nitrene Transfer Reactions. Curr. Opin. Biotechnol. 2017, 47, 102–111. 10.1016/j.copbio.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. 10.1002/anie.201708408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz L. M.; Rosenthal K.; Lütz S. Recent Advances in Heme Biocatalysis Engineering. Biotechnol. Bioeng. 2019, 116, 3469–3475. 10.1002/bit.27156. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Arnold F. H. Navigating the Unnatural Reaction Space: Directed Evolution of Heme Proteins for Selective Carbene and Nitrene Transfer. Acc. Chem. Res. 2021, 54, 1209–1225. 10.1021/acs.accounts.0c00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber-Zucker S.; Mindel V.; Garcia-Ruiz E.; Weinstein J. J.; Alcalde M.; Fleishman S. J. Stable and Functionally Diverse Versatile Peroxidases Designed Directly from Sequences. J. Am. Chem. Soc. 2022, 144, 3564–3571. 10.1021/jacs.1c12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro C.; Kawasaki Y.; Wanyoike G.; Nishioka T.; Yamamoto T.; Snedecor B.; Robinson S. J.; Gosselin F. Engineered Cytochrome P450-Catalyzed Oxidative Biaryl Coupling Reaction Provides a Scalable Entry into Arylomycin Antibiotics. J. Am. Chem. Soc. 2022, 144, 14838–14845. 10.1021/jacs.2c06019. [DOI] [PubMed] [Google Scholar]
- Athavale S. V.; Gao S.; Das A.; Mallojjala S. C.; Alfonzo E.; Long Y.; Hirschi J. S.; Arnold F. H. Enzymatic Nitrogen Insertion into Unactivated C-H Bonds. J. Am. Chem. Soc. 2022, 144, 19097–19105. 10.1021/jacs.2c08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan L.; DeGrado W. F. Characterization of a Helical Protein Designed from First Principles. Science 1988, 241, 976–978. 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- Choma C. T.; Lear J. D.; Nelson M. J.; Dutton P. L.; Robertson D. E.; DeGrado W. F. Design of a Heme-Binding Four-Helix Bundle. J. Am. Chem. Soc. 1994, 116, 856–865. 10.1021/ja00082a005. [DOI] [Google Scholar]
- Robertson D. E.; Farid R. S.; Moser C. C.; Urbauer J. L.; Mulholland S. E.; Pidikiti R.; Lear J. D.; Wand A. J.; DeGrado W. F.; Dutton P. L. Design and Synthesis of Multi-Haem Proteins. Nature 1994, 368, 425–432. 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- Farid T. A.; Kodali G.; Solomon L. A.; Lichtenstein B. R.; Sheehan M. M.; Fry B. A.; Bialas C.; Ennist N. M.; Siedlecki J. A.; Zhao Z.; Stetz M. A.; Valentine K. G.; Anderson J. L. R.; Wand A. J.; Discher B. M.; Moser C. C.; Dutton P. L. Elementary Tetrahelical Protein Design for Diverse Oxidoreductase Functions. Nat. Chem. Biol. 2013, 9, 826–833. 10.1038/nchembio.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koder R. L.; Anderson J. L. R.; Solomon L. A.; Reddy K. S.; Moser C. C.; Dutton P. L. Design and Engineering of an O(2) Transport Protein. Nature 2009, 458, 305–309. 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenner R.; Steventon J. W.; Seddon A.; Anderson J. L. R. A de Novo Peroxidase Is Also a Promiscuous yet Stereoselective Carbene Transferase. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 1419–1428. 10.1073/pnas.1915054117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenner R.; Anderson J. L. R. Chemoselective N-H Insertion Catalyzed by a de Novo Carbene Transferase. Biotechnol. Appl. Biochem. 2020, 67, 527–535. 10.1002/bab.1924. [DOI] [PubMed] [Google Scholar]
- Watkins D. W.; Jenkins J. M. X.; Grayson K. J.; Wood N.; Steventon J. W.; Le Vay K. K.; Goodwin M. I.; Mullen A. S.; Bailey H. J.; Crump M. P.; MacMillan F.; Mulholland A. J.; Cameron G.; Sessions R. B.; Mann S.; Anderson J. L. R. Construction and in Vivo Assembly of a Catalytically Proficient and Hyperthermostable de Novo Enzyme. Nat. Commun. 2017, 8, 358 10.1038/s41467-017-00541-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennist N. M.; Stayrook S. E.; Dutton P. L.; Moser C. C. Rational Design of Photosynthetic Reaction Center Protein Maquettes. Front. Mol. Biosci. 2022, 9, 997295 10.3389/fmolb.2022.997295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennist N. M.; Zhao Z.; Stayrook S. E.; Discher B. M.; Dutton P. L.; Moser C. C. De Novo Protein Design of Photochemical Reaction Centers. Nat. Commun. 2022, 13, 4937 10.1038/s41467-022-32710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins G. H.; Noble C. E. M.; Blackburn H.; Hardy B.; Landau C.; Parnell A. E.; Yadav S.; Williams C.; Race P. R.; Oliveira A. S. F.; Crump M. P.; Berger-Schaffitzel C.; Mulholland A. J.; Ross Anderson J. L.. Precision Design of Single and Multi-Heme de Novo Proteins. bioRxiv 2020, 2020-09-24, 2020.09.24.311514. 10.1101/2020.09.24.311514. [DOI] [Google Scholar]
- Huang P.-S.; Boyken S. E.; Baker D. The Coming of Age of de Novo Protein Design. Nature 2016, 537, 320–327. 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Marcos E.; Basanta B.; Chidyausiku T. M.; Tang Y.; Oberdorfer G.; Liu G.; Swapna G. V. T.; Guan R.; Silva D.-A.; Dou J.; Pereira J. H.; Xiao R.; Sankaran B.; Zwart P. H.; Montelione G. T.; Baker D. Principles for Designing Proteins with Cavities Formed by Curved β Sheets. Science 2017, 355, 201–206. 10.1126/science.aah7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J.; Vorobieva A. A.; Sheffler W.; Doyle L. A.; Park H.; Bick M. J.; Mao B.; Foight G. W.; Lee M. Y.; Gagnon L. A.; Carter L.; Sankaran B.; Ovchinnikov S.; Marcos E.; Huang P.-S.; Vaughan J. C.; Stoddard B. L.; Baker D. De Novo Design of a Fluorescence-Activating β-Barrel. Nature 2018, 561, 485–491. 10.1038/s41586-018-0509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos E.; Chidyausiku T. M.; McShan A. C.; Evangelidis T.; Nerli S.; Carter L.; Nivón L. G.; Davis A.; Oberdorfer G.; Tripsianes K.; Sgourakis N. G.; Baker D. De Novo Design of a Non-Local β-Sheet Protein with High Stability and Accuracy. Nat. Struct. Mol. Biol. 2018, 25, 1028–1034. 10.1038/s41594-018-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishchenko I.; Pellock S. J.; Chidyausiku T. M.; Ramelot T. A.; Ovchinnikov S.; Hao J.; Bafna K.; Norn C.; Kang A.; Bera A. K.; DiMaio F.; Carter L.; Chow C. M.; Montelione G. T.; Baker D. De Novo Protein Design by Deep Network Hallucination. Nature 2021, 600, 547–552. 10.1038/s41586-021-04184-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky B. I. M.; Milles L. F.; Courbet A.; Ragotte R. J.; Dauparas J.; Kinfu E.; Tipps S.; Kibler R. D.; Baek M.; DiMaio F.; Li X.; Carter L.; Kang A.; Nguyen H.; Bera A. K.; Baker D. Hallucinating Symmetric Protein Assemblies. Science 2022, 378, 56–61. 10.1126/science.add1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. L.; Juergens D.; Bennett N. R.; Trippe B. L.; Yim J.; Eisenach H. E.; Ahern W.; Borst A. J.; Ragotte R. J.; Milles L. F.; Wicky B. I. M.; Hanikel N.; Pellock S. J.; Courbet A.; Sheffler W.; Wang J.; Venkatesh P.; Sappington I.; Torres S. V.; Lauko A.; De Bortoli V.; Mathieu E.; Barzilay R.; Jaakkola T. S.; DiMaio F.; Baek M.; Baker D.. Broadly Applicable and Accurate Protein Design by Integrating Structure Prediction Networks and Diffusion Generative Models. bioRxiv 2022, 2022-12-14, 2022.12.09.519842. 10.1101/2022.12.09.519842. [DOI] [Google Scholar]
- Wang J.; Lisanza S.; Juergens D.; Tischer D.; Watson J. L.; Castro K. M.; Ragotte R.; Saragovi A.; Milles L. F.; Baek M.; Anishchenko I.; Yang W.; Hicks D. R.; Expòsit M.; Schlichthaerle T.; Chun J.-H.; Dauparas J.; Bennett N.; Wicky B. I. M.; Muenks A.; DiMaio F.; Correia B.; Ovchinnikov S.; Baker D. Scaffolding Protein Functional Sites Using Deep Learning. Science 2022, 377, 387–394. 10.1126/science.abn2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock S. L.; Crawshaw R.; Basler S.; Levy C.; Baker D.; Hilvert D.; Green A. P. The Road to Fully Programmable Protein Catalysis. Nature 2022, 606, 49–58. 10.1038/s41586-022-04456-z. [DOI] [PubMed] [Google Scholar]
- Park K.; Shen B. W.; Parmeggiani F.; Huang P.-S.; Stoddard B. L.; Baker D. Control of Repeat-Protein Curvature by Computational Protein Design. Nat. Struct. Mol. Biol. 2015, 22, 167–174. 10.1038/nsmb.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle L.; Hallinan J.; Bolduc J.; Parmeggiani F.; Baker D.; Stoddard B. L.; Bradley P. Rational Design of α-Helical Tandem Repeat Proteins with Closed Architectures. Nature 2015, 528, 585–588. 10.1038/nature16191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini A.; Jiang L.; Wollacott A. M.; Cheng G.; Meiler J.; Althoff E. A.; Röthlisberger D.; Baker D. New Algorithms and an in Silico Benchmark for Computational Enzyme Design. Protein Sci. 2006, 15, 2785–2794. 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter F.; Leaver-Fay A.; Khare S. D.; Bjelic S.; Baker D. De Novo Enzyme Design Using Rosetta3. PLoS One 2011, 6, e19230 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.; Zhou G.; Baek M.; Baker D.; DiMaio F. Force Field Optimization Guided by Small Molecule Crystal Lattice Data Enables Consistent Sub-Angstrom Protein-Ligand Docking. J. Chem. Theory Comput. 2021, 17, 2000–2010. 10.1021/acs.jctc.0c01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott M.; Hayashi T.; Mori T.; Mittl P. R. E.; Green A. P.; Hilvert D. A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold. J. Am. Chem. Soc. 2018, 140, 1535–1543. 10.1021/jacs.7b12621. [DOI] [PubMed] [Google Scholar]
- Sowole M. A.; Vuong S.; Konermann L. Interactions of Hemoglobin and Myoglobin with Their Ligands CN(-), CO, and O2 Monitored by Electrospray Ionization-Mass Spectrometry. Anal. Chem. 2015, 87, 9538–9545. 10.1021/acs.analchem.5b02506. [DOI] [PubMed] [Google Scholar]
- Hofbauer S.; Hagmüller A.; Schaffner I.; Mlynek G.; Krutzler M.; Stadlmayr G.; Pirker K. F.; Obinger C.; Daims H.; Djinović-Carugo K.; Furtmüller P. G. Structure and Heme-Binding Properties of HemQ (Chlorite Dismutase-like Protein) from Listeria Monocytogenes. Arch. Biochem. Biophys. 2015, 574, 36–48. 10.1016/j.abb.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Mar G. N.; Davis N. L.; Parish D. W.; Smith K. M. Heme Orientational Disorder in Reconstituted and Native Sperm Whale Myoglobin. Proton Nuclear Magnetic Resonance Characterizations by Heme Methyl Deuterium Labeling in the Met-Cyano Protein. J. Mol. Biol. 1983, 168, 887–896. 10.1016/S0022-2836(83)80080-1. [DOI] [PubMed] [Google Scholar]
- Aojula H. S.; Wilson M. T.; Drake A. Characterization of Haem Disorder by Circular Dichroism. Biochem. J. 1986, 237, 613–616. 10.1042/bj2370613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.; Diwu Z.; Panchuk-Voloshina N.; Haugland R. P. A Stable Nonfluorescent Derivative of Resorufin for the Fluorometric Determination of Trace Hydrogen Peroxide: Applications in Detecting the Activity of Phagocyte NADPH Oxidase and Other Oxidases. Anal. Biochem. 1997, 253, 162–168. 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- Kwon H.; Basran J.; Pathak C.; Hussain M.; Freeman S. L.; Fielding A. J.; Bailey A. J.; Stefanou N.; Sparkes H. A.; Tosha T.; Yamashita K.; Hirata K.; Murakami H.; Ueno G.; Ago H.; Tono K.; Yamamoto M.; Sawai H.; Shiro Y.; Sugimoto H.; Raven E. L.; Moody P. C. E. XFEL Crystal Structures of Peroxidase Compound II. Angew. Chem. Int. Ed. 2021, 60, 14578–14585. 10.1002/anie.202103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmayer M.; Hardy F. J.; Quesne M. G.; Fisher K.; Levy C.; Heyes D. J.; Catlow C. R. A.; de Visser S. P.; Rigby S. E. J.; Hay S.; Green A. P. A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase. JACS Au 2021, 1, 913–918. 10.1021/jacsau.1c00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner C.; Carreira E. M. Cyclopropanation Strategies in Recent Total Syntheses. Chem. Rev. 2017, 117, 11651–11679. 10.1021/acs.chemrev.6b00798. [DOI] [PubMed] [Google Scholar]
- Maguire J. B.; Haddox H. K.; Strickland D.; Halabiya S. F.; Coventry B.; Griffin J. R.; Pulavarti S. V. S. R. K.; Cummins M.; Thieker D. F.; Klavins E.; Szyperski T.; DiMaio F.; Baker D.; Kuhlman B. Perturbing the Energy Landscape for Improved Packing during Computational Protein Design. Proteins 2021, 89, 436–449. 10.1002/prot.26030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwizer F.; Okamoto Y.; Heinisch T.; Gu Y.; Pellizzoni M. M.; Lebrun V.; Reuter R.; Köhler V.; Lewis J. C.; Ward T. R. Artificial Metalloenzymes: Reaction Scope and Optimization Strategies. Chem. Rev. 2018, 118, 142–231. 10.1021/acs.chemrev.7b00014. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Rebelein J. G.; Mallin H.; Trindler C.; Pellizzoni M. M.; Ward T. R. Genetic Engineering of an Artificial Metalloenzyme for Transfer Hydrogenation of a Self-Immolative Substrate in Escherichia Coli’s Periplasm. J. Am. Chem. Soc. 2018, 140, 13171–13175. 10.1021/jacs.8b07189. [DOI] [PubMed] [Google Scholar]
- Natoli S. N.; Hartwig J. F. Noble-Metal Substitution in Hemoproteins: An Emerging Strategy for Abiological Catalysis. Acc. Chem. Res. 2019, 52, 326–335. 10.1021/acs.accounts.8b00586. [DOI] [PubMed] [Google Scholar]
- Kalvet I.De novo heme binder design repository. https://github.com/ikalvet/heme_binder_design.git.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.