Abstract
The family Streptomycetaceae, notably species in the genus Streptomyces, have long been the subject of investigation due to their well-known ability to produce secondary metabolites. The emergence of drug resistant pathogens and the relative ease of producing genome sequences has renewed the importance of Streptomyces as producers of new natural products and resulted in revived efforts in isolating and describing strains from novel environments. A previous large study of the phylogeny in the Streptomycetaceae based on 16S rRNA gene sequences provided a useful framework for the relationships among species, but did not always have sufficient resolution to provide definitive identification. Multi-locus sequence analysis of 5 house-keeping genes has been shown to provide improved taxonomic resolution of Streptomyces species in a number of previous reports so a comprehensive study was undertaken to evaluate evolutionary relationships among species within the family Streptomycetaceae where type strains are available in the ARS Culture Collection or genome sequences are available in GenBank. The results of the analysis supported the distinctiveness of Kitasatospora and Streptacidiphilus as validly named genera since they cluster outside of the phylogenetic radiation of the genus Streptomyces. There is also support for the transfer of a number of Streptomyces species to the genus Kitasatospora as well for reducing at least 31 species clusters to a single taxon. The multi-locus sequence database resulting from the study is a useful tool for identification of new isolates and the phylogenetic analysis presented also provides a road map for planning future genome sequencing efforts in the Streptomycetaceae.
Keywords: MLSA, Streptomyces, Kitasatospora, Streptacidiphilus, Systematics
Introduction
Species within the family Streptomycetaceae, notably those in the genus Streptomyces, have been the subject of investigations for over a century, particularly following the discovery of their prodigious ability to produce antibiotics. The importance and role of Streptomyces in nature and biotechnology have been thoroughly reviewed elsewhere and thus will not be discussed here. However it was noted that 58 new Streptomyces species have been described to date since 2012 and Streptomyces were mentioned more than 22,000 times in the patent literature worldwide during the same period. The emergence of multiple drug resistant pathogens and relative ease of generating draft whole genome sequences from microorganisms has elevated the importance of Streptomyces strains as producers of valuable new natural products and resulted in renewed efforts in isolating and describing strains from new and novel environments. As thoroughly discussed by Labeda et al. (2012) in their study of the 16S rRNA gene phylogeny of the Streptomycetaceae, the systematics and taxonomy of the members of the family have had a long and meandering history, from one based largely on morphological characteristics, through numerical taxonomic analysis of large numbers of physiological (i.e., phenotypic) traits, and finally on to use of molecular genetic methods such as DNA:DNA hybridization and 16S rRNA gene sequencing. The 16S rRNA gene phylogeny provides a useful framework for inferring the relationships between species, but did not always seem to have enough taxonomic resolution to provide definitive identifications in the absence of DNA–DNA relatedness data. The observation that species of the genera Kitasatospora and Streptacidiphilus were found within the phylogenetic radiation of Streptomyces based on 16S rRNA gene sequences led some to question the validity of these genera (Kämpfer 2012). The use of multi-locus sequence analysis (MLSA) has been clearly demonstrated to provide the necessary resolution, comparable to that of DDH, for species discrimination within the Streptomycetaceae (Guo et al. 2008; Rong et al. 2009, 2010; Rong and Huang 2010, 2012, 2014). MLSA has been used recently to clarify the taxonomic position of members of a number of the 16S rRNA gene phylogeny clades including Clade 126, the Streptomyces albus clade (Labeda et al. 2014), and Clade 6, the Streptomyces hirsutus clade (Labeda et al. 2016), as well as a study of the strains within the NRRL Culture Collection identified as Streptomyces scabiei (Labeda 2016). A comprehensive study was therefore undertaken to evaluate the phylogenetic relationship of all taxa within the family Streptomycetaceae whose type strains were currently available in the ARS (NRRL) Culture Collection, as well as those type strains having draft genome sequences available in the public databanks.
Methods
The strains sequenced in the study were obtained from the ARS (NRRL) Culture Collection, Peoria, IL, USA where they had been maintained in long-term storage as lyophilized stocks and are listed in Supplemental Table S1. Strains were cultivated on yeast extract-malt extract agar (YM) ISP-2 medium (Shirling and Gottlieb 1966) at 28 °C for generation of biomass for DNA extraction. Some data were obtained from previous MLSA studies of from genome sequences deposited in NCBI for strains sequenced elsewhere.
Genomic DNA was isolated from all strains using UltraClean®Microbial DNA isolation kits (MoBio Labs, Carlsbad, CA) following the instructions of the manufacturer. Partial sequences of the house-keeping genes atpD (ATP synthase F1, beta subunit), gyrB (DNA gyrase B subunit), rpoB (RNA polymerase beta subunit), recA (recombinase A) and trpB (tryptophan synthetase, beta subunit) were amplified and sequenced using the primers and protocols described previously by Labeda et al. (2014). Amplified products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA), sequenced using BigDye 3.1 on an ABI model 3730 sequencer and assembled using Sequencher version 5.2 (Gene Codes, Ann Arbor, MI). The gene sequences for the 5 house-keeping loci for strains sequenced locally were deposited in Genbank (see Supplemental Table S1).
The draft genome sequences of a number of strains were also determined in the course of the present study. Libraries were prepared with genomic DNA isolated as described above using a Nextera XT DNA library preparation kit (Illumina, San Diego, CA) following the manufacturer’s instructions. The library preparations were sequenced with a MiSeq Desktop Sequencer (Illumina, San Diego, CA) and the resulting short sequence reads were trimmed for quality and removal of adapter sequences, and subsequently de novo assembled with CLC bio Genomics Workbench version 8.0.1 (CLC bio, Waltham, MA). The draft genome sequences have been deposited in the NCBI Whole Genome Shotgun database (See Supplementary Table S2).
The gene sequences for the 5 house-keeping loci for strains sequenced locally were organised using Bacterial Isolate Genomic Sequence Database (BIGSdb) version 1.12.3 (Jolley and Maiden 2010) on the ARS Microbial Genomic Sequence Database server (https:// 199.133.98.43). Where available (see Supplemental Table S2), genome sequences were up-loaded into the sequence bin for the respective strains in the BIGSdb isolate database. The genome sequences were scanned within BIGSdb for house-keeping loci, the specific sequence regions were tagged and the allele sequences and respective allele designations added to the sequence database if new alleles were found. The strain record was then updated with the matching allele id for each locus in the strain database. The sequences for the alleles of the loci for all strains in the study were individually aligned with MAFFT (Katoh and Standley 2013), subsequently concatenated head to tail in-frame, and exported in FASTA format, providing a dataset of 692 strains and 2608 positions. Phylogenetic relationships were constructed from the partitioned gene data in IQ-Tree version 1.41 (Nguyen et al. 2015) using the Maximum Likelihood based on the General Time Reversible model (Nei and Kumar 2000), GTR + F + I + G4, which had been determined to be the optimal model for these data during the model test phase of analysis. The trees were subjected to 1000 ultrafast bootstrap replications (Minh et al. 2013) followed by 1000 replications of assessment of branch supports with single branch tests with SH-like approximate likelihood ratio test (Guindon et al. 2010).
MLSA evolutionary distances were determined using MEGA 6.0 (Tamura et al. 2013) by calculating the Kimura 2-parameter distance (Kimura 1980) and are shown in Supplemental Table S3 for each strain pair. Strain pairs having ≤0.007 MLSA evolutionary distance were considered conspecific based on the guideline empirically determined by Rong and Huang (2012) that this MLSA distance (Kimura 2-paramenter distance) computed from the partial sequences of these house-keeping loci equates to 70% DNA–DNA homology.
Results and discussion
The phylogenetic relationship of species within the Streptomycetaceae based on the partial sequences of the 5 house-keeping loci can be seen in Fig. 1a–j. The node labels also include the taxon’s clade designation from the 16S rRNA gene sequence phylogenetic study of Labeda et al. (2012) as well as the clade assignment in the numerical taxonomic study of Williams et al. (1983), if included in that study. The clades designated on the trees were defined based on an MLSA distance ≤0.007 (shown in Supplemental Table S3), indicating that these represented the same species. Bootstrap values less than 95% or branches having SH-aLRT less than 85% were not shown on the tree as recommended by the developers of IQ-Tree.
Fig. 1 a–j.








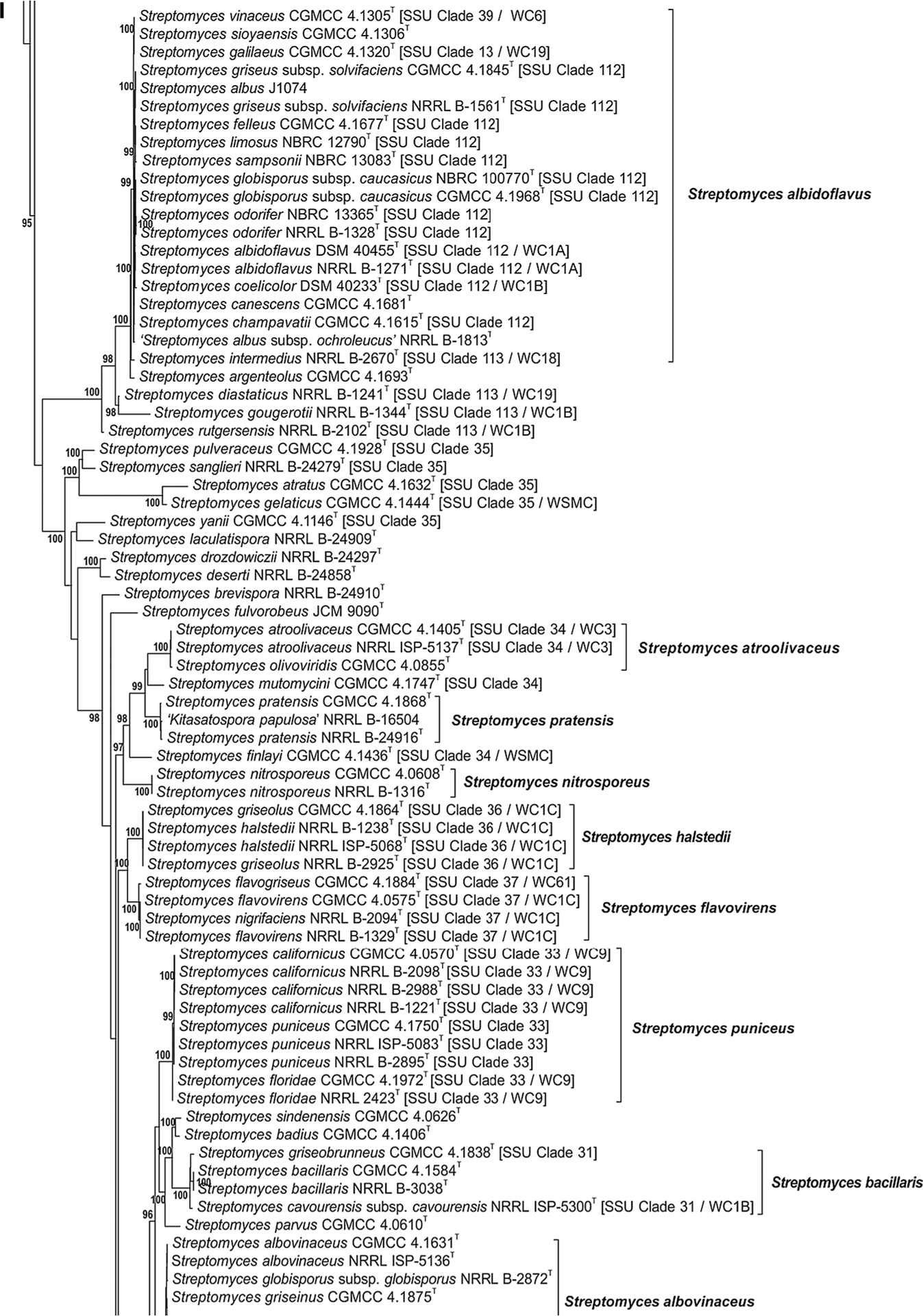
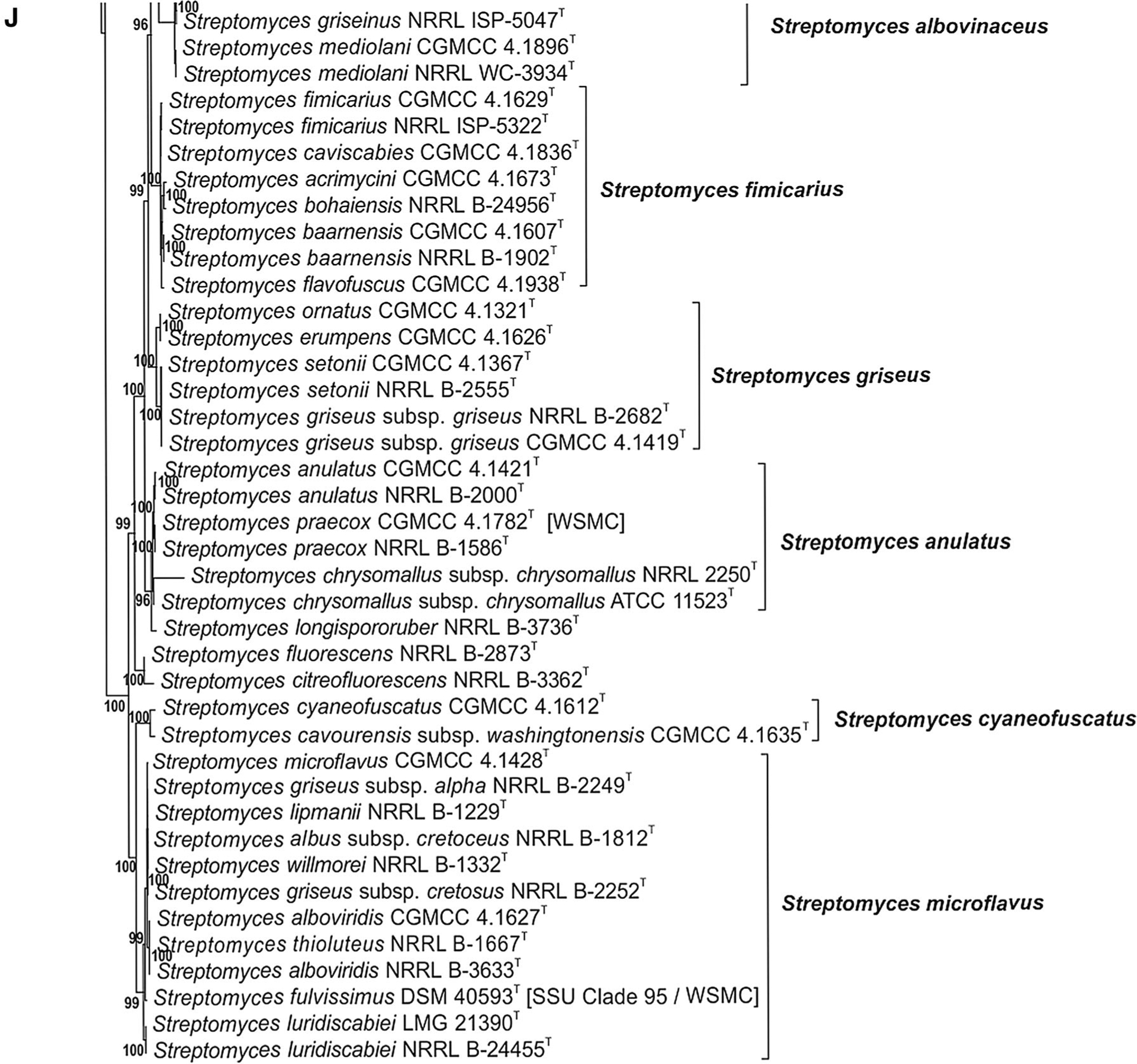
Phylogenetic relationships among strains within the Streptomycetaceae were constructed in IQ-Tree version 1.4.2 (Nguyen et al. 2015) using the Maximum Likelihood based on the full partition General Time Reversible model (Nei and Kumar 2000) including invariate sites plus a discrete Gamma model with 4 rate categories (GTR + F + I + G4) with separate branch lengths and models between partitions, which had been determined to be the optimal model considering the 5 gene partitions of these data during the model test phase of analysis. The trees were subjected to 1000 ultrafast bootstrap replications (Minh et al. 2013) and SH-like average likelihood ratio test replications (SH-aLRT) (Guindon et al. 2010). Bootstrap values less than 95% and/or SH-aLRT less than 85% were omitted as suggested by the IQ-Tree developers. Bar scale reflects number of substitutions per sites
There is generally good correlation between the phylogenetic relationships observed between species in the individual clades (Fig. 1a–j) with those observed in our previous phylogenetic study based on 16S rRNA gene sequences (Labeda et al. 2012). Not surprisingly, there is also fairly good correlation within clades to those assigned in the numerical taxonomic study of Williams et al. (1983), which could have been expected since the MLSA study should correlate well with observed phenotype. A major difference from the phylogenetic tree constructed using 16S rRNA sequence data is that those strains identified as species within the genera Kitasatospora and Streptacidiphilus are clearly observed (Fig. 1a) to fall outside of the radiation of the genus Streptomyces, thus confirming their validity and making the proposal of Kämpfer (2012) that these are genera incertae sedis unlikely. Girard and colleagues previously reported (Girard et al. 2014) evolutionary genetic changes in conserved developmental genes between Kitasatospora and Streptomyces that supported the validity of the genus Kitasatospora and their proposal was confirmed not only by the phylogenetic relationships shown in Fig. 1a but by a scan of the genomes held in the ARS Microbial Genomic Sequence database (http://199.133.98.43) in the present study that demonstrated that the bldB gene locus was absent in strains identified as Kitasatospora or Streptacidiphilus as they suggested. The 5-gene phylogeny shown in Fig. 1a also makes it quite evident that the type strains of many validly named Streptomyces species clearly belong within the genus Kitasatospora including: Streptomyces aburaviensis Nishimura et al. 1957, Streptomyces albolongus Tsukiura et al. 1964, Streptomyces aureofaciens Dugger 1948 (with Streptomyces avellaneus Baldacci and Grein 1966 as a later synonym), Streptomyces chrysomallus subsp. fumigatus Frommer 1959, Streptomyces cinereorectus Terekhova and Preobrazhenskaya 1986, Streptomyces herbaricolor Kawato and Shinobu 1959, Streptomyces misakiensis Nakamura 1961, Streptomyces psammoticus Virgilio and Hengeller 1960, and Streptomyces purpeofuscus Yamaguchi and Saburi 1955. Strain NRRL B-1588T, the type strain of Streptomyces cinnamonensis subsp. cinnamonensis, was also unexpectedly found within the Kitasatospora clade but equivalent type material from another culture collection must be obtained and analysed before any proposal is made to reclassify this strain. It is interesting to note that NRRL B-16,504, a strain deposited into the ARS Culture Collection as a proposed new species ‘Kitasatospora papulosa’ but demonstrated as a member of the species Streptomyces pratensis in this study (Fig. 1i) was found to contain an allele of the bldB locus. Streptacidiphilus species form a distinct, well supported clade that contains all of the taxa for which sequences were available. Strain NRRL B-24555, an isolate from cave soil collected on Jeju Island, Korea and deposited by S. D. Lee as the type strain of a possible new species to be named Streptacidiphilus jeojiense but never published, appears distinct from the other Streptacidiphilus species sequenced and likely represents a new species in this genus but further characterisation of morphological, chemotaxonomic and physiological traits will be necessary to fully describe it.
It is notable that both strains of Streptomyces griseoplanus that were included in the present study, representing the type strain and the International Streptomyces Project (ISP) strain, appear to be phylogenetically distinct from the genera Kitasatospora, Streptacidiphilus while also lacking the gene bldB and outside of the phylogenetic radiation of Streptomyces sensu stricto. Although a draft genome sequence has been determined for the type strain (NRRL B-3064T), further morphological, chemotaxonomic, and physiological study is necessary to formally propose a new genus for these strains. Likewise it will be interesting for future studies to address the relationships of the type species of the recently described genus Allostreptomyces (Huang et al. 2016) to other genera of the family Streptomycetaceae using whole genome sequencing and MLSA.
It is interesting to observe that certain morphological traits of Streptomyces species correlate with phylogenetic relationships observed from MLSA, including the clustering of those taxa having rugose spore surface ornamentation into a single well supported clade (Fig. 1a), while those exhibiting verticillate sporophore structures, originally described as species within the genus Streptoverticillium, are also found within another well-supported clade (Fig. 1c). Both clades are found within the radiation of the genus Streptomyces, as currently defined, and any proposal toward subdividing Streptomyces will require the availability of far more genome sequences for representative type strains for analysis.
Many of the clades presented in Fig. 1a–j illustrate species synonymies for which proposals have already been published. These include: Streptomyces albidoflavus (Rong et al. 2009) with Streptomyces canescens, Streptomyces champavatii, Streptomyces coelicolor, Streptomyces felleus, Streptomyces globisporus subsp. caucasicus, Streptomyces griseus subsp. solvifaciens, Streptomyces limosus, Streptomyces odorifer and Streptomyces sampsonii as later heterotypic synonyms, with the addition of Streptomyces galilaeus CGMCC 4.1320T, Streptomyces sioyaensis CGMCC 4.1306T and Streptomyces vinaceus CGMCC 4.1305T in the present study (Fig. 1a); Streptomyces albovinaceus (Rong and Huang 2010), with Streptomyces griseinus and Streptomyces mediolani as later heterotypic synonyms, now includes S. globisporus subsp. globisporus NRRL B-2872T in the present study (Fig. 1i) which has priority over S. albovinaceus requiring an emended description after genome sequences are obtained for all and DDH determinations are performed; S. albus (Labeda et al. 2012) with Streptomyces almquistii, Streptomyces flocculus, Streptomyces gibsonii and Streptomyces rangoonensis as later heterotypic synonyms (Fig. 1b); Streptomyces anulatus (Rong and Huang 2010) with Streptomyces praecox as a later heterotypic synonym including Streptomyces chrysomallus subsp. chrysomallus NRRL 2250T and ATCC 11523T in the present study (Fig. 1j), confirming the observation of Lanoot et al. (2005); Streptomyces atroolivaceus (Rong and Huang 2010) with Streptomyces olivaceoviridis as a later heterotypic synonym (Fig. 1i); Streptomyces bacillaris (Rong and Huang 2010) with Streptomyces griseobrunneus as a later heterotypic synonym, as well as Streptomyces cavourensis subsp. cavourensis NRRL ISP-5300T in the present study (Fig. 1i); Streptomyces castelarensis (Rong and Huang 2012) with Streptomyces antimycoticus and Streptomyces sporoclivatus as later heterotypic synonyms including Streptomyces mordarskii NRRL B-1346T in the present study (Fig. 1a); Streptomyces cyaneofuscatus (Rong and Huang 2010) with Streptomyces cavourensis subsp. washingtonensis as a later heterotypic synonym, but not Streptomyces fluorescens as proposed by Rong and Huang (2010) (Fig. 1j); Streptomyces ehimensis (Rong and Huang 2012) with Streptomyces luteoverticillatus as a later heterotypic synonym (Fig. 1c); Streptomyces fimicarius (Rong and Huang 2010) with Streptomyces acrimycini, Streptomyces baarnensis, Streptomyces caviscabies and Streptomyces flavofuscus as later heterotypic synonyms, including Streptomyces bohaiensis NRRL B-24956T in the present study (Fig. 1j); Streptomyces flavovirens (Rong and Huang 2010) with Streptomyces flavogriseus as a later heterotypic synonym including Streptomyces nigrifaciens NRRL B-2094T in the present study (Fig. 1i); S. griseus (Rong and Huang 2010) with Streptomyces erumpens, ‘Streptomyces ornatus’ and Streptomyces setonii as later heterotypic synonyms (Fig. 1j); Streptomyces hirsutus (Labeda et al. 2016) with Streptomyces cyanoalbus as a later heterotypic synonym (Fig. 1f); Streptomyces hygroscopicus (Rong and Huang 2012) with Streptomyces demainii, Streptomyces endus and Streptomyces sporocinereus as later heterotypic synonyms (Fig. 1a); Streptomyces javensis (Rong and Huang 2012) with Streptomyces yogyakartensis as a later heterotypic synonym (Fig. 1a); Streptomyces microflavus (Rong and Huang 2010) with Streptomyces alboviridis, S. griseus subsp. alpha, S. griseus subsp. cretosus and Streptomyces luridiscabiei as later heterotypic synonyms including Streptomyces lipmanii NRRL B-1229T, Streptomyces willmorei NRRL B-1332T (confirming the proposals of Lanoot et al. (2005) and Labeda et al. (2014)) and Streptomyces thioluteus NRRL B-1667T in the present study (Fig. 1j); Streptomyces nigrescens (Rong and Huang 2012) with Streptomyces libani subsp. libani as a later heterotypic synonym; Streptomyces prasinopilosus (Labeda et al. 2016) with Streptomyces emeiensis as a later heterotypic synonym (Fig. 1f); Streptomyces prasinus (Labeda et al. 2016) with Streptomyces bambergiensis as a later heterotypic synonym (Fig. 1f); Streptomyces puniceus (Rong and Huang 2010) with Streptomyces californicus and Streptomyces floridae as later heterotypic synonyms (Fig. 1i); Streptomyces rectiverticillatus (Rong and Huang 2012) with Streptomyces aureoversilis as a later heterotypic synonym (Fig. 1c); Streptomyces rhizosphaericus (Rong and Huang 2012) with Streptomyces asiaticus, Streptomyces cangkringensis and Streptomyces indonesiensis as later heterotypic synonyms (Fig. 1a).
This phylogenetic study also supported proposals in previous studies for elevating former subspecies to new species, including Streptomyces glebosus, formerly Streptomyces hygroscopicus subsp. glebosus (Rong and Huang 2012) (Fig. 1b), Streptomyces ossamyceticus, formerly S. hygroscopicus subsp. ossamyceticus (Rong and Huang 2012) (Fig. 1h) and Streptomyces pathocidini, formerly S. albus subsp. pathocidicus (Labeda et al. 2014) (Fig. 1b).
In the present study some potentially misidentified collection strains were identified based on phylogenetic position and an MLSA distance ≤0.007. Streptomyces narbonensis NRRL ISP-5016T is identified as a strain of S. albus (see Fig. 1b) while the original type strain, NRRL B-1680T is found nearest to Streptomyces zaomyceticus NRRL B-2038T (near S. venezuelae in Fig. 1c). As mentioned by Labeda et al. (2012), ‘S. albus’ J1074, whose genome has been sequenced, is actually a strain of Streptomyces albidoflavus and is found in that clade in Fig. 1i. It is also quite evident that Streptomyces lydicus NRRL ISP-5461 is actually a strain of Streptomyces varsoviensis because all share identical alleles for the 5 house-keeping loci (Fig. 1c) and are phylogenetically very distant from Streptomyces lydicus CGMCC 4.1412T (Fig. 1b).
It should also be noted that Streptomyces fulvissimus DSM 40593T (added from a draft genome sequence) is found in the Streptomyces microflavus clade (Fig. 1j), quite distant from Streptomyces fulvissimus NRRL B-1453T (near Streptomyces alboflavus in Fig. 1d) that was sequenced locally. It is not clear in this case which strain represents the actual type of Streptomyces fulvissimus and comparison with the type strain acquired from one or more other culture collections will be necessary to clarify this situation.
The relationships shown in the maximum likelihood phylogenetic tree in Fig. 1a–j, constructed from the alignment resulting from concatenation of the partial sequences of the 5 house-keeping loci, where the observed MLSA distances between clade members is also ≤0.007 suggest that a number of validly named Streptomyces species should be considered synonymous although further studies, including the preparation of draft genome sequences and determination of genomic relatedness, are needed to confirm any merging of these taxa.
To simplify the discussion of the results illustrated in this very long phylogenetic tree (Fig. 1), observations on each page of the figure will be discussed in order.
In Fig. 1a, aside from the previously discussed species to be reassigned to Kitasatospora, it was observed that Streptomyces samsunensis NRRL B-24803T represents a later heterotypic synonym of Streptomyces malaysiensis NRRL B-24313T while Streptomyces cuspidosporus NRRL B-5620T represents a later heterotypic synonym of Streptomyces sparsogenes NRRL 2940T. The equivalent strains of ‘Streptomyces cattleya’ (NRRL 8057 and DSM 46488) that have genome sequences available in the public databanks clearly represent a distinct species but further morphological, chemotaxonomic and phenotypic characterisation is necessary to prepare a formal species description.
In Fig. 1b, the phylogenetic analysis and observed MLSA distance (0.002) confirmed that Streptomyces aminophilus NRRL ISP-5186T is a later heterotypic synonym of Streptomyces cacaoi subsp. cacaoi NRRL B-1220T as proposed earlier by Lanoot et al. (2002) based on SDS-PAGE of proteins. It can also be seen that Streptomyces ochraceiscleroticus CGMCC 4.1096T is equivalent to both S. ochraceiscleroticus NRRL ISP-559T and Streptomyces violens NRRL ISP-5597T but not S. violens CGMCC 4.1786T which will require sequencing of additional type strains of S. violens from other collections to clarify the taxonomic status of this species.
In Fig. 1c, it was confirmed that Streptomyces spitsbergensis NRRL B-24285T is a later synonym of Streptomyces baldaccii NRRL B-3500T as proposed by Hatano et al. (1997) but the present study supports the revival of Streptomyces griseoverticillatus because it does not appear to represent a later synonym of Streptomyces cinnamoneus as Hatano et al. (1997) proposed. Streptomyces sapporonensis DSM 41675T appears to be a later synonym of S. griseoverticillatus DSM 40507T. It can be observed that Streptomyces alboverticillatus NRRL B-24281T has the later synonyms Streptomyces griseocarneus NRRL B-1350T and Streptomyces septatus NRRL ISP-5577T, Streptomyces abikoensis CGMCC 4.1662T has the later synonym Streptomyces kashmirensis NRRL B-3103T and Streptomyces mashuensis DSM 40221T has the later synonym Streptomyces kishiwadensis NRRL B-12326T. Streptomyces cinnamoneus subsp. albosporus NRRL B-5624 appears to represent a distinct species and S. cinnamoneus subsp. lanosus NRRL B-24290 and S. cinnamoneus subsp. sparsus NRRL B-24291 appear to represent a single new species but it is necessary to determine draft genome sequences for all of the Streptomyces cinnamoneus subspecies for genomic DDH comparisons along with morphological and physiological characterisation before these formal descriptions can be prepared.
In Fig. 1d, it was confirmed that Streptomyces laceyi CGMCC 4.1832T and Streptomyces spheroides NRRL 2449T represent later synonyms of Streptomyces niveus NRRL 2466T as proposed by Tamura et al. (2008). It was also confirmed that Streptomyces fradiae NRRL B-1195T is an earlier synonym of Streptomyces roseoflavus NRRL B-2789T as proposed by Lanoot et al. (2004). This section of the tree supports that Streptomyces aureocirculatus NRRL B-3324T (and NRRL ISP-5386T) has the later synonym Streptomyces glomeroaurantiacus NRRL B-3375T, Streptomyces coerulescens CGMCC 4.1597T has the later synonym Streptomyces coeruleofuscus NRRL B-5417T and Streptomyces angustmyceticus CGMCC 4.0207T has the later synonym Streptomyces pluricolorescens CGMCC 4.0236T. The status of strain Streptomyces noursei CGMCC 4.0213T is unclear because two other type strains of S. noursei are found in the tree at the top of Fig. 1c.
In Fig. 1e it was confirmed that Streptomyces flaviscleroticus NRRL B-12173T is a later synonym of Streptomyces minutiscleroticus NRRL B-12202T as proposed by Lanoot et al. (2005). The following observations can also be made: Streptomyces pseudo-griseolus NRRL B-3288T has the later synonyms Streptomyces gancidicus NRRL B-1872T, Streptomyces nashvillensis NRRL B-2606Tand Streptomyces rubiginosus NRRL B-3983T; Streptomyces althioticus NRRL B-3981T has the later synonyms Streptomyces griseorubens NRRL B-3982T, Streptomyces matensis NRRL B-2576T and Streptomyces phaeogriseichromatogenes NRRL 2834T; Streptomyces arabicus NRRL B-1733T has the later synonyms Streptomyces erythrogriseus NRRL B-3808T, Streptomyces griseoincarnatus NRRL B-5313T and Streptomyces variabilis NRRL B-3984T; Streptomyces pilosus NRRL ISP-5097T has the later synonym Streptomyces flavoviridis NRRL ISP-5153T; Streptomyces asterosporus NRRL B-24328T has the later synonym Streptomyces aureorectus NRRL B-24301T; Streptomyces coelescens NRRL B-12348T has the later synonyms Streptomyces anthocyanicus NRRL B-24292T, Streptomyces humiferus NRRL B-3088T, Streptomyces rameus NRRL B-16924T, Streptomyces sannanensis NRRL B-24303T and Streptomyces violaceorectus NRRL B-12181T. It is noted that ‘Streptomyces coelicolor’ JI 1147 and ‘Streptomyces lividans’ TK24 are also strains of S. coelescens. In addition, Streptomyces ennisocaesilis NRRL B-16365T, Streptomyces geysiriensis NRRL B-12102T and Streptomyces vinaceusdrappus NRRL ISP-5169T represent later synonyms of Streptomyces rochei NRRL B-2410T while Streptomyces daghestanicus NRRL B-5418T is a later synonym of Streptomyces griseoviridis NRRL ISP-5229T.
In Fig. 1f it should be noted that the thermophilic Streptomyces species group into a single clade, continued onto Fig. 1g. It was observed that Streptomyces roseodiastaticius CGMCC 4.1788T has the later synonyms Streptomyces tricolor NRRL B-16925T (as proposed by Lanoot et al. 2004) and Streptomyces bangladeshensis NRRL B-24326T. The following observations can also be made: Streptomyces olivaceoviridis NRRL B-12280T has the later synonym Streptomyces corchorusii NRRL B-12289T (also DSM 40340T); Streptomyces murinus NRRL B-2286T has the later synonyms Streptomyces costaricanus NRRL B-16897T and Streptomyces griseofuscus NRRL B-5429T; Streptomyces viridosporus NRRL ISP-5243T has the later synonym Streptomyces ghanaensis NRRL B-12104T (also ATCC 14672T); Streptomyces griseomycini NRRL B-5421T has the later synonyms Streptomyces graminearus NRRL B-16369T and Streptomyces griseostramineus NRRL B-5422T; Streptomyces thermovulgaris NRRL B-12375T has the later synonym Streptomyces thermonitrificans NRRL B-12534T.
In Fig. 1g it can be observed that Streptomyces phaeopurpureus NRRL B-2260T (and DSM 40125T) is confirmed as a later synonym of Streptomyces griseorubiginosus CGMCC 4.1766T (and DSM 40469T) as proposed in Gauze et al. (1983). The following observations can also be made: Streptomyces inusitatus NRRL B-16929T has the later synonym Streptomyces longwoodensis NRRL B-16923T (=DSM 41677T); Streptomyces clavifer CGMCC 4.1064Thas the later synonyms Streptomyces canus NRRL B-3980T (=DSM 40017T) and Streptomyces ciscaucasicus NRRL ISP 5275T (=DSM 40275T).
In Fig. 1h it was observed that Streptomyces goshikiensis NRRL B-5428T has the later synonym Streptomyces sporoverrucosus NRRL B-16379T and Streptomyces melanogenes NRRL B-2072T has the later synonym Streptomyces noboritoensis NRRL B-12152T.
Most relationships in Fig. 1i and 1j have already been discussed earlier with the exception of the observation that Streptomyces griseolus (both CGMCC 4.1864T and NRRL B-2925T) is a later synonym of Streptomyces halstedii NRRL B-1238T and NRRL ISP-5016T) based on presence of identical alleles for all 5 house-keeping loci. It should be noted that Streptomyces graminofaciens CGMCC 4.1359T (bottom of Fig. 1h) is shown not to be a later synonym of Streptomyces halstedii as was suggested by Rong and Huang (2010).
The phylogenetic analyses performed during the course of this study (see Fig. 1a) support the transfer of those Streptomyces species found within the radiation of the genus Kitasatospora to that genus and emended descriptions for these follow below. Although it would be optimal to also have phenotypic properties to support the proposed taxonomic changes, no useful traits have been discovered in past studies and gene sequencing capabilities have become ubiquitous and within the reach of most investigators so a gene phylogeny-based classification should be completely acceptable. The taxonomic status of Streptomyces chrysomallus subsp. fumigatus has been problematic since the Lanoot et al. (2005) proposal that S. chrysomallus subsp. chrysomallus is a later is synonym of S. anulatus which is also supported by the present study (see Fig. 1j). S. chrysomallus subsp. fumigatus is phylogenetically distant from S. chrysomallus subsp. chrysomallus and represents a new species within Kitasatospora for which the name Kitasatospora fumigata is proposed below.
The results presented in this study demonstrate the value of MLSA using partial sequences of single-copy house-keeping genes to provide resolution of taxonomic issues in the Streptomycetaceae and could result in a 116 species reduction from the over 780 Streptomyces species currently listed on the List of Prokaryote Names With Standing in Nomenclature (www.bacterio.net). This method has been applied to over 400 uncharacterised strains in the NRRL Culture Collection to date and at least 20 potentially new species have been discovered among the microbial strains collected since the 1950s (Labeda, unpublished obervations). Although there could be criticism of the taxonomic resolution and accuracy in utilising only the limited set of 5 house-keeping genes in the present study, our experience has shown that these loci provide an excellent assessment of the phylogenetic relationships between species in the Streptomycetaceae. Moreover, the phylogenetic relationships of 170 strains based on maximum likelihood analysis of the sequences of the 5 genes utilised in this study shows good correlation (See Supplemental Figure S1a–d) with that determined utilizing 1487 core genes (1943,267 bp) extracted from the genome sequences of these strains using the Genome Comparator function of BIGSdb, utilising the well-annotated genome sequence (Genbank FN554889) for S. scabiei RL87.22 (=NRRL B-24449) as the reference and with the core gene threshold set at 90%. Although the number of draft or finished genome sequences for strains in the Streptomycetaceae is continually growing, totalling more than 770 at this time, only about 182 of these are from type strains making definitive whole genome or core gene molecular systematics of the entire family Streptomycetaceae not yet within reach. The alleles of the house-keeping genes utilised in this study can be easily discovered within draft or finished genomes and added to the growing sequence database using the genome scanning capability within the BIGSdb software, making it possible to positively and correctly identify incorrectly named genome strains, such as classifying J1074 as a strain of S. albidoflavus rather than S. albus as reported in the genome databases. Furthermore this database can expanded to include all of the relevant single-copy core genes for the family Streptomycetaceae once there are genome sequences available for a representative set of type strains. A major value of the phylogenetic relationships illustrated in Fig. 1a–j, aside from providing taxonomic insight into the genus Streptomyces, is in providing a guide map for selection and prioritising critical species within the Streptomycetaceae for future genome sequencing efforts, as well as highlighting those strains requiring genome sequencing because taxonomic proposals of synonyminity need DDH between strains for confirmation, including many of those identified above. The multigene database has been demonstrated as an important tool for identifying phytopathogenic Streptomyces species when 16 of 43 strains of putative S. scabiei strains in the ARS Culture Collection were confirmed to represent non-pathogenic species while 6 strains possibly represent new phytopathogens (Labeda 2016). We have already demonstrated the usefulness of utilising the genomic sequence database, and the associated phylogeny constructed from the data stored in it, to discover new antibiotic producing strains as well as novel secondary metabolites (Price et al. 2016) in the course of discovery of the new tunicamycin analog, quinvosomycin, and its producing strain. The addition of substantially more Streptomyces genomes and expansion of the gene set used for phylogenetic analysis, aside from providing for a phylogenomic revision of the systematics of the Streptomycetaceae, should make this database an invaluable tool for mining these genomes for hitherto undiscovered natural products.
The ARS Microbial Genomic Sequence Database database is available at http://199.133.98.43/. The new names proposed as a result of this study are described below.
Description of Kitasatospora aburaviensis comb. nov.
Kitasatospora aburaviensis (a.bu.ra.vi.en’sis. N.L. fem. adj. aburaviensis of or belonging to Aburabi, Shiga Prefecture, Japan, the source of the soil from which the microorganism was isolated).
Basonym Streptomyces aburaviensis Nishimura, Kimura, Tawara, Sasaki, Nakajima, Shimaoka, Okamoto, Shimohira and Isono 1957AL.
The description is that reported in Kämpfer (2012).
The type strain is AS 4.1469T, ATCC 23869T, BCRC 11617T, CBS 280.60T, CBS 608.68T, CCRC 11617T, CECT 3315T, CGMCC 4.1469T, DSM 40033T, IFO 12830T, IMET 43031T, IMET 43081T, ISP 5033T, JCM 4170T, JCM 4613T, KACC 20033T, KCTC 9663T, LMG 19305T, NBRC 12830T, Nishimura S-66T, NRRL B-2218T, NRRL-ISP 5033T, VKM Ac-1868T.
Description of Kitasatospora albolonga comb. nov.
Kitasatospora albolonga (al.bo.lon’ga. L. adj. albus white; L. adj. longus long; N.L. fem. adj. albolonga white and long).
Basonym Streptomyces albolongus Tsukiura, Okanishi, Koshiyama, Ohmori, Miyaki and Kawaguchi 1964AL.
The description is that reported in Shirling and Gottlieb (1972).
The type strain is AS 4.1661T, ATCC 27414T, Bristol-Banyu 304R7T, CBS 766.72T, CGMCC 4.1661T, DSM 40570T, IFO 13465T, ISP 5570T, JCM 4716T, KCTC 9676T, KCTC 9749T, NBRC 13465T, NRRL B-3604T, NRRL ISP-5570T, RIA 1426T, VKM Ac-704T.
Description of Kitasatospora aureofaciens comb. nov.
Kitasatospora aureofaciens (au.re.o.fa’ci.ens. L. adj. aureus golden; L. part. adj. faciens producing; L. part. adj. aureofaciens producing golden (referring to pigment produced by the vegetative mycelium of the microorganism).
Basonym Streptomyces aureofaciens Duggar 1948, 177AL; later synonym Streptomyces avellaneus Baldacci and Grien 1966, 195AL.
The descriptions is that reported in Kämpfer (2012).
The type strain is ATCC 10762T, ATCC 23884T, CBS 434.51T, CECT 3206T, CGMCC 4.0568T, CIP 57.11T, DSM 40127T, IFO 12594T, IFO 12843T, IMET 43577T, ISP 5127T, JCM 4008T, JCM 4624T, KACC 20180T, Lederle Labs A-377T, LMG 5968T, NBRC 12594T, NCIB 8234T, NRRL ISP-5127T, RIA 1129T, Waksman 3708T.
Description of Kitasatospora cinereorecta comb. nov.
Kitasatospora cinereorecta (ci.ne.re.o.rec’ta. L. adj. cinereus similar to ashes, ash-colored; L. adj. rectus straight; N.L. fem adj. cinereorecta ash-colored, straight.)
Basonym Streptomyces cinereorectus Terekhova and Preobrazhenskaya 1986, 574AL.
The descriptions is that reported in Kämpfer (2012).
The type strain is AS 4.1622T, ATCC 43679T, CGMCC 4.1622T, DSM 41469T, IFO 15395T, INA 5202T, JCM 6916T, NBRC 15395T, NRRL B-16360T. Type strain AS 4.1589T, CGMCC 4.1589T, DOA 1196T, DSM 41424T, IFO 15394T, JCM 3371T, KCTC 9705T, NBRC 15394T, NRRL B-2289T.
Description of Kitasatospora herbaricolor comb. nov.
Kitasatospora herbaricolor (her.ba.ri’co.lor. L. n. herbarius one skilled in plants, a botanist: L. n. color color; N. L. adj. herbaricolor grass colored green referring to the grass green diffusible pigment produced by the microorganism on chemically defined media).
Basonym Streptomyces herbaricolor Kawato and Shinobu 1959, 114AL.
The descriptions is that reported in Kämpfer (2012).
Type strain AS 4.1849T, AS 4.1887T, ATCC 23922T, CBS 424.61T, CBS 906.68T, CGMCC 4.1849T, CGMCC 4.1887T, DSM 40123T, IFO 12876T, IFO 3838T, IFO 3932T, ISP 5123T, JCM 4138T, JCM 4645T, NBRC 12876T, NBRC 3838T, NBRC 3932T, NCIMB 9837T, NRRL B-3299T, NRRL ISP-5123T, RIA 1126T, RIA 654T, Shinobu 608T, VKM Ac-793T.
Description of Kitasatospora misakiensis comb. nov.
Kitasatospora misakiensis (mi.sa.ki.en’sis. N.L. fem. adj. misakiensis belonging to misaki (referring to Misakicho, Kanagawa Prefecture, Japan, the source of the soil from which the microorganism was isolated).
Basonym Streptomyces misakiensis Nakamura 1961, 86AL.
Description is that reported in Kämpfer (2012).
Type strain AS 4.1437T, ATCC 23938T, CBS 278.65T, CBS 922.68T, CGMCC 4.1437T, DSM 40222T, IFO 12891T, ISP 5222T, JCM 4062T, JCM 4653T, KCTC 19951T, LMG 19369T, NBRC 12891T, NCIB 9852T, NRRL B-2923T, NRRL ISP-5222T, RIA 1166T, VKM Ac-625T.
Description of Kitasatospora psammotica comb. nov.
Kitasatospora psammotica (psam.mo’ti.ca. Gr. n. psammos sand; N.L. fem. adj. psammotica sandy).
Basonym Streptomyces psammoticus Virgilio and Hengeller 1960, 167AL.
Description is that reported in Kämpfer (2012).
Type strain is AS 4.1465T, ATCC 14125T, ATCC 25488T, CBS 175.61T, CBS 299.65T, CBS 916.69T, CGMCC 4.1465T, DSM 40341T, IFO 13076T, ISP 5341T, JCM 4434T, KCTC 19966T, Lepetit Labs C17190T, NRRL B-3291T, NRRL ISP-5341T, RIA 1268T, RIA 832T, VKM Ac-996T.
Description of Kitasatospora purpeofusca comb. nov.
Kitasatospora purpeofusca (pur.pe.o.fus’ca. L. adj. purpureus purple; L. adj. fuscus dark, tawny; N.L. fem. adj. purpeofusca dark purple referring to the color of the vegetative mycelium).
Basonym Streptomyces purpeofuscus Yamaguchi and Saburi 1955, 207AL.
Description is that reported in Kämpfer (2012).
Type strain is ATCC 23952T, CBS 935.68T, CGMCC 4.1767T, CGMCC 4.1999T, DSM 40283T, IFO 12905T, ISP 5283T, JCM 4156T, JCM 4665T, KCTC 19967T, LMG 20283T, NBRC 12905T, NCIMB 9822T, NRRL B-1817T, NRRL ISP-5283T, RIA 1197T, VKM Ac-1825T, Yamaguchi H-5080T.
Description of Kitasatospora fumigata comb. nov.
Kitasatospora fumigata (fu.ma.ga’ta. L. fem. part. adj. fumigata smoked).
Basonym Streptomyces chrysomallus subsp. fumigatus Frommer 1959, 202AL.
Description is that reported in Kämpfer (2012).
Type strain AS 4.1589T, CGMCC 4.1589T, DOA 1196T, DSM 41424T, IFO 15394T, JCM 3371T, KCTC 9705T, NBRC 15394T, NRRL B-2289T.
Supplementary Material
Acknowledgements
The able technical assistance of E. Basehoar in determining the house keeping gene sequences and A. McGovern and H. Walker in determining draft genome sequences is gratefully acknowledged. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer. DPL and the ARS Culture Collection CRIS project was supported by ARS National Program 301. CAD was supported by ARS National Program 206. XR and YH were supported partially by the Specialized Research Fund for State Key Laboratories of China. WWM was supported by the National Institutes of Health (GM PO1 GM077596). JRD was funded by an Institute for Genomic Biology Postdoctoral Fellowship. KSJ was funded by an NIH NRSA Postdoctoral Fellowship (GM100658).
Abbreviation
- DDH
DNA–DNA hybridization
Contributor Information
David P. Labeda, Mycotoxin Prevention and Applied Microbiology Research Unit, National Center for Agricultural Utilization Research, 1815 N, University Street, Peoria, IL 61604, USA
Christopher A. Dunlap, Crop Bioprotection Research Unit, USDA, Agricultural Research Service, National Center for Agricultural Utilization Research, Peoria, IL, USA
Xiaoying Rong, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, People’s Republic of China.
Ying Huang, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, People’s Republic of China.
James R. Doroghazi, Department of Microbiology and Institute for Genomic Biology, University of Illinois at Champaign-Urbana, Urbana, IL, USA
Kou-San Ju, Department of Microbiology and Institute for Genomic Biology, University of Illinois at Champaign-Urbana, Urbana, IL, USA.
William W. Metcalf, Department of Microbiology and Institute for Genomic Biology, University of Illinois at Champaign-Urbana, Urbana, IL, USA
References
- Gauze GF, Preobrazhenskaya TP, Sveshnikova MA, Terekhova LP, Maximova TS (1983) A guide for the determination of actinomycetes. Genera Streptomyces, Streptoverticillium and Chainia. Nauka, Moscow [Google Scholar]
- Girard G, Willemse J, Zhu H, Claessen D, Bukarasam K, Goodfellow M, van Wezel GP (2014) Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie Van Leeuwenhoek 106:365–380 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321 [DOI] [PubMed] [Google Scholar]
- Guo Y, Zheng W, Rong X, Huang Y (2008) A multi-locus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multi-locus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58:149–159 [DOI] [PubMed] [Google Scholar]
- Hatano K, Nishii T, Mordarska H et al. (1997) Streptomyces spitsbergensis Wieczorek et al. 1993 is a later subjective synonym of Streptomyces baldaccii (Farina and Locci 1966) Witt and Stackebrandt 1991. Int J Syst Bacteriol 47:573–574 [DOI] [PubMed] [Google Scholar]
- Huang M-J, Rao MPN, Salam N, Xiao M, Huang H-Q, Li W-J (2016) Allostreptomyces psammosilenae gen. nov., sp. nov., an endophytic actinobacterium isolated from the roots of Psammosilene tunicoides and emended description of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997, emend. Kim et al. 2003, emend. Zhi et al. 2009. Int J Syst Evol Microbiol. doi: 10.1099/ijsem.0.001617 [DOI] [PubMed] [Google Scholar]
- Jolley K, Maiden M (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer P (2012) Family I Streptomycetaceae Waksman and Henrici 1943, 339AL emend. Rainey, Ward-Rainey and Stackebrandt 1197, 486 emend. Kim, Lonsdale, Seong and Goodfellow 2003B, 113 emend. Zhi, Li and Stackebrandt 2009, 600. In: Goodfellow M, Kampfer P, Busse H-J, Trujillo M, Suzuki KE, Ludwig W, Whitman WB(eds) Bergey’s manual of systematic bacteriology, vol 5, 2nd edn. Springer, New York, pp 1446–1447 [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120 [DOI] [PubMed] [Google Scholar]
- Labeda DP (2016) Taxonomic evaluation of putative Streptomyces scabiei strains held in the ARS Culture Collection (NRRL) using multi-locus sequence analysis. Antonie Van Leeuwenhoek 109:349–356 [DOI] [PubMed] [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim S-B, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104 [DOI] [PubMed] [Google Scholar]
- Labeda DP, Doroghazi JP, Ju K-S, Metcalf WW (2014) Taxonomic evaluation of Streptomyces albus and related species using multi-locus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int J Syst Evol Microbiol 64:894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Rong X, Huang Y, Doroghazi JR, Ju K-S, Metcalf WM (2016) Taxonomic evaluation of species in the Streptomyces hirsutus clade using multi-locus sequence analysis and proposals to reclassify several species in this clade. Int J Sys Evol Microbiol 66:2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoot B, Vancanneyt M, Cleenwerck I, Wang L, Li W, Liu Z, Swings J (2002) The search for synonyms among streptomycetes by using SDS-PAGE of whole-cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus and Streptomyces violaceus. Int J Syst Evol Micro-biol 52:823–829 [DOI] [PubMed] [Google Scholar]
- Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert M, Zhang J, Huang Y, Liu Z, Swings J (2004) BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst Appl Microbiol 27:84–92 [DOI] [PubMed] [Google Scholar]
- Lanoot B, Vancanneyt M, Van Schoor A, Liu Z, Swings J (2005) Reclassification of Streptomyces nigrifaciens as a later synonym of Streptomyces flavovirens; Streptomyces citreofluorescens, Streptomyces chrysomallus subsp. chrysomallus and Streptomyces fluorescens as later synonyms of Streptomyces anulatus; Streptomyces chibaensis as a later synonym of Streptomyces corchorusii; Streptomyces flaviscleroticus as a later synonym of Streptomyces minutiscleroticus; and Streptomyces lipmanii, Streptomyces griseus subsp. alpha, Streptomyces griseus subsp. cretosus and Streptomyces willmorei as later synonyms of Streptomyces microflavus. Int J Syst Evol Microbiol 55:729–731 [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NP, Labeda DP, Naumann TA, Vermillion KE, Bowman MJ, Berhow MA, Metcalf WW, Bischoff KM (2016) Quinovosamycins: new tunicamycin-type antibiotics in which the α, β-1",11'-linked N-acetylglucosamine residue is replaced by N-acetylquinovosamine. J Antibiotics 69:637–646 [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y (2010) Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol 60:696–703 [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multi-locus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for the systematics of the whole genus. Syst Appl Microbiol 35:7–18 [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y (2014) Multilocus sequence analysis: taking prokaryote systematics to the next level, Chapter 11. In: Goodfellow M, Sutcliffe I, Chun J(eds) Methods in microbiology, volume 41. New approaches to prokaryote systematics. Elsevier, London, pp 221–251 [Google Scholar]
- Rong X, Gou Y, Huang Y (2009) Proposal to reclassify the Streptomyces albidoflavus clade based on the basis of multi-locus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol 32:314–322 [DOI] [PubMed] [Google Scholar]
- Rong X, Liu N, Ruan J, Huang Y (2010) Multilocus sequence analysis of Streptomyces griseus isolates delineating intraspecific diversity in terms of both taxonomy and biosynthetic potential. Antonie van Leeuenhoek 98:237–248 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Shirling E, Gottlieb DA (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340 [Google Scholar]
- Shirling E, Gottlieb DA (1972) Cooperative description of type strains of Streptomyces. V. Additional descriptions. Int J Syst Bacteriol 22:265–394 [Google Scholar]
- Tamura T, Ishida Y, Otoguro M, Hatano K, Labeda D, Price NP, Suzuki K (2008) Reclassification of Streptomyces caeruleus as a synonym of Actinoalloteichus cyanogriseus and reclassification of Streptomyces spheroides and Streptomyces laceyi as later synonyms of Streptomyces niveus. Int J Syst Evol Microbiol 58:2812–2814 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


