Abstract
The past decade has witnessed remarkable advances in the simultaneous measurement of neuronal activity across many brain regions, enabling fundamentally new explorations of the brain-spanning cellular dynamics that underlie sensation, cognition and action. These recently developed multiregion recording techniques have provided many experimental opportunities, but thoughtful consideration of methodological trade-offs is necessary, especially regarding field of view, temporal acquisition rate and ability to guarantee cellular resolution. When applied in concert with modern optogenetic and computational tools, multiregion recording has already made possible fundamental biological discoveries — in part via the unprecedented ability to perform unbiased neural activity screens for principles of brain function, spanning dozens of brain areas and from local to global scales.
Improved instrumentation for observing the sky markedly transformed our view of the universe. New tools enabled the detection of subtle and sparse, or massive and universal, interrelated phenomena such as black holes and celestial-body dynamics1,2. In neuroscience, we are now experiencing a more rapid but similarly fundamental transformation. Over the past century, we advanced from the invention of tools for measuring electrical activity in single nerve fibres3 and neurons4,5 to an age when we now use electrical and optical tools to routinely obtain simultaneous, high-speed measurements from thousands of neurons in behaving mammals. But these new methods allow us to do more than just sample from larger numbers of neurons in one brain region. We are now able to record synchronously from large neuronal populations that span multiple brain regions — at, or near, single-cell resolution. This multiregion recording capability offers a burgeoning opportunity to see both the emergent whole and the constituent parts (the forest and the trees) of neural computations in a single dataset. Using these reliable new neurotechnologies, we are poised to make swift progress in understanding how cells in interconnected brain areas work together to produce global brain states and behaviour.
These advances in large-scale neural recording technologies are exciting for the field, but the rapid pace of innovation has made it difficult for researchers to learn about each new approach that might be relevant for their work. No single recording modality is best for all applications, and efforts to build ever better tools mean that the most viable approach one year might change in the next. Depending on the experimental question of interest, researchers must make choices, and accept trade-offs, along multiple different axes, such as spatial resolution, field of view, temporal resolution and cost. Different methods are also differentially compatible with optogenetics, freely moving behaviour and molecular phenotyping approaches. Moreover, there are many approaches to analysing and deriving insight from the resulting datasets, with each analysis method offering different perspectives and biases.
Here we summarize advances in multiregion recording and analysis techniques, consider trade-offs among different technical approaches and review key findings resulting from application to neural coding and brain-wide computation. Other recent reviews have focused on complementary issues, such as optical methods specifically6,7, or on conceptual insights that have emerged from the analysis of neuronal population recordings of ever increasing size8. In contrast, here we specifically focus primarily on technology and discoveries relating to large multiregion neural datasets, where simultaneous recordings were obtained from neuronal populations spanning many areas of the brain. We pay particular attention to the rodent literature, which has seen the most dramatic development in this regard in recent years. However, many of the ideas and technologies we discuss are either currently or well on their way to being applied to non-human9–11 and human12 primates.
Why do we care about multiregion recording?
Brains are highly interconnected systems, composed of networks of neurons that span spatially distant regions. Anatomical tracing has identified consistent and diverse brainwide connectivity patterns13, with individual or neighbouring neurons receiving input from or sending output to multiple regions in parallel (for example, in the primary visual cortex14). Research across many decades identified key behavioural and sensory correlates of neuronal activity in particular brain regions, leading to the assignment of primary functions to these regions. However, until the past few decades, most recording of neuronal activity was limited to monitoring a handful of neurons in one region at a time — a situation that has changed dramatically with the development and application of new methods15–17. Because of the interconnectivity and nonlinearity of neural circuits, it is not guaranteed that the conclusions reached by observing a few neurons in one region will also apply to data taken from neurons across many regions. Thus, many questions have persisted, and others have newly emerged, about the extent to which individual brain regions perform individual functions, whether specific computations are local or distributed, and whether brain states and representations are broadcast or confined.
As researchers began to obtain population recording data from multiple brain regions simultaneously, behaviourally relevant neuronal codes were found to be distributed across the brain18–20. For example, motor actions were found to modulate neural activity in many non-motor areas — including in the sensory cortex21–24. Overall, an intriguing picture of planning and outcome processing is beginning to emerge, in which neural computations are distributed, information is distributed or both. Local computations appear to be important, but also should perhaps not be analysed in isolation. We now must investigate why representations of sensation, cognition and action are so widespread, and what role they play in guiding behaviour20. In the past decade, much work has shifted from a focus on the computational properties of single neurons to a population doctrine that is focused on the computations performed by groups of neurons from a given brain region25. In an analogous fashion, as population recording methods continue to scale up from single to multiple regions, perhaps a comparable shift in perspective will emerge from the study of multiregion population dynamics.
A common taxonomy of brain regions
To synthesize findings about multiregion neural dynamics across studies, a common taxonomy of brain regions (or areas; here either word is used interchangeably) is required. Over the past few decades, a small number of rodent brain atlases have become widely adopted26–28. Until recently, atlas borders between areas were primarily delineated on the basis of cytoarchitectural differences (that is, clear differences in the particular arrangement and density of neurons between regions). More recently, a wealth of additional information has become available to enumerate and delineate brain regions, including viral-based connectivity13,29 and gene expression patterns30,31. These disparate data streams have been combined into an updated 3D atlas: the Allen Mouse Brain Common Coordinate Framework version 3 (CCFv3)32. For electrophysiology data, either 2D sections or 3D volumes can be obtained post hoc from fixed experimental brains so as to reconstruct the trajectories of dye tracks left behind by recording electrodes18,33,34, unstained tracks visible from larger probes35,36 or tissue damage left by electrolytic lesions37,38. For optical data, alignment can be achieved using known anatomical or functional landmarks23,39. As a whole, it is encouraging to see that increasingly more rodent studies are aligning their functional brain data to a common reference atlas; this trend is likely to continue along with greater data sharing between research groups. The use of a standard anatomical framework such as CCFv3 may also permit the development of searchable databases that use atlas-registered physiology data to generate summary statistics and perform meta-analyses across thousands of published articles — as has been done for the human neuroimaging community with tools such as NeuroQuery and Neurosynth40,41.
Recording techniques
Three main technical approaches are currently used to record multiregion neuronal activity: one-photon fluorescence imaging, multiphoton fluorescence imaging and electrophysiology. Optical techniques enable cell type-specific recording by leveraging genetically encoded fluorescent activity indicators such as Ca2+ or voltage sensors; cell types are commonly targeted on the basis of genetic expression profiles or by cellular connectivity42–44 or assigned on the basis of post hoc registration to cell type-specific labelling45–47. Some intact-skull optical techniques also have the potential to be minimally invasive and require no craniotomy — if used in tandem with transgenic animals or intravenous gene transfer to drive sensor expression48,49.
Electrophysiology, on the other hand, offers direct recording of cellular action potentials, at sub-millisecond temporal resolution. This high sampling rate is necessary for resolving the shape and timing of individual action potential waveforms. Additionally, electrophysiology is label-free, and therefore does not require any genetic manipulation of the animal, but also does not readily allow detailed anatomical or molecular understanding of the recorded cells. While it is possible, in some situations, to pair electrophysiology with optogenetics to identify specific types of neurons (‘optical tagging’)36,50–53, this approach is of low throughput relative to two-photon imaging, which permits exhaustive characterization of a genetically defined cell type in a local area. Another limitation of extracellular electrophysiology is that, despite recent progress54–57, it remains difficult to track recorded single neurons across multiple days with electrophysiology, in contrast to optical methods with genetically encoded fluorescent indicators, where such an approach is relatively straightforward. However, unlike with most optical imaging approaches that require head fixation, or are limited by the potential photobleaching of a fluorescent sensor, it is possible to obtain continuous electrophysiological recordings for many days57. In this section, we summarize the current state of diverse brainwide recording methods and discuss their strengths and weaknesses for studying multiregion neural computation (FIG. 1 and TABLE 1).
Fig. 1 |. A spectrum of methods for multiregion recording.
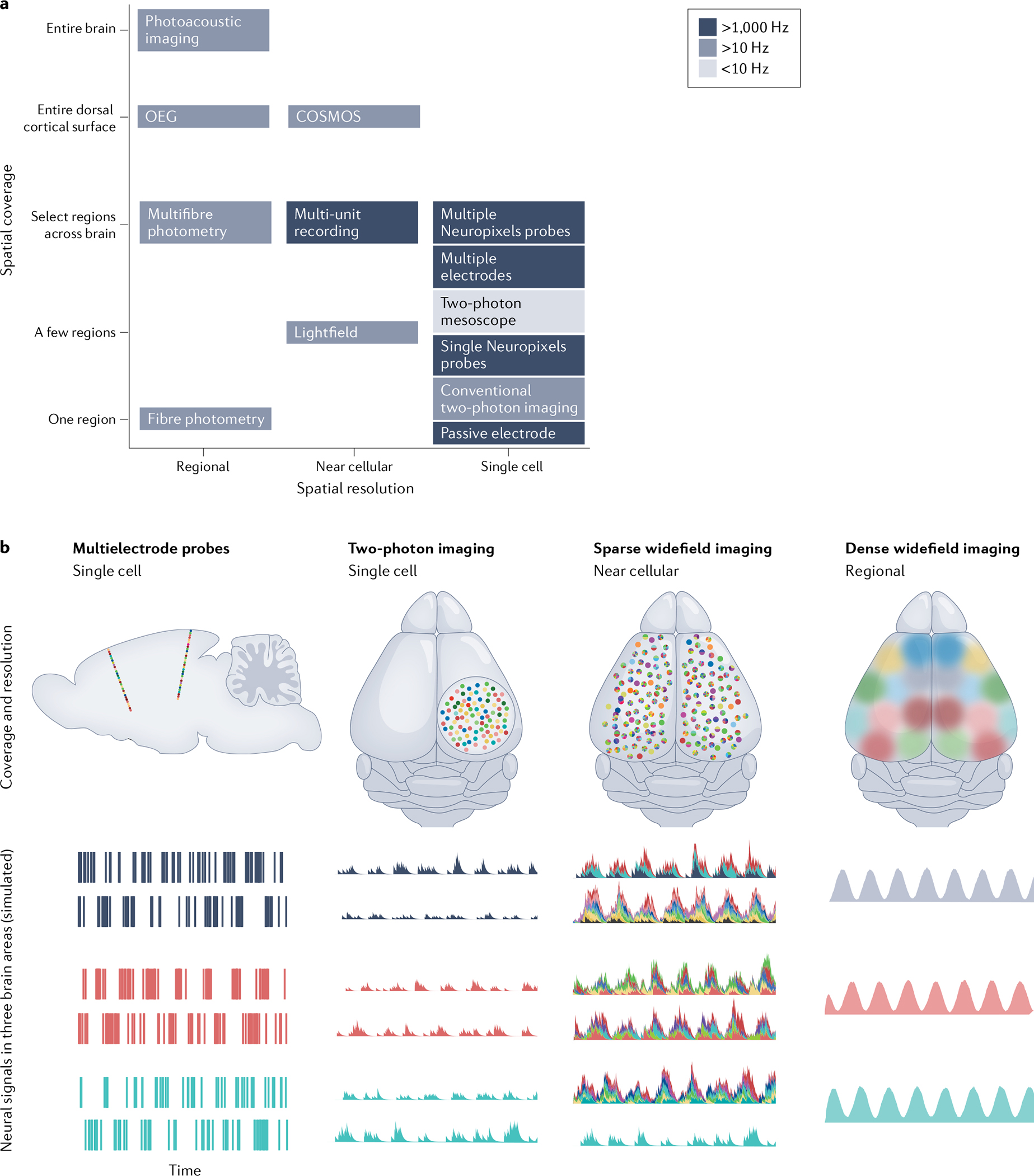
a | Each multiregion recording method exhibits a set of trade-offs among spatial resolution (x-axis; ranging from regional-scale resolution to single-cell resolution), spatial coverage of a simultaneous recording (y-axis; ranging from just one region to the entire brain) and acquisition speed shading. b | As a consequence of the trade-offs illustrated in part a, the spatial and temporal features of the data diverge between different methods. Synthetic data, simulated on the basis of the characteristics of each method, qualitatively illustrate the kinds of data that are produced by different multiregion recording methods — ranging from sparse sampling of neurons around an electrode array (left panel, top) with high-fidelity single-neuron recordings (left panel, bottom), to complete coverage of dorsal cortex (right panel, top) without full cellular resolution (right panel, bottom). Scales on the synthetic neural activity traces are arbitrary. COSMOS, cortical observation by synchronous multifocal optical sampling; OEG, optoencephalography.
Table 1|.
Multiregion recording techniques present distinct trade-offs
| Method | Acquisition speed | Spatial resolution | Spatial coverage | Cost and complexity | Genetic specificity | Freely moving | Example refs. |
|---|---|---|---|---|---|---|---|
| Multiple Neuropixels probes | More than kilohertz | Single cell | Select regions throughout the brain, along multiple linear trajectories | Specialized mount, multiple fragile probes (each ~$1,000), acquisition hardware (<$50,000), relatively simple burr-hole craniotomy surgery | Possible with optotagging | Up to two probes currently, probably more possible | 22,180,224 |
| Two-photon mesoscope | <10 Hz | Single cell | A few adjacent cortical regions (~5-mm diameter) | Custom large microscope objective, high speed optomechanics and detectors, lasers, large optical table, more than hundreds of thousands of dollars, commercial system available, large glass window surgery | Yes, with viral or transgenic reporter expression | Not currently | 138,139,142,143 |
| Multi-objective two photon imaging | >10 Hz | Single cell | A few select regions in the cortex, or accessible with a GRIN lens | Custom optics and optomechanics, lasers, large optical table, more than hundreds of thousands of dollars, multiple glass window or GRIN-lens surgery | Yes, with viral or transgenic reporter expression | Not currently | 136 |
| COSMOS | >10 Hz | A few cells | Entire dorsal cortical surface (~10-mm diameter) | Large-sensor sCMOS camera, lenslet array (<$1,000), minimal alignment, small spatial footprint, total <$50,000, large glass window surgery | Yes, with viral or transgenic reporter expression | Not currently | 23 |
| Single Neuropixels probe | More than kilohertz | Single cell | Select regions throughout the brain, along a single linear trajectory | Fragile probe (each ~$1,000), acquisition hardware (<$50,000), relatively simple burr-hole craniotomy surgery | Possible with optotagging | Yes, including probe reusability | 54,56,151,224 |
| Optoencephalography (widefield imaging) | >10 Hz | Bulk activity of many cells | Entire dorsal cortical surface (~10-mm diameter) | sCMOS camera, minimal alignment, small spatial footprint of hardware system, total <$50,000, simple surgery | Yes, with viral or transgenic reporter expression | Yes | 6,16,48,58,83 |
| Multifibre photometry | >10 Hz | Bulk activity of many cells | Select regions throughout a brain | sCMOS camera, fibre bundle, commercial systems (<$20,000), multiple fibre implant surgery | Yes, with viral or transgenic reporter expression | Yes | 67,111,113 |
| Photoacoustic imaging | >10 Hz | Bulk activity of many cells | Whole brain | High-power pulsed laser, custom ultrasound transducer array, non-invasive preparation with intact skin and skull (~$350,000 total) | Yes, with viral or transgenic reporter expression | Not currently | 133 |
COSMOS, cortical observation by synchronous multifocal optical sampling; GRIN, gradient index; sCMOS, scientific CMOS.
One-photon fluorescence
Regional-scale widefield imaging of the cortex.
One-photon optical fluorescence techniques use short-wavelength (for example, blue) excitation light to elicit fluorescence of a longer wavelength (for example, green). This process is efficient and enables illumination and imaging of an entire field of view, yielding fast recording speeds that can take advantage of modern scientific CMOS image sensors; these sensors can have acquisition rates of hundreds of frames per second with low read noise (less than one electron) and extremely high sensitivity (quantum efficiencies greater than 0.9). One-photon fluorescence is also relatively robust to illumination alignment and detection parameters, and such techniques are therefore easier and cheaper to implement than alternatives such as two-photon techniques — especially over large fields of view. The primary drawback of one-photon illumination is that any fluorophore in the specimen can fluoresce if it absorbs an excitation photon. This effect can lead to out-of-focus background fluorescence that adds noise to the signal measured at the focal plane (originating from fluorescent molecules in the surrounding tissue).
The development of bright genetically encoded fluorescent activity sensors led to the use of widefield one-photon imaging for simultaneously measuring neural activity across multiple regions of the rodent dorsal cortex58. These widefield techniques that combine fast, mesoscopic resolution on the scale of multiple subregions of the brain (even beyond just the cerebral cortex) can be termed ‘optoencephalography’ (OEG), as they offer a perspective reminiscent of electroencephalography, but with the genetic and spatial specificity of optical activity sensors. Early optoencephalographic approaches used synthetic voltage-sensitive dyes applied to exposed cortex16,59,60. More recently, the development of transgenic mice that express sensitive and bright genetically encoded Ca2+ sensors (for example, GCaMP) emerged as a reliable means of obtaining cell type-specific recordings from across the cortex61–63. Additionally, the development of viral capsids that efficiently cross the blood-brain barrier enabled intravenous delivery of desired transgenes (for example, encoding GCaMP) across the brain48,49,64. Of practical importance, the skull can even be made sufficiently transparent to facilitate widefield imaging with no craniotomy, through refractive-index matching by application of optical glue or cement65. However, due to scattering (as well as typically dense neuronal labelling), regional-scale optoencephalographic approaches should be considered as mesoscopic, with information in a typical pixel derived from regional activity on the order of thousands of neurons.
Microscopes suitable for OEG must offer high light collection with good image quality across a large field of view, particularly when imaging more-sensitive but less bright fluorophores such as GCaMP6f. A common approach is inspired by a tandem-lens design with consumer camera lenses that were originally used for intrinsic imaging66. One key consideration is that the optoencephalographic signal can be contaminated by time-varying haemodynamic artefacts, caused by variation in absorption as the amounts of oxygenated and deoxygenated haemoglobin fluctuate in a given area of the brain. This artefact can be corrected by measuring the Ca2+-independent fluctuations, either by using the isosbestic excitation wavelength of GCaMP around 410 nm (REFS.48,67) or by measuring haemodynamic absorbance with reflected green light63,68, and subtracting this from the raw Ca2+-dependent optoencephalographic signal. Failing to correct for haemodynamic artefacts may lead to spurious conclusions and will hinder reproducibility both within and between mice.
Precisely what is being measured by an optoencephalographic imaging technique depends on the specific experimental preparation — and the means of delivering the fluorescent sensor. Specific cell types are commonly targeted using genetically or anatomically delivered recombinases such as Cre, which through recombination enable cell type-specific expression of an indicator gene that is more universally present but in a recombinase-dependent form. Many relevant Cre-driver transgenic rodent lines have been created62,69,70, including as part of the BRAIN Initiative Cell Census Network71,72, along with diverse viral vectors carrying genetically encoded indicators that can even depend on two or three different recombinases for highly specific expression44,73. Targeted cell types can be excitatory (for example, VGLUT1 expressing), inhibitory (GAD2 expressing, somatostatin expressing or parvalbumin expressing) or cortical layer specific. Drivers exist for layer 2/3 (Cux2–Cre), layer 4 (Scnn1a–Tg3-Cre), layer 5 (Rbp4–Cre) and layer 6 (Ntsrl–Cre)74. With less specific expression, for example when an Slc17a7–Cre mouse is used to drive GCaMP expression across all cortical layers, Monte Carlo simulations have suggested that most of the signal should arise from layer 2/3 (REF75), although this may depend on the specific expression pattern, and there is evidence that most of the optoencephalographic signal derives from fluorescence emitted by layer 1 neuropil that may include layer 5 dendrites48. The combinatorial use of different sensors also affords new experimental opportunities. For example, two-colour optoencephalographic imaging has been used to record from excitatory and inhibitory populations48; one recent implementation combined a red fluorescent Ca2+ indicator (jRCaMP1b) with a green fluorescent acetylcholine sensor (ACh3.0)76. Beyond Ca2+ sensors, other genetically encoded fluorescent sensors can be used in conjunction with these same optical methods and Cre lines to enable the brainwide measurement of the release of glutamate, GABA or dopamine77,78. Finally, a new generation of voltage sensors is becoming available that may be more suitable than Ca2+ sensors for some widefield imaging experiments (see ‘Voltage imaging’).
Regarding the use of transgenic animals to express genetically encoded sensors, it is important to be aware that the expression of any non-native protein in the brain (especially during development) may lead to changes in cellular, or even circuit-level, function. For instance, it has been shown that using some strains of transgenic mice to express the Ca2+ sensor GCaMP during development can lead to aberrant cortical activity (reminiscent of seizures) in some mouse lines79. Therefore, it is always important to validate an experimental preparation by performing appropriate control experiments. In this specific case, use of inducible GCaMP lines, where the expression of non-native protein can be delayed until mice have reached adulthood, can mitigate this issue79.
Beyond the cortex, OEG has been successfully applied to other structures along the surface of the brain — namely the cerebellum80 and superior colliculus81,82. Moving forward, it will be exciting to develop new experimental preparations that will enable simultaneous visualization of these structures in addition to the surface of the dorsal cortex. OEG has also recently been extended to freely moving settings, with head-mounted microscope designs for rats83 and mice84. Finally, OEG can be paired with other techniques, such as whole-brain functional MRI85, or use of home-cage systems where mice learn to head-fix themselves for widefield imaging86,87.
Cellular-scale widefield imaging of the cortex.
A number of steps are required to advance beyond the regional-scale (millimetre scale in the mouse) spatial resolution of OEG. First, the quality of optical access to the brain must be improved beyond that afforded by an index-matched clear skull preparation. Among other steps, this improvement requires a large craniotomy, as well as a clear window that is curved to match the curvature of the brain, and made from glass88 or plastic89. It is possible to flatten the brain to some extent, but there is a limit to how large the window can be before significant tissue damage is caused48. Craniotomies have been successfully demonstrated with use of manual surgical techniques or with the semi-automated assistance of a robotic stereotaxic apparatus23.
Second, to address defocused signal and scattering, fluorescent protein expression must be restricted, either to a subset of neurons or to a localized part of each neuron. In one approach, the use of tamoxifen-dependent, layer 2/3/4-restricted expression of Cux2–CreER allowed limitation of GCaMP expression to a sparse set of superficial neurons23. The influence of layer 1 neuropil signal was thereby minimized (relative to less specific expression strategies used for widefield imaging). Other strategies include driving sparse expression using intravascular injections of blood–brain barrier-crossing adeno-associated virus variants such as PHPeB49. Limiting neuropil fluorescence can also be helpful in this respect, most efficaciously and definitively through the use of nuclear-restricted GCaMPs created by histone H2B fusions90–92, although other forms of targeting can include partial restriction of GCaMP to cell bodies through use of peptide tags derived from potassium channels (Kv2.1) or ribosomal subunits93–95.
Third, an imaging system is needed to permit recording across the curved, centimetre-scale extent of the mouse dorsal brain surface, with high light collection and good image quality. For mesoscopic OEG, the curvature of the brain surface is less of an issue because the defocus blur is itself roughly on par with the spatial resolution of the technique. When the goal is cellular or near cellular resolution, however, the defocus becomes a severe limitation on the accessible field of view88. This is due to the curvature of the dorsal surface of the brain, and is therefore an issue regardless of whether the dorsal skull has been replaced with a curved-glass window23,88 or a polymer-based window89,96.
To address this issue, cortical observation by synchronous multifocal optical sampling (COSMOS) uses a bifocal lenslet array and a single camera sensor to simultaneously record in-focus videos of the medial and lateral regions of the cortical surface. This approach has been used to record ~30 Hz signals from thousands of cellular to near cellular resolution neuronal sources simultaneously across the entirety of the mouse dorsal cortex23. A more complex technique (real-time, ultra-large-scale, high-resolution imaging) uses a set of 35 cameras arranged in a 5 × 7 array and a custom objective lens to achieve gigapixel imaging of neuronal dynamics across the curved cortical surface97. Additional possible tactics include use of fast tunable lenses, although this approach is hindered by optical aberrations and a trade-off between the speed of tunability and the size of the optical aperture. By combining high numerical aperture objectives with high-resolution cameras, lightfield98,99 and light-sheet100–102 microscopes can potentially enable truly volumetric multiregion imaging; moreover, eventually these approaches may be miniaturized to the level of applicability in freely behaving rodents. Along these lines, the Computational Miniature Mesoscope used miniaturized lenslet optics in a first step towards head-mounted, cortex-wide, volumetric imaging in freely moving rodents, although considerable development work will still be required to achieve that goal103.
One-photon imaging techniques in scattering mammalian tissue do not guarantee true single-cell resolution. For example, with use of a visual-stimulus assay with comparison with ground-truth high-magnification two-photon data, different neuronal sources computationally extracted from COSMOS data were estimated to be derived from 1–15 neurons23. Similar preparations using different Cre lines, or higher-magnification objectives, may yield results even closer to single-cell resolution104, but all such one-photon microscopy approaches fundamentally lack the axial resolution to guarantee single-neuron resolution. Still, it has been demonstrated that these cellular-scale data occupy a fundamentally different regime of experimental utility, compared with much lower resolution regional-scale widefield approaches. Although one-photon cellular-scale recording techniques should not be generally used to make claims about the response properties of individual cells, these methods permit the study of high-dimensional population coding across large neuronal ensembles23.
Multifibre photometry.
While OEG and COSMOS provide straightforward access to multiple superficial brain areas, these optical approaches are not readily applicable for imaging deep regions. Therefore, to reach areas deep in the brain, it is common to remove tissue or implant a light conduit, taking advantage of the fact that one-photon illumination is easy to transport through a multimode optical fibre. Such fibre photometry105 techniques can be sensitive enough to acquire activity signals arising from axons deep in the living mouse brain, while also being compatible with optogenetics, and enable cell type-specific optical recording access to anywhere in the brain that can be reached by an implanted fibre optic cannula105–109, although they average signal across many neurons in a volume110. Similar principles have been adapted for recording using genetically encoded voltage sensors111, and use of a tapered fibre can even enable depth-resolved recording from along the extent of the fibre112. Importantly, this fibre-based recording approach can be extended to multiple implanted fibres to enable multiregion recording, as demonstrated by frame-projected independent-fibre photometry67, as well as by a large-scale photometry technique that uses high-density arrays of optical fibres to simultaneously target up to 48 brain regions113. Of course, increasing the number of optical fibres inserted into the brain displaces more brain tissue — with greater potential for adverse effects on circuit function and behaviour, a theme common to all forms of brain interfacing, including electrophysiology and microendoscopy (which requires implanted lenses). In each case, the size and the number of neural implants must be balanced against concerns about potential damage; validating relevant baseline behaviour of animal subjects is always important in this regard.
Voltage imaging.
With extracellular electrophysiological recordings (discussed later), it is difficult to know what fraction of the active neurons surrounding an electrode are being sampled — especially when there are many sparsely firing cells. Identifying spatial structure at fine spatial scales is also difficult: electrode arrays do not permit precise localization of units except in very limited scenarios (for example, 2D arrays on organotypic slices or cell cultures). On the other hand, Ca2+ imaging presents different limitations. Ca2+ sensors permit high spatial resolution but provide an indirect, and low-pass-filtered, measure of action potential firing114–116 (also see BOX 1). While models exist to estimate action potential firing rates from Ca2+ measurements, in some situations it would be preferable to simply measure voltage directly. Indeed, cellular resolution, high-speed voltage imaging techniques could represent the best of both worlds — offering genetic specificity in recordings, dense measurements from even sparsely active neurons, an optical readout of action potential waveform shape and even information about subthreshold membrane voltage115,117–121.
Box 1 |. Processing multiregion neural data.
A new generation of mostly automated data-processing pipelines has become available to efficiently analyse multiregion neural datasets of increasing size.
Extracellular electrophysiology and spike sorting
Starting with raw multielectrode voltage data, spike waveform-specific bandpass filters are applied before a spike detection algorithm (often just a threshold) is run to identify candidate action potential times. This leaves the final step of ‘spike sorting’: the process of taking each extracted spike and its waveform shape and sorting them into distinct lists of spikes (called ‘units’) that share similar properties. The resulting units are the starting point for most subsequent analyses. There are now many competing approaches for semi-automated spike sorting56,254–259, but because it is a difficult problem, no single algorithm is appropriate for every experimental scenario257. New pipelines that include many alternative algorithms make it easier to compare many approaches on a single dataset254.
Two-photon Ca2+ imaging and source extraction
Many algorithms114,260–262 now automatically identify the location and shape of each neuron present in a two-photon fluorescence video. Some pipelines also perform a final step, which is to perform ‘spike inference’ and estimate a discrete-time firing rate histogram for each neuron so as to recover information that might have been smoothed out by the slow temporal kinetics of the Ca2+ sensor263–265. A recent study acquired data wherein two-photon Ca2+ imaging was performed before electron microscopy provided a ‘gold standard’ for quantitatively evaluating spatial source extraction algorithm performance266. Benchmarks quantifying spike inference algorithm performance are also available267,268, as are other packages for generating realistic simulated data that can be used for comparing different analysis approaches269.
One-photon Ca2+ imaging and source extraction
Single-photon Ca2+ fluorescence videos can vary dramatically — depending on whether they are obtained from a small field of view endoscope implanted deep in the brain or from a widefield microscope. In either case, the signals observed in a given pixel are likely to have arisen from many neurons stacked on top of each other in space. Recent work has involved the development of statistical inference tools270–272 to separate in-focus neural signals from deeper out-of-focus noise — and to extract a single denoised trace for each detected neuronal source. Manual validation of components extracted with any processing approach here is especially critical, especially if motion artefacts are significant.
Significant progress has been made over the past decade along these dimensions. A host of novel genetically encoded voltage indicators are now available, including the ASAP family119,122, ArcLight118, the QuasAr family121,123–127 and Voltron (which requires the addition of a synthetic Janelia Fluor fluorophore)117. With use of the latest variants of these tools, it is now possible to perform cellular resolution voltage imaging such that both action potentials and subthreshold signals can be measured in single trials, from small ensembles of neurons. Variants of QuasAr are also compatible with optogenetic tools such as CheRiff (a blue-shifted channelrhodopsin) or other newly developed red-shifted opsins such as ChRmine123,125,126,128; this approach could lead to the voltage sensor version of all-optical reading and writing of neural activity into neural ensembles to modulate animal behaviour, as achieved with Ca2+ sensors previously.
Together, these results point to a promising future for voltage imaging. Unfortunately, at the moment, there are also significant challenges that must be overcome before Ca2+ imaging or electrophysiology is displaced — especially for multiregion experiments. First, voltage dynamics are much faster than Ca2+ dynamics, necessitating that high signal-to-noise ratio (SNR) optical signals be measured at kilohertz rates to resolve action potential waveforms, compared with the 2–200-Hz range seen with Ca2+ imaging data. Consequently, genetically encoded voltage indicators must emit far more photons per unit time than Ca2+ sensors to achieve a comparable SNR, because signals can be integrated for far less time per frame. To remedy this issue, most voltage imaging systems use one-photon methods to image small fields of view. The most obvious alternative would be to increase the excitation laser power to levels that might damage tissues of interest, or to use two-photon methods that require novel approaches for fast laser scanning, such as beam multiplexing.
In line with this, recent articles have presented two-photon microscopy approaches called ‘ultrafast local volume excitation’ and ‘free-space angular chirp-enhanced delay, which were reported to permit in vivo measurement of action potentials and subthreshold dynamics with the ASAP3 genetically encoded voltage indicators (but only from 3 and 20 simultaneously recorded neurons, respectively122,129). One-photon approaches can measure activity from more neurons at coarser spatial resolution — but due to constraints of camera acquisition rate, thermal damage and photon-flux concerns, only up to a few dozen neurons can be imaged simultaneously117,120,123. True multiregion population voltage data are, at the moment, attainable only by combining use of the existing genetically encoded voltage indicators with imaging methods that lack cellular resolution, such as OEG and fibre photometry111,130,131. But a recent preprint reports integration of a custom two-photon system, a new voltage sensor (SpikeyGi) and a nonlinear denoising algorithm (DeepVid) that permitted in vivo imaging of approximately 100 neurons for over 1 h (REF.132). While this approach remains to be validated, especially because denoising algorithms rely on difficult-to-characterize supervised deep learning methods, this progress gives reason to believe that within a few years population voltage imaging may become more broadly applicable — at least for measuring single-region neural population activity.
Photoacoustic imaging.
Another approach to brainwide recording of neural signals (versus haemodynamic or structural signals) uses the optoacoustic effect. Here, ultrasound waves are generated by transient light absorption, which can be detected through centimetres of tissue. By the pulsing of bright one-photon excitation, changes in fluorescent indicator absorbance can be measured throughout the entire brain volume133. Unlike functional MRI or intrinsic imaging methods which measure haemodynamic signals, photoacoustic methods using genetically expressed fluorescent indicators directly measure signals emitted from neurons. Ultrasound transducer arrays must be coupled to the brain by water or gel — as with high-resolution optical microscopy methods that rely on immersion objective lenses. But to enable whole-brain tomography, these ultrasound arrays must be coupled over a much larger area. These steric constraints may pose a challenge for application of this method to freely moving rodent preparations, and potentially to even some awake-behaving scenarios. In vivo, photoacoustic methods are also limited by the degree to which blue excitation for GCaMP can travel through the brain without blood absorption. However, application of this technique with red-shifted indicators should increase the depth, and will reduce the impact of haemodynamic-related signals. This approach can be combined with the simultaneous use of other ultrasound-based methods for functional stimulation or haemodynamic recording134.
Two-photon fluorescence
While one-photon methods are straightforward to implement and use, are relatively inexpensive and permit video-rate acquisition from molecularly defined neuronal populations across large fields of view, they generally do not offer unambiguous single-cell resolution in scattering mammalian brain tissue. In contrast, two-photon optical fluorescence techniques use very high intensity excitation light of a longer wavelength (that is, infrared) to elicit fluorescence of a shorter wavelength (that is, green). Two-photon fluorescence depends on the square of the excitation light intensity, because sufficient photon density is required to achieve simultaneous fluorophore excitation by two lower-energy photons. This nonlinear dependence affords two key advantages: optical sectioning, which results from restriction of emission to the focal plane, and robustness to scattering, which results from the increased scattering length of infrared light, as well as the raster-scanned photon counting imaging process135. Disadvantages of two-photon methods include the high cost of the pulsed laser and inherent speed limitations of a raster-scanned approach.
Originally, two-photon microscopy approaches used high-magnification objectives to observe sub-millimetre fields of view. Over the past few years, there have been efforts to extend this technique to record from multiple regions. One approach is to use two high-magnification objective lenses, with separate beam paths136. With careful planning, these objective lenses can be positioned across the brain, potentially in tandem with implanted endoscopes or with optogenetic manipulation of additional regions137.
Another approach is to use a single, large, low-magnification objective lens. Because of the specialized high numerical aperture requirements of two-photon imaging, this approach has required the design of expensive, customized objective lenses. The two-photon random access mesoscope developed by Sofroniew et al. has a 5-mm-diameter field of view, with a numerical aperture of 0.6, near-diffraction-limited performance and a remote focusing module to allow access to multiple focal planes across the imaged volume138. The raster-scan pattern is adjustable, and can image the entire field of view at up to 4.3 frames per second at low resolution or 0.7 frames per second at high resolution.
Higher speeds can be achieved by imaging a few subregions, yielding performance similar to that of multiple objective lens microscopes. The Trepan2p microscope has a 3.5-mm-diameter field of view, with a numerical aperture of 0.43 NA, diffraction-limited performance with a curved field and a tunable lens for volumetric acquisition139. The full field of view could be scanned with one beam at 0.1 frames per second, but the microscope has two separate beam paths to enable simultaneous acquisition of two smaller subregions at 30 frames per second.
Other microscopes have been explicitly designed to image multiple subregions with one objective, for scenarios wherein it would be mechanically difficult to place two objective lenses next to each other, such as when one is imaging the primary and secondary somatosensory cortex140,141. The Diesel2p mesoscope has a 5-mm-diameter field of view, with a numerical aperture of 0.54, and dual independent scan engines for simultaneous imaging of two regions, or from four regions in the Quadroscope version of the microscope142,143. By use of an elongated point spread function, it is possible to scan an entire 4 mm× 4 mm × 100 μm volume, as opposed to just a single focal plane, at 3.2 Hz (REF144). Last, light beads microscopy uses a set of axially separate and temporally distinct foci to record nearly simultaneously from the entire axial imaging range, recording from approximately 5.4 × 6 × 0.5 mm3 volumes at around 2 Hz — potentially enabling cellular resolution recordings from up to one million total neurons145.
Many approaches further increase imaging speed by multiplexing the two-photon beam into many beam-lets that can be scanned in parallel (or remain statically parked on neurons of interest). For example, one microscope with 16 beams and 16 detectors can sample from a 2 × 2 mm2 field of view at up to 17.5 Hz (REF146), while another uses 400 beams with scientific CMOS camera detection to sample a sub-millimetre field of view at kilohertz frame rates147. With two-photon excitation, however, the SNR is reduced for a given laser power due to the focusing of illumination into additional focal spots148. Additionally, scattering-related advantages of two-photon imaging begin to decrease as more focal spots illuminate the specimen.
Two-photon imaging in a small field of view has also been combined with simultaneous OEG, through the use of a prism to enable high-magnification access with relatively little obstruction of the OEG field of view149. Recently developed head-mounted two-photon microscopes suitable for studying freely moving behaviour in mice150 might be productively integrated with OEG to provide broader functional information. High-resolution structural two-photon scans could also be obtained from these same mice under head fixation. Such a multimodal approach would enable registration of freely moving population activity datasets to detailed anatomical and molecular datasets — all at cellular resolution.
Electrophysiology
Unlike optical methods, extracellular electrophysiology directly records electrical activity associated with action potentials on the millisecond timescale of individual spikes; such high acquisition rates facilitate assigning individual spikes to specific neurons, or units, on the basis of the characteristic shape of each neuron’s spike waveform. However, algorithms to perform this task of ‘spike sorting’ are imperfect, require some manual curation and are sensitive to artefacts from animal movement and probe location drift over time. In recent years, advances have been made towards improving and automating these data-processing techniques, but challenges remain (see BOX 1). Other challenges inherent to electrophysiology, compared with imaging, include reduced compatibility with targeting the readout to genetically or anatomically defined cell types, reduced long-term stability of single-cell identification across days and, until recently, recording simultaneously from only a handful of neurons in vivo. However, with the development of high-density, multiple-site electrodes, it has now become possible to simultaneously record from thousands of units, spanning many brain regions (with straightforward access to subcortical regions in mouse), including during optogenetic control18,34,56,151. These advances have been driven primarily by the transition from microwire-based recording systems to silicon-based and polymer-based probes. Microwire tetrode arrays remain a benchmark tool for obtaining stable recordings from single units over many weeks152, but this may change over the next decade as easier-to-manufacture silicon-based and polymer-based probes become available.
The choice to use electrophysiological versus imaging methods presents a number of trade-offs115. For instance, imaging permits dense sampling of neurons along individual planes, whereas electrode-based methods sparsely sample neurons along the depth of each recording probe56,153, or from dispersed points in space where tetrodes have been placed17,152. For a discussion of these considerations and others regarding temporal resolution, spatial sampling, and optogenetic compatibility, see BOX 2.
Box 2 |. Which multiregion recording technique should you use?
Spatial resolution: many multi-unit signals versus fewer unambiguous single units
Cellular resolution data are critical for questions pertaining to single-neuron tuning, and for relating physiology to cell-type identity. But when the goal is to decode information rapidly, to compare activity between regions or to study population codes under different behavioural circumstances, multi-unit spiking data or multineuron imaging techniques such as cortical observation by synchronous multifocal optical sampling may offer many more signals than cellular resolution approaches — and still yield suitable data for many population analyses23,162.
Spatial coverage: dense, slow data versus sparse, fast data
Silicon electrode arrays such as Neuropixels measure activity from a sparse subset of neurons near the probe, with straightforward access to subcortical regions in the mouse. In contrast, a two-photon microscope can see vastly more neurons per unit area. But this increased spatial coverage comes at the expense of temporal resolution: scanning a laser across an entire plane or volume takes many milliseconds, whereas each electrode on an array can be recorded at kilohertz data rates. More generally, scanning a laser over a volume, while remaining at each pixel long enough to measure a useful signal from any neurons that may be present there, enforces a trade-off among temporal resolution, coverage density and area sampled.
Temporal resolution: many neurons at low speeds versus few neurons at high speeds
While Ca2+ imaging permits dense access to cellular resolution activity measurements from genetically defined populations, this comes at a cost. Most existing Ca2+ sensors are unable to reliably measure single action potentials in single trials114–116, independent of the data acquisition rate. Ca2+ sensors report a noisy proxy of neuronal firing passed through a nonlinear filter114,267,268. Despite advances in voltage imaging, electrophysiology remains the ‘gold standard’ for recording with single-spike resolution.
Optogenetic compatibility: writing neural activity without compromising read capability
Shining a light near an electrode array can generate stimulus-locked electrical artefacts via photovoltaic, photoelectrochemical or electromagnetic effects273,274; however, recently designed electrodes appear fairly resilient to this issue151,274, and recently developed ultrasensitive opsins permit the use of reduced light intensities for photostimulation128,275,276. Similarly, new spectral variants of Ca2+ indicators, such as XCaMPs and sRGECO73,277, have increased compatibility with optogenetic tools — particularly in conjunction with red-shifted channelrhodopsins128.
Silicon-shank probes.
Silicon-based probes with tens to thousands of electrical contacts per shank are now widely available — a major increase versus microwire arrays, which typically consist of a few dozen wires at most152. Silicon-shank probes are also significantly narrower than microwire-based probes, therefore reducing tissue damage. More recently, silicon-based probes (termed Neuropixels) were developed with active amplification and digitization on the base of each probe itself151. This design significantly increases the SNR, especially in freely moving settings. These Neuropixels 1.0 probes are manufactured using CMOS nanofabrication and, in their most common configuration, have 960 recording sites across an ~4-mm linear span, with up to 384 sites recordable simultaneously. A single probe thus allows sampling from multiple brain regions, depending on the trajectory of insertion. Multiple successive probe insertions can be used to accumulate asynchronously recorded data from many regions across multiple sessions18,34.
More recently, Neuropixels 2.0 probes56 were described with a geometry similar to in the original probes, but are also available in a four-shank configuration. This means 384 simultaneous channels can now be measured over an area 750 μm wide and to a depth of 720 μm (with the four shanks evenly spaced across this area). A single headstage can also now mount two probes, permitting another four shanks to be inserted within a few hundred microns of the first probe — with spacing limited only by the mounting fixture used, as the probes themselves connect to the headstages with flexible connectors. Building upon previous work with the first-generation probes54,151,154, Neuropixels 2.0 probes are more suitable for long-term (chronic) implantation and for use with freely moving animals owing to their reduced weight (the total weight of two 2.0 probes and a headstage is ~1.1 g versus ~1.3 g for a single 1.0 probe and a headstage; implant weights exclude the weight of the structural materials and cement that must be used to stabilize each probe) and new methods correcting for motion artefacts in acquired data56,153.
Use of multiple probes simultaneously, typically in a head-fixed configuration, permits a substantial increase in the number of regions that can be monitored. Recordings in mice have been performed from up to eight simultaneously inserted Neuropixels probes across the brain22, or in a targeted manner to investigate visual cortical and thalamic regions33. Typically, electrode probes are dipped in lipophilic dyes155 so that the insertion track of each probe can be identified in histological sections, or in three-dimensionally cleared tissue18,33,34, and are then aligned with a reference atlas such as the Allen Mouse Brain CCFv3 (discussed earlier). But limitations remain, including with respect to compatibility with cell type-specific recording methods such as OEG, since silicon-based probes and headstages are not optically transparent. Recording stability and quality also significantly degrade over time (1–2 months), probably as a consequence of issues with biocompatibility and mechanical damage to the surrounding tissue arising from a lack of flexibility.
Flexible polymer-based probes.
In terms of combining multielectrode recordings with OEG or two-photon imaging, flexible or transparent probes may be useful. These properties can be obtained through the development of neural interfaces fabricated on polymer substrates, instead of shanks made of silicon156–158. Polymer-based probes such as the Neuro-FITM probe may be particularly useful for simultaneous imaging158, which is possible but difficult with silicon-based probes159,160. The Neuro-FITM probe is a 32-channel or 64-channel device with electrodes deposited on a flexible polymer, while maintaining a spike SNR comparable with that of Neuropixels probes, and is optically transparent so as to permit simultaneous OEG.
Flexible polymer-based probes may also be valuable for obtaining stable, high-yield recording over many months57,157. Again, obtaining such data is possible with modern silicon-based probes55,56,154 and tetrode arrays152. However, improved biocompatibility relative to microwire bundles and silicon devices may permit increased long-term unit yield, and the flexible nature of polymer-based probes may also be more suitable for recording from larger animals or deep neural structures (such as the brainstem and spinal cord), where the ability of these probes to move and bend with neural tissue compares favourably with the properties of silicon-based probes57,158,161. But for now, the capability of modern silicon-based probes such as Neuropixels 2.0 is quite impressive, and it remains to be seen whether alternative, polymer-based approaches will be widely adopted beyond specific use cases, such as where a transparent or highly flexible probe is required.
Future advances in multiregion recording
Recording methods continue to improve but eventually will encounter physical limits. Will it ever be possible to simultaneously record the action potential firing of every individual neuron in an entire mammalian brain? Theoretical analysis suggests arrays of advanced electrode probes may someday be able to record from most of the neurons in the cortex, or potentially a large fraction of a rodent or primate brain161. Nearer term, the highest-yield recording methods are still based on optical imaging techniques with significant trade-offs between spatial and temporal resolution, such as light beads microscopy, which might simultaneously obtain Ca2+ signals from up to one million neurons but at ~2 Hz (REF145). Multiple optical paths with identical optics could potentially be constructed to simultaneously measure adjacent million neuron-sized fields of view so as to sample densely from most of the neurons in the mouse cortex, albeit still at coarse temporal resolution. More practically, approaches such as COSMOS that approximate cellular resolution but cover huge spatial extents may find increasing utility23. By combination of large field of view OEG methods with electrode arrays159,160, two-photon imaging149 or voltage imaging, many multiregion experiments that require different kinds of information from different brain areas are already feasible. Over the next decade, trade-offs among these rapidly advancing methods will likely become less significant as the field moves ever closer to the goal of comprehensively recording neural activity across the entire brain. But regardless of the ultimate method chosen, the question of how to best analyse multiregion data will remain a pressing concern (given these vast datasets), thus representing another area that has seen many innovations in the past decade23,162.
Analysis techniques
Embedded in the choice of a data analysis method, and of each processing step, is a set of assumptions and biases for looking at the brain in a particular way. In this section, we present a taxonomy of existing neural analysis approaches that apply to cellular-scale datasets spanning multiple brain areas. Unique challenges and opportunities have arisen with the advent of multiregion cellular-scale data streams. Our intention is to articulate how particular analysis strategies are more appropriate for some kinds of questions about multiregion population coding than others. Importantly, selecting an analysis strategy implicitly restricts the hypotheses that can be tested, although this is often not explicitly acknowledged.
Analysis strategies for multiregion data
Historically large-scale neural recording approaches include indirect haemodynamic methods such as functional MRI for imaging and bulk recording methods such as electroencephalography or field recording for electrophysiology. These approaches have permitted interrogation of brainwide circuits and systems — but at spatial and temporal resolution orders of magnitude coarser than for cells and spikes. As discussed earlier, recent advances have changed this landscape, presenting new approaches for relating the information contained in the spike trains of individual, genetically defined neurons to computations performed by brainwide networks.
Recent analyses of multiregion cortical datasets have revealed that many phenomena previously thought to be fairly restricted to specific brain regions are actually present across many areas18,21–23. Our capacity to understand these results is rapidly expanding now that we have access to multiregion data. For instance, the limited field of view of conventional two-photon imaging typically requires asynchronous sampling from distinct brain regions, and thus only trial-averaged responses can be compared between areas. However, such trial-averaged comparisons may bear little resemblance to true moment-to-moment correlations measured using simultaneous multiregion data23. While low spike-train correlations do not necessarily prove the existence of low levels of shared input163,164, such results do suggest that there is much for us to uncover regarding how the downstream actions of a particular neural circuit may yield different behavioural outputs, depending on the context165,166; new analytical approaches will be key for successfully performing large-scale single-trial analyses instead of pooling data across trials and animals.
To analyse large neural datasets and attempt to answer these questions, at least three general approaches are relevant (FIG. 2). First are approaches for localizing information, largely based on computing correlations between recorded neurons and external covariates such as measured behavioural and stimulus features (FIG. 2a). This approach can be applied by fitting predictive models to test specific hypotheses, or by more exploratory analyses ranging from trial averaging based on behavioural structure to the study of neuronal firing across different temporal epochs of a dataset. Second are approaches for identifying population activity patterns — either by examining how neurons fire relative to one another or by examining how groups of neurons fire with respect to coincident behaviour (FIG. 2b). These algorithms enable visualization, description and modelling of the joint activity of groups of thousands of simultaneously recorded neurons. Third are approaches for quantifying network interactions that occur both within and between different brain areas (FIG. 2 c). One increasingly popular approach here is to use new modelling techniques that can be fitted to large neural datasets. The resulting models match many features of the neural data but are more amenable to analysis and understanding. Therefore, these models can be rapidly analysed and experimented on in silico before the predictions are tested in new biological experiments. We discuss each analytical approach in turn.
Fig. 2 |. Approaches for analysing multiregion recording data.
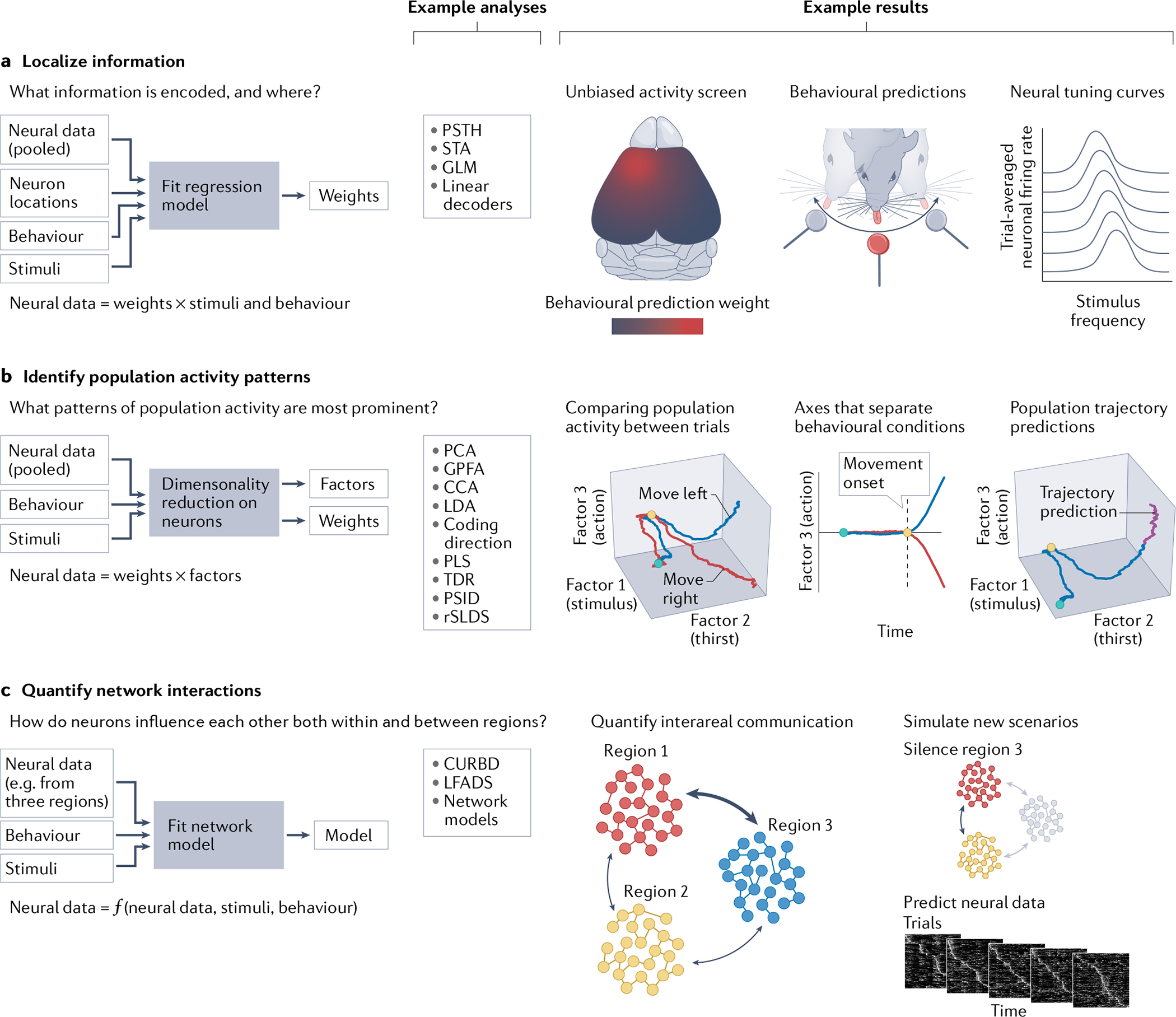
There are many approaches for analysing and interpreting multiregion neural data, each of which can address different types of questions. a | What information is being represented by neurons in a dataset? Questions of this nature can be studied using regression models that try to model neural data as a weighted sum of other variables, including information about animal behaviour, experimental stimuli, information about neuronal identity and position, and signals from other neurons. In the unbiased activity screen example (left), the weights obtained from a model might represent the brain areas that are most useful for predicting information about an animal’s behaviour during a task. In other cases (middle), the product of another set of regression weights and neural data may be used to make behavioural predictions (for example, to predict which waterspout the animal might lick to obtain a reward) or compute a neural tuning curve to show the average response of a neuron to a stimulus (right). The algorithm used to compute these weights is often some form of linear regression, such as a generalized linear model (GLM), and may rely on spike-triggered averaging (STA) to reveal the stimulus that a neuron maximally responds to or may rely on computing a trial-averaged response (often called a ‘peristimulus time histogram’ (PSTH)) to reveal the neural response to a specific stimulus. b | What is the regularity and prominence of different patterns of neural population activity? To study this question, dimensionality reduction techniques can be used to compress the activity of hundreds of neurons into a few prominent factors that can then be used to represent the joint activity of a neural population as a low-dimensional trajectory. These algorithms typically attempt to approximate a data matrix of neurons over time (N × T) as the product of a matrix of neuron weights over factors (N × D) and a matrix of factors over time (D × T), where the number of factors (D) is typically set to be some number less than N (often 2–5). This can be accomplished (in use of many algorithms) with different constraints on the features that must be present in the weights and factors (which are sometimes formulated in slightly different ways and may also incorporate information about behaviour, stimuli and how neural signals evolve over time). These methods, which are described in the main text, include principal component analysis (PCA), Gaussian process factor analysis (GPFA), canonical correlation analysis (CCA), linear discriminant analysis (LDA), coding direction analyses, partial least squares regression (PLS), targeted dimensionality reduction (TDR), preferential subspace identification (PSID) and recurrent switching linear dynamical systems (rSLDS). Projecting the neural data into a low-dimensional space defined by factors can be used to construct neural trajectories that can be visualized, quantified and used to compare the joint activity of a neural population across different trials and behavioural conditions (the green dot represents trial onset, the yellow dot represents movement onset and the red and blue lines represent schematic trial-averaged population activity as a mouse prepares to move right or left, respectively; left panel). Specific factors may be constructed to maximally separate population activity trajectories during different behavioural conditions (middle; format matches left panel). Many of these models can also be used to predict how a neural population trajectory might evolve in the future — in the absence of additional neural data (rightmost panel; general form of trajectory matches left panel). c | How do neurons interact with each other both within and between different brain areas? This can be studied by using algorithms such as latent factor analysis via dynamical systems (LFADS) and current-based decomposition (CURBD) to fit network models to datasets consisting of multiregion neural data, information about animal behaviour and sensory stimuli. The fitted network models can then be used to simulate how neural data might be generated by novel sets of stimuli or to generate unique behavioural outputs. Unlike with a real neural dataset, no element of these models is unobservable. Therefore, direct analysis of these models as a surrogate for the neural data of interest can be used, for example, to quantify the direction and strength of interareal communication between distinct brain regions (left). Communication strengths are indicated by the widths of the arrows between the areas. Use of these models also permits simulation of new experimental scenarios (for example, the behavioural and network-wide effects of silencing a set of neurons; right).
Localize information
A common goal in many of these analyses, whether for large or small datasets, is to relate information about sensory stimuli or behavioural output to recorded neural activity (FIG. 2a). This can be accomplished through a variety of means, but a key feature of many analysis methods is that they are dependent on correlations within the data. The simplest and perhaps most common form of correlational analysis is the notion of a trial-averaged response. This idea dates back more than a century to the original notions of neuronal tuning curves and receptive fields167,168. Beyond this basic idea of plotting average neural responses against stimulus properties169, the more general idea of computing a stimulus-triggered average firing rate is commonly used and is often referred to as the ‘peristimulus time histogram’ or developed into various elaborated forms170,171. These kinds of analyses do not historically treat neural population data any differently than a set of individual neurons. Tuning curves can be simply computed independently for each neuron, or by averaging over multiple neurons. But with new multiregion datasets, understanding unaveraged single-trial responses is of particular interest. Fortunately, a host of modern correlation-based approaches are designed to work in this newer setting.
Many analyses along these lines can be dichotomized into those that attempt to predict neural activity from stimulus and behavioural features (often called ‘encoding models’) and models which predict stimuli and behaviour from neurons (‘decoding models’)18,21,22,34. Traditionally, encoding models attempt to predict the response of a single neuron at a time with different combinations of task features — and are fit using regression algorithms. In contrast, decoding models more obviously lend themselves to the analysis of a whole neuronal population, because a simple regression scheme could be used to predict a single task feature from many simultaneously recorded neurons (potentially spanning multiple areas). But over the past decade multiple techniques have been developed to generalize encoding models to neuronal populations. Fitting encoding regression models to whole populations at once, rather than treating neurons independently, has been found to generate better predictive performance172. In a similar way, the performance of encoding models can be improved by incorporating known information about the structure of a neural circuit and the statistics of spike trains (for example, by using Poisson–generalized linear models (GLM))172,173. Variants of this approach can be specifically constructed to account for interregional connectivity and unknown time lags between neurons in the case of multiregion data174–176.
This process of building, fitting and analysing encoding models can reveal much about the processes that might generate observed patterns of neural activity — and has become increasingly common as available software packages have made such models easier to implement177–179. In the past, these methods were most frequently applied to trial-averaged data, or even measurements pooled between recording sessions or experimental animals. More recently, new recording methods have enabled the acquisition of sufficiently large datasets to perform these analyses on simultaneously measured neurons from single sessions. As a whole, this increase in scale is significant because it enables experimentalists to perform unbiased activity screens, by analogy with unbiased genetic screens that have been so useful in other fields of biology, here to determine which brain regions might be functionally implicated in a behaviour of interest180.
Regardless of the specific analysis approach used, working with large multiregion datasets presents new challenges. For example, if one were to rely on hundreds of pairwise statistical tests to assess whether significant differences might exist between the firing patterns of neurons, it would be necessary to account for the chance of false positive comparisons by using a false discovery rate correction. In a similar way, when one is working with hundreds of slow, time-varying neural signals, a key concern to keep in mind is that of ‘nonsense correlations’181. As many correlation metrics that are commonly used assume each time point is independent of all others (which is obviously false for filtered data), it is often possible to observe strong correlations between time-varying neural signals and even unrelated time-varying variables (for example, the stock market or the price of a cryptocurrency182). Without appropriate control analyses, these kinds of correlations may be erroneously judged as significant. Simple controls where one signal is randomly shuffled to generate a comparative ‘null distribution’ are often insufficient if there is clear time-varying structure imposed in a neural dataset by a stimulus or stereotyped behavioural response. Stronger controls that preserve long-term structure, such as shift permutation or trial and session shuffling, can help mollify such concerns181. The observation that ongoing movements explain a large fraction of the variance in neural activity across the brain further highlights the need for careful behavioural task design — and analysis strategy choice183. In general, both experiments and subsequent analyses should be designed with appropriate controls to ensure that reported results based on correlations do not simply occur due to chance.
Identify population activity patterns
As datasets increased in size over the past decade, new approaches for describing the joint activity of thousands of neurons were developed. In particular, population firing-rate trajectories became increasingly common tools for modelling the joint firing of whole neural populations184. This approach is quite practical, as it permits compression of the joint neural activity from even hundreds of neurons to something that can be visualized on a 3D plot (FIG. 2b, left). The most common means of projecting high-dimensional neural data into a low-dimensional space is principal component analysis (PCA) — which performs linear decomposition on the covariance structure between neurons to define a set of orthogonal axes (often called a ‘latent-variable space’, as the variable representing each axis is inferred, rather than directly observed) where each explains as much variance as possible in the data. This approach is highly effective in neural systems. Across many brain areas and behavioural tasks, most of the variance in the firing of hundreds of neurons can be explained using far fewer dimensions than the number of neurons18,21,185–187. However, there are important limitations to this approach.
First, the process of estimating a smooth ‘neural trajectory’ that represents the evolution of population activity over time requires more specialized methods than standard PCA. One such approach is Gaussian process factor analysis (GPFA) — an algorithm that simultaneously identifies basis vectors and defines a smooth neural trajectory188. In a similar way, methods based on canonical correlation analysis can identify shared neural dimensions between different datasets (which might not share any neurons), such that neural trajectories measured in a brain area could be aligned among different datasets and permit changes in neural dynamics to be tracked for many months or even years189 or to compare trajectories between different areas174. Along these lines, the recurrent switching linear dynamical systems model attempts to decompose neural population trajectories into segments that can be approximated by models with linear dynamics190. This approach has been found to identify states of neural activity that correlate well with manually labelled behavioural states not used to train the model191.
Second, performing dimensionality reduction and then quantification on a resultant trajectory makes two strong assumptions about the data. First, it assumes that the signal of interest is low-dimensional (for example, that the joint activity of 300 neurons can be summarized by three time-varying signals). Second, it assumes that the specific dynamics of the neural trajectory capture relevant features of the data under study192. This second assumption depends on the strong hypothesis that smooth firing rate dynamics, rather than features of precise spike timing, contain the neuronal population codes of interest. We know that this assumption is often at least partially wrong, as single-neuron spike timing codes that have been observed in various experimental settings are eroded by most common methods for smoothing spike trains into firing rates (which often assume Poisson-like spiking statistics that do not necessarily match the data)193,194. Nevertheless, dynamical models that make these assumptions constitute an exciting area of computational neuroscience research because there is much emerging evidence that the evolution of these neural trajectories over time may indeed describe certain neural computations195.
However, interesting structure apparent in neural trajectories based on just a few dimensions need not remain in the full high-dimensional dataset, and other unappreciated features may exist in the data beyond a single trajectory184. One solution to this problem is to use complementary analyses that operate on many more dimensions than were visualized, or on the full-dimensional neural space128,162,185,196. Empirical methods also exist for testing whether novel claims about population codes (for example, based on fitting latent-variable models) are potentially explainable by known features of single-cell response properties197.
So what is the relevant dimensionality of the neural activity in a given brain region? This parameter is likely to depend on the neural structure under study (for example, sensory versus motor) and the complexity of the behaviour186,192, as well as technical details such as the temporal resolution of the data acquired (for example, resampled second-long time bins versus short single-spike time bins). To approach these questions, other dimensionality reduction schemes (besides PCA) may be of use, which quantify the differences in neural activity between distinct experimental conditions — regardless of whether they explain most of the variance in the data (which is the goal of PCA). For example, neurons across the dorsal cortex encode motor-related information such as the current location of a reward, but this explains only a small fraction of the total variance in the data23. In this case, PCA is inappropriate, and using it to draw low-dimensional neural trajectories may not yield any obvious differences in population activity between different experimental conditions.
What alternatives to PCA are available? One approach, linear discriminant analysis, seeks to find an axis (the ‘linear discriminant’) that best separates data points on the basis of some covariate, such as lick direction. This approach works for arbitrary numbers of conditions, but if there are only two conditions to separate in the data (for example, lick left or lick right), the linear discriminant can be approximated by simply computing the vector difference between mean activity under the two conditions. This is sometimes referred to as the ‘coding direction’198. Similarly, an algorithm called ‘partial least squares regression’ is a common approach23,199,200 for jointly achieving two goals: (1) finding a low-dimensional representation of the data that explains much of the variance in the data and (2) using the data in that low-dimensional space to solve a regression problem (that is, to separate trajectories from different experimental conditions). Related algorithms that attempt to jointly find a hidden or ‘latent’ state space that explains variance in the data but also separates the data along experiment-defined conditions include demixed PCA, tensor component analysis and targeted dimensionality reduction201–203. A recent method, preferential subspace identification (PSID), has developed a dynamical model for identifying low-dimensional neural trajectories that also incorporate behavioural information, using measurements of the animal’s behavioural dynamics to aid in the identification of task-relevant neural dynamics204. Interestingly, a nonlinear variant of PSID (which relies upon recurrent neural networks (RNNs) used in deep learning models) performs similarly to the linear variant of the algorithm in mapping cortical activity into a latent space. Only behavioral decoding is significantly improved by the use of a nonlinear model — suggesting that cortical dynamics may be readily explainable by linear dynamics, but that transformations from cortical activity to behavior may be particularly nonlinear205.
More work is required to further generalize these approaches to explicitly multiregion data. One key issue is that many current approaches treat neurons identically, without regard for known differences between neurons residing in different brain areas or those with different gene expression profiles. Alternatively, the neural activity within each region is reduced to just a single signal (or a small number of signals per unit area). This use of a single time-varying scalar for coupling areas makes it easier to know that interregional interactions may be occurring — and could enhance the experimental use of closed-loop interventions in health and disease206 but comes at the expense of knowing what information might actually be transmitted207. But it is likely that the mechanisms that govern information flow between areas in the cortex are not the same everywhere. Thus, multiregion models that treat the neural activity from different brain regions distinctly are necessary — especially since we know there are clear anatomical and physiological distinctions between brain regions. For example, the motor cortex and the spinal cord are coupled via many ascending and descending neural pathways, but it seems unlikely that either region forms the majority of the inputs to the other under any behavioural circumstance; therefore, using only a single set of latent factors to represent a dataset comprising recordings from both areas would make little sense. Developing richer models that can incorporate this kind of information will be critical for building robust brain-machine interfaces and neural decoding algorithms that achieve high performance in complex, real-world scenarios — in addition to guiding us towards a better understanding of the brain.
Quantify network interactions
Beyond quantitatively describing population codes within distinct brain areas, a second-order set of questions seeks to understand how brain areas communicate with each other. New analyses will be important for identifying the mechanisms of interregional communication, and for testing several major hypotheses regarding corticocortical communication208. Three main mechanisms have been proposed. First, correlations between the spike trains of neurons in different areas seem to facilitate information transfer209,210. Second, coherent oscillations, particularly in the gamma band, may enhance information transmission by cortical neurons211,212. Third, interareal communication (both between cortical areas and in cortical–subcortical pathways) can occur within a ‘communication subspace’ such that projection neurons usually fire in a pattern whereby their net effect on a downstream area cancels out (that is, firing in the null space of the postsynaptic area) — except when they are actively broadcasting information213,214.
Each of these mechanisms, in addition to the operation of potential ‘gate’ neurons in pathways beyond the cortex (for example, ‘omnipause’ neurons in the brainstem that gate descending inputs during eye movements), likely plays a role in different behavioural circumstances215, and may now be accessible using multiregion cellular-scale methods. This question of how areas communicate is intertwined with the question of what information is communicated. However, it is still very much an open question whether or how upstream areas ‘command’ downstream areas216 or whether some interareal connections may influence other areas in a subtler manner, for example, via gain regulation217.
Methods from network analysis and topology may be applied to multiregion neural datasets to address these questions. Either simple correlation-based metrics or other, related metrics such as Granger causality, or information theoretic quantities, can be used to quantify directional dependencies between individual neurons or areas, and then network models can be defined using either neurons or brain areas as nodes and with the chosen metric defining functional connections between them. These network models may be useful to develop schemes for controlling the brain, or for better understanding its function218. Indeed, a whole host of network models at different levels of complexity may be applied for better understanding different aspects of interregional communication219.
However, multiregion neural datasets present a particular problem that hinders many kinds of network analyses — the fact that mammalian multiregion recording techniques afford only the ability to incompletely sample from a subset of neurons in a subset of brain areas. One emerging approach is to use these incomplete multiregion neural datasets (which have not measured every relevant activity parameter) for training recurrent neural network (RNN) models that can be then perturbed and analysed in silico (FIG. 2c). While this core idea of building detailed computational simulations of neural circuits can be taken to highly detailed and biophysically realistic levels220, these RNN-based methods usually seek to model features of neural and behavioural responses by using modern deep learning methods, rather than by creating explicit models of biological neurons221–231. For example, in the widely used latent factors analysis via dynamical systems (LFADS) framework, individual artificial units do not represent individual biological neurons221–224. Rather, as Sylwestrak and colleagues recently demonstrated224, LFADS can be used to directly model the underlying dynamical systems corresponding to distinct biological neural populations. Another approach (current-based decomposition) maintains a one-to-one correspondence between biological neurons and artificial units during model fitting, but this procedure is used to generate a 1D time-varying interaction signal between different brain areas (regardless of the number of neurons fitted per area)230. Even in the case of incomplete sampling from the brain, these approaches fit available multiregion neural data to analytically tractable ‘surrogate’ models that can be used to generate testable hypotheses for future experiments.
This approach of building a surrogate model that can generate known neural dynamics is also appealing because RNN models have become increasingly amenable to detailed analysis. For example, specific low-dimensional dynamical motifs can be reliably identified in trained RNN models as learned solutions to many common language processing and neuroscience-inspired tasks225–228. However, some of the analytical tools used to find these motifs are difficult to apply to real neural datasets. For example, dynamical fixed points and basins of attraction may be hard to identify in cortical areas because the area-specific recurrent dynamics are usually happening in the presence of strong external input from other, often unobserved, brain areas or sensory systems229. But surrogate neural models, such as current-based decomposition230, that explicitly model multiple brain areas (which need not be at cellular resolution) may offer a path forward here: a multiregion RNN model can be fit to a large neural dataset and then analysed in situations wherein external inputs to a brain area of interest are disabled, or perturbed in other ways223,231. Optogenetic manipulations could then be used to experimentally validate a tractable set of predictions made by the model.
At a more abstract level, RNN models that generate behavioural data can be used to test different neural analysis strategies. A combined experimental and computational article that validated this analysis paradigm emerged from the study of larval zebrafish, where single-cell neural activity can be measured almost comprehensively from the entire brain and spinal cord45. By the fitting of an RNN to activity measurements from most neurons in the zebrafish brain, distinct changes in the coupling strength between the habenula and raphe nucleus could be seen as fish entered a depression-like state, passively rather than actively coping with a stressor, in the process clearly identifying a circuit previously hypothesized to be involved in depression and passive coping. Importantly, because this approach processes data from all regions across the brain in an identical way, this brainwide analysis was not biased towards any particular answer. At a less comprehensive level, multiregion RNNs have also been used to reproduce interregional network dynamics within both the mammalian cortex and subcortical areas198,232–234. Finally, causal tests will be crucial in validating these models. Recent work experimentally perturbed information flow between two areas of the visual cortex (V1 and LM) by inhibiting activity in one area, at different time lags relative to a visual stimulus235; influence between the areas was observed to vary over time — much analogous to how recently developed models have been used to estimate time-varying ‘currents’, or lagged latent variables, that link brain areas230,236.
Moving forward, a key question is how complete the neural population recordings must be to build a model of the type described here that can accurately recapitulate population dynamics. Explicit incorporation of neuronal cell-type information to delineate subpopulations may be useful in this way. Of similar importance is a clear means of identifying the number of brain areas present in a dataset. Changing the definition of a brain region in this context will certainly influence any measures of interregional communication219. As discussed earlier, using a common reference atlas framework to delineate gross anatomical areas is an important first step. But when one is building an RNN model, is it best to try to learn regional groupings between neurons from the data themselves? Or must granular anatomical labels (for example, cortical layer within an area instead of just areal identity) be applied230,231? To some extent, the answer to these questions will depend on the biological questions of interest, the regions being studied and the behavioural state of the animal. However, it is exciting to consider the idea that as this framework increases in sophistication, single models may be able to accurately model neural activity during complex behaviours, as well as during a variety of perturbations to the relevant neural circuitry (for example, activation or silencing of different brain areas) or behavioural setting (for example, across different environments). If this potential can be realized, studying RNN models as a surrogate for new experimental data will be a tremendously powerful tool for systems neuroscience.
Recent findings and future directions
Initial studies applying multiregion recording methods in different behavioural contexts have begun to demonstrate the types of findings that can result from a broad, brainwide perspective. At least three major themes have emerged (FIG. 3). First, many behavioural features and stimuli have widespread neuronal population representations, and are decodable from neuronal dynamics in seemingly surprising locations across the brain. Second, the location and content of multiregion neural representations and dynamics depend on behavioural context. Third, specific interregional patterns of synchrony and asynchrony appear to be important features of behaviourally relevant neural dynamics.
Fig. 3 |. New perspectives arising from multiregion recording.

a | Example insights thus far into brainwide activity. Widespread nature of state and stimulus representations, here shown as widespread encoding of different actions (left; as in23). Context dependence of interregional dynamics, here shown as different patterns of regional dynamics depending on the behavioural strategy (such as an active or a passive detection strategy, as in243; middle). Roles of synchrony, here shown as desynchronized rhythmic activity between the retrosplenial cortex (red oscillations) that are decoupled from activity in other cortical regions (blue oscillations) that was observed to be elicited by ketamine, a dissociative drug (as in180; right). b | An unbiased activity screen using optoencephalography widefield imaging reveals a ketamine-elicited (50 mg kg−1) rhythm localized to one cortical region, yielding desynchrony between the retrosplenial cortex and other cortical regions. Ketamine’s effect is an important example of the value of multiregion imaging since the uniqueness of the effect seen in the retrosplenial cortex would not have been otherwise appreciated, and also could not have been predicted. This effect is evident in the top row as a sinusoidal pattern of activity in the retrosplenial cortex after infusion (red trace) that is not correlated with the post-infusion activity of other regions. In the bottom row, this is exhibited as a peak at 1 Hz in the spectral power of the activity after infusion (red trace). dF/F is the baseline-corrected change in fluorescence, blue traces are for before ketamine infusion and red traces are for 10 min after ketamine infusion. c | Recordings with multiple Neuropixels silicon-based probe electrodes further reveal ketamine-elicited correlation between the retrosplenial cortex (RSP) and the laterodorsal thalamus (LD), and inverse correlation between the retrosplenial cortex and the anteromedial thalamus (AM). The inset illustrates a Neuropixels silicon-based probe (version 1.0) with a dense arrangement of electrodes that enables recording from many individual neurons across multiple regions of the brain (indicated by colours that correspond to the data shown on the right). See REF180 for further information on recording and analysis details. PSD, power spectral density. Panel b reprinted from REF. 180, Springer Nature Limited. Panel c adapted from REF. 180 and REF. 151, Springer Nature Limited.
Widespread representations
One simple and powerful advantage of multiregion recording lies in surveying activity across many regions, in an unbiased way — thus including areas not expected to be particularly involved in a given behaviour. As a consequence of applying this approach, many recent studies have revealed that neural representations for various behavioural features are not confined to specific individual brain regions. For example, ongoing motor behaviour is represented not only in anterior motor areas — as expected — but also in posterior areas such as the primary visual cortex21,22,237. During a visual task, neurons in nearly all of 42 regions electrophysiologically recorded across the brain were observed to respond non-specifically when mice initiate an action34. Furthermore, even specific history-guided motor plans are encoded widely across the cortex23. Finally, sensory evidence appears to modulate activity in the secondary motor cortex in the absence of movement238. Whether these widespread signals subserve learning, context setting, distributed computation or even no behaviourally relevant purpose at all remains an important question well suited for future causal investigation.
Another important takeaway message from recent analyses of multiregion data is that neurons with similar trial-averaged activity patterns often display very different single-trial combinations of cognitive and movement variables21. For example, in one recent analysis of trial-averaged cortex-wide imaging data, there was no clear dependence of correlation strength over space — that is, pairs of neurons at near and far distances had high correlations. In contrast, single-trial correlations computed on the same dataset exhibited more localized structure23. Similarly, in a different experimental setting, spike-triggered maps (which are inherently trial averaged) from simultaneous electrophysiology and OEG displayed widespread cortical activity motifs related to the activity of individual thalamic or cortical neurons239. Thus, population-level signatures of behaviour are not only widespread; these signatures also manifest themselves differently on analyses of single-trial versus trial-averaged neural data.
What causal role do these widespread representations of behaviour play? Optogenetic interventions have the potential to provide important insight. As with multiregion recording, optogenetic manipulations have progressed to ever wider fields of view128 — even to cortex-wide scales23. Importantly, though, since brainwide activity patterns can arise from activity in localized populations of neurons (for example, sensory neurons, or neuromodulatory neurons that correlate with brain states such as arousal240), investigators can likewise readily generate naturalistic brainwide patterns of activity with even focal optogenetic interventions (if properly targeted). An example is a study using Neuropixels paired with optogenetics in which focal stimulation of input to the neurons of the subfornical organ with a single deep fibre optic triggered brainwide naturalistic internal representations of thirst, and of seeking water when thirsty18. These experiments illustrate how optogenetic interventions operate in ways fundamentally analogous to gain-of-function or loss-of-function genetic interventions (for example, gene knock-in/knockout, RNAi/short hairpin RNA and CRISPR–Cas genome editing) in other realms of biology241, wherein precise highly local perturbations provide insight into the global causal underpinnings of complex system function.
Context dependence
Another key benefit of multiregion investigation is the enhanced ability to compare neural dynamics within different contexts. These contexts can include task difficulty, sensing strategy and behavioural state242. By simultaneously measuring activity across the brain, one can survey the context-dependent involvement and interactions of many regions and ensure that regional differences are not due to uncontrolled differences in context or behaviour that might occur with asynchronous recordings. Moreover, by recording joint activity across behavioural conditions, one can disentangle potentially complex behavioural variables that confound interpretation of population neural activity.
A number of studies have discovered patterns of multiregion activity that distinguish scenarios with similar stimuli or actions but differing higher-level context. For example, the difficulty of a task can alter how identical stimuli are processed, with widespread multiregion activity ramps and decreased correlation across the cortex during a more complicated evidence-accumulation task versus a simpler explicit visual response task233. Moreover, use of optogenetic inactivation to silence activity in single regions across the dorsal cortex influenced performance on the evidence
accumulation task. However, performance on the simpler task only depended on activity in a few visual cortical regions. The representation of a stimulus can also change depending on the strategy used by the animal for sensing the stimulus, as in one example where the locus of short-term memory encoding in the dorsal cortex changed depending on whether the mouse used an active or a passive whisking strategy to identify a texture, and targeted optogenetic inactivation could even induce the mouse to use a different strategy243. The temporal sequence of stimulus presentation can also impact multiregion neural representation, as during a delayed non-match to sample task wherein the secondary somatosensory cortex was sensitive to whether the second stimulus matched the first stimulus and appeared to relay recalled information to primary somatosensory regions141. Last, the degree of agency that an animal has over a stimulus can influence multiregion activity. Using a multiregion brain–machine interface, one study found that when the position of a cursor was controllable, higher visual areas were more active, cursor position was more decodable from population neural activity and units exhibited increased correlation with cortex-wide activity244.
The behavioural state of an animal can also influence multiregion activity. For example, when an animal locomotes, units in the primary visual cortex become more strongly coupled to motor and local visual cortical regions, whereas retrosplenial units become less locally coupled159. Task engagement can also globally influence cortical activity, eliciting desynchronization and persistently decreased low-frequency (3–6-Hz) activity245. Finally, motivational state, such as whether a mouse is thirsty or sated, impacts global activity patterns, leading to a brainwide ‘initial condition’ that influences the transformation of sensory input into behavioural output18.
Synchrony and desynchrony
Perhaps one of the most valuable aspects of simultaneous multiregion recording is the capability to observe the details of correlated activity across the brain. Recording in two regions at the same time, such as the medial prefrontal cortex and hippocampus246–248, medial prefrontal cortex and ventral striatum249, frontal and visual areas34, or secondary motor cortex and posterior parietal cortex250 has already given rise to many synchrony-related insights, including into neuropsychiatric symptoms such as anhedonia249; indeed, synchrony and desynchrony have long been hypothesized to be important in neurobehaviourally important conditions such as schizophrenia, autism, depression and dissociative states. The advances now making it feasible to record from many more than two regions (for example, recordings with six simultaneously deployed Neuropixels probes revealing hierarchical structure in multiregion functional connectivity at the cellular level33, or widefield imaging251) promise an expansion of this perspective, both for basic science understanding and for insight into neuropsychiatric disorders. Further evidence for altered functional connectivity has been observed with multiregion recording in depression-related states, such as during recording from seven regions in a mouse model of stress response, which yielded multiregion activity factors that could serve as signatures for discriminating behavioural conditions252. Similarly, recording from five regions in a model of autism spectrum disorder yielded the discovery of diminished social stimulus-induced increases in coherence between the cingulate cortex, thalamus and nucleus accumbens253. Additionally, multiregion recording led to the discovery of a key role for desynchronized dynamics in the clinically important state of dissociation, whereby administration of dissociative drugs such as ketamine elicited a 1–3-Hz oscillation localized to the retrosplenial cortex (but not other dorsal cortical regions), a brainwide disappearance of most correlations with the retrosplenial cortex and an uncoupling of activity between laterodorsal and anteromedial thalamic regions180. Importantly, the mere presence of a slow oscillation in the retrosplenial cortex was not the distinguishing factor, but rather the spatial restriction of the oscillation and its desynchronization from other cortical regions was the distinguishing factor. Indeed, these multiregion recording observations were critical for informing the design of causal optogenetic and gene knockout experiments that pinpointed the role of the retrosplenial oscillation in dissociation-like behaviour, guiding analysis of multiregion intracranial electrophysiological recordings in the dissociating human brain and the discovery of similar oscillations in the homologous human retrosplenial and deep posteromedial cortical regions.
Looking forward, there are many opportunities for investigating the roles of synchrony in disease states. For example, since altered interregional brainwide communication has been long hypothesized to be relevant to the symptoms of schizophrenia and other psychotic states, it will be interesting to test whether multiregion relationships are altered in preclinical or clinical states with perceptual alterations, including during administration of psychosis-inducing pharmacological agents.
Conclusion
The mammalian brain is a complex system composed of many interdependent parts. In such systems, macroscopic properties emerge from properties and interactions of the individual parts of the system, and the state of each part depends on the state of the others. Neuroscientists may now draw upon new methods to investigate how dynamics of the whole brain and the behaviour of the animal depend on interactions among elemental parts. To advance this goal, here we suggest that it will be crucial to see the parts and the whole at the same time — in particular, by measuring cellular activity in multiple brain regions at once. This integrative approach, encompassing optical, electrophysiological and computational innovations, enables new types of observations, such as measurements of distributed population codes and of interregional synchrony, which are inaccessible to methods that probe one region or cell at a time. Especially when paired with optogenetic control241, multiregion recording provides a vital source of information on naturally occurring brainwide activity patterns that can be screened for in an unbiased fashion180, and then tested for causal significance in physiology and behaviour. Ultimately, by using the experimental and computational approaches discussed here, we have the opportunity to see both the forest and the trees of the brain — emergent brain-spanning states and their constituent cellular dynamics — at the same time.
Acknowledgements
T.A.M. is supported by an NIH NINDS Pathway to Independence Award (K99/NS116122), an A.P Giannini Fellowship and a Stanford School of Medicine Dean’s Fellowship. I.V.K. is a Merck Awardee of the Life Science Research Foundation and a Wu Tsai Stanford Neurosciences Institute Interdisciplinary Scholar. K.D. is supported by NIMH, NIDA, the NIH BRAIN Initiative, the National Science Foundation NeuroNex programme, the NOMIS Foundation, the Else Kröner Fresenius Foundation, the Gatsby Foundation and the AE Foundation. The authors also thank W. Allen, S. Bradbury, M. Inoue, C. Kim, J. Kochalka, B. Midler, A. Mitra, S. Quirin, E. Richman, S. Vesuna and other current and former members of the Deisseroth laboratory for valuable discussions.
Footnotes
Competing interests
The authors declare no competing financial interests
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
References
- 1.The Event Horizon Telescope Collaboration. et al. First M87 event horizon telescope results. I. The shadow of the supermassive black hole. Astrophys. J. Lett. 875, L1 (2019). [Google Scholar]
- 2.Galilei G Sidereus Nuncius (Univ. Chicago Press, 1610). [Google Scholar]
- 3.Adrian ED The Basis of Sensation (WW Norton & Co, 1928). [Google Scholar]
- 4.Brock LG, Coombs JS & Eccles JC The recording of potentials from motoneurones with an intracellular electrode. J. Physiol. 117, 431–460 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodbury JW & Patton HD in Cold Spring Harbor Symposia on Quantitative Biology vol. 17, 185–188 (Cold Spring Harbor Laboratory Press, 1952). [DOI] [PubMed] [Google Scholar]
- 6.Ren C & Komiyama T Characterizing cortex-wide dynamics with wide-field calcium imaging. J. Neurosci. 41, 4160–4168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim T H. & Schnitzer, M. J. Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185, 9–41 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urai AE, Doiron B, Leifer AM & Churchland AK Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 25, 11–19 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Siegel M, Buschman TJ & Miller EK Cortical information flow during flexible sensorimotor decisions. Science 348, 1352–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernández A et al. Procedure for recording the simultaneous activity of single neurons distributed across cortical areas during sensory discrimination. Proc Natl Acad. Sci. USA 105, 16785–16790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández A et al. Decoding a perceptual decision process across cortex. Neuron 66, 300–314 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Paulk AC et al. Large-scale neural recordings with single neuron resolution using Neuropixels probes in human cortex. Nat. Neurosci. 25, 252–263 (2022). [DOI] [PubMed] [Google Scholar]
- 13. Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). This study uses viral anterograde tracing in mice to systematically map mesoscale connectivity between brain regions and produce a foundational brain atlas.
- 14.Han Y et al. The logic of single-cell projections from visual cortex. Nature 556, 51–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown CE, Aminoltejari K, Erb H, Winship IR & Murphy TH In vivo voltage-sensitive dye imaging in adult mice reveals that somatosensory maps lost to stroke are replaced over weeks by new structural and functional circuits with prolonged modes of activation within both the peri-infarct zone and distant sites. J. Neurosci. 29, 1719–1734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferezou I et al. Spatiotemporal dynamics of cortical sensorimotor integration in behaving mice. Neuron 56, 907–923 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Santos L, Opris I, Fuqua J, Hampson RE & Deadwyler SA A novel tetrode microdrive for simultaneous multi-neuron recording from different regions of primate brain. J. Neurosci. Methods 205, 368–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen WE et al. Thirst regulates motivated behavior through modulation of brainwide neural population dynamics. Science 364, eaav3932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel T A. & Steinmetz, N. A. New perspectives on dimensionality and variability from large-scale cortical dynamics. Curr. Opin. Neurobiol. 58, 181–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider DM Reflections of action in sensory cortex. Curr. Opin. Neurobiol. 64, 53–59 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Musall S, Kaufman MT, Juavinett AL, Gluf S & Churchland AK Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 22, 1677–1686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stringer C et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, eaav7893 (2019). This study uses eight simultaneously deployed Neuropixels probes to discover brainwide activity driven by spontaneous facial movements, including in the primary visual cortex.
- 23. Kauvar IV et al. Cortical observation by synchronous multifocal optical sampling reveals widespread population encoding of actions. Neuron 107, 351–367.e19 (2020). This article introduces and applies COSMOS, a method for recording near cellular resolution activity at video rates from thousands of neuronal sources spanning the mouse dorsal cortex.
- 24.Schneider DM, Nelson A & Mooney R A synaptic and circuit basis for corollary discharge in the auditory cortex. Nature 513, 189–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena S & Cunningham J P Towards the neural population doctrine. Curr Opin. Neurobiol. 55, 103–111 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Dong HW The Allen Reference Atlas: A Digital Color Brain Atlas of the C57Bl/6J Male Mouse (Wiley, 2008). [Google Scholar]
- 27.Paxinos G & Franklin KB J. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (Academic, 2019). [Google Scholar]
- 28.Swanson L Brain Maps: Structure of the Rat Brain (Gulf Professional Publishing, 2004). [Google Scholar]
- 29.Zingg B et al. Neural networks of the mouse neocortex. Cell 10.1016/j.cell.2014.02.023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones AR, Overly CC & Sunkin SM The Allen Brain Atlas: 5 years and beyond. Nat. Rev. Neurosci. 10, 821–828 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Wang Q et al. The Allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siegle JH et al. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92 (2021). This study uses six simultaneously deployed Neuropixels probes to establish the hierarchical nature of functional connectivity in the mouse visual system.
- 34.Steinmetz NA, Zatka-Haas P, Carandini M & Harris KD Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafting T, Fyhn M, Molden S, Moser M-B & Moser EI Microstructure of a spatial map in the entorhinal cortex. Nature 436, 801–806 (2005). [DOI] [PubMed] [Google Scholar]
- 36.da Silva JA, Tecuapetla F, Paixão V & Costa RM Dopamine neuron activity before action initiation gates and invigorates future movements. Nature 554, 244–248 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Carus-Cadavieco M et al. Gamma oscillations organize top-down signalling to hypothalamus and enable food seeking. Nature 542, 232–236 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Senzai Y, Fernandez-Ruiz A & Buzsáki G Layer-specific physiological features and interlaminar interactions in the primary visual cortex of the mouse. Neuron 101, 500–513.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couto J et al. Chronic, cortex-wide imaging of specific cell populations during behavior. Nat. Protoc. 16, 3241–3263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dockès J et al. NeuroQuery, comprehensive meta-analysis of human brain mapping. eLife 9, e53385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC & Wager TD Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim CK, Adhikari A & Deisseroth K Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci. 18, 222–235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tervo DGR et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andalman AS et al. Neuronal dynamics regulating brain and behavioral state transitions. Cell 177, 970–985.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovett-Barron M et al. Ancestral circuits for the coordinated modulation of brain state. Cell 171, 1411–1423.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu S et al. Behavioral state coding by molecularly defined paraventricular hypothalamic cell type ensembles. Science 370, eabb2494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen WE et al. Global representations of goal-directed behavior in distinct cell types of mouse neocortex. Neuron 94, 891–907.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lima SQ, Hromádka T, Znamenskiy P & Zador AM PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS ONE 4, e6099 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff SBE et al. Amygdala interneuron subtypes control fear learning through disinhibition. Nature 509, 453–458 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Cohen JY, Haesler S, Vong L, Lowell BB & Uchida N Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature 482, 85–88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herrera CG et al. Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat. Neurosci. 19, 290–298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juavinett AL, Bekheet G & Churchland AK Chronically implanted Neuropixels probes enable high-yield recordings in freely moving mice. eLife 8, e47188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoonover CE, Ohashi SN, Axel R & Fink AJP Representational drift in primary olfactory cortex. Nature 594, 541–546 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Steinmetz NA et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science 372, eabf4588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung JE et al. High-density, long-lasting, and multi-region electrophysiological recordings using polymer electrode arrays. Neuron 101, 21–31.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardin JA, Crair MC & Higley MJ Mesoscopic imaging: shining a wide light on large-scale neural dynamics. Neuron 108, 33–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grinvald A & Hildesheim R VSDI: a new era in functional imaging of cortical dynamics. Nat. Rev. Neurosci. 5, 874–885 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Mohajerani MH et al. Spontaneous cortical activity alternates between motifs defined by regional axonal projections. Nat. Neurosci. 16, 1426–1435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dana H et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE 9, e108697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wekselblatt JB, Flister ED, Piscopo DM & Niell CM Large-scale imaging of cortical dynamics during sensory perception and behavior. J. Neurophysiol. 115, 2852–2866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo ZV et al. Flow of cortical activity underlying a tactile decision in mice. Neuron 81, 179–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ratzlaff EH & Grinvald A A tandem-lens epifluorescence macroscope: hundred-fold brightness advantage for wide-field imaging. J. Neurosci. Methods 36, 127–137 (1991). [DOI] [PubMed] [Google Scholar]
- 67. Kim CK et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat. Methods 13, 325–328 (2016). This article introduces a technique for simultaneously recording cell type-specific neural activity from seven regions throughout the brain.
- 68.Valley MT et al. Separation of hemodynamic signals from GCaMP fluorescence measured with wide-field imaging. J. Neurophysiol. 123, 356–366 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Daigle TL et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerfen CR, Paletzki R & Heintz N GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80, 1368–1383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matho KS et al. Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature 598, 182–187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callaway EM et al. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fenno LE et al. Comprehensive dual- and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals. Neuron 107, 836–853.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris J et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front. Neural Circuits 10.3389/fncir.2014.00076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waters J Sources of widefield fluorescence from the brain. eLife 9, e59841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lohani S et al. Dual color mesoscopic imaging reveals spatiotemporally heterogeneous coordination of cholinergic and neocortical activity. Preprint at bioRxiv 10.1101/2020.12.09.418632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sabatini BL & Tian L Imaging neurotransmitter and neuromodulator dynamics in vivo with genetically encoded indicators. Neuron 108, 17–32 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Shen Y, Nasu Y, Shkolnikov I, Kim A & Campbell RE Engineering genetically encoded fluorescent indicators for imaging of neuronal activity: progress and prospects. Neurosci. Res. 152, 3–14 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Steinmetz NA et al. Aberrant cortical activity in multiple GCaMP6-expressing transgenic mouse lines. eNeuro 4, ENEURO.0207-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heffley W et al. Coordinated cerebellar climbing fiber activity signals learned sensorimotor predictions. Nat. Neurosci. 21, 1431–1441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ackman JB, Burbridge TJ & Crair MC Retinal waves coordinate patterned activity throughout the developing visual system. Nature 490, 219–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, Turan Z & Meister M Functional architecture of motion direction in the mouse superior colliculus. Curr. Biol. 30, 3304–3315.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scott BB et al. Imaging cortical dynamics in GCaMP transgenic rats with a head-mounted widefield macroscope. Neuron 100, 1045–1058.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rynes ML et al. Miniaturized head-mounted microscope for whole-cortex mesoscale imaging in freely behaving mice. Nat. Methods 18, 417–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lake EMR et al. Simultaneous cortex-wide fluorescence Ca2+ imaging and whole-brain fMRI. Nat. Methods 17, 1262–1271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murphy TH et al. High-throughput automated home-cage mesoscopic functional imaging of mouse cortex. Nat. Commun. 7, 11611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy TH et al. Automated task training and longitudinal monitoring of mouse mesoscale cortical circuits using home cages. eLife 9, e55964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim TH et al. Long-term optical access to an estimated one million neurons in the live mouse cortex. Cell Rep. 17, 3385–3394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghanbari L et al. Cortex-wide neural interfacing via transparent polymer skulls. Nat. Commun. 10, 1500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freeman J et al. Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Kim CK et al. Prolonged, brain-wide expression of nuclear-localized GCaMP3 for functional circuit mapping. Front. Neural Circuits 8, 138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vladimirov N et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 11, 883–884 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Chen Y et al. Soma-targeted imaging of neural circuits by ribosome tethering. Neuron 107, 454–469.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shemesh OA et al. Precision calcium imaging of dense neural populations via a cell-body-targeted calcium indicator. Neuron 107, 470–486.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim ST, Antonucci DE, Scannevin RH & Trimmer JS A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron 25, 385–397 (2000). [DOI] [PubMed] [Google Scholar]
- 96.Cramer SW et al. Through the looking glass: a review of cranial window technology for optical access to the brain. J. Neurosci. Methods 354, 109100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan J et al. Video-rate imaging of biological dynamics at centimetre scale and micrometre resolution. Nat. Photonics 13, 809–816 (2019). [Google Scholar]
- 98.Broxton M et al. Wave optics theory and 3-D deconvolution for the light field microscope. Opt. Express 21, 25418–25439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nöbauer T et al. Video rate volumetric Ca2+ imaging across cortex using seeded iterative demixing (SID) microscopy. Nat. Methods 14, 811–818 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Voleti V et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 16, 1054–1062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang C.-P (Jonathan) & Holy TE in Optical Techniques in Neurosurgery, Neurophotonics, and Optogenetics vol. 11629, 20–28 (SPIE, 2021). [Google Scholar]
- 102.Kumar M, Kishore S, Nasenbeny J, McLean DL & Kozorovitskiy Y Integrated one- and two-photon scanned oblique plane illumination (SOPi) microscopy for rapid volumetric imaging. Opt. Express 26, 13027–13041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xue Y, Davison IG, Boas DA & Tian L Single-shot 3D wide-field fluorescence imaging with a Computational Miniature Mesoscope. Sci. Adv. 6, eabb7508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ebrahimi S et al. Emergent reliability in sensory cortical coding and inter-area communication. Nature 10.1038/s41586-022-04724-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gunaydin LA et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cui G et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature 494, 238–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lütcke H et al. Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice. Front. Neural Circuits 4, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schulz K et al. Simultaneous BOLD fMRI and fiberoptic calcium recording in rat neocortex. Nat. Methods 9, 597–602 (2012). [DOI] [PubMed] [Google Scholar]
- 109.Stroh A et al. Making waves: initiation and propagation of corticothalamic Ca2+ waves in vivo. Neuron 77, 1136–1150 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Pisanello M et al. The three-dimensional signal collection field for fiber photometry in brain tissue. Front. Neurosci. 13, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marshall JD et al. Cell-type-specific optical recording of membrane voltage dynamics in freely moving mice. Cell 167, 1650–1662.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pisano F et al. Depth-resolved fiber photometry with a single tapered optical fiber implant. Nat. Methods 16, 1185–1192 (2019). [DOI] [PubMed] [Google Scholar]
- 113.Sych Y, Chernysheva M, Sumanovski LT & Helmchen F High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods 16, 553–560 (2019). [DOI] [PubMed] [Google Scholar]
- 114.Pnevmatikakis EA et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siegle JH et al. Reconciling functional differences in populations of neurons recorded with two-photon imaging and electrophysiology. eLife 10, e69068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei Z et al. A comparison of neuronal population dynamics measured with calcium imaging and electrophysiology. PLoS Comput. Biol. 16, e1008198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Abdelfattah AS et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Jin L et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75, 779–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin MZ & Schnitzer MJ Genetically encoded indicators of neuronal activity. Nat. Neurosci. 19, 1142–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Piatkevich KD et al. Population imaging of neural activity in awake behaving mice. Nature 574, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu Y, Zou P & Cohen AE Voltage imaging with genetically encoded indicators. Curr. Opin. Chem. Biol. 39, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Villette V et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Adam Y et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan LZ et al. All-optical synaptic electrophysiology probes mechanism of ketamine-induced disinhibition. Nat. Methods 15, 823–831 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fan LZ et al. All-optical electrophysiology reveals the role of lateral inhibition in sensory processing in cortical layer 1. Cell 180, 521–535.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hochbaum DR et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Piatkevich KD et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marshel JH et al. Cortical layer–specific critical dynamics triggering perception. Science 365, eaaw5202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu J et al. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 17, 287–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Carandini M et al. Imaging the awake visual cortex with a genetically encoded voltage indicator. J. Neurosci. 35, 53–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Platisa J et al. Voltage imaging using transgenic mouse lines expressing the GEVI ArcLight in two olfactory cell types. Preprint at bioRxiv 10.1101/2020.08.26.268904 (2020). [DOI] [Google Scholar]
- 132.Platisa J et al. High-speed low-light in vivo two-photon voltage imaging of large neuronal populations. Preprint at bioRxiv 10.1101/2021.12.07.471668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gottschalk S et al. Rapid volumetric optoacoustic imaging of neural dynamics across the mouse brain. Nat. Biomed. Eng. 3, 392–401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rabut C et al. Ultrasound technologies for imaging and modulating neural activity. Neuron 108, 93–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Helmchen F & Denk W Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005). [DOI] [PubMed] [Google Scholar]
- 136.Lecoq J et al. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat. Neurosci. 17, 1825–1829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wagner MJ et al. Shared cortex-cerebellum dynamics in the execution and learning of a motor task. Cell 177, 669–682.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sofroniew NJ, Flickinger D, King J & Svoboda K A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, e14472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stirman JN, Smith IT, Kudenov MW & Smith SL Wide field-of-view, multi-region two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 34, 857–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen JL, Voigt FF, Javadzadeh M, Krueppel R & Helmchen F Long-range population dynamics of anatomically defined neocortical networks. eLife 5, e14679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Condylis C et al. Context-dependent sensory processing across primary and secondary somatosensory cortex. Neuron 106, 515–525.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yu C-H, Stirman JN, Yu Y, Hira R & Smith SL Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry. Nat. Commun. 12, 6639 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Clough M et al. Flexible simultaneous mesoscale two-photon imaging of neural activity at high speeds. Nat. Commun. 12, 6638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lu R et al. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat. Methods 17, 291–294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Demas J et al. High-speed, cortex-wide volumetric recording of neuroactivity at cellular resolution using light beads microscopy. Nat. Methods 18, 1103–1111 (2021). This article introduces light beads microscopy, a two-photon method that enables cellular resolution imaging from hundreds of thousands of neurons at rates of a few hertz.
- 146.Rumyantsev OI et al. Fundamental bounds on the fidelity of sensory cortical coding. Nature 580, 100–105 (2020). [DOI] [PubMed] [Google Scholar]
- 147.Zhang T et al. Kilohertz two-photon brain imaging in awake mice. Nat. Methods 16, 1119–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang SJ et al. Extended field-of-view and increased-signal 3D holographic illumination with time-division multiplexing. Opt. Express 23, 32573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barson D et al. Simultaneous mesoscopic and two-photon imaging of neuronal activity in cortical circuits. Nat. Methods 17, 107–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zong W et al. Large-scale two-photon calcium imaging in freely moving mice. Cell 185, 1240–1256.e30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jun JJ et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Voigts J, Newman JP, Wilson MA & Harnett MT An easy-to-assemble, robust, and lightweight drive implant for chronic tetrode recordings in freely moving animals. J. Neural Eng. 17, 026044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Varol E et al. Decentralized motion inference and registration of neuropixel data. In ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 1085–1089 (IEEE, 2021). [Google Scholar]
- 154.Luo TZ et al. An approach for long-term, multi-probe Neuropixels recordings in unrestrained rats. eLife 9, e59716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jensen KHR & Berg RW CLARITY-compatible lipophilic dyes for electrode marking and neuronal tracing. Sci. Rep. 6, 32674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vázquez-Guardado A, Yang Y, Bandodkar AJ & Rogers JA Recent advances in neurotechnologies with broad potential for neuroscience research. Nat. Neurosci. 23, 1522–1536 (2020). [DOI] [PubMed] [Google Scholar]
- 157.Wang X et al. A parylene neural probe array for multiregion deep brain recordings. J. Microelectromech. Syst. 29, 499–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu X et al. Multimodal neural recordings with Neuro-FITM uncover diverse patterns of cortical–hippocampal interactions. Nat. Neurosci. 24, 886–896 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Clancy KB, Orsolic I & Mrsic-Flogel TD Locomotion-dependent remapping of distributed cortical networks. Nat. Neurosci. 22, 778–786 (2019). In this study, the authors simultaneously use single-neuron recordings and OEG to describe how locomotion influences the relationship between single-neuron firing and cortex-wide activity patterns.
- 160.Peters AJ, Fabre JMJ, Steinmetz NA, Harris KD & Carandini M Striatal activity topographically reflects cortical activity. Nature 591, 420–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kleinfeld D et al. Can one concurrently record electrical spikes from every neuron in a mammalian brain? Neuron 103, 1005–1015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Trautmann E et al. Accurate estimation of neural population dynamics without spike sorting. Neuron 103, 292–308.e4 (2019). This article illustrates how many popular population-level analyses of neural activity can successfully be applied to multi-unit electrophysiology data that lack single-cell resolution.
- 163.Ecker AS et al. Decorrelated neuronal firing in cortical microcircuits. Science 327, 584–587 (2010). [DOI] [PubMed] [Google Scholar]
- 164.Renart A et al. The asynchronous state in cortical circuits. Science 327, 587–590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dacre J et al. A cerebellar-thalamocortical pathway drives behavioral context-dependent movement initiation. Neuron 109, 2326–2338.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang W et al. Coordination of escape and spatial navigation circuits orchestrates versatile flight from threats. Neuron 109, 1848–1860.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Adrian ED The impulses produced by sensory nerve endings. J. Physiol. 61, 49–72 (1926). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sherrington C The Integrative Action of the Nervous System (Charles Scribner’s Sons, 1906). [Google Scholar]
- 169.Hubel DH & Wiesel TN Receptive fields of single neurones in the cat’s striate cortex. J. Physiol. 148, 574–591 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chichilnisky EJ A simple white noise analysis of neuronal light responses. Netw. Comput. Neural Syst. 12, 199–213 (2001). [PubMed] [Google Scholar]
- 171.Schwartz O, Pillow JW, Rust NC & Simoncelli EP Spike-triggered neural characterization. J. Vis. 6, 13–13 (2006). [DOI] [PubMed] [Google Scholar]
- 172.Pillow JW et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 454, 995–999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Truccolo W, Eden UT, Fellows MR, Donoghue JP & Brown EN A point process framework for relating neural spiking activity to spiking history, neural ensemble, and extrinsic covariate effects. J. Neurophysiol. 93, 1074–1089 (2005). [DOI] [PubMed] [Google Scholar]
- 174.Kang B & Druckmann S Approaches to inferring multi-regional interactions from simultaneous population recordings. Curr. Opin. Neurobiol. 65, 108–119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Keeley SL, Zoltowski DM, Aoi MC & Pillow JW Modeling statistical dependencies in multi-region spike train data. Curr. Opin. Neurobiol. 65, 194–202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yates JL, Park IM, Katz LN, Pillow JW & Huk AC Functional dissection of signal and noise in MT and LIP during decision-making. Nat. Neurosci. 20, 1285–1292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Benjamin AS et al. Modern machine learning as a benchmark for fitting neural responses. Front. Comput. Neurosci. 12, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Linderman S, Adams RP & Pillow JW Bayesian latent structure discovery from multi-neuron recordings. in Advances in Neural Information Processing Systems Vol. 29 (NIPS, 2016). [Google Scholar]
- 179.Pedregosa F et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 180. Vesuna S et al. Deep posteromedial cortical rhythm in dissociation. Nature 586, 87–94 (2020). Using multiregion recording techniques, the authors reveal a unique oscillatory firing pattern in the retrosplenial cortex that relates to a dissociation-like behavioural state.
- 181.Harris KD Nonsense correlations in neuroscience. Preprint at bioRxiv 10.1101/2020.11.29.402719 (2021). [DOI] [Google Scholar]
- 182.Meijer G Neurons in the mouse brain correlate with cryptocurrency price: a cautionary tale. Preprint at PsyArXiv 10.31234/osf.io/fa4wz (2021). [DOI] [Google Scholar]
- 183.Zagha E et al. The importance of accounting for movement when relating neuronal activity to sensory and cognitive processes. J. Neurosci. 42, 1375–1382 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cunningham JP & Yu BM Dimensionality reduction for large-scale neural recordings. Nat. Neurosci. 17, 1500–1509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Churchland MM et al. Neural population dynamics during reaching. Nature 487, 51–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gao P et al. A theory of multineuronal dimensionality, dynamics and measurement. Preprint at bioRxiv 10.1101/214262 (2017). [DOI] [Google Scholar]
- 187.Stringer C, Pachitariu M, Steinmetz N, Carandini M & Harris KD High-dimensional geometry of population responses in visual cortex. Nature 571, 361–365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yu BM et al. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J. Neurophysiol. 102, 614–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gallego JA, Perich MG, Chowdhury RH, Solla SA & Miller LE Long-term stability of cortical population dynamics underlying consistent behavior. Nat. Neurosci. 23, 260–270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Linderman S et al. Bayesian learning and inference in recurrent switching linear dynamical systems. in Proceedings of the 20th International Conference on Artificial Intelligence and Statistics 914–922 (PMLR, 2017). [Google Scholar]
- 191.Linderman S, Nichols A, Blei D, Zimmer M & Paninski L Hierarchical recurrent state space models reveal discrete and continuous dynamics of neural activity in C. elegans. Preprint at bioRxiv 10.1101/621540 (2019). [DOI] [Google Scholar]
- 192.Humphries MD Strong and weak principles of neural dimension reduction. Preprint at 10.48550/arXiv.2011.08088 (2021). [DOI]
- 193.Gollisch T & Meister M Rapid neural coding in the retina with relative spike latencies. Science 319, 1108–1111 (2008). [DOI] [PubMed] [Google Scholar]
- 194.Maimon G & Assad JA Beyond Poisson: increased spike-time regularity across primate parietal cortex. Neuron 62, 426–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vyas S, Golub MD, Sussillo D & Shenoy KV Computation through neural population dynamics. Annu. Rev. Neurosci. 43, 249–275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ames KC, Ryu SI & Shenoy KV Neural dynamics of reaching following incorrect or absent motor preparation. Neuron 81, 438–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Elsayed GF & Cunningham J P Structure in neural population recordings: an expected byproduct of simpler phenomena? Nat. Neurosci. 20, 1310–1318 (2017). In this study, the authors develop a statistical framework for testing whether population-level structure in neuronal firing patterns is explainable by simpler features of single-neuron responses.
- 198.Li N, Daie K, Svoboda K & Druckmann S Robust neuronal dynamics in premotor cortex during motor planning. Nature 532, 459–464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Geladi P & Kowalski BR Partial least-squares regression: a tutorial. Anal. Chim. Acta 185, 1–17 (1986). [Google Scholar]
- 200.Remedios R et al. Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 550, 388–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kobak D et al. Demixed principal component analysis of neural population data. eLife 5, 614–635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mante V, Sussillo D, Shenoy KV & Newsome WT Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature 503, 78–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Williams AH et al. Unsupervised discovery of demixed, low-dimensional neural dynamics across multiple timescales through tensor component analysis. Neuron 98, 1099–1115.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sani OG, Abbaspourazad H, Wong YT, Pesaran B & Shanechi MM Modeling behaviorally relevant neural dynamics enabled by preferential subspace identification. Nat. Neurosci. 24, 140–149. (2021). [DOI] [PubMed] [Google Scholar]
- 205.Sani OG, Pesaran B & Shanechi MM Where is all the nonlinearity: flexible nonlinear modeling of behaviorally relevant neural dynamics using recurrent neural networks. Preprint at bioRxiv 10.1101/2021.09.03.458628 (2021). [DOI] [Google Scholar]
- 206.Scangos KW, Makhoul GS, Sugrue LP, Chang EF & Krystal AD State-dependent responses to intracranial brain stimulation in a patient with depression. Nat. Med. 27, 229–231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Semedo JD, Gokcen E, Machens CK, Kohn A & Yu BM Statistical methods for dissecting interactions between brain areas. Curr. Opin. Neurobiol. 65, 59–69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kohn A et al. Principles of corticocortical communication: proposed schemes and design considerations. Trends Neurosci. 43, 725–737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hahn G, Ponce-Alvarez A, Deco G, Aertsen A & Kumar A Portraits of communication in neuronal networks. Nat. Rev. Neurosci. 20, 117–127 (2019). [DOI] [PubMed] [Google Scholar]
- 210.Zandvakili A & Kohn A Coordinated neuronal activity enhances corticocortical communication. Neuron 87, 827–839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Buzsáki G & Wang X-J Mechanisms of gamma oscillations. Annu. Rev. Neurosci. 35, 203–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sohal VS, Zhang F, Yizhar O & Deisseroth K Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kaufman MT, Churchland MM, Ryu SI & Shenoy KV Cortical activity in the null space: permitting preparation without movement. Nat. Neurosci. 17, 440–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Semedo JD, Zandvakili A, Machens CK, Yu BM & Kohn A Cortical areas interact through a communication subspace. Neuron 102, 249–259.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Keller EL Participation of medial pontine reticular formation in eye movement generation in monkey. J. Neurophysiol. 37, 316–332 (1974). [DOI] [PubMed] [Google Scholar]
- 216.Kupfermann I & Weiss KR The command neuron concept. Behav. Brain Sci. 1, 3–10 (1978). [Google Scholar]
- 217.Darlington TR & Lisberger SG Mechanisms that allow cortical preparatory activity without inappropriate movement. eLife 9, e50962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bassett DS & Sporns O Network neuroscience. Nat. Neurosci. 20, 353–364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bassett DS, Zurn P & Gold JI On the nature and use of models in network neuroscience. Nat. Rev. Neurosci. 19, 566–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xie ME et al. High-fidelity estimates of spikes and subthreshold waveforms from 1-photon voltage imaging in vivo. Cell Rep. 35, 108954 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Keshtkaran MR et al. A large-scale neural network training framework for generalized estimation of single-trial population dynamics. Preprint at bioRxiv 10.1101/2021.01.13.426570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pandarinath C et al. Latent factors and dynamics in motor cortex and their application to brain-machine interfaces. J. Neurosci. 38, 9390–9401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pandarinath C et al. Inferring single-trial neural population dynamics using sequential auto-encoders. Nat. Methods 15, 805–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Sylwestrak EL et al. Cell type-specific population-dynamics of diverse reward computations. Cell 185, 3568–3587.e27 (2022). In this study, the authors collect multiregion neural recording data during a reward-seeking behaviour, model the data with an RNN model called LFADS and then use the model to describe how a population of genetically defined neurons in the medial habenula integrated reward history over time.
- 225.Aitken K et al. The geometry of integration in text classification RNNs. Preprint at 10.48550/arXiv.2010.15114 (2020). [DOI]
- 226.Maheswaranathan N, Williams AH, Golub MD, Ganguli S & Sussillo D Universality and individuality in neural dynamics across large populations of recurrent networks. Preprint at 10.48550/arXiv.1907.08549 (2019). [DOI] [PMC free article] [PubMed]
- 227.McIntosh L, Maheswaranathan N, Nayebi A, Ganguli S & Baccus S Deep learning models of the retinal response to natural scenes. Adv. Neural Inf. Process. Syst. 29, 1369–1377 (2016). [PMC free article] [PubMed] [Google Scholar]
- 228.Sussillo D & Barak O Opening the black box: low-dimensional dynamics in high-dimensional recurrent neural networks. NeuralComput. 25, 626–649 (2013). [DOI] [PubMed] [Google Scholar]
- 229.Sauerbrei BA et al. Cortical pattern generation during dexterous movement is input-driven. Nature 577, 386–391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Perich MG et al. Inferring brain-wide interactions using data-constrained recurrent neural network models. Preprint at bioRxiv 10.1101/2020.12.18.423348 (2021). This article describes current-based decomposition, an algorithm that uses RNN models to quantify interregional interactions in multiregion neural datasets.
- 231.Perich MG & Rajan K Rethinking brain-wide interactions through multi-region ‘network of networks’ models. Curr. Opin. Neurobiol. 65, 146–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lo C-C & Wang X-J Cortico-basal ganglia circuit mechanism for a decision threshold in reaction time tasks. Nat. Neurosci. 9, 956–963 (2006). [DOI] [PubMed] [Google Scholar]
- 233. Pinto L et al. Task-dependent changes in the large-scale dynamics and necessity of cortical regions. Neuron 104, 810–824 (2019). This study uses OEG and optogenetics to relate cognitive task complexity to cortical engagement — and then analyses this effect in more detail by reproducing qualitative features of the dataset using a multiregion RNN model.
- 234.Hattori R & Komiyama T Context-dependent persistency as a coding mechanism for robust and widely distributed value coding. Neuron 10.1016/j.neuron.2021.11.001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Javadzadeh M & Hofer SB Dynamic causal communication channels between neocortical areas. Preprint at bioRxiv 10.1101/2021.06.28.449892 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gokcen E et al. Disentangling the flow of signals between populations of neurons. Nat. Comput. Sci 2, 512–525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Allen WE et al. Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Orsolic I, Rio M, Mrsic-Flogel TD & Znamenskiy P Mesoscale cortical dynamics reflect the interaction of sensory evidence and temporal expectation during perceptual decision-making. Neuron 109, 1861–1875.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Xiao D et al. Mapping cortical mesoscopic networks of single spiking cortical or sub-cortical neurons. eLife 6, e19976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Carter ME et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 10.1038/nn.2682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deisseroth K From microbial membrane proteins to the mysteries of emotion. Cell 184, 5279–5285 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cardin JA Functional flexibility in cortical circuits. Curr. Opin. Neurobiol. 58, 175–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gilad A, Gallero-Salas Y, Groos D & Helmchen F Behavioral strategy determines frontal or posterior location of short-term memory in neocortex. Neuron 99, 814–828.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 244.Clancy KB & Mrsic-Flogel TD The sensory representation of causally controlled objects. Neuron 109, 677–689.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Jacobs EAK, Steinmetz NA, Peters AJ, Carandini M & Harris KD Cortical state fluctuations during sensory decision making. Curr. Biol. 30, 4944–4955.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Colgin LL Oscillations and hippocampal-prefrontal synchrony. Curr. Opin. Neurobiol. 21, 467–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Park AJ et al. Reset of hippocampal–prefrontal circuitry facilitates learning. Nature 591, 615–619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA & Gordon JA Impaired hippocampal–prefrontal synchrony in a genetic mouse model of schizophrenia. Nature 464, 763–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ferenczi EA et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science 10.1126/science.aac9698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mimica B, Dunn BA, Tombaz T, Bojja VPTNCS & Whitlock JR Efficient cortical coding of 3D posture in freely behaving rats. Science 362, 584–589 (2018). [DOI] [PubMed] [Google Scholar]
- 251.Cramer JV et al. In vivo widefield calcium imaging of the mouse cortex for analysis of network connectivity in health and brain disease. Neuroimage 199, 570–584 (2019). [DOI] [PubMed] [Google Scholar]
- 252.Hultman R et al. Brain-wide electrical spatiotemporal dynamics encode depression vulnerability. Cell 173, 166–180.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wang X et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 7, 11459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Buccino AP et al. SpikeInterface, a unified framework for spike sorting. eLife 9, e61834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Chung JE et al. A fully automated approach to spike sorting. Neuron 95, 1381–1394.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lee J et al. YASS: yet another spike sorter. Preprint at bioRxiv 10.1101/151928 (2017). [DOI] [Google Scholar]
- 257.Magland J et al. SpikeForest, reproducible web-facing ground-truth validation of automated neural spike sorters. eLife 9, e55167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yger P et al. A spike sorting toolbox for up to thousands of electrodes validated with ground truth recordings in vitro and in vivo. eLife 7, e34518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hazan L, Zugaro M & Buzsaki G Klusters, NeuroScope, NDManager: a free software suite for neurophysiological data processing and visualization. J. Neurosci. Methods 155, 207–216 (2006). [DOI] [PubMed] [Google Scholar]
- 260.Giovannucci A et al. CalmAn an open source tool for scalable calcium imaging data analysis. eLife 8, e38173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mukamel EA, Nimmerjahn A & Schnitzer MJ Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pachitariu M et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Preprint at bioRxiv 10.1101/061507 (2017). [DOI] [Google Scholar]
- 263.Greenberg DS Accurate action potential inference from a calcium sensor protein through biophysical modeling. Preprint at bioRxiv 10.1101/479055 (2018). [DOI] [Google Scholar]
- 264.Pnevmatikakis EA, Merel J, Pakman A & Paninski L Bayesian spike inference from calcium imaging data. in Signals, Systems and Computers, 2013 Asilomar Conference on 349–353 (lEEE, 2013). [Google Scholar]
- 265.Vogelstein J et al. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol. 104, 3691–3704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Zhou P et al. EASE: EM-assisted source extraction from calcium imaging data. Preprint at bioRxiv 10.1101/2020.03.25.007468 (2020). [DOI] [Google Scholar]
- 267.Berens P et al. Community-based benchmarking improves spike rate inference from two-photon calcium imaging data. PLoS Comput. Biol. 14, e1006157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rupprecht P et al. A database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. Nat. Neurosci. 24, 1324–1337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Song A, Gauthier JL, Pillow JW, Tank DW & Charles AS Neural anatomy and optical microscopy (NAOMi) simulation for evaluating calcium imaging methods. J. Neurosci. Methods 358, 109173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhou P et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lu J et al. MIN1PIPE: a miniscope 1-photon-based calcium imaging signal extraction pipeline. Cell Rep. 23, 3673–3684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Friedrich J, Giovannucci A & Pnevmatikakis EA Online analysis of microendoscopic 1-photon calcium imaging data streams. PLoS Comput. Biol. 17, e1008565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cardin JA et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2. Nat. Protoc. 5, 247–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kim K et al. Artifact-free and high-temporal-resolution in vivo opto-electrophysiology with microLED optoelectrodes. Nat. Commun. 11, 2063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen R et al. Deep brain optogenetics without intracranial surgery. Nat. Biotechnol. 39, 161–164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kishi KE et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689.e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Inoue M et al. Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell 177, 1346–1360.e24 (2019). [DOI] [PubMed] [Google Scholar]


