Abstract
Autophagy is a highly conserved process that utilizes lysosomes to selectively degrade a variety of intracellular cargo, thus providing quality control over cellular components and maintaining cellular regulatory functions. Autophagy is triggered by multiple stimuli ranging from nutrient starvation to microbial infection. Autophagy extensively shapes and modulates the inflammatory response, the concerted action of immune cells, and secreted mediators aimed to eradicate a microbial infection or to heal sterile tissue damage. Here, we first review how autophagy affects innate immune signaling, cell-autonomous immune defense, and adaptive immunity. Then, we discuss the role of non-canonical autophagy in microbial infections and inflammation. Finally, we review how crosstalk between autophagy and inflammation influences infectious, metabolic, and autoimmune disorders.
Introduction
Autophagy: A process that maintains intracellular homeostasis
Macroautophagy (henceforth referred to as autophagy) is a highly conserved self-degradative process that regulates several vital processes in the cell. Autophagy starts with the formation of the autophagosome, a double membrane-bound structure that sequesters cytoplasmic material and delivers it to lysosomes for degradation 1, 2. The molecular details of this process have been comprehensively reviewed elsewhere3. Once the cellular contents are degraded, energy is released that can buoy cell survival at critical times of nutrient stress. However, autophagy also plays several key housekeeping roles by clearing damaged organelles, aggregated proteins, and pathogens from cells 1, 2, 4. Importantly, autophagy plays a significant role in the regulation of multiple innate and adaptive immune responses, as we review here.
Inflammation: the double-edged sword
Inflammation is the body’s immune response to harmful stimuli, such as pathogens, toxic substances, or physical/physiological stress. Many regulatory pathways are integrated to help mount a balanced and calculated inflammatory response against each stimulus 5. Once the stimuli are suppressed, in most cases the inflammatory response is resolved by a set of regulatory feedback mechanisms. However, genetic defects in mechanisms of resolution can cause acute inflammation to become chronic. This results in tissue and organ damage and can eventually lead to chronic inflammatory disorders 5.
The tissue damage and microbial infection are sensed by pattern-recognition receptors (PRRs) that activate cascades of events resulting in cytokine responses, which are important for the recruitment of adaptive immune cells to the site of injury or infection 6. The integrated and coordinated program of innate and adaptive immune response determines the fate of the threat 6. The four major PRR families include Toll-like receptors (TLRs), NOD-like receptors (NLRs), Retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), and C-type lectin receptors (CLRs). These PRRs sense many different types of ligands and initiate several distinct programs of inflammatory cytokine responses.
Recent evidence suggests that autophagy is a key system induced by inflammatory stimuli to modulate inflammatory responses. Indirectly, autophagy suppresses inflammation by targeting the source of inflammation such as microbes and directly degrades innate immune signaling proteins (inflammophagy) keeping inflammation in check 7. Here, we have focused on the crosstalk that exists between autophagy and immune signaling pathways.
1. Autophagy, Innate immune sensors, and cell death
A. Crosstalk: TLRs and autophagy
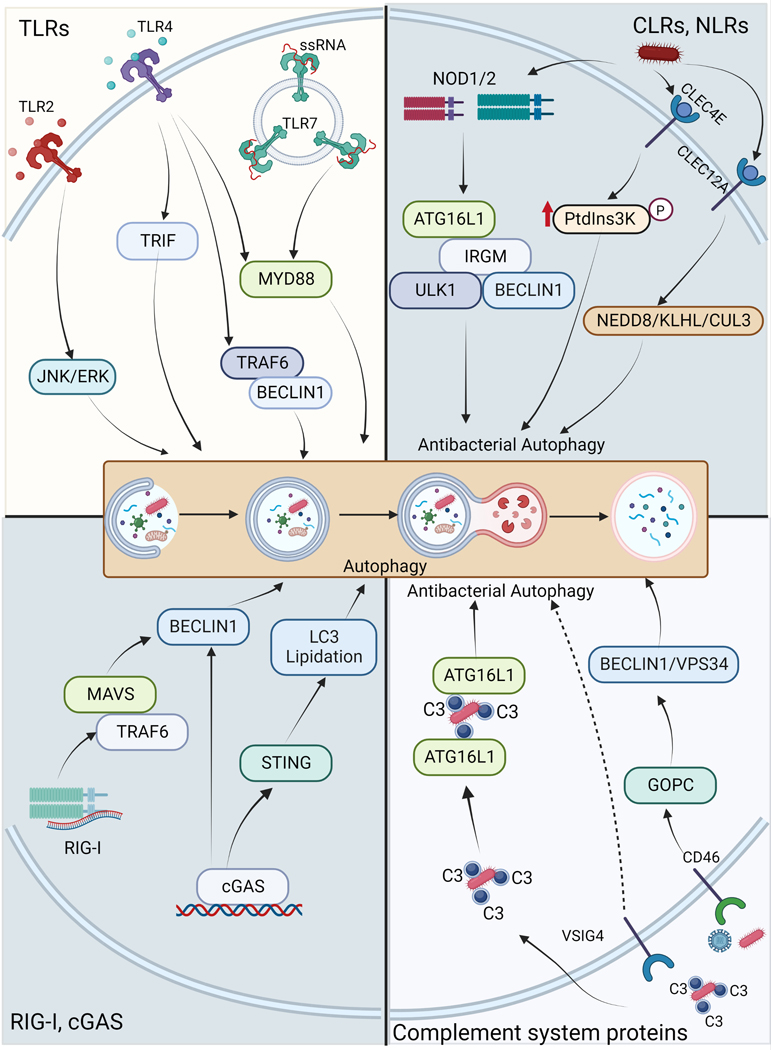
TLRs are germline-encoded receptors that can sense pathogens through the detection of pathogen-associated molecular patterns (PAMPs) that are conserved among large classes of microbes. TLRs localize in the plasma membrane or within endocytic vesicles to allow for the detection of a broad range of lipids, proteins, lipoproteins, and nucleic acids of microbial origin. TLR-associated signaling relies on the recruitment of the adaptor protein myeloid differentiation primary response 88 (MyD88) and/or in some cases, Toll/interleukin-1 receptor domain-containing adaptor inducing interferon-β (TRIF). Downstream events triggered by TLR membrane signaling pathways include activation of the NF-κB, p38 MAPK, JNK, ERK, and IRF pathways culminating in the production of proinflammatory cytokines and type I interferons. Microbes detection by TLR enhances autophagy that greatly contributes to the associated cellular response to infection8 (Figure 1). TLR7 induces LC3 lipidation upon exposure to synthetic single-stranded RNA or during infection with either vesicular stomatitis virus (VSV) or human immunodeficiency virus (HIV) 9, 10. TLR2/1 heterodimer formation enhances autophagy through modulation of the vitamin D receptor signaling during macrophage infection by mycobacteria 11. The signaling associated with bacterial sensing by TLR2 can activate autophagy in macrophages by activating the ERK and JNK pathways 12–14 (Figure 1). TLR4 engagement by LPS activates autophagy by mobilizing signaling pathways associated with both MyD88 and TRIF adaptors 15, 16 (Figure 1). TLR4-TRIF mobilization during Salmonella infection can induce TRAF3-dependent activation of TANK-binding kinase (TBK)1, leading to optimized engagement of optineurin in antibacterial autophagy17. In macrophages, TLR4-induced autophagy involves the modulation of BECLIN1 via ubiquitination 18 (Figure 1). Autophagy activation through BECLIN1 regulation also occurs in the presence of TLR2/4 agonists that induce plasminogen activator inhibitor-2 19. Such regulation often attenuates the BECLIN1-BCL2 interaction that negatively regulates autophagy at a steady state. TLR-signaling induced upon mycobacterial infection can also promote autophagosome formation and maturation through induction of the autophagy modulator DRAM1 20 indicating that TLR engagement can modulate the autophagy flux at distinct steps. At the moment, the mechanisms of autophagy enhancement associated with TLR engagement are incompletely understood and deserve further analysis. What is clear is that multiple TLRs can activate autophagy in various cell types and that the response can be cell-type specific. For instance, unlike in macrophages, TLR7 engagement induces very poor autophagy activation in plasmacytoid dendritic cells (pDC). Because prolonged inflammatory responses can be detrimental to the host, mechanisms exist to dampen such responses when pathogens are being cleared and autophagy contributes to this resolution. Autophagy can thus target components of TLR-associated signaling pathways for degradation 21–24. For example, depending on the context, the TRIF adaptor can be targeted by the autophagy receptors NDP52, TAX1BP1, or p62/SQSTM1 for lysosomal degradation subsequently to TLR3 and/or TLR4 engagement (Figure 2). Such regulatory selective autophagy can involve the participation of factors as varied as TRAF6 [tumor necrosis factor (TNF) receptor-associated factor 6], IRGM [Immunity related GTPase clade M], TRIM32, or ATG16L1 for its completion 21–23, 25 (Figure 2). Thus, TLR signaling and autophagy cross-regulate each other to finetune inflammatory and cell-autonomous immune responses to invaders.
Figure 1. Innate immune signaling pathways activate autophagy.
Upon stimulation with PAMPs, DAMPs, and microbes, several PRRs of the different family including TLRs, CLRs, NLRs, RLRs, and cGAS induces autophagy for anti-microbial and homeostatic functions. The pathways are described in text in detail. Illustrations are made using biorender software.
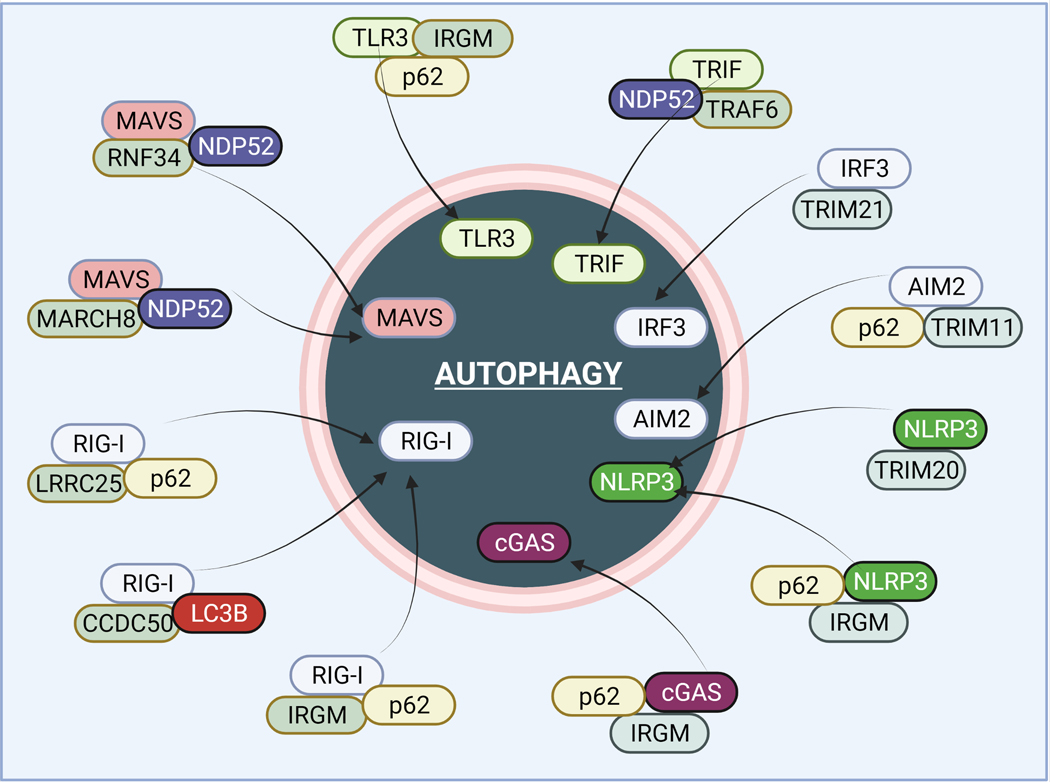
Figure 2. Autophagy degrades several innate immune sensors and intermediary proteins to suppress inflammation.
Upon stimulations, autophagy utilizes different molecular mechanisms to degrade PRR pathways sensors and other vital proteins. Autophagy proteins such as p62/SQSTM1, NDP52, IRGM, and TRIM20/21 play a vital role in the delivery of these inflammatory proteins to autophagosomes for degradation. Several E3 ligases such as RNF34 and MARCH8 play a vital role in the ubiquitination of inflammatory proteins targeted for degradation. The mechanisms are detailed in the text. The illustrations are made using biorender software.
B. C-Type Lectin Receptors (CLRs) and autophagy.
CLRs are pattern recognition receptors expressed predominantly by myeloid cells that participate in immune responses through the sensing of pathogen-associated ligands. Some studies indicate that CLRs can effectively modulate anti-microbial autophagy both positively and negatively. For instance, during Salmonella infection of epithelial and immune cells, CLEC12A can selectively promote autophagy by modulating the activity of the NEDD8-KLHL13-CUL3-KLHL9 E3 ligase complex 26 (Figure 1). CLEC4E can favor antibacterial autophagy as well. In macrophages exposed to Mycobacterium tuberculosis, CLEC4E engagement can synergize with TLR4-associated signaling to support antibacterial autophagy by promoting PtdIns3K phosphorylation27 (Figure 1), whereas CLEC4E in neutrophils is required for the autophagic activity needed to assist the formation of extracellular traps in response to Klebsiella pneumoniae 28. On the other hand, CLRs can also negatively influence anti-microbial autophagy. In macrophages, Dectin-1, a receptor for β-glucan, can down-regulate autophagy in an NF-κB-dependent manner 29. Engagement of DC-SIGN by Kaposi’s sarcoma-associated herpes virus (KSHV) blocks autophagy in dendritic cells due to the stabilized activation of signal transducer and activator of transcription 3 (STAT3) 30. DC-SIGN engagement also negatively regulates the autophagic response of dendritic cells exposed to Porphyromonas gingivalis, which is promoted by TLR2-induced signaling 31.
C. Crosstalk between NLRs and autophagy
Many essential intracellular PRRs belong to the family of NLR proteins, which are classified as proteins with a tripartite domain organization, a hallmark being a centrally localized NACHT domain that serves for oligomerization. The interaction between NLRs and autophagy is complex, and ambivalent, as the activation of certain NLRs by pathogens can induce selective autophagy of invading microbes, while autophagy also acts as a control mechanism both for NLR activation and for the outcome of associated signaling.
C.1. NLR-induced autophagy for debugging
NOD1 and NOD2 were the first NLR-proteins to be identified as intracellular PRRs that activate NF-κB and MAPK pathways by interacting with the kinase RIPK2. They are sensors for bacterial peptidoglycan 32 but more recently have also been shown to respond to cell stress 33, 34, manipulation of F-actin dynamics 35, 36, and viruses 37–39. The first link between NLRs and autophagy was the observation of selective xenophagy of bacterial pathogens following activation of NOD1 and NOD2. The NOD2 ligand muramyl dipeptide (MDP) can induce autophagy by ATG16L1 activation in epithelial and dendritic cells leading to restriction of Salmonella growth 40, 41 (Figure 1). Of note, polymorphisms in both NOD2 and ATG16L1 are associated with Crohn’s disease, a severe inflammatory bowel disease. One possible etiologic mechanism is a reduced anti-bacterial function of NOD2. Crohn’s disease-associated mutations in NOD2 and ATG16L1 render these proteins inactive for autophagy induction 42. Functionally, NOD1 and NOD2 bind ATG16L1 41 and recruit another Crohn’s disease risk factor IRGM that can act as a scaffold to induce autophagosome formation by recruiting core autophagy proteins 42 (Figure 1). In dendritic cells, this NOD2/ATG16L1 dependent autophagy also contributes to MHC class II antigen presentation 43. Induction of autophagy downstream of NOD1/2 is dependent on RIPK2 and its tyrosine kinase activity 40, albeit recruitment of ATG16L1 to membranes by NOD1/2 was reported to be RIPK2 independent in some cell lines 41. Besides this effector function that enhances the anti-bacterial response induced by NOD1/2, autophagy can also affect the NOD1/2-induced proinflammatory response, although the molecular details of this pathway are not well understood. For example, inhibition of ATG16L1 in cell lines can reduce NOD2-induced NF-κB activation 44 but ATG16L1 was also reported to suppress NOD1/2-mediated inflammatory cytokine release by interfering with RIPK2 ubiquitylation 45. This suggests a complex regulatory network of autophagy and NLR innate immune pathway regulation and future research is needed to clarify these differences.
The induction of selective autophagy by NLR proteins is not limited to bacterially induced NOD1/2 activation. Influenza A virus infection and cytosolic delivery of the dsRNA mimetic poly(I:C) also can activate NOD2 and RIPK2 dependent mitophagy via Ulk1 activation 45. This mechanism may reduce immunopathology upon influenza A infection by dampening activation of the NLRP3 inflammasome 45. This suggests a functional link between autophagy and the suppression of immunopathology. NAIP proteins, that were originally identified as susceptibility genes for Legionella infection, can target Legionella pneumophila containing vacuoles for autophagy 46. NAIP proteins form an inflammasome with the NLR protein NLRC4 whereby NAIPs act as sensors for flagellin and components of the bacterial type III secretion system. Functionally, the NLRC4 inflammasome can induce turnover of LC3 at L. pneumophila-containing vesicles to control bacterial growth and prevent pyroptotic cell death to optimize anti-bacterial activity 47.
C.2. Mitophagy and NLR activation
Mitochondrial damage induced by pathogens is a general cellular mechanism. The NLRP3 and NLRC4 inflammasomes can sense this type of danger, resulting in the release of the key inflammatory mediators IL-1β, and IL-18 and the induction of pyroptosis (see below). NLRP3 and NLRC4 activation in macrophages is partly driven by mitochondrial damage and the release of mitochondrial DNA (mtDNA). Autophagy controls the magnitude of this response by sequestering mitochondria and mtDNA. Accordingly, LC3 deficiency leads to enhanced NLRP3-dependent LPS mortality in mice 48 and intracellular Pseudomonas aeruginosa infection activates NLRC4 inflammasomes by the cytosolic release of mtDNA, which is dampened by autophagy 49. Of all human NLRs, only NLRX1 is predominantly localized at mitochondria and has several roles in the control of inflammatory and interferon responses as well as autophagy 50. NLRX1 contains an LC3-interacting region (LIR) and can direct non-canonical, LC3-associated phagocytosis (LAP) and can mediate mitophagy via this process 51 and contributes to LAP upon fungal infection with Histoplasma capsulatum 52 and bacterial infections with Listeria monocytogenes 51. NLRX1 can also downregulate type I interferons by induction of autophagy during viral infection by interaction with Tu translational elongation factor mitochondrial (TUFM) 53. Of note, NLRX1-mediated autophagosomal degradation of interferon pathway components including STING can contribute to immune evasion of papillomavirus transduced cancer cells54. By contrast, the interaction of NLRX1 with BECLIN1 was associated with negative regulation of autophagy of group A streptococci 55.
D. The connections between autophagy and inflammasome activity.
The recognition of PAMPs by PRRs bearing cells such as macrophages triggers the innate immune system including inflammasomes. Inflammasomes are cytosolic signaling complexes that induce inflammation and can activate pyroptotic cell death. They consist of an NLR protein, the adaptor ASC and recruit pro-caspase 1 for autocatalytic activation 56, 57. The most extensively investigated canonical inflammasome uses NLRP3 as a sensor. NLRP3 serves to recognize PAMPs and host-derived damage-associated molecular pattern molecules (DAMPs). NLRP3 inflammasomes require two signals for activation, a priming signal that depends upon NF-κB, and a second signal provided by the inflammasome activator that triggers the assembly of NLRP3 with ASC, and caspase-1. This leads to the cleavage of pro-IL-1β and the release of mature IL-1β and IL-18. Crystal and cryo-EM structures of NLRC4 and a cryo-EM structure of the NLRP3-NEK7 complex have provided insights into the mechanism of inflammasome activation 58–61. Non-canonical inflammasomes depend on caspase-11 (in mice) or caspase-4/5 (in humans) 62. These inflammatory caspases sense intracellular LPS derived from Gram-negative bacteria and together with caspase-1 cleave the pore-forming protein gasdermin-D (GSDMD), which is needed for the cellular release of IL-1 cytokines but also permeabilizes the cell membrane and can trigger pyroptosis. GSDMD pore formation alters ion fluxes and can trigger NLRP3 inflammasome activation. Several recent reviews describe the molecular mechanisms underlying classical and non-classical inflammasome activation 63, 64, which is not dicussed here.
Autophagy and the innate immune system are functionally intertwined. The finding that mouse macrophages lacking an essential autophagy component, ATG16L1, produced excessive amounts of the inflammasome-derived cytokines IL-1β and IL-18 following challenge provided early evidence for this connection 65. Loss of ATG16L1 in hematopoietic cells also rendered mice highly susceptible to dextran sulfate sodium-induced acute colitis, an animal model of inflammatory bowel disease 65. Upon activation, NLRP3 gets ubiquitinated and can recruit the autophagic adaptor p62/SQSTM1, leading to autophagic degradation of the inflammasome, which might be a central mechanism to end inflammasome activation in some cells 66(Figure 2). Subsequent studies have revealed multiple interactions between the pathways that lead to autophagy and those that direct inflammasome assembly. A major mechanism is the autophagic removal of endogenous or exogenous inflammasome activators, thereby reducing the triggers for subsequent inflammasome activity. For example, the ubiquitin (Ub) sensor p62/SQSTM1 binds poly-Ub chains and can direct the removal of damaged mitochondria, which reduces NLRP3 inflammasome activation 67. If autophagy is impaired, damaged mitochondria accumulate, which increases ROS production, mitochondrial DNA release, and subsequent NLRP3 inflammasome assembly 68. The autophagy regulatory protein TRIM20 interacts with CASP1, NLRP1, and NLRP3 targeting them for degradation, hence preventing excessive IL1β- and IL18-mediated inflammation 69(Figure 2). TRIM20 is encoded by the MEFV gene, a known risk locus for the autoinflammatory disease Familial Mediterranean Fever. The autophagy regulatory protein IRGM, which is upregulated in response to PAMPs and DAMPs, suppresses NLRP3 inflammasome activation by inhibiting its assembly. IRGM also mediates selective autophagic degradation of NLRP3 and ASC 70, 71(Figure 2). Supporting the human studies, a mouse ortholog of IRGM (Irgm1) limits gut inflammation in a mouse model of Crohn’s disease 70. TLR4 stimulation of macrophages induces autophagy sequestering pro-IL-1β into autophagosomes leading to its degradation72. AIM2 inflammasomes can also be degraded by ubiquitin-dependent autophagy 73 (Figure 2).
Several studies describe signaling events that regulate both inflammasomes and autophagy. The protein kinase WNK1 inhibits autophagy and also limits IL-1β production following NLRP3 inflammasome stimulation 74. Depletion of WNK1 stimulates class III phosphatidylinositol 3-kinase complex (PI3KC3) activity, which induces autophagy 75, however, a signaling pathway initiated by WNK1 balances intracellular ion concentrations during NLRP3 activation. In macrophages its absence causes intracellular K+ and Cl- levels to excessively decline, which augments macrophage IL-1β production 74. Stimulation of the PI3K/AKT/mTOR pathway in macrophages, which often occurs following pathogen encounters, can limit both autophagy and inflammasome activation 76. Upon activation of this pathway, mTORC1 inhibits ULK1, which phosphorylates the autophagy initiation machinery. AKT can also phosphorylate the N-terminus of NLRP3, limiting its oligomerization and reducing activation 77. A known risk factor for Crohn’s disease, leprosy, and certain types of cancers, autosomal dominant mutations in LRRK2 are the most common genetic cause of familial Parkinson’s disease 78. In mice, Lrrk2 deficiency reduces macrophage caspase-1 activation and IL-1β secretion in response to NLRC4 inflammasome activators. Lrrk2 deficient mice poorly clear Salmonella Typhimurium infections and they exhibit a marked impairment in selective forms of autophagy and lysosomal function, however only a minor defect in nonselective autophagy 79. Understanding the multiple regulatory mechanisms that control autophagy and inflammasomes may help design therapies to counter their manipulation by pathogens.
While initial studies largely focused on autophagy and inflammasomes in the setting of intracellular bacterial infection, the regulation and dysregulation of autophagy and inflammasomes contribute to the pathogenesis of many viral infections 80–82. Autophagy can eliminate invading viruses and foster antiviral responses; however, some enveloped viruses use autophagy-related vesicles for transit and as sites for replication. SARS-CoV-2 has a complex effect on both autophagy and inflammasomes, and their dysregulation may contribute to the pathogenesis of severe, life-threatening diseases 83–85. Several of the SARS CoV-2 encoded proteins manipulate autophagy. E protein, M protein, (open reading frame) ORF3a, and ORF7a all cause an accumulation of autophagosomes, whereas Nsp15 prevents their efficient formation. The viroporin ORF3a is a small, hydrophobic molecule that targets the host cell membrane altering their ion permeability 86. ORF3a localizes to intracellular vesicles, in the endoplasmic reticulum, and at the plasma membrane. It helps viral egress via a lysosomal exocytosis-pathway 87, 88. Conversely, it induces a specialized form of autophagy termed reticulophagy through the high mobility group box 1 (HMGB1)-BECLIN1 pathway 89. Inflammation releases nuclear HMGB1 into the cytosol and extracellular spaces where it helps sustain autophagy and functions as a DAMP. By disrupting ER homeostasis ORF3a induces ER stress and triggers inflammation. Evidence of inflammasome activation in COVID-19 patients has come from studies showing inflammasome ASC specks, active caspase-1, and cleaved GSMD in SARS-CoV-2 bearing monocytes 90. Cytokine release, immune cell recruitment, and positive feedback loops help drive the formation of the highly inflammatory milieu found in critically ill patients. Besides ORF3a the SARS-CoV-2 N protein also activates inflammasomes 91. It directly interacts with NLRP3 promoting binding to ASC and facilitating NLRP3 inflammasome assembly. These and other data indicate that SARS-CoV-2 manipulates the host’s innate immune responses and that targeting these pathways should provide avenues to attenuate viral pathogenicity.
The central role of autophagy in inflammasome regulation is well supported by the fact that some bacterial pathogens manipulate the autophagy response as a subversion strategy. The SpvC effector protein from Salmonella blocks NLRP3 and NLRC4-induced autophagy 92. By contrast, VopQ from Vibrio parahaemolyticus induces autophagy to dampen NLRC4 inflammasome activation 93. Shigella flexneri which activates the NLRC4 inflammasome in macrophages can also induce autophagy to reduce IL-1β release and pyroptotic cell death 94 and Mycobacterium tuberculosis-induced IL-1β release is also negatively controlled by autophagy 95.
The autophagy-dependent regulation of inflammasome responses might be of use for therapeutic intervention for the treatment of hereditary disorders associated with mutations in NLRs. Rapamycin for example was shown to reduce NLRC4-mediated IL-1 cytokine production in myeloid cells expressing a disease-associated hypermorph of NLRC4 96. Future research will help to delineate the whole picture of NLR-autophagy crosstalk. It already emerges that besides NOD1/2, NLRP3 and NLRC4 also other, less well-studied, NLR proteins are linked to autophagy as well. The MHC class I regulator NLRC5, for example, was shown to be degraded by LC3 autophagy in endometrial cancer cells, leading to loss of MHC class I expression that was associated with worse clinical outcomes 97. Besides the NLR discussed above, NLRC4, NLRP3, NLRP4, and NLRP10 were also shown to interact with BECLIN1 and for NLRP4 it was shown that recruitment to Streptococci containing phagosomes leads to the release of BECLIN1 resulting in the local induction of autophagy 98. In the gut, NLRP6 regulates autophagy in goblet cells to control mucus production 99. Taken together, this suggests the potential of NLR proteins as targets to tailor inflammophagy responses and in line with the recent characterization of specific inhibitors for RIPK2 and NLRP3 has clinical potential.
E. Interplay between RLRs and Autophagy
Another family of intracellular PRRs is the RIG-I-like receptors (RLRs). The three ubiquitously expressed RLR members are RIG-I, MDA5, and LGP2 in contrast to the NLRs described above mainly act in anti-viral responses. Upon activation via cytoplasmic viral RNA or processed self RNA, RLRs initiate a series of events that activate interferon regulatory factors (IRFs) 100, 101. IRFs are the key transcription regulators of Interferons (mainly type I and type III IFNs).
RIG-I and MDA5 are the most studied members of the RLR family. Upon sensing PAMPs, RIG-I oligomerizes and interacts with adaptor protein mitochondrial antiviral signaling (MAVS) on the mitochondria. MAVS oligomerizes to form large aggregates and associates with multiple adaptor proteins including TRAF2, TRAF3 TRAF6, TANK, and TRADD to activate Tank-binding kinase-1 (TBK1) and I kappa B kinase epsilon (IKKε) 100, 101. The IRFs (IRF3/7) are phosphorylated by activated TBK1 and IKKε resulting in their homo- or heterodimerization and translocation to the nucleus to promote the expression of IFNs. The secreted interferons are sensed by interferon receptors leading to the activation of JAK-STAT1/2 pathways resulting in the activation of a large number of interferon-stimulated genes (ISGs) that have a myriad of functions in innate immunity 100, 101.
The interplay between RIG-I signaling and autophagy has been documented, however, mechanistically, how RIG-I signaling participates in autophagy modulation is not very clear. On the other hand, how autophagy suppresses RIG-I-MAVS signaling is well understood. The activation of RIG-I-MAVS-TRAF6 signaling by PAMPs and virus induces the interaction between TRAF6 and BECLIN1 resulting in increased K63-linked ubiquitination of BECLIN1 102, which is an important event in autophagy upregulation (Figure 1).
Several autophagy-dependent mechanisms targets RIG-I, MDA5, and MAVS for degradation and suppressing inflammation. In the absence of autophagy, dysfunctional mitochondria accumulate leading to the production of high levels of mtROS that likely activates RLR signaling leading to IFN production 103. The Atg5–Atg12 conjugate can directly interact with the CARD domains of MAVS and RIG-I to suppress the production of type I IFN’s 104. Upon viral cellular invasion, the leucine-rich repeat-containing protein 25 (LRRC25) interacts with activated ISG15-tagged RIG-I and enhances its interaction with autophagy receptor p62/SQSTM1 105. The ISG15-tagged RIG-I is delivered to the autophagosome via p62/SQSTM1 for degradation suppressing type I IFN signaling (Figure 2). Recently, the Coiled-Coil Domain Containing 50 (CCDC50) protein was found to be a new receptor for autophagic degradation of K63-polyubiquitinated RIG-I/MDA5 106(Figure 2). CCDC50 directly interacts with LC3 protein on autophagosome membranes to deliver ubiquitinated RIG-I/MDA5, thereby suppressing the type 1 IFN response. CCDC50 deficient mice exhibited a reduced autophagic degradation of RIG-I/MDA5, a heightened type I IFN response, and enhanced viral resistance106.
MAVS is targeted by several proteins including Tetherin, RNF34, and HFE for autophagic degradation107–109(Figure 2). Tetherin, an ISG and anti-viral protein that recruits MARCH8 to enhance K27-linked ubiquitination of MAVS, is recognized by autophagy receptor protein NDP52107. NDP52 mediates autophagic degradation of MAVS and suppression of type I IFN response. Similarly, RNF34, a ring finger domain-containing E3-ligase, interacts and catalyzes the K27 and K29-linked ubiquitination of MAVS for NDP52-dependent autophagy resulting in reduced type I IFN responses 108.
Several studies have shown genetic and functional linkage of IRGM protein with type 1 interferonopathies and autoimmune diseases 25, 110, 111. The mechanism remained unclear until recently when IRGM was shown to directly interact with RIG-I and MAVS to mediate RIG-I degradation via p62/SQSTM1-dependent selective autophagy (Figure 2). IRGM strongly suppresses type 1 IFN response and reducing its expression induces more than 100 ISGs 25. Similar results were obtained in Irgm1-deficient mice. Downregulation of IRGM/Irgm1 in humans and mice causes mitophagy defects25, 110. The resulting accumulation of defunct mitochondria increased mtROS, mtRNA, and mtDNA fueled RIG-I-MAVS and cGAS-STING signaling for the production of IFN response. These studies delineated the mechanisms by which IRGM protects against type 1 interferonopathies and autoinflammatory diseases. On the flip side, since IRGM is a master negative regulator of the interferon response, its depletion could suppress the replication of a large number of different RNA/DNA viruses112.
Thus, autophagy-dependent mechanisms play a vital role in the suppression of RLRs mediated cytokine response and hence maintain innate immune balance during microbial infection or sterile damage.
F. cGAS-STING signaling and autophagy
Cyclic GMP-AMP synthase (cGAS) senses cytosolic dsDNA to trigger activation of STING (TMEM173) protein to induce both type I interferon and NF-κB responses 113. Extensive functional interactions have been documented between autophagy and the cGAS-STING pathway. Stimulation of the cGAS-STING pathway not only induces type 1 IFN response but also enhances autophagy by several mechanisms to suppress IFN response and thus maintain immune homeostasis. For example, cGAS interaction with autophagy protein BECLIN1 results in reduced cGAMP synthesis and enhanced autophagy-dependent degradation of dsDNA causing suppression of the IFN response 114. Genotoxic stress results in increased micronuclei in the cytosol that are suggested to be sensed by cGAS and induce interferon response. Recently, it was shown that cGAS also acts as a receptor for micronuclei and could target them for autophagy by directly interacting with LC3B on autophagosomes 115. By doing so, cGAS maintains the balance of the IFN response induced against micronuclei.
STING can induce LC3 lipidation and upregulation of autophagy (Figure 1). The cGAMP-induced STING-containing ERGIC (Endoplasmic reticulum–Golgi intermediate compartment) provides a membrane source for LC3 lipidation and autophagosome biogenesis 116. The cGAMP-induced STING-dependent autophagy could clear DNA and viruses from the cytoplasm, possibly to reduces the source of inflammation. Another study confirmed STING-induced LC3 lipidation but reported that LC3 lipidation occurs at single membrane perinuclear vesicles and was mediated by v-ATPase and ATG16L1 117.
cGAS and STING themselves are degraded by p62/SQSTM1-dependent autophagy to maintain the homeostatic balance of the IFN response 25, 118(Figure 2). Interestingly, STING-activated TBK1 can phosphorylate both IRF3 and p62/SQSTM1. Where activated IRF3 induces type I IFN production, phosphorylated p62/SQSTM1 interacts with STING to degrade it and suppress type I IFN response 118. IRGM interacts with cGAS to enhance p62/SQSTM1-dependent autophagic degradation resulting in suppression of type I interferon response 25. Altogether, the cGAS-STING pathway like NLR, TLR, and RLR signaling can induce autophagy and in most cases, this autophagy can suppress inflammation in a feedback loop.
G. Complement-mediated modulation of autophagy.
Complement is a tightly regulated system that critically contributes to the humoral innate immune defense against pathogens promotes both local inflammation, phagocytosis, and lysis of invading microbes. Several components of the complement system can be involved in anti-microbial autophagy (Figure 1). Attenuated measles virus (MeV) infects human cells through CD46, a regulatory factor that prevents complement-mediated cell lysis. MeV entry rapidly activates autophagy through the CD46-Cyt-1 intracytoplasmic splice variant that recruits the scaffold protein GOPC, an interactor of the VPS34-BECLIN1 complex119. This early autophagy resolves spontaneously prior to a second and sustained autophagic phase that depends on virus replication and benefits from it120. The CD46-Cyt-1-GOPC-VPS34-BECN1 axis also activates autophagy during infection by Group A Streptococcus (GAS) 119 and Neisseria gonorrhoeae 121, both bind to CD46. Because CD46 also serves as a receptor for other viruses (HHV-6, BVDV pestivirus, and adenoviruses B/D), this axis is likely activates autophagy upon infection by additional pathogens. Thus, the complement regulatory factor CD46 acts as a pathogen sensor able to initiate autophagy upon infection (Figure 1). V-set and immunoglobulin domain containing 4 (VSIG4) is a surface receptor expressed on phagocytic antigen-presenting cells and that functions as a receptor for the phagocytosis of bacteria opsonized with the complement factor C3. VSIG4 engagement by C3b-coated Listeria monocytogenes promotes the ubiquitination and targeting of cytosolic bacteria to autophagic degradation in both macrophage-like cells and primary macrophages 122. Hence, by engaging the VSIG4 receptor, C3b deposited on L. monocytogenes induces an antibacterial autophagic response in professional phagocytic cells. C3b deposited on L. monocytogenes or adherent-invasive Escherichia coli (AIEC), can also activate anti-microbial autophagy in epithelial cells after reaching the cytosol 123 (Figure 1). This capacity relies on the direct interaction of C3 with ATG16L1 and is altered in cells lacking ATG16L1 or carrying its T300A variant that is associated with Crohn’s disease. The strong anti-microbial potential of this effect is best illustrated by the fact that some bacteria evolved strategies to counteract the C3-ATG16L1 interaction124. For instance, opsonized Shigella flexneri and S. Typhimurium can attenuate autophagy restriction through the shedding of adsorbed C3 soon after infection by engaging proteases of the omptin family124. These studies thus indicate that complement factors can regulate the autophagic response to intracellular microbes through both surface and cytosolic interactions.
2. Interferon-inducible GTPases (IRGs) and autophagy
The Immunity-Related GTPases (IRG; also known as p47 GTPases) and the Guanylate Binding Proteins (GBP, also known as p65 GTPases) are related families of GTPases whose expression is dramatically induced by type I (IFN α/β) and type II (IFNγ) interferons 125, 126. The genes encoding these GTPases were cloned beginning in the late 1990’s as a consequence of genetic searches for IFN-induced genes. The existence of GBP proteins was actually known earlier, as their high levels in cell lysates from IFN-induced cells result in intense, diagnostic spots on two-dimensional protein gels. IRG genes are abundant in rodents with 23 family members found in C57BL/6 mice 127. In contrast, IRGs are more restricted in humans where there are two members - IRGC that is constitutively expressed only in the testis, and IRGM that is more widely expressed but truncated relative to mouse IRGs 127. Genes encoding IRGs are present in small numbers in other species such as dogs, fish etc., 127, 128. These patterns suggest that IRGs have proliferated in rodents under evolutionary pressure from endemic pathogens and play more expansive roles in rodent innate immunity, though important roles in humans exist. The GBP gene families are more equitable between rodents and humans, perhaps suggesting a higher degree of functional conservation.
IRGs/GBPs are expressed in a range of hematopoietic and non-hematopoietic cells following IFN activation. Their activities are essential for normal IFN-induced immune responses to some pathogens 129–131. IRGs and GBPs are thought to function as dynamins, a superfamily of large GTPases that bind cellular membranes and then undergo GTP hydrolysis that induces conformational changes and enables various cellular functions 132. For instance, in Irgb6 conformational changes following GTP hydrolysis allow recognition of a phospholipid binding site on the vacuolar membranes surrounding Toxoplasma gondii 133. IRGs and GBPs bind diverse membrane compartments where they are thought to carry out a collection of activities that support two major cellular functions: cell-autonomous immunity and autophagic regulation.
A. IRGs / GBPs as Regulators of Autophagy of Cell Autonomous Immunity
The first immune function assigned to mouse IRGs was the ability to kill Toxoplasma gondii and Chlamydia trachomatis in host cells (macrophages, astrocytes, and fibroblasts) that have been activated with IFN 134–139. A large family of over 20 mouse IRGs bifurcates into two types: the majority are known as GKS IRGs because they possess a canonical GKS sequence motif in the GTP-binding region, while the less common GMS or IRGM proteins possess a non-canonical GMS motif 128. The GKS IRGs are the major players in this cell-autonomous killing mechanism. They are present in the cytoplasm or on membranes in host cells, but following T. gondii or C. trachomatis infection they rapidly home to the parasitophorous vacuole (PV) that surrounds the pathogens 138, 140–142. They do so in a hierarchical order that ostensibly is required for function, with Irgb6 being a lead protein that recognizes specific phospholipids on the T. gondii PV 143 (Figure 3). Once the GKS IRG complex coats the PV, the T. gondii PV vesiculates extruding the naked pathogen into the cytoplasm, effectively stripping the parasite of its protective niche and leading to its death136, 144 (Figure 3). This vesiculation function is reminiscent of the ability of classical dynamins to contort membranes to form vesicles. The GKS-coated C. trachomatis PVs similarly undergo lytic disintegration 145, suggesting a conserved host defense mechanism executed by mouse IRGs. The mouse IRG family is highly diverse among wild species of mice and has likely been shaped by co-evolution with pathogens like T. gondii that naturally infect rodents 146. IRGM proteins play a more peripheral role in these processes, as they bind endomembranes in the cell and hold the GKS IRGs in a biochemically inactive state in those locations until they are needed to target PVs 141, 147. GMS proteins thus play a critical chaperone role and in their absence, the GKS proteins aggregate and lose their capacity to correctly target to pathogen membranes 141, 148, 149. The central role of this cell-autonomous killing mechanism for pathogen control is illustrated by the fact that T. gondii and C. muridarum, a rodent-adapted pathogen, have both evolved the capacity to circumvent IRG-mediated killing. In T. gondii, specific virulence factors (rhoptry proteins ROP5, ROP17, and ROP18) phosphorylate GKS IRGs to inactivate them150–152. This ability to neutralize IRGs, or not, determines the overall virulence category of T. gondii stains. C. muridarum can also avoid GKS-mediated killing 142 although the molecular basis for this evasion is not yet defined.
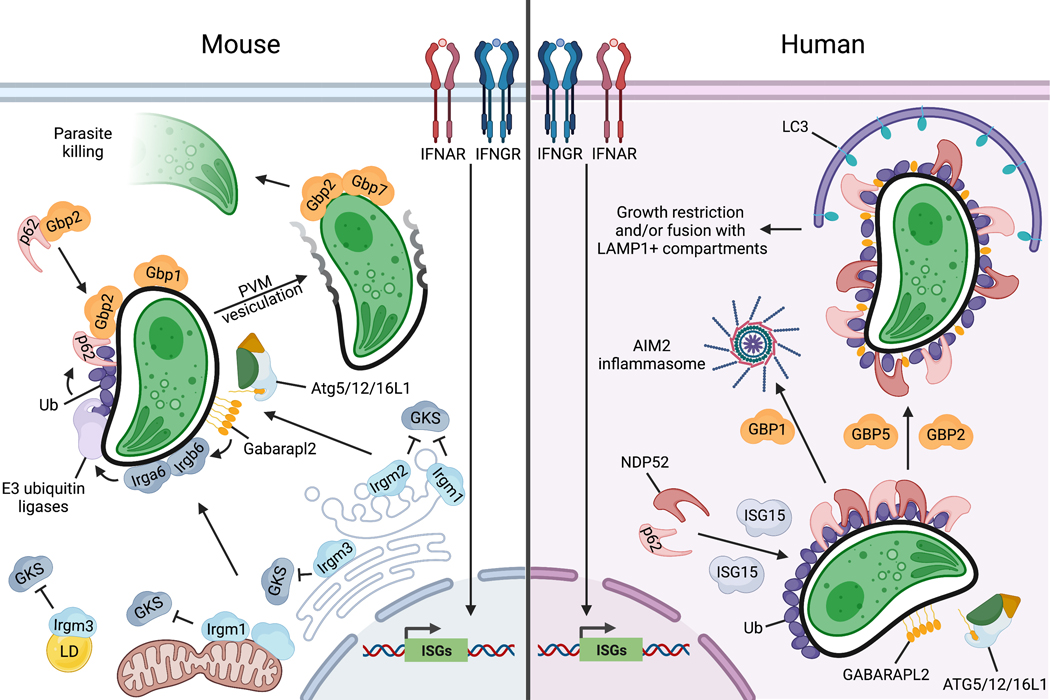
Figure 3. Mechanisms of cell-autonomous immunity to Toxoplasma in mouse and human cells.
Mouse: Activation of IFNAR or IFNGR by type I or II IFNs, respectively, induces expression of >1000 interferon-stimulated genes (ISGs), including IRGs and GBPs. Irgm proteins reside on host organelles including Golgi (Irgm1/m2), ER (Irgm3), mitochondria (Irgm1), and lipid droplets (LDs; Irgm3) and inhibit the binding and activation of cytosolic GKS proteins on those host membranes. Irgm2 promotes the conjugation of GABARAPL2 on the PVM, which promotes GKS and GBP recruitment. Irgm1 and Irgm3 coordinate the targeting of GKS proteins to the Toxoplasma PV, with Irgb6 acting as a pioneer to recruit other GKS proteins such as Irga6. GKS targeting to the PV promotes PV ubiquitination and ubiquitin-dependent recruitment of cytosolic complexes containing p62/SQSTM1 and GBP2, which in turn promotes recruitment of other GBPs including GBP1. Decoration of the PV with host effectors leads to vesiculation of the PVM, and the exposed Toxoplasma is targeted by GBPs including GBP2 and GBP7, and destroyed. Human: IFN stimulation drives the expression of ISGs including GBPs and ISG15 as well as ubiquitination of the Toxoplasma PVM. ISG15 promotes the recruitment of p62/SQSTM1 and NDP52 to ubiquitinated PVs, which facilitates association with LC3+ membrane structures. In some cell types, this leads to restriction of Toxoplasma growth and replication, and in others, the PV is delivered to LAMP1+ lysosomes for degradation. Decoration of the PVM with GABARAPL2 drives parasite clearance by an unknown mechanism. GBP1 is recruited to the Toxoplasma PV in macrophages but not A549 cells and liberates parasite DNA for activation of the AIM2 inflammasome. GBP2 and GBP5 are not recruited to the PV but contribute to the clearance of Toxoplasma.
GBPs also target the T. gondii PV and C. trachomatis inclusion. The GBP family in mouse consists of two clusters one on Chromosome (Chr) 3 (containing Gbp1. Gbp2, Gbp3, Gbp5, Gbp7, and Gbp2ps) and one on Chr 5 (containing Gbp4, Gbp6, Gbp8–11) 153. Deletion of the Chr3 cluster renders mice susceptible to infection with T. gondii 153. Like IRGs, GBPs also home to PVs in an ordered manner with Gbp1 154 and Gbp2 155 initially recruited to the PV surrounding T. gondii, while Gbp2 and Gbp7 are thought to target the parasite membrane for destruction after the vacuole has ruptured 156 (Figure 3). In contrast to IRGs, GBPs may not affect PV vesiculation of C. trachomatis PVs; rather, they are involved in inflammasome activation 145, 157. There is robust cross-regulation among the GBPs and the two IRG subfamilies: IRGM proteins can influence the recruitment of GBPs to PVs 158, and GBPs can affect the recruitment of IRGs 159, 160. Ubiquitination by the E3 ubiquitin ligase TRAF6 and the autophagy adapter protein p62/SQSTM1 are also involved in the process by recruiting GBPs to the PV 145 (Figure 3).
Although autophagy is not required for vesiculation of PVs per se, autophagy and IRGs/ GBPs do cooperate to recognize and eradicate T. gondii in a cell-autonomous manner in murine cells 161–163. This process does not require the initiation steps of autophagy (i.e. Beclin, Atg14) nor the degradative steps (lysosomal fusion), but rather relies on a core set of ATG proteins that has been referred to as non-canonical autophagy 164–166. The autophagy proteins Atg3, Atg7, and the Atg5-Atg12-Atg16L1 complex are recruited directly to the PV, perhaps by recognition of phosphatidylinositols in the PV membrane 143, 163(Figure 3). This non-canonical autophagy protein complex then conjugates LC3 homologs (Atg8 and the GABARAPs) to phosphatidylethanolamine in the membrane. Of the five LC3 orthologs in mice, the major effector is Gabarapl2, while others play lessor roles 167(Figure 3). It is thought that the recruitment of this ATG complex to the PV membrane facilitates the recruitment of IRGs/ GBPs that subsequently carry out vacuole lysis. In the absence of a core set of ATG proteins (i.e. Atg3, Atg7, Atg5-Atg12-Atg16), IRGs and GBPs become activated and aggregate in cytoplasmic clusters and are therefore unable to function in recruitment to pathogen containing vacuoles 165, 168(Figure 3). The lack of ATG proteins thus leads to an absence of IRG/ GBP homing to the PV, essentially undermining the whole cell-autonomous mechanism in mouse cells.
In contrast to murine cells where GKS IRGs play a prominent role, human cells lack this set of effectors and yet still control T. gondii through a process that relies on non-canonical autophagy and GBPs. Human cells express seven GBPs that are found in a single cluster on Chr1 169. It should be noted that in human cells autophagy proteins are dispensable for GBP delivery but essential for the recruitment of GABARAPL2 170(Figure 3), which is required for cell-autonomous restriction of T. gondii, though presumably utilizing different effector proteins 168, 170 In Hela cells, the process is initiated by ubiquitination of unknown targets on the PV followed by recruitment of adaptors p62/SQSTM1, NDP52 and finally conjugation of LC3 168. Unlike the process of xenophagy that restricts intracellular bacteria 171, control of T. gondii in human cells by noncanonical autophagy requires activation with IFN-γ. Autophagy proteins are not normally induced by IFN- γ, rather the link between these two pathways is mediated by induction of ISG15, which facilitates recruitment of the Ub binding adaptors p62/SQSTM1 and NDP52 to the PV surrounding T. gondii 172. The fate of LC3 positive vacuoles differs somewhat based on cell type: in HeLa cells engulfment in LC3 positive membranes stunts parasite growth 168, while in HUVEC cells the parasite is delivered to LAMP1+ compartments 173. In human macrophages, GBP1 performs its role following recruitment to the PV 174, although it acts at a distance in A549 human lung epithelial cells infected with the protists T. gondii or Leishmania donovani 175, 176. GBP1 also contributes to parasite restriction by inducing the AIM2 inflammasome following the release of parasite DNA into the cytosol 177(Figure 3). Additionally, GBP2 and GBP5 have been implicated in parasite control despite not being recruited to the vacuole 178, 179(Figure 3). Both the GTPase functions and lipidation are required for the activities of GBP1, GBP2, and GBP 5 in human monocytes treated with IFN- γ 178. The ability of GBPs to act at a distance contrasts with the role such effectors normally have in targeting the PV membrane or parasite within.177.
Beyond the vesiculation mechanism, IRGs and GBPs can recruit other anti-microbial factors that function on a cell-autonomous level. These factors are numerous and once recruited to vacuoles/phagosomes or bacteria, they act through diverse mechanisms. For instance, Irgm1 and Irgm3 control the translocation of multiple E3 ubiquitin ligases including TRAF6 to T. gondii and C. trachomatis vacuoles 145, 180, which then tags the vacuoles with ubiquitin to allow recognition by other cellular factors. In contrast, Irgm2 controls the targeting of Gabarapl2 to T. gondii phagosomes 181, which then modifies phagosome processing. Regarding GBPs, they have been reported to control the targeting of phagocyte oxidase, antimicrobial peptides, and autophagy effectors to membrane-bound intracellular bacteria to elicit their killing 131. Further, GBP complexes assemble on cytosolic bacteria, where they can recruit caspases that trigger inflammasome activation 182, 183. GBPs are also able to block actin tail-mediated motility of cytosolic bacteria such as Shigella 183–186. Finally, both GBPs and IRGMs modulate the sensing of cytosolic LPS 187–190. The underlying biochemical mechanisms are not yet clear that allow IRGs/ GBPs to recruit these factors; nor is it clear how these activities hinge on the core dynamin-like activity of the proteins.
B. IRGM as Regulators of Autophagy
IRG/GBP proteins regulate autophagic processes that are important for pathogen control and limiting potentially excessive inflammatory cytokine production. This is underscored by polymorphisms in the human IRGM gene that reduce IRGM expression and autophagic activity, and are associated with increases in Crohn’s Disease 191, 192, ankylosing spondylitis193, non-alcoholic fatty liver disease 194, and Mycobacterium tuberculosis infection 195, as well as poor outcomes to sepsis 196. IRGM has been shown to regulate the assembly of the core autophagic machinery 42. It does so by interacting with ULK1 and BECLIN1 to promote their assembly into autophagy initiation complexes. IRGM also interacts with NOD2, which enhances the K63-linked polyubiquitination of IRGM that is required for interactions of the protein with the autophagy complex 42. This ability to promote autophagy impacts immunity and inflammation in various ways. For instance, mouse Irgm1 and human IRGM both stimulate autophagic killing of phagosomal M. tuberculosis 197, 198 and adherent-invasive Escherichia coli 199 in macrophages. Both proteins regulate autophagic degradation of NLRP3 and ASC, which has the effect of blocking NLRP3 inflammasome activation and limiting IL-1β production 70. IRGM also targets viral replication complexes that are subsequently ubiquitinated and thus tagged for autophagic removal 200.
A particularly important aspect of the involvement of Irgm1/IRGM in autophagy may be in their ability to promote autophagic clearance of mitochondria, a process known as mitophagy. When Irgm1 and/or IRGM is absent, mitochondrial homeostasis is perturbed as reflected in an accumulation of defective mitochondria, altered mitochondrial fission/fusion states, and increased presence of intracellular mitochondrial DNA in the cytoplasm 24, 25, 201–204. These alterations in mitochondria result in changes in cellular metabolism that shape immune cell function and increase inflammation 25, 205. The mitochondrial DNA/RNA soiling of the cytoplasm has profound consequences, as it activates intracellular sensors such as cGAS/STING. RIG-I-MAVS, and TLR7 that trigger a cascade of cytokine production 24, 25, 204. This notably drives a type I ‘interferonopathy’ that significantly modulates multiple immune responses and is a key driver of disease.
3. Autophagy and adaptive immunity
A. Autophagy and MHC class II antigen presentation.
Because class II molecules of the major histocompatibility complex (MHC II) constitutively traffic through endo-lysosomal acidic compartments where they get complexed to antigenic peptides (MHC II compartments), autophagosomal cargoes that cross such compartments can influence MHC class II antigen presentation to CD4+ T lymphocytes helper cells. This happens in cell types as various as epithelial cells, melanocytes, and professional antigen-presenting cells, including B cells 206–210. As a consequence, conjugating antigens to LC3 promotes their MHC II presentation due to imposed autophagosomal targeting 210–214,215. Autophagy can promote the MHC II presentation of endogenous (self) antigens of both cytosolic and nuclear origin, including factors of the ATG8 family and autophagy receptors such as TAX1BP1 208,216. In the case of thymic epithelial cells (TECs)217, such an effect participates in the selection of the T cell receptor (TCR) repertoire for both effector and regulatory (Treg) CD4+CD8- thymocytes 211, 218, 219. Accordingly, interfering with TEC autophagy can trigger multi-organ inflammation 218, and fusing antigens to LC3 leads to the intra-thymic deletion of immature CD4+CD8- thymocytes expressing cognate TCRs 211. The role of autophagy in the antigen-presenting function of TECs appears to be regulated by the C-type lectin CLEC16A whose deficiency diminishes the autophagic activity of TECs and attenuates the positive selection of autoreactive thymocytes220. How CLEC16A positively influences autophagy in TECs remains to be dissected as it can also repress autophagy in epithelial cells through activation of the mTOR pathway221. Autophagy also contributes to the MHC II presentation of extracellular antigens, including determinants derived from viruses and bacteria 207,209 222–226. Indeed, the influence of autophagy on the MHC II presentation of microbial antigens can be so efficient that microorganisms evolved means to interfere with it 223, 227–230.
B. Autophagy and MHC I antigen presentation
MHC class I molecules are specialized in the acquisition of antigenic peptides resulting from proteasomal degradation for presentation to CD8+ T lymphocytes. This acquisition occurs within the endoplasmic reticulum (ER) after the importation of cytosolic peptides. Autophagy appears able to influence MHC I-peptide complexation when this classical MHC I pathway is perturbed for instance by viruses that interfere with peptide transport into the ER. In that context, some complexation events can take place in late endosomal compartments that receive contents from autophagosomes. This appears to be the case during infection with the herpes simplex virus (HSV) 1 or human cytomegalovirus (HCMV) for the MHC I presentation of peptides from HSVgB and pUL138 viral antigens, respectively 231, 232. However, autophagy negatively regulates the level of MHC I antigen presentation by promoting cell surface MHC I molecules internalization and lysosomal degradation and thereby, negatively modulating the presentation of viral or tumor antigens to CD8 T cells 233–235. In fact, both MHC I and non-classical MHC I (such as CD1d) molecule expression is augmented on the surface of professional antigen-presenting cells lacking ATG5 or ATG7 factors 226, 233. In this context, the adaptor-associated kinase 1 (AAK1) which regulates the activity of the AP2 complex is not properly recruited to MHC I molecules233, indicating a role for autophagy factors in some forms of endocytosis. In tumor cells, MHC I degradation can involve the autophagy receptor NBR1 that mediates ERphagy 235.
The ability of MHC I molecules to present peptides from exogenous antigens is named cross-presentation. This process, which is very efficient in some subsets of DCs, may depend on the persistence of internalized antigens in endosomes that intersect with recycling endosomes carrying MHC I molecules. In some instances, core autophagy genes may be required for such a phenomenon. Thus, Atg7 is needed for the MHC I cross-presentation of peptides from soluble ovalbumin (OVA) but not that of OVA from apoptotic cell corpses or OVA directed to the DEC205 endocytic receptor 236. In B cells, the cross-presentation of protein antigens can require both autophagy and proteasome activity 237. Cross presentation of some extracellular antigens on MHC I can be affected by the absence of VPS34238. On the donor cell side, functional autophagy can favor the cross-presentation of both viral and tumor antigens by efficiently conditioning exosomal vesicles captured by dendritic cells 239–241. In addition, a contribution of autophagy to the transfer of endocytosed antigenic material into the cytosol before connection to the classical MHC I pathway has been suggested 242, 243, although the exact modalities of the autophagy machinery contribution to exocytosis remain to be elucidated. Hence, while the autophagy machinery limits the presentation of endogenous antigens by regulating MHC I expression level, it can facilitate MHC I presentation of some forms of exogenous antigens by acting both in presenting cells and antigen donor cells.
C. Autophagy in T lymphocyte biology.
Autophagy is instrumental for T lymphocyte homeostasis both during development and their effector functions. It also has a role in immunological memory (Figure 4). Lymphocyte activation via antigen receptor (TCR) engagement triggers multiple biochemical changes that are necessary for clonal expansion and effector cell differentiation. Most notably, lymphocytes increase their glucose uptake, glycolytic activity, and glutamine metabolism to generate ATP and the metabolites required for activation and effector functions, a transition that involves contributions from autophagy. During thymic development, autophagy is important for the CD4-CD8- to CD4+CD8+ transition244 before commitment to either CD4+CD8- or CD4-CD8+ lineages and exit into the periphery (Figure 4). Mice lacking core autophagy factors, such as those involved in the initiation/conjugation steps, display a reduced pool of both immature and mature T cells due to increased cell death245–247 indicating an important role for autophagy in the homeostasis of both developing and mature T cells (Figure 4). Autophagy is also important for the development of invariant NKT cells, a specialized subset of T cells that react to microbial glycolipid in the context of CD1d248.
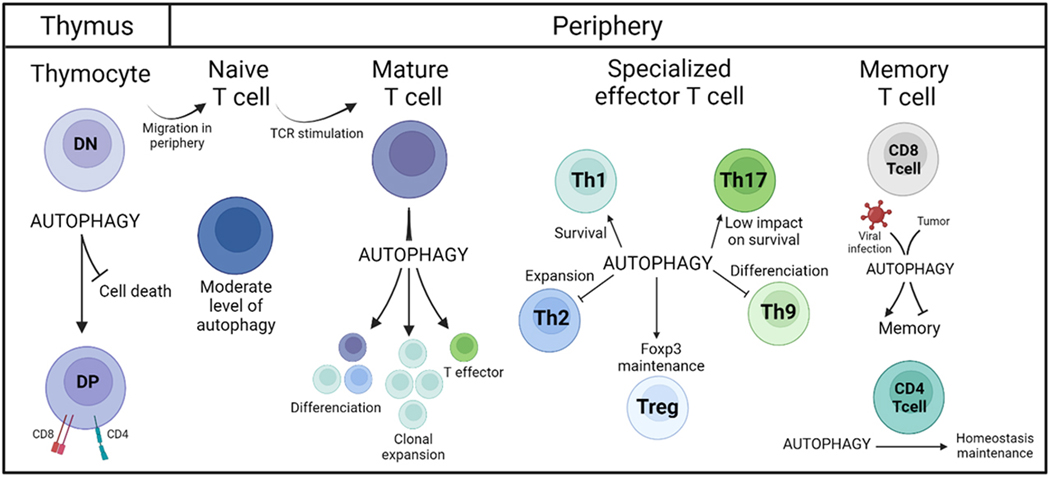
Figure 4. Autophagy in T lymphocyte biology.
Autophagy is critical for T cell homeostasis during their development and their effector functions. In thymocytes, autophagy prevents cell death and allows the transition from double-negative (DN) CD4-CD8- cells to double-positive (DP) CD4+CD8+ cells. After they migrate to the periphery, resting naïve T cells harbor a low level of autophagy. However, upon TCR engagement, autophagy is important for the differentiation, clonal expansion, and the effector function of mature T cells. Autophagy also has a role in the maintenance of memory T cells.
Naïve resting T cells possess a moderate level of constitutive autophagy and depend on oxidative phosphorylation for their energetic needs. Upon T cell activation, the overall autophagy flux is enhanced245, 246, 249 while the autophagic degradation of mitochondria is diminished250, 251. During this process, autophagy protects T cells from a form of functional inactivation called anergy 252. During differentiation into helper (CD4+) or cytotoxic (CD8+) T cell subsets, autophagic and metabolic interconnected adjustments influence the functional potential of the resulting cells. The proinflammatory Th1 subset of differentiated CD4 T cells is more dependent on autophagy than other subsets, such as Th17 cells, for their survival253, 254(Figure 4). In Th2 cells, autophagy appears to negatively regulate their persistence as they further persist/expand in its absence255 253. Along the same line, autophagy deficiency favors the differentiation and function of Th9 CD4 T cells 256. Thus, autophagy differentially impacts the differentiation of naïve CD4 T cells into their specialized effector subsets. In the absence of mTOR, CD4 T cell differentiation is skewed toward regulatory T cells (Treg), which are important for repressing autoimmune T cells, at the expense of conventional CD4 T helper cell subsets. Tregs rely on fatty acid oxidation for energy generation while effector CD4 T cells favor glycolysis to support their function 257–260. Deficiency in ATG16L1, Vps34, ATG5, or ATG7 alters the persistence and function of Tregs while it promotes those of Th2 cells 255, 261, 262 revealing the importance of autophagy in Treg maintenance, especially in terms of transcriptional reprogramming (Figure 4). Alteration of autophagy promotes mTOR activation and glycolytic enzyme expression, which enhances glycolysis and better fits the requirements of conventional T helper cells. In CD8 T cells, antigen receptor activation leads to augmented glycolysis but differs from CD4 T cells as there is a significant role for the pyruvate dehydrogenase 263. Upon viral infection, the importance of functional autophagy in effector CD8 T cells varies according to the involved pathogen 264 265. After contraction of the effector response, some CD8 T cells revert to oxidative phosphorylation and acquire a status of long-lived, antigen-specific, memory cells. As autophagy can be important for such metabolic adjustments it is not surprising that the emergence and persistence of CD8 memory T cells are greatly sensitive to the autophagy status 264–266. For instance, CD8 T deficient in ATG7 after encountering viral antigens were altered in their capacity to generate a memory pool 264, and autophagy was found to promote the maintenance of liver resident memory CD8 T cells 267. In the context of tumor antigens, autophagy represses the function of effector/memory CD8 T cells by controlling histone methylation, glycolysis, and immune response gene expression268, Finally, autophagy can act positively on stemness and survival of antigen-experienced CD8 T cells within the tumor microenvironment 269. In many situations, the metabolic profile of CD8 T cells was modified by the level of cell-autonomous autophagy confirming that autophagy-associated changes in metabolism greatly influence the generation and function of CD8 memory T cells (Figure 4). Within the CD4 T cell lineage, autophagy was found important for the homeostasis of lymphoid organ memory T cells by regulating the mitochondrial pool and lipid load 270.
4. Non-canonical autophagy, infection, and immune response
Non-canonical autophagy has emerged as an essential component of the innate immune system, which serves as a cell-autonomous defense mechanism. Non-canonical autophagy regulates pathogen clearance, inflammation, and antigen presentation. Thus, it plays an indispensable role in protecting against infectious, autoimmune, and inflammatory diseases 271–273.
Unlike the involvement of LC3 with double-membrane autophagosomes in macroautophagy, non-canonical autophagy involves conjugating LC3 family proteins to single-membrane compartments in a ULK1/2 independent manner. For example, LC3 conjugation to the phagosomal membrane (LAPosome) occurs by a process called LC3-associated phagocytosis (LAP), while LC3-associated endocytosis (LANDO) involves conjugation of LC3 to endosomes. Interestingly, Rubicon, a negative regulator of canonical autophagy, positively regulates non-canonical autophagy pathways. Apart from Rubicon, either BECLIN1-VSP34 complex generated PI3P, or PI3P independent LC3 conjugation on the single membrane, requires the involvement of canonical autophagy elongation complex (ATG12-ATG5-ATG16L1) 274–276. The later process of covalent association of LC3 with a bilayer is known as the Conjugation of ATG8 to Single Membranes (CASM). During CASM, in addition to phosphatidylethanolamine (PE), LC3/GABARAP can also be conjugated to phosphatidylserine (PS) 277. While similarities and differences exist between these two autophagy processes, we focus on the crosstalk between non-canonical autophagy (LAP/LANDO) and innate immunity to maintain immune homeostasis. We also highlight the unconventional roles of autophagy proteins beyond their role in autophagy, especially on innate immune regulation.
Douglas R. Green’s group first showed direct and rapid recruitment of LC3 on phagosomal membranes upon TLRs stimulation in mouse macrophages. This was shown to facilitate the maturation of phagosomes resulting in enhanced degradation of engulfed foreign entities during the early stages of infection278. The authors later named this process “LC3-associated phagocytosis (LAP) 279. LAP has been implicated in the clearance of several intracellular pathogens such as Listeria monocytogenes, Streptococcus pneumoniae, Aspergillus fumigatus, Salmonella typhimurium, Mycobacterium tuberculosis, and Influenza A virus (IAV) 280–286. As explained above for CASM, during LAP, LC3 lipidation to a single membrane requires the presence of PI3P, ROS, and ATG16L1. This occurs by the concerted action of Rubicon, NADPH oxidase (NOX), and V-ATPase. While Rubicon is important for the generation of PI3P, NOX is indispensable for producing ROS, and the V-ATPase facilitates the recruitment of ATG16L1 to the phagosome. However, how exactly these events are coordinated, the potential cross-talk, and their regulation is still obscure 287–289. In addition, recent studies suggest that LAP plays a crucial role in antigen presentation 210, 290. Further clarity on the mechanism by which LAP participates in these processes may lie in elucidating the detailed role of V-ATPase as it is not only associated with the recruitment of key proteins but is important for an efficient lysosomal function that effectively generates peptides for antigen presentation.
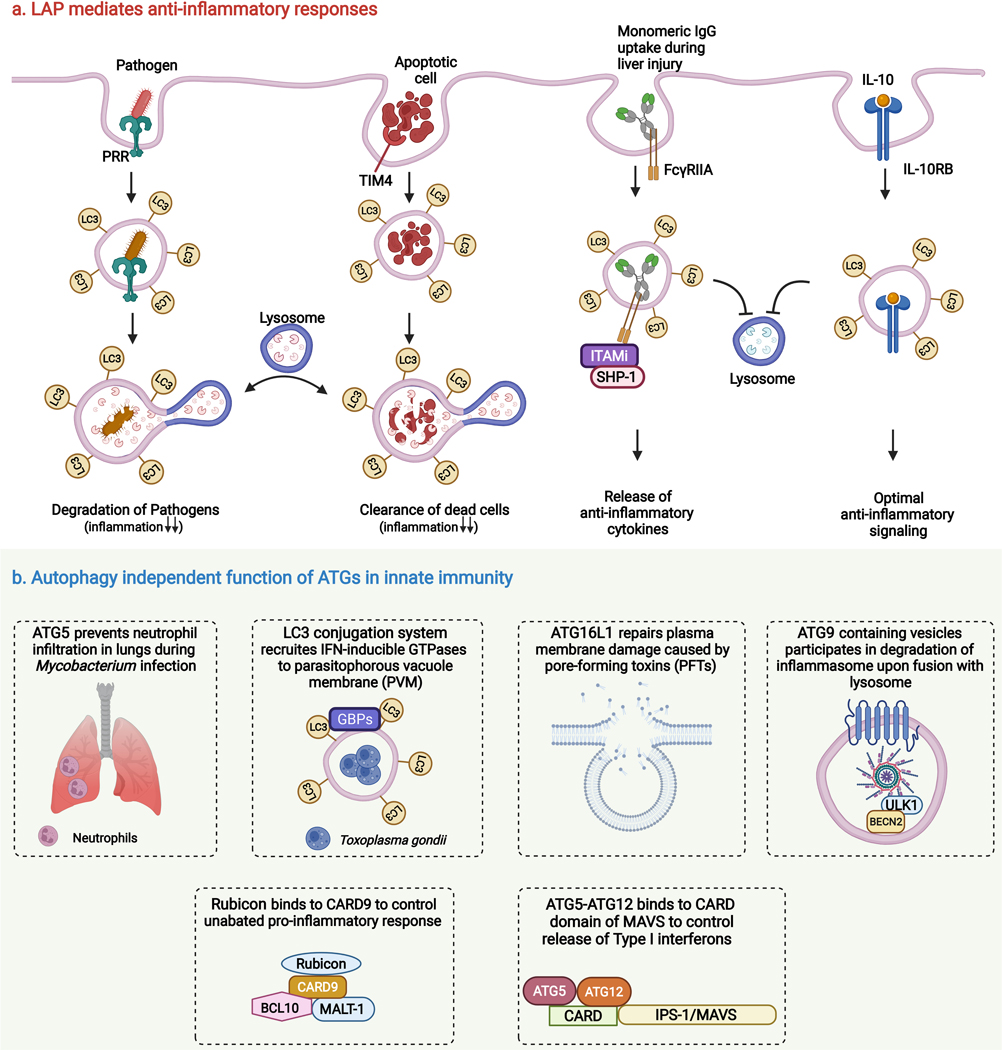
LAP limits pro-inflammatory responses by degrading phagocytosed dead cells. TIM4, a phosphatidylserine receptor essential for this process, induces rapid translocation of LC3 to the phagosome-containing cell corpses (Figure 5a). This promotes phagosome acidification and effective degradation of engulfed apoptotic and necrotic cells 291. In 2016, the same group performed in vivo experiments and showed that LAP-lacking animals display an increased level of inflammation, autoantibodies, kidney damage, and a systemic lupus erythematosus (SLE) like phenotype upon repeated injection of dead cells 292. This highlights the importance of LAP in dampening inflammation. LAP is also a significant contributor to efferocytosis, a process by which dead cells are cleared. Disruption of LAP upon long-term exposure to cigarette smoke (CS) results in failed efferocytosis, thus contributing to Chronic obstructive pulmonary disease (COPD). CS exposure reduces Rubicon levels, which perturbs the LAP pathway, causing severe lung inflammation and damage 293.
Figure 5. Role of unconventional autophagy and autophagy proteins in regulation of innate immune responses.
(a) LC3-associated phagocytosis (LAP) mediates anti-inflammatory responses. LAP contributes to the degradation of intracellular pathogens (via PRR-mediated phagocytosis) and dead cells (via TIM4-mediated engulfment of apoptotic cells) which facilitate to dampen the inflammatory responses. As opposed to its degradative role, LAP also promotes the assembly of several anti-inflammatory signaling complexes (FcγRIIA-SHP-1-ITAMi and IL-10–1L10-RB) to control the levels of inflammation (b) Autophagy independent functions of ATGs in innate immunity. Several core autophagy proteins, independent of their autophagic function, regulate inflammatory responses. ATG5 prevents neutrophil infiltration in the lungs during Mycobacterium infection to control hyper inflammation and induce cell survival. LC3-conjugation system facilitates the targeting of guanylate-binding proteins (GBPs) on the Toxoplasma parasitophorous vacuolar membrane, thereby damaging and destroying the vacuolar niche of the parasite. ATG16L1 contributes to plasma membrane repair upon infection of pore-forming toxins (PFTs) producing bacteria, thus providing resiliency towards PFTs. ATG9 containing vesicles are involved in lysosomal mediated degradation of inflammasome to regulate inflammation, the assembly of inflammasome sensors into atg9 containing vesicles is dependent on Beclin2 and ULK1. Rubicon and ATG5-ATG12 complex control proinflammatory responses by binding to CARD9 and MAVS respectively.
Furthermore, the LAP pathway in monocytes has been reported to reduce inflammation during cirrhosis. However, patients with Acute-on-chronic liver failure (ACLF) showed negligible LAP levels. Patients affected with liver diseases show elevated serum monomeric IgG levels. Surprisingly, the introduction of intravenous monomeric IgG in ACLF patients restores LAP via activation of the FcγRIIA-SHP-1-ITAMi signaling axis to impart anti-inflammatory responses 294 (Figure 5a). However, exactly how LAP facilitates the assembly of anti-inflammatory signaling complex upon exposure to monomeric IgG is yet to be explored.
The C-terminal WD40 domain of ATG16L1, critical for LAP but not for canonical autophagy295, is a key regulator of anti-inflammatory cytokine signaling. Cytokine receptors interact with ATG16L1 WDD via a WDD-binding motif, which facilitates the LC3 lipidated compartmentalization of anti-inflammatory cytokines and their receptors (such as IL-10/IL-10RB) for optimal signaling instead of degradation (Figure 5a). Interestingly, this process is independent of Rubicon 296. Though all the above studies point towards the anti-inflammatory function of LAP in monocytes/macrophages, a small number of studies have also highlighted the pro-inflammatory effects of LAP. FcγR-mediated phagocytosis of DNA-specific IgG autoantibodies (hallmarks of autoimmunity) by plasmacytoid dendritic cells (pDCs) leads to excessive secretion of IFN-α. Henault et al. showed that LAP is essential for IFN-α secretion in response to DNA autoantibodies. LAP drives the trafficking of TLR9 (DNA sensor) containing phagosomes to a mature compartment (late endosomes), wherein the interferon signaling components interacts (TLR9, TRAF3, IRF7, etc.). This results in the pathogenic production of Type I interferons from pDCs, suggesting a role for LAP in autoimmunity 297. Histoplasma capsulatum, a fungal pathogen that causes pulmonary mycosis in immunocompromised individuals, is recognized by the PRR Dectin-1. Dectin-1 activates the downstream effector, Syk, to generate ROS through NADPH oxidase in macrophages. Besides ROS, the NLRX1-TUFM-ATG5-ATG12 axis also promotes LAP induction. Eventually, LAP activates the MAPK-AP-1 pathway and cytokine production in a Rubicon-independent manner, contributing to anti-fungal immunity 298. Although, the mechanistic details are lacking as to how LAP activates the MAPK-AP-1 pathway.
5. Unconventional functions of Atg proteins
Several autophagy proteins have been reported to play role in inflammation regulation without the involvement of the autophagy pathway. Herein we highlight their unconventional roles in the hierarchical order as they participate in different stages in the autophagy pathway (Figure 5b).
Mycobacterium infection in mice depleted of Atg5 in myeloid cells results in aberrant infiltration of neutrophils in the lung, independent of its role in autophagy. This leads to hyper inflammation, bacterial proliferation, and reduced survival 299. However, the molecular mechanism is yet to be discerned for this phenotype. Another study by Choi et al., showed the role of LC3 conjugation machinery in the recruitment of IFN-γ effectors on LC3 coated parasite containing vacuoles independent of their role in degradation. Toxoplasma gondii, a protozoan parasite, survives inside a single membrane parasitophorous vacuole membrane (PVM) by avoiding fusion with lysosomes. Upon IFN-γ stimulation, PVMs of T. gondii conjugate with LC3 by ATG12-ATG5-ATG16L1 complex independently of ULK1/PI3K complex. LC3 conjugation to PVMs results in the recruitment of IFN-inducible GTPases. IFN-inducible GTPases damage PVMs and subsequently restrict parasite replication 300. Pathogens such as Listeria and Streptococcus often use pore-forming toxins (PFTs) to promote dissemination by damaging plasma membrane permeability and epithelial barriers 301. Tan et al. showed that autophagy protein ATG16L1 and its interactors ATG5 and ATG12 are essential for providing resistance to PFTs by repairing plasma membrane independent of their autophagy function. ATG16L1 triggers lysosomal exocytosis and bleb formation to repair PM dependent on cholesterol trafficking towards PM 302.
Rubicon acts as a specific feedback inhibitor of CARD9, an adaptor protein that plays a vital role in providing innate immunity against infection. CARD9 forms a downstream signaling complex with BCL10-MALT-1 upon recognition of PAMPs by PRR and induces the production of proinflammatory cytokines. Rubicon directly interacts with CARD9 to control its unabated activation and eventually inflammation upon specific stimulation (β−1,3-glucan or SeV infection). The Rubicon-CARD9 interaction is functionally and genetically separate from Rubicon’s role in canonical and non-canonical autophagy 303. Another autophagy protein, Atg9a, plays a distinct role in regulating the translocation of STING (cytosolic DNA sensor) and assembly of STING and TBK1 upon dsDNA stimulation. This prevents the uncontrolled release of cytokines 304. BECLIN2 (BECN2) negatively regulates inflammasome activation in an Atg9a and ULK1, not ATG16L1, LC3, and BECLIN1 dependent manner. ULK1 serves as an assembling factor of inflammasome sensors and BECN2 in Atg9a containing vesicles. Further, several SNAREs (SEC22A, STX5, and STX6) drive membrane fusion for inflammasome degradation to control inflammation 305.
Autophagosomes serve as a replicative platform for several RNA viruses 306, 307. RIG-I or MDA5 sense the cytosolic viral RNA, which exposes their two amino-terminal caspase activation and recruitment domains (CARDs). Upon viral RNA detection, RIG or MDA5 interacts with the CARD domain of mitochondrial antiviral signaling protein (MAVS/ IPS-1). This interaction activates the downstream signaling cascade to activate the transcription of type I interferon genes, an essential factor contributing to antiviral host response. Intriguingly, Atg5-Atg12 conjugate associates with the CARD domain of IPS-1 upon RNA virus infection to suppress type I interferon response. RNA viruses have evolved to leverage this interaction, thus sabotaging the host’s innate antiviral immunity for their survival. The authors have also highlighted the importance of this atypical role of Atg5-Atg12 to maintain immune homeostasis under physiological conditions 308. Nevertheless, it remains to be identified which domain or amino acid of Atg5 is required for interaction with IPS-1.
These studies highlight the non-canonical role of autophagy and autophagy proteins in regulating inflammatory response to maintain immune homeostasis. However, a clearer distinction and better mechanistic understanding will be required before unconventional autophagy pathways can be targeted for therapeutic intervention in inflammatory diseases.
6. Autophagy and innate immune crosstalk: disease perspective
As the first line of defense, our innate immune system consists of barrier tissue and its resident immune cells as well as patrolling leukocytes that can be mobilized swiftly to neutralize the threat. Barrier systems can act as a physical wall consisting of cells connected through tight junctions that insulate intruders, which are reinforced by chemical weapons and armor, such as secreted antimicrobials and mucus. Both the structural and immune cells present in barrier tissue activate innate immune responses following stimulation of pattern recognition receptors (PRRs). Activation of signaling cascades downstream of PRRs leads to a series of effector mechanisms such as secretion of soluble factors (cytokines, chemokines, and microbicidal agents), adjustment of barrier permeability, regeneration after injury, intracellular restriction/processing of hazards, triggering cell death and initiation of adaptive immunity. Autophagy, as a cardinal mediator of cellular homeostasis, is fundamental to both the process of PRR signaling and these downstream effector mechanisms. As such, a broad spectrum of diseases has been linked to dysregulated autophagy in innate immunity. In this section, we highlight the autophagy-innate immunity crosstalk from a disease perspective (Figure 6).
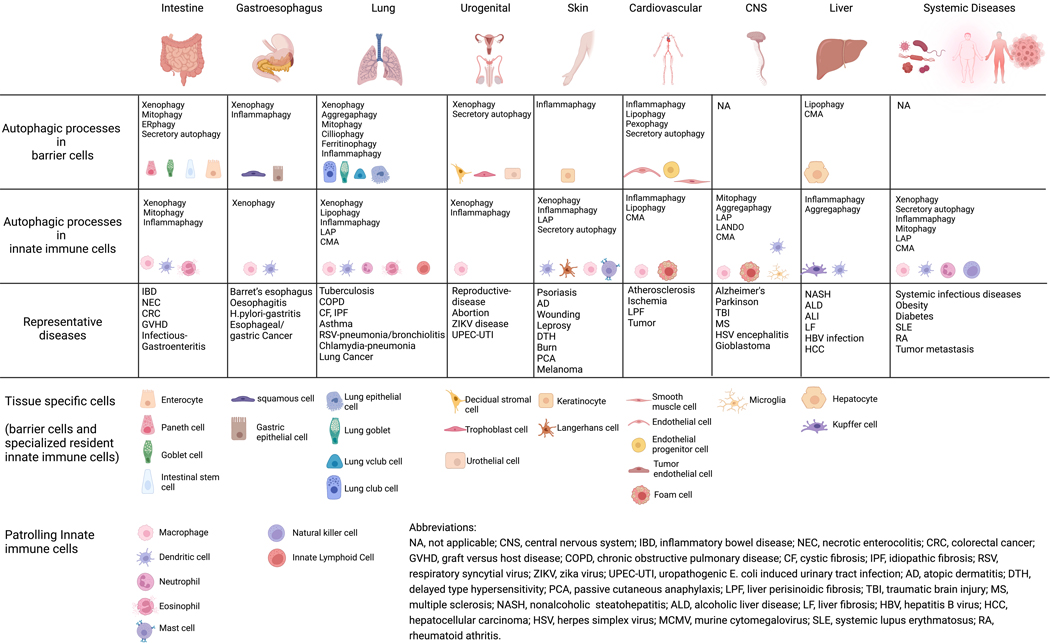
Figure 6. Autophagic processes regulate innate immunity in diseases.
Innate immunity in organs is mediated by autophagic processes occurring in both tissue-specific barrier cells and innate immune cells. Alterations in autophagic processes are linked to a variety of organ-specific or systemic diseases. These autophagic processes include macroautophagy, LC3-associated phagocytosis (LAP), LC3-associated endocytosis (LANDO), secretory autophagy, and chaperone-medidated autophagy (CMA). Macroautophagy processes can be further classified as selective autophagy processes such as xenophagy (targeting intracellular microbes), aggregaphagy (targeting protein aggregates), mitophagy (targeting mitochondria), ERphagy (targeting endoplasmic reticulum), pexophagy (targeting peroxisome), lipophagy (targeting lipids), cilliophagy (targeting cilia), and ferritinophagy (targeting ferritin). Autophagic processes associated with regulating innate immunity, either within barrier cells or specialized innate immune cell subsets, and the representative diseases reviewed in this article are grouped by the organs listed.
A. Autophagy regulation of barrier sites in diseases
Gastrointestinal tract
The gastrointestinal (GI) tract represents an enormous mucosal surface area that requires protection from infectious and non-infectious foreign material ingested along with food and water. The GI tract is also persistently colonized by a massive community of resident microbes, the microbiota. A role for autophagy in supporting a balanced immune reaction at this interface is supported by the association between variants in autophagy genes (NOD2, ATG16L1, IRGM, ULK1, and LRRK2) and inflammatory bowel disease (IBD) 309, a debilitating disease of the GI tract that involves abnormal reactions against microbes. Additionally, autophagy activity is regulated by innate immune signaling at the intestinal barrier. For example, both commensal and invasive bacteria, or bacterial products, can activate PRRs such as NOD2 to induce autophagy or autophagy-related processes by recruiting the core autophagy machinery 41, 44, 310–315. LC3 conjugation occurs constitutively in the epithelium due to the presence of the microbiota and can be further enhanced to control Salmonella enterica Serovar Typhimurium (STm) infection311, 312. Factors associated with damage such as HMGB1, can also induce autophagy 316, 317. Autophagy genes can be transcriptionally regulated, as demonstrated by the upregulation of ATG16L1 by the vitamin D receptor to maintain Paneth cells, an epithelial cell that secretes antimicrobial molecules to protect adjacent intestinal stem cells (ISCs) in the small intestinal crypts 318, 319.
ATG16L1 and autophagy proteins are necessary for the viability and function of Paneth cells. Patients with a major type of IBD called Crohn’s disease who are homozygous for the T300A variant of ATG16L1 display morphologically aberrant Paneth cells, which is reproduced in mice harboring mutations in various autophagy genes320–329. ATG16L1T300A protein is prone to caspase-3 mediated processing and degradation, suggesting that individuals harboring this risk variant may have reduced capacity for autophagy or an autophagy-related pathway under conditions in which caspases are activate330, 331. Paneth cell function is supported by the role of autophagy in countering organelle stress and mediating secretion of the antimicrobial protein lysozyme when the ER-Golgi secretory pathway is disrupted by infections321, 332 or mutations in the unfolded protein response (UPR) genes333–335. Autophagy also prevents cell death of intestinal epithelial cells, and Paneth cells are particularly vulnerable to inflammatory forms of cell death such as necroptosis upon inhibition of autophagy proteins or mitophagy314, 336–339. Thus, deleting autophagy specifically in Paneth cells leads to inflammatory events resembling IBD325.
The function of autophagy proteins in other epithelial cells also contribute to supporting the integrity of the intestinal barrier. In a manner resembling the above role in Paneth cells, autophagy controls ISC homeostasis by limiting toxic ROS production, which is critical for epithelial regeneration after irradiation340. In mucus-secreting goblet cells, the autophagy machinery is mobilized downstream of another NOD-like protein, NLRP6, to mediate the exocytosis of mucin-containing granules, a process that is dependent on FOXO1 and NADPH oxidase activity 99, 341–345. More direct functions of autophagy proteins in supporting the physical barrier include breaking down the tight junction protein Claudin-2346, regulating proliferation347, and maintaining plasma membrane integrity348. The latter involves activating the ATG16L1 complex in response to damage by Listeria monocytogenes through a mechanism independent of an autophagosome. Therefore, autophagy proteins have a major role in supporting the intestinal barrier through autophagy-dependent and -independent processes.
The above processes involved in supporting the viability and function of intestinal epithelial cells are complemented by the role of autophagy in tuning the innate immune response to gut microbes, especially the production of inflammatory mediators such as interferons and interleukins. ATG16L1 deficiency in the epithelium leads to augmented type I interferon responses that enhance resistance to the model enteric pathogen Citrobacter rodentium349, 350 but increases susceptibility to TNF induced cell death339, 351. Similarly, knockout of Epg5, which is involved in autophagosome maturation and mutated in a combined immunodeficiency called VICI syndrome352, 353, increases resistance to enteric virus infection via elevated type III interferon354. Selective autophagy receptors Optineurin and NDP52 affect IBD by regulating proinflammatory cytokines release and TLR-NF-κB activation, respectively. 355–357. In rodent models of necrotizing enterocolitis (NEC), an intestinal inflammatory disorder associated with preterm birth, excessive autophagy downstream of innate immune responses contributes to disease, suggesting a pathologic role for the pathway in this context358–360. Whether autophagy prevents or promotes intestinal malignancies depends on the stage and context of tumorigenesis361. For example, autophagy deficiency suppresses tumor growth in the setting of APC-haploinsuffiency362. However, increased epithelial autophagy activity by colibactin-producing Escherichia coli (CoPEC) is protective in a similar APCmin/+ colorectal cancer (CRC) model363. Also, mitophagy in IECs promotes iron accumulation in lysosomes that trigger lysosomal membrane damage and leakage of proteases into the cytosol, which in turn enhances MHC-I antigen presentation of processed peptides that boost the antitumor CD8 T cell response364.
In addition to events affecting the small intestine and colon, upper GI tract diseases in the esophagus and stomach are associated with autophagy dysregulation. Barret’s esophagus, an inflammatory condition of the esophagus caused by chronic gastric acid reflux, is associated with altered autophagy activity365, 366. Autophagy is important for epithelial cell survival in eosinophilic oesophagitis367, but high autophagic activity is associated with a poor prognosis of esophageal squamous cell carcinoma368, 369. In the context of gastric Helicobacter pylori (H. pylori) infection, autophagy-induced drugs help control infection370 and the autophagy-compromising ATG16L1T300A variant exacerbates inflammation371. However, increased autophagy might contribute to H. pylori-associated gastric cancer via promoting cancer cell stemness 372, 373. These examples highlight how the homeostatic function of autophagy is generally protective but occasionally contributes to disease pathogenesis, especially when persistent inflammation is involved.
Respiratory tract
The respiratory tract represents another major mucosal barrier site where important functions of autophagy have been documented. Autophagy processes, including ciliophagy (targeting airway epithelial cilia)374, 375, ferritinophagy (targeting ferritin)376, mitophagy-mediated necroptosis377 and autophagy induced apoptosis378, 379, have been proposed to contribute to chronic obstructive pulmonary disease (COPD). Particulate matter triggered lung injury also requires autophagy for disease onset380. However, during cystic fibrosis (CF), a disease associated with opportunistic infections and mucus barrier abnormalities due to mutations in cystic fibrosis transmembrane conductance regulator (CFTR), defective autophagy is associated with more severe disease381, 382. Aggregaphagy (autophagy targeting protein aggregates), xenophagy (autophagy targeting intracellular microorganisms), and autophagic restriction of inflammation (inflammophagy) may also safeguard against CF progression383, 384. Similar protective functions of autophagy are found in idiopathic pulmonary fibrosis (IPF) 385, 386, such as PINK1/PARK2-mediated mitophagy in alveolar epithelial cells type 2387–389. In asthma, another major pulmonary disease, impaired autophagy in epithelial cells contributes to eosinophilic inflammation in obese asthmatic mice390. Alveolar epithelial-selective autophagic degradation of the E3 ligase TRIM37 leads to stable levels of its target TRAF6, which mediates NF-κB signaling and chemokine production to recruit neutrophils that promote lung cancer metastasis in response to particulates391. Similar to the intestine, lung epithelial stem-like vClub cells require autophagy for tissue repair under disease conditions such as allergic inflammation392. Secretion of mucus by lung goblet cells is also mediated by autophagy, and excess autophagy activity predisposes mice to type 2 airway diseases393, 394. The secretion of defensive proteins in Club cells is also mediated by autophagy and defective autophagy is associated with COPD395. One interpretation of these collective findings is that autophagy can be either pathologically augmented or reduced during lung disease, and in either case, innate immune mechanisms intrinsic to the epithelium can become dysregulated, such as inappropriate production of soluble inflammatory mediators and mucus.
Urogenital tract
In the urogenital system, autophagy controls inflammation to prevent complications during pregnancy, such as by dampening the NLRP3 inflammasome396, 397. Autophagy in decidual stromal cells is also important to maintain local natural killer cells that are important for preventing spontaneous abortion398. Autophagy in the epithelial barrier is necessary for defense against urogenital pathogens. Likely due to the role of the autophagy machinery in rearranging membranes subverted by viruses for replication, autophagy deficiency in mouse trophoblasts renders the host resistant to Zika virus399. Analogously, deficiency of ATG16L1 or ATG7 in the urothelium lining of the urinary tract, or carriage of the ATG16L1T300A allele, interferes with vesicle trafficking events subverted by uropathogenic E. coli (UPEC) for persistence400.
Skin
Autophagy deficiency leads to the compromised organization of skin barriers because autophagy governs the development and proliferation of epithelial cells, and also mediates the proper inflammatory status in response to purturbations401–404. For example, keratinocyte autophagy is required for suppressing inflammation and facilitating wound healing, and resisting particulate matter-induced dermal fibroblast injury405–408 and psoriasis409–411. In atopic dermatitis (AD) patients, increased autophagy activity was observed in skin epithelial cells, potentially as a compensatory mechanism to mitigate inflammation412.
Endovascular barrier
The endovascular system serves as both a barrier and conduit for the trafficking of immune cells. Autophagy dysfunction in endothelial cells has been shown to instigate a variety of diseases413. For example, the endothelium requires autophagy to maintain integrity via controlling ROS levels in the cell414, promoting angiogenesis415, and secreting blood clotting factors416. Notably, endothelial cell autophagy declines with aging, and decreases in specialized functions of autophagy such as lipophagy are associated with degenerative arterial disease, like atherosclerosis417–420. Targeting of peroxisomes by autophagy (pexophagy) in endothelial cells is important to prevent LPS-induced organ injury421. During Staphylococcus aureus infection, endothelial-intrinsic ATG16L1 is important to tolerate damage caused by a bacterial pore-forming toxin called α-toxin422. Mechanistically, the secretory autophagy pathway mediates the exocytosis of decoy exosomes termed defensosomes that are decorated by the toxin receptor ADAM10, which binds and inhibits α-toxin423. In the blood-brain barrier (BBB), endothelial autophagy has been shown either to prevent ischemia-induced barrier damage by reducing apoptosis424, 425 or to exacerbate brain ischemia BBB damage in diabetic mice by degrading the junction protein claudin-5426, 427, suggesting a context-specific role of endothelial autophagy at this site. Deletion of autophagy genes in liver sinusoidal endothelial cells renders these cells susceptible to increased inflammatory responses in the setting of non-alcoholic steatohepatitis (NASH) 428.
B. Autophagy regulation of innate immune cells in diseases
In addition to directly functioning with barrier cells, autophagy in innate immune cells such as macrophages, dendritic cells (DCs), granulocytes and innate lymphoid cells (ILCs) affects both tissue-specific and systemic innate immune responses.
Gastrointestinal tract
ATG16L1 deficiency in hematopoietic or myeloid cells renders mice more susceptible to experimental colitis due to hyperactivation of pan inflammatory responses including the inflammasome and IL1β/IL18 processing429–432. Excess IL-1 activity also mediates colitis in chronic granulomatous disease patients and the mouse model, which is caused by loss of function mutations in the NADPH oxidase and impaired recruitment of LC3 to the phagosome433. Macrophages in anti-TNF treated IBD patients display increased autophagy activity concomitant with “M2-like” anti-inflammatory features, but this effect is impaired by the ATG16L1T300A variant434. However, this role of autophagy in polarizing macrophages towards an anti-inflammatory state may support CRC by suppressing antitumor immunity435. Nevertheless, therapeutically increasing autophagy including mitophagy can enhance resistance to CRC in the murine model by dampening the inflammasome436, 437. DCs and macrophages without SQSTM1 or TAX1BP1 selective autophagy have amplified TLR-TRIF-IFNβ signaling, which is associated with unresponsiveness to anti-TNF treatment in IBD patients22 23. In myeloid cells, the absence of ATG5 will shunt selective autophagy receptor NBR1 to target IL-12 to late endosomes for secretion, thus instigating inflammation in a murine colitis model438. In addition to controlling inflammation, autophagy proteins in myeloid cells affect intestinal diseases in many other ways. For example, macrophages use the autophagy gene NRBF2 to scavenge apoptotic cells by enhancing phagosome maturation during intestinal inflammation439. Eosinophil autophagy is required for eosinopoiesis but impedes effector function upon C. rodentium challenge440. During STm infection, LAP induced by flagellin-TLR5 activation in zebra fish macrophages delivers antimicrobial activity441, 442. Also, the V-ATPase subunits on STm containing phagosomes mediate xenophagy by promoting phagosome-lysosome fusion, and loss of ATP6V0D2 in macrophages leads to impaired bacterial control in vivo after gastrointestinal infection432. This finding is corroborated by another study that demonstrates that V-ATPase can target ATG16L1 to STm containing vesicles for xenophagy443. Autophagy in intestinal DCs is important for proper interactions with T cells, as impaired autophagy and the ATG16L1-T300A variant prolong their interactions through the immunological synapse leading to hyperactivation of T cells, 444. DC autophagy deficiency leads to enhanced levels of T cell costimulation and exacerbated intestinal damage in a mouse model of graft-versus-host diseases (GVHD)234. In the upper GI tract, H. pylori infection can induce xenophagy in macrophages and DCs which is impaired in PBMCs from individuals with the ATG16L1-T300A variant 445–449.
Urogenital tract
Although mechanistically distinct from how autophagy deficiency in the urothelium is protective, myeloid-specific deletion of ATG16L1 strongly enhances resistance to urinary tract infection by UPEC. This enhanced immunity is dependent on increased NLRP3/IL-1β signaling450. DC depletion of ATG5 increased disease pathology and morbidity in mice during vaginal HSV infection due to impaired T cell priming224.
CNS
Neuronal cell-intrinsic autophagy dysfunction has been well documented to associate with neurodegenerative diseases (including Alzheimer’s disease, AD, and Parkinson’s disease, PD). Experimental models implicate an essential role for autophagy and related processes in removing harmful extra- or intracellular protein aggregates and large organelles and neuronal cell death451–456. In microglia, which are myeloid cells in the CNS, LC3-associated endocytosis (LANDO) is critical for β-amyloid (Aβ) receptor recycling from endosomes to the plasma membrane, which mediates removal of accumulated Aβ in the tissue457, 458. Selective autophagy mediated by SQSTM1 in microglia removes α-synuclein aggregates released by neurons 459. Autophagy and chaperone-mediated autophagy (CMA), which is upregulated after traumatic brain injury or spinal cord injury in neuronal cells including microglia460, 461, both control inflammasome activation to prevent PD-like symptoms in mice462, 463. In glioblastoma, pericytes upregulate their CMA for acquiring an immunosuppressive feature to promote tumor growth464. Autophagy is also important in multiple sclerosis (MS), a chronic inflammatory condition in the CNS465, 466. Myeloid/DC autophagy (including mitophagy, lipophagy) or LAP inhibition reduces experimental autoimmune encephalomyelitis (EAE), a mouse model of MS, through multiple mechanisms467–475. Intriguingly, a gain-of-function variant of the mitophagy receptor NDP52 is associated with improvement in MS patients by reducing proinflammatory cytokines production476, 477. In HSV-induced encephalitis, patients with loss of function mutations in ATG4A and LC3B2 have a defect in autophagy induction, explaining increased viral replication and recurrent clinical symptoms478. These findings support results obtained in animal models implicating autophagy proteins in the control of herpes viruses. As further support of interactions between the virus and autophagy, the HSV encoded factors ICP34.5 and ICP0 interfere with BECLIN1 and SQSTM1/Optineurin, respectively479, 480.
Lung
Mycobacterium tuberculosis (Mtb) infects macrophages, and autophagy has been shown to both limit active Mtb infection and the inflammation in vitro and in vivo 197, 481. Mtb encodes virulence factors ESAT-6 and CpsA that interfere with autophagy and LAP, respectively, to promote survival in macrophages 482. In contrast to macrophages, ATG5 has a unique protective role in mediating the proper turnover of neutrophils independent of other autophagy proteins, which is necessary to limit immunopathology of the lung during Mtb infection 483. During Chlamydia pneumoniae infection, autophagy is required to restrict inflammasome activation to restrict infection in vivo484. Autophagy in ILC2 is required for disease development in a mouse model of allergic asthma; inhibiting ATG5 in activated ILC2s increases apoptosis due to a deficit in lipophagy and a consequent metabolic shift485. In contrast, autophagy in DCs is protective against airway inflammation induced by the respiratory syncytial virus (RSV) by ensuring proper antiviral responses through T cell polarization486, 487. CMA limits lung cancer progression in a mouse model through regulating macrophage activation by degrading ERK3 488.
Skin
Autophagy has been shown to promote antigen presentation by Langerhans cells, a type of macrophage in the skin, to restrict Mycobacterium leprae infection489. LAP in dermal DCs is required for immunosuppression after UV exposure and contact hypersensitivity490. Vitamin D-induced autophagy activity in skin macrophages improves sunburn resolution by promoting wound healing macrophage differentiation491. Skin macrophage autophagy also protects against the treatment of mice with the TLR7 agonist imiquimod, a model of psoriasis, by inhibiting NF-κB492. However, ATG7 and LC3 targeting is necessary for mast cell degranulation and sustains inflammation in passive cutaneous anaphylaxis493. Autophagy inhibition in melanoma cells also renders improved tumor killing by NK cells via upregulating chemokine CCL5 expression, as autophagy deficiency leads to PP2A deactivation and thus promotes JNK-c-Jun-CCL5 axis activity494.
Liver
Patients with chronic HCV infection displayed heightened autophagy activity compared to other liver disease patients495. This upregulation of autophagy, including CMA, is associated with control of lipid storage in hepatocytes496, and resistance to IFN-I and ribavirin treatment due to downregulation of IFNAR1 and nucleoside transporters497–501. During acute liver injuries, autophagy in Kupffer cells, the liver type of macrophages, is induced and chemical inhibition or myeloid ablation of ATG5 had increased disease with elevated inflammatory response502–504. The Kupffer cell autophagy is also protective in alcoholic liver disease505, liver fibrosis506, and hepatocarcinogenesis507 in mice via restraining ROS-induced IL-1α and IL-1β production506. In the context of hepatocellular cancer (HCC), hepatocyte autophagy restricts tumorigenesis by reducing SQSTM1 accumulation, while autophagy in the myeloid compartment is also antitumorigenic through downregulation of immunosuppressive molecules like PD-L1508–511.
Cardiovascular disease
In atherosclerosis, macrophage foam cells use autophagy to mediate intracellular lysosomal cholesterol hydrolysis and efflux512 thereby reducing inflammation and plaque formation/necrosis513, 514. These observations are corroborated by the finding that ectopic overexpression of ATG14 in macrophages decreases plaque formation in atherosclerosis-prone ApoE−/− mice515. Autophagy deficiency in smooth muscle cells leads to increased CCL2 secretion and recruitment of macrophages to increase plaque formation in vivo516. CMA activity is lower in atherosclerosis plaques of patients, and CMA controls inflammasome activation to reduce atherosclerosis in ApoE−/− mice517, 518.
Obesity and diabetes
Impaired autophagy in adipose tissue is associated with an inflammatory state in individuals with obesity 519, 520 and primes insulin resistance in a mouse model521. In line with these observations, autophagy inhibition in macrophages exacerbates metabolic disease in genetic and diet-induced models of obesity and diabetes due to increased inflammasome and ROS production430, 522, 523. Mitophagy is specifically important for staving off obesity524, 525. However, discrepant results have questioned the necessity of macrophage autophagy in adipose tissue lipid metabolism526, 527, perhaps reflecting differences in techniques and models.
Multisystem inflammatory disease
Autophagy gene expression and markers are decreased in patients with systemic lupus erythematous (SLE)528, 529, and the ATG5 genetic locus is associated with disease susceptibility530. These findings could reflect a protective or adverse function of autophagy proteins. SQSTM1 activity in macrophages mediates cell death downstream of TLR7 that coincides with autophagy induction in a mouse model of SLE531, and LAP in DCs is necessary for TLR9 activation by DNA-immune complexes that underly interferon production, an autoimmune disease hallmark297. However, LAP deficient mice develop spontaneous SLE-like disease due to impaired phagocytosis of accumulated apoptotic cell bodies532. The same mechanisms by which autophagy dysfunction can contribute to tissue-specific innate immune hyperactivity could potentially explain multi-organ manifestations. Uveitis, a form of inflammation in the eye, occurs more commonly in IBD patients, and inhibiting autophagy in macrophages causes disease in mice through inflammasome activation and IL1β533. The role of autophagy in antigen-presenting cells could contribute to autoimmune diseases, such as the presentation of the citrullinated peptides underlying rheumatoid athritis534. Although the molecular mechanism is unclear, drugs that upregulate autophagy mitigate lethal lung damage in mouse models of sepsis independently of microbial burden535, highlighting the role of autophagy in tissue resilience to immune-mediated damage.
Tumors in non-barrier sites
Dendritic cell-specific deletion of ATG5 impairs priming of antitumor CD4 and CD8 T cells, which is associated with increased scavenger receptor CD36 and reduced MHCII-tumor antigen presentation536. In an ovarian cancer metastasis model, mitophagy mediates adaptation to oxidative stress in Tim4+ tumor-associated macrophages (TAMs) that interfere with anti-tumor immunity537. LAP supports the tolerogenic nature of TAMs by facilitating the non-inflammatory uptake of apoptotic tumor carcasses, and thus, ablation of LAP in the myeloid compartment leads to increased IFN-I and antitumor immunity538.
7. Autophagy as a pharmacological target
Several studies have highlighted the effectiveness of autophagy regulation via small molecules 539–541. This has opened up considerable interest in the therapeutic potential of pharmacologically active molecules that work through autophagy to have beneficial effects on inflammation. Some of such studies that have used autophagy modulating small molecules in various diseases associated with inflammation have been summarized in Table 1. Although there are not many bonafide autophagy modulators known, several of the molecules, whose mechanism of action and/or targets are known, work indirectly to control autophagy flux (Table 1). Studies have also highlighted the effectiveness of combining such autophagy modulating small molecules with anti-inflammatory drugs 542, 543.
Table 1.
Pharmacological Activators of Autophagy
| Diseases | Autophagy modulator | Mechanism | References |
|---|---|---|---|
| Pathogenic infections | Rapamycin | mTOR inhibition | 544, 545 |
| Torin1 | mTOR inhibition | 546 | |
| AICAR | AMPK activation | 547 | |
| Gefitinib | EGFR inhibition and increase lysosomal biogenesis genes | 548, 549 | |
| AR-12 | Akt kinase inhibition | 550 | |
| Tat–beclin 1 | GAPR-1 inhibition | 551 | |
| Nt-Arg | Induces autophagy via p62 | 552 | |
| Tamoxifen | Estrogen receptor inhibition | 553 | |
| Flubendazole | Inhibits mTOR and releases Beclin1 from Bcl2-Beclin1 complex | 554 | |
| Acacetin | Promotes TFEB nuclear translocation | 555 | |
| Trehalose | Activates MCOLN1 | 556, 557 | |
| Vitamin D | Activates MCOLN3 | 558–560 | |
| Carbamazepine | Depletes cellular myo-inositol, IP3 levels and activates AMPK | 561 | |
| Inflammatory bowel disease (IBD) | Rapamycin/Sirolimus | mTOR inhibition | 562–565 |
| AICAR | AMPK activation | 566 | |
| Celastrol | Suppresses PI3K/Akt/mTOR signaling | 567 | |
| Docosahexaenoic Acid | Inhibition of mTOR signaling pathway | 568 | |
| AMA0825 | ROCK inhibition | 569 | |
| Azathioprine | Inhibits mTOR and activates PERK | 570 | |
| Vitamin D | Activates MCOLN1 | 571, 572 | |
| Lung inflammation | Carbamazepine | Depletes cellular myo-inositol and IP3 levels, activates AMPK | 573, 574 |
| AICAR | AMPK activation | 575 | |
| Tubastatin A | Histone deacetylase 6 inhibition | 576 | |
| Mdivi-1 | Mitophagy inhibition | 577 | |
| Cysteamine | Transglutaminase 2 (TGM2) inhibitor, restores Beclin1 levels | 578, 579 | |
| Epigallocatechin-3-Gallate (EGCG) | Elevates ATG12 expression | 579, 580 | |
| Heart inflammation | Rapamycin | mTOR inhibition | 581–584 |
| Metformin | AMPK activation and mTOR inhibition | 585–587 | |
| Verapamil | Depletes ATP levels | 588 | |
| Liver inflammation | Verapamil | Induces autophagic flux | 589 |
| Micheliolide | Activates PPAR-γ and p-AMPK and inhibits p-mTOR | 590, 591 | |
| Spermine | Increases ATG5 expression | 591 | |
| Epigallocatechin-3-Gallate (EGCG) | Increase phosphorylation of AMPK | 592 | |
| Systemic lupus erythematosus (SLE) | Rapamycin | mTOR inhibition | 593 |
| Hydroxychloroquine | Inhibit lysosomal activity | 594 | |
| Rheumatoid arthritis (RA) | Arsenic Trioxide | Enhances autophagic flux | 595 |
| Metformin | AMPK activation and mTOR inhibition | 596 | |
| Artesunate | Suppresses PI3K/Akt/mTOR signaling | 597 | |
| Triptolide | Inhibits autophagy | 598 | |
| Tomorou | Mechanism unclear | 599 | |
| Oridonin | Inhibits autophagy | 600 | |
| TIPTP | p22phox (subunit of NOX) inhibition | 601 | |
| Hydroxychloroquine | Inhibits lysosomal activity | 543 | |
| Multiple sclerosis (MS) | Rapamycin | mTOR inhibition | 542, 602 |
| 1,25-dihydroxyvitamin D3 | Elevates Beclin1 expression | 603 | |
| Hydroxychloroquine | Inhibits lysosomal activity | 604 | |
| Clioquinol | Induce autophagy | 605 |
Conclusions
Since the hallmark discovery of autophagy as a homeostatic mechanism induced by starvation in yeast by Yoshinori Ohsumi and co-workers, we now realize that autophagy has multiple functions in eukaryotic cells. One important and likely evolutionary conserved one is the contribution to immune defense, discussed here. The obvious possibility of autophagy to clear intercellular microbes has meanwhile been well documented and underlying molecular mechanisms have been worked out in several cases. This contributes to innate immune defense, particularly towards bacterial infection in barrier epithelia. Probably more surprisingly, autophagy also contributes to controlling the magnitude of innate immune responses and to resolve inflammation by targeting cellular components of the inflammatory cascades, and to adaptive immune responses by influencing MHC antigen presentation. The molecular details of these rather newly discovered mechanisms are beginning to emerge, and recent data suggests that several inflammatory disorders can be explained, at least in part, by dysfunctional autophagy processes. The host protective role of inflammation on the other hand is undermined by pathogens that evolved mechanisms to subvert autophagic processes.
With a deeper understanding of the molecular mechanisms and thus identification of druggable targets, we will have the opportunity of developing novel treatment options for both inflammatory and infectious diseases.
Acknowledgements
SC acknowledge the support of DBT EMR grant (BT/PR23942/BRB/10/1808/2019). JC acknowledge the support of National Institutes of Health grants AI139425 and AI103197. JC holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. JHK and CSS acknowledge the support of the Intramural Program, NIAID, NIH. GT acknowledges the support of National Institutes of Health grants AI145929 and AI148243. KC acknowledges the support of NIH grants HL123340, DK093668, AI140754, AI121244, AI130945; Synergy Award from the Kenneth Rainin Foundation, Senior Research Award from the Crohn’s & Colitis Foundation, and a pilot award from the Takeda-Columbia-NYU Alliance. MF and Team is supported by Fondation pour la Recherche Médicale (Label FRM DEQ20170336729) and Association François Aupetit (AFA).
REFERENCES
- 1.Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018; 14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 2009; 335:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawabata T, Yoshimori T. Autophagosome biogenesis and human health. Cell Discov 2020; 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010; 221:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan L, Rao X, Sigdel KR. Regulation of Inflammation in Autoimmune Disease. J Immunol Res 2019; 2019:7403796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805–20. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan S, Jena KK, Mehto S, Chauhan NR, Sahu R, Dhar K, et al. Innate immunity and inflammophagy: balancing the defence and immune homeostasis. FEBS J 2021. [DOI] [PubMed] [Google Scholar]
- 8.Richetta C, Faure M. Autophagy in antiviral innate immunity. Cell Microbiol 2013; 15:368–76. [DOI] [PubMed] [Google Scholar]
- 9.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo j 2008; 27:1110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007; 315:1398–401. [DOI] [PubMed] [Google Scholar]
- 11.Shin DM, Yuk JM, Lee HM, Lee SH, Son JW, Harding CV, et al. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol 2010; 12:1648–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang L, Wu HM, Ding PS, Liu RY. TLR2 mediates phagocytosis and autophagy through JNK signaling pathway in Staphylococcus aureus-stimulated RAW264.7 cells. Cell Signal 2014; 26:806–14. [DOI] [PubMed] [Google Scholar]
- 13.Lu Z, Xie D, Chen Y, Tian E, Muhammad I, Chen X, et al. TLR2 mediates autophagy through ERK signaling pathway in Mycoplasma gallisepticum-infected RAW264.7 cells. Mol Immunol 2017; 87:161–70. [DOI] [PubMed] [Google Scholar]
- 14.Anand PK, Tait SW, Lamkanfi M, Amer AO, Nunez G, Pagès G, et al. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J Biol Chem 2011; 286:42981–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007; 27:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem 2008; 283:33175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy 2010; 6:986–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang SY, Yang CH, Chou CC, Chiang YP, Chuang TH, Hsu LC. TLR-induced PAI-2 expression suppresses IL-1β processing via increasing autophagy and NLRP3 degradation. Proc Natl Acad Sci U S A 2013; 110:16079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected]. Cell Host Microbe 2014; 15:753–67. [DOI] [PubMed] [Google Scholar]
- 21.Inomata M, Niida S, Shibata K, Into T. Regulation of Toll-like receptor signaling by NDP52-mediated selective autophagy is normally inactivated by A20. Cell Mol Life Sci 2012; 69:963–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog 2017; 13:e1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samie M, Lim J, Verschueren E, Baughman JM, Peng I, Wong A, et al. Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat Immunol 2018; 19:246–54. [DOI] [PubMed] [Google Scholar]
- 24.Nath P, Jena KK, Mehto S, Chauhan NR, Sahu R, Dhar K, et al. IRGM links autoimmunity to autophagy. Autophagy 2021; 17:578–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jena KK, Mehto S, Nath P, Chauhan NR, Sahu R, Dhar K, et al. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep 2020; 21:e50051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begun J, Lassen KG, Jijon HB, Baxt LA, Goel G, Heath RJ, et al. Integrated Genomics of Crohn’s Disease Risk Variant Identifies a Role for CLEC12A in Antibacterial Autophagy. Cell Rep 2015; 11:1905–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pahari S, Negi S, Aqdas M, Arnett E, Schlesinger LS, Agrewala JN. Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy 2020; 16:1021–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A, Simonson TJ, Jondle CN, Mishra BB, Sharma J. Mincle-Mediated Neutrophil Extracellular Trap Formation by Regulation of Autophagy. J Infect Dis 2017; 215:1040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Luo H, Ye Y, Chen X, Zou Y, Duan J, et al. β‑glucan, a dectin‑1 ligand, promotes macrophage M1 polarization via NF‑κB/autophagy pathway. Int J Oncol 2019; 54:271–82. [DOI] [PubMed] [Google Scholar]
- 30.Santarelli R, Gonnella R, Di Giovenale G, Cuomo L, Capobianchi A, Granato M, et al. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci Rep 2014; 4:4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Awady AR, Miles B, Scisci E, Kurago ZB, Palani CD, Arce RM, et al. Porphyromonas gingivalis evasion of autophagy and intracellular killing by human myeloid dendritic cells involves DC-SIGN-TLR2 crosstalk. PLoS Pathog 2015; 10:e1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nature reviews Immunology 2014; 14:9–23. [DOI] [PubMed] [Google Scholar]
- 33.Pei G, Zyla J, He L, Moura-Alves P, Steinle H, Saikali P, et al. Cellular stress promotes NOD1/2-dependent inflammation via the endogenous metabolite sphingosine-1-phosphate. EMBO J 2021:e106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chavez-Arroyo A, Tsai AY, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature 2016; 532:394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legrand-Poels S, Kustermans G, Bex F, Kremmer E, Kufer TA, Piette J. Modulation of Nod2-dependent NF-kappa B signaling by the actin cytoskeleton. Journal of Cell Science 2007; 120:1299–310. [DOI] [PubMed] [Google Scholar]
- 36.Bielig H, Lautz K, Braun PR, Menning M, Machuy N, Brügmann C, et al. The cofilin phosphatase slingshot homolog 1 (SSH1) links NOD1 signaling to actin remodeling. PLoS pathogens 2014; 10:e1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, et al. Activation of innate immune antiviral response by NOD2. Nature immunology 2009; 10:1073–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing H, Fang L, Wang D, Ding Z, Luo R, Chen H, et al. Porcine reproductive and respiratory syndrome virus infection activates NOD2-RIP2 signal pathway in MARC-145 cells. Virology 2014; 458–459:162–71. [DOI] [PubMed] [Google Scholar]
- 39.Kapoor A, Forman M, Arav-Boger R. Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PloS one 2014; 9:e92704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. United States: 2010. AGA Institute. Published by Elsevier Inc, 2010:1630–41, 41 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55–62. [DOI] [PubMed] [Google Scholar]
- 42.Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Molecular cell 2015; 58:507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature medicine 2010; 16:90–7. [DOI] [PubMed] [Google Scholar]
- 44.Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE, et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013; 39:858–73. [DOI] [PubMed] [Google Scholar]
- 45.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature immunology 2013; 14:480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cellular microbiology 2005; 7:765–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byrne BG, Dubuisson JF, Joshi AD, Persson JJ, Swanson MS. Inflammasome components coordinate autophagy and pyroptosis as macrophage responses to infection. mBio 2013; 4:e00620–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology 2011; 12:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jabir MS, Hopkins L, Ritchie ND, Ullah I, Bayes HK, Li D, et al. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy 2015; 11:166–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fekete T, Bencze D, Biro E, Benko S, Pazmandi K. Focusing on the Cell Type Specific Regulatory Actions of NLRX1. International journal of molecular sciences 2021; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Yao Y, Qiu X, Wang G, Hu Z, Chen S, et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nature immunology 2019; 20:433–46. [DOI] [PubMed] [Google Scholar]
- 52.Huang JH, Liu CY, Wu SY, Chen WY, Chang TH, Kan HW, et al. NLRX1 Facilitates Histoplasma capsulatum-Induced LC3-Associated Phagocytosis for Cytokine Production in Macrophages. Front Immunol 2018; 9:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei Y, Wen H, Ting JP. The NLR protein, NLRX1, and its partner, TUFM, reduce type I interferon, and enhance autophagy. Autophagy 2013; 9:432–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo X, Donnelly CR, Gong W, Heath BR, Hao Y, Donnelly LA, et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J Clin Invest 2020; 130:1635–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aikawa C, Nakajima S, Karimine M, Nozawa T, Minowa-Nozawa A, Toh H, et al. NLRX1 Negatively Regulates Group A Streptococcus Invasion and Autophagy Induction by Interacting With the Beclin 1-UVRAG Complex. Front Cell Infect Microbiol 2018; 8:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroder K, Tschopp J. The Inflammasomes. Cell 2010; 140:821–32. [DOI] [PubMed] [Google Scholar]
- 57.Rathinam VA, Fitzgerald KA. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 2016; 165:792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 2013; 341:172–5. [DOI] [PubMed] [Google Scholar]
- 59.Hu Z, Zhou Q, Zhang C, Fan S, Cheng W, Zhao Y, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science 2015; 350:399–404. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 2015; 350:404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 2019; 570:338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature 2011; 479:117–21. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Sharif H, Vora SM, Zheng Y, Wu H. Structures and functions of the inflammasome engine. J Allergy Clin Immunol 2021; 147:2021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateman G, Hill B, Knight R, Boucher D. Great balls of fire: activation and signalling of inflammatory caspases. Biochem Soc Trans 2021; 49:1311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 2008; 456:264–8. [DOI] [PubMed] [Google Scholar]
- 66.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature immunology 2012; 13:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 2016; 164:896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakahira K, Haspel JA, Rathinam VAK, Lee S-J, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature Immunology 2011; 12:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 2015; 210:973–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, et al. The Crohn’s Disease Risk Factor IRGM Limits NLRP3 Inflammasome Activation by Impeding Its Assembly and by Mediating Its Selective Autophagy. Mol Cell 2019; 73:429–45 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mehto S, Chauhan S, Jena KK, Chauhan NR, Nath P, Sahu R, et al. IRGM restrains NLRP3 inflammasome activation by mediating its SQSTM1/p62-dependent selective autophagy. Autophagy 2019; 15:1645–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris J, Hartman M, Roche C, Zeng SG, O’Shea A, Sharp FA, et al. Autophagy Controls IL-1β Secretion by Targeting Pro-IL-1β for Degradation. Journal of Biological Chemistry 2011; 286:9587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, et al. TRIM11 Suppresses AIM2 Inflammasome by Degrading AIM2 via p62-Dependent Selective Autophagy. Cell Rep 2016; 16:1988–2002. [DOI] [PubMed] [Google Scholar]
- 74.Mayes-Hopfinger L, Enache A, Xie J, Huang CL, Kochl R, Tybulewicz VLJ, et al. Chloride sensing by WNK1 regulates NLRP3 inflammasome activation and pyroptosis. Nat Commun 2021; 12:4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gallolu Kankanamalage S, Lee AY, Wichaidit C, Lorente-Rodriguez A, Shah AM, Stippec S, et al. Multistep regulation of autophagy by WNK1. Proc Natl Acad Sci U S A 2016; 113:14342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol 2017; 198:1006–14. [DOI] [PubMed] [Google Scholar]
- 77.Zhao W, Shi CS, Harrison K, Hwang IY, Nabar NR, Wang M, et al. AKT Regulates NLRP3 Inflammasome Activation by Phosphorylating NLRP3 Serine 5. J Immunol 2020; 205:2255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahmadi Rastegar D, Dzamko N. Leucine Rich Repeat Kinase 2 and Innate Immunity. Front Neurosci 2020; 14:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu W, Liu X, Li Y, Zhao J, Liu Z, Hu Z, et al. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella Typhimurium infection. J Exp Med 2017; 214:3051–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lupfer C, Malik A, Kanneganti TD. Inflammasome control of viral infection. Curr Opin Virol 2015; 12:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chawla K, Subramanian G, Rahman T, Fan S, Chakravarty S, Gujja S, et al. Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism. Immuno 2022; 2:153–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Viret C, Duclaux-Loras R, Nancey S, Rozières A, Faure M. Selective Autophagy Receptors in Antiviral Defense. Trends Microbiol 2021; 29:798–810. [DOI] [PubMed] [Google Scholar]
- 83.Sargazi S, Sheervalilou R, Rokni M, Shirvaliloo M, Shahraki O, Rezaei N. The role of autophagy in controlling SARS-CoV-2 infection: An overview on virophagy-mediated molecular drug targets. Cell Biol Int 2021; 45:1599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong LR, Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy? Nat Rev Immunol 2022; 22:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vora SM, Lieberman J, Wu H. Inflammasome activation at the crux of severe COVID-19. Nat Rev Immunol 2021; 21:694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kern DM, Sorum B, Mali SS, Hoel CM, Sridharan S, Remis JP, et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol 2021; 28:573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, et al. beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020; 183:1520–35 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen D, Zheng Q, Sun L, Ji M, Li Y, Deng H, et al. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev Cell 2021; 56:3250–63 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X, Yang Z, Pan T, Long X, Sun Q, Wang PH, et al. SARS-CoV-2 ORF3a induces RETREG1/FAM134B-dependent reticulophagy and triggers sequential ER stress and inflammatory responses during SARS-CoV-2 infection. Autophagy 2022:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Junqueira C, Crespo A, Ranjbar S, Lewandrowski M, Ingber J, de Lacerda LB, et al. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. Res Sq 2021. [Google Scholar]
- 91.Pan P, Shen M, Yu Z, Ge W, Chen K, Tian M, et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat Commun 2021; 12:4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou L, Li Y, Gao S, Yuan H, Zuo L, Wu C, et al. Salmonella spvC Gene Inhibits Autophagy of Host Cells and Suppresses NLRP3 as Well as NLRC4. Front Immunol 2021; 12:639019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, Masumoto J, et al. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS pathogens 2013; 9:e1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS pathogens 2007; 3:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleinnijenhuis J, Oosting M, Plantinga TS, van der Meer JW, Joosten LA, Crevel RV, et al. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology 2011; 134:341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barsalou J, Blincoe A, Fernandez I, Dal-Soglio D, Marchitto L, Selleri S, et al. Rapamycin as an Adjunctive Therapy for NLRC4 Associated Macrophage Activation Syndrome. Front Immunol 2018; 9:2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhan L, Zhang J, Zhang J, Liu X, Zhu S, Shi Y, et al. LC3 and NLRC5 interaction inhibits NLRC5-mediated MHC class I antigen presentation pathway in endometrial cancer. Cancer Lett 2021. [DOI] [PubMed] [Google Scholar]
- 98.Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin1. Journal of immunology 2011; 186:1646–55. [DOI] [PubMed] [Google Scholar]
- 99.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014; 156:1045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 2020; 20:537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Loo YM, Gale M Jr., Immune signaling by RIG-I-like receptors. Immunity 2011; 34:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee NR, Ban J, Lee NJ, Yi CM, Choi JY, Kim H, et al. Activation of RIG-I-Mediated Antiviral Signaling Triggers Autophagy Through the MAVS-TRAF6-Beclin-1 Signaling Axis. Front Immunol 2018; 9:2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A 2009; 106:2770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A 2007; 104:14050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du Y, Duan T, Feng Y, Liu Q, Lin M, Cui J, et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J 2018; 37:351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hou P, Yang K, Jia P, Liu L, Lin Y, Li Z, et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res 2021; 31:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, et al. Tetherin Suppresses Type I Interferon Signaling by Targeting MAVS for NDP52-Mediated Selective Autophagic Degradation in Human Cells. Mol Cell 2017; 68:308–22 e4. [DOI] [PubMed] [Google Scholar]
- 108.He X, Zhu Y, Zhang Y, Geng Y, Gong J, Geng J, et al. RNF34 functions in immunity and selective mitophagy by targeting MAVS for autophagic degradation. EMBO J 2019; 38:e100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu J, Wu X, Wang H, Wei J, Wu Q, Wang X, et al. HFE inhibits type I IFNs signaling by targeting the SQSTM1-mediated MAVS autophagic degradation. Autophagy 2021; 17:1962–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rai P, Janardhan KS, Meacham J, Madenspacher JH, Lin WC, Karmaus PWF, et al. IRGM1 links mitochondrial quality control to autoimmunity. Nat Immunol 2021; 22:312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Azzam KM, Madenspacher JH, Cain DW, Lai L, Gowdy KM, Rai P, et al. Irgm1 coordinately regulates autoimmunity and host defense at select mucosal surfaces. JCI Insight 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nath P, Chauhan NR, Jena KK, Datey A, Kumar ND, Mehto S, et al. Inhibition of IRGM establishes a robust antiviral immune state to restrict pathogenic viruses. EMBO Rep 2021; 22:e52948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 2014; 15:228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao M, Wang F, Wu J, Cheng Y, Cao Y, Wu X, et al. CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy 2021; 17:3976–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019; 567:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fischer TD, Wang C, Padman BS, Lazarou M, Youle RJ. STING induces LC3B lipidation onto single-membrane vesicles via the V-ATPase and ATG16L1-WD40 domain. J Cell Biol 2020; 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 2018; 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Joubert PE, Meiffren G, Grégoire IP, Pontini G, Richetta C, Flacher M, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 2009; 6:354–66. [DOI] [PubMed] [Google Scholar]
- 120.Richetta C, Grégoire IP, Verlhac P, Azocar O, Baguet J, Flacher M, et al. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog 2013; 9:e1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim WJ, Mai A, Weyand NJ, Rendón MA, Van Doorslaer K, So M. Neisseria gonorrhoeae evades autophagic killing by downregulating CD46-cyt1 and remodeling lysosomes. PLoS Pathog 2019; 15:e1007495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim KH, Choi BK, Kim YH, Han C, Oh HS, Lee DG, et al. Extracellular stimulation of VSIG4/complement receptor Ig suppresses intracellular bacterial infection by inducing autophagy. Autophagy 2016; 12:1647–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sorbara MT, Foerster EG, Tsalikis J, Abdel-Nour M, Mangiapane J, Sirluck-Schroeder I, et al. Complement C3 Drives Autophagy-Dependent Restriction of Cyto-invasive Bacteria. Cell Host Microbe 2018; 23:644–52.e5. [DOI] [PubMed] [Google Scholar]
- 124.Viret C, Rozieres A, Duclaux-Loras R, Boschetti G, Nancey S, Faure M. Regulation of anti-microbial autophagy by factors of the complement system. Microb Cell 2020; 7:93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coers J, Brown HM, Hwang S, Taylor GA. Partners in anti-crime: how interferon-inducible GTPases and autophagy proteins team up in cell-intrinsic host defense. Curr Opin Immunol 2018; 54:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pilla-Moffett D, Barber MF, Taylor GA, Coers J. Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease. Journal of molecular biology 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol 2005; 6:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hunn JP, Feng CG, Sher A, Howard JC. The immunity-related GTPases in mammals: a fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm Genome 2011; 22:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martens S, Howard J. The interferon-inducible GTPases. Annu Rev Cell Dev Biol 2006; 22:559–89. [DOI] [PubMed] [Google Scholar]
- 130.Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat Rev Immunol 2004; 4:100–9. [DOI] [PubMed] [Google Scholar]
- 131.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 2011; 332:717–21. [DOI] [PubMed] [Google Scholar]
- 132.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 2012; 13:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saijo-Hamano Y, Sherif AA, Pradipta A, Sasai M, Sakai N, Sakihama Y, et al. Structural basis of membrane recognition of Toxoplasma gondii vacuole by Irgb6. Life Sci Alliance 2022; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Butcher BA, Greene RI, Henry SC, Annecharico KL, Weinberg JB, Denkers EY, et al. p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect Immun 2005; 73:3278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Halonen SK, Taylor GA, Weiss LM. Gamma interferon-induced inhibition of Toxoplasma gondii in astrocytes is mediated by IGTP. Infect Immun 2001; 69:5573–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, et al. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathogens 2005; 1:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Finethy R, Coers J. Sensing the enemy, containing the threat: cell-autonomous immunity to Chlamydia trachomatis. FEMS Microbiol Rev 2016; 40:875–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, Meyer TF. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One 2009; 4:e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc Natl Acad Sci U S A 2006; 103:14092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol 2010; 12:939–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, et al. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog 2013; 9:e1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard JC, Taylor GA, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol 2008; 180:6237–45. [DOI] [PubMed] [Google Scholar]
- 143.Lee Y, Yamada H, Pradipta A, Ma JS, Okamoto M, Nagaoka H, et al. Initial phospholipid-dependent Irgb6 targeting to Toxoplasma gondii vacuoles mediates host defense. Life Sci Alliance 2020; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med 2006; 203:2063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, et al. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A 2015; 112:E5628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murillo-Leon M, Muller UB, Zimmermann I, Singh S, Widdershooven P, Campos C, et al. Molecular mechanism for the control of virulent Toxoplasma gondii infections in wild-derived mice. Nat Commun 2019; 10:1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maric-Biresev J, Hunn JP, Krut O, Helms JB, Martens S, Howard JC. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC biology 2016; 14:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J 2008; 27:2495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Henry SC, Daniell XG, Burroughs AR, Indaram M, Howell DN, Coers J, et al. Balance of Irgm protein activities determines IFN-{gamma}-induced host defense. J Leukoc Biol 2009; 85:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Behnke MS, Fentress SJ, Mashayekhi M, Li LX, Taylor GA, Sibley LD. The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog 2012; 8:e1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 2010; 8:484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reese ML, Boothroyd JC. A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J Biol Chem 2011; 286:29366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, et al. A cluster of Interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 2012; 37. [DOI] [PubMed] [Google Scholar]
- 154.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HWt, et al. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Path 2013; 9:e1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Degrandi D, Kravets E, Konermann C, Beuter-Gunia C, Klumpers V, Lahme S, et al. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc Natl Acad Sci U S A 2013; 110:294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Steffens N, Beuter-Gunia C, Kravets E, Reich A, Legewie L, Pfeffer K, et al. Essential Role of mGBP7 for Survival of Toxoplasma gondii Infection. mBio 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, et al. Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun 2015; 83:4740–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, et al. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem 2011; 286:30471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HWt, et al. Guanylate-binding Protein 1 (Gbp1) Contributes to Cell-autonomous Immunity against Toxoplasma gondii. PLoS Pathog 2013; 9:e1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 2012; 37:302–13. [DOI] [PubMed] [Google Scholar]
- 161.Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One 2014; 9:e86684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014; 40:924–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Park S, Choi J, Biering SB, Dominici E, Williams LE, Hwang S. Targeting by AutophaGy proteins (TAG): Targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy 2016; 12:1153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Choi J, Park S, Biering S, Selleck EM, Liu C, Zhang X, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conugation systems of autophagy. Immunity 2014; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008; 4:458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, et al. Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. J Immunol 2014; 192:3328–35. [DOI] [PubMed] [Google Scholar]
- 167.Sasai M, Sakaguchi N, Ma JS, Nakamura S, Kawabata T, Bando H, et al. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat Immunol 2017; 18:899–910. [DOI] [PubMed] [Google Scholar]
- 168.Selleck EM, Orchard RC, Lassen KG, Beatty WL, Xavier RJ, Levine B, et al. A Noncanonical Autophagy Pathway Restricts Toxoplasma gondii Growth in a Strain-Specific Manner in IFN-gamma-Activated Human Cells. MBio 2015; 6:e01157–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 2012; 12:367–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang Z, Gu H, Li Q, Zheng J, Cao S, Weng C, et al. GABARAPL2 Is Critical for Growth Restriction of Toxoplasma gondii in HeLa Cells Treated with Gamma Interferon. Infect Immun 2020; 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol 2014; 12:101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bhushan J, Radke JB, Perng YC, McAllaster M, Lenschow DJ, Virgin HW, et al. ISG15 Connects Autophagy and IFN-gamma-Dependent Control of Toxoplasma gondii Infection in Human Cells. mBio 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Clough B, Wright JD, Pereira PM, Hirst EM, Johnston AC, Henriques R, et al. K63-Linked Ubiquitination Targets Toxoplasma gondii for Endo-lysosomal Destruction in IFNgamma-Stimulated Human Cells. PLoS Pathog 2016; 12:e1006027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fisch D, Clough B, Domart MC, Encheva V, Bando H, Snijders AP, et al. Human GBP1 Differentially Targets Salmonella and Toxoplasma to License Recognition of Microbial Ligands and Caspase-Mediated Death. Cell Rep 2020; 32:108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Johnston AC, Piro A, Clough B, Siew M, Virreira Winter S, Coers J, et al. Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol 2016; 18:1056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haldar AK, Nigam U, Yamamoto M, Coers J, Goyal N. Guanylate Binding Proteins Restrict Leishmania donovani Growth in Nonphagocytic Cells Independent of Parasitophorous Vacuolar Targeting. mBio 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fisch D, Bando H, Clough B, Hornung V, Yamamoto M, Shenoy AR, et al. Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J 2019; 38:e100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fisch D, Clough B, Khan R, Healy L, Frickel EM. Toxoplasma-proximal and distal control by GBPs in human macrophages. Pathog Dis 2022; 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Matta SK, Patten K, Wang Q, Kim BH, MacMicking JD, Sibley LD. NADPH oxidase and guanylate binding protein 5 restrict survival of avirulent Type III strains of Toxoplasma gondii in naive macrophages. MBio 2018; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee Y, Sasai M, Ma JS, Sakaguchi N, Ohshima J, Bando H, et al. p62 Plays a Specific Role in Interferon-gamma-Induced Presentation of a Toxoplasma Vacuolar Antigen. Cell Rep 2015; 13:223–33. [DOI] [PubMed] [Google Scholar]
- 181.Dockterman J, Fee BE, Taylor GA, Coers J. Murine Irgm Paralogs Regulate Nonredundant Functions To Execute Host Defense to Toxoplasma gondii. Infect Immun 2021; 89:e0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wandel MP, Kim BH, Park ES, Boyle KB, Nayak K, Lagrange B, et al. Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat Immunol 2020; 21:880–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kutsch M, Sistemich L, Lesser CF, Goldberg MB, Herrmann C, Coers J. Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J 2020; 39:e104926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kutsch M, Gonzalez-Prieto C, Lesser CF, Coers J. The GBP1 microcapsule interferes with IcsA-dependent septin cage assembly around Shigella flexneri. Pathog Dis 2021; 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Piro AS, Hernandez D, Luoma S, Feeley EM, Finethy R, Yirga A, et al. Detection of Cytosolic Shigella flexneri via a C-Terminal Triple-Arginine Motif of GBP1 Inhibits Actin-Based Motility. MBio 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wandel MP, Pathe C, Werner EI, Ellison CJ, Boyle KB, von der Malsburg A, et al. GBPs Inhibit Motility of Shigella flexneri but Are Targeted for Degradation by the Bacterial Ubiquitin Ligase IpaH9.8. Cell Host Microbe 2017; 22:507–18 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Finethy R, Dockterman J, Kutsch M, Orench-Rivera N, Wallace GD, Piro AS, et al. Dynamin-related Irgm proteins modulate LPS-induced caspase-11 activation and septic shock. EMBO Rep 2020; 21:e50830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Eren E, Planes R, Bagayoko S, Bordignon PJ, Chaoui K, Hessel A, et al. Irgm2 and Gate-16 cooperatively dampen Gram-negative bacteria-induced caspase-11 response. EMBO reports 2020; 21:e50829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A 2014; 111:6046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 2014; 509:366–70. [DOI] [PubMed] [Google Scholar]
- 191.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447:661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet 2007; 39:830–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xia Q, Wang M, Yang X, Li X, Zhang X, Xu S, et al. Autophagy-related IRGM genes confer susceptibility to ankylosing spondylitis in a Chinese female population: a case-control study. Genes Immun 2017; 18:42–7. [DOI] [PubMed] [Google Scholar]
- 194.Lin YC, Chang PF, Lin HF, Liu K, Chang MH, Ni YH. Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J Hepatol 2016; 65:1209–16. [DOI] [PubMed] [Google Scholar]
- 195.King KY, Lew JD, Ha NP, Lin JS, Ma X, Graviss EA, et al. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One 2011; 6:e16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kimura T, Watanabe E, Sakamoto T, Takasu O, Ikeda T, Ikeda K, et al. Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. PLoS One 2014; 9:e91522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753–66. [DOI] [PubMed] [Google Scholar]
- 198.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006; 313:1438–41. [DOI] [PubMed] [Google Scholar]
- 199.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol 2010; 12:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, et al. Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe 2017; 22:74–85 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 2010; 12:1154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. American journal of physiology Gastrointestinal and liver physiology 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Henry SC, Schmidt EA, Fessler MB, Taylor GA. Palmitoylation of the Immunity Related GTPase, Irgm1: Impact on Membrane Localization and Ability to Promote Mitochondrial Fission. PLoS One 2014; 9:e95021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rai P, Janardhan KS, Meacham J, Madenspacher JH, Lin WC, Karmaus PWF, et al. IRGM1 links mitochondrial quality control to autoimmunity. Nat Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schmidt EA, Fee BE, Henry SC, Nichols AG, Shinohara ML, Rathmell JC, et al. Metabolic alterations contribute to enhanced inflammatory cytokine production in Irgm1-deficient macrophages. J Biol Chem 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol 1997; 27:1506–14. [DOI] [PubMed] [Google Scholar]
- 207.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol 2003; 33:1250–9. [DOI] [PubMed] [Google Scholar]
- 208.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A 2005; 102:7922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005; 307:593–6. [DOI] [PubMed] [Google Scholar]
- 210.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2007; 26:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med 2013; 210:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fonteneau JF, Brilot F, Münz C, Gannagé M. The Tumor Antigen NY-ESO-1 Mediates Direct Recognition of Melanoma Cells by CD4+ T Cells after Intercellular Antigen Transfer. J Immunol 2016; 196:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol 2011; 85:6453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Coulon PG, Richetta C, Rouers A, Blanchet FP, Urrutia A, Guerbois M, et al. HIV-Infected Dendritic Cells Present Endogenous MHC Class II-Restricted Antigens to HIV-Specific CD4+ T Cells. J Immunol 2016; 197:517–32. [DOI] [PubMed] [Google Scholar]
- 215.Jin Y, Sun C, Feng L, Li P, Xiao L, Ren Y, et al. Regulation of SIV antigen-specific CD4+ T cellular immunity via autophagosome-mediated MHC II molecule-targeting antigen presentation in mice. PLoS One 2014; 9:e93143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Suri A, Walters JJ, Rohrs HW, Gross ML, Unanue ER. First signature of islet beta-cell-derived naturally processed peptides selected by diabetogenic class II MHC molecules. J Immunol 2008; 180:3849–56. [DOI] [PubMed] [Google Scholar]
- 217.Kasai M, Tanida I, Ueno T, Kominami E, Seki S, Ikeda T, et al. Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J Immunol 2009; 183:7278–85. [DOI] [PubMed] [Google Scholar]
- 218.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 2008; 455:396–400. [DOI] [PubMed] [Google Scholar]
- 219.Niven J, Madelon N, Page N, Caruso A, Harlé G, Lemeille S, et al. Macroautophagy in Dendritic Cells Controls the Homeostasis and Stability of Regulatory T Cells. Cell Rep 2019; 28:21–9.e6. [DOI] [PubMed] [Google Scholar]
- 220.Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity 2015; 42:942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tam RC, Li MW, Gao YP, Pang YT, Yan S, Ge W, et al. Human CLEC16A regulates autophagy through modulating mTOR activity. Exp Cell Res 2017; 352:304–12. [DOI] [PubMed] [Google Scholar]
- 222.Leung CS, Haigh TA, Mackay LK, Rickinson AB, Taylor GS. Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc Natl Acad Sci U S A 2010; 107:2165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr., Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med 2009; 15:267–76. [DOI] [PubMed] [Google Scholar]
- 224.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010; 32:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Thiele F, Tao S, Zhang Y, Muschaweckh A, Zollmann T, Protzer U, et al. Modified vaccinia virus Ankara-infected dendritic cells present CD4+ T-cell epitopes by endogenous major histocompatibility complex class II presentation pathways. J Virol 2015; 89:2698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Keller CW, Sina C, Kotur MB, Ramelli G, Mundt S, Quast I, et al. ATG-dependent phagocytosis in dendritic cells drives myelin-specific CD4(+) T cell pathogenicity during CNS inflammation. Proc Natl Acad Sci U S A 2017; 114:E11228–e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol 2009; 83:12164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 2010; 32:654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gobeil PA, Leib DA. Herpes simplex virus γ34.5 interferes with autophagosome maturation and antigen presentation in dendritic cells. mBio 2012; 3:e00267–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, et al. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol 2016; 1:16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tey SK, Khanna R. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood 2012; 120:994–1004. [DOI] [PubMed] [Google Scholar]
- 233.Loi M, Müller A, Steinbach K, Niven J, Barreira da Silva R, Paul P, et al. Macroautophagy Proteins Control MHC Class I Levels on Dendritic Cells and Shape Anti-viral CD8(+) T Cell Responses. Cell Rep 2016; 15:1076–87. [DOI] [PubMed] [Google Scholar]
- 234.Hubbard-Lucey VM, Shono Y, Maurer K, West ML, Singer NV, Ziegler CG, et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 2014; 41:579–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020; 581:100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mintern JD, Macri C, Chin WJ, Panozza SE, Segura E, Patterson NL, et al. Differential use of autophagy by primary dendritic cells specialized in cross-presentation. Autophagy 2015; 11:906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dasari V, Rehan S, Tey SK, Smyth MJ, Smith C, Khanna R. Autophagy and proteasome interconnect to coordinate cross-presentation through MHC class I pathway in B cells. Immunol Cell Biol 2016; 94:964–74. [DOI] [PubMed] [Google Scholar]
- 238.Parekh VV, Pabbisetty SK, Wu L, Sebzda E, Martinez J, Zhang J, et al. Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8α(+) dendritic cells. Proc Natl Acad Sci U S A 2017; 114:E6371–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li Y, Hahn T, Garrison K, Cui ZH, Thorburn A, Thorburn J, et al. The vitamin E analogue α-TEA stimulates tumor autophagy and enhances antigen cross-presentation. Cancer Res 2012; 72:3535–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ 2009; 16:991–1005. [DOI] [PubMed] [Google Scholar]
- 241.Li Y, Wang LX, Pang P, Cui Z, Aung S, Haley D, et al. Tumor-derived autophagosome vaccine: mechanism of cross-presentation and therapeutic efficacy. Clin Cancer Res 2011; 17:7047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lee Y, Sasai M, Ma JS, Sakaguchi N, Ohshima J, Bando H, et al. p62 Plays a Specific Role in Interferon-γ-Induced Presentation of a Toxoplasma Vacuolar Antigen. Cell Rep 2015; 13:223–33. [DOI] [PubMed] [Google Scholar]
- 243.Fiegl D, Kägebein D, Liebler-Tenorio EM, Weisser T, Sens M, Gutjahr M, et al. Amphisomal route of MHC class I cross-presentation in bacteria-infected dendritic cells. J Immunol 2013; 190:2791–806. [DOI] [PubMed] [Google Scholar]
- 244.Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, et al. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol 2011; 186:2201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol 2006; 177:5163–8. [DOI] [PubMed] [Google Scholar]
- 246.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med 2007; 204:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy 2009; 5:625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, et al. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A 2014; 111:E5678–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol 2010; 185:7349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol 2011; 186:5313–22. [DOI] [PubMed] [Google Scholar]
- 251.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol 2009; 182:4046–55. [DOI] [PubMed] [Google Scholar]
- 252.Mocholi E, Dowling SD, Botbol Y, Gruber RC, Ray AK, Vastert S, et al. Autophagy Is a Tolerance-Avoidance Mechanism that Modulates TCR-Mediated Signaling and Cell Metabolism to Prevent Induction of T Cell Anergy. Cell Rep 2018; 24:1136–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ 2012; 19:144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yang G, Song W, Postoak JL, Chen J, Martinez J, Zhang J, et al. Autophagy-related protein PIK3C3/VPS34 controls T cell metabolism and function. Autophagy 2021; 17:1193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kabat AM, Harrison OJ, Riffelmacher T, Moghaddam AE, Pearson CF, Laing A, et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. Elife 2016; 5:e12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rivera Vargas T, Cai Z, Shen Y, Dosset M, Benoit-Lizon I, Martin T, et al. Selective degradation of PU.1 during autophagy represses the differentiation and antitumour activity of T(H)9 cells. Nat Commun 2017; 8:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 2011; 186:3299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood 2005; 105:4743–8. [DOI] [PubMed] [Google Scholar]
- 259.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 2009; 30:832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Valmori D, Tosello V, Souleimanian NE, Godefroy E, Scotto L, Wang Y, et al. Rapamycin-mediated enrichment of T cells with regulatory activity in stimulated CD4+ T cell cultures is not due to the selective expansion of naturally occurring regulatory T cells but to the induction of regulatory functions in conventional CD4+ T cells. J Immunol 2006; 177:944–9. [DOI] [PubMed] [Google Scholar]
- 261.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagómez D, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol 2013; 190:5086–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 2016; 17:277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Menk AV, Scharping NE, Moreci RS, Zeng X, Guy C, Salvatore S, et al. Early TCR Signaling Induces Rapid Aerobic Glycolysis Enabling Distinct Acute T Cell Effector Functions. Cell Rep 2018; 22:1509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol 2014; 15:1152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Schlie K, Westerback A, DeVorkin L, Hughson LR, Brandon JM, MacPherson S, et al. Survival of effector CD8+ T cells during influenza infection is dependent on autophagy. J Immunol 2015; 194:4277–86. [DOI] [PubMed] [Google Scholar]
- 266.Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife 2014; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Swadling L, Pallett LJ, Diniz MO, Baker JM, Amin OE, Stegmann KA, et al. Human Liver Memory CD8(+) T Cells Use Autophagy for Tissue Residence. Cell Rep 2020; 30:687–98.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.DeVorkin L, Pavey N, Carleton G, Comber A, Ho C, Lim J, et al. Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell Rep 2019; 27:502–13.e5. [DOI] [PubMed] [Google Scholar]
- 269.Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 2019; 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Murera D, Arbogast F, Arnold J, Bouis D, Muller S, Gros F. CD4 T cell autophagy is integral to memory maintenance. Sci Rep 2018; 8:5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR. LC3-Associated Phagocytosis and Inflammation. Journal of molecular biology 2017; 429:3561-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Herb M, Gluschko A, Schramm M. LC3-associated phagocytosis - The highway to hell for phagocytosed microbes. Seminars in Cell and Developmental Biology 2020; 101:68–76. [DOI] [PubMed] [Google Scholar]
- 273.Münz C. Non-canonical functions of autophagy proteins in immunity and infection. Molecular Aspects of Medicine 2021; 82:100987-. [DOI] [PubMed] [Google Scholar]
- 274.Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. The EMBO Journal 2018; 37:e97840-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Heckmann BL, Green DR. LC3-associated phagocytosis at a glance. Journal of cell science 2019; 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Rickman A, Hilyard A, Heckmann B. Dying by fire: noncanonical functions of autophagy proteins in neuroinflammation and neurodegeneration. Neural Regeneration Research 2022; 17:246-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Durgan J, Lystad AH, Sloan K, Carlsson SR, Wilson MI, Marcassa E, et al. Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Molecular Cell 2021; 81:2031–40.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007. 450:7173 2007; 450:1253–7. [DOI] [PubMed] [Google Scholar]
- 279.Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunological Reviews 2009; 227:203–20. [DOI] [PubMed] [Google Scholar]
- 280.Gluschko A, Farid A, Herb M, Grumme D, Krönke M, Schramm M. Macrophages target Listeria monocytogenes by two discrete non-canonical autophagy pathways. Autophagy 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Inomata M, Xu S, Chandra P, Meydani SN, Takemura G, Philips JA, et al. Macrophage LC3-associated phagocytosis is an immune defense against Streptococcus pneumoniae that diminishes with host aging. Proceedings of the National Academy of Sciences of the United States of America 2020; 117:33561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Köster S, Upadhyay S, Chandra P, Papavinasasundaram K, Yang G, Hassan A, et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proceedings of the National Academy of Sciences of the United States of America 2017; 114:E8711–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li T, Kong L, Li X, Wu S, Attri KS, Li Y, et al. Listeria monocytogenes upregulates mitochondrial calcium signaling to inhibit LC3-associated phagocytosis as a survival strategy. Nature microbiology 2021; 6:366-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature cell biology 2015; 17:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 285.Wang Y, Sharma P, Jefferson M, Zhang W, Bone B, Kipar A, et al. Non‐canonical autophagy functions of ATG16L1 in epithelial cells limit lethal infection by influenza A virus. The EMBO Journal 2021; 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, et al. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 2019; 178:552–66.e20. [DOI] [PubMed] [Google Scholar]
- 287.Hooper KM, Jacquin E, Li T, Goodwin JM, Brumell JH, Durgan J, et al. V-ATPase is a universal regulator of LC3-associated phagocytosis and non-canonical autophagy. The Journal of cell biology 2022; 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ulferts R, Marcassa E, Timimi L, Lee LC, Daley A, Montaner B, et al. Subtractive CRISPR screen identifies the ATG16L1/vacuolar ATPase axis as required for non-canonical LC3 lipidation. Cell Reports 2021; 37:109899-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature Cell Biology 2014. 17:7 2015; 17:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 290.Keller CW, Sina C, Kotur MB, Ramelli G, Mundt S, Quast I, et al. ATG-dependent phagocytosis in dendritic cells drives myelin-specific CD4+ T cell pathogenicity during CNS inflammation. Proceedings of the National Academy of Sciences of the United States of America 2017; 114:E11228–E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proceedings of the National Academy of Sciences of the United States of America 2011; 108:17396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, et al. Noncanonical autophagy inhibits the auto-inflammatory, lupus-like response to dying cells. Nature 2016; 533:115-. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 293.Asare PF, Tran HB, Hurtado PR, Perkins GB, Nguyen P, Jersmann H, et al. Inhibition of LC3-associated phagocytosis in COPD and in response to cigarette smoke. Therapeutic Advances in Respiratory Disease 2021; 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wan JH, Weiss E, Mkaddem SB, Mabire M, Choinier PM, Picq O, et al. LC3-associated phagocytosis protects against inflammation and liver fibrosis via immunoreceptor inhibitory signaling. Science Translational Medicine 2020; 12. [DOI] [PubMed] [Google Scholar]
- 295.Florey KF, Rachel U, Elise J, Talitha V, Noor G, Julia MA, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. The EMBO Journal 2018; 37:e97840-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Serramito-Gómez I, Boada-Romero E, Villamuera R, Fernández-Cabrera Á, Cedillo JL, Martín-Regalado Á, et al. Regulation of cytokine signaling through direct interaction between cytokine receptors and the ATG16L1 WD40 domain. Nature Communications 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical Autophagy Is Required for Type I Interferon Secretion in Response to DNA-Immune Complexes. Immunity 2012; 37:986–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Huang JH, Liu CY, Wu SY, Chen WY, Chang TH, Kan HW, et al. NLRX1 Facilitates Histoplasma capsulatum-Induced LC3-Associated Phagocytosis for Cytokine Production in Macrophages. Frontiers in immunology 2018; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in PMN-mediated immunopathology during M. tuberculosis infection. Nature 2015; 528:565-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, et al. The parasitophorous vacuole membrane of toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014; 40:924–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Peraro MD, Van Der Goot FG. Pore-forming toxins: ancient, but never really out of fashion. Nature Reviews Microbiology 2015. 14:2 2015; 14:77–92. [DOI] [PubMed] [Google Scholar]
- 302.Tan JMJ, Mellouk N, Osborne SE, Ammendolia DA, Dyer DN, Li R, et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nature Microbiology 2018. 3:12 2018; 3:1472–85. [DOI] [PubMed] [Google Scholar]
- 303.Yang CS, Rodgers M, Min CK, Lee JS, Kingeter L, Lee JY, et al. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host and Microbe 2012; 11:277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences of the United States of America 2009; 106:20842-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Deng G, Li C, Chen L, Xing C, Fu C, Qian C, et al. BECN2 (beclin 2) Negatively Regulates Inflammasome Sensors Through ATG9A-Dependent but ATG16L1- and LC3-Independent Non-Canonical Autophagy. 10.1080/1554862720211934270 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Choi Y, Bowman JW, Jung JU. Autophagy during viral infection — a double-edged sword. Nature Reviews Microbiology 2018. 16:6 2018; 16:341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Twu WI, Lee JY, Kim H, Prasad V, Cerikan B, Haselmann U, et al. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Reports 2021; 37:110049-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America 2007; 104:14050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020; 16:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 2010; 139:1630–41, 41 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Benjamin JL, Sumpter R Jr., Levine B, Hooper LV. Intestinal Epithelial Autophagy Is Essential for Host Defense against Invasive Bacteria. Cell Host Microbe 2013; 13:723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Conway KL, Kuballa P, Song JH, Patel KK, Castoreno AB, Yilmaz OH, et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology 2013; 145:1347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Gutierrez MG, Saka HA, Chinen I, Zoppino FC, Yoshimori T, Bocco JL, et al. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc Natl Acad Sci U S A 2007; 104:1829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Burger E, Araujo A, Lopez-Yglesias A, Rajala MW, Geng L, Levine B, et al. Loss of Paneth Cell Autophagy Causes Acute Susceptibility to Toxoplasma gondii-Mediated Inflammation. Cell Host Microbe 2018; 23:177–90 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Chang SY, Lee SN, Yang JY, Kim DW, Yoon JH, Ko HJ, et al. Autophagy controls an intrinsic host defense to bacteria by promoting epithelial cell survival: a murine model. PloS one 2013; 8:e81095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Zhu Y, Deng J, Nan ML, Zhang J, Okekunle A, Li JY, et al. The Interplay Between Pattern Recognition Receptors and Autophagy in Inflammation. Adv Exp Med Biol 2019; 1209:79–108. [DOI] [PubMed] [Google Scholar]
- 317.Zhu X, Messer JS, Wang Y, Lin F, Cham CM, Chang J, et al. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J Clin Invest 2015; 125:1098–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lu R, Zhang YG, Xia Y, Sun J. Imbalance of autophagy and apoptosis in intestinal epithelium lacking the vitamin D receptor. FASEB J 2019; 33:11845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy 2016; 12:1057–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456:259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010; 141:1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol 2013; 305:G573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Cadwell K, Patel KK, Komatsu M, Virgin HWt, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 2009; 5:250–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wittkopf N, Gunther C, Martini E, Waldner M, Amann KU, Neurath MF, et al. Lack of intestinal epithelial atg7 affects paneth cell granule formation but does not compromise immune homeostasis in the gut. Clinical & developmental immunology 2012; 2012:278059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Cabrera S, Fernandez AF, Marino G, Aguirre A, Suarez MF, Espanol Y, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy 2013; 9:1188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Shanahan MT, Carroll IM, Grossniklaus E, White A, von Furstenberg RJ, Barner R, et al. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.VanDussen KL, Liu TC, Li D, Towfic F, Modiano N, Winter R, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014; 146:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 2014; 506:456–62. [DOI] [PubMed] [Google Scholar]
- 331.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A 2014; 111:7741–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 2017; 357:1047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013; 503:272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Tschurtschenthaler M, Adolph TE, Ashcroft JW, Niederreiter L, Bharti R, Saveljeva S, et al. Defective ATG16L1-mediated removal of IRE1alpha drives Crohn’s disease-like ileitis. J Exp Med 2017; 214:401–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Diamanti MA, Gupta J, Bennecke M, De Oliveira T, Ramakrishnan M, Braczynski AK, et al. IKKalpha controls ATG16L1 degradation to prevent ER stress during inflammation. J Exp Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017; 214:3687–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Pott J, Kabat AM, Maloy KJ. Intestinal Epithelial Cell Autophagy Is Required to Protect against TNF-Induced Apoptosis during Chronic Colitis in Mice. Cell Host Microbe 2018; 23:191–202 e4. [DOI] [PubMed] [Google Scholar]
- 338.Slowicka K, Serramito-Gomez I, Boada-Romero E, Martens A, Sze M, Petta I, et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun 2019; 10:1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med 2018; 215:2868–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S, et al. Intrinsic Autophagy Is Required for the Maintenance of Intestinal Stem Cells and for Irradiation-Induced Intestinal Regeneration. Cell Rep 2017; 20:1050–60. [DOI] [PubMed] [Google Scholar]
- 341.Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. Embo J 2013; 32:3130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Tiwari S, Begum S, Moreau F, Gorman H, Chadee K. Autophagy is required during high MUC2 mucin biosynthesis in colonic goblet cells to contend metabolic stress. Am J Physiol Gastrointest Liver Physiol 2021; 321:G489–G99. [DOI] [PubMed] [Google Scholar]
- 343.Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016; 352:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Chen Z, Luo J, Li J, Kim G, Chen ES, Xiao S, et al. Foxo1 controls gut homeostasis and commensalism by regulating mucus secretion. J Exp Med 2021; 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Engevik MA, Luk B, Chang-Graham AL, Hall A, Herrmann B, Ruan W, et al. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio 2019; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Nighot PK, Hu CA, Ma TY. Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J Biol Chem 2015; 290:7234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Nagai H, Tatara H, Tanaka-Furuhashi K, Kurata S, Yano T. Homeostatic Regulation of ROS-Triggered Hippo-Yki Pathway via Autophagic Clearance of Ref(2)P/p62 in the Drosophila Intestine. Dev Cell 2021; 56:81–94 e10. [DOI] [PubMed] [Google Scholar]
- 348.Tan JMJ, Mellouk N, Osborne SE, Ammendolia DA, Dyer DN, Li R, et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat Microbiol 2018; 3:1472–85. [DOI] [PubMed] [Google Scholar]
- 349.Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe 2013; 14:216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, et al. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol 2018; 3:1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Matsuzawa-Ishimoto Y, Hine A, Shono Y, Rudensky E, Lazrak A, Yeung F, et al. An intestinal organoid-based platform that recreates susceptibility to T-cell-mediated tissue injury. Blood 2020; 135:2388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 2013; 45:83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Wang Z, Miao G, Xue X, Guo X, Yuan C, Wang Z, et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol Cell 2016; 63:781–95. [DOI] [PubMed] [Google Scholar]
- 354.Lee S, Kalugotla G, Ingle H, Rodgers R, Wu C, Wang Y, et al. Intestinal antiviral signaling is controlled by autophagy gene Epg5 independent of the microbiota. Autophagy 2021:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, et al. Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology 2013; 145:339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Smith AM, Sewell GW, Levine AP, Chew TS, Dunne J, O’Shea NR, et al. Disruption of macrophage pro-inflammatory cytokine release in Crohn’s disease is associated with reduced optineurin expression in a subset of patients. Immunology 2015; 144:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Chew TS, O’Shea NR, Sewell GW, Oehlers SH, Mulvey CM, Crosier PS, et al. Optineurin deficiency in mice contributes to impaired cytokine secretion and neutrophil recruitment in bacteria-driven colitis. Dis Model Mech 2015; 8:817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Maynard AA, Dvorak K, Khailova L, Dobrenen H, Arganbright KM, Halpern MD, et al. Epidermal growth factor reduces autophagy in intestinal epithelium and in the rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 2010; 299:G614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Neal MD, Sodhi CP, Dyer M, Craig BT, Good M, Jia H, et al. A critical role for TLR4 induction of autophagy in the regulation of enterocyte migration and the pathogenesis of necrotizing enterocolitis. J Immunol 2013; 190:3541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Yu Y, Shiou SR, Guo Y, Lu L, Westerhoff M, Sun J, et al. Erythropoietin protects epithelial cells from excessive autophagy and apoptosis in experimental neonatal necrotizing enterocolitis. PLoS One 2013; 8:e69620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. How autophagy controls the intestinal epithelial barrier. Autophagy 2022; 18:86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Levy J, Cacheux W, Bara MA, L’Hermitte A, Lepage P, Fraudeau M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 2015; 17:1062–73. [DOI] [PubMed] [Google Scholar]
- 363.Lucas C, Salesse L, Hoang MHT, Bonnet M, Sauvanet P, Larabi A, et al. Autophagy of Intestinal Epithelial Cells Inhibits Colorectal Carcinogenesis Induced by Colibactin-Producing Escherichia coli in Apc(Min/+) Mice. Gastroenterology 2020; 158:1373–88. [DOI] [PubMed] [Google Scholar]
- 364.Ziegler PK, Bollrath J, Pallangyo CK, Matsutani T, Canli O, De Oliveira T, et al. Mitophagy in Intestinal Epithelial Cells Triggers Adaptive Immunity during Tumorigenesis. Cell 2018; 174:88–101 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kong J, Whelan KA, Laczko D, Dang B, Caro Monroig A, Soroush A, et al. Autophagy levels are elevated in barrett’s esophagus and promote cell survival from acid and oxidative stress. Mol Carcinog 2016; 55:1526–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Verbeek RE, Siersema PD, Vleggaar FP, Ten Kate FJ, Posthuma G, Souza RF, et al. Toll-like Receptor 2 Signalling and the Lysosomal Machinery in Barrett’s Esophagus. J Gastrointestin Liver Dis 2016; 25:273–82. [DOI] [PubMed] [Google Scholar]
- 367.Whelan KA, Merves JF, Giroux V, Tanaka K, Guo A, Chandramouleeswaran PM, et al. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut 2017; 66:1197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Ge J, Li L, Jin Q, Liu YC, Zhao L, Song HH. Functional IRGM polymorphism is associated with language impairment in glioma and upregulates cytokine expressions. Tumour Biol 2014; 35:8343–8. [DOI] [PubMed] [Google Scholar]
- 369.Yamashita K, Miyata H, Makino T, Masuike Y, Furukawa H, Tanaka K, et al. High Expression of the Mitophagy-Related Protein Pink1 is Associated with a Poor Response to Chemotherapy and a Poor Prognosis for Patients Treated with Neoadjuvant Chemotherapy for Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 2017; 24:4025–32. [DOI] [PubMed] [Google Scholar]
- 370.Yang JC, Chien CT. A new approach for the prevention and treatment of Helicobacter pylori infection via upregulation of autophagy and downregulation of apoptosis. Autophagy 2009; 5:413–4. [DOI] [PubMed] [Google Scholar]
- 371.Mommersteeg MC, Simovic I, Yu B, van Nieuwenburg SAV, Bruno IMJ, Doukas M, et al. Autophagy mediates ER stress and inflammation in Helicobacter pylori-related gastric cancer. Gut Microbes 2022; 14:2015238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Rao SV, Solum G, Niederdorfer B, Norsett KG, Bjorkoy G, Thommesen L. Gastrin activates autophagy and increases migration and survival of gastric adenocarcinoma cells. BMC Cancer 2017; 17:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Courtois S, Haykal M, Bodineau C, Sifre E, Azzi-Martin L, Menard A, et al. Autophagy induced by Helicobacter pylori infection is necessary for gastric cancer stem cell emergence. Gastric Cancer 2021; 24:133–44. [DOI] [PubMed] [Google Scholar]
- 374.Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest 2013; 123:5212–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Cloonan SM, Lam HC, Ryter SW, Choi AM. “Ciliophagy”: The consumption of cilia components by autophagy. Autophagy 2014; 10:532–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Yoshida M, Minagawa S, Araya J, Sakamoto T, Hara H, Tsubouchi K, et al. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat Commun 2019; 10:3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest 2014; 124:3987–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One 2008; 3:e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A 2010; 107:18880–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Wu YF, Li ZY, Dong LL, Li WJ, Wu YP, Wang J, et al. Inactivation of MTOR promotes autophagy-mediated epithelial injury in particulate matter-induced airway inflammation. Autophagy 2020; 16:435–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol 2010; 12:863–75. [DOI] [PubMed] [Google Scholar]
- 382.De Stefano D, Villella VR, Esposito S, Tosco A, Sepe A, De Gregorio F, et al. Restoration of CFTR function in patients with cystic fibrosis carrying the F508del-CFTR mutation. Autophagy 2014; 10:2053–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy 2011; 7:1359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zhao X, Wei S, Li Z, Lin C, Zhu Z, Sun D, et al. Autophagic flux blockage in alveolar epithelial cells is essential in silica nanoparticle-induced pulmonary fibrosis. Cell Death Dis 2019; 10:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Mi S, Li Z, Yang HZ, Liu H, Wang JP, Ma YG, et al. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Immunol 2011; 187:3003–14. [DOI] [PubMed] [Google Scholar]
- 386.Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, et al. Autophagy in idiopathic pulmonary fibrosis. PLoS One 2012; 7:e41394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest 2015; 125:521–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Patel AS, Song JW, Chu SG, Mizumura K, Osorio JC, Shi Y, et al. Epithelial cell mitochondrial dysfunction and PINK1 are induced by transforming growth factor-beta1 in pulmonary fibrosis. PLoS One 2015; 10:e0121246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Kobayashi K, Araya J, Minagawa S, Hara H, Saito N, Kadota T, et al. Involvement of PARK2-Mediated Mitophagy in Idiopathic Pulmonary Fibrosis Pathogenesis. J Immunol 2016; 197:504–16. [DOI] [PubMed] [Google Scholar]
- 390.Suzuki Y, Aono Y, Akiyama N, Horiike Y, Naoi H, Horiguchi R, et al. Involvement of autophagy in exacerbation of eosinophilic airway inflammation in a murine model of obese asthma. Autophagy 2022:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Liu J, Li S, Fei X, Nan X, Shen Y, Xiu H, et al. Increased alveolar epithelial TRAF6 via autophagy-dependent TRIM37 degradation mediates particulate matter-induced lung metastasis. Autophagy 2021:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Li K, Li M, Li W, Yu H, Sun X, Zhang Q, et al. Airway epithelial regeneration requires autophagy and glucose metabolism. Cell Death Dis 2019; 10:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Dickinson JD, Alevy Y, Malvin NP, Patel KK, Gunsten SP, Holtzman MJ, et al. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy 2016; 12:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Jiang X, Fang L, Wu H, Mei X, He F, Ding P, et al. TLR2 Regulates Allergic Airway Inflammation and Autophagy Through PI3K/Akt Signaling Pathway. Inflammation 2017; 40:1382–92. [DOI] [PubMed] [Google Scholar]
- 395.Malvin NP, Kern JT, Liu TC, Brody SL, Stappenbeck TS. Autophagy proteins are required for club cell structure and function in airways. Am J Physiol Lung Cell Mol Physiol 2019; 317:L259–L70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Cao B, Camden AJ, Parnell LA, Mysorekar IU. Autophagy regulation of physiological and pathological processes in the female reproductive tract. Am J Reprod Immunol 2017; 77. [DOI] [PubMed] [Google Scholar]
- 397.Navarro-Pando JM, Alcocer-Gomez E, Castejon-Vega B, Navarro-Villaran E, Condes-Hervas M, Mundi-Roldan M, et al. Inhibition of the NLRP3 inflammasome prevents ovarian aging. Sci Adv 2021; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Lu H, Yang HL, Zhou WJ, Lai ZZ, Qiu XM, Fu Q, et al. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy 2021; 17:2511–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Cao B, Parnell LA, Diamond MS, Mysorekar IU. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J Exp Med 2017; 214:2303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Wang C, Bauckman KA, Ross ASB, Symington JW, Ligon MM, Scholtes G, et al. A non-canonical autophagy-dependent role of the ATG16L1(T300A) variant in urothelial vesicular trafficking and uropathogenic Escherichia coli persistence. Autophagy 2019; 15:527–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Yoshihara N, Ueno T, Takagi A, Oliva Trejo JA, Haruna K, Suga Y, et al. The significant role of autophagy in the granular layer in normal skin differentiation and hair growth. Arch Dermatol Res 2015; 307:159–69. [DOI] [PubMed] [Google Scholar]
- 402.Noguchi S, Honda S, Saitoh T, Matsumura H, Nishimura E, Akira S, et al. Beclin 1 regulates recycling endosome and is required for skin development in mice. Commun Biol 2019; 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Lee Y, Shin K, Shin KO, Yoon S, Jung J, Hwang E, et al. Topical application of autophagy-activating peptide improved skin barrier function and reduced acne symptoms in acne-prone skin. J Cosmet Dermatol 2021; 20:1009–16. [DOI] [PubMed] [Google Scholar]
- 404.Liu C, Gu L, Ding J, Meng Q, Li N, Dai G, et al. Autophagy in skin barrier and immune-related skin diseases. J Dermatol 2021; 48:1827–37. [DOI] [PubMed] [Google Scholar]
- 405.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, Lee SH, et al. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol 2011; 186:1248–58. [DOI] [PubMed] [Google Scholar]
- 406.Tian R, Li Y, Yao X. PGRN Suppresses Inflammation and Promotes Autophagy in Keratinocytes Through the Wnt/beta-Catenin Signaling Pathway. Inflammation 2016; 39:1387–94. [DOI] [PubMed] [Google Scholar]
- 407.Yoon SJ, Lim CJ, Chung HJ, Kim JH, Huh YH, Park K, et al. Autophagy Activation by Crepidiastrum Denticulatum Extract Attenuates Environmental Pollutant-Induced Damage in Dermal Fibroblasts. Int J Mol Sci 2019; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Qiang L, Yang S, Cui YH, He YY. Keratinocyte autophagy enables the activation of keratinocytes and fibroblastsand facilitates wound healing. Autophagy 2021; 17:2128–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Mahil SK, Twelves S, Farkas K, Setta-Kaffetzi N, Burden AD, Gach JE, et al. AP1S3 Mutations Cause Skin Autoinflammation by Disrupting Keratinocyte Autophagy and Up-Regulating IL-36 Production. J Invest Dermatol 2016; 136:2251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Johansen MD, Irving A, Montagutelli X, Tate MD, Rudloff I, Nold MF, et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol 2020; 13:877–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Kim JE, Kim HR, Kang SY, Jung MJ, Heo NH, Lee HJ, et al. Aryl Hydrocarbon Receptor and Autophagy-Related Protein Microtubule-Associated Protein Light Chain 3 Expression in Psoriasis. Ann Dermatol 2021; 33:138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Klapan K, Frangez Z, Markov N, Yousefi S, Simon D, Simon HU. Evidence for Lysosomal Dysfunction within the Epidermis in Psoriasis and Atopic Dermatitis. J Invest Dermatol 2021; 141:2838–48 e4. [DOI] [PubMed] [Google Scholar]
- 413.Schaaf MB, Houbaert D, Mece O, Agostinis P. Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ 2019; 26:665–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Patella F, Neilson LJ, Athineos D, Erami Z, Anderson KI, Blyth K, et al. In-Depth Proteomics Identifies a Role for Autophagy in Controlling Reactive Oxygen Species Mediated Endothelial Permeability. J Proteome Res 2016; 15:2187–97. [DOI] [PubMed] [Google Scholar]
- 415.Kim KA, Shin D, Kim JH, Shin YJ, Rajanikant GK, Majid A, et al. Role of Autophagy in Endothelial Damage and Blood-Brain Barrier Disruption in Ischemic Stroke. Stroke 2018; 49:1571–9. [DOI] [PubMed] [Google Scholar]
- 416.Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA, et al. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med 2013; 19:1281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol 2012; 590:3305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Weikel KA, Cacicedo JM, Ruderman NB, Ido Y. Glucose and palmitate uncouple AMPK from autophagy in human aortic endothelial cells. Am J Physiol Cell Physiol 2015; 308:C249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Torisu K, Singh KK, Torisu T, Lovren F, Liu J, Pan Y, et al. Intact endothelial autophagy is required to maintain vascular lipid homeostasis. Aging Cell 2016; 15:187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Vion AC, Kheloufi M, Hammoutene A, Poisson J, Lasselin J, Devue C, et al. Autophagy is required for endothelial cell alignment and atheroprotection under physiological blood flow. Proc Natl Acad Sci U S A 2017; 114:E8675–E84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Vasko R, Ratliff BB, Bohr S, Nadel E, Chen J, Xavier S, et al. Endothelial peroxisomal dysfunction and impaired pexophagy promotes oxidative damage in lipopolysaccharide-induced acute kidney injury. Antioxid Redox Signal 2013; 19:211–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Maurer K, Reyes-Robles T, Alonzo F 3rd, Durbin J, Torres VJ, Cadwell K. Autophagy Mediates Tolerance to Staphylococcus aureus Alpha-Toxin. Cell Host Microbe 2015; 17:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Keller MD, Ching KL, Liang FX, Dhabaria A, Tam K, Ueberheide BM, et al. Decoy exosomes provide protection against bacterial toxins. Nature 2020; 579:260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Li H, Gao A, Feng D, Wang Y, Zhang L, Cui Y, et al. Evaluation of the protective potential of brain microvascular endothelial cell autophagy on blood-brain barrier integrity during experimental cerebral ischemia-reperfusion injury. Transl Stroke Res 2014; 5:618–26. [DOI] [PubMed] [Google Scholar]
- 425.Shi W, Wei X, Wang Z, Han H, Fu Y, Liu J, et al. HDAC9 exacerbates endothelial injury in cerebral ischaemia/reperfusion injury. J Cell Mol Med 2016; 20:1139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Wei N, Yu SP, Gu XH, Chen DD, Whalin MK, Xu GL, et al. The involvement of autophagy pathway in exaggerated ischemic brain damage in diabetic mice. CNS Neurosci Ther 2013; 19:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Liu J, Weaver J, Jin X, Zhang Y, Xu J, Liu KJ, et al. Nitric Oxide Interacts with Caveolin-1 to Facilitate Autophagy-Lysosome-Mediated Claudin-5 Degradation in Oxygen-Glucose Deprivation-Treated Endothelial Cells. Mol Neurobiol 2016; 53:5935–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Hammoutene A, Biquard L, Lasselin J, Kheloufi M, Tanguy M, Vion AC, et al. A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis. J Hepatol 2020; 72:528–38. [DOI] [PubMed] [Google Scholar]
- 429.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264–8. [DOI] [PubMed] [Google Scholar]
- 430.Lee HY, Kim J, Quan W, Lee JC, Kim MS, Kim SH, et al. Autophagy deficiency in myeloid cells increases susceptibility to obesity-induced diabetes and experimental colitis. Autophagy 2016; 12:1390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Zhang H, Zheng L, McGovern DP, Hamill AM, Ichikawa R, Kanazawa Y, et al. Myeloid ATG16L1 Facilitates Host-Bacteria Interactions in Maintaining Intestinal Homeostasis. J Immunol 2017; 198:2133–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Xia Y, Liu N, Xie X, Bi G, Ba H, Li L, et al. The macrophage-specific V-ATPase subunit ATP6V0D2 restricts inflammasome activation and bacterial infection by facilitating autophagosome-lysosome fusion. Autophagy 2019; 15:960–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A 2014; 111:3526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Levin AD, Koelink PJ, Bloemendaal FM, Vos AC, D’Haens GR, van den Brink GR, et al. Autophagy Contributes to the Induction of Anti-TNF Induced Macrophages. J Crohns Colitis 2016; 10:323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Maisonneuve C, Tsang DKL, Foerster EG, Robert LM, Mukherjee T, Prescott D, et al. Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. Cell Rep 2021; 34:108677. [DOI] [PubMed] [Google Scholar]
- 436.Guo W, Sun Y, Liu W, Wu X, Guo L, Cai P, et al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014; 10:972–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Zhao Y, Guo Q, Zhao K, Zhou Y, Li W, Pan C, et al. Small molecule GL-V9 protects against colitis-associated colorectal cancer by limiting NLRP3 inflammasome through autophagy. Oncoimmunology 2017; 7:e1375640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Merkley SD, Goodfellow SM, Guo Y, Wilton ZER, Byrum JR, Schwalm KC, et al. Non-autophagy Role of Atg5 and NBR1 in Unconventional Secretion of IL-12 Prevents Gut Dysbiosis and Inflammation. J Crohns Colitis 2022; 16:259–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Wu MY, Liu L, Wang EJ, Xiao HT, Cai CZ, Wang J, et al. PI3KC3 complex subunit NRBF2 is required for apoptotic cell clearance to restrict intestinal inflammation. Autophagy 2021; 17:1096–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Germic N, Hosseini A, Stojkov D, Oberson K, Claus M, Benarafa C, et al. ATG5 promotes eosinopoiesis but inhibits eosinophil effector functions. Blood 2021; 137:2958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Masud S, Prajsnar TK, Torraca V, Lamers GEM, Benning M, Van Der Vaart M, et al. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy 2019; 15:796–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Masud S, van der Burg L, Storm L, Prajsnar TK, Meijer AH. Rubicon-Dependent Lc3 Recruitment to Salmonella-Containing Phagosomes Is a Host Defense Mechanism Triggered Independently From Major Bacterial Virulence Factors. Front Cell Infect Microbiol 2019; 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, et al. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 2019; 178:552–66 e20. [DOI] [PubMed] [Google Scholar]
- 444.Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, Te Velde AA, et al. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology 2012; 142:1493–503 e6. [DOI] [PubMed] [Google Scholar]
- 445.Wang YH, Wu JJ, Lei HY. When Helicobacter pylori invades and replicates in the cells. Autophagy 2009; 5:540–2. [DOI] [PubMed] [Google Scholar]
- 446.Wang YH, Gorvel JP, Chu YT, Wu JJ, Lei HY. Helicobacter pylori impairs murine dendritic cell responses to infection. PloS one 2010; 5:e10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology 2012; 142:1160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Deen NS, Huang SJ, Gong L, Kwok T, Devenish RJ. The impact of autophagic processes on the intracellular fate of Helicobacter pylori: more tricks from an enigmatic pathogen? Autophagy 2013; 9:639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Castano-Rodriguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM. Autophagy in Helicobacter pylori Infection and Related Gastric Cancer. Helicobacter 2015; 20:353–69. [DOI] [PubMed] [Google Scholar]
- 450.Symington JW, Wang C, Twentyman J, Owusu-Boaitey N, Schwendener R, Nunez G, et al. ATG16L1 deficiency in macrophages drives clearance of uropathogenic E. coli in an IL-1beta-dependent manner. Mucosal Immunol 2015; 8:1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885–9. [DOI] [PubMed] [Google Scholar]
- 452.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880–4. [DOI] [PubMed] [Google Scholar]
- 453.Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J 2021; 40:e108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Fleming A, Bourdenx M, Fujimaki M, Karabiyik C, Krause GJ, Lopez A, et al. The different autophagy degradation pathways and neurodegeneration. Neuron 2022; 110:935–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 2005; 64:113–22. [DOI] [PubMed] [Google Scholar]
- 456.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 2008; 28:6926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, et al. LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 2019; 178:536–51 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Heckmann BL, Teubner BJW, Boada-Romero E, Tummers B, Guy C, Fitzgerald P, et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Sci Adv 2020; 6:eabb9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Choi I, Zhang Y, Seegobin SP, Pruvost M, Wang Q, Purtell K, et al. Microglia clear neuron-released alpha-synuclein via selective autophagy and prevent neurodegeneration. Nat Commun 2020; 11:1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Park Y, Liu C, Luo T, Dietrich WD, Bramlett H, Hu B. Chaperone-Mediated Autophagy after Traumatic Brain Injury. J Neurotrauma 2015; 32:1449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Handa K, Kanno H, Matsuda M, Sugaya T, Murakami T, Prudnikova M, et al. Chaperone-Mediated Autophagy after Spinal Cord Injury. J Neurotrauma 2020; 37:1687–95. [DOI] [PubMed] [Google Scholar]
- 462.Cheng J, Liao Y, Dong Y, Hu H, Yang N, Kong X, et al. Microglial autophagy defect causes parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy 2020; 16:2193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chen J, Mao K, Yu H, Wen Y, She H, Zhang H, et al. p38-TFEB pathways promote microglia activation through inhibiting CMA-mediated NLRP3 degradation in Parkinson’s disease. J Neuroinflammation 2021; 18:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Valdor R, Garcia-Bernal D, Riquelme D, Martinez CM, Moraleda JM, Cuervo AM, et al. Glioblastoma ablates pericytes antitumor immune function through aberrant up-regulation of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 2019; 116:20655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, et al. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy 2009; 5:152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Liang P, Le W. Role of autophagy in the pathogenesis of multiple sclerosis. Neurosci Bull 2015; 31:435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Bhattacharya A, Parillon X, Zeng S, Han S, Eissa NT. Deficiency of autophagy in dendritic cells protects against experimental autoimmune encephalomyelitis. J Biol Chem 2014; 289:26525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Yang G, Song W, Xu J, Postoak JL, Cheng F, Martinez J, et al. Pik3c3 deficiency in myeloid cells imparts partial resistance to experimental autoimmune encephalomyelitis associated with reduced IL-1beta production. Cell Mol Immunol 2021; 18:2024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Yang G, Postoak JL, Song W, Martinez J, Zhang J, Wu L, et al. Dendritic cell PIK3C3/VPS34 controls the pathogenicity of CNS autoimmunity independently of LC3-associated phagocytosis. Autophagy 2022; 18:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Keller CW, Sina C, Kotur MB, Ramelli G, Mundt S, Quast I, et al. ATG-dependent phagocytosis in dendritic cells drives myelin-specific CD4(+) T cell pathogenicity during CNS inflammation. Proc Natl Acad Sci U S A 2017; 114:E11228–E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Keller CW, Kotur MB, Mundt S, Dokalis N, Ligeon LA, Shah AM, et al. CYBB/NOX2 in conventional DCs controls T cell encephalitogenicity during neuroinflammation. Autophagy 2021; 17:1244–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Srimat Kandadai K, Kotur MB, Dokalis N, Amrein I, Keller CW, Munz C, et al. ATG5 in microglia does not contribute vitally to autoimmune neuroinflammation in mice. Autophagy 2021; 17:3566–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Patergnani S, Bonora M, Ingusci S, Previati M, Marchi S, Zucchini S, et al. Antipsychotic drugs counteract autophagy and mitophagy in multiple sclerosis. Proc Natl Acad Sci U S A 2021; 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Haidar M, Loix M, Vanherle S, Dierckx T, Vangansewinkel T, Gervois P, et al. Targeting lipophagy in macrophages improves repair in multiple sclerosis. Autophagy 2022:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Berglund R, Guerreiro-Cacais AO, Adzemovic MZ, Zeitelhofer M, Lund H, Ewing E, et al. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci Immunol 2020; 5. [DOI] [PubMed] [Google Scholar]
- 476.Di Rita A, Strappazzon F. A protective variant of the autophagy receptor CALCOCO2/NDP52 in Multiple Sclerosis (MS). Autophagy 2021; 17:1565–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Di Rita A, Angelini DF, Maiorino T, Caputo V, Cascella R, Kumar M, et al. Characterization of a natural variant of human NDP52 and its functional consequences on mitophagy. Cell Death Differ 2021; 28:2499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Hait AS, Olagnier D, Sancho-Shimizu V, Skipper KA, Helleberg M, Larsen SM, et al. Defects in LC3B2 and ATG4A underlie HSV2 meningitis and reveal a critical role for autophagy in antiviral defense in humans. Sci Immunol 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007; 1:23–35. [DOI] [PubMed] [Google Scholar]
- 480.Waisner H, Kalamvoki M. The ICP0 Protein of Herpes Simplex Virus 1 (HSV-1) Downregulates Major Autophagy Adaptor Proteins Sequestosome 1 and Optineurin during the Early Stages of HSV-1 Infection. J Virol 2019; 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A 2012; 109:E3168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Koster S, Upadhyay S, Chandra P, Papavinasasundaram K, Yang G, Hassan A, et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc Natl Acad Sci U S A 2017; 114:E8711–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 2015; 528:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Crother TR, Porritt RA, Dagvadorj J, Tumurkhuu G, Slepenkin AV, Peterson EM, et al. Autophagy Limits Inflammasome During Chlamydia pneumoniae Infection. Front Immunol 2019; 10:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Galle-Treger L, Hurrell BP, Lewis G, Howard E, Jahani PS, Banie H, et al. Autophagy is critical for group 2 innate lymphoid cell metabolic homeostasis and effector function. J Allergy Clin Immunol 2020; 145:502–17 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Reed M, Morris SH, Jang S, Mukherjee S, Yue Z, Lukacs NW. Autophagy-inducing protein beclin-1 in dendritic cells regulates CD4 T cell responses and disease severity during respiratory syncytial virus infection. J Immunol 2013; 191:2526–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Owczarczyk AB, Schaller MA, Reed M, Rasky AJ, Lombard DB, Lukacs NW. Sirtuin 1 Regulates Dendritic Cell Activation and Autophagy during Respiratory Syncytial Virus-Induced Immune Responses. J Immunol 2015; 195:1637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Jin Y, Pan Y, Zheng S, Liu Y, Xu J, Peng Y, et al. Inactivation of EGLN3 hydroxylase facilitates Erk3 degradation via autophagy and impedes lung cancer growth. Oncogene 2022; 41:1752–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Dang AT, Teles RM, Liu PT, Choi A, Legaspi A, Sarno EN, et al. Autophagy links antimicrobial activity with antigen presentation in Langerhans cells. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Sil P, Suwanpradid J, Muse G, Gruzdev A, Liu L, Corcoran DL, et al. Noncanonical autophagy in dermal dendritic cells mediates immunosuppressive effects of UV exposure. J Allergy Clin Immunol 2020; 145:1389–405. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 491.Das LM, Binko AM, Traylor ZP, Peng H, Lu KQ. Vitamin D improves sunburns by increasing autophagy in M2 macrophages. Autophagy 2019; 15:813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Feng L, Song P, Xu F, Xu L, Shao F, Guo M, et al. cis-Khellactone Inhibited the Proinflammatory Macrophages via Promoting Autophagy to Ameliorate Imiquimod-Induced Psoriasis. J Invest Dermatol 2019; 139:1946–56 e3. [DOI] [PubMed] [Google Scholar]
- 493.Ushio H, Ueno T, Kojima Y, Komatsu M, Tanaka S, Yamamoto A, et al. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol 2011; 127:1267–76 e6. [DOI] [PubMed] [Google Scholar]
- 494.Mgrditchian T, Arakelian T, Paggetti J, Noman MZ, Viry E, Moussay E, et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A 2017; 114:E9271–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Rautou PE, Cazals-Hatem D, Feldmann G, Mansouri A, Grodet A, Barge S, et al. Changes in autophagic response in patients with chronic hepatitis C virus infection. Am J Pathol 2011; 178:2708–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Vescovo T, Romagnoli A, Perdomo AB, Corazzari M, Ciccosanti F, Alonzi T, et al. Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology 2012; 142:644–53 e3. [DOI] [PubMed] [Google Scholar]
- 497.Gunduz F, Aboulnasr FM, Chandra PK, Hazari S, Poat B, Baker DP, et al. Free fatty acids induce ER stress and block antiviral activity of interferon alpha against hepatitis C virus in cell culture. Virol J 2012; 9:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Chandra PK, Bao L, Song K, Aboulnasr FM, Baker DP, Shores N, et al. HCV infection selectively impairs type I but not type III IFN signaling. Am J Pathol 2014; 184:214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Panigrahi R, Chandra PK, Ferraris P, Kurt R, Song K, Garry RF, et al. Persistent hepatitis C virus infection impairs ribavirin antiviral activity through clathrin-mediated trafficking of equilibrative nucleoside transporter 1. J Virol 2015; 89:626–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Kurt R, Chandra PK, Aboulnasr F, Panigrahi R, Ferraris P, Aydin Y, et al. Chaperone-Mediated Autophagy Targets IFNAR1 for Lysosomal Degradation in Free Fatty Acid Treated HCV Cell Culture. PLoS One 2015; 10:e0125962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature 2009; 458:1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Ilyas G, Zhao E, Liu K, Lin Y, Tesfa L, Tanaka KE, et al. Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1beta. J Hepatol 2016; 64:118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Zhu S, Zhang J, Zhang L, Ma W, Man N, Liu Y, et al. Inhibition of Kupffer Cell Autophagy Abrogates Nanoparticle-Induced Liver Injury. Adv Healthc Mater 2017; 6. [DOI] [PubMed] [Google Scholar]
- 504.Yu Z, Xie X, Su X, Lv H, Song S, Liu C, et al. ATRA-mediated-crosstalk between stellate cells and Kupffer cells inhibits autophagy and promotes NLRP3 activation in acute liver injury. Cell Signal 2022; 93:110304. [DOI] [PubMed] [Google Scholar]
- 505.Denaes T, Lodder J, Chobert MN, Ruiz I, Pawlotsky JM, Lotersztajn S, et al. The Cannabinoid Receptor 2 Protects Against Alcoholic Liver Disease Via a Macrophage Autophagy-Dependent Pathway. Sci Rep 2016; 6:28806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Lodder J, Denaes T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, et al. Macrophage autophagy protects against liver fibrosis in mice. Autophagy 2015; 11:1280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Sun K, Xu L, Jing Y, Han Z, Chen X, Cai C, et al. Autophagy-deficient Kupffer cells promote tumorigenesis by enhancing mtROS-NF-kappaB-IL1alpha/beta-dependent inflammation and fibrosis during the preneoplastic stage of hepatocarcinogenesis. Cancer Lett 2017; 388:198–207. [DOI] [PubMed] [Google Scholar]
- 508.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100:15077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Deust A, Chobert MN, Demontant V, Gricourt G, Denaes T, Thiolat A, et al. Macrophage autophagy protects against hepatocellular carcinogenesis in mice. Sci Rep 2021; 11:18809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell metabolism 2011; 13:655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell metabolism 2012; 15:534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 2012; 15:545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Zhang H, Ge S, Ni B, He K, Zhu P, Wu X, et al. Augmenting ATG14 alleviates atherosclerosis and inhibits inflammation via promotion of autophagosome-lysosome fusion in macrophages. Autophagy 2021; 17:4218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Osonoi Y, Mita T, Azuma K, Nakajima K, Masuyama A, Goto H, et al. Defective autophagy in vascular smooth muscle cells enhances cell death and atherosclerosis. Autophagy 2018; 14:1991–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Qiao L, Wang HF, Xiang L, Ma J, Zhu Q, Xu D, et al. Deficient Chaperone-Mediated Autophagy Promotes Lipid Accumulation in Macrophage. J Cardiovasc Transl Res 2021; 14:661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Qiao L, Ma J, Zhang Z, Sui W, Zhai C, Xu D, et al. Deficient Chaperone-Mediated Autophagy Promotes Inflammation and Atherosclerosis. Circ Res 2021; 129:1141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Jansen HJ, van Essen P, Koenen T, Joosten LA, Netea MG, Tack CJ, et al. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 2012; 153:5866–74. [DOI] [PubMed] [Google Scholar]
- 520.Bravo-San Pedro JM, Sica V, Martins I, Pol J, Loos F, Maiuri MC, et al. Acyl-CoA-Binding Protein Is a Lipogenic Factor that Triggers Food Intake and Obesity. Cell Metab 2019; 30:754–67 e9. [DOI] [PubMed] [Google Scholar]
- 521.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010; 11:467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy 2015; 11:271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Kang YH, Cho MH, Kim JY, Kwon MS, Peak JJ, Kang SW, et al. Impaired macrophage autophagy induces systemic insulin resistance in obesity. Oncotarget 2016; 7:35577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Wu H, Wang Y, Li W, Chen H, Du L, Liu D, et al. Deficiency of mitophagy receptor FUNDC1 impairs mitochondrial quality and aggravates dietary-induced obesity and metabolic syndrome. Autophagy 2019; 15:1882–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Ko MS, Yun JY, Baek IJ, Jang JE, Hwang JJ, Lee SE, et al. Mitophagy deficiency increases NLRP3 to induce brown fat dysfunction in mice. Autophagy 2021; 17:1205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Grijalva A, Xu X, Ferrante AW Jr., Autophagy Is Dispensable for Macrophage-Mediated Lipid Homeostasis in Adipose Tissue. Diabetes 2016; 65:967–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Litwinoff EMS, Gold MY, Singh K, Hu J, Li H, Cadwell K, et al. Myeloid ATG16L1 does not affect adipose tissue inflammation or body mass in mice fed high fat diet. Obes Res Clin Pract 2018; 12:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Majai G, Kiss E, Tarr T, Zahuczky G, Hartman Z, Szegedi G, et al. Decreased apopto-phagocytic gene expression in the macrophages of systemic lupus erythematosus patients. Lupus 2014; 23:133–45. [DOI] [PubMed] [Google Scholar]
- 529.Wu J, Singh K, Lin A, Meadows AM, Wu K, Shing V, et al. Boosting NAD+ blunts TLR4-induced type I IFN in control and systemic lupus erythematosus monocytes. J Clin Invest 2022; 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70:1330–7. [DOI] [PubMed] [Google Scholar]
- 531.Li X, Liu F, Zhang X, Shi G, Ren J, Ji J, et al. Notch-Hes-1 axis controls TLR7-mediated autophagic death of macrophage via induction of P62 in mice with lupus. Cell Death Dis 2016; 7:e2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 2016; 533:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 533.Santeford A, Wiley LA, Park S, Bamba S, Nakamura R, Gdoura A, et al. Impaired autophagy in macrophages promotes inflammatory eye disease. Autophagy 2016; 12:1876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med 2011; 208:2625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Figueiredo N, Chora A, Raquel H, Pejanovic N, Pereira P, Hartleben B, et al. Anthracyclines induce DNA damage response-mediated protection against severe sepsis. Immunity 2013; 39:874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Oh DS, Lee HK. Autophagy protein ATG5 regulates CD36 expression and anti-tumor MHC class II antigen presentation in dendritic cells. Autophagy 2019; 15:2091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Xia H, Li S, Li X, Wang W, Bian Y, Wei S, et al. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, et al. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 2018; 175:429–41 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nature Reviews Drug Discovery 2017. 16:7 2017; 16:487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Panda PK, Fahrner A, Vats S, Seranova E, Sharma V, Chipara M, et al. Chemical screening approaches enabling drug discovery of autophagy modulators for biomedical applications in human diseases. Frontiers in Cell and Developmental Biology 2019; 7:38-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Tavakol S, Ashrafizadeh M, Deng S, Azarian M, Abdoli A, Motavaf M, et al. Autophagy Modulators: Mechanistic Aspects and Drug Delivery Systems. Biomolecules 2019; 9:530-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Xu L, Zhang C, Jiang N, He D, Bai Y, Xin Y. Rapamycin combined with MCC950 to treat multiple sclerosis in experimental autoimmune encephalomyelitis. Journal of cellular biochemistry 2019; 120:5160–8. [DOI] [PubMed] [Google Scholar]
- 543.O’Dell JR, Leff R, Paulsen G, Haire C, Mallek J, Eckhoff PJ, et al. Treatment of rheumatoid arthritis with methotrexate and hydroxychloroquine, methotrexate and sulfasalazine, or a combination of the three medications: results of a two-year, randomized, double-blind, placebo-controlled trial. Arthritis and rheumatism 2002; 46:1164–70. [DOI] [PubMed] [Google Scholar]
- 544.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LAM, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host and Microbe 2012; 11:563–75. [DOI] [PubMed] [Google Scholar]
- 545.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, et al. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. 10.4161/auto6246 2008; 4:744–53. [DOI] [PubMed] [Google Scholar]
- 546.Hanson KK, Ressurreicao AS, Buchholz K, Prudencio M, Herman-Ornelas JD, Rebelo M, et al. Torins are potent antimalarials that block replenishment of Plasmodium liver stage parasitophorous vacuole membrane proteins. Proceedings of the National Academy of Sciences of the United States of America 2013; 110:E2838–E47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Kumar A, Singh PK, Giri S. AMP-activated kinase (AMPK) promotes innate immunity and antiviral defense against Zika virus induced ocular infection. The Journal of Immunology 2018; 200. [Google Scholar]
- 548.Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, et al. Identification of Host-Targeted Small Molecules That Restrict Intracellular Mycobacterium tuberculosis Growth. PLOS Pathogens 2014; 10:e1003946-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Sogi KM, Lien KA, Johnson JR, Krogan NJ, Stanley SA. The Tyrosine Kinase Inhibitor Gefitinib Restricts Mycobacterium tuberculosis Growth through Increased Lysosomal Biogenesis and Modulation of Cytokine Signaling. ACS infectious diseases 2017; 3:564-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Chiu HC, Kulp SK, Soni S, Wang D, Gunn JS, Schlesinger LS, et al. Eradication of intracellular Salmonella enterica serovar Typhimurium with a small-molecule, host cell-directed agent. Antimicrobial agents and chemotherapy 2009; 53:5236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy–inducing peptide. Nature 2013; 494:201-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Lee YJ, Kim JK, Jung CH, Kim YJ, Jung EJ, Lee SH, et al. Chemical modulation of SQSTM1/p62-mediated xenophagy that targets a broad range of pathogenic bacteria. 10.1080/1554862720222054240 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Dittmar AJ, Drozda AA, Blader IJ. Drug Repurposing Screening Identifies Novel Compounds That Effectively Inhibit Toxoplasma gondii Growth. mSphere 2016; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Chauhan S, Ahmed Z, Bradfute SB, Arko-Mensah J, Mandell MA, Won Choi S, et al. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nature Communications 2015. 6:1 2015; 6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Ammanathan V, Mishra P, Chavalmane AK, Muthusamy S, Jadhav V, Siddamadappa C, et al. Restriction of intracellular Salmonella replication by restoring TFEB-mediated xenophagy. Autophagy 2020; 16:1584-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Belzile J-P, Sabalza M, Craig M, Clark E, Morello CS, Spector DH. Trehalose, an mTOR-Independent Inducer of Autophagy, Inhibits Human Cytomegalovirus Infection in Multiple Cell Types. Journal of virology 2015; 90:1259–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Sharma V, Makhdoomi M, Singh L, Kumar P, Khan N, Singh S, et al. Trehalose limits opportunistic mycobacterial survival during HIV co-infection by reversing HIV-mediated autophagy block. Autophagy 2021; 17:476–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Dai J, Liang Y, Li H, Zhou W, Wang B, Gong A, et al. Vitamin D enhances resistance to aspergillus fumigatus in mice via inhibition of excessive autophagy. American Journal of Translational Research 2018; 10:381-. [PMC free article] [PubMed] [Google Scholar]
- 559.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, et al. Vitamin D Is Required for IFN-γ–Mediated Antimicrobial Activity of Human Macrophages. Science Translational Medicine 2011; 3:104ra2-ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, et al. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy 2019; 15:707–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Schiebler M, Brown K, Hegyi K, Newton SM, Renna M, Hepburn L, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO molecular medicine 2015; 7:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Massey DCO, Bredin F, Parkes M. Use of sirolimus (rapamycin) to treat refractory Crohn’s disease. Gut 2008; 57:1294–6. [DOI] [PubMed] [Google Scholar]
- 563.Zhong M, Cui B, Xiang J, Wu X, Wen Q, Li Q, et al. Rapamycin is Effective for Upper but not for Lower Gastrointestinal Crohn’s Disease-Related Stricture: A Pilot Study. Frontiers in Pharmacology 2021; 11:2451-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Mutalib M, Borrelli O, Blackstock S, Kiparissi F, Elawad M, Shah N, et al. The use of sirolimus (rapamycin) in the management of refractory inflammatory bowel disease in children. Journal of Crohn’s and Colitis 2014; 8:1730–4. [DOI] [PubMed] [Google Scholar]
- 565.Kellermayer R, Chang A, Patel K. Sirolimus (Rapamycin) Induced Mucosal Healing in Anti-Tumor Necrosis Factor Refractory Pediatric Ulcerative Colitis. JPGN Reports 2022; 3:e183-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Bai A, Ma AG, Yong M, Weiss CR, Ma Y, Guan Q, et al. AMPK agonist downregulates innate and adaptive immune responses in TNBS-induced murine acute and relapsing colitis. Biochemical pharmacology 2010; 80:1708–17. [DOI] [PubMed] [Google Scholar]
- 567.Zhao J, Sun Y, Shi P, Dong JN, Zuo LG, Wang HG, et al. Celastrol ameliorates experimental colitis in IL-10 deficient mice via the up-regulation of autophagy. International immunopharmacology 2015; 26:221–8. [DOI] [PubMed] [Google Scholar]
- 568.Zhao J, Dong JN, Wang HG, Zhao M, Sun J, Zhu WM, et al. Docosahexaenoic Acid Attenuated Experimental Chronic Colitis in Interleukin 10–Deficient Mice by Enhancing Autophagy Through Inhibition of the mTOR Pathway. Journal of Parenteral and Enteral Nutrition 2017; 41:824–9. [DOI] [PubMed] [Google Scholar]
- 569.Holvoet T, Devriese S, Castermans K, Boland S, Leysen D, Vandewynckel YP, et al. Treatment of Intestinal Fibrosis in Experimental Inflammatory Bowel Disease by the Pleiotropic Actions of a Local Rho Kinase Inhibitor. Gastroenterology 2017; 153:1054–67. [DOI] [PubMed] [Google Scholar]
- 570.Hooper KM, Casanova V, Kemp S, Staines KA, Satsangi J, Barlow PG, et al. The Inflammatory Bowel Disease Drug Azathioprine Induces Autophagy via mTORC1 and the Unfolded Protein Response Sensor PERK. Inflammatory Bowel Diseases 2019; 25:1481–96. [DOI] [PubMed] [Google Scholar]
- 571.Gubatan J, Mitsuhashi S, Zenlea T, Rosenberg L, Robson S, Moss AC. Low Serum Vitamin D During Remission Increases Risk of Clinical Relapse in Patients With Ulcerative Colitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 2017; 15:240–6.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Kabbani TA, Koutroubakis IE, Schoen RE, Ramos-Rivers C, Shah N, Swoger J, et al. Association of Vitamin D Level With Clinical Status in Inflammatory Bowel Disease: A 5-Year Longitudinal Study. The American journal of gastroenterology 2016; 111:712–9. [DOI] [PubMed] [Google Scholar]
- 573.Lomia M, Tchelidze T, Pruidze M. Bronchial asthma as neurogenic paroxysmal inflammatory disease: A randomized trial with carbamazepine. Respiratory Medicine 2006; 100:1988–96. [DOI] [PubMed] [Google Scholar]
- 574.Tran I, Ji C, Ni I, Min T, Tang D, Vij N. Role of cigarette smoke-induced aggresome formation in chronic obstructive pulmonary disease-emphysema pathogenesis. American Journal of Respiratory Cell and Molecular Biology 2015; 53:159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Hoogendijk AJ, Pinhanços SS, Van Der Poll T, Wieland CW. AMP-activated Protein Kinase Activation by 5-Aminoimidazole-4-carbox-amide-1-β-d-ribofuranoside (AICAR) Reduces Lipoteichoic Acid-induced Lung Inflammation. Journal of Biological Chemistry 2013; 288:7047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, et al. Histone deacetylase 6–mediated selective autophagy regulates COPD-associated cilia dysfunction. The Journal of Clinical Investigation 2013; 123:5212–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. The Journal of clinical investigation 2014; 124:3987–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Villella VR, Esposito S, Ferrari E, Monzani R, Tosco A, Rossin F, et al. Autophagy suppresses the pathogenic immune response to dietary antigens in cystic fibrosis. Cell Death & Disease 2019. 10:4 2019; 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Tosco A, De Gregorio F, Esposito S, De Stefano D, Sana I, Ferrari E, et al. A novel treatment of cystic fibrosis acting on-target: cysteamine plus epigallocatechin gallate for the autophagy-dependent rescue of class II-mutated CFTR. Cell Death & Differentiation 2016. 23:8 2016; 23:1380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Caution K, Pan A, Krause K, Badr A, Hamilton K, Vaidya A, et al. Methylomic correlates of autophagy activity in cystic fibrosis. Journal of Cystic Fibrosis 2019; 18:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Flynn JM, O’Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging cell 2013; 12:851-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Gao G, Chen W, Yan M, Liu J, Luo H, Wang C, et al. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. International Journal of Molecular Medicine 2020; 45:195-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Gu J, Hu W, Song ZP, Chen YG, Zhang DD, Wang CQ. Rapamycin inhibits cardiac hypertrophy by promoting autophagy via the MEK/ERK/Beclin-1 pathway. Frontiers in Physiology 2016; 7:104-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Quarles E, Basisty N, Chiao YA, Merrihew G, Gu H, Sweetwyne MT, et al. Rapamycin persistently improves cardiac function in aged, male and female mice, even following cessation of treatment. Aging Cell 2020; 19:e13086-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Han Y, Xie H, Liu Y, Gao P, Yang X, Shen Z. Effect of metformin on all-cause and cardiovascular mortality in patients with coronary artery diseases: A systematic review and an updated meta-analysis. Cardiovascular Diabetology 2019; 18:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Kanamori H, Naruse G, Yoshida A, Minatoguchi S, Watanabe T, Kawaguchi T, et al. Metformin Enhances Autophagy and Provides Cardioprotection in δ-Sarcoglycan Deficiency-Induced Dilated Cardiomyopathy. Circulation: Heart Failure 2019; 12. [DOI] [PubMed] [Google Scholar]
- 587.Xie F, Xu S, Lu Y, Wong KF, Sun L, Hasan KMM, et al. Metformin accelerates zebrafish heart regeneration by inducing autophagy. NPJ Regenerative medicine 2021; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Kania E, Pająk B, O’Prey J, Sierra Gonzalez P, Litwiniuk A, Urbańska K, et al. Verapamil treatment induces cytoprotective autophagy by modulating cellular metabolism. The FEBS journal 2017; 284:1370–87. [DOI] [PubMed] [Google Scholar]
- 589.Lai JL, Lian YE, Wu JY, Wang YD, Bai YN. Verapamil induces autophagy to improve liver regeneration in non-alcoholic fatty liver mice. Adipocyte 2021; 10:532–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Zhong J, Gong W, Chen J, Qing Y, Wu S, Li H, et al. Micheliolide alleviates hepatic steatosis in db/db mice by inhibiting inflammation and promoting autophagy via PPAR-γ-mediated NF-кB and AMPK/mTOR signaling. International Immunopharmacology 2018; 59:197–208. [DOI] [PubMed] [Google Scholar]
- 591.Zhou S, Gu J, Liu R, Wei S, Wang Q, Shen H, et al. Spermine alleviates acute liver injury by inhibiting liver-resident macrophage pro-inflammatory response through ATG5-dependent autophagy. Frontiers in Immunology 2018; 9:948-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Zhou J, Farah BL, Sinha RA, Wu Y, Singh BK, Bay BH, et al. Epigallocatechin-3-Gallate (EGCG), a Green Tea Polyphenol, Stimulates Hepatic Autophagy and Lipid Clearance. PLOS ONE 2014; 9:e87161-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Lai ZW, Kelly R, Winans T, Marchena I, Shadakshari A, Yu J, et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. The Lancet 2018; 391:1186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Ponticelli C, Moroni G. Hydroxychloroquine in systemic lupus erythematosus (SLE). Expert opinion on drug safety 2017; 16:411–9. [DOI] [PubMed] [Google Scholar]
- 595.Wang W, Li C, Zhang Z, Zhang Y. Arsenic Trioxide in Synergy with Vitamin D Rescues the Defective VDR-PPAR- γ Functional Module of Autophagy in Rheumatoid Arthritis. PPAR Research 2019; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Yan H, Zhou H-f, Hu Y, Pham CTN. Suppression of experimental arthritis through AMP-activated protein kinase activation and autophagy modulation. Journal of rheumatic diseases and treatment 2015; 1:5-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Feng FB, Qiu HY. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2018; 102:1209–20. [DOI] [PubMed] [Google Scholar]
- 598.Huang G, Yuan K, Zhu Q, Zhang S, Lu Q, Zhu M, et al. Triptolide inhibits the inflammatory activities of neutrophils to ameliorate chronic arthritis. Molecular Immunology 2018; 101:210–20. [DOI] [PubMed] [Google Scholar]
- 599.Jannat A, John P, Bhatti A, Hayat MQ. Tomorou attenuates progression of rheumatoid arthritis through alteration in ULK-1 independent autophagy pathway in collagen induced arthritis mice model. Cell Death Discovery 2019. 5:1 2019; 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.He SD, Huang SG, Zhu HJ, Luo XG, Liao KH, Zhang JY, et al. Oridonin suppresses autophagy and survival in rheumatoid arthritis fibroblast-like synoviocytes. 10.1080/1388020920201711783 2020; 58:146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Kim YR, Kim JS, Gu SJ, Jo S, Kim S, Young Kim S, et al. Identification of highly potent and selective inhibitor, TIPTP, of the p22phox-Rubicon axis as a therapeutic agent for rheumatoid arthritis. Scientific Reports 2020. 10:1 2020; 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Esposito M, Ruffini F, Bellone M, Gagliani N, Battaglia M, Martino G, et al. Rapamycin inhibits relapsing experimental autoimmune encephalomyelitis by both effector and regulatory T cells modulation. Journal of neuroimmunology 2010; 220:52–63. [DOI] [PubMed] [Google Scholar]
- 603.Zhen C, Feng X, Li Z, Wang Y, Li B, Li L, et al. Suppression of murine experimental autoimmune encephalomyelitis development by 1,25-dihydroxyvitamin D3 with autophagy modulation. Journal of neuroimmunology 2015; 280:1–7. [DOI] [PubMed] [Google Scholar]
- 604.Koch MW, Zabad R, Giuliani F, Hader W, Lewkonia R, Metz L, et al. Hydroxychloroquine reduces microglial activity and attenuates experimental autoimmune encephalomyelitis. Journal of the neurological sciences 2015; 358:131–7. [DOI] [PubMed] [Google Scholar]
- 605.Choi BY, Jang BG, Kim JH, Seo JN, Wu G, Sohn M, et al. Copper/zinc chelation by clioquinol reduces spinal cord white matter damage and behavioral deficits in a murine MOG-induced multiple sclerosis model. Neurobiology of disease 2013; 54:382–91. [DOI] [PubMed] [Google Scholar]