Abstract
Hrp1 of Schizosaccharomyces pombe is a member of the CHD protein family, characterized by a chromodomain, a Myb-like telobox-related DNA-binding domain and a SNF2-related helicase/ATPase domain. CHD proteins are thought to be required for modification of the chromatin structure in transcription, but the exact roles of CHD proteins are not known. Here we examine the sub-cellular localization and biochemical activity of Hrp1 and the phenotypes of hrp1Δ and Hrp1-overexpressing strains. Fluorescence microscopy revealed that Hrp1 protein is targeted to the nucleus. We found that Hrp1 exhibited DNA-dependent ATPase activity, stimulated by both single- and double-stranded DNA. Overexpression of Hrp1 caused slow cell growth accompanied by defective chromosome condensation in anaphase resulting in a ‘cut’ (cell untimely torn) phenotype and chromosome loss. The hrp1Δ mutation also caused abnormal anaphase and mini-chromosome loss phenotypes. Electron micrographs demonstrated that aberrantly shaped nucleoli appeared in Hrp1-overexpressing cells. Therefore, these results suggest that Hrp1 may play a role in mitotic chromosome segregation and maintenance of chromatin structure by utilizing the energy from ATP hydrolysis.
INTRODUCTION
In all eukaryotic organisms, DNA is packaged via nucleosomes into highly ordered and dynamic chromatin. During the mitotic cell division cycle, entire chromosomes must first be completely replicated before the newly synthesized sister chromatids condense and are segregated equally to the nuclei of daughter cells in which they decondense in interphase. Following cytokinesis the whole chromosome cycle is ready to be reinitiated. In addition, activation and repression of transcription are frequently accompanied by reorganization of the chromatin structure, facilitating access of the required DNA-binding proteins. These structural changes involve local disruption or reformation of nucleosomes and can be mediated by large multiprotein structures called chromatin remodeling complexes (1,2). These events are highly conserved in all eukaryotes from unicellular yeast to human.
The first remodeling complex identified was the budding yeast 11 subunit SWI/SNF complex, which is required for chromatin derepression and activation of a set of inducible genes (3–5). Other related remodeling complexes were more recently isolated from Drosophila. These complexes, named NURF (nucleosome remodeling factor; 6), CHRAC (chromatin accessibility complex; 7) and ACF (ATP-dependent chromatin assembly and remodeling factor; 8), were found to arrange the histone octamers to promote nucleosome mobility and to stimulate binding of transcription factors. Interestingly, one or more subunits of each complex contain a conserved sequence called the SNF2-related helicase/ATPase domain. The basic function of this domain has been proposed to destabilize protein–DNA interaction by moving along DNA templates (9).
Another class of related proteins implicated in chromatin remodeling is the CHD (chromo-helicase/ATPase DNA-binding protein) family. These proteins contain not only the SNF2-related domain but also two chromodomains (chromatin organization modifier), generally believed to be involved in the formation of repressed and heterochromatic structures (10,11). The chromodomain consists of 30–50 amino acids conserved in several eukaryotic chromatin-binding proteins such as Drosophila HP1 (heterochromatin protein 1), Pc (Polycomb) and Suvar3-9 proteins in Drosophila and mouse (12–14). In fission yeast there are three previously known chromodomain proteins: Swi6p (switching; 15), Clr4p (cryptic loci regulator; 16,17) and Chp1p (chromo domain protein in Schizosaccharomyces pombe; 18). Although the mechanism by which chromodomains interact with chromatin is unclear, synthetic chromodomain peptides are able to self-associate, providing chromodomain-containing proteins the potential to bind to each other and to form complexes among unknown components of the heterochromatin (19).
The S.pombe Hrp1 is a new member of the chromodomain and of the SNF2/SWI2 family, with two chromodomains, a SNF2-related helicase/ATPase domain and a Myb-like telobox-related DNA-binding domain (20). Until now, Hrp1 has been known to contain DNA-binding activity with a preference for (A+T)-rich tracts in double-stranded DNA via interaction with the minor groove. However, like other family members, it exhibits no helicase activity in spite of the presence of the conserved helicase domains (21).
In this report, we show that Hrp1 has a DNA-dependent ATPase activity. We investigate the subcellular localization of Hrp1 and further characterize its cellular functions. We demonstrate that hrp1Δ has a slight mitotic chromosome loss and defective anaphase phenotypes and that overexpression of Hrp1 leads to a severe chromosome decondensation and chromosome segregation defect similar to that of top2 and condensin mutants (22,23). Thus, Hrp1 directly or indirectly affects the sister chromatid structure that is vital for segregation and separation of chromosomes in mitosis.
MATERIALS AND METHODS
Strains, media and techniques for molecular biology and genetics
Schizosaccharomyces pombe strains used in this study are listed in Table 1. Schizosaccharomyces pombe cells were grown in Edinburgh Minimal Medium (EMM) supplemented with appropriate amino acids at 30°C (24). For overexpression, cultures were grown in EMM containing 2 µM thiamine (Sigma) to exponential phase, then washed three times and subsequently grown in medium lacking thiamine for 12–14 h. Transformation of S.pombe was performed by the dimethyl sulfoxide-enhanced lithium method (25). Escherichia coli strain XL2 blue (Stratagene, USA) was used as a host for propagation of plasmids. Western blot analysis was carried out as described by Jin et al. (20).
Table 1. Schizosaccharomyces pombe strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| FY367 | h+ ade6-M210 leu1-32 ura4-D18 | R. Allshire |
| JY746 | h+ ade6-M216 leu1-32 ura4-D18 | M. Yamamoto |
| JYK121 | h– ade6-M216 leu1-32 ura4-D18 hrp1::ura4+ | Y.H. Jin |
| JYK672 | h+ ade6-M216 leu1-32 ura4-D18 hrp1::nmt1-6(His)-hrp1+-ura4+ | Y.H. Jin |
| FY1497 | h+ leu1-32 ura4DS/E ade6-210//Ch16 ade6-216 ura4-tel | R. Allshire |
| FY121 | h+ leu1-32 hrp1::LEU2 ura4DS/E ade6-210//Ch16 ade6-216 ura4-tel | This study |
| FY672 | h+ leu1-32 ura4DS/E ade6-210 hrp1::nmt1-6(His)- hrp1+-LEU2//Ch16 ade6-216 ura4-tel | This study |
Fluorescence microscopy
DNA was visualized by staining with 2.0 µg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) in mounting medium (26). Septa were visualized with 0.2 mg/ml Calcofluor (fluorescent brightener; Sigma). Yeast cells were fixed with 3% (w/v) paraformaldehyde as described by Guthrie and Fink (27). Indirect immunofluorescence microscopy was performed using DAPI, affinity-purified anti-Hrp1 antibody, anti-tubulin TAT1 antibody and FITC-conjugated donkey anti-rabbit and anti-mouse IgG antibody (Jackson ImmunoResearch, USA). Fluorescence was observed with Zeiss Axiophot and Axioskop 2 microscopes with a 100 W light source, Hamamatsu CCD camera and Openlab2 image capturing software (Improvision).
Preparation of 6×His-tagged Hrp1 for ATPase activity assay
The 6×His-tagged Hrp1 was purified from Hrp1-overproducing JYK672 cells as described by Jin et al. (21). For ATPase activity assay, purified Hrp1 protein (40 ng) was incubated at 30°C for 15 min in 20 mM Tris–HCl (pH 7.0), 5% glycerol, 0.05% Tween-20, 30 mM NaCl, 1 mM DTT, 2 mM MgCl2, 0.5 mg/ml BSA, 1 mM ATP, 1 µCi of [γ-32P]ATP (3000 Ci/mmol; Amersham) and 100 ng pBluescript II KS(+) dsDNA. After incubation, an aliquot (1 µl) was spotted onto a polyethyleneimine–cellulose TLC plate (Merck, Germany) and developed in a solution containing 0.5 M LiCl and 1 M formic acid. Radiolabeled ATP and hydrolyzed free 32P were quantitated with a Bio-Imaging Analyzer BAS-1500 and Image Guage v.3.1 software (Fujifilm, Japan). The quantities of ATP hydrolysis (pmol, y-axis) were calculated from the percentage of hydrolyzed 32P out of total ATP {(radioactivity of hydrolyzed 32P/total radioactivity) × ([γ-32P]ATP + ATP)}.
Flow cytometry
Yeast cell fixation and nucleus staining were carried out as described by Sazer and Sherwood (28). Cells (2–3 × 106) from exponentially growing cultures were fixed with 1 ml of cold 70% ethanol. After spinning briefly, the ethanol was removed and cells were rehydrated by washing with 1 ml of 50 mM sodium citrate. The pelleted cells were resuspended in 0.5 ml of 50 mM sodium citrate containing RNase (0.1 mg/ml) and incubated at 37°C for 2 h. For staining, another 0.5 ml of 50 mM sodium citrate containing 10 µg/ml propidium iodide (Sigma) was added to a final concentration of 5 µg/ml. FACS analysis was carried out using a Becton-Dickinson FACStar+ and LYSIS II software.
Measurement of mini-chromosome stability
Determination of mitotic stability of the Ch16 mini-chromosome was carried out as follows. FY1497 (wild-type), FY121 (hrp1Δ) and FY672 (Hrp1-overexpressing) strains carrying a Ch16 mini-chromosome were inoculated into 10 ml of liquid EMM with uracil (75 mg/l), leucine (250 mg/l) and thiamine (2 µM) at 30°C. When cell density reached 2 × 107 cells/ml, each sample was washed and re-inoculated into EMM (plus adenine and uracil) at a concentration of 2 × 106 cells/ml. Then the cells were plated on YE (plus uracil and leucine) at the intervals indicated and incubated at 30°C for 3–5 days followed by an additional 2–3 days at 4°C to allow the red color to deepen. The rate of chromosome loss was measured as the number of half-sectored red and white colonies divided by the total number of white and half-sectored red and white colonies, according to the procedure of Allshire et al. (16).
High pressure freezing and transmission electron microscopy
Cells were prepared for electron microscopy by freeze substitution fixation after rapid high pressure freezing (29). Cells were harvested from liquid cultures grown in the presence or absence of thiamine by centrifugation at 3000 g for 4 min at room temperature. Cells were transferred to a brass hat and frozen with a jet of liquid N2 at a pressure of ~1000 bar within 0.6–0.7 s. Frozen samples were kept in liquid N2 until they were chemically fixed and dehydrated. Fixation and dehydration by freeze substitution was done in methanol containing 2% glutaraldehyde, 0.5% uranyl acetate and 1% OsO4 at –94°C for 8 h, followed by –60°C for 8 h and finally –45°C for 2 h. Samples were then transferred to acetone kept at room temperature for 30 min, whereupon they were gradually embedded in LX112/acetone (1:2 for 3 h, 1:1 overnight and 2:1 for 1 day and, finally, in LX112 for 3–4 days at 60°C). Serial sections of 40–60 nm thickness were cut using a Leica Ultracut E microtome, picked up on formvar-coated carbon-stabilized slot grids and stained with 5% uranyl acetate in 70% methanol followed by 3% lead citrate. Sections were imaged on Kodak 4489 film in a Leo 906 electron microscope operating at 80 kV.
RESULTS
Hrp1 protein is localized in the nucleus
Indirect immunofluorescence microscopy with affinity-purified anti-Hrp1 polyclonal antibodies was used to examine the subcellular distribution of Hrp1 protein in S.pombe cells. The analysis showed that Hrp1 is predominantly localized in the nucleus with an evenly dispersed pattern (compare upper and lower left panels in Fig. 1A). The control experiment (right panels of Fig. 1A) revealed that the faint spots in the cytoplasm shown in the top left panel of Figure 1A were non-specific. In order to analyze the location of the nuclear localization signal (NLS), we made green fluorescent protein (GFP)–Hrp1 fusion constructs. As seen in Figure 1B, amino acids 1–152 were sufficient to direct GFP to the nucleus and deletion of amino acids 122–152 from this construct abolished the nuclear localization. The results suggest that Hrp1 is predominantly a nuclear protein and it is possible that its subcellular localization may be determined by a putative NLS which is within amino acids 122–152.
Figure 1.

Determination of Hrp1 subcellular localization. (A) Wild-type (FY367) and hrp1-deleted cells (JYK121) were stained either with DAPI for DNA or with fluorescein-linked anti-rabbit IgG antibody to anti-Hrp1 antibody. The upper and lower panels show fluorescein and DAPI images, respectively. (B) The pGFP (pREP42 backbone) was used to make the constructs pGFPH1NH, pGFPH1NS, pGFPH1h52 and pGFPH1NT. Each cell containing the construct was grown in EMM (+2 µM thiamine) to OD595 = 0.7 and resuspended in EMM lacking thiamine. The cells were fixed after 12 h incubation and stained with DAPI. The upper images are GFP and the lower are DAPI stained. Scale bar 10 µm.
Hrp1 has a DNA-dependent ATPase activity
Sequence analyses showed that Hrp1 contains a unique ATPase motif, a DNA-binding domain and helicase motifs found in the SWI2/SNF2 protein family. To see if the ATPase motif of Hrp1 has a true function, we purified 6×His-tagged Hrp1 protein and tested it for ATPase activity. As shown in Table 2, ATPase activity of Hrp1 was stimulated up to 9-fold by both dsDNA and ssDNA. Little ATPase activity was observed when S.pombe total RNA was added to the reaction, implying that RNA is a poor cofactor for ATP hydrolysis by Hrp1 (Table 2). At least 100 ng of dsDNA were required for DNA-dependent ATP hydrolysis (Fig. 2A). In vitro ATPase activity of Hrp1 had an optimal pH of 7.0 (Fig. 2B) and required Mg2+, which could be replaced by Ca2+ and Mn2+, but not by Zn2+ (Table 2). As shown in Figure 2C, ATP hydrolytic activity of Hrp1 proceeds in a concentration- and time-dependent manner. We also examined whether other nucleotides could be substrates for Hrp1 ATPase. The enzyme hydrolyzed dATP with similar efficacy and cofactor requirements as with ATP (data not shown). Other NTPs and dNTPs, however, were not significantly hydrolyzed by Hrp1 (data not shown).
Table 2. Requirements of Hrp1 ATPase activity.
| Conditions | Amount added or final concentration | Relative activitya (%) |
|---|---|---|
| Completeb | 100 | |
| No DNA | 11 | |
| Double-stranded DNAc | 100 ng | 100 |
| Single-stranded DNAd | 100 and 240 ng | 92, 100 |
| Schizosaccharomyces pombe total RNA | 100 and 1000 ng | 26, 29 |
| Omit Mg2+ | <1 | |
| Add Mg2+ | 2 mM | 100 |
| Add Ca2+ | 2 mM | 97 |
| Add Mn2+ | 2 mM | 105 |
| Add Zn2+ | 2 mM | 1 |
aThe value of 100% in this experiment corresponded to hydrolysis of 1990 pmol of ATP.
bThe complete reaction contained 40 ng of Hrp1 in the standard reaction mixture described under Materials and Methods.
cThe double-stranded DNA was the circular form of pBluescript II KS(+). The 100 ng of DNA added was equivalent to 2.54 nM molar concentration.
dThe single-stranded DNA used was the circular form of M13mp18. The 100 and 240 ng of DNA added were equivalent to 2.11 and 5.08 nM molar concentrations, respectively.
Figure 2.
Characterization of ATPase activity of Hrp1. ATP hydrolysis was measured as described in Materials and Methods under the following various conditions. (A) Determination of minimal requirement for dsDNA. ATPase activity was assayed by increasing amounts of dsDNA [pBluescript II KS(+), 2.5–1000 ng]. (B) Examination of optimal pH. ATP hydrolysis was measured under various buffer conditions as follows: pH 5.0–6.5, MES (2-[N-morpholino]ethanesulfonic acid); pH 7.0–8.5, Tris–HCl; pH 10, CAPS (3-[cyclohexylamino]-1-propanesulfonic acid). (C) Kinetic analysis of ATPase activity. Reaction mixtures containing 20 ng (closed circle), 40 ng (open circle), 80 ng (closed triangle) and 160 ng (open triangle) of Hrp1were incubated for the period of time indicated. (D) Determination of Km for ATP of Hrp1. The double reciprocal Lineweaver–Burk graph (1/reaction velocity, 1/v (1/mmol of ATP hydrolyzed min–1) versus 1/ATP) was plotted according to results obtained from the standard ATPase assay containing 40 ng Hrp1 with varying concentrations of ATP after incubation for 30 min.
Kinetic analysis of the Hrp1 ATPase was carried out by varying the ATP concentration (100–2000 µM). The Km value, representing the ATP concentration at half maximal velocity, was calculated from a Lineweaver–Burk plot to be 5.5 × 10–4 M (Fig. 2D). These results show that Hrp1 also has a catalytic function driven by the energy from ATP hydrolysis, which is similar to that of other ATPases from the SWI2/SNF2 family.
Hrp1 disrupts mitotic chromosome segregation when overexpressed
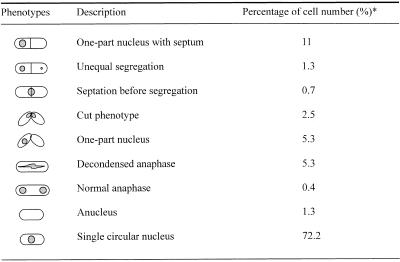
The growth rates of hrp1 deletion (hrp1Δ) and Hrp1-overproducing cells (JYK672) have been previously reported (20). Strain hrp1Δ enters the exponential growth phase slightly faster than the wild-type, whereas JYK672 (overexpressing) cells are unable to grow exponentially. To investigate if the growth defects correlated with structural changes in nuclear morphology, the hrp1Δ and Hrp1-overexpressing cells were examined by fluorescence microscopy. While we observed infrequently altered states of chromatin compaction in hrp1Δ cells, chromosome decondensation and missegregation were readily observed by microscopic examination in JYK672 (Hrp1-overexpressing) cells after DAPI staining (Fig. 3) and altered ploidy was detected by FACS analysis of DNA content (Fig. 4). JYK672 cells were apparently not able to proceed with coordinated mitosis and thus showed various aberrant patterns of chromosomal segregation, as shown in Figure 3. At least 27% of the Hrp1-overexpressing cells (JYK672) showed abnormal segregation of the chromosomes after 12 h in thiamine-depleted (overexpression) conditions (Fig. 3C–F and Table 3). The number of cells showing abnormal mitotic phenotypes was closely correlated with the incubation time under derepressing condition of the nmt1 promoter. Some of the cells had undergone cytokinesis without a prior completion of mitosis, resulting in displacement of the nucleus and anucleation in daughter cells (Fig. 3C) and some showed uneven nuclear segregation (Fig. 3D). Some of septated cells displayed a ‘cut’ (cell untimely torn) phenotype (Figs 3E and F and 5C; 30) due to untimely cleavage of the undivided nucleus by the septum.
Figure 3.

Phenotypic characterization of hrp1-deleted and Hrp1-overexpressing cells with DAPI and Calcofluor. (A) JY746 (wild-type), (B) JYK121 (hrp1Δ) and (C–F) JYK672 (Hrp1-overexpressing) cells were cultured under the appropriate conditions and fixed with paraformaldehyde and stained with DAPI and Calcofluor. Scale bar 5 µm.
Figure 4.
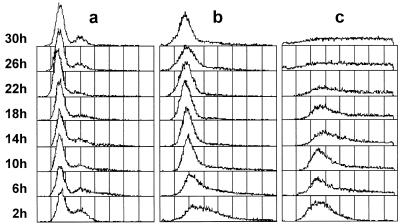
Flow cytometric analysis of (a) JY746 (wild-type), (b) JYK672 cultured with thiamine (Hrp1-repressing) and (c) JYK672 cultured without thiamine (Hrp1-overexpressing). Cells were pre-grown in EMM (+2 µM thiamine), washed and then divided into EMM lacking thiamine and containing thiamine, respectively. After collection at the indicated time points, the cells were fixed with ethanol, stained with propidium iodide and their DNA content was determined by flow cytometry. x-axis, fluorescence (DNA content on an arbitrary scale); y-axis, frequency (cell number).
Table 3. Aberrant phenotypes of Hrp1-overexpressing cells.
*A total of 603 cells was measured
Figure 5.
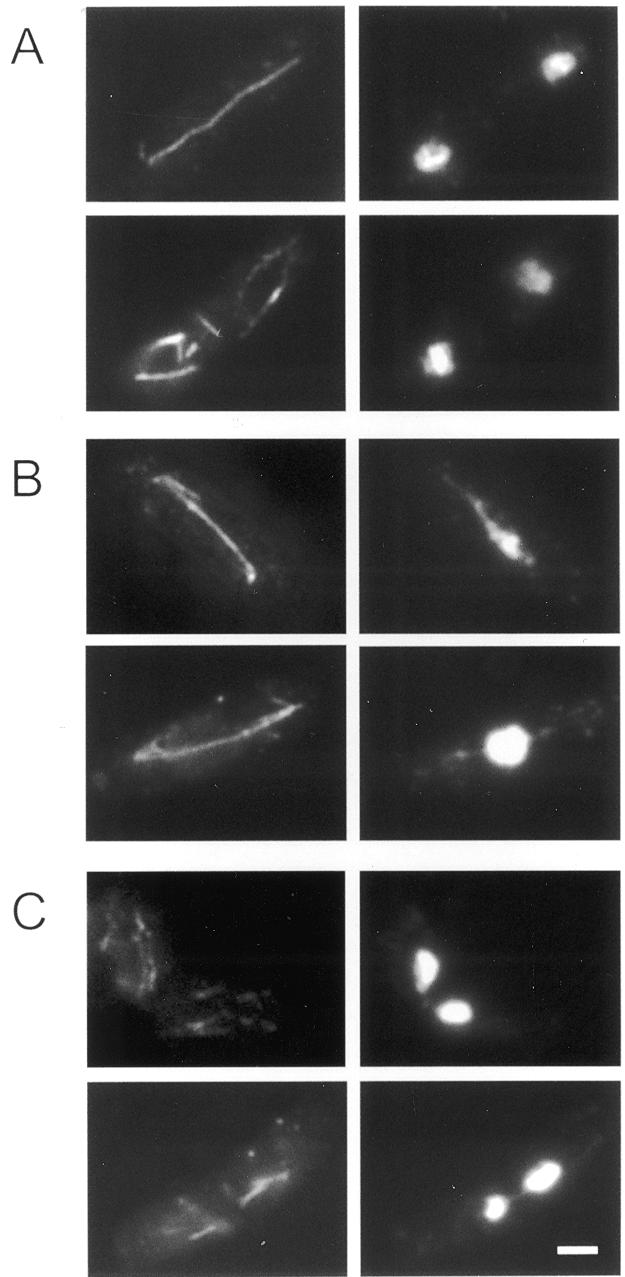
Anaphase decondensation and ‘cut’ phenotype of Hrp1-overexpressing cells. Cells were stained with TAT1 antibody detecting α-tubulin (left) and DAPI detecting chromosomes (right). (A) Wild-type in anaphase (top) and interphase (bottom). (B) Hrp1-overexpressing cells showing decondensed anaphase similar to top2, cut3 and cut14 mutants (22). (C) Hrp1-overexpressing cells showing the ‘cut’ phenotype (30). Scale bar 3 µm.
As shown in Figure 4, FACS analysis revealed an aberrant DNA profile of Hrp1-overexpressing cells. While the DNA profile of the wild-type cells was unchanged even after 30 h in the thiamine-depleted medium, a significant change was observed in the DNA profile of Hrp1-overproducing cells. During the early period of incubation, JYK672 cells cultured both with (Fig. 4, b) and without (Fig. 4, c) thiamine showed a somewhat broader spectrum but their 2C DNA peaks were slightly shifted toward higher DNA contents compared to that of wild-type cells (Fig. 4, a). Prolonged thaimine-depleted incubation of JYK672 cells under the same conditions led to a decrease in the number of cells with 2C DNA content and then the appearance of cells containing higher or lower DNA contents. Incubation >26 h resulted in eventual loss of the 2C peak. Thus, it seems as if overproduction of Hrp1 led to aberrant mitosis and missegregation of chromosomes.
Both deletion of the hrp1 gene and overexpressed Hrp1 cause anaphase decondensation in mitosis
To examine the anaphase defects in hrp1Δ and Hrp1-overexpressing cells, we performed immunofluorescent staining with an anti-tubulin antibody, TAT1 (Fig. 5). Tubulin staining revealed a substantial fraction of abnormal anaphase (12% abnormal anaphase cells, n = 50) in hrp1Δ (JYK121) cells as compared to wild-type (FY367) control cells (<2.0% abnormal anaphase cells, n = 51). The abnormal hrp1Δ cells had lagging chromosomes in the mid zone of late anaphase spindles. Overexpression of Hrp1 caused a more severe defect in anaphase chromosome behavior. Figure 5A shows normal anaphase (top) and normal post-anaphase configurations (bottom) of microtubules and chromosomes. While all wild-type cells found in anaphase showed normal morphologies, most (93%) Hrp1-overexpressing cells in anaphase had abnormal decondensed and entangled chromosomes (Fig. 5B and Table 3). These cells had elongated nuclear structures, as seen in top2 mutants, and their chromatin fibers seemed to extend along the spindle. The phenotype of defective anaphase found in Hrp1-overexpressing cells was similar to those of cut3, cut14 and top2 mutants (22,23).
Hrp1 is required for mitotic mini-chromosome stability
Since the cytological studies indicated that Hrp1-overexpressing cells were likely to be defective in chromosome segregation, we examined the mitotic stability of the Ch16 mini-chromosome in strains FY672 and FY121. These are hrp1-deleted (FY121) and an integrated copy of nmt1-hrp1+ (FY672) strains which contain the Ch16 mini-chromosome carrying an ade6-216 allele that can complement the ade6-210 mutation of the host cell. Hrp1 expression in the nmt1-hrp1 integrant was measured by western blot analysis (Fig. 6). The percentage of cells retaining a mini-chromosome was measured after each culture was incubated on YE plates containing a low concentration of adenine. As shown in Table 4, Ch16 mini-chromosomes were frequently lost in the Hrp1-overexpressing strain. Since full induction of the Hrp1 protein in FY672 cells led to some lethality, the mini-chromosome loss rate was likely to be underestimated. In addition, loss of function of Hrp1 resulted in a 10-fold increase in chromosome loss rate compared to that of wild-type cells (0.03%). This result suggests that a balanced level of Hrp1 is important for proper chromosome segregation.
Figure 6.
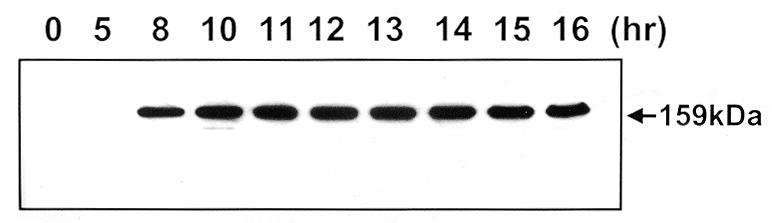
Induction kinetics of Hrp1 protein. Hrp1-overexpressing cells (FY672) grown in EMM with 2 µM thiamine were washed three times with fresh EMM and diluted in EMM to OD595 = 0.1. Cells were further cultured at 30°C and collected at each time point as indicated. After preparation of whole cell extract, expression of Hrp1 was monitored by western blot.
Table 4. Effect of Hrp1 overexpression and hrp1 deletion on Ch16 minichromosome mitotic instability.
| Strain | Background | Induction time (h) | Half-sectored colonies/total colonies | Loss rate (%)a |
|---|---|---|---|---|
| FY1497 | Wild-type | 1/3536 | 0.03 | |
| FY121 | hrp1Δ | 11/3615 | 0.30 | |
| FY672 | nmt1-hrp1 | 0 | 2/477 | 0.40 |
| 4 | 2/457 | 0.40 | ||
| 8 | 3/481 | 0.62 | ||
| 12 | 7/605 | 1.16 | ||
| 16 | 12/624 | 1.92 | ||
| 24 | 10/398 | 2.51 |
aThe loss rate was determined by the method of Allshire et al. (16) as described in Materials and Methods.
Overexpression of Hrp1 induces an aberrant nucleolar structure
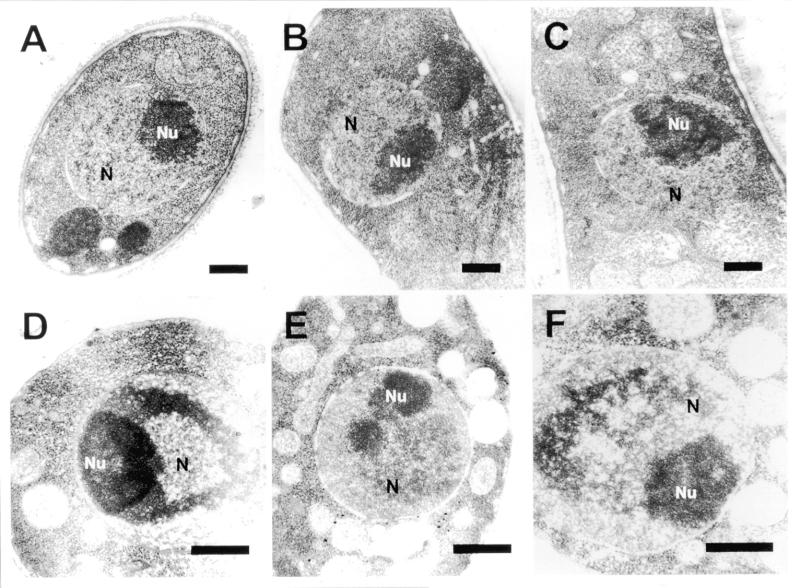
To determine whether abnormal chromatin compaction causes the slow growth and unequal chromatin segregation of Hrp1-overexpressing cells, the cellular morphologies of wild-type (FY367), hrp1-deleted (JYK121) and Hrp1-overexpressing (JYK672) cells were examined by transmission electron microscopy. As shown in Figure 7A–C, intact nuclei and nucleoli were observed in the wild-type, hrp1 deletion mutant and Hrp1-repressed JYK672 cells. However, odd electron-dense materials were found to accumulate in the nucleus of Hrp1-overexpressing cells (Fig. 7D–F). The distinct nucleoli structures found in the cells were classified as follows: (i) horseshoe-shaped nucleoli (Fig. 7D, most frequently found); (ii) two unequally separated nucleoli (Fig. 7E); and (iii) in rare cases, a widely dispersed pattern of dark material in the nucleus as shown in Figure 7F. These results indicate that Hrp1 may also play a role in assembly or maintenance of nucleolar structures.
Figure 7.
Transmission electron micrographs of wild-type (FY367), hrp1Δ (JYK121) and Hrp1-overexpressing (JYK672) cells. (A) Wild-type strain, FY367. (B) hrp1-deleted mutant, JYK121. (C) Hrp1-repressed JYK672 grown in the presence of thiamine. (D–F) Hrp1-overexpressing cells. Nucleolar condensation is seen in the Hrp1-overexpressing strain. N, nucleus, Nu, nucleolus. Scale bar 0.72 µm.
DISCUSSION
In this report, we present the subcellular localization of Hrp1 protein and its enzymatic activities in vitro and describe several interesting phenotypes of hrp1Δ and Hrp1-overexpressing cells with relevance to its in vivo function.
Diffuse sub-nuclear localization of Hrp1
In fission yeast, Swi6p, Clr4p and Chp1p are known to contain a chromodomain. Swi6p and Clr4p are required for formation of heterochromatin structures at centromeres, near telomeres and in the silent mating type region (15,16,31). Since Hrp1 possesses a chromodomain, it is conceivable that this protein also functions as part of heterochromatin structures. Hrp1 also has a Myb-like DNA-binding domain in the C-terminus, a domain which is conserved among several proteins binding with telomere-related sequences (32). We previously reported that Hrp1 is a DNA-binding protein having a preference for (T+A)-rich minor grooves (21). However, our experiments here demonstrate that Hrp1 protein is uniformly localized in the nucleus. Therefore, we suggest that Hrp1p at least is not exclusively localized to heterochromatin, whilst the other chromodomain proteins in fission yeast, Swi6 and Clr4, are localized to heterochromatin (15,31). The CHD1 homologs from mouse (mCHD1), Drosophila (dCHD1) and Saccharomyces cerevisiae (ScCHD1) were also found to be localized in the nucleus (33–35).
DNA-stimulated ATPase activity of Hrp1
Many SNF2/SWI2 proteins have been reported to have DNA-dependent ATPase activity (6,36–39) and mutations in this ATPase domain have been shown to affect the function of the protein complex (40). In other SWI2/SNF2 proteins the energy released from ATP hydrolysis is used for isomerization of the nucleosome structure by alteration of histone–DNA contacts critical for their function. In this study, we show that Hrp1 protein, a new member of the SWI2/SNF2 protein family and CHD1 subfamily, also has a DNA-stimulated ATPase activity in vitro and it is therefore possible that Hrp1 functions in nucleosome remodeling like the other family members.
Defective chromosome segregation
We report an elevated mini-chromosome loss rate for both hrp1Δ cells and Hrp1-overexpressing cells. In general, the missegregation of chromatin could be caused by the defects in chromosome condensation, sister chromatid separation, anaphase-promoting proteolysis and/or spindle formation (41). We found that the hrp1 deletion mutant had a slight defect in mitotic chromosome segregation, detected as an increase in Ch16 mini-chromosome loss, and some aberrant mitosis, detected by staining with TAT1 antibody. Interestingly, mutations in other chromodomain protein genes, clr4 and swi6, cause similar defects, such as lagging chromosomes in late anaphase spindles (31). In contrast, Hrp1-overexpressing cells revealed a relatively high incidence of cytological defects, including a decondensed anaphase and ‘cut’ phenotype, which is very similar to decondensed chromatin in cut3, cut14 and top2 mutants (22,23). Therefore, the chromosome missegregation in Hrp1-overexpressing cells is likely to be caused by interference with chromatin condensation in mitosis.
The electron microscopic analysis shows that nucleolar electron-dense regions were larger when Hrp1 was overexpressed as compared to nucleoli of wild-type cells. These enlarged dark regions also appeared to exhibit unusual horseshoe shapes. These results suggest that overexpression of Hrp1 also causes condensation of the nucleolus. Schizosaccharomyces pombe rDNA is known to consist of 100–150 copies of 10.9 kb tandem head-to-tail repeats in two clusters at both ends of chromosome III (42,43). Thus, it is possible that a higher cellular level of Hrp1 causes unusual assembly of nucleolar structures, including the rDNA clusters.
A direct or indirect role of Hrp1 in chromosome condensation?
The observed defects in chromosome segregation and nucleolar morphology in Hrp1-overexpressing cells may be interpreted by the following hypotheses. (i) Hrp1 may directly or indirectly affect the chromatin structure by virtue of the conserved ATPase/helicase and chromodomains. During the cell cycle, Hrp1 may facilitate and/or counteract the action of condensins. (ii) Hrp1 may directly compete with other proteins such as condensins or topoisomerase II for (A+T)-rich binding sites. (iii) High Hrp1 levels may cause inappropriate heterochromatin formation that prevents normal chromosome segregation. Our previous data showed that Hrp1 prefers to bind an (A+T)-rich sequence (21) and 13S condensin is also proposed to bind a scaffold attachment region containing an (A+T)-rich DNA sequence (44). Thus, Hrp1 may normally play a role in condensation of some specialized regions of chromatin (rDNA, telomere, centromere, etc). Possibly, overexpressed Hrp1 may prevent the binding of some condensin-like proteins and in turn inappropriately condense or decondense non-cognate structures. It is clear that proper condensation is a prerequisite for normal sister chromatid separation (45).
It remains unclear what exact role the Hrp1 protein might play in chromatin remodeling. To understand the exact role of Hrp1, further studies examining the relationship between Hrp1 and other proteins that play a role in chromatin condensation, such as Swi6, Clr4, Top2 and the condensins, remain to be performed. However, this present study establishes that Hrp1 is a nuclear protein that contains ATPase activity like other SNF2/SWI2 protein family members and that it is required for chromosome segregation.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs R. Allshire and M. Yamamoto for providing S.pombe cell strains and Dr Keith Gull for TAT1 antibody. K.E. acknowledges colleagues Dr J. R. McIntosh and E. O’Toole (University of Boulder, Boulder, CO) for generous help in establishing S.pombe HPF in Sweden, as well as local EM support from Dr K. Hultenby (Huddinge). This work was supported in part by grants from the Korea Science and Engineering Foundation through the Research Center for Cell Differentiation (1999G0301-3) and the Ministry of Education (1998-019-D00027), Republic of Korea. Y.K.J., S.H.H. and S.D.P. were also supported in part by BK21 Research Fellowship from the Korean Ministry of Education. Work in the K. Ekwall laboratory was supported by MFR grant 12562 and NFR grant 0990-302. M.T. was the recipient of a Wennergren stipend.
REFERENCES
- 1.Felsenfeld G. (1996) Cell, 86, 13–19. [DOI] [PubMed] [Google Scholar]
- 2.Workman J.L. and Kingston,R.E. (1998) Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 3.Côté J., Quinn,J., Workman,J.L. and Peterson,C.L. (1994) Science, 265, 53–60. [DOI] [PubMed] [Google Scholar]
- 4.Carns B.R., Kim,J.-Y., Sayre,M.H., Laurent,B.C. and Kornberg,R.D. (1994) Proc. Natl Acad. Sci. USA, 91, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson C.L., Dingwall,A. and Scott,M.P. (1994) Proc. Natl Acad. Sci. USA, 91, 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama T. and Wu,C. (1995) Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- 7.Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Nature, 7, 598–602. [DOI] [PubMed] [Google Scholar]
- 8.Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- 9.Pazin M.J. and Kadonaga,J.T. (1997) Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- 10.Aasland R. and Stewart,A.F. (1995) Nucleic Acids Res., 23, 3168–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin E.V., Zhou,S. and Lucchesi,J.C. (1991) Nucleic Acids Res., 23, 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paro R. and Hogness,D.S. (1991) Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh P.B., Miller,J.R., Pearce,J., Kothary,R., Burton,R.D., Paro,R., James,T.C. and Gaunt,S.J. (1991) Nucleic Acids Res., 19, 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aagaard L., Laible,G., Selenko,P., Schmid,M., Dorn,R., Schotta,G., Kuhfittig.S., Wolf,A., Lebersorger,A., Singh,P.B., Reuter,G. and Jenuwein,T. (1999) EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekwall K., Javerzat,J., Lorentz,A., Schmidt,H., Cranston,G. and Allshire,R. (1995) Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- 16.Allshire R.C., Nimmo,E.R., Ekwall,K., Javerzat,J.P. and Cranston,G. (1995) Genes Dev., 9, 218–233. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova A.V., Bonaduce,M.J., Ivanov,S.V. and Klar,A.J.S. (1998) Nature Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- 18.Doe C.L., Wang,G., Chow,C.-M., Fricker,M.D., Singh,P.B. and Mellor,E.J. (1998) Nucleic Acids Res., 26, 222–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell I.G. and Austin,C.A. (1997) Biochim. Biophys. Acta, 1337, 198–206. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y.H., Yoo,E.J., Jang,Y.K., Kim,S.H., Kim,M.J., Shim,Y.S., Lee,J.S., Choi,I.S., Seong,R.H., Hong,S.H. and Park,S.D. (1998) Mol. Gen. Genet., 257, 319–329. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y.H., Yoo,E.J., Jang,Y.K., Kim,S.H., Lee,C.G., Seong,R.H., Hong,S.H. and Park,S.D. (1998) Korean J. Biol. Sci., 2, 539–543. [Google Scholar]
- 22.Saka Y., Sutani,T., Yamashita,Y., Saitoh,S., Takeuchi,M., Nakaseko,Y. and Yanagida,M. (1994) EMBO J., 13, 4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uemura T., Morikawa,K. and Yanagida,M. (1986) EMBO J., 5, 2355–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 133–136.
- 25.Keeney J.B. and Boeke,J.D. (1994) Genetics, 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pringle J.R., Peterson,R.A., Adams,E.M., Stearns,T., Drubin,D.G., Haarer,B.K. and Jones,E.W. (1989) Methods Cell Biol., 31, 357–435. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie C. and Fink,G.R. (1991) Methods Enzymol., 194, 821–823. [Google Scholar]
- 28.Sazer S. and Sherwood,S.W. (1990) J. Cell Sci., 97, 509–516. [DOI] [PubMed] [Google Scholar]
- 29.Ding R., McDonald,K.L. and McIntosh,J.R. (1993) J. Cell Biol., 120, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano T., Funahashi,S., Uemura,T. and Yanagida,M. (1986) EMBO J., 5, 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekwall K., Nimmo,E.R., Javerzat,J., Borgstrøm,B., Egel,R., Cranston,G. and Allshire,R. (1996) J. Cell Sci., 109, 2637–2648. [DOI] [PubMed] [Google Scholar]
- 32.Bilaud T., Koering,C.E., Binet-Brasselet,E., Ancelin,K., Pollice,A., Gasser,S.M. and Gilson,E. (1996) Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes D.G., Tartof,K.D. and Perry,R.P. (1996) Proc. Natl Acad. Sci. USA, 93, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoke D.G. and Perry,R.P. (1995) Mol. Cell. Biol., 15, 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodage T., Basrai,M.A., Baxevanis,A.D., Hieter,P. and Collins,F.S. (1997) Proc. Natl Acad. Sci. USA, 94, 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurent B.C., Treich,I. and Carlson,M. (1993) Genes Dev., 7, 583–591. [DOI] [PubMed] [Google Scholar]
- 37.Cairns B.R., Lorch,Y., Li,Y., Zhang,M., Lacomis,L., Erdjument-Bromage,H., Tempst,P., Du,J., Laurent,B. and Kornberg,R.D. (1996) Cell, 87, 1249–1260. [DOI] [PubMed] [Google Scholar]
- 38.Auble D.T., Hansen,K.E., Mueller,C.G., Lane,W.S., Thorner,J. and Hahn,S. (1994) Genes Dev., 8, 1920–1934. [DOI] [PubMed] [Google Scholar]
- 39.Johnson R.E., Prakash,S. and Prakash,L. (1994) J. Biol. Chem., 269, 28259–28262. [PubMed] [Google Scholar]
- 40.Richmond E. and Peterson,C.L. (1996) Nucleic Acids Res., 24, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanagida M. (1998) Trends Cell Biol., 8, 144–149. [DOI] [PubMed] [Google Scholar]
- 42.Mizukami T., Chang,W.I., Garkavtsev,I., Kaplan,N., Lombardi,D., Matsumoto,T., Niwa,O., Kounosu,A., Yanagida,M., Marr,T.G. and Beach,D. (1993) Cell, 73, 121–132. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh Y. and Laemmli,U.K. (1994) Cell, 76, 609–622. [DOI] [PubMed] [Google Scholar]
- 44.Yin H., Baart,E., Betzendahl,I. and Eichenlaub-Ritter,U. (1998) Mutagenesis, 13, 567–580. [DOI] [PubMed] [Google Scholar]
- 45.Torok T., Harvie,P.D., Buratovich,M. and Bryant,P.J. (1997) Genes Dev., 11, 213–225. [DOI] [PubMed] [Google Scholar]