Fig. 2 |. A cluster of regularly interspersed copies of an 18-bp imperfect palindromic sequence at 11q23 mediates sequence-specific enrichment of EBNA1.
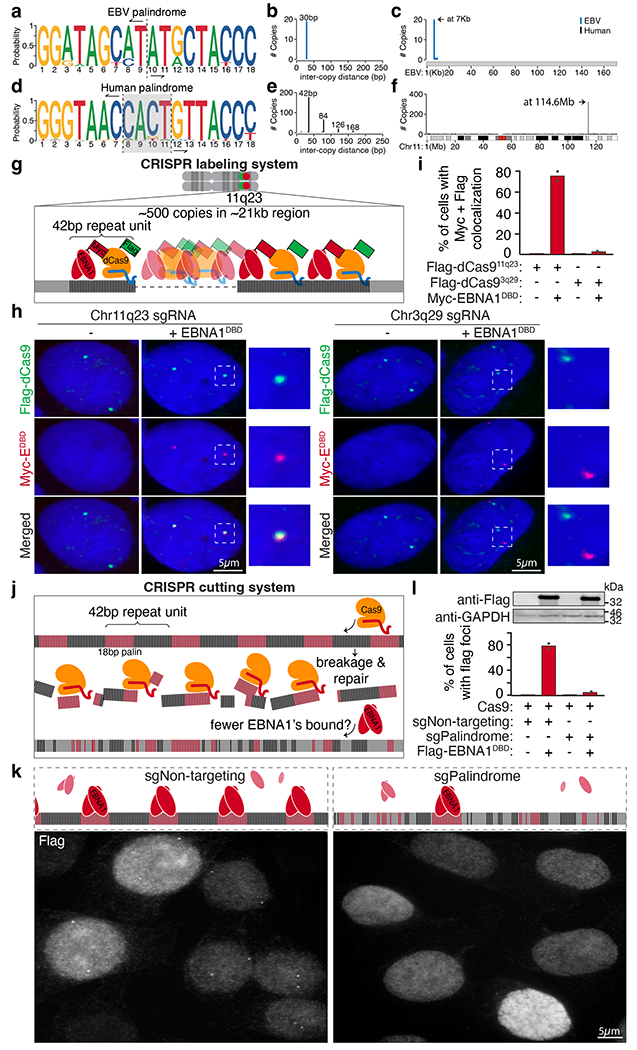
a–c, Consensus motif logo (a), inter-copy distance (b) and position (c) of 18-bp palindromic repeats in the EBV genome. d–f, Consensus motif logo (d), inter-copy distance (e) and position (f) of 18-bp imperfect palindromic repeats with up to 6 variant and 2 asymmetric nucleotides on human chromosome 11. Vertical dashed lines in the consensus motif logos represent the point of symmetry of the palindromes. The grey shaded area highlights the 4-bp non-palindromic core of the human palindrome. g, Schematic representation of the CRISPR labelling system. h, Representative anti-Flag (green) and anti-MYC (red) immunofluorescence micrographs showing localization of Flag–dCas9 directed by the indicated sgRNAs labelling either 11q23 or 3q29 with and without expression of MYC–EBNA1(DBD). Scale bars, 5 μm. i, Quantification of staining in h. A total of 95 cells were counted for Flag–dCas9 labelling 11q23. A total of 110 cells were counted for Flag–dCas9 labelling 3q29. Data are presented as bars representing mean values from two independent experiments. j, Schematic representation of the CRISPR cutting system. k, Representative anti-Flag immunofluorescence micrographs showing localization of Flag–EBNA1(DBD) in cells treated with either non-targeting sgRNA (sgNon-targeting) or palindrome-targeting sgRNA (sgPalindrome). Scale bar, 5 μm. For gel source data, see Supplementary Fig. 1. l, Quantification of staining in k. A total of 410 cells were counted for Cas9-mediated cutting with non-targeting sgRNA. A total of 630 cells were counted for Cas9-mediated cutting with palindrome-targeting sgRNA. Data are presented as bars representing mean values from two independent experiments.
