Abstract
High-precision bioengineering and synthetic biology require fine-tuning gene expression at both transcriptional and posttranscriptional levels. Gene transcription is tightly regulated by promoters and terminators. Promoters determine the timing, tissues and cells, and levels of the expression of genes. Terminators mediate transcription termination of genes and affect mRNA levels posttranscriptionally, e.g., the 3′-end processing, stability, translation efficiency, and nuclear to cytoplasmic export of mRNAs. The promoter and terminator combination affects gene expression. In the present article, we review the function and features of plant core promoters, proximal and distal promoters, and terminators, and their effects on and benchmarking strategies for regulating gene expression.
Introduction
A plant’s DNA sequence can be fragmented into its smallest functional units to generate standardized, functionally interchangeable biological parts (bioparts) or biobricks. Bioparts are the fundamental building blocks in plant synthetic biology for assembling novel functional units or synthetic devices [1,2]. Bioparts include gene promoters, promoter cis-regulatory elements or motifs, exons, introns, terminators, and open reading frames (ORFs). Identification and characterization of individual bioparts and their respective biological interactions with other bioparts allows for improved understanding of transcription and gene expression as a whole. These bioparts are implemented in the design and construction of synthetic devices, which are integrated into plant (and nonplant) biological systems for precise regulation of gene transcription [3].
A plant gene promoter encompasses the DNA sequence flanking the transcription start site(s) (TSSs) of a gene that contains various promoter cis-regulatory elements, to which trans-acting transcription factors (TFs) bind and promote (or repress) initiation of gene transcription. Initiation of transcription is the first step for the designated genetic information of a synthetic device or system to be able to function as expected. Typically, a plant promoter spans the DNA region several hundred to a few thousand base pairs upstream of the TSS, with each promoter containing a core, a proximal, and a distal region based upon the function of the regulatory elements present in each region and their proximity to the TSS. The core promoter directly flanks the gene’s TSS and facilitates the assembly of a pre-initiation complex (PIC) upon the TSS(s), which consists of RNA polymerase (Pol) II and general transcription factors (GTFs). The PIC determines the basal transcription level of a gene, the direction of transcription, and the selection of TSSs, as one plant core promoter may harbor multiple and sometimes mutually exclusive TSSs [4]. The core promoter directs the assembly and recruitment of TFs through its cis-regulatory elements. A plant core promoter may contain conserved, direction-sensitive core promoter motifs such as the TATA-box (for TATA-box-containing promoters), downstream promoter element (DPE; for TATA-less promoters), initiator element (Inr), Y patch, CCAAT-box, and TSS(s) (Fig. 1). The TATA-box is named for its 8-base pair (bp) consensus sequence TATA(A/T)A(A/T)(A/G) and is commonly located 30 to 70 bp upstream of the TSSs in plants [5,6]. Binding of the TATA-binding protein (TBP), a subunit of the transcription factor IID (TFIID), to the TATA-box recruits RNA polymerase II (Pol II) to form the PIC and initiate transcription. In plants, most TATA-box-containing promoters are mainly involved in tissue-specific expression and stress responses [7,8]. Yamamoto et al. [9] found that the TATA-box-containing promoters in Arabidopsis thaliana tend to be associated with the presence of Y-patches and Inr elements, and have high promoter strength with sharp-peaked TSS clusters. In TATA-less promoters such as housekeeping photosynthesis-related gene promoters [7,8], a DPE with the consensus sequence RGWCGTG plays a similar function to that of the TATA-box for TFIID binding. DPEs are usually present approximately 30 bp downstream of the TSSs and are often found in plant promoters controlling stimulus-responsive genes [10]. The Inr element is another important core promoter motif directly covering TSSs [11] with a consensus sequence of PTCA+1NTPP, where A+1 is the initiator and the first base transcribed [11]. The TFIID binds to the Inr cooperatively with the TATA-box or the DPE to initiate recruitment of the PIC [12]. A Y patch is an 8-bp motif consisting mostly of pyrimidine C and T dimers (CT or TC), located 1 to 100 bp upstream of the TSSs, and has a distribution peak around the TSSs [13]. The CCAAT-box is the binding site for the CCAAT-binding factor (CBF) nuclear transcription factor Y (NF-Y). The binding of NF-Y to the CCAAT-box can result in positive or negative posttranslational histone modifications, contributing to activation or repression of gene expression [15]. Regarding the sequences surrounding the TSSs, Yamamoto et al. [13] reported a “YR Rule,” indicating that there is usually a C or T nucleotide 1 bp upstream of the TSSs and an A or G nucleotide 1 bp downstream of the TSSs (i.e., Y, C−1 or T−1; R, A+1 or G+1) for both Arabidopsis and rice.
Fig. 1.
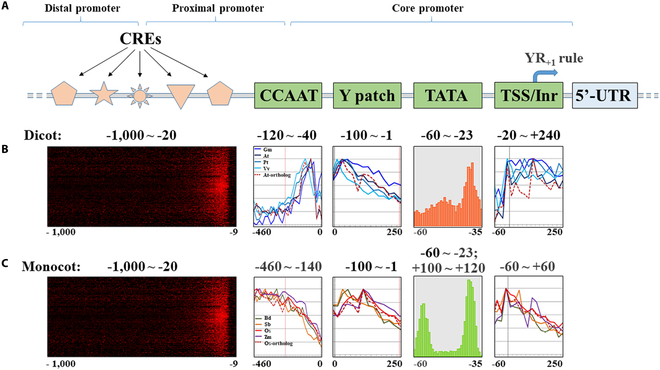
Locations of the cis-regulatory elements (CREs) and core promoter motifs CCAAT, Y patch, TATA-box, and TSS/Inr within dicot and monocot promoters. (A) Representative structure of a plant promoter. (B and C) Local distribution of short sequences (LDSS)-positive octamer CREs, normalized frequency distribution profiles of CCAAT, Y patch, and Inr, and the percentage of TATA-box-containing promoters at the indicated positions on the dicot (B) and monocot (C) promoters. The images of dicot and monocot CREs were adapted from [14] (Arabidopsis) and [13] (rice), respectively. The images of CCAAT, Y-patch, and Initiator (Inr) were adapted from [6] (soybean, Arabidopsis, poplar, grape, Brachypodium distachyon, sorghum, rice, and maize). The images of TATA-box were adapted from [5] (Arabidopsis and maize).
The basal transcription level conferred by core promoters can be greatly increased (or inhibited) by enhancers (or repressors/silencers), which are promoter motifs located upstream of the core promoter regions and define the proximal and distal promoter regions (note: enhancers and repressors can also be located downstream of the TSSs). Cis-regulatory elements interact bidirectionally and in tandem with TFs, cofactors, and chromatin-remodeling complexes to determine the strength and the temporal and spatial expression patterns of a gene during plant growth and development and in response to changing environmental conditions. Studies in which promoters were functionally analyzed for constitutive, tissue-specific, or inducible expression have been extensively reviewed recently [10,16–22] and are not part of this review. Cis-regulatory elements used for plant synthetic promoter engineering have also been reviewed recently [23–28].
The proper combination of a promoter and a terminator is essential for successful gene transcription as the processing of the 3′-end of an RNA transcript is the last essential step in mRNA biogenesis. The terminator is one of the 3′-end regulatory elements and marks the end of an RNA transcript. It assists transcription initiation by interacting with TFs and the C-terminal domain of Pol II, and halts transcription by adding termination signals on the newly synthesized transcript, triggering the release of the transcript from the transcription machinery. Proper mRNA termination promotes the transport of mRNAs from the nucleus to the cytoplasm [29,30] and stabilizes and protects the mRNAs from degradation [31–35]. Thus, proper mRNA termination is necessary for the translation of mRNAs into proteins [31,35–37]. Proper mRNA termination also prevents read-through transcription of downstream sequences [38,39]. Termination factors interact with many RNA processing and degradation enzymes, which in turn define the fate and half-life of the transcripts. There are 2 termination pathways, i.e., transcription termination of protein coding transcripts (i.e., mRNAs) and transcription termination of noncoding RNAs (ncRNAs). Termination of mRNAs leads to the production of stable RNA transcripts that are transported to the cytoplasm, while termination of ncRNAs leads to their sequestration in the nucleus and subsequent degradation. In this review, we will only discuss mRNA transcriptional termination and its relevance for synthetic biology.
Thus, plant core promoters, proximal and distal promoters, and terminators are the starting bioparts regulating gene expression, and it is of crucial importance to determine the best promoter and terminator combination for highly efficient transcription of transgenes in high-precision plant bioengineering.
Plant Core Promoters
Generally, dicot and monocot core promoters perform well when used in respective dicot and monocot species [5]. Core promoter performance is attributed to the nucleotide sequence, position, and number of copies of each core promoter motif, as well as the core promoter’s chromatin configuration. It was reported that 32%, 19%, and 38% genes in Arabidopsis [7], rice (Oryza sativa) [8], and maize (Zea mays) [40], respectively, are TATA-box-containing genes. Kumari and Ware [6] conducted an in silico genome-wide analysis of core promoter motifs in the dicot species Arabidopsis, soybean (Glycine max), poplar (Populus trichocarpa), and grape (Vitis vinifera), and in the monocot species rice, maize, sorghum (Sorghum bicolor), and purple false brome (Brachypodium distachyon). They found that the Inr motifs were located −20 to +240 bp upstream of the TSSs of the promoters in the 4 dicot species, while for the promoters of the 4 monocot species the Inr motifs stretched from −60 to +60 bp and from +100 to +120 bp, suggesting a distinct difference in the genome-wide distribution of the motif between dicot and monocot species. Similarly, the CCAAT-box was mainly positioned −120 to −40 bp in dicot promoters but positioned −460 to −140 bp in monocots [6]. Additionally, it was reported that less than 18% of Arabidopsis promoters and 50% of rice promoters contain Y patches [8,41]. All these distinctions indicate the differences in the functions of the core promoters between monocots and dicots. Thus, it is reasonable to use dicot and monocot core promoters for synthetic promoter engineering in respective species types.
To date, the most widely used core promoter is the minimal CaMV 35S promoter, which has shown functional expression in nearly all dicot species and one monocot species, i.e., rice [42,43] (see Table 1 for a list of representative plant core promoters used for gene expression). While plant core promoter engineering is still in its infancy, a pivotal study was recently published describing plant core promoter analysis and engineering [5]. Jores et al. [5] constructed individual core promoter libraries from Arabidopsis, maize, and sorghum genomes. These libraries included the core promoter regions −165 to +5 nucleotides (nt) (relative to the TSSs) of 18,329, 34,415, and 27,094 protein-encoding and microRNA genes from each of the respective species. Evaluation of the core promoters using tobacco leaf agroinfiltration and maize leaf protoplast expression revealed substantial differences in core promoter strength, indicating that dicot and monocot plants interact with the same core promoter in different manners. Interestingly, core promoter strength was positively associated with gene expression levels only in some of the genes. GC content of the core promoters showed negative correlation with core promoter strength in the tobacco leaf expression system but had no correlation in the maize protoplast expression system. This finding could be attributed to the differences between the GC content of the promoters in these species because tobacco and Arabidopsis promoters are AT-rich, while maize and sorghum promoters tend to be higher in GC content. Moreover, the distribution locations of TATA-box have one peak ~30 bp upstream of the Arabidopsis TSSs, but 2 or 3 peaks upstream of the sorghum (~30 and ~40 bp) and maize (~30, ~55, and ~70 bp) TSSs. They also noticed that maize promoters with TATA-boxes in one of the 3 peaks or closer to the TSSs have higher strength than those with TATA-boxes elsewhere. In addition, TATA-box-containing core promoters exhibited up to 4-fold higher strength than TATA-less core promoters, and interestingly, the presence of Inr elements and Y patches was linked with significantly increased core promoter strength in Arabidopsis, maize, and sorghum [5].
Table 1.
List of representative plant core promoter elements used for gene expression.
| Core promoter element | Consensus sequence | Approximate location | Reference |
|---|---|---|---|
| TATA-box | TATAWAW | −70 to −20 bp | [5,6] |
| Inr | YYA(+1)NWYY | Overlapping +1 | [44] |
| Y patch | CYTCYYCCYC | +20 to +80 | [13] |
| TC motif | YYYYYY | −39 to −26 | [41] |
| GA element | RRRRRRRR | +25 bp to +75 bp | [9] |
| CA element | YMMMMMMM | +1 bp to +30 bp | [9] |
| GC-box | GGGCGG | −70 bp to +250 bp | [6] |
| CAAC | MCMAMCCM | −50 bp to +40 bp | [9] |
| DPE | RGWCGTG | +30 bp | [10] |
| CCAAT-box | CCAAT | −80 bp | [10,15] |
More importantly, Jores et al. [5] conducted plant core promoter engineering by generating random core promoters with average nucleotide frequencies similar to those of Arabidopsis or maize, followed by the addition of TATA-box, Inr, and/or Y patch motifs. As expected, the randomized core promoters had very weak strength, but the addition of the 3 motifs significantly increased the random core promoter activities individually or in combination, with TATA-box and Inr increasing the strength the most and the least, respectively. The strongest synthetic core promoters they developed had strength comparable to the minimal 35S promoter. These results demonstrate that rational design and construction of plant synthetic core promoters with varying strength can be achieved by inserting core promoter motifs into an appropriate nucleotide background. Based on these results, computational models were developed for in silico evolution of synthetic core promoters to predict and improve core promoter strength. The models showed that the developed synthetic core promoters could work independent of the choices of enhancers [5]. Additionally, Jores et al. [5] found that inserting a TATA-box into the previously TATA-less core promoters of Arabidopsis, maize, and sorghum enhanced the core promoter’s strength. This is different from the finding by Nakamura et al. [11], which mutated the TATA-less Inr-containing core promoter of the tobacco psaDb gene to then contain a TATA-box without Inr, and found that the Inr rather than the TATA-box contributed to light-responsive transcriptional regulation of the photosynthesis gene.
It was reported that active (constitutive or housekeeping) core promoters are featured by the presence of nucleosome depletion regions (NDRs) since their nucleosomes are highly dynamic, ensuring the accessibility of the transcription machinery [45,46]. It was also reported that the NDRs in the active core promoters in Arabidopsis, rice, sorghum, and maize have high G/C content [46–48]. Srivastava et al. [49] identified that mutations in TATA-boxes or Inr elements of the Arabidopsis light-regulated promoters could result in the formation of nucleosomal structures, inhibiting gene expression. Oldfield et al. [50] found that the histone-fold domain protein NF-Y maintains the core promoter region in a nucleosome-depleted state in metazoans. Oldfield et al. [50] also found that loss of NF-Y binding to the core promoters disrupts the core promoter’s chromatin architecture, resulting in nucleosome enrichment on the core promoters and repression of gene expression. NF-Y is a ubiquitously expressed, heterotrimeric TF and the key player of TSS selection. Binding of NF-Y to its binding site CCAAT in the Flowering Locus T (FT) gene promoter in Arabidopsis and rice activated FT expression [51]. Still, the key differences in the chromatin landscapes of the dicot and monocot core promoters remain largely unknown.
It is worthwhile to point out that the practical use of plant promoters for gene expression almost always includes the use of 5′-untranslated regions (UTRs) and leader introns that are located within 5′-UTRs since they affect mRNA levels posttranscriptionally [50] and gene function in a length-dependent manner [52]. The average length of 5′-UTRs is 155 and 259 bp in Arabidopsis and rice, respectively [49]. It was reported that the length, GC content, and leader intron number of a 5′-UTR showed significant positive correlation with the expression breadth in various tissues of Arabidopsis, rice, maize, and sorghum [46]. It was also reported that the presence of leader introns enhanced gene expression. Examples include the leader introns of the Arabidopsis ubiquitin3/10/11 (UBQ3, UBQ10, and UBQ11) [53], Mg2+/H+ exchanger (MHX) [54], and cytochrome c oxidase subunit 5c (COX5c) [55], tobacco Ubiquitin.U4 (Ubi.U4) [56] and carnation S-adenosylmethionine decarboxylase9(CSDC9) [57], tomato (Solanum lycopersicum)ascorbate peroxidase20 (APX20) [58], potato (Solanum tuberosum)sucrose synthase3 and 4 (Sus3; Sus4) [59,60], mustard (Brassica juncea) S-adenosylmethionine decarboxylase2 (SAMDC2) [61], rice β-tubulin isotype 6 (Ostub6) [60,62] and 16 (Ostub6) [63], and ubiquitin3 (rubi3) [64]. Leader introns may contain cis-regulatory elements [65] and affect transcription, mRNA stability and export, and tissue-specific gene expression [53,66–74].
Plant Proximal and Distal Promoters
Plant natural promoters contain numerous cis-regulatory elements widely distributed across their proximal and distal promoter regions, with many of them acting as specific regulators of gene expression, i.e., stimulating (enhancers) or repressing (repressors) the basal expression levels conferred by the core promoters. Enhancers and repressors are direction-insensitive and are not typically position-restricted, increasing the likelihood of functioning in both dicot and monocot systems. It was found that about 30 to 50% of 8-bp promoter motifs are conserved between Arabidopsis and rice [13], indicating that transcriptional regulation is relatively conserved between dicots and monocots. As demonstrated by Belcher et al. [75], cis-regulatory elements from a different source (yeast in this case) could be used together with a plant core promoter to develop functional synthetic promoters that work well with plant endogenous transcriptional machinery. Screening of a synthetic promoter library consisting of 5 cis-regulatory elements from yeast and 5 plant core promoters showed that each cis-regulatory element has an independent effect on promoter strength, indicating the orthogonality of the cis-regulatory elements in engineering of synthetic promoters with tunable expression levels in a heterologous plant system.
Various native plant promoters have been functionally characterized and used for constitutive or conditional transgene expression (Table 2; also see the listed plant native promoters in [16,18–22]). Plant promoter databases have also been created for the annotation and curation of the information about functionally active promoters from various plant species. These include PlantProm (http://linux1.softberry.com/berry.phtml?topic=plantprom&group=data&subgroup=plantprom) [117], TransGene Promoters (TGP; http://wwwmgs.bionet.nsc.ru/mgs/dbases/tgp/home.html) [118], Plant Promoter Database (PPDB; https://ppdb.agr.gifu-u.ac.jp/ppdb/cgi-bin/index.cgi#about) [119,120], and Root-associated Genes and Promoters Database (RGPDB; http://sysbio.unl.edu/RGPDB/) [121].
Table 2.
List of representative plant promoters used for gene expression.
| Promoter | Source | Transgenic plant | Expression pattern | References |
|---|---|---|---|---|
| Constitutive promoters | ||||
| Act2 | A. thaliana | A. thaliana | Constitutive | [76] |
| Act-1 | O. sativa | O. sativa | Constitutive | [77] |
| UBQ1 | A. thaliana | N. tabacum | Constitutive | [78] |
| Ubi1, Ubi2 | Panicum virgatum | P. virgatum; O. sativa; N. tabacum | Constitutive | [79] |
| Ubi1 | Zea mays | Z. mays | Constitutive | [80,81] |
| Tissue-specific promoters | ||||
| SlREO | S. lycopersicum | S. lycopersicum | Root | [82] |
| NAC10 | O. sativa | O. sativa | Root | [83] |
| PAT21 | S. tuberosum | S. tuberosum | Tuber | [84] |
| hspr | A. thaliana | A. thaliana | Vascular tissue | [85] |
| Pfn2 | P. virgatum | O. sativa | Vascular tissue | [86] |
| PEPC | Z. mays | Z. mays | Leaf | [87] |
| Lhcb | P. virgatum | O. sativa | Green tissue | [88] |
| TA29 | N. tabacum | N. tabacum | Flower | [89] |
| Lat52 | S. lycopersicum | N. tabacum | Pollen | [90] |
| Zm13 | Z. mays | Tradescantia paludosa; Z. mays | Pollen | [91] |
| Oleosin | A. thaliana | G. max | Seed | [92] |
| Glutenin | T. aestivum | T. aestivum | Seed | [93] |
| D-hordein | T. aestivum | Z. mays | Seed (endosperm) | [94] |
| E8 | S. lycopersicum | S. lycopersicum | Fruit | [95] |
| Abiotic stress-inducible promoters | ||||
| Adh-1 | Z. mays | N. tabacum | Anaerobic conditions | [96] |
| wun1 | S. tuberosum | N. tabacum | Wounding | [97] |
| GBSS | S. tuberosum | S. tuberosum | Sugar | [98] |
| HSP18.2 | A. thaliana | A. thaliana | Heat shock | [99] |
| Rd29 | A. thaliana | A. thaliana; N. tabacum; O. sativa | Drought, cold, salt | [100] |
| SR2 | Phaseolus vulgaris | N. tabacum | Heavy metals | [101] |
| CCA1 | A. thaliana | A. thaliana | Light cycle | [102] |
| UGT71C5 | A. thaliana | A. thaliana | Light | [103] |
| GSE | O. rufipogon | A. thaliana | Light | [104] |
| Biotic stress-inducible promoters | ||||
| win3.12 | Populus sp. hybrid | S. tuberosum | Fusarium solani | [105] |
| R2329 | O. sativa | O. sativa | Magnaporthe grisea | [106] |
| Bs3 | Capsicum annuum | N. benthamiana | Xanthomonas campestris pv. vesicatoria | [107] |
| CaPrx | Coffea arabica | N. tabacum | Meloidogyne incognita | [108] |
| 4×M1.1, 4×M2.3 | G. max | G. max | Heterodera glycines | [107, 109] |
| IFS2 | G. max | G. max | Bradyrhizobium japonicum | [110] |
| SAG12 | A. thaliana | S. lycopersicum | Senescence | [111] |
| SEOF1 | Pisum sativum | N. tabacum | Methyl jasmonate, auxin, abscisic acid (ABA) | [112] |
| Em | T. aestivum | O. sativa | ABA | [113] |
| Rd29 | A. thaliana | A. thaliana, N. tabacum | ABA | [114] |
| SAUR15A | G. max | N. tabacum | Auxin | [115] |
| Chn48 | N. tabacum | N. tabacum | Ethylene | [116] |
The strength and expression patterns of plant native promoters are determined by the presence of combinatorial cis-regulatory elements and their 3-dimensional structures following the binding of their respective TFs [122,123]. Although reports have been published for the 3-dimensional structures of promoters from Escherichia coli [124,125], Xenopus tropicalis [126], and Homo sapiens [127,128], 3-dimensional structures have not been constructed for any plant promoters. Fortunately, cis-regulatory elements can be taken out of their natural context and be implemented as bioparts for synthetic promoter engineering. Known cis-regulatory elements can be identified in a plant promoter by searching against the 3 promoter motif databases PlantCARE [129], PLACE [130], and TRANSFAC [131], while novel cis-regulatory elements can be discovered through promoter serial deletion and de novo motif discovery [26]. It was shown that bioinformatics tools developed for de novo motif discovery have limited prediction efficiency (15 to 25%) of true motifs [132,133], thus making experimental function validation necessary. Liu et al. [134] developed an ensemble approach by focusing on overlapping motif regions detected by multiple de novo motif discovery tools, which improved the prediction efficiency to be 68.8% and led to the discovery of 23 experimentally validated, novel soybean cyst nematode (SCN)-inducible motifs in soybean. Using the same approach, Yang et al. [135,136] identified well-conserved salt- or drought-responsive promoter motifs in hybrid poplar, which were used to generate functionally validated water-deficit stress-, salt stress-, and osmotic stress-inducible synthetic promoters.
With the availability of functionally characterized cis-regulatory elements, it still remains largely undetermined how to rationally and reliably engineer plant synthetic promoters. To date, most plant synthetic promoter engineering studies have implemented a trial-and-error approach by using multiple copies of one or multiple motifs in front of a core promoter. Recently, Cai et al. [137] established a design system for plant synthetic promoter engineering by inserting various cis-regulatory elements between a 19-bp random nucleotide sequence and a TATA-box motif (TATATAA) in front of a 43-bp-long core promoter region and a TSS (Fig. 2). They found that random combinations of different trimerized cis-regulatory elements gave rise to increases in synthetic promoter strength than individual trimers. They also found that changes to the flanking sequences or relative positions of the cis-regulatory elements and moderate increases in spacing contributed negligible effects to promoter strength, implying that the corresponding TFs may work relatively independently other than via direct protein-protein interactions. However, they also reported that the >50-bp distance between the first motif at the 5′-end and the TATA-box significantly affected promoter strength. This raises an interesting nuance regarding motif spacing. The short spacing limit identified here may be pertinent to the specific motifs tested but may not be applicable to other classes of motifs, e.g., stress-inducible motifs that may function in a much less distance-restricted manner [138].
Fig. 2.
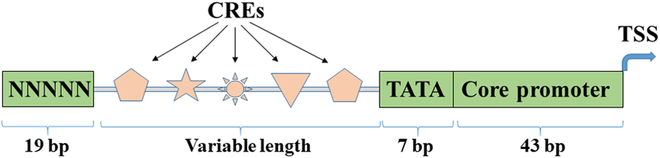
Schematic design of the synthetic promoters by Cai et al. [137], which contain a 19-bp random sequence (NNNNN), a region of variable length to which CREs are added, a TATA-box, and a 43-bp core sequence plus a transcription start site (TSS).
Although it remains largely unknown how fine-tuning gene expression is achieved via a combination of cis-regulatory elements, machine learning algorithms could be applied to build models for predicting synthetic promoter expression patterns by using multiple features, e.g., the presence and absence, location, copy number, and combinatorial relationships of cis-regulatory elements, as input variables. Cis-regulatory information has been implemented to predict biotic and abiotic stress- [139], high salinity- [140], and iron deficiency-responsive [141] transcription of Arabidopsis native promoters. Zou et al. [139] found that the prediction models based on combinatorial relationships (obtained by using a classification algorithm to integrate association rule mining) performed significantly better than those based on only singular traits, i.e., copy number or location.
Plant Terminators
A gene’s terminator sequence mediates transcriptional termination, 3′-end processing, stability, translation efficiency, and nuclear to cytoplasmic export of the mRNA transcripts [142]. A terminator may contain one or more polyadenylation signals (PASs; with consensus sequence of AAUAAA) in the 3′-UTR. Once the poly-A signals are transcribed, a cleavage and polyadenylation specificity factor (CPSF) recognizes the PAS(s) and cleaves the pre-mRNA 10 to 30 bp downstream of the PAS, freeing the mRNA from the Pol II transcription machinery [143]. After the cleavage, a poly(A) tract of ~200 nt in length is added to the mRNA at the cleavage site (CS). Improper termination or unpolyadenylated mRNA transcripts serve as the templates for RNA-dependent RNA polymerase 6 (RDR6) to generate small interfering RNAs (siRNAs), leading to posttranscriptional gene silencing (PTGS) due to homology-based mRNA cleavage or translational repression [144,145].
In plants, 3 cis-termination elements function cooperatively to find the correct PAS(s) in the 3′-UTRs and initiate transcriptional termination. The 3 cis-termination elements include the near upstream element (NUE; AAUAAA-like motifs), the far upstream element (FUE), and the CS [146–149]. NUEs are short (6 to 10 bp) A-rich sequence located approximately 10 to 40 nt upstream of the poly(A) sites. FUEs are also short (6 to 18 bp) sequence with a more diverse nucleotide enrichment (U>A>G) and are typically found starting 30 bp upstream of the poly(A) sites. FUEs enhance processing efficiency at the CSs. The CS is a dinucleotide (CA or UA) typically located within the FUE where polyadenylation takes place. Less than 10% of terminators in Arabidopsis [146,150] and rice [151] contain the canonical PAS (AAUAAA). Genes can also contain multiple PASs [152].
Evidence shows that transgene expression level is significantly affected by the chosen terminators. In Arabidopsis, Pérez-González and Caro [153] demonstrated a comparison between the Arabidopsis heat shock protein 18.2 terminator (tHSP18) and the 35S terminator (t35S). The tHSP18, when used with the firefly luciferase (LUC) reporter gene driven by the 35S promoter (35S:LUC), showed significantly higher protein levels and lower rates of promoter DNA methylation in transgenic Arabidopsis plants than the t35S. Using various promoters, de Felippes et al. [154] observed higher green fluorescent protein (GFP) fluorescence in Agrobacterium-infiltrated tobacco leaves when the Arabidopsis tHSP18 was used, followed by the A. tumefaciens Nopaline synthase terminator (tNos), and then the Arabidopsis RuBisCO small subunit terminator (tRbcS). Differences in selection accuracy between terminator CSs may contribute to the differences in trans(gene) expression by various terminators. de Felippes et al. [154] found that only 40% of the transcripts generated from the overexpressed GFP transgene with tRbcS had the same poly(A) sites, while 76% of the GFP transcripts with tHSP18 were cleaved at the same locations.
Terminators isolated from exogenous sources such as tNos, A. tumefaciens Octopine synthase terminator (tOcs), and t35S are frequently used in plant transgenic experiments. However, evidence shows that the terminator sequences from plant endogenous genes have shown even greater potential to increase transgene expression than exogenous terminators [155] (Table 3). Production of the miraculin (MIR) protein by the 35S promoter was greatly increased (6.5×) in transgenic tomato when the tomato tHSP18 was used over tNos [159]. Similarly, the native MIR terminator showed significantly higher MIR production in transgenic tomato than tNos when the MIR gene was driven by the 35S promoter or the native or a sequence-optimized MIR promoter [165]. Diamos and Mason [156] compared the expression levels of 35S:GFP using one of 20 native terminator sequences from various plant species and found that, in most cases, plant native terminators led to higher GFP protein production than t35S or tNos. Moreover, these plant native terminators showed variable abilities in regulating gene expression. Ingelbrecht et al. [166] studied the effects of a handful of tobacco native terminator sequences, including the terminators from a 2S seed storage protein gene, an RbcS gene, an extensin gene, and a chalcone synthase gene on transcription of the neomycin phosphotransferase II (nptII) gene driven by the 35S promoter (35S:nptII). They found that tRbcS led to the highest mRNA accumulation and protein production, corresponding to 3, 5, 10, and 60 times that of the Ocs, 2S, extensin, and chalcone synthase terminators, respectively. These results suggest that terminators play an important role in posttranscriptional processes including the 3′-end processing efficiency and/or mRNA stability. Interestingly, Nagaya et al. [152] found that Arabidopsis tHSP18 always procured the highest mRNA and protein abundance of the Renilla and firefly luciferase (Rluc and Fluc) reporter genes when driven by the 35S promoter in both Arabidopsis and rice protoplasts, indicating that a plant native terminator could function well in both dicot and monocot species.
Table 3.
List of representative plant terminators used for gene expression.
| Terminator | Terminator source | Length (bp) | Effect a | Transformed species | Expression system | References |
|---|---|---|---|---|---|---|
| tACT3 | N. benthamiana | 617 | 3.9× higher GFP than with tNOS | N. benthamiana | Transient b | [156] |
| 1,044 | ~7.5× higher GFP than with tNOS | Lactuca sativa | ||||
| tACT3 –tRb7MAR | N. benthamiana (tACT3); N. tabacum (tRb7; tTM6) | 2,213 | ~8× higher GFP than with tNOS | N. benthamiana | Transient b | [156] |
| tACT3 –tTM6MAR | 2,243 | ~7× higher GFP than with tNOS | N. benthamiana | Transient b | [156] | |
| tEU | N. tabacum | 732 | 2.5× higher GFP mRNA and 2.5× higher GFP protein than with tVspB3 (Tobacco etch virus (TEV) promoter) | N. benthamiana | Transient b | [157] |
| 1,900 | ~1.5×higher DsRed than with tNOS;~44× higher DsRed than with tNOS | N. benthamiana;L. sativa | Transient b | [157] | ||
| tEU –tTM6MAR | N. tabacum | 1,931 | ~23× higher GFP than with tNOS;~10× higher DsRed than with tNOS | N. benthamiana | Transient b | [156] |
| tEU (intronless) | N. tabacum | 480 | ~10× higher GFP than with tNOS | L. sativa | Transient b | [156,158] |
| 11.9× higher GFP than with tVspB3; | N. benthamiana | |||||
| 2.8× higher GFP than with t35S; | ||||||
| ~15× higher DsRed than with tNOS | ||||||
| tEU (intronless) –tACT3 –tRB7MAR | N. tabacum (tEU; tRB7); N. benthamiana (tACT3) | 2,693 | ~56× higher GFP than with tNOS | N. benthamiana | Transient b | [156] |
| tHSP18 | A. thaliana | 249 | 6.5× or 8.4× higher miraculin protein than with tNOS | S. lycopersicum | Stable c | [159,160] |
| 249 | 4× higher miraculin protein (SlE8 promoter) than with MIR-tNOS | S. lycopersicum | Stable c | |||
| tHSP18 | A. thaliana | N.A. | 1.5× or 2.5× higher GUS or Fluc than with t35S, tOCS or tNOS | A. thaliana; O. sativa | Transient (protoplasts) | [152] |
| 878 | 1.5× higher Rluc or Fluc than with 250 bp tHSP18 | N. benthamiana | Stable c | [161] | ||
| 249 | ~7.5× higher GFP than with tNOS | L. sativa | Transient b | [156] | ||
| 249 | 2.5× higher GFP than with tNOS | N. benthamiana | ||||
| tHSP18 –tEU –tRb7MAR | A. thaliana (tHSP18); N. tabacum (tEU; tRb7) | 1,898 | ~20× higher GFP than with tNOS | N. benthamiana | Transient b | [156] |
| tHSP18 –tACT3 | N. benthamiana (tHSP18; tACT3); N. tabacum (tTM6; tRb7); S. tuberosum (tPINII) | 1,485 | ~25× higher GFP than with tNOS | N. benthamiana | Transient b | [156] |
| tHSP18 –tACT3 –tRb7 | 2,654 | ~50× higher GFP than with tNOS | ||||
| tHSP18 –tPINII –tRb7MAR | 2,580 | ~15× higher GFP than with tNOS | ||||
| tHSP18 –tPINII –tTM6MAR | 2,610 | ~15× higher GFP than with tNOS | ||||
| tHSP18 –tRb7MAR | 1,610 | ~14× higher GFP than with tNOS | N. benthamiana | Transient b | [156] | |
| tProteinase inhibitor II (tPINII) | S. tuberosum | N.A. | 20× higher HBsAg than with tNOS | S. tuberosum | Stable c | [162] |
| 970 | 8.5× higher GFP than with tNOS | N. benthamiana | Transient b | [156] | ||
| trbcS | M. domestica | 582 | 11.1× higher GUS than with tNOS (M. domestica rbcS promoter – 1,679 bp) | N. benthamiana | Transient b | [163] |
| Medicago sativa | 400 | ~1.4× higher GUS than with tNOS (Figwort mosaic virus 35S promoter) | M. sativa | Stable c | [164] | |
| Pisum sativum | 684 | 5.4× higher GFP than with tNOS; ~10× higher DsRed than with tNOS | N. benthamiana | Transient b | [156] |
a The 35S promoter was used except when specified.
b Transient, leaf agroinfiltration.
c Stable, Agrobacterium-mediated transformation.
Plant terminators determine which PASs the gene’s mRNA is processed at. For example, Yang et al. [167] overexpressed a modified house dust mite allergen gene mDerf2 in transgenic rice using the maize Ubiquitin promoter together with different terminators and examined where the transgene mRNA was processed in the seeds and leaves of the transgenic rice plants. They found that the transgene mRNA with the rice glutelin B-1 terminator (tGluB-1) was processed at 2 specific PAS sites in the seed but at 6 sites in the leaf. The 2 PAS sites in the seed were identical to those in the native GluB-1 mRNA. However, when regulated by tNos, the transgene mRNA was processed at 8 PAS sites in the seed and at 6 sites in the leaf.
Plant terminators also affect the length of read-through mRNA transcripts, which is a normal phenomenon of gene transcription. Xing et al. [168] studied the effects of soybean native terminators on transgene expression in transgenic soybean plants. They found that read-through transcription occurred in all the transgenic plants, with ~1% of total transgene mRNA being generated from this process. In addition, Hiwasa-Tanase et al. [165] observed longer read-through mRNA transcripts when the transgene was processed by tNOS rather than tMIR.
Interestingly, studies showed that 2 terminators can be linked together for enhanced transgene expression; perhaps the addition of a second terminator can lead to better detection of read-through transcription and potentially inhibit it [169]. Diamos and Mason [155] combined the Nicotiana benthamiana extensin terminator (tEU) with the terminator of a tobacco Actin (NbACT3) gene to regulate 35S:GFP expression. Transient expression of the reporter gene was observed at levels of 37.7 times greater when the combined terminators were used over tNos. Similarly, Yamamoto et al. [170] measured transient GFP expression in lettuce (Lactuca sativa), tomatoes, eggplants (Solanum melongena), hot peppers (Capsicum frutescens), melons (Cucumis melo), orchids (Phalaenopsis aphrodite), and tobacco by using combined terminators. Combination of Arabidopsis tHSP18 and tobacco tEU rendered the highest expression when compared to each terminator by itself or a double terminator of the same kind. The combined tHSP18-tEU terminator also worked better than a triple terminator made up of one tHSP, one t35S, and one tEU.
The presence of a matrix attachment region (MAR) in a terminator enhances gene expression. MARs are AT-rich DNA sequences that act as epigenetic regulatory sequences by mediating the binding of the chromatin to a protein nuclear matrix and changing chromatin conformation, leading to enhanced gene expression [171]. Matsui et al. [161] compared transient reporter gene expression in tobacco protoplasts when an 878-bp version versus a 250-bp version of the tHSP18 was used. Higher protein abundance of the Rluc and Fluc reporter genes was detected when the longer version was used due to the presence of a MAR in the longer version. It was reported that adding MARs into terminators enhanced GFP mRNA accumulation and protein production in Agrobacterium-infiltrated tobacco leaves [155,158].
Rosenthal et al. [158] tested the effects of the tobacco tEU with and without an intron in tEU on transient 35S:GFP expression in tobacco leaves. They found that the intronless tEU significantly increased transient 35S:GFP expression when compared to the intron-containing tEU. de Felippes et al. [154] also found that introns located close to the terminator can lead to aberrant termination and splicing in a terminator-dependent manner. They found that having a well-defined poly(A) site increased the likelihood that the transcripts were cleaved at the same site, and inefficient termination leads to transgene silencing. Thus, terminators have a great effect on transgene silencing because they affect siRNA production and splicing efficiency.
Utilization of Promoters and Terminators for Plant Biodesign
Promoters control the strength and spatiotemporal expression of a gene, while the terminators mainly contribute to the expression level of a gene [19,154,172]. Based on expression patterns, promoters can be categorized into constitutive promoters, responsive/inducible promoters, and tissue-specific (or cell type-specific) promoters (Table 2). With the advent of plant synthetic biology, elegant genetic circuits are needed for fine-tuning gene expression and high-precision genome engineering in plants. Hence, promoters with various strengths and specific expression patterns are necessary for bioengineering of complex plant systems. In this section, we discuss how promoters have been utilized for plant biodesign, with a focus on some representative examples such as plant transformation, plant metabolic engineering, plant-based biosensing, and genome editing (Fig. 3). Through these cases, we cover multiple technical aspects including expression strength, expression patterns, and terminator and promoter configuration.
Fig. 3.
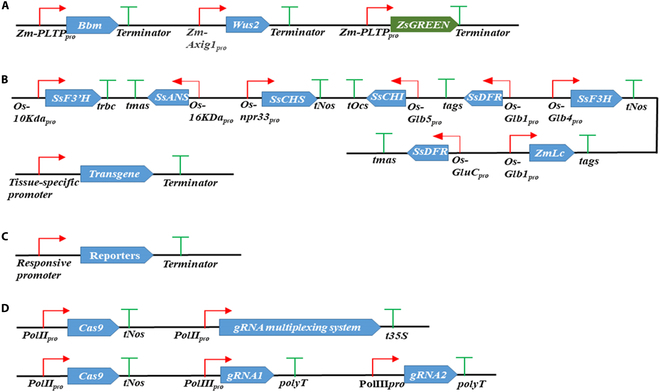
Illustration of construct design for application of promoters in plants. (A) Illustration of the use of promoters for improving plant transformation [174]. (B) Illustration of the use of promoters for metabolic engineering in plants [180]. (C) Illustration of the use of promoters for engineering biosensors in plants. (D) Illustration of the use of promoters for CRISPR/Cas9-based genome editing in plants [185]. Zm-PLTPpro, maize phospholipid transfer protein gene promoter; Zm-Axig1pro, maize auxin-inducible AXIG1 promoter; Os-10Kdapro, rice 10-kDa prolamin promoter; trbc, tobacco RuBisCO terminator; tmas, mannopine synthase gene terminator from A. tumefaciens; Os-npr33pro, rice 13-kDa prolamin promoter; tNos, nopaline synthase terminator from A. tumefaciens; tOcs, octopine synthase terminator from A. tumefaciens; Os-Glb5pro, rice globulin 5 promoter; tags, agropine synthase terminator from A. tumefaciens; Os-Glb4pro, rice globulin 4 promoter; Os-Glb1pro, rice globulin 1 promoter; Os-GluCpro, rice glutelin C promoter; PolIIpro, polymerase II promoter; t35S, 35S terminator; PolIIIpro, polymerase III promoter; polyT, the poly T termination signal for Pol III transcription.
Application of promoters for improving plant transformation
Tissue culture-based plant transformation is a rate-limiting step for plant biodesign. Since 2016, morphogenic regulators have been applied to transform recalcitrant cultivars, increase transformation efficiency, shorten the time for plant regeneration, and generate desirable phenotypes while bypassing the tissue culture process [173–175]. Constitutive expression of the maize morphogenic regulators Baby Boom (BBM) and Wuschel2 (WUS2) promoted direct somatic embryogenesis and thereby increased transformation efficiency in maize [173]. However, this overexpression approach caused side effects, e.g., phenotypic abnormalities and sterility [173], which could be minimized by using tissue-specific or inducible expression of the genes. When the maize leaf- and embryo-specific phospholipid transferase protein (PLTP) gene promoter was used to confer a strong tissue-specific expression of BBM and the maize auxin-inducible AXIG1 promoter was used to drive WUS2 expression, normal transgenic plants were obtained [174] (Fig. 3A).
Application of promoters for metabolic engineering in plants
Expression of partial or complete biosynthetic pathways from one species can be used for plant metabolic engineering in another species. Multiple genes can be stacked into a plant genome via the use of a single T-DNA sequence. For example, a fungal caffeic acid biosynthesis and recycling pathway containing 4 fungal genes plus a LUC reporter gene, with each gene in a constitutive expression cassette, has been used to generate luciferase-based autoluminescent tobacco, tomato, Arabidopsis, periwinkle (Catharanthus roseus), petunia (Petunia hybrida), and rose (Rosa rubiginosa) plants [176,177]. The stability of transgene expression is always a concern for plant metabolic engineering when the identical expression cassettes are used for the expression of multiple genes [178]. Thus, different promoters are preferred to be used in a multi-gene construct for the expression of different genes [179]. For example, 8 different rice endosperm-specific promoters have been used to drive the expression of 8 anthocyanin biosynthesis pathway genes individually to develop purple endosperm rice [180] (Fig. 3B).
Application of promoters for engineering biosensors in plants
Precise control of gene expression is essential for plant responses to various environmental stimuli. Stimulus-responsive promoters ensure accurate timing of gene expression. They can be used to drive the expression of optical (fluorescent proteins/luciferases) or morphological reporter genes to form stimulus-responsive “promoter–reporter” systems (Fig. 3C). Whenever a stimulus signal is present, the reporter gene’s expression is activated by the stimulus-responsive promoter, illuminating the dynamics of the signal in real time. Hence, stimulus-responsive promoters are very useful components in plant biodesign for building transcriptional regulation-based plant biosensors. Several phytohormone-responsive promoters have been generated for monitoring phytohormone signaling pathways. For example, the auxin-responsive DR5 promoter has been used to build biosensors monitoring auxin levels at various developmental stages of Arabidopsis roots [181]. In addition, the abscisic acid (ABA)-responsive DR29 promoter, the cytokinin-responsive TCS promoter, and the salicylic acid (SA)-responsive FLS2 promoter have been used to drive the expression of optical reporters for monitoring the dynamic changes in the levels of their corresponding phytohormones [182–184]. With the availability of rich omics resources, novel cis-regulatory elements can be identified and used to generate synthetic stimulus-responsive promoters by multimerizing cis-regulatory elements upstream of a core promoter sequence [134–136].
Application of promoters for CRISPR/Cas9-based genome editing in plants
CRISPR/Cas9-based genome engineering tools are important for plant synthetic biology. There are 2 functional components in the CRISPR/Cas9 system. The short guide RNA (gRNA) determines the target specificity, while the Cas9 endonuclease generates a double-strand break at each target region [185]. These 2 functional components are usually driven by different expression cassettes. The Cas9 gene is often driven by a strong constitutive promoter, such as the 35S, AtUbi10, and ZmUbi promoters, while the gRNA is usually under the control of a RNA polymerase III (Pol III) promoter such as the U3 or U6 promoter [185–187] (Fig. 3D). One limitation of using Pol III promoters is that a specific nucleotide is required in the first position of each transcript, e.g., the U3 and U6 promoters in plants have discrete TSSs that are adenine (A) and guanine (G), respectively [188]. Compared with Pol III promoters, Pol II promoters can also be used to express a polycistronic gRNA array, which can be processed posttranscriptionally into individual gRNAs by RNA-cleaving enzymes [189,190], providing a flexibility for spatiotemporal control of multiplexed gene editing.
Tissue-specific promoters can be used to improve the efficiency of genome editing in plants. The Arabidopsis meristematic tissue-specific YAO, Egg Cell 1 (EC1), and CLAVATA3 (CLV3) promoters have been used to drive Cas9 expression in Arabidopsis, resulting in enhanced CRISPR/Cas9-induced mutation efficiency [191]. Similarly, the Arabidopsis root cap-specific SOMBRERO (SMB) promoter, stomatal lineage-specific TOO MANY MOUTHS (TMM) and FAMA promoters, and lateral root primordia-specific GATA23 promoter have been utilized to drive root cap-, stomatal lineage-, and lateral root primordia-specific knockouts in Arabidopsis [192,193].
Application of combined promoter-terminator combinations for genetic engineering in plants
Promoters can be used together with terminators to fine-tune transgene expression. For example, a total of 105 promoter–terminator combinations have been evaluated in N. benthamiana leaves and N. tabacum cells, providing a wide range of expression strength [172]. Moreover, Schaart et al. [163] studied the effects of the promoter–terminator combinations on the β-glucuronidase (GUS) reporter gene expression by using the 35S promoter, 2 variants (with different lengths) of the apple (Malus × domestica) RbcS (MdRbcS) promoter, and the tNos and MdRbcS terminator (tMdRbcs). Using transient expression in tobacco leaves, they found that the 35S promoter–tMdRbcs combination generated the highest GUS mRNA and protein levels, followed by the 35S promoter–tNOS, and each MdRbcS promoter–tMdRbcs. The lowest GUS expression was rendered by the MdRbcS promoter–tNos combination. Similarly, Kurokawa et al. [160] found that the transgenic tomato plants expressing the MIR transgene by the tomato fruit-ripening specific E8 promoter–Arabidopsis tHSP18 produced 4 times more MIR protein per fruit fresh weight than the transgenic tomato plants containing 35S:MIR–tNos. Thus, it is important to select an appropriate promoter–terminator combination for optimal transgene expression in plant biodesign [172].
Strategies for Benchmarking Promoters and Terminators for Plant Biodesign
To date, more than 8,000 plant promoter sequences have been identified through transcriptomic and genomic analyses and documented in various plant promoter databases such as PlantProm DB [117], PlantPromoterDB [119], and PlantCARE [129]. However, only a small number of promoters and terminators have been experimentally characterized and validated in plants (see above). Thus, there is high demand to expand the number of functionally characterized promoters and terminators to serve as standard biological parts for plant synthetic biology research and bioengineering. Benchmarking via standardized procedures is an essential step for providing necessary semiquantitative and quantitative information about the performance of promoters and terminators.
Requirements for evaluating promoters and terminators in plants
The features and performance of promoters and terminators can be systematically evaluated in 4 aspects: (a) temporal expression pattern, (b) spatial expression pattern, (c) environmental responses, and (d) cross-species variation (Fig. 4). For each of the 4 aspects, the strength of promoters and promoter–terminator combinations needs to be quantitatively evaluated. Promoters can be explored based on seasonal and diurnal patterns. In terms of spatial expression patterns, numerous tissue- and cell type-specific promoters have been identified in plants including leaf-, stem-, root-, flower-, fruit-, guard cell-, root hair-, and companion cell-specific promoters [194]. Developmental gradient is also one of the aspects that reflect the spatial expression patterns of some promoters. For instance, the developmental gradient in maize leaves has been exploited to redefine the current C4 model and gain new insights into the regulation of C4 photosynthesis [195]. Notably, the performance and the relative usefulness of individual promoters and terminators may be different across plant species. It would be necessary to characterize promoters and terminators in different plant species representing dicots (e.g., Arabidopsis, tobacco, poplar) and/or monocots (e.g., rice, maize, wheat (Triticum aestivum)). Bioengineering of stress tolerance in plants requires the identification and characterization of environmental stress-responsive promoters [196], which can be tested under biotic treatments (e.g., pathogens and beneficial microbes) and abiotic stresses (e.g., drought, salinity, and temperature).
Fig. 4.
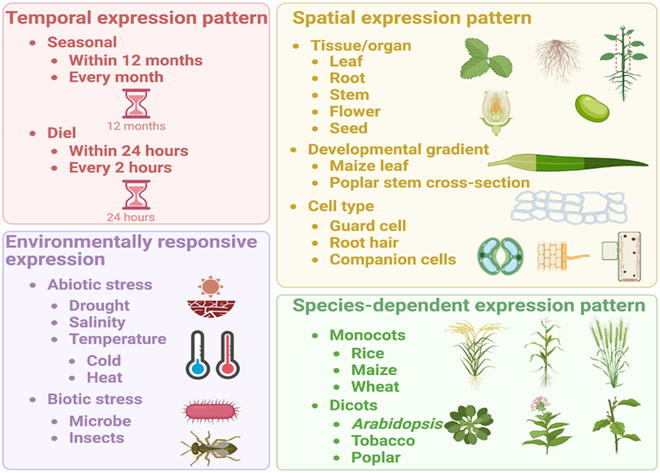
Aspects of promoter and terminator evaluation for plant biodesign.
Methodologies for benchmarking promoters and terminators in plants
Many different methodologies for validating plant promoters and terminators have been developed and applied to various plant species. These methodologies can be classified into 2 main categories, i.e., transient expression approaches and stable expression approaches (Fig. 5A). Transient expression approaches are relatively simple and effective and can be quickly completed in various plant cells and tissue/organs. Examples for transient expression systems include protoplast transfection, leaf agro-infiltration, and hairy root transformation. In contrast, stable expression systems involve complex and lengthy stable plant transformation but provide the most robust information on the function and strength of promoters and terminators. Reporter genes such as GFP, GUS, and LUC are often used to pinpoint the performance of a target promoter and/or terminator in either transient or stable expression systems (Fig. 5A). The reporter assay for the analysis of promoter and/or terminator activity in a transient expression system allows for semiquantitative determination of reporter gene expression using microscope and/or plate reader, leading to variable output due to the use of different cell types and sample preparation methods. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) and droplet digital PCR (ddPCR) analysis of transgenic plant samples can provide more reliable and robust information on promoter and terminator features (Fig. 5B). Moreover, certain features (e.g., environmental response or time-course development) of a target promoter and/or terminator can only be investigated using stable transformed plants or samples.
Fig. 5.
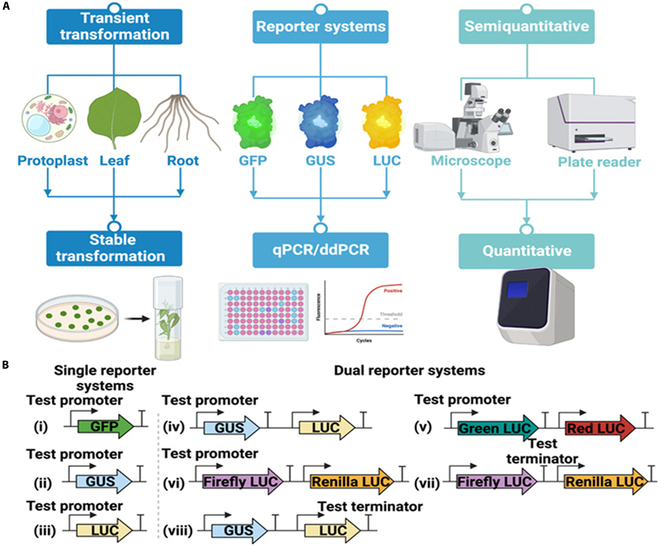
The technologies for benchmarking promoters and terminator in plants. (A) Methods and tools used for performance analysis of plant promoters and terminators. (B) Different architectures of single-reporter and dual-reporter systems. ddPCR, droplet digital PCR; LUC, luciferase.
Both single-reporter and dual-reporter systems have been used to examine promoter and terminator performance in either transient or stable expression systems. A single-reporter system allows reporter gene expression to be imaged and detected in the samples of interest through a microscope and/or a plate reader when compared to the samples of mock treatments (Fig. 5A and B). A dual-reporter system provides the means for normalization of the reporter gene expression when a weak promoter–terminator combination is used to drive the expression of a second reporter gene that serves as a reliable internal standard for normalization. The use of a dual-reporter system permits the elimination of the difference in delivery efficiency (for transient expression systems) or positional effects of the transgene (for stable expression systems). To date, multiple dual-reporter systems have been developed for the quantitative characterization of plant-based promoters (Fig. 5B). For example, a GUS/LUC system, whereby LUC was used to normalize and calculate the relative GUS activities, was used to generate relative promoter activities in wheat protoplasts [197]. A dual-color luciferase ratiometric reporter system with green- and red-emitting luciferases was developed for fast characterization of transcriptional regulatory elements in plants [198]. However, the partial signal overlap between green/LUC and red/LUC hinders the highly precise evaluation of the genetic parts. For more precise promoter characterization, the FLUC/RLUC luciferase reporter system is more sensitive since the assay is based on a chemiluminescence reaction and has markedly reduced interference between the 2 luciferases [172]. The FLUC/RLUC assay was used to examine the strength of combinations of promoters and terminators, revealing a 326-fold difference in the level of reporter gene expression between the strongest and the weakest promoters tested in plants [172]. Using a GUS/LUC reporter system, it was reported that appropriate selection of terminator sequences is an important factor for transgene expression in both monocot and dicot plants [199].
Conclusion and Perspectives
Gene expression is largely under the control of promoters and terminators. Although the selection of promoters and terminators is important for plant bioengineering, efforts to conduct comprehensive sequence analysis of promoters, promoter motifs, and terminators remain limited, hindering high-precision plant bioengineering. The traditional means of identifying regulatory elements in the upstream, intronic, and downstream sequences of genes of interest have limitations. For instance, promoter lengths are frequently chosen in an arbitrary manner (e.g., 1 or 2 kb from the start codons or the TSSs) and regulatory elements at distal sites are sometimes missed. Sequencing-based approaches now permit precise determination of TSSs by mapping nascent transcripts [200]. A recent study used template-switching reverse transcription in conjunction with rolling circle amplification (Smart-Seq2 Rolling Circle to Concatemeric Consensus, or Smar2C2) for global mapping of TSSs in multiple plant species [201]. The growing adoption of ATAC-seq (assay for transposase-accessible chromatin using sequencing) provides a rich resource for mining the regulatory landscape in open chromatin regions, which are genomic sites accessible by TFs. ATAC-seq analysis across multiple plant genomes revealed that the majority of accessible regions fall within 3 kb upstream of the TSSs [202,203]. Those studies also uncovered a substantial number of distal cis-regulatory elements. For instance, more than 10% of ATAC-seq signals are located at >20 kb away from their nearest genes in maize [204]. Integration of RNA-seq and ATAC-seq across a wide range of tissues and increasingly with single-cell resolution should facilitate discovery of tissue-specific cis-regulatory elements for precision gene manipulation. Benchmarking cis-regulatory elements, promoters, and terminators with different expression patterns and strength will provide invaluable insight for characterizing, curating, and constructing genetic circuits and pathways in plant biodesign.
The orthogonality of the key regulatory elements present in the promoters and terminators permits the use of these biobricks in a heterologous system for plant biodesign. However, these regulatory elements (as well as the promoters and terminators themselves) are optimized to function properly in their native contexts, and their regulation in a heterologous system may be different from that in their natural contexts [205]. Thus, whether these biobricks function in an orthogonal manner in a different species must be taken into consideration. Combining a trial-and-error approach with fine-tuning for design, testing, and validation will help obtain the desired functions and properties [1].
Acknowledgments
Funding: The writing of this manuscript was supported by the USDA Floriculture and Nursery Research Initiative (FNRI) grant 8020–21000-071-23S, the USDA National Institute of Food and Agriculture (NIFA) Hatch project 02913, and the Center for Bioenergy Innovation (CBI), which is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. DOE under contract number DE-AC05-00OR22725.
Author contributions: X.Y. and W.L. conceived the idea, and all authors wrote and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Liu W, Stewart CN Jr. Plant synthetic biology. Trends Plant Sci. 2015;20(5):309–317. [DOI] [PubMed] [Google Scholar]
- 2.Rourke M. Access and benefit-sharing DNA componentry for plant synthetic biology: Bioparts expressed in plant chassis. Plants People Planet. 2022;4(1):76–83. [Google Scholar]
- 3.Huang D, Kosentka PZ, Liu W. Synthetic biology approaches in regulation of targeted gene expression. Curr Opin Plant Bio. 2021;63: Article 102036. [DOI] [PubMed] [Google Scholar]
- 4.Schor IE, Degner JF, Harnett D, Cannavò E, Casale FP, Shim H, Garfield DA, Birney E, Stephens M, Stegle O, et al. Promoter shape varies across populations and affects promoter evolution and expression noise. Nat Genet. 2017;49(4):550–558. [DOI] [PubMed] [Google Scholar]
- 5.Jores T, Tonnies J, Wrightsman T, Buckler ES, Cuperus JT, Fields S, Queitsch C. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat Plants. 2021;7(6):842–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari S, Ware D. Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots. PLOS ONE. 2013;8(10): Article e79011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina C, Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6(1): Article 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Civáň P, Švec M. Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y patch promoter elements. Genome. 2009;52(3):294–297. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto YY, Yoshitsugu T, Sakurai T, Seki M, Shinozaki K, Obokata J. Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 2009;60(2):350–362. [DOI] [PubMed] [Google Scholar]
- 10.Porto MS, Pinheiro MP, Batista VG, dos Santos RC, Albuquerque Melo Filho P, de Lima LM.. Plant promoters: An approach of structure and function. Mol Biotechnol. 2014;56(1):38–49. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura M, Tsunoda T, Obokata J. Photosynthesis nuclear genes generally lack TATA-boxes: A tobacco photosystem I gene responds to light through an initiator. Plant J. 2002;29(1):1–10. [DOI] [PubMed] [Google Scholar]
- 12.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72(1):449–479. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto YY, Ichida H, Abe T, Suzuki Y, Sugano S, Obokata J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007;35(18):6219–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8: Article 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18(3):157–166. [DOI] [PubMed] [Google Scholar]
- 16.Baruah I, Baldodiya GM, Sahu J, Baruah G. Dissecting the role of promoters of pathogen-sensitive genes in plant defense. Curr Genomics. 2020;21(7):491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunennvaldt RL, Degenhardt-Goldbach J, Gerhardt IR, Quoirin M. Promoters used in genetic transformation of plants. Res J Biol Sci. 2015;10(1-2):1–9. [Google Scholar]
- 18.Jeong HJ, Jung KH. Rice tissue-specific promoters and condition-dependent promoters for effective translational application. J Integr Plant Biol. 2015;57(11):913–924. [DOI] [PubMed] [Google Scholar]
- 19.Kummari D, Palakolanu SR, Kishor PK, Bhatnagar-Mathur P, Singam P, Vadez V, Sharma KK. An update and perspectives on the use of promoters in plant genetic engineering. J Biosci. 2020;45: Article 119. [PubMed] [Google Scholar]
- 20.Misra S, Ganesan M. The impact of inducible promoters in transgenic plant production and crop improvement. Plant Gene. 2021;27: Article 100300. [Google Scholar]
- 21.Muthusamy SK, Sivalingam PN, Sridhar J, Singh D, Haldhar SM, Kaushal P. Biotic stress inducible promoters in crop plants—A review. J Agric Ecol. 2017;4:14–24. [Google Scholar]
- 22.Singhal P, Jan AT, Azam M, Haq QM. Plant abiotic stress: A prospective strategy of exploiting promoters as alternative to overcome the escalating burden. Front Life Sci. 2016;9(1):52–63. [Google Scholar]
- 23.Ali S, Kim WC. A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front Plant Sci. 2019;10: Article 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biłas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 2016;127:269–287. [Google Scholar]
- 25.Dey N, Sarkar S, Acharya S, Maiti IB. Synthetic promoters in planta. Planta. 2015;242(5):1077–1094. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Stewart CN Jr. Plant synthetic promoters and transcription factors. Curr Opin Biotechnol. 2016;37:36–44. [DOI] [PubMed] [Google Scholar]
- 27.Marand AP, Zhang T, Zhu B, Jiang J. Towards genome-wide prediction and characterization of enhancers in plants. Biochim Biophys Acta Gene Regul Mech. 2017;1860(1):131–139. [DOI] [PubMed] [Google Scholar]
- 28.Shrestha A, Khan A, Dey N. Cis–trans engineering: Advances and perspectives on customized transcriptional regulation in plants. Mol Plant. 2018;11(7):886–898. [DOI] [PubMed] [Google Scholar]
- 29.Vinciguerra P, Stutz F. mRNA export: An assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16(3):285–292. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16(4):1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: Messages from the 3′ end. Curr Opin Genet Dev. 1997;7(2):220–232. [DOI] [PubMed] [Google Scholar]
- 32.Wormington M, Searfoss AM, Hurney CA. Overexpression of poly (A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15(4):900–909. [PMC free article] [PubMed] [Google Scholar]
- 33.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly (A) binding protein is independent of poly (a) and requires translation. Genes Dev. 1998;12(20):3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ford LP, Bagga PS, Wilusz J. The poly (a) tail inhibits the assembly of a 3’-to-5’ exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17(1):398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2(4):237–246. [DOI] [PubMed] [Google Scholar]
- 36.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell. 1997;89(6):831–838. [DOI] [PubMed] [Google Scholar]
- 37.Chekanova JA, Belostotsky DA. MicroRNAs and messenger RNA turnover. MicroRNA Protoc. 2006;342:73–85. [DOI] [PubMed] [Google Scholar]
- 38.Porrua O, Libri D. Transcription termination and the control of the transcriptome: Why, where and how to stop. Nat Rev Mol Cell Biol. 2015;16(3):190–202. [DOI] [PubMed] [Google Scholar]
- 39.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352(6291): Article aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mejía-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, Grotewold E. Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell. 2015;27(12):3309–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernard V, Brunaud V, Lecharny A. TC-motifs at the TATA-box expected position in plant genes: A novel class of motifs involved in the transcription regulation. BMC Genomics. 2010;11: Article 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McElroy D, Brettell RI. Foreign gene expression in transgenic cereals. Trends Biotechnol. 1994;12(2):62–68. [Google Scholar]
- 43.Sah SK, Kaur G, Cheema GS. Genetic transformation of rice: Problems, progress and prospects. Rice Res. 2014;3(1): Article 1000132. [Google Scholar]
- 44.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 2002;16(20):2583–2592. [DOI] [PubMed] [Google Scholar]
- 45.Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19(10):621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das S, Bansal M. Variation of gene expression in plants is influenced by gene architecture and structural properties of promoters. PLOS ONE. 2019;14(3): Article e0212678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu MJ, Seddon AE, Tsai ZT, Major IT, Floer M, Howe GA, Shiu SH. Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res. 2015;25(8):1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh M, Bag SK, Bhardwaj A, Ranjan A, Mantri S, Nigam D, Sharma YK, Sawant SV. Global nucleosome positioning regulates salicylic acid mediated transcription in Arabidopsis thaliana. BMC Plant Biol. 2015;15: Article 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava R, Rai KM, Srivastava M, Kumar V, Pandey B, Singh SP, Bag SK, Singh BD, Tuli R, Sawant SV. Distinct role of core promoter architecture in regulation of light-mediated responses in plant genes. Mol Plant. 2014;7(4):626–641. [DOI] [PubMed] [Google Scholar]
- 50.Oldfield AJ, Henriques T, Kumar D, Burkholder AB, Cinghu S, Paulet D, Bennett BD, Yang P, Scruggs BS, Lavender CA, et al. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat Commun. 2019;10(1): Article 3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaves-Sanjuan A, Gnesutta N, Gobbini A, Martignago D, Bernardini A, Fornara F, Mantovani R, Nardini M. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J. 2021;105(1):49–61. [DOI] [PubMed] [Google Scholar]
- 52.Sun M, Ding J, Li D, Yang G, Cheng Z, Zhu Q. NUDT21 regulates 3′-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410:158–168. [DOI] [PubMed] [Google Scholar]
- 53.Norris SR, Meyer SE, Callis J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol. 1993;21:895–906. [DOI] [PubMed] [Google Scholar]
- 54.Akua T, Shaul O. The Arabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5′ UTR intron. J Exp Bot. 2013;64(14):4255–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curi GC, Chan RL, Gonzalez DH. The leader intron of Arabidopsis thaliana genes encoding cytochrome c oxidase subunit 5c promotes high-level expression by increasing transcript abundance and translation efficiency. J Exp Bot. 2005;56(419):2563–2571. [DOI] [PubMed] [Google Scholar]
- 56.Plesse B, Criqui MC, Durr A, Parmentier Y, Fleck J, Genschik P. Effects of the polyubiquitin gene Ubi. U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol. 2001;45(6):655–667. [DOI] [PubMed] [Google Scholar]
- 57.Kim YJ, Lee SH, Park KY. A leader intron and 115-bp promoter region necessary for expression of the carnation S-adenosylmethionine decarboxylase gene in the pollen of transgenic tobacco. FEBS Lett. 2004;578(3):229–235. [DOI] [PubMed] [Google Scholar]
- 58.Gadea J, Conejero V, Vera P. Developmental regulation of a cytosolic ascorbate peroxidase gene from tomato plants. Mol Gen Genomics. 1999;262:212–219. [DOI] [PubMed] [Google Scholar]
- 59.Fu H, Kim SY, Park WD. High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5’ and 3’ flanking sequences and the leader intron. Plant Cell. 1995;7(9):1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu H, Kim SY, Park WD. A potato Sus3 sucrose synthase gene contains a context-dependent 3’ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell. 1995;7(9):1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu WW, Gong H, Pua EC. The pivotal roles of the plant S-adenosylmethionine decarboxylase 5’ untranslated leader sequence in regulation of gene expression at the transcriptional and posttranscriptional levels. Plant Physiol. 2005;138(1):276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gianì S, Altana A, Campanoni P, Morello L, Breviario D. In trangenic rice, α-and β-tubulin regulatory sequences control GUS amount and distribution through intron mediated enhancement and intron dependent spatial expression. Transgenic Res. 2009;18:151–162. [DOI] [PubMed] [Google Scholar]
- 63.Morello L, Bardini M, Sala F, Breviario D. A long leader intron of the Ostub16 rice β-tubulin gene is required for high-level gene expression and can autonomously promote transcription both in vivo and in vitro. Plant J. 2002;29(1):33–44. [DOI] [PubMed] [Google Scholar]
- 64.Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R. Gene expression enhancement mediated by the 5’ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Gen Genomics. 2008;279(6):563–572. [DOI] [PubMed] [Google Scholar]
- 65.Shi X, Wu J, Mensah RA, Tian N, Liu J, Liu F, Chen J, Che J, Guo Y, Wu B, et al. Genome-wide identification and characterization of UTR-introns of Citrus sinensis. Int J Mol Sci. 2020;21(9): Article 3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vitale A, Wu RJ, Cheng Z, Meagher RB. Multiple conserved 5’ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Mol Biol. 2003;52(6):1135–1151. [DOI] [PubMed] [Google Scholar]
- 67.David-Assael O, Berezin I, Shoshani-Knaani N, Saul H, Mizrachy-Dagri T, Chen J, Brook E, Shaul O. AtMHX is an auxin and ABA-regulated transporter whose expression pattern suggests a role in metal homeostasis in tissues with photosynthetic potential. Funct Plant Biol. 2006;33(7):661–672. [DOI] [PubMed] [Google Scholar]
- 68.De La Torre CM, Finer JJ. The intron and 5’ distal region of the soybean Gmubi promoter contribute to very high levels of gene expression in transiently and stably transformed tissues. Plant Cell Rep. 2015;34(1):111–120. [DOI] [PubMed] [Google Scholar]
- 69.Gallegos JE, Rose AB. An intron-derived motif strongly increases gene expression from transcribed sequences through a splicing independent mechanism in Arabidopsis thaliana. Sci Rep. 2019;9(1): Article 13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grant TNL, De La Torre CM, Zhang N, Finer JJ. Synthetic introns help identify sequences in the 5’-UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta. 2017;245(4):849–860. [DOI] [PubMed] [Google Scholar]
- 71.Kamo K, Kim AY, Park SH, Joung YH. The 5’ UTR-intron of the gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in gladiolus and Arabidopsis. BMC Plant Biol. 2012;12: Article 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laxa M, Müller K, Lange N, Doering L, Pruscha JT, Peterhänsel C. The 5’ UTR intron of Arabidopsis GGT1 aminotransferase enhances promoter activity by recruiting RNA polymerase II. Plant Physiol. 2016;172(1):313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samadder P, Sivamani E, Lu J, Li X, Qu R. Transcriptional and post-transcriptional enhancement of gene expression by the 5’ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Gen Genomics. 2008;279(4):429–439. [DOI] [PubMed] [Google Scholar]
- 74.Sivamani E, Qu R. Expression enhancement of a rice polyubiquitin gene promoter. Plant Mol Biol. 2006;60(2):225–239. [DOI] [PubMed] [Google Scholar]
- 75.Belcher MS, Vuu KM, Zhou A, Mansoori N, Agosto Ramos A, Thompson MG, Scheller HV, Loqué D, Shih PM. Design of orthogonal regulatory systems for modulating gene expression in plants. Nat Chem Biol. 2020;16(8):857–865. [DOI] [PubMed] [Google Scholar]
- 76.An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10(1):107–121. [DOI] [PubMed] [Google Scholar]
- 77.McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2(2):163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Callis J, Raasch JA, Vierstra RD. Ubiquitin extension proteins of Arabidopsis thaliana. Structure, localization, and expression of their promoters in transgenic tobacco. J Biol Chem. 1990;265(21):12486–12493. [PubMed] [Google Scholar]
- 79.Mann DG, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, et al. Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol. 2011;11: Article 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18(4):675–689. [DOI] [PubMed] [Google Scholar]
- 81.Bhattacharyya J, Chowdhury AH, Ray S, Jha JK, Das S, Gayen S, Chakraborty A, Mitra J, Maiti MK, Basu A, et al. Native polyubiquitin promoter of rice provides increased constitutive expression in stable transgenic rice plants. Plant Cell Rep. 2012;31(2):271–279. [DOI] [PubMed] [Google Scholar]
- 82.Jones MO, Manning K, Andrews J, Wright C, Taylor IB, Thompson AJ. The promoter from SlREO, a highly-expressed, root-specific Solanum lycopersicum gene, directs expression to cortex of mature roots. Funct Plant Biol. 2008;35(12):1224–1233. [DOI] [PubMed] [Google Scholar]
- 83.Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153(1):185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jefferson R, Goldsbrough A, Bevan M. Transcriptional regulation of a patatin-1 gene in potato. Plant Mol Biol. 1990;14(6):995–1006. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L, Yang T, Li X, Hao H, Xu S, Cheng W, Sun Y, Wang C. Cloning and characterization of a novel Athspr promoter specifically active in vascular tissue. Plant Physiol Biochem. 2014;78:88–96. [DOI] [PubMed] [Google Scholar]
- 86.Xu W, Liu W, Ye R, Mazarei M, Huang D, Zhang X, Stewart CN. A profilin gene promoter from switchgrass (Panicum virgatum L.) directs strong and specific transgene expression to vascular bundles in rice. Plant Cell Rep. 2018;37(4):587–597. [DOI] [PubMed] [Google Scholar]
- 87.Yanagisawa S, Izui K. MNF1, a leaf tissue-specific DNA-binding protein of maize, interacts with the cauliflower mosaic virus 35S promoter as well as the C 4 photosynthetic phosphoenolpyruvate carboxylase gene promoter. Plant Mol Biol. 1992;19(4):545–553. [DOI] [PubMed] [Google Scholar]
- 88.Liu W, Mazarei M, Ye R, Peng Y, Shao Y, Baxter HL, Sykes RW, Turner GB, Davis MF, Wang ZY, Dixon RA, Stewart CN Jr.. Switchgrass (Panicum virgatum L.) promoters for green tissue-specific expression of the MYB4 transcription factor for reduced-recalcitrance transgenic switchgrass. Biotechnol Biofuels. 2018;11(1): Article 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2(12):1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bate N, Twell D. Functional architecture of a late pollen promoter: Pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol. 1998;37(5):859–869. [DOI] [PubMed] [Google Scholar]
- 91.Hamilton DA, Roy M, Rueda J, Sindhu RK, Sanford J, Mascarenhas JP. Dissection of a pollen-specific promoter from maize by transient transformation assays. Plant Mol Biol. 1992;18(2):211–218. [DOI] [PubMed] [Google Scholar]
- 92.Karunanandaa B, Qi Q, Hao M, Baszis SR, Jensen PK, Wong YH, Jiang J, Venkatramesh M, Gruys KJ, Moshiri F, et al. Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab Eng. 2005;7(5-6):384–400. [DOI] [PubMed] [Google Scholar]
- 93.Lamacchia C, Shewry PR, Di Fonzo N, Forsyth JL, Harris N, Lazzeri PA, Napier JA, Halford NG, Barcelo P. Endosperm-specific activity of a storage protein gene promoter in transgenic wheat seed. J Exp Bot. 2001;52(355):243–250. [PubMed] [Google Scholar]
- 94.Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Perez Conesa D, Ros G, Sandmann G, Capell T, et al. Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci U S A. 2009;106(19):7762–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deikman J, Fischer RL. Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 1988;7(11):3315–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ellis JG, Llewellyn DJ, Dennis ES, Peacock WJ. Maize Adh-1 promoter sequences control anaerobic regulation: Addition of upstream promoter elements from constitutive genes is necessary for expression in tobacco. EMBO J. 1987;6(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siebertz B, Logemann J, Willmitzer L, Schell J. Cis-analysis of the wound-inducible promoter wun1 in transgenic tobacco plants and histochemical localization of its expression. Plant Cell. 1989;1(10):961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Visser RG, Somhorst I, Kuipers GJ, Ruys NJ, Feenstra WJ, Jacobsen E. Inhibition of the expression of the gene for granule-bound starch synthase in potato by antisense constructs. Mol Gen Genomics. 1991;225(2):289–296. [DOI] [PubMed] [Google Scholar]
- 99.Takahashi T, Naito S, Komeda Y. The Arabidopsis HSP18.2 promoter/GUS gene fusion in transgenic Arabidopsis plants: A powerful tool for the isolation of regulatory mutants of the heat-shock response. Plant J. 1992;2(5):751–761. [Google Scholar]
- 100.Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 1993;101(3):1119–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi X, Zhang Y, Chai T. Characterization of a novel plant promoter specifically induced by heavy metal and identification of the promoter regions conferring heavy metal responsiveness. Plant Physiol. 2007;143(1):50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. [DOI] [PubMed] [Google Scholar]
- 103.Liu Z, Luo QH, Wang JM, Li XF, Yang Y. Functional characterization and analysis of the Arabidopsis UGT71C5 promoter region. Genet Mol Res. 2015;14(191):73–83. [DOI] [PubMed] [Google Scholar]
- 104.Xue M, Long Y, Zhao Z, Huang G, Huang K, Zhang T, Jiang Y, Yuan Q, Pei X. Isolation and characterization of a green-tissue promoter from common wild rice (Oryza rufipogon Griff.). Int J Mol Sci. 2018;19(7): Article 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hollick JB, Gordon MP. Transgenic analysis of a hybrid poplar wound-inducible promoter reveals developmental patterns of expression similar to that of storage protein genes. Plant Physiol. 1995;109(1):73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sasaki K, Yuichi O, Hiraga S, Gotoh Y, Seo S, Mitsuhara I, Ito H, Matsui H, Ohashi Y. Characterization of two rice peroxidase promoters that respond to blast fungus-infection. Mol Gen Genomics. 2007;278:709–722. [DOI] [PubMed] [Google Scholar]
- 107.Römer P, Hahn S, Jordan T, Strauß T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318(5850):645–648. [DOI] [PubMed] [Google Scholar]
- 108.Severino FE, Brandalise M, Costa CS, Wilcken SR, Maluf MP, Gonçalves W, Maia IG. CaPrx, a Coffea arabica gene encoding a putative class III peroxidase induced by root-knot nematode infection. Plant Sci. 2012;191:35–42. [DOI] [PubMed] [Google Scholar]
- 109.Sultana S, Mazarei M, Millwood RJ, Liu W, Hewezi T, Stewart N. Functional analysis of soybean cyst nematode-inducible synthetic promoters and their regulation by biotic and abiotic stimuli in transgenic soybean (Glycine max). Front Plant Sci. 2022;13: Article 988048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Subramanian S, Hu X, Lu G, Odelland JT, Yu O. The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol Biol. 2004;54(5):623–639. [DOI] [PubMed] [Google Scholar]
- 111.Swartzberg D, Dai N, Gan S, Amasino R, Granot D. Effects of cytokinin production under two SAG promoters on senescence and development of tomato plants. Plant Biol (Stuttg). 2006;8(5):579–586. [DOI] [PubMed] [Google Scholar]
- 112.Srivastava VK, Raikwar S, Tuteja N. Cloning and functional characterization of the promoter of PsSEOF1 gene from Pisum sativum under different stress conditions using agrobacterium-mediated transient assay. Plant Signal Behav. 2014;9(9): Article e29626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marcotte WR Jr, Bayley CC, Quatrano RS. Regulation of a wheat promoter by abscisic acid in rice protoplasts. Nature. 1988;335(6189):454–457. [Google Scholar]
- 114.Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genomics. 1993;236(2-3):331–340. [DOI] [PubMed] [Google Scholar]
- 115.Li Y, Liu ZB, Shi X, Hagen G, Guilfoyle TJ. An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 1994;106(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shinshi H, Usami S, Ohme-Takagi M. Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol. 1995;27(5):923–932. [DOI] [PubMed] [Google Scholar]
- 117.Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003;31(1):114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smirnova OG, Ibragimova SS, Kochetov AV. Simple database to select promoters for plant transgenesis. Transgenic Res. 2012;21(2):429–437. [DOI] [PubMed] [Google Scholar]
- 119.Hieno A, Naznin HA, Hyakumachi M, Sakurai T, Tokizawa M, Koyama H, Sato N, Nishiyama T, Hasebe M, Zimmer AD, et al. Ppdb: Plant promoter database version 3.0. Nucleic Acids Res. 2014;42(Database issue):D1188–D1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamamoto YY, Obokata J. PPDB: A plant promoter database. Nucleic Acids Res. 2007;36(suppl_1):D977–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moisseyev G, Park K, Cui A, Freitas D, Rajagopal D, Konda AR, Martin-Olenski M, Mcham M, Liu K, Du Q, Schnable JC. RGPDB: Database of root-associated genes and promoters in maize, soybean, and sorghum. Database (Oxford) 2020;2020: Article baaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zuin J, Roth G, Zhan Y, Cramard J, Redolfi J, Piskadlo E, Mach P, Kryzhanovska M, Tihanyi G, Kohler H, et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature. 2022;604(7906):571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsai ZT, Shiu SH, Tsai HK. Contribution of sequence motif, chromatin state, and DNA structure features to predictive models of transcription factor binding in yeast. PLOS Comput Biol. 2015;11(8): Article e1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nerdal W, Hare DR, Reid BR. Three-dimensional structure of the wild-type lac pribnow promoter DNA in solution: Two-dimensional nuclear magnetic resonance studies and distance geometry calculations. J Mol Biol. 1988;201(4):717–739. [DOI] [PubMed] [Google Scholar]
- 125.Polyakov A, Severinova E, Darst SA. Three-dimensional structure of E. coil core RNA polymerase: Promoter binding and elongation conformations of the enzyme. Cell. 1995;83(3):365–373. [DOI] [PubMed] [Google Scholar]
- 126.Tena JJ, Alonso ME, Calle-Mustienes E, Splinter E, De Laat W, Manzanares M, Gómez-Skarmeta JL. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2(1):310. [DOI] [PubMed] [Google Scholar]
- 127.Kerkour A, Marquevielle J, Ivashchenko S, Yatsunyk LA, Mergny JL, Salgado GF. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J Biol Chem. 2017;292(19):8082–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kumari D, Gabrielian A, Wheeler D, Usdin K. The roles of Sp1, Sp3, USF1/USF2 and NRF-1 in the regulation and three-dimensional structure of the fragile X mental retardation gene promoter. Biochem J. 2005;386(2):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hu J, Li B, Kihara D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 2005;33(15):4899–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23(1):137–144. [DOI] [PubMed] [Google Scholar]
- 134.Liu W, Mazarei M, Peng Y, Fethe MH, Rudis MR, Lin J, Millwood RJ, Arelli PR, Stewart CN Jr. Computational discovery of soybean promoter cis-regulatory elements for the construction of soybean cyst nematode-inducible synthetic promoters. Plant Biotechnol J. 2014;12(8):1015–1026. [DOI] [PubMed] [Google Scholar]
- 135.Yang Y, Lee JH, Poindexter MR, Shao Y, Liu W, Lenaghan SC, Ahkami AH, Blumwald E, Stewart CN Jr. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol J. 2021;19(7):1354–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang Y, Shao Y, Chaffin TA, Lee JH, Poindexter MR, Ahkami AH, Blumwald E, Stewart CN Jr. Performance of abiotic stress-inducible synthetic promoters in genetically engineered hybrid poplar (Populus tremula × Populus alba). Front Plant Sci. 2022;13:1011939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cai YM, Kallam K, Tidd H, Gendarini G, Salzman A, Patron NJ. Rational design of minimal synthetic promoters for plants. Nucleic Acids Res. 2020;48(21):11845–11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goldsbrough AP, Albrecht H, Stratford R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 1993;3(4):563–571. [DOI] [PubMed] [Google Scholar]
- 139.Zou C, Sun K, Mackaluso JD, Seddon AE, Jin R, Thomashow MF, Shiu S-H. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108(36):14992–14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Uygun S, Azodi CB, Shiu SH. Cis-regulatory code for predicting plant cell-type transcriptional response to high salinity. Plant Physiol. 2019;181(4):1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schwarz B, Azodi CB, Shiu SH, Bauer P. Putative cis-regulatory elements predict iron deficiency responses in Arabidopsis roots. Plant Physiol. 2020;182(3):1420–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luo Z, Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19(3):943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R. Silencing in Arabidopsis T-DNA transformants: The predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell. 2004;16(10):2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138(3):1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hunt AG, Messenger RNA 3′ end formation in plants. In: Reddy ASN, Golovkin M, editors. Nuclear pre-mRNA processing in plants. Current topics in microbiology and immunology. Berlin, Heidelberg: Springer; 2008; vol. 326; p. 151–177. [PubMed]
- 148.Li Q, Hunt AG. The polyadenylation of RNA in plants. Plant Physiol. 1997;115(2):321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xing D, Li QQ. Alternative polyadenylation and gene expression regulation in plants. RNA. 2011;2(3):445–458. [DOI] [PubMed] [Google Scholar]
- 150.Zhao T, Huan Q, Sun J, Liu C, Hou X, Yu X, Silverman IM, Zhang Y, Gregory BD, Liu CM, et al. Impact of poly (a)-tail G-content on Arabidopsis PAB binding and their role in enhancing translational efficiency. Genome Biol. 2019;20(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ. Genome level analysis of rice mRNA 3’-end processing signals and alternative polyadenylation. Nucleic Acids Res. 2008;36(9):3150–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nagaya S, Kawamura K, Shinmyo A, Kato K. The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 2010;51(2):328–332. [DOI] [PubMed] [Google Scholar]
- 153.Pérez-González A, Caro E. Effect of transcription terminator usage on the establishment of transgene transcriptional gene silencing. BMC Res Notes. 2018;11:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Felippes F, McHale M, Doran RL, Roden S, Eamens AL, Finnegan EJ, Waterhouse PM. The key role of terminators on the expression and post-transcriptional gene silencing of transgenes. Plant J. 2020;104(1):96–112. [DOI] [PubMed] [Google Scholar]
- 155.Diamos AG, Mason HS. Chimeric 3′ flanking regions strongly enhance gene expression in plants. Plant Biotechnol J. 2018;16(12):1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Diamos AG, Mason HS. High-level expression and enrichment of norovirus virus-like particles in plants using modified geminiviral vectors. Protein Expr Purif. 2018;151:86–92. [DOI] [PubMed] [Google Scholar]
- 157.Diamos AG, Rosenthal SH, Mason HS. 5′ and 3′ untranslated regions strongly enhance performance of geminiviral replicons in Nicotiana benthamiana leaves. Front Plant Sci. 2016;7:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rosenthal SH, Diamos AG, Mason HS. An intronless form of the tobacco extensin gene terminator strongly enhances transient gene expression in plant leaves. Plant Mol Biol. 2018;96:429–443. [DOI] [PubMed] [Google Scholar]
- 159.Hirai T, Kurokawa N, Duhita N, Hiwasa-Tanase K, Kato K, Kato K, Ezura H. The HSP terminator of Arabidopsis thaliana induces a high level of miraculin accumulation in transgenic tomatoes. J Agric Food Chem. 2011;59(18):9942–9949. [DOI] [PubMed] [Google Scholar]
- 160.Kurokawa N, Hirai T, Takayama M, Hiwasa-Tanase K, Ezura H. An E8 promoter–HSP terminator cassette promotes the high-level accumulation of recombinant protein predominantly in transgenic tomato fruits: A case study of miraculin. Plant Cell Rep. 2013;32:529–536. [DOI] [PubMed] [Google Scholar]
- 161.Matsui T, Sawada K, Takita E, Kato K. The longer version of Arabidopsis thaliana heat shock protein 18.2 gene terminator contributes to higher expression of stably integrated transgenes in cultured tobacco cells. Plant Biotechnol. 2014;31(2):191–194. [Google Scholar]
- 162.Richter LJ, Thanavala Y, Arntzen CJ, Mason HS. Production of hepatitis B surface antigen in transgenic plants for oral immunization. Nat Biotechnol. 2000;18(11):1167–1171. [DOI] [PubMed] [Google Scholar]
- 163.Schaart JG, Tinnenbroek-Capel IE, Krens FA. Isolation and characterization of strong gene regulatory sequences from apple, malus × domestica. Tree Genet Genomes. 2011;7:135–142. [Google Scholar]
- 164.Weeks JT, Ye J, Rommens CM. Development of an in planta method for transformation of alfalfa (Medicago sativa). Transgenic Res. 2008;17:587–597. [DOI] [PubMed] [Google Scholar]
- 165.Hiwasa-Tanase K, Nyarubona M, Hirai T, Kato K, Ichikawa T, Ezura H. High-level accumulation of recombinant miraculin protein in transgenic tomatoes expressing a synthetic miraculin gene with optimized codon usage terminated by the native miraculin terminator. Plant Cell Rep. 2011;30:113–124. [DOI] [PubMed] [Google Scholar]
- 166.Ingelbrecht IL, Herman LM, Dekeyser RA, Van Montagu MC, Depicker AG. Different 3′ end regions strongly influence the level of gene expression in plant cells. Plant Cell. 1989;1(7):671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang L, Wakasa Y, Kawakatsu T, Takaiwa F. The 3′-untranslated region of rice glutelin GluB-1 affects accumulation of heterologous protein in transgenic rice. Biotechnol Lett. 2009;31:1625–1631. [DOI] [PubMed] [Google Scholar]
- 168.Xing A, Moon BP, Mills KM, Falco SC, Li Z. Revealing frequent alternative polyadenylation and widespread low-level transcription read-through of novel plant transcription terminators. Plant Biotechnol J. 2010;8(7):772–782. [DOI] [PubMed] [Google Scholar]
- 169.Abe H, Abo T, Aiba H. Regulation of intrinsic terminator by translation in Escherichia coli: Transcription termination at a distance downstream. Genes Cells. 1999;4(2):87–97. [DOI] [PubMed] [Google Scholar]
- 170.Yamamoto T, Hoshikawa K, Ezura K, Okazawa R, Fujita S, Takaoka M, Mason HS, Ezura H, Miura K. Improvement of the transient expression system for production of recombinant proteins in plants. Sci Rep. 2018;8(1):4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dolgova AS, Dolgov SV. Matrix attachment regions as a tool to influence plant transgene expression. 3 Biotech .2019;9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tian C, Zhang Y, Li J, Wang Y. Benchmarking intrinsic promoters and terminators for plant synthetic biology research. BioDes Res. 2022;2022:9834989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, et al. Morphogenic regulators baby boom and Wuschel improve monocot transformation. Plant Cell. 2016;28(9):1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W. Rapid genotype “independent” Zea mays L.(maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol Plant. 2018;54:240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38(1):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khakhar A, Starker CG, Chamness JC, Lee N, Stokke S, Wang C, Swanson R, Rizvi F, Imaizumi T, Voytas DF. Building customizable auto-luminescent luciferase-based reporters in plants. eLife. 2020;9: e52786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mitiouchkina T, Mishin AS, Somermeyer LG, Markina NM, Chepurnyh TV, Guglya EB, Karataeva TA, Palkina KA, Shakhova ES, Fakhranurova LI, et al. Plants with genetically encoded autoluminescence. Nat Biotechnol. 2020;38(8):944–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dietz-Pfeilstetter A. Stability of transgene expression as a challenge for genetic engineering. Plant Sci. 2010;179(3):164–167. [Google Scholar]
- 179.Peremarti A, Twyman RM, Gómez-Galera S, Naqvi S, Farré G, Sabalza M, Miralpeix B, Dashevskaya S, Yuan D, Ramessar K, et al. Promoter diversity in multigene transformation. Plant Mol Biol. 2010;73:363–378. [DOI] [PubMed] [Google Scholar]
- 180.Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, et al. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant. 2017;10(7):918–929. [DOI] [PubMed] [Google Scholar]
- 181.Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods. 2015;12(3):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chinnusamy V, Stevenson B, Lee BH, Zhu JK. Screening for gene regulation mutants by bioluminescence imaging. Sci STKE. 2002;2002(140):10. [DOI] [PubMed] [Google Scholar]
- 183.Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161(3):1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Abd El-Halim HM, Ismail IM, Al Aboud NM, Elghareeb D, Metry EA, Hossien AF, Fahmy EM. Evaluation of two promoters for generating transgenic potato plants as salicylic acid biosensors. Biol Plant. 2020;64:535–540. [Google Scholar]
- 185.Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJ, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29(6):1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lowder LG, Zhang D, Baltes NJ, Paul JW III, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169(2):971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu Y, Merrick P, Zhang Z, Ji C, Yang B, Fei SZ. Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnol J. 2018;16(2):381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kor SD, Chowdhury N, Keot AK, Yogendra K, Chikkaputtaiah C, Reddy PS. RNA pol III promoters—Key players in precisely targeted plant genome editing. Front Genet. 2022;13: 989199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hassan MM, Zhang Y, Yuan G, De K, Chen JG, Muchero W, Tuskan GA, Qi Y, Yang X. Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 2021;26(11):1133–1152. [DOI] [PubMed] [Google Scholar]
- 190.Yuan G, Martin S, Hassan MM, Tuskan GA, Yang X. PARA. A new platform for the rapid assembly of gRNA arrays for multiplexed CRISPR technologies. Cells. 2022;11(16):2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wolter F, Klemm J, Puchta H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018;94(4):735–746. [DOI] [PubMed] [Google Scholar]
- 192.Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, Van Isterdael G, Beeckman T, Nowack MK, Jacobs TB. CRISPR-TSKO: A technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell. 2019;31(12):2868–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Singha DL, Das D, Sarki YN, Chowdhury N, Sharma M, Maharana J, Chikkaputtaiah C. Harnessing tissue-specific genome editing in plants through CRISPR/Cas system: Current state and future prospects. Planta. 2022;255(1):28. [DOI] [PubMed] [Google Scholar]
- 194.Mithra SA, Kulkarni K, Srinivasan R. Plant promoters: Characterization and applications in transgenic technology. In: Abdin M, Kiran U, Kamaluddin Ali A, editors. Plant biotechnology: Principles and applications. Singapore: Springer; 2017; p. 117–172. [Google Scholar]
- 195.Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, et al. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell. 2011;23(12):4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ahanger MA, Akram NA, Ashraf M, Alyemeni MN, Wijaya L, Ahmad P. Plant responses to environmental stresses—From gene to biotechnology. AoB Plants. 2017;9(4):plx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fan M, Miao F, Jia H, Li G, Powers C, Nagarajan R, Alderman PD, Carver BF, Ma Z, Yan L. O-linked N-acetylglucosamine transferase is involved in fine regulation of flowering time in winter wheat. Nat Commun. 2021;12(1):2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.González-Grandío E, Demirer GS, Ma W, Brady SM, Landry MP. A ratiometric dual-color luciferase reporter for fast characterization of transcriptional regulatory elements in plants. bioRxiv .2021:2021.05.26.443018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Feike D, Korolev AV, Soumpourou E, Murakami E, Reid D, Breakspear A, Rogers C, Radutoiu S, Stougaard J, Harwood WA, et al. Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots. Plant Biotechnol J. 2019;17(12):2234–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hetzel J, Duttke SH, Benner C, Chory J. Nascent RNA sequencing reveals distinct features in plant transcription. Proc Natl Acad Sci USA. 2016;113(43):12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Murray A, Mendieta JP, Vollmers C, Schmitz RJ. Simple and accurate transcriptional start site identification using Smar2C2 and examination of conserved promoter features. Plant J. 2022;112(2):583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lu Z, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 2017;45(6): e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maher KA, Bajic M, Kajala K, Reynoso M, Pauluzzi G, West DA, Zumstein K, Woodhouse M, Bubb K, Dorrity MW, et al. Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell. 2018;30(1):15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, et al. Widespread long-range cis-regulatory elements in the maize genome. Nat Plants. 2019;5(12):1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Costello A, Badran AH. Synthetic biological circuits within an orthogonal central dogma. Trends Biotechnol. 2021;39(1):59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


