Abstract
Autoproteolysis has been discovered to play key roles in various biological processes, but functional autoproteolysis has been rarely reported for transmembrane signaling in prokaryotes. In this study, an autoproteolytic effect was discovered in the conserved periplasmic domain of anti-σ factor RsgIs from Clostridium thermocellum, which was found to transmit extracellular polysaccharide-sensing signals into cells for regulation of the cellulosome system, a polysaccharide-degrading multienzyme complex. Crystal and NMR structures of periplasmic domains from three RsgIs demonstrated that they are different from all known proteins that undergo autoproteolysis. The RsgI-based autocleavage site was located at a conserved Asn-Pro motif between the β1 and β2 strands in the periplasmic domain. This cleavage was demonstrated to be essential for subsequent regulated intramembrane proteolysis to activate the cognate SigI, in a manner similar to that of autoproteolysis-dependent activation of eukaryotic adhesion G protein–coupled receptors. These results indicate the presence of a unique prevalent type of autoproteolytic phenomenon in bacteria for signal transduction.
A unique type of autoproteolytic phenomenon was discovered in Gram-positive bacteria for transmembrane signal transduction.
INTRODUCTION
Transmembrane signaling is essential for both prokaryotic and eukaryotic cells to survive and communicate with the complex extracellular environment. Autoproteolysis has been reported to play key roles in various biological processes, such as serine protease maturation, eukaryotic signal transduction, and virus penetration (1–8). In prokaryotes, except for various autoproteolytic proteases for maturation (1, 2), functional autoproteolysis has been rarely reported for transmembrane signaling. The best-studied transmembrane signaling mechanisms in bacteria involve the allosteric conformational changes of the transmembrane helical bundle by ligand binding in chemoreceptors and the cascade proteolysis in anti-σ factors (9, 10). In the present study, we discovered an autoproteolytic effect in the distinctive anti-σ factor RsgI family that exists widely in Bacilli and Clostridia (11–13). We demonstrated that the autoproteolytic process is essential for transmembrane signaling, to some degree similar to the well-known autoproteolysis in eukaryotic adhesion G protein–coupled receptors (aGPCRs) (14).
Many bacterial extracytoplasmic function (ECF) σ factors, together with their cognate anti-σ factors cotranscribed in the same gene operon, are responsible for the cellular responses to environmental signals (10). The anti-σ factors sense the extracellular signals, transduce the signals across the membrane by various mechanisms, and lastly release the combined intracellular σ factors, which recruit RNA polymerases to initiate the transcription of specific genes (15, 16). The best-studied transmembrane signaling mechanism by ECF anti-σ factors is the regulated intramembrane proteolysis (RIP) (16). RIP was first described for controlling cholesterol metabolism in mammals (17) and was found in many signaling processes of species from bacteria to mammals (18, 19). The RIP process of bacterial ECF anti-σ factors involves sequential cleavages of the anti-σ factor by cascade proteases (site-1 protease, site-2 protease, etc.) to finally release the bound σ factor (10, 15).
SigI/RsgI is a distinct family of ECF-like σ/anti-σ factors that functions in the heat stress response, virulence, and polysaccharide sensing in Firmicutes (12). The N-terminal part of RsgI is highly conserved and includes a cytoplasmic N-terminal domain (NTD), a transmembrane helix, and a periplasmic domain (PD; Fig. 1A) (13, 20). The NTD is responsible for binding to SigI and inhibiting its activity (11, 13). The PD may potentially participate in the signaling (20), but its structure and function have not yet been elucidated. A previous genetic study showed that multiple proteases, such as RasP and ClpEP in RIP, might be involved in the RsgI-SigI signaling of Bacillus subtilis (21), but the site-1 protease was not identified, and it is still unknown how the RsgI and RIP protease interact.
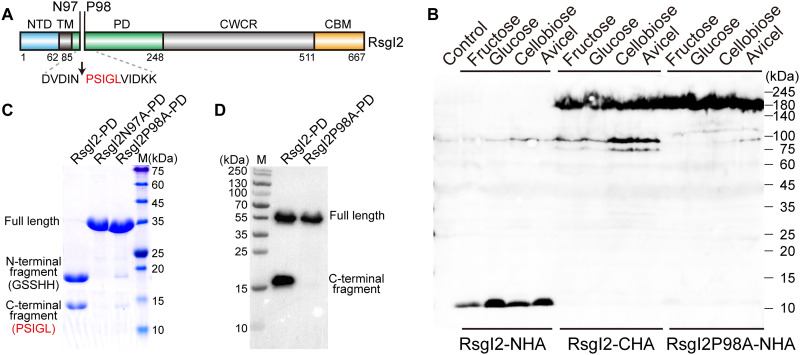
Fig. 1. Identification of the autocleavage phenomenon and the cleavage site of C. thermocellum RsgI2.
(A) Schematic representation of the composition and the location of the autocleavage site of RsgI2. RsgI2 contains an NTD, a transmembrane helix (TM), a PD, a probable cell wall–crossing region (CWCR) and a C-terminal substrate-binding CBM3b domain (CBM). The amino acid sequence around the autocleavage site is shown, and the first five amino acid residues identified by N-terminal protein sequencing are colored in red. (B) Western-blot analysis of RsgI2 expressed in C. thermocellum. The mutant ΔrsgI2 (control), the ΔrsgI2 bearing the plasmids to express RsgI2 with either an N- or C-terminal HA-tag (RsgI2-NHA and RsgI2-CHA), and the P98A mutant RsgI2P98A-NHA were cultured on the different designated carbon sources, and the resultant labeled RsgI2 components were detected by an anti–HA-tag antibody. (C) SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the purified SMT3-RsgI2-PD protein and its N97A and P98A mutants. The results of N-terminal protein sequencing for the two bands of SMT3-RsgI2-PD are denoted parenthetically on the left. (D) RsgI2-PD fused with an N-terminal glutathione S-transferase (GST) and a C-terminal FLAG tag (RsgI2-PD) and its P98A mutant (RsgI2P98A-PD) were expressed using the PURE cell-free transcription/translation system and analyzed by Western blotting using an anti-FLAG antibody.
The C-terminal parts of the RsgIs are not conserved. Multiple paralogous pairs of SigI-RsgIs have been found to regulate the expression of cellulosomes, the multiple enzyme complexes produced by many anaerobic lignocellulolytic bacteria (11, 22–26). The C-terminal part of these RsgIs has a divergent carbohydrate-binding module (CBM), which was proposed to sense different types of polysaccharides (27–29). In Clostridium thermocellum, RsgI1, RsgI2, and RsgI4 contain family 3b CBMs that target cellulose; RsgI3 contains a tandem protective antigen 14 (PA14) motif that recognizes pectin; RsgI5 contains a family 42 CBM that binds to arabinose; RsgI6 contains a family 10 glycosidase hydrolase that targets both xylan and cellulose; RsgI7 and RsgI8 do not contain an obvious CBM, and their functions are currently unknown; and lastly, RsgI9 contains a protease-like domain that is capable of binding cellulose (28–30). After signal transduction across the cell membrane, the RsgIs release their bound cognate SigIs for initiating transcription of a subset of cellulosomal genes, resulting in the dynamic regulation of cellulosome function, which is one of the major reasons for the high efficiency of anaerobic cellulosome-producing bacteria in lignocellulose degradation (11, 24–26). The specific recognition of cellulosomal gene promoters by paralogous SigIs has been extensively studied (25, 26), but how the sensing signals of the CBM are transduced into the cell is largely unknown.
In this work, we found that RsgIs of C. thermocellum undergo an autoproteolysis event to initiate signal transduction. The cleavage site was identified to consist of a pair of conserved Asn and Pro residues located between the first two β strands of the PD in all of the C. thermocellum RsgIs except RsgI8. We further showed that after removing the C-terminal part of the RsgI from the cleavage site, the remaining N-terminal peptide in the PD can activate the SigI following proteolytic cleavage by RseP (a protease in membrane and homolog of RasP), thus defining a distinctive signaling mechanism in bacteria. The residues around the autocleavage site are conserved in the great majority of the RsgIs from both Bacilli and Clostridia, indicating a prevalent autoproteolytic process in bacteria for signal transduction.
RESULTS
RsgIs are cleaved in a manner independent of the cultivation substrate types
To study the signal transduction mechanism of RsgI, we first constructed an RsgI2 inactivation mutant (ΔrsgI2) using the thermotargetron technique (31). Two plasmids for expressing RsgI2 with either an N- or a C-terminal hemagglutinin (HA)–tag (32) (termed RsgI2-NHA and RsgI2-CHA, respectively) were then transformed separately into ΔrsgI2. The expression of HA-tagged RsgI2 in media containing different carbon sources was determined by Western blot analysis using an anti-HA antibody. Unexpectedly, instead of the expected size of 73.5 kDa, major bands of about 180 and 10 kDa were observed for the RsgI2-CHA and RsgI2-NHA expression, respectively (Fig. 1B). The bands were consistently observed whenever the RsgI2-expressing strains were cultured in various substrates, including fructose, glucose, cellobiose, or Avicel (microcrystalline cellulose) as the sole carbon source (Fig. 1B). The unexpectedly large molecular size of RsgI2-CHA in the Western blot was likely caused by the cell wall–crossing region (CWCR) between the PD and the C-terminal CBM3b domain because the CWCR-truncated RsgI2 showed the band with the expected molecular weight (fig. S1, A to C). Further mass spectrometry analysis of the high–molecular weight bands confirmed that one of the bands is RsgI2, but only the peptide fragments from PD and CBM can be detected (fig. S1, D and E), suggesting that the CWCR might be covalently modified by the cell wall. The small size (about 10 kDa) of RsgI2-NHA suggested that the expressed RsgI2-NHA underwent a cleavage at the N-terminal region, independent of the type of cultivation substrate.
Cleavage of RsgI2 was observed at different periods of cell growth (fig. S2A). Considering that the boiling of the samples for SDS–polyacrylamide gel electrophoresis (SDS-PAGE) may cause the cleavage of some peptide bonds (33), we treated the samples with lysozyme and heating at lower temperatures or without heating and found that the cleavage state of RsgI2 was the same under these conditions (fig. S2B). To investigate whether the observed cleavage of RsgI2 is conserved in other RsgIs, we further expressed different RsgIs with an HA-tag in C. thermocellum. Similar to RsgI2, all of the other tested RsgIs, including RsgI1, RsgI3, RsgI4, and RsgI6, showed short N-terminal cleaved fragments (~11 to 17 kDa), independent of the carbon source used in cultivation (fig. S2, C and D). These results indicated that RsgIs have conserved peptide bond cleavage in the N-terminal region, which is independent of the growth stage and carbon source.
The PD of RsgI bears an autocleavage site between a conserved Asn-Pro motif
Considering the size of the observed RsgI N-terminal fragment and the domain organization of RsgI, the cleavage site was considered to reside in the N-terminal part of the RsgI-PD (20). For analysis of the cleavage site, we expressed the N-terminal 6×His-SMT3 tag-fused (34) RsgI2-PD in Escherichia coli. The purified recombinant SMT3-RsgI2-PD showed two bands (~18 and ~14 kDa) in the SDS-PAGE (Fig. 1C), instead of the full-length protein of 32.8 kDa. N-terminal protein sequencing by Edman degradation showed that the N-terminal sequence of the ~18-kDa band is GlySerSerHisHis, which corresponds to the expected N-terminal residues of the 6×His-SMT3 tag, while the N-terminal sequence of the ~14-kDa band is ProSerIleGlyLeu, which comprises the residues after Asn97 of RsgI2-PD (Fig. 1C). Therefore, the cleavage site should be between Asn97 and Pro98 of RsgI2-PD, which agrees with the observed molecular sizes of the two fragments after the cleavage. The mutation of Asn97 or Pro98 to alanine essentially eliminated the cleavage as shown by SDS-PAGE (Fig. 1C), indicating that the two residues are essential for the cleavage. Furthermore, the mutant RsgI2P98A-NHA expressed in C. thermocellum also showed only a single band at about 180 kDa without cleavage in Western blots (Fig. 1B), indicating the same cleavage site in vivo at Asn97-Pro98 of RsgI2 (Fig. 1A).
The same type of cleavage of both RsgI2 in C. thermocellum and recombinant RsgI2-PD in E. coli implies that the protein should undergo autocleavage. To exclude the influence of other proteases in bacteria, we synthesized the RsgI2-PD fused with an N-terminal glutathione S-transferase (GST) and a C-terminal FLAG tag using the PURE cell-free express system (35), which is reconstituted from purified components and is therefore nuclease- and protease-free. As shown in Fig. 1D, the synthesized recombinant RsgI2-PD proteins contained both the full-length protein and the C-terminal fragment detected by Western blotting using an anti-FLAG antibody. The observed full-length protein without cleavage is probably caused by improper folding of RsgI2-PD in this cell-free system that lacks chaperones (36). Furthermore, mutation of Pro98 to Ala in RsgI2-PD did not affect the production of the protein, but no C-terminal fragment was observed, indicating that the mutation completely blocked the cleavage. Therefore, the cleavage of RsgI consists of an autoproteolytic process.
Sequence alignments showed that the two amino acids Asn97 and Pro98 are conserved in most RsgIs from both C. thermocellum (Fig. 2A) and other RsgI-containing bacteria (fig. S3). Western blot analysis of RsgI6P95A-NHA expressed in C. thermocellum showed no band of the cleaved N-terminal fragment (fig. S4), the same as the phenomenon observed for the RsgI2P98A mutant. These results thus indicated that the cleavage between the Asn-Pro motif in the PD is a conserved feature of the RsgIs.
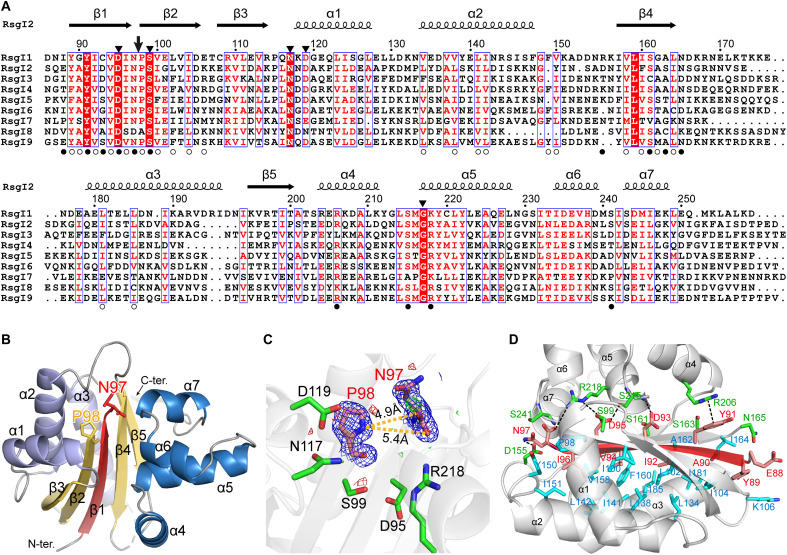
Fig. 2. Structure of RsgI2-PD.
(A) Sequence alignments of the RsgI-PDs from C. thermocellum. The autocleavage site and the surrounding conserved residues are indicated by an arrow and filled triangles, respectively, at the top. The residues involved in the polar and hydrophobic interactions between the disconnected β1 and the other parts of the protein are indicated by filled and open circles, respectively, at the bottom. (B) Ribbon representation of the crystal structure of RsgI2-PD. The disconnected β1 caused by the autocleavage is colored in red, and residues Asn97 and Pro98 are shown as sticks. (C) The conserved residues around the Asn97 and Pro98. The 2mFo-DFc densities for Asn97 and Pro98 are contoured in blue at 1.0σ. The mFo-DFc maps are shown in green and red for positive and negative densities, respectively, at 3.0σ. (D) The interactions of β1 with other parts of the protein. The residues of α helix subdomains involved in the polar and hydrophobic interactions are shown as green and cyan sticks, respectively.
Structure of RsgI-PDs
To provide the structural basis of the autocleavage in RsgI-PD, we solved the crystal structure of RsgI2-PD at 1.85-Å resolution by x-ray crystallography and the solution structures of RsgI2-PD, RsgI1-PD, and RsgI6-PD by nuclear magnetic resonance (NMR; Fig. 2B, fig. S5, and tables S4 and S5). In the crystal structure of RsgI2-PD, the unambiguous electron densities showed that the bond between Asn97 and Pro98 was cleaved (Fig. 2C), with around 5.0-Å distance between the CO of Asn97 and N of Pro98. This agrees with the cleavage site identified in the N-terminal sequencing results (Fig. 1C).
All of the RsgI-PDs present an α/β/α sandwich structure with seven α helices in two subdomains and five β strands forming a central β sheet (Fig. 2B). The first α helix subdomain contains three α helices (α1, α2, and α3), and the second helical subdomain contains four α helices (α4, α5, α6, and α7). The central β sheet is formed in the order β3-β2-β1-β4-β5, and all strands are antiparallel, except for β4 and β5, which are parallel (Fig. 2B). The cleavage between Asn97 and Pro98 in RsgI2-PD breaks the connection between β1 and β2, leaving the N-terminal β1-forming chain with only twelve residues. The β1 strand is in the center of the β sheet and surrounded by the helical subdomains, forming numerous interactions to maintain the stability of PD despite the cleavage between β1 and β2 (Fig. 2D). Besides the hydrogen bonds between β1 and β2/β4, β1 forms hydrophobic interactions with the first helical subdomain and many hydrogen bonds with the second helical subdomain. Furthermore, a π-π stacking interaction between the Tyr91 and Arg206 side chains, as well as electrostatic interactions between Arg218 and Asp95 and the carboxyl of Asn97, were also observed in the structure of RsgI2-PD. These interactions are largely conserved in the three RsgI-PD structures, and the residues involved in the interactions are largely conserved in all of the RsgIs (Fig. 2A), suggesting the conserved structural features of the cleaved N-terminal β1 strand in the RsgIs.
The overall structures of the RsgI-PDs are similar to each other (fig. S5, G and H), with the backbone root mean square deviations of 1.0 to 1.5 Å between them. The major structural differences between RsgI-PDs are the length and orientation of the helices. The search for similar structures in the Protein Data Bank (PDB) using the Dali and secondary-structure matching (SSM) servers gave some α/β/α sandwich structures or α/β half-sandwich structures (fig. S6). However, none of them has the same topology for the β sheet, nor do they contain the cleaved peptide bonds in the protein. Furthermore, these structurally similar proteins have no relationship with anti-σ factors or transmembrane signaling transduction, so they cannot provide valuable functional implications for RsgI-PD. Therefore, the structure-function relationship of RsgI-PD is unique in known protein structures.
According to the sequence alignment, the most conserved residues in RsgI-PDs are located near the cleavage site in the structures (Fig. 2, A and C), implying that the cleavage may have important functions. These conserved residues include several polar residues such as Asp95, Ser99, Asn117, Asp119, and Arg218 in RsgI2-PD, which might play catalytic roles in the autocleavage. However, mutation of each of these surrounding residues did not change the autocleavage of RsgI2-PD. Double mutation was shown to partly reduce the amount of autocleavage (fig. S7), suggesting that multiple residues might be involved in the activation of the cleavage.
RsgI-PD represents a distinct autoproteolysis domain
Although autoproteolysis exists in serine proteases (1, 2), such as the GPCR autoproteolysis-inducing (GAIN) domain of aGPCR (3), the anti-σ factor FoxR (4, 5), and the outer capsid protein VP5/μ1 of reoviruses (6–8) besides the anti-σ factor RsgI, these proteins have divergent structures, autocleavage sites, and catalytic mechanisms. The maturation of serine proteases, such as bacterial subtilisins, is dependent on the autoprocessing of the propeptide domain by the catalytic domain. The N-terminal propeptide domain folds into a compact structure with a long linker connecting the C-terminal catalytic domain (Fig. 3A) (1), and during maturation, the linker is cleaved by the catalytic triad of the enzyme (2).
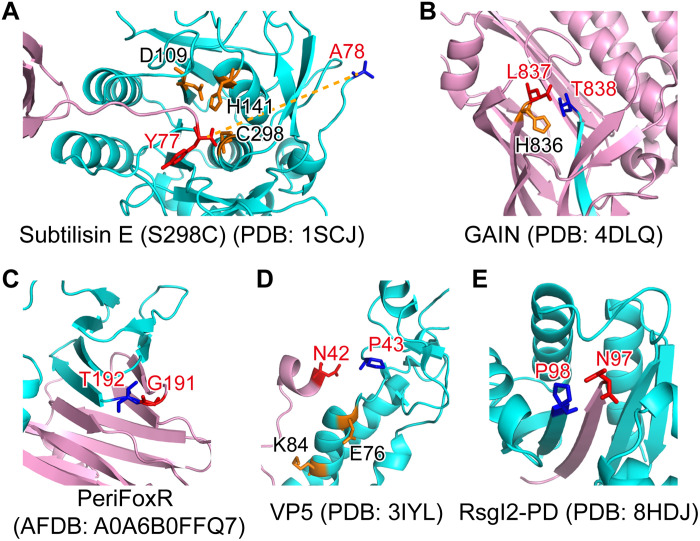
Fig. 3. The structure of RsgI-PD is distinct from other known autocleaved proteins.
The N- and C-terminal parts are colored pink and cyan, respectively. The P − 1 and P + 1 residues of the cleavage site are shown in red and blue sticks, respectively, and other residues probably involved in the catalysis are shown in orange sticks. (A) The structure of the propeptide-subtilisin E (S298C mutant) complex from B. subtilis. (B) The crystal structure of the GAIN domain of the GPCR CL1. (C) The AlphaFold2 structural model of the PD of FoxR (PeriFoxR) from P. aeruginosa. (D) The structure of the VP5 protein of Aquareovirus. (E) The structure of RsgI2-PD from C. thermocellum.
Autoproteolysis in aGPCR GAIN domains is located in the β sandwich subdomain with a conserved HisLeu(Thr/Ser) motif before its C-terminal β strand β13 (Fig. 3B). The conserved histidine or arginine in the motif acts as a general base and extracts a hydrogen from the side chain of the polar threonine (or serine) at the P1′ position, initiating a nucleophilic attack of the carbonyl group of the P1 residue (commonly leucine) (3). After the autocleavage event, the C-terminal β13 strand is left with the transmembrane helices for downstream signaling.
The anti-σ factor FoxR of Pseudomonas aeruginosa undergoes autocleavage between the periplasmic Gly191 and Thr192 residues, located in a turn between two β strands (Fig. 3C), with a proposed N-O acyl rearrangement mechanism, but the autocleavage process is not essential for the signaling function of FoxR (4, 5). This type of autoprocessing mechanism also occurs in the cell wall protein CwpV (37) of Clostridium difficile and the CBM-54 domain (38) of the unique chitinase ChiW in Paenibacillus sp. str. FPU-7. These proteins share similar three-dimensional β helix structures in their cleavage domains, and the intramolecular acyl rearrangement depends on the hydroxyl or sulfhydryl group of the +1 residue of the cleavage site (Thr, Ser, or Cys), which plays the role of a nucleophile, similar to the self-cleavage of inteins (39). The +1 residue of the cleavage site in RsgI, however, is a proline, which does not fulfill the requirement of this N-O acyl rearrangement mechanism.
Both the outer capsid proteins VP5 in Aquareovirus and μ1 in mammalian orthoreovirus exhibit autocleavage between Asn42 and Pro43 located at the C terminus of a short helix (Fig. 3D) (6), and the cleavage event is critical for membrane penetration and cell entry of the virus (7). Asn42 plays a key role in the cleavage process: The nearby carbonyl group of Glu or Asp forms a hydrogen bond with the amide proton on the side chain of Asn42 to polarize the nitrogen of the Asn42 side chain, and the side chain performs a nucleophilic attack on its peptide bond, resulting in the autocleavage (6, 8).
Autocleavage of RsgI also occurs between conserved Asn and Pro residues (Fig. 3E), so it may also use a similar type of chemical mechanism (fig. S8). However, the Asn-Pro motif of RsgI is located in a turn between two β strands, which exhibit completely different structures compared with those of the outer capsid proteins VP5 and μ1 (Fig. 3, D and E).
Autocleavage is involved and essential in the signal transduction of RsgI6
The conserved autocleavage in RsgIs implies its functional significance. We therefore investigated the role of the cleavage in RsgI function in C. thermocellum. Unfortunately, mutant ΔrsgI2 did not exhibit an obvious change in growth phenotype, cellulosome production, or cellulase activity, probably because SigI2/RsgI2 also regulates minor components of cellulosomes according to the previous analyses (26). We therefore decided to use a parallel RsgI to analyze the functional role of autocleavage in the signal transduction mechanism. Considering that SigI6/RsgI6 has been reported to regulate many cellulosomal genes (26) and the cellulosomal xylanases have been confirmed to be up-regulated by the deletion of RsgI6 (40), we thus constructed a similar RsgI6 deletion mutant ΔrsgI6 using the homology recombination method. RsgI6 has a C-terminal GH10 domain, which can bind to xylan and Avicel (27), and the PD of RsgI6 bears the conserved Asn94 and Pro95, which corresponds to Asn97 and Pro98 of RsgI2 (Fig. 4A). Similar to RsgI2-PD, the recombinant RsgI6-PD also undergoes autocleavage, and the P95A mutation can block its autocleavage (Fig. 4B). We found that the culture supernatant of ΔrsgI6 showed two clear additional bands on SDS-PAGE gels (Fig. 4C and table S6), which were identified to be xylanases Xyn10Z and Xyn11B by mass spectrometry (fig. S9). The overexpressed xylanases provided a convenient phenotype for studying the signal transduction mechanism of the RsgIs.
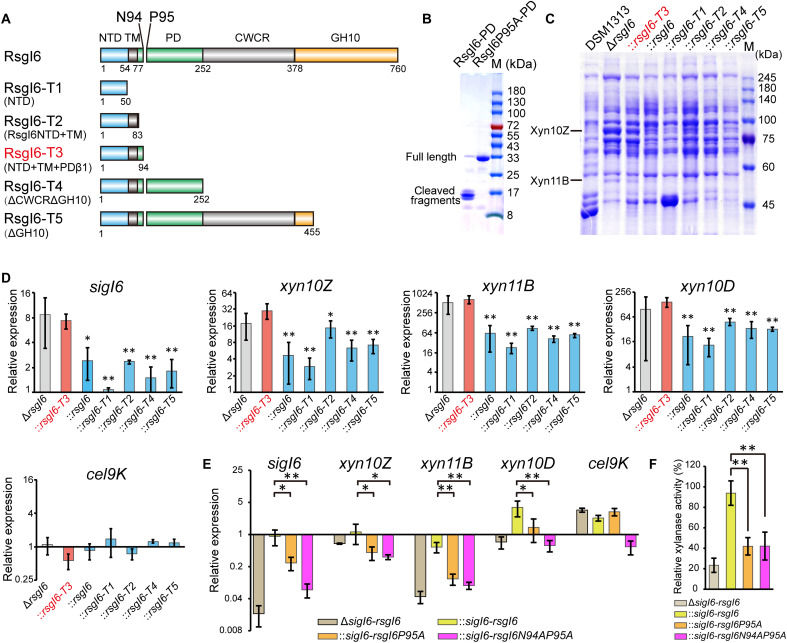
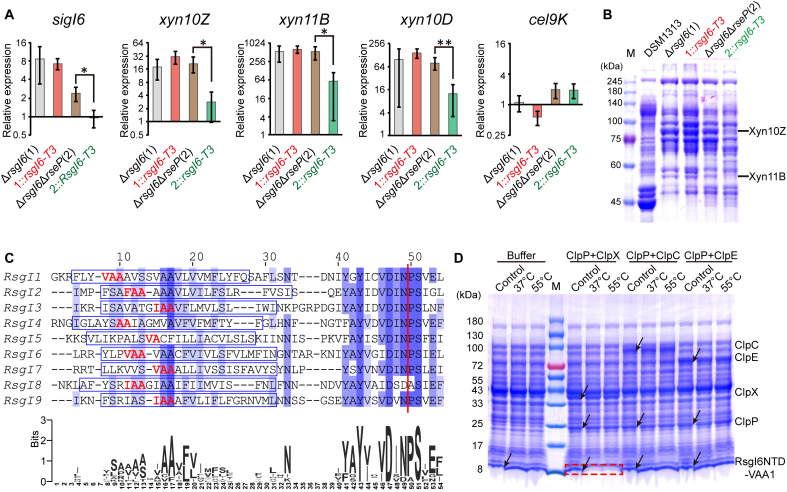
Fig. 4. Analysis of SigI6 and xylanase expression in mutant strains to demonstrate the role of autoproteolysis in signaling.
(A) Schematic representation of the composition of RsgI6 and its truncation mutants. RsgI6 contains an intracellular NTD, a TM, a PD, a proposed CWCR, and a C-terminal substrate-binding GH10 domain (GH10). (B) SDS-PAGE analysis of the purified SMT3-RsgI6-PD protein and its P95A mutant. (C) SDS-PAGE analysis of the extracellular proteins of wild type (DSM1313), ΔrsgI6, and its complemented strains with various RsgI6 truncations of C. thermocellum DSM1313, cultured in media with cellobiose as the carbon source. (D) qRT-PCR analysis of the genes of SigI6 and cellulosomal xylanases in ΔrsgI6 and its complemented strains grown in the cellobiose medium. The relative expression represents the ratio of transcribed mRNA levels in the mutant strains compared to that of DSM1313. The P values were calculated on the basis of three replicates using Student’s t test with ΔrsgI6::rsgI6-T3 as the reference. (E) qRT-PCR analysis of ΔsigI6-rsgI6 and its complemented strains grown in medium with alkali-pretreated wheat straw as carbon source. The relative expression represents the ratio of transcribed mRNA levels in the mutant strains compared to that of DSM1313. (F) Xylanase activity of the culture supernatant. The P values in (D) and (E) were calculated on the basis of three replicates using Student’s t test. *P < 0.05 and **P < 0.01.
We then constructed several plasmids to express different forms of truncated RsgI6, controlled under the promoter of SigI6 in the mutant ΔrsgI6 (Fig. 4A). Because the NTD of RsgI is responsible for binding to and inhibition of SigI (13), the designed RsgI6 truncations contained different N-terminal parts, including truncation RsgI6-T3, which contains the precise autocleaved N-terminal part of RsgI6. The expression levels of Xyn10Z and Xyn11B in the cultural supernatant of these mutants were then analyzed by SDS-PAGE (Fig. 4C and table S6). The results showed that overexpression of Xyn10Z and Xyn11B in ΔrsgI6 was largely impaired by plasmid expression of full-length and truncated RsgI, except that the expression of RsgI6-T3 had little effect on the overexpression of the xylanases. Consequently, the SigI in ΔrsgI6::rsgI6-T3 was still in an activated state, thus indicating that cleavage at Asn94-Pro95 provided a signal to release the SigI if the C-terminal part of RsgI-PD was removed. To further confirm that the phenotype was related to SigI6-controlled gene transcription, the transcription levels of sigI6, xyn10Z, xyn11B, xyn10D, and cel9K in these strains were analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). As expected, the up-regulated transcription of sigI6, xyn10Z, xyn11B, and xyn10D in ΔrsgI6 can be suppressed by plasmid-based expression of RsgI and its truncations, except for RsgI3-T3 (Fig. 4D). As a control, Cel9K, which does not have a SigI6-dependent promoter, did not show suppression of RsgI6-dependent transcription.
These results demonstrated that removal of the C-terminal part of RsgI from the cleavage site can lead to signal transduction for activation of SigI, but it is not clear whether autocleavage is essential for the RsgI6-mediated signal transduction. To solve this problem, we first knocked out the operon of sigI6-rsgI6 in C. thermocellum by homologous recombination. When the ΔsigI6-rsgI6 mutant grew in the medium with alkali-pretreated wheat straw as the carbon source, the transcription levels of sigI6, xyn10Z, xyn11B, and xyn10D (Fig. 4E) and the xylanase activity of the culture supernatant (Fig. 4F) were decreased, which proved that SigI6/RsgI6 was the main regulator responsible for sensing the substrates and expressing the cellulosomal xylanases. Subsequently, we constructed three complemented strains of ΔsigI6-rsgI6 using the respective plasmids that express SigI6-RsgI6, SigI6-RsgI6P95A, and SigI6-RsgI6N94AP95A with the promoter of SigI6. As shown in Fig. 4E, the transcription levels of sigI6 and its regulated cellulosomal xylanase genes were rescued in the complemented strain ΔsigI6-rsgI6::sigI6-rsgI6. However, the phenotype was not rescued in the complemented strains ΔsigI6-rsgI6::sigI6-rsgI6P95A and ΔsigI6-rsgI6::sigI6-rsgI6N94AP95A. A similar result was also observed for the xylanase activity of the culture supernatant of the three complementation mutants (Fig. 4F). Taking these results together, we thus demonstrate that the autocleavage of RsgI6 is essential for its signal transduction.
Therefore, the autocleavage site Asn94-Pro95 in RsgI6 (Asn97-Pro98 in RsgI2) provides a distinct activation mechanism for the cognate SigI: When the N-terminal fragment is separated from the C-terminal fragment of RsgI, the RsgI-bound SigI will be released and activated for downstream transcription. Considering that SigI/RsgI in B. subtilis has been suggested to have a RIP mechanism for signal transduction (21), we speculate that the autocleavage in RsgI provides the peptide recognized by site-2 protease, i.e., the autocleavage mimics the site-1 proteolysis in the classical RIP process.
Proteases RseP and ClpXP are required for the activation of SigI6
A previous study showed that RasP, a homolog of the site-2 protease RseP in the RIP process (41), is involved in the activation of SigI in B. subtilis (21). To investigate whether RseP is involved in the processing of RsgI6 in C. thermocellum, we first constructed an inactivation mutant of RseP (Clo1313_1217) termed ΔrsgI6ΔrseP, on the basis of the ΔrsgI6 strain, using the thermotargetron technique. Gene transcription detected by qRT-PCR in the relevant mutant strains showed that the expression of SigI6 and xylanases in ΔrsgI6ΔrseP was increased and similar to that in ΔrsgI6 and ΔrsgI6::rsgI6-T3, but their overexpression in ΔrsgI6ΔrseP::rsgI6-T3 was significantly reduced (Fig. 5A). These results indicate that activation of SigI through RsgI6-T3 was substantially inhibited when rseP was inactivated. Similarly, SDS-PAGE of the cultural supernatant proteins showed that the expression of Xyn10Z and Xyn11B in ΔrsgI6ΔrseP was also similar to that in ΔrsgI6 and ΔrsgI6::rsgI6-T3, but their expression in ΔrsgI6ΔrseP::rsgI6-T3 was notably reduced (Fig. 5B and table S6). Consequently, the site-2 protease RseP is involved in the process of releasing the bound SigI6 in RsgI6-T3.
Fig. 5. The role of RseP and ClpXP in the signal transduction of RsgI6.
(A) qRT-PCR analysis of SigI6 and xylanase expression in ΔrsgI6, ΔrsgI6ΔrseP, and their rsgI6-T3-complemented strains. The relative expression represents the ratio of the mRNA levels in the mutant strains compared to that of the wild-type DSM1313 strain. The P values were calculated on the basis of three replicates using Student’s t test between ΔrsgI6ΔrseP and ΔrsgI6ΔrseP::rsgI6-T3. *P < 0.05 and **P < 0.01. (B) SDS-PAGE analysis of the extracellular proteins of ΔrsgI6, ΔrsgI6ΔrseP, and their rsgI6-T3–complemented strains cultured with cellobiose as the carbon source. (C) Sequence alignment of the transmembrane helix regions of the RsgIs in C. thermocellum. The transmembrane helices predicted by TMHMM (65) are indicated by blue rectangles. A consensus WebLogo (66) is shown at the bottom. The red vertical line indicates the Asn-Pro cleavage site. The potential ClpP recognition motifs are colored in red. (D) Protease assay using RsgI6NTD-VAA1 as the substrate. The positions of the proteases and substrates in the control (the mixture before the reaction) are indicated by arrows. The degradation of substrate bands is indicated by a red dashed rectangle.
A previous study has shown that ClpEP might be the downstream protease after the transmembrane helix was degraded by RasP in the RsgI-SigI signaling of B. subtilis (21). The site-2 protease RseP/RasP generally cleaves the transmembrane helix at a site after two or more consecutive alanine residues (42, 43). This hydrophobic motif can be recognized as a cryptic proteolytic tag by the cytoplasmic ClpP protease complex [containing a catalytic subunit ClpP and one of Clp adenosine triphosphatases (ATPases)/unfoldases ClpX, ClpA, ClpE, or ClpC] for complete degradation (44, 45). Sequence analysis showed that most RsgIs in C. thermocellum contain a potential proteolytic AA tag in the transmembrane helices, except RsgI5 whose predicted transmembrane helix only contains two nonconsecutive alanine residues (Fig. 5C). Most of the proteolytic tags are in a VAA, IAA, or FAA motif, which are most preferred by ClpP protease recognition (43, 46), suggesting that the product of RsgI cleaved by RseP could be lastly degraded by ClpP. To further clarify which ClpP protease complex is involved in the degradation of RsgI for the final step to release the bound SigI, we performed in vitro protease assays using recombinant ClpP protease and its unfoldases ClpX, ClpE, and ClpC. Two N-terminal fragments of RsgI6, RsgI6NTD-VAA1, and RsgI6NTD-VAA2, truncated at the two VAA sequences, respectively, in the transmembrane helix (Fig. 5C), were expressed in E. coli, and the cell lysates were used as the substates. The protease assays showed that the ClpXP complex can efficiently degrade both substrates but cannot degrade the RsgI6NTD without the VAA motif (Fig. 5D and fig. S10). In contrast, ClpEP and ClpCP failed to degrade any of these substrates. These results indicated that ClpXP is the protease responsible for the final step to degrade RsgI6 and release the bound SigI6 in C. thermocellum, which is different from the final degradation step of RsgI in B. subtilis, where ClpEP was identified as the proteolytic enzyme (21).
Proposed model for RsgI signal transduction
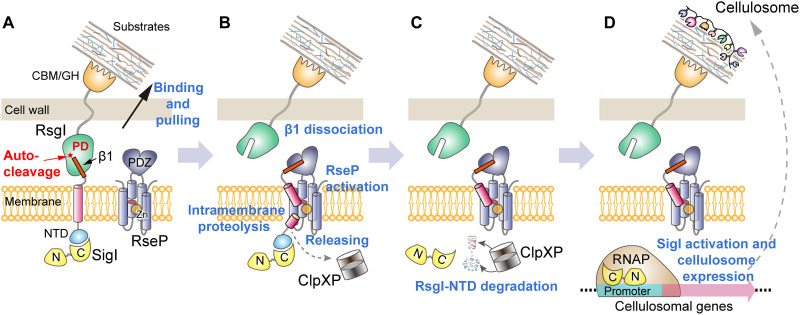
On the basis of the results of this study, we hereby propose a model of RsgI/SigI-mediated signal transduction for cellulosomal gene expression (Fig. 6). The peptide bond between the conserved Asn-Pro motif in the PD of RsgI is autocleaved when RsgI is located on the membrane in the correctly folded state, whereby the β1 strand in the PD of RsgI is noncovalently connected to the C-terminal part of RsgI. When the cells are exposed to the appropriate substrate, the C-terminal CBM domain of RsgI binds to the substrate (Fig. 6A), and an as-yet-undefined process separates the C- and N-terminal parts of RsgI, resulting in dissociation of the N-terminal β1 strand. The β1 fragment and the transmembrane helix are recognized and further cleaved by the RseP protease (Fig. 6B), thereby releasing the N-terminal part of RsgI that contains the NTD and the small fragment of the transmembrane helix. Last, the N-terminal part of RsgI is recognized and degraded by the cytosolic protease ClpP (Fig. 6C), resulting in the release and activation of the bound SigI for the transcription of cellulosomal genes (Fig. 6D).
Fig. 6. Proposed model of transmembrane signal transduction of RsgI in C. thermocellum.
(A) Autocleavage of the conserved Asn-Pro motif and binding of the extracellular RsgI-based CBM to substrate. (B) Recognition and cleavage of the β1 fragment and the transmembrane helix by the RseP protease; release of the NTD. (C) Degradation of the NTD by the cytosolic protease ClpXP. (D) Release of SigI and transcription of cellulosomal genes.
DISCUSSION
Transmembrane signaling is essential for bacterial cells to respond to environmental changes. In this study, we discovered an autoproteolytic process in the signaling of the anti-σ factor RsgI, which is widespread in the Bacilli and Clostridia. The autoproteolysis occurs in the conserved PD whose structure differs from those of other known autoproteolytic proteins. The cleavage leaves a short peptide together with the transmembrane helix of RsgI, which is sufficient to activate the downstream RIP protease RseP and lastly release the bound SigI. Although autoproteolysis has been extensively studied in eukaryote transmembrane signaling processes, it has rarely been reported in bacteria. To the best of our knowledge, the only reported autoproteolytic signaling protein in bacteria is the anti-σ factor FoxR, which responds to ferrioxamine, mainly in Gram-negative Proteobacteria and Bacteroidetes (4). However, the autoproteolysis of FoxR is not essential for its signal transduction (4). The anti-σ factor RsgI thus represents the first autoproteolytic signaling protein described in Gram-positive bacteria, with an essential distinctive conserved cleavage site and overall structure. The proposed model of RsgI signaling in C. thermocellum for substrate sensing and cellulosome regulation retains some of the characteristics of the traditional RIP mechanism, but the signal transduction in the model is initiated by an autocleavage mechanism without the participation of any protease, which substitutes the site-1 cleavage by a protease in classical RIP signaling.
The proposed model of RsgI signaling is, to some degree, similar to the signaling of aGPCR by its GAIN domain (3). In both mechanisms, autocleavage provides a peptide that could be dissociated from the folded domain when the external environment changes, and the peptide provides the initial signal for downstream signal transduction. Despite these similarities, we note that the structure of RsgI-PD and its downstream signaling mechanism differ from those of the GAIN domain. Autoproteolysis of RsgI-PD results in its N-terminal fragment connected to the membrane-spanning helix for subsequent recognition by the downstream protease RseP. In contrast, following autoproteolysis, the C-terminal fragment of the GAIN domain remained, whereby hydrophobic agonist residues in the fragment are rapidly bound to its orthosteric binding pocket within the seven transmembrane helix bundle of aGPCR, a process driven by hydrophobic effects (3). Therefore, the signaling mediated by RsgI-PD currently represents a unique phenomenon in transmembrane signal transduction mechanisms.
RsgIs widely exist in Bacilli and Clostridia and contain PDs with the conserved Asn-Pro residues and structurally adjacent residues, indicating that autoproteolysis of RsgIs is prevalent in these bacteria (fig. S3). However, note that exceptions may exist because RsgI8 (Clo1313_0524) in C. thermocellum has Asp-Ala residues instead of the required Asn-Pro at the prospective autoproteolysis site. However, RsgI8 does not contain a known C-terminal substrate-binding domain, and the promoters recognized by SigI8 have not been identified experimentally, so the function of RsgI8 remains to be elucidated in the future.
In summary, we discovered a bacterial signal transduction mechanism that relies on an autocleavage of the PD of the anti-σ factor RsgI, which is widespread in Firmicutes. Its PD, RsgI-PD, has a unique protein fold and conserved Asn-Pro cleavage site. The cleavage mimics site-1 proteolysis of the RIP process, which is, in a way, similar to aGPCR activation by autoproteolysis of the GAIN domain. This is the first report of a defined autoproteolytic event for transmembrane signal transduction in bacteria, indicating the diversity and complexity of bacterial signaling. Furthermore, the SigI-RsgI system is the main regulator of the intricate multicomponent cellulosome system in many lignocellulolytic bacteria. Therefore, the present study will provide a better understanding of the regulation of this complicated multienzyme complex, which will inspire the engineering of more efficient cellulosome-producing bacteria for a wide range of bioenergy and synthetic biology applications.
MATERIALS AND METHODS
Bacterial strains, growth media, and culture conditions
The strains used in this study are listed in table S1. E. coli strains were routinely cultivated in Luria-Bertani (LB) medium with shaking at 200 rpm at 37°C. C. thermocellum strains were grown anaerobically at 55°C in GS-2 medium (47) with fructose (5 g/liter), glucose (5 g/liter), cellobiose (5 g/liter), or Avicel (5 g/liter; PH-101, Sigma-Aldrich) as the sole carbon source. When required, the media for E. coli and C. thermocellum were supplemented with kanamycin (50 μg/ml), chloramphenicol (30 μg/ml), or thiamphenicol (Tm; 3 μg/ml).
Plasmid construction
All plasmids are described in table S2, and the primers used are listed in table S3. The thermotargetron plasmids pHK-TT1A-RsgI2-9a and pHK-TT1A-RseP-13a were constructed for inactivation of the genes rsgI2 (clo1313_1962) and rseP (clo1313_1217), respectively, through the thermotargetron method (31). The plasmids pHK-HR-RsgI6 and pHK-HR-SigI6-RsgI6 for deletion of the rsgI6 (clo1313_2777) and sigI6-rsgI6 (clo1313_2778 and clo1313_2777), respectively, were constructed to obtain mutants ΔrsgI6 and ΔsigI6-rsgI6 using the homologous recombination method (48). These plasmids contain the endogenous selection marker pyrF, the selection marker tdk (Teth514_0091) from Thermoanaerobacter sp. X514, and ~1-kb upstream and downstream homologous arms of the targeted genes.
The plasmids for expression of full-length RsgIs, RsgI-PDs, the truncated fragments of RsgI6, and SigI6-RsgI6 in C. thermocellum or E. coli were constructed using either a seamless cloning kit (Vazyme Biotech, Nanjing, China) or the traditional restriction enzyme digestion and ligation method. The gene fragments were amplified by PCR from C. thermocellum DSM 1313 genomic DNA and cloned into the expression vector pHK using either the constitutive promoter P2638 of clo1313_2638 (47) or promoter PsigI6 of sigI6 (clo1313_2778). For detection of protein expression, an N-terminal or C-terminal HA-tag (YPYDVPDYA) (32) fused to the protein was designed in the primers. Plasmids pET30a and pET28a-SMT3 (34) were used for protein overexpression in E. coli, and the constructs contained a C-terminal 6×His tag and a cleavable N-terminal 6×His-SMT3 tag, respectively. Mutants of RsgI2, RsgI6, and RsgI2-PD were constructed by the QuikChange method using the designed primers and appropriate templates (table S2). All plasmids were verified by colony PCR and sequencing.
The plasmids for expression of RsgI6-NTD-VAA1 (RsgI residues 1 to 60), RsgI6-NTD-VAA2 (RsgI residues 1 to 63), and ClpP (Clo1313_0329) in E. coli were constructed using the vector pET30a. The plasmids for expression of RsgI6-NTD, the unfoldases ClpX (Clo1313_0330), ClpE (Clo1313_1914), and ClpC (Clo1313_2462) were constructed using the vector pET28a. The resultant recombinant ClpP contains a C-terminal His-tag, while the recombinant RsgI6NTD, ClpX, ClpE, and ClpC contain an N-terminal His-tag.
Construction of C. thermocellum mutants
All plasmids for constructing C. thermocellum mutants were first transformed into E. coli BL21(DE3) to remove Dcm methylation and then transformed into the corresponding parent C. thermocellum cells as previously described (49). After recovering at 51°C, the cells were plated on solid GS-2 medium, supplemented with Tm. Transformants were screened by PCR and sequencing. For the gene inactivation mutants ΔrsgI2 and ΔrsgI6ΔrseP constructed by the thermotargetron technique, the thermotargetron plasmids were cured by continuous inoculation and growth of cells in fresh medium without antibiotic as previously described (49). For the gene deletion mutants ΔrsgI6 and ΔsigI6-rsgI6 constructed by homologous recombination, the appropriate transformants with the pHK-HR-RsgI6 and pHK-HR-SigI6-RsgI6 plasmids were further inoculated into MJ medium and plated onto 5-Fluoro-2'-deoxyuridine (FUDR)–supplemented solid MJ medium to screen for successful recombinant strains as previously described (48).
Recombinant protein expression and purification
Recombinant plasmids for protein expression were transformed into E. coli BL21 (DE3). The cells were cultured in LB medium or M9 minimal media at 37°C with kanamycin sulfate (50 μg/ml). Upon reaching an optical density at 600 nm of 0.8 to 1.0, 0.5 mM isopropyl-β-D-thiogalactopyranoside was added, and the culture was incubated for 4 to 6 hours at 37°C to induce protein expression. The cells were then harvested by centrifugation at 10,000g for 10 min.
The procedure of protein purification of RsgI1-PD (residues 87 to 253), RsgI2-PD (residues 89 to 248), and RsgI6-PD (residues 86 to 252) used for NMR structure determination was the same as described before (20). The purified proteins contained an additional C-terminal His-tag, and the proteins were thus purified initially by affinity chromatography using a HisTrap column (GE Healthcare, Uppsala, Sweden), followed by gel filtration using a Superdex 75 column (GE Healthcare, Uppsala, Sweden). The final NMR sample consisted of ~1.0 mM protein in 50 mM phosphate buffer (pH 6.8) with 90% H2O/10% D2O (v/v), 150 mM NaCl, and 0.02% (w/v) sodium 2,2-dimethylsilapentane-5-sulfonate.
The protein RsgI2-PD (residues 86 to 259) used for crystal structure determination was obtained by protein expression using the plasmid pET28a-SMT3-RsgI2-PD. After initial purification by affinity chromatography, the protein SMT3-RsgI2-PD was further purified by ion-exchange chromatography using a Mono-Q column (GE Healthcare, Uppsala, Sweden). The binding buffers were 50 mM tris-HCl at pH 7.4, and the protein was eluted by adding 1 M NaCl into the corresponding binding buffers. The eluent was pooled, treated with ubiquitin-like-specific protease 1 (ULP1) to cleave the SMT3 tag, and then passed through a HisTrap column to remove the cleaved SMT3 tag. The flow-through target protein RsgI2-PD was concentrated and lastly purified by gel filtration with the buffer containing 10 mM tris-HCl and 100 mM NaCl at pH 7.4.
The recombinant ClpP, RsgI6NTD, ClpX, ClpE, and ClpC were first purified using a HisTrap column (GE Healthcare, Uppsala, Sweden). The eluted proteins from the HisTrap column were changed into the PZ buffer [25 mM Hepes, 200 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, and 10% glycerol (pH 7.6)] (50) by using a desalting column (GE Healthcare, Uppsala, Sweden).
SDS-PAGE and Western blotting
For detection of cellulosomal expression in the extracellular proteins, C. thermocellum strains were cultivated in GS-2 medium with the different carbon sources at 5 g/liter until the late log phase. The cultures were then centrifuged at 7200g for 15 min; the supernatant fluids were filtered through a 0.22-μm syringe filter and concentrated by ultrafiltration (10.0-kDa cutoff). Protein concentration was determined by the Bradford method, and the composition was examined by 10% SDS-PAGE. Individual protein bands were cut from the Coomassie blue–stained gels, digested by trypsin, and identified by LTQ–electrospray ionization tandem mass spectrometry as described before (49).
For Western blot detection of RsgI expression, the pelleted cells of C. thermocellum or E. coli were solubilized in SDS-PAGE sample buffer and boiled for 5 min. Samples were normalized according to the optical density at 600 nm of the culture. After separation by SDS-PAGE with 10 or 15% polyacrylamide resolving gel, proteins were electrotransferred to nitrocellulose membranes (Millipore). Subsequently, the membrane was blocked using skim milk (50 g/liter) in TBST buffer [20 mM tris-HCl, 150 mM NaCl, and 0.05% Tween 20 (pH 7.4)]. Afterward, targeted proteins were detected using the antibody directed against either the HA- or FLAG-tag and horseradish peroxidase (HRP)–labeled anti-tag antibody (Sangon Biotech Co. Ltd., Shanghai) as previously described (49).
Protein N-terminal sequencing
The protein SMT3-RsgI2-PD used for protein N-terminal sequencing was purified by affinity chromatography. The purified SMT3-RsgI2-PD was separated by SDS-PAGE and transferred to presoaked polyvinylidene difluoride membrane (Millipore). The two main bands corresponding to the N-terminal and C-terminal fragments of SMT3-RsgI2-PD were excised from the membrane after staining with Ponceau S dye. The first five N-terminal amino acids of the protein were determined by Edman degradation (Sangon Biotech Co. Ltd., Shanghai).
NMR structure determination
The NMR assignments of RsgI2-PD were reported in a previous study (20), and the same procedures were used to obtain the NMR assignments of RsgI1-PD and RsgI6-PD. All NMR experiments were performed at 298 K on a Bruker Avance III 600-MHz NMR spectrometer equipped with a z-gradient triple-resonance TCI cryoprobe. The NMR structure calculations followed the procedures described before (51). Briefly, the structures were initially calculated by the CYANA software (52) and then refined using the programs SANE (53) and CNS (54), with explicit water refinement protocol implemented in RECOORDScripts (55). Distance restraints derived from the 15N-edited and 13C-edited NOESY-HSQC datasets, dihedral angle restraints derived from the chemical shifts using the program TALOS-N (56), and hydrogen bond restraints according to the secondary structures in the late refinement stages were used in the structure calculations. A set of 20 lowest energy conformers was validated by PROCHECK_NMR (57) and WHAT_CHECK (58) and selected to represent the final ensemble of structures.
Crystal structure determination
The purified protein RsgI2-PD was concentrated to approximately 20 mg/ml. Crystallization conditions of the proteins were screened by using commercial screening kits and then optimized in 24-well crystallization plates at 18°C. The final crystallization condition was 0.2 M lithium sulfate monohydrate (pH 6.0) and 20% (w/v) polyethylene glycol 3350. The crystal used for data collection was cryoprotected by soaking in the well solution supplemented with 20% (v/v) glycerol for 10 s and then flash-cooled in cold liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility, beamlines BL02U1 in a 100-K nitrogen stream. Data indexing, integration, and scaling were conducted using the XDS software (59). The initial structures were determined by molecular replacement using the NMR structure of RsgI2-PD as the search model in the PHENIX program Phaser-MR (60). Refinements of the structures were performed using COOT (61) and PHENIX (60). The search for structure homology was performed using the Dali server (62) and SSM server (63). All of the structure figures were made using PyMOL (www.pymol.org/).
Cell-free protein synthesis
To express RsgI-PD in a cell-free system, the E. coli–based PURE system was used (35). The PCR products encoding RsgI2-PD (residues 89 to 248) or its mutant with an N-terminal GST and a C-terminal FLAG tag were inserted into a prokaryotic expression vector, which contained the T7 promoter, the ribosome-binding site, and the T7 terminator. The proteins were expressed in a 25-μl reaction containing 100 ng of purified plasmids as the template and 20 U of ribonuclease inhibitor (Thermo Fisher Scientific). Reactions were incubated at 37°C for 2 hours and subsequently analyzed by Western blotting as described above.
Quantitative reverse transcription PCR
For RNA extraction, C. thermocellum strains were cultivated to the mid-log phase with cellobiose (5 g/liter) or alkali-pretreated wheat straw (5 g/liter) as the carbon source. The alkali-pretreated wheat straw was obtained by pretreatment with 20% sodium hydroxide in a 1:9 ratio of wheat straw and solution at 160°C for 1 hour. The alkali-pretreated wheat straw contained 59.20% of glucan, 14.47% of hemicellulose, and 5.83% of lignin, determined by the standard two steps of sulfuric acid hydrolysis (64). After centrifugation, the cell pellets were suspended in TRIzol reagent (Invitrogen), and total RNAs were extracted using RNeasy Mini Kit (QIAGEN). To generate cDNA from RNA samples, SuperScript III (Invitrogen) was used according to the manufacturer’s protocol with 3 μg of total RNA and random hexamers. Each qPCR reaction system (20 μl) contained cDNA, reverse and forward primers (10 μM each), and QuantiNova SYBR Green Master Mix (QIAGEN). The primers are listed in table S3. RNA samples were checked to be DNA-free by lack of PCR products when the reverse transcription step was omitted. Experiments were performed in triplicate independently. Data were normalized to RNA levels of the housekeeping gene rps (ribosomal protein S10, clo1313_0444).
Xylanase activity of the culture supernatant
Culture supernatant fluids, using alkali-pretreated wheat straw as the carbon source for C. thermocellum strains, were collected by centrifugation at 7200g for 15 min and then filtered through a 0.22-μm syringe filter. The xylan degradation reaction was initiated by adding 20 mg of beechwood xylan (Megazyme Co. Ltd) into 200 μl of the culture supernatant and then maintained at 55°C for 10 min. Production of reducing sugar was determined by the 3,5-dinitrosalicylic acid (DNS) method, and protein concentrations of the supernatant fractions were determined by the Bradford method. One unit of xylanase activity was defined as the amount of supernatant protein that releases 1 μmol of reducing sugar (xylose equivalent) per minute.
Statistical analysis
All data of expression level and activity are the average with an error bar of SD for three independent biological replicates. Original data values are listed in table S7. Statistical analysis was performed using Student’s t test method in Microsoft Excel.
ClpP protease assay
In vitro protease assays were performed with reference to the previous literature (50). The reaction was carried out in the PZ buffer, containing an adenosine triphosphate (ATP) regeneration mix [4 mM ATP, 16 mM creatine phosphate, and creatine kinase (20 U/ml); Shanghai Yuanye Bio-Technology Co. Ltd.]. ClpP (3 μM) and its ATPases ClpX, ClpE, or ClpC (3 μM) were first added into the buffer and preincubated at 30°C for 15 min. The substrate (cell lysate of RsgI6NTD-VAA1 or RsgI6NTD-VAA2, or purified RsgI6NTD) was then added and reacted at 37° or 55°C for 10 min. The quantity of added substrates was estimated according to the visible band on a preparative SDS-PAGE gel. The degradation of the bands of the substrate proteins was detected by SDS-PAGE.
Acknowledgments
We thank the staff from BL02U1 beamline of the Shanghai Synchrotron Radiation Facility for assistance during x-ray diffraction data collection.
Funding: This work was supported by the National Natural Science Foundation of China (grant nos. 32070125 to Y.F., 31800022 to C.C., 32170051 to Q.C., 32171203 to S.D., 32070028 to Y.-J.L., and 32130063 to J. Lin), Shandong Energy Institute (SEI) (grant nos. SEI S202106 to Q.C. and SEI I202106 to Y.F.), Qingdao Independent Innovation Major Project (grant no. 21-1-2-23-hz to Y.-J.L.), and Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA21060201 to Q.C.).
Author contributions: Y.F. conceived and supervised the study. C.C., S.D., J. Li, and X.D. performed protein expression and purification. S.D. and Y.F. performed crystallization and crystal structure determination. C.C. and Y.F. performed NMR experiments and solution structure determination. C.C., Y.Q., and R.L. performed the genetic operation. C.C., S.D., and Y.Q. performed the Western blotting and N-terminal protein sequencing. Z.Y. and J. Lin performed the cell-free protein synthesis. C.C. performed the qPCR and xylanase activity measurements. C.C. wrote the original manuscript. E.A.B., Y.-J.L., Q.C., and Y.F. analyzed the data and revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Atomic coordinates and NMR restraints (for solution structures) or structure factors (for crystal structure) have been deposited in the PDB under accession numbers 8HDJ (crystal structure of RsgI2-PD), 8HEP (solution structure of RsgI1-PD), 8HEQ (solution structure of RsgI2-PD), and 8HER (solution structure of RsgI6-PD). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S7
REFERENCES AND NOTES
- 1.S. C. Jain, U. Shinde, Y. Li, M. Inouye, H. M. Berman, The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 Å resolution. J. Mol. Biol. 284, 137–144 (1998). [DOI] [PubMed] [Google Scholar]
- 2.H. Ikemura, M. Inouye, In vitro processing of pro-subtilisin produced in Escherichia coli. J. Biol. Chem. 263, 12959–12963 (1988). [PubMed] [Google Scholar]
- 3.A. Vizurraga, R. Adhikari, J. Yeung, M. Yu, G. G. Tall, Mechanisms of adhesion G protein-coupled receptor activation. J. Biol. Chem. 295, 14065–14083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. C. Bastiaansen, J. R. Otero-Asman, J. Luirink, W. Bitter, M. A. Llamas, Processing of cell-surface signalling anti-sigma factors prior to signal recognition is a conserved autoproteolytic mechanism that produces two functional domains. Environ. Microbiol. 17, 3263–3277 (2015). [DOI] [PubMed] [Google Scholar]
- 5.K. C. Bastiaansen, P. van Ulsen, M. Wijtmans, W. Bitter, M. A. Llamas, Self-cleavage of the Pseudomonas aeruginosa cell-surface signaling anti-sigma factor FoxR occurs through an N-O acyl rearrangement. J. Biol. Chem. 290, 12237–12246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.X. Zhang, L. Jin, Q. Fang, W. H. Hui, Z. H. Zhou, 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Liemann, K. Chandran, T. S. Baker, M. L. Nibert, S. C. Harrison, Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108, 283–295 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. J. Taylor, J. E. Johnson, Folding and particle assembly are disrupted by single-point mutations near the autocatalytic cleavage site of Nudaurelia capensis ω virus capsid protein. Protein Sci. 14, 401–408 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. J. Falke, G. L. Hazelbauer, Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. de Dios, E. Santero, F. Reyes-Ramírez, Extracytoplasmic function σ factors as tools for coordinating stress responses. Int. J. Mol. Sci. 22, 3900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.H. Kahel-Raifer, S. Jindou, L. Bahari, Y. Nataf, Y. Shoham, E. A. Bayer, I. Borovok, R. Lamed, The unique set of putative membrane-associated anti-σ factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol. Lett. 308, 84–93 (2010). [DOI] [PubMed] [Google Scholar]
- 12.O. Ramaniuk, M. Převorovský, J. Pospíšil, D. Vítovská, O. Kofroňová, O. Benada, M. Schwarz, H. Šanderová, J. Hnilicová, L. Krásný, σI from Bacillus subtilis: Impact on gene expression and characterization of σI-dependent transcription that requires new types of promoters with extended −35 and −10 elements. J. Bacteriol. 200, e00251–e00218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Z. Wei, C. Chen, Y.-J. Liu, S. Dong, J. Li, K. Qi, S. Liu, X. Ding, L. Ortiz de Ora, I. Muñoz-Gutiérrez, Y. Li, H. Yao, R. Lamed, E. A. Bayer, Q. Cui, Y. Feng, Alternative σI/anti-σI factors represent a unique form of bacterial σ/anti-σ complex. Nucleic Acids Res. 47, 5988–5997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. Nieberler, R. J. Kittel, A. G. Petrenko, H. H. Lin, T. Langenhan, Control of adhesion GPCR function through proteolytic processing. Handb. Exp. Pharmacol. 234, 83–109 (2016). [DOI] [PubMed] [Google Scholar]
- 15.M. S. Paget, Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 5, 1245–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Sineva, M. Savkina, S. E. Ades, Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 36, 128–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.X. Wang, R. Sato, M. S. Brown, X. Hua, J. L. Goldstein, SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62 (1994). [DOI] [PubMed] [Google Scholar]
- 18.J. O. Ebinu, B. A. Yankner, A RIP tide in neuronal signal transduction. Neuron 34, 499–502 (2002). [DOI] [PubMed] [Google Scholar]
- 19.T. Danyukova, K. Schöneck, S. Pohl, Site-1 and site-2 proteases: A team of two in regulated proteolysis. BBA-Mol. Cell. Res. 1869, 11 (2022). [DOI] [PubMed] [Google Scholar]
- 20.X. Ding, C. Chen, Q. Cui, W. Li, Y. Feng, Resonance assignments of the periplasmic domain of a cellulose-sensing trans-membrane anti-sigma factor from Clostridium thermocellum. Biomol. NMR Assign. 9, 321–324 (2015). [DOI] [PubMed] [Google Scholar]
- 21.T. Y. Liu, S. H. Chu, Y. N. Hu, J. J. Wang, G. C. Shaw, Genetic evidence that multiple proteases are involved in modulation of heat-induced activation of the sigma factor SigI in Bacillus subtilis. FEMS Microbiol. Lett. 364, fnx054 (2017). [DOI] [PubMed] [Google Scholar]
- 22.L. Artzi, E. A. Bayer, S. Moraïs, Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15, 83–95 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Y. Feng, Y.-J. Liu, Q. Cui, Research progress in cellulosomes and their applications in synthetic biology. Synth. Biol. J. 3, 138–154 (2022). [Google Scholar]
- 24.Y. Nataf, L. Bahari, H. Kahel-Raifer, I. Borovok, R. Lamed, E. A. Bayer, A. L. Sonenshein, Y. Shoham, Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18646–18651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.I. Muñoz-Gutiérrez, L. Ortiz de Ora, I. Rozman Grinberg, Y. Garty, E. A. Bayer, Y. Shoham, R. Lamed, I. Borovok, Decoding biomass-sensing regulons of Clostridium thermocellum alternative sigma-I factors in a heterologous Bacillus subtilis host system. PLOS ONE 11, e0146316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.L. Ortiz de Ora, R. Lamed, Y.-J. Liu, J. Xu, Q. Cui, Y. Feng, Y. Shoham, E. A. Bayer, I. Muñoz-Gutiérrez, Regulation of biomass degradation by alternative σ factors in cellulolytic clostridia. Sci. Rep. 8, 11036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L. Bahari, Y. Gilad, I. Borovok, H. Kahel-Raifer, B. Dassa, Y. Nataf, Y. Shoham, R. Lamed, E. A. Bayer, Glycoside hydrolases as components of putative carbohydrate biosensor proteins in Clostridium thermocellum. J. Ind. Microbiol. Biotechnol. 38, 825–832 (2011). [DOI] [PubMed] [Google Scholar]
- 28.O. Yaniv, G. Fichman, I. Borovok, Y. Shoham, E. A. Bayer, R. Lamed, L. J. Shimon, F. Frolow, Fine-structural variance of family 3 carbohydrate-binding modules as extracellular biomass-sensing components of Clostridium thermocellum anti-σI factors. Acta Crystallogr. D Biol. Crystallogr. 70, 522–534 (2014). [DOI] [PubMed] [Google Scholar]
- 29.I. R. Grinberg, O. Yaniv, L. O. de Ora, I. Muñoz-Gutiérrez, A. Hershko, O. Livnah, E. A. Bayer, I. Borovok, F. Frolow, R. Lamed, M. Voronov-Goldman, Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum. Proteins 87, 917–930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. J. Mahoney, A. Takayesu, A. Zhou, D. Cascio, R. T. Clubb, The structure of the Clostridium thermocellum RsgI9 ectodomain provides insight into the mechanism of biomass sensing. Proteins 90, 1457–1467 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.G. Mohr, W. Hong, J. Zhang, G. Z. Cui, Y. Yang, Q. Cui, Y. J. Liu, A. M. Lambowitz, A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLOS ONE 8, e69032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Field, J. Nikawa, D. Broek, B. Macdonald, L. Rodgers, I. A. Wilson, R. A. Lerner, M. Wigler, Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8, 2159–2165 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. Lamed, R. Kenig, E. Morag, S. Yaron, Y. Shoham, E. A. Bayer, Nonproteolytic cleavage of aspartyl proline bonds in the cellulosomal scaffoldin subunit from Clostridium thermocellum. Appl. Biochem. Biotechnol. 90, 67–73 (2001). [DOI] [PubMed] [Google Scholar]
- 34.C. Chen, Z. Cui, Y. Xiao, Q. Cui, S. P. Smith, R. Lamed, E. A. Bayer, Y. Feng, Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation. J. Struct. Biol. 188, 188–193 (2014). [DOI] [PubMed] [Google Scholar]
- 35.H. Ohashi, T. Kanamori, Y. Shimizu, T. Ueda, A highly controllable reconstituted cell-free system-a breakthrough in protein synthesis research. Curr. Pharm. Biotechnol. 11, 267–271 (2010). [DOI] [PubMed] [Google Scholar]
- 36.T. Niwa, B. W. Ying, K. Saito, W. Jin, S. Takada, T. Ueda, H. Taguchi, Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 4201–4206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Dembek, C. B. Reynolds, N. F. Fairweather, Clostridium difficile cell wall protein CwpV undergoes enzyme-independent intramolecular autoproteolysis. J. Biol. Chem. 287, 1538–1544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Itoh, T. Hibi, F. Suzuki, I. Sugimoto, A. Fujiwara, K. Inaka, H. Tanaka, K. Ohta, Y. Fujii, A. Taketo, H. Kimoto, Crystal structure of chitinase ChiW from Paenibacillus sp. str. FPU-7 reveals a novel type of bacterial cell-surface-expressed multi-modular enzyme machinery. PLOS ONE 11, e0167310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. D. Clarke, A proposed mechanism for the self-splicing of proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 11084–11088 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Sand, E. K. Holwerda, N. M. Ruppertsberger, M. Maloney, D. G. Olson, Y. Nataf, I. Borovok, A. L. Sonenshein, E. A. Bayer, R. Lamed, L. R. Lynd, Y. Shoham, Three cellulosomal xylanase genes in Clostridium thermocellum are regulated by both vegetative SigA (σA) and alternative SigI6 (σI6) factors. FEBS Lett. 589, 3133–3140 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Y. Hizukuri, T. Oda, S. Tabata, K. Tamura-Kawakami, R. Oi, M. Sato, J. Takagi, Y. Akiyama, T. Nogi, A structure-based model of substrate discrimination by a noncanonical PDZ tandem in the intramembrane-cleaving protease RseP. Structure 22, 326–336 (2014). [DOI] [PubMed] [Google Scholar]
- 42.J. M. Flynn, I. Levchenko, R. T. Sauer, T. A. Baker, Modulating substrate choice: The SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 18, 2292–2301 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Parrell, Y. Zhang, S. Olenic, L. Kroos, Bacillus subtilis intramembrane protease RasP activity in Escherichia coli and In vitro. J. Bacteriol. 199, e00381-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Zellmeier, W. Schumann, T. Wiegert, Involvement of Clp protease activity in modulating the Bacillus subtilis σW stress response. Mol. Microbiol. 61, 1569–1582 (2006). [DOI] [PubMed] [Google Scholar]
- 45.D. Frees, K. Savijoki, P. Varmanen, H. Ingmer, Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63, 1285–1295 (2007). [DOI] [PubMed] [Google Scholar]
- 46.M. Bramkamp, L. Weston, R. A. Daniel, J. Errington, Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62, 580–591 (2006). [DOI] [PubMed] [Google Scholar]
- 47.K. Qi, C. Chen, F. Yan, Y. Feng, E. A. Bayer, A. Kosugi, Q. Cui, Y.-J. Liu, Coordinated β-glucosidase activity with the cellulosome is effective for enhanced lignocellulose saccharification. Bioresour. Technol. 337, 125441 (2021). [DOI] [PubMed] [Google Scholar]
- 48.J. Zhang, S. Liu, R. Li, W. Hong, Y. Xiao, Y. Feng, Q. Cui, Y.-J. Liu, Efficient whole-cell-catalyzing cellulose saccharification using engineered Clostridium thermocellum. Biotechnol. Biofuels 10, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.W. Hong, J. Zhang, Y. Feng, G. Mohr, A. M. Lambowitz, G. Z. Cui, Y.-J. Liu, Q. Cui, The contribution of cellulosomal scaffoldins to cellulose hydrolysis by Clostridium thermocellum analyzed by using thermotargetrons. Biotechnol. Biofuels 7, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. Gatsogiannis, D. Balogh, F. Merino, S. A. Sieber, S. Raunser, Cryo-EM structure of the ClpXP protein degradation machinery. Nat. Struct. Mol. Biol. 26, 946–954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.X. Yao, C. Chen, Y. Wang, S. Dong, Y.-J. Liu, Y. Li, Z. Cui, W. Gong, S. Perrett, L. Yao, R. Lamed, E. A. Bayer, Q. Cui, Y. Feng, Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules. Sci. Adv. 6, eabd7182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.T. Herrmann, P. Guntert, K. Wuthrich, Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002). [DOI] [PubMed] [Google Scholar]
- 53.B. M. Duggan, G. B. Legge, H. J. Dyson, P. E. Wright, SANE (Structure Assisted NOE Evaluation): An automated model-based approach for NOE assignment. J. Biomol. NMR 19, 321–329 (2001). [DOI] [PubMed] [Google Scholar]
- 54.A. T. Brunger, P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, G. L. Warren, Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. Biol. Crystallogr. 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 55.A. J. Nederveen, J. F. Doreleijers, W. Vranken, Z. Miller, C. A. Spronk, S. B. Nabuurs, P. Guntert, M. Livny, J. L. Markley, M. Nilges, E. L. Ulrich, R. Kaptein, A. M. Bonvin, RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Y. Shen, A. Bax, Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 1260, 17–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. A. Laskowski, J. A. Rullmannn, M. W. MacArthur, R. Kaptein, J. M. Thornton, AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 58.R. W. Hooft, G. Vriend, C. Sander, E. E. Abola, Errors in protein structures. Nature 381, 272 (1996). [DOI] [PubMed] [Google Scholar]
- 59.W. Kabsch, XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.L. Holm, P. Rosenström, Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E. Krissinel, K. Henrick, Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 64.S. Liu, Y.-J. Liu, Y. Feng, B. Li, Q. Cui, Construction of consolidated bio-saccharification biocatalyst and process optimization for highly efficient lignocellulose solubilization. Biotechnol. Biofuels 12, 35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Moller, M. D. R. Croning, R. Apweiler, Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653 (2001). [DOI] [PubMed] [Google Scholar]
- 66.G. E. Crooks, G. Hon, J. M. Chandonia, S. E. Brenner, WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S7