Abstract
Fluorescein angiography is a crucial examination in ophthalmology to identify retinal and choroidal pathologies. However, this examination modality is invasive and inconvenient, requiring intravenous injection of a fluorescent dye. In order to provide a more convenient option for high-risk patients, we propose a deep-learning-based method to translate fundus photography into fluorescein angiography using Energy-based Cycle-consistent Adversarial Networks (CycleEBGAN) We propose a deep-learning-based method to translate fundus photography into fluorescein angiography using CycleEBGAN. We collected fundus photographs and fluorescein angiographs taken at Changwon Gyeongsang National University Hospital between January 2016 and June 2021 and paired late-phase fluorescein angiographs and fundus photographs taken on the same day. We developed CycleEBGAN, a combination of cycle-consistent adversarial networks (CycleGAN) and Energy-based Generative Adversarial Networks (EBGAN), to translate the paired images. The simulated images were then interpreted by 2 retinal specialists to determine their clinical consistency with fluorescein angiography. A retrospective study. A total of 2605 image pairs were obtained, with 2555 used as the training set and the remaining 50 used as the test set. Both CycleGAN and CycleEBGAN effectively translated fundus photographs into fluorescein angiographs. However, CycleEBGAN showed superior results to CycleGAN in translating subtle abnormal features. We propose CycleEBGAN as a method for generating fluorescein angiography using cheap and convenient fundus photography. Synthetic fluorescein angiography with CycleEBGAN was more accurate than fundus photography, making it a helpful option for high-risk patients requiring fluorescein angiography, such as diabetic retinopathy patients with nephropathy.
Keywords: fluorescein angiographs, fundus photographs, generative adversarial network, paired translation
1. Introduction
Fundus photography is a safe, fast, and noninvasive ophthalmologic modality that allows for visualization of the details of the retina and provides a bird eye view of all retinal layers. This method enables early diagnosis of several diseases, including diabetes and hypertensive retinopathy. The clinical accessibility of fundus photography is continuously improving with the development of modern tabletop, handheld, and smartphone fundus cameras.[1]
Fluorescein angiography (hereafter abbreviated as “angiography”) is an imaging modality that involves taking photographs of the retina and its blood vessels after the injection of a fluorescent dye.[2] It is an essential examination modality in ophthalmological practice that is widely used to identify retinal and choroidal pathologies. Hyperfluorescences observed during angiography are classified as window defects, abnormal vessels, and leakage. Among these, leakage is a pathognomonic sign of retinal vascular hyperpermeability and is subdivided into pooling and staining. Many retinal diseases exhibit characteristic pooling and staining patterns depending on their pathophysiology. Therefore, angiography is important for diagnosing and treating retinal diseases.[2,3]
However, angiography is an invasive and inconvenient modality that requires the intravenous injection of a fluorescent dye. Side effects ranging from mild (such as nausea and vomiting) to severe (such as renal failure and, rarely, anaphylaxis and death) have been reported.[4] Additionally, only tabletop angiography cameras have been developed, making it impossible to perform angiography on bedridden patients. Therefore, angiography can only be performed in specialized medical centers, and necessary angiography may be delayed or impossible under restrictive conditions.
Tavakkoli et al[5] attempted to generate angiography from fundus photography using Generative Adversarial Networks (GAN) and achieved satisfactory results. However, their GAN model required precisely aligned images for proper training. Their real fundus photographs and real angiographs had almost identical shapes and sizes, and the optic discs and retinal vessels were located in precisely identical locations. As 3-dimensional concave retinal images are rarely identically matched between tests, it is difficult to construct a consistent image set. Therefore, an improved method is needed to translate non-identical paired image sets.
Cycle-consistent adversarial networks (CycleGAN)[6] is a well-known deep learning unpaired image translation method introduced in 2017 that can translate images from 1 domain to another without a one-to-one mapping between input and target images. CycleGAN can efficiently translate images even if the mapping between domains is nonlinear. Several studies have used CycleGAN to convert medical modality images into another modality.[7] For example, some studies have shown excellent performance in converting magnetic resonance imaging (MRI) images to computed tomography (CT) images,[8] and cone-beam CT images to MRI images.[9]
However, CycleGAN cannot learn the specific features of each image because of the inherent limitations of unpaired methods, and therefore has limited clinical utility. Several studies have proposed pairing modifications of CycleGAN.[8,10–14] but these methods were specifically modified for their datasets and were not easily applicable to other datasets.
To address this limitation, we proposed a more generalized method for effectively translating the detailed features of paired input and target images. We developed Energy-based Cycle-consistent Adversarial Networks (CycleEBGAN), a conceptual combination of CycleGAN[6] and Energy-based Generative Adversarial Networks (EBGAN).[15] This method has the advantage of translating corresponding but non-identical image pairs. In this study, we synthesized angiographs using the CycleEBGAN method and examined their clinical usefulness.
2. Methods
2.1. Study design
This retrospective study was approved by the institutional review board of Gyeongsang National University Changwon Hospital (GNUCH 2021-01-001) and followed the principles of the Declaration of Helsinki. Informed consent was waived by the institutional review board due to the retrospective nature of the study.
2.2. Image acquisition protocol
For image acquisition, an expert examiner took fundus photographs using a digital retinal camera (CR-2; Canon, Tokyo, Japan). The same examiner performed fluorescein angiography using a Heidelberg retina angiograph (Heidelberg Engineering, Heidelberg, Germany). Angiography was initiated by injecting 4 mL of fluorescein (Fluorescite; Alcon, Fort Worth, TX) into the cephalic vein. After video recording during the first 30 seconds, the examiner captured images of both eyes alternately. We selected late-phase angiographs taken after 5 minutes as the target images. Fundus photographs taken on the same day were selected as the input images.
2.3. Image preprocessing
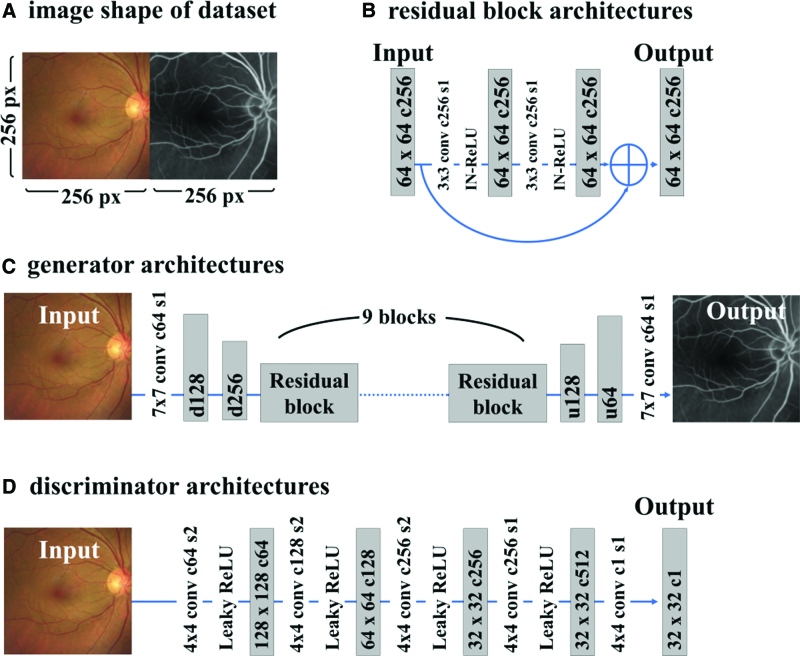
We acquired the stored images in an uncompressed BMP format in order to minimize image distortion. We adjusted the image sizes to be consistent in order to reduce training errors. For image preprocessing, we trimmed the images to make them square in shape and resized them to 256 × 256 pixels using OpenCV and NumPy.[16] The initial size of the fundus photographs was 1113 × 1112 pixels, which were trimmed to 786 × 786 pixels and then resized to 256 × 256 pixels. The initial size of the angiographs was 768 × 768 pixels, which were trimmed to 542 × 542 pixels and then resized to 256 × 256 pixels. The fundus photograph and angiograph were merged horizontally (Fig. 1A). Since we did not perform alignment for the paired fundus photographs and angiography, the positions of the disc and macula varied slightly between the paired images (Figs. 2–4).
Figure 1.
Schematic of the architecture of CycleEBGAN. (A) The images constituting the dataset were 512 × 256 pixels in size, and fundus photographs and angiographs taken on the same day were arranged horizontally. (B) The residual block repeated the convolutional layer, instance normalization, and ReLU activation twice, and the output was summed with the input. (C) The generator repeated the downsampling twice to reduce the size to 1 quarter of the original size. The residual block was repeated 9 times to translate the image from 1 domain to another. Upsampling was repeated twice to restore the image to its original size. (D) The discriminator repeated the convolution of stride 2 3 times and the convolution of stride 1 twice. Finally, a tensor of 32 × 32 × 1 pixels was generated. c = channel, conv = convolutional layer, CycleEBGAN = energy-based cycle-consistent adversarial networks, d = downsampling layer, IN-ReLU = instance normalization and ReLU activation, px = pixel size, s = stride, u = upsampling layer.
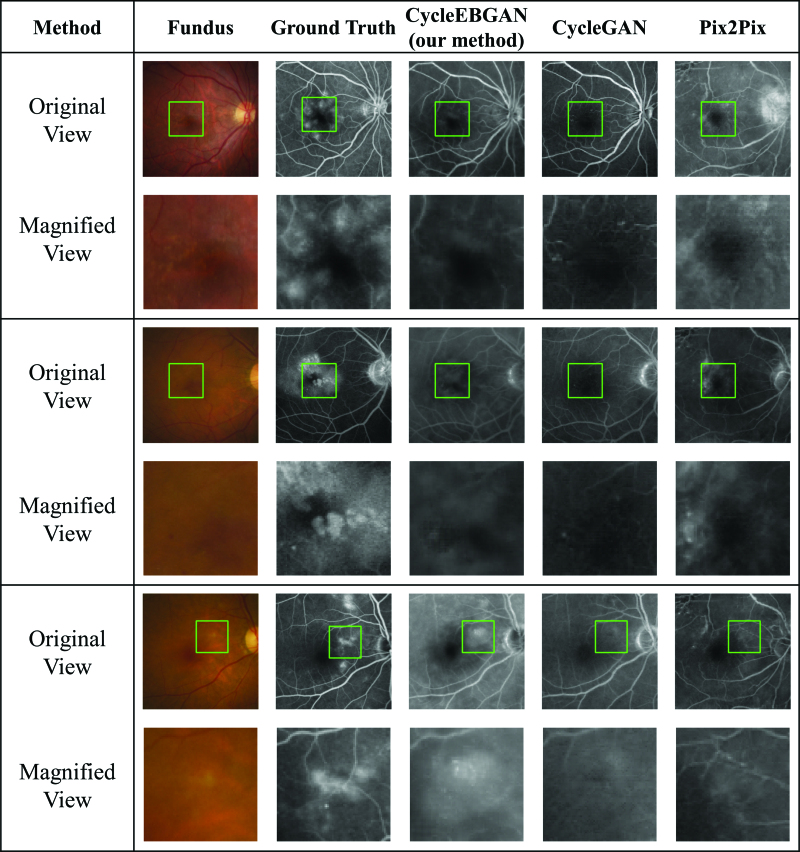
Figure 2.
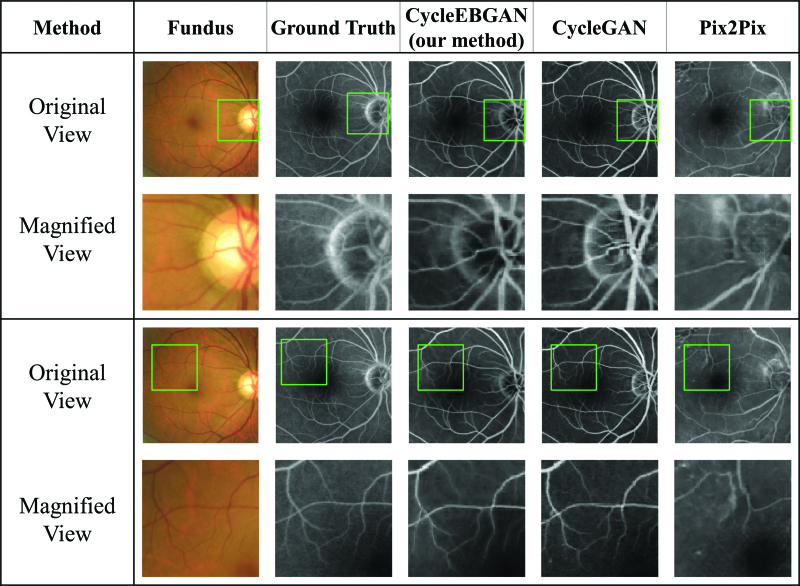
Representative synthetic result of normal fundus photographs and angiographs. The size and position of the green square were adjusted to match the magnified view. Note that as the paired fundus photograph and the angiograph are not aligned, the positions of the green rectangle are not identical. Looking at the magnified view, both CycleEBGAN and CycleGAN produced good synthesis results that matched the angiography. In contrast, Pix2Pix generated abnormal disc and vessels. Compared to Pix2Pix, both CycleGAN and CycleEBGAN show robust results in dealing with the misalignment of paired images. CycleEBGAN = energy-based cycle-consistent adversarial networks, CycleGAN = cycle-consistent adversarial networks.
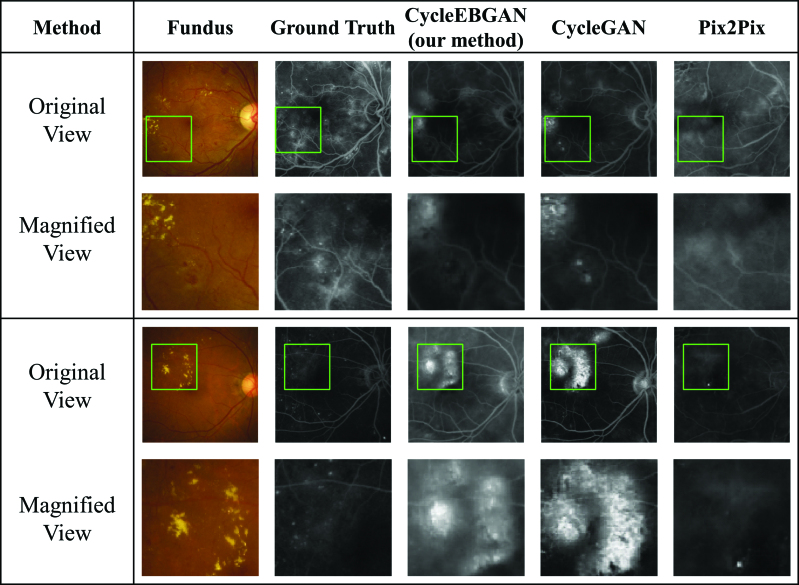
Figure 4.
Representative synthetic result of obviously abnormal fundus photographs and angiographies. Both CycleEBGAN and CycleGAN converted hard exudates into hyperfluorescence, whereas microaneurysms and microaneurysms were ignored. The simulated images generated by the 2 models are almost identical. While Pix2Pix showed excellent performance in transforming vascular leaking, it generated abnormal optic discs and vessels. CycleEBGAN = energy-based cycle-consistent adversarial networks, CycleGAN = cycle-consistent adversarial networks.
2.4. Architecture of CycleEBGAN
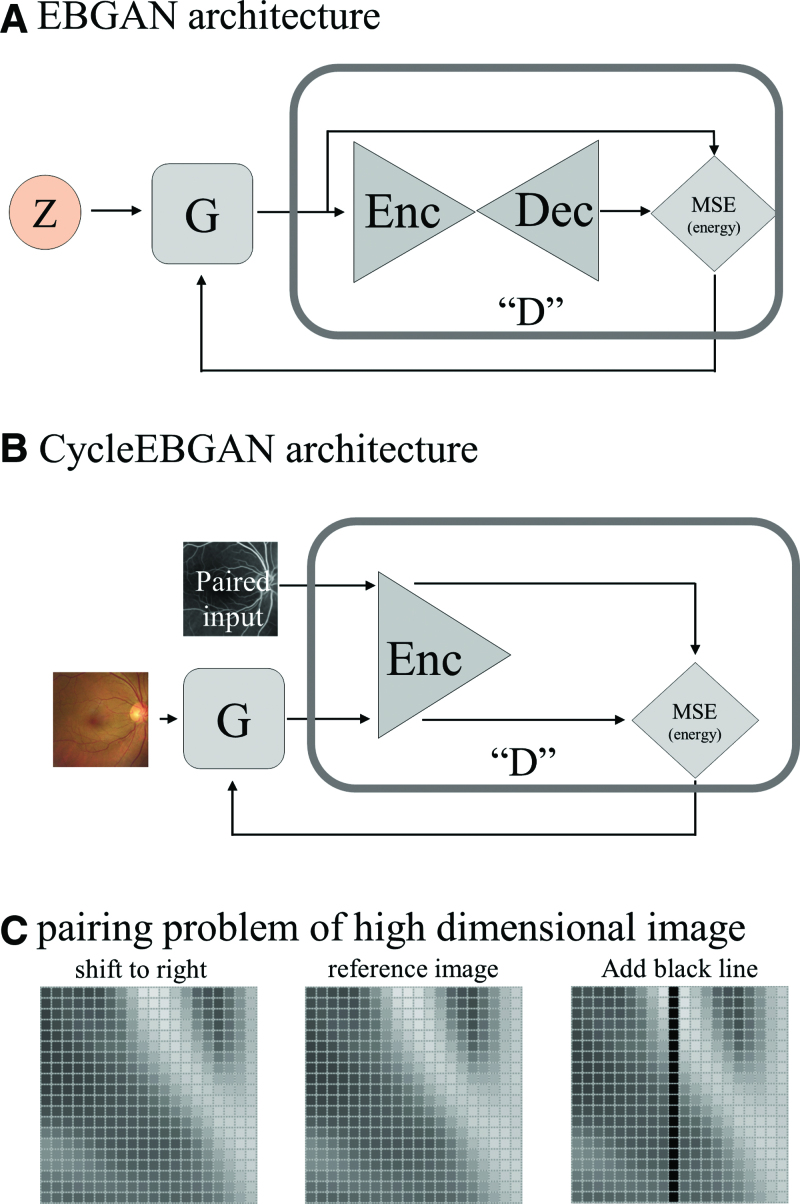
The architecture of CycleEBGAN is almost identical to that of CycleGAN but differs in 2 respects. First, while CycleGAN was trained using unpaired datasets, CycleEBGAN was trained using paired images as in the Pix2Pix method[17] (Fig. 1A). Second, we modified the adversarial loss of the generators by redefining it as an energy function, which will be explained after describing the discriminators.
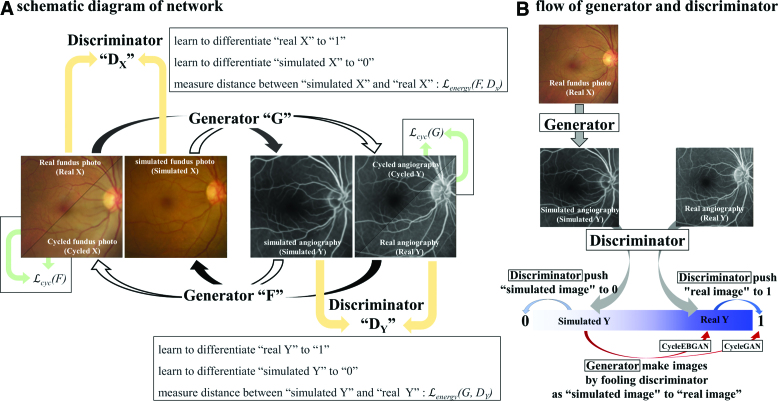
The generator of CycleEBGAN had an identical architecture to that proposed in CycleGAN[6] and consisted of 3 parts: downsampling layers, residual blocks, and upsampling layers (see Fig. 1C). Two downsampling layers downsized the images from 256 × 256 to 64 × 64 pixels while increasing the number of channels from 3 to 256. The images then passed through 9 residual blocks without changing size. Two upsampling layers restored the image size to 256 × 256 × 3 pixels. We applied reflection padding, instance normalization, and ReLU activation. The residual blocks[18] had the same architecture as that proposed in CycleGAN (see Fig. 1B). The data flow in the generators is shown in Figure 5A. Paired real fundus photographs and angiographs were analyzed by Generators G and F, respectively. Generator G translated the real fundus photograph into a simulated angiograph, and Generator F translated the real angiograph into a simulated fundus photograph. Generator F also translated the simulated angiograph into a cycled fundus photograph, and Generator G translated the simulated fundus photograph into a cycled angiograph.
Figure 5.
Schematic diagram of CycleEBGAN. Real and cycled fundus photographs were converted into angiographs by G and F, respectively. Real and cycled angiographs were converted into simulated fundus photographs by F and G, respectively. The discriminators DX and DY were trained to distinguish between the real and simulated images. The generators were trained to “fool” the discriminator to make the simulated image look like a real image. We calculated the L1 loss so that the real and cycled images were identical. In addition, in CycleEBGAN, L2 loss led to the constraint of identical real and simulated images (red arrows). The main difference between CycleEBGAN and CycleGAN is that CycleGAN measures the distance between the simulated image and 1, whereas CycleEBGAN measures the distance between the simulated and paired real images (red arrows). CycleEBGAN = energy-based cycle-consistent adversarial networks, CycleGAN = cycle-consistent adversarial networks.
For the discriminator, we used 70 × 70 PatchGANs (Fig. 1D).[17,19] The discriminator analyzed and compressed simulated and real images into 32 × 32 × 1-sized tensors. Each tensor had a total of 1024 (32 × 32 × 1) elements, which could have values between “0” and “1.” The discriminators were trained to distinguish tensors from simulated images as “0” and those from real images as “1” (Fig. 5B, gray arrows).
In CycleGAN, the generators had 2 types of loss: cycle consistency loss and adversarial loss. Cycle consistency loss is based on the similarity between the real and cycled images (Fig. 5A, green arrows). Adversarial loss measures how realistic the simulated image looks. Adversarial loss is the element-wise distance between the tensor of the simulated image and “1” (Fig. 5B, red arrow).
In CycleEBGAN, we modified the adversarial loss and measured the distance between the tensor of the simulated image and that of the real image (Fig. 5B, red arrow). We performed the following calculations to compare the adversarial losses of CycleGAN and CycleEBGAN:
We trained the models with 200 epochs and a learning rate of 0.0002. Weights were initialized with a Gaussian distribution (mean = 0, standard deviation = 0.02).
2.5. Objective of CycleEBGAN
Our objective is defined as:
where domain X is the input image and domain Y is the target image. G is the generator that converts domain X to Y, and F is the generator that converts domain Y to X. Discriminator DY distinguishes between real and simulated Y, and DX distinguishes between real and simulated X. λ determines the relative importance of adversarial and cycle consistency loss. We set λ to 10, as in CycleGAN.[6] The generator attempts to generate images similar to real images, whereas the discriminator aims to distinguish simulated and real images. The generator aims to minimize the objective function against the adversarial discriminator that attempts to maximize this function. Finally, we aimed to solve:
2.6. Energy function (corresponding to adversarial loss)
We modified the adversarial loss and redefined it as the energy function. A common problem with GAN is that the generator tends to generate samples clustered in one or a few high data density regions.[15] Thus, the images generated by the GAN look similar to each other and have low diversity. We devised an energy function to translate the paired images, as described in the architecture section. The generators were trained to transform real images into simulated images by minimizing the adversarial losses. To calculate the adversarial losses, the discriminators converted the simulated and paired real images into 32 × 32 × 1-sized tensors. Then, we compared the element-wise distance of the tensors. The mean square error (MSE) between the 2 tensors is defined as the energy (Fig. 5B, red arrow):
2.7. Cycle consistency loss
The cycle consistency loss of CycleEBGAN was identical to that of CycleGAN (Fig. 5B, green arrow):
2.8. Simulated image quality evaluation
A small number of images were used as the test set. After training the models, the generator G of CycleGAN and CycleEBGAN converted the test set fundus photographs into simulated angiographs. We evaluated the image quality by analyzing the hyperfluorescence patterns of the simulated images.
Two retinal specialists interpreted the images to determine whether the simulated images were clinically useful. First, the 2 retinal specialists determined whether real angiographs had leakage spots. Then, they predicted the leakage on fundus photographs and simulated angiographs. The area under the receiver operating characteristic curve (AUC) was computed to evaluate the image quality of simulated angiographs. The agreement between the 2 ophthalmologists’ readings was evaluated using Pearson correlation.
2.9. Software
In this study, we used Python version 3.7.9. The CNN model was built using TensorFlow 2.4.1, Keras 2.4.3, OpenCV 4.5.1.48, and NumPy 1.19.5. The performance of each CNN model was given by the accuracy of the test set results. The CPU used to train the CNN model was an Intel Core i9-10980XE, and the GPU was a GeForce RTX 3090 Graphics Card. We analyzed the results for the test set using SPSS for Windows software (version 24.0; SPSS Inc., Chicago, IL). In all analyses, P < .05 was considered to indicate statistical significance.
3. Results
3.1. Baseline characteristics
The medical charts of patients who visited Gyeongsang National University, Changwon Hospital, between February 2016 and December 2020 were reviewed retrospectively. A total of 3359 eyes of 1691 patients were included in this study. In total, 3859 angiographs were included in the study; 193 images were excluded because the macula and optic disc could not be identified due to severe cataracts, vitreous hemorrhage, corneal opacity, etc. In addition, 1061 angiographs were excluded as matched fundus photographs were unavailable. Finally, 2605 image pairs were included in the study. The models were trained with 2555 of these paired images; the remaining 50 paired images were used as the test set.
3.2. Simulated angiographs generated by CycleEBGAN and CycleGAN
The test set consisted of 50 pairs of fundus photographs and angiographs. After training, CycleGAN and CycleEBGAN converted the fundus photographs into simulated angiographs. A total of 200 images were evaluated by CycleGAN and CycleEBGAN: 50 real angiographs, 50 fundus photographs, and 2 sets of 50 simulated angiographs (Figs. 2–4).
CycleEBGAN and CycleGAN generated realistic simulated images similar to the real angiographs. Figure 2 shows normal image pairs. Both models accurately translated normal fundus photographs and did not produce false features. Figure 4 shows the obvious pathological features in both paired fundus photographs and angiographs. Both models converted hard exudates into hyperfluorescent patterns, whereas microaneurysms were ignored. In normal and obviously abnormal image pairs, the models generated almost identical simulated images.
However, CycleEBGAN and CycleGAN showed distinguishable performance in terms of translating the subtle features of fundus photographs. Figure 3 shows abnormal fundus photographs and angiographs with subtle abnormal features. Trained ophthalmologists could identify the features, including retinal thickening, low transparency, and decreased foveal reflex. CycleGAN ignored the features and generated simulated images similar to normal ones. However, CycleEBGAN captured the subtle features and simulated hyperfluorescent patterns similar to the real paired angiographs.
Figure 3.
Representative synthetic result of subtle abnormal fundus photographs and angiographs. Image pairs of fundus photographs with subtle abnormal changes and angiographies with leakage spots. CycleEBGAN learned subtle features from fundus photographs better than CycleGAN, and generated images similar to real angiographs. While Pix2Pix showed excellent performance in transforming these subtle features, it generated abnormal optic discs and vessels. CycleEBGAN = energy-based cycle-consistent adversarial networks, CycleGAN = cycle-consistent adversarial networks.
3.3. Evaluation of image quality by retinal specialists
Two retinal specialists determined that 31 of 50 (62%) real angiographs had leakage spots (Table 1). We compared the readings for real angiography with those for the other images. When the ophthalmologists predicted leakage based on fundus photographs, the AUCs were 0.857 and 0.855, respectively. The AUCs of CycleEBGAN were 0.941 and 0.925, respectively, and those of CycleGAN were 0.899 and 0.903, respectively. CycleEBGAN showed the best AUC, but it was not statistically significantly superior (shown by the overlapping confidence interval).
Table 1.
Image quality evaluation by retinal specialists.
| Examiner | Number of images | AUC (95% CI) | Pearson correlation (P value) | ||
|---|---|---|---|---|---|
| Leakage (+) | Leakage (−) | ||||
| Fluorescein angiography | A & B | 31 (62%) | 19 (38%) | ||
| Fundus photography |
A | 30 (60%) | 20 (40%) | 0.857 (0.739–0.974) | 0.707 (<.001) |
| B | 22 (44%) | 28 (56%) | 0.855 (0.750–0.960) | ||
| CycleEBGAN | A | 30 (60%) | 20 (40%) | 0.941 (0.864–1.000) | 0.877 (<.001) |
| B | 29 (58%) | 21 (42%) | 0.925 (0.841–1.000) | ||
| CycleGAN | A | 30 (60%) | 20 (40%) | 0.899 (0.798–1.000) | 0.735 (<.001) |
| B | 25 (50%) | 25 (50%) | 0.903 (0.816–0.991) | ||
AUC = area under the receiver operating characteristic curve, CI = confidence interval, CycleEBGAN = energy-based cycle-consistent adversarial networks, CycleGAN = cycle-consistent adversarial networks.
The agreement between ophthalmologists was evaluated using Pearson correlation. The coefficients for fundus photography, CycleEBGAN, and CycleGAN were 0.707, 0.877, and 0.735, respectively. CycleEBGAN generated simulated angiographs with the most definite features.
4. Discussion
Angiography is an essential imaging modality for diagnosing and treating retinal and choroidal diseases.[3] However, angiography is invasive and can cause life-threatening anaphylaxis and fatal kidney damage.[4] In addition, angiography cannot be performed in bedridden patients because the examination can only be performed while sitting. It would be beneficial to avoid angiography in high-risk patients. As patients examined with angiography usually also undergo fundus photography, fundus photographs and fluorescein angiographs could be paired. Therefore, we attempted to convert fundus photographs into angiographs using machine learning methods.
Tavakkoli et al[5] utilized a method known as the conditional Generative Adversarial Network (GAN) to generate angiographs from fundus photographs, using a set of 70 well-matched images. Their findings differed from ours, as Tavakkoli et al discovered that the conditional GAN yielded superior results compared to the traditional CycleGAN, and CycleGAN pretended to produce false features. Another research for creating angiographs involves segmenting retinal vessels from fundus photographs. Son et al[20] developed a GAN network using the U-Net architecture, demonstrating its superior performance compared to conventional methods. They showed that their GAN could detect subtle features of tiny blood vessels in the fovea.
The datasets employed in the previous 2 studies were public and differed in characteristics from our own. Tavakkoli dataset was composed of 70 images, each measuring 512 × 512 pixels, whereas Son dataset comprised 110 images, each 640 × 640 pixels. Compared to our dataset, their dataset had a smaller sample size and larger image size, and the paired images were well-aligned. In contrast, our dataset was significantly larger (2605 images), and image size was small (256 × 256 pixels). Additionally, our paired images were not well-aligned, which could result in the generation of false features.[5] Therefore, conventional methods of previous studies may not be suitable for our dataset.
First, we attempted to apply translation methods to paired images. The well-known U-net based pix2pix conditional GAN (pix2pix) maps the input image to a corresponding output image.[17] Pix2pix directly compares pixels constituting synthesized and target images. A significant limitation of pix2pix is that it assumes that objects are in identical locations in images. Therefore, direct comparison of the pixels would generate significant error due to image shifting (Fig. 6C). Thus, the retinal vessels and optic disc in pix2pix were often corrupted, and false-positive artifacts were common (Figs. 2–4). A previous study also reported misrepresentations when generating angiographs from fundus photographs.[5] Because the patient posture and ocular position changed slightly between taking fundus photographs and angiography, it was not possible to match the images perfectly. Moreover, the retina is a 3-dimensional concave surface, so identical images could not be obtained; the retinal images were always slightly different.
Figure 6.
Comparison of EBGAN and CycleEBGAN. (A) In EBGAN, generator G takes a random vector (z) and synthesizes G(z). The discriminator takes G(z) and transforms it. The discriminator estimates the energy value (E) from the MSE of the G(z) and transformed G(z). (b) In CycleEBGAN, the generator (G) synthesized and simulated an angiograph from a real fundus photograph. The discriminator pairs the simulated angiograph and real angiograph. The discriminator estimates the energy value (E) from the MSE of the 2 images. (c) The central image is a reference image; the left image is the reference image moved to the right by 1 pixel; the right image is the reference image to which a black line has been added. The left and central images look similar. However, the distance (Lp-norm) between them is significantly larger than that between the right and central images. The black line in the right image induces a slight difference. CycleEBGAN = energy-based cycle-consistent adversarial networks, D = discriminator, Dec = decoder, E = energy, EBGAN = energy-based generative adversarial networks, Enc = encoder, G = generator, MSE = mean square error, x = real image, z = latent vector.
CycleGAN, a well-known unpaired image translation method, has been used in several studies to convert medical images, such as MRI, CT, cone-beam CT, and positron emission tomography (PET)-CT.[7,14,21–34] As shown in Figures 2 to 4, CycleGAN could successfully translate fundus photographs into angiographs. CycleGAN could capture hard exudates, soft exudates, and depigmentation from fundus photographs. It showed excellent performance in normal and obviously abnormal images (Figs. 2 and 4). CycleGAN generated similar simulated images and ignored subtle features, such as curved vessels, retinal thickening, retinal non-transparency, and decreased foveal reflex (Fig. 3). Ophthalmologists often derive important information from these subtle features; therefore, it was necessary to devise a model to learn these subtle features from paired images.
CycleGAN generated samples clustered in regions with high data density and ignored sparse samples.[15] The generator of CycleGAN tried to generate simulated images that the discriminator might interpret as “1.” We realized that “1” referred to the common characteristics of real images. The simulated images looked alike as the generator was trained with common characteristics. We believe that the interpreted value should not only be “1” but have different values. We reviewed many published models and concluded that EBGAN was suitable to overcome our problem.
The energy function, which is a crucial part of EBGAN, quantifies the compatibility between the model parameters and the observed data. The main concept of EBGAN is that if an autoencoder (discriminator) can compress and restore data effectively, it considered low energy (real). On the other hand, if there are larger errors, it considered high energy (fake). Unlike the traditional GAN that derives results within a range of 0 to 1, the error from the compression and restoration process in the autoencoder can have a wider range of values, which allows the model to learn from a richer set of data. EBGAN propose to view the discriminator as an energy function (or a contrast function) without explicit probabilistic interpretation.[15]
We observed that CycleGAN tended to overlook subtle features, resulting in the production of similar images (Fig. 3). We hypothesized that this was due to the generated images clustering together. To overcome this limitation, we explored training the generator with a wider range of scalar values other than “1.” Our idea was that this approach might lead the generator to create more diverse and less clustered output. As a result, this could enable better preservation and transformation of the subtle features present in the images.
EBGAN views the discriminator as an energy function that associates low energies with regions near the data manifold and higher energies with other regions.[15] In addition, the generator is trained to produce samples with minimal energy. The generator takes a random vector z and synthesizes G(z). The discriminator takes G(z) and generates transformed G(z) with the autoencoder. The discriminator calculates the MSE (L2-norm) of G(z) and transformed G(z). The MSE is the energy (E) in EBGAN. The discriminator learns various scalar energy values other than “1” (Fig. 6A). These values prevent clustering of generated images and reflect the real image manifold.[15]
At the time of abstracting the input image information, discriminators of CycleGAN functionally correspond to autoencoder of EBGAN. The discriminators of EBGAN and CycleGAN had different structures. The EBGAN discriminators consist of a combination of an encoder and decoder, whereas that of CycleGAN consists only of an encoder (Fig. 6A and B). Despite the structural disparity, the discriminators of CycleGAN compress image information into a tensor. The tensor compiles the input image and compresses the spatial information. We redefined the discriminator of CycleGAN as an energy function that associates low energies with regions near the data manifold. A tensor transformed from a paired real image is considered a low-energy manifold. Thus, the energy function is the MSE of tensors from simulated and real paired images (Fig. 5B red arrows). The generators are trained to generate simulated images with minimal energy, and the discriminators distinguish real images (“1”) from simulated images (“0”). Through repetition of this adversarial learning, the generator could generate an image with the characteristics of a paired real image.
We devised CycleEBGAN by modifying CycleGAN, as mentioned previously. After this modification, the generators and discriminators could learn the manifold of paired images. CycleEBGAN could learn subtle features and showed better performance than CycleGAN (Fig. 3). The interpretations of the retinal specialists were better than those of CycleGAN. The agreement for simulated image readings also improved. CycleEBGAN could generate simulated images closer to real fluorescein angiographs than CycleGAN.
The main advantage of CycleEBGAN over CycleGAN in previous studies[8,10–14] was that it overcomes the pairing problem of high-dimensional images. Training the model using one-to-one image mapping can result in a large error even if images are only slightly spatially mismatched. For example, Figure 6C shows 400-dimensional images (20 × 20). The central image is a reference image, and the left image depicts the reference image moved to the right by one pixel; in the right image, a black line is added to the reference image. Although the left image appears similar to the middle image, when the distance (Lp-norm) is measured, the left image shows a much larger difference than the right one.[35] Previous studies used complex methods to overcome the mismatch of image pairs.[8,10–14] For example, they split the input and target images into small patches and then merged them to restore the original size.[10] In addition, they used 1.5 as the Lp-norm, whereas standard machine learning models use values of 1 or 2. They also introduced the concept of gradient magnitude distance to reduce the large error seen for non-identical paired images.
Autoencoder is a well-known and effective solution to deal with the pairing problem of high-dimensional space. The autoencoder consists of an encoder that reduces dimensionality and a decoder that restores it. In the reduced dimensional space, Lp-norm could better reflect the similarity.[35] CycleEBGAN performs dimensionality reduction using the discriminator. CycleGAN includes discriminators performing dimensionality reduction, compressing 14,700 numbers (70 × 70 × 3) into 1 float number. In this study, a 256 × 256 × 3 dimensional image was reduced to a 32 × 32 × 1-sized tensor. CycleEBGAN calculates the L2-norm in the reduced dimensional space. In this way, we could easily solve the problem of high-dimensional space that affected previous studies.
Some experiments showed L1-norm is better than MSE. EBGAN uses L2-norm, whereas in CycleGAN, use combination of L1 and L2 (cycle loss: L1-norm, discriminator loss: L2-norm). Joseph Harms has also employed the L1.5 loss in an effort to harness the benefits of both the L1 and L2. The L1-norm is more robust to outliers, but its constant gradient when differentiated can sometimes lead to non-convergence during training. We tried both of L1 and L2 loss as energy function (discriminator loss). Our experiments with using the L1 demonstrated that the loss diverge after approximately 80 to 100 epochs. When training CycleEBGAN using the L1-norm with various learning rates, we noticed the discriminator loss initially decreased before it started bouncing and increasing. Therefore, the L2-norm proved to be more effective as an energy function.
This study has the limitation of being conducted on images acquired from a single device at a single medical center. It is believed that this could be improved by learning from additional data. Particularly considering the rapidly advancing image generation GAN methodologies, there is a high potential for improvement in the future. CycleEBGAN had several advantages. First, it rarely generated features absent in the input image (i.e., had a low false-positive rate; Fig. 2). The models generated simulated angiographs that only included the features found in the fundus photographs. However, CycleEBGAN could not generate images by capturing features that ophthalmologists could not detect in fundus photographs. In cases of central serous chorioretinopathy with no pathological change on fundus photographs, CycleEBGAN showed disappointing performance. Also, the CycleEBGAN does not actually find neovascularization, but rather it identifies various features in the fundus that are caused by neovascularization. For example, it detects features such as hard exudates or a decrease in retinal transparency and interprets these as leakage in the angiograph. Therefore, CycleEBGAN cannot completely replace angiography. Thus, it is important to obtain additional information for screening and diagnosis. Second, image synthesis was affected by the images constituting the training set. Only late-phase angiographs were included in this study. Therefore, hard exudate was accurately converted into hyperfluorescence, whereas microaneurysms were ignored (Fig. 4). In contrast, if the training set consisted of early phase fluorescein angiographs without leakage spots, hard exudates tended to be ignored, and microaneurysms were transformed into hyperfluorescence.
Generative adversarial networks (GAN) has demonstrated impressive performance in image translation. For instance, GAN could generate traditional retinal fundus photographs using ultra-widefield images. As we structured image pairs to generate late-phase angiography, thereby enabling more effective learning of leakage. However, when paired with early-phase angiography, GAN could also effectively identify microaneurysms and retinal hemorrhages. In combination with early ICG images, GAN emphasized the choroidal vessels. By altering the paired image for a single fundus photograph, we could identify various features. In ophthalmology, various modalities can be used to measure the eye. Since these examinations have the potential to be transformed into one another, GAN could be employed to complement existing examinations.
Most medical imaging modalities cannot reproduce identical images due to subtle changes in the patient posture and movement of the internal organs. CycleEBGAN effectively translated such inconsistently paired images. In addition, it has a much simpler structure than other paired CycleGAN methods. CycleEBGAN would be helpful for disease screening and data augmentation. This study showed that CycleEBGAN could generate angiographs from fundus photographs, which would be helpful in high-risk patients who require fluorescein angiography, such as diabetic retinopathy patients with nephropathy.
Author contributions
Conceptualization: Taeseen Kang, Kilhwan Shon, Yong Seop Han.
Data curation: Taeseen Kang, Bum Jun Kim, Yong Seop Han.
Formal analysis: Taeseen Kang, Bum Jun Kim, Yong Seop Han.
Investigation: Taeseen Kang, Yong Seop Han.
Methodology: Taeseen Kang, Yong Seop Han.
Project administration: Taeseen Kang, Yong Seop Han.
Resources: Taeseen Kang, Yong Seop Han.
Software: Taeseen Kang, Kilhwan Shon, Sangkyu Park, Yong Seop Han.
Supervision: Taeseen Kang, Sangkyu Park, Yong Seop Han.
Validation: Taeseen Kang, Kilhwan Shon, Sangkyu Park, Woohyuk Lee, Bum Jun Kim, Yong Seop Han.
Visualization: Taeseen Kang, Sangkyu Park, Woohyuk Lee, Yong Seop Han.
Writing – original draft: Taeseen Kang, Kilhwan Shon, Yong Seop Han.
Writing – review & editing: Taeseen Kang, Kilhwan Shon, Woohyuk Lee, Yong Seop Han.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- CT
- computed tomography
- CycleEBGAN
- energy-based cycle-consistent adversarial networks
- CycleGAN
- cycle-consistent adversarial networks
- EBGAN
- energy-based generative adversarial networks
- GAN
- generative adversarial networks
- MRI
- magnetic resonance imaging,
- MSE
- mean square error
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
How to cite this article: Kang TS, Shon K, Park S, Lee W, Kim BJ, Han YS. Translation of paired fundus photographs to fluorescein angiographs with energy-based cycle-consistent adversarial networks. Medicine 2023;102:27(e34161).
Contributor Information
Tae Seen Kang, Email: tskang85@naver.com.
Kilhwan Shon, Email: rlfghks84@gmail.com.
Sangkyu Park, Email: skpark@keei.re.kr.
Woohyuk Lee, Email: lwhyuk@naver.com.
Bum Jun Kim, Email: deankim21c@gmail.com.
References
- [1].Panwar N, Huang P, Lee J, et al. Fundus photography in the 21st century—a review of recent technological advances and their implications for worldwide healthcare. Telemed J E Health. 2016;22:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bennett TJ, Quillen DA, Coronica R. Fundamentals of fluorescein angiography. Curr Concepts Ophthalmol. 2001;9:43–9. [PubMed] [Google Scholar]
- [3].Ffytche T, Shilling J, Chisholm I, et al. Indications for fluorescein angiography in disease of the ocular fundus: a review. J R Soc Med. 1980;73:362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kornblau IS, El-Annan JF. Adverse reactions to fluorescein angiography: a comprehensive review of the literature. Surv Ophthalmol. 2019;64:679–93. [DOI] [PubMed] [Google Scholar]
- [5].Tavakkoli A, Kamran SA, Hossain KF, et al. A novel deep learning conditional generative adversarial network for producing angiography images from retinal fundus photographs. Sci Rep. 2020;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu J-Y, Park T, Isola P, et al. Unpaired image-to-image translation using cycle-consistent adversarial networks. 2017:2223–32.
- [7].Wang T, Lei Y, Fu Y, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. 2021;22:11–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lei Y, Harms J, Wang T, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. 2019;46:3565–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lei Y, Wang T, Tian S, et al. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI. Phys Med Biol. 2020;65:035013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Harms J, Lei Y, Wang T, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46:3998–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jin C-B, Kim H, Liu M, et al. DC2Anet: generating lumbar spine MR images from CT scan data based on semi-supervised learning. Appl Sci. 2019;9:2521. [Google Scholar]
- [12].Dong X, Wang T, Lei Y, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dong X, Lei Y, Tian S, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother Oncol. 2019;141:192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lei Y, Dong X, Wang T, et al. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Phys Med Biol. 2019;64:215017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao J, Mathieu M, LeCun Y. Energy-based generative adversarial network. arXiv preprint arXiv:160903126. 2016.
- [16].Harris CR, Millman KJ, Van Der Walt SJ, et al. Array programming with NumPy. Natur. 2020;585:357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Isola P, Zhu J-Y, Zhou T, et al. Image-to-image translation with conditional adversarial networks. 2017:1125–34.
- [18].He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. 2016:770–8.
- [19].Larsen ABL, Sønderby SK, Larochelle H, et al. Autoencoding beyond pixels using a learned similarity metric. PMLR. 2016:1558–66. [Google Scholar]
- [20].Son J, Park SJ, Jung K-H. Towards accurate segmentation of retinal vessels and the optic disc in fundoscopic images with generative adversarial networks. J Digit Imaging. 2019;32:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kurz C, Maspero M, Savenije MH, et al. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol. 2019;64:225004. [DOI] [PubMed] [Google Scholar]
- [22].Liu Y, Lei Y, Wang T, et al. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy. Med Phys. 2020;47:2472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sorin V, Barash Y, Konen E, et al. Creating artificial images for radiology applications using generative adversarial networks (GANs)–a systematic review. Acad Radiol. 2020;27:1175–85. [DOI] [PubMed] [Google Scholar]
- [24].Lei Y, Dong X, Tian Z, et al. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network. Med Phys. 2020;47:530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jin C-B, Kim H, Liu M, et al. Deep CT to MR synthesis using paired and unpaired data. Sensors. 2019;19:2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fu Y, Lei Y, Wang T, et al. Deep learning in medical image registration: a review. Phys Med Biol. 2020;65:20TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dong X, Lei Y, Wang T, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys Med Biol. 2020;65:055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang T, Lei Y, Tian Z, et al. Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy. J Med Imaging. 2019;6:1–043504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu Y, Lei Y, Wang Y, et al. Evaluation of a deep learning-based pelvic synthetic CT generation technique for MRI-based prostate proton treatment planning. Phys Med Biol. 2019;64:205022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liang X, Chen L, Nguyen D, et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64:125002. [DOI] [PubMed] [Google Scholar]
- [31].Shafai-Erfani G, Lei Y, Liu Y, et al. MRI-based proton treatment planning for base of skull tumors. Int J Part Ther. 2019;6:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu Y, Lei Y, Wang T, et al. MRI-based treatment planning for liver stereotactic body radiotherapy: validation of a deep learning-based synthetic CT generation method. Br J Radiol. 2019;92:20190067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McKenzie EM, Santhanam A, Ruan D, et al. Multimodality image registration in the head-and-neck using a deep learning-derived synthetic CT as a bridge. Med Phys. 2020;47:1094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kida S, Kaji S, Nawa K, et al. Visual enhancement of cone-beam CT by use of CycleGAN. Med Phys. 2020;47:998–1010. [DOI] [PubMed] [Google Scholar]
- [35].Doersch C. Tutorial on variational autoencoders. arXiv preprint arXiv:160605908. 2016.