Abstract
Malignant bone neoplasms can be represented by osteosarcoma (OS), which accounts for 36% of all sarcomas. To reduce tumor malignancy, extensive efforts have been devoted to find an ideal target from numerous candidates, among which RNA-binding proteins (RBPs) have shown their unparalleled competitiveness. With the special structure of RNA-binding domains, RBPs have the potential to establish relationships with RNAs or small molecules and are considered regulators of different sections of RNA processes, including splicing, transport, translation, and degradation of RNAs. RBPs have considerable significant roles in various cancers, and experiments revealed that there was a strong association of RBPs with tumorigenesis and tumor cell progression. Regarding OS, RBPs are a new orientation, but achievements in hand are noteworthy. Higher or lower expression of RBPs was first found in tumor cells compared to normal tissue. By binding to different molecules, RBPs are capable of influencing tumor cell phenotypes through different signaling pathways or other axes, and researches on medical treatment have been largely inspired. Exploring the prognostic and therapeutic values of RBPs in OS is a hotspot where diverse avenues on regulating RBPs have achieved dramatical effects. In this review, we briefly summarize the contribution of RBPs and their binding molecules to OS oncogenicity and generally introduce distinctive RBPs as samples. Moreover, we focus on the attempts to differentiate RBP's opposite functions in predicting prognosis and collect possible strategies for treatment. Our review provides forwards insight into improving the understanding of OS and suggests RBPs as potential biomarkers for therapies.
1. Introduction
Osteosarcoma (OS), the most frequent occurrence in bone sarcomas, is the primary malignant neoplasm of mesenchymal tissue that occurs in the long bone metaphysis, especially in the distal femur and proximal tibia [1]. Stimulating data have concluded the significant impact of age on tumorigenesis and gender preference, in which boys above 13 years old have more preference to attack [2]. People in the sixth decade have a similar risk of affecting OS as those in the 20th, but surprisingly, elderly people have the same survival rates as the young [2]. Similar to other cancer cells, OS cells have a strong tendency in hematogenous metastasis, and the lung is the optimal organ for cells to regrow, which brings a sharp drop to a 5-year event-free survival (EFS). Ground-breaking achievements have been made by the combination of surgical operation, neoadjuvant and adjuvant chemotherapy, and radiotherapy in maintaining 5-year EFS to more than 70% with OS cells localized [3, 4]. Nevertheless, the growth of EFS reached a plateau in the last 30 years [5], and there are abundant challenges in improvement; in particular, less than half of patients who have metastatic tumors will not be disturbed by tumor recurrence in 5 years [2]. Under this circumstance, new strategies to improve the prognosis of OS are urgently needed.
Current studies focusing on the basic mechanism of cancer cell phenotypes, including proliferation, invasion, apoptosis, and epithelial-mesenchymal transcription (EMT), have gradually helped us uncover the mystery of cancer, and during the course, a vast number of proteins and molecules have been reported to have potential functions in influencing cancer cells. Among these proteins and molecules, RNA-binding proteins (RBPs) with multiple regulatory RNAs are considered participants impacting the cancer cell phenotypes and have become a new hotspot in this research area [6, 7]. Stimulating evidence has proven that aberrant expression of RBPs is linked to tumorigenesis [8, 9], and regulating RBP expression are possible approaches to cancer therapy. Herein, we conclude abnormal expression of RBPs, emphasize the combination of RBPs and multiple molecules in regulating cell phenotypes, and present prognostic and therapeutic values, suggesting that RBPs may be possible avenues in OS treatment.
2. Interactions of RBPs and RNAs
2.1. RBPs Bind to RNAs through RNA-Binding Domains
Identified mostly in plants, animals, and microbial species [10], RBPs were originally thought of as proteins binding to coding and noncoding RNAs through RNA-binding domains (RBDs) and formed a stable secondary structure. The classical viewpoint stated that RBDs taking part in ribonucleoprotein complexes were all well-defined, such as the RNA recognition motif (RRM), K homology (KH), DEAD box helicase domains, and others [11–15]. The consensus had been reached for a long time, but new findings that searched out the structure of large ribonucleoproteins consisting of RBPs with unknown RBDs forcefully challenged this point [16]. Advanced studies revealed that previous well-defined RBDs are classified as canonical RBDs and account for just a quarter of total RBDs, whereas other noncanonical RBDs are in the process of searching and concluding their structures and functions. For example, prion-like domain (PrLD) leads to liquid–liquid phase transitions but has a preference for RBP misfolding, which causes neurodegenerative disease [15]. Both canonical and noncanonical RBDs are reliable tools to connect with RNAs and naturally affect RNA metabolism.
2.2. RBPs Regulate RNAs at the Posttranscriptional Level
Apart from its incredible function in forming ribonucleic protein complexes, RBPs act as regulators in posttranscriptional processes, including alternative splicing, alternative polyadenylation, capping, modification, mRNA stabilization, and mRNA localization [17]. Alternative splicing is the major regulation that promotes protein diversity and mRNA stability, and RBPs are determinants for alternative splicing. Abnormal RBPs are partly due to the disturbed splicing process and induce tumor growth [18]. Alternative polyadenylation is utilized to explain the work of generating transcripts and modifying the coding sequence to impact the protein [19]. RNA stability is closely related to its poly(A) tail at the 3′ end, and some RBPs can stabilize the RNA structure by capping the RNA poly(A) tail with an N7 methylated guanine (m7GpppN) [20]. RBPs can also recognize cis motifs or zip codes in the 3′-untranslated region (3′-UTR) of RNA to regulate the localization of RNA in cells. RBPs are necessary to activate RNA translation in all stages of the progress [21]. More collective and sufficient evidence can be found in the previous review [6].
3. RBPs as Hub Regulators in Diverse Cancers
Aberrant expression or disorders of RBPs affect RNA metabolic processing and alter gene expression patterns, driving serious diseases or even cancers. To obtain more information on the fundamental mechanism, much attention has been given to this field. Ectopic expression of RBPs was first noticed in various tumor tissues, and among these tissues, elevated expression was more easily observed. Human RBP La has been demonstrated to be overexpressed in lung, cervical, head and neck, and chronic myelogenous leukemia, leading to various cancers by supporting proliferation, mobility, invasion, and maintaining the survival of cancer cells that may cause chemotherapeutic resistance [22]. Musashi (MSI) proteins were highly expressed in colorectal, lung, and pancreatic cancers, glioblastoma, and several leukemias by operating crucial oncogenic signaling pathways [23]. In addition to those highly expressed RBPs, low expression may also indicate high malignancy. One of the examples is polyC-RNA-binding protein 1 (PCBP1). As a member of the PCBP family proteins, PCBP1 is reported to be reduced in lung cancer, cervical cancer, breast cancer, colon cancer, and liver cancer [24–27], suggesting that altering PCBP1 expression may become a possible therapeutic strategy. Moreover, previous studies have elucidated the identification of RBPs as prognostic factors and treatment targets in cancers. For example, high expression of RNA-binding motif protein 38 (RBM38) represents longer overall survival in ovarian cancer, breast cancer, and glioma [28] but implies poor prognosis in breast cancer [29]. The opposite meanings indicate the dual role of RBM38 in predicting prognosis, and more roles of RBPs in the prognosis and potential therapy of cancers deserve to be explored in future studies.
4. Dysregulation of RBPs in OS
To date, RBPs and cancers are hotspots in investigating the primary mechanism of cancer progression, and OS is one of those investigated cancers that shows a considerable relationship with RBPs. Summarized from several studies, we identified several dysregulated RBPs in OS, and the correlated molecules were also collected (Table 1). From the expression level, four RBPs (RBM10, PUM2, QKI2, and TARBP2) were downregulated in tumor tissues compared with normal tissues, while other identified RBPs were consistently upregulated in OS cells.
Table 1.
Summary of RBPs regulating OS.
| Sarcoma types | RBD | RBPs | Expression in OS | Protein function | Targets | Signaling pathway or axis | Cell lines | Reference |
|---|---|---|---|---|---|---|---|---|
| Osteosarcoma | RRM | RBM10 | – | Alternative splicing, 3′- end processing of some pre-mRNAs, p53 stabilization, cell cycle arrest, and antiviral reactions | Bcl-2, caspase-3, and TNF-α | N | U2OS | [30] |
| HuR | + | mRNAs stability, translation, and nucleus-to-cytoplasm shuttling | AGO2, lncRNA xist | lncRNA XIST/AGO2 | hFOB1.19 | [31] | ||
| YAP | N | HEK293T, SaOS2, 143B, U2OS, MG63 | [32] | |||||
| LncRNA B4GALT1-AS1 | B4GALT1-AS1/HuR/YAP axis | MG63, U2OS, Saos2, 143B | [33] | |||||
| DUXAP10, SOX18 | N | U2OS, SAOS2, HOS, MG63, hFOB1.19 | [34] | |||||
| HMGA1 | miR 142 3p/HMGA1 axis | MG63 | [35] | |||||
| MSI1 | + | Regulation of early asymmetric divisions | p21 and p27 | N | MG63, HOS | [36] | ||
| MSI2 | + | Asymmetric division, hematopoietic, and intestinal systems, self-renew | miR-149, LncRNA DANCR | miR-149/MSI2 axis | hFOB1.19, Saos-2 | [37] | ||
| AUF1 | + | mRNA stability and mRNA translation | miR-141 and miR-146b-5p | N | U2OS, HOS, MG63, 143B, SaOS2, HFSN1 | [38] | ||
| HIF-1α and VEGF-A | N | U2OS, HOS, MG63, 143B | [39] | |||||
| PTBP1 | + | mRNA splicing, translation, stability, and localization | LncRNA HOTTIP | Wnt/β-catenin signaling pathway | hFOB 1.19, U2OS, MG63, Saos-2 | [40] | ||
| Pumilio family | PUM2 | – | mRNA repression; mRNA degradation | miR-590-3p and miR-9 | RhoA/Rock pathway | MG63, Saos2 | [41] | |
| KH domain | hnRNPK | + | RNA splicing, mRNA stabilization, translation, chromatin restruction, and DNA damage response | DDX3 | N | U2OS | [42] | |
| QKI2 | – | Facilitating spliceosomal complex formation, mRNA stability, and localization | miR-17-92 cluster | miR-17-92 cluster/QKI2/β-catenin axis | hFOB 1.19, HOS, 143B, HEK-293TN | [43] | ||
| miR-20a | N | 293TN, 143B, hFOB 1.19 | [44] | |||||
| RRM&KH domain | IGF2BP1 | + | mRNA stability, translatability, or localization | miR-150 | miR-150- IGF2BP1 axis | N | [45] | |
| IGF2BP2 | + | RNA localization, stability, and translation, abate RNAs endonucleases or microRNA-mediated degradation | lncRNA HCG11 | N | MG63, U2OS, HOS, 143B, Saos-2, hFOB1.19 | [46] | ||
| DDX11-AS1 | N | SaOS-2, MG-63, HOS, U2OS, hFOB1.19 | [47] | |||||
| IGF2BP3 | + | mRNA stability, localization | Igf2 | N | AXT, AX | [48] | ||
| dsRBD | TARBP2 | – | miRNA processing and maturation | let-7f-5p | Wnt signaling pathway | Hfob, U2OS, HOS, Saos | [49] | |
| PrLD | FUS | + | RNA splicing | miR-141-3p,LDHB | N | U2OS, Saos-2, 143B, MG-63, HOS, hFOB | [50] | |
| EWSR1 | + | Transcription and RNA splicing | miR-199a-5p, Sox2 | miR-199a-5p/Sox2 axis | MG63 | [51] | ||
| hnRNPA1 | + | Pre-mRNA processes | Fatty acid synthase | Fatty acid synthase /hnRNPA1 | hFOB1.19, 143B, HOS, U2OS | [52] | ||
| DEAD-box | DDX5 | + | Translation, RNA decay, and miRNA processing | miR-671-5p | miR-671-5p/DDX5 axis | hFOB1.19, HOS, MG63, U2OS, and Saos-2 | [53] | |
| miR-214-5p | circ-XPR1/miR-214-5p/DDX5 axis | hFOB1.19, U2OS, U2OS/MTX300, HOS, MG63, 143B, ZOS, and ZOSM | [54] | |||||
| Cap-binding | eIF4E | + | RNA export, translation, RNA stability. and/or sequestration | miR-496 | N | Saos-2, HOS, U-2OS, SOSP-9607, MG63, hFOB1.19 | [55] | |
| La module | LARP1 | + | mRNA stability and mRNA translation | miR-129-5p | miR-129-5p/LARP1 axis | HFOB1.19, HOS, MG63, 143B,U2OS | [56] | |
| RRM&Zinc finger domain | CPEB1 | + | mRNA translation | miR-320a | N | 143B, U2OS, hFOB | [57] | |
| miR-377-3p | miR-377-3p/CPEB1 axis, | N | [58] | |||||
| CPEB4 | + | Polyadenylation, mRNA translation | lncRNA RP11-361F15.2, miR-30c-5p | RP11-361F15.2/miR-30c-5p/CPEB4 loop | MG-63, U2OS, HOS, 13B, hFOB 1.19, RAW264.7 | [59] |
“+” represents upregulated. “−” represents downregulated. “N” not detected.
5. RBPs Act as Oncogenic Proteins or Anticancer Proteins to Regulate OS Cells
Dysregulation of RBPs in OS is the most typical and exterior characteristic of all considerable functions in promoting or suppressing tumor cells, and the mechanism of RBP expression need further explorations. As shown in the stimulating data, RBPs have a significant impact on the junction with OS cell phenotypes, such as proliferation, migration and invasion, EMT, and apoptosis. We cartoon different roles of RBPs in OS progression (Figure 1). Of note, IGF2 mRNA binding protein 3 (IGF2BP3) is consistently regarded as an oncofoetal protein [60], and this assertion was strongly proven in murine OS cells [48]. Abundance analyses revealed that the expression of IGF2BP3 was higher in AXT cells than in AX cells, which show less malignancy than the former, and contributed to activating tumors and promoting tumor cell growth in vivo and in vitro. Human antigen R (HuR) was elucidated to be tightly correlated with OS cell migration and invasion, lying in supporting cells to abandon epithelial cell-like characteristics. However, when HuR was knocked down, this effect was attenuated [32]. In addition, DDX3 interacting with heterogeneous nuclear ribonucleoprotein K (hnRNPK) via its C-terminal region contributed to cell apoptosis, and further studies demonstrated that modified DDX3 lost the ability to initiate DNA damage progression due to unfavorable contact with hnRNPK, which emphasizes the credible efficiency of DDX3-hnRNPK to induce apoptosis [42].
Figure 1.
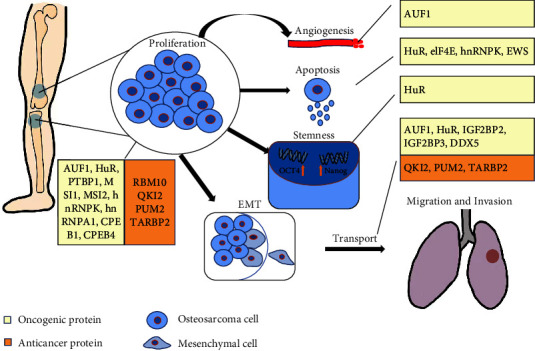
Diverse function of RBPs in OS progression.
RBPs and their binding molecules are regulators of each other, and the combinations act directly on tumor cell progression. Evidence below have shown that regulations on RBPs or their binding molecules can bring out wonderful effects to reduce tumor malignancy. To more specifically amplify the roles of RBPs in suppressing or supporting OS cell progression, conclusions on the interaction between RBPs and molecules will be shown via the classification of molecule types.
5.1. Interaction with Noncoding RNAs
5.1.1. RBPs and MicroRNA (miRNAs)
miRNAs are highly conserved small noncoding RNAs that have been identified as suppressive effectors in regulating mRNAs and are critical for cellular progress and developmental pathways [61]. Collective analyses indicated that RBPs play irreplaceable roles in the canonical and noncanonical pathways to mature miRNAs. The double-stranded RBP DGCR8 and its cooperator RNase III-type enzyme DROSHA were observed to guide primary miRNA (pri-miRNA) into precursors miRNA (pre-miRNA) in the nucleus, which eventually turned into mature miRNA with the help of DICER in the cytoplasm. In the other pathways, miRNA expression was greatly influenced by RBPs in positive or negative effects through binding to the terminal loop of pri-miRNAs and pre-miRNAs [62]. Cooperation of miRNAs and RBPs has a considerable impact on regulating diseases and malignant tumors. For example, upregulated fused in sarcoma (FUS) and downregulated miR-138-5p were correlated with the regulation of angiogenesis with the mediator circ_002136 in glioma cells [63]. In this part, we gather the studies involving the interplay of miRNAs and RBPs in OS to emphasize the favorable effect on cancer regulation.
HuR, also termed ELAVL1, is the only antigen to be expressed in all human tissues among all ELAVL family members [64] containing two tandem RRMs, a hinge region, and a third RRM [65], and the abundant proof was collected to completely explain the regulatory mechanism. Pan et al. [35] found that HuR could harbor high-mobility group AT-hook 1 (HMGA1) and that miR-142-3p could directly bind to the HMGA1 3′ untranslated region. With the characteristics of reducing miR-142-3p expression and increasing HMGA1 expression, HuR notably promotes OS cell progression by suppressing the miR-142-3p/HMGA1 axis (Figure 2(a)).
Figure 2.
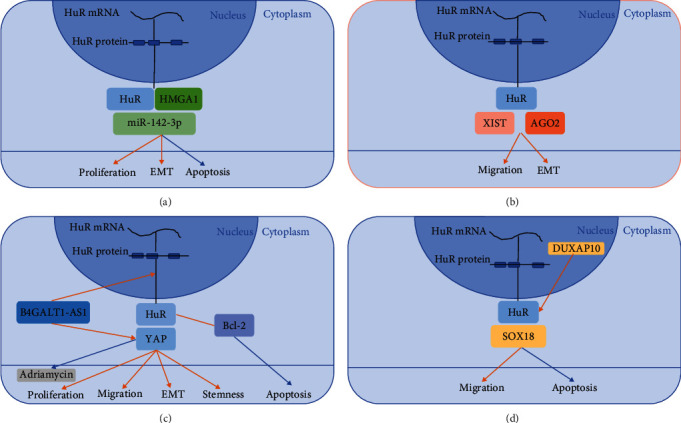
Mechanisms of HuR in regulating OS progression. Red arrows represent positive regulation, while blue ones are negative. (a) HuR binds to HMGA1 and miR-142-3p to promote OS cell proliferation and EMT while reduce cell apoptosis at the same time. (b) LncRNA XIST combined AGO2 and HuR to promote EMT and migration of OS. (c) HuR was positively related to Bcl-2 overexpression and thus reduce apoptosis. B4GALT1-AS1 was contributed to the delivery of HuR from nucleus to cytoplasm and upregulated YAP expression that promotes OS cell proliferation, migration and invasion, EMT, and stemness. The effect of Adriamycin was observed attenuated when YAP overexpressed. (d) DUXAP10 was largely expressed in nucleus and was demonstrated to bind HuR and SOX18 to promote migration and invasion and reduce apoptosis of OS cells.
With two RRMs, AU-binding factor 1 (AUF1) is an RBP rooted in alternative premessenger RNA (pre-mRNA) splicing [66] and is involved in mRNA stability and mRNA translation [64]. In particular, AUF1 is mostly upregulated in cancers, including breast, skin, thyroid, and liver cancers [67, 68], as well as in OS. AUF1 was found to be strongly modulated by miR-141 and miR-146b-5p. The relationship between regulators and the effector was sensitive. AUF1 was shown to be correlated with the proliferation, migration, invasion, and mesenchymal features of OS cells but was reduced by miR-141 and miR-146b-5p [38].
PUM2 is a Pumilio (PUM) protein encoded by PUM genes on chromosome 2 containing a C-terminal highly conserved PUF domain that binds to several protein cofactors to repress direct mRNA and lead to degradation [69]. However, in another aspect, PUM2 has an effort to develop mammalian neural stem cells, epilepsy, and human germ cell progression [70–72], and accumulating evidence proving that PUM2 is correlated with cancer progression [73, 74]. As a tumor suppressor, PUM2 is typically attenuated in OS tissue. Hu et al. [41] designed a series of experiments to determine the significant repression of overexpressed PUM2 on OS cell proliferation, migration, and progression in vivo. Subsequent studies investigated whether PUM2 could regulate and interact with its potential target STARD13 by binding to the STARD13 3′-UTR with miR-590-3p and miR-9. With this binding, PUM2 exerts an inhibitory effect on OS cells.
Quaking (QKI), one of the signal transduction and activation of RNA (STAR) protein family and hnRNPK homology type family [75], is involved in posttranscriptional mRNA processing, including facilitating spliceosomal complex formation, mRNA stability, and localization [76–78]. There are backlogging certifications disclosing that QKI serves as a tumor suppressor gene in different types of cancers [79–81]. QKI2 is one of the isoforms of QKI, showing its surprising role in restraining OS cell progression. Yang et al. [43] demonstrated that the decrease in QKI2 protein expression was regulated by the miR-17-92 cluster in OS and led to a decrease in β-catenin protein levels soon afterward. Based on the discovery, they concluded that the miR-17-92 cluster/QKI2/β-catenin axis promotes OS progression. A few years later, they noticed positive characteristics of miR-20a in OS cell proliferation, migration, and invasion and found that QKI2 is a direct target of miR-20a [44]. QKI2 mRNA can be inhibited by aberrantly increased miR-20a and thus promote OS cell progression, implying that QKI2 has a potential suppressive influence in OS cells.
As a critical regulator of tumor and stem cells, the IGF2 mRNA-binding protein family (IGF2BPs) are oncogenic proteins in cancer [82] containing RRM and KH domains. In the consistent research, a consensus has been reached that IGF2BP1 has a strong conversed potential in cancer-derived cell lines [83, 84]. Mostly expressed in fetal tissues, while in a few adult tissue cases, IGF2BP1 is important in embryogenesis, carcinogenesis, and chemoresistance by influencing the stability, translatability, or localization of direct mRNAs [85, 86]. Consistently, IGF2BP1 is a prognostic factor in abundant human cancers with high expression in tumor tissue. Wang et al. [87] detected the expression levels of miR-150 and IGF2BP1 at the mRNA and protein levels and evaluated the associations of miR-150 and/or IGF2BP1 protein expression. With data analysis, they elucidated the imbalance of the miR-150- IGF2BP1 axis, which contributed to the development, progression, and prognosis of OS, leaving the mechanism problem of the miR-150- IGF2BP1 axis unsolved. Later, miR150 modulated with mesenchymal stem cell-derived exosomes was also verified to downregulate IGF2BP1 and suppress OS development [45].
Similar to the prion domain in yeast, PrLD is characterized by low complexity and exists in nearly 70 RBPs in humans [15]. FUS is an RNA- or DNA-binding protein that was originally detected in human liposarcoma [88] characterized with PrLD. Coupled with splicing regulators or precursor mRNAs, FUS is capable of regulating RNA splicing [89]. Aberrant mutation of FUS can frequently cause amyotrophic lateral sclerosis, a well-known neurodegenerative disease, and FUS aggression has been identified ubiquitously in other neurodegenerative diseases, such as frontotemporal lobar degeneration, polyglutamine diseases, essential tremor and Parkinson's disease [90–93]. Currently, the property of FUS in stabilizing the high expression of messenger RNA (mRNA) lactate dehydrogenase B (LDHB) was illustrated by Wang [50]. LDHB is unfavorably overexpressed in OS cells and can promote tumor malignancy. In a later study, upstream of FUS, miR-141-3p was identified to be downregulated in OS cells, and its upregulation was correlated with the abatement of FUS and LDHB at both the mRNA and protein levels.
The Ewing sarcoma breakpoint region one gene, also known as the Ewing sarcoma breakpoint region 1 (EWSR1) gene, is a member of the FET gene family (together with FUS/TLS and TAF15) and consists of a low-complexity PrLD and an SYGQ-rich N-terminal transactivation domain [94]. EWS was identified as having a critical impact on the pathogenesis of Ewing's sarcoma, and its regulation in OS was uncovered in later research. He and Ding [51] discovered that upregulated EWS was typically characterized in OS and was related to tumor size, advanced stage, and metastasis. The corresponding factor Sox2 was adjusted by EWS to induce OS cell proliferation, colony formation, and apoptosis. Subsequent analysis proved that miR-199a-5p could directly bind to the Sox2 3′-UTR and negatively affect Sox2 biological function in MG63 cells, indicating that EWS regulates OS cells through the miR-199a-5p/Sox2 axis.
DDX5 is typically recognized with an Asp-Glu-Ala-Asp (DEAD) motif [64] and belongs to the largest helicase family. Previous studies elucidated that DDX5 was highly expressed in OS and that multiple factors were involved in the modulation. Chen et al. [53] found that miR-671-5p was able to attenuate DDX5 expression and downregulate long noncoding RNA (lncRNAs) DLEU1 expression to deplete OS cell proliferation and migration. Subsequently, Mao et al. [54], inspired by previous research, identified miR-214-5p as having similar effects on DDX5, which was sponged by circ-XPR1 to induce OS cell progression.
RBPs of eukaryotic translation initiation factor 4E (eIF4E) are characterized by cap-binding domains and are well known to induce tumorigenesis and cancer progression. Qi et al. [55] inferred that eIF4E was negatively influenced by overexpressed miR-496, which was pervasively expressed at low levels in OS cells. Evidence has shown that miR-496 acts as an inhibitor of OS cell proliferation, migration and invasion, and suppresses tumor growth in vivo by depressing the expression of eIF4E, suggesting a potential therapeutic target for OS treatment.
Cytoplasmic polyadenylation element-binding protein (CPEB) is a series of binding proteins that regulate mRNA translation via the 3′-UTR of mRNAs [95], and CPEB1 has been considered a positive factor in tumorigenesis. Wang et al. [57] showed that CPEB1 was upregulated in OS and had a close relationship with OS cell proliferation and metastasis. In structure, the target gene miR-320a binds directly to the 3′-UTR of CPEB1 to indirectly inhibit the biological function of CPEB1 by downregulating its expression. In addition, Zhou et al. [58] enumerated another target gene, miR-377-3p, that binds to both CPEB1 and circ_0003732 and has meaningful effects on suppressing CPEB1 effectiveness.
5.1.2. RBPs and lncRNAs
lncRNAs are a large group of RNAs that are candidates for multiple modifications of cancers through binding molecules. lncRNAs were demonstrated to sponge miRNAs and combine with RBPs to induce cancer progression [96] and control stem cell self-renewal and differentiation [97]. However, well-defined lncRNAs represent only a minor part of all lncRNAs, and more explorations of the foundations of lncRNA regulation remain to be carried out. In this section, we summarize the interplay of lncRNAs and diverse RBPs and/or miRNAs in modulating OS cell phenotypes and reveal the mechanism of RBPs and lncRNAs as much as possible.
The oncogenic role of the lncRNA XIST in OS was previously uncovered [98], but the exact mechanism remains unexplored. Liu et al. [31] verified the linkage of lncRNA XIST and RBP HuR as possible regulators of OS cell progression. Then, they discovered that AGO2 was positively related to mRNA to facilitate the activation of XIST-HuR on EMT and migration in OS cells, indicating that silencing AGO2 may become a possible way to reduce HuR [31] (Figure 2(b)). lncRNA B4GALT1-AS1 has a function inverse to B4GALT1 and was verified as a suppression factor for hepatic gluconeogenesis and lipogenesis [99]. Li et al. [33] discovered the process of HuR translocation from the nucleus to the cytoplasm by virtue of B4GALT1-AS1 and thus upregulated YAP expression, explaining the regulatory effect of B4GALT1-AS1 in promoting OS proliferation, migration, and cell stemness (Figure 2(c)). lncRNA double homeobox A pseudogene 10 (DUXAP10) is especially overexpressed in OS tissues and acts as a promotor of OS cells in vivo and in vitro. Wang et al. [34] demonstrated the correlation between DUXAP10 and HuR, and SOX18 acted as a downstream factor of DUXAP10 to affect OS cell progression (Figure 2(d)).
MSI2 is a translational repressor participating in regulating asymmetric division, hematopoietic, and intestinal systems, self-renewal, etc. [22, 100–103]. MSI2 is generally considered an oncogenic factor in promoting cancer cell proliferation, invasion, migration, and metastasis and predicting prognosis [104–106]. In addition, the function of MSI2 in regulating OS cells has been uncovered based on a deep comprehension of lncRNAs. Zhang et al. [37] proved the tumorigenesis influence of lncRNA anti-differentiating noncoding RNA (DANCR) in OS. Inspired by the correlation of DANCR and the miR-149/MSI2 axis in bladder cancer, they verified the combination of DANCR and miR-149 and the inverse correlation between MSI2 and miR-149. Thus, they concluded that DANCR can target OS cells by regulating the miR-149/MSI2 axis.
Polypyrimidine tract-binding protein 1 (PTBP1) participates in all aspects of the mRNA cascade, and its combination with lncRNAs was verified. Yao et al. [40] demonstrated that lncRNA HOPPIT had a positive correlation in booming PTBP1 expression and regulated KH-type splicing regulatory protein via PTBP1 to activate OS cell proliferation, migration, and invasion but left the limitation on therapy value for further research.
IGF2BP2 is generally considered a participant in the localization, stability, and translation of RNAs and is also capable of abating RNA endonucleases or miRNA-mediated degradation [107, 108]. Recent studies revealed that IGF2P2 was beneficial for inducing OS, suggesting an association between the dysregulation of IGF2BP2 and cancer progression. Gu et al. [46] verified the hypothesis that IGF2BP2 was recruited by lncRNA HCG11 to stabilize p27 Kip1 mRNA and helped HCG11 suppress human OS growth.
CPEB4 belongs to the CPEBs and serves as a promoter on OS because of its high expression in all types of OS cell lines [59]. Yang et al. [59] provided an assumption that the role of lncRNA RP11-361F15.2 was important in regulating OS cell progression by connecting and interacting with its target genes miR-30c-5p and CPEB4. The later experiments proved the inverse correlation of antitumour gene miR-30c-5p and oncogenes CPEB4 or lncRNA RP11-361F15.2, which exploited updated approaches to repress OS cell growth and invasion.
La-related protein 1 (LARP1) is a member of the LARP family with a highly conserved La module. It is doubtless that LARP1 is correlated with high malignancy and poor prognosis in most cancers. Zhang et al. [56] found that miR-129-5p can directly bind to LARP1 and lncRNA KCNQ1OT1 and inhibit the progression of cell proliferation, invasion, and drug resistance when KCNQ1OT1 expression is suppressed.
5.2. Interaction with mRNA
Among all kinds of RNAs, mRNAs are the most common corresponding factors that bind with RBPs through 3′ or 5′-UTRs or coding sequences and take part in posttranscriptional regulation [109]. The emerging roles of mRNA-RBP complexity in regulating malignant cancers show a bright new way to treat cancer, and the discoveries of complexity in OS are summarized below.
As confirmed in 1995 for the first time [110], RNA-binding motif protein 10 (RBM10) is an RBP containing two RRMs and is located at p11.23 on the X chromosome. RBM10 is regarded as a tumor suppressor [111–113], but recent studies have shown that in small cell lung cancer, RBM10 has a reversed exertion with endogenous RBM5 deletion [114]. The dual state makes RBM10 an intricate therapeutic target, and its mechanism in OS is unknown. Han et al. [30] investigated the role of RBM10 in OS and found that the efficiency of RBM10 upregulation was a feasible avenue for inhibiting OS growth. During the exploration of material inhibition, downregulated Bcl-2 and upregulated caspase-3, and TNF-α mRNA were selected as potential regulators to induce cell apoptosis.
Xu et al. [32] affirmed that HuR binds directly to YAP and showed its promotor role by YAP expression. Further study detected the sensitivity development of adriamycin in OS cells by HuR knockout and YAP dependence (Figure 2(c)).
The MSI RBPs were first found in Drosophila by Nakamura et al. [115], and humans have two homologs, Musashi-1 (MSI1) and Musashi-2 (MSI2), consisting of two RRMs [65]. MSI1 is a kind of stem cell-related gene that is involved in early asymmetric divisions generating differentiated cells from neural stem cells or progenitor cells. MSI1 can be upregulated in multiple cancers [116–118] and was elucidated as a prognostic factor in breast cancer and ovarian cancer [119, 120]. Niu et al. [36] obtained evidence showing the appearance of increased expression of MSI1 in OS and proved the inhibition of MSI1 knockdown on proliferation, apoptosis, tumor formation, and cell cycle arrest of OS cells. Then, they found the relationship between knockdown MSI1 and increment of p21 and p27 protein that indicated the effect of MSI1 on cell cycle arrest.
Al-Khalaf and Aboussekhra [39] hypothesized that the combination of AUF1 with VEGF-A mRNA and HIF-1α mRNA was correlated with controlling angiogenesis. AUF1 was highly expressed in OS cells and was demonstrated to induce VEGF-A and HIF-1α, which are typical factors in hypoxic conditions. Thus, antiangiogenic therapies targeting AUF1 could provide effective methods for treating OS.
IGF2BP3 shares 73% amino acid sequence identity with IGF2BP1 and is particularly interesting in tumorigenesis and tumor progression. Compared to human tissue, the expression of IGF2BP3 is highly elevated in stomach adenocarcinoma, skin cutaneous melanoma, lung adenocarcinoma, etc. [121]. Previously, IGF2BP3 was shown to predict metastasis and angiogenic potential in human OS [122, 123]. In a subsequent study, IGF2BP3 could regulate mRNA lgf2 to affect cell tumorigenicity in vivo in murine AXT cells [48], but data analyses refused to affirm the single contribution of lgf2 in regulation, thus leaving further experiments to uncover the mechanism.
TARBPs are a group of RBPs with typical double-stranded RNA-binding domains (dsRBDs). The dsRBD consists of 70–90 amino acids and a characterized αβββα protein folding pattern to recognize dsRNA [124, 125] and has RNA interference, localization, editing, and translocation abilities [65]. TARBP2, the branch of TARBP, acts as an RNA-binding subunit on the RNA-induced silencing complex and is capable of influencing miRNA processing and maturation [126, 127]. Notably, overexpressed or downregulated TARBP2 was found in various cancers [128–130], and the assumption of improvement in tumor angiogenesis and metastasis was verified through dysregulating miRNAs in lung, breast, and liver cancer cell lines [131]. In OS cells, TARBP2 was observed to have decreased expression and target the anticancer gene let-7f-5p [49]. The regulation between TARBP2 and let-7f-5p is negative, where TARBP2 can activate let-7f-5p, and let-7f-5p will also downregulate TARBP2 in return. In particular, researchers have pointed out a feedback loop consisting of TARBP2 and let-7f-5p that strongly modulates the two genes to a low level of expression and is induced by hypoxia. The feedback loop was demonstrated to hamper the activation of the Wnt signaling pathway so that the proliferation and invasion of OS cells were largely promoted.
The expression of lncRNA DDX11 antisense RNA 1 (DDX11-AS1) was found to be elevated in OS cells, and Zhang et al. [47] demonstrated the contribution of DDX11-AS1 to OS cell progression by regulating the RBP DDX11. IGF2BP2 was consistent with its oncogenic role in OS and bound to DDX11-AS1 to upregulate DDX11 expression, whereas the anticancer gene miR-873-5p was attenuated by DDX11-AS1 to reach the same goal.
5.3. Interaction with Other Types of Molecules
hnRNPK is considered a DNA/RNA binding protein as well as a coregulator of p53 participating in RNA splicing, mRNA stabilization, translation, chromatin restructuring, and the DNA damage response [132, 133]. In particular, high expression of hnRNPK is observed in several cancers, and its promoting effect on cell metastasis and its significance in poor prognosis implicate hnRNPK as a cancer promotor [134]. However, arginine-methylated hnRNPK has a negative impact on OS cells by hampering the interaction between DDX3 and hnRNPK. Similarly, inhibition of DDX3 helicase promoted cell apoptosis, and this characteristic belonged to RK-33 [42]. Research gives us a bright new decoy to OS cell apoptosis.
Heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is the hub factor involved in pre-mRNA processing and RNA splicing [135]. FASN is a lipogenic enzyme that contains two polypeptides. Both of these polypeptides are characterized as oncogenes that promote cancer cell progression in several cancers. An initial study elucidated that FANS activates the PI3K/Akt signaling pathway to induce OS cell migration and invasion [136], but the latest research [52] validated that hnRNPA1 was the downstream target of FANS and was positively related to FANS expression. Elevated hnRNPA1 expression was associated with poor prognosis in OS and proliferation, migration, and invasion of OS cells. The therapeutic value of hnRNPA1 deserves to be explored in further studies.
6. Therapeutic Value of RBPs in OS
Generally, cancers are too complex to explore the complete reason why we have no specific treatments for curing various cancers at present. OS is a troublesome challenge that hurts doctors all over the world due to its rarity, complexity, and difficulty. Clinical therapies have significantly improved thanks to the amazing inspiration of combining surgical operation and radiotherapy with neoadjuvant and adjuvant chemotherapy but have failed to renew for a long time. Many articles about sensitive biomarkers have manifested their favorable function in distinguishing the molecular differences in specific tumors and are beneficial to adjust the present treatment strategy. RBPs, with enrichment in biological processes, have been shown to have prognostic and pharmacological value in OS in recent years.
6.1. Evidence of RBPs as Prognostic Factors in OS
The prognostic value of RBPs has been recognized in recent years when RBPs and their associated molecules were considered simultaneously, which will help us renew the understanding of different tumor recoveries. The IGFBP family plays irreplaceable roles in this section (Figure 3). Statistical data from 100 patients with OS showed that miR150/IGF2BP1 expression (P = 0.01, for overall/disease-free survival, Cox proportional hazard model) was a symbolic target for poor response in OS therapy. Kaplan–Meier and log-rank tests suggested that the expression levels of two targets resulted in four degrees of recovery, in which low miR-150 and high IGF2BP1 expression were the worst [87].
Figure 3.
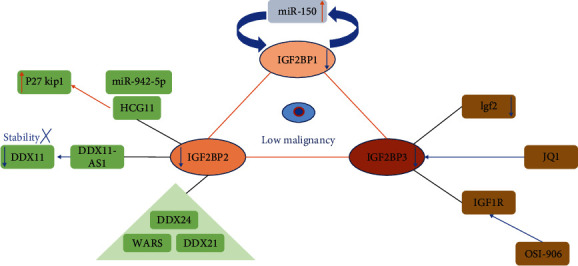
Multiple functions of IGF2BPs in reducing OS malignancy: regulatory mechanism, medicine therapy, and prognosis. Red arrows represent positive regulation, while blue ones are negative.
In addition to predicting patients' recoveries by means of both RBPs and their correlated molecules, systematic analysis is another high-efficiency strategy to select target RBPs. In the beginning of 2021, Li et al. [137] published the results that four key RBPs (DDX24, DDX21, WARS, and IGF2BP2) were identified as prognostic factors in OS. GSE33382, a GEO dataset of OS, first revealed 38 RBPs aberrantly expressed from the comparison between tumor and normal samples, and then enrichment analysis and PPI network analysis were performed to predict the potential functions and connections of these RBPs. Subsequently, univariate and multiple stepwise Cox regression analyses selected DDX24, DDX21, WARS, and IGF2BP2 as hub RBPs, among which WARS was closely correlated with tumor immune infiltration. A risk score and a prognostic model were constructed, and the calculation suggested that both have the expected properties. At the end of 2021, Zhang et al. [138] developed and validated another 10 RBPs (TDRD6, TLR8, NXT2, EIF4E3, RPS27L, CPEB3, RBM34, TERT, RPS29, and ZC3HAV1) as novel predictive factors in OS. With the analyses, the risk assessment model was prominent in connection with metastasis, and the nomogram is the dependable strategy to predict patient survival, indicating that the prognostic function of RBPs has a great influence in the future.
6.2. RBPs in Medicine Therapy
Apart from the prognostic value in predicting OS malignancy, what cannot be ignored is the applications of RBPs in pharmacology existing underlying remedies. High expression of IGF2BP3 can be decreased by the inhibitor JQ1 [139], but low expression of IGF2BP3/IGF1R is sensitive to the inhibitor OSI-906, which specifically targets IGF1R but not IGF2BP3 [48]. Not only inhibitors but also medicines that act on RBPs make researchers continually update pharmacologic ways to reduce tumor cell viability. Some medicines were certified to adapt RBP expression. Knockdown of DDX5 has been reported to be able to moderate camptothecin (CPT)-induced DNA damage, and the interaction protein NONO was observed to coimmunoprecipitate with DDX5. Thus, camptothecin can restore the cpt resistance of OS by breaking down the bond of DDX5 and NONO at the protein level [140]. Adriamycin was either observed to be sensitive to OS cells with HuR or HuR-related lncRNA knockdown [32, 33]. In summary, RBPs have been extensively used in exploring therapeutic strategies in the pharmacology of OS.
7. Perspectives
RBPs are critical regulators that participate in OS cell progression by controlling mRNAs, lncRNA-miRNA complexes, and RBPs or impacting other signaling pathways. However, as the understanding of RBPs in OS gradually accumulates, research on the therapeutic value of RBPs is still too preliminary to certify its contribution in prognosis and therapy strategies. Compared to the numerous RBPs identified, only some of them were validated to have an abnormal expression in cancers, let alone rare tumors such as OS. Therefore, updated certifications on new oncogenic or nononcogenic abilities of RBPs deserve more attention. Additionally, the conclusions in Table 1 are consistent with the notion that RBPs exert positive or negative effects on OS; thus, inhibitors of molecules or techniques, such as the CRISPR-Cas9 system, directly regulating RBP expression or inhibiting RBP function indirectly can be possible ways to suppress OS cell progression. In medicine therapy, drug-resistance cannot be avoided, but the key to solve the problem can be another succedaneum or specific structure to improve the efficiency. Similar to adriamycin, verteporfin is another YAP-TEAD binding inhibitor that can repress OS cell progression by reducing YAP expression and activation in transcription, suggesting its possible application in curing adriamycin-resistant patients, but more reliable data need to be explored in future studies. Estrogen-related receptor alpha was found to employ IGF2BP1 to facilitate mRNA stability and rescue the efficiency of adriamycin in chemotherapy-resistant OS cells [141]. At present, attempts to uncover the characteristics of RBPs in DNA damage and radiotherapy broaden our insights into OS treatment. DNA damage is one method to stimulate cell apoptosis, and RBM10 [30] and hnRNPK with arginine methylation [42] were proven to play support or suppression roles in this progress. Previous studies have identified five correlating RBPs (RUNX2, CDC5L, MDM2, RECQL4, and CDK4) as new clues in pediatric OS for chemotherapy-negative response [142], but further experiments are needed to explain the underlying principles. Radiotherapy uses radiation or X-rays to kill tumor cells, and the efficacy is largely correlated with the oxygen level. Hypoxia fails to stabilize radiation to produce DNA radicals and causes DNA damage that prevents tumor cells from death [143]. Additionally, hypoxia was found to have a positive influence on RBP overexpression. Decreased expression of the interaction loop of TARBP2 and let-7f-5p was verified under hypoxic conditions [49]. To prevent hypoxia and improve radiotherapy effects, bacteria have the potential as a medium. As probiotics, bacteria benefit the host, and many studies have attested to their ability to enhance the radiotherapy effect under hypoxia and protect normal tissue [144]. Taken together, we suggest the development potential of RBPs in OS treatment.
8. Conclusions
RBPs are proteins that bind to multiple RNAs and have been identified as potential targets in cancers by modulating cancer cell proliferation, migration and invasion, apoptosis, and EMT. RBPs have identified complex characteristics in various cancers, and dual effects in regulating OS have recently been discovered. Summarizing previous studies, we naturally concluded that HuR, AUF1, PTBP1, and IGF2BPs were confirmed to promote OS cell progression, while TARBP2, PUM2, QKI2, and RBM10 acted as suppression factors. Of note, the close relationship between RBPs and binding molecules helped us comprehend the mechanism of RBP regulation. mRNA, miRNAs, and lncRNAs have gradually manifested considerable roles in OS by combining and influencing RBPs. Anticancer miRNAs and oncogenic lncRNAs are canonical couples in regulating cancers, and RBPs have dramatic joint mediation on the couples in OS, thus suggesting an indirect way to affect RBP function. Additionally, accumulating evidence has shown the prognostic value of RBPs that are able to distinguish tumor malignancy, and inhibitors or medicines targeting RBPs exert favorable efficiency on OS treatment. However, the existing findings are too small to completely explain the whole mechanism. What can be ascertained is that new avenues to enrich our knowledge of OS are visible in studying RBPs, and new methods to attenuate tumor malignancy are feasible through modulating RBPs or their binding. The fruitful achievements we can see in this regard are forthcoming.
Abbreviations
- OS:
Osteosarcoma
- RBPs:
RNA-binding proteins
- EFS:
Event-free survival
- EMT:
Epithelial-mesenchymal transcription
- RBDs:
RNA-binding domains
- RRM:
RNA recognition motif
- KH:
K homology
- PrLD:
Prion-like domain
- m7GpppN:
N7 methylated guanine
- 3′-UTR:
3′-Untranslated region
- MSI:
Musashi
- PCBP1:
PolyC-RNA-binding protein 1
- RBM38:
RNA-binding motif protein
- IGF2BP3:
IGF2 mRNA-binding protein 3
- HuR:
Human antigen R
- hnRNPK:
Heterogeneous nuclear ribonucleoprotein K
- miRNAs:
MicroRNAs
- AUF1:
AU-binding factor 1
- pre-mRNA:
Premessenger RNA
- PUM:
Pumilio
- QKI:
Quaking
- STAR:
Signal transduction and activation of RNA
- FUS:
Fused in sarcoma
- ALS:
Amyotrophic lateral sclerosis
- LDHB:
Lactate dehydrogenase B
- EWSR1:
Ewing sarcoma breakpoint region 1
- DEAD:
Asp-Glu-Ala-Asp
- eIF4E:
Eukaryotic translation initiation factor 4E
- CPEB:
Cytoplasmic polyadenylation element-binding protein
- lncRNAs:
Long noncoding RNAs
- DUXAP10:
Double homeobox A pseudogene 10
- DANCR:
Anti-differentiating noncoding RNA
- PTBP1:
Polypyrimidine tract-binding protein 1
- LARP1:
La-related protein 1
- RBM10:
RNA-binding motif protein 10
- MS1-1:
Musashi-1
- MSI-2:
Musashi-2
- dsRBDs:
Double-stranded RNA-binding domains
- RISC:
RNA-induced silencing complex
- DDX11-AS1:
DDX11 antisense RNA 1
- hnRNPA1:
Heterogeneous nuclear ribonucleoprotein A1
- CPT:
Camptothecin
- ERRα:
Estrogen-related receptor alpha.
Data Availability
Data are available upon reasonable request to the corresponding author.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
ZYQ: study conception and design, data collection, and draft manuscript preparation; NW and SYL: study conception and design and manuscript editing; KY, TL: study conception and design, data collection, manuscript writing, and visualization. All authors reviewed and approved the final version of the manuscript. Kang Yang and Ziyuan Que these authors contributed equally.
References
- 1.Enneking W. F., Spanier S. S., Goodman M. A. The classic: a system for the surgical staging of musculoskeletal sarcoma. Clinical Orthopaedics and Related Research . 2003;415:4–18. doi: 10.1097/01.blo.0000093891.12372.0f. [DOI] [PubMed] [Google Scholar]
- 2.Schuetze S. M. Chemotherapy in the management of osteosarcoma and Ewing’s sarcoma. Journal of the National Comprehensive Cancer Network . 2007;5(4):449–455. doi: 10.6004/jnccn.2007.0039. [DOI] [PubMed] [Google Scholar]
- 3.Meyers P. A., Schwartz C. L., Krailo M., et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. Journal of Clinical Oncology . 2005;23(9):2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 4.Chou A. J., Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Review of Anticancer Therapy . 2006;6(7):1075–1085. doi: 10.1586/14737140.6.7.1075. [DOI] [PubMed] [Google Scholar]
- 5.Harrison D. J., Geller D. S., Gill J. D., Lewis V. O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Review of Anticancer Therapy . 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 6.Qin H., Ni H., Liu Y., et al. RNA-binding proteins in tumor progression. Journal of Hematology & Oncology . 2020;13 doi: 10.1186/s13045-020-00927-w.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Sun Z., Lei Z., Zhang H.-T. RNA-binding proteins and cancer metastasis. Seminars in Cancer Biology . 2022;86, Part 2:748–768. doi: 10.1016/j.semcancer.2022.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Zou C., Wan Y., He L., et al. RBM38 in cancer: role and mechanism. Cellular and Molecular Life Sciences . 2021;78:117–128. doi: 10.1007/s00018-020-03593-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiße J., Rosemann J., Krauspe V., et al. RNA-binding proteins as regulators of migration, invasion and metastasis in oral squamous cell carcinoma. International Journal of Molecular Sciences . 2020;21(18) doi: 10.3390/ijms21186835.6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keene J. D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proceedings of the National Academy of Sciences . 2001;98(13):7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nature Reviews Genetics . 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cléry A., Blatter M., Allain F. H.-T. RNA recognition motifs: boring? Not quite. Current Opinion in Structural Biology . 2008;18(3):290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Valverde R., Edwards L., Regan L. Structure and function of KH domains. FEBS Journal . 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 14.Linder P., Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nature Reviews Molecular Cell Biology . 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 15.Harrison A. F., Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochemical Journal . 2017;474(8):1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentze M. W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nature Reviews Molecular Cell Biology . 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 17.Choi S. K., Park C., Kim K. E., Kim K. K. An in vitro technique to identify the RNA binding-site sequences for RNA-binding proteins. BioTechniques . 2017;63(1):28–33. doi: 10.2144/000114567. [DOI] [PubMed] [Google Scholar]
- 18.Fu X.-D., Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nature Reviews Genetics . 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkon R., Ugalde A. P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature Reviews Genetics . 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 20.Hai Y., Kawachi A., He X., Yoshimi A. Pathogenic roles of RNA-binding proteins in sarcomas. Cancers . 2022;14(15) doi: 10.3390/cancers14153812.3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W., Deng X., Chen J. RNA-binding proteins in regulating mRNA stability and translation: roles and mechanisms in cancer. Seminars in Cancer Biology . 2022;86, Part 2:664–677. doi: 10.1016/j.semcancer.2022.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommer G., Heise T. Role of the RNA-binding protein La in cancer pathobiology. RNA Biology . 2021;18(2):218–236. doi: 10.1080/15476286.2020.1792677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudinov A. E., Karanicolas J., Golemis E. A., Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clinical Cancer Research . 2017;23(9):2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai M. R., Chacko P., Kesari L. A., Jayaprakash P. G., Jayaram H. N., Antony A. C. Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. Journal of Clinical Pathology . 2003;56(8):569–574. doi: 10.1136/jcp.56.8.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur S., Nakamura T., Calin G., et al. Regulation of BRCA1 transcription by specific single-stranded DNA binding factors. Molecular and Cellular Biology . 2003;23(11):3774–3787. doi: 10.1128/MCB.23.11.3774-3787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H., Vardy L. A., Tan C. P., et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell . 2010;18(1):52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T., Huang X.-H., Dong L., et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Molecular Cancer . 2010;9 doi: 10.1186/1476-4598-9-72.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldstein O., Ben-Hamo R., Bashari D., Efroni S., Ginsberg D. RBM38 is a direct transcriptional target of E2F1 that limits E2F1-induced proliferation. Molecular Cancer Research . 2012;10(9):1169–1177. doi: 10.1158/1541-7786.MCR-12-0331. [DOI] [PubMed] [Google Scholar]
- 29.Chin K., DeVries S., Fridlyand J., et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell . 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Han L.-P., Wang C.-P., Han S.-L. Overexpression of RBM10 induces osteosarcoma cell apoptosis and inhibits cell proliferation and migration. Med Sci (Paris) . 2018;34:81–86. doi: 10.1051/medsci/201834f114. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Zhang Y., Zhang J., et al. Silencing of HuR inhibits osteosarcoma cell epithelial-mesenchymal transition via AGO2 in association with long non-coding RNA XIST. Frontiers in Oncology . 2021;11 doi: 10.3389/fonc.2021.601982.601982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W., Chen C., Xu R., et al. Knockdown of HuR represses osteosarcoma cells migration, invasion and stemness through inhibition of YAP activation and increases susceptibility to chemotherapeutic agents. Biomedicine & Pharmacotherapy . 2018;102:587–593. doi: 10.1016/j.biopha.2018.03.098. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Wang Y., Hu R., Xu R., Xu W. LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Proliferation . 2018;51(6) doi: 10.1111/cpr.12504.e12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G., Zhang Q., Wang Q., et al. Long non-coding RNA DUXAP10 exerts oncogenic properties in osteosarcoma by recruiting HuR to enhance SOX18 mRNA stability. Human Cell . 2022;35:1939–1951. doi: 10.1007/s13577-022-00772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan W., Pang J., Ji B., et al. RNA binding protein HuR promotes osteosarcoma cell progression via suppressing the miR-142-3p/HMGA1 axis. Oncology Letters . 2018;16(2):1475–1482. doi: 10.3892/ol.2018.8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu J., Zhao X., Liu Q., Yang J. Knockdown of MSI1 inhibited the cell proliferation of human osteosarcoma cells by targeting p21 and p27. Oncology Letters . 2017;14(5):5271–5278. doi: 10.3892/ol.2017.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W., Li J.-Z., Tai Q.-Y., Tang J.-J., Huang Y.-H., Gao S.-B. LncRNA DANCR regulates osteosarcoma migration and invasion by targeting miR-149/MSI2 axis. European Review for Medical and Pharmacological Sciences . 2020;24(12):6551–6560. doi: 10.26355/eurrev_202006_21639. [DOI] [PubMed] [Google Scholar]
- 38.Al-Khalaf H. H., Aboussekhra A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. Journal of Biological Chemistry . 2014;289(45):31433–31447. doi: 10.1074/jbc.M114.593004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Khalaf H. H., Aboussekhra A. AUF1 positively controls angiogenesis through mRNA stabilization-dependent up-regulation of HIF-1α and VEGF-A in human osteosarcoma. Oncotarget . 2019;10:4868–4879. doi: 10.18632/oncotarget.27115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao X.-Y., Liu J.-F., Luo Y., Xu X.-Z., Bu J. LncRNA HOTTIP facilitates cell proliferation, invasion, and migration in osteosarcoma by interaction with PTBP1 to promote KHSRP level. Cell Cycle . 2021;20(3):283–297. doi: 10.1080/15384101.2020.1870820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu R., Zhu X., Chen C., Xu R., Li Y., Xu W. RNA-binding protein PUM2 suppresses osteosarcoma progression via partly and competitively binding to STARD13 3’UTR with miRNAs. Cell Proliferation . 2018;51(6) doi: 10.1111/cpr.12508.e12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C.-C., Yang J.-H., Fu S.-L., Lin W.-J., Lin C.-H. Arginine Methylation of hnRNPK Inhibits the DDX3-hnRNPK Interaction to Play an Anti-Apoptosis Role in Osteosarcoma Cells. International Journal of Molecular Sciences . 2021;22(18) doi: 10.3390/ijms22189764.9764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H., Peng Z., Liang M., et al. The miR-17-92 cluster/QKI2/β-catenin axis promotes osteosarcoma progression. Oncotarget . 2018;9(38):25285–25293. doi: 10.18632/oncotarget.23935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H., Li Y., Peng Z., Wang Y. Overexpression of miR-20a promotes the progression of osteosarcoma by directly targeting QKI2. Oncology Letters . 2019;18(1):87–94. doi: 10.3892/ol.2019.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z., Zhou X., Wu J., et al. Mesenchymal stem cell-derived exosomes carrying microRNA-150 suppresses the proliferation and migration of osteosarcoma cells via targeting IGF2BP1. Translational Cancer Research . Sep. 2020;9(9):5323–5335. doi: 10.21037/tcr-20-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu J., Dai B., Shi X., et al. lncRNA HCG11 suppresses human osteosarcoma growth through upregulating p27 Kip1. Aging (Albany NY) . 2021;13(17):21743–21757. doi: 10.18632/aging.203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Lin J., Chen J., et al. DDX11-AS1 contributes to osteosarcoma progression via stabilizing DDX11. Life Sciences . 2020;254 doi: 10.1016/j.lfs.2020.117392.117392 [DOI] [PubMed] [Google Scholar]
- 48.Ueki A., Shimizu T., Masuda K., et al. Up-regulation of Imp3 confers in vivo tumorigenicity on murine osteosarcoma cells. PLOS ONE . 2012;7(11) doi: 10.1371/journal.pone.0050621.e50621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen G., Gu H., Fang T., Zhou K., Xu J., Yin X. Hypoxia-induced let-7f-5p/TARBP2 feedback loop regulates osteosarcoma cell proliferation and invasion by inhibiting the Wnt signaling pathway. Aging (Albany NY) . 2020;12(8):6891–6903. doi: 10.18632/aging.103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L. MiR-141-3p overexpression suppresses the malignancy of osteosarcoma by targeting FUS to degrade LDHB. Bioscience Reports . 2020;40(6) doi: 10.1042/BSR20193404.BSR20193404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He P., Ding J. EWS promotes cell proliferation and inhibits cell apoptosis by regulating miR-199a-5p/Sox2 axis in osteosarcoma. Biotechnology Letters . 2020;42:1263–1274. doi: 10.1007/s10529-020-02859-4. [DOI] [PubMed] [Google Scholar]
- 52.Fu D., Liu S., Liu J., et al. iTRAQ-based proteomic analysis of the molecular mechanisms and downstream effects of fatty acid synthase in osteosarcoma cells. Journal of Clinical Laboratory Analysis . 2021;35(3) doi: 10.1002/jcla.23653.e23653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X., Zhang C., Wang X. Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis. Artificial Cells, Nanomedicine, and Biotechnology . 2019;47(1):3322–3328. doi: 10.1080/21691401.2019.1648285. [DOI] [PubMed] [Google Scholar]
- 54.Mao X., Guo S., Gao L., Li G. Circ-XPR1 promotes osteosarcoma proliferation through regulating the miR-214-5p/DDX5 axis. Human Cell . 2021;34:122–131. doi: 10.1007/s13577-020-00412-z. [DOI] [PubMed] [Google Scholar]
- 55.Qi N.-N., Tian S., Li X., Wang F.-L., Liu B. Up-regulation of microRNA-496 suppresses proliferation, invasion, migration and in vivo tumorigenicity of human osteosarcoma cells by targeting eIF4E. Biochimie . 2019;163:1–11. doi: 10.1016/j.biochi.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Cai W., Zou Y., Zhang H. Knockdown of KCNQ1OT1 inhibits proliferation, invasion, and drug resistance by regulating miR-129-5p-mediated LARP1 in osteosarcoma. BioMed Research International . 2020;2020:10. doi: 10.1155/2020/7698767.7698767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Yang J., Chen P., et al. MicroRNA-320a inhibits invasion and metastasis in osteosarcoma by targeting cytoplasmic polyadenylation element-binding protein 1. Cancer Medicine . 2020;9(8):2833–2845. doi: 10.1002/cam4.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Z., Liu T., Li Z., Wang L. Circ_0003732 promotes osteosarcoma progression through regulating miR-377-3p/CPEB1 axis and Wnt/β-catenin signaling pathway. Anti-Cancer Drugs . 2022;33(1):e299–e310. doi: 10.1097/CAD.0000000000001206. [DOI] [PubMed] [Google Scholar]
- 59.Yang D., Liu K., Fan L., et al. LncRNA RP11-361F15.2 promotes osteosarcoma tumorigenesis by inhibiting M2-Like polarization of tumor-associated macrophages of CPEB4. Cancer Letters . 2020;473:33–49. doi: 10.1016/j.canlet.2019.12.041. [DOI] [PubMed] [Google Scholar]
- 60.Yaniv K., Yisraeli J. K. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene . 2002;287(1-2):49–54. doi: 10.1016/S0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 61.Ebert M. S., Sharp P. A. Roles for microRNAs in conferring robustness to biological processes. Cell . 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michlewski G., Cáceres J. F. Post-transcriptional control of miRNA biogenesis. RNA . 2019;25(1):1–16. doi: 10.1261/rna.068692.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Z., Ruan X., Liu X., et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. Journal of Experimental & Clinical Cancer Research . 2019;38 doi: 10.1186/s13046-019-1065-7.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lukong K. E., Chang K.-W., Khandjian E. W., Richard S. RNA-binding proteins in human genetic disease. Trends in Genetics . 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 65.Mohibi S., Chen X., Zhang J. Cancer the‘RBP’eutics–RNA-binding proteins as therapeutic targets for cancer. Pharmacology & Therapeutics . 2019;203 doi: 10.1016/j.pharmthera.2019.07.001.107390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White E. J. F., Matsangos A. E., Wilson G. M. AUF1 regulation of coding and noncoding RNA. Wiley Interdisciplinary Reviews: RNA . 2017;8(2) doi: 10.1002/wrna.1393.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zucconi B. E., Wilson G. M. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Frontiers in Bioscience-Landmark . 2011;16(6):2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdelmohsen K., Tominaga-Yamanaka K., Srikantan S., Yoon J.-H., Kang M.-J., Gorospe M. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Research . 2012;40(22):11531–11544. doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldstrohm A. C., Tanaka Hall T. M., McKenney K. M. Post-transcriptional regulatory functions of mammalian pumilio proteins. Trends in Genetics . 2018;34(12):972–990. doi: 10.1016/j.tig.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zahr S. K., Yang G., Kazan H., et al. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron . 2018;97(3):520–537.e6. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- 71.Follwaczny P., Schieweck R., Riedemann T., et al. Pumilio2-deficient mice show a predisposition for epilepsy. Disease Models & Mechanisms . 2017;10(11):1333–1342. doi: 10.1242/dmm.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ginter-Matuszewska B., Kusz K., Spik A., et al. NANOS1 and PUMILIO2 bind microRNA biogenesis factor GEMIN3, within chromatoid body in human germ cells. Histochemistry and Cell Biology . 2011;136:279–287. doi: 10.1007/s00418-011-0842-y. [DOI] [PubMed] [Google Scholar]
- 73.Zhang L., Chen Y., Li C., et al. RNA binding protein PUM2 promotes the stemness of breast cancer cells via competitively binding to neuropilin-1 (NRP-1) mRNA with miR-376a. Biomedicine & Pharmacotherapy . 2019;114 doi: 10.1016/j.biopha.2019.108772.108772 [DOI] [PubMed] [Google Scholar]
- 74.Xu L., Zhang B., Li W. Downregulated expression levels of USP46 promote the resistance of ovarian cancer to cisplatin and are regulated by PUM2. Molecular Medicine Reports . 2021;23(4) doi: 10.3892/mmr.2021.11902.263 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Darbelli L., Richard S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. WIREs RNA . 2016;7(3):399–412. doi: 10.1002/wrna.1344. [DOI] [PubMed] [Google Scholar]
- 76.Edenfeld G., Volohonsky G., Krukkert K., et al. The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron . 2006;52(6):969–980. doi: 10.1016/j.neuron.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 77.Doukhanine E., Gavino C., Haines J. D., Almazan G., Richard S. The QKI-6 RNA binding protein regulates actin-interacting protein-1 mRNA stability during oligodendrocyte differentiation. Molecular Biology of the Cell . 2010;21(17):3029–3040. doi: 10.1091/mbc.e10-04-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanchard J., Wanka L., Tung Y.-C., et al. Pharmacologic reversal of neurogenic and neuroplastic abnormalities and cognitive impairments without affecting Aβ and tau pathologies in 3xTg-AD mice. Acta Neuropathologica . 2010;120:605–621. doi: 10.1007/s00401-010-0734-6. [DOI] [PubMed] [Google Scholar]
- 79.Li Z. Z., Kondo T., Murata T., et al. Expression of Hqk encoding a KH RNA binding protein is altered in human glioma. Japanese Journal of Cancer Research . 2002;93(2):167–177. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ichimura K., Mungall A. J., Fiegler H., et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene . 2006;25:1261–1271. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen A.-J., Paik J.-H., Zhang H., et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes & Development . 2012;26(13):1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Müller S., Bley N., Glaß M., et al. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Research . 2018;46(12):6285–6303. doi: 10.1093/nar/gky229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gutschner T., Hämmerle M., Pazaitis N., et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology . 2014;59(5):1900–1911. doi: 10.1002/hep.26997. [DOI] [PubMed] [Google Scholar]
- 84.Fakhraldeen S. A., Clark R. J., Roopra A., et al. Two isoforms of the RNA binding protein, coding region determinant-binding protein (CRD-BP/IGF2BP1), are expressed in breast epithelium and support clonogenic growth of breast tumor cells. Journal of Biological Chemistry . 2015;290(21):13386–13400. doi: 10.1074/jbc.M115.655175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahaira L. G., Katsara O., Pappou E., et al. IGF2BP1 expression in human mesenchymal stem cells significantly affects their proliferation and is under the epigenetic control of TET1/2 demethylases. Stem Cells and Development . 2014;23(20):2501–2512. doi: 10.1089/scd.2013.0604. [DOI] [PubMed] [Google Scholar]
- 86.Yuan P., Meng L., Wang N. SOX12 upregulation is associated with metastasis of hepatocellular carcinoma and increases CDK4 and IGF2BP1 expression. European Review for Medical and Pharmacological Sciences . 2017;21(17):3821–3826. [PubMed] [Google Scholar]
- 87.Wang L., Aireti A., Aihaiti A., Li K. Expression of microRNA-150 and its target gene IGF2BP1 in human osteosarcoma and their clinical implications. Pathology & Oncology Research . 2019;25:527–533. doi: 10.1007/s12253-018-0454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crozat A., Åman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature . 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 89.Alliegro M. C., Alliegro M. A. A nuclear protein regulated during the transition from active to quiescent phenotype in cultured endothelial cells. Developmental Biology . 1996;174(2):288–297. doi: 10.1006/dbio.1996.0074. [DOI] [PubMed] [Google Scholar]
- 90.Neumann M., Roeber S., Kretzschmar H. A., Rademakers R., Baker M., Mackenzie I. R. A. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathologica . 2009;118:605–616. doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doi H., Koyano S., Suzuki Y., Nukina N., Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neuroscience Research . 2010;66(1):131–133. doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 92.Merner N. D., Girard S. L., Catoire H., et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. The American Journal of Human Genetics . 2012;91(2):313–319. doi: 10.1016/j.ajhg.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Labbé C., Rayaprolu S., Soto-Ortolaza A., et al. Investigating FUS variation in Parkinson’s disease. Parkinsonism & Related Disorders . 2014;20, Supplement 1:S147–S149. doi: 10.1016/S1353-8020(13)70035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riggi N., Suvà M. L., Stamenkovic I. Ewing’s Sarcoma. The New England Journal of Medicine . 2021;384:154–164. doi: 10.1056/NEJMra2028910. [DOI] [PubMed] [Google Scholar]
- 95.Caldeira J., Simões-Correia J., Paredes J., et al. CPEB1, a novel gene silenced in gastric cancer: a Drosophila approach. Gut . 2012;61(8):1115–1123. doi: 10.1136/gutjnl-2011-300427. [DOI] [PubMed] [Google Scholar]
- 96.Ferrè F., Colantoni A., Helmer-Citterich M. Revealing protein–lncRNA interaction. Briefings in Bioinformatics . 2016;17(1):106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao C., Xie W., Zhu H., et al. LncRNAs and their RBPs: How to influence the fate of stem cells? Stem Cell Research & Therapy . 2022;13 doi: 10.1186/s13287-022-02851-x.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang W., Shen H., Cao G., Huang J. Long non-coding RNA XIST predicts poor prognosis and promotes malignant phenotypes in osteosarcoma. Oncology Letters . 2019;17(1):256–262. doi: 10.3892/ol.2018.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J., Yang W., Chen Z., et al. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes . 2018;67(4):581–593. doi: 10.2337/db17-0799. [DOI] [PubMed] [Google Scholar]
- 100.Park S.-M., Deering R. P., Lu Y., et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. Journal of Experimental Medicine . 2014;211(1):71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wuebben E. L., Mallanna S. K., Cox J. L., Rizzino A. Musashi2 is required for the self-renewal and pluripotency of embryonic stem cells. PLOS ONE . 2012;7(4) doi: 10.1371/journal.pone.0034827.e34827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang S., Li N., Yousefi M., et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nature Communications . 2015;6 doi: 10.1038/ncomms7517.6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He L., Zhou X., Qu C., et al. Musashi2 predicts poor prognosis and invasion in hepatocellular carcinoma by driving epithelial–mesenchymal transition. Journal of Cellular and Molecular Medicine . 2014;18(1):49–58. doi: 10.1111/jcmm.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang C., Zhang W., Wang L., et al. Musashi-2 promotes migration and invasion in bladder cancer via activation of the JAK2/STAT3 pathway. Laboratory Investigation . 2016;96(9):950–958. doi: 10.1038/labinvest.2016.71. [DOI] [PubMed] [Google Scholar]
- 105.Yang K., Du J., Shi D., et al. Knockdown of MSI2 inhibits metastasis by interacting with caveolin-1 and inhibiting its ubiquitylation in human NF1-MPNST cells. Cell Death & Disease . 2020;11 doi: 10.1038/s41419-020-2703-x.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ito T., Kwon H. Y., Zimdahl B., et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature . 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dai N., Rapley J., Angel M., Yanik M. F., Blower M. D., Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes & Development . 2011;25:1159–1172. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai N. The diverse functions of IMP2/IGF2BP2 in metabolism. Trends in Endocrinology & Metabolism . 2020;31(9):670–679. doi: 10.1016/j.tem.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 109.Di Liegro C. M., Schiera G., Di Liegro I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review) International Journal of Molecular Medicine . 2014;33(4):747–762. doi: 10.3892/ijmm.2014.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nagase T., Seki N., Tanaka A., Ishikawa K.-I., Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Research . 1995;2(4):167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 111.Jung J. H., Lee H., Zeng S. X., Lu H. RBM10, a new regulator of p53. Cells . 2020;9(9) doi: 10.3390/cells9092107.2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hernández J., Bechara E., Schlesinger D., Delgado J., Serrano L., Valcárcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biology . 2016;13(4):466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamada H., Tsutsumi K., Nakazawa Y., Shibagaki Y., Hattori S., Ohta Y. Src family tyrosine kinase signaling regulates FilGAP through association with RBM10. PLOS ONE . 2016;11(1) doi: 10.1371/journal.pone.0146593.0146593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loiselle J. J., Roy J. G., Sutherland L. C. RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5. PLOS ONE . 2017;12(6) doi: 10.1371/journal.pone.0180258.e0180258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakamura M., Okano H., Blendy J. A., Montell C. Musashi, a neural RNA-binding protein required for drosophila adult external sensory organ development. Neuron . 1994;13(1):67–81. doi: 10.1016/0896-6273(94)90460-X. [DOI] [PubMed] [Google Scholar]
- 116.Sanchez-Diaz P. C., Burton T. L., Burns S. C., Hung J. Y., Penalva L. O. F. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer . 2008;8 doi: 10.1186/1471-2407-8-280.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan L.-F., Dong W.-G., Jiang C.-Q., Xia D., Liao F., Yu Q.-F. Expression of putative stem cell genes Musashi-1 and β1-integrin in human colorectal adenomas and adenocarcinomas. International Journal of Colorectal Disease . 2010;25:17–23. doi: 10.1007/s00384-009-0791-2. [DOI] [PubMed] [Google Scholar]
- 118.Wang X.-Y., Yu H., Ilona Linnoila1 R., et al. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget . 2013;4(5):739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X.-Y., Penalva L. O. F., Yuan H., et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Molecular Cancer . 2010;9 doi: 10.1186/1476-4598-9-221.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen P.-X., Li Q.-Y., Yang Z. Musashi-1 expression is a prognostic factor in ovarian adenocarcinoma and correlates with ALDH-1 expression. Pathology & Oncology Research . 2015;21:1133–1140. doi: 10.1007/s12253-015-9943-6. [DOI] [PubMed] [Google Scholar]
- 121.Mancarella C., Scotlandi K. IGF2BP3 from physiology to cancer: novel discoveries, unsolved issues, and future perspectives. Frontiers in Cell and Developmental Biology . 2020;7 doi: 10.3389/fcell.2019.00363.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Do S.-I., Kim Y. W., Park H.-R., Park Y.-K. Expression of insulin-like growth factor-II mRNA binding protein 3 (IMP3) in osteosarcoma. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics . 2008;17(6):269–272. doi: 10.3727/096504008786991639. [DOI] [PubMed] [Google Scholar]
- 123.Chen P., Wang S.-J., Wang H.-B., et al. The distribution of IGF2 and IMP3 in osteosarcoma and its relationship with angiogenesis. Journal of Molecular Histology . 2012;43:63–70. doi: 10.1007/s10735-011-9370-2. [DOI] [PubMed] [Google Scholar]
- 124.Bycroft M., Grünert S., Murzin A. G., Proctor M., St Johnston D. NMR solution structure of a dsRNA binding domain from drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. The EMBO Journal . 1995;14(14):3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gleghorn M. L., Maquat L. E. ‘Black sheep’ that don’t leave the double-stranded RNA-binding domain fold. Trends in Biochemical Sciences . 2014;39(7):328–340. doi: 10.1016/j.tibs.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Daniels S. M., Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiology and Molecular Biology Reviews . 2012;76(3):652–666. doi: 10.1128/MMBR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gatignol A., Lainé S., Clerzius G. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology . 2005;2 doi: 10.1186/1742-4690-2-65.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Caramuta S., Lee L., Özata D. M., et al. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocrine-Related Cancer . 2013;20(4):551–564. doi: 10.1530/ERC-13-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin X., Wu M., Liu P., et al. Up-regulation and worse prognostic marker of cytoplasmic TARBP2 expression in obstinate breast cancer. Medical Oncology . 2014;31 doi: 10.1007/s12032-014-0868-9.868 [DOI] [PubMed] [Google Scholar]
- 130.Kim M. S., Oh J. E., Kim Y. R., et al. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. The Journal of Pathology . 2010;221(2):139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 131.Zhou M., Lu W., Li B., Liu X., Li A. TARBP2 promotes tumor angiogenesis and metastasis by destabilizing antiangiogenic factor mRNAs. Cancer Science . 2021;112(3):1289–1299. doi: 10.1111/cas.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barboro P., Ferrari N., Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Letters . 2014;352(2):152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 133.Wang Z., Qiu H., He J., et al. The emerging roles of hnRNPK. Journal of Cellular Physiology . 2020;235(3):1995–2008. doi: 10.1002/jcp.29186. [DOI] [PubMed] [Google Scholar]
- 134.Gao R., Yu Y., Inoue A., Widodo N., Kaul S. C., Wadhwa R. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. Journal of Biological Chemistry . 2013;288(21):15046–15056. doi: 10.1074/jbc.M113.466136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roy R., Huang Y., Seckl M. J., Pardo O. E. Emerging roles of hnRNPA1 in modulating malignant transformation. WIREs RNA . 2017;8(6) doi: 10.1002/wrna.1431.e1431 [DOI] [PubMed] [Google Scholar]
- 136.Liu Z. L., Mao J. H., Peng A. F., et al. Inhibition of fatty acid synthase suppresses osteosarcoma cell invasion and migration via downregulation of the PI3K/Akt signaling pathway in vitro. Molecular Medicine Reports . 2013;7(2):608–612. doi: 10.3892/mmr.2012.1220. [DOI] [PubMed] [Google Scholar]
- 137.Li B., Fang L., Wang B., Yang Z., Zhao T. Identification of prognostic RBPs in osteosarcoma. Technology in Cancer Research & Treatment . 2021;20 doi: 10.1177/15330338211004918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang J., Miao X., Wu T., Jia J., Cheng X. Development and validation of Ten-RNA binding protein signature predicts overall survival in osteosarcoma. Frontiers in Molecular Biosciences . 2021;8 doi: 10.3389/fmolb.2021.751842.751842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mancarella C., Pasello M., Manara M. C., et al. Insulin-like growth factor 2 mRNA-binding protein 3 influences sensitivity to Anti-IGF system agents through the translational regulation of IGF1R. Frontiers in Endocrinology . 2018;9 doi: 10.3389/fendo.2018.00178.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao X., Bao M., Zhang F., Wang W. Camptothecin induced DDX5 degradation increased the camptothecin resistance of osteosarcoma. Experimental Cell Research . 2020;394(2) doi: 10.1016/j.yexcr.2020.112148.112148 [DOI] [PubMed] [Google Scholar]
- 141.He Q., Hao P., He G., et al. IGF2BP1-regulated expression of ERRα is involved in metabolic reprogramming of chemotherapy resistant osteosarcoma cells. Journal of Translational Medicine . 2022;20 doi: 10.1186/s12967-022-03549-7.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Martin J. W., Chilton-MacNeill S., Koti M., van Wijnen A. J., Squire J. A., Zielenska M. Digital expression profiling identifies RUNX2, CDC5L, MDM2, RECQL4, and CDK4 as potential predictive biomarkers for neo-adjuvant chemotherapy response in paediatric osteosarcoma. PLOS ONE . 2014;9(5) doi: 10.1371/journal.pone.0095843.e95843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harris A. L. Hypoxia—a key regulatory factor in tumour growth. Nature Reviews Cancer . 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 144.Kouhsari E., Ghadimi-Daresajini A., Abdollahi H., et al. The potential roles of bacteria to improve radiation treatment outcome. Clinical and Translational Oncology . 2018;20:127–139. doi: 10.1007/s12094-017-1701-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request to the corresponding author.


