Abstract
The molecular mechanisms of angiogenesis have been intensely studied, but many genes that control endothelial behavior and fate still need to be described. Here, we characterize the role of Apold1 (Apolipoprotein L domain containing 1) in angiogenesis in vivo and in vitro. Single-cell analyses reveal that - across tissues - the expression of Apold1 is restricted to the vasculature and that Apold1 expression in endothelial cells (ECs) is highly sensitive to environmental factors. Using Apold1−/− mice, we find that Apold1 is dispensable for development and does not affect postnatal retinal angiogenesis nor alters the vascular network in adult brain and muscle. However, when exposed to ischemic conditions following photothrombotic stroke as well as femoral artery ligation, Apold1−/− mice display dramatic impairments in recovery and revascularization. We also find that human tumor endothelial cells express strikingly higher levels of Apold1 and that Apold1 deletion in mice stunts the growth of subcutaneous B16 melanoma tumors, which have smaller and poorly perfused vessels. Mechanistically, Apold1 is activated in ECs upon growth factor stimulation as well as in hypoxia, and Apold1 intrinsically controls EC proliferation but not migration. Our data demonstrate that Apold1 is a key regulator of angiogenesis in pathological settings, whereas it does not affect developmental angiogenesis, thus making it a promising candidate for clinical investigation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10456-023-09870-z.
Keywords: Angiogenesis, Endothelial cell, EC proliferation, Hindlimb ischemia, Stroke, Tumor
Introduction
Angiogenesis, the formation of new blood vessels from existing ones, is crucial for normal embryonic development and thus essential for life [1–3]. In healthy tissues, however, angiogenesis only occurs in the muscle [4] and in specific, highly activated brain areas following exercise training [5] and in the uterus where it ensures endometrial growth during the menstrual cycle [6, 7]. Angiogenesis is a strictly timed, multi-step process that is tightly controlled by an intimate interplay between several pro- and anti-angiogenic growth factors that regulate the activity of complex cellular signaling networks [8]. Those ultimately orchestrate changes in the function and fate of endothelial cells (ECs), the main cell type composing the vascular wall. Decades of intense research, mostly using models of developmental angiogenesis, have led to the discovery of a plethora of angiogenic master regulators [2, 3]. But undoubtedly, many genes that control EC behavior, fate, and/or EC interactions with other cells to ensure the generation of functional blood vessels still need to be described [9].
In many pathological conditions, angiogenesis plays a crucial role. On the one hand, angiogenesis secures tissue viability and recovery during and after injuries often associated with hypoxia, such as in wounds, upon ischemia, or after stroke, thereby ensuring tissue repair [10–12]. On the other hand, the “angiogenic switch” in cancer fuels tumor growth and malignancy [13, 14]. In pathological conditions, angiogenesis often relies on the reactivation of developmental pathways that control physiological angiogenesis [15, 16]. For example, many tumors activate the expression of vascular endothelial growth factor (VEGF), a master regulator of physiological angiogenesis, in response to environmental signals, such as hypoxia or following oncogenic (or loss of tumor suppressor function) mutations. As a result, anti-angiogenic drugs targeting VEGF, but also other crucial regulators of developmental angiogenesis, have been extensively tested for anti-cancer treatment. Despite some successes, their efficacy to slow down tumor progression and improve patient survival remains limited due to serious side effects, toxicity, and acquired resistance [13, 17–19]. Therefore, identifying novel molecular targets that are redundant for physiological angiogenesis and endothelial survival, but specifically regulate pathological angiogenesis, holds great therapeutic potential.
Apold1 (Apolipoprotein L domain containing 1, also known as Verge) is a gene that is thought to be selectively expressed in ECs [20]. Apold1 expression rapidly increases under metabolically demanding conditions, e.g., in the rat heart after vigorous physical activity [21], in human muscle after acute aerobic and resistance exercise [22, 23], and in the rodent brain in response to activating/stressful stimuli [24, 25] and seizures [20]. It appears that this response is driven by increased oxygen demand, as Apold1 mRNA increases in ischemic brains [20, 26] and under hypoxic conditions in various tissues [27–30]. Angiogenic growth factors such as FGF2 and (to a lesser extent) Angiopoietin-2 also activate Apold1 [20]. But while in vitro studies suggested that Apold1 controls vascular permeability [20, 31] and the secretion of Weibel–Palade bodies [31], little is known about the functional role of Apold1 in vivo. In this respect, a mutation in Apold1 was recently described in a family of patients with a novel inherited bleeding disorder [31], yet Apold1 knockout mice (Apold1−/−) present with higher platelet reactivity and a prothrombotic phenotype [32]. In a mouse model of neonatal stroke, Apold1−/− pups showed reduced angiogenesis after stroke and impaired long-term functional recovery [26]. However, in adult mice, where Apold1 expression is much lower, acute stroke caused similar size lesions and comparable functional impairment in Apold1−/− mice and wild-type controls [33]. Importantly, the role of Apold1 in recovery from stroke, including angiogenesis and revascularization, was not assessed. Thus, whether Apold1 contributes to angiogenesis in adulthood is not known.
Here we investigated the role of Apold1 in angiogenesis in vivo and in vitro. We report that Apold1 is dispensable for developmental angiogenesis, but it is necessary for functional revascularization during recovery from ischemia in the central nervous system and in the periphery. We also find that human tumor ECs express strikingly higher levels of Apold1 and that Apold1 deletion in mice reduces tumor growth by limiting EC proliferation.
Methods
Animals
All experiments were conducted in accordance with the Swiss federal guidelines for the use of animals in research and under licenses ZH153-18, ZH050-21, and ZH115-19. Mice were housed in groups of 4–5 per cage in a temperature- and humidity-controlled facility on a 12-h light–dark cycle, with food and water ad libitum and used for experiments at the age of 8–15 weeks. Only for behavior experiments the light cycle was reversed so that after two weeks of adaptation mice were tested during the active phase of the light cycle. Experiments were performed in male and female Apold1 knockout mice (C57BL/6 J-Apold1tm1(LacZ)Pfw, here referred to as Apold1−/−), in heterozygous mice (Apold1−/−), and in wild-type (WT) control C57Bl6/J. In the Apold1−/− mice, the open reading frame was replaced with a beta-gal/neo cassette [33]. By breeding heterozygous (Apold1±) males and females, littermates with different genotypes were used in in vivo experiments. Genotyping was performed by PCR as described previously [32], using the following primers: F2 5′-CTCTAGCCTAGGGCAGCAAC-3′; wtR1 5′-GAGAGAGGTCGGACGTGATG-3′; and LacZR 5′-GGCGATTAAGTTGGGTAACG-3′. PCR cycling was performed at 95 °C for 5 min followed by 45 cycles at 95 °C for 30 s, 64 °C for 30 s, 72 °C for 30 s, and finally 72 °C for 5 min.
RNA sequencing and data analysis
Sequencing
For the RNA sequencing of samples from FACS-sorted mECs incubated under angiogenic conditions for 16 h, the following protocol was used: The TruSeq-stranded RNA kit (Illumina Inc.) was used according to the manufacturer’s protocol. The mRNA was purified by polyA selection, chemically fragmented, and transcribed into cDNA before adapter ligation. Single-end (100nt) sequencing was performed with Illumina Novaseq 6000. Samples were all run on the same lane and demultiplexed. A sequencing depth of ~ 20 M reads per sample was used.
For RNA sequencing of FACS-sorted mECs without prior incubation, cDNA conversion was performed using the Smart-seq2 protocol [34] using 800-pg mRNA as input. Library preparation and paired-ended sequencing (150 bp) were performed by Novogene (Cambridge, UK) on an Illumina Novaseq 6000 machine. A sequencing depth of ~ 20-M read pairs per sample was achieved.
Analysis
Adapters were trimmed using cutadapt [35] (v 1.15) with a maximum error rate of 0.05 and a minimum length of 15. Kallisto [36] (v0.44.0) was used for pseudoalignment of reads on the transcriptome level using the genecode.vM17 assembly with 30 bootstrap samples. For single-end samples an estimated fragment length of 200 ± 20 was used. For differential gene expression (DGE) analysis we aggregated reads of protein coding transcripts on the gene level and used R (v. 4.0.3) with the package “edgeR” (v 3.32.1) [37] for analysis. A filter was used to remove genes with low expression prior to DGE analysis. EdgeR was then used to calculate the normalization factors (TMM method) and estimate the dispersion (by weighted likelihood empirical Bayes). For two-group comparisons the genewise exact test was used. For multiple testing correction the Benjamini–Hochberg false discovery rate (FDR) method was used. GO analyses on significant genes vs. the background of all tested genes were performed using the R package “topGO” (v 2.42.0) with a node size of 10 and a fisher p-value cutoff of 0.05 as determined by the algorithm “weight01.” The mus musculus mapping “org.Mm.eg” was used across all three categories (biological processes, molecular function, and cellular component).
Analyses of Apold1 expression and HIF binding based on Mimura et al. [38]
Microarray data produced following both a hypoxic time series and siRNA knockdowns were obtained from the NCBI gene expression omnibus (accession: GSE35932) published by Mimura et al. [38]. To determine the relative expression values, “RMA” was used for background correction, “qspline” for normalization, and “liwong” as a summary method. Probes corresponding to APOLD1 and VEGFA were identified using the hgu133plus2.db R package (version: version 3.2.3). Four probes were identified for VEGFA from which the mean values were calculated to represent the relative expression level of the gene.
Raw Chip-seq data for the identification of HIF1-binding sites in HUVECs for both normoxic and hypoxic conditions were obtained via the accession GSE39089 [38]. Reads were trimmed using trimmomatic [39] and aligned to the GRCh38 genome using bowtie2 [40] with default parameters. Coverage tracks were normalized using Counts Per Million (CPM) reads mapped, and visualization was made with the epiwraps package.
Analyses of single-cell RNA-seq data from muscle tissue
Single-cell RNA-seq data of mononuclear cells from hindlimb skeletal muscle in wild-type mice was obtained from the GEO database via the accession GSE110878 [41]. All single-cell analyses and visualization were carried out using Seurat (version 3.1.0). The two data sets available for uninjured wild-type mice were integrated using Seurat’s standard workflow. Integration anchors were identified following canonical correlation analysis for dimension reduction. T-SNE was applied to the integrated dataset for visualization and cluster identification. The makers used to assign cell-type identities to each cluster are shown in Table 1. These labels were chosen as to reflect those findings in the original Giordani paper and to distinguish APOLD1 expressing cell types [41].
Table 1.
Overview of cell types and their respective markers used for cell annotation
| Cell type | Markers |
|---|---|
| T cells | Cd3d, Cd3e, Cd3g, Cd8a, Cd4, Ptprc |
| B cells | Cd19, Cd22, Ms4a1, Ptprc |
| Neutrophils | Itgam, Cd14, S100a8, S100a9m |
| Scx + cells | Scx, Tnmd, Col1a1, Tnc |
| FAPs | Ly6a, Ly6e, Pdgfra, Dcn |
| Glial cells | Plp1, Kcna1, S100b, Mbp |
| MuSCs | Pax7, Myod1, Vcam1, Sdc4 |
| SMCs | Acta1, Pdgfrb, Myl9, Acta2 |
| Pericytes | Pdgfrb, Notch3, Cspg4, Kcnj8 |
| Endothelial cells | Pecam1, Cdh5 |
Analyses of brain single-cell RNA-seq datasets:
For the dataset from Wu et al., 2017 [42] (Fig. 1J–K), the re-analysis described in Floriou-Servou et al., 2021 [43] was used. For the dataset from von Ziegler et al., 2022 [44] (Fig. 1L–M), the processed data were obtained from the github repository of the original publication, and vascular cells were reassigned to more specific categories in a supervised fashion. The counts of markers for SMCs, ECs, and pericytes (see Table 1) were summed for each cell type, and cells which did not have at least twice as much signal for markers of one cell type as for the others were excluded as ambiguous cells. Cells were then assigned to the cell type with the maximum signal.
Fig. 1.
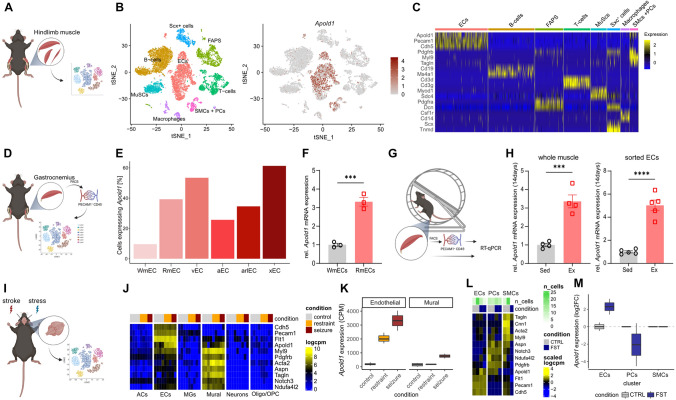
Apold1 gene expression in muscle and brain is restricted to the vasculature, including endothelial cells, pericytes, and smooth muscle cells and triggered by environmental stimuli. A Experimental design [41]. B t-Stochastic neighbor embedding (tSNE) plots of distribution of main muscle-resident populations reanalyzed [41] (ECs endothelial cells; PCs pericytes; SMCs smooth muscle cells). C RNA expression heat map for given cell populations (column) and genes (row) sorted by population specific gene expression reanalyzed [41]. D Experimental design [52]. E Percentage of Apold1 RNA expression in all endothelial populations (WmEC white mECs; RmEC red mECs; vEC venous ECs; aEC arterial ECs; arlEC arteriolar ECs; xEC unknown ECs) reanalyzed [52]. F Relative Apold1 mRNA expression in WmECs and RmECs (n (WmECs/RmECs−) = 3/3). G Experimental design. H Relative Apold1 mRNA expression in whole muscle and sorted ECs after 14 days of voluntary wheel running (n (Sed/Ex) = 4/4). I Experimental design [42, 44]. J RNA expression heat map for control, restraint stress, and seizure conditions (column) and genes (rows) reanalyzed [42]. K Apold1 RNA expression in control, restraint stress, and seizure conditions in ECs and mural cells in the mice brain [42]. L RNA expression heat map of ECs, PCs, and SMCs in mice brain exposed to control (CTRL) and forced swim test conditions (FST) [44]. M Apold1 RNA expression upon forced swim test (FST) in ECs, PCs, and SMCs of the brain compared to control conditions [44]. Student’s t test in F and H (***p < 0.001, **** = p < 0.0001). The data shown are mean ± SEM
For the dataset from Zeisel et al., 2018 [45] (Fig. S2), we downloaded the L5 dataset from http://mousebrain.org/adolescent/downloads.html and used the TaxonomyRank4. To perfect the vascular cell types, we isolated cells that had been assigned to cell types containing any of the words smooth muscle, pericytes, endothelial, or vascular and reanalyzed them separately using scran 1.18.7 [46]. We used 20 components of a PCA based on the 2000 most expressed genes, followed by Louvain clustering on the KNN graph. Clusters were then manually linked to cell types using the markers from Table 1. For other cell types, the original annotation was used.
Postnatal retinal angiogenesis model
To assess postnatal retinal angiogenesis, WT and Apold1−/− pups were sacrificed at P6. Thereafter, eyes were harvested for retinal dissection and analysis of blood vessel outgrowth. Different parameters were automatically quantified with the angiogenesis plug in tool of ImageJ (NIH).
Stroke induction
Mice were anesthetized using isoflurane (5% induction, 1.5–2% maintenance; Attane, Provet AG). Analgesic (Novalgin, Sanofi) was administered 1 d prior to the start of the procedure via drinking water. A photothrombotic stroke to unilaterally lesion the sensorimotor cortex was induced on the right hemisphere, as previously described [47, 48]. Briefly, animals were placed in a stereotactic frame (David Kopf Instruments), the surgical area was sanitized, and the skull was exposed through a midline skin incision. A cold light source was positioned over the right forebrain cortex (anterior/posterior: − 1.5 −/ + 1.5 mm and medial/lateral 0 −/ + 2 mm relative to Bregma). 5 min prior to illumination, Rose Bengal (10 mg/ml, in 0.9% NaCl, Sigma) was injected intraperitoneally and the region of interest was subsequently illuminated through the intact skull for 8 min (mild stroke) or 10.5 min (severe stroke). To restrict the illuminated area, an opaque template with an opening of 3 × 1.5 mm (mild stroke) or 3 × 4 mm (severe stroke) was placed directly on the skull. The wound was closed using a 6/0 silk suture and animals were allowed to recover. For postoperative care, all animals received analgesics (Novalgin, Sanofi) for at least 3 days after surgery.
Stroke volume quantification
Stroke volume was calculated from coronal brain sections stained with NeuroTrace 640/660 (ThermoFischer). Brain sections at six defined landmarks (2.5, 1.5, 0.5, − 0.5, − 1.5, − 2.5 mm in relation to bregma) were analyzed for depth of the cortical lesion. The dorso-ventral, medio-lateral, and anterior–posterior stroke extent were then used to modulate a precise ellipsoid with the coordinates relative to Bregma.
RNA extraction
RNA extraction of brain tissue was carried out using Qiagen RNeasy kit according to the manufacturer’s recommendations. qPCR was performed using SYBR green kit (iTaq Universal SYBR Green Supermix from Bio-rad) containing 0.5 µM of each primer with the following cycling conditions (hold stage: 95 °C, 10 min, 1 cycle; PCR stage (95 °C, 15 s, 60 °C 1 min; 95 °C 15 s, 40 cycles; Melting curve (95 °C, 15 s, 60 °C, 1 min).
EdU analysis
To label proliferating vascular endothelial cells mice received three consecutive i.p. injections of 5-ethynyl-2’-deoxyuridine (EdU, 50 mg/kg body weight, ThermoFischer) on day 6, 7, and 8 after stroke. EdU incorporation was detected 21 days after stroke using the Click-it EdU Alexa Fluor 647 Imaging Kit (ThermoFischer) on 40-µm free floating coronal sections. We quantified the number of EDU+ PECAM1+ cells per mm2 stroked brain tissue.
Vascular analysis
Vascular quantification including vessel area fraction, length, and branching was performed based on an automated script previously established [49, 50]. Briefly, for area fraction, the area covered by PECAM1 was quantified using ImageJ after applying a constant threshold. The vascular length was quantified using the “skeleton length” tool and number of branches was assessed using the “analyze skeleton” tool. For the vascular length and branches, results were normalized per mm2 of brain tissue.
Behavioral characterization
Open-field testing
Open-field testing took place inside sound insulated ventilated multiconditioning chambers (TSE Systems Ltd, Germany). The open-field arena (45 cm (l), 45 cm (w), 40 cm (h)) consisted of four transparent Plexiglas walls and a light gray PVC floor. Animals were tested under yellow light (4 Lux across the floor of the open field) with 60–65 dB of white noise playing through the speakers of each box. An infrared light also illuminated the boxes so that an infrared camera could be used to record the tests. Prior to testing each animal, the entire open-field arena was cleaned using 10-ml/l detergent (For, Dr. Schnell AG). The room housing the multiconditioning chambers was illuminated with red LED lights (637 nm). Animals were removed from their home cage by the tail and placed directly into the center of the open field. The doors of the conditioning chamber were then swiftly closed. Tracking/recording was initiated upon first locomotion grid beam break. All open-field tests were 10 min in duration.
Light–Dark box
Light–Dark box testing took place inside sound insulated ventilated multiconditioning chambers (TSE Systems Ltd, Germany). The Light–Dark box (internal dimensions: 42.5 cm (l), 29.5 cm (w), 24.5 cm (h) (dark compartment 15 cm (l) 29.5 cm (w) with a centered square opening 6 cm x 6 cm) consisted of both transparent and infrared permeable black Plexiglas walls and a light gray PVC floor. Animals were tested under white light (200 Lux across the floor of the light compartment) with 60–65 dB of white noise playing through the speakers of each box. An infrared light also illuminated the boxes so that an infrared camera could be used to record the tests. Prior to testing each animal, the entire arena was cleaned using 10-ml/l detergent (For, Dr.Schnell AG). The room housing the multiconditioning chambers was illuminated with red LED lights (637 nm). Animals were removed from their home cage by the tail and placed directly into the center of the light compartment. The doors of the conditioning chamber were then swiftly closed. Tracking/recording was initiated upon first locomotion grid beam break. All Light–Dark box tests were 10 min in duration.
Hindlimb ischemia model
Hindlimb ischemia experiments were performed as described before [51]. Briefly, mice were anesthetized with isoflurane, the hindlimb was shaved, and following a small incision in the skin, both the proximal end of the femoral artery and the distal portion of the saphenous artery were ligated. The artery and all side branches were dissected free; after this, the femoral artery and attached side branches were excised. Immediately after surgery, perfusion was measured by Laser Doppler Imaging of plantar regions of interest (Moor Instruments Ltd, Axminster, Devon, England) and calculated as ratio of left (ligated) versus right (unligated) values. To label proliferating cells, an intraperitoneal injection of 5-ethynyl-2’-deoxyuridine (EdU) (E10187, Thermo Fischer Scientific) solution (5 mg/ml in saline, 10 μg per gram body weight) was performed 7 h before sacrificing the mice.
Isolation of primary muscle endothelial cells (mECs)
mEC isolation for mRNA analysis was performed as described before [52]: Mice were euthanized and calf muscles from different groups were immediately dissected and superficial big vessels were carefully removed. Muscles were minced in a petri dish on ice with a scalpel until a homogeneous paste-like mash was formed. Thereafter, the mashed muscle was enzymatically digested in digestion buffer containing 2-mg/ml Dispase II (D4693, Sigma-Aldrich, Steinheim, Germany), 2-mg/ml Collagenase IV (17104019, Thermo Fisher Scientific, Zurich, Switzerland), 2-mM CaCl2, and 2% BSA in PBS at 37 °C for 10 min, with gentle shaking every 3 min. The reaction was stopped by immediately adding an equal volume of ice cold HBSS containing 20% FBS and the suspension was passed through a 70-μm cell strainer (#431751, Corning, New York, USA) and then 40-μm cell strainer (#431750, Corning, New York, USA) to remove tissue debris. Cell suspension was centrifuged at 500 g for 5 min at 4 °C and then the pellet was washed with ice cold HBSS (+ 20% FBS) followed by a centrifugation at 400 g for 5 min in 4 °C. Next, the cell pellet was re-suspended in antibody medium (EGM2 CC-3162, Lonza, Basel, Switzerland) with anti-mouse CD31 (PECAM1) PE antibody (1:400) (553373, BD Biosciences, Basel, Switzerland) and anti-mouse CD45 PerCP antibody (1:400) (557235 BD Biosciences, Basel, Switzerland) and placed on ice for 20 min in the dark. Before sorting, the cell suspension was washed in FACS buffer (1xPBS + 1%BSA) and centrifuged at 400 g for 5 min, 4 °C and then the washed cell pellet was re-suspended in FACS buffer containing cell viability dye, SYTOX™ blue (1:1000) (S34857, Thermo Fischer Scientific, Zurich, Switzerland). Viable endothelial cells (PECAM1+, CD45−, SYTOX™ blue−) were sorted by a FACS Aria III (BD Bioscience) sorter. 200,000 events (ECs) were directly sorted (70-μm nozzle) into 700-μl RNA lysis buffer, and RNA extraction was performed by RNeasy Plus Micro Kit (74034 QIAGEN). mEC isolation for culturing: Whole skeletal muscle tissues from hindlimb were dissected and digested as described above. After a series of centrifugation and washing steps, the heterogeneous cell population was re-suspended in EC culture medium and seeded in collagen type I (125–50, Sigma)-coated plates. Due to the higher expression of P-glycoprotein in ECs compared to other skeletal muscle cells, mECs were selected by adding 4-μg/ml puromycin (P8833, Sigma-Aldrich, St. Louis, USA) to the medium overnight. After 7 days in culture, the purity of mECs was determined by PECAM1 fluorescence staining and only cultures containing at least 85% of the cells positive for PECAM1 were used for further experiments.
In situ hybridization:
For in situ hybridization, the RNAscope® Multiplex Fluorescent Assay v2 Kit (Cat.# 323110; Advanced cell diagnostics, Newark, United States) and probes (Table 2) were employed. The instructions were followed in accordance to the manufacturer’s manual. The procedure briefly described: Tissue preparation for RNAscope. Tissue were dissected and left in 4% PFA until the tissue sunk to the bottom. Same procedure was repeated in 10%, 20%, and 30% sucrose (Cat.# 573113; MilliporeSigma, Burlington, United States). Samples were frozen in the fume of liquid nitrogen and stored at − 80 °C. Before sectioning (14 µm on Superfrost microscope slides), samples acclimatized in the cryostat (Leica CM1950 cryostat) at − 20 °C for 60 min and were embedded in OCT. RNAscope pre-treatment. The sections were first washed in 1 × PBS for 5 min to remove OCT and treated with RNAscope Hydrogen Peroxide for 10 min at RT and washed in distilled water twice. For target retrieval, the slides were placed into slightly boiling (90–95 °C) RNAscope 1 × Target Retrieval Reagents for 5 min. Slides were washed in distilled water and 100% ethanol and air-dried at RT. After marking the slides with a hydrophobic pen RNAscope protease III was applied to the sections and incubated in the HybEZ Oven at 40 °C for 30 min. Slides were washed in distilled water. RNAscope. The hybridization step occurred by mixing pre-warmed probes (Table2; water bath at 40 °C) as followed and adding the mix to the cover slides and incubated at 40 °C for two hours:
Table 2.
Overview of RNAscope probes
| Channel | Gene | Species | Relative volume | Cat.# |
|---|---|---|---|---|
| 1 | Apold1 | Mm | 50 x | 418161 |
| 2 | Pecam1 | Mm | 1 x | 316721-C2 |
| 3 | Acta2 | Mm | 1 x | 319531-C3 |
Listed are all employed probes purchased from advanced cell diagnostics and their according gene, the species, the relative volume used, and their Cat.#
Slides were washed twice with 1 × wash buffer for 2 min. Excess liquid was removed and the RNAscope Multiplex FL v2 Amp1 was added to the sections and incubated for 30 min. After washing the slides twice for 2 min with 1 × washing buffer, RNAscope Multiplex FL v2 Amp2 was added and again incubated at 40 °C for 30 min. Same washing step occurred and RNAscope Multiplex FL v2 Amp 3 was applied to the slides and incubated at 40 °C for 15 min. The slides were washed twice in 1 × wash buffer for 2 min and excess liquid was removed. RNAscope Multiplex Fl v2 Hrp-C1 was added to slides and incubated for 15 min at 40 °C. Meanwhile, each opal dye in DMSO (520 nm, 570 nm, 650 nm; Cat#. NEL810001KT; PerkinElmer, Waltham, United States) was mixed 1:2000 in TSA buffer. After another washing step, opal 520 was applied to the sections and incubated for 30 min at 40 °C. Once more, the slides were washed in 1 × washing buffer and blocked with RNAscope Multiplex FL v2 HRP blocker for 15 min at 40 °C. Slides were washed twice in 1 × washing buffer. This procedure was repeated for RNAscope Multiplex FL v2 HRP-C2 and Opal 570, followed by RNAscope Multiplex FL v3 HRP-C3 and Opal 650. Lastly, sections were incubated with DAPI for 30 s, mounted with DAKO, and dried overnight in the dark at RT. Slides were stored at 4 °C in the dark.
Cell culture
Isolated primary mouse skeletal muscle endothelial cells (mECs) and commercially purchased human umbilical vein endothelial cells (HUVECs) from pooled donors (C-12203, PromoCell, Heidelberg, Germany) were routinely cultured in a 1:1 ratio of M199 ((11150059, Thermo Fisher Scientific) supplemented with 20% fetal bovine serum (FBS) (10270–106, Thermo Fisher Scientific), 30-mg/L endothelial cell growth factor supplements (EGCS) (E2759, Sigma-Aldrich), 10-U/ml heparin (H3149 Sigma-Aldrich), and 1% Penicillin–Streptomycin (10,000 U/ml) (15140122, Thermo Fisher Scientific) and Endopan 3 (P04-0010 K, PAN BIOTECH, Aidenbach, Germany) (denoted as M + E). Murine B16 (F10) cells were purchased from ATCC® (CRL-6475™, Molsheim, France) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher, 41965039, Zurich, Switzerland) containing 10% fetal bovine serum (FBS) and 1% pen strep (PS) (100-IU/ml penicillin and 100-μg/ml streptomycin). Primary mECs were only used until passage (P)1, HUVECs were used between P1 and P5. Cells were routinely maintained in 5% CO2 and 95% air at 37 °C and regularly tested for mycoplasma.
RNA extraction and quantitative RT-PCR
RNA of cultured mECs and HUVECs was extracted using PureLink™ RNA Mini Kit (12183020, Thermo Fischer Scientific). For RNA isolation from muscle tissues, 100-mg muscles were quickly dissected and homogenized in 1-ml Trizol, after 5-min incubation, and 200 µl of chloroform was added and spined down at 1,200 × g for 15 min at 4 °C. Then transfer the transparent upper phase to a new tube and add equal volume of 70% ethanol. Transfer to an RNeasy Mini spin column and process as described above. RNA purity and concentration were assessed via a spectrophotometer (Spark 10 M, Tecan). RNA was reverse transcribed to cDNA by High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, 43-688-13). A SYBR Green-based master mix (Thermo Fisher Scientific, A25778) was used for real-time qPCR analysis with primers listed in Table 3. To compensate for variations in RNA input and efficiency of reverse transcription, 18S was used as a housekeeping gene. The delta–delta CT method was used to normalize the data.
Table 3.
Forward and reverse primer sequences for murine (m) and human (h) primers used for RT-qPCR
| Species | Gene | Forward | Reverse |
|---|---|---|---|
| m | 18S | AGTCCCTGCCCTTTGTACACA | CGATCCGAGGGCCTCACTA |
| m | Apold1 | ACCTCAGGCTCTCCTTCCATCAT | ACCCGAGACAAAGCACCAATGC |
| h | Apold1 | CTACTTCATCGTCTTCTTTGGCT | TGAATCTTGGCCTTCAGCAC |
| h | Vegfa | TTCATGGATGTCTATCAGCG | CATCTCTCCTATGTGCTGGC |
| h | Glut1 | GACCCTGCACCTCATAGGC | GATGCTCAGATAGGACATCCAGG |
| h | Redd1 | CGCACTTGTCTTAGCAGTTCTC | TAGGCATGGTGAGGACAGAC |
| h | Bnip3 | CGCAGACACCACAAGATACC | TCATCTTCCTCAGACTGTGAGC |
| m | Apelin | GTTGCAGCATGAATCTGAGG | CTGCTTTAGAAAGGCATGGG |
| m | Angpt2 | GGAAGCCCAAGTACTAAACC | GCTTGTTTATTTCACTGGTCTG |
| m | Pgf | GTGTGCCGATAAAGACAGCC | TCCAGAATAGGTCTGCATTCAC |
| m | Ramp2 | GGAAGATGGAAGACTACGAAACAC | CTGTAATGCCTGCTAATCAAAGTC |
| m | Edn1 | GGAAACTACGAAGGTTGGAGG | CTCTGTAGTCAATGTGCTCGG |
| m | Adm | GATGTTATTGGGTTCACTCGCT | CCACTTATTCCACTTCTTTCGG |
Subcutaneous tumor model
Mice at the age of 7–10 weeks were subcutaneously injected cultured murine B16 (F10) melanoma cells (1 million cells per mouse) proximate under the left forelimb. The mice were monitored over the next 15 days to detect and quantify tumor growth. Tumor diameters were measured by caliper and the size is calculated using the formula: Tumor volume (mm3) = d2x D/2, where d and D are the shortest and longest diameter in mm, respectively. After two weeks, mice were anesthetized and sacrificed by transcardial perfusion with 2% paraformaldehyde (PFA, Cat.# J61984 Alfa Aesar, Haverhill, United States). Tumors were dissected and fixed with 4% PFA for 24–48 h and frozen in the fume of liquid nitrogen. Samples were stored at − 80 °C. Tumor vessel perfusion was quantified on tumor cryosections following intravenous injection of 0.05-mg fluorescein-labeled Lycopersicon esculentum (Tomato) lectin (FL-1171, Vector laboratories, Brussels, Belgium) in tumor-bearing mice (tumors were harvested 10 min after injection). The perfused area was defined as the lectin+ PECAM1+ area expressed as a percentage of the total PECAM1+ area (NIH ImageJ software). Tumor perfusable area was analyzed by measuring the total vessel lumen area (i.e., sum of the lumen area of all vessels) and expressing it as a percentage of the total tumor area.
Knockdown and lentiviral particle production
Multiple GIPZ Lentiviral shRNAs target human APOLD1 (V2LHS_117002; V2LHS_117004; V2LHS_117217) were purchased from Dharmacon (Horizon Discovery; Waterbeach, United Kingdom). A nonsense scrambled shRNA sequence was used as control. Lentiviral particles were generated by transfection of HEK 293 cells (Cat.# ACC635; DSMZ, Braunschweig, Germany) with Pmd2 (AddGene, Plasmid #12259), lentiviral envelope plasmid psPAX2 (AddGene, Plasmid #12260), and the plasmid containing scramble or against APOLD1 gene sequences. Lentiviral particles were generated by transfection of HEK 293 cells with the respective plasmid and pLenti-C-mGFP-P2A-Puro Lentiviral Gene Expression Vector (Cat. #PS100093, Origene). Lipofectamine 2000 (Cat.# 11668030; Thermo Fisher Scientific) was used for transfection. Viral particles were collected at least 48 h after incubation. Cells were transduced 24 h in the presence of 8-μg/ml polybrene and re-fed with fresh medium the next day. Apold1-KD HUVECs were used in functional assays at least 5-day post-transduction.
Immunohistochemistry and histology
Muscle samples were harvested and embedded in Tissue-Tek and frozen in liquid N2-cooled isopentane. Frozen muscle cross-Sects. (7–10 μm) were made using a cryostat (Leica CM 1950) and collected on Superfrost Ultra Plus slides (Thermo Fischer Scientific). Cross-sections were permeabilized in PBS with 0.5% Triton X-100 and blocked for 1 h at room temperature in PBS with 0.05% Tween-20, 1% BSA (BS). Primary antibody incubations were performed at 4 °C overnight. Slides were subsequently washed in PBS and incubated for 1 h in blocking buffer with the appropriate secondary antibodies. Nuclei were stained with Hoechst. The following primary and secondary antibodies were used: anti-CD31 (PECAM1) (1:250, AF3628, R&D Systems), anti-α smooth muscle actin Cy3™ conjugated (αSMA) (1:500, C6198, Sigma-Aldrich), isolectin B4, (IB4, 1:500, I32450, Thermo Fisher Scientific), Alexa Fluor 594 donkey anti-goat IgG (H + L) (1:250, A-11058, Thermo Fisher Scientific), Alexa Fluor 568 donkey anti-rat IgG (H + L) (1:250, ab175475, abcam), Alexa Fluor 350 donkey anti-sheep IgG (H + L) (1:250, A-21097, Thermo Fisher Scientific), Alexa Fluor 488 donkey anti-rabbit IgG (H + L) (1:250 dilution, A-32790, Thermo Fisher Scientific), Alexa Fluor 488 donkey anti-goat IgG (H + L) (1:250, A-32814, Thermo Fisher Scientific), and Alexa Fluor 350 donkey anti-rabbit IgG (H + L) (1:250 dilution, A- 10039, Thermo Fisher Scientific).
Fiber-type staining combined with isolectin B4 staining was performed in two steps: First, fiber-type staining was performed as described [53]. In short, sections were dried and washed for 5 min in PBS supplemented with 0.05% Triton X-100 (PBST) and subsequently blocked for 60 min in PBST + 10% goat serum (16210064, Thermo Fisher Scientific). Afterward a primary antibody cocktail diluted in PBST + 10% goat serum was applied for 120 min against MyHC-I (1:50 dilution, BA-F8 from hybridoma, Iowa City, IA, USA), MyHC-IIa (1:200 dilution, SC-71 from hybridoma), and MyHC-IIb (1:100 dilution, BF-F3 from hybridoma). After washing 3 times for 5 min, a secondary antibody cocktail, diluted in PBST + 10% goat serum, was applied for 60 min. Secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG2B (1:250 dilution, Thermo Fisher Scientific), Alexa Fluor 350 goat anti-mouse IgG1 (1:250 dilution, A-21120, Thermo Fisher Scientific), and Alexa Fluor 568 goat anti-mouse IgM (1:250 dilution, A-21043, Thermo Fisher Scientific). After a 3 × 5 min wash, muscle sections were briefly fixed in 2% PFA for 5 min and washed with PBS. For additional IB4 vascular staining, muscle sections were incubated overnight at 4 °C with Isolectin GS-IB4 Alexa Fluor™ 647 (IB4, 1:500, I32450, Thermo Fisher Scientific) diluted in blocking buffer 0.1% Triton X-100. For EdU detection combined with ERG and/or PECAM1/IB4, EdU was first visualized using the EdU Click-iT™ Cell Reaction Buffer Kit (C10269, Thermo Fisher Scientific) according to manufacturer’s instructions and subsequently incubated for 1 h in blocking buffer (PBS with 1% BSA) at room temperature (RT). Thereafter, sections were incubated overnight at 4 °C with Isolectin GS-IB4 Alexa Fluor™ 568 (1:500, I21412, Thermo Fischer Scientific), goat anti-Mouse/Rat CD31 (PECAM1) antibody (1:250, 3628, R&D Systems), and ERG antibody (#97249, Cell Signaling, 1:200) diluted in blocking buffer with 0.1% Triton X-100.
Images were taken with a Zeiss Axio observer Z.1 or an Olympus confocal microscope (FV1200). All images were captured at the same exposure time for one experiment. Composite images were stitched together using the tiles module in the ZEN 2011 imaging software (Zeiss). Fiber-type composition, vascular density (% IB4/PECAM1 + area), and ERG numbers were quantified using ImageJ software.
Permeability assay
Mice were intravenously injected with 200 µl of 0.5% Evans blue (E2129, Sigma-Aldrich) in PBS. Thirty min later, mice were euthanized and perfused with PBS. Kidney, gastrocnemius, and brain were dissected and weighted. Tissues were incubated in 500-µl formamide (#75-12-7, AppliChem) at 55 °C for 24 h, centrifuged, and the absorbance of the supernatant was measured by a spectrophotometer (Spark 10 M, Tecan) at 610 nm. Relative absorbance unit per mg tissue was calculated.
Blood–brain barrier permeability
Briefly, animals were injected with the small-molecule tracer sodium fluorescein (NaF, 376 Da) in 0.9% saline (120 mg/kg, 30 mg/ml, i.p.). 10 min afterward, mice were anesthetized with an injection of Esconarkon (i.p.) and ~ 700-µl blood were taken from the right ventricle before perfusion with 50 ml of ice cold 0.9% saline. The brain was rapidly extracted, and the hippocampus, cerebellum, and cortex were dissected and collected. The cerebral extraction ratio for sodium fluorescein was determined as described in detail in Roszkowski & Bohacek 2016 [54].
In vitro analysis of EC function
Proliferation: Cultured primary mouse mECs (no longer than 8 days, P1) or HUVECs were incubated in growth medium containing 10-μM 5-ethynyl-2’-deoxyuridine (EdU) for 15 h. As a measure of proliferation, incorporation EdU was assessed using the Click-iT™ Cell Reaction Buffer Kit (C10269, Thermo Fisher Scientific), according to the manufacturer’s instructions. Briefly, after EdU incorporation, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized for 20 min in 0.5% Triton X-100 with 3% BSA in PBS, followed by reaction with the Click-iT reaction cocktail for 45 min in dark at room temperature. Thereafter, cells were washed briefly and counterstained with Hoechst (#62249, Thermo Fisher Scientific,1:2000) and antibodies against CD31 (PECAM1) (AF3628, R&D Systems, 1:250) or ERG (#97249, Cell Signaling, 1:250). Cells were imaged using a Zeiss Axio Observer.Z1 fluorescence microscope (Carl Zeiss, Oberkochen, Germany). EdU+ cells and ERG+ ECs were counted in at least 5 random fields and the percentage of EdU+ cells in ECs was calculated. Scratch wound assay: A scratch wound was applied on confluent EC monolayers (pre-treated with 1-µg/ml mitomycin C for 24 h where indicated) using a 200 µl tip. After scratch wounding (T0) and photography using a Leica DMI6000 B inverted microscope (Leica Microsystems, Mannheim, Germany), the cultures were further incubated in growth medium and fixed with 4% PFA 24 h after first scratch (T24). Cells were photographed again (T24) and gap areas at both time points were measured using the Fiji software package (https://fiji.sc) to calculate the percentage of wound closure using the following expression: (1 − (T24gap area/T0gap area)) × 100. Spheroid capillary sprouting assay: Spheroids were prepared as previously described [55] with minor modifications. Briefly, spheroids containing 1000 HUVECs or mECs per 25-µl droplet were plated overnight as hanging drops in a 20% methylcellulose (9004-67-5, Sigma-Aldrich) in EGM2 mixture. The next day, spheroids were collected in 10% FBS in PBS, concentrated using several centrifugation steps, and embedded in a Fibrinogen gel (5-mg/ml fibrinogen (F8630, Sigma-Aldrich) dissolved in EGM2 plus 1U/ml thrombin (T4648, Sigma-Aldrich). To assess tip cell competition, cells were mixed at the indicated ratio. Growth medium (with or without Mitomycin C) was pipetted on top of the gel to induce sprouting. 24 h later, spheroids were fixed with 4% PFA at room temperature and photographed using a Leica DM IL LED microscope (Leica Microsystems GmbH, Wetzlar, Germany). Contact inhibition: HUVECs were seeded in 50% EGM2 / 50% full M199 medium at a density of 15,000 cells/cm2 and were further cultured for 3 days until contact inhibition was reached. To generate the corresponding proliferative control, contact-inhibited cells were trypsinized and cultured for 24 h to re-initiate proliferation. VEGFA stimulation: HUVECs were starved with DMEM medium without any growth factors for 4 h. One group was treated with VEGF (Cat.# 450–32; PeproTech; London, United Kingdom) (end conc.: 30 ng/ml) for two hours, while the other group was starved for another two hours. RNA was extracted as described above. Assessment of autophagy: Cultured primary muscle endothelial cells were incubated in either growth medium or medium lacking amino acids for 16 h. Cells were collected and lysed with [50-mM Tris–HCl pH 7.0, 270-mM sucrose, 5-mM EGTA, 1-mM EDTA, 1-mM sodium orthovanadate, 50-mM glycerophosphate, 5-mM sodium pyrophosphate, 50-mM sodium fluoride, 1-mM DTT, 0.1% Triton X-100, and a complete protease inhibitor tablet (C755C25, Roche Applied Science)]. Lysates were centrifuged at 10,000 g for 10 min at 4 °C. Supernatant was collected and protein concentration was measured using the DC protein assay kit (5000116, Bio-rad). Total protein (5–10 ug) was loaded in 15 well pre-casted gradient gel (456-8086, Bio-Rad). After electrophoresis, a picture of the gel was taken under UV-light to determine protein loading using stain-free technology. Proteins were transferred onto a PVDF membrane (Bio-rad, 170-4156) with a semi-dry system. Membrane was blocked for 1 h at room temperature with 5% BSA in 0.1% TBS tween. Membranes were incubated in primary antibodies (1:500, NB100-2220, Novus Biologicals; 1:500, ab109012, Abcam) overnight. Anti-rabbit IgG HRP-linked secondary antibody (1:500, 70748, Cell Signaling Technology) was used for chemiluminescent detection of proteins. Membranes were scanned with a Chemidoc imaging system (Bio-rad) and quantified using Image Lab software (Bio-rad).
Flow cytometry
For endothelial cell analysis in muscles, muscles were dissected, separated, and enzymatically digested as described above, and cells were incubated in dark for 30 min with anti-mouse CD31 (PECAM1) PE antibody (553373, BD Biosciences) and anti-mouse CD45 PerCP antibody (557235 BD Biosciences) (1:400 diluted in FACS buffer (1xPBS + 1% FBS)). Cells were washed with FACS buffer before loading. For EdU proliferation experiments in B16-F10 melanoma, tumors were dissected after seven hours labeling with EdU (i.p. 5 mg/ml in saline, 10 μg per gram body weight), then the dissociated cells were briefly fixed with 2% PFA, and processed with the click-iT plus EdU Alexa Fluor® 647 Flow Cytometry Assay Kit (C10634, ThermoFischer Scientific) according to the manufacturer’s instructions. Subsequently, they were incubated in the dark for 30 min with CD31 (PECAM1) PE antibody (553373, BD Biosciences) and Alexa Fluor® 488 anti-mouse CD45 Antibody (1:400, 103122, BioLegend). Cells were analyzed using SONY SH800S cell sorter. Data were analyzed using FlowJo 10 software (Tree Star).
Quantification and statistical analysis
The images presented in the manuscript are representative of the data (quantification of image is approximately the group average) and the image/staining quality. All data represent mean ± SEM. GraphPad Prism software (version 8.0.0) was used for statistical analyses. Investigators were always blinded to group allocation. Unless otherwise indicated, when comparing two group means, Student’s t test was used in an unpaired two-tailed fashion. For more than two groups, one-way ANOVA with Tukey’s multiple comparisons test was used and for experimental set-ups with a second variable, two-way ANOVA with Sidak’s multiple comparisons test and two-way ANOVA with Tukey’s multiple comparison test were used. The statistical method used for each experiment is indicated in each figure legend. Asterisks in figure legends denote statistical significance. No experiment-wide multiple test correction was applied. p > 0.05 is considered non-significant (n.s.). p < 0.05 is considered significant (*).
Results
Apold1 is expressed in endothelial cells and regulated by metabolic demand
Apold1 was originally reported to be an immediate early gene expressed in endothelial cells (ECs), primarily during embryogenesis [20]. To assess Apold1 expression in adult animals, we first interrogated “Tabula Muris,” a large single-cell RNA sequencing (scRNAseq) compendium covering 20 mouse organs [56]. Across organs, Apold1 expression was largely restricted to ECs (Fig. S1A-C). When narrowing the analysis to individual, highly vascularized tissues, Apold1 was highly expressed in ECs in the brain (Fig. S1D), muscle (Fig. S1E), and liver (Fig. S1F). As ECs differ substantially between tissues [57], we took a closer look at Apold1 mRNA in both brain and muscle, two tissues that differ radically in cellular complexity and metabolic profile. Reanalysis of a scRNAseq dataset from mouse adult skeletal hindlimb muscle [41] confirmed that Apold1 expression is restricted to vascular cells (Fig. 1A–C), with abundant expression in ECs, but also to a lesser extent in pericytes and smooth muscle cells (Fig. 1B, C). To better understand the distribution of Apold1 within the different types of ECs within muscle, we evaluated Apold1 expression in FACS-sorted PECAM1+CD45− ECs that were collected from the mouse gastrocnemius muscle [52] (Fig. 1D). Apold1 was widely distributed across the different EC populations ranging from arterial to capillary and venous ECs (Fig. 1E). Interestingly, its expression in capillary ECs derived from white, glycolytic muscle (WmECs), which have low angiogenic potential, was noticeably lower when compared to capillary ECs derived from red, oxidative muscle (RmECs) with high angiogenic potential (Fig. 1E). To confirm this, we isolated WmECs (from EDL) and RmECs (from Soleus) using FACS (Pecam1+CD45−) and showed with RT-qPCR that Apold1 was enriched in RmECs compared to WmECs (Fig. 1F). We then tested whether exercise, an intervention known to activate RmECs, would affect Apold1 expression (Fig. 1G). Following two weeks after daily voluntary wheel running, we again isolated Pecam1+CD45− ECs using FACS from calf muscle. RT-qPCR showed a marked increase in Apold1 expression in whole muscle as well as in sorted mECs after exercise (Fig. 1H).
We then assessed Apold1 expression in the brain and turned to an extensive single-cell resource on the mouse nervous system [45], which confirmed that Apold1 expression is restricted to vascular cells including ECs and to a lesser extent mural cells (pericytes and smooth muscle cells) (Fig. S2). It had previously been reported that metabolically demanding environmental insults like seizures, stress, or exercise increase Apold1 expression in brain tissue [20, 21, 24, 25]. To explore in which cell types this regulation occurs, we used a scRNAseq study that compared mice that had been injected with the seizure-inducing agent pentylenetetrazole (PTZ, which triggers strong pathological levels of neuronal activity), exposed to immobilization stress (which triggers high but physiological levels of neuronal activity), or received no treatment [42] (Fig. 1I). This dataset contained several biological replicates per group, allowing us to quantitatively compare multi-condition scRNAseq datasets [58]. We found that Apold1 expression was again restricted to vascular cells, including ECs and mural cells (Fig. 1J), and that seizures triggered robust Apold1 expression mainly in ECs, although we also observed a small increase in mural cells (Fig. 1K). In addition, immobilization stress triggered Apold1 expression only in ECs but not in mural cells (Fig. 1K), suggesting that Apold1 expression in ECs is particularly sensitive to environmental stimuli. To further examine whether Apold1 regulation is indeed more sensitive in ECs than in mural cells, we turned to a single-nucleus RNA sequencing experiment (snRNAseq), in which we had assessed the transcriptomic response of mice to an acute swim stress exposure [44]. Indeed, Apold1 was expressed in ECs and pericytes, but the stress-induced increase in Apold1 expression was restricted to ECs (Fig. 1L, M). These analyses demonstrate that Apold1 expression across the organism is restricted to vascular cells and that Apold1 expression is readily stimulated by environmental challenges, particularly in ECs.
Loss of Apold1 does not impair normal development
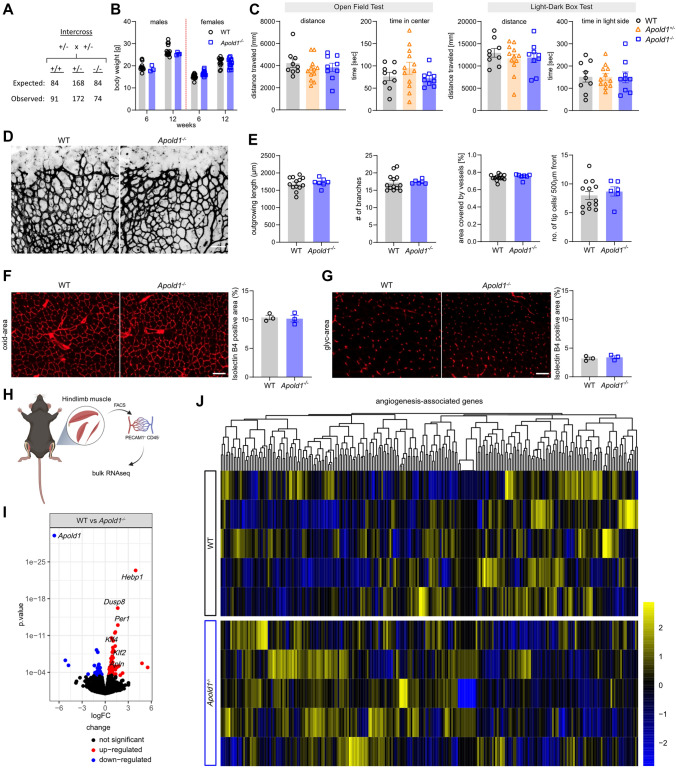
To assess whether Apold1 plays a role in EC function in vivo, we turned to Apold1 knockout (Apold1−/−) mice. Because of the key role of angiogenesis during development [59] and because Apold1 expression is high during development [20], we first assessed whether Apold1−/− mice display developmental defects. We set up several heterozygous (Apold1±) breeding pairs and observed the expected Mendelian distribution of offspring genotypes with very similar distributions in males and females (Fig. 2A). Also, Apold1−/− females were able to produce normal numbers of healthy pups when mated with wild-type (WT) or Apold1−/− males, and we noticed no increased incidence of natural deaths in Apold1−/− relative to wild-type WT mice (data not shown). Moreover, mice had similar body weight (Fig. 2B). Apold1−/− mice as well as heterozygous Apold1± mice appeared developmentally normal and were behaviorally indistinguishable from WT littermates in terms of locomotor activity, exploratory behaviors, and emotionality in the open-field test and the light–dark box test (Fig. 2C). To investigate whether loss of Apold1 leads to more subtle changes in developmental angiogenesis, we subsequently used the postnatal retina, which allows the characterization of distinct changes in EC fate [60, 61]. At postnatal day 5 (P5), whole-mount isolectin-B4-stained retinas showed that vascular parameters are normal in Apold1−/− mice, with normal vascular outgrowth, number of branch points, total vessel area, and number of tip cells in Apold1−/− mice when compared to their WT littermates (Fig. 2D–E). Further, the analysis of the vasculature in the oxidative as well as the glycolytic part of the gastrocnemius muscle of adult animals showed no alterations in terms of vessel area (Fig. 2F–G). Because previous work had raised the possibility of changes in vessel permeability [20, 31], we performed a vascular permeability assay by injecting Evans Blue intravenously before sacrificing the animals and analyzed the distribution of the dye within the different tissues. We did not observe any differences in vascular permeability in kidney, muscle, and brain (Fig. S3A). We assessed the integrity of the blood–brain barrier in more detail using a dye with smaller molecular weight (sodium fluorescein). We again found that WT and Apold1−/− animals displayed similar permeability in multiple brain regions (Fig. S3B).
Fig. 2.
Apold1 deficiency does not affect physiological angiogenesis A Expected and observed Mendelian distribution of heterozygous breedings (Chi-square test: 0.9585; p = 0.6193). B Quantification of body weight at 6 and 12 weeks of age in females and males between genotypes (n = 3–14 per group). C Behavioral analysis of WT and Apold1−/− in the open-field test and light–dark box measured by distance traveled and time in center/time in light side (n (WT/Apold1±/Apold1−/−) = 9/12/9). D Representative images of isolectin-B4-stained (black) retinal vessels of postnatal day 5 (P5) pups. Scale bar, 100 µm. E Quantification of outgrowing vessel length, number of branches, percentage of area covered by vessels, and number of tip cells in 500 µm front of the retina (n (WT/Apold1−/−) = 13/6). F Representative images and percentage positive area of blood vessels stained with isolectin B4 (red) in the oxidative area of the gastrocnemius (oxid-area) (n (WT/Apold1−/−) = 3/3). Scale bar, 200 μm. G Representative images and percentage of positive area of blood vessels stained with isolectin B4 in the glycolytic area of the gastrocnemius (glyc-area) (n (WT/Apold1−/−) = 3/3). Scale bar 200 µm. H Experimental design. I Volcano plot of significantly differentially expressed genes by bulk RNAseq of WT vs Apold1−/− mECs (n (WT/Apold1−/−) = 5/5). J RNA expression heat map of all angiogenesis-associated genes (n(WT/Apold1−/−) = 5/5). Student’s test in B, E, F, and G; one-way ANOVA with Tukey’s multiple comparison test in C. The data shown are mean ± SEM
Screening of the transcriptomic profile of WT and Apold1−/−-isolated muscle ECs by bulk RNAseq revealed minor transcriptional differences, as only 89 genes were differentially expressed (Fig. 2H–I). Among those were Klf2 and Klf4, known regulators of vascular integrity by shear stress [62, 63], but activation of their downstream targets was not obvious (Fig. 2I; see list of differentially expressed genes in Supplementary Table1/Supplementary file 8). We also observed an increase in Apln (a gene with pleiotropic functions in ECs, reported to promote angiogenesis [64] (Fig. 2I), but we could not detect any concerted regulation of angiogenesis-related genes (Fig. 2J). Altogether, these data show that loss of Apold1 does not lead to any obvious developmental deficits in overall health and that Apold1 is dispensable for developmental angiogenesis.
Loss of Apold1 impairs recovery from stroke
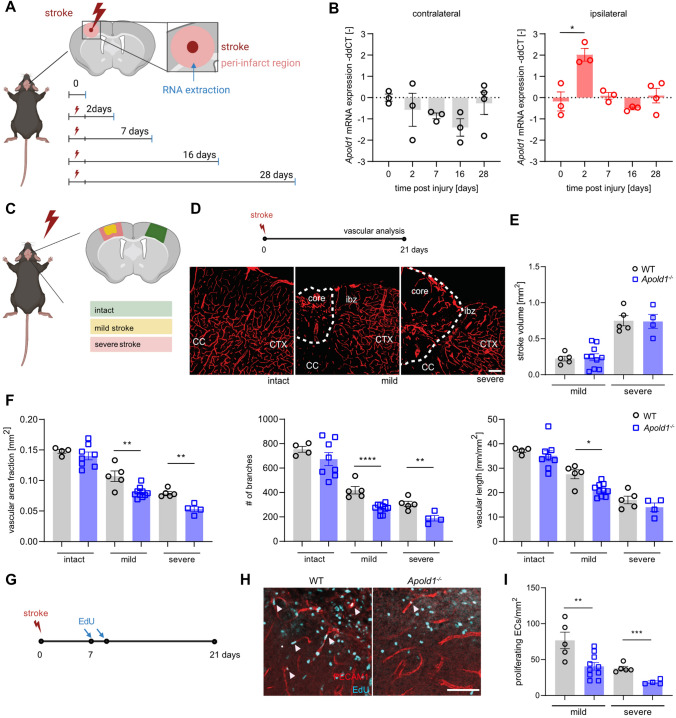
To subsequently study the role of Apold1 during angiogenesis in pathological settings, we turned to a mouse model of stroke. We first tested whether photothrombotic (i.e., microthrombotic focal) strokes in the sensorimotor cortex of adult mice would lead to changes in Apold1 mRNA expression in the ischemic border region, where angiogenesis is induced [47] (Fig. 3A). As expected, Apold1 expression increased in the microdissected peri-infarct tissue 2 days after stroke and returned to baseline levels within a week (Fig. 3A, B, right). Apold1 expression did not change in the intact contralateral cortex (Fig. 3A, B, left). Then, we induced photothrombotic strokes in Apold1−/− mice and WT littermates, using either a weaker or a stronger paradigm to generate milder and more severe strokes (Fig. 3C). Stroke volume was similar in Apold1−/− and WT mice (Fig. 3E), indicating that loss of Apold1 does not impact the severity of an ischemic injury, in agreement with previous findings [33]. Also, the non-injured (contralateral) cortex of Apold1−/− mice had a normal vascular network (vascular area fraction, number of branches, and vascular length), consistent with normal vascular development of Apold1−/− mice (Fig. 3D, F). However, 21 days after the stroke, we observed a reduction in vascular area, branch numbers, and total length of the vascular network in the peri-infarct border zone of Apold1−/− mice as compared to the corresponding region of WT mice (Fig. 3F). Labeling of proliferating cells one week after stroke using 5-ethynyl-2’-deoxyuridine (EdU) injections combined with immunofluorescent labeling of PECAM1+ ECs showed that Apold1−/− ECs within the ischemic border zone proliferate less (Fig. 3G, I). Thus, Apold1−/− mice show impaired angiogenesis in the ischemic stroke border zone during the three-week recovery period.
Fig. 3.
Apold1 is required for angiogenesis and revascularization after photothrombotic stroke. A Experimental design. B Time course of relative Apold1 mRNA expression on contralateral and ipsilateral cortex (n = 3–4 per timepoint). C Illustration of stroke size and location. D Experimental design and representative images of intact and injured PECAM1+ vasculature. E Stroke volumes 21 days following injury (n = 4–10 per group; Scale bar, 100 µm). F Quantitative analysis of vascular density, number of branches, and length of blood vessels in the ischemic border regions (n = 4–8 per group). G Experimental design. H Representative images of newly formed vascular cells by PECAM1+/EdU+ co-staining in WT and Apold1−/− animals. Scale bar, 50 µm. I Quantification of proliferating ECs (n = 4–10 per group). Two-way ANOVA with Tukey’s multiple comparisons test in B; Student’s t test in E, F, and G (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001). The data shown are mean ± SEM
Apold1 is required for revascularization after hindlimb ischemia
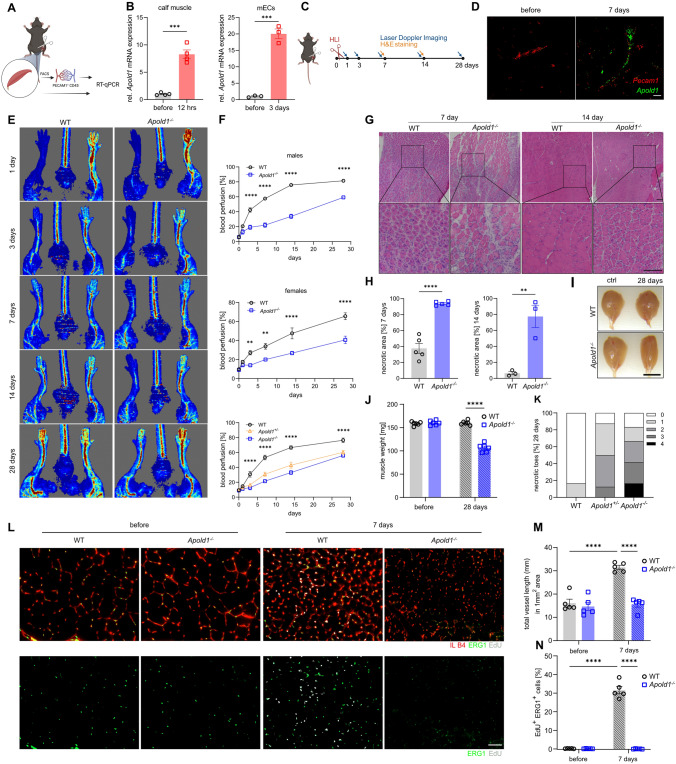
Because the regenerative capacity of peripheral tissues is much greater than that of the central nervous system (CNS) and because Apold1 is abundantly expressed in ECs from non-CNS vascular beds (see Figs. 1 and S1), we asked whether Apold1−/− mice also show impaired angiogenesis in the regeneration of peripheral tissue. We used hindlimb ischemia, where ligation of the femoral artery strongly reduces perfusion of the hindlimb and causes severe hypoxia followed by a robust angiogenic response [65, 66]. Analysis of Apold1 mRNA showed a strong (eightfold) increase in whole tissue isolated from the ipsilateral (ischemic) muscle compared to the contralateral side 12 h after femoral artery ligation (Fig. 4A; 4B, left). Isolation of muscle endothelial cells (mECs) from the ligated hindlimb 3 days after induction of ischemia revealed a nearly 20-fold increase in Apold1 mRNA content when compared to the non-ligated contralateral side (Fig. 4B, right). In situ hybridization confirmed a strong increase in Apold1 expression after ischemia, colocalized with Pecam1 (Fig. 4D). Laser Doppler imaging showed that blood flow was similarly reduced upon femoral artery ligation in both WT and Apold1−/− mice, but while it gradually recovered in WT mice during the 4-week follow-up period, reperfusion remained severely impaired in Apold1−/− mice (Fig. 4E, F). This impairment was observed in both sexes, and the deficit was similarly profound in Apold1−/− and in mice haplo-deficient for Apold1 (Apold1−/+) (Fig. 4F).
Fig. 4.
Impaired revascularization, angiogenesis, and EC proliferation after hindlimb ischemia in Apold1−/− mice. A Experimental design. B Apold1 expression in qRT-PCR in whole calf muscle tissue 12 h after hindlimb ischemia (left) and in muscle ECs (right) 3 days after hindlimb ischemia in WT mice (n = 3–4 per group). C Experimental design. D Representative image of in situ hybridization for Apold1 7 days after ischemia in the ischemic muscle (ipsilateral) and the contralateral control muscle. Scale bar, 20 μm. E Representative images of blood perfusion measured by laser Doppler imaging (LDI) 1, 3, 7, 14, and 28 days after hindlimb ischemia in Apold1−/− and WT mice. F Time course quantification of blood perfusion across 28 days after hindlimb ischemia comparing recovery in Apold1−/− vs. WT males (n (WT/Apold1−/−) = 6/6), females (n (WT/Apold1−/−) = 6/6), and in a separate experiment in Apold1−/−, Apold1±, and WT males (n (WT/Apold1±/Apold1−/−) = 6/6/4). G Representative hematoxylin–eosin (H&E) staining images of the triceps surae muscles at 7 and 14 days after hindlimb ischemia. Scale bar, 100 μm. H Quantification of necrotic area 7 and 14 days after hindlimb ischemia in WT and Apold1−/− mice (n (WT/Apold1−/−) = 6/6). I Photographs of whole calf muscle isolated from control and ischemic leg of WT and Apold1−/− mice. Scale bar, 7 mm. J Whole calf muscle weight 28 days after hindlimb ischemia (n = 6 per group). K Measurement of number of necrotic toes 28 days after hindlimb ischemia (n = 8–12 per group). L Representative images of isolectin B4 (IL B4, red), Erg1 (green), and EdU (white) immunofluorescent images on gastrocnemius muscle cross-sections of control and ischemic leg 7 days after hindlimb ischemia in WT and Apold1−/− mice. Scale bar, 50 μm. M Total vessel length in 1 mm2 area of muscle cross-sections before and 7 days after hindlimb ischemia in WT and Apold1−/− mice (n = 5 per group). N Percentage of EdU+ Erg1+ proliferating mECs at 7 days after induction of hindlimb ischemia (n = 5 per group). Student’s t test in B, H, and J; Two-way ANOVA with Sidak’s multiple comparison test in F; Two-way ANOVA with Tukey’s multiple comparison test in M and N (**p < 0.01; ***p < 0.001; and ****p < 0.0001). The data shown are mean ± SEM
Histological examination of H&E-stained muscle sections revealed that Apold1−/− triceps surae muscles had much larger necrotic areas at 7-day post-ischemia (Fig. 4G, H). Fourteen days after ischemia Apold1−/− mice still had large areas containing necrotic fibers, while WT muscles had almost completely recovered (Fig. 4G, H). In addition, while WT mice already showed clear signs of regeneration (evidenced by centrally nucleated fibers) at 7 days, Apold1−/− muscle only had low numbers of regenerating fibers, even at 14 days after ischemia (Fig. 4G). Further, impaired muscle recovery resulted in lower muscle weight in Apold1−/− mice 28 days after ischemia (Fig. 4I, J). We also noticed a higher frequency of necrotic toes in Apold1−/− mice and in haplo-deficient Apold1± mice (Fig. 4K). Finally, we measured vascular density in ischemic and (contralateral) control muscles on day 7 after hindlimb ischemia using isolectin B4 (IB4). While ischemic muscles in WT mice showed the typical increase in total vessel length, this response was completely abolished in Apold1−/− mice (Fig. 4L, M). Consistent with our observations in stroke, EdU labeling of proliferating mECs at 7 days following induction of ischemia revealed that Apold1-deficient mECs were unable to proliferate in response to ischemia (Fig. 4L, N).
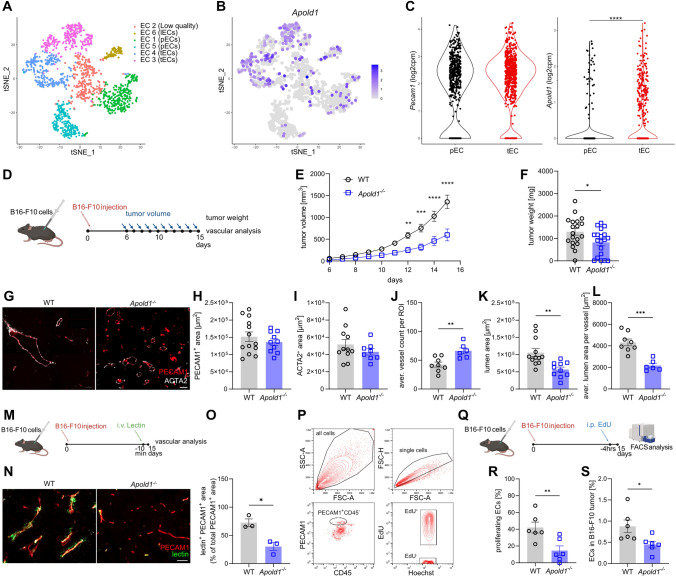
Apold1 controls tumor angiogenesis
Because of the profound role of Apold1 in regulating angiogenesis in pathological settings, we next asked whether Apold1 controls tumor angiogenesis. We first used a published scRNAseq dataset and quantified Apold1 expression in ECs of normal versus malignant lung tissue resected from 5 patients with untreated, non-metastatic lung tumors (Fig. 5A, B) [67]. Strikingly, we found that within ECs, Apold1 expression was enriched in tumor ECs (tECs) compared to pulmonary ECs (pECs) (Fig. 5C). We confirmed these findings by re-analyzing a second scRNAseq dataset, where freshly isolated human tECs and peritumoral non-tumor pECs (collected from the same individual) were analyzed from 1 large cell carcinoma, 4 squamous cell carcinomas, and 3 adenocarcinoma of treatment-naive patients [68]. Again, tECs expressed more Apold1 than pECs (Fig. S4A). To functionally test the role of Apold1 in tumorigenesis, we implanted syngeneic B-16-F10 melanoma cells in WT and Apold1−/− mice and followed tumor growth over time (Fig. 5D). While WT mice showed exponential tumor growth, tumor growth was retarded in Apold1−/− mice leading to lower tumor volume and tumor weight at end stage (Fig. 5E, F). Combined PECAM1 and ACTA2 staining showed similar vessel (PECAM1+) area with similar coverage by pericytes (ACTA2+) in the tumor tissue of both WT and Apold1−/− mice (Fig. 5G,H, I). However, closer investigation of the tumor vasculature showed that Apold1−/− mice had more vessels (Fig. 5J), but these vessels had a much smaller lumen (Fig. 5K), resulting in a dramatic reduction in lumen area per vessel in Apold1−/− mice (Fig. 5L). Functional assessment of vessel perfusion following intravenous injection with fluorescein-labeled Lycopersicon esculentum (tomato) lectin confirmed these data (Fig. 5 M–O). Finally, we injected EdU four hours before sacrificing the mice and subsequently FACS-sorted PECAM1+/CD45− tumor ECs (Fig. 5P, Q). We found that loss of Apold1 reduced the percentage of proliferating (Edu+) ECs leading to a lower fraction of ECs inside the tumor (Fig. 5R, S).
Fig. 5.
Apold1 is enriched in tumor ECs and loss of Apold1 slows tumor growth. A t-Stochastic neighbor embedding (tSNE) plots of distribution of ECs in lung tissue resected from 5 patients with untreated, non-metastatic lung tumors reanalyzed [67]. B t-Stochastic neighbor embedding (tSNE) plots of distribution of Apold1 in ECs in lung tissue reanalyzed [67]. C Apold1 expression in tumor ECs (tECs) compared to pulmonary ECs (pECs) reanalyzed [67]. D Experimental design. E In vivo measurement of tumor volume in Apold1−/− and WT mice after injection of B16-F10 melanoma cells (n (WT/Apold1−/−) = 11/11). F Weight of isolated tumors 15 days after injection (n (WT/Apold1−/−) = 19/19). G Representative images of the vasculature in tumors isolated from WT and Apold1−/− mice stained for PECAM1 (red) and ACTA2 (white). Scale bar, 100 µm. H Quantification of PECAM1+ area (n (WT/Apold1−/−) = 13/10) and I ACTA2+ area (n (WT/Apold1−/−) = 13/10). Scale bar, 100 µm. (J) Quantification of average vessel count per region of interest (ROI) (n (WT/Apold1−/−) = 8/6), K lumen size (n (WT/Apold1−/−) = 12/10), and L average lumen area per vessel (n (WT/Apold1−/−) = 8/6). M Experimental design. N Representative image of perfusion stained by injected fluorescein-labeled Lycopersicon esculentum (tomato) lectin (green) in PECAM1+ (red) in tumors isolated from WT and Apold1−/− mice. O Percentage of lectin+PECAM1+ area of total PECAM1+ area (n (WT/Apold1−/−) = 3/3). P Representative flow cytometric analysis of ECs (PECAM1+CD45−) and proliferating ECs (EdU+) in B16-F10 melanoma. Q Experimental design. R Percentage of proliferating ECs (EdU+) (n (WT/Apold1−/−) = 6/6) and S percentage of ECs (PECAM1+CD45+) in B16-F10 melanoma (n (WT/Apold1−/−) = 6/6). Student’s t test in C, F, H, I, J, K, L, O, R, and S. Two-way ANOVA with Sidak’s multiple comparison test in E (*p < 0.05; **p < 0.01; and ***p < 0.001). The data shown are mean ± SEM
Altogether, these results demonstrate a critical role for Apold1 in angiogenesis and revascularization after hypoxic injury in the CNS and in the periphery, as well as during tumor growth. This is particularly striking when considering that disruption of many vascular genes involved in angiogenesis leads to severe developmental deficits, whereas Apold1-deficient mice are developmentally normal, suggesting that Apold1 plays a role primarily during angiogenesis in pathological settings.
Apold1 controls EC proliferation
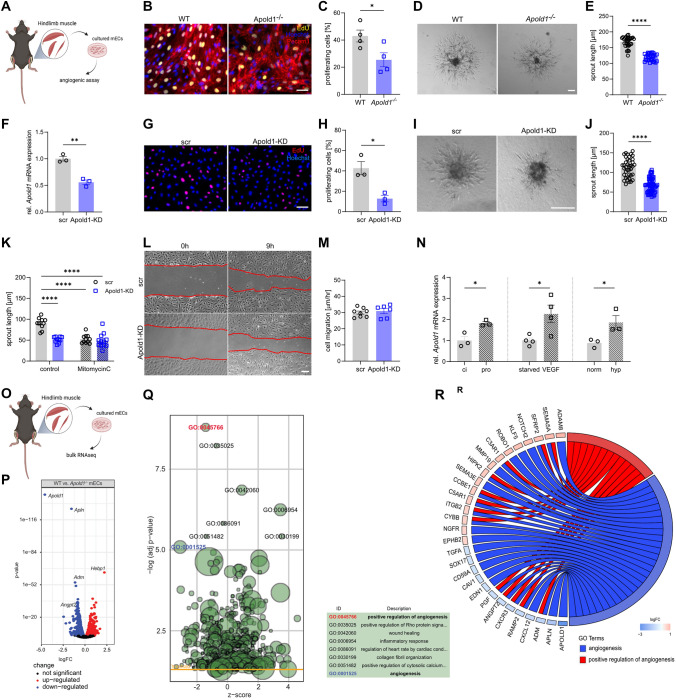
To evaluate whether the ability of Apold1 to control angiogenesis occurs via EC intrinsic mechanisms, we isolated and cultured primary ECs from Apold1−/− mice and wild-type littermates (Fig. 6A). We observed that ECs from Apold1−/− mice proliferated less in highly angiogenic cell culture medium (Fig. 6B, C). Moreover, spheroids created from ECs of Apold1−/− mice showed less sprouting than those from wild-type mice (Fig. 6D, E). To test whether Apold1 is also required for cell proliferation in human umbilical vein endothelial cells (HUVECs), we knocked down Apold1 using lentiviral transfection with short hairpin RNAs (shRNAs), which reduced Apold1 expression by > 50% compared to scrambled controls (Fig. 6F). Similar to our findings in Apold1−/− mECs, Apold1 knockdown (Apold1-KD) in HUVECs reduced proliferation (Fig. 6G, H), and it also dramatically reduced sprouting length in a spheroid assay (Fig. 6I, J). This impairment was dependent on proliferation, since inhibiting proliferation using Mitomycin C lowered sprouting similar to Apold1-KD and prevented any further decrease in sprouting after Apold1-KD (Fig. 6K). Indeed, scratch assays confirmed that migration into the scratch was not impaired after Apold1-KD (Fig. 6L, M).
Fig. 6.
Angiogenesis is impaired in Apold1-deficient cells in vitro. A Experimental design. B, C Representative immunofluorescent images and quantification of four independent experiments of EdU (yellow) labeling of proliferating ECs isolated from skeletal muscle of WT and Apold1−/− mice, co-stained with Hoechst (blue) (n (WT/Apold1−/−) = 4/4. Scale bar, 100 μm). D, E Representative bright-field images and morphometric quantification of average sprout length of sprouting spheroids of mECs isolated from WT and Apold1−/− mice (n (WT/Apold1−/−) = 30/33. Scale bar, 100 μm). F Apold1 knockdown efficiency in HUVECs treated with scrambled (scr) shRNAs or shRNAs against Apold1 (Apold1-KD). G, H Representative immunofluorescent images and quantification of percentage of EdU (red)-labeled proliferating HUVECS to all nuclei stained with Hoechst (blue) in scr and Apold1-KD HUVECs (n (WT/Apold1-KD) = 3/3. Scale bar, 100 μm). I, J Representative image and quantification of sprout length of spheroids of scr and Apold1-KD HUVECs (n (scr/Apold1-KD) = 30/33. Scale bar, 100 μm). K Quantification of sprout length in scr and Apold1-KD HUVECs treated in control conditions or Mitomycin C (n (scr/Apold1-KD) = 10/10). L, M Representative images and quantification of cell migration in scratch assay (n (scr/Apold1-KD) = 7/6). N Relative Apold1 expression in contact-inhibited (ci) and proliferative conditions (pro), after VEGF treatment and in hypoxic condition (0.1% O2) (n (scr/Apold1-KD) = 3-4/3-4). O Experimental design. P Volcano plots with red and blue dots showing significantly changed genes within 5% false discovery rate (FDR) determined by bulk RNAseq of cultured WT vs Apold1−/− mECs (n (WT/Apold1−/−) = 3/3). Q Gene ontology analysis of bulk RNAseq of cultured and WT vs Apold1−/− mECs (n (WT/Apold1−/−) = 3/3). R Overview of genes significantly differentially expressed within the GO terms angiogenesis (blue) and positive regulation of angiogenesis (red) in cultured WT and Apold1−/− mECs. Student’s test in C, E, F, H, J, M, and N; Two-way ANOVA with Tukey’s multiple comparison test in K (*p < 0.05; **p < 0.01; ****p < 0.0001). The data shown are mean ± SEM
Consistent with our in vivo data, we found that Apold1 expression was higher under proliferating angiogenic conditions versus contact inhibition, when ECs are quiescent (Fig. 6N, left). Also, Apold1 increased upon stimulation with VEGF (Fig. 6N, middle), while previous work already showed increased expression upon FGF2 [20]. Furthermore, since hypoxia is a known driver of angiogenesis, we confirmed that Apold1 expression is increased in response to hypoxia in ECs [20, 30, 69, 70] (Fig. 6N, right). In fact, revisiting a published HIF1 ChIP-seq screen in HUVECs under normoxia or after 24 h of hypoxia [38] revealed that hypoxia increases HIF binding to Apold1 very similarly to the well-described HIF1 binding to VEGF (Fig. S5A). Additional analysis of this dataset showed that HIF1a knockdown abolished hypoxia-induced Apold1 expression, further supporting Apold1 regulation by HIF-1α (Fig. S5B). On the other hand, knocking down Apold1 did not affect the regulation of HIF-responsive genes under hypoxia (Fig. S5C). Thus, Apold1 can be activated by angiogenic growth factors as well as hypoxia. Altogether, Apold1 is required to increase EC proliferation in response to a variety of angiogenic stimuli.
To subsequently explore how Apold1 affects angiogenesis, we initially focused on autophagy, since it is a known regulator of endothelial function under pathological conditions [71] and because other apolipoproteins have been linked to autophagy before [72]. Moreover, autophagy was recently reported to be impaired upon Apold1 knockdown in human dermal blood ECs, but we could not find any differences in LC3B nor p62 expression during normal culture conditions, nor under amino acid deprivation, a condition known to activate autophagy (Fig. S6A, B). We next performed RNA sequencing on primary ECs derived from Apold1−/− or WT ECs, which were briefly (16 h) cultured in angiogenic medium (Fig. 6O). We found 2255 genes to be differentially expressed between Apold1−/− and WT mECs (1041 up, 1246 down; FDR adj.p < 0.05; Fig. 6P; see list of differentially expressed genes in Supplementary Table2/Supplementary file 9). We confirmed differential expression of several top regulated genes in muscle 24 h after ischemia induction (Fig. S7A). These results are in striking contrast to the subtle gene expression differences observed under baseline conditions (Fig. 2I) and suggest that pro-angiogenic conditions unmask a profound functional impairment in Apold1-deficient ECs. In support of our findings, GO analysis showed that main top downregulated pathways were linked to angiogenesis and positive regulation of angiogenesis (Fig. 6Q). In fact, most genes included in those GO terms which were affected upon loss of Apold1 all crucially control vascular development and many, when genetically removed, are associated with severe developmental phenotypes or even lethality. These data underscore the role of Apold1 as a regulator of angiogenesis under pathological conditions.
Discussion
In this study, we report that the vascular gene Apold1 is dispensable for developmental angiogenesis but that it crucially contributes to ischemia-induced revascularization of the brain (stroke) and muscle (femoral artery ligation) as well as tumor angiogenesis by controlling EC proliferation. Despite the large phenotypes observed under such conditions, we did not observe a role for Apold1 during developmental angiogenesis. Apold1−/− mice were born at the expected Mendelian frequencies, showed normal development, and showed normal retinal angiogenesis. Furthermore, we did not find any differences in vascular density of adult brain nor muscle, and Apold1−/− mice behaved similar to their WT littermates. These data are remarkable since the constitutive deletion of angiogenic regulators often leads to severe developmental phenotypes or even lethality. In fact, there are only a few genes such as Placental Growth Factor [73], Vegfb [74], Ang2 [75], or Robo4 [76] that upon constitutive deletion do not impair development but only affect angiogenesis in pathological conditions.
It is unclear why Apold1 does not affect developmental angiogenesis while strongly restricting angiogenesis in several pathological settings. We showed that Apold1 expression is activated by VEGF, which is known to activate an immediate early gene response [77], as well as by hypoxia. A previous RNA-seq screen already identified Apold1 to be part of a HIF-dependent angiogenic response downstream of YAP/TAZ in bone ECs [69]. We also found higher Apold1 levels in muscle ECs during hindlimb ischemia and in the brain upon stroke. In those conditions, deleting Apold1 impaired angiogenesis. Previous reports however also observed higher Apold1 expression during development [20], a condition not affected by Apold1 deletion. Notably, Apold1 is also rapidly activated under specific conditions, such as stress, which are not commonly associated with angiogenesis. This might indicate that the role of Apold1 is not restricted to angiogenesis but contributes to the adaptive, homeostatic response to stress, consistent with its original description as an early immediate gene [20]. In such a scenario, Apold1 might fine tune the normal angiogenic response to quickly restore tissue homeostasis.
Given the high expression of Apold1 during development, a previous report evaluated loss of Apold1 in a mouse model of neonatal stroke. Apold1-deficient pups showed reduced angiogenesis after stroke and had impaired long-term functional recovery [26]. However, in adult mice, where Apold1 expression is much lower, acute stroke caused similar size lesions and comparable functional impairment (24–72 h after stroke) in Apold1−/− mice and wild-type controls [33]. In agreement with those observations, we found that stroke size was not different between genotypes, but, importantly, revascularization at the ischemic border was highly reduced in Apold1−/− mice. Because revascularization of the ischemic area is crucial for functional recovery in preclinical mouse models and patients after stroke [78–80], we speculate that loss of Apold1 might impair functional recovery in stroke.
Although little is known about the possible role of Apold1 in cancer, two studies have reported dysregulation of Apold1 expression through DNA methylation. One study reported hypermethylation of Apold1 in testicular germ cell tumors and in testicular embryonal carcinoma NT2 cells, lower Apold1 expression in two types of testicular tumor (seminoma and embryonal carcinoma), and increased Apold1 expression in response to treatment with the demethylating agent 5-azacytidine [81]. In contrast, another study found that Apold1 DNA is hypomethylated and its expression strongly increased in two independent cohorts of patients with colorectal cancer [82]. Vascular heterogeneity, determined by tumor origin and type, is a hallmark feature of cancer and might explain these differences in Apold1 regulation [14]. Therefore, we used single-cell datasets to evaluate Apold1 expression in human cancer. We found that Apold1 expression is restricted to ECs and is higher in tumor ECs when compared to normal ECs [67, 68], confirming its relevance in humans. In a mouse model of melanoma (B16-F10), we found that subcutaneous tumor growth was reduced in Apold1−/− mice. Vessels were smaller, with very small lumens, leading to impaired tumor perfusion. Further experiments showed that ECs proliferated much less, leading to a lower fraction of ECs in the tumor. While these data require confirmation in other, more chronic tumor models, they suggest that inhibiting Apold1 could prevent uncontrolled endothelial proliferation in pathological settings.
Interrogating several mouse as well as human single-cell sequencing datasets, we consistently found that Apold1 expression was restricted to vascular cells, predominantly ECs. Some datasets also revealed expression in pericytes and/or SMCs, but those cells were much less responsive to metabolic challenges, while endothelial Apold1 was highly activated under such conditions. Another manuscript also reported that Apold1 is expressed in platelets, but while Apold1-deficient platelets exhibit increased reactivity to collagen [32], human data currently do not support a role for Apold1 in platelets [31]. Since we used constitutive Apold1−/− mice in our experiments, we cannot rule out a contribution of perivascular cells nor platelets to the observed phenotypes nor can we exclude that Apold1 controls angiogenesis by affecting the crosstalk between ECs and other cells, such as immune cells. Pathway analysis of our sequencing experiments under angiogenic conditions already showed the activation of ‘inflammatory response’ and ‘wound healing’ upon loss of Apold1 which could indicate a potential immune regulatory function. Also, a potential role for APOLD1 in regulating endothelial permeability could affect immune responses during pathological conditions, but while we did not observe vascular leakiness under basal conditions in vivo, previous work using cultured ECs yielded conflicting data [31]. Future work will need to further address these outstanding questions.
Our ex vivo and in vitro experiments all confirmed a strong cell-intrinsic role of Apold1 inhibition leading to reduced endothelial cell proliferation, a key feature of angiogenic ECs. It is noteworthy that Apold1 haploinsufficiency already led to a profound impairment in angiogenesis after ischemia, mimicking the effect of the full knockout and that a 50–70% knockdown of Apold1 in HUVECs in vitro also reproduced the profound impairment in proliferation and sprouting observed in knockout ECs. This suggests that normal levels of Apold1 expression are critical for proper EC function under pathological conditions. How Apold1 controls proliferation remains to be investigated. We found a concerted downregulation of many pro-angiogenic genes and growth factors in Apold1−/− conditions. It is not clear though whether this is directly linked to Apold1 function or secondary to the impaired proliferative capacity downstream of APOLD1.
Previous work has localized APOLD1 to the membrane (presumably focal adhesion sites) [20, 31] or at Weibel–Palade bodies [31]. Unfortunately, we did not find any commercially available antibody that specifically labeled APOLD1 in WT versus Apold1−/− ECs, nor upon the use of overexpression/knockdown approaches in human ECs using Western blot as well as immunohistochemistry (data not shown). In addition, we did not observe changes in autophagy. Likely, the role of APOLD1 is contextual and is dependent on the endothelial source, the growth state of the cell, the stimulus that induces APOLD1 function, and the specific culture conditions. For instance, Stritt et al. used confluent monolayers to evaluate the role of APOLD1 [31], while our experiments specifically aimed to study APOLD1 under angiogenic (non-confluent), growth factor-stimulated conditions.
Taken together, we here show that Apold1 is dispensable for developmental angiogenesis, but that it controls ischemia-induced revascularization of the brain (stroke) and muscle (femoral artery ligation) and regulates pathological tumor angiogenesis, probably largely by controlling EC proliferation. The absence of a developmental phenotype, the observation that Apold1 is activated under ischemic and pro-angiogenic conditions, and the fact that its expression is restricted to vascular endothelial cells could make it an interesting target for future therapeutic interventions to either enhance vascular repair or to restrict tumor vascularization.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
KDB is endowed by the Schulthess foundation. The lab of KDB is funded by the ETH Zurich and SNSF Grant 31003A_208041. The lab of JB is funded by the ETH Zurich, the ETH Project Grant ETH-20 19-1, SNSF Grant 310030_172889, Swiss 3R Competence Center, Botnar Research Center for Child Health, and Multi-Investigator Project. Apold1-/- mice were generously provided by Prof. Paul Worley. We thank Han-Yu Lin for skilled technical assistance and Prof. Sonia Tugues Solsona for thoughtful discussions. We thank Laura Hinte and Prof. Ferdinand von Meyenn for helpful discussions. We thank the ETH Zurich Flow Cytometry Core Facility for the assistance.
Author contributions
ZF, RA, JB, and KDB contributed to the study conception and design. ZF and RA performed and analyzed most experiments. AAB, LVZ, AEM, and PLG performed bioinformatic analysis of RNA-seq datasets. RR, RWe, and Rwa performed and analyzed stroke experiments. RWa helped with RNAseq. HD performed and analyzed retina experiments. JZ and TD assisted with hindlimb ischemia surgeries and analysis. OS performed behavior experiments. MR performed blood–brain barrier permeability experiments. MES, JB, and KDB supervised the experiments and provided funding. The manuscript was written by ZF, RA, JB, and KDB. All authors read and approved the final manuscript.
Funding
Open access funding provided by Swiss Federal Institute of Technology Zurich.
Data availability
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession codes GSE217694.
Declarations
Conflict of interest
The authors disclose no financial or non-financial interests related to this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zheng Fan and Raphaela Ardicoglu equally contributed.
Contributor Information
Johannes Bohacek, Email: johannes.bohacek@hest.ethz.ch.
Katrien De Bock, Email: katrien-debock@ethz.ch.
References
- 1.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 3.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 4.Gorski T, De Bock K. Metabolic regulation of exercise-induced angiogenesis. Vasc Biol. 2019;1:H1–H8. doi: 10.1530/VB-19-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishijima T, Torres-Aleman I, Soya H. Chapter 11 - Exercise and cerebrovascular plasticity. In: Masamoto K, Hirase H, Yamada K, editors. Progress in Brain Research. Elsevier; 2016. pp. 243–268. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds LP, Killilea SD, Redmer DA. Angiogenesis in the female reproductive system. FASEB J. 1992;6:886–892. doi: 10.1096/fasebj.6.3.1371260. [DOI] [PubMed] [Google Scholar]
- 7.Augustin HG. Angiogenesis in the female reproductive system. Mech Angiogenesis. 2005 doi: 10.1007/3-7643-7311-3_3. [DOI] [PubMed] [Google Scholar]
- 8.Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A, Zakeri P, Mousa M, et al. Angiogenesis goes computational - the future way forward to discover new angiogenic targets? Comput Struct Biotechnol J. 2022;20:5235–5255. doi: 10.1016/j.csbj.2022.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeltsch M, Leppänen V-M, Saharinen P, Alitalo K. Receptor tyrosine kinase-mediated angiogenesis. Cold Spring Harb Perspect Biol. 2013;5:a009183. doi: 10.1101/cshperspect.a009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza GL. Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol. 2010;30:648–652. doi: 10.1161/ATVBAHA.108.181644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 13.De Bock K, Mazzone M, Carmeliet P. Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nat Rev Clin Oncol. 2011;8:393–404. doi: 10.1038/nrclinonc.2011.83. [DOI] [PubMed] [Google Scholar]
- 14.De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17:457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 15.Chung AS, Lee J, Ferrara N. Targeting the tumour vasculature: insights from physiological angiogenesis. Nat Rev Cancer. 2010;10:505–514. doi: 10.1038/nrc2868. [DOI] [PubMed] [Google Scholar]
- 16.Wälchli T, Wacker A, Frei K, et al. Wiring the vascular network with neural cues: a CNS perspective. Neuron. 2015;87:271–296. doi: 10.1016/j.neuron.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 17.Augustin HG. Translating angiogenesis research into the clinic: the challenges ahead. BJR. 2003;76:S3–S10. doi: 10.1259/bjr/68078705. [DOI] [PubMed] [Google Scholar]
- 18.Jain RK. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26:605–622. doi: 10.1016/j.ccell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comunanza V, Bussolino F. Therapy for cancer: strategy of combining anti-angiogenic and target therapies. Front Cell Dev Biol. 2017;5:101. doi: 10.3389/fcell.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regard JB, Scheek S, Borbiev T, et al. Verge: a novel vascular early response gene. J Neurosci. 2004;24:4092–4103. doi: 10.1523/JNEUROSCI.4252-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonsen ML, Alessio HM, White P, et al. Acute physical activity effects on cardiac gene expression. Exp Physiol. 2010;95:1071–1080. doi: 10.1113/expphysiol.2010.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catoire M, Mensink M, Boekschoten MV, et al. Pronounced effects of acute endurance exercise on gene expression in resting and exercising human skeletal muscle. PLoS One. 2012;7:e51066. doi: 10.1371/journal.pone.0051066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickinson JM, D’Lugos AC, Naymik MA, et al. Transcriptome response of human skeletal muscle to divergent exercise stimuli. J Appl Physiol. 2018;124:1529–1540. doi: 10.1152/japplphysiol.00014.2018. [DOI] [PubMed] [Google Scholar]
- 24.Floriou-Servou A, von Ziegler L, Stalder L, et al. Distinct proteomic, transcriptomic, and epigenetic stress responses in dorsal and ventral hippocampus. Biol Psychiatry. 2018;84:531–541. doi: 10.1016/j.biopsych.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Roszkowski M, Manuella F, von Ziegler L, et al. Rapid stress-induced transcriptomic changes in the brain depend on beta-adrenergic signaling. Neuropharmacology. 2016;107:329–338. doi: 10.1016/j.neuropharm.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 26.Mirza MA, Capozzi LA, Xu Y, et al. Knockout of vascular early response gene worsens chronic stroke outcomes in neonatal mice. Brain Res Bull. 2013;98:111–121. doi: 10.1016/j.brainresbull.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keenan SW, Hill CA, Kandoth C, et al. Transcriptomic responses of the heart and brain to anoxia in the western painted turtle. PLoS One. 2015;10:e0131669. doi: 10.1371/journal.pone.0131669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma P, Bansal A, Sharma PC. RNA-seq-based transcriptome profiling reveals differential gene expression in the lungs of Sprague-Dawley rats during early-phase acute hypobaric hypoxia. Mol Genet Genomics. 2015;290:2225–2240. doi: 10.1007/s00438-015-1064-0. [DOI] [PubMed] [Google Scholar]
- 29.Trollmann R, Rehrauer H, Schneider C, et al. Late-gestational systemic hypoxia leads to a similar early gene response in mouse placenta and developing brain. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1489–R1499. doi: 10.1152/ajpregu.00697.2009. [DOI] [PubMed] [Google Scholar]
- 30.Zhou L, Wang LM, Song HM, et al. Expression profiling analysis of hypoxic pulmonary disease. Genet Mol Res. 2013;12:4162–4170. doi: 10.4238/2013.October.7.2. [DOI] [PubMed] [Google Scholar]
- 31.Stritt S, Nurden P, Nurden AT, et al. APOLD1 loss causes endothelial dysfunction involving cell junctions, cytoskeletal architecture, and Weibel-Palade bodies, while disrupting hemostasis. Haematologica. 2022 doi: 10.3324/haematol.2022.280816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Cañestro C, Bonetti NR, Wüst P, et al. Apold1 deficiency associates with increased arterial thrombosis in vivo. Eur J Clin Invest. 2020;50:e13191. doi: 10.1111/eci.13191. [DOI] [PubMed] [Google Scholar]
- 33.Liu F, Turtzo LC, Li J, et al. Loss of vascular early response gene reduces edema formation after experimental stroke. Exp Transl Stroke Med. 2012;4:12. doi: 10.1186/2040-7378-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 35.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 36.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mimura I, Nangaku M, Kanki Y, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible Factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giordani L, He GJ, Negroni E, et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell. 2019;74:609–621.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 42.Wu YE, Pan L, Zuo Y, et al. Detecting activated cell populations using single-cell RNA-Seq. Neuron. 2017;96:313–329.e6. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 43.Floriou-Servou A, von Ziegler L, Waag R, et al. The acute stress response in the multiomic era. Biol Psychiat. 2021;89:1116–1126. doi: 10.1016/j.biopsych.2020.12.031. [DOI] [PubMed] [Google Scholar]
- 44.von Ziegler LM, Floriou-Servou A, Waag R, et al. Multiomic profiling of the acute stress response in the mouse hippocampus. Nat Commun. 2022;13:1824. doi: 10.1038/s41467-022-29367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeisel A, Hochgerner H, Lönnerberg P, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with bioconductor. F1000Research. 2016 doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rust R, Grönnert L, Gantner C, et al. Nogo-A targeted therapy promotes vascular repair and functional recovery following stroke. Proc Natl Acad Sci. 2019;116:14270–14279. doi: 10.1073/pnas.1905309116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber RZ, Grönnert L, Mulders G, et al. Characterization of the blood brain barrier disruption in the photothrombotic stroke model. Front Physiol. 2020;11:586226. doi: 10.3389/fphys.2020.586226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rust R, Kirabali T, Grönnert L, et al. A practical guide to the automated analysis of vascular growth, maturation and injury in the brain. Front Neurosci. 2020 doi: 10.3389/fnins.2020.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rust R, Grönnert L, Dogançay B, Schwab ME. A revised view on growth and remodeling in the retinal vasculature. Sci Rep. 2019;9:3263. doi: 10.1038/s41598-019-40135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Muri J, Fitzgerald G, et al. Endothelial lactate controls muscle regeneration from ischemia by inducing M2-like macrophage polarization. Cell Metab. 2020;31:1136–1153.e7. doi: 10.1016/j.cmet.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Z, Turiel G, Ardicoglu R, et al. Exercise-induced angiogenesis is dependent on metabolically primed ATF3/4+ endothelial cells. Cell Metab. 2021;33:1793–1807.e9. doi: 10.1016/j.cmet.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masschelein E, D’Hulst G, Zvick J, et al. Exercise promotes satellite cell contribution to myofibers in a load-dependent manner. Skeletal Muscle. 2020;10:21. doi: 10.1186/s13395-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roszkowski M, Bohacek J. Stress does not increase blood-brain barrier permeability in mice. J Cereb Blood Flow Metab. 2016;36:1304–1315. doi: 10.1177/0271678X16647739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Bock K, Georgiadou M, Schoors S, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154:651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 56.Schaum N, Karkanias J, Neff NF, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalucka J, de Rooij LPMH, Goveia J, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 58.Crowell HL, Soneson C, Germain P-L, et al. Muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat Commun. 2020;11:6077. doi: 10.1038/s41467-020-19894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerhardt H, Golding M, Fruttiger M, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- 62.Sangwung P, Zhou G, Nayak L, et al. KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight. 2017 doi: 10.1172/jci.insight.91700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dekker RJ, Boon RA, Rondaij MG, et al. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 64.Helker CS, Eberlein J, Wilhelm K, et al. Apelin signaling drives vascular endothelial cells toward a pro-angiogenic state. eLife. 2020;9:e55589. doi: 10.7554/eLife.55589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JJ, Arpino J-M, Yin H, et al. Systematic interrogation of angiogenesis in the ischemic mouse hind limb. Arterioscler Thromb Vasc Biol. 2020;40:2454–2467. doi: 10.1161/ATVBAHA.120.315028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Limbourg A, Korff T, Napp LC, et al. Evaluation of postnatal arteriogenesis and angiogenesis in a mouse model of hind-limb ischemia. Nat Protoc. 2009;4:1737–1746. doi: 10.1038/nprot.2009.185. [DOI] [PubMed] [Google Scholar]
- 67.Lambrechts D, Wauters E, Boeckx B, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 68.Goveia J, Rohlenova K, Taverna F, et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37:21–36.e13. doi: 10.1016/j.ccell.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Sivaraj KK, Dharmalingam B, Mohanakrishnan V, et al. YAP1 and TAZ negatively control bone angiogenesis by limiting hypoxia-inducible factor signaling in endothelial cells. eLife. 2020;9:e50770. doi: 10.7554/eLife.50770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartoszewski R, Moszyńska A, Serocki M, et al. Primary endothelial cell–specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. FASEB J. 2019;33:7929–7941. doi: 10.1096/fj.201802650RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schaaf MB, Houbaert D, Meçe O, Agostinis P. Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ. 2019;26:665–679. doi: 10.1038/s41418-019-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pays E. The function of apolipoproteins L (APOLs): relevance for kidney disease, neurotransmission disorders, cancer and viral infection. FEBS J. 2021;288:360–381. doi: 10.1111/febs.15444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carmeliet P, Moons L, Luttun A, et al. Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med. 2001;7:575–583. doi: 10.1038/87904. [DOI] [PubMed] [Google Scholar]
- 74.Zhang F, Tang Z, Hou X, et al. VEGF-B is dispensable for blood vessel growth but critical for their survival, and VEGF-B targeting inhibits pathological angiogenesis. Proc Natl Acad Sci USA. 2009;106:6152–6157. doi: 10.1073/pnas.0813061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 76.Jones CA, London NR, Chen H, et al. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat Med. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanki Y, Muramatsu M, Miyamura Y, et al. Bivalent-histone-marked immediate-early gene regulation is vital for VEGF-responsive angiogenesis. Cell Rep. 2022;38:110332. doi: 10.1016/j.celrep.2022.110332. [DOI] [PubMed] [Google Scholar]
- 78.Nih LR, Gojgini S, Carmichael ST, Segura T. Dual-function injectable angiogenic biomaterial for the repair of brain tissue following stroke. Nat Mater. 2018;17:642–651. doi: 10.1038/s41563-018-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rust R, Grönnert L, Weber RZ, et al. Refueling the ischemic CNS: guidance molecules for vascular repair. Trends Neurosci. 2019;42:644–656. doi: 10.1016/j.tins.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 80.Krupinski J, Kaluza J, Kumar P, et al. Role of angiogenesis in patients with cerebral ischemic stroke. Stroke. 1994;25:1794–1798. doi: 10.1161/01.str.25.9.1794. [DOI] [PubMed] [Google Scholar]
- 81.Cheung HH, Lee TL, Davis AJ, et al. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Øster B, Linnet L, Christensen LL, et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer. 2013;132:2303–2315. doi: 10.1002/ijc.27921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession codes GSE217694.