Abstract
MicroRNAs (miRNAs) are clinical biomarkers for various human diseases, including cancer. They have been found in liquid biopsy samples, including various bodily fluids. They often play an important role in the early diagnosis and prognosis of cancer, and the development of simple and effective analytical methods would be of pivotal importance for the entire community. The determination of these targets may be affected by the different physicochemical parameters of the specimen of interest. In this work, an electrochemical detection platform for miRNA based on a screen-printed gold electrode was developed. In the present study, miRNA-21 was selected as a model sequence, due to its role in prostate, breast, colon, pancreatic, and liver cancers. A DNA sequence modified with methylene blue (MB) was covalently bound to the electrochemical strip and used to detect the selected target miRNA-21. After optimization of selected parameters in standard solutions, including the study of the effect of pH, the presence of interferent species, and NaCl salt concentration in the background, the application of square-wave voltammetry (SWV) technique allowed the detection of miRNA-21 down to a limit in the order of 2 nM. The developed device was then applied to several urine samples. In this case too, the device showed high selectivity in the presence of the complex matrix, satisfactory repeatability, and a limit of detection in the order of magnitude of nM, similarly as what observed in standard solutions.
Keywords: miRNA, Screen-printed, Electroanalysis, Cancer, Urine
Introduction
Prostate cancer (PCa) has an incidence of more than 1.2 million new cases and 350,000 deaths yearly [1]. Statistics report that PCa is the second most common cause of cancer mortality in men globally [2]. The chances of occurrence increase significantly with age, with > 85% of newly diagnosed individuals are over 60 years old [3]. The outlook for distant metastatic PCa (mPCa) is poor, and the overall 5-year survival rate is only 31% [1]. Therefore, diagnosis in the initial stage of the disease, prior to the development of metastatic sites, is vital to reduce mortality and to the successful onset of treatment.
Regular monitoring of prostate-specific antigens (PSA) and digital rectum examinations (DREs) are the gold standard screening technique for prostate cancer in clinical practice [4–6]. However, the precision of these approaches for detecting PCa is limited. PSA analysis has low specificity, resulting in a high risk of false positives or negatives. Biopsies are unpleasant processes that can result in infections, incontinence, and erectile dysfunction. Moreover, it has not been possible to this date to specify a precise range of PSA levels in mPCa [7].
Additionally, DREs are subjective tests and the degree of precision depends on the examiner's experience [8]. In a meta-analysis, DRE has shown a sensitivity and a positive predictive value (PPV) of 17.8% for PCa [6]. Recently, there has been an upsurge in liquid biopsies that use circulating tumor cells, exosomes, DNA, RNA, or microRNAs (miRNA) as biomarkers. Liquid biopsies are gaining ground as routine, minimally invasive tests for regular monitoring, early diagnostics, and monitoring of chronic and recurring diseases [9–14]. Research shows that miRNAs play an important role in the development of various diseases, including oncogenesis and metastasis [15–17]. Some type of miRNA control gene expression post-transcriptionally by repressing or degrading mRNA transcripts [18]. The presence of specific miRNA in various bodily fluids makes them circulating biomarkers of high interest for the research in the field of liquid biopsy [19].
Recent observations indicate that a “signature” of determined groups of microRNA widely influence pathways associated with tumor development, progression, and response to therapy [20]. MiRNAs can thus act as fingerprints to detect disease, making them a viable biomarker. For example, ongoing studies focused on lung cancer, blood miRNAs associated with individual risk stratification by low-dose computed tomography (LDCT) are currently object of a large clinical trial (> 4000 participants) at the Multicentric Italian Lung Detection (MILD). The study demonstrates that the combination of a prespecified circulating miRNA signature and LDCT improves the prediction of individual lung cancer incidence and mortality over LDCT alone [21]. In the field of breast cancer, large trials have been designed using multi-gene expression to personalize the therapies, including OncotypeDX, TAILORx, and RxPONDER [22–24]. These trials could allow in future to address specific markers of response, facilitating tumor eradication and limiting excessive therapies. In addition, the scientific community is now considering the relevance of miRNA (both tumor and circulating) to be added to the markers’ list for triple-negative breast cancer, as reported in different studies [24]. The effectiveness of circulating miRNAs as diagnostic biomarkers for the precise and early diagnosis of PCa has been reported in multiple studies [25–27]. Even if not validated by massive clinical trials, many recent studies have reported promising findings relative to using miRNAs for future diagnostic/prognostic applications.
So far, considerable effort has been made to develop sensitive and selective analytical methods for detecting miRNA. The main traditional assays developed to analyze miRNA molecules include real-time polymerase chain reaction (RT-PCR) [28], Northern blot [29], in situ hybridization (ISH) with locked nucleic acid probes [30], next-generation sequencing (NGS) [31], fluorescence in situ hybridization (FISH) [32], and microarrays [33]. These techniques tend to have limitations such as the requirement of qualified manpower, being time-consuming, and the need to access to laboratory facilities with costly high-end instruments [34]: those may include the isolation of target, gel denaturation, transfer to external/solid support (blotting), and the use of specific equipment and tools. A cost in the range of ca. 1000 Eur is associated to these approaches, and this does not include the cost of equipment. In order to overcome limitations caused by laboratory-bound approaches, a wide range of sensor- and biosensor-based techniques can be used, based on colorimetry, fluorescence, based on cantilevers, and electrochemical. For example, Choi and Seo reported a hairpin structure-mediated diagnostic method for the simple and rapid colorimetric detection of miRNA with high sensitivity (LOD = 132 aM) and high selectivity [35]. Nemati and Hosseini detected miRNA by an enzyme-free fluorescence probe with a 5 pM detection limit [36]. Recently, Andrade et al. reported a nanomechanical genosensor based on an atomic force microscope (AFM) cantilever functionalized with complementary strands of miRNA-203 and miRNA-205 which are potential biomarkers of metastasis in head and neck squamous cell carcinomas (HNSCC) [37]. Although these methods showed an acceptable range of accuracy and precision in detecting miRNA in biological samples, electrochemical architectures are promising, due do their miniaturization potential, simple experimental setup, and direct application towards colored matrices, where colorimetric methods often meet their main limitation. In addition, they can be designed for multiplexed measurements [38, 39]. The adoption of miniaturized electrochemical sensors, exploiting the advantage of microfabrication, and the use of hybrid nanoprobes have demonstrated to be satisfactorily applied towards the detection of clinically relevant molecules, i.e., exosomes, protein, nucleic acids, and enzymes [40]. Several different electrochemical architectures have been reported to detect circulating miRNAs: for example, miRNA-492 has been detected in serum samples through the nanoengineering of a paper-based electrode with a peptide nucleic acid (PNA) probe that was coupled to an external ruthenium-based redox probe, producing a current signal [41]. A more sophisticated approach was developed exploiting DNA nanotechnology, with a novel concept of dynamic nanomachine, defined as “walking and rolling.” This approach is powered by two enzymes, namely nuclease and nicking endonuclease, and the presence of pH-controlled triplex structures, which allows the reversible use of the nanomachine. The main drawback of this approach is that several steps are necessary. Authors achieved an ultra-low detection limit in the order of 10−18 M [42]. A different strategy, based on the formation of a miRNA/DNA assembly was used to detect miRNA sequences, that was electrochemically quantified by observing the hydrogen peroxide reduction in the presence of HRP-labeled p53 and hydroquinone [43]. Another novel and sophisticated strategy to detect miRNA-141 uses the RNA-triggered copper ion reduction method. Authors reported the possibility to detect a miRNA target in the presence of DNA recognition strand, dithiothreitol, and reducing agents: briefly, the presence of target miRNA induces the reduction of Cu2+ with a subsequent change in electrochemical signals generated from the remaining Cu2+ [44]. Although different approaches have been developed, it should be noted that some of these are based on complex and time-consuming procedures, often involving the use of external reagents to carry out the assay. In addition to this, the reported application to real samples is scarce. Test in real complex biological media are of high relevance to understand the drawbacks of a technique and evaluate troubleshooting approaches. Relatively to prostate cancer, a recent preliminary study highlighted the possibility of discriminating prostate cancer patients from healthy controls by detecting a signature of miRNAs, including miRNA-21 [45], and urine seems to be a promising matrix to be investigated [46]. The electrochemical platform herein reported (to be also adapted to other circulating miRNAs) represents an easy-to-use and quick cheap device that does not require skilled technicians and/or equipment like for the RT-qPCR. In particular, the adoption of RT-qPCR is also not suitable for portable screening tool, considering RNA is first transcribed into complementary DNA by reverse transcriptase, and the use of primers and proprietary instruments are necessary for the quantification.
According to these findings, we report in the present study the development of a screen-printed electrochemical platform for the determination of miRNA-21 in real urine samples. The portable sensing platform, to be used in decentralized settings, has been developed on gold-based screen-printed electrodes. The herein reported work describes the detailed fabrication procedure of the disposable platform, which uses a methylene blue tagged immobilized DNA probe, providing an integrated sensing platform without the necessity of additional/time consuming tasks to carry out the determination. The study focuses in particular on the effect of pH and salt concentration and their effect on the sensitivity of the platform. The optimized probes and salt concentrations were then applied to a preliminary study in real urine samples. According to the results, miRNA-21 has been detected down to a low nanomolar range with high selectivity. The reported results open the possibility of developing multiplex screen-printed devices for miRNA signature determination in heterogenous urine samples and the use of such samples for liquid biopsy purposes.
Experimental section
Materials and apparatus
Sodium chloride (NaCl), sodium hydrogen phosphate (Na2HPO4), magnesium chloride (MgCl2), 6-Mercapto-1-hexanol (MCH, C6H14OS), and tris(2-carboxyethyl)phosphine (TCEP; C9H15O6P) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The MB-DNA probe selective to miRNA-21 (5′thiol C6 SS-TCAACATCAGTCTGATAAGCTA- methylene blue 3′) and the miRNA-21 target (5′UAGCUUAUCAGACUGAGUUGA 3′) were purchased from LGC Biosearch technologies, Hoddesdon, UK. Thin-film gold electrodes (ref. DRP-PW-AU10) were purchased from Metrohm DropSens. Deionized water (DI) was used to perform the experiments. All solvents and chemicals were of analytical grade. The urine samples have been collected directly from volunteers working at CSEM SA—Center Landquart. Electrochemical measurements were performed with a potentiostat MultiPalmSens 4, 8 channels (PalmSens, the Netherlands), and data were recorded with potentiostat MultiPalmSens software PsTrace interfaced with a laptop.
Sensor fabrication on screen-printed gold electrodes
Before the immobilization of the probe, the working electrode surface was cleaned by following a protocol reported by the research group of Plaxco [47].To do this, we first performed a series of cyclic voltammetry (CV) cycles in 0.5 M H2SO4, scanning from 0 to + 1.5 V (step of 1 mV at a scan rate of 1 V/s), until the gold peak became stable (approximately 20 scans). After thorough rinsing with DI water, the active area of the electrode was dried, avoiding contact with the three-electrode system. After this, the immobilization of the oligonucleotide probe was carried out in the following three main steps [48]: (1) the 100 μM MB-DNA probe, dissolved in 50 mM phosphate buffer pH = 7, 250 mM NaCl, 10 mM MgCl2, was first reduced in the presence of 0.01 M TCEP for 1 h prior to being immobilized; (2) the resulting solution was diluted in 50 mM phosphate buffer to the chosen concentration (in the nanomolar range) for immobilization on the screen-printed working electrode. To do this, 6 μL of the diluted solution was added onto the working electrode for 1 h in a humidity chamber at room temperature. Subsequently, the working electrode surface was carefully rinsed with DI water. Finally, (3) the working electrode was incubated with 6 μl of 2 mM MCH in a humidity chamber, to passivate the remaining active sites on the working electrode and to define the orientation of the immobilized MB-DNA probes. Before being used, the electrode was rinsed with DI water to eliminate the unbound MCH. The main steps for modification are reported in Fig. 1A.
Fig. 1.
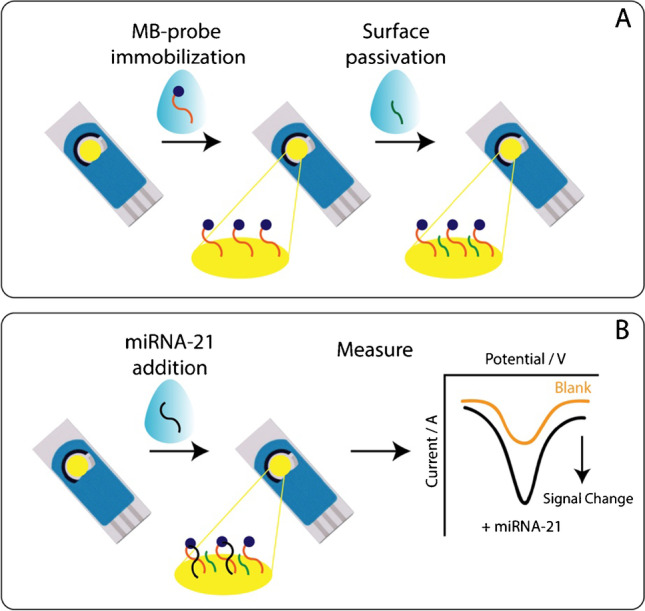
A Schematic representation of electrochemical device fabrication and B principle of miRNA-21 detection
Measurement of miRNA target
Our electrochemical measurements for the miRNA target were performed at room temperature with eight electrodes inserted into a multi-eight potentiostat connected to a laptop. Square wave voltammetry (SWV) was performed using the following parameters (T equilibration = 5 s, E begin = 0.1 V, E end = − 0.4 V, E step = 0.001 V, amplitude = 0.04 V, frequency = 5–1000 Hz). Briefly, the sensing architecture is based on the affinity between the miRNA-21 and the immobilized probe. When the target is recognized at the modified surface of the electrode, the signal due to the electron transfer, mediated by methylene blue tag, is affected. The signal change is correlated with the presence of miRNA-21 that is hybridized with the complementary DNA probe that is immobilized on top of the working electrode. The signal change is based on changes in electron transfer as a function of reporter separation from the electrode surface. From a physical point of view, two populations of probes exist on the electrode with different rates of electron transfer, one slower than the other. Because the rates are different, the time constants, i.e., half-life, of the current decay after pulsing the voltage will be different for each of the two populations. By adjusting the frequency of the square wave, the frequency of measurements is tuned to the time constant of the electron transfer process for the populations. When the high frequency is tuned to the time constant of the folded population, it increases upon target binding, and the consequence is the possibility to have both ON and OFF responses. With hybridization systems, the oligo-duplex may be so rigid that is not possible to experimentally match the time constant of the electron transfer event (low frequency). To our knowledge, in literature the existence of signal OFF and signal ON frequencies has been described towards the detection of the target analyte [49].
The modified electrode was stabilized for 30 min in a buffer solution before detecting the target. After 30 min, the signal of the immobilized probe was considered stable and the background signal (blank) was registered (Fig. 1B). Following the background detection, the sample solution containing the miRNA target in the nanomolar range was added to the surface of the sensor, and after 20 min, the signal was recorded. For in the determination of the calibration curve, the % signal change I% was plotted against the concentration of the target analyzed. With the use of the following equation, we were able to calculate the percentage of signal change for a given frequency:
| 1 |
where Itarget represents the current signal obtained in presence of miRNA-21 and I0 represents the current signal obtained in the absence of target.
Results and discussion
Optimization of experimental parameters in standard solution
All the experimental parameters have been optimized to develop a biosensor adapted to detect miRNA-21 in real urine matrix with minimal sample preparation. The electroanalytical platform is based on a binding-induced conformational change of an immobilized oligonucleotide probe containing a redox reporter, in the present case methylene blue. When the target is present in the solution, it leads to a change in electron transfer that is quantified by using square-wave voltammetry. The optimization was performed in 50 mM phosphate buffer solutions pH = 7 (containing 10 mM MgCl2) and different concentrations of sodium chloride, depending on the experimental conditions. The adjustment of ionic strength was achieved by variation of sodium chloride concentration. The signal change in function of the frequency has been calculated from the data collected by varying the square-wave frequency between 5 and 1000 Hz. These electrochemical architectures can be divided into “signal on” and “signal off,” respectively. The presence of the target increases and decreases the peak current [50–52]. In the present case, we observe a”signal on” behavior.
It should be noted that an electrostatic repulsion exists between negatively charged target and probe. In such case, high ionic strength is required for the successful binding of the two molecules [53]. To perform these studies, the final configuration of the working electrode was obtained by modifying it with a 250-nM probe solution that gave the best compromise in terms of sensitivity and signal-to-noise ratio. In fact, 125 and 500 nM were also tested for the manufacturing of the device, but the former was consistent with a low amount of immobilized probes, giving low current, and the latter resulted in a too high concentration of probe on the sensor surface, creating a steric hindrance which affected probe-target recognition and resulting in low signal change.
For these experiments, 50 mM phosphate buffer at pH equal to 7 was used as the working solution to which the following concentrations of NaCl were added: 100, 250, 500, and 1000 mM. For all the experiments, a concentration of 60 nM miRNA-21 was used. As shown in Fig. 2, the device is characterized by a “signal on” change, and a signal change increase can be observed as the ionic strength increases. This is ascribable to the higher sodium concentration, which promotes probe-target recognition. In addition, the higher signal change is associated with low square-wave frequency in the 10–30 Hz. Among these, the signal at 15 Hz represented a good compromise in terms of sensitivity and repeatability. The frequency tests have been performed up to 1000 Hz; however from 100 to 1000 Hz, signal change was not significant. By consequence, the frequency of 15 Hz was chosen to analyze various concentrations of miRNA-21 in buffer solutions and in urine, as reported in Figs. 3, 4, and 5.
Fig. 2.
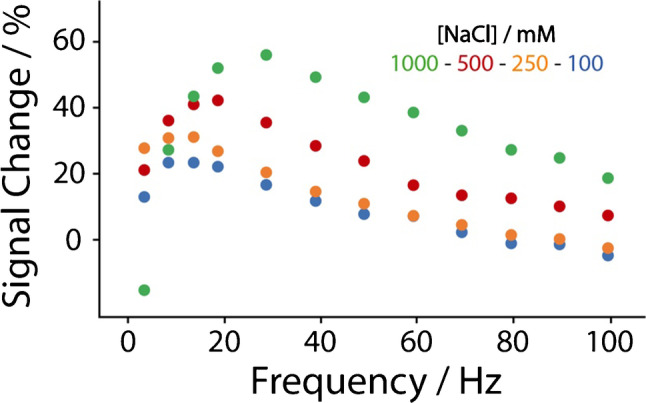
Evaluation of the correlation between square-wave frequency and signal change using different concentrations of sodium chloride in 50 mM phosphate buffer pH = 7 (containing 10 mM MgCl2), namely 100 (blue dots), 250 (orange dots), 500 (red dots), and 1000 mM (green dots). All the studies have been performed in triplicate, the reported value is the mean of measurements (standard deviation < 10%, not shown), and the tested concentration of miRNA-21 was of a nominal value of 60 nM
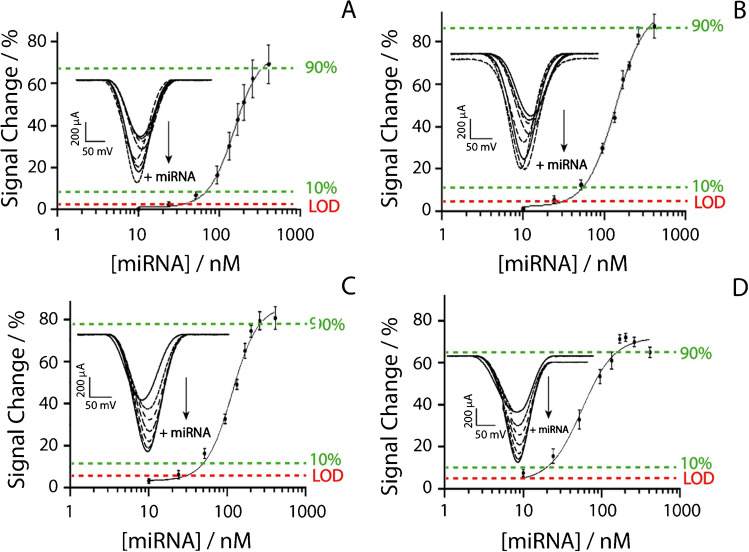
Fig. 3.
Binding curves obtained at 15 Hz in presence of miRNA-21 ranging from 10 to 410 nM in 50 mM phosphate buffer solution spiked with A 100, B 250, C 500, and D 1000 mM of sodium chloride. Insets of each figures display the “signal on” voltametric curves responding to the increase of miRNA-21. All the studies have been performed in triplicate
Fig. 4.
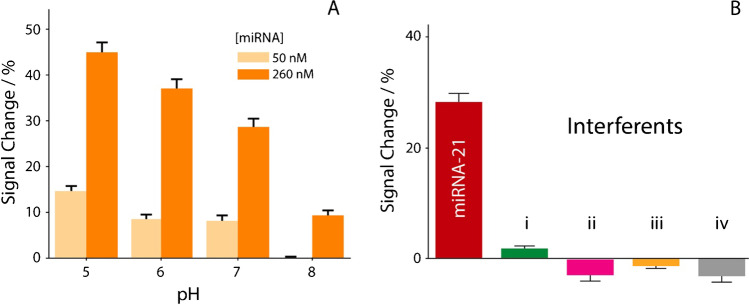
A Evaluation of the effect of pH in buffer solutions spiked with 250 mM sodium chloride. The effect of pH was investigated ranging from 5 to 8, using two fixed level of miRNA-21, namely 50 nM (light bars) and 260 nM (dark bars). B Selectivity studies in buffer solutions, comparing the signal intensities obtained in the presence of 50 nM miRNA-21 (red bar) and in the presence of two RNA single strands (i, ii) and two DNA single strands (iii, iv). All the studies have been performed in triplicate
Fig. 5.
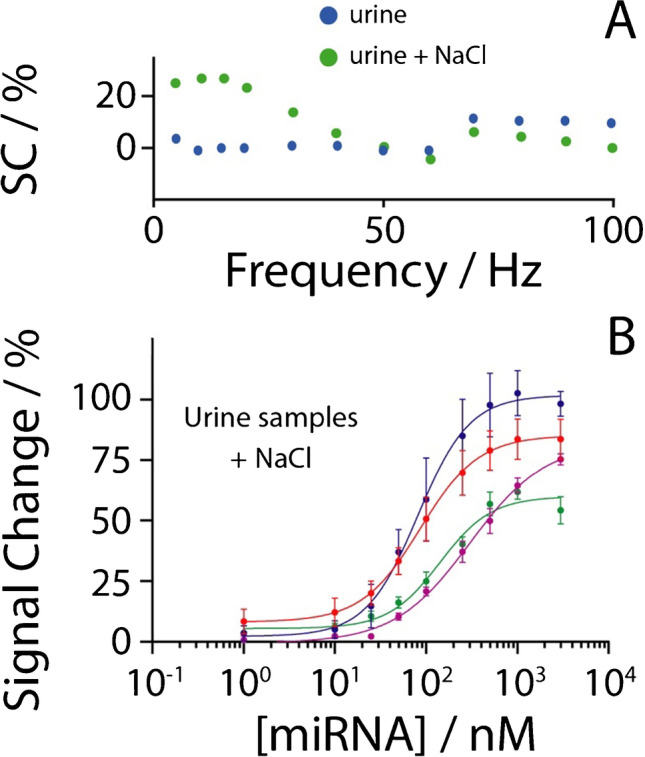
A Effect of the addition of 250 mM sodium chloride in urine sample application. B Binding curves in the presence of miRNA-21 ranging from 10 to 3000 nM in four different urine samples with the addition of sodium chloride. All the studies have been performed in triplicate
All the experiments have been carried out in the presence of miRNA-21 ranging from 10 to 410 nM, utilizing 15 Hz as the square-wave frequency and varying the concentration of sodium chloride from 100 to 1000 mM. Depending on the different concentrations of sodium chloride, each platform was able to detect miRNA-21 down to 34, 31, 30, and 8 nM, respectively, adding 100, 250, 500, and 1000 mM to the buffer solution. The dynamic ranges were calculated for each set of parameters as the target concentration which produced a signal change comprised between 10 and 90% of the maximum: the use of 100, 250, 500, and 1000 mM sodium chloride produced the dynamic ranges of 70–345 nM, 50–340 nM, 50–270 nM, 20–160 nM, respectively. Interestingly, when 250 and 500 mM sodium chloride were added, the curves showed very similar performance in terms of detection limit and repeatability (ca. 10%, calculated as the relative standard deviation). All the analytical features have been included in following Table 1.
Table 1.
Analytical performance of the developed e-platform for miRNA-21 detection
| NaCl level (mM) | LOD (nM) | Dynamic range (nM) | CV (%) |
| 100 | 34 | 70–345 | 15 |
| 250 | 31 | 50–340 | 8 |
| 500 | 30 | 50–270 | 9 |
| 1000 | 8 | 20–160 | 12 |
| Recovery studies | |||
| Added (nM) | Founded (nM) | Recovery (%) | |
| 50 | 45 | 90 | |
| 100 | 105 | 105 | |
Considering that the physiological range of sodium chloride in the urine ranges in general from 20 to 250 mM, and based on the abovementioned results, it was decided to proceed with the analysis in urine by supplementing the raw samples with an additional amount of sodium chloride to reach exogenous 250 mM, in order to reach a NaCl level comprised between 250 and 500 mM.
Evaluation of the pH effect and interferents
The effect of the variation of pH was investigated, because for real matrix application in urine, the pH might be diverse among the various samples, varying in general from pH 4.6 to pH 8 [54]. In particular, a high pH value (generally > 8.5–9) can lead to the denaturation of the probe/target double helix. In contrast, a low value of pH (< 3) can lead to the hydrolysis of the nucleic acid strands [55]. The effect of the pH on the sensing architecture was evaluated by varying the pH of the sensing buffer solution between 5 and 8, as reported in Fig. 4A.
All the experiments were performed using the optimized experimental conditions, and for each pH value, the concentrations of miRNA-21 interrogated were 50 and 260 nM. pH 5 gave the best results in signal variation. The next step of investigations was to evaluate the response of the platform in the presence of interference. For this study, the device was interrogated in the presence of four interferents, as reported in inset of Fig. 4B. The selectivity of the platform was tested in the presence of the following single-stranded RNA and DNA targets: (i) 5′-AAACUUUUGGGGAUGACGA-3′, (ii) 5′-ACACUUCCGCGCGAUGACGCC-3′, (iii) 5′-TATCCCATTTAGACTACTA-3′ and 5′-TATCCATTAGACTACTACGCA-3′. All the experiments were performed in the presence of 50 nM of the sequences described above, and they were compared with the detection of 50 nM of the entirely complementary miRNA-21 target (5′-UAGCUUAUCAGACUGAGUUGA-3′). The presence of interferents was only responsible for neglectable signal changes, lower than 10%.
Urine samples monitoring of miRNA-21
The following steps of the work were focused on the analysis of miRNA-21 in urine samples. From the results of the studies in standard solution, it was defined to adjust the salt content of real urine samples by an additional 250 mM NaCl. As shown in Fig. 5A, the square-wave frequency studies (up to 100 Hz) highlighted how, for these samples, the addition of 250 mM of sodium chloride was effective in inducing a detectable and reproducible signal change, producing a similar trend as obtained in standard solutions.
Subsequently, four different urine samples were spiked with miRNA-21 ranging from 1 to 3000 nM for a preliminary evaluation of the applicability of the platform when in use with this complex medium. For each urine sample, as resulting from the optimization tests, a 250-mM NaCl solution was added, in order to achieve a background total concentration of sodium chloride ranging between 250 and 500 mM. The pH was measured prior to perform miRNA-21 quantification. As displayed in Fig. 5B, four sigmoidal curves were obtained for the four urine samples tested. It should be noted that the pH value was slightly different, namely 6.38, 6.22, 6.50, and 7.20, respectively. The adoption of exogenous sodium chloride allowed satisfactory results in the presence of miRNA-21 in the range between 10 and 3000 nM, as displayed in Fig. 5D, reaching detection limits down to 10, 34, 2, and 24 nM. It should be noted that all the real urine samples have not been treated, causing the different LOD values that have been calculated; however, the analysis of different pH- and ionic strength samples might be mitigated by the use of the standard addition method, thus reducing the effect due to the heterogeneity of untreated samples. These values are in the same order of magnitude with respect to the results obtained in buffer solutions. In addition, with respect to other electrochemical systems reported for miRNA-21 detection in biological samples, Table 2 has been included for a quick comparison, focusing on the platform, the analytical features, and the experimental setup.
Table 2.
Comparison of the proposed miRNA-21 platform with other electrochemical-based approaches
| Platform | LOD | Matrix | Required tasks | Ref |
|---|---|---|---|---|
| LNA-integrated nucleic acid hairpin probe, biotin-labeled bridge DNA–AuNPs–bio-barcode, enzymatic signal amplification | 6 fM (buffer) | Human hepatocarcinoma cells extract | Addition of HRP, H2O2, hydroquinone | [56] |
| ssDNA/pSAM/ITO recognized RNA, and ALP-conjugated JAZ binds RNA–DNA hybrid | 30 fM | tenfold diluted serum | Cell stored for 10’ at 37 °C in TZ buffer with Ru(NH3)63+, HQDP, TCEP | [57] |
| Sulfonamide-bound antisense hybridization | 20 fM (buffer) | Urine after proteinase K digestion by spin-filtration | 30’ shaking at 50 °C, addition of external electrochemical probe (ferricyanide) | [58] |
| Magnetosensing platform using p19 viral protein as receptor | 0.45 nM (buffer) | Breast cancer cells Extract | Magnetic beads, HRP, H2O2, 75’ | [59] |
| miRNA-DNA-MB hybridization onto AuNPs | 2 nM | Urine | Addition of NaCl | This work |
LNA, locked nucleic acid; ssDNA, single stranded DNA; pSAM, polymeric self-assembled monolayer; ITO, indium − tin oxide; ALP: alkaline phosphatase; JAZ, just another zinc finger protein ZNF346; HRP, horseradish peroxidase
With respect to other existing methodologies, it should be noted that the approach which involves the use of a MB-modified recognizing probe represents a very easy-to-use approach, based on the fact that all the reagents are already incorporated on top of the working electrode. Although other systems are characterized by lower detection limit, down to fM level (often based on measurements carried in buffer solutions), it should be highlighted how these approaches are based on both laborious manufacturing and measuring tasks, for instance, involving the addition of external electrochemical mediators, enzyme–substrate, magnetic beads for pre-concentration, and in some cases heating steps are performed. These systems are majorly developed using traditional/bulk electrodes, and the adoption of such disposable screen-printed platform makes the proposed device very interesting for following application, also considering the unique application in un-treated urine.
Conclusion
Timely diagnosis and effective therapies are essential to improve the chances of survival for PCa patients. To enable point-of-care testing, analytical devices need to satisfy the ASSURED principle established by the World Health Organization. Herein, we reported the development of an electrochemical printed platform to determine miRNA-21 making use of a DNA probe. After optimization of the experimental parameters in standard solutions, the feasibility of analysis of miRNA-21 in real urine samples has been evaluated. The present study was launched with the target of finding novel solutions for the design of portable health screening and diagnostics methods for the application to real urine samples. We demonstrated the potential of the method for simple detection assays in complex bodily fluids. The developed devices detected miRNA-21 down to 2 nM in real urine samples. When real matrices are interrogated, the major drawback is represented by the heterogeneity among patients; with this study we evaluated various strategies to compensate for it. The impact of multiple treatment procedures on the detection of the target, i.e., salt concentration and pH adjustment, was discussed. The results of this study indicate that the present approach represents a valid starting point toward the future development of multi-sensory platforms, possibly in combination with multivariate analysis/artificial intelligence, to detect validated miRNA signatures for ambulatory health screening tests and monitoring for early detection of tumor or its recurrence. In addition, these results could be implemented in future by combining the fully integrated printed platform with additional steps, i.e., magnetic beads, aimed to improve the sensitivity of the method, even if these steps would be consistent with an increase of the complexity of the assay.
Acknowledgements
Mr. Stefano Del Giovane is acknowledged for the assistance to W. C. during her stay at CSEM. Prof. Netz Arroyo (Johns Hopkins University School of Medicine) is acknowledged for the helpful discussions.
Funding
Open access funding provided by Università degli Studi di Napoli Federico II within the CRUI-CARE Agreement. The research leading to these results has received funding from AIRC under MFAG 2022 - ID. 27586 project – P.I. Cinti Stefano. Sima Singh also acknowledges Fondazione Umberto Veronesi for Postdoctoral Fellowship 2022.
Declarations
Ethics approval
All urine samples used in this research project have been collected and stored anonymously, without any type of list of donors or correspondence table. Moreover, as they have been artificially reinforced, it further increases the difficulty to identify anyone from the results. As this research therefore involves anonymized biological material, it falls out of the scope of the Swiss Federal Act on Research involving Human Beings (article 2) and thus does not require to be submitted to an ethics committee.
Conflict of interest
The authors declare no competing interests.
Footnotes
Published in the topical collection Young Investigators in (Bio-)Analytical Chemistry 2023 with guest editors Zhi-Yuan Gu, Beatriz Jurado-Sánchez, Thomas H. Linz, Leandro Wang Hantao, Nongnoot Wongkaew, and Peng Wu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wanda Cimmino and Davide Migliorelli equally contributed.
References
- 1.Numan A, Singh S, Zhan Y, Li L, Khalid M, Rilla K, Ranjan S, Cinti S. Advanced nanoengineered—customized point-of-care tools for prostate-specific antigen. Microchim Acta. 2022;189:27. doi: 10.1007/s00604-021-05127-y. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Prostate cancer Nat Rev Dis Prim. 2021;7:8. doi: 10.1038/s41572-021-00249-2. [DOI] [PubMed] [Google Scholar]
- 4.Walsh AL, Considine SW, Thomas AZ, Lynch TH, Manecksha RP. Digital rectal examination in primary care is important for early detection of prostate cancer: a retrospective cohort analysis study. Br J Gen Pract. 2014 doi: 10.3399/bjgp14X682861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Leeuwen PJ, Hayen A, Thompson JE, Moses D, Shnier R, Böhm M, Abuodha M, Haynes AM, Ting F, Barentsz J, Roobol M, Vass J, Rasiah K, Delprado W, Stricker PD. A multiparametric magnetic resonance imaging-based risk model to determine the risk of significant prostate cancer prior to biopsy. BJU Int. 2017 doi: 10.1111/bju.13814. [DOI] [PubMed] [Google Scholar]
- 6.Mistry K, Cable G. Meta-analysis of prostate-specific antigen and digital rectal examination as screening tests for prostate carcinoma. J Am Board Fam Pract. 2003 doi: 10.3122/jabfm.16.2.95. [DOI] [PubMed] [Google Scholar]
- 7.Pinto FG, Mahmud I, Harmon TA, Rubio VY, Garrett TJ. Rapid prostate cancer noninvasive biomarker screening using segmented flow mass spectrometry-based untargeted metabolomics. J Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00006. [DOI] [PubMed] [Google Scholar]
- 8.Pinto RA, Corrêa Neto IJF, Nahas SC, Froehner Junior I, Soares DFM, Cecconello I. Is the physician expertise in digital rectal examination of value in detecting anal tone in comparison to anorectal manometry? Arq Gastroenterol. 2019 doi: 10.1590/s0004-2803.201900000-04. [DOI] [PubMed] [Google Scholar]
- 9.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 10.Mizuno R, Oya M. Genitourinary cancers. Nippon rinsho Japanese J Clin Med. 2010;68(Suppl 8):485–488. doi: 10.1007/978-3-662-05225-9_11. [DOI] [PubMed] [Google Scholar]
- 11.Pantel K, Alix-Panabières C. The potential of circulating tumor cells as a liquid biopsy to guide therapy in prostate cancer. Cancer Discov. 2012;2:974–975. doi: 10.1158/2159-8290.CD-12-0432. [DOI] [PubMed] [Google Scholar]
- 12.Singh S, Arshid N, Cinti S. Electrochemical nano biosensors for the detection of extracellular vesicles exosomes: from the benchtop to everywhere? Biosens Bioelectron. 2022;216:114635. doi: 10.1016/j.bios.2022.114635. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67(1):129–39. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 14.Liang H, Gong F, Zhang S, Zhang CY, Zen K, Chen X. The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip Rev RNA. 2014;5:285–300. 10.1002/wrna.1208 [DOI] [PubMed]
- 15.Farazi TA, Spitzer JI, Morozov P, Tuschl T. MiRNAs in human cancer. J Pathol. 2011;223:102–115. 10.1002/path.2806 [DOI] [PMC free article] [PubMed]
- 16.Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol. 2020;72:1732–49. 10.1111/jphp.13351 [DOI] [PubMed]
- 17.Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J, Fang X, Gong C. Mirnas and cancer: Key link in diagnosis and therapy. Genes (Basel). 2021;12:1289. 10.3390/genes12081289 [DOI] [PMC free article] [PubMed]
- 18.Stuopelyte K, Daniunaite K, Bakavicius A, Lazutka JR, Jankevicius F, Jarmalaite S. The utility of urine-circulating miRNAs for detection of prostate cancer. Br J Cancer. 2016 doi: 10.1038/bjc.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010 doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed]
- 21.Pastorino U, Boeri M, Sestini S, Sabia F, Milanese G, Silva M, Suatoni P, Verri C, Cantarutti A, Sverzellati N, Corrao G, Marchianò A, Sozzi G. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann Oncol. 2022 doi: 10.1016/j.annonc.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004 doi: 10.1056/nejmoa041588. [DOI] [PubMed] [Google Scholar]
- 23.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Dees EC, Goetz MP, Olson JA, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, Ellis MJ, Paik S, Wood WC, Ravdin PM, Keane MM, Gomez Moreno HL, Reddy PS, Goggins TF, Mayer IA, Brufsky AM, Toppmeyer DL, Kaklamani VG, Berenberg JL, Abrams J, Sledge GW. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018 doi: 10.1056/nejmoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinsky K, Barlow WE, Meric-Bernstam F, Gralow JR, Albain KS, Hayes D, Lin N, Perez EA, Goldstein LJ, Chia S, Dhesy-Thind S, Rastogi P, Alba E, Delaloge S, Martín M, Gil MG, Arce-Salinas C, Brain E, Park IH, Pierga J-Y, Lluch AH, Vasquez MR, Borrego MR, Jung KH, Ferrero J-M, Schott A, Shak S, Sharma P, Lew DL, Miao J, Tripathy D, Hortobagyi G, Pusztai L. Abstract GS3-00: first results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET) +/- chemotherapy (CT) in patients (pts) with 1–3 positive nodes, hormone receptor-positive (HR+) and HER2-negative (HER2-) breast cancer. Cancer Res. 2021 doi: 10.1158/1538-7445.sabcs20-gs3-00. [DOI] [Google Scholar]
- 25.Lee J, Kwon MH, Kim JA, Rhee WJ. Detection of exosome miRNAs using molecular beacons for diagnosing prostate cancer. Artif Cells Nanomed Biotechnol. 2018 doi: 10.1080/21691401.2018.1489263. [DOI] [PubMed] [Google Scholar]
- 26.Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21:126–44. 10.1007/s12094-018-1910-8. [DOI] [PubMed]
- 27.Endzeliņš E, Melne V, Kalniņa Z, Lietuvietis V, Riekstiņa U, Llorente A, Line A. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: A systematic review. Mol Cancer. 2016;15:1–13. 10.1186/s12943-016-0523-5. [DOI] [PMC free article] [PubMed]
- 28.Fang C, Zhao J, Li J, Qian J, Liu X, Sun Q, Liu W, Tian Y, Ji A, Wu H, Yan J. Massively parallel sequencing of microRNA in bloodstains and evaluation of environmental influences on miRNA candidates using realtime polymerase chain reaction. Forensic Sci Int Genet. 2019 doi: 10.1016/j.fsigen.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Koscianska E, Starega-Roslan J, Sznajder LJ, Olejniczak M, Galka-Marciniak P, Krzyzosiak WJ. Northern blotting analysis of microRNAs, their precursors and RNA interference triggers. BMC Mol Biol. 2011 doi: 10.1186/1471-2199-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arata H, Komatsu H, Han A, Hosokawa K, Maeda M. Rapid microRNA detection using power-free microfluidic chip: coaxial stacking effect enhances the sandwich hybridization. Analyst. 2012 doi: 10.1039/c2an16154k. [DOI] [PubMed] [Google Scholar]
- 31.Su YJ, Lin IC, Wang L, Lu CH, Huang YL, Kuo HC. Next generation sequencing identifies miRNA-based biomarker panel for lupus nephritis. Oncotarget. 2018 doi: 10.18632/oncotarget.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renwick N, Cekan P, Bognanni C, Tuschl T. Multiplexed miRNA fluorescence in situ hybridization for formalin-fixed paraffin-embedded tissues. Methods Mol Biol. 2014 doi: 10.1007/978-1-4939-1459-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Zhao B, Jin Y, Ruan K. Development of a low-cost detection method for miRNA microarray. Acta Biochim Biophys Sin (Shanghai) 2010 doi: 10.1093/abbs/gmq017. [DOI] [PubMed] [Google Scholar]
- 34.El Aamri M, Yammouri G, Mohammadi H, Amine A, Korri-Youssoufi H. Electrochemical biosensors for detection of MicroRNA as a cancer biomarker: pros and cons. Biosensors. 2020;10:186. 10.3390/bios10110186. [DOI] [PMC free article] [PubMed]
- 35.Choi MH, Seo YJ. Rapid and highly sensitive hairpin structure-mediated colorimetric detection of miRNA. Anal Chim Acta. 2021 doi: 10.1016/j.aca.2021.338765. [DOI] [PubMed] [Google Scholar]
- 36.Nemati F, Hosseini M. Fluorescence turn-on detection of miRNA-155 based on hybrid Ce-MOF/ PtNPs /graphene oxide serving as fluorescence quencher. J Photochem Photobiol A Chem. 2022;429:113943. doi: 10.1016/j.jphotochem.2022.113943. [DOI] [Google Scholar]
- 37.Andrade MA, Leite FL, Rodrigues LF, Ierich JCM, Melendez ME, Carvalho AL, De Carvalho AC, Steffens C, Abdalla FC, Oliveira ON. A nanomechanical genosensor using functionalized cantilevers to detect the cancer biomarkers miRNA-203 and miRNA-205. IEEE Sens J. 2020 doi: 10.1109/JSEN.2019.2948506. [DOI] [Google Scholar]
- 38.Xie X, Liu J, Ding M, Yang X, Peng Y, Zhao Y, Ouyang R, Miao Y. Recent development of the electrochemical sensors for miRNA detection. Int J Electrochem Sci. 2021 doi: 10.20964/2021.04.35. [DOI] [Google Scholar]
- 39.Singh S, Numan A, Cinti S. Point-of-care for evaluating antimicrobial resistance through the adoption of functional materials. Anal Chem. 2022;94:26–40. doi: 10.1021/acs.analchem.1c03856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh S, Wang J, Cinti S. Review—an overview on recent progress in screen-printed electroanalytical (bio)sensors. ECS Sensors Plus. 2022;1:023401. doi: 10.1149/2754-2726/ac70e2. [DOI] [Google Scholar]
- 41.Miao P, Tang Y. DNA walking and rolling nanomachine for electrochemical detection of miRNA. Small. 2020 doi: 10.1002/smll.202004518. [DOI] [PubMed] [Google Scholar]
- 42.Miao P, Tang Y. DNA walking and rolling nanomachine for electrochemical detection of miRNA. Small. 2020;16:1–7. doi: 10.1002/smll.202004518. [DOI] [PubMed] [Google Scholar]
- 43.Moccia M, Caratelli V, Cinti S, Pede B, Avitabile C, Saviano M, Imbriani AL, Moscone D, Arduini F. Paper-based electrochemical peptide nucleic acid (PNA) biosensor for detection of miRNA-492: a pancreatic ductal adenocarcinoma biomarker. Biosens Bioelectron. 2020 doi: 10.1016/j.bios.2020.112371. [DOI] [PubMed] [Google Scholar]
- 44.Kim HY, Song J, Park HG, Kang T. Electrochemical detection of zeptomolar miRNA using an RNA-triggered Cu2+ reduction method. Sensors Actuators B Chem. 2022 doi: 10.1016/j.snb.2022.131666. [DOI] [Google Scholar]
- 45.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018 doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghorbanmehr N, Gharbi S, Korsching E, Tavallaei M, Einollahi B, Mowla SJ. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine—promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019;79:88–95. doi: 10.1002/pros.23714. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Y, Lai RY, Plaxco KW. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat Protoc. 2007 doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 48.Cinti S, Proietti E, Casotto F, Moscone D, Arduini F. Paper-based strips for the electrochemical detection of single and double stranded DNA. Anal Chem. 2018 doi: 10.1021/acs.analchem.8b04052. [DOI] [PubMed] [Google Scholar]
- 49.Dauphin-Ducharme P, Yang K, Arroyo-Currás N, Ploense KL, Zhang Y, Gerson J, Kurnik M, Kippin TE, Stojanovic MN, Plaxco KW. Electrochemical aptamer-based sensors for improved therapeutic drug monitoring and high-precision, feedback-controlled drug delivery. ACS Sens. 2019;10:2832–2837. doi: 10.1021/acssensors.9b01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White RJ, Plaxco KW. Exploiting binding-induced changes in probe flexibility for the optimization of electrochemical biosensors. Anal Chem. 2010;82:73–76. doi: 10.1021/ac902595f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dauphin-Ducharme P, Plaxco KW. Maximizing the signal gain of electrochemical-DNA sensors. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b03227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cinti S, Cinotti G, Parolo C, Nguyen EP, Caratelli V, Moscone D, Arduini F, Merkoci A. Experimental comparison in sensing breast cancer mutations by signal on and signal off paper-based electroanalytical strips. Anal Chem. 2020 doi: 10.1021/acs.analchem.9b02560. [DOI] [PubMed] [Google Scholar]
- 53.Fan C, Plaxco KW, Heeger AJ. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michael R. Clarkson CNM, BMB. Pocket companion to Brenner and Rector’s the kidney. 2011. 10.1016/C2010-0-65550-7.
- 55.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Effect of pH on the overstretching transition of double-stranded DNA: evidence of force-induced DNA melting. Biophys J. 2001;80:874–881. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin H, Zhou Y, Chen C, Zhu L, Ai S. An electrochemical signal ‘off–on’sensing platform for microRNA detection. Analyst. 2012;137:1389–1395. doi: 10.1039/C2AN16098F. [DOI] [PubMed] [Google Scholar]
- 57.Fang CS, Kim KS, Yu B, Jon S, Kim MS, Yang H. Ultrasensitive electrochemical detection of miRNA-21 using a zinc finger protein specific to DNA–RNA hybrids. Anal Chem. 2017;89:2024–2031. doi: 10.1021/acs.analchem.6b04609. [DOI] [PubMed] [Google Scholar]
- 58.Smith DA, Newbury LJ, Drago G, Bowen T, Redman JE. Electrochemical detection of urinary microRNAs via sulfonamide-bound antisense hybridisation. Sensors Actuators B. 2017;253:335–341. doi: 10.1016/j.snb.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torrente-Rodríguez RM, Campuzano S, López-Hernández E, Granados R, Sánchez-Puelles JM, Pingarrón JM. Direct determination of miR-21 in total RNA extracted from breast cancer samples using magnetosensing platforms and the p19 viral protein as detector bioreceptor. Electroanalysis. 2014;26:2080–2087. doi: 10.1002/elan.201400317. [DOI] [Google Scholar]