Highlights
-
•
Isolation and formulation of porcine spinal cord-derived ECM spinning solution.
-
•
Production of highly directional ECM fiber scaffolds by integrating electrospinning into 3D bioprinter.
-
•
Adhesion, proliferation and differentiation of neural cells into mature neurons supported by ECM fiber scaffolds.
-
•
Enhanced migration of mature neurons on aligned ECM fiber scaffolds after lesion underlining superior cell guiding capacities of ECM by combination of biochemical and topographical cues.
Keywords: Neural cell guiding, Neuronal migration, Electrospinning, Extracellular matrix, Decellularized scaffolds
Abstract
Traumatic injury to the spinal cord (SCI) causes the transection of neurons, formation of a lesion cavity, and remodeling of the microenvironment by excessive extracellular matrix (ECM) deposition and scar formation leading to a regeneration-prohibiting environment. Electrospun fiber scaffolds have been shown to simulate the ECM and increase neural alignment and neurite outgrowth contributing to a growth-permissive matrix. In this work, electrospun ECM-like fibers providing biochemical and topological cues are implemented into a scaffold to represent an oriented biomaterial suitable for the alignment and migration of neural cells in order to improve spinal cord regeneration. The successfully decellularized spinal cord ECM (dECM), with no visible cell nuclei and dsDNA content < 50 ng/mg tissue, showed preserved ECM components, such as glycosaminoglycans and collagens. Serving as the biomaterial for 3D printer-assisted electrospinning, highly aligned and randomly distributed dECM fiber scaffolds (< 1 µm fiber diameter) were fabricated. The scaffolds were cytocompatible and supported the viability of a human neural cell line (SH-SY5Y) for 14 days. Cells were selectively differentiated into neurons, as confirmed by immunolabeling of specific cell markers (ChAT, Tubulin ß), and followed the orientation given by the dECM scaffolds. After generating a lesion site on the cell-scaffold model, cell migration was observed and compared to reference poly-ε-caprolactone fiber scaffolds. The aligned dECM fiber scaffold promoted the fastest and most efficient lesion closure, indicating superior cell guiding capabilities of dECM-based scaffolds. The strategy of combining decellularized tissues with controlled deposition of fibers to optimize biochemical and topographical cues opens the way for clinically relevant central nervous system scaffolding solutions.
Graphical abstract

1. Introduction
Spinal cord injuries (SCI), caused by trauma such as accidents, sport injuries or acute or chronic diseases, temporarily and permanently change the function of the spinal cord, resulting in decreased sensitivity or mobility, long-term social and financial consequences, and decreased lifespan [1,2]. Lesioned sensory and motor neurons, representing the primary cells responsible for signal transduction between the central nervous system (CNS) and the peripheral nervous system (PNS), fail to regenerate spontaneously. Once transected, neurons neither grow beyond the lesion site nor reconnect into functional neuronal networks in mature CNS. Among the potential reasons for poor tissue repair, the limited intrinsic growth ability of mature CNS neurons, the presence of external inhibitory factors related to fibrotic and astrocytic scarring, and the timing and sequence of growth-promoting factors are considered to be the most problematic [3], [4], [5], [6].
To date there is no treatment for complete recovery or regeneration of motor and sensory function after SCI. The current clinical strategies mainly focus on neuroprotective treatments, such as surgeries for decompression and stabilization of the injury site, or pharmaceutical interventions for pain and inflammation control [7]. The traditional cell-based approach for neuroregeneration in CNS has been engrafting neural stem cells. However, this procedure often leads to immune rejection of transplanted externally reprogrammed cells and poor functional integration into the host tissue [8,9]. Alternative cell-based approaches include in vivo reprogramming of endogenous glial cells into neurons avoiding adverse immune responses. This methods, but encounters issues regarding long-term survival of neurons after reprogramming, generation of specific neuronal subtypes and rebuilding of functional circuits [10,11]. Compared to cell-based strategies, scaffold-based approaches are cell-free and focus on bridging the lesion site, providing biocompatible substrates and guiding platforms for neural cells. This has the advantage that specific biophysical and biochemical cues can be provided, while no additional cells are introduced in a lesion site, which has already been infiltrated by activated cells (reactive astrocytes, microglia, and fibroblasts) due to the injury. Traditionally, scaffolds have been produced either from hydrogels or fibers. Hydrogels display tissue-matching mechanical strength and degradation rates, while fibers are characterized by large surface area, submicron diameter and high porosity, mimicking the 3D structure of the native ECM [12].
Electrospinning is one of the techniques commonly used to generate fiber scaffolds [13], [14], [15]. Synthetic polymers can serve as spinning solutions, however their lack of physiologically relevant biochemical properties can lead to poor cell adhesion, impaired proliferation, and inflammatory reactions [16], [17], [18], [19]. Therefore, materials based on naturally occurring ECM constituents, like collagen [7,15,20] have been employed for guidance of neural cells. Collagen-based scaffolds, however, typically include only one or two isoforms and cannot fully represent the diversity of proteins found in the native tissue. dECM-derived materials can also be used as a 3D-network of macromolecules that offers tissue-like properties [21,22]. Decellularization of several tissues, including heart, muscle, or peripheral nerves, has been reported, envisioning the application of the matrices in regenerative medicine [23], [24], [25], [26], [27], [28]. The preservation of the complex biochemical cues specific to the native microenvironment is essential to the biological success of these matrices [29], but also a major challenge of tissue decellularization. Tissue decellularization techniques must achieve the elimination of contaminating nucleic acids down to a final double-stranded DNA (dsDNA) content < 50 ng per mg of native tissue [25], while maintaining the biochemical characteristics required to promote cell attachment and ultimately tissue regeneration. The first reported biomaterial composed of decellularized CNS tissue was based on hydrogels of dECM isolated from porcine brain [26], which supported the differentiation of human induced pluripotent stem cells (iPSC) into neurons. Since then, several other CNS-based scaffolds have been proposed [23,[29], [30], [31], [32], [33], [34]], also in form of fiber-based scaffolds. To our knowledge, the use of aligned dECM CNS fiber scaffolds, combining the topology and biochemical environment of native spinal cord for the support of tissue regeneration, has not yet been reported.
In this work, we suggest a novel strategy for the generation of cell-friendly, anisotropic scaffolds for the promotion of neuronal regeneration in SCI. We propose an approach based on decellularization techniques to produce tissue-specific materials combined with well-established tissue engineering concepts, such as nanofiber scaffolds for cell guidance. The data demonstrate that scaffolds generated by electrospinning of dECM provide biochemical and topological cues, induce the alignment of neurons in vitro and enhance neuronal migration across a lesion inflicted to the cell cultures. These scaffolds may lead to improved neuronal re-connectivity forming neuronal networks and restoring damaged sensory and locomotive functions.
2. Matrials and methods
2.1. Spinal cord decellularization
Porcine spinal cord was received frozen from the butcher shop Braunwalder (Wohlen, Switzerland). The dura was removed after thawing the tissue. The spinal cord was either directly used for decellularization or frozen at -196 °Cin liquid N2 in 10 mM Trizma (Sigma, T2194) solution supplemented with 5 mM ethylenediaminetetraacetic (EDTA, Sigma, 03610). For decellularization, we adapted the protocol described by Crapo et al. [23]. Briefly, the spinal cord was first thawed at room temperature and again frozen in liquid nitrogen (repeated 3×). Subsequently, the tissue was mechanically disrupted with a scalpel into small pieces and immersed in deionized water at 4 °C for 24 h. Afterwards, the tissue was incubated in 0.02% Trypsin/0.05% EDTA (Sigma Aldrich, 9002–07–7) at 60 rpm on an orbital shaker (Major Science, MS-NOR-30) and 37 °C for 1 h, followed by 1.0% Triton X-100 (Sigma Aldrich, 9002-93–1) for 1 h, 1 M Sucrose (Sigma Aldrich, 57–50–1) for 15 min, deionized water for 30 min, 1.0% Deoxycholic acid (Sigma Aldrich, 302–95–4) for 1 h, 0.1% Peracetic acid (Sigma Aldrich, 79–21–0) in 4% (v/v) ethanol (Sigma, 64–17–5) for 2 h. The final steps included washing in PBS (Sigma Aldrich, 25,777–14–4) for 15 min, twice with deionized water for 15 min and in PBS for 15 min. If not noted otherwise all steps were carried out at room temperature and a shaking speed of 180 rpm. The extracted dECM was lyophilized under -87 °C at 0.01 mbar for 24 h.
2.2. Characterization of dECM
After each decellularization and washing step, the tissue was weighed (Mettler Toledo, PG5002-S). To quantify the remaining dsDNA, the native and decellularized tissues were lysed with proteinase K for 2 h at 56 °C, the dsDNA was extracted, washed, and purified (Qiagen DNeasy blood & tissue kit 69,504). The purified dsDNA was diluted 1/20 in Qubit working solution and the amount of dsDNA in native and decellularized samples quantified by fluorescence intensity (Qubit Fluorometer4, Thermo Fisher). For histological stainings, native and decellularized tissues were fixed for 24 h at RT in 4% formaldehyde solution. After rinsing, the tissue was dehydrated in increasing ethanol series, followed by m-xylene (Sigma, 108–38–3) incubation and embedding in paraffin (Carl Roth, CN 49) overnight. The embedded tissue blocks were cut with a microtome (SLEE Medical, Microtome CUT 6062) in 5 µm slices, collected on glass slides (Corning, 2948) and the paraffin removed at 80 °C. Prior to the staining, the tissue slides were rehydrated in m-xylene, a decreasing ethanol series and water. For Hematoxylin-Eosin (H-E) staining, the tissue slides were incubated for 8 min in Mayer's hemalum solution (Merck, 109,249), followed by insertion in ethanol with 0.3% hydrochloric acid (Sigma, 30,721), water wash and 2 min in 0.5% aqueous eosin solution (Merck, 109,844). For PAS-Alcian blue staining the tissue slides were first incubated for 5 min in Alcian blue solution (Merck, 101,647) and after rinsing with water for 10 min in 0.5% periodic acid solution (Merck, 100,482). After another rinse with water, the tissue slides were incubated for 15 min in Schiff's reagent (Sigma, 3,952,016) and three times 2 min in sulphite water (0.5% Sodium disulfite, Sigma 31,448 in 0.05 M hydrochloric acid). After another dehydration series, all stained tissue slides were mounted with DPX (Merck, 10,059) and imaged using an optical microscope (Olympus IX73) [33,35,36].
2.3. Production of spinning solutions
The lyophilized decellularized spinal cord was milled with a cryomill (Retsch) in a 50 mL stainless steel container with 2 stainless steel balls (15 mm diameter) using automatic pre-freezing and 3 milling cycles of 5 min milling at 5 Hz and −196 °C each. The dECM powder was dissolved in precooled hexafluoroisopropanol (Merck, 920–66–1) to 5% w/v solutions and homogenized with a vortex at 3000 rpm in 4 cycles of each 1 min at RT. 0.1% w/v of polyethylenglycol (Sigma, 182,028) was added and all dissolved at 20 rpm over 48 h at 4 °C. Prior to spinning, the dECM dispersion was centrifuged at 300xg for 5 min at room temperature.
Polycaprolactone (PCL, Sigma, 440744) was dissolved as a 10% w/w solution in a 70/30% v/v acetic acid/acetone (Sigma, 695092/Sigma, 34850).
2.4. Electrospinning of fiber scaffolds
To produce dECM-based fiber scaffolds, a RegenHU bio printer (RegenHU, Discovery) was equipped with an electrospinning head, which was connected to a high voltage source, and a grounded collector plate. The voltage was set to 10 kV with a polymer flow of 0.05 mm/s with a 24 G needle. The distance between needle tip and collector was 45 mm. PCL-based fiber scaffolds were typically spun with 15 kV, a flow rate of 0.001 mm/s, 24 G needle and 45 mm distance. For aligned fiber scaffolds, dECM and PCL-based, 2 additional zinc wires were placed 2 cm apart on the collector plate and the spinning head in the printer operated with a speed of 7.0 mm/s in a linear movement within the 2 cm of zinc wires. The electrospun fibers were collected on glass cover slips (Huberlab, 10.0360.52). The scaffolds were kept at RT for 1 h after spinning to ensure complete removal of highly volatile HFIP and stored sealed at RT in the dark until use. All scaffolds were sterilized with 70% ethanol and then washed three times in PBS to remove any entrapped solvent prior to use in cell culture experiments.
2.5. Cell culture of neural stem cells
The human neuroblastoma cell line SH-SY5Y was cultured in proliferation medium composed of DMEM high glucose (Gibco, 11960044) supplemented with 10% FBS (Gibco, 26140) and 1% penicillin-streptomycin (Gibco, 15140122). For all in vitro experiments, cells at passages 10–20 were used. Throughout the cultivation period, the cells were incubated at 37 °C and 5% CO2 and the medium was exchanged every two or three days. Typically, cells were seeded on poly-l-lysine (PLL, Sigma, P4707)-coated (1 h, RT) tissue culture polystyrene cell culture flasks (TCPS) and were detached with Accutase™ (Millipore Cat. No. SCR005) for 2-3 min at 37 °C. For experiments with dECM and PCL fiber scaffolds and TCPS reference the cell seeding density was kept constant. Cells were seeded on the scaffolds with a cell density of 4500 cells/cm [2], resulting in a final cell seeding density of 5000 cells/scaffold.
Neuronal differentiation was induced by changing the medium to “RA differentiation medium” consisting of DMEM (high glucose) supplemented with 10% FBS, 1% penicillin-streptomycin and 10 mM retinoic acid (RA, Sigma, R2625). After 5 days, the medium composition was changed to DMEM (high glucose) supplemented with 1% penicillin-streptomycin and 50 ng/mL brain derived neural growth factor (BDNF, Sigma, B3795) for the next 9 days [37], [38], [39], [40].
2.6. Characterization of cell adhesion, proliferation and differentiation
The adhesion of SH-SY5Y cells on the fiber scaffolds and TCPS was analyzed by optical microscopy (Olympus IX73) and images taken 24 h after seeding with different magnifications. The proliferation of cells was observed at days 1, 5, 7, 11 and 14 after cell seeding as a function of the metabolic activity with an Alamar Blue assay (Thermo Fisher, DAL1025). After 2 h of incubation with Alamar blue solution, the fluorescence was measured at 560 nm.
For immunofluorescence staining after 1, 5-, 7-, 14- and 16-days cells were fixed at room temperature in 4% PFA solution (EM-grade, EMS, 15,712), permeabilized with 0.2% Triton X-100 (Sigma, 94,326) and blocked with 3% BSA (Sigma, A4503). After washing thrice with PBS, the cells were incubated with the primary antibody overnight at 4 °C followed by the secondary antibody at room temperature for 2 h. SH-SY5Y cells were probed with a primary anti-tubulin antibody (Sigma, T8578, 1/500) detected by an anti-mouse IgG Alexa Fluor 488 secondary antibody (Thermo Fisher, A11017, 1/1000). As a proliferation marker Ki-67 was marked with a primary rabbit antibody (Abcam, 15,580, 1/1000) detected by an anti-rabbit IgG Alexa Fluor 546 (Thermo Fisher, A11071, 1/1000). After three washing steps with PBS, the cells were incubated at room temperature for 5 min with 4′6-diamidino-2-phenylindole (DAPI, Invitrogen, 1:1000) to stain the nuclei. For imaging with a confocal microscope (Olympus, FV3000) the cells were mounted on the scaffolds with antifade mounting medium (Thermo Fisher, P36982).
2.7. Quantitative PCR
RNA was extracted from cells at day 16 of the experiment, 48 h after induction of the lesion on dECM and PCL scaffolds each in random and aligned configuration and on TCPS reference. Cells on scaffolds without any lesion and on TCPS have served as reference. For mRNA isolation, the cells were washed with PBS and lysed with Trizol (Qiagen 79,306, 5 min, RT). Chloroform (Sigma, C7559) was added 1/5 to each sample, the tubes were shaken vigorously by hand for 15 s and incubated for 3 min at RT. After centrifuging at 12′000×g for 15 min at 4 °C, the aqueous upper phase (about 200 µl) was pipetted into a new tube and 2 µl of 5 mg/mL RNase-free glycogen (Thermo Fisher, AM9510) and isopropanol (Sigma, I9516, 1/2 v of trizol) were added to each sample. The mixture was incubated for 10 min at RT, followed by centrifuging at 12′000×g for 10 min at 4 °C. The supernatant was removed, and the RNA pellet washed with 75% EtOH (Sigma, 51,976). After brief vortexing and centrifuging at 7′500×g for 5 min at 4 °C, the pellets were resuspended in 25 µL RNase-free water (Qiagen, 129,112). For reverse transcription, mRNA was incubated for 5 min at 70 °C with 0.57 µg/mL PolydT primer (Qiagen, 79,237) in RNase-free water in a thermocycler (Biometra, trio). Reverse transcriptase mix containing 1× buffer (Promega, M531A), 2 mM dNTP (Solids biodyne, 02–31–00,100) and 200 U/µL reverse transcriptase (Promega, M170B) in RNase-free water was added and incubated for 1 h at 37 °C. As a negative control non-RT mix was prepared similarly but excluding the reverse transcriptase. The cDNA was mixed with a master mix containing 1× TaqMan probe master (Sigma, 04,673,433,001) and 1× PTK2 (Thermo fisher, 1× Hs01056457_m1) in a pipetting robot (QIAgility, 015,790). qPCR was measured with the LightCycler 480 II (Roche). The housekeeping gene was B2M (Thermo Fisher, Hs0187842_m1). Data are expressed as fold change versus cells cultured on TCPS using the 2−ΔΔCt method.
2.8. Scratch assay
For scratch assays, dECM and PCL fiber scaffolds in random and aligned orientation and TCPS as reference were used. The lesion was induced with a 200 µL pipette tip (Eppendorf, 0,030,076,320) over the scaffolds. For experiments with undifferentiated neural cells, the scratch was induced on day 5, and for experiments with differentiated neurons the scratch was induced on day 14. The cell migration and lesion closure were followed with live microscopy (Olympus, IXplore) directly after lesioning and during 48 h, with images taken every 30 min. The acquired images were analyzed using the wound healing macro on ImageJ, typically using parameters variance radius 10, threshold value 100 and% of saturated pixel 0.001.
2.9. Statistical analysis
dECM was isolated from three different spinal cords (N = 3) and each dECM preparation analyzed for dsDNA content and presence of biochemical cues with histology in triplicates. From each dECM preparation six fiber scaffolds were produced per condition (random and aligned). Six PCL scaffolds for random and aligned orientation were fabricated from three different PCL solutions. Each lesion closure was measured in triplicates for each fiber scaffold condition. Data were analyzed using GraphPad Prism 9 (GraphPad Software, Version 9.3.0) and expressed as mean values ± SD. The student's unpaired t-test was used for analysis of decellularization efficiency. Two-way and one-way ANOVA were used for statistical analysis of metabolic activity, expression of proliferation marker, gene expression of PTK2 with multiple fiber scaffold conditions. Differences among means of p < 0.05 were considered to be significant (**, p ≤ 0.005; ****, p ≤ 0.0001).
3. Results
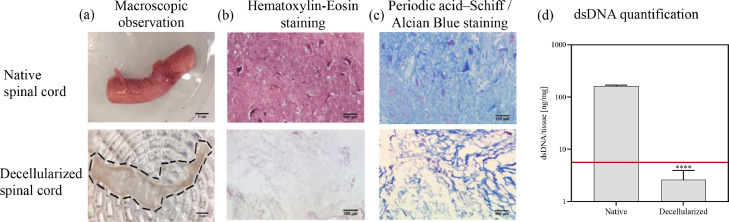
The dECM for production of the spinning solution was isolated from porcine spinal cord as described in Material and Methods. Porcine material was chosen as it closely resembles the physiology and anatomy of human spinal cord. After removal of the dura, cellular components, fat and blood were removed from the finely chopped spinal cord by sequential incubation in a series of different solutions containing enzymes or chemicals. The decellularization process was followed macroscopically by a change in color resulting in a white fibrous tissue and a loss in weight of > 90 w% (Fig. 1a). Further, successful decellularization was confirmed by histological staining and DNA quantification (Fig. 1b and d). The amount of isolated and purified dsDNA in native spinal cord was 161 ng/mg tissue and was significantly decreased by a factor of 60 to 2.6 ng/mg tissue. In the hematoxylin-eosin staining of formalin fixed tissue slices, cell nuclei of neurons and glia cells in dark violet were only found in native spinal cord, while the decellularized spinal cord was free of cell nuclei. With the periodic acid Schiff/alcian blue staining, glycosaminoglycans and polysaccharides were stained in blue both in native and decellularized tissue, indicating the preservation of ECM components after decellularization (Fig. 1c).
Fig. 1.
Characterization of native and decellularized porcine spinal cord. (a) Macroscopic observation; (b) Histological stainings with hematoxylin-eosin and (c) periodic acid Schiff-alcian blue; (d) dsDNA quantification of native and decellularized spinal cord; bar graphs represent means ± SD; statistical analysis was based on Student's unpaired t-test; ****, p ≤ 0.0001; number of spinal cords n = 3.
To formulate a spinning solution applicable for electrospinning, the dECM was first lyophilized to remove all remaining water. To improve the dissolution rate and homogenize the sample, the dECM was ground into a fine powder at very low temperature to minimize degradation of dECM. Lastly, the dECM was dissolved in hexafluoroisopropanol (HFIP) as a 5% w/v solution with the addition of 0.1% polyethylene glycol (PEG) to improve the viscosity and enhance spinnability of the spinning solution.
This solution was used to produce dECM-based electrospun fiber scaffolds in two different settings for random and aligned fiber deposition, named “dECM aligned” and “dECM random” for aligned and random scaffolds, respectively. The electrospinning process was optimized by adjusting the distance between collector and needle to 45 mm and the high voltage to 10 kV with a slow flow rate of 0.05 mm/s, resulting in bead-free fibers (Supplementary data, Fig. 1). The fibers had a uniform size distribution with a diameter in both settings in the sub-micrometer range with an average length of 925 ± 96 nm for aligned and 875 ± 80 µm for random fibers, closely resembling the size of ECM fibers found in native tissue [41]. To control the deposition of the fibers into an aligned pattern, two conductive wires were placed on the collector plate as indicated by the two green arrows depicted in Fig. 2a.
Fig. 2.
Electrospun ECM fiber scaffolds. (a) Spinning set-up at RegenHu bioprinter for random and aligned fiber deposition; (b) Random and aligned fiber scaffolds spun at 10 kV, with flow rate of 0.05 mm/s and 7.0 mm/s for aligned deposition; (c) Histograms of dECM fibers in random and aligned deposition, alignment was determined by ImageJ (directionality macro); six scaffolds were analyzed per orientation (n = 6).
In order to enhance the aligned deposition of the fibers, the printhead additionally moved in between the two wires in a linear movement. This resulted in highly aligned fiber scaffolds with fibers oriented parallel to each other, with more than 95% of the fibers orienting parallel to the main axis (0 ± 7°) (Fig. 2b and c). In contrast, the wire-free electrospinning set-up resulted in randomly distributed fiber scaffolds without recognizable alignment. The histogram depicted in Fig. 2c shows fibers distributed without a preferred main angle, with a minor preference for deposition at around 15°
Once the scaffolds were generated in two different topological orientations using dECM and PCL, we set out to analyze the effect of the fiber alignment on the neuronal cells. To this end, we assessed the behavior of neuronal precursor cells on the dECM scaffolds. Undifferentiated SH-SY5Y cells, a human neuroblastoma cell line, were seeded on the dECM scaffolds to test the biocompatibility of the scaffold, and the ability of the cells to attach to the scaffold (adhesion), and to proliferate. As shown in Fig. 3a, undifferentiated SH-SY5Y cells successfully attached to dECM scaffolds (both random and aligned) and presented the typical cellular phenotype with a round cell body and short neurite extensions. SH-SY5Y cells cultured in the same conditions on the PCL fiber scaffolds and TCPS reference, displayed phenotypic morphology and proliferation, and in case of PCL fibers followed the fiber pattern (Supplementary data, Fig. 3). After 5 days, the metabolic capacity of the cultures increased compared to day 1, indicating that the scaffold is not cytotoxic and supports the proliferation of healthy, undifferentiated cells. In addition, cells grown on aligned dECM scaffolds displayed a cellular orientation that followed the alignment cues presented by the scaffold (Fig. 3a and Supplementary data, Fig. 2).
Fig. 3.
Undifferentiated SH-SY5Y cells adhere, proliferate and align on dECM fiber scaffolds. (a) SH-SY5Y cells on random (top row) and aligned (bottom row) dECM scaffolds at day 1 and day 5, scale bar = 100 µm; (b) Optical micrographs of SH-SY5Y cells on random and aligned dECM scaffolds, images taken with live microscope at 0 h, 24 h and 48 h post lesion; (c) Graphical representation of scratch closure of SH-SY5Y cells on random and aligned dECM scaffolds during the first 48 h post lesion (n = 3).
In a simplified injury model, a confluent cell layer grown on random and aligned dECM scaffolds was damaged by scratching it with a pipette tip (Fig. 3b). On both random and aligned scaffolds, the cells were able to occupy the 200 µm gap and close the lesion within 48 h. Due to the inherent proliferative behavior of undifferentiated SH-SY5Y cells, the cells replicated, migrated and filled the gap. As depicted in Fig. 3c, the kinetics of the lesion closure is very similar for both scaffold configurations and the pre-alignment of cells, as induced by the aligned dECM scaffold, does not have a significant influence on the speed or density of the lesion closure (as assessed for three independent scaffolds).
The behavior of differentiated neurons showing a lower proliferative rate and perhaps more representative of a SCI injury site was also evaluated. Differentiated neurons were generated through incubation of SH-SY5Y under specific differentiation conditions for 9 days (see Material and Methods) and were also able to attach to the dECM-based scaffolds. In addition, neurons also adhered to PCL-scaffolds serving as comparator and providing a purely topographical reference, without any intrinsic physiological biochemical properties. dECM and PLC-based scaffolds were produced using similar electrospinning protocols leading to comparable spatial dispositions. Random PCL (PCL random) and aligned PCL (PCL aligned) scaffolds were topologically comparable to dECM-based scaffolds (dECM random or dECM aligned) based on fiber diameter, density, and orientation. SH-SY5Y cells were seeded on all four scaffold types, cultured for 24 h in proliferation medium allowing proper cell adhesion to the scaffold prior to differentiation into neurons by adapting the culture conditions as described in Materials and Methods. The differentiation into mature neurons was monitored by means of optical microscopy, cell viability assays and immunofluorescence stainings.
The differentiation of SH-SY5Y cells into neurons was studied directly on the fiber scaffolds. Undifferentiated SH-SY5Y cells did not express tubulin βIII (cytoskeletal protein mainly found in neurons) or ChAT (enzyme mainly expressed in the neuronal body), both markers of a neuronal phenotype. In contrast, expression of both markers was observed after 14 days in culture under differentiation conditions, indicating the successful differentiation of SH-SY5Y cells into neurons on dECM fiber-based scaffolds (Fig. 4a). Furthermore, the cell morphology changed over the differentiation period from cells with a more rounded on day 1 into cells with a more elongated soma displaying typical neurites extending from the cell body on days 7–14 (Supplementary data, Figs. 4 and 5 for PCL fibers and TCPS reference). Concordant to the progression of differentiation, the cellular metabolism determined by Alamar blue (surrogate for number of viable cells) increased significantly during the first week in culture on fiber scaffolds. The subsequent stagnation of metabolic activity on fiber scaffolds (Fig. 4b) indicates a switch from a proliferative to a differentiated phenotype. Furthermore, the number of proliferating cells expressing Ki-67 was significantly lower after 14 days culture under neuronal differentiation conditions than upon seeding (Fig. 4c). Comparison of metabolic activity and proliferation index dECM fiber scaffolds and TCPS did not show any significant difference, suggesting that the dECM fiber scaffolds are not cytotoxic. The differentiation into neurons does not seem to be significantly changed between both fiber scaffold types (dECM and PCL), indicating negligible influence on the differentiation by the additional biochemical components provided by the dECM fibers.
Fig. 4.
Differentiation of SH-SY5Y cells into neurons on dECM scaffolds. (a) Expression of neuronal differentiation marker choline acetyltransferase (left column, green) and tubulin βIII (right column, green), counterstained with DAPI (cyan) in SH-SY5Y (before differentiation) and neurons (after 14 days differentiation) on dECM scaffold; (b) Cellular metabolism determined by Alamar blue of SH-SY5Y cells during differentiation process into neurons on dECM scaffolds and PCL scaffolds with TCPS as reference (day 1, 7, 11); c) Expression of proliferation marker Ki67 of SH-SY5Y cells during differentiation process into neurons on dECM and PCL scaffolds with TCPS as reference (day 1, 14); bar graphs represent means ± SD (b, c); statistical analysis was based on two-way ANOVA (b, c); ****, p ≤ 0.0001; n = 3.
Fig. 5.
Lesioned neurons migrate on dECM and PCL fiber scaffolds. (a) Optical micrographs of neurons on random and aligned dECM (top row) and PCL (bottom row) scaffolds, images taken with live microscope at 0 h, 24 h and 48 h post lesion; (b) Graphical representation of scratch closure of SH-SY5Y cells on random and aligned dECM and PCL scaffolds within 48 h post lesion; (c) Expression of proliferation marker Ki67 of neurons post lesion on aligned dECM scaffolds at day 14 before and post lesion, day 1 as reference; (d) Gene expression of PTK2 in neurons on random and aligned dECM and PCL scaffolds 48 h post lesion, expressed as 2-ΔΔCt of cells differentiated on TCPS reference; bar graphs represent means ± SD (c, d); statistical analysis was based on one-way ANOVA (c) and two-way ANOVA (d); **, p ≤ 0.05, ****, p ≤ 0.0001; n = 3.
Once determined that the cells could attach and differentiate on the generated dECM scaffolds, we set off to investigate if the biochemistry and/or the topology of the scaffolds would influence the cell migration and injury repair. For this, we induced a lesion on differentiated neurons (at day 14) and observed the kinetic of the wound closure using live microscopy for 48 h. As depicted in Fig. 5a, all four tested scaffolds promoted the cell migration into the gap (Supplementary data, figure 6 for TCPS reference). There were, however remarkable differences in the kinetics: after 48 h, only neurons grown on dECM aligned scaffolds closed the gap to nearly 100%, whereas the other scaffolds (dECM random, PCL aligned and PCL random) only achieved a partial closure of approximately 40–50%. The difference noted with the dECM aligned is particularly pronounced from 24 h post lesion on (Fig. 5b). Not only the scaffold influenced the speed of gap closure, but also the differentiation status. As depicted in Fig. 3c, the gap closure was slower with differentiated neurons than with the undifferentiated SY-SH5Y, probably due to the observed lower proliferation rate in differentiated cells. In the reference PCL scaffolds the lesion closure seems to be independent of the orientation of the PCL fibers, since the difference between closure speed and amount is not significant between aligned and random scaffolds as visualized in Fig. 5b.
The combination of biochemical and topographical cues, as constructed in the aligned dECM fiber scaffold, promoted the fastest migration of neurons. Additional to the biochemical characteristics, dECM and PCL fiber scaffolds possess different elastic properties. Nanoindentation measurements showed a significantly lower young's modulus for dECM scaffolds (random dECM of 326 ± 48 Pa, aligned dECM of 464 ± 18 Pa compared to random PCL of 12.13 ± 1.0 kPa and aligned PCL of 12.60 ± 0.8 kPa) as visualized in Supplementary data, Fig. 7.
To verify the migratory behavior of neurons, and exclude a lesion induced activation of cell proliferation, the proportion of proliferating neurons was assessed immediately after the lesion by quantifying the proliferation marker Ki67 (Fig. 5c). Compared to day 1, prior to differentiation when cells were highly proliferative, the level of Ki67 decreased significantly in differentiated neurons. Expression of Ki67 remained low after the lesion, indicating that the cell proliferation was not reactivated. Also, the transcriptional induction of PTK2 (focal adhesion kinase; FAK) support a positive effect of aligned dECM scaffolds on neuronal migration and it was highest in neurons cultured in aligned dECM (Fig. 5d). This was associated with the faster and more efficient lesion closure after 48 h and indicated the superior guiding properties of aligned dECM fiber scaffolds by the combination of biochemical (dECM) and topological (fiber) cues.
4. Discussion
In this work, we sought to investigate whether the type and/or the topology of the scaffolds may promote the healing of an in vitro injury as a model for spinal cord injury. For this purpose, we focused on scaffolds produced using either a biomaterial (dECM) or a polymer (PCL) in different configurations. Moreover, we used the scaffolds to evaluate the wound closing ability of an undifferentiated neuroblastoma cell line (SH-SY5Y) and on neurons derived from this cell line following well described differentiation protocols.
For the production of dECM-based scaffolds, the first crucial step was the successful decellularization of the tissue, native spinal cord from pig. The results show excellent decellularization efficiency as indicated by a final dsDNA concentration of 2.6 ± 1.3 ng/mg compared to starting concentration of 161.5 ± 7.4 ng/mg and compared to the 37.9 ± 7.7 ng/mg in spinal cord dECM reported by the Crapo and other decellularization protocols, using similar reagents [30,33]. Compared to the original protocol of Crapo [23], in our adapted method the concentrations of deoxycholate and Triton x-100 were reduced to 1.0% for both reagents from 4% and 3%, respectively. For single reagent methods, e.g. purely SDS-based protocols [26,32], the amount of residual dsDNA/mg tissue can achieve comparable results for porcine brains, but require a much longer incubation time. Hence, our method requires less time and less reagents for a better outcome in terms of dsDNA content. Possible reasons for the good performance of method described here can be the tissue preparation steps with physical and mechanical disruption, including repeated freeze-thawing cycles and a sophisticated mechanical dissection, which result in smaller tissue pieces. The optimized tissue disruption probably offers higher surface to volume ratio allowing a faster reaction in the subsequent chemical/enzymatic decellularization. Achieving a low dsDNA content implicates the removal of large quantities of cellular and genetic material from the tissue potentially impacting also the surrounding dECM. However here the elimination of dsDNA did not negatively affect the preservation of biochemical cues as shown in the histological stainings with H-E and AB-PAS; the decellularized dECM maintained glycosaminoglycans, including chondroitin sulfate proteoglycans, such as neurocan or versican. The main challenges of producing an dECM-based fiber scaffold are the dECM's batch-to-batch variation due to its non-synthetic origin. This causes difficulties to form stable polymer jets for controlled electrospinning. Such issues can be circumvented by mixing the finely grinded dECM of several different decellularized spinal cords homogeneously followed by the addition of 0.1% PEO before solubilizing and spinning. Inspired by pioneering work of the Machluf lab, who paved the way in dECM-based electrospinning with primary focus on cardiac-based dECM spinning solutions [27,42,43], the dECM is solubilized in HFIP enabling adequate charge distribution and consistent fiber spinning conditions. With this dECM-spinning solution the produced fibers consist only of dECM since PEO is easily washed away in aqueous solutions. This renders them completely tissue-specific in contrast to the first CNS-derived fiber scaffolds which are based on 9% gelatin with 1% dECM [30]. The produced fiber scaffold is stable under physiological conditions, as shown by an intact scaffold structure after 14 days incubation in cell culture medium at 37 °C. This was achieved without the need of additional crosslinking, thus reducing the post-production steps and costs and avoiding the use of potentially cytotoxic chemicals for crosslinking [44]. PCL served as a reference material, with fiber scaffolds providing purely topographical cues. Due to its high biocompatibility, inflammation reducing effects and ability to align neural cells it is a common choice in neural tissue engineering [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]. The diameter of the dECM and PCL fibers are sub-micron (925 ± 96 nm, 875 ± 80 nm), recreating the fiber diameter found in the native tissue of e.g. collagen fibers with a diameter of 20–500 nm [41,[55], [56], [57]], and in the same size range than dECM fibers reported previously [30,42,43]. Besides the topographical and biochemical properties of the fibers, the scaffolds’ elastic characteristics could also influence differentiation and alignment of neurons. The young's moduli determined for dECM and PCL fiber scaffolds are in accordance with previously described results [51]. Electrospun skin-derived dECM fiber scaffolds have been reported with comparable elasticity properties of 456 ± 62 Pa by Weigel et al. [58]. As described in literature, the differentiation and migration behavior can be directed by the elasticity of the surrounding tissue with neurons preferring softer tissues [59]. In order to produce a scaffold with a defined architecture, the control of the spinning procedure is crucial, and we achieved this by utilizing the bioprinter's ability of controlled movement in depositing fibers in random or aligned fashion. The electrospinning of porcine spinal cord-derived dECM into an aligned fiber scaffold, has to the best of our knowledge not been reported yet. The fibers are deposited randomly with densities similar as reported for other dECM formulations [28,30,43], and as an anisotropically aligned scaffold [43]. Using a bioprinter, offers the advantage of creating structures of higher complexity without being bound to a rotating drum or gap-based electrospinning set-up mainly generating aligned structures [44].
The viability assays and optical micrographs demonstrated the high biocompatibility and bioactivity of dECM scaffolds. Further, these dECM-based scaffolds supported the adhesion, proliferation, and differentiation of SH-SY5Y cells. This human immortalized cell line could be differentiated into mature functional neurons as described before. Due to their neuronal morphology and functionality, differentiated SH-SY5Y cells have been used as an in vitro model for spinal cord development and in neurite outgrowth studies [38,40,[60], [61], [62]]. The alignment of differentiated neurons on the dECM scaffolds was successfully achieved indicating that the fiber diameter and fiber density in the engineered scaffolds represented topographical cues suitable for alignment as reported in literature [54,63].
To simulate an injury of the spinal cord, the cells grown on the scaffolds were lesioned and a gap with injured neurons on both sides generated. This experiment was based on well-established migration assays, but modified by implementing the engineered fiber scaffolds as cell substrates creating an oriented 3D-like environment [64]. The rate and area of lesion closure was significantly different when comparing biological (dECM) to synthetic (PCL) fiber scaffolds. The higher migration rate and as a result the faster and more complete scratch closure on dECM scaffolds could be associated to the positive effect of dECM on migration, outgrowth and network formation [65]. Moreover, an aligned scaffold results in faster closure, which could be speculated to be due to the pre-alignment of the cells [66]. This pre-alignment would facilitate a directed migration and in turn lead to faster lesion closure. Further, the results of the migration assay indicate that the lesion closure is primarily due to neuronal migration, rather than proliferation. In our model no activation of proliferation or change of the neuron-like phenotype back to undifferentiated neural cell was detected by immunostainings after lesion. This is particularly of interest, because one main contribution to the inhibitory environment after SCI is the activation of cell proliferation and the secondary injury cascade inducing excessive dECM secretion into the lesion site [[67], [68], [69]]. The concept of inducing migration and recruitment of neural cells to close the lesion site has been shown to improve motor functions and new synaptic connections in animal models post injury [67,70]. Neural stem cells, either endogenous or grafted on scaffolds, have been shown to migrate towards the lesion site. However, it is crucial to direct the differentiation into neuronal lineage to avoid further contribution to the (astrocytic) scar formation, and enhance the reconnection of neurons into functional circuits [[71], [72], [73], [74], [75], [76]]. The bioengineered aligned dECM scaffolds could actively support neuronal migration in lesion environment and thus promote the formation of neuronal relays and regeneration. One potential molecular mechanism involved is the induction of the expression of PTK2 by the aligned dECM. This is a cytoplasmic protein tyrosine kinase which is found concentrated in the focal adhesions that form between cells growing in the presence of extracellular matrix constituents [77]. It may be an important early step in cell growth and signal transduction triggered in response to cell interactions with the extracellular matrix [78,79]. The increased expression of PTK2 in cells grown on aligned dECM support the effect of the scaffold on the cellular phenotype. Altogether, our results showcase that the use of aligned dECM-based scaffolds deliver biochemical cues that support the topographical cues provided by synthetic polymer-based scaffolds. With this, the generated scaffolds influence cellular behavior, enhance neuronal migration, and can promote wound healing. This could be implemented as a potential strategy to address SCI with specific scaffolds derived of dECM.
5. Conclusion
A dECM-fiber scaffold offering anisotropical cues was developed to study the alignment and migration of neurons. The ECM was efficiently isolated from porcine spinal cord, achieving low dsDNA content, while preserving ECM proteins. The concentration of detergents and incubation times could be reduced without decreasing the decellularization efficiency due to improved tissue preparation. The integration of an electrospinning printhead into a 3D bioprinter enabled the production of randomly and highly aligned nanofiber scaffolds. Parallel orientated spinal cord-derived dECM fiber scaffolds have not been reported previously. On aligned dECM scaffolds mature neurons migrated into the lesion site most efficiently, showing a faster and more complete wound closure efficiency compared to random dECM or any PCL-based fiber scaffolds. The combination of topographical and biochemical cues have thus a direct influence on cell phenotype. The results of this work improve the understanding of combined effects of topographical and biochemical signals on neural behavior. Further, the fabrication of dECM-based spinning solutions bring us one step closer to bioengineer scaffolds with tissue-specific native cues, which represents a way forward for future in vivo studies for the treatment of SCI.
Author contributions
The research was designed by L.M and A.T and discussed with J.R and L.S-D; L.M performed the experimental work; R.N helped in the preparation of decellularized spinal cord; C.G. supported the confocal microscopy studies; L.M prepared the manuscript in consultation with A.T, J.R and L.S-D. All authors have given approval to the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the funding of FHNW HLS to conduct this study. The authors are grateful to Prof. Johannes Mosbacher for the kind donation of SH-SY5Y cells.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbiosy.2023.100081.
Appendix. Supplementary materials
References
- 1.Ahuja C.S., et al. Traumatic spinal cord injury. Nat. Rev. Dis. Primers. 2017;3:17018. doi: 10.1038/nrdp.2017.18. [DOI] [PubMed] [Google Scholar]
- 2.Krause J.S. Mortality after spinal cord injury: an ll-year prospective study. Arch. Phys. Med. Rehabil. 1997;78:7. doi: 10.1016/s0003-9993(97)90193-3. [DOI] [PubMed] [Google Scholar]
- 3.Liu K., Tedeschi A., Park K.K., He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu. Rev. Neurosci. 2011;34:131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- 4.Klapka N., Müller H.W. Collagen matrix in spinal cord injury. J. Neurotrauma. 2006;23:422–435. doi: 10.1089/neu.2006.23.422. [DOI] [PubMed] [Google Scholar]
- 5.Silver J., Miller J.H. Regeneration beyond the glial scar. Nat. Rev. Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 6.Sun F., et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu W., et al. Polymer-based scaffold strategies for spinal cord repair and regeneration. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.590549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortazavi M.M., et al. Engraftment of neural stem cells in the treatment of spinal cord injury. Transl. Res. Anat. 2015;1:11–16. [Google Scholar]
- 9.Dulin J.N., Lu P. Bridging the injured spinal cord with neural stem cells. Neural Regen. Res. 2014;9:229–231. doi: 10.4103/1673-5374.128212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Chen G. In vivo reprogramming for CNS repair: regenerating neurons from endogenous glial cells. Neuron. 2016;91:728–738. doi: 10.1016/j.neuron.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinrich C., Spagnoli F.M., Berninger B. In vivo reprogramming for tissue repair. Nat. Cell Biol. 2015;17:204–211. doi: 10.1038/ncb3108. [DOI] [PubMed] [Google Scholar]
- 12.Rashid T.U., Gorga R.E., Krause W.E. Mechanical properties of electrospun fibers—a critical review. Adv. Eng. Mater. 2021;23 [Google Scholar]
- 13.Xie J.W., MacEwan M.R., Schwartz A.G., Xia Y.N. Electrospun nanofibers for neural tissue engineering. Nanoscale. 2010;2:35–44. doi: 10.1039/b9nr00243j. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal S., Wendorff J.H., Greiner A. Use of electrospinning technique for biomedical applications. Polymer. 2008;49:5603–5621. (Guildf) [Google Scholar]
- 15.Ding J., et al. Electrospun polymer biomaterials. Prog. Polym. Sci. 2019;90:1–34. [Google Scholar]
- 16.Park S.B., Lih E., Park K.S., Joung Y.K., Han D.K. Biopolymer-based functional composites for medical applications. Prog. Polym. Sci. 2017;68:77–105. [Google Scholar]
- 17.Hussey G.S., Dziki J.L., Badylak S.F. Extracellular matrix-based materials for regenerative medicine. Nat. Rev. Mater. 2018;3:159–173. [Google Scholar]
- 18.Chan G., Mooney D.J. New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol. 2008;26:382–392. doi: 10.1016/j.tibtech.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Zhu M., et al. In vivo engineered extracellular matrix scaffolds with instructive niches for oriented tissue regeneration. Nat. Commun. 2019;10:4620. doi: 10.1038/s41467-019-12545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., et al. Electrospun collagen fibers with spatial patterning of SDF1α for the guidance of neural stem cells. Adv. Healthc. Mater. 2015;4:1869–1876. doi: 10.1002/adhm.201500271. [DOI] [PubMed] [Google Scholar]
- 21.Morris A.H., Stamer D.K., Kyriakides T.R. The host response to naturally-derived extracellular matrix biomaterials. Semin. Immunol. 2017;29:72–91. doi: 10.1016/j.smim.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Badylak S., Freytes D., Gilbert T. Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Crapo P.M., et al. Biologic scaffolds composed of central nervous system extracellular matrix. Biomaterials. 2012;33:3539–3547. doi: 10.1016/j.biomaterials.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou J.L., et al. Peripheral nerve-derived matrix hydrogel promotes remyelination and inhibits synapse formation. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 25.Keane T.J., Swinehart I.T., Badylak S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods. 2015;84:25–34. doi: 10.1016/j.ymeth.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 26.DeQuach J.A., Yuan S.H., Goldstein L.S.B., Christman K.L. Decellularized porcine brain matrix for cell culture and tissue engineering scaffolds. Tissue Eng. Part A. 2011;17:2583–2592. doi: 10.1089/ten.tea.2010.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishtul S., Baruch L., Machluf M. Processed tissue–derived extracellular matrices: tailored platforms empowering diverse therapeutic applications. Adv. Funct. Mater. 2019 doi: 10.1002/adfm.201900386. [DOI] [Google Scholar]
- 28.Kim Y.S., Majid M., Melchiorri A.J., Mikos A.G. Applications of decellularized extracellular matrix in bone and cartilage tissue engineering. Bioeng. Transl. Med. 2018;4:83–95. doi: 10.1002/btm2.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volpato F.Z., Führmann T., Migliaresi C., Hutmacher D.W., Dalton P.D. Using extracellular matrix for regenerative medicine in the spinal cord. Biomaterials. 2013;34:4945–4955. doi: 10.1016/j.biomaterials.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 30.Baiguera S., et al. Electrospun gelatin scaffolds incorporating rat decellularized brain extracellular matrix for neural tissue engineering. Biomaterials. 2014;35:1205–1214. doi: 10.1016/j.biomaterials.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 31.Medberry C.J., et al. Hydrogels derived from central nervous system extracellular matrix. Biomaterials. 2013;34:1033–1040. doi: 10.1016/j.biomaterials.2012.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granato, A.E.C. et al. A Novel Decellularization Method to Produce Brain Scaffolds. http://biorxiv.org/lookup/doi/10.1101/680702 (2019) doi: 10.1101/680702. [DOI] [PubMed]
- 33.Guo S.Z., Ren X.J., Wu B., Jiang T. Preparation of the acellular scaffold of the spinal cord and the study of biocompatibility. Spinal Cord. 2010;48:576–581. doi: 10.1038/sc.2009.170. [DOI] [PubMed] [Google Scholar]
- 34.Ren, T., van der Merwe, Y. & Steketee, M.B. Developing extracellular matrix technology to treat retinal or optic nerve injury. eNeuro 2, ENEURO.0077-15.2015 (2015). [DOI] [PMC free article] [PubMed]
- 35.Goto N. Discriminative staining methods for the nervous system: luxol fast blue–periodic acid-Schiff–hematoxylin triple stain and subsidiary staining methods. Stain Technol. 1987;62:305–315. doi: 10.3109/10520298709108015. [DOI] [PubMed] [Google Scholar]
- 36.Fischer A.H., Jacobson K.A., Rose J., Zeller R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc. 2008;2008 doi: 10.1101/pdb.prot4986. pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- 37.Janesick A., Wu S.C., Blumberg B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 2015;72:1559–1576. doi: 10.1007/s00018-014-1815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Constantinescu R., Constantinescu A.T., Reichmann H., Janetzky B., Gerlach M., Deckert J., Double K., Koutsilieri E. Neuropsychiatric Disorders An Integrative Approach. Springer; 2007. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y; pp. 17–28. [DOI] [PubMed] [Google Scholar]
- 39.Påhlman S., Ruusala A.I., Abrahamsson L., Mattsson M.E.K., Esscher T. Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 1984;14:135–144. doi: 10.1016/0045-6039(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 40.Kovalevich J., Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes C.P., Sell S.A., Boland E.D., Simpson D.G., Bowlin G.L. Nanofiber technology: designing the next generation of tissue engineering scaffolds. Adv. Drug Deliv. Rev. 2007;59:1413–1433. doi: 10.1016/j.addr.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 42.Efraim Y., et al. 3D structure and processing methods direct the biological attributes of ECM-based cardiac scaffolds. Sci. Rep. 2019;9:5578. doi: 10.1038/s41598-019-41831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoen B., et al. Electrospun extracellular matrix: paving the way to tailor-made natural scaffolds for cardiac tissue regeneration. Adv. Funct. Mater. 2017;27 [Google Scholar]
- 44.Smoak M.M., et al. Fabrication and characterization of electrospun decellularized muscle-derived scaffolds. Tissue Eng. Part C Methods. 2019;25:276–287. doi: 10.1089/ten.tec.2018.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jakobsson A., et al. Three-dimensional functional human neuronal networks in uncompressed low-density electrospun fiber scaffolds. Nanomed. Nanotechnol. Biol. Med. 2017;13:1563–1573. doi: 10.1016/j.nano.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 46.Lee J.Y., Bashur C.A., Gomez N., Goldstein A.S., Schmidt C.E. Enhanced polarization of embryonic hippocampal neurons on micron scale electrospun fibers. J. Biomed. Mater. Res. 2009;9999A doi: 10.1002/jbm.a.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrudo F.F.F., et al. The effect of electrospun scaffolds on the glycosaminoglycan profile of differentiating neural stem cells. Biochimie. 2021;182:61–72. doi: 10.1016/j.biochi.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMurtrey R.J. Patterned and functionalized nanofiber scaffolds in three-dimensional hydrogel constructs enhance neurite outgrowth and directional control. J. Neural Eng. 2014;11 doi: 10.1088/1741-2560/11/6/066009. [DOI] [PubMed] [Google Scholar]
- 49.Donoghue P.S., et al. The development of a ɛ-polycaprolactone scaffold for central nervous system repair. Tissue Eng. Part A. 2013;19:497–507. doi: 10.1089/ten.TEA.2012.0382. [DOI] [PubMed] [Google Scholar]
- 50.Li Z., Wang Q., Hu H., Zheng W., Gao C. Research advances of biomaterials-based microenvironment-regulation therapies for repair and regeneration of spinal cord injury. Biomed. Mater. 2021;16 doi: 10.1088/1748-605X/ac1d3c. [DOI] [PubMed] [Google Scholar]
- 51.Mungenast L., et al. Directional submicrofiber hydrogel composite scaffolds supporting neuron differentiation and enabling neurite alignment. Int. J. Mol. Sci. 2022;23:11525. doi: 10.3390/ijms231911525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi J., et al. Topographic orientation of scaffolds for tissue regeneration: recent advances in biomaterial design and applications. Biomimetics. 2022;7:131. doi: 10.3390/biomimetics7030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y.A., et al. Nanoimprinted anisotropic topography preferentially guides axons and enhances nerve regeneration. Macromol. Biosci. 2018;18 doi: 10.1002/mabi.201800335. [DOI] [PubMed] [Google Scholar]
- 54.Vedaraman S., et al. Anisometric microstructures to determine minimal critical physical cues required for neurite alignment. Adv. Healthc. Mater. 2021;10 doi: 10.1002/adhm.202100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ushiki T., Ide C. Three-dimensional organization of the collagen fibrils in the rat sciatic nerve as revealed by transmission- and scanning electron microscopy. Cell Tissue Res. 1990;260:175–184. doi: 10.1007/BF00297503. [DOI] [PubMed] [Google Scholar]
- 56.Siadat S.M., Silverman A.A., DiMarzio C.A., Ruberti J.W. Measuring collagen fibril diameter with differential interference contrast microscopy. J. Struct. Biol. 2021;213 doi: 10.1016/j.jsb.2021.107697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buttafoco L., et al. Electrospinning of collagen and elastin for tissue engineering applications. Biomaterials. 2006;27:724–734. doi: 10.1016/j.biomaterials.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 58.Weigel T., et al. Fully synthetic 3D fibrous scaffolds for stromal tissues—replacement of animal-derived scaffold materials demonstrated by multilayered skin. Adv. Mater. 2022;34 doi: 10.1002/adma.202106780. [DOI] [PubMed] [Google Scholar]
- 59.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 60.Li Q., Yang C., Zhang B., Guo Z., Lin J. Wnt3a ectopic expression interferes axonal projection and motor neuron positioning during the chicken spinal cord development. J. Mol. Neurosci. 2018;64:619–630. doi: 10.1007/s12031-018-1060-z. [DOI] [PubMed] [Google Scholar]
- 61.Hwang K., et al. Glial cell line-derived neurotrophic factor-overexpressing human neural stem/progenitor cells enhance therapeutic efficiency in rat with traumatic spinal cord injury. Exp. Neurobiol. 2019;28:679–696. doi: 10.5607/en.2019.28.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park K.W., Lin C.Y., Benveniste E.N., Lee Y.S. Mitochondrial STAT3 is negatively regulated by SOCS3 and upregulated after spinal cord injury. Exp. Neurol. 2016;284:98–105. doi: 10.1016/j.expneurol.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omidinia-Anarkoli A., Ephraim J.W., Rimal R., De Laporte L. Hierarchical fibrous guiding cues at different scales influence linear neurite extension. Acta Biomater. 2020;113:350–359. doi: 10.1016/j.actbio.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 64.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 65.Lam D., et al. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-40128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrie R.J., Doyle A.D., Yamada K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Echeverry S., Shi X.Q., Zhang J. Characterization of cell proliferation in rat spinal cord following peripheral nerve injury and the relationship with neuropathic pain. PAIN®. 2008;135:37–47. doi: 10.1016/j.pain.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Liddelow S.A., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wanner I.B., et al. Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via stat3-dependent mechanisms after spinal cord injury. J. Neurosci. 2013;33:12870–12886. doi: 10.1523/JNEUROSCI.2121-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou S. Relay strategies combined with axon regeneration: a promising approach to restore spinal cord injury. Neural Regen. Res. 2014;9:1177. doi: 10.4103/1673-5374.135322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meletis K., et al. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 2008;6:e182. doi: 10.1371/journal.pbio.0060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., et al. Cetuximab modified collagen scaffold directs neurogenesis of injury-activated endogenous neural stem cells for acute spinal cord injury repair. Biomaterials. 2017;137:73–86. doi: 10.1016/j.biomaterials.2017.05.027. [DOI] [PubMed] [Google Scholar]
- 73.McDonough A., Martínez-Cerdeño V. Endogenous proliferation after spinal cord injury in animal models. Stem Cells Int. 2012;2012 doi: 10.1155/2012/387513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lacroix S., et al. Central canal ependymal cells proliferate extensively in response to traumatic spinal cord injury but not demyelinating lesions. PLoS One. 2014;9:e85916. doi: 10.1371/journal.pone.0085916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mothe A.J., Tator C.H. Proliferation, migration, and differentiation of endogenous ependymal region stem/progenitor cells following minimal spinal cord injury in the adult rat. Neuroscience. 2005;131:177–187. doi: 10.1016/j.neuroscience.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 76.Zai L.J., Wrathall J.R. Cell proliferation and replacement following contusive spinal cord injury. Glia. 2005;50:247–257. doi: 10.1002/glia.20176. [DOI] [PubMed] [Google Scholar]
- 77.Nikolic M. The molecular mystery of neuronal migration: FAK and Cdk5. Trends Cell Biol. 2004;14:1–5. doi: 10.1016/j.tcb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Katoh K. FAK-dependent cell motility and cell elongation. Cells. 2020;9:192. doi: 10.3390/cells9010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan C.L., et al. Integrin activation promotes axon growth on inhibitory chondroitin sulfate proteoglycans by enhancing integrin signaling. J. Neurosci. 2011;31:6289–6295. doi: 10.1523/JNEUROSCI.0008-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.