Abstract
Introduction
Harmonization protocols that address batch effects and cross‐site methodological differences in multi‐center studies are critical for strengthening electroencephalography (EEG) signatures of functional connectivity (FC) as potential dementia biomarkers.
Methods
We implemented an automatic processing pipeline incorporating electrode layout integrations, patient–control normalizations, and multi‐metric EEG source space connectomics analyses.
Results
Spline interpolations of EEG signals onto a head mesh model with 6067 virtual electrodes resulted in an effective method for integrating electrode layouts. Z‐score transformations of EEG time series resulted in source space connectivity matrices with high bilateral symmetry, reinforced long‐range connections, and diminished short‐range functional interactions. A composite FC metric allowed for accurate multicentric classifications of Alzheimer's disease and behavioral variant frontotemporal dementia.
Discussion
Harmonized multi‐metric analysis of EEG source space connectivity can address data heterogeneities in multi‐centric studies, representing a powerful tool for accurately characterizing dementia.
Keywords: AD, automatic harmonization, bvFTD, dementia classification, EEG, inverse solution methods, multi‐centric studies, whole‐brain functional connectivity
1. INTRODUCTION
Alzheimer's disease (AD) and behavioral variant frontotemporal dementia (bvFTD) are two of the most common and impactful forms of dementia globally. As a result, AD and bvFTD are life‐limiting conditions that represent an enormous burden on developing countries and across underrepresented populations, where socioeconomic and educational inequities compromise access to traditional dementia biomarkers and undermine the timely diagnosis of the disease. 1 , 2 Efforts to overcome these challenges include the development of biomarkers derived from high‐density electroencephalography (hd‐EEG), a cost‐effective, scalable, and portable technology that allows for the accurate assessment of brain oscillatory dynamics and network disintegration. 3 , 4 In this regard, functional connectivity (FC) analyses are particularly relevant because FC is associated with cognitive deficits and neuronal dysfunction in neurodegeneration. 5 , 6
Two major challenges underpin the development of dementia biomarkers derived from EEG connectivity. First, progress in this field relies on large‐scale multi‐center studies addressing geographic and sociodemographic heterogeneities. These studies require harmonization protocols to bring heterogeneous data together into a unified analytical space, thereby minimizing batch effects. 7 , 8 Second, various conceptual frameworks comprising different connectivity metrics and estimation procedures have been used to assess EEG connectivity. 9 , 10 To comprehensively characterize abnormal FC in dementia, composite connectivity metrics that provide insight into various types of functional interactions are needed. 10 , 11 Joint analyses of FC benefit from EEG source localization methods, 12 , 13 since they provide topographic information on brain functional interactions and reduce spurious effects commonly observed in sensor‐space connectivity analyses. 14
This study presents a novel framework for dementia characterization, in which joint analyses of EEG source space connectivity in site‐harmonized data serve as input for gradient‐boosting machine learning classifiers tuned by Bayesian optimization. 15 , 16 The tool considers four essential stage of harmonization that follow recommendations for assesing EEG connectivity in neurodegenerative diesasese (the ConnEEGtome), 11 the tool incorporates four critical harmonization stages: (a) adoption of standards for data storage that facilitate information exchange 17 ; (b) adequate reference schemes by implementing average reference or Reference Electrode Standardization Technique (REST) 18 , 19 ; (c) integration of different electrode layouts 20 ; and (d) EEG rescaling using patient–control normalizations. 11 , 15
We hypothesized that (1) spline interpolations of re‐referenced EEG allow for the integration of different electrode layouts, (2) a more comprehensive representation of the EEG source space connectomics is obtained after the patient–control normalization of the EEG data using Z‐score transformations, and (3) multi‐metric analyses of source space connectivity allow for the accurate classification of dementia subtypes. The following sections describe the workflow for data harmonization and classification. The validity of the harmonization procedures and the use of the multi‐metric approach to source space EEG connectivity for dementia characterization is illustrated. The study is accompanied by a repository where the code, a user guide, and source space connectivity matrices are available (https://github.com/PavelPrado/EEG-Harmonization).
2. METHODS
2.1. Data set for validation (AD, bvFTD, and HCs)
Because the resting state EEG (rsEEG) is the ideal acquisition protocol for data harmonization in EEG multicentric studies, 11 the pipeline's functioning is described using this EEG modality. To illustrate the different steps of the processing pipeline, we used eyes‐closed rsEEG acquired from participants enrolled in two centers of the Multi‐Partner Consortium to Expand Dementia Research in Latin America (ReDLat), 2 a regional effort aimed at the harmonization of participant enrollment and neurocognitive assessment in multi‐centric studies. Therefore, the pipeline's validation was carried out using data from a well‐described cohort that was recruited and assessed for cognitive abilities using standardized procedures. Consequently, inter‐site variability was restricted predominantly to the EEG acquisition.
Participants were patients with AD (n = 35), patients with bvFTD (n = 19), and healthy control (HCs, n = 46). All patients were in the early/mild stages of the disease and had no proven track of substance abuse, primary language deficits, and neurological or psychiatric disorders. The demographic information of the sample (Table A1), the diagnosis criteria, and the neurophysiological examinations are presented in Appendix A. The EEG acquisition is also described in Appendix A. The study was approved by the institutional review board at each recruitment site. A signed informed consent was provided by all participants, following the Declaration of Helsinki.
2.2. Analysis workflow
The pipeline runs with a default workflow that can be modified following instructions provided in the user guide (Appendix B). Additional technical and practical information is provided elsewhere. 21 The pipeline has a modular organization in which a module can offer different processing alternatives, for example, seven data normalizations, three methods for EEG source space estimation, and 22 possible joint analyses of connectivity (Table C1, Appendix C). Therefore, more than 462 different processing strategies can be implemented.
2.2.1. Data structure
The pipeline (Figure 1) is implemented in Matlab as a Command Line Interface and built upon pre‐existing tools. Data (rsEEG or task‐related activity) is organized according to the EEG‐BIDS format, an extension of the brain imaging data structure (BIDS) that addresses the heterogeneity of data organization by following the FAIR principles of findability, accessibility, and interoperability. 17
FIGURE 1.
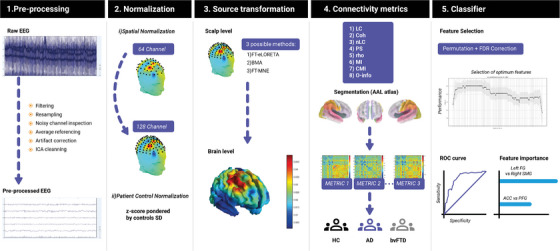
Flowchart of the pipeline for dementia classification based on multi‐metric analyses of electroencephalography (EEG) source space connectivity. From left to right, the figure presents the five modules of the pipeline. Traditional reprocessing steps are indicated in module 1. This is followed by the normalization stage, where spatial harmonization and data rescaling are conducted (module 2). Source reconstruction (module 3) assessing the inverse problem in EEG is implemented for joint analyses of functional whole‐brain functional connectivity in Alzheimer's disease (AD) and behavioral variant frontotemporal dementia patients (bvFTD) (module 4), alongside parameters describing the performance of machine learning classification of each dementia subtype (module 5).
2.2.2. Preprocessing
In the preprocessing module (Figure 1A), filtering (default cut‐off: 0.5 and 40 Hz) and resampling (default frequency: 512 Hz) are executed automatically via EEGLAB. 22 These steps are followed by a bad channel inspection that incorporates a built‐in graphical user interface (GUI) to facilitate the manual identification of noisy channels. Next, data are referenced using the average reference (AVE, the average time series of all channels) or REST. 18 , 19 Artifact removal can be carried out with (1) ICLabel, a classification method that labels EEG‐independent components into signals and different categories of noise, 23 (2) EyeCatch, a tool containing a database of manually identified eye‐related ICA scalp maps, 24 and (3) BLINKER, a toolbox for blinking extraction and quantification of ocular indices. 25 Subsequently, channels marked as noisy are replaced by spherical interpolation of neighboring channels 26 (Figure 1A).
RESEARCH IN CONTEXT
Systematic review: Authors reviewed the literature using traditional sources (e.g., PubMed) regarding functional connectivity (FC) with electroencephalography (EEG) for dementia and harmonization protocols to target cross‐site variability and batch effects in multi‐center studies. In addition, integrative connectivity analysis in the EEG source space were reviewed. No previous work has proposed a harmonized, multi‐centric and multi‐metric approach to EEG source connectivity in dementia research.
Interpretation: Findings indicate the effectiveness of critical harmonization steps to reduce cross‐site methodological differences. The high accuracy of classifications based on joint analysis of theoretical information metrics of FC highlights the relevance of integrative approach to assess complex connectivity patterns that profit the characterization and classification of dementia subtypes.
Future directions: Results strengthen global strategies for the assessment of dementia based on scalable and cost‐effective technologies. Future studies need to explore harmonization steps to control the effect of demographic covariates, and the test–retest reliability of the composite metrics for connectivity.
2.2.3. Spatial normalization
In this stage, EEG acquired with different electrode layouts is registered into a common topographical space using a modified version of the EEGLAB function “headplot,” 26 , 27 which projects the original EEG into a 6067‐point mesh head model and projects the signal back into the desired layout (Figure 1B). Currently the spatial normalization step supports two EEG layouts: Biosemi128 and Biosemi64 (international 10/20 system).
2.2.4. Patient–control normalization
This data rescaling aims to reduce cross‐site (centers) variability. The normalization is carried out separately by each recruitment center and consists of computing a constant for rescaling (weighting factor) from the data belonging to HCs (Figure 1B). Subsequently the EEG of all individuals is rescaled using the same weighting factor. Seven options of normalization are included, where the default option is the Z‐score transformation. 15 , 16 The latter metric describes the position of a raw score in terms of its distance from the mean when measured in standard deviation units. Other rescaling factors included in the pipeline are (1) the robust standard deviation of all data, (2) the robust standard deviation per channel, (3) the Huber mean of robust standard deviation per channel, (4) the Huber mean of the robust standard deviation per subject, (5) the mean of robust standard deviation per subject, and (6) the L‐2 norm of the robust standard deviation per subject. 20
2.2.5. EEG source space estimation
The pipeline includes three methods for solving the EEG inverse problem. The default method is the exact Low‐Resolution Electromagnetic Tomography (eLORETA), 28 which is a distributed, linear, weighted minimum norm inverse solution method providing exact localization to test seeds, albeit with a high correlation between neighboring generators. Alternatively, EEG sources can be localized using the Bayesian model averaging (BMA) method. 29 The BMA method considers anatomic constraints to address the model uncertainty, thereby allowing for the accurate estimation of deep EEG generators. The second alternative is the minimum‐norm estimation (MNE) method, which provides the EEG inverse solution that best fits the sensor data with minimum overall amplitude of brain activity. 30
2.2.6. Estimation of FC
Whole‐brain FC is computed using 82 anatomic compartments of the Automated Anatomical Labeling Atlas (AAL atlas) 31 (Table C2, Appendix C), and therefore comprises 3321 pair‐wise functional interactions. 32 , 33 The set of connectivity metrics comprises four frequency‐domain connectivity metrics (Table 1), which in turn considers instantaneous, lagged, and total connectivity in eight EEG frequency bands: delta (δ: 1.5–4 Hz), theta (θ: 4–8 Hz), alpha1 (α1: 8–10 Hz), alpha2 (α2: 10–13 Hz), beta1 (β1: 13–18 Hz), beta2 (β2: 18–21 Hz), beta3 (β3: 21–30 Hz), and gamma (γ: 30–40 Hz). This results in 96 frequency‐domain connectivity metrics (96 representations of whole‐brain FC). In addition, five time‐domain connectivity metrics can be computed (Table 1). Therefore, it is possible to conduct joint analyses of up to 101 connectivity metrics. 34 The FC metrics and the technical parameters used to compute the FC matrices are presented in Prado et al. 34 and in Appendix C (sections Connectivity metrics and Feature selection, respectively).
TABLE 1.
Connectivity metrics for the analysis of whole‐brain functional connectivity.
| Functional connectivity | ||
|---|---|---|
| Frequency‐domain | Time‐domain | |
| Linear metrics | Linear connectivity | Pearson's correlation |
| Coherence () | ||
| Nonlinear metrics | Nonlinear connectivity | Mutual information |
| Phase synchronization | Weighted mutual information | |
| Conditional mutual information | ||
| O‐information | ||
2.3. Multi‐feature analyses for classification
A threefold multi‐feature analysis 16 , 35 , 36 is implemented using the entire set (or a predefined subset) of connectivity metrics. First, the most relevant EEG features for binary classifications (i.e., AD vs HC and bvFTD vs HC) are identified by combining statistical tests and progressive feature elimination in the training sets, which consisted of 80% of the total samples (one set containing ADs and HCs, the other containing bvFTD and HCs). In this stage, statistical criteria are used to reduce the dimension of the connectivity matrices to those connections with statistically significant differences between the dementia subtype (AD or bvFTD) and HCs (Feature filter selection section). Once optimum features are selected, the machine learning classifier is used on new out‐of‐sample data. Finally, we run a feature importance analysis to gain insights into which features played a more significant role in classifying the testing sample (machine learning algorithm section).
2.3.1. Filtering for feature selection
The dimension of the connectivity matrices is reduced using group‐level statistics. 34 This step is conducted independently for each connectivity metric by comparing the whole‐brain connectivity maps of the groups being assessed (a given dementia subtype and HCs, for example) using nonparametric permutation tests (α = 0.05; 5000 randomizations). 37 To control the false discovery rate (FDR), the Benjamini and Hochberg FDR method was utilized. 38 This filter method 39 avoids adaptation to the data set specificities (overfitting) and therefore increases generalizability. 40 Furthermore, this approach allows for a more direct interpretation of the results than other alternatives of dimensionality reduction, such as principal component analysis, 41 it is computationally inexpensive, and does not consider the classifier performance. 39 Connections with statistically significant differences between groups (e.g., HCs‐AD, and HCs‐bvFTD) are selected for the subsequent analysis steps.
2.3.2. Machine learning algorithm
Machine learning classifiers utilize connections that vary significantly between groups as features to distinguish between dementia subtypes and HCs, using the procedure described in Moguilner et al. 16 Random divisions of the data set are obtained using an 8:2 split ratio for training and testing, respectively, without using the testing data set during the validation phase for out of k‐folds (k = 10) predictions. Stratified cross‐validation is used during the training phase to tune Bayesian hyperparameters and ensure equal sample distribution among the folds, thereby reducing potential biases in classification due to imbalanced sample sizes.
In addition, regularized boosting helps to reduce overfitting. The classification accuracy values (F1 score) are reported along with the receiver‐operating characteristic (ROC) curve and the SHapley Additive exPlanations (SHAP), which allow identifying the set of parameters that provide a compressive explanation of the classification. Further details of the classification algorithm are provided in Section 8.2, Appendix B.
3. RESULTS
Validation of the pipeline approached critical steps for data harmonization (spatial normalization and Z‐transformation for EEG rescaling) and using a composite connectivity metric to classify AD and bvFTD.
3.1. Spatial normalization for harmonization of EEG data
We quantitatively evaluated the spatial normalization by transforming the EEG (128 channels, Biosemi acquisition system) to a 64‐channel 10/20 layout, using the 6067‐point head model, and repeating the process in the opposite direction (Figure 2A). The validity of the normalization was expressed as the absolute error of the transformation, that is, the absolute difference between the original and recreated signals. For all electrode locations, the transformation error was always two orders of magnitude lower than the original signal (Figure 2A). Furthermore, regression analyses showed that the recreated signal was accurately predicted from the original voltages. In this scenario, the spatial normalization resulted in moderate underestimations of voltages in distal frontotemporal scalp locations (Figure 2B). This distortion in the EEG topology resulted in phantom activities estimated in inferior portions of frontolateral, temporal, and occipitotemporal cortical regions (Figure 2B). Nevertheless, the maximum error in the estimation of cortical activities (differences of the current density maps derived from the original and recreated EEG topologies) was also two orders of magnitude lower than the original cortical activity, which confirmed the validity of the spatial normalization.
FIGURE 2.
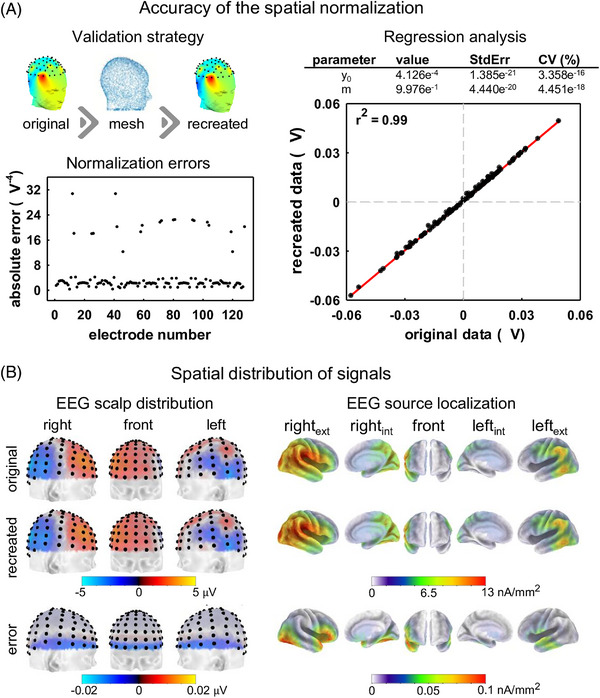
Spatial normalization of electroencephalography (EEG). (A) A scheme of the validation strategy (comparison of original and estimated voltage samples of the 128 Biosemi electrode layout) is illustrated in the left, top panel. The absolute error between the original and estimated voltages recorded in one healthy control is presented for each electrode in the left, inferior panel. Original and estimated voltages, as the theoretical line representing the relationship between original and predicted voltages, are plotted in the right panel. The parameters of the regression analysis are presented in the table. (B) Scalp distribution of voltages of the original and recreated EEG, as well as their corresponding brain generators estimated using the eLORETA method. Errors (differences between the original and recreated voltages) are provided.
3.2. Effect of data rescaling on FC
Examples of time courses of EEG at different scalp locations, along with the corrections derived from the Z‐score transformation, are illustrated in Figure 3A. The rescaling caused only slight variations in the EEG topology. At the source space level, the spatial normalization did not disturb the topographical distribution of the EEG generators. Nevertheless, the current density of some cortical areas varied (Figure 3B). In this illustrative example, the latter is reflected in the reduced activity of the left inferior frontal gyrus, and the left inferior temporal gyrus obtained after normalization (Figure 3B).
FIGURE 3.
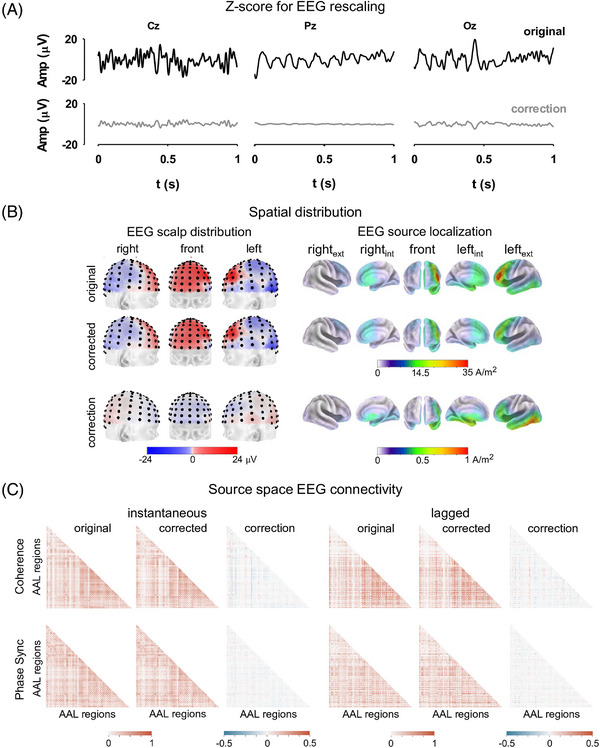
Electroencephalography (EEG) rescaling using the Z‐score transformation. (A) Illustrative examples of EEG time series acquired at different scalp locations, from an elderly healthy control. Original signals are presented along with their corresponding transformations. (B) Scalp distribution of voltages of the original and rescaled EEG, as well as their corresponding brain generators estimated using the eLORETA method. Different views of the brain are presented. (C) Whole‐brain functional connectivity estimated in the low alpha EEG frequency band with different frequency‐domain connectivity metrics. Functional connectivity matrices computed from original and rescaled EEG. The differences between the connectivity matrices are illustrated. For clarity, only inferior triangles of the connectivity matrices are presented. ext, external; in, internal; phase sync., phase synchronization.
Furthermore, rescaling induced modest corrections of the EEG source space connectivity maps (Figure 3C). To illustrate the above, the 20 strongest connections in the EEG alpha band were estimated before and after applying the Z‐score transformation. Connectivity maps were computed with two frequency‐domain metrics: coherence and phase synchronization (Figure 4). The EEG rescaling resulted in brain FC with reduced hemispheric asymmetry (lateralization). This was reflected by the emergence of interhemispheric connections and long‐range connections within a particular hemisphere, accompanied by a reduction of short‐range interactions (Figure 4).
FIGURE 4.
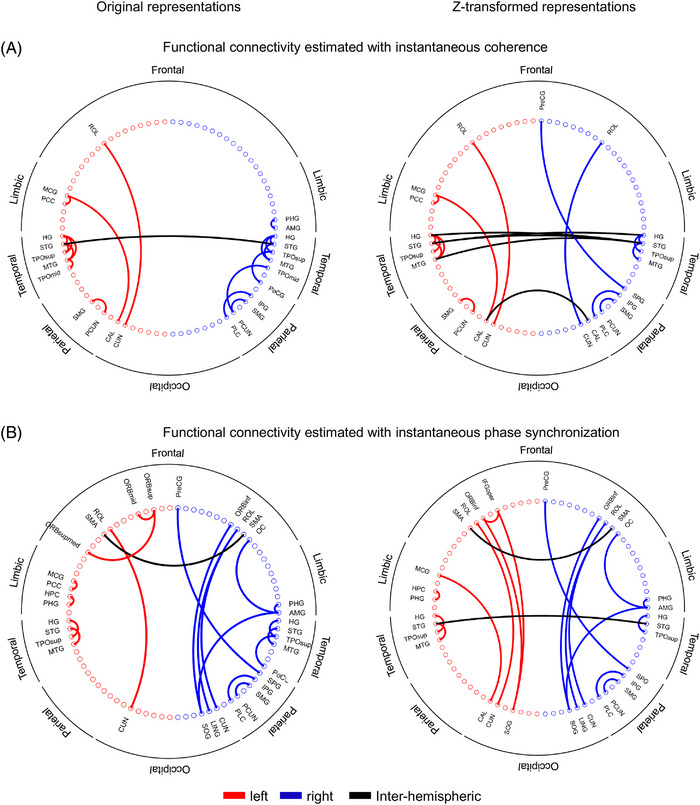
Topographic distribution of the 20 strongest brain functional connections estimated in the electroencephalography (EEG) low alpha frequency band with (A) instantaneous coherence and (B) instantaneous phase synchronization. The EEG source space connectivity was estimated from the original time series (Left panels: Z‐transformed representations). Left, right, and interhemispheric connections are illustrated with different colors.
3.3. Classification of dementia subtypes using a composite metric of FC
Figure 5 illustrates how joint analyses of FC help characterize dementia. In this case, AD and bvFTD were classified by integrating information theoretic metrics (MI: mutual information, CMI: conditional mutual information, and O info: Organizational information). A comprehensive analysis of the integration of frequency‐domain connectivity metrics is reported elsewhere. 34
FIGURE 5.
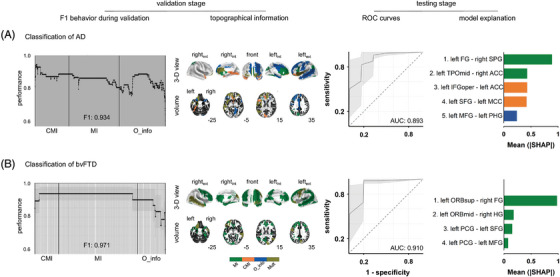
Dementia classification based on source localized resting state EEG (rsEEG) connectivity estimated with theoretic‐information metrics. (A) Classification of Alzheimer's disease (AD). (B) Classification of behavioral variant frontotemporal dementia (bvFTD). For each classification, the performance of the classification (F1 during validation) is presented as a function of the number of features (functional connections) that were sequentially included in the analyses. The performance (F1) obtained with the optimum set of features is provided. The topographical information of the optimum set of features of the classification is shown (topographical information). Regions denoted by “mult” in the color bar belong to atypical connections captured by different connectivity metrics. The z coordinate of the horizontal plane of the brain is presented in the right low portion of each volume view. The receiver‐operating characteristic (ROC) curves are presented. The area under the curve (AUC) is indicated for each case. ROC curves are presented as the mean (thick line) and the 95% confidence interval (shadows), which were obtained by bootstrapping (5000 times). Connections with the highest contributions to the predictive model are presented (model explanation) in descendent order of relevance (SHAP values). ACC: anterior cingulate cortex; AD, Alzheimer's disease; AUC, area under the curve; bvFTD, behavioral variant frontotemporal dementia; CMI, conditional mutual information; FG, fusiform gyrus; HG, Helsch gyrus; IFGoper, opercular part of the inferior frontal region; MFG, middle frontal gyrus; MI: mutual information; O_info: Organizational information; ORBmid: orbital part of the middle frontal gyrus; ORBsup: orbital part of the superior frontal gyrus; PCG, precentral gyrus; PHG, parahippocampal gyrus; ROC, receiver operating characteristic curves; SFG, superior frontal gyrus; SHAP, SHapley Additive exPlanations; SMG, supramarginal gyrus, TPOmid, middle temporal pole.
The number of features used for the classification (which resulted from the feature filter selection step, section 2.3) is presented in Table C3 (Appendix C). The location of these features within the connectivity matrices is illustrated in Figure SC1 (Appendix C), which is complemented by the list of AAL regions provided in Table C4 (Appendix C). The complete list of connections used for the classifications is provided in Tables C4 (AD vs HCs classification) and C5 (bvFTD vs HCs classification) (Appendix C). Finally, how features were sorted to input the classifier is illustrated in Figure C1 (Appendix C).
During the validation stage, the maximum performance of the classification (F1) was 0.93 (for AD vs HCs) and 0.97 (for bvFTD vs HCs) (Figure 5). The complete list of optimum features is presented in Table C7 (Appendix C). It is noteworthy that the optimum set of features comprised connections captured by different metrics (Table C2). Furthermore, these sets of features varied for the AD and bvFTD classifications. The classifications resulted in ROC curves of 0.89 and 0.91 for AD and bvFTD, respectively. The complete set of performance metrics is presented in Table C8 (Appendix C).
Regions with atypical connectivity included the superior middle and inferior frontal gyri, the parahippocampal gyrus and the anterior cingulate cortex, the middle temporal pole, the supramarginal gyrus, and the fusiform gyrus (Figure 5A, model explanation). For bvFTD, atypical connectivity was restricted mainly to areas in the frontal and temporal lobes (Figure 5B, topographical information). The areas identified included the precentral gyrus, the superior and middle frontal gyri, the Heschl gyrus, and the supramarginal gyrus (Figure 5A, model explanation).
The analyses above were repeated in a matched subsample with an equal number of subjects to ensure that the results were not driven by the differences in sample size or demographics (Table C9, Appendix C). The results (Table C10 and C11, Appendix C) confirmed the classifications obtained with the whole sample.
4. DISCUSSION
An automatic pipeline supporting multi‐centric studies of EEG source space connectomics was implemented. The pipeline incorporates standards for data storage and critical harmonization stages, including integrating different electrode layouts and rescaling algorithms for patient–control normalizations. Results show that joint analyses of connectivity allow for assessing complex connectivity patterns not evident in traditional single metric approaches, thereby improving the characterization of dementia subtypes. The automatic processing step sequence facilitates the analysis of large data volumes with minimal supervision. The pipeline's strengths rely on storage, transparency, data set reusability, and reproducibility of the results.
4.1. Harmonization steps
The four essential elements of data harmonization are discussed below. The first relates to adopting BIDS standards for storage and data exchange. 17 The second aspect is incorporating uniform criteria for EEG re‐referencing, which is achieved by using the AVE reference and the infinite reference estimated with REST. These two reference choices provide consistent results in EEG connectivity studies and outperform single electrode (Cz) and linked mastoid references. 18 , 19 The third element is the integration of different electrode layouts, which had been addressed previously using assignment/replacement procedures based on the closest equivalent electrodes and minimum electrode distance. 20 , 42 In this study, electrode layouts are integrated by generating virtual sensors computed from topographic interpolation transforms. The high correlation between the EEG topographies constructed from the spherically splined EEG field map and that resulting from the original signals reflect the efficacy of this method of spatial harmonization (Figure 2A).
The fourth aspect, the patient–control normalization using the Z‐score transform, has been applied successfully in multi‐centric studies on neurodegeneration 15 , 16 and has been crucial to developing quantitative EEG norms 8 that increase the diagnostic accuracy of brain dysfunction. In addition, the Z‐score transformation results in FC matrices with higher bilateral symmetry than that obtained with unharmonized EEG (Figure 4). This method promotes the emergence of interhemispheric connections and long‐range connections within a particular hemisphere while reducing short‐range interactions (Figure 4). The latter is particularly relevant because short‐range connections can be interpreted as spurious interactions that result from head volume conduction effects on the estimation of FC. 43
4.2. A multi‐metric analysis of FC for dementia classification
EEG source space connectivity analyses provide topographic representations of brain functional interactions 14 with increased test–retest reliability, compared to sensor‐space estimates. 44 FC analyses provide better classification performance of dementia in comparison with spectral descriptors. 45 , 46 Likewise, joint analyses of connectivity result in a more robust classification of dementia than single metric approaches. 45 , 47 The heterogeneous set of features underlying AD and bvFTD classifications (Figure 5A) confirms the relevance of this integrative approach and the utility of joint analyses of EEG spectral measures and FC descriptors. 9 , 46 , 47
The atypical connectivity observed in AD supports the hypothesis that AD is a disconnection syndrome, mainly affecting posterior brain regions. 9 , 48 Nodes of the default mode network (DMN) are significantly affected by this condition. 49 , 50 In bvFTD, the anterior cingulate and the insula (hubs of the salience network), 51 the dorsolateral prefrontal cortex, and the orbitofrontal cortex are critically impaired. 50 , 52 Other frontotemporal regions with atypical connectivity have been associated with diverse symptoms in bvFTD. 52
It is noteworthy that the multi‐metric approach to dementia implemented in this study is boosted by machine learning algorithms. Critical elements are recursive feature reduction (from thousands to dozen) to prevent overfitting, and Bayesian optimization for tuning model hyper‐parameters on datasets consisting of several features. As a result, the performance of dementia classifications based on this multi‐metric and multi centric framework 16 , 46 approaches those obtained using advanced biomarkers.
4.3. Limitations and future directions
The main limitation of this study resides in the lack of steps to control the effects of demographic covariates (age, sex, and ages of formal education) 8 , 53 on dementia classification (but see the confirmation results using matched samples in Appendix C). Future pipeline implementations will allow for the inclusion of demographics in the classification, as is done elsewhere. 16 , 35 , 36 Likewise, future studies must address statistical approaches to validate the best pipelines created by the synergy of available choices (options for spatial normalization, EEG source reconstruction, connectivity metrics, and classification algorithms). Although this study focused on integrating three time‐domain connectivity metrics, network topologies across EEG frequency bands and dementia classifications based on integrating frequency‐domain connectivity metrics have been provided. 34 Nevertheless, the accuracy of classifications based on integrating frequency‐ and time‐domain metrics needs to be explored. Furthermore, although joint connectivity analyses benefit dementia classification even without controlling for metric crosswalk, 34 a better classification accuracy may be achieved by limiting the analysis to uncorrelated metrics. 54
To confirm if the advantages of spatial normalization mentioned in this research are relevant to electrode arrangements offered by other suppliers, additional testing is required. Likewise, EEG inverse solution methods that reduce distortions in connectivity due to the leakage effect 55 and the use of individual head models based on electrode digitalization and anatomic MRI information should be further considered. Future research should include additional validation by comparing normalized versus non‐normalized data and resting‐state fMRI. In addition, the benefits of using simultaneous recording of EEG and functional near‐infrared spectroscopy could be explored. Furthermore, conceptualizing short‐range connections as potentially spurious is a complex issue requiring further investigation, as well as the enhanced bilateral symmetry and long‐range connectivity that resulted from spatial normalization. To ensure accuracy and validity, it is important to gather diverse and sizable samples from both mainstream and underrepresented populations. 15 , 16 It is also essential to test the reliability of connectivity analyses through repeated testing. 56 Doing so will help address geographic and socioeconomic variability and validate classification strategies with worldwide impact.
5. CONCLUSIONS
We present a robust workflow for harmonizing multi‐metric analysis of source space EEG connectivity and developing composite metrics of FC for the classification of dementia subtypes. Findings emphasize the importance of spatial and patient–control normalizations to control cross‐site variance. In addition, the study highlights the relevance of joint analyses of information‐theoretic metrics of EEG connectivity to accurately classify dementias. This multi‐metric approach may allow for a more comprehensive characterization of functional brain interactions in neurodegeneration, supporting the development of neuroimaging biomarkers of dementia based on scalable and cost‐effective technologies.
CONFLICT OF INTEREST STATEMENT
Agustín Ibáñez is partially supported by grants of Takeda CW2680521; FONCYT PICT (2017‐1818, 2017‐1820); ANID/FONDECYT Regular (1210195, 1210176, 1220995); ANID/FONDAP (15150012); FONDEF ID20I10152, ID22I10029; ANID/PIA/ANILLOS ACT210096; and the Multi‐Partner Consortium to Expand Dementia Research in Latin America (ReDLat), funded by the National Institutes of Aging of the National Institutes of Health under award number R01AG057234, an Alzheimer's Association grant (SG‐20‐725707‐ReDLat), the Rainwater Foundation, and the Global Brain Health Institute (GBHI). Pavel Prado, Jhony A. Mejía, Agustín Sainz‐Ballesteros, Agustina Birba, Sebastian Moguilner, Rubén Herzog, Mónica Otero, Jhosmary Cuadros, Lucía Zepeda, Daniel Franco‐O'Byrne, and Mario Parra have nothing to disclose. The content of the article is solely the responsibility of the authors and does not represent the official views of these institutions. The authors thank the ReDLat participants and their families for their commitment to our study. Author disclosures are available in the supporting information.
CONSENT STATEMENT
Before enrolling, a signed informed consent was provided by all the participants following the Declaration of Helsinki.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
The authors thank the ReDLat participants and their families for their valuable time and commitment to our study. AI is supported by Takeda Grant CW2680521; CONICET; FONCYT‐PICT (2017‐1818, 2017‐1820); ANID/FONDECYT Regular (1210195, 1210176, 1220995); ANID/FONDAP (15150012); ANID/PIA/ANILLOS ACT210096; ANID/FONDEF ID20I10152, ID22I10029; and the Multi‐Partner Consortium to Expand Dementia Research in Latin America (ReDLat), funded by the National Institutes of Aging of the National Institutes of Health under award number R01AG057234, an Alzheimer's Association grant (SG‐20‐725707‐ReDLat), the Rainwater Foundation, and the Global Brain Health Institute. SF is an Atlantic Fellow for Equity in Brain Health at the Global Brain Health Institute (GBHI) and is supported with funding from GBHI, BrainLat, ANID/FONDEF ID22I10029, and CONICET. MO is supported by ANID/FONDECYT Postdoctorado 3210508. The content is solely the responsibility of the authors and does not represent the official views of these institutions.
Open access funding provided by IReL.
Prado P, Mejía JA, Sainz‐Ballesteros A, et al. Harmonized multi‐metric and multi‐centric assessment of EEG source space connectivity for dementia characterization. Alzheimer's Dement. 2023;15:e12455. 10.1002/dad2.12455
REFERENCES
- 1. LAC‐CD . Parra MA, Garcia AM, Ibanez A. Addressing dementia challenges through international networks: evidence from the Latin American and Caribbean Consortium on Dementia (LAC‐CD). Alzheimers Dement. 2021;17(S8):e055106. doi: 10.1002/alz.055106 [DOI] [PubMed] [Google Scholar]
- 2. Ibanez A, Yokoyama JS, Possin KL, et al. The multi‐partner consortium to expand dementia research in Latin America (ReDLat): driving multicentric research and implementation science. Front Neurol. 2021;12:631722. doi: 10.3389/fneur.2021.631722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babiloni C, Arakaki X, Azami H, et al. Measures of resting state EEG rhythms for clinical trials in Alzheimer's disease: recommendations of an expert panel. Alzheimers Dement. 2021;17(9):1528‐1553. doi: 10.1002/alz.12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goriely A, Kuhl E, Bick C. Neuronal oscillations on evolving networks: dynamics, damage, degradation, decline, dementia, and death. Phys Rev Lett. 2020;125(12):128102. doi: 10.1103/PhysRevLett.125.128102 [DOI] [PubMed] [Google Scholar]
- 5. Bhattarai A, Chen Z, Chua P, et al. Network diffusion model predicts neurodegeneration in limb‐onset amyotrophic lateral sclerosis. PLoS One. 2022;17(8):e0272736. doi: 10.1371/journal.pone.0272736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large‐scale human brain networks. Neuron. 2009;62(1):42‐52. doi: 10.1016/j.neuron.2009.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pavlov YG, Adamian N, Appelhoff S, et al. Investigating the replicability of influential EEG experiments. Cortex. 2021;144:213‐229. doi: 10.1016/j.cortex.2021.03.013 [DOI] [PubMed] [Google Scholar]
- 8. Li M, Wang Y, Lopez‐Naranjo C, et al. Harmonized‐Multinational qEEG norms (HarMNqEEG). Neuroimage. 2022;256:119190. doi: 10.1016/j.neuroimage.2022.119190 [DOI] [PubMed] [Google Scholar]
- 9. Briels CT, Schoonhoven DN, Stam CJ, De Waal H, Scheltens P, Gouw AA. Reproducibility of EEG functional connectivity in Alzheimer's disease. Alz Res Therapy. 2020;12(1):68. doi: 10.1186/s13195-020-00632-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohanty R, Sethares WA, Nair VA, Prabhakaran V. Rethinking measures of functional connectivity via feature extraction. Sci Rep. 2020;10(1):1298. doi: 10.1038/s41598-020-57915-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prado P, Birba A, Cruzat J, et al. Dementia ConnEEGtome: towards multicentric harmonization of EEG connectivity in neurodegeneration. Int J Psychophysiol. 2022;172:24‐38. doi: 10.1016/j.ijpsycho.2021.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Babiloni C, Del Percio C, Lizio R, et al. Abnormalities of resting‐state functional cortical connectivity in patients with dementia due to Alzheimer's and Lewy body diseases: an EEG study. Neurobiol Aging. 2018;65:18‐40. doi: 10.1016/j.neurobiolaging.2017.12.023 [DOI] [PubMed] [Google Scholar]
- 13. San‐Martin R, Fraga FJ, Del Percio C, et al. Classification of patients with Alzheimer's disease and dementia with Lewy bodies using resting EEG selected features at sensor and source levels: a proof‐of‐concept study. CAR. 2021;18(12):956‐969. doi: 10.2174/1567205018666211027143944 [DOI] [PubMed] [Google Scholar]
- 14. Thompson PM, Jahanshad N, Schmaal L, et al. The enhancing neuroimaging genetics through meta‐analysis consortium: 10 years of global collaborations in human brain mapping. Hum Brain Mapp. 2022;43(1):15‐22. doi: 10.1002/hbm.25672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birba A, Santamaría‐García H, Prado P, et al. Allostatic‐Interoceptive overload in frontotemporal dementia. Biol Psychiatry. 2022;92(1):54‐67. doi: 10.1016/j.biopsych.2022.02.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moguilner S, Birba A, Fittipaldi S, et al. Multi‐feature computational framework for combined signatures of dementia in underrepresented settings. J Neural Eng. 2022;19(4):046048. doi: 10.1088/1741-2552/ac87d0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pernet CR, Appelhoff S, Gorgolewski KJ, et al. EEG‐BIDS, an extension to the brain imaging data structure for electroencephalography. Sci Data. 2019;6(1):103. doi: 10.1038/s41597-019-0104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dong L, Li F, Liu Q, et al. MATLAB toolboxes for Reference Electrode Standardization Technique (REST) of Scalp EEG. Front Neurosci. 2017;11:601. doi: 10.3389/fnins.2017.00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu S, Lai Y, Valdes‐Sosa PA, Bringas‐Vega ML, Yao D. How do reference montage and electrodes setup affect the measured scalp EEG potentials? J Neural Eng. 2018;15(2):026013. doi: 10.1088/1741-2552/aaa13f [DOI] [PubMed] [Google Scholar]
- 20. Bigdely‐Shamlo N, Touryan J, Ojeda A, Kothe C, Mullen T, Robbins K. Automated EEG mega‐analysis II: cognitive aspects of event related features. Neuroimage. 2020;207:116054. doi: 10.1016/j.neuroimage.2019.116054 [DOI] [PubMed] [Google Scholar]
- 21. Ballesteros AS, Prado P, Ibanez A, Perez JAM, Moguilner S, A pipeline for large‐scale assessments of dementia EEG connectivity across multicentric settings. OFS Preprints, 22 Feb. 2023. doi: 10.31219/osf.io/h2wgv [DOI]
- 22. Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single‐trial EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134(1):9‐21. doi: 10.1016/j.jneumeth.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 23. Pion‐Tonachini L, Kreutz‐Delgado K, Makeig S. ICLabel: an automated electroencephalographic independent component classifier, dataset, and website. Neuroimage. 2019;198:181‐197. doi: 10.1016/j.neuroimage.2019.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bigdely‐Shamlo N, Kreutz‐Delgado K, Kothe C, Makeig S, EyeCatch: data‐mining over half a million EEG independent components to construct a fully‐automated eye‐component detector. 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2013:5845‐5848. doi: 10.1109/EMBC.2013.6610881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleifges K, Bigdely‐Shamlo N, Kerick SE, Robbins KA. BLINKER: automated extraction of ocular indices from EEG enabling large‐scale analysis. Front Neurosci. 2017;11:12. doi: 10.3389/fnins.2017.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kothe CA, Makeig S. BCILAB: a platform for brain–computer interface development. J Neural Eng. 2013;10(5):056014. doi: 10.1088/1741-2560/10/5/056014 [DOI] [PubMed] [Google Scholar]
- 27. Melnik A, Legkov P, Izdebski K, et al. Systems, subjects, sessions: to what extent do these factors influence EEG Data? Front Hum Neurosci. 2017;11:150. doi: 10.3389/fnhum.2017.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pascual‐Marqui RD, Lehmann D, Koukkou M, et al. Assessing interactions in the brain with exact low‐resolution electromagnetic tomography. Phil Trans R Soc A. 2011;369(1952):3768‐3784. doi: 10.1098/rsta.2011.0081 [DOI] [PubMed] [Google Scholar]
- 29. Trujillo‐Barreto NJ, Aubert‐Vázquez E, Valdés‐Sosa PA. Bayesian model averaging in EEG/MEG imaging. Neuroimage. 2004;21(4):1300‐1319. doi: 10.1016/j.neuroimage.2003.11.008 [DOI] [PubMed] [Google Scholar]
- 30. Hämäläinen MS, Ilmoniemi RJ. Interpreting magnetic fields of the brain: minimum norm estimates. Med Biol Eng Comput. 1994;32(1):35‐42. doi: 10.1007/BF02512476 [DOI] [PubMed] [Google Scholar]
- 31. Rolls ET, Joliot M, Tzourio‐Mazoyer N. Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage. 2015;122:1‐5. doi: 10.1016/j.neuroimage.2015.07.075 [DOI] [PubMed] [Google Scholar]
- 32. Herzog R, Rosas FE, Whelan R, et al. Genuine high‐order interactions in brain networks and neurodegeneration. Neurobiol Dis. 2022;175:105918. doi: 10.1016/j.nbd.2022.105918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cruzat J, Herzog R, Prado P, et al. Temporal irreversibility of large‐scale brain dynamics in Alzheimer's disease. J Neurosci. 2023;43(9):1643‐1656. doi: 10.1523/JNEUROSCI.1312-22.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prado P, Moguilner S, Mejía JA, et al. Source space connectomics of neurodegeneration: one‐metric approach does not fit all. Neurobiol Dis. 2023;179:106047. doi: 10.1016/j.nbd.2023.106047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santamaría‐García H, Baez S, Aponte‐Canencio DM, et al. Uncovering social‐contextual and individual mental health factors associated with violence via computational inference. Patterns. 2021;2(2):100176. doi: 10.1016/j.patter.2020.100176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maito MA, Santamaría‐García H, Moguilner S, et al. Classification of Alzheimer's disease and frontotemporal dementia using routine clinical and cognitive measures across multicentric underrepresented samples: a cross sectional observational study. Lancet Reg Health Am. 2023;17:100387. doi: 10.1016/j.lana.2022.100387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manly BFJ. Randomization, Bootstrap, and Monte Carlo Methods in Biology. 3rd ed. Chapman & Hall/CRC; 2007. [Google Scholar]
- 38. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289‐300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 39. Kassraian‐Fard P, Matthis C, Balsters JH, Maathuis MH, Wenderoth N. Promises, pitfalls, and basic guidelines for applying machine learning classifiers to psychiatric imaging data, with autism as an example. Front Psychiatry. 2016;7:177. doi: 10.3389/fpsyt.2016.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller AC, Guido S. Introduction to Machine Learning with Python: A Guide for Data Scientists. 1st ed. O'Reilly media, INC. 2016. [Google Scholar]
- 41. Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. Neuroimage. 2009;45(1):S199‐S209. doi: 10.1016/j.neuroimage.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farzan F, Atluri S, Frehlich M, et al. Standardization of electroencephalography for multi‐site, multi‐platform and multi‐investigator studies: insights from the Canadian biomarker integration network in depression. Sci Rep. 2017;7(1):7473. doi: 10.1038/s41598-017-07613-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colclough GL, Smith SM, Nichols TE, et al. The heritability of multi‐modal connectivity in human brain activity. eLife. 2017;6:e20178. doi: 10.7554/eLife.20178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pourmotabbed H, Jongh Curry AL, Clarke DF, Tyler‐Kabara EC, Babajani‐Feremi A. Reproducibility of graph measures derived from resting‐state MEG functional connectivity metrics in sensor and source spaces. Hum Brain Mapp. 2022;43(4):1342‐1357. doi: 10.1002/hbm.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blinowska KJ, Rakowski F, Kaminski M, et al. Functional and effective brain connectivity for discrimination between Alzheimer's patients and healthy individuals: a study on resting state EEG rhythms. Clin Neurophysiol. 2017;128(4):667‐680. doi: 10.1016/j.clinph.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 46. Hughes LE, Henson RN, Pereda E. Biomagnetic biomarkers for dementia: a pilot multicentre study with a recommended methodological framework for magnetoencephalography. Alzheimers Dement. 2019;11(1):450‐462. doi: 10.1016/j.dadm.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alonso JF, Poza J, MÁ Mañanas, Romero S, Fernández A, Connectivity HorneroRMEG. Analysis in patients with Alzheimer's disease using cross mutual information and spectral coherence. Ann Biomed Eng. 2011;39(1):524‐536. doi: 10.1007/s10439-010-0155-7 [DOI] [PubMed] [Google Scholar]
- 48. Yu M, Gouw AA, Hillebrand A, et al. Different functional connectivity and network topology in behavioral variant of frontotemporal dementia and Alzheimer's disease: an EEG study. Neurobiol Aging. 2016;42:150‐162. doi: 10.1016/j.neurobiolaging.2016.03.018 [DOI] [PubMed] [Google Scholar]
- 49. Hafkemeijer A, Möller C, Dopper EGP, et al. Resting state functional connectivity differences between behavioral variant frontotemporal dementia and Alzheimer's disease. Front Hum Neurosci. 2015;9:474. doi: 10.3389/fnhum.2015.00474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J, Greicius MD, Gennatas ED, et al. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease. Brain. 2010;133(5):1352‐1367. doi: 10.1093/brain/awq075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seeley WW. The Salience Network: a Neural System for Perceiving and Responding to Homeostatic Demands. J Neurosci. 2019;39(50):9878‐9882. doi: 10.1523/JNEUROSCI.1138-17.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Connor CM, Landin‐Romero R, Clemson L, et al. Behavioral‐variant frontotemporal dementia: distinct phenotypes with unique functional profiles. Neurology. 2017;89(6):570‐577. doi: 10.1212/WNL.0000000000004215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parra MA. Barriers to Effective Memory Assessments for Alzheimer's Disease. J Alzheimers Dis. 2022;90(3):981‐988. doi: 10.3233/JAD-215445 [DOI] [PubMed] [Google Scholar]
- 54. Dauwels J, Vialatte F, Musha T, Cichocki A. A comparative study of synchrony measures for the early diagnosis of Alzheimer's disease based on EEG. Neuroimage. 2010;49(1):668‐693. doi: 10.1016/j.neuroimage.2009.06.056 [DOI] [PubMed] [Google Scholar]
- 55. Gonzalez‐Moreira E, Paz‐Linares D, Areces‐Gonzalez A, et al. Caulking the leakage effect in MEEG source connectivity analysis. Published online December 1, 2019. Accessed May 29, 2023. http://arxiv.org/abs/1810.00786
- 56. Rolle CE, Narayan M, Wu W, et al. Functional connectivity using high density EEG shows competitive reliability and agreement across test/retest sessions. J Neurosci Methods. 2022;367:109424. doi: 10.1016/j.jneumeth.2021.109424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information


