Abstract
Introduction
Several studies have emphasized the potential of artificial intelligence (AI) and its subfields, such as machine learning (ML), as emerging and feasible approaches to optimize patient care in oncology. As a result, clinicians and decision-makers are faced with a plethora of reviews regarding the state of the art of applications of AI for head and neck cancer (HNC) management. This article provides an analysis of systematic reviews on the current status, and of the limitations of the application of AI/ML as adjunctive decision-making tools in HNC management.
Methods
Electronic databases (PubMed, Medline via Ovid, Scopus, and Web of Science) were searched from inception until November 30, 2022. The study selection, searching and screening processes, inclusion, and exclusion criteria followed the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines. A risk of bias assessment was conducted using a tailored and modified version of the Assessment of Systematic Review (AMSTAR-2) tool and quality assessment using the Risk of Bias in Systematic Reviews (ROBIS) guidelines.
Results
Of the 137 search hits retrieved, 17 fulfilled the inclusion criteria. This analysis of systematic reviews revealed that the application of AI/ML as a decision aid in HNC management can be thematized as follows: (1) detection of precancerous and cancerous lesions within histopathologic slides; (2) prediction of the histopathologic nature of a given lesion from various sources of medical imaging; (3) prognostication; (4) extraction of pathological findings from imaging; and (5) different applications in radiation oncology. In addition, the challenges in implementation of AI/ML models for clinical evaluations include the lack of standardized methodological guidelines for the collection of clinical images, development of these models, reporting of their performance, external validation procedures, and regulatory frameworks.
Conclusion
At present, there is a paucity of evidence to suggest the adoption of these models in clinical practice due to the aforementioned limitations. Therefore, this manuscript highlights the need for development of standardized guidelines to facilitate the adoption and implementation of these models in the daily clinical practice. In addition, adequately powered, prospective, randomized controlled trials are urgently needed to further assess the potential of AI/ML models in real-world clinical settings for the management of HNC.
Keywords: Head and neck cancer, Artificial intelligence, Machine learning, Systematic review
Key Summary Points
| Several studies have emphasized the potentials of artificial intelligence/machine learning (AI/ML) for the improved management of head and neck cancer (HNC). |
| Researchers, clinicians, and healthcare decision-makers are faced with the challenge of summarizing these studies in HNC management |
| We analyzed all the systematic reviews relating to the application of AI/ML for the management of HNC. |
| The applications of AI/ML for head and neck oncology can be thematized into: (1) precancerous and cancerous lesions detection within histopathologic slides; (2) prediction of the histopathologic nature of a given lesion from various sources of medical imaging; (3) prognostication; (4) extraction of pathological findings from imaging; and (5) different applications in radiation oncology. |
| Standardized guidelines are warranted to facilitate the adoption and implementation of these models in everyday clinical practice. |
Introduction
Head and neck cancer (HNC) comprises a heterogenous group of cancers in terms of etiology, behavior, and outcome, with squamous cell carcinoma representing the most common histology [1]. In recent decades, there have been considerable advancements in the therapeutic repertoire for the management of HNC [2]. However, HNC mortality rates have not significantly improved [2], as the majority of these tumors are still diagnosed at an advanced stage, which reduces survival rate even after curative-intent treatment [3, 4]. Therefore, different methods and strategies have been explored for early detection of HNC to improve treatment outcome. In recent years, machine learning (ML) and deep learning (DL) techniques, which are subfields of artificial intelligence (AI), have shown promising results in various efforts of outcome prognostication in HNC due to their ability to learn complex relationships between datasets. The method of learning relationships is used to classify different patterns to more effectively predict treatment outcome [5, 6].
Several studies have utilized AI techniques on various forms of medical data, such as clinical, videoendoscopic, histologic, pathologic, genetic, radiologic, metabolic, or a combination of these, to improve clinical decision-making or to speed up novel drug discovery. In addition, recent technological advancements in computer science, availability of large medical imaging datasets, and improved ML/DL algorithms have further enhanced the potential for application of AI in oncology. As a result, several promising studies emphasizing the diagnostic and prognostic potentials of AI models as an assistant decision-making tool have been reported during the last decade [7–9]. Subsequently, clinicians and decision-makers are now faced with a plethora of reviews summarizing the evidence for the application of AI in HNC management.
This article aims to address the research question: what is the current status and what are the limitations of the application of AI platforms as adjunctive decision-making tools in HNC management? Several articles have been published emphasizing the promising potential of AI (ML/DL) models as an ancillary tool for HNC management. As a result, several reviews have been published to summarize these articles. However, these reviews significantly vary in quality and scope. Thus, a systematic analysis of these reviews is essential to appraise, summarize, present, compare, and contrast separate contributions in a single study [10]. Here, we systematically examined all the existing systematic review articles regarding the application of AI in HNC management.
Methods
Search of Databases and Study Period
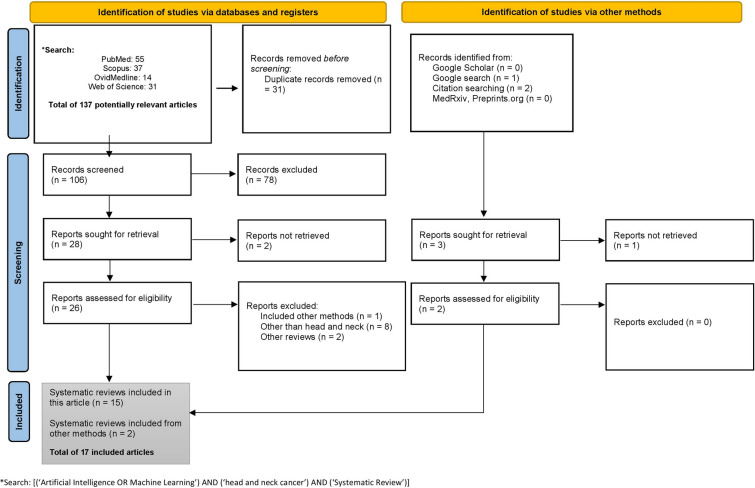
Medline via Ovid, PubMed, Scopus, and Web of Science databases were systematically searched from inception until 30 November, 2022, to retrieve all systematic review articles that examined the application of AI or ML in HNC (Fig. 1). To reduce research waste and to maximize grey literature, Google Scholar was searched for potentially relevant systematic reviews. Research Ethics Committee approval was not needed for this systematic literature search.
Fig. 1.
The PRISMA flowchart
Search Terms
The potentially relevant articles were retrieved by combining search keywords: [(‘Artificial Intelligence OR Machine Learning’) AND (‘head and neck cancer’) AND (‘Systematic Review’)].
Search Analysis
All the retrieved potentially relevant articles were exported to Endnote for further analysis. The hits were analyzed for possible duplicates and irrelevant studies. The inclusion and exclusion criteria were defined based on the study-specific research questions.
Inclusion Criteria
All studies that had systematically reviewed articles that examined the application of AI or its subfields in HNC. To minimize inadvertent omissions, the reference lists of all the potentially systematic reviews were manually searched to ensure that all the relevant systematic reviews were adequately included. The potential reviews were further analyzed based on the PICO model (Population, Intervention, Comparison, and Outcome) prior to inclusion in this review (Table 1).
Table 1.
Inclusion and exclusion using modified PICO model
| Selection criteria | Inclusion criteria | Exclusion criteria |
|---|---|---|
| P: population | Review or systematic review that examined the application of deep learning (DL) or its subfields such as machine learning in head and neck cancer (HNC) | Reviews that examine other subsites other than HNC. Reviews in other languages than English |
| I: intervention | Artificial intelligence and its subfield such as deep learning (DL) or machine learning (ML) | Traditional statistical method |
| C: comparison | Reviews that compared several articles using PRISMA guidelines | Non-systematic reviews (i.e. narrative reviews) |
| O: outcome | Examined the application of DL or ML for either diagnosis, prognosis, or both | General overview, editorial, narrative studies, and comments |
Exclusion Criteria
All studies that reviewed the application of AI or its subfields in any of the subsites of HNC were excluded. Comments, opinions, perspectives, guidelines, editorials, articles other than systematic reviews, and papers in languages other than English were excluded.
Search Reporting and Screening
Two independent researchers performed the screening of potentially relevant articles. The screening was done in two phases. In the first phase, the review titles and abstracts were examined in relation to the research objective of this study. In the second phase, a comprehensive full-text assessment of the potential reviews identified in the first phase was further analyzed. A data extraction sheet was used to minimize the omission of possible eligible studies. The same two independent researchers discussed to resolve possible discrepancies. The inter-observer reliability between these researchers was measured using Kappa Cohen’s coefficient (). All eligible studies to be included are summarized in Table 2. The entire process of literature search, screening, inclusion and exclusion, and reporting of the potentially relevant studies followed the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) (Fig. 1).
Table 2.
Extracts of the main findings from the included studies
| Study, Year, Country (Cancer type/subtypes) |
Review objective | Number of databases searched | Number of studies included | Conclusion |
|---|---|---|---|---|
|
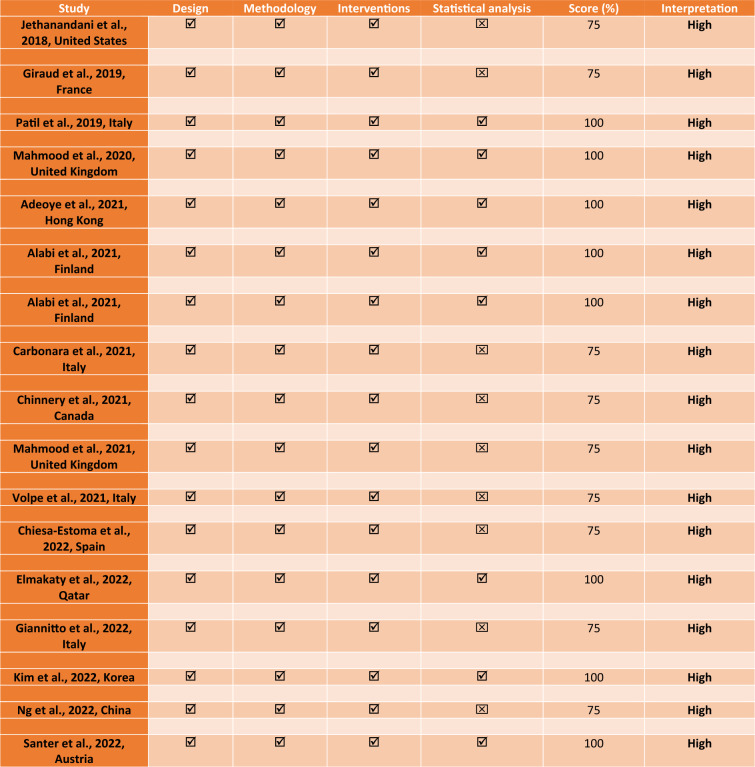
Jethanandani et al., 2018, United States (HNC, Radiomics) |
Application of radiomics in MRI of HNC | 5 | 16 | MRI radiomic applications demonstrate predictive potential in analyzing diverse HNC outcomes. However, methodological variances affect the accurate interpretation of results |
|
Giraud et al., 2019, France (HNC, Radiomics) |
Review the application of radiomics and ML in HNC | 1 | 8 | Radiomics can be explored by ML for decision making in oncology |
|
Patil et al., 2019, Italy (HNC, Genomic) |
Access the application of ML on genomic data in HNC | 4 | 7 | ML techniques can play a significant role in the prognostic prediction of HNC |
|
Mahmood et al., 2020, United Kingdom (Oral, WSI) |
Detection and grading of potentially malignant (pre-cancerous) and cancerous head and neck lesions using whole slide images (WSI) | 3 | 11 | AI has the potential as a diagnostic aid for some oral potentially malignant and malignant lesions |
|
Adeoye et al., 2021, Hong Kong (Oral cavity, Tabular data) |
To determine the current status of the application of these learning models as adjunctive decision-making tools in oral cavity cancer management | 8 | 27 | Machine learning algorithms have a satisfactory to excellent accuracy for predicting three of four oral cavity cancer outcomes i.e., malignant transformation, nodal metastasis, and prognosis |
|
Alabi et al., 2021, Finland (OSCC, Tabular data) |
A review of diagnostic and prognostic application of machine learning in oral squamous cell carcinoma (OSCC) and highlights some of the limitations and concerns of clinicians towards the implementation of machine learning-based models for daily clinical practice | 5 | 41 | The main limitations and concerns can be grouped as either the challenges inherent to the science of machine learning or relating to the clinical implementations |
|
Alabi et al., 2021, Finland (OSCC, Imaging modality) |
The application of deep learning for OSCC | 5 | 34 | Deep learning has great potential in the prognostication of OSCC |
|
Carbonara et al., 2021, Italy (HNC, Radiomics [radiation oncology]) |
Examined the potential of radiomics combined with ML methods in the prediction and assessment of radiation-induced toxicities | 2 | 8 | Studies showed promising results, but these require further validation to improve decision-making processes in preventing and managing radiation-induced toxicities |
|
Chinnery et al., 2021, Canada (HNC) |
To summarize the growing trend in the field of radiomics for AI | - | 24 | Predictive models for oncologic outcomes, treatment toxicity, and pathological findings have been reported to aid clinical decision making |
|
Mahmood et al., 2021, United Kingdom (HNC, Imaging modalities) |
The application and diagnostic accuracy of AI methods for detection and grading of potentially malignant (pre-cancerous) and cancerous head and neck lesions using WSI | 3 | 11 | The quality of evidence suggesting the use of AI for detection and grading of potentially malignant and cancerous HNC is low |
|
Volpe et al., 2021, Italy (HNC, Radiation oncology) |
To illustrate the potential and limitations of ML in solving everyday clinical issues in HNC and RT | 3 | 48 | ML is poised to revolutionize head and neck radiation oncology |
|
Chiesa-Estomba et al., 2022, Spain (OCSCC) |
To explore the application of ML for the management of OCSCC | 4 | 8 | ML will improve and democratize the application of algorithms to improve the prediction of cancer prognosis and its management |
|
Elmakaty et al., 2022, Qatar (OSCC) |
Meta-analysis to explore the potential of AI in detecting OSCC | 6 | 16 | AI can assist in the detection of OSCC |
|
Giannitto et al., 2022, Italy (HNC) |
ML to detect lymph node metastases in HNC | 3 | 7 | ML can assist in the detection of lymph node metastases in HNC |
|
Kim et al., 2022, Korea (Oral cancerous lesions) |
Evaluation of the potential of AI for discriminating oral cancerous lesions from normal mucosa | 6 | 14 | AI has the potential to be used as a clinical tool for the early diagnosis of pathological lesions |
|
Ng et al., 2022, China (NPC) |
To explore the unique application and implementation direction of AI in the management of NPC | 3 | 60 | AI has been used in auto-contouring, diagnosis, prognosis, and miscellaneous planning such as radiotherapy planning. Therefore, AI is poised to contribute to the routine management of NPC |
|
Santer et al., 2022, Austria (HNSCC) |
Review of studies that specifically explored the role of AI to classify lymph nodes in locally advanced HNSCC | 3 | 13 | AI showed a promising potential as a diagnostic support tool for lymph node classification in HNSCC |
MRI magnetic resonance imaging, HNC head and neck cancer, HNSCC head and neck squamous cell carcinoma, NPC nasopharyngeal cancer, OCSCC oral cavity squamous cell carcinoma, OSCC oral squamous cell carcinoma, WSI whole slide image
Data Extraction
For each eligible systematic review, the first author’s name, year of publication, country, area of application of the review, review objectives, number of databases searched, number of included studies, and conclusion from the systematic review were reported (Table 2). Based on the conclusion from the included reviews, the various applications of AI were summarized. The limitations mentioned in these reviews were noted. This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors. Therefore, a research ethics board approval was not applicable.
Quality Appraisal
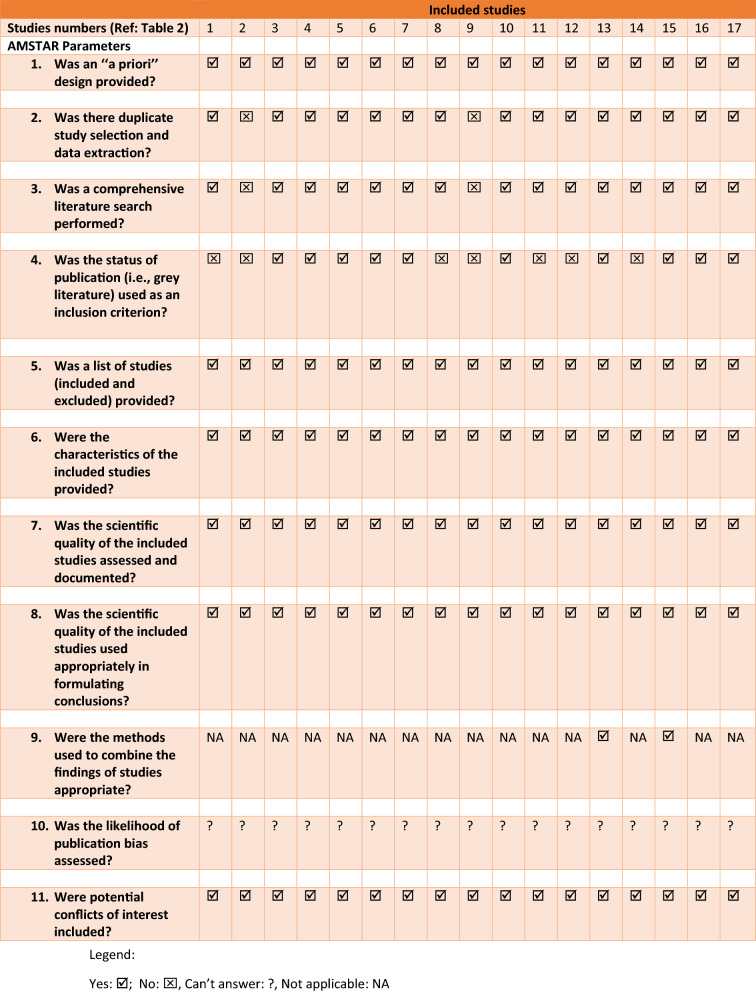
The quality appraisal of the included systematic reviews was done using two different quality assessment tools: a modified version of the National Institute of Health Quality Assessment tools and the Assessment of Multiple Systematic Reviews (AMSTAR-2) tool. Similarly, the risk of bias of the included studies was analyzed using the Risk of Bias in Systematic Reviews (ROBIS) tool (Sect. 2.9). Following the extraction using the PRISMA guideline, a preliminary assessment of the quality of the included studies was done using a modified version of the National Institute of Health Quality Assessment tools [11]. The modification was warranted considering the nature of this study as a review of systematic reviews. The modification includes design (systematic review), methodology (electronic databases were systematically searched), interventions (AI and its subfields were applied), and statistical analysis (summary of the performance metrics and conclusion from the included studies) (Table 3) [6]. For each criterion, a corresponding score was assigned (. Studies that met the minimum quality threshold were subjected to the main quality assessment using the revised version of the AMSTAR-2 tool (Table 4) [12].
Table 3.
The quality appraisal of the included systematic reviews
Modification includes design (review or systematic review), methodology (electronic databases were systematically searched), interventions (AI and/or its subfield were applied), and statistical analysis (conclusion from the included studies)
Table 4.
Assessment of the quality of the included studies using modified AMSTAR tool
Risk of Bias Analysis
Assessing the risk of bias of the included systematic reviews ensures that their quality is reliable. The risk of bias of the included reviews was assessed using the ROBIS tool. The details of the bias analysis and the corresponding results from each examined bias are given in Table 5.
Table 5.
Presentation of the ROBIS results
| Reviews | Phase 2 | Phase 3 | |||
|---|---|---|---|---|---|
| 1. Study eligibility criteria | 2. Identification and selection of studies | 3. Data collection and study appraisal | 4. Synthesis and findings | Risk of bias in the review | |
| #1 | |||||
| #2 | NA | ||||
| #3 | |||||
| #4 | |||||
| #5 | |||||
| #6 | |||||
| #7 | |||||
| #8 | |||||
| #9 | NA | ||||
| #10 | |||||
| #11 | |||||
| #12 | |||||
| #13 | |||||
| #14 | |||||
| #15 | |||||
| #16 | |||||
| #17 | |||||
Low risk = , high risk = , unclear = NA
Results
Results of the Database Search
A total of 137 hits were retrieved. After deleting duplicates (n = 31), and irrelevant papers (n = 81), we found 17 studies eligible to be included in this review as shown in Fig. 1 [1, 5, 7, 13–26].
Characteristics of Relevant Studies
All the articles included in this review were published in English. Of the 17 included systematic reviews [1, 5, 7, 13–26], 11 were conducted in Europe [1, 5, 7, 14, 15, 17–19, 21, 22, 24] while 4 were conducted in Asia [16, 23, 25, 26] and 2 in the United States [13, 20] (Table 2). All but one of the included systematic reviews showed high-quality appraisal and low risk of bias (Tables 3, 4). Seven of the systematic reviews were conducted in the year 2021 [1, 5, 16–20], 6 in 2022 [21–26], and the remaining 4 before the year 2021 [7, 13–15].
Current Status of AI in HNC Oncology
The findings of the published systematic reviews (Table 1) suggest that the application of AI and its subfields in HNC can be summarized in 5 distinct fundamental themes: (1) detection of precancerous and cancerous lesions in histopathologic slides [7, 18]; (2) prediction of histopathologic nature of a given lesion from imaging [1, 5, 13–15, 17, 21, 22, 24–26]; (3) prognostication [5, 17, 18, 20, 21, 23]; (4) extraction of pathological findings from imaging [15, 16, 20]; and (5) applications in radiation oncology [15, 17, 19].
Theme 1: Detection of Precancerous and Cancerous Lesions in Histopathologic Slides
The included studies produced ML models with an average accuracy ranging between 79 and 100% for the detection and grading of potentially malignant (precancerous) and cancerous head and neck lesions using whole-slide images (WSI) of human tissue slides [7]. The average dataset used ranged between 40 and 270 unicentric WSI. Thus, with this promising accuracy, ML models are poised to act as a diagnostic aid for detection and grading of oral potentially malignant and malignant lesions [7, 18], especially as ML accuracy can improve as more datasets are utilized.
Theme 2: Prediction of Histopathologic Nature of a Given Lesion from Imaging
ML models act as a diagnostic aid for the HNC detection using a range of imaging modalities such as histologic WSI of hematoxylin and eosin (H&E)-stained tissue sections (as detailed above), radiologic data (MRI, CT, PET/CT, and plain film intraoral radiographs), hyperspectral imaging (HSI), videoendoscopic/clinical examinations, and multimodal optical imaging. For instance, the application of ML models for predicting the histopathologic nature of a given lesion from endoscopic or radiologic images includes the detection of oral squamous cell carcinoma (OSCC) with an average sensitivity of 92% and specificity of 91.9% [25]. Similarly, AI models have shown an average sensitivity of 90.4% and specificity of 88.4% in discriminating between oral precancerous and cancerous lesions from normal mucosa by means of clinical pictures [26]. Furthermore, radiomics-based ML has been employed to identify occult involvement of cervical lymph nodes in HNSCC [21, 22, 24] and to aid in the assessment and evaluation of/or differentiation between oral potentially malignant disorders and OSCC [1, 7]
This approach has been reported to be useful for region of interest (ROI) segmentation methods, image pre-processing, and feature extraction [13, 17]. ML models have been reported to be used for the classification of the HPV status of oropharyngeal SCC and the identification of nasopharyngeal SCC [1, 15]. Moreover, they have also been used to detect oral, nasopharyngeal, oropharyngeal, and laryngeal cancers using videoendoscopic/clinical images. HSI has been used by AI/ML models for early detection and diagnosis of OSCC, differentiation between normal and cancerous tongue tissue, and multispectral wide-field optical imaging to distinguish between oral cancer/precancer and non-neoplastic mucosa [1].
Theme 3: Prognostication
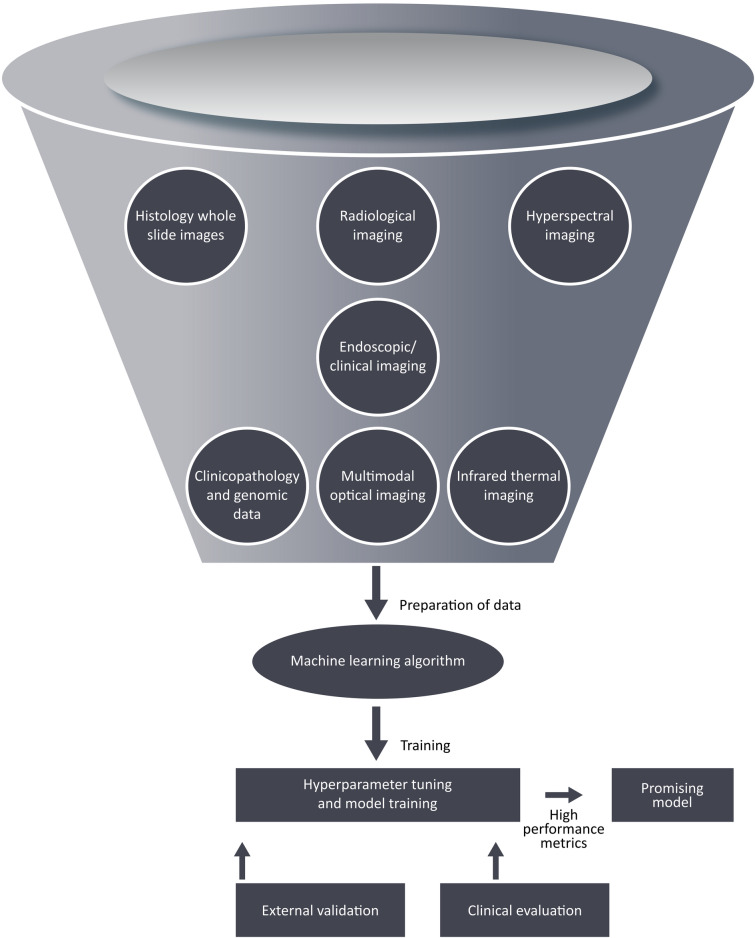
AI techniques have been utilized to explore vital information contained in clinicopathologic and genomic data to aid in cancer management (Fig. 2). For genomic data, ML models have been used for prognostic prediction by identifying and classifying patterns for the discovery of new biomarkers, drug targets, and a better identification of critical cancer genes in HNC management [14]. In recent years, radiomics-based ML approaches have been used for predicting oncologic outcomes based on tumor characteristics associated with overall survival in multiple cohorts of patients with HNC [20]. ML models have been generated and used to predict other oncologic outcomes such as progression-free survival, local–regional relapse, and occurrence of distant metastases [17, 20, 23]. These models, which aid the prediction of survival, provide a step closer to achieve personalized risk-based treatment selection, which may be used to escalate or de-escalate treatment intensity in a patient-tailored fashion [5, 18]. Furthermore, HNC patients may be stratified into risk groups for effective treatment planning [17, 20, 21]. It should be emphasized that the suggested escalating and de-escalating treatment regimens should be comprehensively investigated in clinical trials before incorporating them in therapeutic guidelines and protocols.
Fig. 2.
Workflow of ML model development for outcome prediction
Theme 4: Pathological Findings Based on Imaging
AI/ML models can be used to guide clinical decision-making through the analysis of pathological findings, such as the number, location, and size of lymph node involvement, malignant transformation of precancerous lesions, evaluation of lympho-vascular invasion, depth of tumor invasion, perineural invasion, and presence of extra-nodal extension [15, 16, 20], based on imaging alone.
Theme 5: Applications for Radiation Oncology
A rRadiomics-based ML approach can assist radiotherapy treatment planning by automation of organs at risk delineation, determining the probability of complications to normal tissues, and predicting of radiation-induced toxicities to guide and facilitate adaptive radiotherapy [15, 17, 19].
Limitations of AI Studies in the Field of Head and Neck Oncology
The observed limitations in the studies include the lack of standardized data collection [10], methodological variations in AI model development and generation [10, 13], low quality of evidence on model performance [13], lack of adequate validation [12, 13, 17], and lack of regulatory framework [16]. Furthermore, the methodological differences in terms of the acquisition of clinical images have prohibited proper evaluation of model accuracy, data interpretation, and external validation with new imaging data [13]. The quality of evidence in terms of the accuracy of these models so far seems low [7].
Discussion
This study highlights the current status and limitations of the application of AI and its subfields as adjunctive decision-making tools in HNC management. We present a summary of all the systematic reviews on the application of AI in HNC management in a logical manner with the findings of separate reviews to be compared and contrasted. This review provides various stakeholders including clinical researchers and decision-makers, hospital management, government agencies, and entrepreneurs with the evidence and future directions with regards to the application of AI in the field of head and neck oncology.
The adoption of these ML models in the daily clinical practice has so far been limited due to several factors [13, 18]. For example, a significant variation exists in data collection methods [6]. Data collection largely consists of data acquisition and labeling, as well as the improvement of existing data [27]. For instance, various centers and databases have different approaches for parameter labeling. In addition, treatment protocols may vary significantly across countries and geographic regions. This prevents the combination of various sources of data for robust model training using relatively large training data, and independent geographic external validation. For image data, the quality varies from one center to the other due to variations for example in tissue fixation, quality, mounting and staining of sections, scanning procedures, unstandardized image digitization methods, and suboptimal image magnification [28]. These variations affect the performance of the model when geographically validating data which are different from the data used for model development [6, 28]. These factors will affect proper data interpretation and the performance metrics from the model training process. Also, the model development varies significantly [7]. These variations usually include the size of the dataset for model training, type of machine learning algorithm, training methodology (data division paradigm), performance metrics for model evaluation, model evaluation on geographic external validation, model reporting, and adherence to AI model checklists [18]. Several efforts have thus been taken in recent years to build guidelines for model development and evaluation [29, 30]. Standardized guidelines for structured data registration and collection and model development are thus warranted and further data are necessary for validation studies which would facilitate the implementation of AI models in daily clinical practices [7, 15, 19]. Another limitation to the adoption of these algorithms for clinical evaluation is that their majority have not been independently or externally validated. In a few studies with performed external validations, there were concerns relating to this process in terms of external dataset similarity, minimum required dataset for external validation, acceptable performance metrics, and the procedure itself used for such a validation (independent or not). Hence, a modular regulatory framework considering the five important and closely related aspects of AI/ML (i.e., data collection, model development, performance metrics, external validation, and reporting), to facilitate the recommendation of these models for clinical evaluation is necessary [18]. Ethical and legal frameworks should be initiated to facilitate the adoption of these models in healthcare in order to prevent their misuse in terms of, for example, self-diagnosis and obtaining treatment recommendations [31].
The learning paradigm of the present AI techniques may be considered as a retrospective learning while it uses existing data resources and assumes that these will apply for the future settings. This approach has been criticized for not being a truly intelligent system [32]. Therefore, besides addressing the aforementioned limitations in this study, the future potential of AI in healthcare should also be considered from the natural intelligence perspective, where AI-based systems can use prospectively collected data for model development [32]. Therefore, a prospective learning paradigm will need to utilize different resources. A significant number of promising results reported on the use of AI in pathology have so far relied on retrospective data obtained from tissue biopsies. This continues to form the cornerstone for efficient AI model development and the training, validation, and assessment of model correctness. In turn, the model may possibly serve as an assistant tool in enabling low-cost and time-saving benefits for increased productivity and decision-making.
In recent years, the application of AI in healthcare has been touted to use natural language processing (NLP), which is a subfield of AI for differential diagnosis, self-triage, or self-treatment in the form of symptoms and clinical sign checkers [33]. Recent trends have shown these AI-assisted symptom checkers being integrated as a free web-based (such as the Isabel Symptom Checker) [34] or AI-powered chatbot system [33, 35, 36]. Therefore, necessary regulations are needed to provide clear guidance for the misuse or unauthorized use of AI. Studies are emerging on the potential of NLP as an approach to automatically transform clinical text in the hospital charts into structured data for various research purposes or improved clinical decision-making [35–41, 43]. More importantly, in these recent applications of AI and its subfield, the roles of clinicians remain important in evaluating the results.
Several studies have emphasized the potential of AI to augment image quality, segmentation, tumor characterization and prognostication, and treatment response evaluation [5, 6, 35]. Our review of the previously published systematic reviews demonstrates that AI has been suggested to play a prominent role in the identification of head and neck precancerous and cancerous lesions in histopathological slides [7, 18], prediction of the histopathologic nature of a given lesion from various sources of medical imaging [1, 5, 13–15, 17, 21, 22, 24–26], prognostication [5, 17, 18, 20, 21, 23], extraction of pathological findings from imaging [15, 16, 20], and different applications in radiation oncology [15, 17, 19].
In HNC, histopathological assessment remains the gold standard for providing prognostic information, but improvement/novel strategies are desirable. AI models may assist in effecting these and also serve as ancillary tools for risk stratification and management guidance [7, 18]. Information on precise location and size of HNC, presence of human papilloma virus (HPV), PDL-1 status/calculation of combined positive score (CPS), depth of invasion, perineural and lymphovascular invasion, number/size of metastases in lymph nodes, and the presence of extra-nodal extension have been reported to be useful prognosticators influencing management and AI models are reasonably expected to effect an efficient and standardized assessment of those parameters.
The application of AI to aid cancer diagnosis has formed the cornerstone of digital pathology. One of the issues affecting effective management of HNC cancer is delayed diagnosis and detection at an advanced stage [38]. It has been reported that early diagnosis of HNC can improve treatment and survival outcomes remarkably [7]. With the current advancements in computational capacity and improvements in various subfields of AI, digital pathology has significantly evolved from using static images to whole slide images (WSI) [39], thus enhancing pathological workflow and quantifying a number of parameters for defining the tumor and its microenvironment [39]. A high-resolution of WSI of human tissue is isolated into regions of clinical significance. This process is followed by pathology extraction (deconstruction of the WSI into smaller images) [40]. The use of AI to analyze WSI can also help in the detection, differentiation, and grading of potentially malignant (precancerous) and cancerous head and neck lesions [7]. Using AI in cancer pathology may refine or even redefine the histopathologic subtypes of different tumors altogether, as current definition of these is based on human visual recognition, interpretation and classification of images differently than in AI methods.
Technological advancements have enhanced the production and availability of medical data in different formats. In recent years, imaging data have become a budding source of interest for diagnostic and prognostic purposes, especially in the area of the quantitative image feature approach. Radiomics (i.e., the conversion of medical images into quantitative high-dimensional data) emerges as a potential tool in clinical practice to effect quick, cost-effective, and non-invasive diagnosis and prognostication [41, 42]. Data thus extracted from clinical imaging can provide specific information on tumor heterogeneity, texture, and morphology [43, 44]. In turn, the combination of AI and radiomics may lead to novel insights into the fundamental pathobiology of tumors, inferring the histomorphology, grading, metabolism, and, eventually, patient survival [43]. This has the potential to aid in clinical decision-making for personalized and precision medicine targeted at improving patient outcomes [41].
Despite the advances in medical care and both surgical and radiotherapy techniques, successful treatment of HNC may be associated with treatment-related late toxicities, such as masticatory, airway, speech, and swallowing impairments, all of which significantly reduce patient-reported quality of life [45]. Therefore, it is important to strike a balance between cancer treatment intensity and the risk of such toxicities. In this context, AI has been reported to show an insightful and efficient method of achieving personalized treatment planning [38, 46]. ML-based algorithms have been used to stratify patients into risk groups (patient-specific selection) for targeted treatment intensity [38, 46]. A personalized risk-based therapeutic approach before starting treatment is an important step towards improved survival and functional outcomes. However, the employment of such a risk stratification into treatment strategies should be followed by clinical trials, as intense treatment may increase toxicity with no prognostic benefit, whereas de-intensified treatment may reduce toxicity with a prognostic disadvantage.
Admittedly, methodological limitations influenced the present study. Firstly, not all the included reviews reported the average performance of the models in terms of any of the widely used performance metrics. Therefore, we could not present summarized performance metrics for each of the highlighted themes. Secondly, not all those systematic reviews assessed the risk of bias of the included studies, and this negatively affects our study. In addition, a systematic literature search always has a time frame limit. This means that the present analysis may miss any important studies that were reported after the 17 reviews were published.
In conclusion, we provided an informative analysis of all the systematic reviews during the selected time period on the evolving status of AI/ML approaches in head and neck oncology. For future studies, it would be desirable to perform an examination of systematic reviews for each of the AI/ML application themes presented here. Finally, although the use of AI/ML-based models for HNC management is a promising and rapidly expanding field, standardized international guidelines are warranted to overcome the limitations of the widespread use and implementation of these models. Thereafter, it is of utmost importance to validate these clinical applications in the management of HNC, as the methodology is progressing rapidly in many specialties.
Acknowledgements
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. Sigrid Jusélius Foundation, Finska Läkaresällskapet, State Research Funding for the Helsinki University Hospital and for the Turku University Hospital funded this review. No funding or sponsorship was received for the publication of this article.
Authorship
All mentioned authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
The study was conceived and designed by Antti Mäkitie. Rasheed Omobolaji Alabi and Antti Mäkitie performed the literature review. Antti Mäkitie and Rasheed Omobolaji Alabi drafted the manuscript. Sweet Ping Ng, Robert P. Takes, K. Thomas Robbins, Ohad Ronen, Ashok R. Shaha, Patrick J Bradley, Nabil F Saba, Sandra Nuyts, Asterios Triantafyllou, Cesare Piazza, Alessandra Rinaldo, and Alfio Ferlito were involved in commenting, revising, and quality assessment of the manuscript. All authors approved the final version.
Disclosures
Antti A. Mäkitie, Rasheed Omobolaji Alabi, Sweet Ping Ng, Robert P. Takes, K. Thomas Robbins, Ohad Ronen, Ashok R. Shaha, Patrick J Bradley, Nabil F Saba, Sandra Nuyts, Asterios Triantafyllou, Cesare Piazza, Alessandra Rinaldo, and Alfio Ferlito all have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
This article was written by members and invitees of the International Head and Neck Scientific Group (www.IHNSG.com).
References
- 1.Mahmood H, Shaban M, Rajpoot N, Khurram SA. Artificial Intelligence-based methods in head and neck cancer diagnosis: an overview. Br J Cancer. 2021 doi: 10.1038/s41416-021-01386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Svider PF, Blasco MA, Raza SN, Shkoukani M, Sukari A, Yoo GH, et al. Head and neck cancer: underfunded and understudied? Otolaryngol Head Neck Surg. 2017;156:10–13. doi: 10.1177/0194599816674672. [DOI] [PubMed] [Google Scholar]
- 3.Pai SI, Westra WH. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol Mech Dis. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muto M, Nakane M, Katada C, Sano Y, Ohtsu A, Esumi H, et al. Squamous cell carcinoma in situ at oropharyngeal and hypopharyngeal mucosal sites. Cancer. 2004;101:1375–1381. doi: 10.1002/cncr.20482. [DOI] [PubMed] [Google Scholar]
- 5.Alabi RO, Bello IO, Youssef O, Elmusrati M, Mäkitie AA, Almangush A. Utilizing deep machine learning for prognostication of oral squamous cell carcinoma—a systematic review. Front Oral Health. 2021 doi: 10.3389/froh.2021.686863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alabi RO, Almangush A, Elmusrati M, Mäkitie AA. Deep machine learning for oral cancer: from precise diagnosis to precision medicine. Front Oral Health. 2022;2:794248. doi: 10.3389/froh.2021.794248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahmood H, Shaban M, Indave BI, Santos-Silva AR, Rajpoot N, Khurram SA. Use of artificial intelligence in diagnosis of head and neck precancerous and cancerous lesions: a systematic review. Oral Oncol. 2020;110:104885. doi: 10.1016/j.oraloncology.2020.104885. [DOI] [PubMed] [Google Scholar]
- 8.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.EhteshamiBejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith V, Devane D, Begley CM, Clarke M. Methodology in conducting a systematic review of systematic reviews of healthcare interventions. BMC Med Res Methodol. 2011;11:15. doi: 10.1186/1471-2288-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Institute of Health. Study quality assessment tools. United States: n.d.
- 12.Shea BJ, Hamel C, Wells GA, Bouter LM, Kristjansson E, Grimshaw J, et al. AMSTAR is a reliable and valid measurement tool to assess the methodological quality of systematic reviews. J Clin Epidemiol. 2009;62:1013–1020. doi: 10.1016/j.jclinepi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Jethanandani A, Lin TA, Volpe S, Elhalawani H, Mohamed ASR, Yang P, et al. Exploring applications of radiomics in magnetic resonance imaging of head and neck cancer: a systematic review. Front Oncol. 2018;8:131. doi: 10.3389/fonc.2018.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil S, Habib Awan K, Arakeri G, Jayampath Seneviratne C, Muddur N, Malik S, et al. Machine learning and its potential applications to the genomic study of head and neck cancer—a systematic review. J Oral Pathol Med. 2019;48:773–779. doi: 10.1111/jop.12854. [DOI] [PubMed] [Google Scholar]
- 15.Giraud P, Giraud P, Gasnier A, El Ayachy R, Kreps S, Foy J-P, et al. Radiomics and machine learning for radiotherapy in head and neck cancers. Front Oncol. 2019;9:174. doi: 10.3389/fonc.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adeoye J, Tan JY, Choi S-W, Thomson P. Prediction models applying machine learning to oral cavity cancer outcomes: a systematic review. Int J Med Inform. 2021;154:104557. doi: 10.1016/j.ijmedinf.2021.104557. [DOI] [PubMed] [Google Scholar]
- 17.Volpe S, Pepa M, Zaffaroni M, Bellerba F, Santamaria R, Marvaso G, et al. Machine learning for head and neck cancer: a safe bet?—a clinically oriented systematic review for the radiation oncologist. Front Oncol. 2021;11:772663. doi: 10.3389/fonc.2021.772663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alabi RO, Youssef O, Pirinen M, Elmusrati M, Mäkitie AA, Leivo I, et al. Machine learning in oral squamous cell carcinoma: current status, clinical concerns and prospects for future—a systematic review. Artif Intell Med. 2021;115:102060. doi: 10.1016/j.artmed.2021.102060. [DOI] [PubMed] [Google Scholar]
- 19.Carbonara R, Bonomo P, Di Rito A, Didonna V, Gregucci F, Ciliberti MP, et al. Investigation of radiation-induced toxicity in head and neck cancer patients through radiomics and machine learning: a systematic review. J Oncol. 2021;2021:1–9. doi: 10.1155/2021/5566508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinnery T, Arifin A, Tay KY, Leung A, Nichols AC, Palma DA, et al. Utilizing artificial intelligence for head and neck cancer outcomes prediction from imaging. Can Assoc Radiol J. 2021;72:73–85. doi: 10.1177/0846537120942134. [DOI] [PubMed] [Google Scholar]
- 21.Chiesa-Estomba CM, Graña M, Medela A, Sistiaga-Suarez JA, Lechien JR, Calvo-Henriquez C, et al. Machine learning algorithms as a computer-assisted decision tool for oral cancer prognosis and management decisions: a systematic review. ORL. 2022;84:278–288. doi: 10.1159/000520672. [DOI] [PubMed] [Google Scholar]
- 22.Giannitto C, Mercante G, Ammirabile A, Cerri L, De Giorgi T, Lofino L, et al. Radiomics-based machine learning for the diagnosis of lymph node metastases in patients with head and neck cancer: systematic review. Head Neck. 2022 doi: 10.1002/hed.27239. [DOI] [PubMed] [Google Scholar]
- 23.Ng WT, But B, Choi HC, de Bree R, Lee AW, Lee VH, et al. Application of artificial intelligence for nasopharyngeal carcinoma management—a systematic review. CMAR. 2022;14:339–366. doi: 10.2147/CMAR.S341583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santer M, Kloppenburg M, Gottfried TM, Runge A, Schmutzhard J, Vorbach SM, et al. Current applications of artificial intelligence to classify cervical lymph nodes in patients with head and neck squamous cell carcinoma—a systematic review. Cancers. 2022;14:5397. doi: 10.3390/cancers14215397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmakaty I, Elmarasi M, Amarah A, Abdo R, Malki MI. Accuracy of artificial intelligence-assisted detection of oral squamous cell carcinoma: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2022;178:103777. doi: 10.1016/j.critrevonc.2022.103777. [DOI] [PubMed] [Google Scholar]
- 26.Kim J-S, Kim BG, Hwang SH. Efficacy of artificial intelligence-assisted discrimination of oral cancerous lesions from normal mucosa based on the oral mucosal image: a systematic review and meta-analysis. Cancers. 2022;14:3499. doi: 10.3390/cancers14143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roh Y, Heo G, Whang SE. A survey on data collection for machine learning: a big data—AI integration perspective. IEEE Trans Knowl Data Eng. 2021;33:1328–1347. doi: 10.1109/TKDE.2019.2946162. [DOI] [Google Scholar]
- 28.Chu CS, Lee NP, Ho JWK, Choi S-W, Thomson PJ. Deep learning for clinical image analyses in oral squamous cell carcinoma: a review. JAMA Otolaryngol Head Neck Surg. 2021 doi: 10.1001/jamaoto.2021.2028. [DOI] [PubMed] [Google Scholar]
- 29.Collins GS, Dhiman P, Andaur Navarro CL, Ma J, Hooft L, Reitsma JB, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11:e048008. doi: 10.1136/bmjopen-2020-048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabitza F, Campagner A, Soares F, García de Guadiana-Romualdo L, Challa F, Sulejmani A, et al. The importance of being external. Methodological insights for the external validation of machine learning models in medicine. Comput Methods Programs Biomed. 2021;208:106288. doi: 10.1016/j.cmpb.2021.106288. [DOI] [PubMed] [Google Scholar]
- 31.Alabi RO, Tero V, Mohammed E. Machine learning for prognosis of oral cancer: what are the ethical challenges? CEUR-Workshop Proceedings 2020.
- 32.Vogelstein JT, Verstynen T, Kording KP, Isik L, Krakauer JW, Etienne-Cummings R, et al. Prospective Learning: Back to the Future 2022. 10.48550/ARXIV.2201.07372.
- 33.Meyer AND, Giardina TD, Spitzmueller C, Shahid U, Scott TMT, Singh H. Patient perspectives on the usefulness of an artificial intelligence–assisted symptom checker: cross-sectional survey study. J Med Internet Res. 2020;22:e14679. doi: 10.2196/14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isabel Healthcare. Isabel Symptom Checker—The One the Patients Use https://symptomchecker.isabelhealthcare.com/isabel-tool-page. AI-Assisted Symptom Checker 2018. https://symptomchecker.isabelhealthcare.com/. Accessed 28 Feb 2023.
- 35.Pham N, Ju C, Kong T, Mukherji SK. Artificial intelligence in head and neck imaging. Semin Ultrasound CT MRI. 2022;43:170–175. doi: 10.1053/j.sult.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Montenegro JLZ, da Costa CA, da Rosa RR. Survey of conversational agents in health. Expert Syst Appl. 2019;129:56–67. doi: 10.1016/j.eswa.2019.03.054. [DOI] [Google Scholar]
- 37.Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7:e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alabi RO, Elmusrati M, Sawazaki-Calone I, Kowalski LP, Haglund C, Coletta RD, et al. Machine learning application for prediction of locoregional recurrences in early oral tongue cancer: a Web-based prognostic tool. Virchows Arch. 2019;475:489–497. doi: 10.1007/s00428-019-02642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassani S, Santonicco N, Eccher A, Scarpa A, Vianini M, Brunelli M, et al. Artificial intelligence in head and neck cancer diagnosis. J Pathol Inform. 2022;13:100153. doi: 10.1016/j.jpi.2022.100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 41.Fh T, Cyw C, Eyw C. Radiomics AI prediction for head and neck squamous cell carcinoma (HNSCC) prognosis and recurrence with target volume approach. BJR|Open. 2021;3:20200073. doi: 10.1259/bjro.20200073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajgor AD, Patel S, McCulloch D, Obara B, Bacardit J, McQueen A, et al. The application of radiomics in laryngeal cancer. BJR. 2021;94:20210499. doi: 10.1259/bjr.20210499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1:207–226. doi: 10.1080/23808993.2016.1164013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao L-J, Hsu W-L, Lo W-C, Cheng P-W, Shueng P-W, Hsieh C-H. Health-related quality of life and utility in head and neck cancer survivors. BMC Cancer. 2019;19:425. doi: 10.1186/s12885-019-5614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alabi R, Almangush A, Elmusrati M, Leivo I, Mäkitie AA. An interpretable machine learning prognostic system for risk stratification in oropharyngeal cancer. Int J Med Inform. 2022;168:104896. doi: 10.1016/j.ijmedinf.2022.104896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.