Abstract
Transcription factor IIIA (TFIIIA) activates 5S ribosomal RNA gene transcription in eukaryotes. The protein from vertebrates has nine contiguous Cys2His2 zinc fingers which function in nucleic acid binding, and a C-terminal region involved in transcription activation. In order to identify protein partners for TFIIIA, yeast two-hybrid screens were performed using the C-terminal region of Xenopus TFIIIA as an attractor and a rat cDNA library as a source of potential partners. A cDNA clone was identified which produced a protein in yeast that interacted with Xenopus TFIIIA but not with yeast TFIIIA. This rat clone was sequenced and the primary structure of the human homolog (termed TFIIIA-intP for TFIIIA-interacting protein) was determined from expressed sequence tags. In vitro interaction of recombinant human TFIIIA-intP with recombinant Xenopus TFIIIA was demonstrated by immunoprecipitation of the complex using anti-TFIIIA-intP antibody. Interaction of rat TFIIIA with rat TFIIIA-intP was indicated by co-chromatography of the two proteins on DEAE-5PW following fractionation of a rat liver extract on cation, anion and gel filtration resins. In a HeLa cell nuclear extract, recombinant TFIIIA-intP was able to stimulate TFIIIA-dependent transcription of the Xenopus 5S ribosomal RNA gene but not TFIIIA-independent transcription of the human adenovirus VA RNA gene.
INTRODUCTION
Transcription of 5S ribosomal RNA genes by RNA polymerase III requires three transcription factors, TFIIIA, TFIIIB and TFIIIC, whereas transcription of tRNA and adenovirus VA RNA genes requires only TFIIIB and TFIIIC (1–3). TFIIIB and TFIIIC, but not TFIIIA, associate in vivo with polymerase III forming a ‘holoenzyme’ (4). These observations demonstrate the TFIIIIA dependence of polymerase III transcription of 5S genes and indicate that the transcription mechanism is distinct from that of tRNA and adenovirus VA genes. Understanding the biochemical details of vertebrate 5S RNA gene transcription will require elucidation of the structure and function of the protein factors involved in this mechanism. Factor IIIA is a single protein whereas factors IIIB and IIIC contain multiple protein species (5–11). Little information is available on the structure of RNA polymerase III initiation complexes in vertebrates whereas more information is available on the structure of these complexes in yeast (12,13). In the case of TFIIIA and 5S gene transcription, the yeast and vertebrate systems are not interchangeable in vitro and a number of proteins are not shared (8,9). The Xenopus and human 5S gene transcription systems are interchangeable in vitro (14).
Considerable structural and functional information is available concerning TFIIIA isolated from Xenopus laevis oocytes. This 38 kDa protein was the first gene regulatory factor discovered to contain zinc and possess repetitive Cys2His2 zinc binding domains referred to as zinc fingers (15–18). These fingers bind the 50 bp internal control region (ICR) of the 5S gene in three groups of three: the N-terminal finger group contacts the 3′ ICR C-box, the middle group contacts the ICR intermediate element and the C-terminal finger group contacts the A-box 5′ ICR sequences (19–24). Xenopus TFIIIA zinc fingers are also involved in binding the 5S ribosomal RNA transcript (22,23). The binding orientation of the TFIIIA zinc fingers on the ICR positions the C-terminal region of the protein pointing upstream toward the start site of 5S RNA transcription. This C-terminal region is conserved in amphibians and human TFIIIA and is essential for transcription activation (25–28). The structure of yeast TFIIIA differs significantly to that of Xenopus and all other known vertebrate TFIIIAs, sharing only ∼20% amino acid sequence identity with frog TFIIIA (29,30). The transcription activation region of yeast TFIIIA does not resemble that of Xenopus or human TFIIIA and is located between fingers VIII and IX (31,32).
The identification and characterization of proteins that interact with the C-terminal portion of TFIIIA will aid in the elucidation of the 5S gene transcription activation mechanism in vertebrates. The yeast two-hybrid screen is a useful experimental tool for identifying cDNA clones from a tissue library that code for proteins that can physically interact with a protein of interest (33). In the present study, the yeast two-hybrid system was used to identify proteins which interact with the C-terminal portion of Xenopus TFIIIA. A 70.6 kDa protein previously not associated with RNA polymerase III transcription was identified that interacts with TFIIIA in vitro and in cell extracts. This protein stimulates TFIIIA-dependent transcription of the 5S RNA gene in vitro but not TFIIIA-independent transcription of the adenovirus VA gene.
MATERIALS AND METHODS
Construction of pAS1-TFIIIA fusions and yeast transformations
Yeast plasmid pAS1 (34) containing the yeast GAL4 DNA binding domain was kindly provided by Steven Elledge. The cDNA sequence coding for zinc fingers VII–IX (starting at amino acid 189, QDLAV) and the C-terminal region of Xenopus TFIIIA (ending at KLTIQ, amino acid 344) was amplified using as template the 1.5-kb X.laevis TFIIIA cDNA clone (15) kindly provided by Robert Roeder. Upstream and downstream primers contained BamHI and SalI sites respectively to allow in-frame cloning into pAS1. A similar strategy was used in the sub-cloning of the yeast TFIIIA insert containing zinc fingers V–IX (starting at amino acid 187, DPEVE) and the C-terminal tail (ending at SAGDK, amino acid 428). Saccharomyces cerevisiae genomic DNA obtained from Promega Life Sciences (Madison, WI) was used for the PCR template. Yeast transformation was performed by modification of a previously described procedure (35). Yeast strain Y190 (containing β-galactosidase and His reporter genes) was grown to about 1 × 107 cells/ml in YTDA media. Cells were pelleted and resuspended in 1 ml of 100 mM lithium acetate and 10 mM Tris–HCl, pH 7.6, 1 mM EDTA. An aliquot of 200 µl of this cell suspension was mixed with 2–4 µg plasmid DNA, 200 µg carrier DNA, 1.2 ml 40% PEG and shaken at 30°C for 30 min. Cells were placed at 42°C for 15 min, pelleted, resuspended in sterile H2O and plated overnight.
Yeast lysate preparation and western blotting
Cells containing pAS1-TFIIIAfrog or TFIIIAyeast were pelleted from 10 ml cultures, resuspended in 200 µl of 16.6% pre-chilled trichloroacetic acid and vortexed repeatedly with siliconized glass beads. The suspension was centrifuged to pellet the protein, washed with 100% ethanol and resuspended in 100 µl SDS–PAGE loading buffer; the pH was adjusted to 6–7 with 1 M Tris–HCl, pH 10. Samples were electrophoresed through a 12% polyacrylamide SDS gel and transferred to a 0.45 µm nitrocellulose filter using an IDEA Scientific trans-blot cell. The filter was blocked with 3% non-fat dry milk in 10 mM Tris–HCl, pH 7.4, 140 mM NaCl and incubated in the same solution overnight with the appropriate antibody. The blot was rinsed and incubated with secondary antibody for 1 h. For alkaline phosphatase-conjugated secondary antibody, the blot was developed with NBT (1 µg/ml) and BCIP (0.5 µg/ml) in 20 mM Tris–HCl, pH 9.5; for horseradish peroxidase antibody, development was performed using the Renaissance Chemiluminescence kit from New England Nuclear (Boston, MA).
Library screening, β-galactosidase filter assay, yeast plasmid isolation and characterization
Yeast strain Y190 transformed with pAS1-TFIIIAfrog was grown in SC media minus tryptophan and transformed with a rat brain cDNA pACT library (36) kindly provided by Ian Macara. The transformation mix was distributed on SC plates lacking tryptophan, leucine and histidine but including 25 mM 3-aminotriazole (3-AT, Sigma Chemical Co., St Louis, MO; A8056) and incubated for 4–5 days at 30°C. A total of 12 × 106 transformants were assayed for Trp+, Leu+, His+ growth in four independent screens. To assay for β-galactosidase activity, Ahlstrom grade 95 paper filters were exposed to the plate surfaces followed by freezing in liquid nitrogen and then thawing at room temperature. The filter was placed in a Petri dish and 1 ml of 1 mM MgSO4, 10 mM KCl, 5.5 g/l NaH2PO4, 8.5 g/l Na2HPO4, 40 µl of X-gal (20 mg/ml) was added followed by incubation at 37°C overnight. Positive colonies were reassayed for β-galactosidase activity and then grown in Leu– Trp+ media to remove the pAS1-TFIIIA plasmids. The remaining pACT plasmids were purified from 5 ml yeast cultures by vortexing cell pellets with glass beads in 100 mM Tris–HCl, pH 8.0, 50 mM EDTA, 1% SDS followed by phenol/chloroform extraction and ethanol precipitation. Plasmid DNA was transformed into yeast cells containing pAS1-Gal4 fusions to either SNF1, lamin or the yeast TFIIIA and assayed for β-galactosidase activity.
Sequence analysis, expression and purification of recombinant proteins
Rat pACT which tested positive in the two-hybrid screen with Xenopus TFIIIA but not with yeast TFIIIA was digested with XhoI; the cDNA insert was cloned into pTrcHis (Invitrogen, Carlsbad, CA) for expression and into pBluescript (Stratagene, La Jolla, CA) for sequencing (37). An open reading frame (ORF) was observed that displayed 85% identity to a human expressed sequence tag (EST) clone from an infant brain library (clone ID 38702; 5′ and 3′ sequence accession numbers, R36308 and R49240). Another EST clone (ID 757436; 5′ and 3′ sequence accession numbers, AA442279 and AA437216) from a human testis cDNA library contained an additional 1000 nt. These two EST clones shared a common BbsI restriction site which was used to splice them together. This procedure yielded a 3-kb cDNA fragment which was cloned into the pET vector 28a(+) (Novagen, Madison, WI) using SacI and NotI sites. The protein coded by this construct, termed TFIIIA-intP for TFIIIA-interacting protein, was expressed in BL21(DE3) cells by induction with IPTG. Cells were pelleted, suspended in 40 mM Tris–HCl, pH 7.4, 6 M guanidine–HCl, 0.5 M NaCl, 10% glycerol, 20 mM imidazole (pH 8.0), broken in a French press and centrifuged at 30 000 r.p.m. in a Beckman Ti45 rotor. The supernatant was loaded on a ProBond Resin column (Invitrogen) and eluted with the same buffer containing 0.3 M imidazole. The eluate was dialyzed against this buffer minus the guanidine/imidazole and stored at –80°C. Recombinant Xenopus TFIIIA was expressed from plasmid pT7-TFIIIA in a similar fashion and cells were broken in a French press in 40 mM Tris–HCl, pH 7.4, 100 mM KCl, 5 M urea, 1 mM dithiothreitol (DTT), 50 µM Zn2SO4 and 10% glycerol. The extract was centrifuged and loaded on a SP Sepharose column equilibrated in the breaking buffer. TFIIIA, eluting in 0.3 M KCl breaking buffer, was dialyzed against breaking buffer minus the urea, and stored at –80°C. Recombinant protein amounts were determined by the dye-binding method (38).
Affinity purification of anti-TFIIIA intP antibody and immunoprecipitation
Rat cDNA for TFIIIA-intP cloned in pTrcHis was expressed and purified as described above. Purified protein was electrophoresed on a 12% polyacrylamide SDS gel and rabbit polyclonal antiserum was developed against the TFIIIA-intP PAGE band at Cocalico Biologicals (Reamstown, PA). Thirty-four milligrams of rat TFIIIA-intP was reacted with Affi-Gel 10 resin according to the manufacturer’s directions (Bio-Rad, Hercules, CA). Unconjugated resin was blocked with 0.1 M ethanolamine and rabbit antiserum was applied to the resin and washed repeatedly. The bound antibody was eluted with 200 mM glycine (pH 2.0), 100 mM NaCl and neutralized with 1.0 M Tris–HCl, pH 10. This antibody reacted with the recombinant human TFIIIA-intP at a 1:5000 dilution in western blotting. Immunoprecipitation of the Xenopus TFIIIA–TFIIIA-intP complex was performed using the Immuno Catcher kit (CytoSignal, Irvine, CA). Fifty microliters of the purified TFIIIA intP antibody (3 µg protein) were incubated for 30 min at 23°C with 30 µl bed volume of protein A/G binding resin; unbound antibody was washed from the resin and then 10 µg of recombinant human TFIIIA-intP was bound for 1.5 h. The resin was washed twice with 20 mM HEPES, pH 7.9, 0.1 M KCl, 10% glycerol followed by addition of 6 µg of recombinant Xenopus TFIIIA; the reaction was incubated in the same buffer for 2 h at 23°C. The resin was washed six times with 20 mM HEPES, pH 7.9, 0.3 M KCl, 10% glycerol; the bound protein was eluted with SDS–PAGE sample buffer and electrophoresed through a 12% polyacrylamide SDS gel.
Chromatographic fractionation of rat and human cell extracts
A rat liver cytosol was prepared and fractionated by (NH4)2SO4 precipitation as described (39). The 0–38% fraction containing TFIIIA-intP (assayed in this and subsequent fractions by western blot) was resuspended in 20 mM HEPES–NaOH, pH 7.9, 0.5 mM EDTA, 1 mM DTT, 10% glycerol (buffer A), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg/ml of antipain and leupeptin. Following centrifugation at 28 000 g, the supernatant was fractionated on phosphocellulose (P11; Whatman, Clifton, NJ) pre-equilibrated in buffer A containing 100 mM KCl and 0.5 mM PMSF. The flow-through containing TFIIIA-intP was fractionated on Toyopearl DEAE-650M (TosoHaas, Montgomeryville, PA) pre-equilibrated in buffer C (40 mM Tris–HCl, pH 7.9, 0.5 mM EDTA, 1 mM DTT, 10% v/v glycerol), 80 mM KCl. The 1000 mM KCl fraction containing TFIIIA-intP was dialyzed against buffer A containing 100 mM KCl and then concentrated by (NH4)2SO4 precipitation. The pellet was resuspended in 10 ml buffer A and dialyzed against buffer A containing 100 mM KCl. Following centrifugation at 12 000 g, the supernatant was applied to an Ultrogel AcA34 gel filtration column pre-equilibrated with buffer A containing 400 mM KCl. Column fractions containing TFIIIA-intP were diluted with an equal volume of buffer C containing no salt and applied to a TSK DEAE-5PW column pre-equilibrated in buffer C containing 200 mM KCl; column contents were eluted with a 10 column volume gradient to 1000 mM KCl. For transcription assays, HeLa nuclear extract (Promega) was loaded on a 1 ml P11 column pre-equilibrated in buffer A containing 100 mM KCl. The contents on the column were washed with buffer A containing 100 mM KCl and then eluted with buffer A containing 800 mM KCl; aliquots were frozen at –80°C.
Transcription assays
In vitro transcription of the Xenopus 5S ribosomal gene and the human adenovirus VA RNA gene (kindly provided by Guang Jer Wu, 40) was performed using the phosphocellulose 0.8 M KCl fraction of the human nuclear extract as a source of RNA polymerase III and associated factors. Transcription assays (25 µl) contained 10 mM HEPES, pH 7.9, 50 mM KCl, 1.5 mM MgCl2, 0.25 mM DTT, 0.1 mM EDTA, 10% glycerol, 400 µM (GTP, ATP, UTP), 1.6 µM CTP, 10 µCi [α-32P]CTP, 3 µg phosphocellulose fraction; plasmid and recombinant protein additions are described in the legend to the Figure 7. Plasmids and recombinant proteins were incubated in the transcription buffer with the phosphocellulose fraction for 30 min at 30°C in the absence of nucleotides and then for 1 h in the presence of nucleotides. The reactions were quenched with 100 µl SDS buffer (0.5% SDS, 0.3 M Tris–HCl, pH 7.4, 0.3 M sodium acetate, 2 mM EDTA, 3 µg/ml yeast tRNA), phenol–chloroform extracted, and ethanol precipitated. Samples were resuspended in 10 mM EDTA, pH 8.0, 98% deionized formamide, 0.04% xylene cyanol/bromophenol blue and heated at 65°C. Samples were electrophoresed through a 7% polyacrylamide gel containing 7 M urea and 89 mM Tris, 110 mM boric acid, 1 mM EDTA, pH 8.0. Gels were subjected to autoradiography overnight using Kodak XAR5 film.
Figure 7.
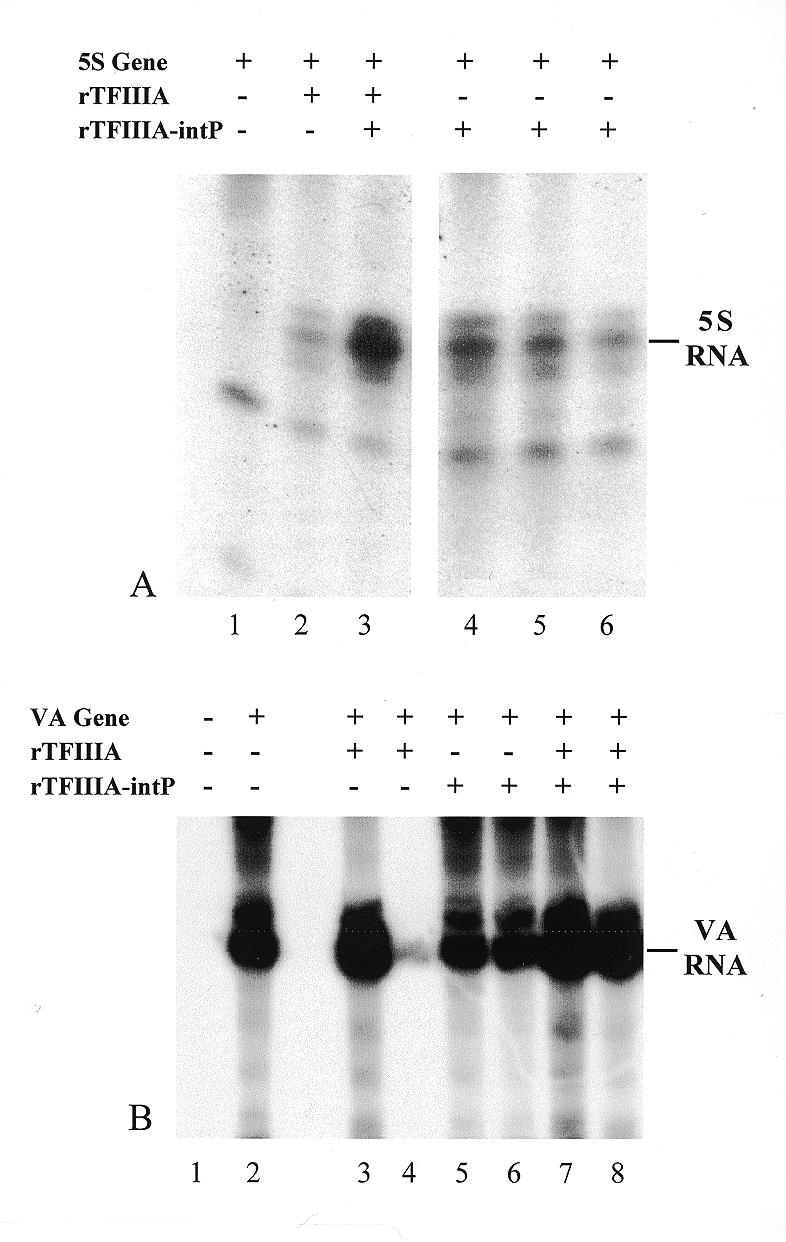
Effect of TFIIIA-intP on 5S RNA gene or adenovirus VA gene transcription in vitro. Preparation of recombinant proteins, transcription assays and gel electrophoresis were performed as described in Materials and Methods. Transcription reactions electrophoresed in lanes 1–6 (A) or 2–8 (B) contained 0.5 µg 5S gene or VA gene plasmid respectively. 5S and VA RNA transcripts are demarcated in the right margins. Reactions in lanes 2 and 3 (A) contained 0.2 µg recombinant Xenopus TFIIIA and lane 3 additionally contained 0.1 µg recombinant human TFIIIA-intP. Reactions in lanes 4–6 (A) contained 0.1, 0.2 or 0.4 µg recombinant human TFIIIA-intP. In (B), transcription reactions electrophoresed in lanes 3 and 4 contained 0.2 and 0.3 µg recombinant Xenopus TFIIIA; reactions in lanes 5 and 6 contained 0.1 and 0.2 µg recombinant human TFIIIA-intP; reactions in lanes 7 and 8 contained 0.2 and 0.3 µg recombinant Xenopus TFIIIA respectively and 0.1 and 0.2 µg recombinant human TFIIIA-intP respectively. The reaction electrophoresed in lane 1 contained no VA gene.
RESULTS
Two-hybrid screen for TFIIIA-interacting proteins
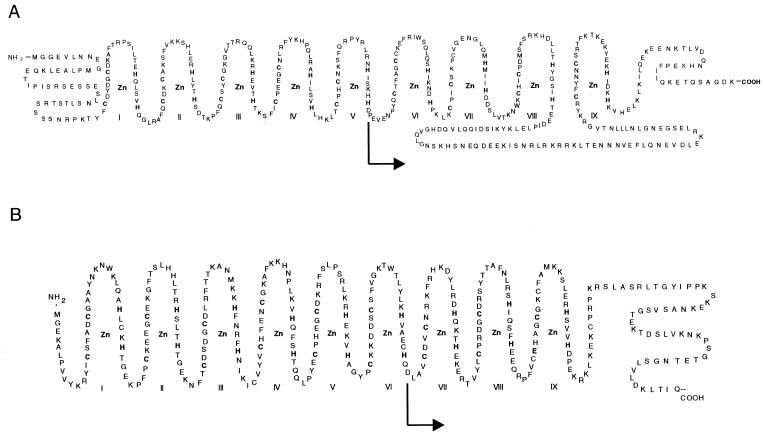
Figure 1 exhibits the polypeptide regions of yeast (A) and Xenopus (B) TFIIIA used to construct the GAL4 DNA binding domain fusions. Rather than clone just the yeast transcription activation region located between fingers XIII and XI into the GAL4 construct, the surrounding fingers beginning at finger VI (Fig. 1A, arrow) and extending to the C-terminus were used for the construction. This was done to possibly help maintain the conformational stability of the activation domain. The same approach was taken with Xenopus TFIIIA where the C-terminal region was cloned along with the C-terminal fingers VII, VIII and IX (Fig. 1B, arrow) into the GAL4 fusion. When expressed in yeast cells, the yeast and Xenopus GAL4 constructs yielded fusion proteins of the appropriate size when analyzed by western blotting using either an antibody against the GAL4 DNA binding domain (Fig. 2, lanes 1–4) or against Xenopus TFIIIA (lanes 7 and 8). The larger molecular weight of the yeast TFIIIA fusion (∼10K, lanes 3 and 4) reflects the additional amino acids present.
Figure 1.
TFIIIA amino acid structures used in two-hybrid plasmid constructs. I–IX indicate the order of the nine Cys2His2 zinc fingers from the N- to C-terminal direction. An arrow marks the site where the TFIIIA polypeptide was fused to the C-terminus of the Gal4 DNA binding domain; the entire polypeptide from the arrow to the C-terminus was used in the construct. (A) and (B), amino acid sequences from S.cerevisiae and X.laevis TFIIIA.
Figure 2.

Protein expression in yeast from TFIIIA-Gal4 fusion plasmids. Plasmid construction, yeast transformation, lysate preparation, antibodies, SDS–PAGE and western blotting are described in Materials and Methods. Dashes on the left margin of gel blot mark approximate electrophoretic migration distances for proteins of the indicated molecular weights (×10–3). Lysates from yeast cells expressing the Xenopus TFIIIA or yeast TFIIIA-Gal4 fusions were electrophoresed in lanes 1–4 and 5–8 respectively. The blots were probed with either anti-Gal4 DNA binding domain antibody (1–4) or anti-Xenopus TFIIIA antibody (5–8) and alkaline phosphatase-conjugated secondary antibody.
For the two-hybrid screen, a rat pACT cDNA expression library was used as a source for potential Xenopus TFIIIA-interacting proteins. This was done because of the availability of this high-complexity library for our studies and because we previously demonstrated that Xenopus TFIIIA promoted transcription of the Xenopus 5S RNA gene to a high degree in a rat in vitro transcription system (28). When this rat library was transformed into yeast expressing the Xenopus TFIIIA-GAL4 construct, a number of positive clones were observed among four independent screens. Four of the clones that were consistently positive and that were negative when retransformed into yeast cells lacking the TFIIIA-GAL4 fusion were additionally screened against the yeast TFIIIA construct as well as the SNF1 or lamin constructs (Fig. 3). Of these four, two were strongly positive for β-galactosidase activity (clones 3 and 4) and two were weakly positive (clones 1 and 2) when assayed against the Xenopus-GAL4 construct (Fig. 3A). Of the two strongly positive ones, clone 4 was also strongly positive when assayed with Yeast TFIIIA (Fig. 3B), yeast SNF1 (Fig. 3C) or lamin (Fig. 3D). The protein coded by this clone may be interacting with the GAL4 DNA binding domain. Significantly, the other strongly interacting clone for Xenopus TFIIIA (clone 3) did not interact with yeast TFIIIA, SNF1 or lamin. These results suggest that this clone may code for a protein that interacts with the C-terminal portion of Xenopus TFIIIA. This putative protein was termed TFIIIA-intP for ‘TFIIIA-interacting protein’.
Figure 3.
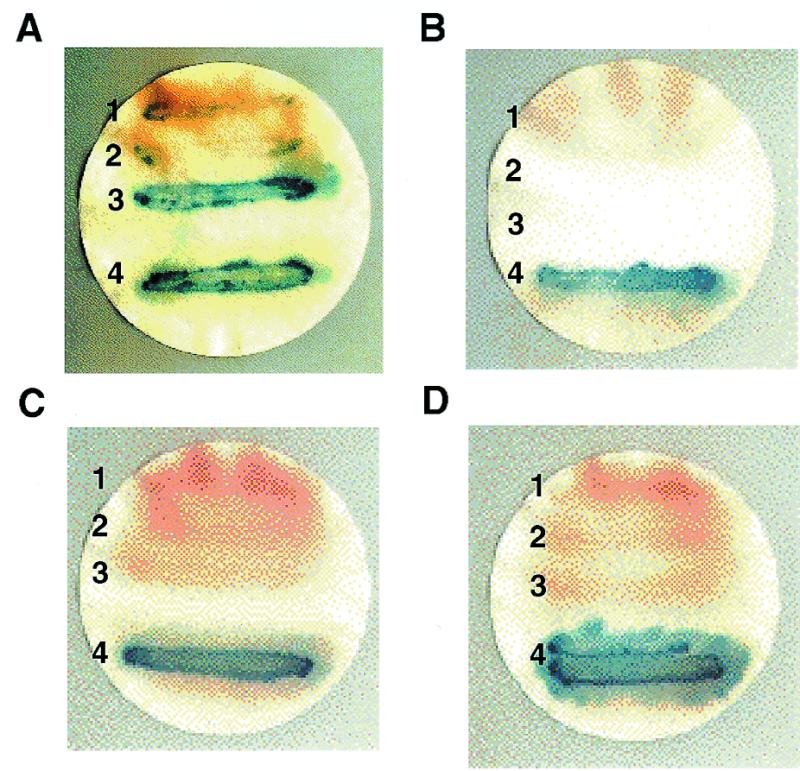
Binding specificity of candidate TFIIIA-interacting protein in yeast. The yeast β-galactosidase filter assay was performed as described in Materials and Methods. Cells streaked on filters (A), (B), (C) and (D) contained Gal4 DNA binding domain-Xenopus TFIIIA, yeast TFIIIA, yeast SNF1 or lamin fusion plasmids respectively. Individual yeast clones 1, 2, 3 and 4 also contained different pACT rat cDNAs fused to Gal4 activation domains and were obtained from the two-hybrid screens.
Sequence analysis of the TFIIIA-intP two-hybrid clone
Clone number 3 from Figure 3 was sequenced and found to contain an ORF coding for 343 amino acids (Fig. 4, rat TFIIIA-intP). No N-terminal methionine was observed so this sequence is likely from an incomplete cDNA. This rat sequence was found to share 85% identity with a number of human ESTs (see Materials and Methods), including 100% identity over a leucine-rich, 36 amino acid stretch. Using these ESTs along with confirmatory sequencing, a complete amino acid sequence for the apparent human homolog for this protein was assembled (Fig. 4), yielding a protein of 617 amino acids with a molecular weight of 70 600. The human sequence has numerous leucine zipper and other leucine-rich regions that are present and conserved with the rat sequence. The sequence contains a potential Cys2HisCys zinc finger at residues 553–582. The protein is rich in glutamate (14.7%) and has an acidic pI. EST database searches in other organisms identified a pig sequence (AU059594) with 92% sequence identity to this human sequence. Searches of the completed yeast and Caenorhabditis elegans genomes revealed no apparent homologies to the TFIIIA-intP sequence.
Figure 4.
Amino acid sequence alignment of the human and rat TFIIIA-intP. DNA sequences were obtained as described in Materials and Methods. Dark and light shaded boxes indicate amino acid sequence identities and similarities respectively between the human rat TFIIIA-intP sequences; dashes indicate amino acid deletions. The shared consensus sequence is given below the rat TFIIIA-intP sequence.
Interaction of TFIIIA-intP and TFIIIA recombinant proteins
Although an in vivo interaction between Xenopus TFIIIA and the proposed rat TFIIIA-intP is indicated by the positive β-galactosidase assay for clone 3 in Figure 3, it was necessary to demonstrate the interaction in vitro. In order to demonstrate an in vitro interaction between Xenopus TFIIIA and the human homolog, recombinant forms of both proteins were expressed, purified, and incubated together to allow potential interactions to occur. The binding reaction was subjected to immunoprecipitation using anti-TFIIIA-intP antibody followed by IgG selection with protein A/G resin. After selection, complexes were subjected to SDS–PAGE and western blotting with anti-Xenopus TFIIIA antibody (Fig. 5). Electrophoresed in lane 1 is the recombinant TFIIIA protein; two bands of equal intensity are observed, one at ∼40 and the other at ∼30K. The 40K band is the intact Xenopus TFIIIA protein containing the nine zinc fingers (30K) and the non-finger C-terminal tail (10K). The 30K band migrating below the 40K band is possibly the nine zinc-finger region since the C-terminal region of Xenopus TFIIIA is protease sensitive (41,42) and protease inhibitors were not used in the preparation of the recombinant protein. Lanes 2 and 3 exhibit the protein A/G selected complexes from TFIIIA binding reactions containing or lacking the TFIIIA-intP protein respectively. In the absence of the TFIIIA-intP (lane 3), very little recombinant TFIIIA protein is precipitated, demonstrating that complex selection is dependent on TFIIIA-intP. Markedly more TFIIIA (40K band) is selected in the presence of TFIIIA-intP (lane 2) demonstrating that a complex was formed between Xenopus TFIIIA and the human TFIIIA-intP. The affinity purified anti-TFIIIA-intP antibody does not immunoprecipitate TFIIIA in the absence of TFIIIA-intP (not shown). It is noted that very little of the 30K band was found in the TFIIIA–TFIIIA-intP complex (lane 2); this could be explained by lack of interaction between TFIIIA-intP and any TFIIIA lacking the C-terminal region.
Figure 5.

Immunoprecipitation of the TFIIIA–TFIIIA-intP complex. Preparation of recombinant Xenopus TFIIIA and human TFIIIA-intP, binding reaction conditions, immunoprecipitations and western blotting using horseradish peroxidase-conjugated secondary antibody were performed as described in Materials and Methods. Binding reaction components are indicated above lanes 1–3.
Interaction of TFIIIA and TFIIIA-intP from a rat liver extract
An experiment that would lend physiological significance to the interaction of TFIIIA and TFIIIA-intP observed with recombinant proteins would be to identify an interaction between the native proteins from a cell extract. One way to identify such an interaction would be to demonstrate co-chromatography of rat TFIIIA-intP and rat TFIIIA after several distinct chromatographic separations of the original cell extract. These separations should take advantage of disparate physical characteristics of the proteins—e.g., TFIIIA-intP is a 70K acidic protein and TFIIIA is a 38K basic protein. Such co-chromatography would be indicative of the presence of the complex in the cell extract and its maintenance during column fractionation. The separation protocol used in this experiment is depicted in Figure 6A. A rat liver cytosol was prepared and fractionated on phosphocellulose. Rat TFIIIA-intP was found in the flow-through (because of its acidic nature) when assayed with the anti-TFIIIA-intP antibody (Fig. 6B, lane 1). This flow-through was then fractionated on a DEAE 650 column where the rat TFIIIA-intP fraction eluted at 1 M KCl. This fraction was then subjected to gel filtration; the rat TFIIIA-intP peak was identified and fractionated on DEAE-5PW whose western blot profile is exhibited in Figure 6C (top panel). Significantly, this chromatographic profile is the same as the chromatographic profile for rat TFIIIA on this column as detected by the Xenopus TFIIIA antibody (bottom panel). These results indicate that rat TFIIIA and rat TFIIIA-intP proteins co-fractionated as a complex on this column since TFIIIA alone does not bind to DEAE (not shown). The result is also consistent with the complex being present in the original rat cell extract and with its survival through phosphocellulose, DEAE (1 M KCl) and gel filtration chromatography.
Figure 6.
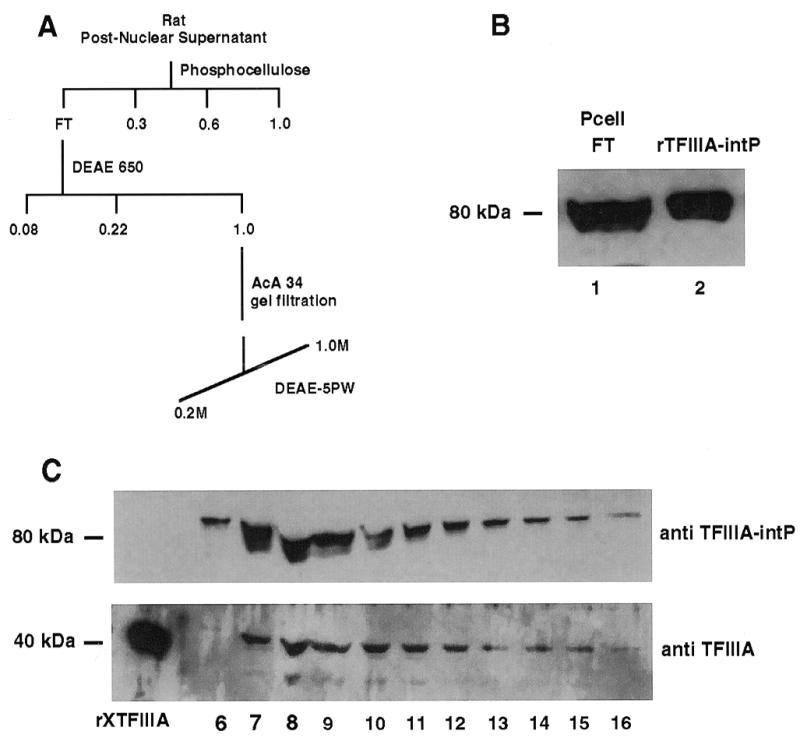
Co-chromatography of TFIIIA and TFIIIA-intP from a rat liver extract. Rat liver cell extracts, chromatographic separations and western blotting using horseradish peroxidase-conjugated secondary antibody were performed as described in Materials and Methods. (A) Chromatography protocol; (B) western blot of phosphocellulose flow-through (lane 1) and recombinant human TFIIIA-intP (lane 2) probed with TFIIIA-intP antibody; (C), western blot of DEAE-5PW column fractions probed with TFIIIA-intP antibody (top) or TFIIIA antibody (bottom).
Stimulation of 5S gene transcription by recombinant TFIIIA-intP
Evidence is presented in the previous experiments indicating that a physical interaction can take place between TFIIIA and TFIIIA-intP. In order to demonstrate whether there is any functional significance to this interaction, in vitro transcription assays (using the 0.8 M KCl phosphocellulose fraction from a HeLa cell nuclear extract) were performed to examine for effects of TFIIIA-intP on 5S RNA gene or adenovirus VA RNA gene transcription by RNA polymerase III (Fig. 7). Lanes 1 and 2 in Figure 7A are 5S gene-containing transcription reactions minus or plus recombinant Xenopus TFIIIA. A 5S RNA signal is observed in the plus recombinant TFIIIA lane and little if any signal is observed in the minus lane. Longer autoradiogram exposures do reveal a weaker 5S RNA band in the minus TFIIIA lane (not shown) indicating that some human TFIIIA is present in this 0.8 M KCl fraction; human TFIIIA was previously observed in this KCl phosphocellulose fraction by others (14). Importantly, addition of the TFIIIA-intP to the TFIIIA-dependent reaction (lane 2) markedly stimulated 5S RNA gene transcription (lane 3). This stimulation by TFIIIA-intP occurs in the absence of exogenously added TFIIIA as is exhibited in lanes 4–6. Also exhibited in these lanes is the diminution of the 5S RNA signal upon increasing amounts of the TFIIIA-intP. Such diminution could be caused by a TFIIIA-intP squelching phenomenon with the limiting amounts of endogenous TFIIIA present in the 0.8 M KCl fraction. Any potential effect of TFIIIA-intP on adenovirus VA transcription is exhibited in Figure 7B. Lanes 1 and 2 exhibit VA RNA transcription in this phosphocellulose column fraction in the absence and presence of the VA RNA gene; lanes 3 and 4 exhibit the transcription product in the presence of the VA gene and recombinant TFIIIA. The lack of signal in the no. 4 TFIIIA lane is due to loss of the ethanol precipitate. Lanes 5 and 6 exhibit the transcription products in the presence of the recombinant human TFIIIA-intP and lanes 7 and 8 exhibit the products in the presence of both recombinant TFIIIA and TFIIIA-intP. Little overall change in signal is observed in these lanes indicating that the TFIIIA-intP does not stimulate VA transcription.
DISCUSSION
Although a great deal of structural and nucleic acid binding data are available on Xenopus TFIIIA, little is known about how it activates 5S gene transcription and even less is known about proteins with which it interacts. In this study, a yeast two-hybrid screen was performed using the C-terminal portion of Xenopus TFIIIA as an attractor and a rat cDNA library as a source of potential partners. One rat cDNA clone was chosen for further study because its protein product in the yeast assay did interact with the Xenopus TFIIIA attractor but did not interact with the C-terminal portion of yeast TFIIIA or other non-TFIIIA proteins. The complete sequence of the human homolog for this rat protein was assembled from ESTs and found to code for a 70.6 kDa protein (Fig. 4). As assayed by immunoprecipitation, this recombinant human protein (termed TFIIIA-intP) was able to form a complex in vitro with recombinant Xenopus TFIIIA (Fig. 5). Rat TFIIIA and rat TFIIIA-intP co-chromatographed on DEAE-5PW after three distinct chromatographic separations of a rat cell extract. This observation is consistent with these two proteins being present in a complex on DEAE-5PW as well as in the original cell extract.
With respect to possible structure(s) in TFIIIA-intP that may interact with the C-terminal portion of TFIIIA, the 70.6K human protein has several motif-rich regions of potential use for protein–protein interactions. The N-terminal quarter of the protein has potential leucine zipper regions and acidic amino acid-rich regions. The large middle portion of the protein also has several leucine-rich zipper regions. The C-terminal portion of TFIIIA-intP has a potential Cys2HisCys zinc finger, a variant where a cysteine replaces the second histidine in the Cys2His2 zinc coordination sphere (43). Because the original rat clone identified in the two-hybrid screen lacked both these N- and C-terminal portions (Fig. 4), the TFIIIA-interacting region is likely within the central leucine-rich central domain. A hydrophobic nature for the TFIIIA–TFIIIA-intP interaction is suggested by the observation that the complex of the two proteins (as evidenced by co-chromatography) is stable at high salt (Fig. 6).
Recombinant human TFIIIA-intP stimulated TFIIIA-dependent 5S gene transcription in vitro but not TFIIIA-independent human adenovirus VA gene transcription (Fig. 7), a result consistent with TFIIIA specificity in the TFIIIA-intP reaction. What possible functions could TFIIIA-intP have in 5S gene transcription stimulation? Although the protein contains a potential zinc finger in its C-terminal region, we were not able to demonstrate DNA binding by this protein to the 5S gene or non-specific plasmid DNA (not shown). The potential zinc finger in its C-terminal region may have a role in 5S RNA binding like some of the TFIIIA fingers and we have yet to explore that possibility. If its does bind 5S RNA, it does not appear to do this in a protective manner as increasing amounts of the protein in vitro reduce the amount of 5S RNA product (Fig. 7). This could be due to squelching of a limiting amount of endogenous TFIIIA in the fractionated nuclear extract.
TFIIIA-intP could have a non-DNA binding role in augmenting TFIIIA interaction to the IIIC complex and/or IIIB and/or pol III. Such a role might be analogous to the co-activators of pol II transcription. As a modulator, it could have a role in stimulating a basal rate of 5S gene transcription depending on changing cellular requirements. TFIIIA-intP could also have a role in altering the conformation of TFIIIA from an inactive to an active state. A previous study indicated that the protein we identified as a TFIIIA-intP was found to interact with the adenovirus E3 14.7 kDa protein in a yeast two-hybrid screen and was termed FIP-2 (44). As assayed by fluorescence microscopy, FIP-2 was found in the cell nucleus and the authors suggested that this protein may be a transcription factor because of the numerous leucine zipper-like motifs. Adenovirus is known to enhance RNA polymerase III transcription including 5S RNA synthesis in human cells (45). There may be a mechanistic connection between adenovirus infection and potential FIP-2/TFIIIA-intP stimulation of 5S gene transcription.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Michael Conrad, Anna Dominguez, James Hocker, Takumi Kamura, Martha Ogilvie and John Wickham for technical assistance. This work was supported by the Oklahoma Medical Research Foundation (M.D), National Institutes of Health (B.R.), Howard Hughes Medical Institute (J.W.C., Associate Investigator), National Institutes of Health (R.C.C.) and Department of Defense (J.H.).
REFERENCES
- 1. Segall J., Matsui,T. and Roeder,R.G. (1980) J. Biol. Chem., 255, 11986–11991. [PubMed] [Google Scholar]
- 2. Shastry B.S., Ng,S.Y. and Roeder,R.G. (1982) J. Biol. Chem., 257, 12979–12986. [PubMed] [Google Scholar]
- 3. Lassar A.B., Martin,P.L. and Roeder,R.G. (1983) Science, 222, 740–748. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z., Luo,T. and Roeder,R.G. (1997) Genes Dev., 11, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 6. Yoshinaga S.K., Boulanger,P.A. and Berk,A.J. (1987) Proc. Natl Acad. Sci. USA, 84, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fradkin L.G., Yoshinaga,S.K., Berk,A.J. and Dasgupta,A. (1989) Mol. Cell. Biol., 9, 4941–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. L’Etoile N.D., Fahnestock,M.L., Shen,Y., Aebersold,R. and Berk,A.J. (1994) Proc. Natl Acad. Sci. USA, 91, 1652–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lagna G., Kovelman,R., Suegawa,J. and Roeder,R.G. (1994) Mol. Cell. Biol., 14, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sinn E., Wang,Z., Kovelman,R. and Roeder,R.G. (1995) Genes Dev., 9, 675–685. [DOI] [PubMed] [Google Scholar]
- 11. Wang Z. and Roeder,R.G. (1995) Proc. Natl Acad. Sci. USA, 92, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaussivert N., Conesa,C., Shaaban,S. and Sentenac,A. (1995) J. Biol. Chem., 270, 15353–15358. [DOI] [PubMed] [Google Scholar]
- 14. Moorefield B. and Roeder,R.G. (1994) J. Biol. Chem., 269, 20857–20865. [PubMed] [Google Scholar]
- 15. Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Cell, 39, 479–489. [DOI] [PubMed] [Google Scholar]
- 16. Hanas J.S., Hazuda,D.J., Bogenhagen,D.F., Wu,F.W. and Wu,C.W. (1983) J. Biol. Chem., 258, 14120–14125. [PubMed] [Google Scholar]
- 17. Miller J., McLachlan,A.D. and Klug,A. (1985) EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown R.S., Sander,C. and Argos,P. (1985) FEBS Lett., 186, 271–274. [DOI] [PubMed] [Google Scholar]
- 19. Pieler T., Hamm,J. and Roeder,R.G. (1987) Cell, 48, 91–100. [DOI] [PubMed] [Google Scholar]
- 20. Hanas J.S., Littell,R.M., Duke,A.L. and Zebrowski,R. (1989) Nucleic Acids Res., 17, 9861–9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayes J.J. and Clemens,K.R. (1992) Biochemistry, 31, 11600–11605. [DOI] [PubMed] [Google Scholar]
- 22. Clemens K.R., Wolf,V., McBryant,S.J., Zhang,P., Liao,X., Wright,P.E. and Gottesfeld,J.M. (1993) Science, 260, 530–533. [DOI] [PubMed] [Google Scholar]
- 23. Theunissen O., Rudt,F., Guddat,U., Mentzel,H. and Peiler,T. (1992) Cell, 71, 679–690. [DOI] [PubMed] [Google Scholar]
- 24. Nolte R.T., Conlin,R.M., Harrison,S.C. and Brown,R.S. (1998) Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vrana K.E., Churchill,M.E.A., Tullius,T.D. and Brown,D.D. (1989) Mol. Cell. Biol., 8, 1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaskins C.J., Smith,J.F., Ogilvie,M.K. and Hanas,J.S. (1992) Gene, 120, 197–206. [DOI] [PubMed] [Google Scholar]
- 27. Mao X. and Darby,M.K. (1994) Mol. Cell. Biol., 13, 7496–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ogilvie M.K. and Hanas,J.S. (1997) Gene, 203, 103–112. [DOI] [PubMed] [Google Scholar]
- 29. Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 30. Woychik N.A. and Young,R.A. (1992) Proc. Natl Acad. Sci. USA, 89, 3999–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Milne C.A. and Segall,J. (1993) J. Biol. Chem., 268, 11364–11371. [PubMed] [Google Scholar]
- 32. Rowland O. and Segall,J. (1998) Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chien C.T., Bartel,P.L., Sternglanz,R. and Fields,S. (1991) Proc. Natl Acad. Sci. USA, 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- 35. Schiestl R.H. and Gietz,R.D. (1989) Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- 36. Brondyk W.H., Mckiernan,C.J., Fortner,K.A., Stabila,P., Holz,R.W. and Macara,I.G. (1995) Mol. Cell. Biol., 15, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bodenteich A., Chissoe,S., Wang,Y.F. and Roe,B.A. (1993) In Venter,J.C. (ed.), Automated DNA Sequencing and Analysis Techniques, Academic Press, London, pp. 42–50.
- 38. Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 39. Conaway R.C., Reines,D., Garrett,K.P., Powell,W. and Conaway,J.W. (1996) Methods Enzymol., 273, 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu G.J. and Cannon,R.E. (1986) J. Biol. Chem., 261, 12633–12642. [PubMed] [Google Scholar]
- 41. Smith D.R., Jackson,I.J. and Brown,D.D. (1984) Cell, 37, 645–652. [DOI] [PubMed] [Google Scholar]
- 42. Hanas J.S., Duke,A.L. and Gaskins,C. (1989) Biochemistry, 28, 4083–4088. [DOI] [PubMed] [Google Scholar]
- 43. Ogilvie M.K., Smith,J.F. and Hanas,J.S. (1993) Nucleic Acids Res., 21, 4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y., Kang,J. and Horwitz,M.S. (1998) Mol. Cell. Biol., 18, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoeffler W.K. and Roeder,R.G. (1985) Cell, 41, 955–963. [DOI] [PubMed] [Google Scholar]