Abstract
SARS-CoV-2 is still wreaking havoc all over the world with surging morbidity and high mortality. The main protease (Mpro) is essential in the replication of SARS-CoV-2, enabling itself an active target for antiviral development. Herein, we reported the design and synthesis of a new class of peptidomimetics-constrained α, γ-AApeptides, based on which a series of aldehyde and ketoamide inhibitors of the Mpro of SARS-CoV-2 were prepared. The lead compounds showed excellent inhibitory activity in the FRET-based Mpro enzymatic assay not only for the Mpro of SARS-CoV-2 but also for SARS-CoV and MERS-CoV, along with HCoVs like HCoV-OC43, HCoV-229E, HCoV-NL63 and HKU1. The X-ray crystallographic results demonstrated that our compounds form a covalent bond with the catalytic Cys145. They also demonstrated effective antiviral activity against live SARS-CoV-2. Overall, the results suggest that α, γ-AApeptide could be a promising molecular scaffold in designing novel Mpro inhibitors of SARS-CoV-2 and other coronaviruses.
Keywords: Broad-spectrum activity, COVID-19, Mpro inhibitor, SARS-CoV-2, X-Ray structure
Graphical Abstract
A series of constrained α, γ-AApeptides served as a novel scaffold to design aldehydes and ketoamides to inhibit the Mpro of the SARS-CoV-2. They show excellent inhibitory activity against SARS-CoV-2 and broad-spectrum activity against different other HCoVs. The X-ray structure of Mpro with inhibitors demonstrated the mechanism of covalent inhibition of Mpro.
Introduction
The coronavirus disease (COVID-19) pandemic has caused 6.7 million deaths among 667 million confirmed diseases until January 07, 2023. The causative agent of COVID-19 is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which poses a great challenge to public health and the global economy.[1] Due to the scarcity of antiviral agents and the emergence of variants of concern and variants of interest,[2] there is still a pressing need for developing new antiviral drugs to combat SARS-CoV-2. Belonging to the β coronavirus genus, SARS-CoV-2 is a single-stranded positive-sense RNA virus.[3] Two known proteolytic enzymes are involved in the replication of SARS-CoV-2, including main protease and papain-like protease. Two viral polyproteins of SARS-CoV-2, pp1a and pp1ab, play a crucial role in the life cycle of SARS-CoV-2, as their proteolytic cleavage products of 16 non-structural proteins (nsps) are used for the replication of subgenomic RNAs,[4] In the processing of these two polyproteins, the main protease, also called 3-chymotrypsin-like protease (3CLpro), cleaves the polyproteins pp1a and pp1ab, making itself one of the most essential viral enzymes among the coronaviruses. It is currently known that Mpro is a cysteine protease and can digest the polyprotein at more than 11 conserved sites including the Leu-Gln↓ (Ser, Ala, Gly) sequence to achieve the coronavirus replication process,[5] As such, Mpro is an active target for the development of inhibitors of SARS-CoV-2. In addition to SARS-CoV-2, six coronaviruses (CoVs) are also known to infect humans: HCoV-229E, HCoV-OC43, SARS-CoV, HCoV-NL63, HKU1, and MERS-CoV.[6] It is found that Mpro of the SARS-CoV-2 shows high sequence similarities among the CoVs group, suggesting inhibitors for Mpro of the SARS-CoV-2 could become broad-spectrum inhibitors to combat a range of coronaviruses in the future.[7]
There has been great progress in developing Mpro inhibitors,[8] most of which are covalent inhibitors, although non-covalent inhibitors were recently reported as well.[9] Among the tremendous achievements, PAXLOVID™, which is a combination of Mpro inhibitor nirmatrelvir and the metabolic enhancer ritonavir, received FDA approval.[8b] However, resistance against nirmatrelvir has been reported.[10] Therefore, developing new inhibitors with novel scaffolds is a viable strategy to combat SARS-CoV-2 and other coronaviruses in the long term.
γ-AApeptide (Figure 1), oligomers of γ-substituted-N-acylated-N-aminoethyl amino acids,[11] is a class of sequence-specific peptidomimetics with functional diversity, remarkable enzymatic stability, and biocompatibility.[12] Whereas α, γ-AApeptides (Figure 1) have not been reported by us previously, we speculated that constrained α, γ-AApeptide scaffold bearing either a 2-pyrrolidone and 2-piperidinone (Figure 1) may closely fit in the pocket of active sits of Mpro of SARS-CoV-2. As such, we envisioned a class of Mpro inhibitors could be developed based on this molecular scaffold. Herein, we reported the rational design of covalent Mpro inhibitors, including aldehydes and ketoamides derived from the constrained α, γ-AApeptide backbone, to target SARS-CoV-2. Both α, γ-AApeptide-based aldehydes and ketoamides showed excellent activity against Mpro of SARS-CoV-2 and exhibited broad-spectrum activity to other coronaviruses in the enzymatic assay. One of the most potent compounds M-1-6 demonstrated a Mpro inhibitory activity of 0.08 μM (IC50). As anticipated, the X-ray structures of compounds with the Mpro of SARS-CoV-2 revealed that the warheads of our compounds form a covalent bond with Cys145, a mechanism of inhibition similar to the reported Mpro inhibitors. The lead compound was subsequently found to effectively inhibit SARS-CoV-2 live virus infection in cell culture. The findings suggested that the molecular scaffold of α, γ-AApeptide is a promising platform to develop novel inhibitors for Mpro of SARS-CoV-2 and other coronaviruses.
Figure 1.
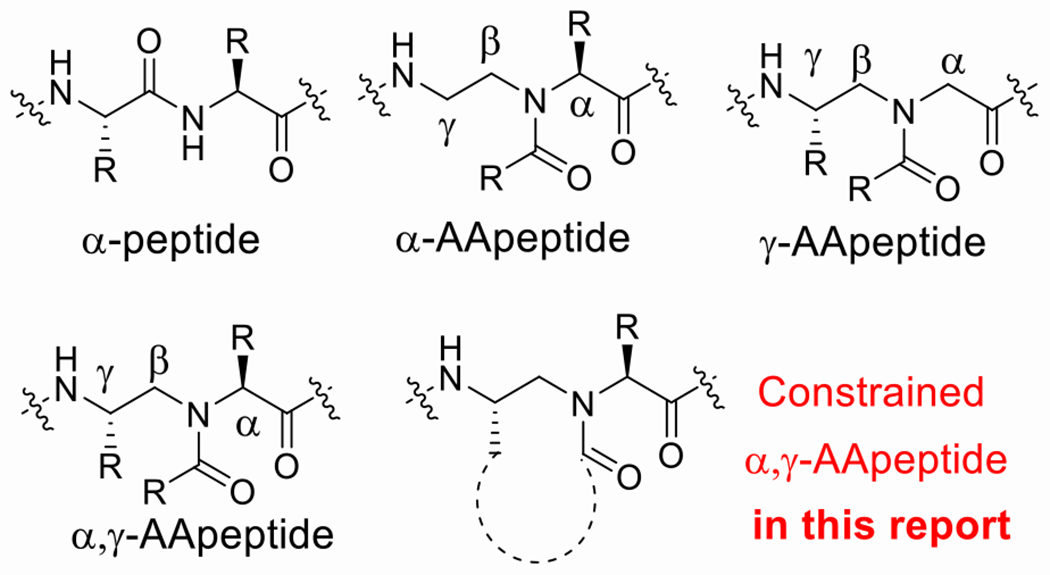
Structures of α-peptide, α-AApeptide, γ-AApeptide, α, γ-AApeptide, constrained α, γ-AApeptide.
Results and Discussion
Structure insight of Mpro.
The X-ray structure of the Mpro of SARS-CoV-2 reveals that Mpro is a homodimer consisting of two protomers (known as “A” and “B”) which orient themselves at the right angle. Each protomer comprises three domains, antiparallel β-barrel domain I (residues 8–101) and domain II (residues 102–184), as well as Domain III (residues 201–303) responsible for the dimerization of Mpro.[13] There are four pockets (S1’, S1, S2, S4) in the active sites of the Mpro enzyme, which can accommodate four groups of peptidomimetic inhibitors (P1′, P1, P2, and P3, respectively) (Figure 2). The S1’ pocket is a catalytic site that crucially relies on Cys145 and His41 residues (Figure 2A). His41 can provide the required pH condition for the thiol group (−SH) in Cys145 to be activated to enable a nucleophilic attack on the substrate.[14] Interestingly, the Gln residue is always preferred at the P1 position of the substrate (Figure 2B). This makes Mpro as an ideal viral target to design inhibitors with minimal side effects.[8a, 15]
Figure 2.
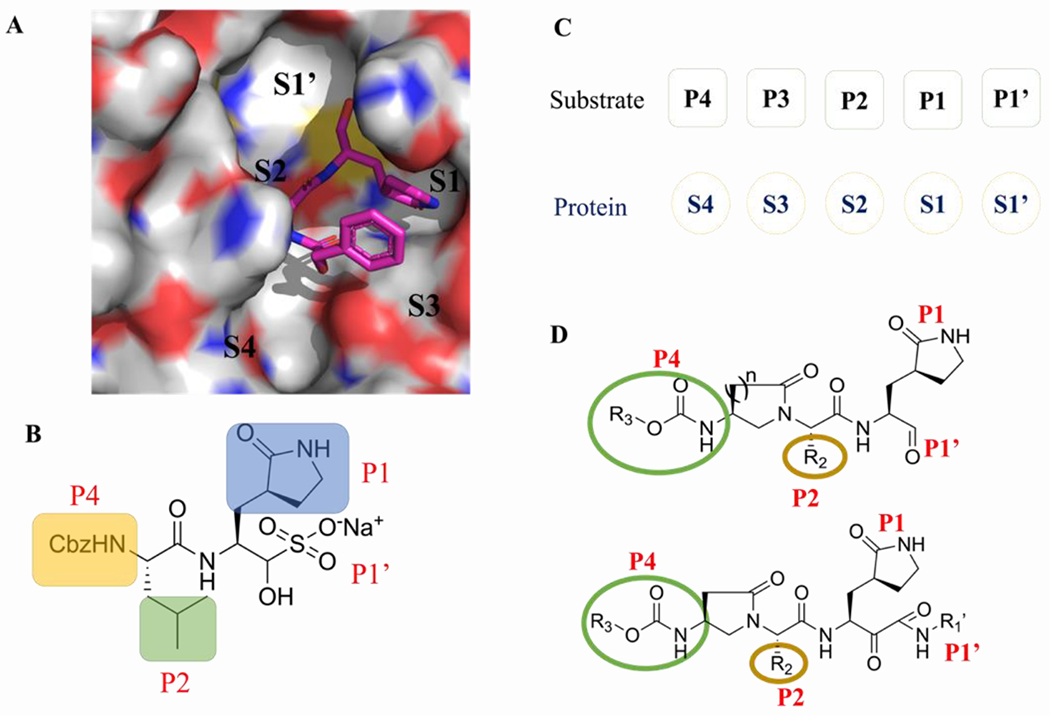
(A) Surface representation of substrate-binding pocket of SARS-CoV-2 Mpro and Molecular recognition of GC-376 toward Mpro of SARS-CoV-2.[5c] (B) Chemical structure of GC-376. (C) The substrate binding sites of the protein (S1’-S4) and substrate residues (P1’-P4). (D) The design strategy of SARS-CoV-2 Mpro inhibitors based on the constrained α, γ-AApeptide scaffolds.
Design of the aldehydes and ketoamides.
Based on the binding mode of reported Mpro inhibitors (such as GC-376) of SARS-CoV-2 (Figure 2), we hypothesized that an α, γ-AApeptide-based scaffold could serve as the substrate mimic (Figure 2D). The intramolecular constraints were expected to increase the molecular rigidity. Conjugation with active-site-reacting warheads such as aldehydes and ketoamides could lead to new classes of Mpro inhibitors. To this end, we started the synthesis of both classes of molecules.
Synthesis of the aldehydes and ketoamides.
The synthesis of the constrained α, γ-AApeptide based aldehydes and ketoamides could be accomplished with commercially available Cbz-protected glutamic acid and aspartic acid 1 (Scheme 1). Firstly, the carboxylic acid group of 1 was reduced to the alcohol to provide intermediate 2, which was subjected to OTs esterification to afford 3. The following SN2 reaction with the methyl ester of an amino acid led to the formation of the secondary amine 4, which was converted to 5 upon intramolecular self-cyclization. Next, the hydrogenation of 5 gave rise to 6, which was subsequently converted to isocyanates 7 and then 8 bearing different N-terminal functional groups. The removal of methyl ester led to compound 9, which reacted with commercially available pyrrolidone to give 10. The following reduction and Dess-Marten oxidation gave rise to the desired aldehydes M-1. To obtain ketoamide inhibitors, M-1 was subject to a nucleophilic addition with isocyanide under the acidic condition to give the intermediate 12, which was followed by the removal of the acetyl group and oxidation to give rise to the desired α-ketoamides M-2.
Scheme 1.
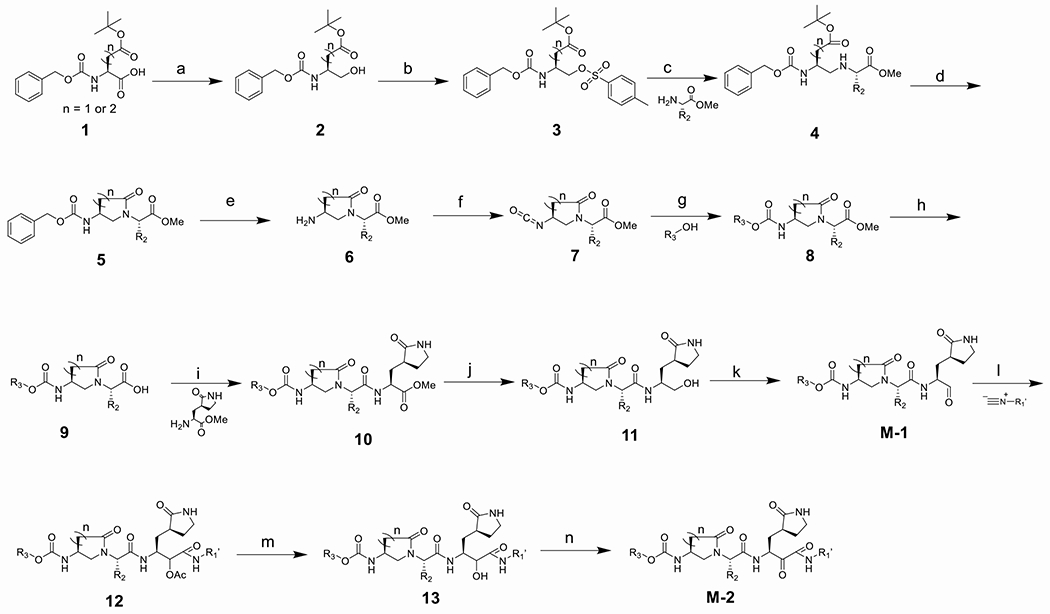
Synthesis of α, γ-AApeptide based aldehydes and ketoamides. Reaction conditions: (a) isobutyl chloroformate, NMM, NaBH4, THF; (b) TosCl, TEA, DCM; (c) K2CO3, NaI, ACN; (d) TFA, DCM, HOBT, DIC, DIPEA, DMF; (e) Pd/C, MeOH; (f) trichloromethyl chloroformate, dioxane; (g) R3-OH, TEA, ACN; (h) LiOH, THF/H2O; (i) DIPEA, HATU, DMF; (j) NaBH4, MeOH; (k) Dess-Martin periodinane (DMP), NaHCO3, DCM; (l) isocyanide, AcOH, DCM; (m) LiOH, THF/H2O; (n) DMP, NaHCO3, DCM
The activity of the aldehydes and ketoamides toward Mpro.
We aimed to develop a new series of aldehydes and ketoamides as SARS-CoV-2 inhibitors based on the constrained α, γ-AApeptide scaffold. Both aldehydes and ketoamides are known to form covalent inhibitors by forming a covalent bond with catalytic Cys145 of Mpro of SARS-CoV-2.[8d, 15] It is generally observed that the aldehyde inhibitors exhibit more potent inhibitory activity than α-ketoamides, however, ketoamides seemed to demonstrate better activity in vivo and clinical applications. We, therefore, decided to explore the activity of both classes of inhibitors. In the reported inhibitors for the Mpro of SARS-CoV and SARS-CoV-2,[8, 14a] it is recognized that the (S)-γ-lactam ring in the P1 position is preferred because of its suitable occupation in the S1 site.[15] As such, we put the lactam ring in the P1 position. As to the P2 position, the side chains of Leu, Phe, as well as other side chains such as Cyclohexyl, Cyclopropyl and Fluorobenzyl side chains were introduced to fit the hydrophobic pocket S2 in Mpro. The activity of the compounds was determined with FRET-based Mpro enzymatic assay.[16] As shown in Table 1, all designed molecules show moderate-to-excellent activity in inhibiting the enzymatic activity of SARS-CoV-2 Mpro. The most potent aldehyde inhibitors are M-1-3 and M-1-6 with IC50 values of 0.08 μM, which is comparable to GC376 (IC50 = 0.03 μM), a previously reported well-known inhibitor.[5c] The results reveal that the compounds with P2 Leu and Cyclopropyl residues (M-1-3, M-1-6) are more potent than those compounds with Cyclohexyl and Fluorobenzyl residues (M-1-4, M-15) against Mpro of SARS-CoV-2. This is similar to the observation for the ketoamides, as the compounds M-2-5 and M-2-8 with P2 Leu and Cyclopropyl residues are also more potent (IC50 = 3.58 μM and 2.14 μM, respectively) than compounds M-2-6 and M-2-7 (with IC50 values of 4.43 μM and 3.99 μM) bearing Cyclohexyl and Fluorobenzyl residues in the P2 position. At the P4 substitution, different hydrophobic groups were introduced to occupy the S4 pocket. The results reveal that the compound M-1-3 with the Cbz group shows slightly better activity than compounds M-1-7 (0.19 μM) and M-1-8 (0.29 μM) which have di-Fluorocyclohexyl and 3,5-di-Fluorobenzyl group at the P4 position. Interestingly, in the ketoamides, the results seem to be different. The compounds M-2-9 and M-2-10 with IC50 values of 1.92 μM and 2.76 μM, respectively, are more potent than M-2-5 (3.58 μM), suggesting that the choice of groups at the P4 position could be related to the groups at the other positions. In addition, the 2-pyrrolidone and 2-piperidinone, which are designed to bridge the P2-P4 position also played an important role in the activity. For instance, compound M-1-3 is more potent than M-1-1 (0.51 μM), suggesting 2-piperidinone is more suitable for the development of inhibitors. Finally, it is noted that all the compounds are highly selective as they did not exhibit any cytotoxicity under the tested condition.
Table 1.
Inhibitory activity of aldehydes and ketoamides against Mpro.
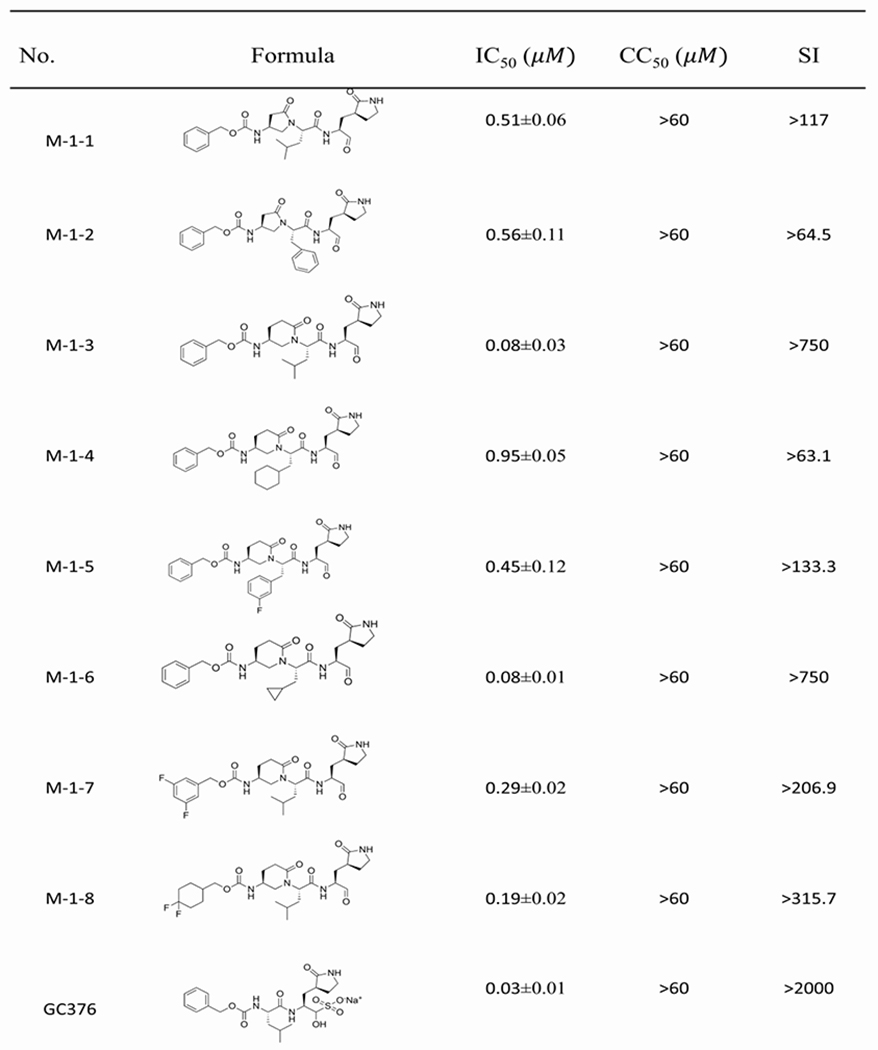
|
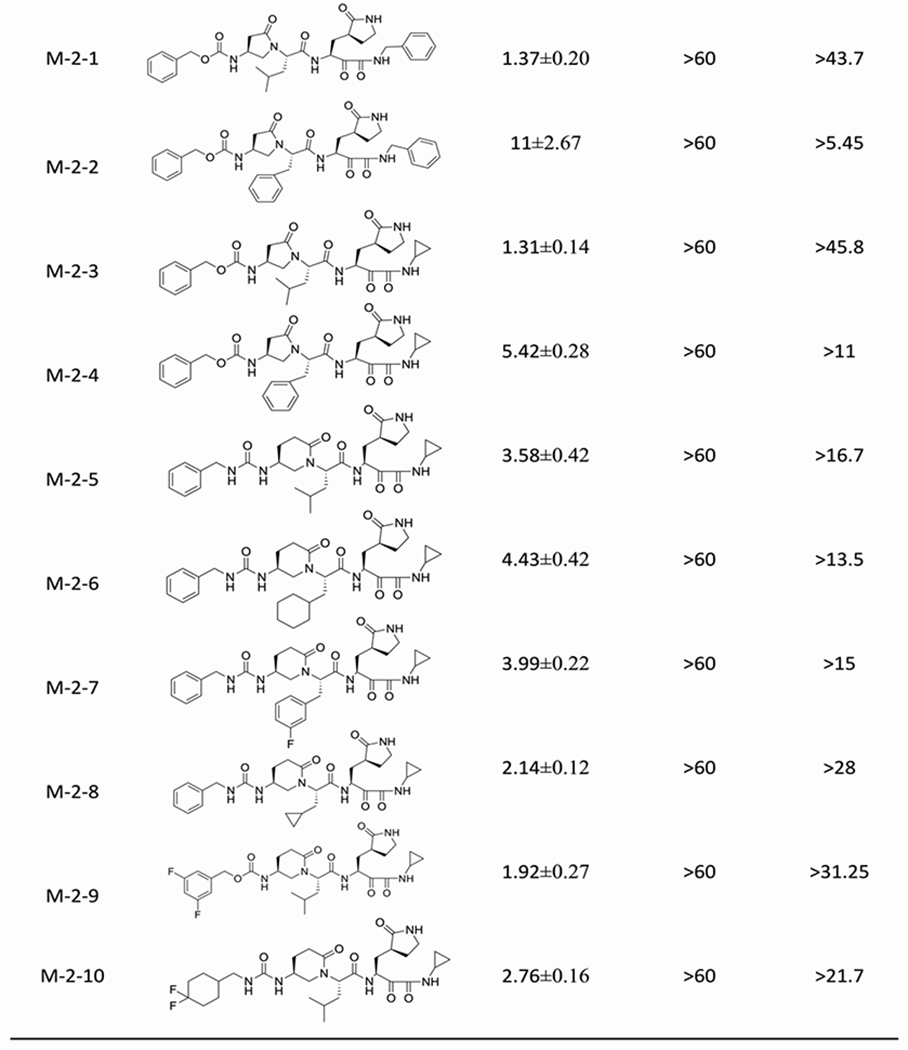
|
Broad-spectrum activity of the aldehydes and ketoamides.
Based on the SARS-CoV-2 Mpro inhibitory activity and the cell cytotoxicity results, as well as the fact that all the Mpros of different coronavirus possess high sequence similarities, we speculated that these constrained α, γ-AApeptide-based compounds should demonstrate broad-spectrum inhibitory activity toward other coronaviruses. To test our hypothesis, we chose the lead compounds (M-1-3, M-1-4 , M-1-6, M-2-3 and M-2-8) and tested their broad-spectrum activity against Mpros of several members of the coronavirus family including SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E, and HKU1 (Table 2).[17] Among them, β-HoVs, including SARS-CoV and MERS-CoV. SARS-CoV, in which Mpro show the most similarities with SARS-CoV-2, could be effectively inhibited by compounds M-1-3, M-1-4 and M-1-6 with IC50 values of 0.25 μM, 0.16 μM and 0.73 μM, respectively (Table 2). Compared to the aldehydes, two ketoamides, M-2-3 and M-2-8 also inhibited Mpro of SARS-CoV with IC50 values of 1.31 μM and 3.88 μM. For MERS-CoV, due to less sequence similarity with SARS-CoV-2 than SARS-CoV, the IC50 values of all five compounds to inhibit Mpro varied from 0.41 μM to > 20 μM. Similarly, all the compounds showed good inhibitory activity against α-HoVs, including HCoV-OC43, HCoV-NL63, HCoV-229E and HUK1. Collectively, all selected five compounds could largely inhibited Mpro of both for α-HCoVs and β-HCoVs, suggesting that the compounds we designed are broad-spectrum inhibitors against different coronaviruses.
Table 2.
Broad-spectrum inhibitory activity of lead compounds against a panel of human CoVs. (IC50, μM)
| Bota coronavirus | Alpha coronavirus | ||||||
|---|---|---|---|---|---|---|---|
| No | SARS-CoV-2 IC50 (μM) | SARS-CoV IC50 (μM) | MERS-CoV IC50 (μM) | HCoV-OC43 IC50 (μM) | HKU1 IC50 (μM) | HCoV-229E IC50 (μM) | HCoV-NL63 IC50 (μM) |
| M-1-3 | 0.08±0.01 | 0.16±0.01 | 0.41±0.05 | 0.71±0.02 | 0.033±0.01 | 0.062±0.01 | 0.89±0.01 |
| M-1-4 | 0.95±0.04 | 0.73±0.10 | >20 | 3.67±0.63 | 1.21±0.20 | 0.61±0.09 | 9.58±1.79 |
| M-1-6 | 0.08±0.01 | 0.25±0.02 | 0.85±0.21 | 0.18±0.01 | 0.12±0.02 | 0.18±0.02 | 0.48±0.06 |
| M-2-3 | 1.31±0.10 | 2.26±0.39 | 8.02±2.07 | 0.12±0.01 | 0.39±0.03 | 0.69±0.08 | 0.83±0.10 |
| M-2-8 | 2.14±0.09 | 3.88±0.67 | >20 | 2.60±0.45 | 2.23±0.46 | 5.67±0.49 | 10.84±2.21 |
| GC376 | 0.03±0.01 | 0.079±0.01 | 0.11±0.02 | 0.011±0.01 | 0.012±0.03 | 0.069±002 | 0.12±0.02 |
Antiviral activity of SARS-CoV-2 Mpro inhibitors and cellular protease inhibitory activity in the Flip-GFP Mpro assay.
All the inhibitors that showed good enzymatic activity in the FRET assay were further tested for cytotoxicity with Vero E6 cells. All the compounds including aldehydes and ketoamides were well tolerant in the Vero E6 cells with their CC50 values greater than 60 μM (data not shown). Previous literature reported a poor correlation between FRET enzyme activity with antiviral activity because of some problems such as cell permeability, off-targeted binding, and so on.[18] Therefore, we used another cell-based assay called Flip-GFP assay (Figure 3) which can remove the effect of the inhibitor’s cytotoxicity and cell permeability. In the Flip-GFP assay, two fragments, the β10–11 fragment of GFP protein was separated from the β1–9 template; while the β10 strand and β11 strand were linked with Mpro cleavage sequences (AVLQ↓SGFR) and were locked in inactive orientation with coiled-coil E5/K5 sequence. Upon protease cleavage, the β11 strand can be freed from restrain and flipped into the opposite direction as the β10 strand and fit into the β1–9 template and generate active GFP protein. This reporter plasmid also included mCherry red fluorescence protein as a transfection control. When 293T cells were co-transfected with the Flip-GFP reporter plasmid and Mpro protease expressing plasmid, the normalized ratio of green fluorescence signal over red fluorescence signal is proportional to the enzymatic activity. [13b, 16] We used GC-376 as the positive control, with the calculated EC50 value of 1.66 μM, which is close to the previous reported value. [5c] Among all the inhibitors that we screened, compound M-1-4 showed the best activity, giving the EC50 value of 18 μM. Another two compounds, M-1-5 and M-1-7 were less active with EC50 values of 49.57 μM and 27.30 μM, respectively. Surprisingly, compounds M-1-3 and M-1-6, which show the best activity in the FRET enzymatic assay, do not display observable activity (EC50> 60 μM) in this assay. To further evaluate these compounds, we chose compounds M-1-3, M-1-4 and M-1-6 to do the cellular antiviral activity test of SARS-CoV-2 (USA-WA1/2020) both in Vero E6 and Caco2-hACE2 cells. As we anticipated, compound M-1-4 showed some activity in the antiviral test with an EC50 value of 19 μM in Caco2-hACE2 cells (Figure 4), which is comparable to EC50 (2.9 μM) of GC376. [9b] Compound M-1-6, which did not inhibit the Mpro of SARS-CoV-2 in the Flip-GFP assay, showed weak antiviral activity. The EC50 value is about 38.25 μM. However, compound M-1-3 is not active in both Vero E6 and Caco2-hACE2 cells, which may be due to poor cell permeability. Overall, compound M-1-4 shows promising antivirus activity in the antiviral activity test.
Figure 3.
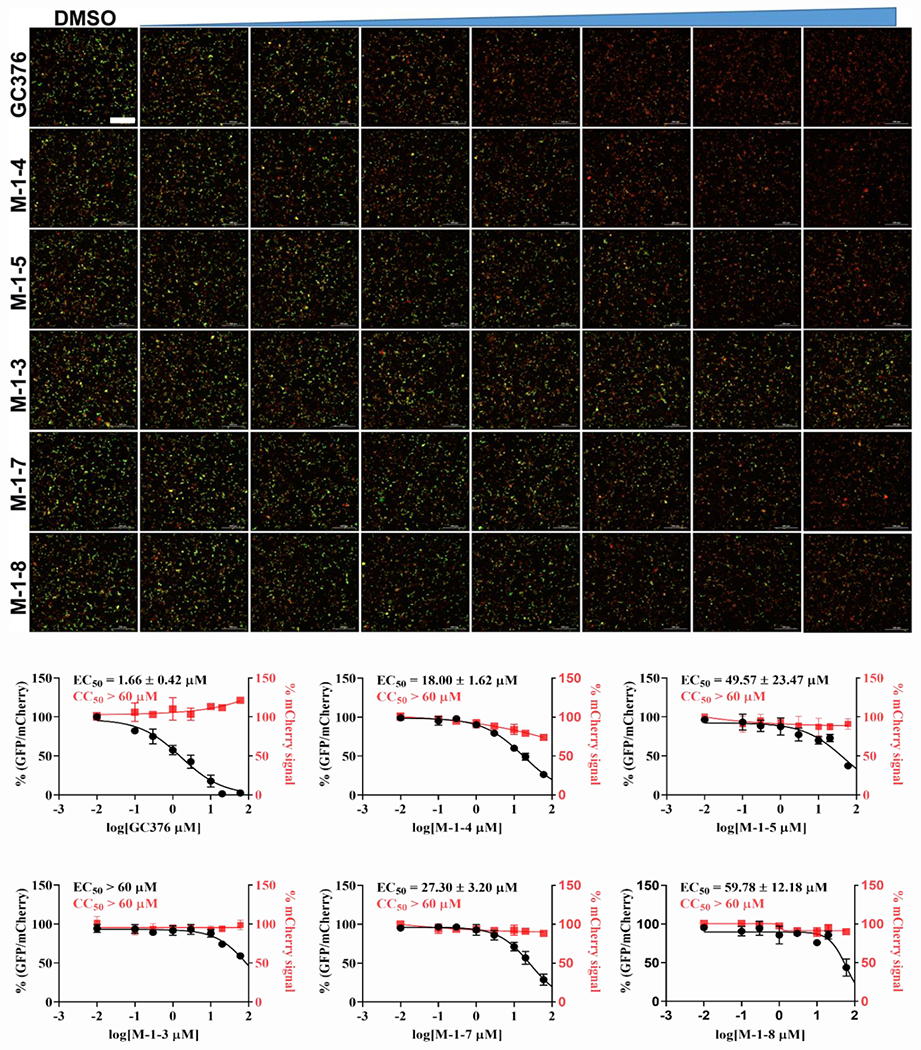
Flip-GFP assay characterization of the inhibition of the cellular enzymatic activity of SARS-CoV-2 Mpro by the four lead compounds (Scale bar is 300 μm).
Figure 4.
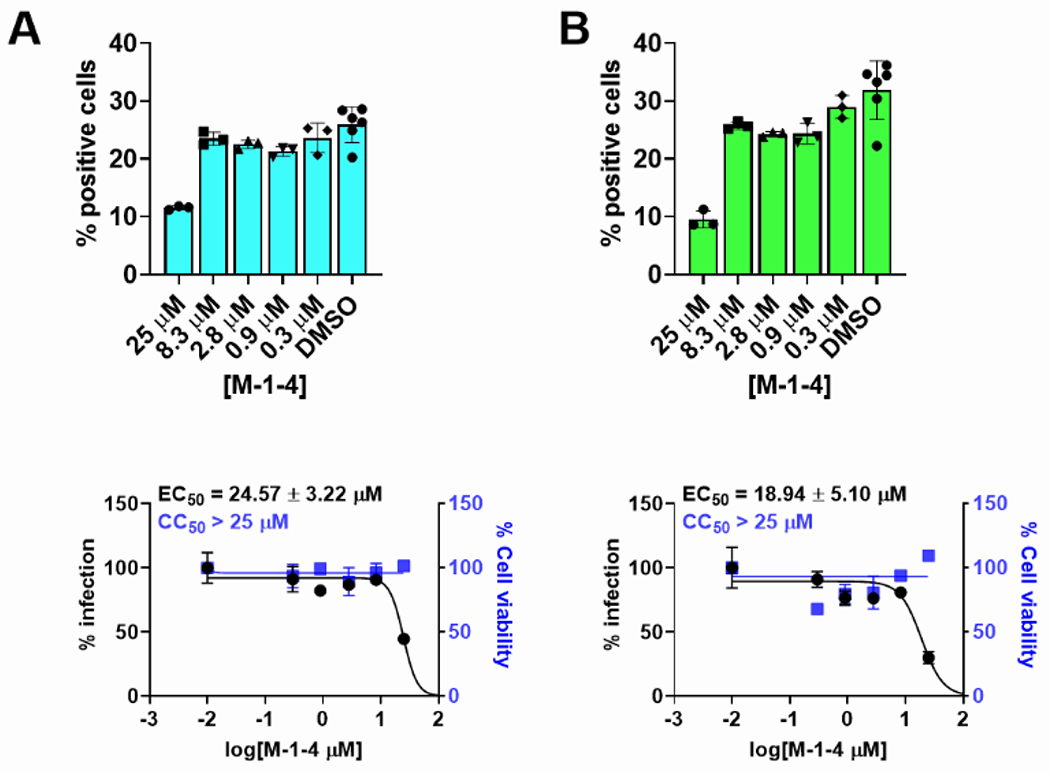
Antiviral activity of compound M-1-4 against SARS-CoV-2 in different cell lines. (A) Antiviral activity against SARS-CoV-2 in Vero E6 cells. (B) Antiviral activity against SARS-CoV-2 in Caco2-hACE2 cells.
X-ray crystal structure of SARS-CoV-2 Mpro in complex with compounds M-2-3 and M-2-9.
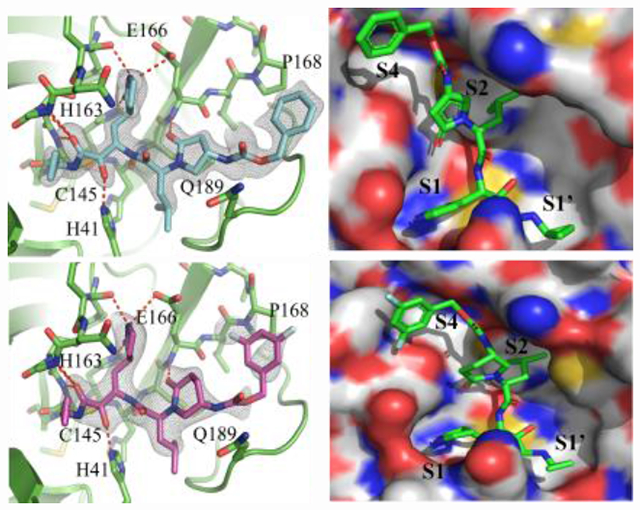
Using X-ray crystallography we solved the complex structures of SARS-CoV-2 Mpro with inhibitors M-2-3 (1.8 Å, PDB 8DZB) and M-2-9 (2.2 Å, PDB 8DZC) (Figure 5). As shown in Figure 5A and 5B, the thiohemiketal forms via nucleophilic attack of catalytic Cys145 onto the α-keto group of M-2-3 and M-2-9, which is clearly observed in the electron density map. Covalent bond formation positions the ketoamide optimally, allowing it to form hydrogen bonds with several catalytic core residues: Gly143, Ser144, and His41. Both M-2-3 and M-2-9 have a similar chemical structure and retain a similar binding pose (Figure 5A–C). The P1 γ-lactam moiety of M-2-3 and M-2-9, designed to mimic the glutamine residue of the native SARS-CoV-2 Mpro substrate,[8, 14a] is deeply inserted in the S1 pocket of the protease. The P1 γ-lactam nitrogen donates a hydrogen bond to the Glu166 sidechain and the backbone of Phe140. The lactam carbonyl oxygen also accepts a hydrogen bond from the imidazole sidechain of His163. The P2 isoleucine side chain of M-2-3 and M-2-9 fits tightly in the S2 subsite, deeply embedding itself into this hydrophobic pocket. As shown in Figures 5A and 5B, the 2-pyrrolidone and 2-piperidinone occupy the P2-P4 position where oxygens of 2-pyrrolidone and 2-piperidinone accept a hydrogen bond with the backbone amide of Glu166 to further stabilize their pose in the active site. Distal to the catalytic core are the Cbz and 3, 5 difluoro phenyl moieties of M-2-3 and M-2-9. However, no apparent intermolecular interactions are observed, suggesting this position is not critical for inhibition. Taken together, compounds M-2-3 and M-2-9 were suitably embedded in the substrate channel of SARS-CoV-2 Mpro and demonstrated excellent interactions with critical regions of the active site. The electron density from these co-crystals provides unambiguous evidence that M-2-3 and M-2-9 form a covalent bond with Cys145, contributing to the potent inhibition of this enzyme.
Figure 5.
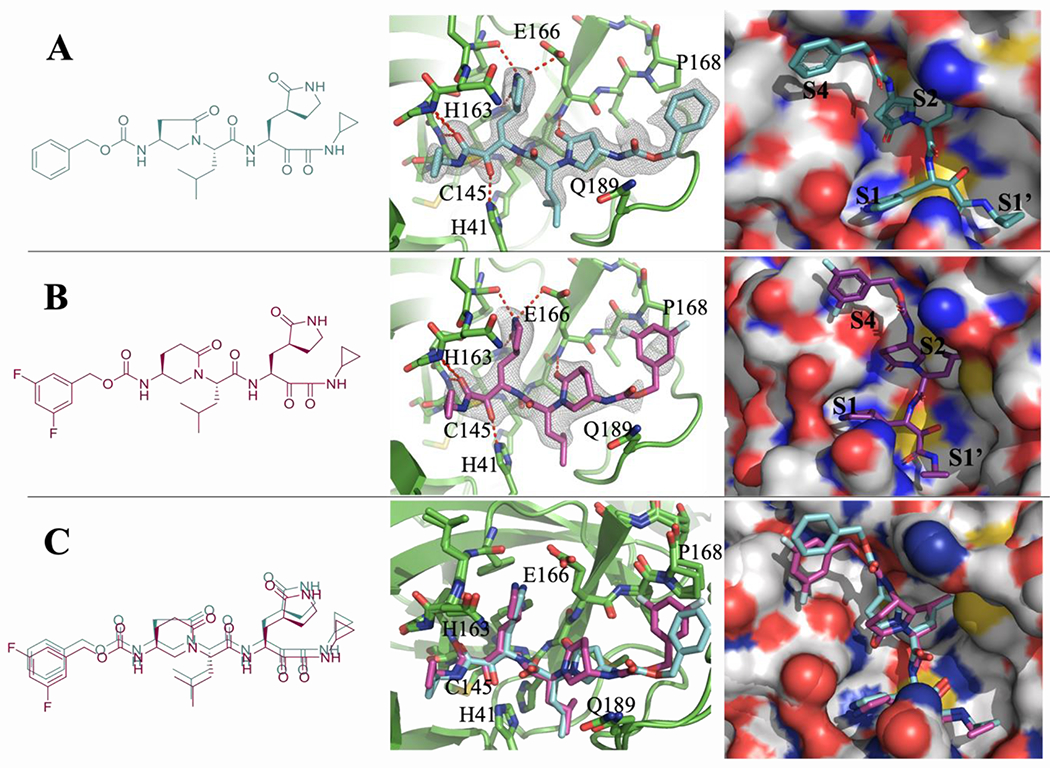
(A) X-ray crystal structure of SARS-CoV-2 Mpro with compound M-2-3. (B) X-ray crystal structure of SARS-CoV-2 Mpro with compound M-2-9. (C) Superimposed binding poses of compounds M-2-3 (green) and M-2-9 (magenta) on SARS-CoV-2 Mpro.
Discussion:
The COVID-19 pandemic caused a serious adverse impact on the public health and the global economy in many countries, and the causative SARS-CoV-2 is still evolving rapidly. In the past two decades, we have experienced three coronavirus infections in different regions around the globe,[19] including SARS-CoV, MERS-CoV and SARS-CoV-2. The COVID-19 pandemic is a timely reminder that broad-spectrum antiviral agents against different CoVs are urgently needed. Nirmatrelvir is the first FDA-approved oral COVID-19 drug. However, its suboptimal pharmacokinetic (PK) properties necessitate the co-administration of ritonavir.[8b] Although ritonavir prolongs the in vivo half-life of nirmatrelvir, its inhibition of cytochrome P450 3A4 leads to drug-drug interactions, which limits its use in people with underlying diseases.[20] Furthermore, drug resistance against nirmatrelvir is a pressing concern, and recent studies have found that resistant mutations can emerge either from natural variants or under the drug selection pressure.[10b, 10c] Therefore, additional Mpro inhibitors with improved PK properties and higher resistance barriers are in dire need. In this work, we designed and synthesized a series of constrained peptidomimetics-α, γ-AApeptides, which could serve as a new molecular scaffold for the next generation of Mpro inhibitors with broad-spectrum activity. A number of compounds exhibited excellent activity in the FRET-based Mpro enzymatic assay. The most potent compounds M-1-3 and M-1-6 inhibited SARS-CoV-2 Mpro with IC50 values of 0.08 μM. We also assessed broad-spectrum activity against Mpro of different coronavirus such as HCoVS like SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E and HUK1. The results suggested that α, γ-AApeptides are promising candidates as broad-spectrum antiviral candidates. Subsequently, we evaluated the activity of the all the inhibitors in the cell-based Flip-GFP Mpro assay. Compound M-1-4 had an EC50 value of 18 μM in the Flip-GFP Mpro assay and it was selected for antiviral assay. M-1-4 inhibited SARS-CoV-2 replication in both Vero E6 and Caco2-hACE2 cells with EC50 values of 18.94 and 24.57 μM, respectively. Further modification to improve the cellular antiviral activity is ongoing.
Conclusion
In conclusion, we have reported the design and synthesis of a new class of peptidomimetics - constrained α, γ-AApeptides, which served as the scaffold to design aldehydes and ketoamides to bind Mpro of the SARS-CoV-2. Certain compounds exhibited potent and broad-spectrum inhibitory activity against infection by SARS-CoV-2 and other HCoVs, as well as bat SARS-CoVs. X-ray crystal structures of SARS-CoV-2 Mpro in complex with these inhibitors revealed that they adopted a similar inhibitory mode as previously reported Mpro inhibitors. Our findings demonstrated that the constrained α, γ-AApeptide scaffold may be adopted to develop new antiviral agents for SARS-CoV-2 and other viruses.
Experimental Section
Materials
All chemicals and solvents were purchased and directly used without any purification from Fisher Scientific, Sigma-Aldrich, or Oakwood. Cbz-protected amino acids were purchased from AK Scientific. On silica gel with a mesh size ranging from 230 to 430, flash column chromatography was carried out. F-254 (0.2mm thickness) was used for analytic thin-layer chromatography (TLC). 1H NMR, 13C NMR spectra were recorded on Bruker Avance NEO-600 MHz spectrometers. Chemical shifts were reported relative to CDCl3 (δ 7.26 ppm) for 1H and CDCl3 (δ 77.00 ppm) for 13C. the mass of each compound was confirmed by high-resolution mass spectrometry detected by Agilent 6220 using electrospray ionization time-of-flight (ESI-QTOF). All the compounds were purified by the Waters Breeze 2 HPLC system. The purity of all compounds tested biologically was ≥ 95. The compounds were dissolved in H2O/MeCN and analyzed on a Waters Alliance HPLC system with a flow rate of 1 ml/min and a 5 to 100% linear gradient of solvent B (0.1% TFA in MeCN) in solvent A (0.1% TFA in H2O) over a duration of 50 min. The ultraviolet detector was set to 215 nm.
Cells and viruses
Vero E6 cells were obtained from ATCC and then cultured in DMEM containing 5% FBS in a 37 °C incubator in a 5% CO2 atmosphere. Caco-2 cells (ATCC HTB-37) were maintained in Eagle’s minimum essential medium with 10% FBS and 1% penicillin-streptomycin antibiotics. Human rhabdomyosarcoma (RD, ATCC CCL-136), Vero C1008 (ATCC CRL-1586), Huh-7 (University of Pittsburgh), and HEK293T expressing ACE2 (293T-ACE2, BEI resources, NR-52511) cell lines were maintained in DMEM. The human fibroblast Cell Line, MRC-5 (ATCC CCL-17) was cultured in EMEM. The media were all treated with 10% FBS and 1% penicillin-streptomycin antibodies and kept in an incubator (humidified, 5% CO2/95% air, 37 °C). The following reagents were obtained through BEI Resources, SARS-CoV-2, isolate USA-WA1/2020 (NR-52281), propagated once on VERO E6 cells before it was used for this study.
Protein expression and purification
SARS-CoV-2 Mpro (Mpro or 3CLpro) gene from strain BetaCoV/Wuhan/WIV04/2019 and SARS-CoV main protease (accession no.: 6W79_A), MERS-CoV main protease (accession no.: 5C3N_B), HCoV-NL63 main protease (accession no.: 5GWY_A), HCoV-HKU1 main protease (accession no.: 3D23_D), HCoV-OC43 main protease (accession no.: QDH43723), HCoV-229E main protease (accession no.: P0C6X1), with E-coli codon optimization and inserted into pET29a(+) plasmid were obtained from GenScript (Piscataway, NJ). The Mpro genes were then subcloned into the pE-SUMO vector as described previous publication10. The expression and purification of all Mpros were described in detail in the previous paper17.
Enzymatic assay
The enzymatic assay was established and performed as in the previous study.[18] To get the IC50 value of the testing compounds against different Main proteases, inside of 100 uL reaction buffer (20 mM HEPES, pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT, and 20% glycerol), 100 nM SARS-CoV-2, 100 nM SARS-CoV, 250 nM MERS-CoV, 50 nM HCoV-229E, 100 nM HCoV-OC43, 100 nM HCoV-NL63 or 50 nM HCoV-HUK1 were incubated with a serial concentration of the testing compounds at 30 °C. After 30 min, the enzymatic reaction was recorded in Cytation 5 imaging reader (Thermo Fisher Scientific) with filters for excitation at 360/40 nm and emission at 460/40 nm for 1 h by adding 1 uL of 1 mM substrate peptide. The initial velocity of the enzymatic reaction with and without testing compounds was calculated by linear regression for the first 15 min of the kinetic progress curves. The IC50 values were calculated by plotting the initial velocity against serial concentrations of the testing compounds using a dose-response curve in prism 8.
Cellular-Based FlipGFP Mpro Assay
The cellular-based FlipGFP Mpro assay of testing compounds was tested according to a previous study.[17b] Briefly, 50 ng pcDNA3-flipGFP-T2A-mCheery plasmid with TEV, Mpro cleavage site and 50 ng protease expression plasmid pcDNA3.1 SARS-CoV-2 Mpro were transfected into each well with 70-90% confluency 293T cells with transfection reagent Transit-293 (Mirus catalog No. MIR 2700) according to the manufacture’s protocol. 3 hrs post transfection, 1 uL of the testing compound was directly added into each well without changing any media. Two days later, images were taken with Cytation 5 imaging reader (Biotek) by using GFP and mCherry channels via 10 × objective lenses and were analyzed with Gen5 3.10 software (Biotek). mCherry signal alone was used to evaluate the compound cytotoxicity.
Antiviral Assays
The antiviral activity of testing compounds was assessed both in the Vero E6 cells and Caco-2 cells via CPE assay, as described elsewhere.[16] Plates with 384 wells contain Vero E6 and Caco-2 cells (ATCC) that have been cultured in Minimal Eagles Medium with the addition of 1% non-essential amino acids, 1% penicillin/streptomycin, and 10% FBS. The following day, 50 nL of the medication suspended in DMSO is added as an 8-pt dosage response with triplicate tests done with 3-fold dilutions between test concentrations starting at a 40 M final concentration. On each assay plate there were a positive control (10 M Remdesivir, n = 32) and a negative control (DMSO, n = 32). Prior to infection, cells were pretreated with controls and testing substances (in triplicate) for 2 hours. SARS-CoV-2 (isolate USA-WA1/2020) was diluted in serum-free growing media and placed to plates in BSL-3 containment to reach an MOI of 0.5. Cells were incubated with compounds and the SARS-CoV-2 virus for 48 h. For automated microscopy, cells were fixed, anti-dsRNA (J2) immunostained, and the nuclei were counterstained with Hoechst 33342. The number of cells per well (toxicity) and the proportion of infected cells (dsRNA+ cells/cell number) are both quantified by automated image analysis. SARS-CoV-2 infection was standardized to aggregated DMSO plate control wells and expressed as a percentage-of-control for each medication concentration: POC Z % Infectionsample/Avg % InfectionDMSO cont. A non-linear regression curve fit analysis (GraphPad Prism 8) of POC Infection and cell viability versus the log10 transformed concentration values to calculate EC50 values for Infection and CC50 values for cell viability.
X-Ray crystallization
As previously described,[9b] a final concentration of 2 mM of compound M-2-3 or M-2-9 was added to 20 mg/mL SARS-CoV-2 Mpro, and the mixture was then incubated for an overnight period at 4 °C. To remove precipitate, samples were centrifuged at 13000 g for 1 minute. Protein was combined in an identical amount with crystallization buffer (25% PEG 3350, 0.2 M AmSO4, and 0.1 M HEPES 7.5) in a vapor diffusion, hanging drop system to generate crystals. The crystals were transferred to a cryoprotectant solution of 30 % PEG 3350, 0.2 M AmSO4, 0.1 M HEPES 7.5, and 15 % glycerol for 5 seconds before being flash-frozen in liquid nitrogen once they had reached their full size, which usually took a few days.
Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and the Structural Biology Center (SBC) 19-ID beamlines at the Advanced Photon Source (APS) in Argonne, IL, then indexed and processed using the CCP4 versions of iMosflm 39. Structural refinement was carried out using REFMAC53340 and COOT41. SARS-CoV-2 Mpro with compound M-2-3 and compound M-2-9 complex structures have been deposited in the Protein Data Bank with accession ID 8DZB and 8DZC, respectively.
Characterization of compounds.
Benzyl ((S)-1-((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl) amino) pentan-2-yl)-5-oxopyrrolidin-3-yl) carbamate (M-1-1):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.54, 9.46 (s, 1H), 8.51, 7.84 (d, J = 7.3 Hz, 1H), δ 7.35 (m, J = 8.3, 6.8, 4.2 Hz, 5H), 6.72 (s, 1H), 6.45 (s, 1H). 5.09 (s, 2H), 4.57 (d, J = 29.8 Hz, 3H), 4.01 – 3.82 (m, 1H), 3.68 – 3.55 (m, 1H), 3.38 – 3.29 (m, 2H), 2.81 (m, J = 39.2, 17.4, 8.3 Hz, 1H), 2.55 – 2.26 (m, 3H), 2.04 – 1.36 (m, 6H), 0.99 – 0.80 (m, 6H). 13C NMR (151 MHz, CDCl3, isomers) δ 199.59, 181.80, 180.89, 171.42, 155.93, 136.10, 128.64, 128.61, 128.38, 128.24, 128.18, 66.99, 53.89, 44.97, 44.84, 41.06, 38.57, 37.30, 31.96, 29.42, 28.14, 24.80, 24.75, 22.99, 22.92, 21.69, 21.59. HRMS (ESI) C25H34N4O6 [M + H] + calcd = 489.569; found [M + H] + =489.2645.
Benzyl ((S)-5-oxo-1-((S)-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl) amino)-3-phenylpropan-2-yl) pyrrolidin-3-yl)carbamate (M-1-2):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.57, 9.43 (s, J = 1.3 Hz, 1H), 8.86, 8.73 (d, 1H δ 7.39 – 7.27 (m, 6H), 7.25 – 7.13 (m, 4H), 6.23 (s, 1H), 5.96 (s, 1H). 5.11 – 4.98 (m, 2H), 4.54 – 4.01 (m, 3H), 3.47 – 2.99 (m, 6H), 2.81 – 2.36 (m, 3H), 2.34 – 2.12 (m, 2H), 1.97 – 1.72 (m, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 201.37, 174.87, 170.36, 156.53, 136.09, 135.93, 128.85, 128.76, 128.67, 128.59, 128.44, 127.19, 67.00, 58.87, 56.89, 56.48, 55.66, 44.52, 41.23, 38.95, 37.68, 32.91, 29.93, 27.76. HRMS (ESI) C28H32N4O6 [M + H] + calcd = 521.586; found [M + H] + =521.2375.
Benzyl ((S)-1-((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl)amino)pentan-2-yl)-6-oxopiperidin-3-yl)carbamate (M-1-3):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.47, 9.29 (s, 1H), 8.55, 8.02 (d, 1H), δ 7.34 (m, J = 16.3, 13.2, 6.8 Hz, 5H), 6.70 (s, J = 8.8 Hz, 1H), 5.97 (s, 1H), 5.23 – 5.03 (m, 2H), 4.60 – 3.93 (m, 3H), 3.53 – 3.05 (m, 7H), 2.64 – 2.34 (m, 3H), 2.17 (s, 1H), 1.86 (m, J = 9.6, 4.7 Hz, 2H), 1.67 – 1.37 (m, 3H), 1.00 – 0.81 (m, 6H). 13C NMR (151 MHz, CDCl3, isomers) δ 199.40, 180.58, 170.90, 160.03, 155.06, 135.66, 128.63, 128.55, 128.47, 128.33, 128.27, 128.19, 66.79, 53.91, 49.12, 40.86, 38.85, 37.83, 36.46, 35.57, 28.69, 25.09, 24.72, 23.24, 22.23, 21.49. HRMS (ESI) C26H36N4O6 [M + H] + calcd = 501.596; found [M + H] + =501.2653.
Benzyl ((S)-1-((S)-3-cyclohexyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl)amino)propan-2-yl)-6-oxopiperidin-3-yl)carbamate (M-1-4):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.46, 9.31 (s, 1H), 8.65, 8.57 (d, 1H), δ 7.35 (m, J = 26.6, 13.8, 7.5, 6.9 Hz, 5H), 6.71 (m, J = 18.4 Hz, 1H), 6.07 (s, 1H), 5.17 – 5.01 (m, 2H), 4.26 – 4.01 (m, 2H), 3.78 (s, 3H), 3.21 (m, J = 40.5, 17.8, 9.8 Hz, 2H), 2.65 – 2.24 (m, 4H), 2.06 – 1.79 (m, 5H), 1.76 – 1.49 (m, 7H), 1.26 – 1.07 (m, 4H), 1.04 – 0.83 (m, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 199.48, 180.81, 170.89, 160.11, 156.00, 136.26, 129.03, 128.61, 128.52, 128.26, 128.24, 67.18, 66.56, 59.47, 53.25, 48.56, 45.61, 40.86, 37.82, 34.18, 33.84, 33.68, 32.65, 32.16, 28.77, 27.89, 26.38, 26.27, 26.02. HRMS (ESI) C29H40N4O6 [M + H] + calcd = 541.661; found [M + H] + =541.3061.
Benzyl ((S)-1-((S)-3-(3-fluorophenyl)-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl) amino) propan-2-yl)-6-oxopiperidin-3-yl)carbamate (M-1-5):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.35, 8.62 (s, 1H), 8.01 (d, J = 7.9, 1.2 Hz, 1H), δ 7.41 – 7.28 (m, 5H), 7.24 – 7.15 (m, 1H), 7.04 – 6.83 (m, 3H), ), 6.39 (s, J = 8.6 Hz, 1H), 6.14 (s, 1H), 5.18 – 4.99 (m, 2H), 4.27 – 3.98 (m, 2H), 3.48 (dt, J = 17.2, 6.3 Hz, 1H), 3.37 – 3.08 (m, 3H), 2.99 – 2.86 (m, 1H), 2.42 – 2.22 (m, 3H), 1.95 – 1.51 (m, 5H), 1.28 (d, J = 26.7 Hz, 2H), 0.86 – 0.78 (m, 1H). 13C NMR (151 MHz, CDCl3, isomers) δ 198.53, 183.23, 169.81, 163.61, 155.95, 141.60, 139.59, 136.29, 130.05, 128.91, 128.55, 128.41, 128.30, 127.98, 124.51, 115.71, 113.33, 66.57, 58.77, 56.33, 49.08, 44.49, 41.67, 38.82, 33.25, 29.71, 28.81, 27.93, 25.47. HRMS (ESI) C29H33FN4O6 [M + H] + calcd = 553.603; found [M + H] + =553.2369.
Benzyl ((S)-1-((S)-3-cyclopropyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl) amino) propan-2-yl)-6-oxopiperidin-3-yl)carbamate: (M-1-6):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.45, 9.34 (s, 1H), 8.88, 8.50 (d, 1H), δ 7.35 (m, J = 26.0, 13.5, 12.5, 7.4 Hz, 5H), δ 6.54 (d, J = 8.5 Hz, 1H), 6.27 (s, 1H), 5.15 – 5.02 (m, 2H), 4.87 (s, 2H), 4.19 – 4.08 (m, 1H), 3.63 – 3.07 (m, 4H), 2.80 – 2.27 (m, 4H), 2.13 – 1.55 (m, 7H), 0.69 – 0.37 (m, 3H), 0.11 (m, J = 18.5, 16.1, 7.9 Hz, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 200.19, 182.50, 173.17, 161.09, 155.71, 137.51, 128.76, 128.62, 128.55, 128.37, 127.77, 67.76, 56.12, 48.36, 43.81, 41.04, 32.29, 31.72, 28.85, 28.72, 25.54, 7.75, 5.07, 4.86, 4.29, 4.17. HRMS (ESI) C26H34N4O6 [M + H] + calcd = 499.580; found [M + H] + =499.2508.
3,5-difluorobenzyl ((S)-1-((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl) propan-2-yl)amino)pentan-2-yl)-6-oxopiperidin-3-yl)carbamate (M-1-7):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.47, 9.40 (s, 1H).δ 8.46 (d, J = 61.9, 21.3 Hz, 1H), 6.93 – 6.67 (m, 3H), 6.18 (s, J = 38.8, 17.3 Hz, 1H), 5.13 – 4.95 (m, 2H), 4.30 – 3.97 (m, 4H), 3.52 – 3.17 (m, 3H), 2.72 – 2.32 (m, 3H), 2.22 – 1.19 (m, 9H), 0.95 (m, J = 13.5, 12.5, 7.8 Hz, 6H). 13C NMR (151 MHz, CDCl3, isomers) δ 200.08, 182.04, 172.05, 171.62, 165.40, 163.01, 160.12, 156.15, 110.42, 103.54, 103.17, 65.81, 59.47, 54.04, 48.46, 41.04, 39.32, 38.80, 35.57, 28.81, 26.25, 24.93, 24.81, 23.28, 23.09, 21.49. HRMS (ESI) C26H34F2N4O6 [M + H] + calcd = 537.577; found [M + H] + =537.2460.
Benzyl ((S)-1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-4-methyl-1-oxopentan-2-yl)-5-oxopyrrolidin-3-yl)carbamate (M-2-1):
1H NMR (600 MHz, Chloroform-d) δ 8.80 (s, 1H), 7.39 – 7.24 (m, 10H), 6.43 (s, 1H), 5.89 (s, J = 7.3 Hz, 1H), 5.16 – 5.05 (m, 2H), 4.62 – 4.25 (m, 4H), 3.81 (m, J = 10.8, 6.6 Hz, 1H), 3.39 – 3.15 (m, 3H), 3.10 – 2.75 (m, 3H), 2.55 (m, J = 8.1 Hz, 1H), 2.35 (m, J = 66.6, 15.6, 5.7 Hz, 2H), 2.07 – 1.64 (m, 5H), 1.46 (m, J = 6.6 Hz, 1H), 0.90 (m, J = 10.2, 5.3 Hz, 6H). 13C NMR (151 MHz, CDCl3, isomers) δ 194.93, 180.48, 173.48, 170.69, 159.63, 155.86, 136.82, 136.18, 128.82, 128.63, 128.34, 128.24, 127.90, 127.83, 66.93, 54.60, 53.43, 51.88, 44.81, 43.38, 40.88, 39.54, 38.27, 37.07, 31.19, 28.65, 24.68, 22.99, 21.68. HRMS (ESI) C33H41N5O7 [M + H] + calcd = 620.719; found [M + H] + =620.2889.
Benzyl ((S)-1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-1-oxo-3-phenylpropan-2-yl)-5-oxopyrrolidin-3-yl)carbamate (M-2-2):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.23 (s, 1H), 8.84 (d, J = 6.1 Hz, 1H), δ 7.40 – 7.10 (m, 15H), 5.99 (s, 1H), 5.80 (s, 1H), 5.17 – 4.95 (m, 2H), 4.58 – 4.34 (m, 2H), 4.21 – 4.02 (m, 1H), 3.39 – 2.97 (m, 5H), 2.64 – 2.03 (m, 7H), 1.90 – 1.72 (m, 1H), 1.27 (d, J = 17.0 Hz, 1H). 13C NMR (151 MHz, CDCl3, isomers) δ 194.72, 180.68, 173.79, 169.72, 159.91, 155.78, 137.67, 136.88, 136.74, 136.30, 136.08, 129.01, 128.88, 128.84, 128.75, 128.69, 128.43, 127.93, 127.84, 126.98, 66.40, 59.08, 54.42, 44.80, 43.42, 41.03, 40.00, 39.27, 33.43, 30.76, 29.15, 27.61. HRMS (ESI) C36H39N5O7 [M + H] + calcd = 654.736; found [M + H] + =654.2845.
Benzyl ((S)-1-((S)-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-4-methyl-1-oxopentan-2-yl)-5-oxopyrrolidin-3-yl)carbamate (M-2-3):
1H NMR (600 MHz, Chloroform-d) δ 8.52 (m, 1H), 8.02 (m, J = 8.0, 1.1 Hz, 0H), 7.93 (m, J = 7.8, 1.7 Hz, 0H), δ 7.43 – 7.31 (m, 5H), 7.01 (s, J = 6.3 Hz, 1H), 5.10 (m, J = 12.0, 10.9 Hz, 2H), 4.59 – 4.36 (m, 2H), 3.90 – 3.78 (m, 1H), 3.44 – 3.24 (m, 3H), 2.99 – 2.59 (m, 3H), 2.53 – 1.64 (m, 8H), 1.46 (m = 18.7, 11.8 Hz, 1H), 0.91 (m, J = 13.3, 6.6 Hz, 6H), 0.86 – 0.79 (m, 2H), 0.61 (m, J = 11.4, 9.8, 5.5 Hz, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 194.55, 181.26, 170.64, 160.93, 156.46, 141.56, 132.91, 131.52, 128.64, 128.37, 128.21, 127.97, 66.70, 55.67, 52.48, 44.89, 41.11, 37.48, 37.46, 37.08, 31.31, 28.91, 28.27, 24.69, 23.01, 22.89, 22.48, 21.61, 21.44. HRMS (ESI) C36H39N5O7 [M + H] + calcd = 570.659; found [M + H] + =570.2796.
Benzyl ((S)-1-((S)-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-1-oxo-3-phenylpropan-2-yl)-5-oxopyrrolidin-3-yl)carbamate (M-2-4):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.18 (s, 1H), 8.80 (s, 1H), 7.37 (m, 5H), 7.21 – 6.97 (m, 5H), 6.52 (s, 1H), 6.06 (s, 1H), 5.06 (m, J = 36.4, 30.6, 17.6 Hz, 3H), 4.40 – 4.07 (m, 2H), 3.36 (m, J = 13.8 Hz, 2H), 3.28 – 3.20 (m, 2H), 3.17 – 3.10 (m, 2H), 2.70 (m, J = 83.0 Hz, 3H), 2.39 – 2.03 (m, 4H), 1.84 (d, J = 38.6 Hz, 1H), 0.83 (s, 2H), 0.62 (m, J = 13.1, 12.2 Hz, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 194.89, 175.10, 171.37, 161.16, 160.68, 155.78, 137.41, 136.26, 136.05, 128.95, 128.76, 128.66, 128.58, 128.38, 128.14, 127.24, 127.02, 66.94, 55.04, 44.78, 44.39, 41.08, 40.82, 39.09, 38.70, 33.58, 30.85, 29.91, 28.97, 22.05, 6.01. HRMS (ESI) C32H37N5O7 [M + H] + calcd = 604.676; found [M + H] + =604.2747.
Benzyl ((S)-1-((S)-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-4-methyl-1-oxopentan-2-yl)-6-oxopiperidin-3-yl) carbamate (M-2-5):
1H NMR (600 MHz, Chloroform-d, isomers) δ 8.90 (s, J = 4.5 Hz, 1H), 7.43 – 7.27 (m, 5H), 7.03 – 6.95 (m, 1H), 6.78 (s, J = 8.9 Hz, 1H), 6.34 (s, 1H), 5.44 (s, J = 10.7, 5.0 Hz, 1H), 5.14 – 5.02 (m, 2H), 4.37 – 4.09 (m, 1H), 3.62 – 3.06 (m, 4H), 2.83 – 2.48 (m, 4H), 2.28 (dt, J = 14.3, 7.6 Hz, 1H), 2.10 – 1.32 (m, 9H), 0.91 (p, J = 8.6, 7.2 Hz, 6H), 0.81 (t, J = 9.1 Hz, 2H), 0.67 – 0.52 (m, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 195.02, 180.99, 171.06, 160.84, 156.14, 137.11, 132.11, 128.69, 128.59, 128.44, 128.27, 128.19, 66.70, 54.22, 53.47, 48.39, 43.85, 41.07, 39.59, 35.34, 30.21, 28.84, 27.62, 25.52, 24.80, 24.59, 23.30, 22.41, 21.56, 6.33. HRMS (ESI) C30H41N5O7 [M + H] + calcd = 584.686; found [M + H] + =584.3008.
Benzyl ((S)-1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-1-oxopropan-2-yl)-6-oxopiperidin-3-yl) carbamate (M-2-6):
1H NMR (600 MHz, Chloroform-d, isomers) δ 8.89 (s, J = 3.9 Hz, 1H), 7.40 – 7.28 (m, 5H), 6.94 (m, J = 31.7, 3.9 Hz, 1H), 6.75 (s, J = 8.9 Hz, 1H), 5.30 (s, 1H), 5.05 (m, J = 33.8 Hz, 2H), 4.38 – 4.01 (m, 2H), 3.43 – 2.97 (m, 7H), 2.85 – 2.71 (m, 2H), 2.64 – 2.40 (m, 3H), 2.13 – 1.88 (m, 4H), 1.86 – 1.76 (m, 3H), 1.69 – 1.60 (m, 5H), 1.27 – 1.04 (m, 5H), 0.85 – 0.78 (m, 2H), 0.66 – 0.54 (m, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 195.12, 180.11, 171.37, 170.36, 162.18, 156.49, 136.44, 128.69, 128.59, 128.44, 128.25, 128.16, 66.65, 55.18, 52.65, 48.76, 45.09, 43.98, 40.96, 39.60, 34.32, 34.18, 34.01, 33.82, 33.70, 32.63, 32.22, 30.32, 28.92, 27.79, 26.25, 26.00, 22.41. HRMS (ESI) C33H45N5O7 [M + H] + calcd = 624.751; found [M + H] + =624.3337.
Benzyl ((S)-1-((S)-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-3-(3-fluorophenyl)-1-oxopropan-2-yl)-6-oxopiperidin-3-yl) carbamate (M-2-7):
1H NMR (600 MHz, Chloroform-d, isomers) δ 9.00 (s, 1H), 7.30 (m, J = 52.7, 27.8, 10.8, 6.4 Hz, 6H), 7.03 – 6.82 (m, 3H), 6.63 (s, J = 8.9 Hz, 1H), 6.17 (s, 1H), 5.75 (s, J = 10.2, 6.1 Hz, 1H), 5.17 – 5.00 (m, 2H), 4.28 – 4.04 (m, 1H), 3.90 (m, J = 65.9 Hz, 1H), 3.47 – 3.22 (m, 3H), 3.07 (m, J = 62.8, 24.0, 11.1 Hz, 2H), 2.91 – 2.61 (m, 5H), 2.53 – 2.24 (m, 3H), 1.99 – 1.63 (m, 3H), 1.50 (m, J = 20.9, 10.0 Hz, 1H), 0.82 (m, J = 6.6 Hz, 2H), 0.71 – 0.48 (m, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 195.16, 184.84, 181.37, 170.15, 161.71, 161.30, 155.33, 136.38, 129.94, 128.80, 128.60, 128.46, 124.53, 116.15, 115.72, 113.68, 67.44, 65.95, 54.31, 48.51, 43.97, 40.94, 39.53, 32.91, 30.36, 29.34, 25.58, 22.86, 22.45, 6.34. HRMS (ESI) C33H38FN5O7 [M + H] + calcd = 636.693; found [M + H] + =636.2766.
Benzyl ((S)-1-((S)-3-cyclopropyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl) butan-2-yl) amino)-1-oxopropan-2-yl)-6-oxopiperidin-3-yl)carbamate (M-2-8):
1H NMR (600 MHz, Chloroform-d, isomers) δ 8.88 (s, 1H), 7.98 (m, J = 46.6, 7.8 Hz, 1H), 7.45 – 7.24 (m, 5H), 7.04 – 6.88 (m, 1H), 6.68 (s, J = 8.7 Hz, 1H), 5.51 – 5.29 (m, 1H), 5.27 – 5.08 (m, 2H), 5.03 (s, 1H), 4.39 – 4.02 (m, 1H), 3.58 – 3.02 (m, 3H), 2.89 – 2.29 (m, 4H), 2.21 – 1.50 (m, 6H), 0.80 (m, J = 7.4 Hz, 2H), 0.51 (m, J = 78.2, 12.0 Hz, 4H), 0.10 (m, J = 10.7, 5.2 Hz, 2H). 13C NMR (151 MHz, CDCl3, isomers) δ 195.01, 170.81, 168.59, 160.88, 156.20, 141.54, 132.86, 131.51, 128.59, 128.47, 128.25, 127.97, 68.72, 57.08, 54.14, 49.77, 43.58, 41.64, 39.60, 31.64, 30.42, 28.66, 27.17, 25.66, 23.21, 22.46, 10.53, 6.33, 4.77, 4.27. HRMS (ESI) C30H39N5O7 [M + H] + calcd = 582.670; found [M + H] + =582.2834.
Supplementary Material
Acknowledgements
The work was supported by NIH R01AI152416 (JC) and NIH R01AI149852 (JC). We thank the beamline scientists at SBC and SER-CAT for their help with data collection. SBC-CAT is operated by UChicago Argonne LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. SER-CAT is supported by its member institutions and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References:
- [1].a) Looi MK, BMJ 2020, 371, m4113; [DOI] [PubMed] [Google Scholar]; b) Smith P, BMJ 2020, 370, m3350. [DOI] [PubMed] [Google Scholar]
- [2].a) Sacco MD, Hu YM, Gongora MV, Meilleur F, Kemp MT, Zhang X, Wang J, Chen Y, Cell Res. 2022, 32, 498–500; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Artese A, Svicher V, Costa G, Salpini R, Di Maio VC, Alkhatib M, Ambrosio FA, Santoro MM, Assaraf YG, Alcaro S, Ceccherini-Silberstein F, Drug Resist. Updat 2020, 53, 100721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Gorbalenya AE, Baker SC, Baric RS, Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J, Microbiol. 2020, 5, 536–544; [Google Scholar]; b) Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL, Nature 2020, 579, 270–273; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ, Nature 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Ren Z, Yan L, Zhang N, Guo Y, Yang C, Lou Z, Rao Z, Protein Cell 2013, 4, 248–250; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ramajayam R, Tan KP, Liang PH, Biochem. Soc. Trans 2011, 39, 1371–1375. [DOI] [PubMed] [Google Scholar]
- [5].a) Sacco MD, Ma CL, Lagarias P, Gao A, Townsend JA, Meng XZ, Dube P, Zhang X, Hu YM, Kitamura N, Hurst B, Tarbet B, Marty MT, Kolocouris A, Xiang Y, Chen Y, Wang J, Sci. Adv. 2020, 6, eabe0751; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fu LF, Ye F, Feng Y, Yu F, Wang QS, Wu Y, Zhao C, Sun H, Huang BY, Niu PH, Song H, Shi Y, Li XB, Tan WJ, Qi JX, Gao GF, Nat. Commun 2020, 11, 4417; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ma CL, Sacco MD, Hurst B, Townsend JA, Hu YM, Szeto T, Zhang XJ, Tarbet B, Marty MT, Chen Y, Wang J, Cell Res. 2020, 30, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Su S, Wong G, Shi WF, Liu J, Lai ACK, Zhou JY, Liu WJ, Bi YH, Gao GF, Trends Microbiol 2016, 24, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Ma CL, Sacco MD, Xia ZL, Lambrinidis G, Townsend JA, Hu YM, Meng XZ, Szeto T, Ba M, Zhang XJ, Gongora M, Zhang FS, Marty MT, Xiang Y, Kolocouris A, Chen Y, Wang J, ACS Cent. Sci 2021, 7, 1245–1260 ; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bar-On YM, Flamholz A, Phillips R, Milo R, Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Zhang LL, Lin DZ, Sun XYY, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R, Science 2020, 368, 409–412; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Owen DR, Allerton CM, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Science 2021, 374, 1586–1593; [DOI] [PubMed] [Google Scholar]; c) Dai WH, Zhang B, Jiang XM, Su HX, Li J, Zhao Y, Xie X, Jin ZM, Peng JJ, Liu FJ, Li CP, Li Y, Bai F, Wang HF, Cheng X, Cen XB, Hu SL, Yang XN, Yang J, Liu X, Xiao GF, Jiang HL, Rao ZH, Zhang LK, Xu YC, Yang HT, Liu H, Science 2020, 368, 1331–1335; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Qiao JX, Li YS, Zeng R, Liu FL, Luo RH, Huang C, Wang YF, Zhang J, Quan BX, Shen CJ, Mao X, Liu XL, Sun WN, Yang W, Ni XC, Wang K, Xu L, Duan ZL, Zou QC, Zhang HL, Qu W, Long YHP, Li MH, Yang RC, Liu XL, You J, Zhou YL, Yao R, Li WP, Liu JM, Chen P, Liu Y, Lin GF, Yang X, Zou J, LI LL, Hu YG, Lu GW, Li WM, Wei YQ, Zheng YT, Lei J, Yang SY, Science 2021, 371, 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Rossetti GG, Ossorio MA, Rempel S, Kratzel A, Dionellis VS, Barriot S, Tropia L, Gorgulla C, Arthanari H, Thiel V, Mohr P, Gamboni R, Halazonetis TD, Sci. Rep 2022, 12, 2505; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ma CL, Xia ZL, Sacco MD, Hu YM, Townsend JA, Meng XZ, Choza J, Tan HZ, Jang J, Gongora MV, Zhang XJ, Zhang FS, Xiang Y, Marty MT, Chen Y, Wang J, J. Am. Chem. Soc 2021, 143, 20697–20709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Joyce RP, Hu VW, Wang J, Med. Chem. Res 2022, 31, 1637–1646; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hu YM, Lewandowski EM, Tan HZ, Zhang XM, Morgan RT, Zhang XJ, Jacobs LMC, Butler SG, Gongora MV, Choy J, Deng XF, Chen Y, Wang J, bioRxiv 2022; [Google Scholar]; c) Iketani S, Mohri H, Culbertson B, Hong SJ, Duan Y, Luck MI, Annavajhala MK, Guo Y, Sheng Z, Uhlemann AC, Goff SP, Sabo Y, Yang H, Chavez A, Ho DD, Nature 2023, 613, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sang P, Shi Y, Huang B, Xue SY, Odom T, Cai JF, Acc. Chem. Res 2020, 53, 2425–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Abdulkadir S, Li CP, Jiang W, Zhao X, Sang P, Wei LL, Hu Y, Li Q, Cai JF, J. Am. Chem. Soc 2022, 144, 270–281; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xue SY, Wang XL, Wang L, Xu W, Xia S, Sun LJ, Wang SH, Shen N, Yang ZQ, Huang B, Li SH, Cao CH, Calcul L, Sun XM, Lu L, Cai JF, Jiang SB, Cell Discov. 2022, 8, 88; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sang P, Zhou ZH, Shi Y, Lee C, Amso Z, Huang D, Odom T, Nguyen-Tran VT, Shen WJ, Cai JF, Sci. Adv 2020, 6, eaaz4988; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zheng MM, Li CP, Zhou M, Jia R, Cai G, She FY, Wei LL, Wang SH, Yu J, Wang DY, Calcul L, Sun XM, Luo XM, Cheng F, Li Q, Wang Y, Cai JF, J. Med. Chem 2021, 64, 11219–11228; [DOI] [PubMed] [Google Scholar]; e) Xue SY, Wang L, Cai JF, Chembiochem 2022, 23, e202200298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Yang HT, Yang MJ, Ding Y, Liu YW, Lou ZY, Zhou Z, Sun L, Mo LJ, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z, Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Qamar MTU, Alqahtani SM, Alamri MA, Chen LL, J. Pharm. Anal 2020, 10, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R, Science 2003, 300, 1763–1767; [DOI] [PubMed] [Google Scholar]; b) Citarella A, Scala A, Piperno A, Micale N, Biomolecules 2021, 11, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang LL, Lin DZ, Kusov Y, Nian Y, Ma QJ, Wang J, Brunn AV, Leyssen P, Lanko K, Neyts J, Wilde AD, Snijder EJ, Liu H, Hilgenfeld R, J. Med. Chem 2020, 63, 4562–4578. [DOI] [PubMed] [Google Scholar]
- [16].Ma C, Tan H, Choza J, Wang Y, Wang J, Acta Pharm. Sin. B 2022, 12, 1636–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Hu YM, Ma CL, Szeto T, Hurst B, Tarbet B, Wang J, ACS Infect. Dis 2021, 7, 586–597; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xia ZL, Sacco MD, Hu YM, Ma CL, Meng XZ, Zhang FS, Szeto T, Xiang Y, Chen Y, Wang J, ACS Pharmacol. Transl. Sci 2021, 4, 1408–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Ma CL, Wang J, Proc. Natl. Acad. Sci. USA 2021, 118, e2024420118;33568498 [Google Scholar]; b) Steuten K, Kim H, Widen JC, Babin BM, Onguka O, Lovell S, Bolgi O, Cerikan B, Neufeldt CJ, Cortese M, Muir RK, Bennett JM, Geiss-Friedlander R, Peters C, Bartenschlager R, Bogyo M, ACS Infect. Dis 2021, 7, 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ, Infez. Med 2020, 28, 174–184. [PubMed] [Google Scholar]
- [20].N. I. o. Health, 2022, September 26.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


