Abstract
Purpose of review:
Mutations in the E3 ubiquitin ligase scaffold cullin 3 (CUL3) cause the disease Familial Hyperkalemic Hypertension (FHHt) by hyperactivating the NaCl cotransporter (NCC). The effects of these mutations are complex and still being unraveled. This review discusses recent findings revealing the molecular mechanisms underlying the effects of CUL3 mutations in the kidney.
Recent findings:
The naturally occurring mutations that cause deletion of exon 9 (CUL3-Δ9) from CUL3 generate an abnormal CUL3 protein. CUL3-Δ9 displays increased interaction with multiple ubiquitin ligase substrate adaptors. However, in vivo data show that the major mechanism for disease pathogenesis is that CUL3-Δ9 promotes degradation of itself and KLHL3, the specific substrate adaptor for an NCC-activating kinase. CUL3-Δ9 displays dysregulation via impaired binding to the CSN and CAND1, which cause hyperneddylation and compromised adaptor exchange, respectively. A recently discovered CUL3 mutant (CUL3-Δ474–477) displays many similarities to CUL3-Δ9 mutations but some key differences that likely account for the milder FHHt phenotype it elicits. Furthermore, recent work suggests that CUL3 mutations could have unidentified complications in patients and/or a predisposition to renal injury.
Summary:
This review summarizes recent studies highlighting advances in our understanding of the renal mechanisms by which CUL3 mutations modulate blood pressure in FHHt.
Keywords: Cullin 3, kidney, Familial Hyperkalemic Hypertension, KLHL3
Introduction
Familial hyperkalemic hypertension (FHHt, also known as Gordon syndrome or Pseudohypoaldosteronism Type II) is a rare monogenic disease characterized by high blood pressure and plasma potassium, and hyperchloremic metabolic acidosis. The phenotype ultimately results from an increase in sodium chloride reabsorption through the NaCl cotransporter (NCC) expressed in the distal convoluted tubule (DCT) of the kidney. Hyperkalemia is secondary to effects on the connecting segment/cortical collecting duct (1), and metabolic acidosis is secondary to effects on the proximal tubule (2). Thiazide diuretic treatment, therefore, alleviates most electrolyte imbalances. Although NCC is upregulated, mutations that cause FHHt are not found in the gene encoding NCC (SLC12A3) itself, but in genes encoding proteins that regulate NCC activity. With No Lysine [K] kinases (WNKs) initiate a phosphorylation cascade that ends with NCC phosphorylation, activating the cotransporter. WNK4, the predominant active WNK in the DCT (3), phosphorylates the intermediate kinases SPAK and OSR1; both of which then directly phosphorylate NCC (Figure 1) (4, 5). WNKs are regulated by the ubiquitin proteasome system via cullin-RING ligases (CRLs), the largest group of E3 ubiquitin ligases (Figure 1). CRLs are modular complexes consisting of a cullin scaffold protein attached to a RING subunit, and an interchangeable adaptor and substrate receptor that confers specificity to one or a small group of substrates (Figure 2). There are 8 known cullins in humans: cullins 1, 2, 3, 4A, 4B, 5, 7, and 9/PARC (6). Cullin 3 (CUL3) is unique in that it interacts with substrate adaptors called BTB (Broad complex, Tramtrack, Bric-a-brac) proteins that serve the role of both the adaptor and substrate receptor (7). WNKs interact with the CUL3 CRL specifically via the substrate adaptor kelch-like 3 (KLHL3). FHHt mutations have been in found in WNK1 (expressed ubiquitously throughout the nephron), WNK4 and KLHL3 (expressed predominately in the DCT), and CUL3 (ubiquitously expressed in all cells) (8-11). A tremendous amount of knowledge was unveiled by studying these disease-causing mutations. The disease onset and severity depend on the causative genetic mutation, with CUL3 mutations being the most severe, and having the earliest onset (8, 12). Boyden and colleagues found that CUL3 mutations that cause FHHt are dominant and de novo (8). All CUL3 mutations initially identified cluster to sites associated in splicing of exon 9, causing deletion of the entire exon; thus, the mutant will be referred to here as CUL3-Δ9. Exon 9 encodes amino acids 403–459 which contains the entire 4HB domain (Figure 3A). More recently, an FHHt-causing mutation was identified in exon 10 (13). This mutation causes deletion of 12-base pairs, resulting in loss of 4 amino acids (Δ474–477) located in the α/β1 domain (Figure 3A). Although there has only been one known case, the CUL3-Δ474–477 mutant showed a less severe phenotype, with functional and mechanistic differences compared to CUL3-Δ9, discussed below. WNK1, WNK4, and KLHL3 mutations impair their interaction with other subunits in the CRL complex, preventing ubiquitin ligase attachment to WNKs (11, 14-17). Additionally, there are intronic WNK1 mutations that cause ectopic WNK1 expression (9, 18). FHHt-causing CUL3 mutations lead to a complex modulation of CRL structure and function that have been studied extensively over the past decade, however, the precise molecular mechanism has been difficult to elucidate. This review highlights recent findings that have unraveled the molecular mechanisms underlying the effects of CUL3 mutations on the kidney that contribute to FHHt.
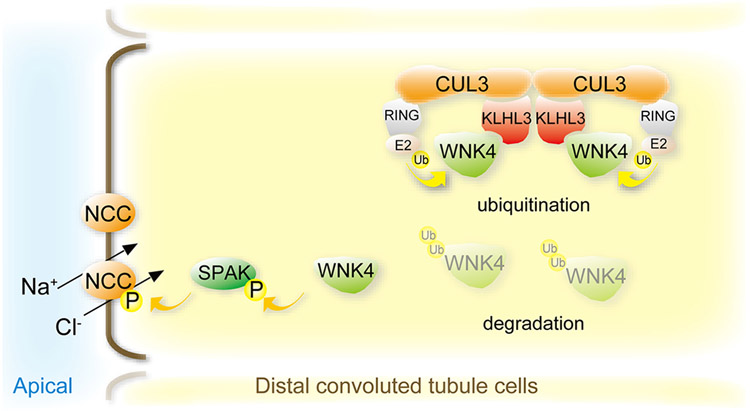
Figure 1. Regulation of NCC activity by the CUL3-WNK4-SPAK pathway.
In normal conditions, With-No-Lysine (K) kinase 4 (WNK4) activates SPAK, which in turn phosphorylates the Na+-Cl− cotransporter (NCC). The cullin ring ligase (CRL) consisting of the scaffold Cullin 3 (CUL3), the substrate adaptor Kelch-like 3 (KLHL3), and a Ring ubiquitin ligase determines abundance of WNK4 via the ubiquitin proteasome system.
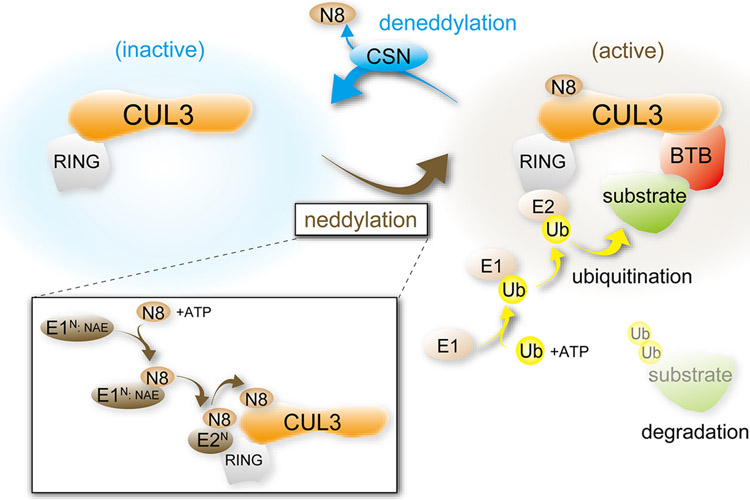
Figure 2. Assembly and activation of the Cullin 3 Ring Ligase (CRL).
Ubiquitination is mediated by sequential actions of ubiquitin (Ub) activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin ligase E3. The scaffold CUL3 assembles with a BTB substrate-binding adaptor, a RING ubiquitin ligase, and E2 to form a CRL, facilitating substrate ubiquitination and subsequently degradation. Stability and activity of the CRL depends upon addition (neddylation) and removal (deneddylation) of the ubiquitin-like protein NEDD8 (N8). NEDD8 is conjugated to CUL3 in an ATP-dependent manner by serial reactions catalyzed by the neddylation-specific enzymes E1n (NAE) and E2n to activate the CRL. Deneddylation is mediated by the COP9 signalosome (CSN).
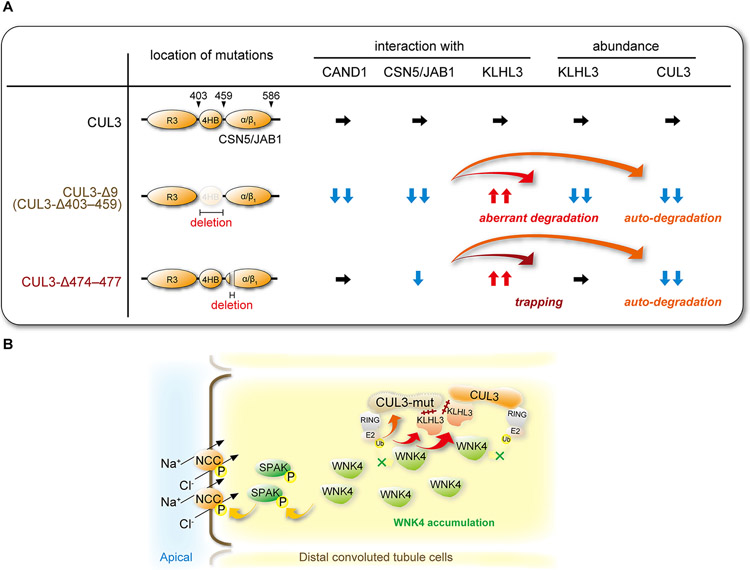
Figure 3. Dysregulation of NCC in Familial Hyperkalemic Hypertension (FHHt) caused by CUL3-Δ9 and CUL3Δ474-477.
A) Most FHHt-causing CUL3 mutations are located in sites associated with splicing of exon 9, resulting in deletion of exon 9 (CUL3-Δ9). CUL3 exon 9 encodes amino acids 403–459 which contains the entire 4HB domain. A recently identified mutation in exon 10 leads to deletion of four amino acids (CUL3-Δ474–477) in the α/β1 domain that interacts with CSN5/c-Jun activation domain-binding protein-1 (JAB1). Based on in vitro findings, CUL3-Δ9 displays dramatically impaired interaction with CSN5/JAB1, which catalyzes deneddylation, and CAND1, which is important for adaptor exchange. This causes hyperneddylation, leading to CUL3-Δ9 autoubiquitination and aberrant degradation of KLHL3, the adaptor for WNKs. In contrast, CUL3-Δ474–477 only mildly impairs interaction with CSN5/JAB1, does not affect CAND1 interaction, and causes autoubiquitination. Both mutants display enhanced affinity for KLHL3, trapping the adaptor in dysfunctional complexes, but only CUL3-Δ9 appears to promote KLHL3 degradation. B) FHHt-causing CUL3 mutants (CUL3-mut) autoubiquitinate and cause their own degradation, and trap KLHL3, resulting in WNK4 accumulation and NCC hyperactivation. Inappropriate KLHL3 degradation is only observed in CUL3-Δ9, as shown in (A), exacerbating WNK4 accumulation and hyperactivation.
CRL ubiquitination, neddylation, and deneddylation
E3 ubiquitination is a multi-step process (Figure 2). First, an E1 ubiquitin activating enzyme activates ubiquitin in an ATP-dependent manner and then transfers the protein to an E2 ubiquitin conjugating enzyme. The ubiquitin-charged E2 binds to the RING subunit where it transfers ubiquitin to the target substrate. Before the ubiquitination process can begin, the CRL must be activated. CRL activity is maintained through a cycling process in which the cullin undergoes modification via covalent attachment of NEDD8, a small ubiquitin like protein (19-21) (Figure 2). This process (neddylation) is similar to ubiquitination but uses neddylation-specific enzymes (E1: NAE; E2: UBE2M, also known as UBC12, and UBE2F) (22). NEDD8 attachment near the RING subunit facilitates recruitment of ubiquitin-charged E2s and initiates ubiquitin transfer by altering the cullin-RING structure, bringing the ubiquitin charged E2 closer to the substrate (23, 24). Ultimately, the neddylation process “turns on” the CRL complex. NEDD8 removal (deneddylation) is mediated by the COP9 signalosome (CSN) (Figure 2). The CSN binds to the CRL complex and the CSN5/JAB1 subunit catalyzes the cleavage of NEDD8, “turning off” ubiquitin ligase activity (25, 26). Thus, neddylation activates the CRL, initiating ubiquitination, whereas deneddylation inactivates the CRL. Cycling between neddylation and deneddylation is important for normal CRL function. This cycling process in necessary for rapid turnover of adaptor proteins allowing CRLs to adapt to the available pool of substrates. When in the unneddylated state CRLs are able to interact with cullin-associated NEDD8-dissociated protein 1 (CAND1); a regulatory protein involved in recycling adaptors and substrates. CAND1 has also helps catalyze the binding of adaptor proteins, after which it releases from the complex, allowing activation via neddylation (27, 28).
CUL3 mutants display enhanced interaction with BTB adaptors
Initial in vitro results published by the Ellison and McCormick groups in JCI in 2014, showed that CUL3-Δ9 displayed increased binding and enhanced degradation of the adaptor KLHL3 compared to WT CUL3 (29). Analysis of both CUL3-Δ474–477 and CUL3-Δ9 mutants has shown an increased interaction not only with KLHL3 (Figure 3A), but with multiple BTB adaptors. We first showed that CUL3-Δ9 had increased binding to KLHL3, BTBD1, and KCTD6 (29). Sigmund and colleagues followed with findings that CUL3-Δ9 had enhanced interaction with Bacurd1 and RhoBTB. However, unlike its effect on KLHL3, the enhanced interaction of CUL3-Δ9 with these adaptors did not lead to degradation, suggesting that sequestration or trapping of adaptors by CUL3-Δ9 could be a key molecular mechanism (30). Two more recent studies using mass spectrometry identified multiple BTB adaptors with preferential binding to CUL3-Δ9 (13, 31) and CUL3-Δ474–477 (13) compared to WT-CUL3. The work by Kouranti et al. further assessed the enhanced binding of CUL3-Δ9 and adaptors (31). They analyzed a small set of BTB adaptor proteins using bioluminescence resonance energy transfer (BRET) concluding that the preferential binding to CUL3-Δ9 was not due to an increase in affinity. However, stable isotope labeling by amino acids in cell culture (SILAC) combined with dynamic pulse-chase experiments showed that CUL3-Δ9 had an impairment in adaptor exchange, supporting the theory that the impact of CUL3-Δ9 on BTB adaptors was to trap/sequester them rather than cause enhanced degradation.
CAND1 is important in CRL adaptor exchange and cannot bind to neddylated CRLs. Kurz and colleagues found that CUL3-Δ9 did not interact with CAND1 (Figure 3A) (32), and Kouranti et al. showed that the neddylation inhibitor MLN4924 did not restore this interaction (31), suggesting that the impaired interaction is caused by structural changes caused by exon 9 deletion. Together the results suggest that impaired CAND1 binding may be an important mechanism for the CUL3-Δ9 mutants. Chatrathi et al., however, showed that the novel CUL3-Δ474–477 mutant did not have impaired binding to CAND1 (Figure 3A). Furthermore, their mass spectrometry data demonstrated that a large number of adaptors had an even higher interaction with CUL3-Δ474–477 compared to CUL3-Δ9 (13). The seemingly contradictory results may shine a light on the molecular mechanisms for CUL3 FHHt mutations as well as the difference in severity between the two mutants (explained in more detail below). The conflicting data demonstrate the need for continued examination of the effect of CUL3 mutations on adaptors and downstream effects on their substrates.
CUL3 FHHt mutants display impaired deneddylation
Our initial findings showed that the CUL3-Δ9 mutant was hyperneddylated, indicating impaired deneddylation (29). Neddylation inhibition via the compound MLN4924 attenuated the reduction in KLHL3 abundance we had observed, suggesting that hyperneddylation causes the aberrant adaptor degradation. The effect of neddylation on KLHL3 sequestration was also explored more recently by Kouranti et al. using BRET which showed that the neddylated form of CUL3 had an enhanced affinity to KLHL3, and neddylation inhibition with MLN4924 reduced the affinity (31). It was also reported that CUL3-Δ9 displays impaired binding to multiple CSN subunits (13, 31-33), including CSN4 (31), CSN5/JAB1, and CSN8 (32), suggesting impaired interaction with the CSN as a cause for the hyperneddylation. Enhanced neddylation of the novel CUL3-Δ474–477 mutant was also demonstrated, but its binding to CSN5/JAB1 was affected to a lesser extent than CUL3-Δ9 (Figure 3A) (13).
To examine the effects of impaired interaction with the CUL3 FHHt mutants the Ellison group generated an inducible, renal tubule-specific CSN5/JAB1 null mouse (KS-Jab1−/−) (34). The mice were examined three-weeks after induction of Jab1 deletion which led to an increase in the neddylated form of CUL3, but lower CUL3 abundance overall. The reduction in CUL3-Δ9 abundance was first reported by Kurz and colleagues (32). They found evidence of enhanced ubiquitination of CUL3-Δ9 in vitro, suggesting aberrant autoubiquitination leading to degradation (Figure 3A). In vivo, their CUL3-Δ9 knockin mice showed reduced CUL3 abundance, with almost undetectable levels of CUL3-Δ9. In addition, the CUL3-Δ474–477 mutant had enhanced autoubiquitination and decreased CUL3 abundance in patient-derived fibroblasts (Figure 4) (13). The KS-Jab1−/− mice also displayed lower KLHL3, and higher abundances of WNK1, WNK4, phosphorylated WNK4, full-length SPAK, and phosphorylated SPAK and OSR1 (34). However, total NCC was lower in these mice due to the effects of impaired deneddylation along the entire nephron, but the ratio of phosphorylated NCC to total NCC was increased. These data indicate that Jab1 deletion and subsequent hyperneddylation of CUL3 causes activation of the WNK-SPAK-NCC pathway. The mice, however, did not develop an FHHt phenotype due to the development of progressive kidney injury (35).
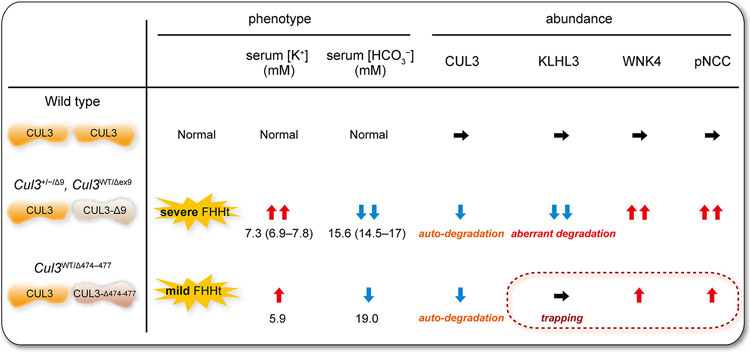
Figure 4. FHHt severity differs between CUL3-Δ9 and CUL3-Δ474–477.
(Left) In patients with FHHt caused by CUL3-Δ9 or CUL3-Δ474–477, serum K+ and HCO3− levels are increased but CUL3-Δ474–477 mutation has a milder effect on electrolyte abnormalities than the majority of CUL3-Δ9-generating mutations. Serum K+ and HCO3− data were obtained from (8,12,13) and represent median (quartile 1–quartile 3). (Right) In CUL3 heterozygous mice expressing CUL3-Δ9 from a transgene (Cul3+/−/Δ9) and knock-in mice with exon 9 deletion (Cul3WT/Δex9), CUL3-Δ9 causes auto-ubiquitination and degradation, and promotes KLHL3 degradation. WNK4 accumulates leading to excessive NCC phosphorylation (pNCC) and hence FHHt. Mice expressing CUL3-Δ474–477 have not been generated yet, but the milder patient phenotype suggests a mild increase in WNK4 and pNCC abundance compared with CUL3-Δ9, which may be explained by a lack of effect of CUL3-Δ474–477 on KLHL3 degradation, based on in vitro findings (Fig. 3A).
Dominant negative effects of CUL3-Δ9 on KLHL3 and CUL3 expression
We reported that CUL3-Δ9 caused KLHL3 degradation in vitro (Figure 3A) and hypothesized that this was the main cause of NCC activation via increased WNK4 abundance (Figure 3B) (29). However, others reported that in newly generated CUL3-Δ9 mouse models KLHL3 was not degraded (32, 36). Rather CUL3-Δ9 caused degradation of itself (32, 36, 37). Thus, they suggested that the mechanism for FHHt was functional CUL3 haploinsufficiency. This was supported by evidence that mice heterozygous for Klhl3 did not cause FHHt (38), but mice heterozygous for Cul3 also did not develop an FHHt phenotype (39), contradicting this hypothesis. Data from two novel FHHt mouse models further supported our KLHL3 degradation hypothesis. The first was a KLHL3 knockout mouse (38), which phenocopied FHHt, and was also used to validate KLHL3 antibody specificity. The second mouse model was an independently generated CUL3-Δ9 knockin mouse (40). Using the KLHL3 knockout mice validated antibody, Sohara and colleagues showed an approximately 70% decrease in KLHL3 in CUL3-Δ9 knockin mice compared to wildtype mice (40). This reduction was specific to KLHL3, as two other BTB proteins, Keap1 and KLHL2, showed no change in protein expression. We developed several novel mouse models to further expand on these results. Using an inducible, renal tubule-specific CUL3-Δ9 mutant mouse model, we expressed CUL3-Δ9 in mice with a CUL3 knockout background (41). Cul3 deletion led to a large increase in KLHL3 protein expression, but when the CUL3-Δ9 mutant was simultaneously expressed with Cul3 deletion, KLHL3 was drastically reduced, demonstrating that CUL3-Δ9 indeed has increased ubiquitin ligase activity toward the adaptor. When the CUL3-Δ9 mutant was expressed in mice heterozygous for Cul3 the results showed an FHHt phenotype, with a decrease in KLHL3 shown using the knockout mouse validated antibody (Figures 3B and 4) (41).
Thus, CUL3-Δ9 causes a decrease in both CUL3 and KLHL3 protein abundance (Figure 4), however, when these proteins are decreased in mice separately, they do not produce an FHHt phenotype. We therefore hypothesized that a combined decrease in both proteins is necessary for the disease. We tested this by generating inducible mice heterozygous for both Cul3 and Klhl3. Therefore, after induction, the mice would have only 50% expression of CUL3 and KLHL3. These mice showed an FHHt-like phenotype with two-fold increases in WNK4 and phosphorylated NCC, higher plasma potassium levels, and an increase in blood pressure when challenged with a high sodium, low potassium diet. The results indicate that degradation of both CUL3 and KLHL3 by CUL3-Δ9 is a key mechanism for the disease (Figure 4) (41).
The hypothesis was further validated using KS-Jab1−/− mice. Examination of the mice early on after induction showed that NCC was lower to the degree observed at the later time point (34). Acute Jab1 deletion caused a decrease in both CUL3 and KLHL3 abundances which led to an increased WNK4 abundance and increased phosphorylation of NCC (41). The results suggest that hyperneddylation of CUL3-Δ9 due to the impaired interaction for the CSN causes the reduction in CUL3 and KLHL3 protein leading to FHHt.
Differential effects of CUL3-Δ474-477 and CUL3 CUL3-Δ9
The FHHt patient with the novel CUL3-Δ474–477 mutation reported by Chatrathi et al. presented with a less severe phenotype compared to patients with CUL3-Δ9 mutations, with a milder elevation in serum potassium and reduction in serum bicarbonate (Figure 4) (13). The CUL3-Δ474–477 mutant showed similar effects as CUL3-Δ9 in vitro including hyperneddylation, enhanced CUL3 degradation, and increased interaction with BTB proteins. There were some notable differences, however, including intact interaction with CAND1 and a lack of KLHL3 degradation (Figure 3A). Furthermore, some of the effects, when compared directly to CUL3-Δ9, were either enhanced or reduced to different extents. For CSN5/JAB1, interaction with CUL3-Δ9 was more impaired than interaction with CUL3-Δ474–477, whereas interactions with BTB adaptor proteins were more enhanced for CUL3-Δ474–477 than for CUL3-Δ9.
The CUL3-Δ474–477 mutant did not promote degradation of KLHL3 in vitro (Figure 3A). This result seems to contradict our KLHL3 degradation hypothesis for CUL3-mediated FHHt; however, one limitation is that they also showed a lack of CUL3-Δ9 mediated KLHL3 degradation in their system, which we have reproduced both in vitro and in vivo (29, 33, 41). If there is indeed a lack of KLHL3 degradation by CUL3-Δ474–477 this may be explained by the intact interaction with CAND1, partially intact CSN5/JAB1 interaction, or the less severe effects of the 4 amino acid deletion on CUL3 structural flexibility. This lack of effect on KLHL3 abundance could explain the less severe electrolyte imbalances observed in the patient carrying this mutation (Figure 4), with vascular effects contributing more to the development of hypertension (36, 42).
Possible additional effects of CUL3 mutations in the kidney
FHHt is primarily considered a disease of high blood pressure and electrolyte abnormalities caused by dysfunction in the kidney and vasculature. However, FHHt patients with CUL3 mutations can also suffer from other maladies, including growth retardation and short stature, atelectasis, facial dysmorphia, and speech dyspraxia, indicating additional effects outside the kidney (12, 13). CUL3 regulates the degradation of proteins involved in an array of cellular functions including cell cycle regulation and oxidative stress response. As described above, multiple BTB adaptors are affected by CUL3 FHHt mutations, which could impair degradation of many different proteins involved in important molecular processes expressed in the kidney. This raises the possibility that FHHt-causing CUL3 mutations could cause more extensive effects in the kidney.
Further important roles for both wildtype CUL3 and the effects of CUL3-Δ9 beyond NCC regulation were revealed using renal tubule-specific CUL3 knockout mice. These mice display proximal tubule injury and fibrosis (29, 43, 44), possibly through effects on the cell cycle (43). The mice also developed polyuria with a reduction in aquaporin 2 (29, 44) and injury to the collecting duct (44). The CUL3 substrates Nrf2 (involved in oxidative stress response) and cyclin E (involved in cell cycle regulation) showed increased abundance after Cul3 deletion (43, 44). Interestingly, the induction of CUL3-Δ9 expression on the knockout background did not rescue the CUL3 knockout phenotype (44), including an inability to degrade Nrf2 and high molecular weight cyclin E, providing further evidence that CUL3-Δ9 does not have normal ubiquitin ligase function toward substrates (44). Additionally, the abundance of Nrf2 in CUL3-Δ9 FHHt mice was increased (44), which was also previously shown in vitro (33). Furthermore, in patient-derived fibroblasts the CUL3-Δ474–477 mutation caused an increase in cyclin E and increased proliferation compared to controls (13). The results suggest the possibility of unidentified complications in FHHt patients, such as an impaired response to oxidative stress, or a predisposition to kidney injury.
Impairing the regulation of cullin deneddylation also caused renal damage. Nephron-specific deletion of the CSN5/JAB1 CSN subunit caused progressive kidney injury that led to severe fibrosis (34, 35). KIM-1, a proximal tubule injury marker, and Ki-67, a proliferation marker, were expressed at high levels in the medulla early on after deletion and this progressed into the cortex over time. Long-term deletion of CSN5/JAB1 led to an increase in high molecular weight cyclin E abundance and remodeling of the distal nephron, with a shortening of the DCT and a large reduction in DCT1-specific proteins. Interestingly, although CSN5/JAB1 was deleted along the whole nephron, only the distal nephron was remodeled. Thus, disruption of CRL activity or regulation leads to renal injury.
Conclusion
The full effects of the FHHt-causing CUL3 mutations are still being determined. The sheer number of BTB adaptors (and therefore substrates) influenced by CUL3 mutations would suggest effects throughout the body, but the main effect is a renal (and vascular) disease. The fact that CUL3-Δ9 seems to have a unique effect on KLHL3 to cause degradation, and since expression of this adaptor predominates in the DCT of the kidney suggests that KLHL3 degradation is an important mechanism in the severity of the disease. Our recent data demonstrating enhanced Nrf2 activity suggest additional effects may occur in the kidney. It is not fully understood why CUL3-Δ9 degrades KLHL3 but CUL3-Δ474–477 does not, but the increased structural flexibility of the CUL3-Δ9 mutation could be a cause. The mechanistic differences between the CUL3-Δ9 and CUL3-Δ474–477 mutants may cause differences in severity of the disease with the CUL3-Δ474–477 mutant being more analogous to KLHL3 and WNK mutations. CUL3-Δ9 mutations are generally more severe, but some patients with the CUL3-Δ9 mutation have less severe electrolyte abnormalities (8, 12). There are multiple CUL3 mutations that cause alternative splicing leading to exon 9 deletion, but in some cases exon 9 deletion is not 100% penetrant (8). Thus, the ratio of WT-CUL3 to CUL3-Δ9 may be higher, potentially increasing the amount of functional CUL3, leading to a less severe phenotype. Future studies both in vitro and in vivo, will help resolve these remaining mechanistic questions. In the DCT, the FHHt-causing mutations are in multiple genes which lead to varying disease severity. This shows similarities with the disease Bartter syndrome in the thick ascending limb. There are 5 types of Bartter syndrome caused by mutations in 5 different genes that ultimately lead to increased sodium-wasting by reducing activity of the furosemide-sensitive Na+-K+−2Cl− Cotransporter 2 (NKCC2); however, the phenotype varies depending on the location of the mutation. In both circumstances, the discovery of these mutations and subsequent research has led to a substantial understanding of the underlying mechanisms of these transport pathways.
Key points.
Significant progress in determining the mechanisms by which CUL3 mutations cause the disease FHHt with the effects on dysregulation via the CSN and CAND1 may likely be central to the downstream effects on BTB adaptor sequestration and degradation.
Degradation of both CUL3 and KLHL3 is central to the disease pathogenesis of FHHt caused by CUL3 mutations leading to skipping of exon 9.
Examining the differences between CUL3-Δ9 mutations and the novel CUL3-474–477 mutant could lead to a better understanding of the molecular mechanisms of FHHt.
Analysis of both CUL3-Δ9 and CUL3-474–477 mutations suggest additional effects to other CRL substrates potentially affecting other cellular processes and organ systems. This could explain the extrarenal manifestations seen in patients with CUL3 mutant-mediated FHHt, such as growth retardation and short stature, and raises the possibility that other renal effects occur.
Financial support and sponsorship:
Y.M. received a postdoctoral award from the Uehara Foundation; R.J.C. is funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant Mentored Research Scientist Career Development Award DK120790; J.A.M. is funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK098141.
Footnotes
Conflicts of interest: none
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Grimm PR, Coleman R, Delpire E, Welling PA. Constitutively Active SPAK Causes Hyperkalemia by Activating NCC and Remodeling Distal Tubules. J Am Soc Nephrol. 2017;28(9):2597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris AN, Grimm PR, Lee HW, et al. Mechanism of Hyperkalemia-Induced Metabolic Acidosis. J Am Soc Nephrol. 2018;29(5):1411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi D, Mori T, Nomura N, et al. WNK4 is the major WNK positively regulating NCC in the mouse kidney. Biosci Rep. 2014;34(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm PR, Taneja TK, Liu J, et al. SPAK isoforms and OSR1 regulate sodium-chloride co-transporters in a nephron-specific manner. J Biol Chem. 2012;287(45):37673–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391(Pt 1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Song J, Ye D. CRL3s: The BTB-CUL3-RING E3 Ubiquitin Ligases. Adv Exp Med Biol. 2020;1217:211–23. [DOI] [PubMed] [Google Scholar]
- 8.Boyden LM, Choi M, Choate KA, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson FH, Disse-Nicodeme S, Choate KA, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293(5532):1107–12. [DOI] [PubMed] [Google Scholar]
- 10.Louis-Dit-Picard H, Barc J, Trujillano D, et al. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet. 2012;44(4):456–60, S1-3. [DOI] [PubMed] [Google Scholar]
- 11.Louis-Dit-Picard H, Kouranti I, Rafael C, et al. Mutation affecting the conserved acidic WNK1 motif causes inherited hyperkalemic hyperchloremic acidosis. J Clin Invest. 2020;130(12):6379–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hureaux M, Mazurkiewicz S, Boccio V, et al. The variety of genetic defects explains the phenotypic heterogeneity of Familial Hyperkalemic Hypertension. Kidney Int Rep. 2021;6(10):2639–52. • This study expanded on the original findings of Boyden (8) regarding the relationship between FHHt severity and the causative gene mutation, confirming that CUL3 mutations that generate CUL3-Δ9 cause the most severe form.
- 13. Chatrathi HE, Collins JC, Wolfe LA, et al. Novel CUL3 Variant Causing Familial Hyperkalemic Hypertension Impairs Regulation and Function of Ubiquitin Ligase Activity. Hypertension. 2022;79(1):60–75. •• Chatrathi identified the FHHt-causing CUL3-Δ474–477 mutation. An extensive molecular characterization was performed, and comparisons were made with CUL3-Δ9 that suggest differences in the mechanisms through which each causes the disease.
- 14.Susa K, Sohara E, Rai T, et al. Impaired degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3 knock-in mice. Hum Mol Genet. 2014;23(19):5052–60. [DOI] [PubMed] [Google Scholar]
- 15.Wakabayashi M, Mori T, Isobe K, et al. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep. 2013;3(3):858–68. [DOI] [PubMed] [Google Scholar]
- 16.Shibata S, Zhang J, Puthumana J, et al. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A. 2013;110(19):7838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta A, Schumacher FR, Mehellou Y, et al. The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J. 2013;451(1):111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaloy C, Elvira-Matelot E, Clemessy M, et al. Deletion of WNK1 first intron results in misregulation of both isoforms in renal and extrarenal tissues. Hypertension. 2008;52(6):1149–54. [DOI] [PubMed] [Google Scholar]
- 19.Osaka F, Saeki M, Katayama S, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19(13):3475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podust VN, Brownell JE, Gladysheva TB, et al. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97(9):4579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275(41):32317–24. [DOI] [PubMed] [Google Scholar]
- 22.Zheng YC, Guo YJ, Wang B, et al. Targeting neddylation E2s: a novel therapeutic strategy in cancer. J Hematol Oncol. 2021;14(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N, Schulman BA, Song L, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–9. [DOI] [PubMed] [Google Scholar]
- 24.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyapina S, Cope G, Shevchenko A, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382–5. [DOI] [PubMed] [Google Scholar]
- 26.Cope GA, Suh GS, Aravind L, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–11. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Yang X, Harrell JM, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10(6):1519–26. [DOI] [PubMed] [Google Scholar]
- 28.Goldenberg SJ, Cascio TC, Shumway SD, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119(4):517–28. [DOI] [PubMed] [Google Scholar]
- 29.McCormick JA, Yang CL, Zhang C, et al. Hyperkalemic hypertension-associated cullin 3 promotes WNK signaling by degrading KLHL3. J Clin Invest. 2014;124(11):4723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibeawuchi SR, Agbor LN, Quelle FW, Sigmund CD. Hypertension-causing Mutations in Cullin3 Protein Impair RhoA Protein Ubiquitination and Augment the Association with Substrate Adaptors. J Biol Chem. 2015;290(31):19208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kouranti I, Abdel Khalek W, Mazurkiewicz S, et al. Cullin 3 Exon 9 Deletion in Familial Hyperkalemic Hypertension Impairs Cullin3-Ring-E3 Ligase (CRL3) Dynamic Regulation and Cycling. Int J Mol Sci. 2022;23(9). • This study extensively characterized CUL3-Δ9 interactions with various substrate adaptors and culling ring ligase regulators. Evidence is provided that CUL3-Δ9 adaptor trapping results from impaired adaptor exchange due to hyperneddylation rather than a direct effect on adaptor affinity.
- 32.Schumacher FR, Siew K, Zhang J, et al. Characterisation of the Cullin-3 mutation that causes a severe form of familial hypertension and hyperkalaemia. EMBO Mol Med. 2015;7(10):1285–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornelius RJ, Zhang C, Erspamer KJ, et al. Dual gain and loss of cullin 3 function mediates familial hyperkalemic hypertension. Am J Physiol Renal Physiol. 2018;315(4):F1006–F18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornelius RJ, Si J, Cuevas CA, et al. Renal COP9 Signalosome Deficiency Alters CUL3-KLHL3-WNK Signaling Pathway. J Am Soc Nephrol. 2018;29(11):2627–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cornelius RJ, Nelson JW, Su XT, et al. COP9 signalosome deletion promotes renal injury and distal convoluted tubule remodeling. Am J Physiol Renal Physiol. 2022;323(1):F4–F19. • Renal tubule-specific disruption of CSN5/JAB1 caused DCT injury leading to hypoplasia. This counteracted the elevation in WNK4 that would be expected to activate NCC, so no FHHt phenotype was observed.
- 36. Abdel Khalek W, Rafael C, Loisel-Ferreira I, et al. Severe Arterial Hypertension from Cullin 3 Mutations Is Caused by Both Renal and Vascular Effects. J Am Soc Nephrol. 2019;30(5):811–23. • Generation of mouse models with ubiquitous or vascular smooth muscle-specific CUL3-Δ9 expression demonstrated that the severe hypertension is due to both renal and vascular effects.
- 37.Araki Y, Rai T, Sohara E, et al. Generation and analysis of knock-in mice carrying pseudohypoaldosteronism type II-causing mutations in the cullin 3 gene. Biol Open. 2015;4(11):1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki E, Susa K, Mori T, et al. KLHL3 Knockout Mice Reveal the Physiological Role of KLHL3 and the Pathophysiology of Pseudohypoaldosteronism Type II Caused by Mutant KLHL3. Mol Cell Biol. 2017;37(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferdaus MZ, Miller LN, Agbor LN, et al. Mutant Cullin 3 causes familial hyperkalemic hypertension via dominant effects. JCI Insight. 2017;2(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida S, Araki Y, Mori T, et al. Decreased KLHL3 expression is involved in the pathogenesis of pseudohypoaldosteronism type II caused by cullin 3 mutation in vivo. Clin Exp Nephrol. 2018;22(6):1251–7. [DOI] [PubMed] [Google Scholar]
- 41. Maeoka Y, Ferdaus MZ, Cornelius RJ, et al. Combined Kelch-like 3 and Cullin 3 Degradation is a Central Mechanism in Familial Hyperkalemic Hypertension in Mice. J Am Soc Nephrol. 2022;33(3):584–600. •• This study demonstrated that KLHL3 is degraded by CUL3-Δ9 in vivo, including in a model of FHHt. Reducing levels of CUL3 and KLHL3 in the absence of CUL3-Δ9 expression phenocopied FHHt. This showed that the combination of autoubiquitination and KLHL3 degradation is a central mechanism in CUL3-Δ9-mediated FHHt.
- 42.Agbor LN, Ibeawuchi SC, Hu C, et al. Cullin-3 mutation causes arterial stiffness and hypertension through a vascular smooth muscle mechanism. JCI Insight. 2016;1(19):e91015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saritas T, Cuevas CA, Ferdaus MZ, et al. Disruption of CUL3-mediated ubiquitination causes proximal tubule injury and kidney fibrosis. Sci Rep. 2019;9(1):4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maeoka Y, Cornelius RJ, Ferdaus MZ, et al. Cullin 3 mutant causing familial hyperkalemic hypertension lacks normal activity in the kidney. Am J Physiol Renal Physiol. 2022;323(5):F564–F76. • CUL3-Δ9 expression was unable to prevent renal fibrosis and polyuria In Cul3 knockout mice, showing that CUL3-Δ9 lacks normal activity in kidney. This study also showed that CUL3-Δ9 exerts effects on NRF2 activity raising the possibility of additional renal defects in FHHt.