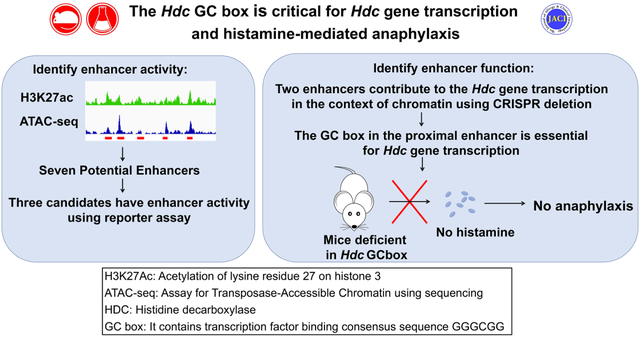
Abstract
Background:
Histamine is a critical mediator of anaphylaxis, a neurotransmitter, and a regulator of gastric acid secretion. Histidine decarboxylase is a rate-limiting enzyme for histamine synthesis. However, in vivo regulation of Hdc, the gene that encodes histidine decarboxylase is poorly understood.
Objective:
We sought to investigate how enhancers regulate Hdc gene transcription and histamine synthesis in resting conditions and in a mouse model of anaphylaxis.
Methods:
H3K27 acetylation histone modification and chromatin accessibility were used to identify candidate enhancers; The enhancer activity of candidate enhancers was measured in a reporter gene assay; and the function enhancers were validated using CRISPR deletion.
Results:
Deletion of the GC box, which binds to zinc finger transcription factors, in the proximal Hdc enhancer, reduced Hdc gene transcription and histamine synthesis in the mouse and human mast cell lines. Mast cells, basophils, brain cells, and stomach cells from GC box-deficient mice transcribed the Hdc gene much less than similar cells from wild-type mice and Hdc GC box-deficient mice failed to develop anaphylaxis.
Conclusion:
Our results demonstrate that the HDC GC box within the proximal enhancer in the mouse and human HDC gene is essential for Hdc gene transcription, histamine synthesis, and histamine-mediated anaphylaxis in vitro and in vivo.
Keywords: Histamine, anaphylaxis, Hdc gene enhancers, GC box
Graphical Abstract
Capsule summary
Our results demonstrate that the Hdc GC box within the proximal enhancer of the mouse and human HDC gene is essential for Hdc gene transcription and histamine synthesis in mast cells. The mouse Hdc GC box is required for histamine-mediated anaphylaxis in the mouse model of anaphylaxis.
Introduction
Anaphylaxis is a life-threatening allergic reaction that rapidly affects the skin, gastrointestinal tract, respiratory system, and cardiovascular system1. Anaphylaxis can be caused by allergy to foods, insect venoms, medications, and other agents1. Food-induced anaphylaxis has risen dramatically in developed countries during the past several decades and continues to rise2–4. Understanding the molecular regulation of anaphylactic shock should promote effective prevention and treatment.
Mast cells (MCs) play a critical role in mediating anaphylaxis by releasing large amounts of histamine and other mediators. MCs express FcεRI, the high-affinity receptor for IgE, a heterotetramer composed of one IgE-binding α subunit, one membrane-tetraspanning β subunit, and a disulfide-linked γ subunit dimer. MCs can be activated by antigen cross-linking of antigen-specific IgE bound to FcεRI. Antigen cross-linking of the IgE/FcεRI complex activates MCs to release histamine that is pre-made and stored in MC granules.
Histamine belongs to a family of biogenic amines that includes neurotransmitters such as serotonin and dopamine and hormones such as adrenaline. Biogenic amines that contain one or more amine groups are formed mainly by decarboxylation of amino acids. Histamine is a monoamine synthesized from the amino acid histidine through a reaction catalyzed by the enzyme histidine decarboxylase (HDC), which removes the carboxyl group from histidine. The Hdc gene encodes HDC protein with a molecular mass of 74 kDa, a proenzyme with little or no enzyme activity. However, it becomes active once the proenzyme is cleaved at the site near its c-terminus, presumably by Caspase-9. HDC is the primary enzyme that catalyzes histamine synthesis.
Knowledge of how the human and mouse Hdc genes are transcribed is limited5. The Mouse Hdc gene is located on chromosome 26. It contains 12 exons. Hdc mRNA is expressed in basophils, MCs, and enterochromaffin-like cells (ECL) of the stomach and is 86% homologous with the human gene. The human HDC gene is located in the 15q21.2 region of chromosome 15 and also contains 12 exons7. Most previous work has concentrated on analyzing the Hdc promoter region using reporter gene assays in transformed MC cell lines. Deletional analysis of Hdc promoter-driven luciferase reporter gene transcription demonstrated that the transcription factor SP1 binds to a GC box (GGGGCGGGG) found in both the human and mouse Hdc gene promoters6, 8. Several promoter elements have been reported to regulate Hdc gene transcription negatively. For example, the transcription factors YY1 and KLF4 negatively regulate the Hdc gene by suppressing SP1 in a gastric cancer cell line9, 10. Our group demonstrated positive regulation of Hdc gene expression by the transcription factor (TF) GATA binding protein 2 (GATA2), a member of the GATA family of TFs and by Microphthalmia-associated TF (MITF)11.
Enhancers are segments of DNA located in the noncoding regions of genes. They activate gene transcription by delivering important accessory factors to the promoter12, 13. Decoding enhancers has been a longstanding goal in the field of gene transcription14. Enhancers and other cis regulatory elements work from a distance in eukaryotes. Transcriptional codes hidden in gene distal regions are often required for full transcription. Locating critical enhancers can be difficult because they can be located up to 100 kb from the transcription start sites in noncoding regions, which constitute 99% of the genome. Enhancers contain clusters of TF binding motifs that act as transcriptional codes that are read by TFs. The binding of TFs and co-factors to enhancers creates accessible regions within enhancers that recruit histone-modifying enzymes to add acetyl or methyl groups to histones. Monomethylation of lysine four on histone 3 (H3K4me1) marks genes that are poised to be transcribed, whereas acetylation of lysine 27 on histone 3 (H3K27ac) identifies genes that are actively being transcribed. DNA regions associated with increased histone modification often coincide with chromatin accessibility, which can be measured by ATAC-seq. The combined presence of histone modifications and chromatin accessibility predicts enhancer activity15–19 and promotes a looping mechanism that approximates TF- and co-factor-bound distal and proximal enhancers to core promoters via a huge protein complex called mediator20; this approximation then facilitates gene transcription. By measuring H3K4me1 and H3K27ac modifications using next generation DNA sequencing, our group has identified a −8.8 kb Hdc enhancer that increased minimal Hdc promoter activity in a luciferase reporter gene transcription assay11.
In this study, by combing H3K27ac modification and chromatin accessibility measurement, we have identified several additional potential enhancers in the 34 kb region of the mouse Hdc gene. We have functionally tested the potential enhancers in the context of chromatin in mouse and human mast cell lines and in mice using an improved CRISPR method. Our work demonstrates a critical role for the HDC proximal enhancer GC box in human HDC gene transcription and histamine synthesis and for murine histamine-mediated anaphylaxis.
METHODS
Mice
sgRNA (TTTATGGGGGCGGGGC) was designed and injected into BALB/cByJ zygotes along with ssDNA HDR template. A more detailed description of mice used is included in the Methods section in this article’s Online Repository. All animal experiments were conducted according to protocols approved by the National Jewish Health Institutional Animal Care and Use Committee (Denver, CO).
BMMC and LAD2 cell culture
BMMCs were cultured from bone marrow cells of BALB/c mice. Human mast cell line LAD2 was provided by Drs. Dean D. Metcalfe and Arnold S. Kirshenbaum (National Institutes of Health, Bethesda, MD). See the Methods section in this article’s Online Repository for further details.
H3K27ac CUT&Tag-seq and Omni-ATAC-seq
The amplified libraries were purified, size-selected, and the quality and quantity of libraries were assessed with an Agilent Technologies TapeStation. The pair-ended sequencing of DNA libraries was performed with an Illumina NovaSEQ6000 platform. Data can be accessed at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo, GSE223388). See the Methods section in this article’s Online Repository for further details about libraries preparation and NGS data analysis.
Luciferase reporter constructs and assay
CFTL-15 cells (5 × 106) were electroporated with plasmids using a Bio-Rad Gene Pulser. Luciferase activities were measured by using an InfiniteMlOOO® microplate reader and the Dual-Luciferase reporter assay system. See the Methods section in this article’s Online Repository for further details.
Deletion of GC boxes using four bicistronic sgRNA guides
The sgRNA sequences targeting Hdc enhancers or transcription factor exons were designed using the online CRISPick tool from Broad Institute. Each of four sgRNA sequences targeting the same enhancer was cloned into vector using the BsmBI cloning site. CFTL-15 cells were either transfected with Lenti plasmids or transduced with lentivirus containing the bicistronic sgRNA guides. The sorted cells were subjected to DNA analysis of GC box deletion. The intensities of DNA fragments were measured using Imagej (NIH, MD). See the Methods section in this article’s Online Repository for further details.
Passive systemic anaphylaxis (PSA)
Mice were sensitized with 10 μg of IgE anti-TNP antibody and challenged with 10 μg of TNP-BSA. Rectal temperatures of the challenged mice were measured before challenge (0 min) and at 5, 10, 15, 20, 25, 30, 40, 60, 80, 100 and 120 minutes after challenge as described previously11.
Histamine and MMCP1 measurements
Plasma was collected 5 minutes after challenge for histamine measurement, using a histamine EIA kit (Beckman Coulter, IM2015). Sera were collected one hour after challenge for determination of serum levels of mucosal mast cell protease (MMCP)1, using an MMCP1 EIA kit (Thermo Fisher Scientific).
Statistical analysis
The nonparametric Mann-Whitney U test or two-tailed student’s t test was used to determine significant differences between two samples. The body temperature changes at various time points before and after PSA challenges were analyzed using a two-way repeated measure ANOVA.
RESULTS
The proximal Hdc enhancer is essential for Hdc gene transcription
Previously, we used the histone modifications H3K4me1 and H3K27ac to identify potential enhancers in the Hdc gene and analyzed the distal −8.8 enhancer in detail. In this study, we used H3K27ac CUT&Tag-seq and Omni-ATAC-seq to perform a higher resolution analysis of potential Hdc enhancers in a 34 kb region of the Hdc gene. In addition to the previously defined Hdc E−8.8, we identified proximal enhancer (PE), E+8.5, E+12.3, E+15.8, E+18.8, and E+20.8 noncoding DNA regions that showed increased H3K27ac modification and increased chromatin accessibility (Fig. 1). To determine which of the DNA regions possess enhancer activity, we constructed a series of luciferase reporter genes in which the PE, E+8.5, E+12.3, E+15.8, E+18.8, or E+20.8 was linked to the minimal Hdc promoter (MP) (−42 to +1 relative to the TSS). We included a DNA region that was not associated with H3K27ac modification or chromatin accessibility as a negative non-enhancer (NE) control (Fig. 1). The constructs were transfected into the CFTL-15 mast cell line, which we previously used to show Hdc E−8.8 enhancer activity11. We found that, in addition to Hdc E−8.8, the Hdc PE and E+18.8 showed enhancer activities relative to the MP promoter (E−8.8, 40.0-fold; PE, 13.4-fold; E+18.8, 12.7-fold) (Fig. 2A). As expected, the NE region did not show enhancer activity (Fig. 2A). These results provide evidence that the Hdc PE, E−8.8, and E+18.8 DNA regions contain sites that can bind to cognate transcription factors in CFTL-15 mast cells to promote the transcription of the luciferase reporter gene.
Figure 1. Identification of the putative Hdc enhancers.
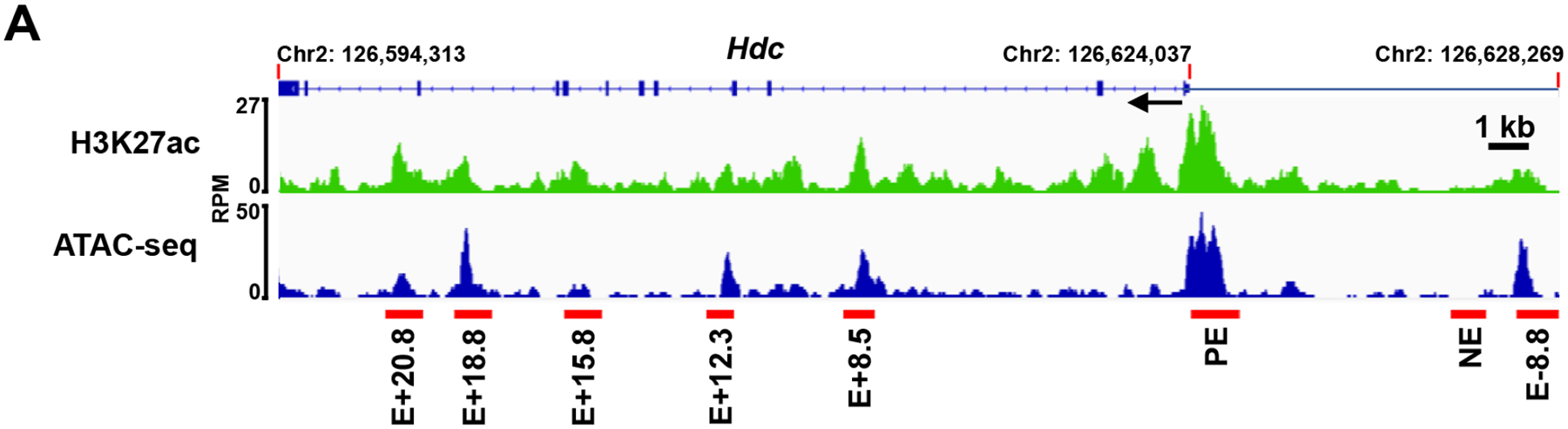
Representative Integrated Genome Viewer (IGV) tracks from H3K27ac CUT&Tag-seq and Omni-ATAC-seq. BMMCs were used for H3K27ac CUT&Tag-seq and Omni-ATAC-seq. Red bars indicate putative Hdc enhancers that show increased H3K27ac modifications and chromatin accessibility. RPM: reads per million mapped reads; E: enhancer; PE: proximal enhancer. The numbers following E indicate the distance (kb) of enhancer to the TSS of the Hdc gene; + means the enhancers are located downstream of the TSS, and − means the enhancers are located upstream of the TSS. One IGV track was from one biological sample, representing two (Omni-ATAC-seq) or three (CUT&Tag-seq) biological samples with similar patterns.
Figure 2. Hdc E−8.8, PE, and E+18.8 possess enhancer activity.
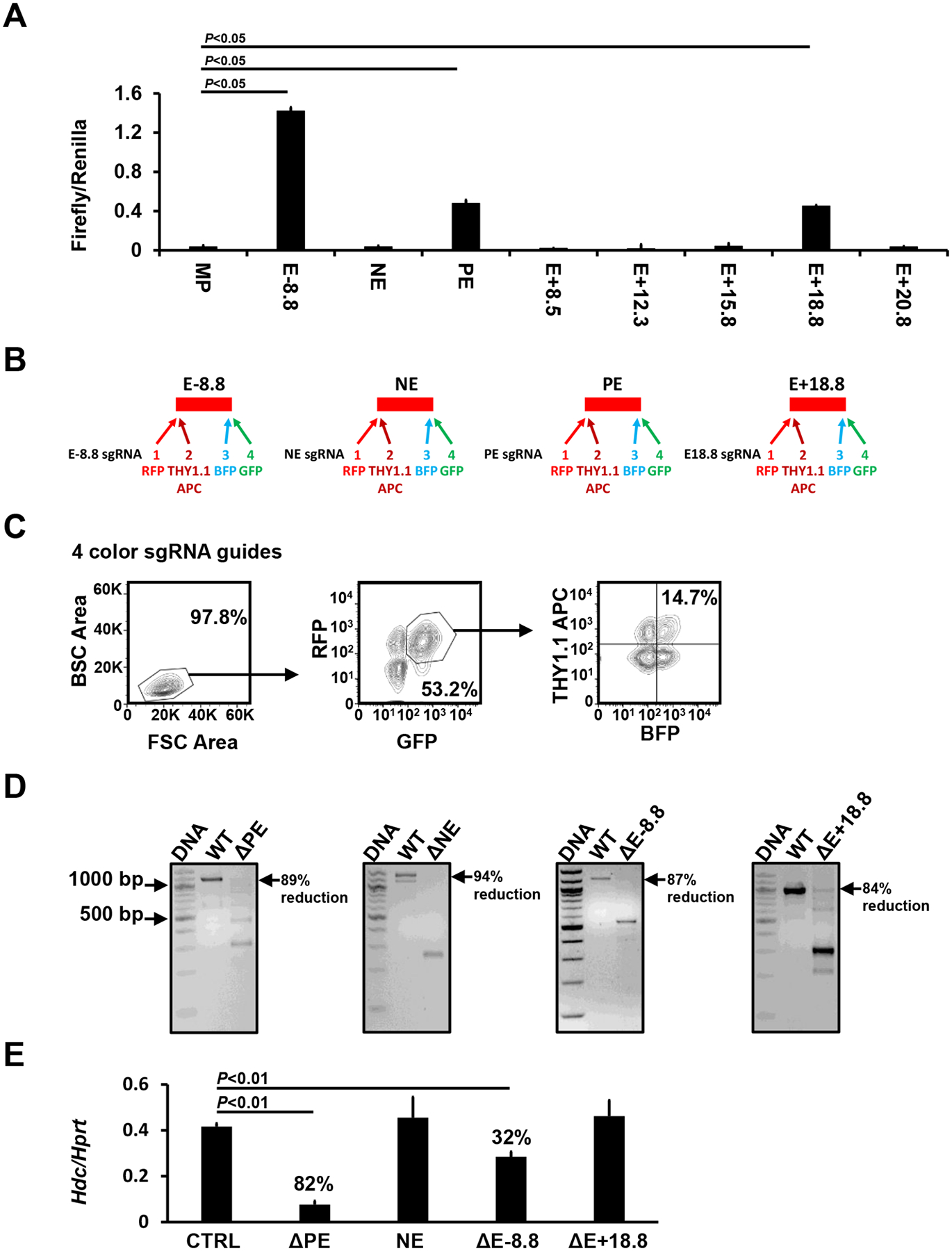
A. Luciferase reporter analysis of the Hdc enhancer activities in CFTL-15 mast cells. Error bars represent mean ± SEM, n=4 transfectants from 2 independent experiments. B. sgRNA guide constructs that co-expressed bicistronic BFP, GFP, RFP, and Thy1.1 genes. C. FACS sorting gates. CFTL15 cells were transduced lentivirus containing four bicistronic sgRNA guides. BFP+, GFP+, RFP+, and Thy1.1+ transduced cells were FACS-sorted using sorting gates indicated. D. DNA deletion efficiency analysis. WT and deleted DNA fragments in FACS-sorted BFP+, GFP+, RFP+, and Thy1.1+ cells were analyzed with PCR. E. Hdc mRNA expression in the FACS-sorted cells was measured by qPCR (mean ± SEM, n=6 transfectants from 3 independent experiments). The percentages indicate the percent reduction in Hdc mRNA expression relative to CTRLs. P values were calculated using the Mann-Whitney U test.
However, the enhancer luciferase reporter assay does not assess enhancer activity in the context of chromatin. To determine whether the Hdc PE, E−8.8, and/or E+18.8 enhancers play a role in Hdc gene transcription in the context of chromatin, we used CRISPR/Cas9 technology to delete these enhancers. We used the NE as a negative control. The CRISPR deletion method using two sgRNA guides often resulted in deletions occurring in one chromosome, creating heterozygous deletion that does not have a phenotype. To overcome this technical challenge, we targeted one enhancer with four sgRNA guides, each expressing a bicistronic gene encoding for fluorescence protein BFP, GFP, RFP, or for Thy1.1 (Fig. 2B). CFTL-15 cells were transduced with lentiviruses containing the four bicistronic sgRNA guides. Cells expressing BFP, GFP, RFP, and Thy1.1 (7.6% of transfected cells were positive for four colors) were FACS-sorted, using gates shown in Fig. 2C. This method achieved 84–94% homozygous deletion of Hdc enhancers in bulk (Fig. 2D). Deletion of the PE enhancer resulted in the greatest reduction (82%) in Hdc mRNA expression (Fig. 2E), while deletion of the E-8.8 enhancer resulted in a 32% reduction in Hdc mRNA expression (Fig. 2E). In contrast, deletion of the NE or Hdc E18.8 enhancer did not affect Hdc mRNA expression in the context of chromatin (Fig. 2E). These data demonstrate that Hdc PE is critical in Hdc gene transcription, while the Hdc E−8.8 contributes to Hdc gene transcription in the context of chromatin.
The GC box in the proximal Hdc enhancer is essential for Hdc gene transcription in the CFTL-15 mast cell line
To analyze transcription factor binding sites in the Hdc PE enhancer, we performed a deletional analysis of the −1401 bp long Hdc PE enhancer using a luciferase reporter gene assay. We found that most PE enhancer activity was driven by sequences located within the −48 bp fragment (Fig. 3A). Transcription factor binding site analysis revealed that a GC box is located within this −48 bp fragment. Mutation in the GC box (GGGCGG, −43 to −48) resulted in a complete loss of enhancer activity in the luciferase reporter gene assay (Fig. 3B). Furthermore, CRISPR-mediated deletion of the GC box in CFTL-15 mast cells reduced Hdc mRNA expression by 78% (Fig. 3C). These data demonstrate that the GC box in the proximal Hdc PE enhancer is important for Hdc gene transcription in a mast cell line.
Figure 3. The GC box in the Hdc PE is required for Hdc gene transcription.
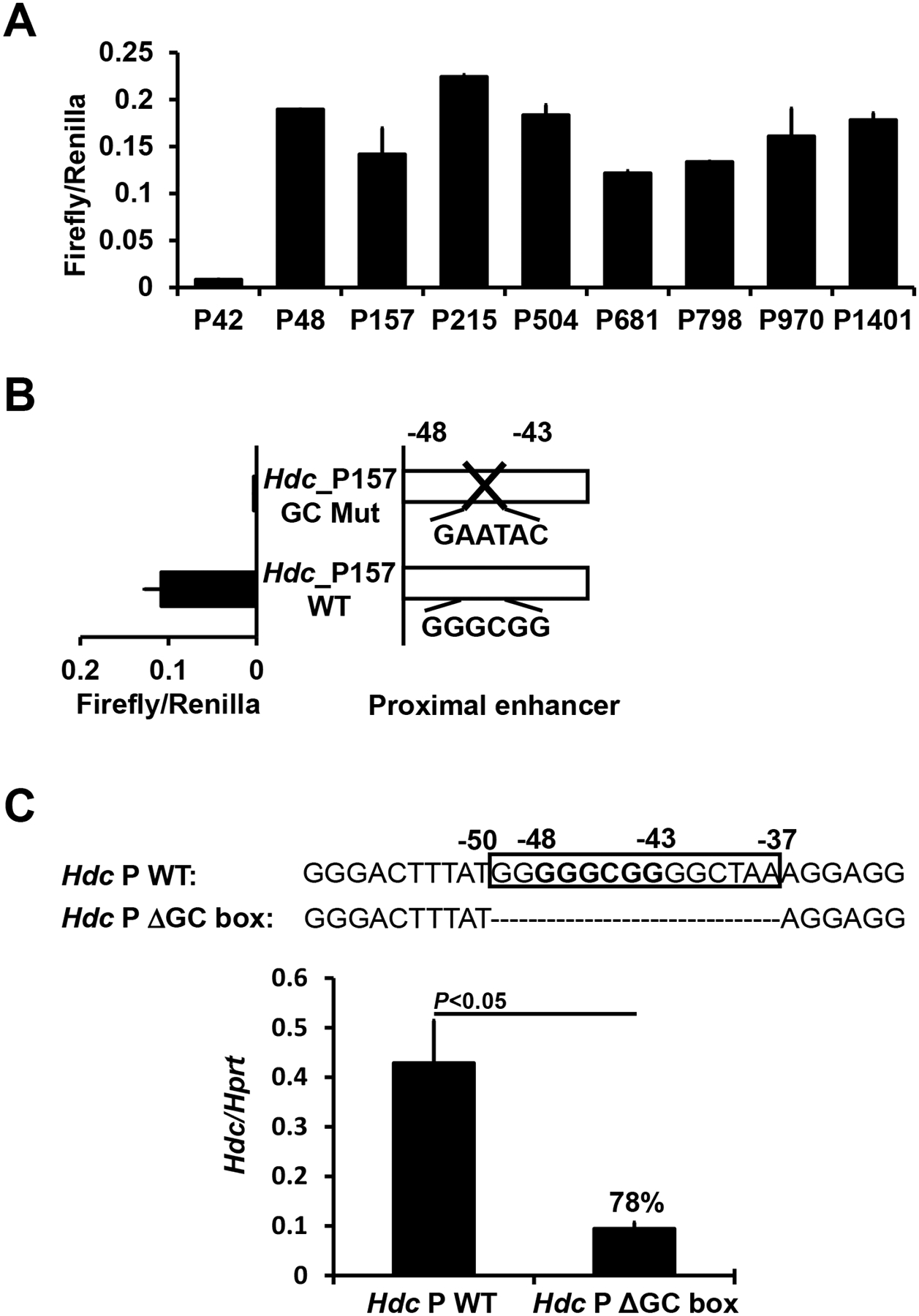
A. Deletional analysis of the Hdc PE in CFTL-15 mast cells. Luciferase reporter genes were transfected into CFTL-15 mast cells. Error bars represent mean ± SEM, n=4 transfectants from 2 independent experiments. B. Mutational analysis of the GC box in the −157 Hdc PE using a luciferase reporter gene. A luciferase reporter gene containing the GC box mutation was transfected into CFTL-15 mast cells. Mean ± SEMs were calculated from 3 transfectants from 3 independent experiments. C. qPCR analysis of Hdc mRNA expression in GC box deleted CFTL15 mast cell clones (mean ± SEM, n=3 colonies). CFTL15 mast cells were transfected with the four bicistronic RNA guides. Clones derived from single transfected cells were obtained. The percentages indicate the extent of reduction in Hdc mRNA expression. P values were calculated using the Mann-Whitney U test.
Mice deficient in the Hdc GC box have greatly reduced synthesis of histamine and IgE-mediated anaphylaxis after antigenic challenge.
To validate the findings obtained using the IL-3-dependent non-transformed mast cell line CFTL-15, we generated mice deficient in the Hdc GC box using a single sgRNA guide as shown in Fig. 4A to make double-stranded breaks and a homology-directed repair template that lacks the GC box to repair the breaks. A founder strain of mice with a six-nucleotide deletion of the Hdc GC box was verified by sequencing (Fig. 4A). Mice deficient in the Hdc GC box (HdcGCbox−/−) tended to have slightly reduced numbers of peritoneal mast cells (Fig. E1A and B). Surprisingly, FcεRIα expression, but not cKit expression, on the peritoneal mast cells in the HdcGCbox−/− mice tended to be elevated compared to control HdcGCbox+/+ mice (Fig. E1C). To determine whether the tendency towards increased peritoneal mast cell FcεRIα expression was a direct or indirect consequence of the Hdc GC box deletion, we compared FcεRIα and cKit expression by BMMCs derived from HdcGCbox+/+ and HdcGCbox−/− mice in vitro, where the indirect extrinsic interactions that occur in vivo are reduced. We found a noticeable difference in FcεRIα expression but not cKit expression on HdcGCbox−/− BMMCs relative to HdcGCbox+/+ BMMCs. A small population of BMMCs expressing relatively low levels of FcεRIα was observed in HdcGCbox+/+ but not HdcGCbox−/− BMMCs (Fig. E2). The difference may be noticeable but it is not statistically significant. In contrast, Hdc mRNA expression was considerably and significantly reduced in both BMMCs (90% reduction) and basophils (77% reduction) (Fig. 4B). To determine whether the Hdc GC box only regulates the transcription of the Hdc gene, we examined mRNA expression of a neighboring gene Gabpb1, which is located just 10 kb upstream of the Hdc gene. Deletion of the Hdc GC box did not affect Gabpb1 mRNA expression, supporting that the GC box in the PE of the Hdc gene acts explicitly on the transcription of the Hdc gene (Fig. 4B).
Figure 4. The GC box in the Hdc PE is critical for histamine synthesis and IgE/mast cell-mediated anaphylaxis.
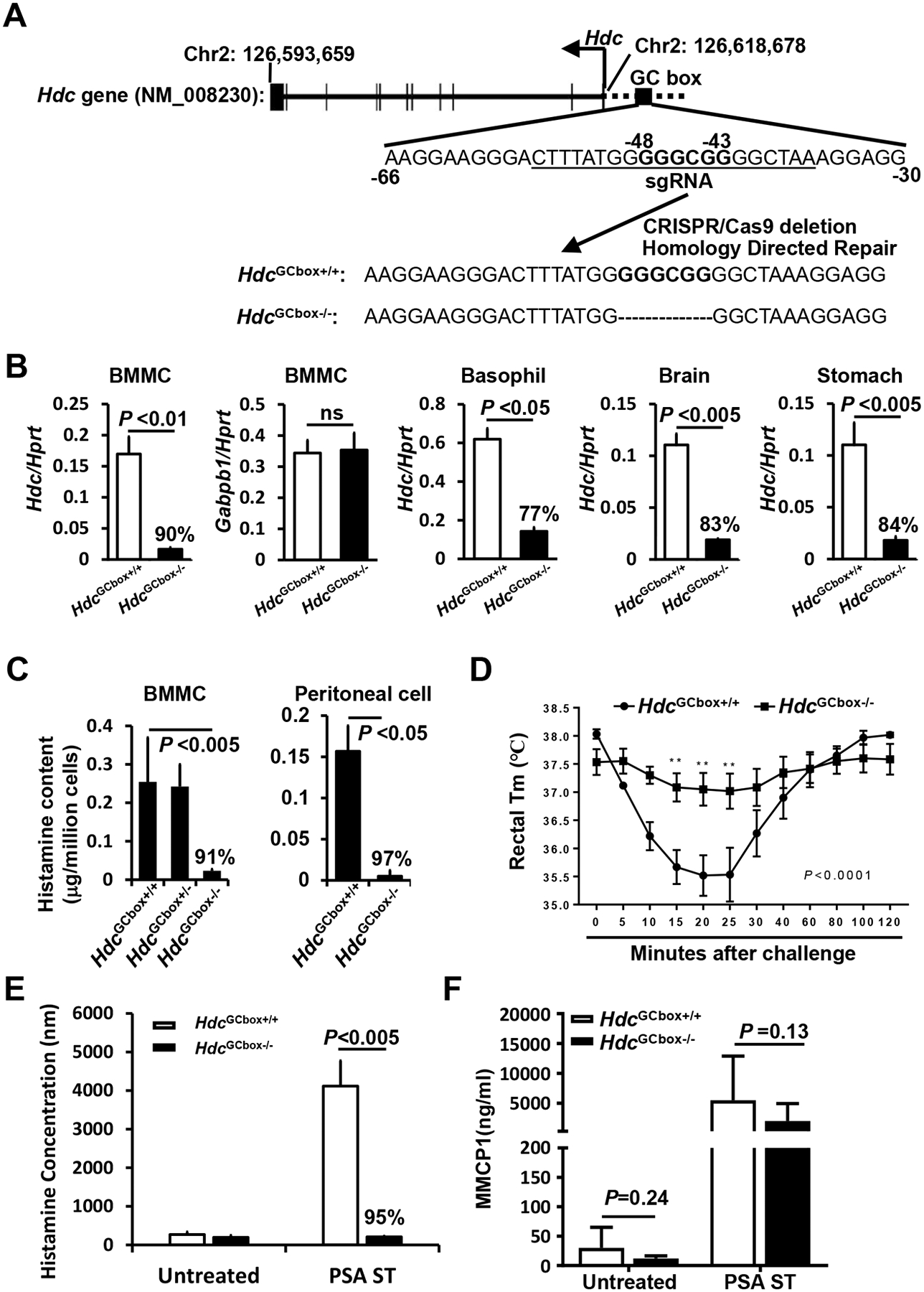
A. mice deficient in GC box (GGGCGG) were generated by a single sgRNA (underlined) targeting the GC box located from−48 to −43 in the proximal promoter of the Hdc gene. B. qPCR analysis of Hdc and Gabpb1 mRNA expression in cells isolated from HdcGCbox+/+ and HdcGC−/− mice (n=4, pooled from two experiments). C. histamine contents in BMMCs derived from HdcGCbox+/+ and HdcGC−/− mice (n=6, two experiments). D. PSA of HdcGCbox+/+ and HdcGC−/− mice (n=6, two experiments). E. histamine release in the plasma of HdcGCbox+/+ and HdcGC−/− mice that were treated with IgE anti-TNP antibody followed by challenge with BSA-TNP. Blood was drawn five minutes after challenge (n=6, two experiments). F. MMCP1 concentrations in the sera of HdcGCbox+/+ and HdcGC−/− mice that were treated with IgE anti-TNP antibody and BSA-TNP for one hour (n=6, two experiments). P values were calculated using the Mann-Whitney U test.
FIG E1. Mice deficient in the Hdc GC box had slightly reduced numbers and expressed higher FcεRIα and cKit expression levels in peritoneal mast cells.
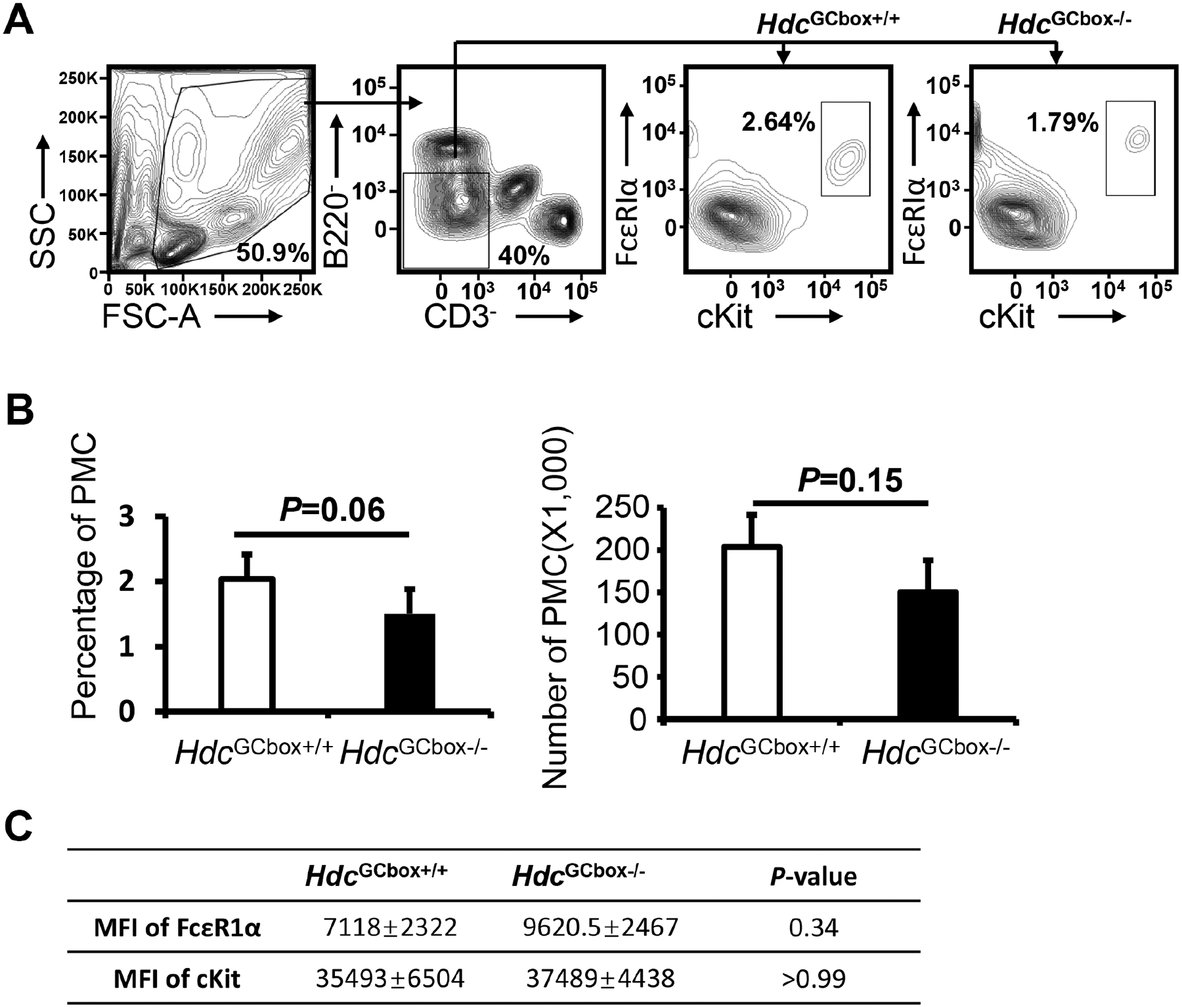
A. Peritoneal cells were collected from WT and Hdc GC box (HdcGCbox−/−) mice and analyzed by FACS, DAPI− CD3ε− B220− FcεRIα + c-Kit+ peritoneal mast cells were shown in the gates. B. Percentage and total numbers of peritoneal mast cells in the WT and Hdc GC box (HdcGCbox−/−) mice (mean ± SEM, n=4 from 2 independent experiments). C. MFI of FcεRIα and cKit of the peritoneal mast cells in the WT and Hdc GC box (HdcGCbox−/−) mice (mean ± SEM, n=4 from 2 independent experiments). P values were calculated using the Mann-Whitney U test.
FIG E2. Mice deficient in the Hdc GC box lack a population of BMMCs that have low FcεRI expression.
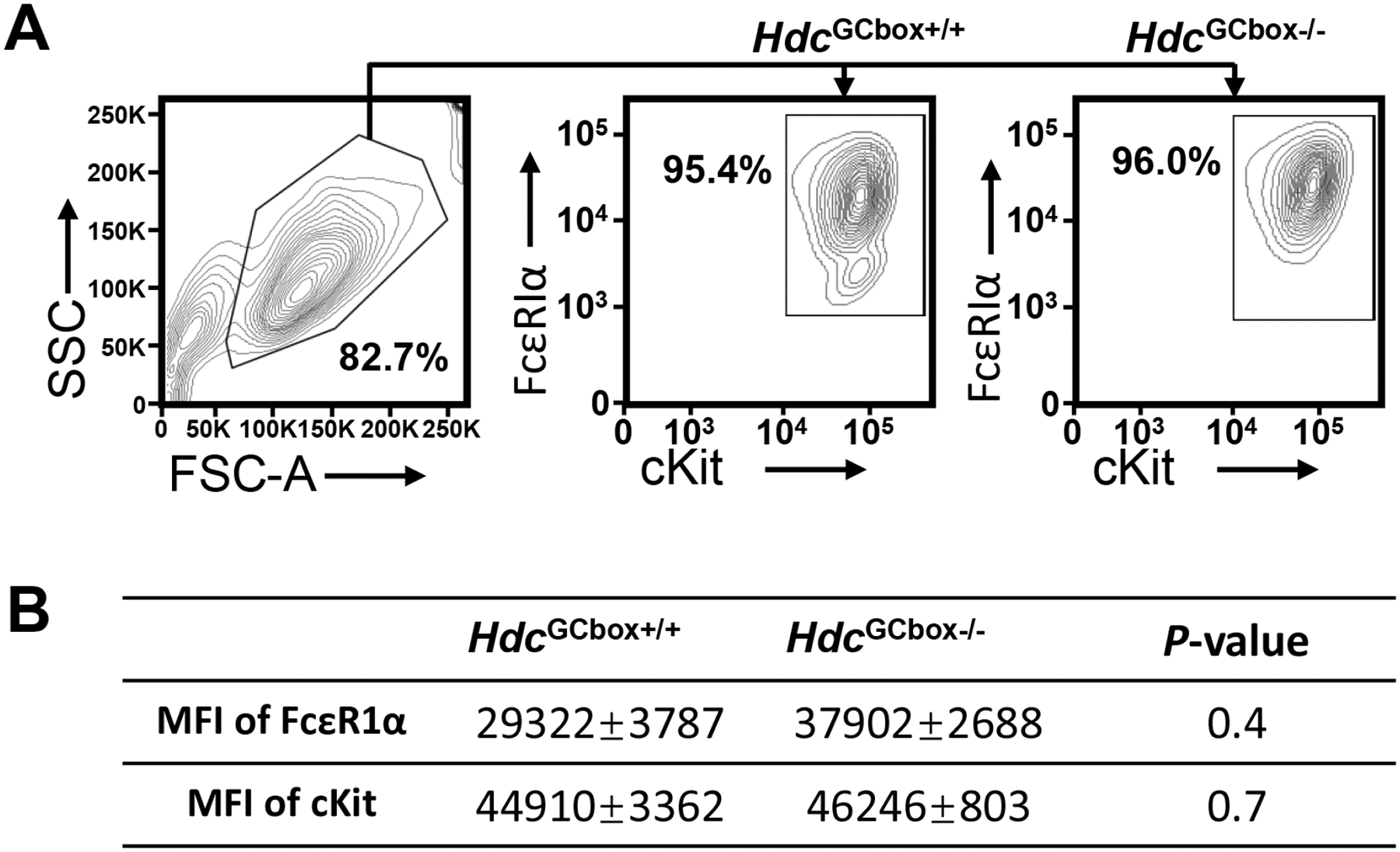
A. BMMCs were cultured from WT and Hdc GC box (HdcGCbox−/−) mice and analyzed by FACS using gates shown. B. MFI of FcεRIα and c-Kit on WT and HdcGCbox−/− BMMCs (mean ± SEM, n=3 from 2 independent experiments). P values were calculated using the Mann-Whitney U test.
In addition to its activity as an allergic mediator, histamine is a neurotransmitter and a regulator of gastric acid secretion5. Histaminergic neurons in the basal ganglia of the brain express Hdc mRNA and synthesize histamine. In the stomach, enterochromaffin-like cells express Hdc mRNA and synthesize histamine, which stimulates parietal cells in the stomach to secrete stomach acid21, 22. We found that the levels of Hdc mRNA were significantly decreased in brain cells (83% reduction) and stomach cells (84% reduction) in the HdcGCbox−/− mice (Fig. 4B). Consistent with the reduction of Hdc mRNA in the HdcGCbox−/− mice, histamine contents in BMMCs and peritoneal mast cells derived from the HdcGCbox−/− mice were dramatically decreased (91% and 97% reduction, respectively, Fig. 4C).
Histamine plays a critical role in IgE/mast cell-mediated anaphylaxis. We found that the HdcGCbox−/− mice developed little or no passive systemic anaphylaxis (Fig. 4D) and released little histamine (Fig. 4E) after the injection of IgE anti-TNP antibody and challenge with TNP-BSA (95% reduction in histamine release relative to HdcGCbox+/+ mice). In contrast, mast cell protease MMCP-1 release to the blood was reduced moderately, if at all, in HdcGCbox−/− mice compared to HdcGCbox+/+ mice (Fig. 4F), indicating that the Hdc GC box deletion primarily affects histamine synthesis. These data demonstrate that the GC box in the proximal Hdc enhancer is critical for histamine synthesis and histamine-mediated anaphylaxis in vivo.
The GC box plays a minor role in regulating chromatin modification and chromatin accessibility at the distal enhancers of the Hdc gene
Because distal enhancers can interact with proximal enhancers and core promoters through a looping mechanism, it is possible that the lack of an Hdc GC box in the proximal enhancer could affect enhancer structures of distal enhancers16. For this reason, we investigated whether the Hdc GC box regulates chromatin structure at the distal enhancers. We performed Omni-ATAC-seq and H3K27ac ChIP-seq in HdcGCbox+/+ and HdcGCbox−/− BMMCs. Chromatin accessibility at the PE enhancer near the GC box was notably reduced in HdcGCbox−/− BMMCs. Reads at two out of three peaks (peak one and two) near the deleted GC Box were reduced by 65% and 43%, respectively (Fig. 5A). However, H3K27ac histone modification was not reduced by the absence of the Hdc GC box. Instead, the H3K27ac peak at the −8.8 position was elevated (Fig. 5B), indicating that the Hdc GC box plays a minor role in chromatin accessibility at the PE enhancer and does not contribute to chromatin accessibility at the distal enhancers. These data demonstrate that the Hdc GC box plays a minor role in regulating chromatin modification and chromatin accessibility at distal enhancers of the Hdc gene.
Figure 5. GC box only promotes chromatin accessibility, but not H3K27ac modification, at the Hdc PE.
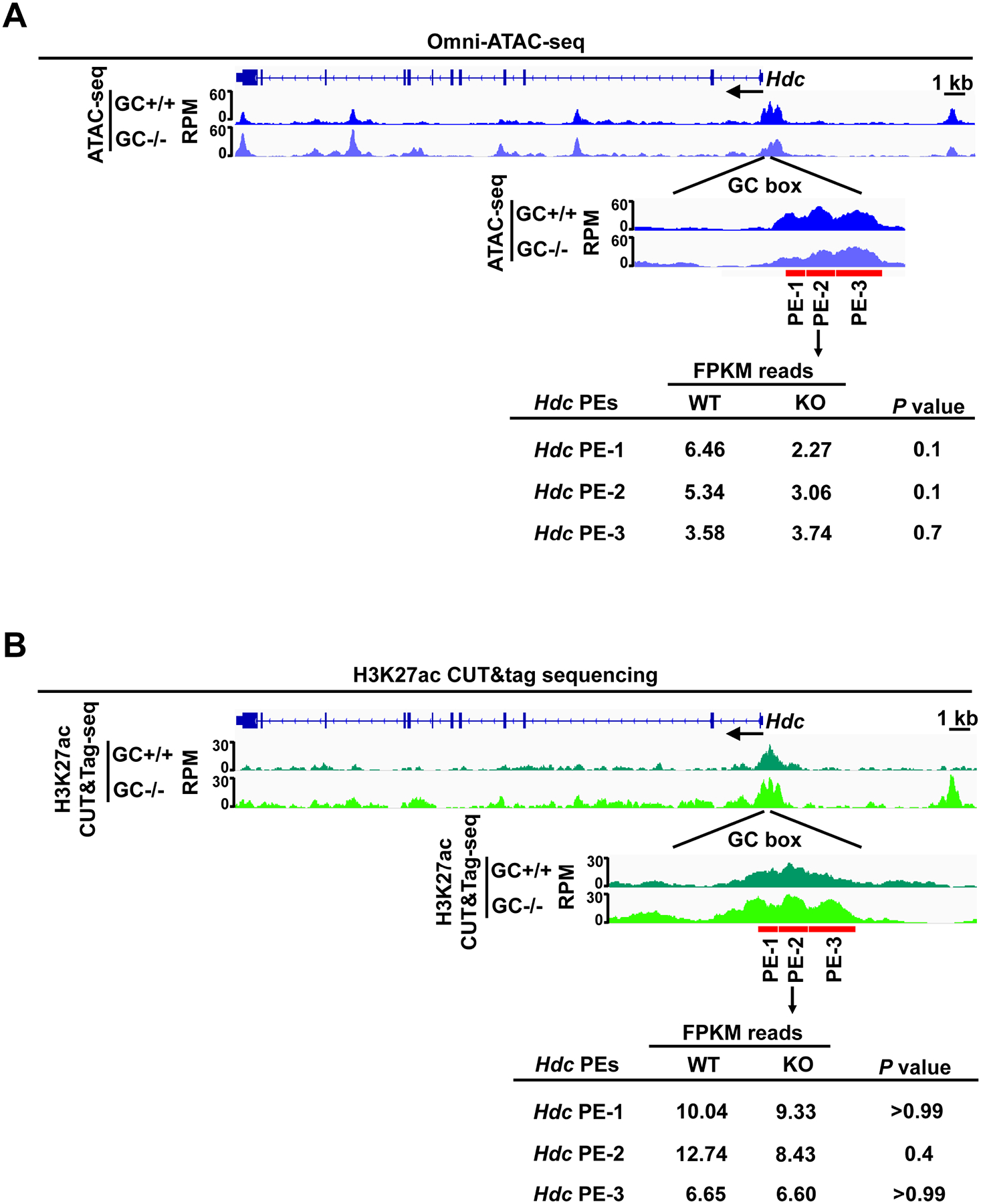
A. Representative Integrated Genome Viewer (IGV) tracks from Omni-ATAC-seq (n=3). B. Representative Integrated Genome Viewer (IGV) tracks from H3K27ac CUT&Tag-seq representing three biological samples (n=3). Statistical significance was analyzed with a two-tailed student’s t test.
The GC box is required for HDC gene transcription and histamine synthesis in human mast cells
To determine whether human HDC is also regulated by the GC box in the context of chromatin, we designed four bicistronic RNA guides targeting the human HDC GC box in the proximal enhancers (−153 to −495, Fig. 6 A). Human mast cell line LAD2 cells were transduced with lentivirus containing the four bicistronic sgRNA guides. LAD2 cells expressing BFP, GFP, RFP, and Thy1.1 (17.6% of transfected cells were positive for 4 colors) were FACS-sorted, using gates shown in Fig. 6B. We achieved 91% homozygous deletion of the GC box (Fig. 6C). Deletion of the HDC GC box resulted in 77% reduction in HDC mRNA expression (Fig. 6D). Consistent with this, histamine content in the GC box-deleted LAD2 cells was reduced by 86% (Fig. 6E). These results demonstrate that the human HDC GC box is as critical for HDC gene transcription in human MCs as in mouse MCs.
Figure 6. The HDC GC box in the human HDC gene is critically important for HDC gene transcription and histamine synthesis.
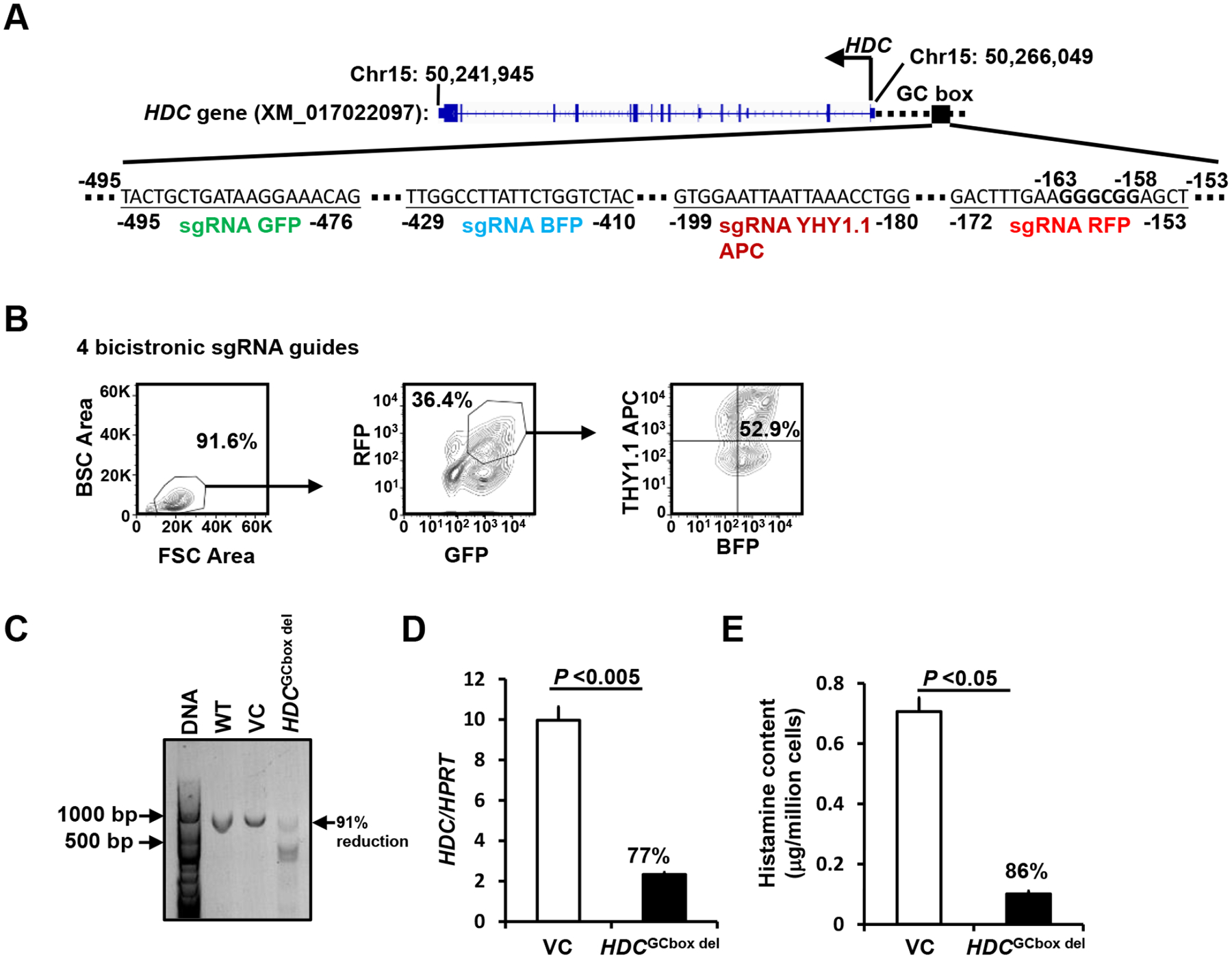
A. Bicistronic BFP, GFP, RFP, and Thy1.1 sgRNA guides targeting the human HDC GC box located in −153 to −495. B. FACS sorting gates. LAD2 cells were transduced with lentivirus containing four bicistronic sgRNA guides. BFP+, GFP+, RFP+, and Thy1.1 APC+ transfectant cells were FACS-sorted using the sorting gates indicated (Fig. 2A). C. DNA deletion efficiency analysis. GC box fragments from untransduced WT, or FACS-sorted BFP+, GFP+, RFP+, and Thy1.1APC+ cells transduced with lenti vector without sgRNA guide vector controls (VCs) and with four sgRNA guides targeting the human HDC GC box were analyzed with PCR. Percentage of reduction was calculated by reduction in density of the WT band. D. HDC mRNA expression in the FACS-sorted BFP+, GFP+, RFP+, and Thy1.1APC+ cells was measured by qPCR (mean ± SEM, n=4 from 2 independent experiments). E. Histamine content in the FACS-sorted BFP+, GFP+, RFP+, and Thy1.1APC+ cells was measured by ELISA (mean ± SEM, n=4 from 2 independent experiments). The percentages indicate the percent reduction in HDC mRNA expression and histamine content relative to VCs. P values were calculated using the Mann-Whitney U test.
DISCUSSION
The work described in this paper contributes to the study and understanding of eukaryote gene transcription in general and Hdc gene transcription in particular in several ways. It: (1) moves beyond in vitro studies with luciferase reporter genes to in vivo studies that incorporate the effects of chromatin; (2) reveals and validates an improved method for CRISPR-mediated homozygous deletion of a gene segment; (3) demonstrates a critical role for the proximal enhancer GC box in both mouse and human HDC gene expression despite its lack of influence on the chromatin structure of Hdc distal enhancers; and (4) provides a novel and useful mouse model in which nearly absent Hdc transcription blocks histamine synthesis and IgE-mediated systemic anaphylaxis with little or no reduction in mast cell number, maturation, or degranulation.
Genes are composed of coding and noncoding DNA regions, spanning tens of kbs in length. Enhancers are noncoding DNA regions that contain cis regulatory regions that are bound by transcription factors and co-factors, such as histone modifying enzymes that can add and remove acetylated or methylated lysines in the histones. Enhancers are often surrounded by chromatin that has histones with acetylated lysines that make the enhancers more accessible to transcription factors and co-factors. The traditional method of analyzing enhancer activity transfects cell lines with reporter genes; although this is a powerful approach for predicting enhancer activity, it has the limitation of ignoring the role of chromatin in enhancer function. The remarkable progress that has been made in CRISPR gene editing technology now enables analysis of enhancer activity in the context of chromatin by deleting enhancers in situ. However, the typical CRISPR deletion method, which uses two sgRNA guides, often deletes the targeted gene in only one chromosome, producing heterozygotes that lack a phenotype. To overcome this technical challenge, we have used four sgRNA guides, each expressing a bicistronic gene that encodes Thy1.1 or a fluorescence protein (BFP, GFP, or RFP), to target an enhancer. By coupling this with fluorescence activated cell sorting to select for cells that express all four marker genes, we achieved 84% to 94% enhancer deletion at both chromosomes without having to perform single-cell cloning. This technical improvement has allowed us to analyze mouse and human HDC gene transcription in the context of chromatin. Prior to the development of this technique, we used a luciferase reporter gene assay with the murine CFTL-15 MC cell line to demonstrate that the distal Hdc E−8.8 enhancer contributes to Hdc gene transcription. Now, using a novel sgRNA technique, we have demonstrated that this enhancer contributes to mouse Hdc gene transcription in the context of chromatin. Furthermore, our work has revealed a critical role of the GC box in both mouse and human HDC gene in the context of chromatin.
The GC box has a consensus sequence of GGGCGG and is often located near the promoter of eukaryotic genes. Members of the 3-zinc finger TF family, which includes SP1, Krox/Egr, Wilms’ tumor, MIGI, and CREA, can bind to the GC box (GGGCGG)23, where they function similarly to TFs that bind to more distal enhancers. There is only a single GC box in the mouse and human HDC promoters, although most promoters that express the GC box express it multiple times24. Remarkably, deletion of the single GC box in the mouse or human HDC proximal enhancer in the context of chromatin profoundly reduces Hdc gene transcription. The importance of the mouse Hdc GC box in allergy is demonstrated further by our study of its role in a mouse model of anaphylaxis. This validation of the GC box function in mice adds rigor and greater biologic relevance to previously published conclusions, which depended on studies with a single cell line that had been generated from a single mouse strain and used a mast cell line that had been cultured for a long time. Our in vivo validation allows broader generalization of our finding that the Hdc GC box is required for Hdc gene transcription, histamine synthesis, and histamine-mediated anaphylaxis. Currently, it is not clear which members of 3-zinc finger TF family bind to the mouse and human HDC GC box under the resting and antigen-stimulated conditions.
The functioning of distal enhancers, proximal enhancers, and core promoters through a looping mechanism20 makes it possible that modifying or eliminating the GC box in proximal enhancer could result in profound changes in enhancer structures at the distal enhancers. However, despite its importance in Hdc gene transcription, absence of the GC box in the proximal Hdc enhancer slightly alters and does not reduce the chromatin structures of distal enhancers of the mouse Hdc gene. This is consistent with the view that other transcription factors, such as GATA2, control Hdc gene chromatin accessibility11. The GC box thus might function as an enhancer to promote the assembly of the transcriptional complex.
In contrast to the existing Hdc gene knockout mice25, the HdcGCbox−/− mice that we have generated show only minor effects on mast cell maturation. The number of peritoneal cavity mast cells, which are considered mature mast cells, appears to be slightly, but not significantly reduced. Surprisingly, cKIT and FcεRIα on HdcGCbox−/− peritoneal cavity mast cells were expressed at normal or higher levels than in HdcGCbox+/+ mast cells. Consistent with the notion that the Hdc GC box primarily regulates Hdc gene transcription, serum mast cell protease MMCP1 levels in naive HdcGCbox−/− mice and HdcGCbox−/− mice that had undergone passive systemic anaphylaxis were reduced moderately, if at all. Any reduction might have been caused by a proportionate reduction in the number of mature mast cells in HdcGCbox−/− mice. Because HdcGCbox−/− mice have relatively normal mast cell differentiation, these mice are more suitable than global Hdc-deficient mice for studying the importance of the Hdc gene in mast cell/IgE-mediated anaphylaxis. Additionally, because HdcGCbox−/− mice also have reduced Hdc gene transcription in stomach and brain tissues, these mice can be used to study how the Hdc gene is regulated in neurons and in gastric enterochromaffin-like cells.
METHODS
Mice
sgRNA (TTTATGGGGGCGGGGC) was designed using the online CRISPick tool from Broad Institute (https://portals.broadinstitute.org/) and was injected into BALB/cByJ zygotes along with ssDNA HDR template (CAAATCAGAAAGGACGGGGACAAGAGAGGCTGAAATTAAGCCTGAAGGAAGGGACTTTATGGGGCTAAAGGAGGGGAATAGGGAACTCCTTAAATAAGAGATGCACTGGCTGCCAGGGAGTGCAC) as describedE1. BALB/cByJ mice (Stock No. 001026) were purchased from the Jackson Laboratory (Bar Harbor, ME). The six-nucleotide deletion (GGGCGG) in HdcGCbox+/−mice was verified by sequencing. Two out of eleven founders that have an exact GGGCGG six-nucleotide deletion were selected for further breeding. Mice deficient in the GC box were backcrossed to BALB/c mice for one generation and designated as Balb/c-Hdcem1huhu/J mice. These mice are available from the Jackson Laboratory (Stock No. 037608). Genotyping was carried out using the following primers: forward, TCAAATCAGAAAGGACGGGGA; reverse, ATTTAAGGAGTTCCCTATTCCCCTC.
BMMC and LAD2 cell culture
Bone marrow-derived mast cells (BMMCs) were cultured from bone marrow cells of BALB/c mice in Iscove’s DMEM (10016CV, Corning™ cellgro™, Manassas, VA) plus 10% FBS,100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM β-mercaptoethanol in the presence of 20 ng/mL IL-3 for four weeks. Over 99% of BMMCs were mast cells had a mast cell phenotype by FACS analysis (FcεRIα+ c-Kit+). Human mast cell line LAD2 was provided by Drs. Dean D. Metcalfe and Arnold S. Kirshenbaum (National Institutes of Health, Bethesda, MD) and cultured in AIM V™ Medium (Cat#31035025, Thermo Fisher Scientific, Waltham, MA) plus 2 mM beta-mercaptoethanol, 2 mM L-Glutamine (Cat#25005CI, Thermo Fisher Scientific, Waltham, MA), 100 units/ml penicillin, and 100 μg/ml streptomycin in the presence of 100 ng/ml recombinant human SCF (Cat#300–07, Peprotech, Cranbury, NJ).
H3K27ac CUT&Tag-seq
The Cleavage Under Targets and Tagmentation (CUT&Tag) assay for H3K27ac modification was performed according to the published protocolE2. Omni-ATAC-seq was performed as previously describedE3, 4. BMMCs (1 × 105) were treated with concanavalin A-coated beads (Bangs Laboratories, BP531) and incubated with rabbit anti-H3K27ac antibody (Abcam, ab4729) at 4 °C overnight. The cells were then incubated with guinea pig anti-rabbit antibody (Antibodies online, ABIN101961) at room temperature for 30 minutes. The cells were washed and incubated with pA-Tn5 adapter complex (Diagenode, C01070001) at room temperature for 1 hour on a rotator. The cell pellet was resuspended in 300 μl tagmentation buffer and incubated at 37 °C for 1 hour. The reaction was stopped by adding 10 μL 0.5M EDTA, 3 μL 10% SDS, and 2.5 μL 20 mg/mL Proteinase K, and cells were digested at 37 °C overnight. Then, DNA was extracted using phenol chloroform and diluted in 25 μl TE buffer with 1/400 RNAse A. The tagmented DNA was enriched by PCR amplification using Universal i5 primer and uniquely barcoded i7 primers under the following conditions: 72 °C for 5 min, and 13 cycles: 98 °C for 30 sec, 98 °C for 10 sec, 63 °C for 10 sec, 72°C for 1 min and hold at 8 °C. The PCR products were cleaned up and size-selected using Ampure XP beads (Beckman Coulter, A63880). The quality and quantity of libraries were analyzed with the Agilent TapeStation 4200 system. Pair-ended sequencing of DNA libraries was performed on an Illumina NovaSEQ6000 platform.
Omni-ATAC-seq
BMMCs (5 × 104) either untreated or activated by IgE receptor crosslinking, were spun down and washed once with cold PBS. The cells were resuspended in 50 μl cold ATAC-RSB-lysis buffer (10mM Tris-HCl pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% NP-40, 0.1% Tween-20 and 0.01% digitonin) and incubated for 3 minutes. The lysis buffer was immediately washed out with 1 mL ATAC-RSB buffer (10mM Tris-HCl pH 7.4, 10mM NaCl, 3mM MgCl2 and 0.1% Tween-20). The cell pellet was resuspended in 50 μl transposition mix (25 μl 2X TD buffer, 2.5 μl transposase (Illumina, FC-121-1030), 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl H2O) and incubated at 37 °C for 30 minutes. The reaction was stopped by adding 2.5 μl of 0.5M EDTA, pH 8, and transposed DNA was purified with a Qiagen MiniElute PCR purification kit (Qiagen). Purified DNA was amplified using the following conditions: 72°C for 5 min, 98 °C for 30 s, and 7 cycles: 98 °C for 10 s, 63 °C for 30 s, 72 °C for 1 min. The amplified libraries were purified, size-selected, and the quality and quantity of libraries were assessed with an Agilent Technologies 2100 Bioanalyzer. The pair-ended sequencing of DNA libraries was performed with an Illumina NovaSEQ6000 platform.
NGS data analysis
Raw sequencing reads (average 118.2 million reads, 2 biological replicates) were adaptor trimmed by Trimmomatic 0.33 and the quality of sequenced data was analyzed by FastQC. The trimmed reads were aligned to the mm10 reference genome using Bowtie2 with -very-sensitive and -x 2000 parameters. The read alignments were filtered using SAMtools (version 1.7) to remove mitochondrial genome and PCR duplicates. Peaks were identified by MACS2 with the q-value cut-off of 0.05 and the sequencing data was displayed using IGV. The representative ATAC-seq IGV tracks showed in the figures were generated from one biological sample, representing two or three biological samples with similar pattens.
Luciferase reporter constructs and assay
CFTL-15 cells (5 × 106) were electroporated with 10 μg of luciferase plasmid and 0.5 μg of Renilla vector at 450 V and 500 microfarads using a Bio-Rad Gene Pulser. Twenty hours after electroporation or transfection, cells were collected, and luciferase activities were measured by using an InfiniteMlOOO® microplate reader (Tecan Systems, Inc., San Jose, CA) and the Dual-Luciferase reporter assay system (E1960, Promega). Transcriptional activity was normalized as the ratio of luciferase activity divided by Renilla activity.
Deletion of GC boxes using four bicistronic sgRNA guides
The sgRNA sequences targeting Hdc enhancers or transcription factor exons were designed using the online CRISPick tool from Broad Institute (https://portals.broadinstitute.org/). Each of four sgRNA sequences targeting the same enhancer was cloned into LentiCRISPRv2GFP (Addgene, Plasmid # 82416), LentiCRISPRv2-mCherry (Addgene, Plasmid #99154), LentiCRISPRv2BFP, or LentiCRISPRv2THY1.1 vectors. LentiCRISPRv2THY1.1 vector was modified by replacing the Gfp gene in LentiCRISPRv2GFP with the gene encoding THY1.1, using the BsmBI cloning site. CFTL-15 cells were either transfected with Lenti plasmids or transduced with lentivirus containing the bicistronic sgRNA guides. For transduction, lentiviruses were prepared using method describe in our publication. CFTL-15 mast cells were transduced using spin infections as described; For transfection, CFTL-15 cells (10 × 106) were electroporated with 10 μg of each CRISPR plasmid using a Bio-Rad Gene Pulser at 450 V and 500 microfarads. Twenty-four hours after electroporation, the cells that expressed BFP, GFP, RFP, and Thy1.1 were FACS-sorted. The sorted cells were subjected to DNA analysis of GC box deletion. The intensities of DNA fragments were measured using Imagej (NIH, MD).
Key messages:
The HDC GC box within the proximal enhancer of the mouse and human HDC gene is essential for Hdc gene transcription and histamine synthesis in mouse and human mast cells.
The mouse Hdc GC box is required for histamine-mediated anaphylaxis in vivo.
Acknowledgements
We thank lab members for thoughtful discussions. We thank Ms. Marlene Gallegos-Sanchez, Kirby Motsinger for technical assistance. We are grateful to Dr. Melissa Brown of Northwestern University for providing us with the CFTL-15 mast cell line; to Drs. Dean Metcalfe and Arnold S. Kirshenbaum of National Institute of Allergy and Infectious Diseases for providing us with the human mast cell line LAD2. We thank Dr. Jennifer Matsuda and her staff in the Regional Mouse Genetics Core Facility at National Jewish Health for generating mice deficient in GC box. We thank the staff at the Animal Care Facility for animal care and other technical assistance. We thank Josh Loomis and Shirley Sobus of the Cytometry Core cell sorting and FACS analysis. We thank staff at genomic core facility at University of Colorado and the Gene and Environment Center at National Jewish Health for NGS sequencing.
Funding
Supported by grants from the National Institutes of Health R01AI107022 and R01AI083986 (H.H.), R01AI145991 (F.D.F.).
Abbreviations used:
- BMMCs
Bone marrow-derived mast cells
- ATAC-seq
Assay for Transposase-Accessible Chromatin using sequencing
- CUT&Tag-seq
The Cleavage Under Targets and Tagmentation
- H3K27Ac
Acetylation of lysine residue 27 on histone 3
- FACS
Fluorescence activated cell sorting
- GC box
It contains transcription factor binding consensus sequence GGGCGG
- HDC
Histidine decarboxylase
- PCMCs
Peritoneal cavity mast cells
- MMCP1
Mucosal mast cell protease-1
- PSA
Passive systemic anaphylaxis
- TNP
2, 4, 6-Trinitrophenyl
- TSS
Transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflict of interest: The authors declare no conflicts of interest.
References
- 1.Simons FE. Anaphylaxis. J Allergy Clin Immunol 2010; 125:S161–81. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol 2010; 125:S116–25. [DOI] [PubMed] [Google Scholar]
- 3.Hogan SP, Wang YH, Strait R, Finkelman FD. Food-induced anaphylaxis: mast cells as modulators of anaphylactic severity. Semin Immunopathol 2012; 34:643–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vickery BP, Chin S, Burks AW. Pathophysiology of food allergy. Pediatr Clin North Am 2011; 58:363–76, ix–x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Li Y, Liang J, Finkelman FD. Molecular Regulation of Histamine Synthesis. Front Immunol 2018; 9:1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki-Ishigaki S, Numayama-Tsuruta K, Kuramasu A, Sakurai E, Makabe Y, Shimura S, et al. The mouse L-histidine decarboxylase gene: structure and transcriptional regulation by CpG methylation in the promoter region. Nucleic Acids Res 2000; 28:2627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunami K, Ohtsu H, Tsuchikawa M, Higuchi T, Ishibashi K, Shida A, et al. Structure of the L-histidine decarboxylase gene. J Biol Chem 1994; 269:1554–9. [PubMed] [Google Scholar]
- 8.Hirasawa N, Torigoe M, Kano K, Ohuchi K. Involvement of Sp1 in lipopolysaccharide-induced expression of HDC mRNA in RAW 264 cells. Biochem Biophys Res Commun 2006; 349:833–7. [DOI] [PubMed] [Google Scholar]
- 9.Ai W, Liu Y, Wang TC. Yin yang 1 (YY1) represses histidine decarboxylase gene expression with SREBP-1a in part through an upstream Sp1 site. Am J Physiol Gastrointest Liver Physiol 2006; 290:G1096–104. [DOI] [PubMed] [Google Scholar]
- 10.Ai W, Liu Y, Langlois M, Wang TC. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem 2004; 279:8684–93. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Liu B, Harmacek L, Long Z, Liang J, Lukin K, et al. The transcription factors GATA2 and MITF regulate Hdc gene expression in mast cells and are required for IgE/mast cell-mediated anaphylaxis. J Allergy Clin Immunol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaffner W. Enhancers, enhancers - from their discovery to today’s universe of transcription enhancers. Biol Chem 2015; 396:311–27. [DOI] [PubMed] [Google Scholar]
- 13.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 2019; 20:437–55. [DOI] [PubMed] [Google Scholar]
- 14.Vaishnav ED, de Boer CG, Molinet J, Yassour M, Fan L, Adiconis X, et al. The evolution, evolvability and engineering of gene regulatory DNA. Nature 2022; 603:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470:279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311–8. [DOI] [PubMed] [Google Scholar]
- 18.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010; 107:21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012; 13:613–26. [DOI] [PubMed] [Google Scholar]
- 20.Soutourina J. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 2018; 19:262–74. [DOI] [PubMed] [Google Scholar]
- 21.Hersey SJ, Sachs G. Gastric acid secretion. Physiol Rev 1995; 75:155–89. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka S, Hamada K, Yamada N, Sugita Y, Tonai S, Hunyady B, et al. Gastric acid secretion in L-histidine decarboxylase-deficient mice. Gastroenterology 2002; 122:145–55. [DOI] [PubMed] [Google Scholar]
- 23.Lundin M, Nehlin JO, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol 1994; 14:1979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blake MC, Jambou RC, Swick AG, Kahn JW, Azizkhan JC. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol Cell Biol 1990; 10:6632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsu H, Tanaka S, Terui T, Hori Y, Makabe-Kobayashi Y, Pejler G, et al. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS Lett 2001; 502:53–6. [DOI] [PubMed] [Google Scholar]
REFERENCES
- E1.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153:910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019; 10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017; 14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Li Y, Gao J, Kamran M, Harmacek L, Danhorn T, Leach SM, et al. GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super-enhancers. Nat Commun 2021; 12:494. [DOI] [PMC free article] [PubMed] [Google Scholar]


