Abstract
PHD fingers comprise a large and well-established family of epigenetic readers that recognize histone H3. A typical PHD finger binds to the unmodified or methylated amino-terminal tail of H3. This interaction is highly specific and can be regulated by posttranslational modifications in H3 and other domains present in the protein. However, a set of PHD fingers have recently been shown to bind non-histone proteins, H3 mimetics, and DNA. In this review, we highlight the molecular mechanisms by which PHD fingers interact with ligands other than the amino terminus of H3 and discuss similarities and differences in engaging with histone and non-histone binding partners.
Keywords: PHD, histone, DNA, binding mechanism
PHD fingers: readers of histone H3
The plant homeodomain (PHD) finger is found in eukaryotic nuclear proteins involved in the regulation of gene transcription and chromatin remodeling (see Glossary). This small, ~65-residue evolutionarily conserved cysteine-rich domain can be distinguished by its canonical C4HC3 motif that coordinates two zinc ions in a cross-braced topology. The typical PHD finger folds into a short double-stranded anti-parallel β sheet, one or two small α helices, and several variable-length loops connecting the zinc coordinating residues. Often, the PHD finger is present in a protein as a single domain or in multiple copies of structurally and functionally independent domains, but it can also form integrated modules, coupling to a zinc-knuckle or to another PHD finger. The assembly of two PHD fingers create a double PHD finger (DPF) domain, and the assembly of two PHD fingers linked through a zinc-knuckle form a PZP domain.
Extensive functional and biochemical studies in the past decade revealed diverse biological roles of the PHD fingers. In 2006-2007, two subsets of PHD fingers were identified: one subset is capable of recognizing trimethylated lysine 4 of histone H3 (H3K4me3) and another binds to unmodified H3 [1-6]. The highly specific interactions with histones mediate many fundamental cellular processes, particularly gene expression, DNA damage repair, nucleosome dynamics, and nuclear signaling (reviewed in [7-14]). Dysregulation of these processes is associated with human diseases, including cancer, immunodeficiency, and neurological disorders.
The molecular mechanism for the recognition of H3K4me3 is conserved in dozens of PHD fingers and can be discerned by the hallmark contacts with three amino acids of the histone tail, i.e. A1, R2 and K4me3. The H3K4me3 tail typically forms an anti-parallel β-strand, pairing with the double-stranded β-sheet of the PHD finger. The extended side chain of K4me3 occupies a binding pocket consisting of one to four aromatic residues and named the ‘aromatic cage’. The aromatic rings in the cage are positioned roughly perpendicular to the protein surface and to each other and are engaged in cation-π, hydrophobic and van der Waals interactions with K4me3. R2 of H3K4me3 is bound in the adjacent to the aromatic cage groove containing negatively charged residues that restrain the guanidinium moiety of R2 via hydrogen bonds and ionic interactions. The primary amino group of A1 of H3K4me3 must be unprotected and is hydrogen bonded to one or more backbone carbonyls of the PHD finger.
The PHD fingers specific toward unmodified H3 show a similar mode of the engagement with A1 and R2 of the histone tail but they lack the aromatic cage. Instead, these PHD fingers contain a set of negatively charged residues that form hydrogen bonds and salt bridges with the positively charged ammonium group of K4 of H3.
In addition to their well-characterized function as readers of histone H3, several PHD fingers have been shown to recognize non-histone proteins and DNA. These new binding activities have expanded the role of PHD fingers as versatile transcriptional regulators and signaling factors. In this review, we summarize the mechanisms by which PHD fingers select for non-histone proteins and DNA and discuss similarities and differences in engaging histone and non-histone ligands. The ability of PHD fingers to form complexes with distinctly different biological ligands highlights the remarkable functional plasticity of this structurally conserved module.
H3 mimetics capture the canonical binding pocket
A number of histone H3 mimetics have been found in eukaryotes and viruses. These proteins can compete with the H3 tail for the same histone readers and hijack or disturb gene transcription and other vital cellular programs [15-20]. Searching the human proteome, the Bedford group identified seven H3 mimetic proteins and using in vitro microarrays showed that they are targeted by histone readers, including PHD fingers [21]. Particularly, the PHD fingers of PHF2, TAF3 and KDM7A associate with both VRK1 (PRVKme3A sequence) and BCL11B (SRRKme3Q sequence) proteins when their H3-like sequences are trimethylated at the K4 position (-K4me3), and the PHD fingers of ING3 and TAF3 associate with TSHZ1-K4me3 (PRRKme3Q sequence). The VRK1-K4me3 is specifically recognized by the PHD fingers of DIDO1, ING1/2 and MLL5, whereas BCL11B-K4me3 is specifically recognized by the PHD fingers of ING3 and PHF23. Some of these proteomics and microarray screening interactions were subsequently chosen for validation, and all were confirmed by pulldown experiments [21].
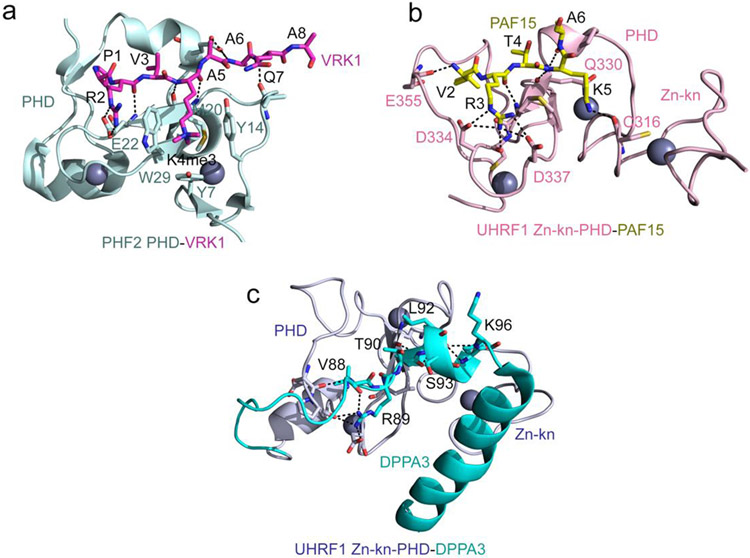
The crystal structure of the PHF2 PHD finger in complex with VRK1-K4me3 provides molecular insight into the binding mechanism [21] (Fig. 1a). The structure of the PHF2 PHD:VRK1-K4me3 complex superimposes to the structure of the PHF2 PHD:H3K4me3 complex with an rmsd of 0.5 Å, indicating a similar mode for the recognition of both ligands [22]. Like H3K4me3, VRK1-K4me3 is bound in a β-strand conformation, with K4me3 occupying the aromatic cage created by three aromatic residues, Y7, Y17 and W29 and a hydrophobic M20, and R2 being restrained through the interaction with the negatively charged carboxyl group of E22 [21]. The difference arises in the coordination of the first residue, which is alanine in H3K4me3 but proline in VRK1-K4me3. P1 does not form hydrogen bonds with two backbone carbonyl groups of the PHD finger as A1 does, and instead is involved in the hydrophobic interactions with I21 and I45, which may explain a three-fold increase in binding affinity of this PHD finger to VRK1-K4me3 compared to its affinity to H3K4me3. The presence of the catalytic Jumonji domain adjacent to the PHD finger is essential for binding of PHF2 to VRK1-K4me3. The crystal structure of the PHD-Jumonji cassette in complex with VRK1-K4me3 peptide shows that the peptide is sandwiched between the two domains [23]. Three negatively charged residues, one from PHD and two from the Jumonji domain constrain R2 of the peptide, whereas K4me3 of the peptide is caged in the aromatic cage, formed by three and two aromatic residues of PHD and the Jumonji domain, respectively. The engagement of VRK1-K4me3 with both domains results in the formation of a very tight complex, characterized by a Kd of 42 nM [23].
Figure 1: H3 mimetics occupy the canonical histone binding pocket.
(a) A ribbon diagram of the crystal structure of the PHF2 PHD finger in complex with the histone-mimicking peptide derived from the VRK1 protein (PDB ID 7M10). The VRK1 peptide is shown as magenta sticks, and the zinc ions are grey spheres. Intermolecular hydrogen bonds are indicated by black dash lines. (b) The crystal structure of the zinc-knuckle-PHD finger of UHRF1 in complex with PAF15 peptide (PDB ID 6IIW). The PAF15 peptide is shown as yellow sticks, and intermolecular hydrogen bonds are indicated by black dash lines. (c) The solution NMR structure of the zinc-knuckle-PHD finger of UHRF1 linked with DPPA3 (Stella) (PDB ID 7XGA). Hydrogen bonds between the UHRF1 (light blue) and DPPA3 (cyan) residues are indicated by black dash lines.
The PHD finger of UHRF1 is a well-established reader of unmodified H3 tail (A1RTK sequence). Two studies from the Nakanishi and Arita groups now demonstrate that this PHD finger can also bind to the mammalian proteins PAF15 (V2RTK sequence) and DPPA3 (V88RTL sequence) [24, 25]. The structure of the PAF15-bound UHRF1 PHD finger demonstrates that the replacement of A1 in H3 with V2 in PAF15 has essentially no effect on the binding mechanism [24, 26] (Fig. 1b). In both complexes of the UHRF1 PHD finger with either H3 or PAF15, R and K of the peptides are similarly bound in the highly negatively charged binding pockets and are involved in electrostatic and hydrogen bonding interactions, whereas the unprotected amino terminal (H3N+) groups of A1 or V2 are restrained via hydrogen bonds with the backbone carbonyl groups of the PHD finger. Comparable binding affinity of the UHRF1 PHD finger toward H3 and PAF15 (~2 μM) supports the structural data [24] and suggests that not only alanine but other small hydrophobic residues in the first position, such as valine, can be recognized by the PHD fingers. However, the presence of lysine in the fourth position is essential, because binding of the UHRF1 PHD finger to the DPPA3 peptide, containing unprotected (H3N+) V88 in the first position but leucine instead of lysine in the fourth position, is reduced by ~15-fold [25]. The structure of the UHRF1 PHD:DPPA3 complex was obtained using a linked construct, in which V88 is not the first residue and therefore does not have the positively charged amino terminus as A1 in H3 and V2 in PAF15 do [25] (Fig. 1c). It will be interesting to explore in future studies whether binding of the UHRF1 PHD finger to DPPA3 with V88 positioned in the middle of the sequence is reduced. Nevertheless, this interaction is physiologically relevant, and overexpression of DPPA3 (also known as Stella) facilitates the disassociation of UHRF1 from chromatin and DNA demethylation [27].
Interactions with non-histone mimetic proteins
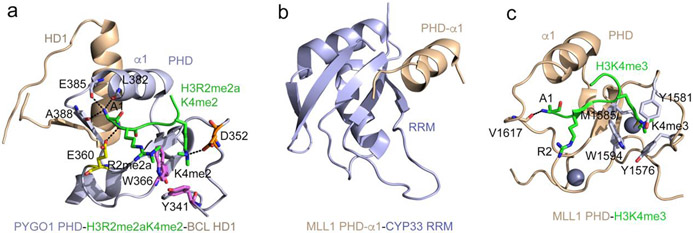
Pygopus (PYGO) and a co-factor BCL9 are components of the Wnt signaling pathway. The PHD finger of PYGO1/2 concurrently associates with H3K4me2/3 and the homology domain 1 (HD1) of BCL9 [28]. The crystal structure of the PYGO1 PHD finger in complex with H3R2me2aK4me2 and HD1 shows that the two ligands bind to the opposite sides of the PHD finger (Fig. 2a). The H3R2me2aK4me2 peptide lays in the canonical histone binding site of the PHD finger, being restrained through the common PHD-histone contacts described above. The C-terminal α-helix of the HD1 domain is packed against the α1 helix connecting zinc-coordinating C6 and C7 in the PHD finger, whereas the N-terminal region of HD1 folds into a β strand and pairs with a β strand of the PHD finger, forming a short parallel double stranded β sheet. Binding of HD1 allosterically shifts a short loop following α1 and opens the binding cavity for A1 of the histone peptide [28]. This allosteric effect leads to a ~2-fold enhancement in binding of the PYGO1 PHD finger to H3K4me3.
Figure 2: Structural bases of non-histone recognition by PHD fingers.
(a) The ternary complex of the PHD finger of PYGO1 (light blue) with H3R2me2aK4me2 peptide (green) and HD1 (wheat) (PDB ID: 2VPG). Residues of the PHD finger involved in the contact with methylated R2 and K4 of the histone peptide are shown as sticks, labeled and colored magenta, orange and yellow. Hydrogen bonds are indicated by black dash lines. (b) The solution NMR structure of the CYP33 RRM domain (light blue) fused with the α1-helix of the MLL1 PHD3 finger (wheat) (PDB: 2KU7). (c) A ribbon diagram of the structure of the MLL1 PHD finger (wheat) in complex with H3K4me3 peptide (green) (PDB: 3LQJ). The aromatic cage residues are shown as sticks, colored light blue and labeled.
The same α1 helix in the third PHD (PHD3) finger of the methyltransferase MLL1 is involved in the interaction with the RNA recognition motif (RRM) of nuclear cyclophilin Cyp33 [29, 30] (Fig. 2b). Interestingly, the MLL1 PHD3 finger also recognizes H3K4me3, a product of the enzymatic activity of MLL1 [29, 31, 32] (Fig. 2c). A remarkably complex mechanism for the PHD3-H3K4me3-RRM assembly and regulation by Cyp33 is described by the Patel group [29]. The structure of the PHD3 finger linked to bromodomain of MLL1 in the apo-state shows that the two domains are in close contact, and the RRM-binding site of the PHD3 finger is blocked by bromodomain. In the apo-state a proline residue located in the linker between the PHD3 finger and bromodomain adopts a cis conformation. Cyp33, a peptidyl-prolyl isomerase, catalyzes isomerization of this proline from the cis to trans conformation, which leads to flattening the linker and the disruption of the PHD3-bromodomain contacts. Consequently, this results in the release of the occluded Cyp33 RRM-binding site, allowing the PHD3 finger to interact with RRM [29]. The RRM-PHD3 binding interface is adjacent to but apparently does not overlap with the H3K4me3-binding site, and the two ligands can be engaged with the PHD3 finger simultaneously, though they influence each other to a degree [29, 32].
The first and fourth PHD fingers (PHD1 and PHD4) of MLL1 cooperate in mediating intramolecular interactions between the proteolytically processed N- and C-terminal regions of MLL1, though it remains to be explored whether these PHD fingers physically interact [33]. The second PHD finger (PHD2) of MLL1 is involved in homo-dimerization [34] and shows E3 ubiquitin ligase activity in the presence of the E2-conjugating enzyme CDC34 [35]. The E3 ubiquitin ligase activity toward ERK2 was observed for the PHD finger of MEKK1 [36]. The PHD finger of KAP1/TRIM28 is also reported to function as a intramolecular E3 ligase for sumoylation of the adjacent bromodomain [37]. Not only the KAP1/TRIM28 PHD finger is in close contact with the bromodomain, it also binds to Ubc9, a SUMO E2 enzyme, and many amide resonances in NMR spectra of the PHD finger are perturbed upon binding of Ubc9 [37, 38]. The tandem of the PHD finger and another zinc finger (PHD-C5HCH) from the methyltransferases NSD1/2/3 has been shown to interact with a zinc-finger of the transcriptional corepressor Nizp1 [39]. Structural and docking studies reveal that the zinc finger of Nizp1 occupies the elongated groove at the interface of PHD-C5HCH [39, 40].
Binding to DNA
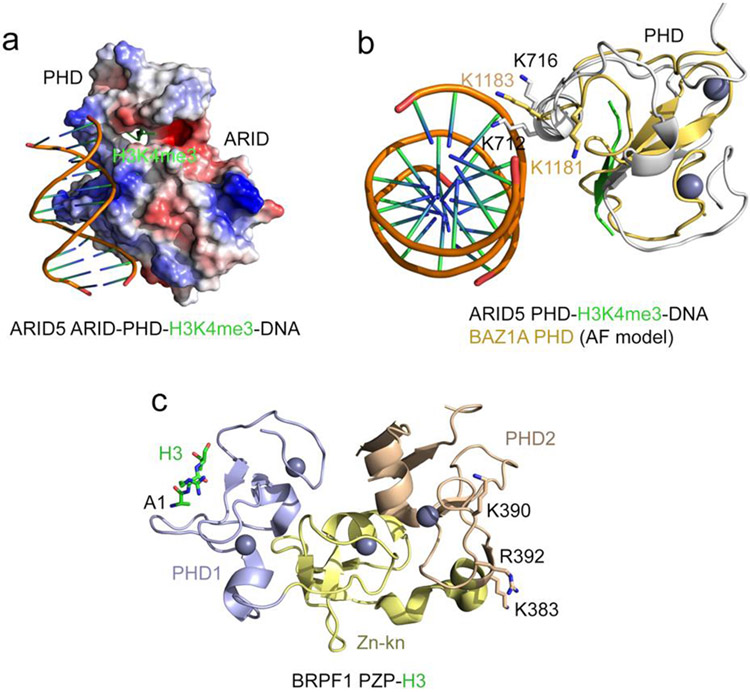
The presence of multiple positively charged lysine and arginine residues on the surface of chromatin binding modules, including zinc fingers, PHD fingers, DPF, and PZP domains, often points to a possible association with DNA. Indeed, the positively charged surfaces of the ARID and PHD domains of ARID5 in the ternary complex with H3K4me3 peptide and DNA interact with DNA, aligning it along one side of the ARID-PHD cassette [41] (Fig. 3a). The DNA-binding interface of the ARID5 PHD finger is far from the binding site for H3K4me3, which is bound by the PHD finger through the typical histone binding mechanism. Interestingly, the α1 helix, which is involved in binding of PHD fingers to non-histone mimetics (Fig. 2), is also implicated in binding of the ARID5 PHD finger to DNA (Fig. 3a). Two lysine residues of α1 in particular, K712 and K716, are in direct contact with DNA. Mutations of lysine residues in this region in the PHD finger of BAZ1A abrogate its binding to DNA [42] (Fig. 3b). The extended PHD finger of PHF6, consisting of a zinc knuckle and PHD, associates with DNA but does not bind histone tails [43].
Figure 3: DNA binding function of PHD fingers.
(a) Electrostatic surface potential of the ARID5 ARID-PHD cassette in complex with H3K4me3 peptide and DNA (PDB: 6LQF). Blue and red colors represent surface positive and negative charges, respectively. The H3K4me3 peptide is depicted as green ribbon. (b) Overlay of the structure of the ARID5 ARID-PHD cassette (grey) in complex with H3K4me3 peptide (green) and DNA (PDB: 6LQF) and the AlphaFold model of the PHD finger of BAZ1 (light orange) (UniProt ID: Q9NRL2). (c) A ribbon diagram of the crystal structure of the BRPF1 PZP domain in complex with H3 (PDB ID: 6U04). The H3 tail is N-terminally linked to PZP. The H3 tail is shown as green sticks, the zinc ions are grey spheres, and the positively charged residues of PHD2 involved in binding to DNA are shown as sticks and labeled.
The distinct, non-overlapped DNA and histone binding interfaces are identified in the PZP domains of BRPF1 and AF10. While the first PHD finger in these PZP domains is a canonical histone H3 reader and along with the zinc knuckle forms an additional binding site for the distal part of H3 tail, the second PHD finger has a DNA binding activity [44-48] (Fig. 3c). The DPF domain of the histone acetyltransferase MORF is capable of binding both acylated K14 of H3 and DNA [49]. Here, the histone and DNA binding sites are in close proximity, which provides a unique mechanism to fine-tune electrostatic contacts with the nucleosome. The dual interaction of the PZP and DPF domains with histone tails and DNA is necessary for tight binding of these domains to chromatin.
Concluding Remarks
The presence of a large number of PHD finger containing proteins in the human genome points to a pivotal role of this module in cell biology. These proteins act in a multifaceted manner and are involved in or mediate fundamental processes such as transcriptional activation and repression, cell growth, differentiation, division and survival. However, only a fraction of PHD fingers have been characterized as histone readers, and biological functions of the majority of this zinc finger family members remains unknown (see Outstanding Questions).
Outstanding questions.
Dozens of PHD fingers have been characterized as readers of histone H3, but many PHD fingers do not contain histone recognizing residues. What are the biological functions of these domains?
Various nuclear proteins contain multiple histone readers with selectivities for distinct posttranslational modifications (PTMs). How does the combinatorial reading of continuously changing epigenetic landscape impact functions of these proteins?
PHD fingers and other readers are also present in many enzymes and scaffolding proteins that link multisubunit enzymatic complexes to chromatin to further modify it by removing PTMs or adding new marks. What are the mechanisms that dictate biological outcomes associated with these chains of the reactions?
Misreading of epigenetic marks by readers, including PHD fingers, has been shown to underlie a host of diseases. However, the PHD finger is known to be a challenging drug target. What are the most efficient strategies to develop chemical probes to disrupt histone binding activities of these readers?
Studies over the past few years reveal the ability of some PHD fingers to bind non-histone proteins either through the histone-recognizing mechanism or via an independent mechanism, and the pace of discoveries of novel binding partners is expected to accelerate considerably. High-throughput screening approaches, including phage display and modified two-hybrid assays, mass spectrometry proteomics analysis and DNA and protein microarrays coupled with computational methods offer tremendous opportunities to identify and validate non-histone ligands of PHD fingers. Furthermore, we anticipate that the development of novel in cellulo and in vivo high-throughput tools and artificial intelligence (AI)-assisted approaches will further speedup the discovery and characterization of physiologically relevant interactors.
Another pressing objective is to study binding activities of PHD fingers in the context of full-length host proteins and their complexes to the intact nucleosome and posttranslational modifications (PTMs)-containing nucleosome arrays [44, 46, 50-53]. The link between aberrant functions of PHD fingers and diseases, including those triggered by viral proteins, suggest great therapeutic potential, and efforts have been put forth to develop inhibitors of PHD fingers, including small molecules and peptidomimetics [14, 54-58]. However, to fully understand and exploit this potential, it is vital to continue deciphering the underlying mechanisms at the atomic-resolution level.
Highlights.
Plant homeodomain (PHD) finger is a major epigenetic reader that recognizes methylated and unmodified histone H3 tail.
A set of PHD fingers capable of binding to proteins other than histone H3 and DNA has been identified.
PHD fingers can bind to the H3-like sequences of eukaryotic and viral proteins, named histone mimetics, and these interactions can disturb normal cell signaling programs.
Dual interaction with histone and DNA enhances the association of zinc finger modules with chromatin.
Acknowledgements
Research in the Kutateladze laboratory is funded by the NIH, HL151334, CA252707, GM125195, GM135671 and AG067664.
Glossary
- Aromatic cage
an arrangement of aromatic rings of two to four aromatic residues to form a cage like cavity on the surface of a protein. The aromatic rings are positioned roughly perpendicular (or parallel) to each other and are involved in cation-π interactions with methylated lysine bound in the aromatic cage
- Cross-braced topology
a structural configuration of eight zinc-coordinating cysteine/histidine residues found in PHD, FYVE and RING zinc fingers. First, second, fifth and sixth zinc-binding residues coordinate one zinc ion, whereas third, fourth, seventh and eighth zinc-binding residues coordinate another zinc ion
- Chromatin
a complex of DNA and DNA-associated proteins
- Nucleosome
a fundamental unit of chromatin, also known as the nucleosome core particle (NCP). It consists of an octamer of four histone proteins, H2A, H2B, H3 and H4 around which double stranded DNA wraps almost twice
- H3 mimetics
proteins containing the histone H3-like sequence, ARTK(me)Q
- H3K4me3
an epigenetic mark enriched in promoters of actively transcribed genes. This mark is added or ‘written’ by specific methyltransferases and is removed or ‘erased’ by demethylases. It is recognized or ‘read’ by readers, including PHD fingers
- Posttranslational modifications (PTMs)
functional groups, covalently (often enzymatically) added to the side chain of specific amino acids in a protein. Acetylation (or acylation in general) of lysine residues, methylation of lysine and arginine residues, and phosphorylation of serine and threonine residues are the most common PTMs
- Transcriptional regulators
proteins that associate with chromatin and activate or repress transcription
- Zinc-knuckle
a small zinc finger domain containing one zinc-binding cluster of four cysteine and/or histidine residues that adopt a tetrahedral geometry to coordinate the zinc ion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
References
- 1.Li H et al. (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442 (7098), 91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pena PV et al. (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442 (7098), 100–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442 (7098), 96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wysocka J et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442 (7098), 86–90. [DOI] [PubMed] [Google Scholar]
- 5.Lan F et al. (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448 (7154), 718–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooi SK et al. (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448 (7154), 714–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musselman CA and Kutateladze TG (2011) Handpicking epigenetic marks with PHD fingers. Nucleic acids research 39 (21), 9061–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taverna SD et al. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14 (11), 1025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruthenburg AJ et al. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25 (1), 15–30. [DOI] [PubMed] [Google Scholar]
- 10.Musselman CA et al. (2012) Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19 (12), 1218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews FH et al. (2016) Insights into newly discovered marks and readers of epigenetic information. Nature Chemical Biology 12 (9), 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musselman CA and Kutateladze TG (2009) PHD fingers: epigenetic effectors and potential drug targets. Mol Interv 9 (6), 314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez R and Zhou MM (2011) The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36 (7), 364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black JC and Kutateladze TG (2023) Atypical histone targets of PHD fingers. Journal of Biological Chemistry online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marazzi I et al. (2012) Suppression of the antiviral response by an influenza histone mimic. Nature 483 (7390), 428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin S et al. (2014) Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1. Nat Commun 5, 3952. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y et al. (2019) MORC3 Is a Target of the Influenza A Viral Protein NS1. Structure 27 (6), 1029–1033 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sampath SC et al. (2007) Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Molecular cell 27 (4), 596–608. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y et al. (2011) MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun 2, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarakhovsky A and Prinjha RK (2018) Drawing on disorder: How viruses use histone mimicry to their advantage. J Exp Med 215 (7), 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J et al. (2021) Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions. Biochem J 478 (10), 1943–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen H et al. (2010) Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem 285 (13), 9322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton JR et al. (2023) A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2. J Biol Chem 299 (2), 102862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishiyama A et al. (2020) Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat Commun 11 (1), 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hata K et al. (2022) Structural basis for the unique multifaceted interaction of DPPA3 with the UHRF1 PHD finger. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arita K et al. (2012) Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A 109 (32), 12950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du W et al. (2019) Stella protein facilitates DNA demethylation by disrupting the chromatin association of the RING finger-type E3 ubiquitin ligase UHRF1. J Biol Chem 294 (22), 8907–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiedler M et al. (2008) Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell 30 (4), 507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z et al. (2010) Pro Isomerization in MLL1 PHD3-Bromo Cassette Connects H3K4me Readout to CyP33 and HDAC-Mediated Repression. Cell 141 (7), 1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hom RA et al. (2010) Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain. J Mol Biol 400 (2), 145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang PY et al. (2010) Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol 400 (2), 137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S et al. (2010) The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression. Biochemistry 49 (31), 6576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama A et al. (2011) Proteolytically cleaved MLL subunits are susceptible to distinct degradation pathways. J Cell Sci 124 (Pt 13), 2208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fair K et al. (2001) Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol 21 (10), 3589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J et al. (2012) A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J Biol Chem 287 (52), 43410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Z et al. (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell 9 (5), 945–56. [DOI] [PubMed] [Google Scholar]
- 37.Ivanov AV et al. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28 (5), 823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng L et al. (2008) Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol 15 (6), 626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berardi A et al. (2016) Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: implications for Sotos syndrome. Nucleic Acids Res 44 (7), 3448–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berardi A et al. (2022) Nizp1 is a specific NUP98-NSD1 functional interactor that regulates NUP98-NSD1-dependent oncogenic programs. FEBS J. [DOI] [PubMed] [Google Scholar]
- 41.Tan LM et al. (2020) Dual Recognition of H3K4me3 and DNA by the ISWI Component ARID5 Regulates the Floral Transition in Arabidopsis. Plant Cell 32 (7), 2178–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oppikofer M et al. (2017) Non-canonical reader modules of BAZ1A promote recovery from DNA damage. Nat Commun 8 (1), 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z et al. (2014) Structural and functional insights into the human Borjeson-Forssman-Lehmann syndrome-associated protein PHF6. J Biol Chem 289 (14), 10069–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein BJ et al. (2016) Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res 44 (1), 472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein BJ et al. (2021) The role of the PZP domain of AF10 in acute leukemia driven by AF10 translocations. Nat Commun 12 (1), 4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein BJ et al. (2020) Molecular Basis for the PZP Domain of BRPF1 Association with Chromatin. Structure 28 (1), 105–110 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S et al. (2015) The PZP Domain of AF10 Senses Unmodified H3K27 to Regulate DOT1L-Mediated Methylation of H3K79. Mol Cell 60 (2), 319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vann KR et al. (2021) Mechanistic similarities in recognition of histone tails and DNA by epigenetic readers. Curr Opin Struct Biol 71, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein BJ et al. (2019) Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat Commun 10 (1), 4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatchalian J et al. (2017) Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 8 (1), 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison EA et al. (2018) The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tallant C et al. (2015) Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC. Structure 23 (1), 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tencer AH et al. (2017) Covalent Modifications of Histone H3K9 Promote Binding of CHD3. Cell Rep 21 (2), 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang MY et al. (2022) Covalent labeling of a chromatin reader domain using proximity-reactive cyclic peptides. Chem Sci 13 (22), 6599–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen HF et al. (2014) Inhibition of histone binding by supramolecular hosts. Biochem J 459 (3), 505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali M et al. (2015) Molecular Insights into Inhibition of the Methylated Histone-Plant Homeodomain Complexes by Calixarenes. J Biol Chem 290 (38), 22919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amato A et al. (2018) Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach. ACS Chem Biol 13 (4), 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller TC et al. (2014) Competitive binding of a benzimidazole to the histone-binding pocket of the Pygo PHD finger. ACS Chem Biol 9 (12), 2864–74. [DOI] [PMC free article] [PubMed] [Google Scholar]