Abstract
Neuroligin-4 (NLGN4) loss-of-function mutations are associated with monogenic heritable autism spectrum disorder (ASD) and cause alterations in both synaptic and behavioral phenotypes. Microglia, the resident CNS macrophages, are implicated in ASD development and progression. Here we studied the impact of NLGN4 loss in a mouse model, focusing on microglia phenotype and function in both male and female mice. NLGN4 depletion caused lower microglia density, less ramified morphology, reduced response to injury and purinergic signaling specifically in the hippocampal CA3 region predominantly in male mice. Proteomic analysis revealed disrupted energy metabolism in male microglia and provided further evidence for sexual dimorphism in the ASD associated microglial phenotype. In addition, we observed impaired gamma oscillations in a sex-dependent manner. Lastly, estradiol application in male NLGN4−/− mice restored the altered microglial phenotype and function. Together, these results indicate that loss of NLGN4 affects not only neuronal network activity, but also changes the microglia state in a sex-dependent manner that could be targeted by estradiol treatment.
Keywords: Microglia, Purinergic signaling, Autism spectrum disorder, Gamma oscillation, Sex difference
1. Introduction
Autism Spectrum Disorders (ASD) are a range of neurodevelopmental and behavioral disorders, characterized by changes in social interaction, repetitive and restrictive behavior/interest, and differing sensory responses as a result of altered signal processing in the brain (Miles, 2011; El-Kordi et al., 2013; Lord et al., 2020). Loss-of-function in the Neuroligin-4 (NLGN4) locus was identified as one of the main monogenic causes of ASD (Jamain et al., 2003; Maćkowiak et al., 2014). Neuroligin 4 is a postsynaptic cell adhesion protein required for synaptic organization and remodeling (Hoon et al., 2011; Hammer et al., 2015; Zhang et al., 2018). It has been shown that NLGN4 knock-out (NLGN4−/−) mice demonstrate major behavioral changes that have been linked to ASD, including increased repetitive behavior and altered social interaction, and that these changes are more severe in males than in females (Jamain et al., 2008; El-Kordi et al., 2013; Ju et al., 2014). These data coincide with epidemiologic data showing a higher incidence of ASD in men (Trabzuni et al., 2013). NLGN4 is highly expressed at various mouse brain regions such as the brainstem, retina and in all subregions of the adult hippocampus (Jamain et al., 2008; Hammer et al., 2015). Specifically, NLGN4−/− causes a reduction in GABA (γ-aminobutyric acid) -ergic synaptic transmission along with disrupted gamma (γ) oscillations in the CA3 region of the mouse hippocampus (Hammer et al., 2015). Gamma oscillations in CA3 are thought to play an important role for cognition and memory (Csicsvari et al., 2003; Shinohara et al., 2013). Moreover, gamma oscillations have been shown to modulate morphology and function of microglia – the resident immune cells of the CNS- in an animal model of Alzheimer’s Disease (Iaccarino et al., 2016), whereas stimulation of interferon gamma (IFN-γ) accelerates changes in microglia states that are capable of reducing neural information processing (Ta et al., 2019). Microglia perform various functions including synaptic pruning, phagocytosis of pathogens and regulation of homeostasis and inflammation in the healthy brain (Kettenmann et al., 2011). Many studies suggest that neuroinflammation in different brain regions of ASD patients correlates with activation of microglia (Vargas et al., 2005; Edmonson et al., 2016; O’Loughlin et al., 2017; Salter and Stevens, 2017). Altered microglial structure, function and transcriptional expression are evident in patients with ASD and relevant animal models. Given the sex bias in ASD (Napolitano et al., 2022) and a known sexual dimorphism of microglia under physiological conditions and other disorders (Bordt et al., 2020), it is of high interest to elucidate the involvement of microglia in ASD, as well as characterize how this involvement varies across sexes. Microglial differences have previously been demonstrated in male mice expressing an ASD-relevant gene mutation (NL3R451C) (Matta et al., 2019, 2020). However, there is a lack of knowledge about a possible sexual dimorphism in microglia in ASD. Since mutations in NLGN4 are amongst the common monogenic causes of ASD, we utilized NLGN4−/− mice to investigate sex differences in microglia morphology, phenotype, and function in the hippocampus. We first investigated the chemotactic movement of microglial processes controlled by extracellular ATP signaling towards a laser lesion in acute brain slices. We further investigated phagocytosis by measuring the in vivo uptake of apoptotic neurons by microglia. Proteomics were used to gain a broader overview about microglial phenotypes associated with the NLGN4−/− model. In addition, we tested whether the microglial state can be modulated by 17-beta estradiol in male mice.
2. Materials and methods
2.1. Animals
All mice of both sexes used for the present study had a C57BL/6J genetic background, and animals were handled according to government and internal rules and regulations. The NLGN4−/− mice were a kind gift of Prof. Nils Brose (Max-Planck-Institute for Experimental Medicine, Göttingen). The NLGN4−/− strain was first backcrossed for 8 generations. For the electrophysiological and laser-lesion in vitro experiments, C57BL/6J mice that express EGFP under the Csf1R promotor (B6.Cg-Tg (Csf1r-EGFP)Hume/Jackson Stock No:018549) were used to allow detection of microglial cells by their fluorescence. We crossed NLGN4−/− with the Csf1R-EGFP strain for 4 generations before we maintained heterozygote breeding of NLGN4−/+xCsfR-EGFPtg/tg. For the generation of NLGN4−/− mice, we performed the very first experiments using the HETxHET breeding strategy, but the strategy was later altered because of reported interaction between NLGN3−/− and WT animals resulting in deficits in sociability in both genotypes when raised together (Kalbassi et al., 2017). To avoid such possible interaction in our NLGN4−/− colony, we changed the breeding strategy to KOxKO and WTxWT. The offspring were separated after weaning according to sex and genotype. All breeding was performed in the same room of the same animal SPF facility. Animals were housed in individually ventilated cages (Typ II Blue Line, 536 cm2) in groups of 3–5 animals per cage including igloos and paper rolls for enrichment. The mice were kept on a 12 h light/dark cycle with food and water supplied ad libitum.
Two cohorts of NLGN4−/− male mice were treated with 17β-estradiol according to a previously described protocol (Fernandez and Frick, 2004). Briefly, 1 μM 17β-estradiol was administered to the mice via the drinking water. The drinking water containing 17β-estradiol was replaced with fresh water containing 17β-estradiol every second day. Treatments began at the 14-week-old age and lasted for 6 weeks (Fig. 6A).
Fig. 6. ß-Estradiol Treatment rescued microglia structure and function in NLGN4−/− male mice.

(A) Schematic of experimental layout for 5 weeks of beta-estradiol (E2) treatment of NLGN4−/− male mice (NLGN4−/−-E2) and aged-matched untreated male WT and NLGN4−/− mice. (B) Representative photomicrographs from WT and NLGN4 treated and untreated male brain samples after laser lesion (microglia in green EGFP, scale bar 10 μm). Quantitative analysis of lesion-induced, microglial process movement expressed in arbitrary units (AU) in 20-week-old male WT, NLGN4−/− and NLGN4−/−-E2 mice. Impaired process extension in male NLGN4−/− mice are rescued by estradiol treatment (right graph, ordinary one-way ANOVA following Bonferroni’s multiple comparisons test: number of sections (and mice): male WT, 12 (3); male NLGN4−/−, 12 (3); male NLGN4−/−-E2, 18 (4)). We performed 3 independent experiments. (C) Average current density to voltage relationships of microglial ATP-induced metabotropic purinergic responses (P2YR12) in the hippocampus of 20-week-old WT, NLGN4−/− and NLGN4−/−-E2 mice (left graph). Summary of the outward conductance’s, which were calculated from ATP (10 μM)-evoked currents at potentials between + 20 mV and + 60 mV (right graph). Both datasets suggested that beta-estradiol treatment reversed the effect of loss of NLGN4 in male microglia (Ordinary one-way ANOVA following Bonferroni’s multiple comparisons test; number of cells (and mice): male WT, 20 (5); male NLGN4−/−, 18 (3); male NLGN4−/−-E2, 22 (4)). We performed 3 independent experiments. (D) The percentage of AnnexinV positive microglial cells is reduced to the level of WT after the treatment in NLGN4−/− male (2 independent experiments, ordinary one-way ANOVA following Bonferroni’s multiple comparisons test: male: n(WT) = 38, n(NLGN4−/−) = 14, n(E2- NLGN4−/−) = 20). (E) Representative photomicrographs of microglia morphology from WT, NLGN4−/− treated and untreated males (scale bar 5 μm, Iba-1 staining for microglia). Microglia morphology analysis shows that the total number of processes, the average process length and the number of branch points per cell in CA3 was significantly altered between male WT and NLGN4−/− microglia in 20 weeks old animals, and beta estradiol treatment normalized the morphology (2 independent experiments, ordinary one-way ANOVA following Bonferroni’s multiple comparisons: n(WT) = 5, n(NLGN4−/−) = 5, and n(E2- NLGN4−/−) = 6). Asterisk represents significant levels as *p < 0.05; **p < 0.01; ***p < 0.0001. All mice used for the study were on a C57BL/6J genetic background.
2.2. Ethics statement
All procedures involving the handling of living animals were performed in strict accordance with the German Animal Protection Law and were approved by the Regional Office for Health and Social Services in Berlin (Landesamt für Gesundheit und Soziales, Berlin, Germany). Adult mice were sacrificed by cervical dislocation, intraperitoneal injection of pentobarbital (Narcoren, Merial GmbH, Hallbergmoos, Germany) or isoflurane anesthesia followed by decapitation. Moreover, mice were euthanized by CO2 inhalation at Harvard according to the Institutional Animal Care and Use Committee at Harvard Medical School’s approved protocol. All efforts were made to minimize suffering.
2.3. Immunohistochemistry and confocal microscopy
Under deep anesthesia, animals were perfused with phosphate buffered saline (PBS) followed by 4% paraformaldehyde in PBS, decapitated and brains were sectioned in the coronal plane at 40 μm thickness as described earlier in Guneykaya et al., 2018. Both dorsal and ventral hippocampal sections were used to analyze CA3 region of the hippocampus. Ki67 + based proliferation analysis was performed as described by Mattei et al., 2014. The following primary antibodies were used in the indicated dilution: goat anti-Iba1 (ab5076/Abcam, Cambridge, UK) 1:250, and rabbit anti-Ki67 (ab16667/Abcam) 1:100 and samples were incubated overnight at 4 °C. Secondary antibodies were also prepared in tris buffered saline (TBS) at the following dilutions: donkey anti-goat Alexa Fluor 488 (705-545-147/Dianova, Hamburg, Germany) 1:200, donkey anti-rabbit Cy3 (Dianova) 1:200 and samples were incubated with secondary antibodies for one hour at room temperature. Finally, the cell nucleus was stained using 4′,6-diamidino-2-phenylindole (DAPI, Dianova) 1:500. For microglia and Ki67 cell density, images were acquired with a Leica SPE confocal microscope (equipped with a 10X objective using LASAF software Leica microsystems, Germany). Images were taken throughout the entire thickness of the section by Z stack analysis (8 μm z-step size, 10 steps), covering the surface of the respective region. The number of microglia cells per/mm2 were quantified using Fiji ImageJ software (NIH, Maryland, USA). A total of four sections per animal were analyzed.
2.4. Microglia morphology analysis
40 μm dorsal and ventral coronal brain sections were collected from NLGN4−/− and WT male and female brains and stained with anti-Iba1 (Abcam, Cambridge, UK) antibody (see section 2.3 for details). Imaging of the CA3 region of the hippocampus was performed on an LSM 880 NLO Airyscan microscope (Zeiss, Oberkochen, Germany) using a 20x objective. Z stacks were defined as 1.44 μm steps, and 1,024 × 1,024-pixel resolution were recorded. 3D reconstructed images were analyzed using IMARIS software (Bitplane, Zurich, Switzerland) with semi-automated filament analysis. Briefly, the surface was rendered based on the threshold and small, unattached cell extensions were filtered using the surface module. After surface rendering, filament analysis was performed (Largest diameter size: 2.5 μm) to determine processes number and structure applying the filament module. 18–20 cells from 4 different sections were reconstructed per mouse.
2.5. Microglia apoptotic neuron phagocytosis
We followed the protocol as described (Krasemann et al., 2017). Briefly, apoptotic neurons were prepared from a E18 primary neuron culture. Mice were anesthetized by i.p injection, using a mixture of Ketamine (100 mg kg – 1), Xylazine (10 mg kg – 1), and a total of 80,000 cells were injected into hippocampus and cortex bilaterally using stereotactic equipment. After 16 h post-surgery, mice were euthanized and microglia isolation was performed accordingly (Butovsky et al., 2014). Cells were stained with Ly-6C-PerCP Cy5.5, CD11b-PeCy7 (Thermo Fisher Scientific), and 4D4-APC (provided by Oleg Butovsky). Apoptotic neurons were labeled by Alexa Fluor™ 488 NHS Ester (Succinimidyl Ester, Thermo Fisher Scientific), and phagocytic versus non-phagocytic were further sorted from the 4D4 + CD11b + -population by detection of Alexa 488 fluorescence. Cells were immediately acquired using FACS Aria II (BD Biosciences), and data analysis was performed using FlowJo v10 software (Tree Star).
2.6. Flow cytometry analysis
Under deep anesthesia, animals were transcardially perfused with ice-cold PBS. After decapitation, the brains were removed, and the hippocampus was dissected in Hanks balanced salt solution (HBSS). Single cell suspension from the hippocampus was homogenized in dissection buffer (1x HBSS (Thermo Fisher Scientific, Waltham, USA), 45 % glucose, 1 M HEPES) and the pellets were resuspended in ice-cold Percoll solution composed of Percoll (GE Healthcare, Little Chalfont, Buckinghamshire, UK) and myelin gradient buffer. A layer of PBS was applied on top and centrifuged at 950 g, 4 °C with no brake and full acceleration. The cell pellet was stained with dye-coupled monoclonal anti-mouse antibodies: CD45, CD11b (eBioscience, San Diego, USA) for 20 min at 4 °C. After incubation, apoptotic cells were detected using the manufacturer’s recommended protocols (FITC AnnexinV Apoptosis Detection Kit I (RUO), Thermo Fisher Scientific). The same isolation method was used to analyze MHCI, MHCII and CD54 expression (eBioscience, San Diego, USA), with 1:100 concentration per million isolated cells. Cells were immediately acquired on a LSRII flow cytometer (BD Bioscience, San Jose, USA) and data was analyzed using FlowJo v10 software (Tree Star, Orlando, USA).
2.7. Acute brain slice preparation and microglia electrophysiological recordings
Acute cortical brain slices were prepared as previously described (Boucsein et al., 2003). In brief, mice were euthanized by cervical dislocation, and their brains removed and cooled in ice-cold artificial cerebrospinal fluid (aCSF) containing (in mM): 230 sucrose, 2.5 KCl, 10 MgSO4, 0.5 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 D-glucose, pH7.4; gassed with 95% O2/ 5% CO2. Brains were then mounted on a vibratome (HM650V, Thermo Scientific, Massachusetts, USA), and 250 μm thick coronal brain samples were generated and kept at room temperature for experiments for up to 5 h in gassed ACSF containing (in mM): 134 NaCl, 2.5 KCl, 1.3 MgCl2, 2 CaCl2, 1.26 K2HPO4, 26 NaHCO3, and 10 D-glucose (pH 7.4). Acute brain slices were used for patch clamp recordings and in situ 2-photon live-cell imaging.
A conventional patch-clamp amplifier was used (EPC9, HEKA Elektronik, Lambrecht, Germany). Acute 250 μm coronal brain sections were prepared from NLGN4−/− and wild type mice generated on a MacGreen background for microglia identification by fluorescence on an epifluorescence microscope. Patch pipettes were pulled from borosilicate glasses and had resistances of 3–8 MOhm. The following intracellular solution was used (in mM): KCl,120; MgCl2; 2; CaCl2,0.5; Na-ATP,2; EGTA,5; HEP10 and sulforhodamine 101,0.01 (Sigma Aldrich) and had an osmolarity of 280–290 mOsm/L adjusted to a pH of 7.3 with KOH (~20 mM). The extracellular solution contained (in mM): NaCl,134; KCl,2.5; MgCl2,1.3; CaCl2,2; K2HPO4,1.25; NaHCO3,26; D-glucose,10; pH7.4; 310–320 mOsm/L and was gassed with carbogen (95% O2/ 5% CO2). Experiments with series resistances<100MOhm were used for data analysis. All experiments were performed in the voltage-clamp configuration. To obtain current–voltage curves during continuous recordings, the membrane was clamped every 5 s from a holding potential of −70 or −20 mV (before and during the ATP response, respectively) to a series of de- and hyperpolarizing voltages ranging from −140 mV to 60 mV with 20 mV increments, 100 ms in duration. Membrane currents were averaged for quantification between 30 and 45 ms after clamping the membrane to a given value from the resting potential. Membrane capacitance was quantified based on an exponential fit of the current decay in response to a 10 mV test pulse. The same pulse was used to quantify series resistance from the peak amplitude of the membrane capacitance currents. Comparisons of membrane currents between different groups were always normalized to the membrane capacitance.
2.8. Two-photon live cell imaging and laser lesion
A custom-built two-photon laser-scanning microscope (Till Photonics, Gräfelfing, Germany) was used for live imaging of microglial processes. Coronal brain sections (250 μm) were obtained as described above (Boucsein et al., 2003). EGFP was excited by a Chameleon Ultra II laser (Coherent, Dieburg, Germany) tuned to 940 nm. A 40x water-immersion objective (NA 0.8, Olympus, Hamburg, Germany) was used for scanning 60 μm thick z-stacks with a step size of 1 μm, covering a field of 320 × 320 μm2. The laser lesion was set focusing the laser beam to 20 μm depth in the selected imaging volume in the hippocampal region dorsal CA3, at a wavelength of 810 nm and the maximum power. The tissue was scanned until autofluorescence of the injured tissue was visible, and the final lesion size was around 20 μm. For data analysis, we used an in house-written algorithm based on IGOR Pro 6.37 using IGOR Pro 6.37 (Lake Oswego, USA) as in Davalos et al., 2005. Briefly, the sequences of 3D image stacks were converted into sequences of 2D images by a maximum intensity projection algorithm. Grayscale images were first converted into binary form using a threshold. Microglial response to focal lesion was quantified by counting EGFP + pixels (associated with microglial processes) in a proximal circular region of 45 μm around the lesion site over time (Rx(t)). Distal fluorescence of the first time point was determined within a diameter of 45 μm to 90 μm around the lesion site for normalization (Ry(0)). The microglial response was represented as R(t)=(Rx(t)-Rx(0))/Ry(0).
2.9. Microglia cell isolation and magnetic cell sorting (MACS) for proteomics
Hippocampi from 3 animals were pooled and digested into a single cell suspension by using an Adult Brain Dissociation Kit for mouse and rat (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany) according to the manufacturer’s manual. After Percoll separation, the cells were washed in ice-cold MACS-buffer (PBS, 0.5% bovine serum albumin, 2 mM EDTA) and stained with anti-mouse CD11b magnetic beads (Miltenyi Biotech) at 4 °C for 20 min. Due to limitation of cell number, six hippocampi from 3 animals were pooled and total cells were passed through large-sized MACS columns (Miltenyi Biotech). The flow through was discarded and the cells were flushed out of the column in MACS buffer, pulled down by centrifugation and resuspended in RIPA buffer (Thermo Fisher Scientific) for Western blotting or 2M urea buffer for microglia specific proteomics, and stored at −80 °C.
2.10. Western blotting
Under deep anesthesia, animals were transcardially perfused with ice-cold PBS. After decapitation, the brain was removed, and the hippocampus total protein extraction was performed with RIPA buffer. Microglia cells were isolated with MACS from hippocampus samples. Total protein concentration of hippocampi and microglia samples w determined by using a BCA assay (Pierce). Proteins were denatured in Laemmli buffer at 95 °C for 5 min, and 20 μg of protein per sample was loaded into a 10% SDS gel. Following SDS-PAGE, proteins were transferred onto PVDF membrane (Immobilon-FL) activated in methanol at 100 V for 2 h by wet transfer. The membrane was blocked in Odyssey blocking buffer (LiCor Biosciences) for 3 h and incubated with the primary antibody (Abcam rabbit monoclonal anti-NLGN4:1/500, mouse anti-GAPDH:1/2500 in Odyssey blocking buffer and 0.2% Tween 20) overnight at 4 °C.Following three washing steps in TBS-T, the membrane was incubated with a secondary antibody (Invitrogen; 1/2500 goat IRDye800 anti-mouse, 1/2500 goat IRDye680 anti-rabbit in Odyssey blocking buffer, 0.2% Tween20 and 0.01% SDS) for 3 h at RT with slow shaking. The membrane was washed three times in TBS-T and proteins were detected by using Odyssey Imaging Systems (LiCor Biosciences).
2.11. Sample preparation for mass spectrometry and proteomics data analysis
Proteomic changes were addressed by label-free quantitation. Samples were subjected to urea-based lysis and tryptic in-solution digest as described (Mertins et al., 2018). After desalting, samples were measured by LC-MS/MS on an Orbitrap Fusion mass spectrometer (Thermo) using a 110-min gradient. Cells were isolated in 3 different cohorts, and mass spectrometry was run in a single experiment. For database search MaxQuant version 1.6.3.4 (Cox and Mann, 2008) was consulted. An FDR of 0.01 was applied for peptides and proteins and database search was performed using a mouse Uniprot database (July 2018) including isoforms. MS intensities were normalized by the LFQ algorithm whilst enabling the match-between-runs feature (Cox et al., 2014). Further data analysis was done in R. Our proteomics analysis was performed in hippocampal microglia only.
A number of at least two peptide IDs per protein group and at least three valid values in any experimental group were required for quantitation. Missing values were imputed using a column-wise Gaussian distribution with a width of 0.2 and a downshift of 1.8. Significance calling among experimental groups was achieved by applying a moderated t-test from the limma package (Ritchie et al., 2015). Gene ontology analysis was done using Metascape (metascape.org (Zhou et al., 2019)). Protein interaction networks were analyzed with STRING using experiments and databases as interaction sources and medium confidence. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD024325.
2.12. Hippocampal gamma oscillation measurements
Mice were anesthetized with isoflurane and then decapitated. Their brains were removed from the skull and immerged in an ice-cold carbogenated (95% O2/5% CO2) sucrose-based solution containing (in mM): NaCl, 80; NaHCO3 25; NaH2PO4 1.25; KCl, 2.5; glucose, 25; sucrose, 85; Cacl2 0.5; MgCl, 3. The brain was cut into 400 μm thick, horizontal, hippocampal sections with a vibratome (DSK microslicer DTK-1000, Dosaka, Japan) and immediately transferred to an interfacetype recording chamber perfused with carbogenated, warm (36 ± 0.2 °C) artificial cerebrospinal fluid (ACSF) of the following composition (in mM): NaCl, 129; NaH2PO4, 1.25; NaHCO3, 21; glucose, 10; MgSO4, 1.8; CaCl2, 1.6; KCl, 3. Sections were incubated for at least one hour before starting recordings. Local field potentials (LFPs) were recorded with glass pipettes filled with ACSF in the stratum pyramidale in both dorsal and ventral CA3b of the hippocampus. Recordings were amplified by an EXT-02B amplifier (npi, Tamm, Germany), low-pass filtered at 1 kHz and sampled by a CED 1401 interface (Cambridge Electronic Design, Cambridge, UK) at 5 kHz. For the induction of gamma oscillations, acetylcholine (10 μM, Ach, Sigma Aldrich, Taufkirchen, Germany) and physostigmine (PING, 2 μM, Tocris Bioscience, Bristol, UK) were administered by bath application for 120 min. We choose cholinergic gamma oscillations over gamma oscillations evoked by kainate (KA, used by Hammer et al., 2015) since this type is very similar to gamma oscillations in awake behaving animals, especially during exploratory behavior where the acetylcholine concentration is the highest. This models the septal cholinergic inputs into the hippocampus known to play key roles in learning and memory. This PING gamma oscillation is also maximally dependent on perisomatic inhibition but indeed might involve a different circuit compared to KA-induced ones. Gamma peak power was calculated between 110 and 120 min of induction. After this period, the induction was stopped for 40 min and the oscillation was then re-induced at 160 min and recorded for 60 min (Lemercier et al., 2016; Meier et al., 2020). Power values were normalized to the period 110–120 min before interruption, re-induced gamma oscillations were represented as normalized gamma power after 50 min re-induction. Log-normally distributed absolute power values were log-transformed before using parametric statistical tests.
2.13. Statistical analysis
We performed a 2-way ANOVA followed by Bonferroni’s multiple comparisons if not stated differently in the respective material and methods section. The statistical tests are mentioned in the figure legends.
3. Results
3.1. Microglia do not express Neuroligin 4 (NLGN4) protein
Since previous studies suggested that loss of NLGN4 causes significant defects on the network activity and changes in the GABAergic synapses in the hippocampal CA3 region (El-Kordi et al., 2013; Hammer et al., 2015), we aimed to investigate microglia structure, function and proteome profile and their related readouts in both sexes of wildtype (WT) and NLGN4−/− 13-week-old mice, specifically in the CA3 region of the hippocampus (see experimental design Fig. 1A).
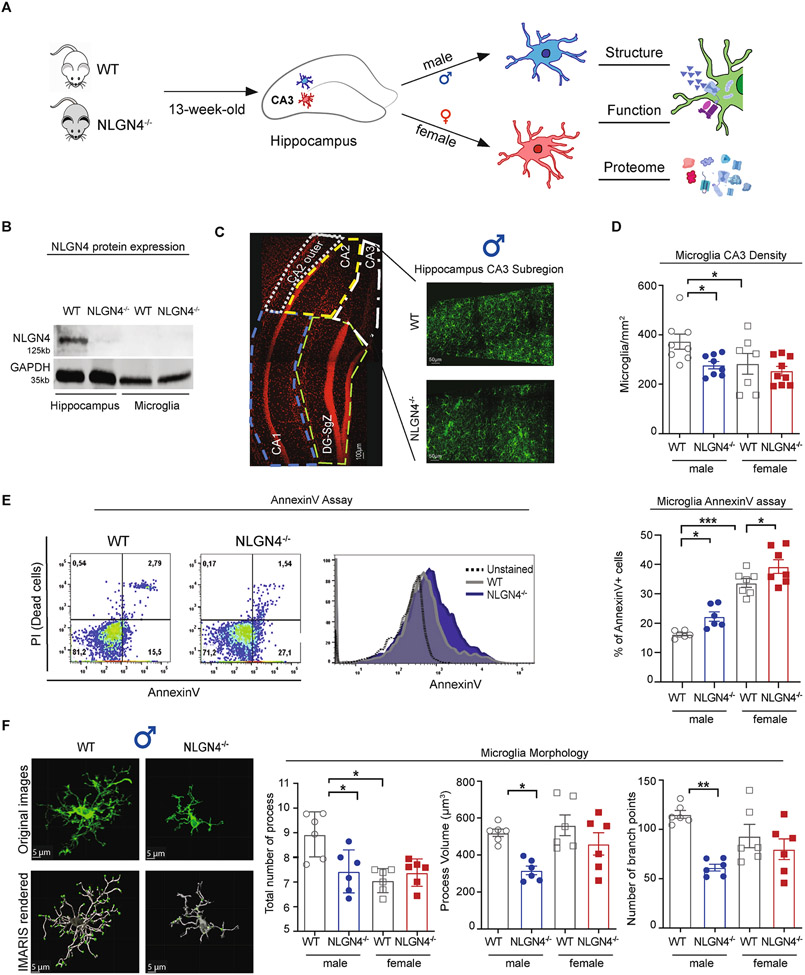
Fig. 1. Loss of NLGN4 results in changes in microglia structure in male hippocampus.
(A) Graphical illustration of the study design. (B) Western blotting plots for NLGN4 protein. Microglia clearly showed no expression of NLGN4 protein. WT hippocampus homogenate was used as positive control, and NLGN4−/− hippocampus homogenate was used as negative control. (C) Representative hippocampus DAPI image to show the definition of hippocampal subregions with cornu ammonis (CA)1-CA3 and dentate gyrus-subgranular zone (DG-SgZ) (left). Representative fluorescence images of Iba1± labeling to identify microglia in coronal slices from 13-week-old WT and NLGN4−/− male CA3 region of the hippocampus (right). (D) Quantitative analysis in the CA3 region (right graph) revealed a lower density of female WT vs. male WT. Loss of NLGN4−/− reduced microglia density in males, and no significant changes were observed between WT and NLGN4−/− female mice (2-way ANOVA followed by Bonferroni’s multiple comparisons, male (WT) n = 8, (NLGN4−/−) n = 8, female (WT) n = 7, (NLGN4−/−) n = 9). We performed 3 independent experiments. (E) Gating strategy for AnnexinV based apoptosis analysis by FACS shown as density plots. The cell populations were gated according to CD45+ CD11b+ expression (microglia), following PI and AnnexinV staining to detect apoptotic state. FSC, forward scatter; SSC, sideward scatter; PI: Promodium iodide (left). The percentage of AnnexinV positive microglia cells indicated higher apoptotic microglia percentage in both male and female NLGN4−/− hippocampi (right, 2-way ANOVA followed by Bonferroni’s multiple comparisons; male n(WT) = 6n(NLGN4−/−) = 6, female: n(WT) = 6n(NLGN4−/−) = 7). We performed 4 independent experiments. (F) Representative photomicrographs of male WT and NLGN4−/− microglia confocal and IMARIS-rendered images from hippocampus CA3 region (scale bars represent 5 μm). 3D-reconstructed IMARIS-based semi-automatic quantification was performed to analyze microglia morphology: the total number and length of processes and the number of branch points of individual cells in CA3. The length of the branches was lower in female WT microglia compared to male WT. All parameters were significantly altered between male WT and NLGN4−/− microglia, whereas no changes were observed between female WT and NLGN4−/− microglia. Each symbol represents one mouse with 18 measured cells per mouse (2-way ANOVA followed by Bonferroni’s multiple comparisons; n = 6 per group). We performed 2 independent experiments. Asterisk indicates significant differences *p < 0.05; **p < 0.01; ***p < 0.001. All mice used for the study were on a C57BL/6J genetic background. All male and female comparison can be found in the supplementary Table S2.
We first examined NLGN4 expression in microglia cells. We performed western blotting analysis from hippocampus homogenate and CD11b+ cells from hippocampus for NLGN4 protein. We could not detect NLGN4 protein expression in CD11b+ cells derived from hippocampus (Fig. 1B). Therefore, we hypothesize that changes in microglia from NLGN4−/− mice are potentially caused by factors in the microenvironment.
3.2. Loss of NLGN4 results in lower density and less ramified morphology in male CA3-hippocampal microglia
Microglia display sex differences in their expression profiles and function under physiological conditions (Guneykaya et al., 2018; Hanamsagar et al., 2017). In addition, the prevalence of ASD is strongly male-biased. Based on the prevalence of ASD in males and sex-dependent structural and functional differences in microglia in several models of clinically relevant disorders (Hanamsagar et al., 2017; Yanguas-Casás et al., 2018; Lynch, 2022), we aimed to investigate changes in structure and function in both male and female microglia from WT and NLGN4−/− mice.
Microglia density and morphology have been extensively studied to define microglia states in disease and different environmental challenges (Haynes et al., 2006; Butovsky et al., 2014). Therefore, we next analyzed microglial density and morphology in the CA3 region of the hippocampus in male and female NLGN4−/− (Fig. 1C). WT males had higher microglia density than females in accordance with our previous data (Guneykaya et al., 2018, Fig. 1D). However, only in male NLGN4−/− mice we found significantly lower density of microglia compared to WT (Fig. 1D, p = 0.0497, t = 2.549).
To further investigate decreased microglia density in CA3 region of the hippocampus, we next analyzed microglia proliferation using Ki67 + Iba1 co-staining (Fig. S1A). We found no difference between WT and NLGN4−/− in either sex (Fig. S1B). In addition, we evaluated apoptotic microglia ex vivo using a flow cytometry based AnnexinV apoptosis detection assay. AnnexinV is a Ca2+-dependent phospholipid-binding protein and commonly used to detect apoptotic cells. Loss of NLGN4 resulted in a significant increase in AnnexinV-positive cells in both sexes. Female WT microglia derived from hippocampus showed a significantly higher proportion of AnnexinV-positive cells compared to male WT (Fig. 1E, male: p = 0.0482, t = 2.91; female: p = 0.0362 t = 3.038; male vs. female (WT): p= <0.0001, t = 6.544). Neither proliferation nor ex vivo apoptosis rates could explain the sex-specific difference in density in the context of NLGN4−/−.
Morphology of microglia is an established indicator for the surveillance and interaction with other cell types via purinergic signaling (Carvalho-Paulo et al., 2021). We next analyzed microglial morphology with IMARIS based semi-automatic quantitative three-dimensional measurements focusing on the CA3 region of the hippocampus. Only NLGN4−/− male microglia had a significantly lower number of processes, branching points, and smaller process volume compared to WT male (Fig. 1F, left to right: p = 0.0119, t = 3.55; p = 0.022 t = 3.288; p = 0.0011, t = 4.582). Although WT female had lower numbers of processes compared to WT male, there was no significant difference in morphology between female WT and NLGN4−/− microglia (Fig. 1F).
3.3. Phagocytosis of apoptotic neurons is impaired in NLGN4−/− microglia
Microglia are brain phagocytes that clear apoptotic or necrotic cells (Green et al., 2016). In a Rett-syndrome-like mouse model (Mecp2-null), it has been shown that the ability of microglia to phagocytose apoptotic neurons is crucial for disease progression (Derecki et al., 2012). Therefore, we stereotactically injected apoptotic neurons into the hippocampus and cortex to evaluate in vivo phagocytosis capacity of microglia using a flow cytometry-based assay as described previously (Krasemann et al., 2017, Fig. 2A). Microglial phagocytosis of apoptotic neurons was significantly reduced in both NLGN4−/− male and female mice compared to their controls (Fig. 2B, male: p = 0.0499 t = 3.067; female: p = 0.0015 t = 3.993).
Fig. 2. Microglia phagocytosis was reduced in both sexes, but MHCI expression was reduced only in male NLGN4−/−.
(A) Gating strategy to identify the microglia that phagocytosed apoptotic neurons. We first gated live cells followed by single cell gating. We next excluded Ly6C expressing cells and used CD11b+4D4+ to gate for the microglia population. Microglia that phagocytosed apoptotic neurons were identified by 4D4 and FITC co-expression. Representative flow cytometry plots from WT and NLGN4−/− both male and female indicated the reduction of phagocyting apoptotic neurons in NLGN4−/− microglia. (B) Percentage of phagocytic microglia was reduced both in NLGN4−/− male and female as compared to WT by flow cytometry (2-way ANOVA followed by Bonferroni’s multiple comparisons, n = 5 per group, gating strategy in S2G). We performed 2 independent experiments. (C) Expression analysis was done by measuring mean fluorescence intensity of CD45+CD11b+. The histogram exemplifies the shift in MHCI expression. (D) Flow cytometry analysis of MHCI, MHCII, and CD54 (ICAM-1) mean fluorescence intensity (MFI) as proxy for antigen presentation. MHCI and CD54 were significantly reduced in WT female compared to WT male. NLGN4−/− lead to a decrease of MHCI and CD54 expression levels compared to WT in male microglia only (2-way ANOVA followed by Bonferroni’s multiple comparisons; n = 9 per each group). We performed 3 independent experiments. Asterisk indicates significant differences *p < 0.05; **p < 0.01; ***p < 0.001. All mice used for the study were on a C57BL/6J genetic background. All male and female comparison can be found in the supplementary Table S2.
3.4. Male NLGN4−/− microglia express lower levels of activation markers
Microglia also serve as professional antigen presenting cells, and MHC expression is vital for interaction with other immune cells (Malo et al., 2018). As reported earlier, MHCI expression levels were higher in WT males compared to WT females both in hippocampus and cortex (Guneykaya et al., 2018, Fig. 2D). MHCI and CD54 expression levels were significantly decreased in NLGN4−/− male microglia compared to WT, and no difference was observed between female NLGN4−/− and WT microglia from hippocampus (Fig. 2D, gating strategy in Fig. S1C, MHCI: male: p = 0.0064 t = 3.721; male vs female: p = 0.0390 t = 2.981; CD54: male: p = 0.0186 t = 3.287; male vs female: p = 0.0414 t = 2.955)). There was no difference in MHCII expression between groups (Fig. 2D).
3.5. Loss of NLGN4 results in decreased microglia purinergic signaling only in NLGN4−/− male mice
Purinergic signaling regulates crucial microglia functions, such as microglial processes movement to a site of injury and phagocytosis (Davalos et al., 2005; Nimmerjahn et al., 2005; Schulz et al., 2012). We tested microglia process movement measured in male and female acutely prepared hippocampal CA3 brain sections by time-lapse two-photon microscopy in response to a laser lesion (Fig. 3A, Fig. S1D). We observed that WT male microglia responded significantly faster than WT female microglia. However, microglia process movement was significantly reduced in NLGN4−/− male compared to WT, while no changes were observed in female samples (Fig. 3B).
Fig. 3. Microglial purinergic signaling is disrupted only in male NLGN4−/− hippocampus.
(A) Representative pictures of laser-induced process movement of EGFP-positive microglia in the hippocampus of acute brain slices from 13-week-old male WT and NLGN4−/− mice. Asterisk indicates laser lesion location. White appearing centers of the cells reflect the over-saturation of the GFP signal. (B) Quantitative analysis of the lesion-induced microglial process movement in arbitrary units (AU) in male and female WT and NLGN4−/− mice from CA3 sections over 30 min observation time. WT female showed lower levels of process movement compared to WT males. NLGN4−/− male microglia significantly reduced the process movement towards the lesion as compared to WT, whereas no changes were observed between females (n = 5 per group, 2-way ANOVA following Bonferroni’s multiple comparisons test was performed for each time point). Multiple (40) p and t values are available upon request. We performed 3 independent experiments. (C) Average current density to voltage relationships of microglial ATP-induced (10 μM) metabotropic purinergic membrane currents (P2YR12) of microglia from 13-week-old female and male WT and NLGN4−/− hippocampus. Current densities were calculated by subtraction of membrane currents before and during ATP application (2-way ANOVA followed by Bonferroni’s multiple comparisons, number of cells (and mice): female WT, 15 (4); female NLGN4−/−, 11 (3); male WT, 18 (5); male NLGN4−/−, 14 (3)). Please note the typical microglial response to 10 μM ATP with activation of currents in the outward direction in all groups. K + current amplitudes in microglia from male NLGN4−/− hippocampus were significantly reduced. (D) Summary of the outward conductances which were calculated from ATP (10 μM)-evoked currents between + 20 mV and + 60 mV. Only in males the outward current is lower in NLGN4−/− compared to WT. (E) Average current density to voltage relationships of ATP (1 mM) -induced ionotropic purinergic membrane currents (P2XR7) from 13-week-old female and male WT or NLGN4−/− microglia (2-way ANOVA followed by Bonferroni’s multiple comparisons, number of cells (and mice): female WT, 13 (4); female NLGN4−/−, 12 (3); male WT, 21 (5); male NLGN4−/−, 15 (3)). We performed 3 independent experiments. (F) Summary of the inward conductances which were calculated from ATP (1 mM)-evoked currents between 120 mV and − 160 mV. No differences are detected between the groups. Asterisk indicates significant differences between WT and NLGN4−/− mice *P < 0.05; **P < 0.01). All mice used for the study were on a C57BL/6J genetic background. All male and female comparisons can be found in the supplementary Table S2.
We have previously demonstrated that microglial membrane current responses upon application of low (10 μM) ATP concentrations largely depend on P2RY12 activation (Elmadany et al., 2020). We then analyzed purinergic P2RY12 responses of microglia in acute hippocampal slices from 13-week-old male and female NLGN4−/− mice using whole-cell patch clamp techniques in the voltage-clamp configuration. Membrane properties of hippocampal microglia and reversal potential did not differ between WT and NLGN4−/− in both sexes (Fig. S1E, F). Consistent with previous studies, WT microglia responded to 10 μM ATP with the induction of an outwardly rectifying current that reversed close to the equilibrium potential for potassium (Guneykaya et al., 2018; Elmadany et al., 2020). The conductance of microglial ATP-evoked potassium currents was similar for female and male WT microglia (Fig. 3C, D). Interestingly, these microglial purinergic responses were only reduced in male NLGN4−/− microglia whereas responses of female NLGN4−/− microglia were not different from WT females, indicating a sex-specific impairment of P2YR12 signaling in the setting of a NLGN4−/− (Fig. 3C,D, male WT vs. NLGN4−/−:6.3(2.3/10.5) pS/pF p = 0.0477, t = 2.758; female WT vs. NLGN4−/−: p > 0.9999 t = 0.9715).
To analyze P2X receptor signaling, we measured microglial membrane currents evoked by the application of 1 mM ATP, a concentration sufficient to activate P2RX7, the most common P2X receptor isoform in microglia (Butovsky et al., 2014; Ousingsawat et al., 2015). There was no significant difference between the four tested groups (Fig. 3E, F, male WT vs. NLGN4−/−: p > 0.9999, t = 0.6789, female WT vs. NLGN4−/−: p > 0.9999, t = 0.3414). Thus, we speculate that the P2RX7 response is unaffected in the NLGN4−/− animals, while the P2Y12 dependent response is impaired in male NLGN4−/− only.
These data collectively suggest that male microglia derived from the NLGN4−/− hippocampus might fail to perform their first responder role in the CNS more than female NLGN4−/− microglia, reflected by aberrant morphology, reduced antigen presentation potential, and impaired P2YR12 signaling. However, microglial proliferation, apoptotic state and apoptotic neuron phagocytosis appeared to be similar in in both sexes of NLGN4−/− compared to WT.
3.6. Protein profile of microglia shows increased sexual dimorphism in the NLGN4−/− context
To determine the proteome profile of NLGN4−/− microglia, we assessed protein composition by mass spectrometry (MS) label-free quantification (LFQ) of CD11b+ MACS sorted cells from the hippocampus. We identified>10,000 peptide sequences for each sample and performed the quantification using Max Quant algorithms with normalized intensity values (LFQ intensity). Analysis of hippocampal MS data from WT and NLGN4−/− microglia samples resulted in 3433 proteins for statistical quantification (Supp. Table S1). We analyzed the abundance of cell type-specific markers (provided in Zhang et al., 2014) in our samples and found microglia-specific proteins (16 proteins) clearly exceeding markers for the other cell types (median 3.5; Fig. 4B) (Zhang et al., 2014). Overall, principal component analysis (PCA) revealed a difference between male WT and NLGN4−/− (Fig. 4C). In males, 235 proteins were differentially expressed between WT and NLGN4−/− microglia, while only 31 proteins were altered in female NLGN4−/− microglia compared to WT (Fig. 4A).
Fig. 4. Proteomics profile of microglia revealed unique phenotypes for male and female NLGN4−/−.
(A) The heatmap shows all proteins that were significantly different between WT and NLGN4−/− microglia in the male and female group at an FDR (False discovery rate) cutoff of 10%. A row-wise z-score was applied. (B) Brain cell type analysis based on Zhang et al. 2017 revealed a high microglial abundance in the proteomics dataset. (C) Principal component analysis of WT and NLGN4−/− microglial proteomics clusters indicated a different impact of NLGN4loss on male and female microglia. (D-E) Volcano plots highlight the most important proteins WT over NLGN4−/− separately in male (D) and female (E). Proteins of interest are indicated with blue (downregulated in NLGN4−/−), and yellow (upregulated in NLGN4−/−). We applied a cut off as 0.1 FDR (False discovery rate) and a row-wise z-score. Cells were isolated from 3 different cohorts, and mass spectrometry was run in single experiment to avoid a batch effect. (F-G) Gene ontology analysis was done using Metascape (metascape.org; (Zhou et al., 2019)). Metascape biological process analysis of male (F) and female (G) significantly different regulated proteins between WT and NLGN4−/− (n = 3 per group). All mice used for the study were on a C57BL/6J genetic background.
We found that Gmfb, Myh9, Ctss, Ctsh, Grn proteins -important for microglia development and homeostasis- were expressed higher in male NLGN4−/− microglia, but proteins that are important for neuronal clearance and phagocytosis function (Rcn1,Dynll2) were downregulated in NLGN4−/− male microglia compared to WT. Notably, expression of mitochondrial proteins that contribute to energy metabolism and anti-oxidative defense (Cox7a2, Cox6c, Hadha/b, Ndufs, Atp5a1, Nnt) were lower in male NLGN4−/− microglia (Fig. 4D). Ergic1, Itga3, and Nptn were expressed higher in female NLGN4−/− microglia, which have been shown to be upregulated in AD-associated microglia (Krasemann et al., 2017; Schedin-Weiss et al., 2017). Moreover, Lamp2 and Enpp2, components of the cell death pathway, were also expressed higher in female NLGN4−/− microglia (Fig. 4E). Metascape analysis of male proteome suggested changes in metabolic, immune system process, and related pathways such as mitochondrial metabolism, and response to activation between male WT and NLGN4−/− microglia (Fig. 4F, G). Taken together, the microglial proteome profile highlighted the importance of sexual dimorphism in relation to NLGN4 loss and pointed towards disruption in energy metabolism.
3.7. Sex differences are prevalent in hippocampal gamma oscillations in the NLGN4−/− model
Gamma oscillations can modulate purinergic signaling, as well as microglia morphology and function (Guzman and Gerevich, 2016; Iaccarino et al., 2016). Our data collectively suggest a sex difference in purinergic signaling and morphology of microglia in CA3. Since perturbed gamma oscillations were previously only studied in male P20 NLGN4−/− mice (Hammer et al., 2015), we next measured gamma oscillations by cholinergic stimulation in the CA3 area of the hippocampus in acute brain slices from adult males and females.
The power of the oscillations during the first induction was lower in female NLGN4−/− compared to WT whereas no difference could be seen between male WT and NLGN4−/− animals (Fig. 5A). The frequency of gamma oscillation remained unchanged between WT and NLGN4−/− in both sexes, but WT males displayed higher frequencies as compared to WT females (Fig. 5B, m(WT) vs f(WT) = p = 0.0005, t = 4.277). After the induction of the oscillations, we washed out the cholinergic stimulation and re-induced the oscillations after 40 minutes (Lemercier et al., 2016; Meier et al., 2020) (Fig. 5C). We observed significantly slower reinduction of gamma oscillations in male NLGN4−/− animals compared to WT, whereas no such difference was seen in female mice (Fig. 5D, male: p = 0.0439 t = 2.064).
Fig. 5. Hippocampal gamma oscillation is disrupted only in male NLGN4−/− hippocampus.
(A) Representative power spectra of gamma oscillations during the first induction in WT and NLGN4−/− female mice (left graph). Quantification of the peak gamma power shows that power of gamma oscillations is reduced in female NLGN4−/− mice compared to WT during the first induction. Geometric mean ± geometric SD, 3 independent experiments (right graph, n(WT) = 15, n(NLGN4−/−) = 16, female: n(WT) = 17, n(NLGN4−/−) = 9). (B) Quantification of the peak power of the oscillations shows significantly lower gamma oscillation frequency in WT females compared to WT males and no effect of NLGN4−/− (2-way ANOVA, n(WT) = 15, n(NLGN4−/−) = 16, female: n(WT) = 17, n(NLGN4−/−) = 9). (C) The first induction of gamma oscillations in the CA3 region of hippocampus was not different in WT and NLGN4−/− males (left graph). The second induction (re-induction) of gamma oscillations was slower in male NLGN4−/− mice compared to WT (middle graph) while no difference was seen between WT and NLGN4−/− females during re-introduction (right graph) (n(WT) = 15, n(NLGN4−/−) = 16, female: n(WT) = 17, n(NLGN4−/−) = 9). (D) Quantification of re-induced gamma oscillation power normalized to the first induction period shows reduced power only in male NLGN4−/− (2-way ANOVA followed by Bonferroni’s multiple comparisons, male: n(WT) = 15, n(NLGN4−/−) = 16, female: n(WT) = 17, n(NLGN4−/−) = 9)). We performed 3 independent experiments. Asterisk indicates significant differences between WT and NLGN4−/− mice *P < 0.05; **P < 0.01). All mice used for the study were on a C57BL/6J genetic background. All male and female comparison can be found in the supplementary Table S2.
3.8. β-Estradiol treatment rescued microglia phenotype in male NLGN4−/− mice
Altered levels of estrogen receptors have been reported in patients with ASD and schizophrenia, and estrogen therapy has been suggested as a possible treatment candidate for schizophrenia (Gogos et al., 2015; Crider and Pillai, 2017). We found NLGN4 loss to affect male mice more severely in microglial analysis. Therefore, we examined whether estrogen treatment could be beneficial for NLGN4−/− male mice. We supplemented 17ß-estradiol (E2) in the drinking water for 6 weeks by following a previously published protocol (Fernandez and Frick, 2004), and analyzed hippocampal microglia function in male mice at the age of 20-weeks (Fig. 6A). We included untreated WT and NLGN4−/− into the analysis at the same age to control for the age effect since all other data were obtained using 13-week-old animals.
We first examined the purinergic signaling related microglia features. Injury-induced process extensions towards a laser lesion in male NLGN4−/− hippocampal samples were significantly decreased like shown before in slices from 13-week-old mice (Fig. 3B). 17ß-estradiol in vivo treatment recovered NLGN4−/− microglia process movement in male brain sections towards the WT levels (Fig. 6B). Along the same line, also in samples derived from 20-week-old male mice, the P2RY12-dependent response to ATP of male WT microglia was significantly larger than that of male NLGN4−/− microglia (Fig. 6C). The NLGN4−/−-related decrease in P2RY12-dependent purinergic responses was fully rescued by the in vivo 17ß-estradiol treatment (Fig. 6C, WT vs. NLGN4−/−: p = 0.0141 t = 3.294, NLGN4−/− vs. NLGN4−/−-E2: p = 0.0236 t = 2.549). We also observed higher number of AnnexinV-positive microglia cells in untreated 20-week-old NLGN4−/− mice, and we were able to reduce the number of AnnexinV-positive cells in the hippocampus by E2 treatment of NLGN4−/− male animals (Fig. 6D, WT vs. NLGN4−/− p = 0.0089 t = 3.081, NLGN4−/− vs. NLGN4−/−E2: p < 0.0001 t = 4.786). Furthermore, we analyzed microglia morphology in the CA3 region of E2 treated NLGN4−/− male animals. In parallel to process movement, E2 treatment rescued total number of processes, process volume, and distance from the origin in microglia from male NLGN4−/− mice (Fig. 6E).
4. Discussion
Microglia do not express NLGN4 protein (Fig. 1B, (Zhang et al., 2014)). This raises the question: why is the microglia state so strikingly different in a mouse model where a synaptic protein is missing? We propose that disrupted gamma oscillations in the NLGN4−/− model leads to unique alterations of microglia structure, proteome profile, and function in a sexually dimorphic manner. Most of the differences observed between WT and NLGN4−/− mice were present in male mice, but absent in females with a few exceptions, indicating multiple sex-specific impairments in microglial function of NLGN4−/− mice. These impairments include altered male microglial morphology such as decreased number of processes, branch points, and cell volume, reduced antigen presentation potential, impaired P2YR12 signaling, reduced phagocytosis and disrupted microglial energy metabolism. Overall, these findings suggest a unique sex-dependent microglia state, which could be a potential signature of ASD.
The 18 kDa translocator protein (TSPO) is widely studied in many functional MRI studies as an indicator of neuro-inflammation with glial activation, since its density is low in the healthy brain under physiological conditions (Zürcher et al., 2021; Nutma et al., 2021b; Van Camp et al., 2021; Simpson et al., 2022). TSPO expression is evident in many different cell types including microglia, astrocytes, and vascular endothelial cells (Nutma et al., 2021a). Current PET studies from young adult ASD patients show a significant reduction in TSPO density levels in the temporal lobe with functional MR imaging (Simpson et al., 2022; Zürcher et al., 2021). In our study, we observed reduced microglia density, less ramification, and decreased microglial activation in the CA3 region in male NLGN4−/− mice. Since TSPO binding is discussed as a surrogate marker for microglia activation (Enache et al., 2019), and it is lower expressed in ASD patients, one could speculated, that microglia are less responsive in ASD patients.
Purinergic receptors play an important role in variety of microglia functions such as proliferation, phagocytosis, migration, immune response and synaptic pruning (Davalos et al., 2005; Nimmerjahn et al., 2005; Schulz et al., 2012). Among them, the P2 receptor family has 2 main subclusters: ionotropic P2X receptors and metabotropic P2Y receptors. P2Y12 receptor was shown to regulate morphology, cell motility and surveillance of microglia in a healthy brain, as an indicator of homeostatic brain environment (Haynes et al., 2006; Butovsky et al., 2014). We observed less ramified morphology, impaired P2YR12-dependent ATP response and process movement exclusively in male NLGN4−/− microglia compared to WT, which hints at a divergence from the homeostatic microglia state. This is in line with a recent report in which ATP-induced P2Y-mediated membrane currents and P2RY12-dependent laser lesion-induced accumulation of microglial processes were only impaired in male, but not female Nf1+/− microglia (Elmadany et al., 2020). Other studies have shown a lower basal and stimulated migratory capacity of female microglia compared to male (Villapol et al., 2017; Lively and Schlichter, 2018; Yanguas-Casás et al., 2018). It is worth mentioning that in another ASD animal model (Fmr1 knockout), a therapy targeting purinergic signaling restored the microglia-enriched proteins such as C1q and TDP43, and a clinical trial using this therapeutic concept has recently begun (Naviaux et al., 2015; Naviaux, 2018).
Recent studies revealed baseline differences in structure, function and gene expression between male and female microglia (Hanamsagar et al., 2017; Guneykaya et al., 2018; Yanguas-Casás et al., 2018; Elmadany et al., 2020). We observed higher MHCI and CD54 expression in WT male microglia compared to WT females. NLGN4 loss decreased surface expression of these immune activation-related proteins in males, leading to similar expression levels in both sexes of NLGN4−/−. In summary, this suggests a baseline sex difference in WT, and sex-dependent impact of NLGN4 loss. To what extent the loss of NLGN4 alters existing sex differences depends clearly on specific microglia function.
Proteomics data revealed that proteins related to energy metabolism are downregulated in NLGN4−/− male microglia. Pathway analysis indicated changes in metabolic processes, response to stimulus, and immune system processes which point to an exhausted and stressed microglia state in NLGN4−/− male mice.
In addition, gamma oscillation in the CA3 region of hippocampus can be modulated by both ionotropic and metabotropic purinergic receptors (Schulz et al., 2012; Guzman and Gerevich, 2016). Hammer et al., 2015 indicated that kainate-induced gamma oscillations were strongly perturbed in the CA3 region in 15–26 days old male NLGN4−/− mice, but females were not studied. Here we show that loss of NLGN4 results in slower re-introduction of gamma oscillations in 13-week-old NLGN4−/− male mice, whereas no changes were observed in female mice.
In females, we have seen changes in microglial phagocytosis, and AnnexinV binding between WT and NLGN4−/− mice. Nevertheless, the microglia phenotype is rather mildly affected in NLGN4−/− females compared to the males as reflected by smaller changes in the proteome profile and no differences in the purinergic signaling and ATP response towards injury. We hypothesized that female microglia might be protected from the effect of NLGN4 loss because of the higher abundance of estrogen in females. Indeed, in vivo E2 treatment was able to rescue male microglia features and functions related to purinergic signaling, response to injury, morphology and AnnexinV binding. Our findings are in line with other reports about the anti-inflammatory effect of E2 treatment on microglia in vitro and in vivo (Loiola et al., 2019; Wu et al., 2016).
Sex differences in microglia structure and function are the focus of significant attention because of the potential impact of cognitive outcomes in neuropsychiatric and neurodevelopmental disorders. Investigating the biological basis by which sex differences are established will assist in understanding the mechanisms contributing to dysregulated processes in ASD. It is therefore important to consider the value of findings that implicate microglia signaling molecules as critical contributors to male and female brain development in the manifestation of ASD cognitive symptoms. Thus, sex-dependent targeted therapies to modulate microglia function might provide beneficial outcomes for precision medicine. The jury is still out on how microglia function and properties might impact behavior directly (Schwarz and McCarthy, 2021). In our model we propose a mixture of effects that stem from synaptic transmission impairments directly and microglia functional impairments. We show here, in a proof-of-principle approach that there is a beneficial effect of 17-beta estradiol treatment in male NLGN4−/− mice. However, before any translational implication for clinical use could be suggested, more in depth analysis of this effect is needed.
We propose that loss of NLGN4 has a sex-dependent impact on gamma oscillation that in turn affects the microglia state predominantly in males. Slower re-introduction of gamma oscillations in male mice impacting the purinergic signaling may explain the effect of synaptic changes on microglia state. It has been shown previously that altered microglia function affects synaptic wiring and neuronal performance (Schafer et al., 2012; Paolicelli et al., 2014; Ta et al., 2019). We propose that in turn, synaptic dysfunction impacts microglia function. NLGN4-associated microglia state might contribute to further ASD progression in a sex-specific manner and may be targeted by 17-beta estradiol treatment.
Supplementary Material
Acknowledgements
We thank Regina Piske, Nadine Scharek, and Michaela Seeger-Zografski for technical assistance. We thank the Advance Light Microscopy core facility of the Max-Delbrueck-Center (MDC-ALM) and Konstantin Grohmann for technical assistance with confocal microscopy and IMARIS-based data analysis. Authors also thank FACS facility for technical assistance. We thank Dr. Angie Michaiel, Emma Robinson and Maya Jay for proofreading the manuscript.
Funding
This work was supported by the DFG (WO 1418/6-1) (S.A.W.), NIH/NINDS R01 NS088137 (OB), R21 NS104609 (OB), R21 NS101673 (OB); NIH/NIA R01 AG051812 (OB) and R01 AG054672 (OB); BrightFocus Foundation 2020A016806 (OB); the Cure Alzheimer’s Fund (O.B.); Shenzhen Key Laboratory of Neuroimmunomodulation for Neurological Diseases ZDSYS20220304163558001(H.K.).
Footnotes
CRediT authorship contribution statement
Dilansu Guneykaya: Conceptualization, Software, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Bilge Ugursu: Software, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Francesca Logiacco: Formal analysis, Investigation, Writing – review & editing. Oliver Popp: Software, Formal analysis, Investigation, Writing – review & editing. Maria Almut Feiks: Formal analysis, Investigation, Writing – review & editing. Niklas Meyer: Formal analysis, Investigation, Writing – review & editing. Stefan Wendt: Formal analysis, Investigation, Writing – review & editing. Marcus Semtner: . Fatma Cherif: Formal analysis, Investigation. Christian Gauthier: Software, Formal analysis, Investigation. Charlotte Madore: Formal analysis, Investigation, Writing – review & editing. Zhuoran Yin: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Özcan Çınar: Formal analysis, Investigation, Writing – review & editing. Taner Arslan: Software. Zoltan Gerevich: Writing – review & editing, Supervision. Philipp Mertins: Writing – review & editing, Supervision. Oleg Butovsky: Writing – original draft, Writing – review & editing, Supervision. Helmut Kettenmann: Writing – original draft, Writing – review & editing, Funding acquisition, Supervision. Susanne A. Wolf: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbi.2023.03.023.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD024325. All other data will be made available on request.
References
- Bordt EA, Ceasrine AM, Bilbo SD, 2020. Microglia and sexual differentiation of the developing brain: A focus on ontogeny and intrinsic factors. Glia 68, 1085–1099. 10.1002/glia.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucsein C, Zacharias R, Färber K, Pavlovic S, Hanisch U-K, Kettenmann H, 2003. Purinergic receptors on microglial cells: Functional expression in acute brain slices and modulation of microglial activation in vitro. Eur. J. Neurosci 17 (11), 2267–2276. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu LiPing, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL, 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci 17 (1), 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Paulo D, Bento Torres Neto J, Filho CS, de Oliveira TCG, de Sousa AA, dos Reis RR, dos Santos ZA, de Lima CM, de Oliveira MA, Said NM, Freitas SF, Sosthenes MCK, Gomes GF, Henrique EP, Pereira PDC, de Siqueira LS, de Melo MAD, Guerreiro Diniz C, Magalhães N.G.d.M., Diniz JAP, Vasconcelos P.F.d.C., Diniz DG, Anthony DC, Sherry DF, Brites D, Picanço Diniz CW, 2021. Microglial Morphology Across Distantly Related Species: Phylogenetic, Environmental and Age Influences on Microglia Reactivity and Surveillance States. Front. Immunol 12 10.3389/fimmu.2021.683026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M, 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13 (9), 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M, 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26 (12), 1367–1372. [DOI] [PubMed] [Google Scholar]
- Crider A, Pillai A, 2017. Estrogen signaling as a therapeutic target in neurodevelopmental disorders. J. Pharmacol. Exp. Ther 360 (1), 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csicsvari J, Jamieson B, Wise KD, Buzsáki G, 2003. Mechanisms of gamma oscillations in the hippocampus of the behaving rat. Neuron 37 (2), 311–322. [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y.i., Jung S, Littman DR, Dustin ML, Gan W-B, 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci 8 (6), 752–758. [DOI] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SBG, Guyenet PG, Kipnis J, 2012. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 484 (7392), 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson CA, Ziats MN, Rennert OM, 2016. A non-inflammatory role for microglia in autism spectrum disorders. Front. Neurol 10.3389/fneur.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kordi A, Winkler D, Hammerschmidt K, Kästner A, Krueger D, Ronnenberg A, Ritter C, Jatho J, Radyushkin K, Bourgeron T, Fischer J, Brose N, Ehrenreich H, 2013. Development of an autism severity score for mice using Nlgn4 null mutants as a construct-valid model of heritable monogenic autism. Behav. Brain Res 251, 41–49. [DOI] [PubMed] [Google Scholar]
- Elmadany N, de Almeida Sassi F, Wendt S, Logiacco F, Visser J, Haage V, Hernandez DP, Mertins P, Hambardzumyan D, Wolf S, Kettenmann H, Semtner M, 2020. The VGF-derived Peptide TLQP21 Impairs Purinergic Control of Chemotaxis and Phagocytosis in Mouse Microglia. J. Neurosci 40 (17), 3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enache D, Pariante CM, Mondelli V, 2019. Markers of central inflammation in major depressive disorder: A systematic review and meta-analysis of studies examining cerebrospinal fluid, positron emission tomography and post-mortem brain tissue. Brain. Behav. Immun 81, 24–40. 10.1016/j.bbi.2019.06.015. [DOI] [PubMed] [Google Scholar]
- Fernandez SM, Frick KM, 2004. Chronic oral estrogen affects memory and neurochemistry in middle-aged female mice. Behav. Neurosci 118 (6), 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos A, Sbisa AM, Sun J, Gibbons A, Udawela M, Dean B, 2015. A Role for Estrogen in Schizophrenia: Clinical and Preclinical Findings. Int. J. Endocrinol 2015, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Oguin TH, Martinez J, 2016. The clearance of dying cells: Table for two. Cell Death Differ. 23 (6), 915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guneykaya D, Ivanov A, Hernandez DP, Haage V, Wojtas B, Meyer N, Maricos M, Jordan P, Buonfiglioli A, Gielniewski B, Ochocka N, Cömert C, Friedrich C, Artiles LS, Kaminska B, Mertins P, Beule D, Kettenmann H, Wolf SA, 2018. Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Rep. 24 (10), 2773–2783.e6. [DOI] [PubMed] [Google Scholar]
- Guzman SJ, Gerevich Z, 2016. P2Y Receptors in Synaptic Transmission and Plasticity: Therapeutic Potential in Cognitive Dysfunction. Neural Plast. 2016, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Krueger-Burg D, Tuffy L, Cooper B, Taschenberger H, Goswami S, Ehrenreich H, Jonas P, Varoqueaux F, Rhee J-S, Brose N, 2015. Perturbed Hippocampal Synaptic Inhibition and γ-Oscillations in a Neuroligin-4 Knockout Mouse Model of Autism. Cell Rep. 13 (3), 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, Bilbo SD, 2017. Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65 (9), 1504–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan W-B, Julius D, 2006. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci 9 (12), 1512–1519. [DOI] [PubMed] [Google Scholar]
- Hoon M, Soykan T, Falkenburger B, Hammer M, Patrizi A, Schmidt K-F, Sassoè-Pognetto M, Löwel S, Moser T, Taschenberger H, Brose N, Varoqueaux F, 2011. Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc. Natl. Acad. Sci. U. S. A 108 (7), 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, Adaikkan C, Canter RG, Rueda R, Brown EN, Boyden ES, Tsai L-H, 2016. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540 (7632), 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg C, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T, Nydén A, Philippe A, Cohen D, Chabane N, Mouren-Siméoni MC, Brice A, Sponheim E, Spurkland I, Skjeldal OH, Coleman M, Pearl PL, Cohen IL, Tsiouris J, Zappella M, Menchetti G, Pompella A, Aschauer H, Van Maldergem L, 2003. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet 34, 27–29. 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Frahm J, Fischer J, Bourgeron T, Ehrenreich H, Brose N, 2008. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc. Natl. Acad. Sci. U. S. A 105 (5), 1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju A, Hammerschmidt K, Tantra M, Krueger D, Brose N, Ehrenreich H, 2014. Juvenile manifestation of ultrasound communication deficits in the neuroligin-4 null mutant mouse model of autism. Behav. Brain Res 270, 159–164. [DOI] [PubMed] [Google Scholar]
- Kalbassi S, Bachmann SO, Cross E, Roberton VH, and Baudouin SJ. 2017. Male and female mice lacking neuroligin-3 modify the behavior of their wild-type littermates. eNeuro. doi: 10.1523/ENEURO.0145-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A, 2011. Physiology of microglia. Physiol. Rev 91, 461–553. 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, Tjon E, Mazaheri F, Hartmann K, Madi A, Ulrich JD, Glatzel M, Worthmann A, Heeren J, Budnik B, Lemere C, Ikezu T, Heppner FL, Litvak V, Holtzman DM, Lassmann H, Weiner HL, Ochando J, Haass C, Butovsky O, 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47 (3), 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier CE, Schulz SB, Heidmann KE, Kovács R, Gerevich Z, 2016. Dopamine D3 Receptors Inhibit Hippocampal Gamma Oscillations by Disturbing CA3 Pyramidal Cell Firing Synchrony. Front. Pharmacol 10.3389/fphar.2015.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively S, Schlichter LC, 2018. Microglia responses to pro-inflammatory stimuli (LPS, IFNγ+TNFα) and reprogramming by resolving cytokines (IL-4, IL-10). Front. Cell. Neurosci 10.3389/fncel.2018.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiola RA, Wickstead ES, Solito E, McArthur S, 2019. Estrogen Promotes Proresolving Microglial Behavior and Phagocytic Cell Clearance through the Actions of Annexin A1. Front. Endocrinol. (Lausanne) 10.3389/fendo.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, Jones EJH, Jones RM, Pickles A, State MW, Taylor JL, and Veenstra-VanderWeele J. 2020. Autism spectrum disorder. Nat. Rev. Dis. Prim doi: 10.1038/s41572-019-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MA, 2022. Exploring Sex-Related Differences in Microglia May Be a Game-Changer in Precision Medicine. Front. Aging Neurosci 14 10.3389/fnagi.2022.868448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maćkowiak M, Mordalska P, Wędzony K, 2014. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol. Rep 66 (5), 830–835. [DOI] [PubMed] [Google Scholar]
- Malo CS, Jin F, Hansen MJ, Fryer JD, Pavelko KD, and Johnson AJ. 2018. MHC class I expression by microglia is required for generating a complete antigen-specific CD8 T cell response in the CNS. J. Immunol [Google Scholar]
- Matta SM, Hill-Yardin EL, and Crack PJ. 2019. The influence of neuroinflammation in Autism Spectrum Disorder. Brain. Behav. Immun 79:75–90. doi: 10.1016/j.bbi.2019.04.037. [DOI] [PubMed] [Google Scholar]
- Matta SM, Moore Z, Walker FR, Hill-Yardin EL, Crack PJ, 2020. An altered glial phenotype in the NL3R451C mouse model of autism. Sci. Rep 10 10.1038/s41598-020-71171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei D, Djodari-Irani A, Hadar R, Pelz A, de Cossío LF, Goetz T, Matyash M, Kettenmann H, Winter C, Wolf SA, 2014. Minocycline rescues decrease in neurogenesis, increase in microglia cytokines and deficits in sensorimotor gating in an animal model of schizophrenia. Brain. Behav. Immun 38, 175–184. [DOI] [PubMed] [Google Scholar]
- Meier MA, Lemercier CE, Kulisch C, Kiss B, Lendvai B, Adham N, Gerevich Z, 2020. The novel antipsychotic cariprazine stabilizes gamma oscillations in rat hippocampal slices. Br. J. Pharmacol 177 (7), 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, Petyuk VA, Thomas SN, Mani DR, Mundt F, Moore RJ, Hu Y, Zhao R, Schnaubelt M, Keshishian H, Monroe ME, Zhang Z, Udeshi ND, Mani D, Davies SR, Townsend RR, Chan DW, Smith RD, Zhang H, Liu T, Carr SA, 2018. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc 13 (7), 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JH, 2011. Autism spectrum disorders-A genetics review. Genet. Med 13 (4), 278–294. [DOI] [PubMed] [Google Scholar]
- Napolitano A, Schiavi S, La Rosa P, Rossi-Espagnet MC, Petrillo S, Bottino F, Tagliente E, Longo D, Lupi E, Casula L, Valeri G, Piemonte F, Trezza V, Vicari S, 2022. Sex Differences in Autism Spectrum Disorder: Diagnostic, Neurobiological, and Behavioral Features. Front. Psychiatry 13. 10.3389/fpsyt.2022.889636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, 2018. Antipurinergic therapy for autism—An in-depth review. Mitochondrion 43, 1–15. [DOI] [PubMed] [Google Scholar]
- Naviaux JC, Wang L, Li K, Bright A, Alaynick WA, Williams KR, Powell SB, Naviaux RK, 2015. Antipurinergic therapy corrects the autism-like features in the Fragile X (Fmr1 knockout) mouse model. Mol. Autism 6 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F, 2005. Neuroscience: Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308 (5726), 1314–1318. [DOI] [PubMed] [Google Scholar]
- Nutma E, Ceyzériat K, Amor S, Tsartsalis S, Millet P, Owen DR, Papadopoulos V, Tournier BB, 2021a. Cellular sources of TSPO expression in healthy and diseased brain. Eur. J. Nucl. Med. Mol. Imaging 49, 146–163. 10.1007/s00259-020-05166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutma E, Gebro E, Marzin MC, van der Valk P, Matthews PM, Owen DR, Amor S, 2021b. Activated microglia do not increase 18 kDa translocator protein (TSPO) expression in the multiple sclerosis brain. Glia 69, 2447–2458. 10.1002/glia.24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin E, Pakan JMP, Yilmazer-Hanke D, McDermott KW, 2017. Acute in utero exposure to lipopolysaccharide induces inflammation in the pre- and postnatal brain and alters the glial cytoarchitecture in the developing amygdala. J. Neuroinflamm 14 (1) 10.1186/s12974-017-0981-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousingsawat J, Wanitchakool P, Kmit A, Romao AM, Jantarajit W, Schreiber R, and Kunzelmann K. 2015. Anoctamin 6 mediates effects essential for innate immunity downstream of P2X7 receptors in macrophages. Nat. Commun doi: 10.1038/ncomms7245. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bisht K, Tremblay MÈ, 2014. Fractalkine regulation of microglial physiology and consequences on the brain and behavior. Front. Cell. Neurosci 10.3389/fncel.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, Stevens B, 2017. Microglia emerge as central players in brain disease. Nat. Med 23 (9), 1018–1027. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B, 2012. Microglia Sculpt Postnatal Neural Circuits in an Activity and Complement-Dependent Manner. Neuron 74, 691–705. 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedin-Weiss S, Inoue M, Hromadkova L, Teranishi Y, Yamamoto NG, Wiehager B, Bogdanovic N, Winblad B, Sandebring-Matton A, Frykman S, Tjernberg LO, 2017. Monoamine oxidase B is elevated in Alzheimer disease neurons, is associated with γ-secretase and regulates neuronal amyloid β-peptide levels. Alzheimer’s Res. Ther 9 (1) 10.1186/s13195-017-0279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz SB, Klaft Z-J, Rösler AR, Heinemann U, Gerevich Z, 2012. Purinergic P2X, P2Y and adenosine receptors differentially modulate hippocampal gamma oscillations. Neuropharmacology 62 (2), 914–924. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, and McCarthy MM. 2021. Microglia, Hormones, and Behavior. 207–228. [Google Scholar]
- Shinohara Y, Hosoya A, and Hirase H. 2013. Experience enhances gamma oscillations and interhemispheric asymmetry in the hippocampus. Nat. Commun doi: 10.1038/ncomms2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson D, Gharehgazlou A, Da Silva T, Labrie-Cleary C, Wilson AA, Meyer JH, Mizrahi R, Rusjan PM, 2022. In vivo imaging translocator protein (TSPO) in autism spectrum disorder. Neuropsychopharmacology 47, 1421–1427. 10.1038/s41386-022-01306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta TT, Dikmen HO, Schilling S, Chausse B, Lewen A, Hollnagel JO, Kann O, 2019. Priming of microglia with IFN-γ slows neuronal gamma oscillations in situ. Proc. Natl. Acad. Sci. U. S. A 116, 4637–4642. 10.1073/pnas.1813562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, Hardy J, and Ryten M. 2013. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp N, Lavisse S, Roost P, Gubinelli F, Hillmer A, and Boutin H. 2021. TSPO imaging in animal models of brain diseases. Eur. J. Nucl. Med. Mol. Imaging 49: 77–109. doi: 10.1007/s00259-021-05379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA, 2005. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol 57 (1), 67–81. [DOI] [PubMed] [Google Scholar]
- Villapol S, Loane DJ, Burns MP, 2017. Sexual dimorphism in the inflammatory response to traumatic brain injury. Glia 65 (9), 1423–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, and Hermjakob H. 2013. The Proteomics Identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chen YW, Tsai SF, Wu SN, Shih YH, Jiang-Shieh YF, Yang TT, Kuo YM, 2016. Estrogen ameliorates microglial activation by inhibiting the Kir2.1 inward-rectifier K+ channel. Sci. Rep 10.1038/srep22864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanguas-Casás N, Crespo-Castrillo A, de Ceballos ML, Chowen JA, Azcoitia I, Arevalo MA, Garcia-Segura LM, 2018. Sex differences in the phagocytic and migratory activity of microglia and their impairment by palmitic acid. Glia 66 (3), 522–537. [DOI] [PubMed] [Google Scholar]
- Zhang Y.e., Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ, 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34 (36), 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.o., Gokce O, Hale WD, Brose N, Südhof TC, 2018. Autism-associated neuroligin-4 mutation selectively impairs glycinergic synaptic transmission in mouse brainstem synapses. J. Exp. Med 215 (6), 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher NR, Loggia ML, Mullett JE, Tseng C, Bhanot A, Richey L, Hightower BG, Wu C, Parmar AJ, Butterfield RI, Dubois JM, Chonde DB, Izquierdo-Garcia D, Wey HY, Catana C, Hadjikhani N, McDougle CJ, Hooker JM, 2021. [11C]PBR28 MR–PET imaging reveals lower regional brain expression of translocator protein (TSPO) in young adult males with autism spectrum disorder. Mol. Psychiatry 26 (5), 1659–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD024325. All other data will be made available on request.