Abstract
BH3-mimetics are used as an efficient strategy to induce cell death in several blood malignancies, including acute myeloid leukemia (AML). Venetoclax, a potent BCL-2 antagonist, is used clinically in combination with hypomethylating agents for the treatment of AML. Moreover, MCL-1 or dual BCL-2/BCL-xL antagonists are under investigation. Yet, resistance to single or combinatorial BH3-mimetics therapies eventually ensues. Integration of multiple genome-wide CRISPR/Cas9 screens revealed that loss of mitophagy modulators sensitizes AML cells to various BH3-mimetics targeting different BCL-2 family members. One such regulator is MFN2, whose protein levels positively correlate with drug resistance in patients with AML. MFN2 overexpression is sufficient to drive resistance to BH3-mimetics in AML. Insensitivity to BH3-mimetics is accompanied by enhanced mitochondria-endoplasmic reticulum interactions and augmented mitophagy flux which acts as a pro-survival mechanism to eliminate mitochondrial damage. Genetic or pharmacologic MFN2 targeting synergizes with BH3-mimetics by impairing mitochondrial clearance and enhancing apoptosis in AML.
Statement of significance
AML remains one of the most difficult to treat blood cancers. BH3-mimetics represent a promising therapeutic approach to eliminate AML blasts by activating the apoptotic pathway. Enhanced mitochondrial clearance drives resistance to BH3-mimetics and predicts poor prognosis. Reverting excessive mitophagy can halt BH3-mimetics resistance in AML.
Keywords: BH3-mimetics, resistance, mitophagy, MFN2, AML
Introduction
Acute myeloid leukemia (AML) is an aggressive blood cancer with dismal clinical outcomes exemplified by a 28% five-year overall survival rate1. Targeting the BCL-2 family-regulated apoptotic pathway is a novel means of treating multiple forms of cancer, including AML2-4. Venetoclax, a selective BCL-2 antagonist, was FDA-approved in 2018 for the treatment of AML in combination with hypomethylating agents (HMA), azacytidine and decitabine. Despite promising early responses of AML patients to venetoclax/HMA, intrinsic or acquired drug resistance ensues after prolonged treatment, highlighting the urgency for a better understanding of the underlying molecular mechanisms5,6.
Accumulating evidence links TP53 aberrations, FLT3 kinase activation or activation of other survival kinase signaling pathways, as well as acquired loss of function mutations in BAX to adaptive resistance to venetoclax-based therapies in patients with AML7-11. Recent studies from our laboratories and others revealed mitochondrial and metabolic adaptations as modes of resistance to BCL-2 antagonism, including alterations in mitochondrial cristae shape and dependency on fatty acid metabolism12-14. Targeting of mitochondrial structure12, translation15, mitochondrial complex I16, protein degradation17,18, or fatty acid oxidation13 sensitizes AML cells to venetoclax-based treatments. Not surprisingly, upregulation or variation on the dependencies on the anti-apoptotic BCL-2 family proteins, including MCL-1, BCL-xL, and BCL2A17,9,12,19, are among the most recurrent determinants of venetoclax resistance in AML.
Concomitant targeting of MCL-1 with BCL-2 has been proven to be an attractive way to overcome resistance to BCL-2 inhibition. Several compounds antagonizing MCL-1 (AMG17620; AZD599121,22; VU66101323; and S6384524) have been developed and have been tested in Phase I/II clinical trials as monotherapy or in combination with venetoclax in patients with hematologic malignancies, including AML (NCT02675452, NCT03218683, NCT03672695, NCT04629443). Moreover, CDK inhibitors, such as CYC065 and voruciclib, which indirectly repress MCL-1, are also under clinical development for the treatment of AML25-27 (NCT04017546, NCT03547115). Yet, resistance is anticipated to arise upon MCL-1 suppression, and most importantly AML cells may eventually escape even combinatorial BH3-mimetics treatments. Thus, there is an imperative need to understand the mechanisms of resistance to these combinatorial treatments and to develop additional therapeutic means to bypass the multi-modal BH3-mimetics resistance.
To gain insight into common principles of resistance to various BH3-mimetics, we integrated loss-of-function CRISPR/Cas9 screens in human AML cells treated with: a) MCL-1 inhibitors (MCL1i) as a monotherapy, b) MCL1i combined with venetoclax, and, c) venetoclax plus azacytidine. Loss of BAX, BAK1, and TP53 were common determinants of drug resistance, while deficiency of MFN2 and MARCH5, genes mediating mitochondrial autophagy (mitophagy), sensitized AML cells to all treatment types. Our work revealed that isogenic human AML cells resistant to BH3-mimetics have increased mitochondrial clearance through mitophagy upon induction of mitochondrial stress compared to their sensitive counterparts, suggesting that mitophagy acts as a pro-survival cellular mechanism to eliminate damaged mitochondria and evade apoptosis. Accordingly, blockage of autophagy or specific targeting of MFN2 potentiates BH3-mimetics action in eliminating leukemic cells in vitro and in preclinical AML patient-derived models.
Results
Integration of CRISPR/Cas9 loss-of-function screens identifies prevailing mechanisms of resistance and synthetic lethality to BH3-mimetics in AML.
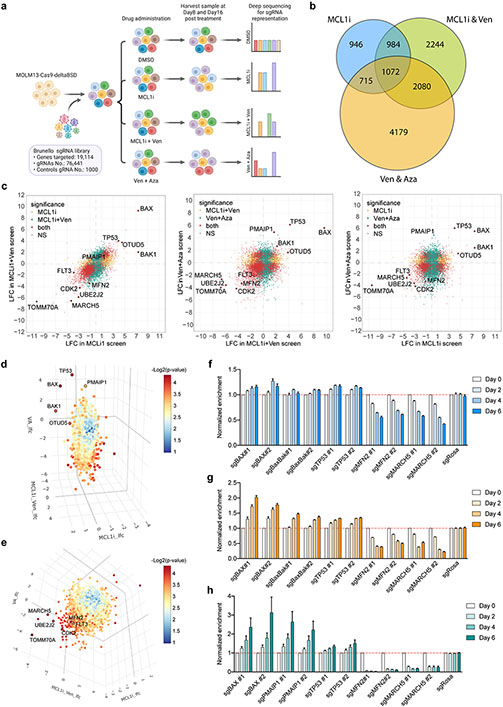
To systematically identify common determinants of resistance to various BH3-mimetics treatments, we performed genome-wide CRISPR/Cas9 loss-of-function screens in human AML cells treated with: a) MCL1i, as a single agent, b) MCL1i combined with venetoclax (MCL1i+Ven), and c) venetoclax plus azacytidine (Ven+Aza), the FDA-approved regimen. After transducing MOLM-13 cells stably expressing Cas9 with the Brunello sgRNA library28 and selecting for the infected cells, cells were treated with BH3-mimetics and their combinations for 16 days (Fig. 1a). Our strategy allowed us to pinpoint the genes whose ablation significantly conferred resistance to or sensitized cells to all the tested BH3-mimetics combinations (Fig. 1b; Suppl. Table 1). Loss of TP53, BAX, or BAK were recurrent primary factors to evade apoptosis induced by MCL1i, MCL1i+Ven, or Ven+Aza regimens (Fig. 1c-d; Fig. S1a). Additionally, one of the top hits in the positive arm of the screens was OTUD5, which encodes a deubiquitinase that stabilizes p5329. This result is in agreement with previous studies showing that perturbations in the p53 pathway and BAX mutations led to venetoclax-based treatment failure in patients with AML7,8,10-12. Consistently, our findings further highlight the challenge to overcome the p53- or BAX-mediated resistance and the urgency to explore alternative combinatorial therapeutic strategies.
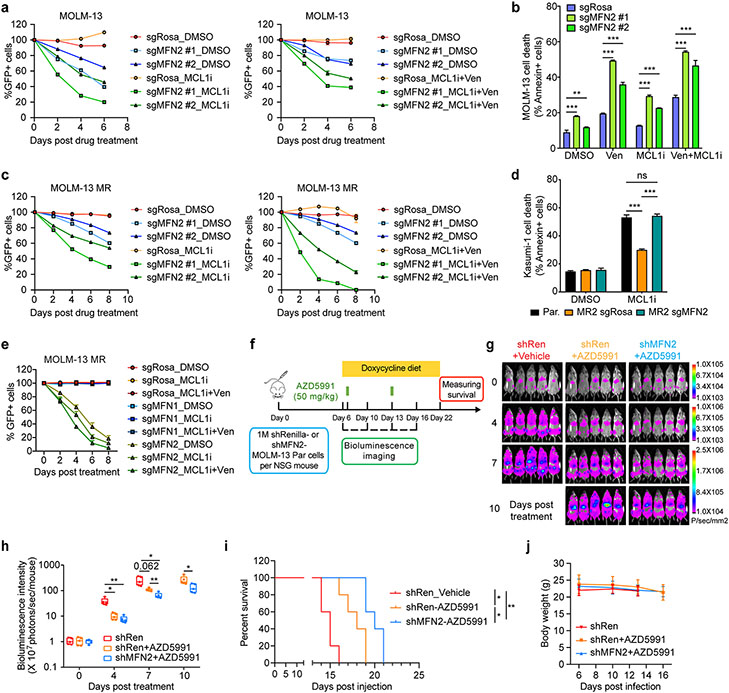
Figure 1. Genome-wide CRISPR/Cas9 loss-of-function screens uncover genes whose ablation synergizes or confers resistance to MCL1i- and Venetoclax- based treatments.
a. Schematic of the screens.
b. Venn diagram depicting the intersection of commonly identified hits in all three screens.
c. Scatter plots showing comparisons of significantly selected genes in MCL1i and MCL1i+Ven screens (left), MCL1i+Ven and Ven+Aza screens (middle), and MCL1i and Ven+Aza screens (right). Genes highlighted in yellow and green are significantly selected in one out of the two indicated screens, while in red are genes significantly selected in both screens shown in each plot. Axes denote the log2 fold change (LFC) of sgRNA representations in drug-treated groups relative to DMSO control. Significance was defined as p < 0.05 in the screen.
d-e. Three-dimensional plots combining the significantly selected hits from all three screens. In panel d, top-ranking positive hits are highlighted, while in e top-scoring negative hits are shown.
f-h. Validation of selected genes using competition-based survival assays in MOLM-13 cells treated with MCL1i (f), MCL1i+Ven (g) or Ven+Aza (h) (n=3, mean ± SD). Normalized enrichment scores were calculated as described in the methods.
On the opposite arm of the screens, amongst the common top-scoring genes whose ablation sensitized the cells to the BH3-mimetics combinations, we identified genes that encode outer mitochondrial membrane proteins, including MFN2, MARCH5, and TOMM70A (Fig. 1c, e; Fig. S1a). All these gene products are involved in mitochondrial dynamics, mitochondrial import, and the selective degradation of mitochondria by autophagy, termed mitophagy30-37. In addition, CDK2 and FLT3 were identified as recurrent negative hits (Fig. 1c, e). CDK2 inhibition has been previously proposed to act synergistically with BH3-mimetics in leukemias, due to CDK2 function in phosphorylating and stabilizing MCL-138,39. The synergy of FLT3 inhibition with MCL-1 targeting has been reported in AML cells carrying FLT3-ITD mutations40,41. In addition, we recently reported a pronounced synergy between FLT3 and BCL-2 inhibition in a clinical trial42, further validating our screen design and underscoring the potential of these combinatorial treatments in human AML.
KEGG pathway analyses of the pairwise intersections of our screens identified key cellular pathways enriched in our screening strategies, including the p53 signaling pathway, fatty acid metabolism, and TCA cycle, in line with previous studies on venetoclax resistance8,13. Core essential processes associated with the ribosome, RNA degradation, and cell cycle were also identified, as well as less common processes like mitophagy and autophagy (Fig. S1b-e). Additional Gene Ontology (GO) analysis of the common hits from all three screens highlighted the relevance of pathways involved in mitochondrial cytochrome c release, response to endoplasmic reticulum (ER) stress, mitochondrial fusion, and mitochondrial organization in the acquisition of drug resistance (Fig. S1f).
Competition-based viability assays for our top-scoring hits in cells treated with MCL1i, MCL1i+Ven, or Ven+Aza, additionally validated the findings of our screens (Fig. 1f-h; Fig. S1g-i). Dose-response curves confirmed that BAX depletion leads to apoptosis resistance upon MCL1i or Ven+Aza treatments (Fig. S1j). Similarly, p53 ablation increases the IC50 of MCL1i, MCL1i+Ven, and Ven+Aza in human AML cells (Fig. S1k). Interestingly, a murine AML model expressing a p53R172H mutant, which is prevalent in patients with AML43, exhibited resistance not only to single MCL1i treatment but also to the combination MCL1i+Ven (Fig. S1l). Hence, although combining BH3-mimetics, such as MCL1i with Ven, is an attractive therapeutic strategy for AML, still resistance develops, underscoring the need for concomitant targeting of alternative cellular pathways together with the apoptosis-inducing agents to competently eliminate leukemic cells.
BH3-mimetics resistance is accompanied by MFN2 upregulation and mitochondria-ER tethering adaptations.
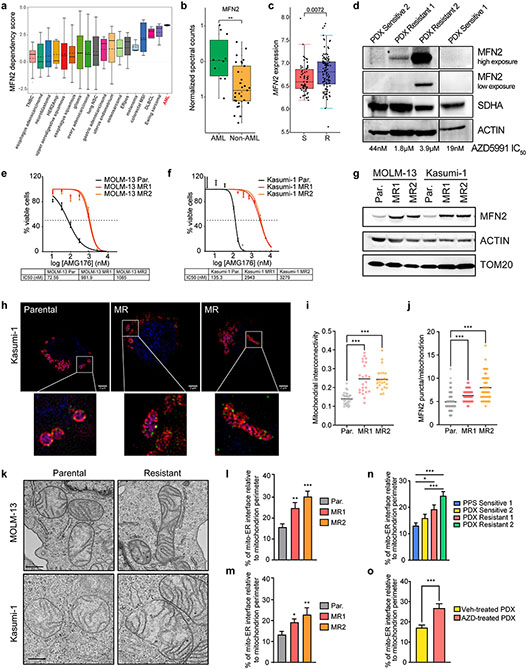
Our top-scoring candidate was Mitofusin-2 (MFN2), which encodes a GTPase that localizes in both the outer mitochondrial membrane and the surface of the ER. According to the DepMap database, MFN2 expression is preferentially required for AML cell survival compared to other tumor types (Fig. 2a). Moreover, MFN2 protein levels are significantly elevated in human AML cell lines relative to other hematologic malignancies44 (Fig. 2b), highlighting the specific relevance of this protein in this blood neoplasm.
Figure 2. MFN2 and mitochondria-ER interactions participate in the acquisition of BH3-mimetics resistance.
a. MFN2 dependency scores across a diverse panel of 563 cancer cell lines from DepMap. A positive score signifies a stronger gene dependency.
b. Quantification of normalized MFN2 spectral counts in AML and other hematologic malignancies based on mass spectrometry-based data from Cancer Cell Line Encyclopedia44.
c. MFN2 expression in BeatAML cohorts with Sensitive (S, IC50 < 0.25 μM) vs Resistant (R, IC50 < 0.25 μM) patients to venetoclax.
d. Western blotting in total cell lysates from patient-derived primary or xenografted AML blasts. MCL1i (AZD5991) IC50 values of each patient sample were calculated after 48 hrs ex vivo treatment.
e-f. IC50 curves of MCL1i (AMG176) in parental (Par.) or MCL1i-resistant (MR) AML cell lines after 48 hrs treatment (MOLM-13; e, Kasumi-1; f) (n=3, mean ± SD).
g. Western blotting in total cell lysates from Par. and MR AML cell lines.
h. Representative z-stack projection (upper) and single z-stack (lower) images from super-resolution microscopy of Parental or MR MOLM-13 and Kasumi-1 cells stained with mitoTracker (magenta), TOM20 (red), MFN2 (green) and DAPI (blue). Scale bars represent 2 μm.
i. Quantification of mitochondrial interconnectivity (area/perimeter) in Kasumi-1 cells in (h).
j. Measurement of MFN2 puncta per individual mitochondrion in Kasumi-1 cells in (h).
k. Representative electron micrographs of mitochondria and their interactions with ER from Par. or MR MOLM-13 and Kasumi-1 cells. Scale bar represents 0.5 μm.
l-m. Quantification of the mitochondria-ER contacts in experiments as in (k), in MOLM-13 (l) and Kasumi-1 (m) cells. Graphs depict the percentage of mitochondria-ER interface relative to the mitochondrial perimeter (n=30, mean ± SEM).
n. Graph depicting the percentage of mitochondria-ER interface relative to the mitochondrial perimeter as quantified in electron micrographs from patient AML blasts (n=45, mean ± SEM).
o. Quantification of the percentage of mitochondria-ER interface relative to the mitochondrial perimeter in electron micrographs from PDX Resistant 1 cells harvested from mice treated weekly with vehicle or 100 mg/kg AZD5991 for 28 days (n=30, mean ± SEM).
The p values were determined by unpaired two-tailed t-test.
Importantly, based on the BeatAML database45, high MFN2 expression in patients with AML positively correlates to high ex vivo IC50 values for a number of drug therapies, including venetoclax (Fig. 2c; Suppl. Table 2). To comprehensively investigate the mechanisms of BH3-mimetics resistance, we, firstly, generated and utilized human AML patient-derived xenografts (PDX) with different mutational backgrounds (Suppl. Table 3). These PDXs were tested ex vivo for their responsiveness to AZD5991. We found that two of them were sensitive (IC50 < 50 nM for 48hr treatment) and the other two PDXs were resistant to AZD5991 (IC50 > 1 μM for 48hr treatment). Notably, the resistant primary patient sample 2 (PDX Resistant 2) was purified from a patient who was refractory to Ven+Aza treatment. Immunoblotting of lysates from these patients’ samples revealed elevated MFN2 protein levels in the resistant blasts compared to the sensitive ones, with the highest levels in the refractory PDX Resistant 2 (Fig. 2d). In addition, we generated human AML clones highly resistant to MCL-1 inhibition, by growing the parental MOLM-13 (expressing MLL-AF9 and FLT3-ITD) and Kasumi-1 (that is AML1-ETO+; KIT, RAD21, TP53 mutant) cells in increasing doses of the MCL1i (AMG176) for over eight weeks. IC50 analyses confirmed a significant increase of MCL1i concentration that is required to induce 50% cell death in the MCL1i-resistant (MR) cell lines with respect to the parental clones (Par) (Fig. 2e-f). The cells displayed resistance also to other compounds that antagonize MCL-1, including AZD5991 (Fig. S2a). Consistently, BH3-profiling also revealed that the resistant cell lines display significantly reduced apoptotic priming compared to the parental blasts (Fig. S2b). Of note, MCL1i resistance was sustained even after a drug holiday (Fig. S2c). Notably, we observed a significant increase in the protein levels of MFN2 in all the MR cell lines relative to the parental clones (Fig. 2g). These findings strongly suggest a role of MFN2 in the acquisition of BH3-mimetics resistance in human AML.
Mechanistically, MFN2 plays critical roles in mitochondrial fusion46, tethering mitochondria to the ER34, while its phosphorylation flags damaged mitochondria for clearance via mitophagy35. Intrigued by the role of MFN2 in mitochondrial dynamics, we initially employed 3D structured illumination microscopy (SIM) to visualize mitochondrial morphology in fixed AML cells stained with mitoTracker (mitochondrial matrix), TOM20 (outer mitochondrial membrane), and MFN2 and observed a significant enlargement of mitochondria of the MCL1i-resistant cells relative to the parental clones (Fig. 2h). Morphometric analysis on z-stack projections revealed that the apoptosis-resistant mitochondria exhibit higher interconnectivity (average area/perimeter) (Fig. 2i; Fig. S2d). In addition, quantification of the number of MFN2 puncta per mitochondrion demonstrated a higher abundance of MFN2 on the surface of individual mitochondria of the resistant clones relative to the parental ones (Fig. 2h, j; Fig. S2d).
Next, to determine potential alterations in mitochondria-ER juxtaposition, we performed electron microscopy-based quantifications (Fig. S2e). We demonstrated that the percentage of mitochondria-ER interface relative to the mitochondrial perimeter was increased in the MR cells, indicating a higher abundance of mitochondria-ER associated membranes (MAMs) (Fig. 2k-m). This is in line with the upregulation of the protein-tether MFN2. Similar results were obtained also in Ven+Aza (VAR) resistant MOLM-13 and MV4-11 cells, even after a drug holiday (Fig. S2f-g, h-i). Likewise, cells from AML patient samples that are resistant to MCL1 inhibition exhibited more MAMs than the sensitive PDXs (Fig. 2n; Fig. S2j). We also transplanted patient-derived cells (PDX Resistant 1) into recipient mice, which were then treated with AZD5991 for three weeks. Electron microscopy in PDX blasts harvested from the bone marrow of the mice showed an increase in mitochondria-ER juxtaposition in AZD5991-treated versus the vehicle-treated xenografts (Fig. 2o). All the above findings indicate that alterations in the mitochondria-ER contact sites are a consistent cellular adaptation mechanism to chronic treatment with BH3-mimetics in human AML.
Cells resistant to BH3-mimetics display high rates of mitophagy.
Besides its role in mitochondrial dynamics, MFN2 is also a mediator of the autophagic clearance of mitochondria through serving as a receptor for PARKIN onto the damaged mitochondria and by supplying autophagosome membranes35,47,48. To study the cellular process of mitophagy, we firstly transduced AML cells with the pH-sensitive fluorescent tag of mitochondria, mitoKeima, and measured the mCherry fluorescence that corresponds to mitochondria inside the acidic lysosomes (pH 4.5) during mitophagy49,50. Using the mitoKeima-stably expressing cells, we verified that sublethal doses of MCL1i or Ven+Aza increased mitophagy in a dose-dependent manner in AML cells (Fig. S3a-b). This finding was also validated using the ratiometric FUGW-PK-hLC3 reporter to monitor autophagic flux51, in experiments demonstrating elevated numbers of autophagic cells upon treatment with BH3-mimetics (Fig. S3c).
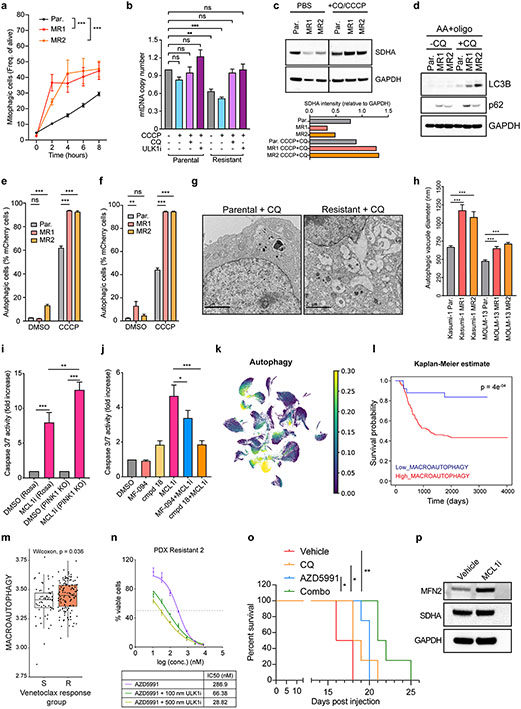
Since the mitophagy receptor MFN2 is upregulated during the acquisition of MCL1-resistance, we asked whether the MR AML cells have higher mitophagy flux compared to the parental AML cells. Under steady state, we observed no significant changes, however, upon an insult provided by the mitochondrial uncoupler CCCP, the resistant cells were more readily undergoing mitophagy compared to the parental clones (Fig. 3a; Fig. S3d). Similar results were also obtained in the VAR AML cells (Fig. S3e-f). In line with this result, we observed that the resistant cells display significantly lower mitochondrial biomass at steady state and in response to CCCP treatment compared to the parental cells using the quantification of mtDNA as a readout. Strikingly, pharmacologic inhibition of autophagy using chloroquine (CQ) or ULK1-inhibition (ULK1i) dramatically increased mtDNA content in the resistant cells to levels that are comparable with the baseline of the parental cells (Fig. 3b). Taken all together our findings suggest that the resistant cells contain fewer mitochondria than the sensitive cells due to enhanced mitochondrial autophagy. Interestingly, when compared to healthy hematopoietic stem and progenitor cells (CD34+ cord blood cells), the AML cell lines have more mitochondrial mass (Fig. S3g), consistent with the previously described mitochondrial dependency of AML cells52. Yet, the BH3-mimetics resistant clones have a reduction in mtDNA content (Fig. S3g). The reduced mitochondria content in the resistant cells is also verified in experiments using mitoTracker green staining (Fig. S3h). To further corroborate these results, we performed western blotting in parental and resistant cells treated with CCCP and CQ and we stained for SDHA, an inner mitochondrial protein that serves as a mitochondrial marker. Notably, we observed that resistant blasts have lower levels of SDHA at baseline than parental cells. Moreover, inhibition of autophagy dramatically increased SDHA levels in resistant cells, while the same conditions had a minimum effect on SDHA levels in parental cells (Fig. 3c). These data further confirm that resistant cells have enhanced mitochondrial clearance via mitophagy relative to the parental clones. Despite the reduced mitochondrial biomass, mitochondrial respiration appears largely unaltered in the resistant clones, suggesting that the increased mitophagy in the resistant clones maintains a healthier and more functional mitochondrial population compared to the parental cells (Fig. S3i-j). This result could possibly explain the reduced sensitivity to apoptosis in the resistant clones as a subsequent mitochondrial adaptation due to increased clearance of damaged organelles.
Figure 3. AML cells resistant to BH3-mimetics have higher capacity for mitophagy.
a. Frequencies of mitophagic cells in a time-course of CCCP treatment (15 μM) in parental or resistant mitoKeima-expressing Kasumi-1 cells treated with 15 μM CCCP or PBS. (n=3, mean ± SD, unpaired two-tailed t-test).
b. Quantification of mtDNA content in Parental and Resistant cells after treatment as indicated (n=6, mean ± SEM). Pretreatment with 100 μM CQ or 200 nM ULK1 inhibitor (ULK1i) for 3 hrs; CCCP 10 μM for 30 min. Statistics were calculated using one-way ANOVA.
c. Western blotting in whole cell lysates from parental and resistant MOLM-13 cells. Where indicated, cells were pretreated with CQ for 3 hrs and CCCP for 30 min (top). Quantitative densitometric analysis of SDHA compared to GAPDH in Western Blots (bottom).
d. Immunoblotting for autophagy markers, LC3B and p62, in MOLM-13 treated with antimycin A (AA, 1 μM) and oligomycin (oligo, 1 μM) with or without chloroquine (CQ, 50 μM) for 19 hrs.
e-f. Bar graphs depicting the frequencies of autophagic cells (mCherry positive) from alive FUGW-PK-hLC3-expressing Kasumi-1 (e) or MOLM-13 (f) cells treated with 10 μM CCCP for 21 hrs (n=3, mean ± SEM, two-way ANOVA).
g. Representative electron micrographs of Kasumi-1 cells treated with 50 μM CQ for 19 hrs.
h. Quantification of autophagic vacuole diameter in experiments as in (g) in Kasumi-1 and MOLM-13 cells (at least 45 autophagosomes from 10 cells per condition; mean ± SEM; unpaired two-tailed t-test).
i. Caspase 3/7 assay in Kasumi-1 Rosa and Kasumi-1 PINK1 KO cells treated with AZD5991 (MCL1i, 1 μM, 8 hr). Data represent mean ± SEM of four independent biological replicates.
j. Caspase 3/7 assay in MOLM-13 parental cells treated with AZD5991 (MCL1i, 1 μM, 8 hr), MF094 (10 μM, 29 hr), and compound 18 (cmpd 18, 20 μM, 29 hr). Data represent mean ± SEM of three independent biological replicates.
k. UMAP colored by the usage of non-negative matrix factorization determined gene expression profile (GEP) containing genes enriched for autophagy-related processes.
l. Kaplan-Meier curves split by high/low mean gene expression of ‘Macroautophagy’ GO pathway genes overlapping with autophagy GEP using overall survival (OS) as an endpoint in TARGET-AML cohort. Split points were determined using rpart. Log-rank test was used to determine significance.
m. Boxplot of ‘Macroautophagy’ scores in venetoclax sensitive and resistant patients from the BeatAML cohort. Wilcoxon rank sum test was used to determine significance. Box plots represent the median with the box bounding the interquartile range (IQR) and whiskers showing the most extreme points within 1.5× IQR.
n. Dose-response curves of AZD5991 and ULK1i from patient-derived xenograft (PDX resistant 2). Treatments were performed for 24 hrs (PDXs) before viability measurements using Cell Titer Glo. Data represent mean ± SD (n=3).
o. Kaplan-Meier survival curves of the Kasumi-1 leukemia recipient mice described in (Fig. S5e). The p-values were determined using the Log-rank Mantel-Cox test.
p. Immunoblotting of sorted GFP+ MOLM-13 leukemic cells from the bone marrow of animals treated with AZD5991 (MCL-1) or vehicle.
Next, to validate the role of mitochondrial autophagy in BH3-mimetics resistance, we harvested AML cells upon mitochondrial stress induction (using antimycin A and oligomycin) or mitochondria depolarization (CCCP) and autophagy inhibition (using CQ) and immunoblotted against LC3B and p62. These experiments demonstrated elevated lipidation of LC3B and increased p62 levels in the MR cells relative to the parental clones, further verifying enhanced mitophagy rates (Fig. 3d, Fig. S3k). Furthermore, we employed the mCherry-GFP-LC3 reporter in MOLM-13 and Kasumi-1 MR cells to monitor autophagic activity upon treatment with CCCP. Flow cytometry demonstrated higher autophagy rates after mitochondrial depolarization in the resistant cells compared to the parental (Fig. 3e-f). These findings were further verified using confocal microscopy, in which the colocalizing events between LC3B puncta and TOM20 (mitochondrial marker) were increased in the MR cells relative to the parental, after mitophagy induction with CCCP (Fig. S3l). Moreover, electron microscopy of cells treated overnight with the autophagy inhibitor CQ revealed a striking phenotype characterized by larger and more autophagic vacuoles in the resistant cells compared to the parental (Fig. 3g-h), again suggesting that BH3-mimetics resistance is accompanied by high autophagic flux, and specifically increased rates of mitochondrial clearance.
To investigate the contribution of mitophagy in the responsiveness of AML cells to BH3-mimetics, we depleted a critical mitophagy regulator, PINK1, in our resistant AML clones. Caspase 3/7 activation experiments showed that PINK1 loss sensitizes resistant cells to BH3-mimetics (Fig. 3i). Next, we took advantage of two commercially available compounds (MF094 and cmpd18) that can increase mitophagy by selectively inhibiting USP3053. USP30 is a deubiquitinase that acts as a brake on mitochondrial autophagy by opposing the Parkin-mediated ubiquitination of several mitochondrial proteins including MFN254,55. Noteworthy, both MF094 and cmpd18 rendered parental AML cells more resistant to apoptosis induced by BH3-mimetics (Fig. 3j). These findings strongly suggest that an increase in mitophagy per se is adequate to alter the AML cell response to BH3-mimetics.
Macroautophagy signatures predict poor survival and low responsiveness to BH3-mimetics in AML patients.
After discovering mitochondrial autophagy as a mechanism of resistance in human AML, we delved into recent single-cell transcriptomic profiling of patients with AML, performed in our laboratory56. Single-cell RNA sequencing (scRNAseq) was performed on bone marrow aspirates from 20 adult AML patients and 5 healthy donors (Controls). Uniform manifold approximation and projection (UMAP) of the malignant AML patient cells and the healthy counterpart hematopoietic stem/progenitor (HSPC) and myeloid cells showed AML patient cells clustering separately from healthy counterparts and by their annotated cell types (Fig. S4a). Thereafter, to uncover common gene signatures that differentiate malignant cells from HSPC and myeloid cells, we employed non-negative matrix factorization (NMF). Remarkably, one of the top gene signatures prevalent in primary bone marrow AML cells was enriched in autophagy-related pathways (macro-autophagy and the process of utilizing autophagy) (Fig. 3k; Fig. S4b). MFN2 and other mitophagy regulators ranked in the top 3% of these signatures (Suppl. Table 4). Strikingly, we found significantly inferior overall survival of AML patients with high mean autophagy gene expression in two independent patients’ cohorts (TARGET and TCGA) (Fig. 3l; Fig. S4c-d).
Next, to associate the abovementioned gene signatures with responsiveness to BH3-mimetics treatment in patients with AML, we initially explored the BeatAML dataset45. Interestingly, high expression of autophagy signatures positively correlates with low responsiveness to venetoclax in AML patients (Fig. 3m; Fig. S4e). Consistent with the above results, gene enrichment pathway analysis of bulk transcriptomics in a different cohort of AML patients showed that autophagy-related gene signatures are enriched in low responders to venetoclax relative to high responders16 (Fig. S4f). In addition, integrated transcriptomics analysis in AML cell lines uncovered that the top differentially expressed pathways in the MCL1i-resistant cells are involved in the autophagic process, again underlying the role of autophagy in the acquisition of BH3-mimetics resistance (Fig. S4g). Importantly, enhancing autophagy through mTOR inhibition with rapamycin treatment renders parental AML cells less susceptible to MCL1i-induced caspase activation and apoptosis (Fig. S4h).
Given that autophagy is a general degrading mechanism comprised of several arms that degrade selectively diverse components which range from peptides to organelles (e.g. mitophagy), we set out to precisely dissect the contribution of mitophagy from the other forms of macroautophagy in the development of resistance to BH3-mimetics. Thus, we performed bioinformatics analysis of the scRNAseq of AML blasts from two PDX models (TUH69 and TUH07) and revealed an upregulation of several mitophagy-related genes upon venetoclax treatment16 (Fig. S4i). Importantly, these data reveal a prognostic significance of autophagy, and specifically mitophagy for the overall survival and drug responsiveness in human AML.
Pharmacologic inhibition of autophagy sensitizes AML to BH3-mimetics.
As a proof of principle for the role of mitochondrial clearance by the process of autophagy in BH3-mimetics resistance, we examined whether chemically targeting of macroautophagy enhances the BH3-mimetics-induced apoptosis in AML models. To test this, we used autophagy inhibitors that were previously shown to increase/rescue mitochondrial biomass in the resistant AML cells (Fig. 3b). First, we utilized an ULK1 inhibitor (ULK1i) that specifically targets the key component of autophagy initiation57,58. Dose-response curves and caspase activation experiments verified the synergism between ULK1i with BH3-mimetics in our resistant AML cell lines and PDXs (Fig. 3n; Fig. S5a-c).
Besides ULK1i, we also tested alternative ways to inhibit the degradation of mitochondria through macroautophagy by chemically blocking the fusion of autophagosomes to the lysosomes using CQ (Fig. 3b). Survival curves in human AML cells demonstrated that CQ acts synergistically with MCL1i even in a p53-mutated background (Kasumi-1) (Fig. S5d). We next assessed the in vivo efficacy of the combination treatment in AML progression. Initially, we transplanted Kasumi-1 cells expressing firefly luciferase and GFP into immune-deficient recipient mice. After tumor engraftment, we treated the animals with MCL1i (AZD5991) weekly and CQ every two-three days (Fig. S5e). In accordance with our in vitro findings, concomitant administration of CQ together with MCL-1 inhibition resulted in a significant synergism. Bioluminescence imaging and quantification showed a marked delay in AML progression in animals treated with both drugs (Fig. S5f). The efficacy of the combinatory treatment was also evident by the enhanced survival of these mice compared to the single-agent treated and untreated mice (Fig. 3o). These results were also verified in an additional human AML preclinical model in vivo (luciferase and GFP-expressing MOLM-13, Fig. S5g). In the aforementioned in vivo experiments, we harvested animals’ bone marrow at late stages of tumor progression and isolated the tumor cells by GFP selection. Western blotting analysis uncovered that surviving AML cells isolated from mice treated with AZD5991 display significantly elevated MFN2 protein levels at advanced stages of the disease compared with those from vehicle-treated mice (Fig. 3p). These findings demonstrate that MFN2 upregulation also occurs in vivo as a mechanism of relapse to the drug.
Next, we examined whether CQ can re-sensitize high MFN2-expressing resistant AML cells to BH3-mimetics. Notably, the addition of CQ was able to re-sensitize MR cells to MCL-1 inhibition as shown by dose-response curves (Fig. S5h). This synergism was also extended when using the dual inhibitor of BCL-2 and BCL-xL (AZD4320)59 as demonstrated by the survival experiments (Fig. S5i). Furthermore, CQ reduced the IC50 of MCL-1 or BCL-2 antagonists in multiple human AML PDXs ex vivo (Fig. S5j-m), suggesting a general combinatory effect of CQ with BH3-mimetics in pre-clinical AML models. To investigate the in vivo synergism of CQ with MCL1i in a BH3-mimetics-resistant AML model, we transplanted cells from the PDX resistant 2 into NSGS recipients and monitored the peripheral blasts every week (Fig. S5n). We demonstrated that this drug combination led to the most significant reduction of AML blasts in the peripheral blood (Fig. S5o), highlighting its therapeutic potential to overcome BH3-mimetics resistance.
To demonstrate that the synergy between the BH3-mimetics and the autophagy inhibitors derives from the on-target effects of the autophagy inhibitors, we genetically ablated ULK1, as well as other main regulators of autophagosome formation, including ATG5, ATG7 and BECLIN160-62, in resistant, high-MFN2 expressing AML cells. Targeting any of these autophagy genes results in the sensitization of the resistant AML clones to BH3-mimetics-induced apoptosis (Fig. S5p-s).
MFN2 overexpression drives resistance to BH3-mimetics by modulating mitophagy.
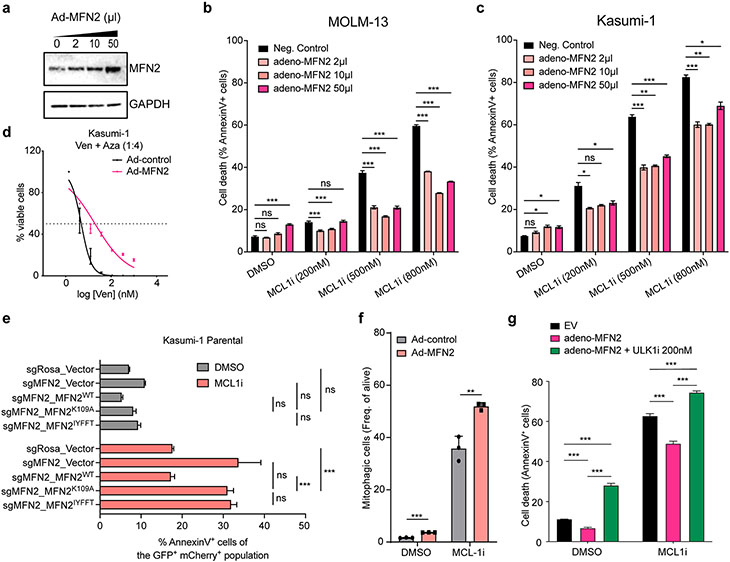
Given the specific upregulation of MFN2 in all previously examined AML models of BH3-mimetics resistance, we next asked whether MFN2 overexpression is sufficient to reduce sensitivity to BH3-mimetics. Ectopic adenoviral overexpression of MFN2 in MOLM-13 and Kasumi-1 cells, verified by western blotting (Fig. 4a), led to decreased cell death upon MCL-1 inhibition and increased the IC50 of Ven+Aza compared to the control group (Fig. 4b-d). To precisely dissect whether the GTPase activity or the subcellular localization of MFN2 are crucial for MFN2 function in BH3-mimetics responsiveness in AML, we generated constructs expressing (1) the full-length, wild type (WT) MFN2, (2) a GTPase-defective MFN2K109A (K109), and (3) an ER-MFN2 construct where the C-terminal domain of MFN2 has been replaced by the stretch of hydrophobic amino acids (IYFFT) that targets the protein exclusively in the ER63. These three constructs were infected in AML cells lacking MFN2 (sgMFN2) and subsequently treated with MCL1i to measure apoptosis after 24 hr. As expected, we found that MFN2-depleted cells are more prone to MCL1i-induced apoptosis. Importantly, overexpression of WT MFN2 completely rescued the cell death sensitization, confirming the specificity of our guides and vectors and that the observed effects are specific to MFN2. On the other hand, we demonstrated that neither of the mutants can protect from MCL1i-induced apoptosis (Fig. 4e). Taken all together our data show that the GTPase activity is necessary for the effects of MFN2 in the BH3-mimetics resistance in AML cells. This result comes in agreement with previous reports showing that the MFN2 GTPase activity is important for mitochondrial fusion63,64, and MAMs formation34. Moreover, the abovementioned data indicate that mitochondria localized MFN2 is indispensable for AML cells to become resistant to apoptosis, possibly due to its effects in recruiting PARKIN on defective mitochondria and, thus, regulating mitophagy.
Figure 4. MFN2 overexpression reduces sensitivity of AML cells to BH3-mimetics.
a. Immunoblotting in lysates from MOLM-13 cells adenovirally transduced with MFN2 (Ad-MFN2) or scramble control.
b-c. Annexin V staining in MOLM-13 (b) or Kasumi-1 (c) cells ectopically expressing MFN2 and subsequently treated with MCL1i (AMG176) for 24 hrs (n=3, mean ± SEM). Statistics were calculated using one-way ANOVA.
d. Dose-response curves of Ven+Aza from Kasumi-1 cells ectopically overexpressing MFN2 or control vector.
e. Annexin V assays of Kasumi-1 cells transduced with control or MFN2-targeting sgRNAs, followed by overexpressing vector control or indicated MFN2 constructs. Cells were treated with 500 nM AMG176 for 24 hrs before staining for Annexin V and DAPI.
f. Bar graphs depicting the frequencies of alive mitophagic cells in mitoKeima-expressing MOLM-13 cells transduced with MFN2 (Ad-MFN2) or scramble control treated with 1 μM MCL1i (AMG176) for 16 hrs (n=3, mean ± SD).
g. Annexin V staining in MOLM-13 cells ectopically expressing MFN2, after treatment with 500 nM AMG176 and 200 nM ULK1i for 24 hrs (n=3, mean ± SD). Statistics were calculated using two-way ANOVA.
Next, we wished to inspect whether MFN2-mediated MCL1i resistance is due to alterations in mitophagy rates. Treatment of mitoKeima-expressing cells with MCL1i leads to induction of mitophagy in AML cells. Interestingly, overexpression of MFN2 results in a further increase of cells undergoing mitophagy, linking MFN2 expression with mitophagy in the context of BH3-mimetics resistance in AML (Fig. 4f). Remarkably, autophagy blockage using ULK1i was able to completely reverse the protective effects of MFN2 overexpression in AML cells, as shown by annexin-V staining assays (Fig. 4g). Collectively, these results indicate that excessive mitochondrial clearance governed by MFN2 overexpression results in acquired resistance to BH3-mimetics and place MFN2-mediated mitophagy as a pro-survival mechanism in response to BH3-mimetics treatment in AML.
Targeting MFN2 activity impairs mitophagy and sensitizes AML cells to BH3-mimetics.
Our previous results prompted us to test whether MFN2 deficiency can elevate BH3-mimetics-induced cell death. Growth competition assays demonstrated that MFN2 loss synergizes with MCL1i, MCL1i+Ven, and Ven+Aza treatments in human AML cells (Fig. 5a; Fig. S6a-b). Cell death experiments further confirmed that MFN2 knockout enhances the BH3-mimetics-induced apoptosis in different human AML cell lines in vitro (Fig. 5b; Fig. S6c). Given that MFN2 is highly expressed in resistant AML cells, we sought to validate the sensitization effect of MFN2 ablation in the resistant cells. Similar to the results observed in the parental clones, MFN2 depletion significantly re-sensitized MR cells to the drug treatments (Fig. 5c; Fig. S6d). In fact, MFN2 loss completely restores the sensitivity of the resistant clones to BH3-mimetics comparable to the levels of the parental cells (Fig. 5d). Noteworthy, MFN1 deletion has no effect on the response to BH3-mimetics in AML (Fig. 5e), suggesting that the specific functions of MFN2 on mitochondria-ER tethering and mitophagy are crucial for the development of resistance in AML cells. Besides MFN2, we also deleted MARCH5, another top hit in our screens and a protein that post-translationally regulates the activity and oligomerization of MFN232,33,37. MARCH5 ablation conferred competitive disadvantages in the viability of both sensitive and resistant AML cell lines through enhancing the cell death induced by BH3-mimetics (Fig. S6e-g). The latter finding is in accordance with a recent study that also proposed that MARCH5 repression enhances venetoclax efficacy in AML65.
Figure 5. Knockout of MFN2 sensitizes AML cells to BH3-mimetics.
a. Competition-based viability assays in MOLM-13 parental cells treated with MCL1i (left) or MCL1i+Ven (right). Y-axis denotes the GFP+ percentage relative to day 0 of treatment (n=3, mean ± SD).
b. Annexin V staining in MOLM-13 cells transduced with sgRNAs targeting MFN2 or control (Rosa), after treatment with BH3 mimetics or DMSO for 24 hrs (n=3, mean SEM, unpaired two-tailed t-test).
c. Competition-based viability assays in MOLM-13 MR cells treated with MCL1i (left) or MCL1i+Ven (right). Y-axis denotes the GFP+ percentage relative to day 0 of treatment.
d. Annexin V staining in Kasumi-1 resistant cells transduced with sgRNAs targeting MFN2 or control (Rosa), after treatment with 600 nM AZD5991 or DMSO for 24 hrs (n=3, mean ± SD). Statistics were calculated using two-way ANOVA.
e. Competition-based viability assays in MOLM-13 MR cells transduced with sgRNAs targeting MFN1, MFN2 or control (Rosa) treated with DMSO, AMG176 (MCL1i) or AMG176 (MCL1i)+Ven. Y-axis denotes the GFP+ percentage relative to day 0 of treatment (n=3, mean ± SD).
f. Schematic outline of the in vivo experiment validating the synergistic effects of MFN2 ablation and AZD5991 treatment using MOLM-13 xenografts.
g. Bioluminescence images of the MOLM-13 leukemia recipient mice described in (f). The same mice are depicted at each time point (n=5 mice per group). Days post-drug administration are shown.
h. Quantification of bioluminescence emitted from the whole body of each mouse described in (f) at the indicated time points (mean ± SD).
i. Kaplan-Meier survival curves of the MOLM-13 leukemia recipient mice described in (f). X-axis denotes days post-transplantation. The p-values were determined using the Log-rank Mantel-Cox test.
j. Graph depicting changes in the animals’ body weight throughout the experiment described in (f).
We then examined the synergistic effects of MFN2 deletion with BH3-mimetics in vivo using pre-clinical animal models. Specifically, NOD SCID gamma (NSG) mice were transplanted with AML (MOLM-13) cells expressing doxycycline-inducible Renilla- or MFN2-targeting shRNAs (shRenilla or shMFN2). After tumor engraftment, the mice were fed with a diet containing doxycycline to induce MFN2 silencing, and the next day we initiated weekly intravenous administration of AZD5991 or vehicle (Fig. 5f). Bioluminescence imaging showed that the reduction of MFN2 levels synergized with MCL1 inhibition in vivo (Fig. 5g-h). This led to prolonged survival of the mice bearing MFN2 knockdown tumors and being treated with MCL1i (Fig. 5i). No side effects were noticed after MCL1i treatment and MFN2 deletion, including no loss in animals’ body weight (Fig. 5j).
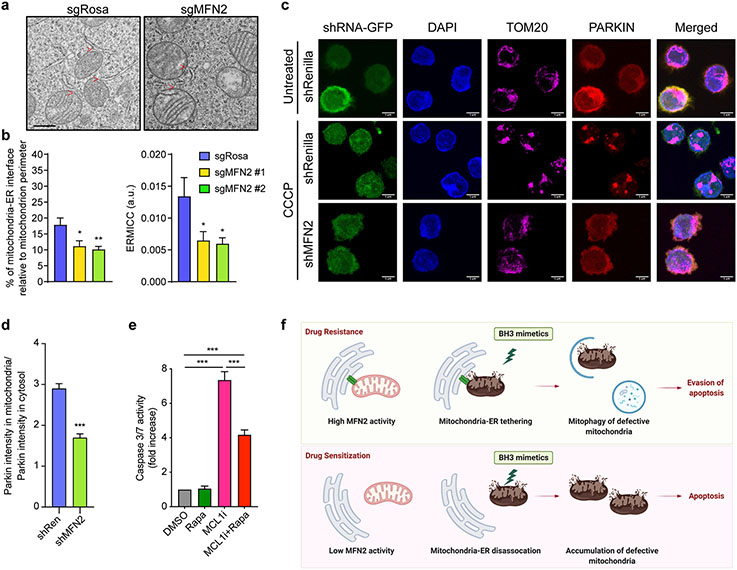
From a mechanistic point of view, electron microscopy in the AML line MOLM-13 verified that MFN2 ablation led to a reduction of mitochondria-ER contact sites (Fig. 6a-b). Like MFN2 loss, MARCH5-depleted AML cells also displayed fewer mitochondria-ER interactions as determined by morphometric analyses of electron micrographs (Fig. S7a-b). To understand how MFN2 loss impacts mitophagy, we stably expressed PARKIN-mCherry in MOLM-13 and HeLa cells, a cytosolic protein that translocates onto mitochondria upon cellular insults i.e., mitochondrial uncoupling by CCCP, and subsequently promotes the clearance of depolarized mitochondria. Confocal microscopy and colocalization analysis in TOM20 stained cells revealed that MFN2 deletion leads to defective PARKIN translocation upon stress, indicating mitophagy impairment (Fig. 6c-d; Fig. S7c-d). Likewise, MARCH5 knockdown in HeLa and MOLM-13 cells led to inadequate PARKIN translocation to mitochondria after CCCP treatment (Fig. S7e-h). In addition, we performed immunoblotting against LC3B and p62 in AML cells treated with mitochondrial stressors (oligomycin and antimycin A) as well as CQ and demonstrated reduced LC3B lipidation and p62 accumulation in cells lacking MFN2 or MARCH5 (Fig. S7i-n). Rapamycin administration rendered MFN2 knockout cells less prone to apoptosis induced by MCL1i (Fig. 6e), further indicating that MFN2 regulates sensitivity to BH3-mimetics at least partially through the control of mitochondrial autophagy. These results suggest that targeting MFN2 or MARCH5 sensitizes AML cells to BH3-mimetics by diminishing mitophagy (Fig. 6f).
Figure 6. Knockout of MFN2 decreases mitochondrial clearance through autophagy.
a. Representative electron micrographs of mitochondria from Cas9-expressing MOLM-13 infected with sgRNA targeting MFN2 or Rosa. Scale bar represents 0.5 μm, red arrows indicate mitochondria-ER interactions.
b. Quantification of the mitochondria-ER contacts in experiments as in (a). The percentage of the mitochondria-ER interface relative to the mitochondrial perimeter (left) and ER-mitochondrial contact coefficient (ERMICC; right) are shown (n = 21, mean ± SEM). ERMICC = interface length / (mitochondrial perimeter × mitochondria-ER distance)94.
c. Representative z-stack projections of confocal 3D images of PARKIN-mCherry-overexpressing MOLM-13 cells transduced with the indicated shRNAs (GFP+), treated with 10 μM CCCP or DMSO for 2 hrs, and stained for TOM20 (magenta; mitochondria). Scale bars represent 5 μm.
d. Quantification of PARKIN translocation onto mitochondria (colocalization index) relative to the cytosolic PARKIN in experiments described in (c) in CCCP-treated cells.
e. Caspase 3/7 assay in MFN2 KO Kasumi-1 cells treated with AZD5991 (MCL1i, 1 μM, 8 hrs) and rapamycin (Rapa, 250 nM, 26 hrs). Data represent mean ± SEM of three independent biological replicates.
f. Schematic diagram of the study.
Pharmacologic targeting of MFN2 potentiates BH3-mimetics activity in AML.
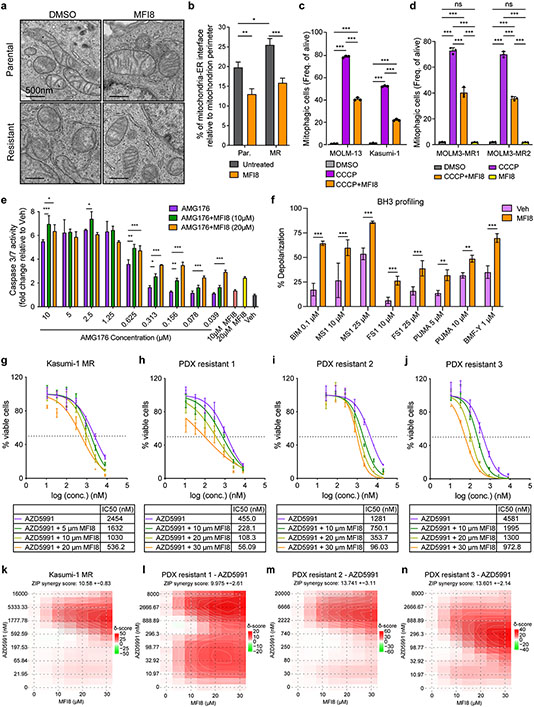
Next, we employed a recently developed small molecule (MFI8) that selectively blocks the tethering activity of mitofusins66 to chemically target mitochondrial quality control and mitophagy in AML cells. Firstly, we validated MFI8 function in AML cells, by performing electron microscopy. MFI8 reduced the mitochondria-ER interactions in parental and resistant MOLM-13 cells (Fig. 7a-b). In addition, we assessed the role of MFI8 treatment in the regulation of mitophagy in AML. MFI8 significantly reduced CCCP-induced mitophagy events in parental and resistant AML cells expressing mitoKeima (Fig. 7c-d).
Figure 7. Pharmacologic targeting of MFN2 sensitizes AML cells to BH3-mimetics.
a. Representative electron micrographs of mitochondria and their interactions with ER from MOLM-13 parental or MR (resistant) cells treated for 6 hr with 20 μM MFI8. Scale bars represent 500 nm.
b. Quantification of MAMs in experiments in MOLM-13 cells as in (a). Graphs depict the percentage of mitochondria-ER interface relative to the mitochondrial perimeter (n=40 mitochondria, mean ± SEM).
c. Bar graphs depicting the mitophagic events in mitoKeima-expressing MOLM-13 and Kasumi-1 cells treated with 10 μM CCCP and 10 μM MFI8 for 16 hrs, as quantified by flow cytometry.
d. Bar graphs depicting the mitophagic events in mitoKeima-expressing MOLM-13-MR1 and MOLM-13-MR2 cells treated with 10 μM CCCP and 10 μM MFI8 for 16 hrs, as quantified by flow cytometry.
e. Caspase 3/7 activation in MOLM-13 cells treated with increasing concentrations of AMG176 and MFI8 16 hrs post-treatment (n=3, mean ± SD).
f. BH3 profiling in MOLM-13 MR cells following MFI8 treatment (6 hrs pre-treatment and 3 hrs during the course of the experiment; 20 μM). The percentage of mitochondrial membrane depolarization was measured (n=4, mean ± SD).
g-n. Dose-response curves (g-j) and matrices (k-n) of AZD5991 and mitofusin inhibitor (MFI8) from indicated cell lines and xenografts. Treatments were performed for 24 hrs (PDXs) or 48 hrs (cell line) before viability measurements using Cell Titer Glo. Data represent mean ± SD (n=3).
The statistical analysis was performed using two-way ANOVA.
Subsequently, and based on our previous experimental data, we decided to examine if pharmacological inhibition of MFN2 can sensitize AML cells to MCL-1 antagonists. Caspase activation assays verified that co-treatment of MFN2 inhibitor with MCL1i results in enhanced apoptosis in vitro compared to the single MCL1i treatment (Fig. 7e). To gain more insight into the role of MFI8 in apoptotic signaling, we performed BH3-profiling67 in MOLM-13 MR cells pretreated with MFI8. Notably, MFI8 primed the resistant cells to apoptotic stimulation and sensitized them to low doses of various peptides (PUMA, FS1, BAD, BMF-Y) with the most remarkable effects upon treatment with the MS1 peptide (MCL-1 antagonist) (Fig. 7f). In accordance with these findings, our viability experiments revealed that MFI8 and AZD5991 act synergistically in eliminating resistant AML cells, as well as AML PDXs ex vivo (Fig. 7g-n). These studies introduce a novel lead compound for targeting AML growth and further support our main hypothesis that inhibition of MFN2 activity and mitochondrial autophagy enhances the efficacy of BH3-mimetics and can overcome drug resistance in AML.
Discussion
Our extensive CRISPR/Cas9 screening strategy allowed us to identify novel determinants of drug resistance that are shared among various BH3-mimetics combinations in AML. Initially, we verified that loss of TP53, BAX, or BAK are recurrent primary factors to evade apoptosis induced by MCL1i, MCL1i+Ven, or Ven+Aza regimens in AML, further highlighting the challenge to overcome the p53- or BAX- mediated resistance and the urgency to explore alternative combinatory therapeutic strategies. Our screening approach prominently uncovered genes involved in mitochondrial dynamics, including MFN2 and MARCH5, as synthetic lethal targets with BH3-mimetics in human AML. Mitochondria actively modify their morphology to respond to the various cellular demands. Specifically, early during apoptosis, mitochondria fragment68. Interestingly, our comprehensive biochemical and morphometric analyses using super-resolution microscopy uncovered alterations in mitochondrial morphology, coupled with MFN2 upregulation, in BH3-mimetics-resistant human AML cells. Increased mitochondrial fusion in the resistant cells might act as a quality control mechanism to buffer the organelles’ damage induced by BH3-mimetics and to resist apoptotic fragmentation. In addition, MFN2 can act as a recruiter for PARKIN to initiate the autophagic clearance of “injured” mitochondria35. Hence, a high abundance of MFN2 onto resistant mitochondria might contribute to an enlarged surface and more receptors for instantaneous mitophagy initiation upon an insult, such as BH3-mimetics. Furthermore, MARCH5, a ubiquitin ligase that was recently shown to regulate MFN2 activity32, PARKIN-mediated mitophagy33, and be essential in AML65, possibly contributes to BH3-mimetics resistance by regulating MFN2-mediated mitophagy and mitochondrial quality control.
MFN2 and MARCH5 also participate in the tethering of mitochondria with the ER32,34. Interestingly, mitochondria-ER interactions, which we found them to be more abundant in BH3-mimetics-resistant cell lines and primary patient AML samples compared to the sensitive clones, have multiple cellular functions, including Ca2+ signaling, lipid synthesis, and cholesterol metabolism69. Notably, metabolic changes have been previously reported in venetoclax resistance, characterized by increased dependency on fatty acid metabolism13, which might be mediated by alterations in MAMs’ abundance and architecture. Moreover, a recent study pinpointed the role of MAMs in lipophagy, energy production, and survival of AML cells70. Noteworthy, MAMs are the sites of BCL-2 family members’ action during cell death71, where MFN2 and MARCH5 localize32, and operate as the platforms for the autophagosome formation47,48,72.
Overall, our study provides mechanistic insights into the role of MFN2 and MARCH5 in mitochondrial autophagy and MAMs in the context of resistance to BH3-mimetics in AML. Importantly, genetic ablation of MFN2, MARCH5, or other key autophagy regulators re-sensitized the resistant AML cells to apoptotic-inducing agents. These data corroborate the implication of mitochondrial clearance through mitophagy in the apoptosis regulation and sensitivity to BH3-mimetics.
The interplay between autophagy and apoptosis has been previously investigated and BCL-2 family members are centered on this crosstalk73,74. Interestingly, autophagy has been implicated in chemoresistance in AML75, while mitophagy has been shown to play a role in leukemia stem cell survival76. In this study, we demonstrated that BH3-mimetics-resistant AML cells have increased capacity of mitochondrial autophagy upon challenge, which possibly acts as an additional cytoprotective mechanism by quickly eliminating the damaged mitochondria that accumulate during BH3-mimetics treatment. We cannot exclude the possibility that also a general enhancement in macroautophagy contributes to the acquisition of resistance, as MFN2 loss has been linked with global autophagy defects77-79.
Multiple ongoing clinical trials examine the effectiveness of several drug candidates in combination with the FDA-approved anti-malarian, chloroquine, for the treatment of various solid tumors80. In our study, we proved that blocking autophagy and subsequently mitophagy with the “off-the-shelf” chloroquine enhances the efficacy of BH3-mimetics (including MCL-1, BCL-2, BCL-xL antagonists, and their combinations) and could overcome drug resistance in vivo in human AML PDX and cell line models by rescuing mitochondrial degradation. Yet, to avoid off-target effects, specific targeting of autophagy regulators like ULK1 with ULK1i58 or mitophagy regulators like MFN2 using MFI866, which we showed to exhibit synergistic effects with BH3-mimetics in AML xenografts ex vivo, might be a more attractive approach to further explore in the future for the treatment of hematologic malignancies. Future studies will focus on optimizing the MFN2-inhibiting compounds, but also examining the effects of MFN2 targeting in healthy cells and tissues to avoid mitochondrial dysfunction similar to what is observed in patients with Charcot-Marie-Tooth disease type 2A (CMT2A) that harbor mutations in MFN2 64,81.
Collectively, our work introduces novel modes of BH3-mimetics resistance, including mitochondrial quality control mechanisms through adaptations in mitochondria-ER communication and mitophagy regulation. All these pathways lead to the elimination of mitochondrial defects and evasion of programmed cell death induced by BH3-mimetics. Blocking MFN2 activity and the specific clearance of damaged mitochondria through mitophagy represents a powerful approach to overcome drug resistance in hematologic malignancies and other tumors.
Methods
Cell lines and cell culture
All human and mouse leukemia cells were cultured in recommended media, typically RPMI medium with 10% FBS and 1% penicillin/streptomycin. KL974 (p53R172H/Δ) primary mouse cell line was cultured with RPMI, 10% FBS (Gibco) supplemented with recombinant mouse SCF (50 ng/ml), IL3 (10 ng/ml) and IL6 (10 ng/ml). The adherent cell lines HeLa and HEK293T cells were grown in DMEM medium with 10% FBS and 1% penicillin/streptomycin. Cas9-expressing cell lines transduced with lentiviral-Cas9-2A-Blast (Addgene, plasmid no. 73310) were selected with blasticidin (InvivoGen) 48 hrs after transduction.
For the generation of the MCL1i-resistant cell lines (MR), MOLM-13 and Kasumi-1 cells were cultured in media containing increasing doses of AMG176 (MedChemExpress, Cat#HY-101565; starting from 30 nM to 1.6 μM) for more than 8 weeks to achieve complete resistance to the drug. For the generation of the Venetoclax/Azacytidine-resistant cell lines (VAR), MOLM-13 and MV4-11 cells were cultured in media containing increasing doses of venetoclax (Selleckchem, Cat#S8048) + azacytidine (Selleckchem, Cat#S1782) (venetoclax:azacytidine=1:4 with concentration ranged from 5:20 to 200:800 nM) for more than 8 weeks to achieve complete resistance to the drug.
Cell lines information is as follows:
MOLM-13, DSMZ, ACC 554
Kasumi-1, ATCC, CRL-2724
MV4–11, ATCC, CRL-9591
HEK293T, ATCC, CRL-1573
HeLa, ATCC, CCL-2
MLL-AF9 NrasG12D (RN2) Cas9, Zuber et al, 201182
KL974 (p53R172H/Δ), Loizou et al, 201943.
All the cell lines were determined negative for mycoplasma, as indicated by the latest Mycoplasma test (February 2022) using the LookOut Mycoplasma PCR Detection Kit (Sigma). Cells were used for experiments within 15 to 20 passages from thawing.
Primary AML cells and patient-derived xenografted cells
Primary AML cells from peripheral blood of patients were collected during routine diagnostic procedures at NYU Langone Health and analyzed with hematopathology at the NYU Langone’s Department of Pathology. The AML patient-derived xenograft (PDX) Sensitive 2 model (MDAM-72403) and Resistant 3 model (MDAM-19692) were purchased from PRoXe.org. The AML PDX Resistant 1 model (I15-4072) was established in NSGS mice using a primary bone marrow (BM) sample (93% blasts), kindly provided by Dr. Giorgio Inghirami (Weil Cornel Medicine). For short term in vitro culture, primary and PDX cells isolated from the mouse BM (90% blast) were maintained in OPTI-MEM (Gibco), 15% FBS, 1% penicillin/streptomycin, 1x b-mercaptoethanol supplemented with human SCF (50 ng/ml), FLT3L (50 ng/ml), IL3 (10 ng/ml) and IL6 (10 ng/ml).
For single cell RNA sequencing experiments, cryopreserved bone marrow aspirates from newly diagnosed adult AML patients were obtained from the OSU Leukemia Tissue Bank (adult AML samples). Cryopreserved primary human bone marrow mononuclear cells were obtained from STEMCELL Technologies (Cat#70001) or from StemExpress (Cat#BMMNC025C).
Ethical reporting
This study complies with all relevant ethical regulations and was approved by the Institutional Review Boards of New York University, entitled Heme Research Biorepository Protocol, Ohio State University, and Weil Cornell Medicine. Informed written consent was obtained from each subject or each subject's guardian, according to the Declaration of Helsinki to use their tissue for studies, and in accordance with the regulations of the institutional review boards of all participating institutes.
Animal Experiments
C57BL/J (Cat#000664), NOD.Cg-PrkdcscidIl2rgtm1WjlTg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSGS, Cat#013062) and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG, Cat#00557) were produced using breeders bought from the Jackson Laboratory. Mice were bred and maintained in individual ventilated cages and fed with autoclaved food and sterile water at NYU School of Medicine Animal Facility.
For drug combination in vivo experiments, 0.5 x 106 GFP and luciferase-expressing Kasumi-1 or MOLM-13 cells were intravenously injected into recipient NSG or NSGS mice. Recipient mice were treated with vehicle or AZD5991 (50 mg/kg, Chemietek) every 6 days by tail vein from day 6 to day 18. AZD5991 was prepared in 30% 2-hydroxypropyl-beta-cyclodextrin (HBPBC; Sigma). Chloroquine (CQ; Sigma) was reconstituted in PBS and administrated to the mice every 2-3 days from day 6 to day 18. Whole-body bioluminescent imaging was performed at indicated time points by intraperitoneally injection of Luciferin (Gold-bio, Cat#LUCK-500) at a 50 mg/kg concentration and imaging was performed after 5 min using an IVIS imager. Bioluminescent signals (radiance) were quantified using Living Image software with standard regions of interests (ROI) rectangles. Peripheral blood of recipient NSG/NSGS mice was collected at the indicated time points after transplantation, and leukemic cells (GFP+) were analyzed by flow cytometry. Kaplan-Meier survival curves were compared using the Wilcoxon Rank-Sum test via GraphPad Prism. Following disease onset, moribund mice were sacrificed with bone marrow (BM) processed. Specifically, both tibia and femurs were flushed using centrifugation and subsequent red cell lysis.
For the in vivo MFN2 knockdown experiment, NSG mice were transplanted with 1 x 106 luciferase-expressing MOLM-13 cells transduced with shRNA targeting MFN2 (shMFN2) or Renilla control (shRen). Doxycycline diet was initiated at day 6 and maintained throughout the experiment. Mice were administrated with vehicle or AZD5991 (AZD) on day 7 and day 13 post transplantation. Bioluminescent imaging was performed at indicated time points to measure tumor burden. Survival was measured in the end of the experiment.
To generate patient-derived xenografts (PDX) from primary peripheral blood or bone marrow, red blood cells were removed using PharmLyse (BD, Cat#555899). CD3+ T cells were depleted using CD3 microbeads (Miltenyi Biotec, Cat#130-050-101). After cell counting, 1-8 x 106 viable cells were resuspended in 150 μl sterile PBS per mouse for intravenous injections. The recipient mice were 4-8 weeks old females. If the mice were 8-10 weeks old, they were irradiated at 200cGy within 24 hr prior to injection. Xenografted mice were clinically assessed daily for physical signs of illness, including lethargy, weight loss, hind limb paralysis, and/or poor grooming. Engraftment of primary AML cells in the animals was assessed by retro-orbital blood collection every two weeks. The peripheral blood was stained with anti-hCD45, anti-hCD34, anti-hCD3, and anti-mCD45 and subsequently analyzed by flow cytometry to determine the fraction of circulating blast (hCD45+, hCD3−). When the circulating disease was > 20% in the peripheral blood, mice were euthanized, and human leukemic cells were harvested from the bone marrow and the spleen. For the bone marrow isolation, tibias, femurs, iliac crests, and spines were dissected and crushed using pestle and mortar. Spleens were minced, homogenized, and resuspended in ACK (ThermoFisher) to lyse the red blood cells. After cell counting, bone marrow samples and splenocytes were frozen in FBS with 10% DMSO. A portion of bone marrow cells and splenocytes were labelled with anti-hCD45, anti-hCD34, anti-hCD3, and anti-mCD45, to determine the percentage of the human blast in the mouse tissues.
For the combinatorial treatment of PDX Resistant 2, 0.6M cells were transplanted in NSGS mice and the treatment started after 4 weeks when the peripheral blast was 0.1-1%. AZD5991 was administered intravenously at 100 mg/kg once per week and CQ was administered intraperitoneally at 75 mg/kg every other day. For the treatment of PDX Resistant 1, once the xenografted cells were engrafted (in approximately 8 weeks post-transplantation), mice were treated weekly with AZD5991 (100mg/kg, intravenously) or vehicle for 28 days. Disease progression was monitored every week by retro-orbital blood collection and flow cytometry analysis.
All animal experiments were performed in accordance with protocols approved by the New York University Institutional Animal Care and Use Committee (IACUC, ID: IA16-00008_TR1), according to national and institutional guidelines.
Genome-wide CRISPR screens
Cas9-expressing MOLM-13 were transduced with the Brunello sgRNA library 28 virus at a low MOI (~0.3). On day 2 post-transduction, GFP+ percentage was assessed to determine infection efficiency and sgRNA coverage (~1000X). Then, puromycin (Sigma Aldrich, Cat#P7255, 1 μg/ml) was added for 5 days to select infected (GFP+) cells. After selection, viable infected cells were isolated by Histopaque 1077 (Sigma Aldrich, Cat#10771) and grown without antibiotics for 2 days. After recovery, 100 x 106 infected cells were cultured with AMG176 (MedChemExpress, Cat#HY-101565), AMG176+Venetoclax, Venetoclax (Selleckchem, Cat#S8048) +Azacytidine (Selleckchem, Cat#S1782) or DMSO. Another 100 x 106 cells were used for genomic DNA (gDNA) extraction and served as an initial reference (day 0 of drug treatment). The concentration of each drug and their combinations were increased during the screen to avoid spontaneous gain of drug resistance. For each passage, 100 x 106 cells were placed back into culture. gDNA of cells containing ~1000X coverage was harvested on day 16 post drug treatment using Qiagen DNA kit (Cat#51306) according to manufacturer’s protocol. For library construction, 300 μg of gDNA were amplified for 25 cycles using EX-Taq (Takara Bio, Cat#RR001B) and primer pairs that contain barcodes. PCR products were size-selected using AMPure XP beads (Beckman Coulter). Barcoded libraries were then sequenced using Next-Seq instrument (single-end, 80 cycles).
Competition-based survival assay
For the competition-based survival assay, Cas9-expressing cells were transduced with the indicated sgRNAs (Suppl. Table 5). Cells were cultured for 8 days to allow complete CRISPR/Cas9 editing. Increasing amounts of BH3 mimetics were then added to the medium every other day. GFP+ percentages were assessed by flow cytometry at 48 hrs upon each drug administration. Normalized enrichment was calculated as the ratio: Enrichment (sgRNA target) / Enrichment (sgRNA control). Enrichment was defined as the ratio: %GFP+ in drug treatment / %GFP+ in DMSO. Details on the construction of sgRNAs can be found in the Supplemental Methods.
Drug treatment and IC50 measurement
Cells were plated in 96-well plates and exposed to AZD5991 (Chemietek Cat#CT-A5991 or Astrazeneca), AMG176 (MedChemExpress), AZD4320 (Astrazeneca), Venetoclax (Selleckchem), Azacytidine (Selleckchem), Venetoclax+Azacytidine (Ven:Aza = 1:4), Chloroquine (Sigma Aldrich, Cat#C6628) or ULK1i (MRT68921, Selleckchem, Cat#S7949) with a minimum of three technical replicates per concentration per cell line. Cell viability was measured with the CellTiter-Glo 2.0 reagent (Promega, Cat#G9243) according to manufacturer’s instructions. Absolute viability values were converted to percentage viability versus DMSO control treatment, and then non-linear fit of log(inhibitor) versus response (three parameters) was performed in GraphPad Prism v8.0 to obtain the IC50 values. For multi-drug combination profiling data, SynergyFinder (version 2.0) was used (https://synergyfinder.fimm.fi/). The expected drug combination responses were calculated based on ZIP reference model using SynergyFinder83. Deviations between observed and expected responses with positive and negative values denote synergy and antagonism respectively. For estimation of outlier measurements, cNMF algorithm84 implemented in SynergyFinder was utilized.
Frozen human BM mononuclear cells preparation
Frozen human bone marrow samples were thawed and transferred into 50mL conical tubes containing PBS + 2% FBS. Cell suspensions were centrifuged at 350 x g for 5 minutes at 4°C, and the supernatant was discarded. Samples were then subjected to dead cell depletion, using a dead cell removal kit (Miltenyi Biotec, Cat#130-090-101), or stained with DAPI (Sigma Aldrich, Cat#D9542, 0.5 μg/mL) and sorted for live cells (DAPIlow), using a FACSAria IIu SORP cell sorter (BD Biosciences). For cell sorting, all samples were gated based on forward and side scatter, followed by doublet exclusion, and then gated on DAPIlow for viable cells. Samples were sorted into 5mL poly-propylene tubes containing 300μL ice-cold PBS + 2% FBS. Following cell sorting, samples were centrifuged at 350 x g for 5 minutes at 4°C.
For CITE-seq, enriched live cells were first tagged with cell-hashing oligo-tagged antibodies (Biolegend) according to the manufacturer’s instructions. Samples were than counted, and a maximum of 105 cells for each sample was pooled together and stained with a CITE-Seq antibody cocktail (Biolegend) according to the manufacturer’s instructions.
Libraries were prepared using the Chromium Single Cell 3’ Reagent Kits (v3 and v3.1, CITE-Seq). Hashtag and antibody-derived tag (ADT) libraries were prepared according to the New York Genome Center CITE-Seq and hashing protocol (https://citeseq.files.wordpress.com/2019/02/cite-seq_and_hashing_protocol_190213.pdf). Libraries were run on an Illumina NovaSeq 6000.
Single cell RNA/CITE sequencing pre-processing
Raw sequencing reads were converted to FASTQ format using Illumina bcl2fastq software. We used Cell Ranger Single Cell Gene Expression Software (version 5.0, 10x Genomics) to demultiplex and align raw 3’ library reads to GRCh38 (version 2020-A). All following downstream analysis was performed using the Seurat R package (version 3.2.2)85, and all visualizations were generated using ggplot2 (version 2_3.3.3). We excluded cells with less than 400 or more than 6000 unique feature counts, as well as cells with more than 15% transcripts originating from mitochondrial genes to filter low-quality cells and droplets that may have captured multiple cells. Details on the subsequent steps can be found in Supplemental Information.
Mitochondria visualization using super-resolution microscopy
Sterile 22 mm square #1.5 thickness coverglasses were precoated with 0.1 mg/ml Poly-L-Lysine (SIGMA) for 5 min, washed with PBS and drained at room temperature overnight. Cells were stained with 200 nM MitoTracker Deep Red (Thermo Scientific, Cat#M22426) in 1x HBSS with 20 nM HEPES for 20 min at 37°C. After the staining, 3.5 x 105 cells were washed with prewarmed PBS and fixed on top of the coverslips with freshly prepared, 3.7% formaldehyde in PBS for 30 min. Next, cells were washed, permeabilized at room temperature with 0.5% Triton X-100 in PBS for 20 min, washed again, and incubated with blocking buffer (0.5% BSA in PBS) for 30 min. After blocking, cells were stained with primary antibodies against TOM20 (1:100; Santa Cruz - clone F10, RRID:AB_628381) and MFN2 (1:100; Cell Signaling – clone D2D10, RRID:AB_2716838) diluted in blocking buffer for 2 hrs at room temperature. The cells were then washed with PBS and incubated with anti-mouse AlexaFluor 568 (1:1000; Invitrogen, RRID:AB_2534072) and anti-rabbit Alexa Fluor 488 (1:1000; Invitrogen, RRID:AB_143165) for 2 hrs at room temperature. After washing, cells were stained with 1 μg/μl DAPI for 1 min and then coverslips were mounted with 10 μl of SlowFade Glass Soft-set antifade mountant (Invitrogen, Cat#S36917) onto slides.
Transmission electron microscopy
Cultured and primary AML cells were fixed in 0.1 mol/L sodium cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde and 2% paraformaldehyde for 2 hrs and post-fixed with 1% osmium tetroxide and 1% potassium ferrocyanide for 1 hr at 4°C. Then, block was stained in 0.25% aqueous uranyl acetate, processed in a standard manner, and embedded in EMbed 812 (Electron Microscopy Sciences). Ultrathin sections (60 nm) were cut, mounted on copper grids, and stained with uranyl acetate and lead citrate. Stained grids were examined under Philips CM-12 electron microscope and photographed with a Gatan (4k × 2.7k) digital camera. For morphometric analysis of mitochondrial-ER contacts, mitochondrial perimeter, mitochondria-ER distance, and length of mitochondria-ER interface were measured using freehand tool in ImageJ (NIH) as shown in Fig. S2e.
FACS analysis of mitophagy
To generate AML cell lines stably expressing mitoKeima, a cassette containing mitoKeima CDS (Addgene, plasmid no. 56018) followed by ires-EGFP-P2A-PURO was cloned into the Lentiviral vector pCDH-EF1s (Addgene, plasmid no. 72484). AML cells were then transduced with the pCDH-EF1s-mitoKeima-ires-EGFP-P2A-PURO construct through spin infections. mitoKeima-expressing AML cells were selected using puromycin. Cells stably expressing mitoKeima were analyzed with flow cytometer BD LSR Fortessa equipped with a 405-nm and 561-nm laser. The Qdot605 signals were obtained using the violet laser (405 nm) in combination with the 610 ± 10 nm filter, while the mCherry signals were measured using the yellow-green laser (561 nm) with the 610 ± 10 nm filter. Events detected with Qdot605 correspond to mitochondrial fluorescence, while events detected with mCherry detector correspond to lysosomal mitochondria. The ratio of the mean intensities of mCherry vs Qdot605 represents mitophagy rate.
BH3 profiling assay
Parental, MR and MR treated with MFI8 (20 μM) AML cells were subjected to BH3 profiling. BIM, BID, PUMA, MS1, FS1, BAD, HRK or BMF-γ peptides; CCCP (10 μM) were added to JC1-MEB staining solution (150 mM mannitol, 10 mM HEPES-KOH, 50 mM KCl, 0.02 mM EGTA, 0.02 mM EDTA, 0.1% BSA, 5 mM succinate, pH 7.5) in a black 384-well plate. Cell suspensions were prepared in JC1-MEB buffer as previously described86. After adding the cells to the 384-well plate, 4 × 104 cells/well, fluorescence was measured at 590 nm emission 545 nM excitation using the M1000 microplate reader (TECAN) at 30 °C every 15 min for a total of 3 h. MFI8 was co-treated with the peptides over the course of 3 hr. Percentage of depolarization was calculated by normalization of the AUC of every peptide to the solvent-only control DMSO (0% depolarization) and the positive control CCCP (100% depolarization), as previously described87.
Caspase-3/7 activation assay
Parental and MR AML cells (4 x 103 cells/well) were seeded in a 384-well white plate and treated with MFI8, MF094 (MedChemExpress, Cat# HY-112438), cmpd18 (MedChemExpress, Cat# HY-141659), rapamycin, ULK1i (MRT68921), AMG176, and AZD5991. Caspase 3/7 activation was measured by addition of the Caspase-Glo 3/7 reagent according to the manufacturer’s protocol (Promega). Luminescence was detected by a F200 PRO microplate reader (TECAN). Caspase assays were performed in at least triplicate and the data normalized to vehicle-treated control wells. Dilutions of the drugs were performed using a TECAN D300e Digital Dispenser from 10 mM stocks.
Quantification and statistical analysis
CRISPR Screen Analysis
Based on the sequence read fastq files, we removed the adapter sequence (5’ adapter CGAAACACCG) and counted the reads for each sgRNAs. We then removed sgRNAs with less than 50 reads at day 0 post-treatment. The differential analysis was performed using MAGecCK (version 0.5.9.2) and genes with good sgRNAs < 3 were removed from all downstream analyses. All scatter plots were generated for different pairs of conditions using R package ggplot2 (version 3.3.4). The significance levels were defined as pvalue < 0.05 and LFCs were center scaled for each sample. Venn diagrams were plotted using R package VennDiagram (version 1.6.20). KEGG pathway analyses were done with R package clusterProfiler (version 4.0.0). Our computational analysis ensured that the “positive” sgRNAs are the ones that are enriched in the drug(s) treated groups compared to the DMSO treated populations and correspond to the genes whose loss confers resistance to the drug treatment. Conversely, “negative” sgRNAs are defined as hits whose abundance is reduced in the treatment(s) relative to the DMSO control and represent the genes whose ablation sensitizes cells to the drug.
Gene essentiality and Vizome
Depmap essentiality matrices were downloaded from https://score.depmap.sanger.ac.uk/. Here we plotted the scaledBayesianfactors against different cancer subtypes. BeatAML data were obtained from Tyner et al. Nature, 2018 (http://vizome.org/aml/), which include both the gene expression levels and IC50 values for different inhibitors. Pearson correlation scores were calculated for different inhibitors with MFN2 expression. Heatmap of correlation score was then plotted for different inhibitors with R package pheatmap (version 1.0.12).
Proteomics
Normalized protein expression levels were downloaded from the article of Nusinow and colleagues44. Expression level of MFN2 was plotted for AML subjects against all other hematological cancer types, with R package ggplot2 (version 3.3.4).
Analysis of single cell RNA/CITE sequencing data
Clustering and visualization
To visualize RNA expression similarities between cells in two-dimensional space, we used the scaled data matrix to perform principal component analysis (PCA) on the 2,000 most variable genes. We ran uniform manifold approximation and projection (UMAP)88 on the first 30 principal components with 25 nearest neighbors defining the neighborhood size and a minimum distance of 0.3. We constructed a shared nearest neighbor (SNN) graph using 25 nearest neighbors and clustered the graph using a range of resolution from 0.1-10 to explore the clusters – resolution 2, which yielded 85 clusters, was used for subsequent broad cell type annotation and occupancy scoring analysis. Details on the annotations can be found in Supplemental Information.
Non-negative matrix factorization
To identify common gene expression profiles in the malignant cells of the adult AML patients, we subset malignant and healthy counterpart HSPC/myeloid populations and performed non-negative matrix factorization (NMF) using cNMF (v1.1)89. We included genes expressed in at least 50 cells and used only the top 2,000 overdispersed genes to define common gene expression profiles (GEP). In order to identify the most stable and accurate number of components (k) within the range of 20 to 35, we used silhouette score and Frobenius reconstruction error as implemented in cNMF over 25 iterations. K = 26 emerged as the smallest most stable solution. The consensus solution was determined over 250 iterations using a density threshold of 0.1 to exclude outlier solutions. UMAP visualization of malignant and healthy counterpart HSPC/myeloid cells was based on the same 2000 overdispersed genes used in the NMF analysis. Cell type related GEPs were excluded by evaluating GEP usage in healthy control cells. We identified 10 cell type specific GEPs, 4 patient-specific profiles and 12 commonly used GEPs across the malignant cells. Marker genes for each GEP were identified using multiple least squares regression of normalized z-scored gene expression against the consensus GEP usage matrix as implemented in cNMF. Positively associated genes were used for subsequent GO analysis as in 89. One commonly expressed GEP, GEP 4, was enriched for autophagy related pathway genes. We utilized the genes within the autophagy related pathways ‘macroautophagy’ and ‘process utilizing autophagic mechanism’ to generate signatures. All analysis was performed in Python (v3.7.0) using scanpy (v1.6.0) and seaborn for visualizations (v0.11.2).
Data analysis of public scRNAseq
Patient-derived xenograft single-cell RNA sequencing data of pre- and post-treatment of venetoclax was downloaded from GEO database (GSE178912). Briefly, in the previous study16, the authors established a patient-derived xenograft model using NOD/LtSz-SCID/IL-2Rγchain null (NSG) mice and treated venetoclax for a week. Viable human CD45+CD33+ AML cells before and after the treatment were used for single-cell RNA sequencing with the Chromium Single Cell 3’ Reagent kit (v3 Chemistry, 10X Genomics). The data was aligned by CellRanger v.3.0.2 and we annotated cell types using the SingleR package90 with the pre-defined `BlueprintEncodeData` labels and visualized the log-transformed gene expression levels of mitophagy related genes using the Seurat package91.
RNA sequencing analysis
Sequence alignment was performed using STAR (version 2.7.3a) using GRCh37.p13 (June 2013) human assembly (https://gdc.cancer.gov/about-data/gdc-data-processing/gdc-reference-files) and differential analysis was done using DESeq2 (version 1.32.0). KEGG and GO pathway analyses were done with R package clusterProfiler (version 4.0.0).
AML bulk RNA-sequencing cohorts
Bulk RNA sequencing data of xenografted AML patient cells treated in vivo with venetoclax to correlate response to the gene expression at diagnosis was downloaded from GEO database (GSE183329). We performed gene set enrichment analysis (GSEA) for both Reactome and GO biological process gene sets (downloaded from MSigDB). GSEA analysis was based on the rank order of log fold change (LFC) from our differential gene list with R package clusterProfiler (v4.0.2). Enriched gene set were then plotted using R package enrichplot (v1.12.2).
The Cancer Genome Atlas (TCGA) Acute Myeloid Leukemia (LAML)92 and Therapeutically Applicable Research To Generate Effective Treatments (TARGET)93 AML RNA sequencing data as well as clinical and survival annotations were downloaded from UCSC GDC Xena Hub (https://gdc.xenahubs.net). To generate ‘macroautophagy’ and ‘process utilizing autophagic mechanism’ gene signatures, we added one to the expression values, and used the average of the log2 transformed values as the gene set score.
The association of the scores with overall survival was addressed with Kaplan-Meier estimation where the continuous score was dichotomized by recursive partitioning method in the rpart R package. The association of the scores with venetoclax resistance was evaluated in the Beat AML cohort45, where patients with IC50 below 0.25 μM were called "Sensitive" and otherwise "Resistant" to venetoclax.
Statistical analysis
No statistical methods were used to predetermine the sample size. Kaplan-Meier survival curve p-values were computed using Logrank test. For statistical comparison, we performed unpaired Student’s t-test. Two-way ANOVA was used to compare among experiments. Data were plotted using GraphPad Prism v9.0 software as mean values, with error bars indicating standard deviation. *p < 0.05, **p < 0.01 and ***p < 0.001, respectively, unless otherwise specified.
Supplementary Material
Acknowledgments
We thank all members of the Aifantis laboratory for discussions throughout this project; A. Heguy and the NYU Genome Technology Center (GTC) RRID: SCR_017929 for expertise with sequencing experiments and the Applied Bioinformatics Laboratory (ABL) RRID: SCR_019178 for computational analyses (the GTC and ABL are shared resources partially supported by the Cancer Center Support Grant from National Institutes of Health (NIH)/National Cancer Institute (NCI) P30CA016087 at the Laura and Isaac Perlmutter Cancer Center). We also thank the NYU Langone Health DART Microscopy Laboratory RRID: SCR_017934 (grants NCI P30CA016087, S10 RR023704, S10 RR024708) for assistance with confocal microscopy and image analysis, and Alice Liang, Chris Petzold, Joseph Sall, and Kristen Dancel-Manning for their assistance with TEM work; the NYU Cytometry and Cell Sorting facility RRID: SCR_019179 for expert cell sorting (partially supported by P30CA016087 from NIH/NCI); the NYU Experimental Pathology Research Laboratory RRID: SCR_017928 (P30CA16087-31) for assistance with histology. In addition, we thank the MD Anderson Cancer Center Advanced Microscopy Core for assistance with super-resolution microscopy using OMX Blaze V4 SIM Super-Resolution Microscope (Leica), which is supported by NIH S10 grant RR029552. We thank Giorgio Inghirami (Weill Cornell Medicine) and Ann-Kathin Eisfeld (Ohio State University) for kindly providing us with the primary AML patient samples, as well as Richard N. Kitsis and Yun Chen (Albert Einstein College of Medicine) for the adenovirus overexpressing MFN2. Moreover, we thank Evangelia Loizou and Scott Lowe (Memorial Sloan Kettering Cancer Center) for kindly sharing with us the murine p53R172H mutant AML cells. We also thank Elena Ziviani and Luca Scorrano (University of Padua, Italy) for fruitful discussions. C.G. is supported by the NIH/NCI 1K99CA252602 grant and is a Special Fellow of The Leukemia & Lymphoma Society. E.G. is supported by the NIH/NCI grants (R01CA178394, R01CA223243, P30CA013330). I.A. is supported by the NIH/NCI (R01CA216421, R01CA173636, R01CA228135, P01CA229086, R01CA242020) and AstraZeneca.
Footnotes
Conflict of interest
The Aifantis laboratory has received funding from AstraZeneca (Pre-Clinical & Retrospective Translational AZ Solicited Research). E.G and E.Z. have filled a patent application 63/286,264 submitted by Albert Einstein College of Medicine that protects compositions and methods for inhibiting mitofusins for the treatment of cancer and other diseases. The authors declare no additional competing interests.
Data availability
All bulk RNA-seq data have been deposited and are available under Gene Expression Omnibus (GEO) accession number GEO: GSE182401. Source data are provided for all experiments.
Details on additional methods used in this study are described in Supplemental Information.
References
- 1.Siegel RL, Miller KD, and Jemal A (2018). Cancer statistics, 2018. CA Cancer J Clin 68, 7–30. 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Cerrano M, and Itzykson R (2019). New Treatment Options for Acute Myeloid Leukemia in 2019. Curr Oncol Rep 21, 16. 10.1007/s11912-019-0764-8. [DOI] [PubMed] [Google Scholar]
- 3.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. (2016). Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov 6, 1106–1117. 10.1158/2159-8290.CD-16-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD, and Daver N (2020). Advances in the Treatment of Acute Myeloid Leukemia: New Drugs and New Challenges. Cancer Discov 10, 506–525. 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 5.Pollyea DA (2020). Venetoclax in AML: Where We Are and Where We Are Headed. Clin Lymphoma Myeloma Leuk 20 Suppl 1, S25–S26. 10.1016/S2152-2650(20)30450-X. [DOI] [PubMed] [Google Scholar]
- 6.Pollyea DA, Amaya M, Strati P, and Konopleva MY (2019). Venetoclax for AML: changing the treatment paradigm. Blood Adv 3, 4326–4335. 10.1182/bloodadvances.2019000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan R, Ruvolo V, Mu H, Leverson JD, Nichols G, Reed JC, Konopleva M, and Andreeff M (2017). Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell 32, 748–760 e746. 10.1016/j.ccell.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. (2019). The TP53 Apoptotic Network Is a Primary Mediator of Resistance to BCL2 Inhibition in AML Cells. Cancer Discov 9, 910–925. 10.1158/2159-8290.CD-19-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Nakauchi Y, Kohnke T, Stafford M, Bottomly D, Thomas R, Wilmot B, McWeeney SK, Majeti R, and Tyner JW (2020). Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer 1, 826–839. 10.1038/s43018-020-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. (2020). Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 135, 791–803. 10.1182/blood.2019003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moujalled DM, Brown FC, Chua CC, Dengler MA, Pomilio G, Anstee NS, et al. (2023). Acquired mutations in BAX confer resistance to BH3-mimetic therapy in acute myeloid leukemia. Blood 141, 634–644. 10.1182/blood.2022016090. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Glytsou C, Zhou H, Narang S, Reyna DE, Lopez A, et al. (2019). Targeting Mitochondrial Structure Sensitizes Acute Myeloid Leukemia to Venetoclax Treatment. Cancer Discov 9, 890–909. 10.1158/2159-8290.CD-19-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens BM, Jones CL, Pollyea DA, Culp-Hill R, D'Alessandro A, Winters A, et al. (2020). Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat Cancer 1, 1176–1187. 10.1038/s43018-020-00126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabe Y, Saitoh K, Yang H, Sekihara K, Yamatani K, Ruvolo V, et al. (2018). Inhibition of FAO in AML co-cultured with BM adipocytes: mechanisms of survival and chemosensitization to cytarabine. Sci Rep 8, 16837. 10.1038/s41598-018-35198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharon D, Cathelin S, Mirali S, Di Trani JM, Yanofsky DJ, Keon KA, Rubinstein JL, Schimmer AD, Ketela T, and Chan SM (2019). Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci Transl Med 11. 10.1126/scitranslmed.aax2863. [DOI] [PubMed] [Google Scholar]
- 16.Bosc C, Saland E, Bousard A, Gadaud N, Sabatier M, Cognet G, et al. (2021). Mitochondrial inhibitors circumvent adaptive resistance to venetoclax and cytarabine combination therapy in acute myeloid leukemia. Nature Cancer 2, 1204–1223. 10.1038/s43018-021-00264-y. [DOI] [PubMed] [Google Scholar]
- 17.Ishizawa J, Kojima K, Chachad D, Ruvolo P, Ruvolo V, Jacamo RO, et al. (2016). ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci Signal 9, ra17. 10.1126/scisignal.aac4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishizawa J, Zarabi SF, Davis RE, Halgas O, Nii T, Jitkova Y, et al. (2019). Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality. Cancer Cell 35, 721–737 e729. 10.1016/j.ccell.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatt S, Pioso MS, Olesinski EA, Yilma B, Ryan JA, Mashaka T, et al. (2020). Reduced Mitochondrial Apoptotic Priming Drives Resistance to BH3 Mimetics in Acute Myeloid Leukemia. Cancer Cell 38, 872–890 e876. 10.1016/j.ccell.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caenepeel S, Brown SP, Belmontes B, Moody G, Keegan KS, Chui D, et al. (2018). AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies. Cancer Discov 8, 1582–1597. 10.1158/2159-8290.CD-18-0387. [DOI] [PubMed] [Google Scholar]
- 21.Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, et al. (2018). Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun 9, 5341. 10.1038/s41467-018-07551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter BZ, Mak PY, Tao W, Warmoes M, Lorenzi PL, Mak D, Ruvolo V, Tan L, Cidado J, Drew L, and Andreeff M (2022). Targeting MCL-1 dysregulates cell metabolism and leukemia-stroma interactions and resensitizes acute myeloid leukemia to BCL-2 inhibition. Haematologica 107, 58–76. 10.3324/haematol.2020.260331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, et al. (2018). A Novel MCL1 Inhibitor Combined with Venetoclax Rescues Venetoclax-Resistant Acute Myelogenous Leukemia. Cancer Discov 8, 1566–1581. 10.1158/2159-8290.CD-18-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. (2016). The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482. 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 25.Luedtke DA, Su Y, Ma J, Li X, Buck SA, Edwards H, et al. (2020). Inhibition of CDK9 by voruciclib synergistically enhances cell death induced by the Bcl-2 selective inhibitor venetoclax in preclinical models of acute myeloid leukemia. Signal Transduct Target Ther 5, 17. 10.1038/s41392-020-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogenberger J, Whatcott C, Hansen N, Delman D, Shi CX, Kim W, et al. (2017). Combined venetoclax and alvocidib in acute myeloid leukemia. Oncotarget 8, 107206–107222. 10.18632/oncotarget.22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibes R, and Bogenberger JM (2019). Transcriptional Silencing of MCL-1 Through Cyclin-Dependent Kinase Inhibition in Acute Myeloid Leukemia. Front Oncol 9, 1205. 10.3389/fonc.2019.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Lu Z, Lu X, Chen L, Cao J, Zhang S, Ling Y, and Zhou X (2013). OTUD5 regulates p53 stability by deubiquitinating p53. PLoS One 8, e77682. 10.1371/journal.pone.0077682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato H, Lu Q, Rapaport D, and Kozjak-Pavlovic V (2013). Tom70 is essential for PINK1 import into mitochondria. PLoS One 8, e58435. 10.1371/journal.pone.0058435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto H, Fukui K, Takahashi H, Kitamura S, Shiota T, Terao K, et al. (2009). Roles of Tom70 in import of presequence-containing mitochondrial proteins. J Biol Chem 284, 31635–31646. 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, et al. (2013). MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell 51, 20–34. 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 33.Koyano F, Yamano K, Kosako H, Tanaka K, and Matsuda N (2019). Parkin recruitment to impaired mitochondria for nonselective ubiquitylation is facilitated by MITOL. J Biol Chem 294, 10300–10314. 10.1074/jbc.RA118.006302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Brito OM, and Scorrano L (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, and Dorn GW 2nd (2013). PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475. 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, and Harper JW (2013). Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376. 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barazzuol L, Giamogante F, Brini M, and Cali T (2020). PINK1/Parkin Mediated Mitophagy, Ca(2+) Signalling, and ER-Mitochondria Contacts in Parkinson's Disease. Int J Mol Sci 21. 10.3390/ijms21051772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary GS, Tat TT, Misra S, Hill BT, Smith MR, Almasan A, and Mazumder S (2015). Cyclin E/Cdk2-dependent phosphorylation of Mcl-1 determines its stability and cellular sensitivity to BH3 mimetics. Oncotarget 6, 16912–16925. 10.18632/oncotarget.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittaker SR, Barlow C, Martin MP, Mancusi C, Wagner S, Self A, et al. (2018). Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor. Mol Oncol 12, 287–304. 10.1002/1878-0261.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnik JL, Kikushige Y, et al. (2009). FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood 114, 5034–5043. 10.1182/blood-2008-12-196055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter BZ, Mak PY, Ruvolo V, Tao WJ, Hughes P, Chen XY, Morrow PK, and Andreeff M (2020). Targeting Mcl-1 Enhances the Activity of Tyrosine Kinase Inhibitor Gilteritinib in FLT3 Mutated AML. Blood 136. 10.1182/blood-2020-141399. [DOI] [Google Scholar]
- 42.Maiti A, DiNardo CD, Daver NG, Rausch CR, Ravandi F, Kadia TM, et al. (2021). Triplet therapy with venetoclax, FLT3 inhibitor and decitabine for FLT3-mutated acute myeloid leukemia. Blood Cancer J 11, 25. 10.1038/s41408-021-00410-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loizou E, Banito A, Livshits G, Ho YJ, Koche RP, Sanchez-Rivera FJ, et al. (2019). A Gain-of-Function p53-Mutant Oncogene Promotes Cell Fate Plasticity and Myeloid Leukemia through the Pluripotency Factor FOXH1. Cancer Discov 9, 962–979. 10.1158/2159-8290.CD-18-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, et al. (2020). Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402 e316. 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. (2018). Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531. 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santel A, and Fuller MT (2001). Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114, 867–874. [DOI] [PubMed] [Google Scholar]
- 47.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 48.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, and Lippincott-Schwartz J (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667. 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun N, Malide D, Liu J, Rovira II, Combs CA, and Finkel T (2017). A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc 12, 1576–1587. 10.1038/nprot.2017.060. [DOI] [PubMed] [Google Scholar]
- 50.Um JH, Kim YY, Finkel T, and Yun J (2018). Sensitive Measurement of Mitophagy by Flow Cytometry Using the pH-dependent Fluorescent Reporter mt-Keima. J Vis Exp 138, 58099. 10.3791/58099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanida I, Ueno T, and Uchiyama Y (2014). A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS One 9, e110600. 10.1371/journal.pone.0110600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. (2015). AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 125, 2120–2130. 10.1182/blood-2014-08-594408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kluge AF, Lagu BR, Maiti P, Jaleel M, Webb M, Malhotra J, Mallat A, Srinivas PA, and Thompson JE (2018). Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg Med Chem Lett 28, 2655–2659. 10.1016/j.bmcl.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 54.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, and Sheng M (2014). The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375. 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 55.Ordureau A, Paulo JA, Zhang J, An H, Swatek KN, Cannon JR, Wan Q, Komander D, and Harper JW (2020). Global Landscape and Dynamics of Parkin and USP30-Dependent Ubiquitylomes in iNeurons during Mitophagic Signaling. Mol Cell 77, 1124–1142 e1110. 10.1016/j.molcel.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lasry A, Nadorp B, Fornerod M, Nicolet D, Wu H, Walker CJ, et al. (2023). An inflammatory state remodels the immune microenvironment and improves risk stratification in acute myeloid leukemia. Nat Cancer 4, 27–42. 10.1038/s43018-022-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zachari M, and Ganley IG (2017). The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61, 585–596. 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, and Ganley IG (2015). Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem 290, 28726. 10.1074/jbc.A114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balachander SB, Criscione SW, Byth KF, Cidado J, Adam A, Lewis P, et al. (2020). AZD4320, A Dual Inhibitor of Bcl-2 and Bcl-x(L), Induces Tumor Regression in Hematologic Cancer Models without Dose-limiting Thrombocytopenia. Clin Cancer Res 26, 6535–6549. 10.1158/1078-0432.CCR-20-0863. [DOI] [PubMed] [Google Scholar]
- 60.Gelmetti V, De Rosa P, Torosantucci L, Marini ES, Romagnoli A, Di Rienzo M, Arena G, Vignone D, Fimia GM, and Valente EM (2017). PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 13, 654–669. 10.1080/15548627.2016.1277309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menon MB, and Dhamija S (2018). Beclin 1 Phosphorylation - at the Center of Autophagy Regulation. Front Cell Dev Biol 6, 137. 10.3389/fcell.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehrpour M, Esclatine A, Beau I, and Codogno P (2010). Overview of macroautophagy regulation in mammalian cells. Cell Res 20, 748–762. 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 63.Rojo M, Legros F, Chateau D, and Lombes A (2002). Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci 115, 1663–1674. 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- 64.Detmer SA, and Chan DC (2007). Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol 176, 405–414. 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S, Larrue C, Scheidegger NK, Seong BKA, Dharia NV, Kuljanin M, et al. (2022). An In Vivo CRISPR Screening Platform for Prioritizing Therapeutic Targets in AML. Cancer Discov 12, 432–449. 10.1158/2159-8290.CD-20-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zacharioudakis E, Agianian B, Kumar Mv V, Biris N, Garner TP, Rabinovich-Nikitin I, et al. (2022). Modulating mitofusins to control mitochondrial function and signaling. Nat Commun 13, 3775. 10.1038/s41467-022-31324-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Del Gaizo Moore V, and Letai A (2013). BH3 profiling--measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett 332, 202–205. 10.1016/j.canlet.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giacomello M, Pyakurel A, Glytsou C, and Scorrano L (2020). The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol 21, 204–224. 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 69.Csordas G, Weaver D, and Hajnoczky G (2018). Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol 28, 523–540. 10.1016/j.tcb.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bosc C, Broin N, Fanjul M, Saland E, Farge T, Courdy C, et al. (2020). Autophagy regulates fatty acid availability for oxidative phosphorylation through mitochondria-endoplasmic reticulum contact sites. Nat Commun 11, 4056. 10.1038/s41467-020-17882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang N, Wang C, Zhao H, He Y, Lan B, Sun L, and Gao Y (2021). The MAMs Structure and Its Role in Cell Death. Cells 10. 10.3390/cells10030657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mizushima N (2018). A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol 20, 521–527. 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 73.Malik SA, Shen S, Marino G, BenYounes A, Maiuri MC, and Kroemer G (2011). BH3 mimetics reveal the network properties of autophagy-regulatory signaling cascades. Autophagy 7, 914–916. 10.4161/auto.7.8.15785. [DOI] [PubMed] [Google Scholar]
- 74.Marino G, Niso-Santano M, Baehrecke EH, and Kroemer G (2014). Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15, 81–94. 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Piya S, Andreeff M, and Borthakur G (2017). Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy 13, 214–215. 10.1080/15548627.2016.1245263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, et al. (2018). AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell 23, 86–100 e106. 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu Y, Chen H, Zhang L, Lin X, Li X, Zhuang H, et al. (2021). The AMPK-MFN2 axis regulates MAM dynamics and autophagy induced by energy stresses. Autophagy 17, 1142–1156. 10.1080/15548627.2020.1749490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munoz JP, Ivanova S, Sanchez-Wandelmer J, Martinez-Cristobal P, Noguera E, Sancho A, et al. (2013). Mfn2 modulates the UPR and mitochondrial function via repression of PERK. EMBO J 32, 2348–2361. 10.1038/emboj.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sebastian D, Sorianello E, Segales J, Irazoki A, Ruiz-Bonilla V, Sala D, et al. (2016). Mfn2 deficiency links age-related sarcopenia and impaired autophagy to activation of an adaptive mitophagy pathway. EMBO J 35, 1677–1693. 10.15252/embj.201593084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mulcahy Levy JM, and Thorburn A (2020). Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ 27, 843–857. 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feely SM, Laura M, Siskind CE, Sottile S, Davis M, Gibbons VS, Reilly MM, and Shy ME (2011). MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology 76, 1690–1696. 10.1212/WNL.0b013e31821a441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, et al. (2011). An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25, 1628–1640. 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ianevski A, Giri AK, and Aittokallio T (2020). SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 48, W488–W493. 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ianevski A, Giri AK, Gautam P, Kononov A, Potdar S, Saarela J, Wennerberg K, and Aittokallio T (2019). Prediction of drug combination effects with a minimal set of experiments. Nat Mach Intell 1, 568–577. 10.1038/s42256-019-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, et al. (2015). Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 160, 977–989. 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryan J, and Letai A (2013). BH3 profiling in whole cells by fluorimeter or FACS. Methods 61, 156–164. 10.1016/j.ymeth.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 89.Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, and Sabeti PC (2019). Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife 8. 10.7554/eLife.43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172. 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancer Genome Atlas Research, N., Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074. 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. (2016). Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res 76, 2197–2205. 10.1158/0008-5472.CAN-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Naon D, Zaninello M, Giacomello M, Varanita T, Grespi F, Lakshminaranayan S, et al. (2016). Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc Natl Acad Sci U S A 113, 11249–11254. 10.1073/pnas.1606786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All bulk RNA-seq data have been deposited and are available under Gene Expression Omnibus (GEO) accession number GEO: GSE182401. Source data are provided for all experiments.
Details on additional methods used in this study are described in Supplemental Information.