Abstract
Purpose of review:
Acute kidney injury (AKI) occurs in approximately 10–15% of patients admitted to hospital and is associated with adverse clinical outcomes. Despite recent advances, management of patients with AKI is still mainly supportive, including the avoidance of nephrotoxins, volume and hemodynamic management and renal replacement therapy. A better understanding of the renal response to injury is the prerequisite to overcome current limitations in AKI diagnostics and therapy.
Recent findings:
Single cell technologies provided new opportunities to study the complexity of the kidney and have been instrumental for rapid advancements in the understanding of the cellular and molecular mechanisms of AKI.
Summary:
We provide an update on single cell technologies and we summarize the recent discoveries on the cellular response to injury in proximal tubule cells from the early response in AKI, to the mechanisms of tubule repair and the relevance of maladaptive tubule repair in the transition to chronic kidney disease (CKD).
Keywords: Acute kidney injury, Single cell RNA sequencing, Proximal tubule, Kidney repair, Maladaptive repair
Introduction
Since the first description of RNA sequencing in individual cells in 2009 (1), single cell technologies have evolved rapidly. In less than 10 years, these approaches are widely employed in biomedical research (2). Single cell technologies offer unprecedent data granularity to advance our understanding of biology and disease by analyzing the transfer of genetic information from genome to protein in individual cells. Single cell technologies have proven to be particularly useful in unravelling complex biological systems, such as the kidney, and dynamic processes, such as acute kidney injury (AKI). Here, we provide an update and some future perspectives on single cell technologies, as applied to the kidney, with a particular interest for the biology of proximal tubule cells in kidney disease.
Single cell technologies
The basics, the wet-lab techniques, the hardware, the protocols, the analytical methods, and the practical challenges related to the application of single cell technologies have been comprehensively reviewed by others (3, 4). The main challenges in isolating single cells, capturing picogram amounts of template (such as DNA or RNA) and amplifying this to obtain enough material for high-throughput sequencing without introducing sample and technical biases, limited the initial application of single cell technologies to highly specialized labs. However, the potential impact was recognized early on, when single cell sequencing was selected as the “Method of the Year 2013” (5).
Remarkably, in a few years, major technological advances, including improvements in throughput, accuracy, and automation, enabled the commercialization and widespread accessibility to single cell technologies where nucleic acid sequence is the technological readout (Table 1). An initial interest for single cell DNA and RNA sequencing was now expanded to include single cell epigenome sequencing (such as scATAC-seq, scHi-C, scChIP-seq)(6, 7), single-cell lineage tracing (by combining CRISPR/Cas9 genome editing with single-cell transcriptomics or by taking advantage of endogenous modifications of genome and mitochondrial DNA) (8, 9), single cell spatial transcriptomics (based on seqFISH or in situ sequencing / capturing strategies) (10–12) and third generation sequencing, opening the door to systematic analysis of alternative splicing, DNA rearrangements and extra-chromosomal circular DNAs (13–15). Technologies have become more integrated, enabling the analysis of multiple parameters in individual cells (16): e.g. CITE-seq combines RNA sequencing with the detection of cell surface proteins at single cell level (17), whereas ASAP-seq pairs scATACseq with protein and mitochondrial DNA detection (18). Such multimodal analyses significantly enhance the power of single cell technologies but their applicability is still limited by the need of specific tissue fixation and processing.
Table 1.
Summary of some relevant single cell technologies.
| Name | Abbreviation | Description |
|---|---|---|
| Single cell DNA sequencing | scDNA-seq | Detection of DNA in individual cells or nuclei. |
| Single cell transposase-accessible chromatin sequencing | scATAC-seq | Mapping of chromatin accessibility across the genome in individual cells. |
| Single cell chromatin immunoprecipitation sequencing | scChIP-seq | Epigenome sequencing technology in individual cells. |
| Single cell Hi-C | scHi-C | Mapping of 3D genome organization in individual cells. |
| Single cell RNA sequencing (Single nucleus RNA sequencing) | scRNA-seq (snRNA-seq) | Detection and quantitative analysis of mRNA in individual cells (or nuclei). |
| Cellular indexing of transcriptomes and epitopes | CITE-seq | Simultaneous quantification of cell surface protein markers and mRNA in individual cells. |
| ATAC with select antigen profiling by sequencing | ASAP-seq | Simultaneous profiling of cell surface/intracellular proteins markers and chromatin accessibility in combination with mitochondrial DNA analysis for clonal tracking. |
Improving quality of single cell studies
Coupled to the expanded use of single cell technologies, data collection and computational strategies evolved to manage the data output, extremely large sets of DNA sequence data. However, several challenges remain to be solved: Data reproducibly and comparability, always a challenge, plays a special role in the context of large consortia in which multiple data obtained in different laboratories, using a variety of technologies, is brought together to illuminate a shared research focus. Several recent studies have attempted to benchmark different protocols and identify the most reliable technologies, so that researchers may determine which method is best applied to a specific purpose (19). Comparative analyses have highlighted substantial differences in protocol performance, with major concerns around sample processing and batch-effects (20–22). Recent developments are expected to overcome those limitations.
First, at the computational level, methods have been developed for data integration and batch-effect correction, including Seurat 3 and Harmony (23–25), which have been shown in comparative studies to manage inherent problems of variability amongst the data (22). Second, technological advances leading to throughput improvement and cost reduction allow for further increases in the number of samples and cells per sample. Appropriate power calculations will become standard as in other areas of biomedical research (26). Third, smart strategies were developed to minimize batch effects: e.g. the sci-RNA-seq3 approach employs a combinational indexing strategy to introduce a high multiplexing capacity. This opens the possibility to analyze many samples and millions of cells in a single experiment (27, 28). The introduction of “gold standard reference samples” can benchmark data quality and improve data comparability. The availability of these data, and the incorporation of existing data into new analyzes, is providing important new insight, building on an existing knowledge-base and avoiding costly repetition of data collection. Improvements in reproducibility, integration and sharing of data, will only enhance the broadest use of single cell datasets to the benefit of scientific discovery, hypothesis building, and study design across disciplines (29, 30).
Interrogating the kidney cell by cell
The application of single cell technologies in kidney research rapidly advanced from first studies in mouse and human kidney development, anatomy and physiology (31–36) to experimental models of acute and chronic kidney disease (37, 38) and investigations on clinical samples (39–41). First studies on disease models were mainly focused on proximal tubule cells (PTCs). The proximal tubule response to ischemia reperfusion injury has been extensively investigated in the last decades and, since PTCs are the most abundant cell type in the kidney, and standard kidney biopsies from patients sample the kidney cortex, PTC signatures predominate in bulk RNAseq experiments (42, 43). Single cell technologies have substantially increased the depth of our understanding of PTC biology, in the normal, diseased, and injured kidney.
Kidney injury (from multiple insults) significantly impacts PTCs resulting in the dedifferentiation or suspension of the differentiated PTC state, proliferation of these cells and their return to normal mature PTCs (44). Single cell technologies were instrumental to characterize how PTC respond to injury by modifying their epigenetic and transcriptional profiles and thereby entering specific cell states (Figure 1). Moreover, we begin to understand the mechanisms determining the dynamic cell state switches along the transition from injury to repair (or failed repair) (Figure 1). (37, 38). Interestingly, different experimental models and first clinical studies suggest the presence of common fundamental molecular mechanisms driving this complex biological process, but those seem to overlap with multiple dimensions of heterogeneity related to the anatomical localization (along the segments of the tubule and in different areas of the kidney defining a local microenvironment), the type of injury, the species and other factors (28, 45).
Figure 1.
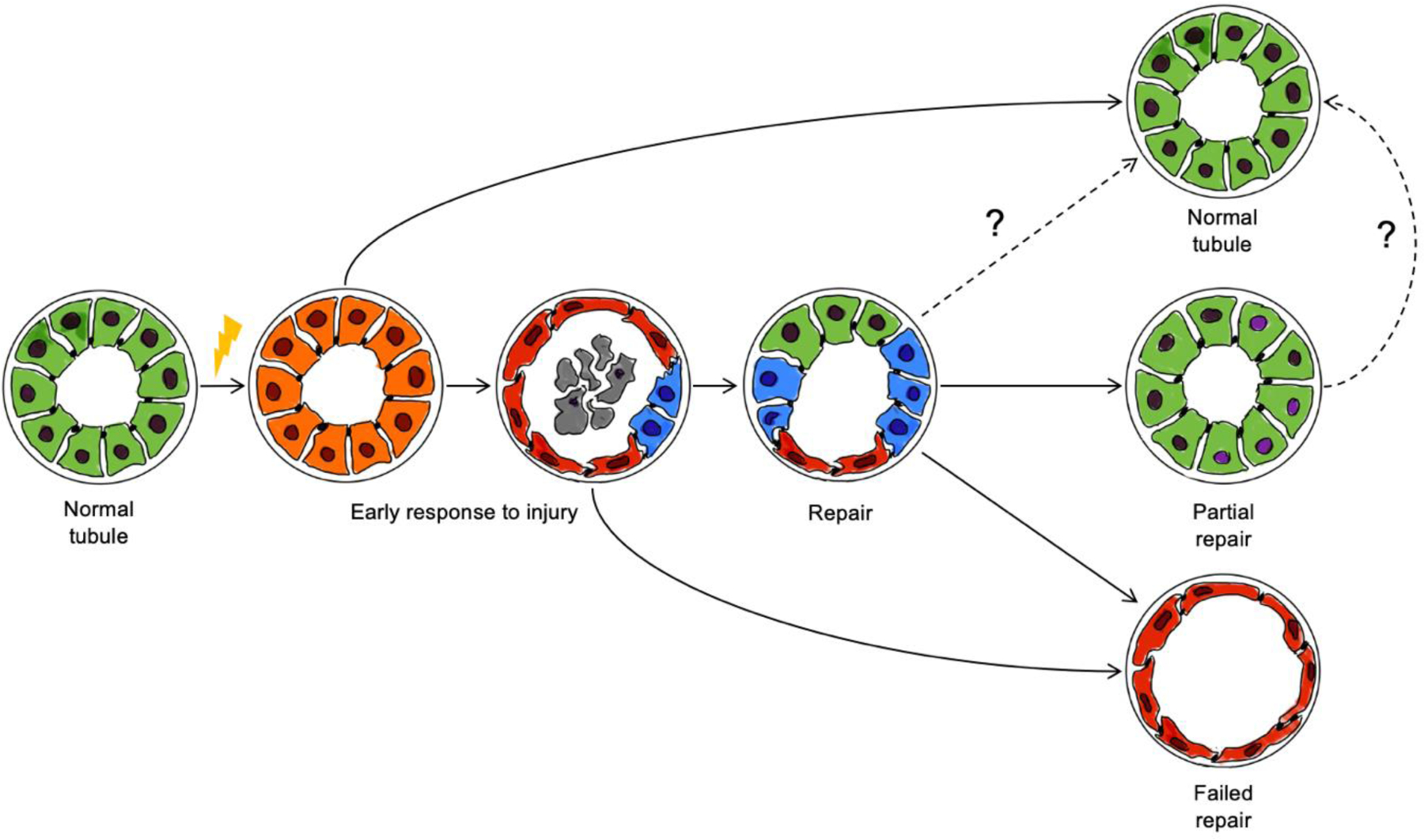
Schematic model of the main cell state transitions in proximal tubule cells following ischemia-related AKI. Almost all PTCs display a transcriptional response to AKI in the first hours/days after ischemia/reperfusion injury (orange). Dead cells accumulate in the tubule lumen (debris, grey). Injured cells dedifferentiate and acquire a squamous epithelium morphology (red). Some cells enter cell cycle and activate developmental pathways to accomplish the repair program. Dedifferentiated squamous epithelial cells can persist (failed repair) or be replaced by reparative tubule cells. The tubule repair program can restore tubule morphology and function, but in some cases PTCs with apparent normal morphology retain an altered epigenetic signature (partial repair).
Early response to injury in proximal tubule cells
The early response to injury in PTCs is characterized by the loss of normal gene expression and cessation of proximal tubule function. Critical to this stage is the down-regulation of regulons controlled by key transcriptional regulators of PTC differentiation and function such as HNF4A and HNFG, and the de novo activation of genes involved in cell adhesion, migration, and proliferation by new transcriptional processes activated by the injury insult (38). Injured cells acquire a simple squamous epithelium morphology. In parallel, some cells within the damaged tubule enter the cell cycle and likely activate a reparative program. Previous studies highlighted a pivotal role for SOX9 in this process (46, 47), preceded by a wave of transcriptional responses to stress, that is highly conserved in the mouse and human kidney (42, 43). The mechanisms triggering and coupling downstream responses are still not well understood and the control of this process and the relationship to developmental pathways leading a full repair of the injured tubule are not clear.
Increasing evidence supported by single cell sequencing suggests a central role for energy metabolism (48). A high metabolic rate is required to maintain the physiological functions of PTCs. In quiescent PTCs this rely on oxidative phosphorylation driven mainly by fatty acid oxidation (FAO) in mitochondria (49). In response to various types of kidney injury, mitochondrial function and therefore the ability of the PTCs to use FAO is impaired and PTCs switch from FAO to anaerobic glycolysis (50). The Ca2+-dependent mitochondria stress response activates a complex signaling response to the nucleus, including the activation of NFκB (51). In parallel, a HIF1A-driven metabolic adaptation likely maintains cell viability and – in analogy with studies in cancer – contributes to major cell state changes and cell proliferation (48). Importantly, different types of acute and chronic kidney injury elicit distinct metabolic alterations and can interfere with the mito-nuclear communication regulating fundamental cellular processes in homeostasis and in stress conditions (51, 52). This opens the opportunity for novel therapeutic options in AKI, e.g. by boosting nicotinamide adenine dinucleotide (NAD+) synthesis (53, 54).
The metabolic rewiring in response to injury also impairs gluconeogenesis, an often neglected, but fundamental function of the kidney, with PTCs processing primarily lactate to provide ca. 40% of endogenous gluconeogenesis in the post-absorptive phase (55). Impaired PTC gluconeogenesis in patients with AKI contributes to systemic metabolic disturbances, was associated with adverse prognosis, and might represent a therapeutic target to modulate kidney repair and the systemic consequences of AKI (50). High-spatial-resolution metabolomics in combination with multiplex immunofluorescence was recently applied to the kidney and might provide more detailed information about the role of cell-cell interactions and of the microenvironment on metabolic perturbations (56). Thus, the application of single cell technologies has facilitated the characterization of the initial tubular response to injury and contributed to identify potential therapeutic targets, but several pieces of the puzzle are still missing to fully understand this complex process, its dynamic, reversibility and systemic relevance.
Altered proximal tubule cells in failed tubule repair
As a result of the repair process and depending on the severity of injury, most PTCs recover by completing the process of re-differentiation (Figure 1). Recent studies have compared cells undergoing normal replicative repair to uninjured PTCs, identifying residual changes in gene activity and epigenetic signatures in cells commonly regarded as having undergone effective repair (57). More dramatically, some cells adopt a markedly altered state of failed repair. These cells form persistent squamous epithelium following AKI, with accompanying inflammatory-associated fibrosis accompanying unresolved tubular injury.
Single cell technologies have allowed for a resolution analysis of these cell types. Further, the application of computational approaches to study cell state transitions (e.g. pseudotime analysis) and cell lineage tracing experiments have begun to delineate the molecular processes determining the shift from the early injury response to adaptive/maladaptive repair. Several groups identified and characterized altered proximal tubule cell states, which were not present in the kidney of normal young mice but persisted in the late states after AKI after normalization of kidney function and in experimental models of chronic kidney disease (28, 37, 38, 57, 58). Late injured cells persistently lacked the expression of terminal differentiation markers (such as Slc34a1 or Slc5a12) and were marked by Vcam1 and Ccl2 and by the activation of the NF-κB and TNF-α pathways (37, 38). The expression of genes associated to inflammation and fibrosis implicated altered proximal tubule cells as central drivers of AKI to CKD transition. Consistently, similar proximal tubule cell states, marked by VCAM1 and NF-κB activation, were identified in patients with CKD; notably however, the same cells were also found in people without apparent kidney disease, suggesting an accumulation of altered PTC in association with aging and possibly in response to subclinical episodes of AKI (35, 39).
Increasing evidence indicates that VCAM1+ PTCs represent the common final step of a failed-repair process independently of the initial type of injury and that different biological processes might contribute to the establishment of this likely irreversible cell state (28, 45). Results obtained in distinct experimental and clinical conditions suggest again a pivotal role for mitochondrial function and fatty acid metabolism along the transition from early injury to failed repair: a delicate balance in the accumulation of lipid droplets in damaged tubule cells might promote tubule repair or conversely contribute to a vicious circle of lipotoxicity (28, 48, 59). The proinflammatory and profibrotic features of failed-repair PTCs is in good agreement with the senescence- associated secretory phenotype (SASP) reported in aged and injured non-kidney tissues (38). Cellular senescence is involved in the pathogenesis of chronic kidney disease and senescence-targeting interventions displayed beneficial role in animal models of kidney aging and disease (60–63). However, the role of some features of cellular senescence in other systems has been controversial in the biology of failed-repair, in particular the role of cell cycle arrest. Conclusions based on cycling proteins suggested epithelial cell cycle arrest in G2/M was associated with kidney fibrosis, but scRNAseq studies fail to observe a pronounced G2/M arrest signature in the gene expression data (37, 38, 64). This might reflect a lack of strict cycle-stage specificity in the antibody determinant studies or a dual role for cyclin G1 in the regulation of the cell cycle and of PTC dedifferentiation, which becomes uncoupled in the context of AKI (65).
Several lines of evidence suggest the accumulation of altered proximal tubule cells following AKI impacts the long term transition to CKD. First, the loss of terminal differentiation genes reduces the amount of functional renal tissue. As an example, impaired renal gluconeogenesis, secondary to reduced proximal tubule function, is a hallmark of chronic kidney disease and was associated with adverse clinical outcomes (66). Second, recent studies on fibrosis highlighted the role of cell-cell and cell-matrix interactions in a well-defined microenvironment referred as the fibrogenic niche in kidney scarring (67). Many questions related to the biology of the fibrogenic niche remains to be solved, but the main cells, structural elements and secreted factors (e.g. TGFβ, WNTs) have been identified. Damaged proximal tubule cells are considered one of the most important drivers in the formation and the maintenance of the niche, stimulating adjacent stroma cell to become scar-forming myofibroblasts (39) and generating a inflammatory microenvironment (68). Third, altered PTC in response to AKI have been linked to the development of papillary renal cell carcinoma (69). Thus, single cell analyses contributed to a better understanding of the intrinsic cellular properties of altered proximal tubule cells in kidney repair, their relevance in the local microenvironment of the damaged kidney and in the pathophysiology of AKI and CKD, and to the identification of pharmacological targets to modulate their generation, accumulation and (dys)function.
Future perspectives
Despite substantial recent advances, our understanding of the fascinating complexity of the kidney remains incomplete and the cellular understanding of kidney diseases is still limited to some general mechanisms and mainly focused on the proximal tubule. The mechanisms determining the cell state switch in the different phases of kidney injury and repair remain to be elucidated (Figure 1). Moreover, we still do not know how the activation of injury and reparative pathways is differentially regulated in response to injury to determine the fate of individual cells in the repair program. Relevant differences between mouse and human might need to be considered here. The recent application of single cell technologies on lineage-traced reparative cells raises new question about the late phase of the repair process, on the possible spread of injury to secondary sites not associated with a primary injury response and on the clinical relevance of an epigenetic signature of injury in repaired tubules (57). Moreover, the power of single cell technologies need to be expanded to other cell types (such as endothelial cells, podocytes or immune cells). Understanding the interaction in the renal microenvironment will provide more comprehensive models, define new areas of research and provide important information for different application in the field of regenerative nephrology. Failed repair PTCs like SASP-exhibiting cells in other organ systems are potential targets for senolytic therapies (70). However, this begs the question of the long-term outcomes for this altered PTC-type.
The application of single cell multi-omics technologies will be instrumental to fully elucidate the dynamic processes of kidney injury and repair. Recent data points to epigenetic alterations persisting after tubular recovery (57). How such changes might impact human kidney function over many years is an open question. Changes in organelle function, notably mitochondria, might not be detectable by transcriptomics analysis but may determine an increased susceptibility to repeated injuries. This is a clinically relevant aspect, in consideration of the multifactorial pathogenesis of CKD. In fact, the integration of multiple level of complexity from the genetic background to the clinical history, down to a multiomics analysis of the cellulome of an individual patient remains a major challenge to be solved in order to translate the power of single cell technologies towards clinical applications. In this setting the analysis of a limited number of patient samples by single cell will not be sufficient to correctly evaluate the inevitable variability of clinical samples and to fully understand the peculiarities the human kidney. New technological and computational advances will determine how single cell analyses will be applicable in the clinic. Improving data reproducibility, comparability and sharing will be instrumental for clinical applications.
Conclusion
Single cell technologies opened the opportunity to study the kidney at a higher level of resolution. Their application is rapidly moving from experimental to clinical samples, revealing new aspects of kidney biology and disease, including the complex cellular mechanisms of tubule injury and repair.
Key points.
Single cell technologies contributed to a more detailed understanding of the cellular mechanism of kidney injury and repair.
The accumulation of altered tubule cells after failed repair and in aging contributes to kidney dysfunction and fibrosis.
The molecular definition of kidney injury patterns is expected to define specific pharmacological targets in the future.
Acknowledgements
Financial support:
Work in PEC’s laboratory is supported by a grant from the Swiss National Foundation (Sinergia, CRSII5_202302) and by the Balli and the Gianella foundations. Work in APM’s laboratory is supported by grants from the National Institutes of Health (UC2 DK126024, R01 DK054364, R01 DK121409, and R01 DK126925).
Footnotes
Conflicts of interest: none.
References
- 1.Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge S, Teichmann SA. Single cell transcriptomics comes of age. Nat Commun. 2020;11(1):4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adil A, Kumar V, Jan AT, Asger M. Single-Cell Transcriptomics: Current Methods and Challenges in Data Acquisition and Analysis. Front Neurosci. 2021;15:591122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Method of the year 2013. Nat Methods. 2014;11(1):1. [DOI] [PubMed] [Google Scholar]
- 6.Cusanovich DA, Hill AJ, Aghamirzaie D, et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell. 2018;174(5):1309–24 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Wen Y, Liang Y, et al. A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility. Nat Protoc. 2021;16(8):4084–107. [DOI] [PubMed] [Google Scholar]
- 8.Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet. 2020;21(7):410–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig LS, Lareau CA, Ulirsch JC, et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell. 2019;176(6):1325–39 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng CL, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568(7751):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriques SG, Stickels RR, Goeva A, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melo Ferreira R, Gisch DL, Eadon MT. Spatial transcriptomics and the kidney. Curr Opin Nephrol Hypertens. 2022;31(3):244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen L, Tang F. Recent advances in single-cell sequencing technologies. Precis Clin Med. 2022;5(1):pbac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta I, Collier PG, Haase B, et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat Biotechnol. 2018. [DOI] [PubMed] [Google Scholar]
- 15.Sedlazeck FJ, Rescheneder P, Smolka M, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20(5):257–72. [DOI] [PubMed] [Google Scholar]
- 17.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mimitou EP, Lareau CA, Chen KY, et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat Biotechnol. 2021;39(10):1246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dance A Which single-cell analysis tool is best? Scientists offer advice. Nature. 2022;612(7940):577–9. [DOI] [PubMed] [Google Scholar]
- 20.Mereu E, Lafzi A, Moutinho C, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020;38(6):747–55. [DOI] [PubMed] [Google Scholar]
- 21.Tian L, Dong X, Freytag S, et al. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat Methods. 2019;16(6):479–87. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Zhao Y, Chen X, et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat Biotechnol. 2021;39(9):1103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran HTN, Ang KS, Chevrier M, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid KT, Hollbacher B, Cruceanu C, et al. scPower accelerates and optimizes the design of multi-sample single cell transcriptomic studies. Nat Commun. 2021;12(1):6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Dixon EE, Wu H, Humphreys BD. Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis. Cell Metab. 2022;34(12):1977–98 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreibing F, Kramann R. Mapping the human kidney using single-cell genomics. Nat Rev Nephrol. 2022;18(6):347–60. [DOI] [PubMed] [Google Scholar]
- 30.Naved BA, Bonventre JV, Hubbell JA, et al. Kidney repair and regeneration: perspectives of the NIDDK (Re)Building a Kidney consortium. Kidney Int. 2022;101(5):845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J, Shrestha R, Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ransick A, Lindstrom NO, Liu J, et al. Single-Cell Profiling Reveals Sex, Lineage, and Regional Diversity in the Mouse Kidney. Dev Cell. 2019;51(3):399–413 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindstrom NO, Guo J, Kim AD, et al. Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol. 2018;29(3):806–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung JJ, Goldstein L, Chen YJ, et al. Single-Cell Transcriptome Profiling of the Kidney Glomerulus Identifies Key Cell Types and Reactions to Injury. J Am Soc Nephrol. 2020;31(10):2341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muto Y, Wilson PC, Ledru N, et al. Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat Commun. 2021;12(1):2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lake BB, Chen S, Hoshi M, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun. 2019;10(1):2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.*.Kirita Y, Wu H, Uchimura K, et al. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci U S A. 2020;117(27):15874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first study to characterize injured proximal tubule cells by single cell RNAseq in a mouse model of AKI.
- 38.Gerhardt LMS, Liu J, Koppitch K, et al. Single-nuclear transcriptomics reveals diversity of proximal tubule cell states in a dynamic response to acute kidney injury. Proc Natl Acad Sci U S A. 2021;118(27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.*.Kuppe C, Ibrahim MM, Kranz J, et al. Decoding myofibroblast origins in human kidney fibrosis. Nature. 2021;589(7841):281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a single cell characterization of kidney fibrosis at single cell resolution in patients with CKD
- 40.Wu H, Malone AF, Donnelly EL, et al. Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol. 2018;29(8):2069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson PC, Wu H, Kirita Y, et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A. 2019;116(39):19619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Kumar S, Dolzhenko E, et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight. 2017;2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cippa PE, Sun B, Liu J, et al. Transcriptional trajectories of human kidney injury progression. JCI Insight. 2018;3(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang-Panesso M, Humphreys BD. Cellular plasticity in kidney injury and repair. Nat Rev Nephrol. 2017;13(1):39–46. [DOI] [PubMed] [Google Scholar]
- 45.Gerhardt LMS, McMahon AP. Identifying Common Molecular Mechanisms in Experimental and Human Acute Kidney Injury. Semin Nephrol. 2022;42(3):151286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Liu J, Pang P, et al. Sox9 Activation Highlights a Cellular Pathway of Renal Repair in the Acutely Injured Mammalian Kidney. Cell Rep. 2015;12(8):1325–38. [DOI] [PubMed] [Google Scholar]
- 47.Kang HM, Huang S, Reidy K, et al. Sox9-Positive Progenitor Cells Play a Key Role in Renal Tubule Epithelial Regeneration in Mice. Cell Rep. 2016;14(4):861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Rijt S, Leemans JC, Florquin S, et al. Immunometabolic rewiring of tubular epithelial cells in kidney disease. Nat Rev Nephrol. 2022;18(9):588–603. [DOI] [PubMed] [Google Scholar]
- 49.Tang C, Cai J, Yin XM, et al. Mitochondrial quality control in kidney injury and repair. Nat Rev Nephrol. 2021;17(5):299–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.*.Legouis D, Ricksten SE, Faivre A, et al. Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat Metab. 2020;2(8):732–43. [DOI] [PubMed] [Google Scholar]; This study demonstrates the pivotal role of reduced gluconeogenesis on altered proximal tubule cells in patients with AKI.
- 51.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17(4):213–26. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Hepokoski M, Gu W, et al. Targeting Mitochondria and Metabolism in Acute Kidney Injury. J Clin Med. 2021;10(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katsyuba E, Mottis A, Zietak M, et al. De novo NAD(+) synthesis enhances mitochondrial function and improves health. Nature. 2018;563(7731):354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poyan Mehr A, Tran MT, Ralto KM, et al. De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat Med. 2018;24(9):1351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Legouis D, Faivre A, Cippa PE, de Seigneux S. Renal gluconeogenesis: an underestimated role of the kidney in systemic glucose metabolism. Nephrol Dial Transplant. 2022;37(8):1417–25. [DOI] [PubMed] [Google Scholar]
- 56.Wang G, Heijs B, Kostidis S, et al. Analyzing cell-type-specific dynamics of metabolism in kidney repair. Nat Metab. 2022;4(9):1109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.*.Gerhardt LMS, Koppitch K, van Gestel J, et al. Lineage Tracing and Single-Nucleus Multiomics Reveal Novel Features of Adaptive and Maladaptive Repair after Acute Kidney Injury. J Am Soc Nephrol. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study combines lineage tracing and single nucleus multiomics to characterized the fate of proximal tubule cells in kidney repair.
- 58.Wu H, Gonzalez Villalobos R, et al. Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metab. 2022;34(7):1064–78 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rinaldi A, Lazareth H, Poindessous V, et al. Impaired fatty acid metabolism perpetuates lipotoxicity along the transition to chronic kidney injury. JCI Insight. 2022;7(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang W, Hickson LJ, Eirin A, et al. Cellular senescence: the good, the bad and the unknown. Nat Rev Nephrol. 2022;18(10):611–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.*.Mylonas KJ, O’Sullivan ED, Humphries D, et al. Cellular senescence inhibits renal regeneration after injury in mice, with senolytic treatment promoting repair. Sci Transl Med. 2021;13(594). [DOI] [PubMed] [Google Scholar]; This study highlight the role of cellular senescence and in kidney injury and repair and support further invetigations on senolytics after AKI.
- 62.Tan H, Xu J, Liu Y. Ageing, cellular senescence and chronic kidney disease: experimental evidence. Curr Opin Nephrol Hypertens. 2022;31(3):235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker DJ, Wijshake T, Tchkonia T, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L, Besschetnova TY, Brooks CR, et al. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med. 2010;16(5):535–43, 1p following 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taguchi K, Elias BC, Sugahara S, et al. Cyclin G1 induces maladaptive proximal tubule cell dedifferentiation and renal fibrosis through CDK5 activation. J Clin Invest. 2022;132(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verissimo T, Faivre A, Rinaldi A, et al. Decreased Renal Gluconeogenesis Is a Hallmark of Chronic Kidney Disease. J Am Soc Nephrol. 2022;33(4):810–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Fu H, Liu Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat Rev Nephrol. 2022;18(9):545–57. [DOI] [PubMed] [Google Scholar]
- 68.Cippa PE, Liu J, Sun B, et al. A late B lymphocyte action in dysfunctional tissue repair following kidney injury and transplantation. Nat Commun. 2019;10(1):1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peired AJ, Antonelli G, Angelotti ML, et al. Acute kidney injury promotes development of papillary renal cell adenoma and carcinoma from renal progenitor cells. Sci Transl Med. 2020;12(536). [DOI] [PubMed] [Google Scholar]
- 70.van Deursen JM. Senolytic therapies for healthy longevity. Science. 2019;364(6441):636–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


