Abstract
Background & aims:
Visceral smooth muscle cells (SMCs) are an integral component of the gastrointestinal (GI) tract that regulate GI motility. SMC contraction is regulated by post-translational signaling and the state of differentiation. Impaired SMC contraction is associated with significant morbidity and mortality, but the mechanisms regulating SMC-specific contractile gene expression, including the role of long non-coding RNAs (lncRNAs), remain largely unexplored. Herein, we reveal a critical role of Carmn (Cardiac mesoderm enhancer-associated noncoding RNA), a SMC-specific lncRNA, in regulating visceral SMC phenotype and contractility of the GI tract.
Methods:
GTEx and publicly available single-cell RNA sequencing (scRNA-seq) datasets from embryonic, adult human and mouse GI tissues were interrogated to identify SMC-specific lncRNAs. The functional role of Carmn was investigated using novel GFP knock-in (KI) reporter/knockout (KO) mice. Bulk RNA sequencing (RNA-seq) and single nucleus RNA sequencing (snRNA-seq) of colonic muscularis were used to investigate underlying mechanisms.
Results:
Unbiased in silico analyses and GFP expression patterns in Carmn GFP KI mice revealed that Carmn is highly expressed in GI SMCs in human and mouse. Premature lethality was observed in global Carmn KO (gKO) and inducible SMC-specific KO (iKO) mice due to GI pseudo-obstruction, severe distension of the GI tract with dysmotility in cecum and colon segments. Histology, GI transit and muscle myography analysis revealed severe dilation, significantly delayed GI transit and impaired GI contractility in Carmn KO versus control mice. Bulk RNA-seq of GI muscularis revealed that loss of Carmn promotes SMC phenotypic switching as evidenced by up-regulation of extracellular matrix genes and down-regulation of SMC contractile genes, including Mylk, a key regulator of SMC contraction. snRNA-seq further revealed SMC Carmn KO not only compromised myogenic motility by reducing contractile gene expression but also impaired neurogenic motility by disrupting cell-cell connectivity in the colonic muscularis. These findings may have translational significance as silencing CARMN in human colonic SMCs significantly attenuated contractile gene expression, including MYLK, and decreased SMC contractility. Luciferase reporter assays showed that CARMN enhances the transactivation activity of the master regulator of SMC contractile phenotype, myocardin, thereby maintaining the GI SMC myogenic program.
Conclusions:
Our data suggest that Carmn is indispensable for maintaining GI SMC contractile function in mice, and that loss of function of CARMN may contribute to human visceral myopathy. To our knowledge this is the first study showing an essential role of lncRNA in the regulation of visceral SMC phenotype.
Keywords: Carmn, lncRNA, Smooth muscle, Contractility, Pseudo-obstruction
Lay Summary
The muscle cells of our GI tract provide the force to move food. Our understanding of muscle cell function has been based on the actions of proteins, but in the current study we found that a special type of RNA, a lncRNA called Carmn that is found in muscle cells is critical for muscle contraction, GI movement and survival.
Graphical Abstract

Introduction
The gastrointestinal (GI) tract is a hollow organ responsible for the movement, digestion and absorption of food as well as the removal of waste. These critical functions rely on the contraction of smooth muscle cells (SMCs) to generate the peristaltic forces that provide motility 1. Loss of GI motility underlies several diseases, including chronic intestinal pseudo-obstruction (CIPO) 2. Smooth muscle-motility disorders can result from loss of function mutation of SMC-contractile genes such as MYH11 3, MYLK 4, LMOD1 5, ACTG2 6, ACTA2 7 and MYL9 8 or inactivation of upstream regulatory transcription factors, such as Srf 9–11 and Myocd 12 which bind to CArG elements within SMC-contractile gene loci 13. The etiology of CIPO is multifactorial, and the importance of epigenetic regulators, such as long non-coding RNAs (lncRNAs), in the regulation of GI motility remains largely unexplored.
LncRNAs are an under-investigated class of RNA transcripts that are defined as having lengths exceeding 200 nucleotides, but no apparent protein-coding potential. Most evidence to date suggests that lncRNAs act as non-coding regulatory molecules that have important roles in a variety of physiological and pathological conditions 14. Although several lncRNAs have been shown to contribute to SMC biology 15, these studies have exclusively focused on vascular SMCs. Visceral SMCs contribute to GI motility through coordinated contractions of single units which is distinct from vascular SMCs that contract individually as multi-units 16–18. To the best of our knowledge, our study is the first to investigate the expression and functional role of lncRNAs in visceral SMCs.
Using a data mining approach to interrogate multiple independent omics datasets, we found a novel lncRNA, CARMN, that was the most abundantly expressed lncRNA in visceral SMCs from both human and mouse. CARMN was initially identified as an important regulator of cardiac differentiation in vitro 19, 20. Recently, we showed that CARMN is a highly abundant and conserved SMC-specific lncRNA that plays a critical role in maintaining a contractile phenotype of vascular SMCs 21–23. Unexpectedly, we found that both germline and SMC-specific inducible deletion of Carmn in mice resulted in premature lethality due to GI pseudo-obstruction. These data suggest that Carmn is indispensable for GI function and to the best of our knowledge, is the first lncRNA to have an important role in regulating GI motility. These data further suggest that loss of function mutations or downregulation of CARMN may contribute to CIPO in humans.
Material and Methods
Generation of Carmn global KO (gKO) mice and SMC-specific inducible Carmn KO (iKO) mice
Carmn KO/GFP KI reporter mice have been recently described 21. Carmn gKO mice were generated by intercrossing Carmn flox mice with mice harboring a ubiquitously expressed Cre (CMV-Cre) 24. SMC-specific, inducible Carmn KO mice (iKO) and SMC-lineage tracing control mice were generated as previously described 21. Carmn iKO studies were restricted to male mice due to the presence of the SMC-specific Myh11-CreERT2 transgene on the Y chromosome 25. All mice were maintained on the C57BL/6J background. The use of experimental animals has been approved by the Institutional Animal Care and Use Committee and Biosafety Committee at Augusta University in accordance with NIH guidelines.
Bulk RNA-seq and single nucleus RNA-seq (snRNA-seq)
Colon or jejunum muscularis were isolated from Carmn iKO and control mice (at 30 days post the 1st tamoxifen injection) for total RNA extraction using the TRIzol reagent. Total RNA was then subjected to whole transcriptome RNA-seq analysis to assess differential gene expression. For snRNA-seq, the colon muscularis was isolated from Carmn iKO and control mice, snap frozen in liquid N2 and nuclei were isolated according to the protocol provided by 10X Genomics. Nuclei were subsequently processed using a 10X Chromium single cell 3’ protocol. Completed 10X libraries were sequenced using an Illumina NextSeq-500/550 high output kit v2. Transcriptomic data including both bulk and snRNA-seq datasets have been deposited in the Sequences Read Archive (NCBI) under the accession # GSE199157.
A detailed description of methods is provided in the Online Supplemental Material.
Results
Identification of CARMN as a SMC-specific lncRNA in the human and mouse GI tract.
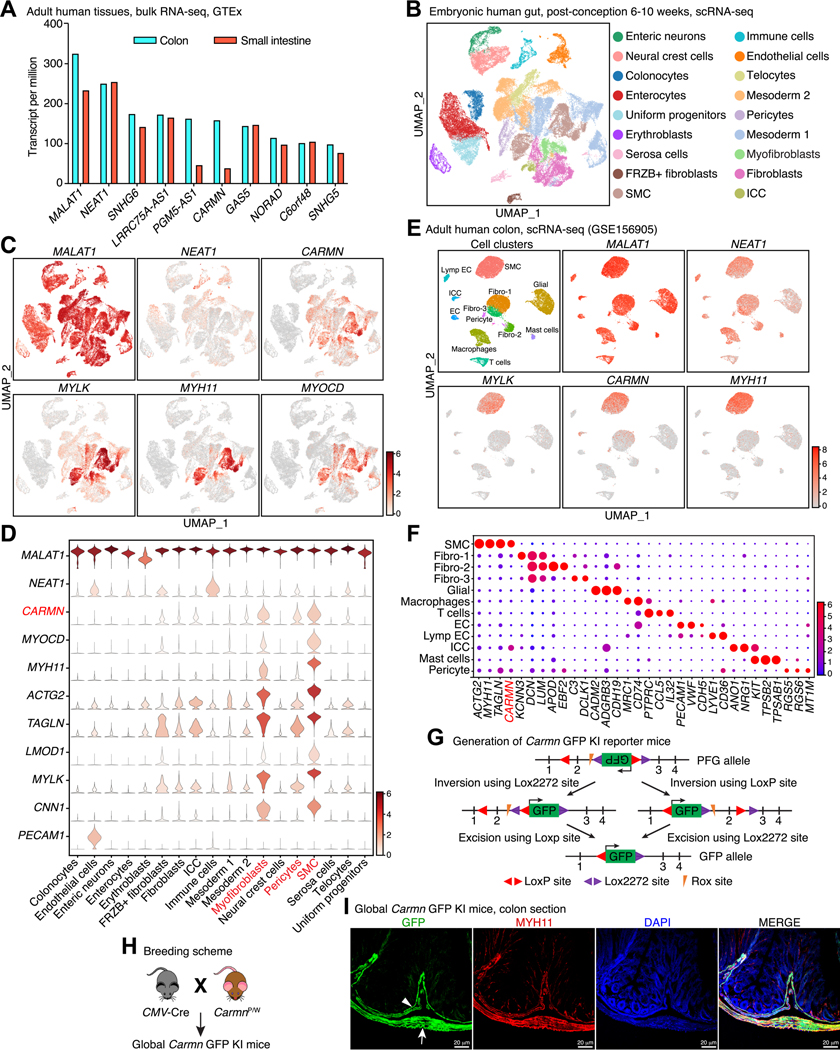
To identify novel lncRNAs in the GI tract, we first screened the most abundant lncRNAs expressed in human colon and small intestine tissues using the GTEx database 26. We identified the 10 highly enriched lncRNAs in adult human GI tissues (Figure 1A) and found that CARMN is the most abundantly expressed lncRNA in SMC-enriched tissues (Figure S1A). Further analysis at the single cell level was performed using a transcriptome dataset of 62,849 cells isolated from embryonic human duodenum, jejunum, ileum, and colon 27. Based on cell specific gene signatures, cells were divided into 43 clusters from 18 cell types (Figure 1B and Figure S1B). UMAP visualization revealed that CARMN is the most abundantly expressed lncRNA in SMCs, with an invariable expression pattern that was consistent with canonical SMC markers, such as MYH11, MYLK, ACTG2, LMOD1, TAGLN and CNN1, and the SMC-specific transcription cofactor MYOCD 28 (Figure 1C-D). CARMN was also detected in myofibroblasts and pericytes, which have SMC like properties (Figure 1D). We next de novo re-analyzed scRNA-seq datasets from human adult colon 29 and fetal intestine 30, and also from adult mouse ileum and colon 31. UMAP visualization revealed that CARMN expression is mostly enriched in SMCs although it can be detected at a lower level in related cell types such as myofibroblasts, pericytes and possibly in interstitial cells of Cajal (ICC) (Figure 1E-F and Figure S1C-D). Although bulk RNA-seq also indicated that the lncRNAs MALAT1, NEAT1 and SNHG6 are highly expressed in the adult human GI tract (Figure 1A), none of them displayed a SMC-specific expression pattern (Figure 1C & E).
Figure 1. Identification of CARMN as a SMC-specific lncRNA in human and mouse GI tissues.
(A) Analysis of the GTEx database showing the top 10 most abundant lncRNAs in human colon and intestinal tissues. (B) UMAP plot of cell populations based on scRNA-seq analysis of developing human gut tissues including duodenum, jejunum, ileum and colon. (C) UMAP visualization of the distribution of gene expression for NEAT1, CARMN, MYLK, MYH11 and MYOCD in embryonic human gut based on scRNA-seq analysis. (D) Violin plot showing the expression of selected genes in the different cell types identified in embryonic human gut. CARMN is found to be specifically expressed in SMCs, myofibroblasts and pericytes (in red). (E) UMAP plot showing cell types and the expression of selected genes in adult human colon as revealed by scRNA-seq analysis. (F) Dot plot showing the marker genes used to identify the cell types that comprise the adult human colon. (G) Strategy used to generate Carmn GFP KI mice. (H) Breeding strategy used to generate global Carmn GFP KI reporter mice. (I) Direct visualization of GFP and immunostaining of MYH11 to determine the cellular distribution of Carmn in relation to SMCs in colon dissected from K/W mice. Arrow and arrowhead denote muscularis externa SMCs and mucosa muscularis SMCs, respectively.
To visualize the distribution of Carmn expression in mice, we generated a novel Carmn KO/GFP KI mouse model 21 (Figure 1G). Cre-mediated inversion of a reversed GFP cassette (PFG to GFP) and deletion of exon 2 in the Carmn gene, results in GFP expression driven by the endogenous Carmn promoter and loss of Carmn expression. Female CarmnPFG/WT (CarmnP/W) mice were intercrossed with male global Cre mice to invert the GFP cassette (Figure 1H). Co-localization of GFP and immuno-fluorescent signals from the SMC-marker, MYH11 in K/W GFP reporter mice, revealed that Carmn is abundantly expressed in MYH11+ SMCs of the colon (Figure 1I) and jejunum (Figure S1E). Whole mount staining of the colon muscularis revealed that the GFP signal completely overlaps with that of MYH11, but rarely with the ICC marker, KIT, confirming that Carmn expression is SMC-specific (Figure S2A-B). Moreover, we found that CARMN is also specifically expressed in SMCs of the uterus and bladder in both human and mouse (Figure S3A-F). Collectively, data from unbiased informatics and our novel Carmn KI GFP reporter mouse model support the concept that the lncRNA, CARMN, is abundantly expressed in visceral SMCs of the GI and genitourinary tracts of both humans and mice.
Global deletion of Carmn in mice results in premature death due to lethal GI pseudo-obstruction.
To investigate a functional role of Carmn, we generated Carmn global KO (gKO) mice via intercross of female K/W and male K/W mice. Resulting WT and heterozygous (Het) littermates served as controls (Figure 2A). Unexpectedly, we observed that approximately 60% of pregnant dams develop a dystocia phenotype, suggesting that Carmn haploinsufficiency is sufficient to compromise uterine contraction and successful parturition (Figure 2B-C). Carmn gKO mice were born at the expected Mendelian ratio without gross abnormalities. However, upon weaning, they developed an unhealthy appearance with reduced body size (Figure 2D) and body weight compared to control mice (Figure 2E). Further, the switch to a normal chow diet resulted in acute lethality (within 1 week) of approximately 60% of gKO mice, with the remainder dying within 15 months (Figure 2F). gKO mice displayed an enlarged abdomen that was caused by a dilated duodenum, jejunum and colon and the accumulation of gas and stool (Figure 2G). Closer examination of gKO mice revealed that the cecum and colon had the greatest distension and accumulation of feces (Figure 2H), suggesting lethal GI pseudo-obstruction. HE histological analysis revealed a severe dilation of the colon and jejunum in gKO mice along with the thinning of muscle layers (Figure 2I-J, Figure S4A-B). Ultrastructural examination by transmission electron microscopy revealed that SMCs, in both the colon and jejunum of gKO mice, had markedly abnormal structures. The morphology of the endoplasmic reticulum was striking with dramatically dilated lumens and massive accumulation of membrane laminated vacuoles with embedded membranes and organelles such as mitochondria, suggesting a stressed and degenerative SMC phenotype and the presence of autophagosomes (Figure 2K and Figure S4C). Collectively, these data demonstrate that global deletion of Carmn causes lethal GI pseudo-obstruction and dystocia, suggesting that Carmn is indispensable for GI and uterine function in mice.
Figure 2. Global deletion of Carmn in mice causes lethal GI pseudo-obstruction.

(A) Breeding strategy used to generate global Carmn KO mice via intercross of Carmn Het (K/W) mice. (B) Representative picture of a uterus from a Carmn Het female mouse in labor under bright field (upper panel) or GFP channel (bottom panel) to show the dystocia phenotype. Arrows denote trapped embryos in the uterine tract. Arrowhead denotes the bladder. (C) The incidence of dystocia in female WT and Carmn Het mice crossed with Carmn Het male mice. (D) Gross pictures of littermates of WT, Het and Carmn gKO mice reveal smaller body sizes of gKO mice on postnatal day (P) 29. (E) The relative body weights of littermate WT, Het and Carmn gKO mice on postnatal P19–31. N = 15; *p < 0.05; One-way ANOVA. (F) Kaplan-Meier survival analysis of WT, Het and Carmn gKO mice. N = 18; *p < 0.05; Log-rank (Mantel-Cox) test. (G) Carmn gKO mice exhibit abdominal distension (top panel) and dramatically enlarged intestines that have accumulated air (arrow), as compared to WT and Het littermates. The arrowhead points to the bladder. (H) Gross pictures of GI tracts isolated from WT, Het and Carmn gKO mice. The GI segments from Carmn gKO mice reveal dramatically different cecum (arrowhead) and colon (arrow). (I) Hematoxylin and eosin (HE) staining on the transverse sections of colon from WT, Het and Carmn gKO mice. The boxed area is magnified on the bottom of each respective panel. (J) The relative thickness of the muscularis layers of the colon (double-head arrows shown in “I”) was measured and plotted. N = 6–8; *p < 0.05; One-way ANOVA. (K) Representative transmission electron microscopy images of control and Carmn gKO colon show the enlarged endoplasmic reticulum lumen (asterisk), autophagic vesicles with lamination (arrow) in gKO mouse colon. The boxed area is magnified on the right.
Inducible SMC-specific deletion of Carmn in adult mice recapitulates the lethal phenotype of Carmn gKO mice.
To exclude roles in other cell types and to establish a functional role of Carmn specifically in adult SMCs, we generated inducible, SMC-specific Carmn KO mice through intercross of tamoxifen (TAM)-inducible SMC-specific Cre mice (Myh11-CreERT2) with conditional CarmnPFG/PFG mice (iKO mice, Figure 3A and Figure S5A). To control for any unanticipated effects of ectopic expression of GFP and Cre in SMCs of SMC-specific Carmn iKO mice, SMC-lineage tracing mice (referred to as WT control mice) were generated by intercrossing Myh11-CreERT2 mice with Rosa26-mTmG dual fluorescence reporter mice (Figure 3B). A separate cohort of mice, in which a single Carmn allele was deleted in SMCs, was used as an additional control (referred to as iHet hereafter, Figure S5A). TAM was administered to 8–10-week-old male mice and body weight was monitored (Figure 3C). At day 62 after the first TAM injection, the body weights of iKO mice were significantly reduced compared to those of WT and iHet mice. We observed increased mortality of iKO mice starting at day 33, with none surviving beyond 130 days (Figure 3D). Autopsy of iKO mice at day 30 revealed a pathology very consistent with that seen in gKO mice. This included dramatic GI tract distention and excess gas and feces that were observed predominantly in the cecum and colon (Figure 3E-F). Histological analysis of iKO colon and jejunum was similar to that seen in gKO mice, with pronounced lumen dilation and thinning of the muscular layers (Figure 3G-H and Figure S5B-C). Similarly, ultrastructural images of both the colon and jejunum from iKO mice also revealed a degenerative SMC phenotype with autophagosomes in the muscularis, consistent with findings in gKO mice (Figure 3I and Figure S5D). To examine whether the loss of Carmn has a broader impact on organs outside of the GI, we performed an extensive phenotypic analysis on brain, ascending and thoracic aorta, bladder, ureter, and uterus in gKO or/and iSM KO mice (Figure S6). We found that there was no major pathological changes in these organs at baseline, except for a thinning of the ascending aortic wall in both KO mouse models (Figure S6C & G). These results collectively suggest that GI pseudo-obstruction is the dominant phenotype resulting from Carmn deficiency and is responsible for the premature deaths of both global and SMC-specific inducible Carmn KO mice.
Figure 3. SMC-specific deletion of Carmn in adult mice causes lethal GI pseudo-obstruction and phenocopies Carmn gKO mice.

(A-B) Schematic diagram of the strategy used to generate SMC-specific Carmn KO mice. Inducible, SMC-specific deletion of Carmn in adult mice (iKO) was achieved by intraperitoneal injections of tamoxifen (TAM) in adult male Myh11-CreERT2+; CarmnPFG/PFG mice and (B) lineage tracing control mice (WT) were created by TAM injection in adult male Myh11-CreERT2+; R26-mTmG+/− mice, respectively. (C) Body weights of the WT, iHet and iKO mice were measured before and after TAM injection at the times indicated. N = 6–8; *p < 0.05; 2-way analysis of variance, followed by post hoc testing. (D) Kaplan-Meier survival analysis of WT, iHet and iKO mice after two rounds of injection with TAM. N = 10–12; *p < 0.05; Log-rank (Mantel-Cox) test. (E) Carmn iKO mice exhibited abdominal distension and dramatic intestinal enlargement (arrow) compared to WT and iHet mice. Arrowhead denotes the bladder. Pictures were acquired using a dissecting scope with bright field (top and middle panels, before and after opening abdominal cavity, respectively) and GFP channel (bottom panel after opening the abdominal cavity). (F) Pictures of the GI tract isolated from WT, iHet and iKO mice on day 30 post the first TAM injection. The cecum (arrowhead) and proximal colon (arrow) were the most dilated parts of the GI tract of Carmn iKO mice. (G) HE staining of transverse sections of colon from WT, iHet and iKO mice. The boxed area is magnified on the bottom. (H) The thicknesses of muscular layers of the colon were measured (double-head arrows in “G”) and plotted. N = 4–7; *p < 0.05; One-way ANOVA. (I) Representative transmission electron microscopy images demonstrate the formation of significant autophagic vesicles (arrow) and a large vesicle containing several small vesicles (asterisk) in Carmn iKO colon SMCs. The boxed area is magnified on the bottom.
Carmn deficiency impairs GI motility and colonic contractility in mice.
Impaired SMC contraction in the GI tract is responsible for GI pseudo-obstruction and CIPO in humans 4–8. To indirectly assess contractile function of the GI tract of mice in vivo, whole-gut transit time from food intake to excretion was measured. Whole-gut transit time was significantly increased in both Carmn KO mice (gKO and iKO) versus respective controls (Figure 4A-B). Using a fluorophore-based spatial GI transit assay, FITC-dextran was found concentrated in the proximal small intestine in iKO mice versus the distal small intestine and cecum of control mice, one hour after gavage (Figure 4C). iKO mice issued larger diameter stools versus controls (Figure 4D-E). These data suggest that Carmn is necessary for efficient GI motility. We next measured the contractile function of the colon using myography. Spontaneous contractions in both Carmn gKO and iKO mice were significantly attenuated (Figure 4F-G and Figure S7A-B). In addition, the amplitude of contraction in response to depolarization with KCl or the muscarinic receptor agonist carbachol, which use different signaling pathways to promote SMC contraction through MYLK-dependent phosphorylation of myosin regulatory light chain (RLC) 32, was significantly impaired in Carmn gKO and iKO mice (Figure 4H-O and Figure S7C-F). These data indicate that SMC Carmn has a major role in regulating GI motility and contractility.
Figure 4. Carmn deficiency impairs GI motility and colonic contractility.
(A) Whole-gut transit time in WT and Carmn gKO mice. N = 7–8; *p < 0.05; Unpaired Student t test. (B) Whole-gut transit time in WT and Carmn iKO mice before (day 0) or after TAM injection at day 45. N = 5; *p < 0.05; 2-way analysis of variance followed by post hoc testing within day. (C) Distribution of intestinal FITC in WT and Carmn iKO mice after gavage with FITC labelled dextran for 1 hour. N=3–5. (D-E) Stools were collected from WT and iKO mice on day 30 post the first TAM injection. Stool diameter was then measured and plotted. N = 9–13; *p < 0.05; Unpaired Student t test. (F-G) Representative recordings of spontaneous contractions in colonic rings from (F) Carmn gKO or (G) iKO mice and their respective control mice. (H-K) Representative recordings of contractions induced by 60 mM KCl on colonic rings from (H) Carmn gKO or (J) iKO mice and their respective control mice. Quantitative analysis of peak force induced by KCl treatment on colonic rings from Carmn gKO or iKO mice are shown in “I” or “K”, respectively. N = 6–7; *p < 0.05; Unpaired Student t test. (L-O) Representative recordings of contraction induced by 1 μM Cch (Carbachol, a muscarinic receptor agonist) on colonic rings from (L) Carmn gKO or (N) iKO mice and their respective WT control mice. The peak force induced by Cch treatment on Carmn gKO or iKO mouse colonic rings are shown in “M” or “O”, respectively. N = 6; *p < 0.05; Unpaired Student t test.
Carmn deficiency induces visceral SMC phenotypic switching.
To investigate the molecular mechanisms underlying the GI pseudo-obstruction phenotype in Carmn deficient mice, bulk RNA-sequencing (RNA-seq) was performed on the muscularis isolated from the colon and jejunum of Carmn iKO and control mice (Figure 5A). Loss of Carmn in iKO mice resulted in 318 significantly down-regulated genes and 243 up-regulated genes in colon muscularis (Figure 5B and Supplemental Table 1), and 561 down-regulated genes and 635 up-regulated genes in jejunum muscularis (Figure 5C and Supplemental Table 1). Gene ontology (GO) analysis in colon muscularis revealed that Carmn deficiency impaired the expression of genes contributing to SMC homeostasis, contraction, development and differentiation (Figure 5D & F). In contrast, the up-regulated genes in colon muscularis have roles in tissue repair, matrix remodeling, wound healing, and inflammatory processes including the response to lipopolysaccharide (Figure 5E-F). In jejunum muscularis, Carmn KO down-regulated genes involved in metabolism (Figure S8A), while up-regulated genes were involved in immune cell infiltration and inflammation (Figure S8B), consistent with changes in gene expression observed in the colon. To better understand the tissue-dependent variability of genes regulated by Carmn, we compared differentially expressed genes in both colon and jejunum muscularis. As shown by the Venn diagram, overlap between Carmn iKO colon and jejunum muscularis was seen with 136 downregulated and 128 upregulated genes (Figure 5G). Strikingly, GO analysis of the overlapping downregulated genes revealed roles in muscle contraction and development. These data suggest that down-regulation of contractile genes is a primary cause of gut hypomotility in Carmn deficient mice (Figure 5H and Figure S8C). GO analysis of the upregulated genes revealed common roles in inflammatory responses and matrix remodeling (Figure S8D-E), implicating possible secondary effects that result from the visceral myopathy. We next analyzed the expression of core genes maintaining contractile SMC homeostasis and found that Mylk, the principal regulator of the myosin II molecular motor that is essential for gastrointestinal motility 33, was the only common gene (Figure 5I). Collectively, the integrative analysis of bulk RNA-seq datasets from the muscular layers of both colon and jejunum reveals that Carmn deficiency disrupts the expression of SMC contractile genes such as Mylk, while increasing expression of genes involved in wound healing, inflammation, and extracellular matrix.
Figure 5. Carmn deficiency down-regulates the expression of genes involved in muscle contraction while increasing the expression of genes regulating extracellular matrix remodeling.
(A) The workflow of bulk RNA-seq of colon and jejunum muscularis isolated from WT and iKO Carmn KO mice. (B) Volcano plots depicting significantly down-regulated (in green) and up-regulated genes (in red) between Carmn iKO and WT mouse colon and (C) jejunum muscularis. FDR: False Discovery Rate. FC: Fold Change. (D) GO analysis showing that the 318 down-regulated genes in Carmn iKO colon muscularis are significantly enriched in biological processes related to muscle contraction and differentiation (E) while the significantly up-regulated genes are enriched in the functional categories involved in the wound healing and extracellular matrix remodeling. The top 10 most significant GO biological processes are shown. (F) Heat map showing the differential expression of select genes in colon involved in muscle contraction and extracellular matrix organization. (G) Venn diagrams showing the overlap of down-regulated (left panel) or up-regulated (right panel) genes between colon and jejunum muscularis in Carmn iKO mice. (H) GO analysis showing that the 136 overlapping genes down-regulated in both colon and jejunum muscularis of Carmn iKO mice are significantly enriched in biological processes related to muscle contraction. The top 10 most significant GO biological processes are shown. (I) Venn diagram showing the identification of overlapping down-regulated genes involved in the regulation of muscle function.
Carmn deficiency disrupts homeostatic SMC contractile gene programs and compromises cell-cell communication in the colonic muscularis.
To obtain a deeper understanding of the transcriptional impact of Carmn on SMCs at the single cell level, cell nuclei were isolated from the colonic muscularis of Carmn iKO and control mice and snRNA-seq performed (Figure S9A). UMAP analysis of 5,072 nuclei revealed 17 distinct clusters with 13 cell types, as defined by cell type-specific transcriptomes (Figure 6A and Figure S9B-E). In control mice, Carmn was specifically detected in SMCs expressing well-established SMC markers, Myh11 and Mylk, and the SMC-specific transcription co-factor of SRF, Myocd (Figure 6A and Figure S9E). Cell composition analysis revealed that the percentage of SMCs, glial cells, Cftr+ cells and Goblet cells was decreased in Carmn iKO colon, while the percentages of fibroblast, fibroblast-like, macrophage and T cells were increased (Figure 6B). We next conducted a differential gene analysis, specifically within SMC clusters, and identified 123 downregulated and 265 up-regulated genes in Carmn iKO SMCs (Figure 6C and Supplemental Table 2). GO analysis of down-regulated genes revealed that Carmn deficiency significantly decreased the expression of genes involved in regulating muscle contraction and synapse organization (Figure S9F). The major up-regulated genes contribute to extracellular matrix organization and adhesion (Figure S9G). Consistent with our bulk RNA-seq data, genes that are involved in muscle contraction like Mylk and Tpm1, and the muscarinic receptor Chrm2, were significantly downregulated. In contrast, genes encoding extracellular matrix proteins such as Ccn2, Fn1 and Eln, were significantly increased in iKO SMCs, consistent with SMC phenotypic switching from a contractile to a synthetic phenotype (Figure S9H). Analysis of the overlap differentially expressed genes (DEGs) between colon muscularis (bulk RNA-seq) and SMC clusters (snRNA-seq) revealed 29 co-downregulated genes and 34 co-upregulated genes, respectively (Figure 6D). GO analysis revealed that these genes are involved in pathways regulating muscle contraction and extracellular matrix organization, respectively (Figure 6E-F). Collectively, the use of bulk and snRNA-seq approaches reveals that deletion of Carmn in SMCs leads to phenotypic switching through the down-regulation of contractile genes and the up-regulation of genes involved in extracellular matrix remodeling.
Figure 6. Identification of transcriptomic changes in SMC sub-populations from the colon muscularis of Carmn iKO mice using snRNA-seq.
(A) UMAP visualization of Myh11 and Carmn expression in the cell types of the colon muscularis using snRNA-seq. (B) Percentage of each cell type in the colon muscularis from WT and Carmn iKO mice. (C) Volcano plot showing differentially expressed genes (DEGs) specifically in SMCs between Carmn iKO and WT colon muscularis as determined by snRNA-seq. (D) Venn diagrams showing the overlap of down-(upper panel) or up-regulated (bottom panel) genes between DEGs in SMC clusters as revealed by snRNA-seq data of colon muscularis and DEGs as revealed by bulk RNA-seq of colon muscularis from Carmn iKO and WT mice. (E) GO analysis of the top 4 GO biological processes using the common genes either down- or (F) up-regulated as shown in “D”. (G) UMAP plot showing 7 distinct SMC sub-clusters (SCs) in mouse colonic muscularis. (H) Heatmap showing the top 5 enriched genes in each SMC SC. (I) Percentage of each SMC SC in both Carmn iKO and WT mouse colonic SMCs. (J) Violin plots showing the expression level of a subset of genes in each SC of SMCs. (K) Pseudotime trajectory analysis showing the branching SMC fates (upper panel) and the fate transition of each SMC SC along the trajectory. (L) Cell-cell connectivity analysis showing cell-cell interactions among all cell types identified in WT and Carmn iKO mouse colon muscularis. (M) Heatmaps showing the overall (both outgoing and incoming) signaling patterns of each cell type mediated by individual ligand-dependent signaling axis in both WT (left panel) and Carmn iKO (right panel) mouse colon muscularis.
To investigate the diversity of SMC sub-populations in mouse colon in greater detail, we compared the respective transcriptomes of 1,529 SMCs identified in iKO and control samples. Unsupervised Seurat-based clustering revealed 7 distinct SMC sub-populations as defined by cluster-specific gene markers (Figure 6G-H). Interestingly, we found that Carmn iKO SMCs have reduced representation of sub-cluster (SC) 4 and higher proportions of SC6 (Figure 6I). Although all 7 SMC subpopulations are Myh11 positive, expression of Myh11 is lower, whereas Mylk is nearly absent, in SC4 of Carmn iKO SMCs. In contrast, the most significantly enriched genes in SC4 SMCs, but depleted in iKO SMCs, were Nrg3 (Neuregulin3, a ligand binding ERBB-family receptors) and Rbfox1 (RNA binding protein, fox-1 homolog) (upper row in Figure 6J). Both are neuron-enriched genes (Figure S9E). In contrast, SC6 was the only subset of SMCs positive for the fibroblast-specific gene, Fbn2. Furthermore, the most significantly upregulated genes in iKO SMC from subpopulation SC6 are Ccn1, Ccn2 and Thbs1, all of which are fibroblast-specific genes and key mediators of SMC extracellular matrix homeostasis (bottom row in Figure 6J) 34. To better understand the functional characteristics of genes enriched in SMC SC4 and 6, GO analysis revealed pathways regulating axonogenesis and matrix remodeling, respectively (Figure S10A-B). Therefore, we classify the SMC subpopulation, SC4 as neuron-like SMCs and those in SC6 as fibroblast-like SMCs. To investigate the transitional potential of these SMC subtypes, we applied the Monocle 2 algorithm to perform single-cell, pseudo time trajectory analysis 35. This analysis revealed that SMCs progressively branch out into two trajectories, culminating in SMC SC4 and 6 (Figure 6K and Figure S10C). We hypothesize that the higher representation of fibroblast-like SMC subpopulation contribute to tissue repair, while the reduction in neuron-like SMC subpopulation in Carmn iKO mice impairs neurogenic motility and communication between cell types 36. To test this hypothesis, we performed cell-cell communication analysis using CellChat 37. The SC4 SMC subpopulation in Carmn iKO cells has impaired cell-cell communication with other contractile SMCs, glial cells, ICC and myenteric neurons (Figure 6L and Figure S10D). The overall signaling patterns show that compared to WT cells, Carmn iKO SMCs have greater activity of pathways promoting SMC phenotypic switching, such as IGF, THBS and TGFβ, which closely correlate with the increased proportions of fibroblast-like, SC6 SMC subpopulation in Carmn iKO mice (Figure 6M). In contrast, the neuron-like SC4 SMC subpopulation loses neurexin (NRXN), SEMA3 and neuregulin (NRG) signals while gaining FN1 ligand-mediated signals (Figure 6M and Figure S10E). Together, snRNA-seq analysis reveals that Carmn deficiency in SMCs compromises myogenic motility, not only by reducing the expression of contractile genes essential for SMC contraction, but also by disrupting cell-cell connectivity for the interactions with neurons, thereby collectively contributing to the GI pseudo-obstruction phenotype.
Deletion of Carmn attenuates the expression of Mylk which is required for SMC contraction.
Validation of bulk RNA-seq and snRNA-seq data using qRT-PCR and Western blotting confirmed that MYLK is consistently down-regulated in colon and jejunum muscularis from both gKO and iKO mice, suggesting that Mylk is a major target gene regulated by Carmn (Figure S11A-F and Figure S12A-B). Carmn deficiency did not significantly affect the expression of the master transcription factor, Srf, nor its cofactor Myocd (Figure S11A & D). IF staining further shows that MYLK expression was markedly reduced in colon and jejunum muscularis of both gKO and iKO mice (Figure S11G-H and Figure S12C-D). These data further support a critical role of Carmn in maintaining the expression of Mylk, an essential gene for SMC contraction.
CARMN is critical for contraction of human colonic SMCs by regulating MYLK gene expression.
We next determined the translational relevance of CARMN in human colonic smooth muscle cells (HuCoSMCs). qRT-PCR and Western blot analysis revealed that silencing CARMN in HuCoSMCs significantly increased expression of the proliferative gene, PCNA, and the matrix gene, CCN2, while attenuating the expression of almost all contractile genes examined, except MYH11, at both the mRNA and protein levels (Figure 7A-C). Since MYLK-mediated phosphorylation of RLC is a prerequisite for the initiation of SMC contraction, we examined KCl and Cch-mediated MYLK-pMLC signaling by Western blotting in HuCoSMCs. CARMN knockdown robustly attenuated MYLK expression, thereby preventing MYLK-mediated phosphorylation of RLC (pMLC) (Figure 7D-E). To assess the contractile competence of human SMCs, we performed collagen gel contraction assays using HuCoSMCs. We found that there was a ~50% decrease in contractile activity in CARMN-depleted HuCoSMCs versus controls (Figure 7F-G). The binding of a transcriptional complex, comprised of MYOCD and SRF, to CArG elements in the promoter regions of SMC contractile genes, is a fundamental mechanism that maintains SMC contractile phenotype 13. Our recent study showed that Carmn directly binds to MYOCD, but not SRF, to potentiate MYOCD transcription 21. Since MYLK is an established SRF target gene 38 and its SMC-specific expression profile overlaps with that of CARMN (Figure 1D), we hypothesized that MYLK is a novel target of the CARMN/MYOCD/SRF complex in visceral SMCs. To test this, we performed luciferase assays using a WT or CArG mutant Mylk gene reporter, with or without co-transfection of Carmn and Myocd. Similar assays were performed with a Lmod1 gene reporter as it is a CARMN- (Figure 7A-C) and SRF/MYOCD-dependent gene and mutations in LMOD1 gene cause an intestinal hypoperistalsis syndrome in mice and humans 5. We found that Carmn alone had no effect on baseline activity of Mylk and Lmod1 promoters, but significantly enhanced MYOCD-mediated transactivation in a CArG-dependent manner (Figure 7H). In summary, our data supports a model where the lncRNA, Carmn, contributes to the efficient contraction of GI SMC through the in trans regulation of a SRF/MYOCD complex to facilitate the expression of SMC genes, MYLK and LMOD1. Global or SMC-specific deletion of Carmn yields congruent phenotypes in mice resulting in attenuated expression of MYLK and comprised cell-cell connectivity. Loss of Carmn significantly impairs SMC contractility, progressively slowing GI motility to the point of premature lethality due to pseudo-obstruction (Figure 7I).
Figure 7. Depletion of CARMN impairs the contractile phenotype of human colonic SMCs.
(A) CARMN or control phosphorothioate-modified ASO were transduced into human colonic SMCs (HuCoSMCs) for 48 hours and then total RNA was harvested for qRT-PCR analysis. The relative mRNA levels were quantified in CARMN deficient cells and compared relative to silencing control cells (set to 1). N = 3; *p < 0.05; Unpaired Student t test. (B) CARMN or control phosphorothioate-modified ASO were transduced into HuCoSMCs for 72 hours and then total protein was harvested for Western blot analysis. (C) Densitometric analysis of relative protein levels as shown in “B”. N = 6; *p < 0.05; Unpaired Student t test. (D-E) CARMN or control phosphorothioate-modified ASO were transduced into HuCoSMCs for 72 hours, followed by treatment with (D) 60 mM KCl or (E) the muscarinic agonist Carbachol (Cch, 1 μM) for the indicated times. Total protein was then harvested for Western blot analysis. (F) HuCoSMCs were transfected with control or CARMN phosphorothioate-modified ASO for 48 hours and then seeded onto 24-well plates for collagen contractility assays. (G) Quantitative analysis of collagen contractility as shown in “F”. N = 4; *p < 0.05; unpaired Student t test. (H) Luciferase reporter assays were performed to examine the role of Carmn in regulating MYOCD-induced promoter activity of SMC-specific Lmod1 and Mylk gene reporters which harbor wild-type (WT) or mutated CArG boxes. N = 3; *p < 0.05; 2-way analysis of variance. (I). Schematic diagram summarizing the major findings of this study.
Discussion
CIPO is a rare, but life-threatening disease characterized by severe intestinal dysmotility. Functional and histopathologic studies have shown that GI dysmotility in CIPO patients results from multiple mechanisms, including defects in neurons, SMCs or ICC 39. Mutations affecting the expression and function of proteins of the SMC contractile apparatus are the most common cause of myopathic CIPO 2. To the best of our knowledge, our study is the first to report that lncRNAs have vital roles in maintaining visceral SMC contractility and GI motility. The loss of Carmn expression, as seen with global deletion during development and SMC-specific inducible ablation in adulthood, resulted in a severe GI motility disorder with a lethal visceral myopathy, resembling SMC dysfunction-mediated CIPO in humans. Our findings raise the possibility that polymorphisms in the CARMN gene, causing loss of expression and/or function, could be responsible for myopathic CIPO in humans. Future studies are needed to examine whether reduced CARMN expression levels or loss-of-function mutations in CARMN can be linked to patients with CIPO.
A comprehensive snRNA-seq analysis of GI muscularis revealed that Carmn deficiency affects NRXNs and NRGs-mediated neuronal signaling. NRXNs and NRGs are synaptic cell-adhesion molecules that connect pre- and post-synaptic neurons, mediate trans-synaptic signaling, and shape neural network properties by specifying synaptic functions 40. How Carmn expression in SMCs can influence NRXN and NRG signaling remains undefined. Moreover, SNPs in NRXN and NRG3 gene loci are associated with the susceptibility to Hirschsprung’s disease 41, 42. We propose that CARMN may play a broader role, not only in SMCs per se, but also in regulating the cell-cell communications that are crucial for coordinating GI motility. Moreover, the possible expression of CARMN in ICC also suggests a potential role of CARMN in regulating SMC contraction initiated by pacemaker cells.
In addition to the effect on GI motility, we also found that pregnant Carmn heterozygous mice develop a dystocia phenotype, likely due to impaired uterine contractions. Notably, starting from day 21 when the pups are weaned from milk to a regular chow diet, 60% of Carmn gKO mice died within 2 weeks, suggesting that Carmn is vital for the force overload response in both uterine and GI SMCs. A limitation of our study is that we were unable to assess the specific functional role of Carmn in postnatal visceral SMCs because the inducible Myh11-CreERT2 transgene we used is active in both vascular and visceral SMCs and only in male mice 25. Thus, the creation of a new visceral SMC-specific Cre mouse model in which Cre is active in both sexes of mice is required to address this question.
Previous studies have shown that both the SRF transcription factor and its cofactor, MYOCD, are necessary for the development and maintenance of the contractile phenotype in visceral SMCs. Targeted deletion of either SRF or MYOCD in SMCs of adult mice results in severe gut dysmotility, characterized by weak peristalsis and dilation of the digestive tract 9, 10, 12. Notably, the SMC-specific inducible Carmn KO mice used in our study exhibited remarkable similarities to the functional, gross appearance, microscopic and molecular changes in SMC-specific inducible Myocd and Srf KO mice 9, 10 11, 12. While this was not completely surprising, given that Carmn binds to and potentiates the function of the MYOCD/SRF complex 21, the extent of the phenotypic regulation of GI SMC by Carmn is remarkable. In addition to actions in the GI tract, SMC-specific inducible Myocd KO mice also develop significant phenotypes in the aorta and genitourinary tracts 12. In contrast, Carmn KO mice only exhibited the lethal GI obstructive phenotype, suggesting that Carmn may have MYOCD-independent mechanisms in SMCs. However, a caveat is that the lethal GI myopathy phenotype resulting from activity of the Myh11 promoter-driven Cre has precluded us from assessing a long-term effect of Carmn deficiency on vascular SMCs in vivo.
We identified that Carmn deficiency disrupts homeostasis of the colon muscularis and impairs colonic motility mainly through down-regulation of an array of SMC contractile genes, and especially the Mylk gene. Consistent with previous studies showing that MYLK is an SRF/MYOCD target gene 38, we found that Carmn-facilitated transcriptional activation of the MYLK gene in a CArG-dependent manner. It is well accepted that SMC RLC phosphorylation by MYLK is a prerequisite for SM contraction and critical for maintaining the physiological movements of hollow organs 33, 43. We found that deletion of Carmn in both murine visceral SMCs in vivo and human colonic SMCs in vitro, not only significantly down-regulated Mylk gene expression, but also attenuated KCl and Cch-induced RLC phosphorylation. Our data is consistent with a previous study showing that inducible SMC-specific deletion of Mylk in mice results in markedly reduced RLC phosphorylation, impaired gut motility and premature lethality 33. The ability of CARMN to strongly upregulate MYLK expression in visceral SMCs suggests that reduced MYLK is, at least in part, responsible for the functional deficits seen in Carmn KO mice. However, we cannot rule out the possibility that other Carmn-dependent genes, such as Dmpk and Chrm2, may play additional functional roles since these genes are also SRF/MYOCD targets genes and have been shown to be important for smooth muscle contraction 11, 44. Interestingly, a recent computational study suggested that Carmn could regulate the expression of downstream genes including MYLK via formation of RNA-DNA-DNA triplexes 45. Although this idea is provocative, whether Carmn regulates the expression of SMC-contractile genes via RNA-DNA interactions awaits experimental validation. Moreover, based on data showing a reduction in a novel population of neuron-like SMCs in Carmn KO mice, our snRNA-seq data advance the exciting concept that Carmn also regulates cellular connectivity between SMCs and other cell types. In addition to the ability of Carmn to regulate SMC gene expression including Mylk, we propose that cell-cell decoupling may contribute to the decreased contractility and motility observed in the GI tracts of Carmn KO mice.
In summary, our study has uncovered a novel role of the lncRNA, CARMN, as a major regulator of gastrointestinal motility and viability through actions in visceral SMCs that serve to maintain contractile function and intercellular connectivity. A greater understanding of the GI roles of lncRNAs such as CARMN expands our tool kit in helping to understand and diagnose human visceral myopathy diseases such as CIPO.
Supplementary Material
What You Need to Know.
BACKGROUND AND CONTEXT
The movement of food inside our gastrointestinal (GI) tract is controlled by the coordinated contraction of smooth muscle cells (SMCs). Impaired function of SMCs can slow and even halt the movement of food thereby causing a disease called chronic intestinal pseudo-obstruction (CIPO). Previously, the behavior of SMCs has been shown to be regulated by proteins controlling DNA transcription as well as contractile proteins that provide SMC tone. The functional role of long non-coding RNAs (lncRNAs) in regulating GI SMCs and motility has largely been unexplored.
NEW FINDINGS
We found that a lncRNA called Carmn is highly concentrated in SMCs of the GI tract of mice and humans. Loss of Carmn expression in mice resulted in GI pseudo-obstruction and premature death by reducing the motility of the GI tract. Using cell specific analysis, we found that Carmn is critical for the expression of SMC contractile proteins in the GI tract and cell: cell connectivity.
LIMITATIONS
Our single cell analysis shows that in the GI tract of mice there are multiple subpopulations of SMCs. Loss of Carmn reduced subpopulations of SMCs committed to expressing contractile proteins as well as neuron-like SMC subpopulations that might contribute to coordinated contractions. A limitation of our study is the characterization of these neuron-like SMC subpopulations and the translational impact of CARMN in human CIPO.
CLINICAL RESEARCH RELEVANCE
Our study suggests that loss of CARMN expression or function may contribute to human visceral myopathies such as CIPO.
BASIC RESEARCH RELEVANCE
Carmn is a SMC-enriched lncRNA that is indispensable for maintaining the contractile function of GI SMCs in mice. To the best of our knowledge, this is the first study showing an essential role of lncRNAs in the regulation of visceral SMC phenotype and GI function.
Source of funding:
The work at the J.Z. laboratory is supported by grants from the National Heart, Lung, and Blood Institute, NIH (HL157568 and HL149995). J.Z. is a recipient of Established Investigator Award (17EIA33460468) and Transformational Project Award (19TPA34910181) from American Heart Association. X.H. and K.D. are supported by a postdoctoral fellowship (836341) and a Career Development Award (938570), respectively, from American Heart Association.
Footnotes
Conflict of interest statement: The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability statement:
Original data, analytic methods, and materials will be made available to other researchers upon reasonable requests. The transcriptomics data including both bulk and snRNA-seq datasets generated in this study have been deposited in the Sequences Read Archive at the NCBI under accession # GSE199157.
Reference
- 1.Sanders KM, Koh SD, Ward SM. Interstitial cells of cajal as pacemakers in the gastrointestinal tract. Annu Rev Physiol 2006;68:307–43. [DOI] [PubMed] [Google Scholar]
- 2.Hashmi SK, Ceron RH, Heuckeroth RO. Visceral myopathy: clinical syndromes, genetics, pathophysiology, and fall of the cytoskeleton. Am J Physiol Gastrointest Liver Physiol 2021;320:G919–G935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong W, Baldwin C, Choi J, et al. Identification of a dominant MYH11 causal variant in chronic intestinal pseudo-obstruction: Results of whole-exome sequencing. Clin Genet 2019;96:473–477. [DOI] [PubMed] [Google Scholar]
- 4.Halim D, Brosens E, Muller F, et al. Loss-of-Function Variants in MYLK Cause Recessive Megacystis Microcolon Intestinal Hypoperistalsis Syndrome. Am J Hum Genet 2017;101:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halim D, Wilson MP, Oliver D, et al. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. Proc Natl Acad Sci U S A 2017;114:E2739–E2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorson W, Diaz-Horta O, Foster J 2nd, et al. De novo ACTG2 mutations cause congenital distended bladder, microcolon, and intestinal hypoperistalsis. Hum Genet 2014;133:737–42. [DOI] [PubMed] [Google Scholar]
- 7.Milewicz DM, Østergaard JR, Ala-Kokko LM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A 2010;152a:2437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno CA, Sobreira N, Pugh E, et al. Homozygous deletion in MYL9 expands the molecular basis of megacystis-microcolon-intestinal hypoperistalsis syndrome. Eur J Hum Genet 2018;26:669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mericskay M, Blanc J, Tritsch E, et al. Inducible mouse model of chronic intestinal pseudo-obstruction by smooth muscle-specific inactivation of the SRF gene. Gastroenterology 2007;133:1960–70. [DOI] [PubMed] [Google Scholar]
- 10.Angstenberger M, Wegener JW, Pichler BJ, et al. Severe intestinal obstruction on induced smooth muscle-specific ablation of the transcription factor SRF in adult mice. Gastroenterology 2007;133:1948–59. [DOI] [PubMed] [Google Scholar]
- 11.Lee MY, Park C, Ha SE, et al. Serum response factor regulates smooth muscle contractility via myotonic dystrophy protein kinases and L-type calcium channels. PLoS One 2017;12:e0171262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Wang T, Wright AC, et al. Myocardin is required for maintenance of vascular and visceral smooth muscle homeostasis during postnatal development. Proc Natl Acad Sci U S A 2015;112:4447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miano JM. Myocardin in biology and disease. J Biomed Res 2015;29:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed ASI, Dong K, Liu J, et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc Natl Acad Sci U S A 2018;115:E8660–E8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhl L, Mocci G, Pietila R, et al. A single-cell transcriptomic inventory of murine smooth muscle cells. Dev Cell 2022;57:2426–2443 e6. [DOI] [PubMed] [Google Scholar]
- 17.Davis MJ, Hill MA. Signaling mechanisms underlying the vascular myogenic response. Physiol Rev 1999;79:387–423. [DOI] [PubMed] [Google Scholar]
- 18.Kuriyama H, Kitamura K, Itoh T, et al. Physiological features of visceral smooth muscle cells, with special reference to receptors and ion channels. Physiol Rev 1998;78:811–920. [DOI] [PubMed] [Google Scholar]
- 19.Ounzain S, Micheletti R, Arnan C, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol 2015;89:98–112. [DOI] [PubMed] [Google Scholar]
- 20.Plaisance I, Perruchoud S, Fernandez-Tenorio M, et al. Cardiomyocyte Lineage Specification in Adult Human Cardiac Precursor Cells Via Modulation of Enhancer-Associated Long Noncoding RNA Expression. JACC Basic Transl Sci 2016;1:472–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong K, Shen J, He X, et al. CARMN Is an Evolutionarily Conserved Smooth Muscle Cell-Specific LncRNA That Maintains Contractile Phenotype by Binding Myocardin. Circulation 2021;144:1856–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vacante F, Rodor J, Lalwani MK, et al. CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice. Circ Res 2021;128:1258–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni H, Haemmig S, Deng Y, et al. A Smooth Muscle Cell-Enriched Long Noncoding RNA Regulates Cell Plasticity and Atherosclerosis by Interacting With Serum Response Factor. Arterioscler Thromb Vasc Biol 2021;41:2399–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995;23:5080–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth A, Benyo Z, Lukasova M, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med 2008;14:64–8. [DOI] [PubMed] [Google Scholar]
- 26.The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elmentaite R, Ross ADB, Roberts K, et al. Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev Cell 2020;55:771–783.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Chang PS, Wang Z, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 2001;105:85162. [DOI] [PubMed] [Google Scholar]
- 29.Wright CM, Schneider S, Smith-Edwards KM, et al. scRNA-Seq Reveals New Enteric Nervous System Roles for GDNF, NRTN, and TBX3. Cell Mol Gastroenterol Hepatol 2021;11:1548–1592.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao J, O’Day DR, Pliner HA, et al. A human cell atlas of fetal gene expression. Science 2020;370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drokhlyansky E, Smillie CS, Van Wittenberghe N, et al. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 2020;182:1606–1622.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno Y, Isotani E, Huang J, et al. Myosin light chain kinase activation and calcium sensitization in smooth muscle in vivo. Am J Physiol Cell Physiol 2008;295:C358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He WQ, Peng YJ, Zhang WC, et al. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology 2008;135:610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin W, Kim HT, Wang S, et al. Fibrillin-2 is a key mediator of smooth muscle extracellular matrix homeostasis during mouse tracheal tubulogenesis. Eur Respir J 2019;53. [DOI] [PubMed]
- 35.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huizinga JD, Chen JH. The myogenic and neurogenic components of the rhythmic segmentation motor patterns of the intestine. Front Neurosci 2014;8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin F, Hoggatt AM, Zhou J, et al. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am J Physiol Cell Physiol 2006;290:C1599–609. [DOI] [PubMed] [Google Scholar]
- 39.Antonucci A, Fronzoni L, Cogliandro L, et al. Chronic intestinal pseudo-obstruction. World J Gastroenterol 2008;14:2953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008;455:903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Duan S, Zhong R, et al. Exome sequencing identified NRG3 as a novel susceptible gene of Hirschsprung’s disease in a Chinese population. Mol Neurobiol 2013;47:957–66. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Liu H, Dong Y. Significance of neurexin and neuroligin polymorphisms in regulating risk of Hirschsprung’s disease. J Investig Med 2018;66:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamm KE, Stull JT. The function of myosin and myosin light chain kinase phosphorylation in smooth muscle. Annu Rev Pharmacol Toxicol 1985;25:593–620. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Rippe C, Hansson O, et al. Regulation of the Muscarinic M3 Receptor by Myocardin-Related Transcription Factors. Front Physiol 2021;12:710968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Yang X, Huang W, et al. Single-cell profiling of long noncoding RNAs and their cell lineage commitment roles via RNA-DNA-DNA triplex formation in mammary epithelium. Stem Cells 2020. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data, analytic methods, and materials will be made available to other researchers upon reasonable requests. The transcriptomics data including both bulk and snRNA-seq datasets generated in this study have been deposited in the Sequences Read Archive at the NCBI under accession # GSE199157.