Abstract
DNA aptamers that bind to cholic acid were previously isolated by an in vitro selection method. Secondary structural prediction and deletion-mutant experiments suggested that the cholic-acid binding regions of 19 sequenced clones could form three-way-junction structures. In this article, the secondary structures of the sequenced clones and the structural requirements for binding to cholic acid were evaluated. A course of mutational-analysis and chemical-modification experiments provided strong support for the predicted secondary structure and also indicated that the binding site is located at the branching point of the three-way junction. Sequence analysis revealed that the sequences of the three base pairs flanking the junction of the three stems are highly conserved among selected clones. The evaluation of the relative binding of several bile acids and structurally related steroids with the aptamer was also carried out. The results revealed a broad range of selectivity and preference for hydrophobic steroids rather than for cholic acid upon binding, indicating that the binding is driven by a hydrophobic interaction. The experimental results reported here allowed us to propose a structural model of a binding site formed by three Watson–Crick base pairs.
INTRODUCTION
DNA and RNA aptamers have been previously selected by in vitro selection for various small molecules including organic dyes (1,2), ATP (3,4), arginine (5,6) and N-methylmesoporphyrin I (7). The sequences of binding to these small target molecules can generally fold into secondary and tertiary structures such as stem–loops (8), pseudo-knots (9) and G-quartets (10). These structural motifs form binding sites specific to the small target molecules (11). Structural studies of these motifs isolated by in vitro selection may provide general information about small molecule–nucleotide interaction.
Recently, we have isolated DNA aptamers that bound to cholic acid, a bile-acid component, from an initial single-stranded DNA (ssDNA) library with ~9 × 1014 independent sequences after 13 rounds of in vitro selection (12). All 19 sequenced clones with a length of 95–100 nt were unique. Subsequently, we defined the regions required for binding to cholic acid in all clones and predicted their secondary structures. A common feature of their sequences was that they consisted of two sets of possible stem–loop regions and a pair of complementary sequences flanking the stem–loop regions at both ends (Table 1a). They bound to cholic acid with Kd values of 5.0–67.5 µM. The sequences and lengths of the binding regions seemed fully variant, but they can be modeled as a secondary structure having a three-way junction, in which each stem generally consists of more than three base pairs and each junction is flanked by two or three G–C base pairs. A three-way junction which can be folded in these 19 clones appeared to dominate as a binding motif in the library throughout 13 rounds of in vitro selection.
Table 1. Sequences of the intact cholic-acid binding regions of 19 sequenced clones (a) and sequences of three ssDNAs that can fold into a three-way junction (b).
(a) The type of junction included in each clone is indicated. (b) Three sets of stem regions are shown in underlined, bold and outlined letters, respectively. Letters in italics denote mismatches or wobble base pairs in stems.
There is considerable interest in the structure and properties of three-way DNA junctions (13–15), primarily in connection with the four-way Holliday junction occurring in genetic recombination (16). Three-way DNA junctions can provide information about the general principle of the folding and stabilization of branched DNA as the simplest DNA junction. They are also known to be involved in certain recombination events (17). In addition to their biological importance and utility as structural models, they can also serve as components in nano-construction (18). Interactions of three-way and four-way DNA junctions with intercalating drugs have been previously reported and applied to probe the structures of junctions (19,20), whereas no small molecules specifically bound to three-way junctions have been found. A characterization of the elements required for the observed binding may provide a new insight into the structure, properties and biological roles of three-way DNA junctions.
In this article, the secondary structure of sequenced clones and the structural requirements for binding to cholic acid were evaluated by a course of mutational analysis. Chemical-modification experiments were done to further evaluate the secondary structure and probe the bases relevant to the interaction. We have also carried out ligand-binding experiments to reveal the binding selectivity of selected aptamers. The experimental results and a molecular-modeling study allowed us to propose a structural model of the binding site.
MATERIALS AND METHODS
Materials
Cholic acid, lithocholic acid, cholesterol and β-estradiol were purchased from Wako Pure Chemical (Osaka, Japan). Chenodeoxycholic acid, 5β-pregnan-3α-ol-20-one, trans-androsterone, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and cholic acid–agarose were obtained from Sigma (St Louis, MO). Digoxigenin hydrate and cholic acid methyl ester were from Aldrich (Milwaukee, WI) and Tokyo Chemical Industry (Tokyo, Japan), respectively. All ssDNAs, including fluorescein isothiocyanate (FITC)-labeled ones, were synthesized by standard phosphoramidite chemistry and purified by reversed-phase high-performance liquid chromatography.
Mutational analysis
Prior to use, all DNA samples were denatured in a binding buffer (50 mM Tris–HCl, 300 mM NaCl, 30 mM KCl, 5 mM MgCl2, pH 7.6) at 95°C for 5 min and then allowed to cool to room temperature for 30 min. The relative binding efficiency of aptamers and their mutants with cholic acid was determined by the equilibrium-filtration method (21). Each DNA sample (50 µM) was incubated with 50 µM of cholic acid in 200 µl of the binding buffer for 5 min at 25°C. The mixture was then placed in a Microcon 10 filtration device (Millipore, Bedford, TX) and centrifuged for 15 min at 850 g. The concentrations of cholic acid in 20–30 µl of filtrate were determined by a diagnostic kit (Wako Pure Chemical) for the quantitative enzymatic determination of total bile acids. For each DNA sample, the concentration of bound cholic acid was determined by the difference between cholic-acid concentration in the filtrate and retentate. The relative binding efficiency of mutant aptamers was reported as a percentage of bound cholic acid normalized to that for wild-type aptamers.
Ligand specificity
The relative elution of 5′-FITC-labeled ch9-48 bound to cholic acid-immobilized agarose matrix (2 µmol cholic acid/g gel) by various competitors was determined to study the specificity of ligand binding. 5′-FITC-labeled ch9-48 (50 pmol) was mixed with the 50 µl of cholic acid–agarose matrix in 200 µl of the binding buffer. After being equilibrated for 15 min, the matrix was washed with binding buffer extensively to remove unbound DNA. Then, 1 ml of the binding buffer containing 50 µM of each competitor compound and 1% methanol were added to the matrix and incubated for 15 min at room temperature. After centrifuging for 5 min at 850 g, 100 µl of supernatants were transferred to a 96-well microtiter plate. The fluorescence intensities of the supernatants were measured using FluorImager 595 (Molecular Dynamics, Sunnyvale, CA) to scan a laser beam (λ = 488 nm) over the sample surface and monitor the resulting fluorescence using a 530 ± 15 nm band-pass filter. The fluorescence images were analyzed using software (ImageQuaNT v4.2a) provided with the FluorImager. The relative ability of different competitors used at 50 µM concentrations to elute 5′-FITC-labeled ch9-48 was expressed as a percentage of cholic-acid effectiveness.
Osmium-tetroxide modification
3′-FITC-labeled ch9-48 and ch9-48-C6 (5 µM) were incubated with 1 mM osmium tetroxide and 3% pyridine in 100 µl of the binding buffer, with or without 5 mM cholic acid, for 15 min at 20°C (70°C for denatured control). The reactions were stopped by two sequential ethanol precipitations. The aptamers were cleaved at the sites of modification by treatment with 15 µl of 1 M piperidine at 90°C for 30 min. After addition of 15 µl of a formamide loading buffer, the cleaved products were run on 19% polyacrylamide gels (19:1 monomer/bis ratio) containing 7 M urea. The buffer system consisted of 90 mM Tris-borate (pH 8.0) and 2 mM EDTA. The gels were scanned and analyzed using FluorImager 595 as described above.
RESULTS
Mutational analysis on ch2-40
To verify the predicted three-way junction structure in 19 clones isolated by the in vitro selection and to define the structural elements important for binding to cholic acid, we performed mutational analysis on ch2-40, a deletion mutant including the intact three-way-junction region in clone 2 (Table 1a). A series of mutants of ch2-40 with single-base substitutions (each base was substituted for its counterpart of the Watson–Crick base pairing) and base-pair compensatory substitutions (each Watson–Crick base pair was substituted for another one with a conserved purine–pyrimidine arrangement) was synthesized, and the affinity of these mutants to cholic acid was evaluated by the equilibrium-filtration method. Several single-base deletions and insertions were also tested.
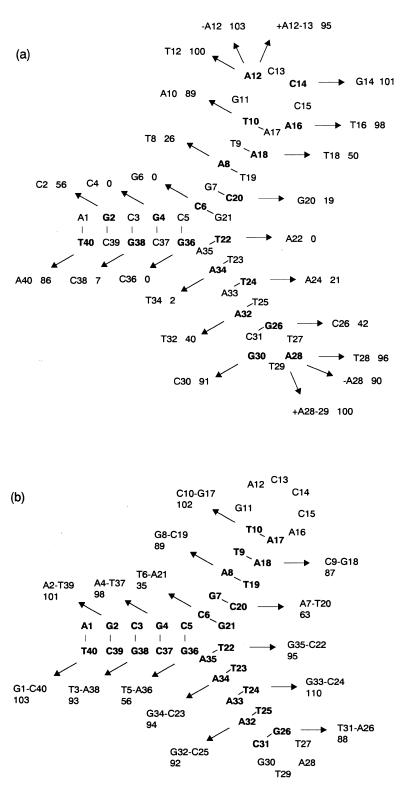
As shown in Figure 1a, several single-base substitutions near the junction of the predicted secondary structure lost binding affinity completely (G4 to C, C6 to G, A22 to T and G36 to C). In contrast, pairwise compensatory substitutions in the possible three stems were tolerated for binding even though changes from the G–C to the A–T base pair near the junction moderately decreased the relative binding affinity (Fig. 1b). With regard to the influence of single-base and pairwise substitutions on the binding affinity, the former, by which Watson–Crick base pairs in the predicted secondary structure were disrupted, reduced binding more significantly than the latter in each base pair. This result provides strong support for the predicted three-way junction structure and indicates that the formation of the structure is crucial for the observed binding. Base pairs at the branching point seemed to play critical roles as compared with others for the binding and would be directly involved in binding or in proper folding of a binding site. Single-base substitutions, deletions and insertions in the two loop regions hardly affected the binding. Thus, two loops may act only as linkers of two sets of complementary sequences included in the three-way junction.
Figure 1.
Mutational analysis on ch2-40. The results are mapped on the predicted secondary structure including a three-way junction. Single-base substitutions, deletions, insertions (a) and base-pair compensatory substitutions (b) were introduced into ch2-40. Replaced nucleotides are shown in bold letters. The arrows indicate introduced nucleotides with the percentage of relative binding efficiency of mutants. –A12 and –A28 denote that A12 and A28 are deleted, respectively. +A12–13, +A28–29 indicate that an adenosine is inserted between A12 and C13, A28 and T29, respectively.
Mutational analysis at the branching point
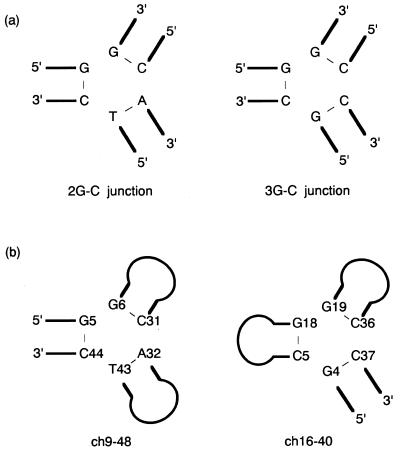
Subsequently, we focused on the sequences of three base pairs flanking the junction of the 19 isolated clones. The junctions were flanked by either three G–C or by two G–C and one A–T base pairs (identified as the 3G–C and the 2G–C junction, respectively). Of the 19 clones selected, 10 clones included the 3G–C junction, while others included the 2G–C junction (Table 1a). Comparing the sequences of the three base pairs flanking the 3G–C junctions, 9 out of 10 clones were identical (Fig. 2a). Six of nine clones with the 2G–C junction also shared a conserved sequence, even though each of the other three clones varied. On the other hand, sequences at more remote positions than the branching point seemed to have no constraints.
Figure 2.
(a) Conserved sequences of three base pairs at the branching point of 2G–C and 3G–C junction. The sequences at the branching point of 2G–C junction were conserved in clones 3, 5, 9, 10, 14 and 17, while those of 3G–C junctions were conserved in clones 1, 4, 6, 8, 11, 12, 13, 15 and 16. (b) Sequences of three base pairs flanking the junctions of ch9-48 and ch16-40. Line drawings denote stem and loop regions.
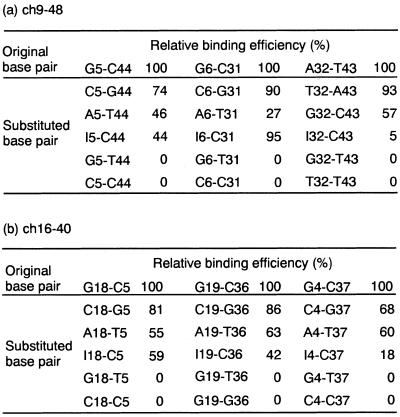
To evaluate the precise structural requirements for binding at the branching point, base-pair substitutions including inosine–cytosine (I–C) base pairs were introduced at the branching point of ch9-48 and ch16-40 (Fig. 2b), and then the binding of these to cholic acid was tested (Table 2). The deletion mutants, ch9-48 and ch16-40, included the 2G–C and the 3G–C junction, respectively. Their interactions with cholic acid had the lowest Kd value in each conserved class (5.0 and 6.4 µM, respectively) (12). With a few exceptions on substitutions of the I–C base pairs, changes to Watson–Crick base pairs were tolerated for binding, whereas all changes to mismatch and wobble base pairs (G–T) eliminated binding completely. Several A–T and G–C base pairs were also substituted for penultimate base pairs from the junctions. Similarly to the pairwise substitutions on ch2-40, none of them lost the binding (data not shown). Consequently, virtually all substitutions of A–T and G–C base pairs near the junctions were tolerated for binding even though the binding affinity was decreased. This result suggests that no bases near the branching point provide base-specific interactions with either cholic acid or other bases and, furthermore, that the binding is distinctively dependent on the formation of the particular secondary structure, the three-way junction. A specific tertiary structure commonly appearing in the three-way junctions may be formed by several nucleotides as a result of the assembly of Watson–Crick base pairs, and the structure must be responsible for the binding. Two conserved sequences at the branching point may offer a more suitable tertiary structure for binding to cholic acid.
Table 2. Mutational analysis on the three base pairs flanking the junction of ch9-48 (a) and ch16-40 (b), respectively.
Relative binding efficiency was evaluated by the equilibrium-filtration method and is indicated for each mutant of ch9-48 and ch16-40.
To further clarify the contribution of the three-way junction to the binding, we prepared three individual ssDNAs (Table 1b, identified as 3S-1, 2 and 3, respectively) forming an intermolecular three-way junction (identified as the 3S junction) (22). Each stem of the 3S junction consists of 10 bp, and the sequences of the three base pairs flanking the junction are consistent with those conserved in the 2G–C junctions. The efficiency of the binding was evaluated by the equilibrium-filtration method described in Materials and Methods. The concentration of cholic acid bound to the 3S junction was 29.5 µM, which was comparable to that for ch2-40 (27.8 µM). The binding ability of the 3S junction was completely lost by the introduction of a mismatch at the branching point (G10 of 3S-1 to C). Both inter- and intra-molecular three-way junctions bound to cholic acid with high affinity, suggesting that the geometry of the global structure around the branching point of the three-way junction in the aptamers is nearly consistent with that of the 3S junction.
Ligand specificity
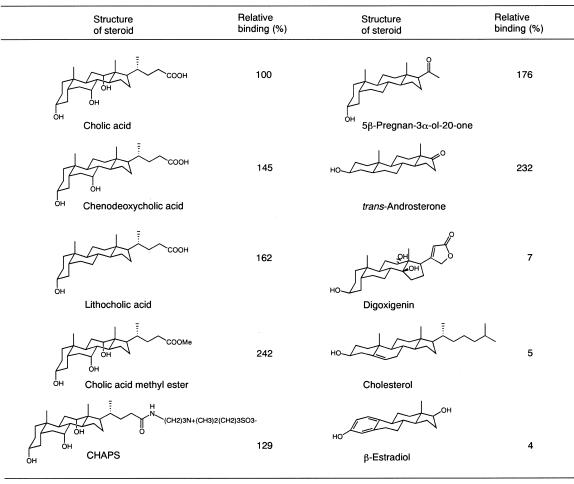
In an effort to study the specificity of ligand–aptamer interaction, the ability of various competitors to elute ch9-48 bound to cholic acid–agarose was observed (Fig. 3). Chenodeoxycholic acid and lithocholic acid bound more tightly to ch9-48 than cholic acid. Moreover, cholic acid methyl ester, which lacks a negative charge at the fatty acid side chain, was more effective as an affinity ligand than other bile acids tested. More hydrophobic bile acids appeared to offer stronger binding. Despite its large polar side chain, CHAPS, a zwitterionic cholic-acid derivative, was also effective. 5β-pregnan-3α-ol-20-one, which has the same steroidal tetracyclic ring with lithocholic acid but lacks the fatty acid side chain, also bound more tightly than cholic acid. These results suggest that the aptamer does not accommodate the side chain of bile acids in the binding site. Surprisingly, trans-androsterone having trans-formed ring A, bound more strongly than cholic acid. On the other hand, neither digoxigenin, including cis-formed ring D and unsaturated γ-lactone at the C-17 position, nor two unsaturated steroids, cholesterol and β-estradiol, were effective. Furthermore, 1-adamantane acetic acid, an alicyclic compound smaller than steroid, and decahydro-2-naphthol, which can be potentially considered as a fragment of cholic acid, did not compete with cholic acid at all (data not shown). The observed structure-affinity relationship suggests that the binding is dominantly governed by the hydrophobic interaction. Steroids appeared to be discriminated by their size and shapes and not by the specific interactions between polar functional groups of steroids and bases.
Figure 3.
Competitive binding analysis with ch9-48. The ability of various steroids to elute 5′-FITC-labeled ch9-48 from cholic acid–agarose was assessed.
Chemical modification
We carried out chemical-modification studies on ch9-48 using oxidizing agent osmium tetroxide to further evaluate the secondary structure of the aptamer and the bases relevant to the interaction. Osmium tetroxide selectively reacted with thymine (T) in the single-stranded state in the presence of pyridine (23). This selectivity is believed to result from the ability of osmium tetroxide to attack C-5, 6 double bond of T out-of-plane when it is unstacked on one side (24). The sites of modification can be cleaved by alkaline treatment. Chemical probing of the structure of three-way DNA junctions formed by three individual ssDNAs revealed that the Ts flanking the junction were highly reactive to osmium tetroxide, while those at more remote positions than the branching point in the three stems were slightly reactive, suggesting that the three stems are not in regular B-form (13).
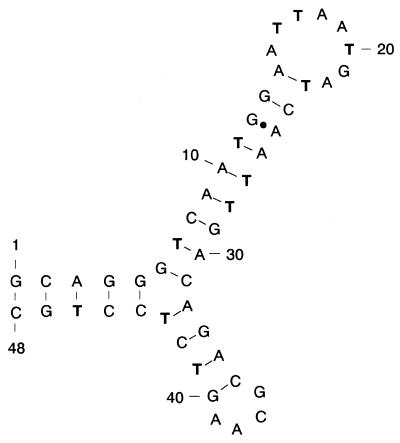
The pattern of modifications on ch9-48 (Fig. 4) under the folding condition (i.e., 20°C and the binding buffer) is highly consistent with the proposed secondary structure (Fig. 5). A strong modification was observed at T43 flanking the junction, while the Ts in the loop regions were also reactive. The Ts in the stem regions were efficiently protected except for T23 and T46: the former should be reactive because it is positioned at the end of the stem region, and the latter might be so because of the potential instability of the short stem lacking the loop. We prepared ch9-48-C6, a mutant of ch9-48, which lacks the ability to bind to cholic acid, by changing G6 to C and compared the modification pattern with ch9-48. In the presence of cholic acid, the degree of modification of T43 at the branching point of ch9-48 was reduced to <40%. In contrast, the reactivity of T43 in ch9-48-C6 was suppressed by cholic acid only slightly. Thus, T43 in ch9-48 may be directly involved in contact with cholic acid. Considering the proposed reaction mechanism of osmium tetroxide (24), cholic acid may sit in the binding site perpendicularly to the planar T43 ring.
Figure 4.
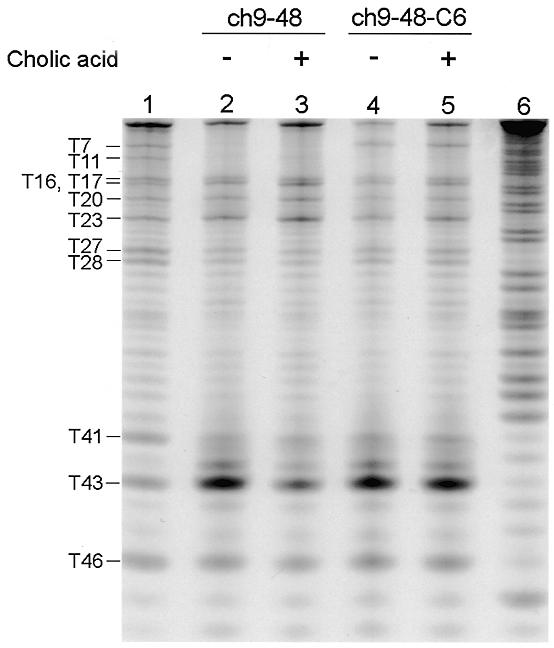
Osmium-tetroxide modification of 3′-FITC-labeled ch9-48 and ch9-48-C6. Nucleotides are numbered from the 5′-end and are indicated at the left. Lane 1, denatured control; lanes 2 and 3, modified ch9-48 in the absence and presence of cholic acid; lanes 4 and 5, modified ch9-48-C6 in the absence and presence of cholic acid; lane 6, Maxam–Gilbert A+G ladder of ch9-48.
Figure 5.
Proposed secondary structure of ch9-48. Originally, the secondary structure was predicted by the MacDNASIS Pro v1.0 program (HITACHI Software Engineering). Several base pairs on a boundary of the stem and loop region were realigned by adopting a G–A wobble base pair so that the stem would extend as long as possible. Thymines are shown in bold letters. The bold dot indicates a wobble base pair.
DISCUSSION
The results of mutational analysis on both ch2-40 and ch9-48 were highly consistent with the predicted secondary structures of the aptamers, in which the three-way junctions are included as essential parts. The three-way junction differs from the secondary and tertiary structural motifs of previously characterized DNA and RNA aptamers in that the binding site consists of only Watson–Crick base pairs. The binding does not require a sequence-specific tertiary structure but proper folding of the secondary structure. Chemical modification using osmium tetroxide provided further evidence for the formation of the three-way junction and also suggests that cholic acid interacts with the three-way junction at the branching point.
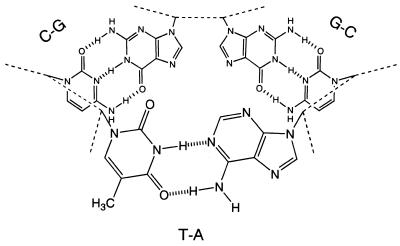
A Corey–Pauling–Koltun (CPK) space filling molecular model of the three base pairs flanking the junction allowed us to address the tertiary structure of the binding site. We found that a cavity lined by three base pairs can be formed at the branching point (Fig. 6). The hyper-reactivity of the Ts at the branching point against osmium tetroxide supports the existence of the cavity. The inner diameter of the cavity is estimated to be ~10–15 Å, which is variable according to the conformation of deoxyribose moieties and phosphodiester bonds. The size of the cavity is likely to be well suited to accommodate a steroidal tetracyclic ring whose width is 12 Å at most, as estimated from CPK models. This model can account for the tolerance of the binding for base-pair changes at the branching point because any combination of base pairs offers the formation of the cavity. It can also account for a broad range of binding selectivity of the aptamers. The cavity could be lipophilic in character, since aromatic bases surround it. Hydrophobic desolvation might provide the major driving force for the binding of various suitably shaped steroids. The conservation of sequences of three base pairs at the branching point can be explained if appropriate shape or increased stability of the cavity is provided.
Figure 6.
Structural model of three base pairs flanking the 2G–C junction. Thin dotted lines denote sugar–phosphate backbones of DNA. A minor groove side of the three-way junction is in the upper part of the figure.
The global structure of the three-way DNA junction formed by three individual ssDNAs has been characterized previously. Stühmeier and co-workers suggested that the global structure had a trigonal pyramidal geometry (22). Therefore, the cavity may have a geometry in which a rim on one side of the cavity is wider than another, although we cannot conclude if it is on a major or a minor groove side of the three-way junction.
The X-ray crystal structures of steroid complexes with antibodies, protein receptors and enzymes revealed that the large apolar steroidal tetracyclic ring is best bound in deep, highly preorganized apolar cavities formed by aromatic amino-acid side chains (25). Their binding mode is very similar to that of the aptamers proposed in this article. Further evaluation of the binding mode utilized by the cholic-acid binding aptamers may enable us to understand the general principle about the mechanism of steroid–macromolecule interactions.
Acknowledgments
ACKNOWLEDGEMENTS
T.K. thanks the Japan Society for the Promotion of Science (JSPS) for a fellowship. This work was supported by research for the future program of JSPS.
REFERENCES
- 1.Ellington A.D. and Szostak,J.W. (1990) Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 2.Ellington A.D. and Szostak,J.W. (1992) Nature, 355, 850–852. [DOI] [PubMed] [Google Scholar]
- 3.Sassanfar M. and Szostak,J.W. (1993) Nature, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- 4.Huizenga D.E. and Szostak,J.W. (1995) Biochemistry, 34, 656–665. [DOI] [PubMed] [Google Scholar]
- 5.Connell G.J., Illangesekare,M. and Yarus,M. (1993) Biochemistry, 32, 5497–5502. [DOI] [PubMed] [Google Scholar]
- 6.Harada K. and Frankel,A.D. (1995) EMBO J., 14, 5798–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Geyer,C.R. and Sen,D. (1996) Biochemistry, 35, 6911–6922. [DOI] [PubMed] [Google Scholar]
- 8.Cho J., Hamasaki,K. and Rando,R.R. (1998) Biochemistry, 37, 4985–4992. [DOI] [PubMed] [Google Scholar]
- 9.Wilson C., Nix,J. and Szostak,J. (1998) Biochemistry, 37, 14410–14419. [DOI] [PubMed] [Google Scholar]
- 10.Wilson C. and Szostak,J.W. (1998) Chem. Biol., 5, 609–617. [DOI] [PubMed] [Google Scholar]
- 11.Hsiung C. and Patel,D.J. (1996) Nat. Struct. Biol., 3, 1046–1050. [DOI] [PubMed] [Google Scholar]
- 12.Kato T., Takemura,T., Yano,K., Ikebukuro,K. and Karube,I. (2000) Biochim. Biophys. Acta, in press. [DOI] [PubMed] [Google Scholar]
- 13.Duckett D. and Lilley,D.M.J. (1990) EMBO J., 9, 1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q., Lu,M., Churchill,M.E.A. and Kallenbach,N.R. (1990) Biochemistry, 29, 10927–10934. [DOI] [PubMed] [Google Scholar]
- 15.Lu M., Guo,Q. and Kallenbach,N.R. (1991) Biochemistry, 30, 5815–5820. [DOI] [PubMed] [Google Scholar]
- 16.Orr-Weaver T.L., Szostak,J.W. and Rothstein,R.J. (1981) Proc. Natl Acad. Sci. USA, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minagawa T., Murakami,A., Ryo,Y. and Yamagishi,H. (1983) Virology, 126, 183–193. [DOI] [PubMed] [Google Scholar]
- 18.Seeman N.C. (1998) Angew. Chem. Int. Ed., 37, 3220–3238. [DOI] [PubMed] [Google Scholar]
- 19.Guo Q., Seeman,N.C. and Kallenbach,N.R. (1989) Biochemistry, 28, 2355–2359. [DOI] [PubMed] [Google Scholar]
- 20.Guo Q., Seeman,N.C. and Kallenbach,N.R. (1990) Biochemistry, 29, 3407–3412. [DOI] [PubMed] [Google Scholar]
- 21.Jenison R.D., Gill,S.C., Pardi,A. and Polisky,B. (1994) Science, 263, 1425–1429. [DOI] [PubMed] [Google Scholar]
- 22.Stühmeier F., Welch,J.B., Murchie,A.I.H., Lilley,D.M.J. and Clegg,R.M. (1997) Biochemistry, 36, 13530–13538. [DOI] [PubMed] [Google Scholar]
- 23.Furlong J.C., Sullivan,K.M., Murchie,A.I.H., Gough,G.W. and Lilley,D.M.J. (1989) Biochemistry, 28, 2009–2017. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen P.E. (1990) J. Mol. Recog., 3, 1–25. [DOI] [PubMed] [Google Scholar]
- 25.Wallimann P., Marti,T., Fürer,A. and Diederich,F. (1997) Chem. Rev., 97, 1567–1608. [DOI] [PubMed] [Google Scholar]