Abstract
Mutations in Fms-like tyrosine kinase 3 (FLT3) are common drivers in acute myeloid leukemia (AML) yet FLT3 inhibitors only provide modest clinical benefit. Prior work has shown that inhibitors of lysine-specific demethylase 1 (LSD1) enhance kinase inhibitor activity in AML. Here we show that combined LSD1 and FLT3 inhibition induces synergistic cell death in FLT3-mutant AML. Multi-omic profiling revealed that the drug combination disrupts STAT5, LSD1, and GFI1 binding at the MYC blood super-enhancer, suppressing super-enhancer accessibility as well as MYC expression and activity. The drug combination simultaneously results in the accumulation of repressive H3K9me1 methylation, an LSD1 substrate, at MYC target genes. We validated these findings in 72 primary AML samples with the nearly every sample demonstrating synergistic responses to the drug combination. Collectively, these studies reveal how epigenetic therapies augment the activity of kinase inhibitors in FLT3-ITD AML.
Keywords: combination therapy, epigenetic inhibitors, mechanism of synergy, MYC super-enhancer, FLT3-ITD acute myeloid leukemia
INTRODUCTION
Mutations in Fms-like tyrosine kinase 3 (FLT3) occur in nearly a third of all patients with acute myeloid leukemia (AML) and are associated with an inferior overall survival (1). The most frequent mutation in FLT3 is the internal tandem duplication (ITD) of the juxta-membrane domain (2). While small molecule inhibitors of FLT3 kinase produce higher overall response rates and improved survival compared to salvage chemotherapy in patients with relapsed or refractory FLT3-ITD positive AML, FLT3 inhibitor monotherapy is rarely curative and responses are short-lived (3–5). There is a clinical need for approaches to deepen the initial response to FLT3 inhibitors, enabling longer-lasting clinical responses.
An approach to improving responses to FLT3 inhibitors in AML is to simultaneously target aberrant FLT3 activity and its downstream mediators. A major driver of mutant-FLT3-dependent oncogenesis is the MYC proto-oncoprotein (6–8) (6,7). MYC, a critical regulator of proliferation and differentiation, is over-expressed in the vast majority of patients with AML (8). Reactivation of MYC-controlled oncogenic networks by the bone marrow microenvironment promotes FLT3 inhibitor resistance (8,9). These findings suggest that improved responses to FLT3 inhibitors may be achieved with combination strategies that target MYC-dependent proliferative programs.
Direct inhibition of MYC has been an objective of anti-cancer therapeutic development for over the last twenty years. However, MYC has been considered undruggable due to its intrinsically disordered nature and lack of enzymatic activity (10). Another approach is to instead disrupt the molecular mechanisms that drive MYC over-expression. In blood cells, MYC expression is regulated by a blood-specific super-enhancer complex (BENC), which is bound by numerous transcription factors and global chromatin activators (11,12). Recent studies in AML cell lines have demonstrated that small molecule inhibitors targeting these activating chromatin complexes, including BRD4, resulted in a loss of MYC expression and leukemia cell death (13–16). However, initial clinical trials have only shown modest clinical activity and substantial toxicity (17).
An alternate approach is to simultaneously target two factors that regulate MYC gene expression. The chromatin regulatory protein lysine specific demethylase 1 (LSD1) is a well-established regulator of MYC gene expression (18–20). LSD1 regulates gene expression by removing activating methylation marks on lysine 4 of histone 3 (H3K4) and repressive methylation marks on lysine 9 of histone 3 (H3K9) or by recruiting repressive complexes to gene promoters (21,22). Inhibitors of LSD1 have been shown to decrease MYC abundance and activity in AML cell lines and primary samples (18–20). Our prior work and that of other groups shows that LSD1 inhibition augments the efficacy of kinase inhibitors in AML (19,20,23). However, the extent to which synergy exists between LSD1 and FLT3 inhibition and the underlying mechanism of drug synergy has not been investigated.
Here we report ex vivo drug screening data on a cell line model of FLT3-ITD positive AML and primary FLT3-ITD positive AML samples demonstrating that LSD1 inhibition potentiates the efficacy of FLT3 inhibition. Using high-sensitivity epigenetic profiling, we establish that dual FLT3/LSD1 inhibition disrupts regulatory factor binding at the MYC BENC, resulting in a loss of MYC expression. Using short-term ex vivo culture, we confirm that these transcriptional and epigenetic responses to combined FLT3/LSD1 inhibition occur in primary FLT3-ITD positive leukemic blasts. Collectively, this data reveals how epigenetic therapies augment the activity of kinase inhibitors in FLT3-ITD AML.
MATERIALS AND METHODS
Cell and Patient Sample Culture
Cell Lines
MOLM13 cells (DSMZ) were cultured in RPMI (Gibco) supplemented with 10% fetal bovine serum (FBS, HyClone), 2 mM GlutaMAX (Gibco), 100 units/mL Penicillin, and 100 μg/mL Streptomycin (Gibco). MV4;11 and K562 cells (ATCC) were cultured in IMDM supplemented with 20% fetal bovine serum (FBS, HyClone), 2 mM GlutaMAX (Gibco), 100 units/mL Penicillin, and 100 μg/mL Streptomycin (Gibco). All cells were cultured at 5% CO2 and 37°C. Cell lines were tested for mycoplasma concentration at the time of freezing as well as monthly for any cell lines in culture. Cell lines were authenticated by the OHSU Cell Line Authentication service, which uses Promega’s GenePrint 10 system to confirm the identity of human cell lines by short tandem repeat analysis. All cell clines were maintained below fifteen passages.
Patient Samples
All patients gave written informed consent to participate in this study, which was conducted in accordance with the Declaration of Helsinki and had the approval and guidance of the institutional review boards at Oregon Health & Science University (OHSU), University of Utah, University of Texas Medical Center (UT Southwestern), Stanford University, University of Miami, University of Colorado, University of Florida, National Institutes of Health (NIH), Fox Chase Cancer Center and University of Kansas. Samples were sent to the coordinating center (OHSU; IRB#9570; #4422; NCT01728402) where they were coded and processed. Mononuclear cells were isolated by Ficoll gradient centrifugation from freshly obtained bone marrow aspirates or peripheral blood draws. Clinical, prognostic, genetic, cytogenetic, and pathologic lab values as well as treatment and outcome data were manually curated from patient electronic medical records. Genetic characterization of the leukemia samples included results of a clinical deep-sequencing panel of genes commonly mutated in hematologic malignancies (Sequenome and GeneTrails [OHSU]; Foundation Medicine [UT Southwestern]; Genoptix; and Illumina). Patient samples were cultured in RPMI with 10% FBS and 10% HS-5 conditioned media (ATCC) or SFEMII supplemented with 1x StemSpan CD34+ Expansion Media and 1 μM UM729 (StemCell Technologies).
Colony Assay
Whole bone marrow was obtained (AllCells) and CD34+ cells were selected using CD34 MicroBead Kit (Miltenyi Biotec) according to manufacturer’s instructions. For the colony assay, 500 CD34+ cells were used per replicate and plated in MethoCult™ H4435 Enriched (StemCell Technologies). The four groups were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or DMSO. Plates were incubated for 14 days in 5% CO2 and 37°C. Samples were imaged using STEMvision (StemCell Technologies) and blinded prior to counting by another investigator by assigning letters randomly. ImageJ (NIH) was used to count colonies after blinding.
Drug Synergy
Drug synergy was assessed using an 8 × 8 matrix of drug concentrations. Cells were treated for 72 hours prior to MTS assay to evaluate viability. Cell viability was used to calculate drug synergy with SynergyFinder based on the ZIP reference model (24).
RNA Interference
Two SMARTvector Inducible short hairpin RNAs (shRNAs) for Human SPI1 (V3IHSHER_10431275, V3IHSHER_10642739), two for STAT5A (V3IHSHEG_6691183, V3IHSHEG_4988581) two for STAT5B (V3IHSHER_4778243, V3IHSHER_6411380) and two for GFI1 (V3IHSHER_5266412 and V3IHSHER_5697821) in a hEF1a-TurboRFP or hEF1a-TurboGFP (STAT5A) backbone were obtained from Horizon Discovery. Both SPI1 shRNA constructs showed effective knockdown of SPI1. STAT5A V3IHSHEG_6691183 was specific for STAT5A, STAT5A V3IHSHEG_4988581 was ineffective against either STAT5A or STAT5B, STAT5B V3IHSHER_6411380 was selective for STAT5B, and V3IHSHER_4778243 knocked down both STAT5A and STAT5B. GFI1 V3IHSHER_5266412 produced effective GFI1 knockdown while V3IHSHER_5697821 was ineffective. Lentivirus was produced by transfecting Lenti-X 293T cells (Clontech) with FuGENE (Promega #E2311) and Opti-MEM (ThermoFisher #31985062) as well as the SMARTvector transfer plasmid and packaging/pseudotyping plasmids. psPAX2 was a gift from Didier Trono (Addgene plasmid #12260) and pMD2.G was a gift from Didier Trono (Addgene plasmid #12259). The supernatants containing lentivirus was collected after 48 hours of culture and filtered with a 0.45 um filter. MOLM13 cells were transduced with virus via spinnoculation in the presence of polybrene. Transduced cells were selected with 1 μg/mL puromycin to produce a stable cell line.
MYC Over-expression
For human MYC over-expression, pDONR223_MYC_WT was a gift from Jesse Boehm & Matthew Meyerson & David Root (Addgene plasmid #82927) and cloned into pCW57.1, a gift from David Root (Addgene plasmid #41393). Lentiviral particles were generated as above and MOLM13 cells were selected after viral transduction with 1 μg/mL puromycin. After selection, cells were treated with 1 μg/mL doxycycline for 48 hours prior to experiments.
Stat5a1*6 Over-expression
For Stat5a1*6 over-expression, pMXs-IRES-Puro (pMX Empty) was acquired from Cell Biolabs Inc. (#RTV-014) and pBABE-Stat5a1*6 was acquired from Addgene (#130668). Retroviral particles were generated by transfecting 293T17 cells (ATCC Number #CRL-3216) with FuGENE (Promega #E2311) and Opti-MEM (ThermoFisher #31985062) as well as pCMB-VSV-G (Addgene plasmid #8454), pUMCV (Addgene plasmid #8449), and the appropriate transfer plasmid. The supernatants containing retrovirus was collected after 48 hours of culture and filtered with a 0.45 um filter. MOLM13 cells were transduced with virus via spinnoculation in the presence of polybrene. Transduced cells were selected with 1 μg/mL puromycin to produce a stable cell line.
LSC FACS
CD34+ cells were isolated from patient samples using CD34 MicroBead Kit (Miltenyi Biotec) according to manufacturer’s instructions. Cells were subsequently stained with Calcein Violet (BioLegend #425203) as well as the following antibodies: CD45RA FITC (BD Pharmigen #55488), IL1-RAP PE (R&D Systems #FAB676P), CD123 PE (BD Pharmigen #554529), CD38 APC (BD Pharmigen #555462), and CD34 APC-Cy7 (BioLegend #343513). Stained cells were analyzed and sorted into cytokine-enriched methylcellulose (MethoCult StemCell #H4435) using a Sony SH800S. 40,000 cells were sorted from each patient sample per replicate (n=3) per drug condition (n=2). The methylcellulose was treated with 500 nM quizartinib and 500 nM GSK-2879552 or an equivalent volume of DMSO. Plates were incubated for 10 days in 5% CO2 and 37°C. Samples were imaged using STEMvision (StemCell Technologies) and blinded prior to counting by another investigator by assigning letters randomly. FIJI (NIH) was used to count colonies after blinding.
Sequencing Methods
Bulk RNA-Seq
MOLM13 cells were treated with 1 nM quizartinib, 100 nM GSK-2879552, the combination, or equal volume of DMSO for 24h. Total RNA was isolated using a RNeasy Plus Mini Kit (Qiagen). BGI performed the library preparation and sequencing with 50 base pair (bp) single-end (SE) sequencing. Patient samples were cultured in 10% HS-5 CM/RPMI with 20% FBS and treated with 500 nM quizartinib, 500 nM GSK-2879552, the combination, or the equivalent volume of DMSO for 24 hours. Total RNA was isolated with RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions. Libraries were prepared using the NEBNext Low Input RNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s instructions. Libraries were sequenced by the OHSU Massively Parallel Sequencing Shared Resource (MPSSR) using 100 bp SE sequencing on an Ilumina NovaSeq S1 flow cell.
Bulk ATAC-Seq
MOLM13 cells were treated with 1 nM quizartinib, 100 nM GSK-2879552, the combination, or an equivalent volume of DMSO for 24 hours. After treatment, 50,000 cells per replicate were harvested for Fast-ATAC sequencing performed as previously described (25). In brief, cells were resuspended in cold PBS and tagmentation master mix (25 μL of 2x tagmentation buffer, 2.5 μL of TDE1 [Illumina], 0.5 μL of 1% digitonin; 2x tagmentation buffer: 66 mM Tris-Acetate, pH 7.8, 132 mM potassium acetate, 20 mM magnesium acetate, 32% v/v N,N-Dimethylformamide) was added. Samples were incubated at 37°C for 30 minutes. DNA was purified using Zymo Clean and Concentrator 5 Kit (Zymo). Transposed DNA was amplified and purified as described previously with adapted primers (26,27). Samples were quantified using Qubit dsDNA HS Assay Kit (Invitrogen), pooled, and sequenced by BGI using 50 bp paired-end (PE) sequencing.
CUT&Tag
MOLM13 cells were treated with 1 nM quizartinib, 100 nM GSK-2979552, the combination, or an equal volume of DMSO for 2 or 6 hours. Benchtop CUT&Tag was performed as previously described (28). In brief, cells were counted, harvested, and centrifuged for 5 min at 300xg at room temperature. Cells were washed 2X in 1.5 mL wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1x Protease inhibitor cocktail). Concanavalin A magnetic coated beads (Bangs Laboratories) were activated in binding buffer by washing 2X (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2). Washed cells were separated into 100,000 cell aliquots and 10 μL of activated beads were added to each sample. Samples were rotated end-over-end for 7 minutes at room temperature. A magnetic stand was used to separate beads and the supernatant was removed. Primary antibody was diluted 1:50 in antibody buffer (20 mM HEPES pH 7.5, 150mM NaCl, 0.5 mM Spermidine, 1× Protease inhibitor cocktail, 0.05% digitonin, 2 mM EDTA, 0.1% BSA). The following antibodies were diluted 1:100 in dig-300 buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM Spermidine, 1× Protease inhibitor cocktail, 0.01% digitonin) and added to samples as previously described: H3K27ac (Abcam #ab4729), H3K4me1 (CST #5326), H3K4me3 (CST #9751), RBP1 (CST #2629), H3K9me1 (Diagenode #C15410065), H3K9ac (Diagenode #C15410004), CEBPA (CST #8178), and Normal Rabbit IgG (CST #2729) (28). Samples incubated for 1 hour at room temperature on nutator. Samples were washed 2X with dig-300 buffer then resuspended in tagmentation buffer (dig-300 buffer with 1 mM MgCl2). Samples were incubated at 37°C for 1 hour. DNA was extracted with phenol:chloroform extraction. Samples were amplified by PCR using custom Nextera primers at 400 nM and NEBNext (29). PCR conditions were set to: 72°C for 5 minutes, 98°C for 30 seconds, 14 cycles of 98°C for 10 sec, 63°C for 10 sec, and 72°C for 1 minute. Libraries were purified with AMPure Beads (Beckman) and sequenced by the OHSU MPSSR on an Ilumina NovaSeq using 50 bp SE sequencing or NextSeq 500 using 37 bp PE sequencing.
CUT&RUN
MOLM13 cells were treated with 1 nM quizartinib, 100 nM GSK-2979552, the combination, or an equal volume of DMSO 24 hours. CUT&RUN was performed as previously described (30). Briefly, concanavalin A magnetic coated beads (Bangs Laboratories) were washed 2x in binding buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2). 500,000 cells per replicate were washed 2x with wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1x Protease inhibitor cocktail). Cells were bound to beads by nutating for 10 minutes at room temperature. Cells were permeabilized and incubated overnight at 4°C on nutator with primary antibody in antibody buffer (wash buffer, 0.001% digitonin, 3 mM EDTA). The following antibodies were used at 1:50 PU.1 (Invitrogen #MA5–15064), GFI1 (Abcam #ab21061), and normal rabbit IgG (CST #2729). Bead slurry was washed 2x with dig wash buffer (wash buffer, 0.001% dig) and resuspended with dig wash buffer and 1x pAG-MNase (Epicypher). Cell were incubated for 10 minutes on nutator at room temperature then washed 2x with dig wash buffer followed by resuspension in pAG-MNase reaction mix (dig wash buffer, 2 mM CaCl2). Bead slurry was incubated for 2 hours at 4°C on nutator. STOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 50 μg/mL RNase A, 50 μg/mL glycogen, 0.02% dig) was then added, then tubes were incubated at 37°C for 10 minutes. DNA was extracted using phenol:cholorform extraction. Libraries were prepared using NEBNext Ultra II DNA Library Prep Kit (NEB), modified for CUT&RUN as previously described (31). After adapter ligation fragments were cleaned up with 1.75x AMPure beads (Beckman). Following PCR amplification, libraries were purified 2x with 1.2x AMPure beads to rid of adaptor fragments. Libraries were quantified on the 2100 Bioanalyzer instrument (Agilent) with the High Sensitivity DNA Analysis Kit (Agilent). Libraries were pooled and sequenced by MPSSR on a NextSeq 500 sequencer (Illumina) using 37 bp PE sequencing.
ChIP-Seq
ChIP-seq was performed using the SimpleChIP plus Enzymatic Chromatin IP Kit (Cell Signaling Technology). For each replicate, 20 million cells were fixed in 4% formaldehyde (Sigma-Aldrich) for 10 minutes at room temperature then quenched with glycine, washed and stored at −80°C until use. Nuclei were extracted according to the manufactures instructions and treated with 1.25 μL MNase in 500 μL Buffer B at 37°C for 20 minutes. Samples were sonicated on a Qsonic sonicator at 50% amplitude for 5 cycles of 15 sec on 15 sec off on ice. Crosslinks were reversed on a small aliquot of extracted chromatin quantified by OD260. A total of 5 μg of chromatin was used for each immunoprecipitation. The following antibodies were used: LSD1 (Abcam #ab17721), MYC (CST #13987), RUNX1 (Abcam #ab23980), STAT5 (CST #94205S) and rabbit IgG (CST #2729). After overnight incubation, complexes were captured using protein G beads. Crosslinks were reversed and libraries prepped using an NEBNex Ultra II for DNA Library Prep kit. Libraries were sequenced by the OHSU MPSSR. The STAT5 libraries were sequenced by Genewiz using a HiSeqX and 150 bp PE sequencing.
Reverse Phase Protein Array (RPPA)
MOLM13 cells were treated for 24 hours with 1 nM quizartinib, 100 nM GSK-2979552, the combination. Cells were washed 2x in PBS then flash frozen. Cell pellets were lysed and processed by the University of Texas MD Anderson Cancer Center Functional Proteomics RPPA Core Facility.
Single Cell ATAC-Seq
Patient samples were treated with 500 nM quizartinib and 500 nM GSK-2879552 or an equal volume of DMSO for 24 hours. Nuclei were prepared using the demonstrated protocol for primary cell nuclei extraction from 10x Genomics. ATAC libraries were prepared using Chromium Single Cell ATAC Library and Gel Bead kit v1.1 (10x Genomics, 1000176). Libraries were sequenced with 50 bp PE sequencing by the OHSU MPSSR.
Primary AML Blast Dataset
Gene mutation, drug response, and gene expression data from primary AML blasts was accessed through the BeatAML database (1). Samples from collected from patients who were in remission or had a history of myelodysplastic syndrome at the time of collection were excluded from downstream analysis. Samples with quizartinib/GSK-2979552 single and dual agent drug response data were selected and stratified by FLT3-ITD mutation status. Gene expression dataset was downloaded in the form of reads per kilobase of exon per million reads mapped (RPKM).
Quantification and Statistical Analysis
Values are represented as the mean and error bars are the SEM unless otherwise stated. Prism software (version 9.1; Prism Software Corp.) or R was used to perform statistical analyses. Significance was tested using Student’s t-test or two-way ANOVA followed by Holm-Šidák post-test correction unless otherwise stated. For differential analysis of RNA-seq, CUT&Tag, CUT&RUN, and ATAC-seq p-values were adjusted for repeated testing using the Benjamini-Hochberg method.
Data Availability
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE190785.
For more information, please see Supplementary Materials & Methods.
RESULTS
Combined FLT3/LSD1 inhibition synergistically represses MYC transcriptional programs, while activating PU.1 transcriptional programs
Prior work from our lab and others suggest that the combination of kinase and LSD1 inhibition may be an effective therapeutic strategy in multiple molecular subtypes of AML (18–20,23). To establish whether this approach is effective for FLT3-ITD AML, we treated FLT3-ITD-positive (MOLM13 and MV4;11) and FLT3-ITD-negative (K562) cell lines with multiple FLT3/LSD1 inhibitor combinations. We observed potent synergy between the FLT3 inhibitors and LSD1 inhibitors in the FLT3-ITD-positive cell lines but not in the FLT3-ITD-negative cell lines, suggesting that this drug combination has specificity for FLT3-ITD-positive AML (Fig. 1A–B; Supplementary Fig. 1). In addition, we observed that the drug combination increased both early (Annexin V+/PI−) and late (Annexin V+/PI+) apoptosis populations with minimal toxicity to healthy CD34+ cells (Supplementary Fig. 2A–C). These results indicate that synergy exists between FLT3/LSD1 inhibition in FLT3-ITD-positive AML.
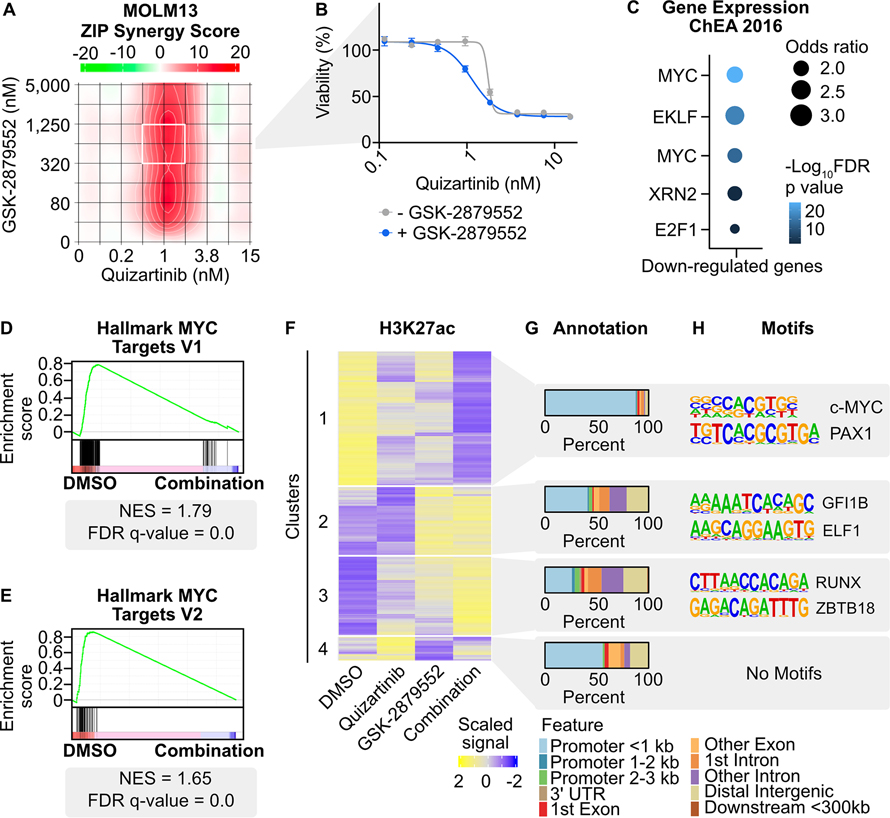
Fig. 1: Transcriptional and chromatin dynamics in response to combined FLT3/LSD1 inhibition in FLT3-ITD-positive AML.
A, MOLM13 cells were treated in triplicate with an 8×8 dose matrix of quizartinib and GSK-2879552 for 72 hours prior to viability assessment by CellTiter Aqueous colorimetric assay. Zero interaction potency (ZIP) synergy scores were calculated on the average values for each drug dose. The white box indicates the quizartinib and GSK-2879552 concentrations corresponding to maximal synergy. B, Quizartinib response curves with and without GSK-2879552 (638 nM, which is the concentration corresponding to maximal synergy in (A)). The GSK-2879552 response curve with and without quizartinib is shown in Supplementary Fig. 1. C, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO vehicle for 24 hours prior to RNA sequencing. Analysis was performed on genes with decreased expression with the drug combination relative to DMSO. D, E, GSEA was performed comparing the drug combination to DMSO. F, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO vehicle for 2 hours prior to CUT&Tag for H3K27ac. Unsupervised hierarchical clustering of regions with differential signal following drug treatment. G, Annotation of regions in clusters from (F). H, Motif enrichment of regions with differential H3K27ac signal. Top two de novo motifs with p-value <10−12 are shown.
To understand the mechanism of synergy, we performed RNA-seq on MOLM13 cells treated with quizartinib, GSK-2879552, or the combination for 24 hours. The majority of the 1,716 differentially expressed genes (80%; 1,374 of 1,716 genes) following the drug combination were not differentially expressed following single agent therapies (Supplementary Fig. 2D). Unsupervised clustering of differentially expressed genes revealed clusters of genes that were either up-regulated or down-regulated by the drug combination (Supplementary Fig. 2E; Supplementary Table 1). Transcription factor target gene enrichment analysis of the down-regulated genes revealed an enrichment of MYC target genes (Fig. 1C; Supplementary Fig. 2F; Supplementary Table 2). To understand the potential impact of these down-regulated genes on cell viability, we investigated essential genes nominated from genome-wide CRISPR dropout screens performed in MOLM13 cells (32). There were 74 genes, including MYC, that were differentially down-regulated by the drug combination and identified as depleting genes (Supplementary Fig. 2G). Notably, these genes were also enriched for MYC target genes (Supplementary Fig. 2H; Supplementary Table 3). In contrast, the genes up-regulated by the drug combination were enriched for SPI1/PU.1 targets. The SPI1 gene encodes the transcription factor PU.1, which is critical for coordinating myeloid differentiation (33). To further corroborate these gene expression profiles, gene set enrichment analysis (GSEA) was performed, which revealed depletion of MYC target genes in cells treated with the drug combination (Fig. 1D and E; Supplementary Table 4). Collectively, we observed that the combination activates an anti-proliferative and pro-differentiative transcriptional program with repression of MYC target genes and activation of PU.1 target genes.
Combined FLT3/LSD1 inhibition disrupts chromatin dynamics at distinct genomic loci
A key component of LSD1 inhibitor activity has been ascribed to displacement of GFI1/CoREST from chromatin and re-activation of enhancers associated with differentiation (34). Therefore, to characterize the early chromatin dynamics following combined FLT3/LSD1 inhibition, we utilized cleavage under targets and tagmentation (CUT&Tag) (35). We used CUT&Tag to assess changes in acetylation of histone 3 lysine 27 (H3K27ac), a marker of transcriptional activation, in MOLM13 cells 2 hours following drug treatment. Unsupervised clustering of the regions with differential H3K27ac signal revealed four clusters (Fig. 1F; Supplementary Table 5). The regions in cluster 1 were associated with repressed H3K27ac signal by the drug combination and were primarily localized to promoters (Fig. 1G). The cluster 1 regions were also enriched for MYC motifs, consistent with the findings of decreased gene expression of MYC target genes (Fig. 1H). An example of a down-regulated region is observed at the PVT1 promoter, a known regulator of MYC expression (Supplementary Fig. 2I) (36). Cluster 2 contained regions with increased H3K27ac signal largely driven by LSD1 inhibition. Cluster 2 regions were nearly equally distributed at promoter and non-promoter elements and were enriched for GFI1/GFI1B motifs. Cluster 3 regions were also localized at promoter and non-promoter elements and showed enrichment for RUNX motifs. RUNX1 is a critical regulator of myeloid differentiation and potentiates the transcriptional activation activity of PU.1 (37). An example of up-regulated non-promoter regions from clusters 2 and 3 were observed upstream of the lysozyme promoter, which is expressed in mature granulocytes (Supplementary Fig. 2J). These data collectively show that the drug combination alters the chromatin landscape at distinct genomic loci. Furthermore, based on pathway analysis and motif enrichment, MYC, GFI1, RUNX1, and PU.1 transcription factors are candidate regulators of these chromatin dynamics.
Chromatin segmentation reveals that MYC-, STAT5-, and PU.1-driven molecular programs underlie the response to combined FLT3/LSD1 inhibition
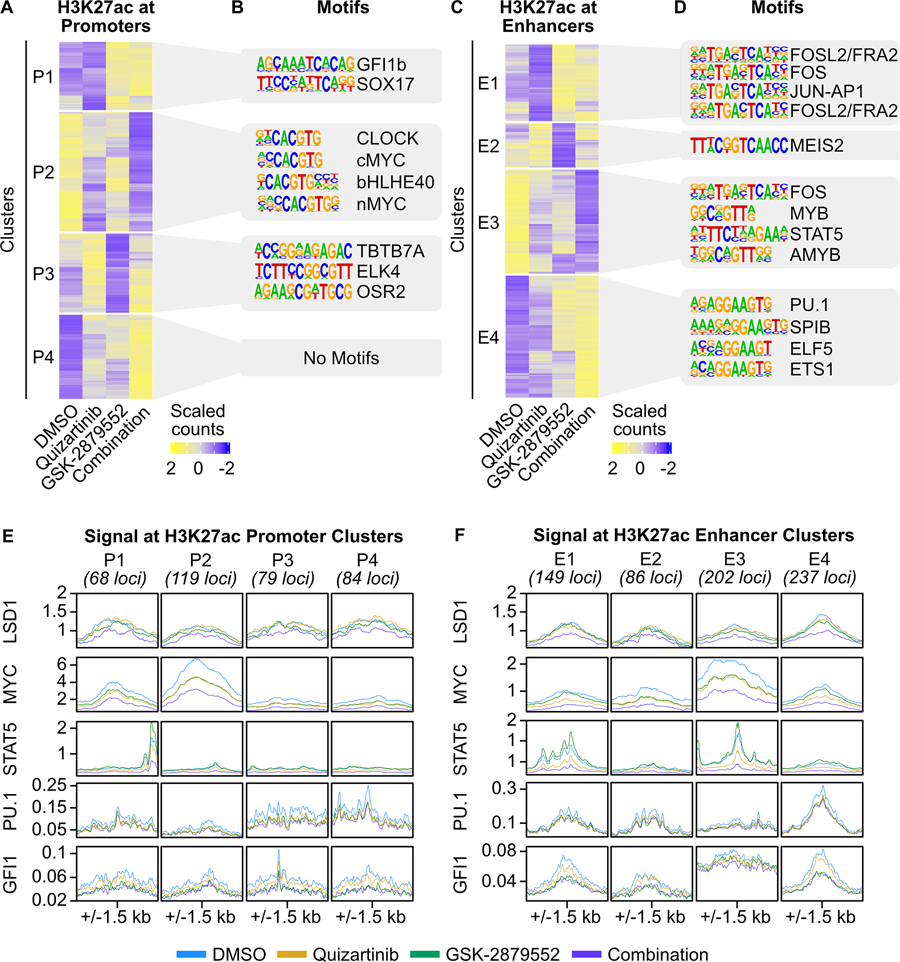
Our CUT&Tag results revealed substantial changes in histone acetylation at both promoters and outside promoters in response to combined FLT3/LSD1 inhibition, arguing that both types of regulatory elements have distinct roles in the drug response. We therefore profiled a series of covalent histone marks in MOLM13 cells 6 hours following drug treatment. These marks enabled the segmentation of chromatin into promoters and enhancers. Trimethylation of histone 3 lysine 4 (H3K4me3) is primarily localized at promoters, whereas monomethylation of histone 3 lysine 4 (H3K4me1) is predominantly at enhancers (38). We also profiled H3K27ac at this same time point to understand transcriptional activation at prompters and enhancers (35). Following LSD1 inhibition, we observed regions of increased H3K4me3 and H3K4me1 signal consistent with the known demethylase activity of LSD1 for H3K4me1/3 (Supplementary Fig. 3A–D) (34,39). Unsupervised clustering of regions with differential H3K27ac signal at promoters and enhancers revealed multiple patterns of regulation (Supplementary Table 6). Similar to the global 2-hour acetylation CUT&Tag data, we observed a large cluster of repressed H3K27ac signal at promoters (cluster P2) that were enriched for MYC motifs as well as a cluster of increased H3K27ac signal (cluster P1) enriched for GFI1/GFI1B motifs (Fig. 2A and B). GO analysis revealed that cluster P2 regions are associated with cell cycle and proliferation (Supplementary Fig. 3E; Supplementary Table 7). At enhancers, we identified a cluster of suppressed H3K27ac signal (cluster E3) associated with STAT5 motifs along with a cluster of increased H3K27ac signal (cluster E4) enriched for SPI1/PU.1 motifs (Fig. 2C and D; Supplementary Fig. 3F). This data suggests that combined FLT3/LSD1 inhibition simultaneously activates GFI1/GFI1B-bound promoters and represses the activation of MYC-bound promoters. In parallel, the drug combination suppresses STAT5-bound enhancers and activates PU.1-bound enhancers.
Fig. 2: Discrete components of the response to FLT3/LSD1 inhibition are mediated by promoters and enhancers.
A, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO vehicle for 6 hours prior to CUT&Tag for H3K27ac, H3K4me1, and H3K4me3. On the basis of these marks, chromatin was segmented into promoters and enhancers. Unsupervised hierarchical clustering of differential H3K27ac signal at promoters. B, Motif enrichment of promoters with differential H3K27ac signal. Top four de novo motifs with p-value <10−12 are shown. C, D, Same analyses as (A) and (B) were performed at enhancers. E, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO for 6 hours. LSD1, MYC, and STAT5 binding was assessed by ChIP-seq. PU.1 and GFI1 binding was assessed by CUT&RUN. Transcription factor binding profiles at promoters with differential H3K27ac identified in (A). F, Transcription factor profiles at enhancers with differential H3K27ac identified in (C).
To better characterize the transcription factors driving the response to combined FLT3/LSD1 inhibition, we profiled the genome-wide binding of multiple candidate transcription factors following single or dual drug treatment using chromatin immunoprecipitation sequencing (ChIP-seq), cleavage under targets and release using nuclease (CUT&RUN), and CUT&Tag. We then examined the signal of these factors at each cluster, identifying promoters and enhancers with differential H3K27ac signal following drug treatment (Fig. 2E and F). We observed modest enrichment of LSD1 at all acetylated promoter and enhancer regions. MYC signal was most pronounced at promoters (cluster P2) and enhancers (cluster E3) associated with H3K27ac signal suppressed by the combination. In both clusters, a greater loss of MYC signal was observed with combined FLT3/LSD1 inhibition compared with no drug or single drug controls. STAT5 binding was localized to enhancers with differential H3K27ac signal that were down-regulated by quizartinib and/or enriched for STAT5 motifs (clusters E1 and E3), which is consistent with studies demonstrating that STAT5 is a primary downstream target of FLT3 inhibitors (40). Notably, combined FLT3/LSD1 inhibition resulted in a global depletion of STAT5 signal, including at clusters E1 and E3 (Supplementary Fig. 3G). PU.1 and GFI1 showed specific enrichment at enhancers with increased differential H3K27ac signal following LSD1 inhibition (clusters E1 and E4). LSD1 inhibition led to a loss GFI1 signal at these clusters, consistent with previously reported displacement of GFI1 from chromatin upon LSD1 inhibition (34). While RUNX1 and CEBPA were enriched at both promoters and enhancers with differential H3K27ac signal, they did not demonstrate appreciable changes in signal following drug treatment (Supplementary Fig. 3H and I). Collectively these results implicate MYC, STAT5, PU.1, and GFI1 in the synergistic cytotoxicity of combined FLT3/LSD1 inhibition.
Loss of MYC expression is critical for the response to combined FLT3/LSD1 inhibition
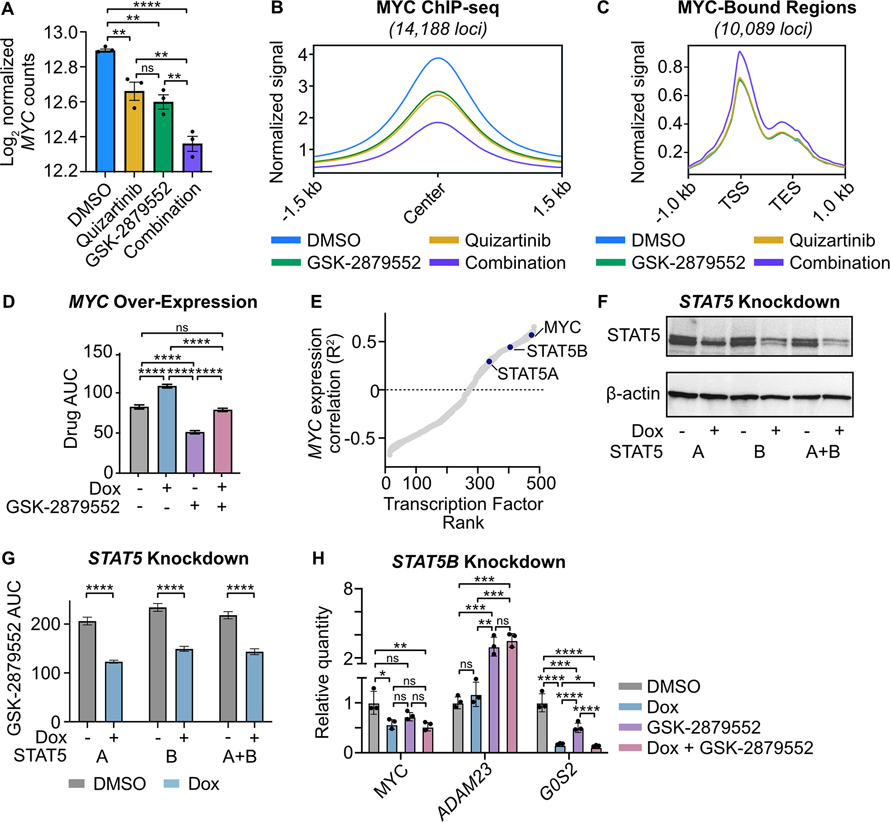
Our transcriptomic and epigenetic analyses nominated MYC as a key driver of the molecular responses to combined FLT3/LSD1 inhibition. We observed that combined FLT3/LSD1 inhibition results in the suppression of MYC transcript abundance and in a genome-wide decrease in MYC binding (Fig. 3A and B). We confirmed that the transcriptional suppression of MYC was associated with a decrease in MYC protein abundance (Supplementary Fig. 4A). Modulation of MYC target gene expression can be influenced both by changes in MYC gene abundance and activity. MYC regulates the transcription of cell cycle proteins through the recruitment of pause-released factors to poised RNA Polymerase II (RNA PolII) (41). We observed that the drug combination increased RNA PolII signal at MYC-bound gene transcription start site (TSS; Fig. 3C). Although the drug combination also results in increased RNA PolII signal at gene bodies, the RNA-PolII pause index shows that the accumulation of paused RNA PolII at gene TSS exceeds the amount present at gene bodies (Supplementary Fig. 4B and C; Supplementary Table 8). Overall, this data indicates that the drug combination disrupts the ability of MYC to promote RNA PolII pause release at its target. In addition, we observed an increase in TP53 protein levels and an enrichment of a phosphoprotein network controlled by TP53 following the drug combination, consistent with repression of MYC-dependent cell cycle regulation (Supplementary Fig. 4D; Supplementary Table 9). Together, these findings suggest a mechanism of combined FLT3/LSD1 that suppresses MYC expression and activity.
Fig. 3: MYC expression is suppressed by combined FLT3/LSD1 inhibition and is associated with STAT5 regulatory activity.
A, Normalized MYC counts from RNA-seq presented in Fig. 1. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. B, MYC binding profile at consensus peaks from MOLM13 ChIP-seq presented in Fig. 2. C, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO vehicle for 6 hours prior to CUT&Tag for RBP1. RBP1 binding profile at RBP1 and MYC co-bound regions. D, MOLM13 cells were transduced with lentiviral particles harboring a doxycycline-inducible MYC expression vector. Cells were treated with doxycycline (1 μg/mL) or DMSO for 48 hours and then plated in an 8×8 matrix of quizartinib and GSK-2879552 for 72 hours prior to viability assessment by CellTiter Aqueous colorimetric assay. AUC data from the 311 nM GSK-2979552 isoline (the concentration corresponding to maximal synergy in the MYC over-expressed MOLM13 cells) is shown. Dose responses and synergy over the entire drug matrix is shown in Supplementary Fig. 4. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. E, Spearman’s correlation of normalized MYC gene counts and predicted transcription factor activity scores. Activity scores were inferred from baseline RNA-seq performed on patients in the BeatAML cohort. Transcription factors are ranked by goodness of fit (R2). F, MOLM13 cells were transduced with lentiviral particles harboring a doxycycline-inducible STAT5 short hairpin RNA (shRNA) knockdown vector. Western blot for STAT5 and β-actin following treatment with doxycycline (1 μg/mL) or DMSO for 48 hours. G, GSK-2879552 AUC of MOLM13 STAT5 knockdown cells treated with doxycycline (1 μg/mL) or DMSO for 72 hours. The GSK-2879552 response curves are shown in Supplementary Fig. 5. Statistical significance was determined by Student’s t-test. H, qPCR assessment of gene expression in MOLM13 cells expressing a doxycycline-inducible STAT5B shRNA. Cells were treated with doxycycline (1 μg/mL) for 48 hours prior to the addition of GSK-2879552 (100 nM) for 24 hours. Expression was normalized to GUSB as an endogenous control. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001, TSS = transcription start site, TES = transcription end site.
To evaluate the importance of MYC expression to the mechanism of dual FLT3/LSD1 inhibition, we derived a MOLM13 cell line with a doxycycline-inducible MYC expression construct (Supplementary Fig. 4E and F). MYC over-expression resulted in decreased sensitivity to the drug combination (Fig. 3D; Supplementary Fig. 4G–I). In addition, MYC over-expression attenuated the induction of apoptosis by the drug combination (Supplementary Fig. 4J). These data suggest that forced expression of MYC partially abrogates the effect of combined FLT3/LSD1 inhibition.
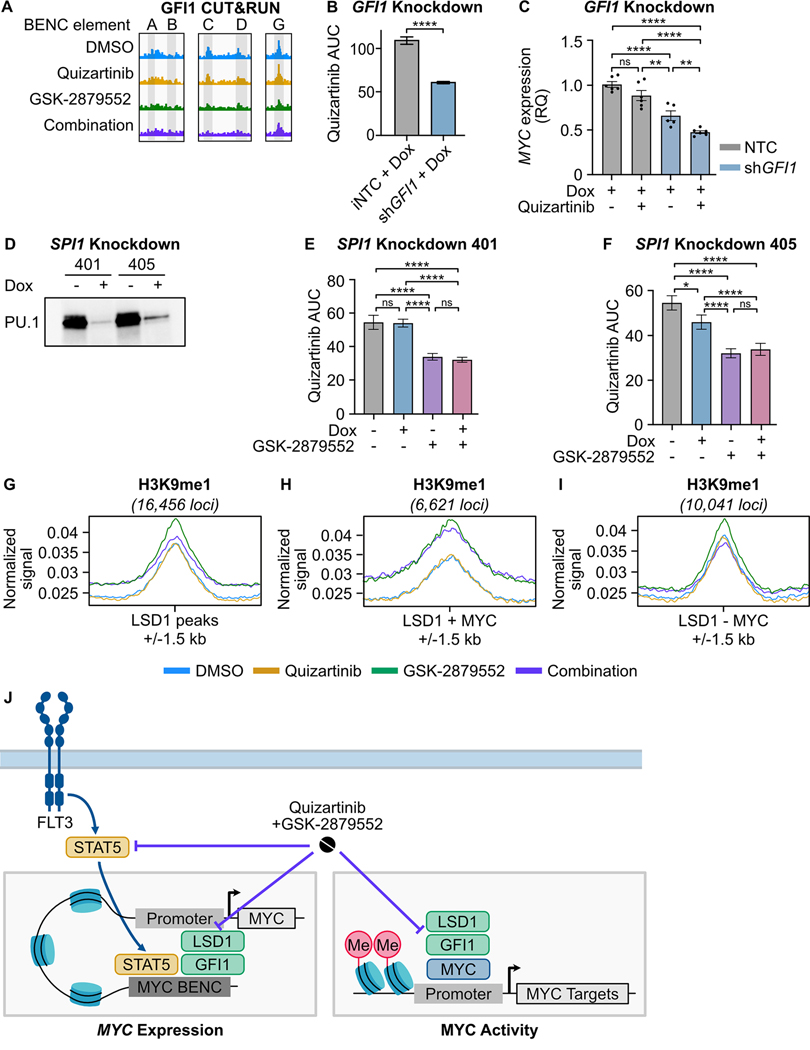
To identify potential regulators of MYC gene expression in AML that may mediate the response to the combination, we analyzed transcription factor activity from RNA-seq on 681 primary AML samples. We generated scores for each sample that reflected the predicted activity of 468 different transcription factors. Correlation of these transcription factor activity scores with MYC gene expression revealed a strong positive correlation with STAT5 transcription factor activity (Fig. 3E; Supplementary Table 10). As FLT3 is a known activator of STAT5, we generated MOLM13 cell lines with perturbed STAT5 activity to evaluate its role in the response to combined FLT3/LSD1 inhibition (40). Knockdown of STAT5A and/or STAT5B resulted in increased sensitivity to GSK-2879552 (Fig. 3F and G; Supplementary Fig. 5A–F). Furthermore, we observed synergy between STAT5 knockdown and GSK-2879552, demonstrating that a loss of STAT5 activity is sufficient to recapitulate a portion of the quizartinib effect (Supplementary Fig. 5G–J). STAT5 knockdown resulted in reduced expression of MYC as well as dysregulated expression of drug combination response markers, ADAM23 and G0S2, which were nominated from our RNA-seq analysis (Fig. 3H; Supplementary Fig. 5K and L). To evaluate whether a decreased STAT5 activity is necessary for drug combination efficacy, we created MOLM13 cells stably expressing Stat5a1*6 (Supplementary Fig. 6A and B). Mutant Stat5a1*6 harbors two point mutations, rendering it constitutively active (42,43). Stat5a1*6 expression markedly attenuated synergy between FLT3 and LSD1 inhibition, demonstrating that STAT5 is a critical target of the drug combination (Supplementary Fig. 6C–F). Collectively, these results show that the STAT5-MYC axis plays a major role in the response to dual FLT3/LSD1 inhibition.
FLT3 inhibition suppresses STAT5 binding to the MYC blood super-enhancer
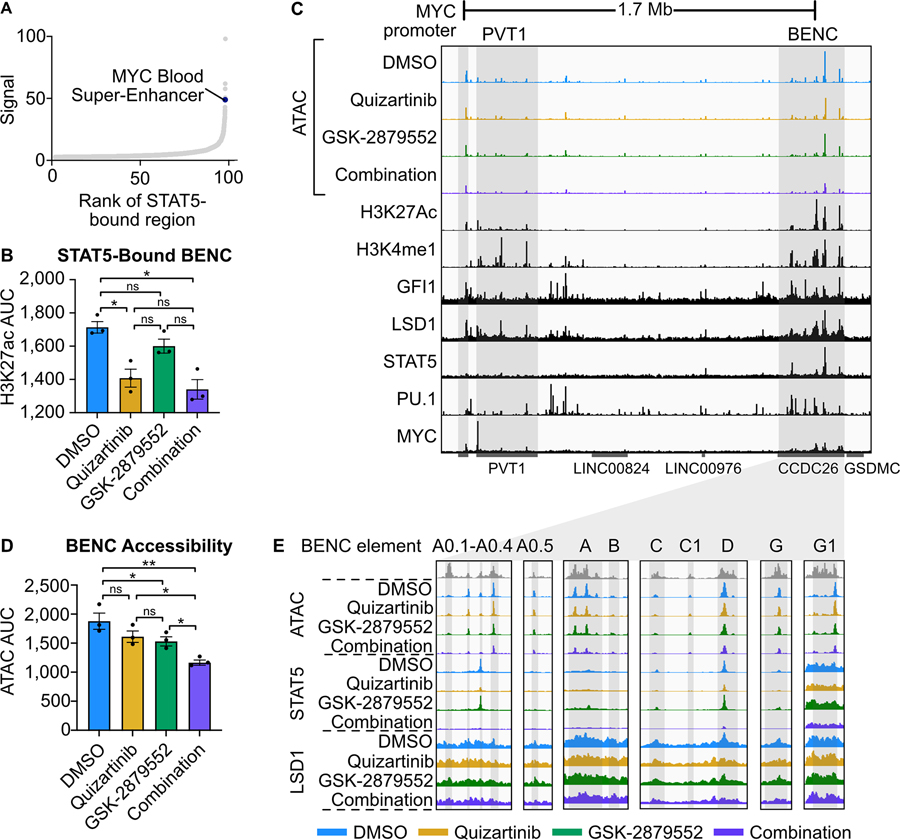
To identify the mechanism by which STAT5 regulates MYC expression in FLT3-ITD AML, we examined our STAT5 ChIP-seq data. While we did not identify a STAT5 binding event at the MYC promoter, ranking STAT5 peaks by normalized signal revealed a strong binding event at the MYC BENC consistent with previous findings (Fig. 4A; Supplementary Tables 11, 12) (11,44). Notably, the MYC BENC was among the cluster of enhancers associated with suppressed H3K27ac signal and STAT5 signal depletion following combined FLT3/LSD1 inhibition (cluster E3). STAT5-bound elements within the MYC BENC showed a significant decrease in H3K27ac signal after treatment with quizartinib or the drug combination (Fig. 4B). To characterize the changes in accessibility of the MYC BENC we performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) (36). This analysis revealed a loss of accessibility across all modules in response to drug combination treatment (Fig. 4C–E). Evaluation of STAT5 binding and H3K27ac signal revealed several sub-modules that also display dynamic behavior in response to drug treatment but have not been previously characterized (A0.1–0.5, C1, G1). Collectively, our findings nominate the MYC BENC as a crucial locus for down-regulation of MYC gene expression by dual FLT3/LSD1 inhibition.
Fig. 4: FLT3-Inhibition represses MYC expression through a loss of STAT5 binding to the MYC blood super-enhancer cluster.
A, STAT5-bound regions from Fig. 2 ranked by ChIP-seq signal. B, H3K27ac CUT&Tag signal AUC at STAT5-bound BENC elements. H3K27ac signal data is from MOLM13 cells in Fig. 1 whereas the STAT5 signal data is from MOLM13 cells in Fig. 2. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. C, ATAC-seq was performed on MOLM13 cells quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO for 24 hours. Representative histone modification and transcription factor tracks (from DMSO conditions in Fig. 2) shown at the extended MYC locus. D, ATAC signal AUC at all MYC BENC modules. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. E, ATAC, STAT5 ChIP-seq, and LSD1 ChIP-seq signal at twelve BENC modules. ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
LSD1 inhibition represses the expression of MYC and its target genes by altering GFI1 and histone modification dynamics
Our data demonstrates that dual FLT3/LSD1 inhibition suppresses MYC gene expression by displacement of STAT5 from the MYC BENC. However, the MYC BENC is bound by many other transcription factors, indicating that drug combination efficacy may be dependent on interruption of MYC BENC-bound factors in addition to STAT5. Prior studies have shown that LSD1 inhibitor monotherapy decreases MYC expression (18–20). A critical component of LSD1-inhibitor efficacy is the disruption of LSD1 scaffolding of GFI1 from the CoREST transcription repressor complex (34). Examination of our GFI1 CUT&RUN data confirmed that GFI1 is bound to the MYC BENC, but is disrupted by the drug combination at module C (Fig. 5A; Supplementary Fig. 7A–E). To evaluate the importance of GFI1 in the response to combined FLT3/LSD1 inhibition, we generated MOLM13 cell lines with doxycycline-inducible knockdown of GFI1 (Supplementary Fig. 7F and G). We found that GFI1 knockdown increased sensitivity to FLT3 inhibition and enhanced FLT3-inhibitor-dependent repression of MYC and its target genes (Fig. 5B and C; Supplementary Fig. 7H and I). Collectively, this data indicates that displacement of GFI1 binding by LSD1 inhibition reduces MYC expression and is important to combined FLT3/LSD1 inhibitor response.
Fig. 5: LSD1 inhibition disrupts GFI1 binding at the MYC BENC and induces a gain of H3K9me1 binding at MYC-bound promoters.
A, GFI1 CUT&RUN signal from Fig. 2 at five BENC modules. B, MOLM13 cells were transduced with lentiviral particles harboring a doxycycline-inducible non-targeting codon (NTC) or GFI1 shRNA knockdown vector. Quizartinib AUC of cells treated with doxycycline (1 μg/mL) or DMSO for 72 hours. Substantial knockdown was observed in the absence of doxycycline treatment, so only doxycycline-treated samples were compared. The quizartinib response curves are shown in Supplementary Fig. 7. Statistical significance was determined by Student’s t-test. C, qPCR assessment of gene expression in cells treated with doxycycline (1 μg/mL) for 48 hours prior to the addition of quizartinib (1 nM) for 24 hours. Expression was normalized to GUSB as an endogenous control. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. D, MOLM13 cells were transduced with lentiviral particles harboring a doxycycline-inducible SPI1 shRNA knockdown vector. Western blot for PU.1, which is encoded by SPI1, in cells treated with doxycycline (1 μg/ml) or an equivalent volume of DMSO for 48 hours. E, F, Cells were treated with doxycycline (1 μg/mL) or DMSO for 48 hours and then plated in an 8×8 matrix of quizartinib and GSK-2879552 for 72 hours prior to viability assessment. AUC data from the 311 nM GSK-2979552 isoline (the concentration corresponding to maximal synergy in the SPI1 knockdown MOLM13 cells) is shown. Dose responses and synergy over the entire drug matrix is shown in Supplementary Fig. 9. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. G-I, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), or the combination for 6 hours prior to CUT&Tag for H3K9me1. Normalized signal for H3K9me1 at LSD1-bound regions, LSD1 and MYC co-bound regions, and at regions bound by LSD1 but not MYC. J, Schematic describing the drug combination mechanism. ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Our data demonstrates that dual FLT3/LSD1 inhibition exerts a portion of its activity via a STAT5- and GFI1-dependent decrease in MYC gene expression. However, MYC over-expression only partially reverses the impact of combination treatment, suggesting the involvement of additional mechanisms. Prior work has shown that LSD1 inhibitor efficacy is also dependent on activation of PU.1-bound enhancers and subsequent induction of myeloid differentiation (45). Our transcriptional and epigenetic data shows that combined FLT3/LSD1 inhibition results in the activation of PU.1 target genes and acetylated enhancers enriched for PU.1 motifs. Therefore, we evaluated whether this transcriptional signal resulted in immunophenotypic differentiation of AML blasts. Drug treatment did result in a modest increase in CD11b expression, however the majority of treated cells remained CD11b negative (Supplementary Fig. 8). To evaluate whether PU.1-dependent activation of this differentiation-associated gene expression program was necessary for drug effect, we evaluated the impact of SPI1 (gene coding for PU.1) knockdown. PU.1-deficient cells demonstrated no significant reduction in drug synergy, arguing that the PU.1-driven pro-differentiation gene expression program is dispensable for the cytotoxic drug effect (Fig. 5D–F; Supplementary Fig. 9A–F).
In other cell types, LSD1 plays a role in gene activation via removal of repressive mono- and demethylation of histone 3 lysine 9 (H3K9me1/2) (46). To evaluate this possible mechanism, we profiled the genome-wide distribution of H3K9me1 using CUT&Tag. LSD1 inhibition, with or without FLT3 inhibition, resulted in an accumulation of H3K9me1 at MYC target genes co-bound with LSD1 (Fig. 5G–I). This was accompanied by a loss of the reciprocal activating mark acetylated histone 3 lysine 9 (H3K9ac), consistent with the observed decrease in the expression of MYC target genes (Supplementary Fig. 9G). These findings suggest that dual inhibition of FLT3/LSD1 exerts locus-specific effects on the chromatin landscape by interrupting STAT5 and GFI1/CoREST transcriptional regulation as well as altering the balance of repressive H3K9 marks at MYC-bound promoters (Fig. 5J).
Efficacy of combined FLT3/LSD1 inhibition in primary AML samples
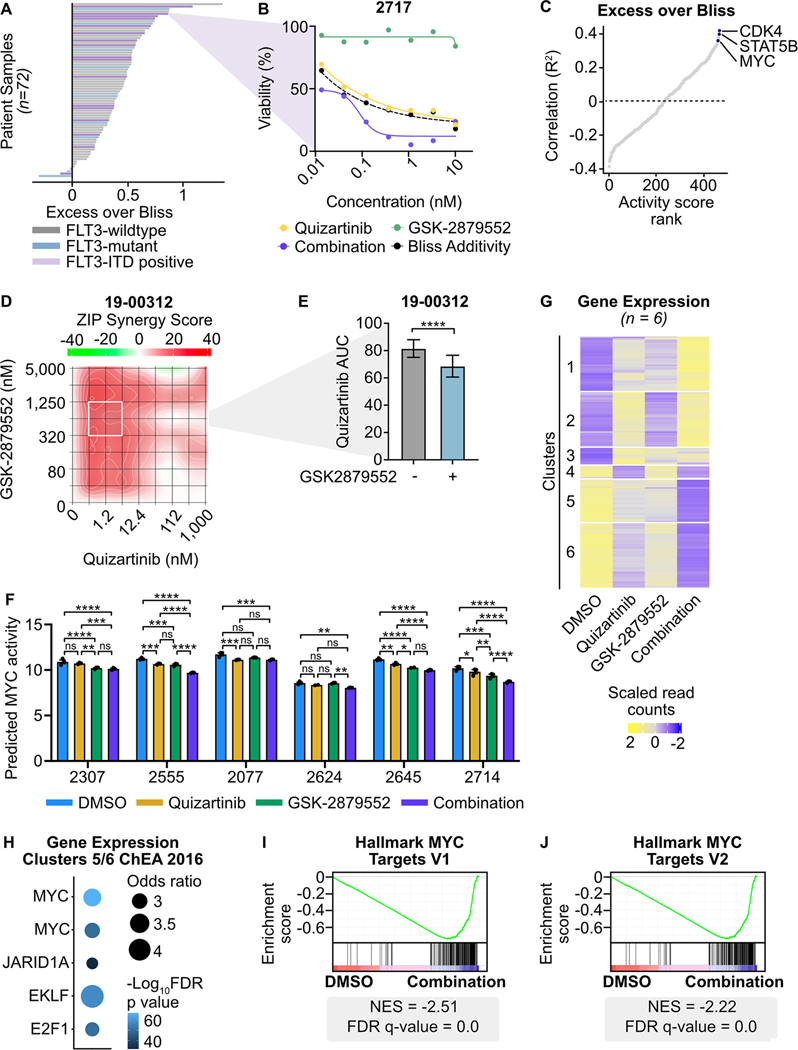
To understand the activity of dual FLT3/LSD1 inhibition in primary AML patients, we performed a 3-day ex vivo drug assay on 72 primary AML samples. Nearly every sample (94%; 68 of 72 samples) demonstrated a synergistic increase in efficacy of dual agent therapy over single agents alone (Fig. 6A and B; Supplementary Fig. 10A; Supplementary Table 13). Although synergy was observed regardless of FLT3 mutation status, the mean quizartinib AUC was lower in FLT3-ITD-positive samples (140.1) as compared to FLT3-wildtype samples (172.4) or samples harboring a FLT3 mutation other than FLT3-ITD (168.2; Supplementary Fig. 10B). To characterize the determinants of response to combined FLT3/LSD1 inhibition, we generated transcription factor activity scores on baseline RNA sequencing performed on the cohort (Supplementary Table 14). Predicted MYC and STAT5B transcription factor activity were among the strongest correlates with the degree of combination synergy (Fig. 6C). CDK4, a known transcriptional target of MYC, was the strongest correlate (47). These findings reveal that AML samples with high baseline MYC activity have the greatest sensitivity to combined FLT3/LSD1 inhibition.
Fig. 6: Combined FLT3/LSD1 inhibition drives synergistic cell death by repressing a MYC-dependent transcriptional network in primary AML blasts.
A, Primary AML blasts from 72 total samples (18 FLT3-ITD-positive) were cultured for 72 hours along a 7-point curve with either quizartinib, GSK-2879552, or equimolar amounts of the drug combination. Cell viability was assessed by CellTiter Aqueous colorimetric assay. Excess over Bliss was calculated using cell viability at corresponding drug concentrations. Each bar represents the mean excess over Bliss across all concentrations. Bar color indicates FLT3 mutation status. B, Dose response curves for quizartinib, GSK-2879552, and the drug combination in a FLT3-ITD-positive AML sample from (A). C, Spearman’s correlation of excess over Bliss and predicted transcription factor activity. Transcription factors were ranked by goodness of fit (R2). D, Primary blasts from a FLT3-ITD-positive AML sample were treated in triplicate with an 8×8 dose matrix of quizartinib and GSK-2879552 for 72 hours prior to viability assessment by CellTiter Aqueous colorimetric assay. ZIP synergy scores were calculated on the average values for each drug dose. E, AUC data from the 628 nM GSK-2979552 isoline (the concentration corresponding to maximal synergy in (D)) is shown. Statistical significance was determined by Student’s t-test. F, Bulk RNA-seq was performed on six FLT3-ITD-positive patient samples treated in triplicate with 500 nM quizartinib, 500 nM GSK-2879552, both drugs in combination, or an equivalent volume of DMSO for 24 hours. MYC transcription factor activity was inferred from RNA-seq. Statistical significance was determined by two-way ANOVA with a Holm-Šidák post-test correction. G, Unsupervised hierarchical clustering of differentially expressed genes following drug treatment. H, Transcription factor target enrichment from clusters in (G). I, J, GSEA was performed comparing the drug combination to DMSO. ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
To characterize the response to combined FLT3/LSD1 inhibition in patient samples, we performed drug sensitivity studies and RNA-seq on 6 FLT3-ITD-positive patient samples treated with single or dual agent therapy. Similar to the pattern of synergy observed in cell lines, we found drug synergy across a broad range of doses (Fig. 6D and E). In addition, the drug combination synergistically induced apoptosis in primary AML blasts (Supplementary Fig. 10C and D). MYC expression and predicted activity was down-regulated in all samples by the drug combination, although differing patterns of individual drug effect were observed (Fig. 6F; Supplementary Fig. 10E). Unsupervised hierarchical clustering of differentially expressed genes across all 6 samples revealed a similar pattern to what was observed in MOLM13 cells (Fig. 6G; Supplementary Table 15). Nearly a quarter of the differentially expressed genes in the patient samples were also identified in the MOLM13 cells (26.0% of up-regulated genes and 18.4% of down-regulated genes; Supplementary Fig. 10F and G). Transcription factor target analysis revealed suppression of MYC target genes and activation of SPI1/PU.1 target genes following combined FLT3/LSD1 inhibition (Fig. 6H; Supplementary Fig. 10H; Supplementary Table 16). Finally, GSEA revealed the drug combination decreased expression of MYC target genes and increased expression of differentiation-associated genes (Fig. 6I and J; Supplementary Fig. 10I; Supplementary Table 17). Collectively, this data confirms the findings of MYC gene expression and transcription factor activity from AML cell lines in primary AML blasts.
Dual FLT3/LSD1 inhibition suppresses MYC super-enhancer accessibility in primary AML blasts
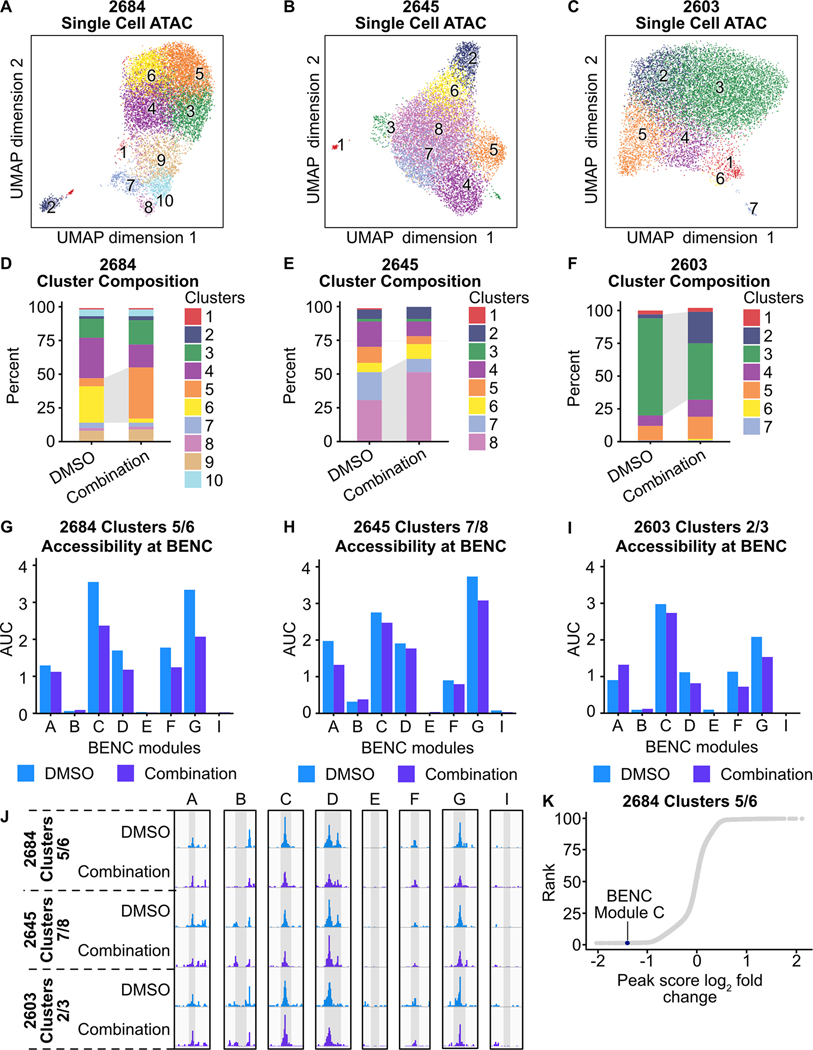
Our initial investigations in AML cell lines demonstrate that suppression of chromatin accessibility at the MYC BENC is an important mechanism of cytotoxicity produced by combined FLT3/LSD1 inhibition. To validate this mechanism in primary AML, we performed single cell ATAC-seq on primary AML blasts from three FLT3-ITD-positive patients 24 hours after treatment with combined quizartinib and GSK-2879552 or DMSO. Cell type identification using a reference dataset and predicted CD34 expression revealed that the samples were largely of GMP-like myeloid blasts (Supplementary Fig. 11A–F; Supplementary Table 18). Clustering revealed population shifts in response to treatment (Fig. 7A–F; Supplementary Fig. 11G–I). Assessment of accessibility at the MYC BENC showed that these population shifts were associated with decreased accessibility at most BENC modules (Fig. 7G–J). However, each sample showed a distinct pattern of change. Sample 2684 showed a decrease in the majority of modules, 2645 demonstrated the most prominent decrease in modules A and G, and sample 2603 exhibited the strongest decrease in modules F and G. To understand the regions that are associated decreased accessibility following drug combination treatment, we calculated the peak score fold change among peaks identified in each condition. In sample 2684, ranking by log2 peak score fold change revealed that MYC BENC module C was amongst the regions with the greatest decrease in accessibility (Fig. 7K). Collectively, these results demonstrate that suppression of MYC BENC accessibility is a conserved feature of the response to dual FLT3/LSD1 inhibition in primary AML samples. However, patient-to-patient heterogeneity does exist, suggesting diversity in the regulatory factors that sustain MYC BENC accessibility in primary AML samples.
Fig. 7: Dual FLT3/LSD1 inhibition results in a shift from a MYC super-enhancer-high to a MYC super-enhancer-low cell state in primary AML blasts.
A-C, Single cell ATAC-seq was performed on three AML patient samples following treatment with quizartinib (500 nM) and GSK-2879552 (500 nM) or an equivalent volume of DMSO for 24 hours. UMAP of DMSO-treated and drug-treated cells colored by cluster. D-F, Percent of cells assigned to each cluster. Dynamic clusters were identified as the populations that shift between DMSO-treated and drug-treated conditions. Dynamic clusters are highlighted with gray shading between bars. G-I, AUC of accessibility at each BENC module. J, Psuedo-bulked accessibility at the MYC BENC modules separated by treatment condition. K, Peak score fold change was calculated between peaks in DMSO-treated and combination-treated cells within dynamic clusters. Peaks are ranked by log2(peak score fold change).
Combined FLT3/LSD1 inhibition disrupts human LSC colony formation
Our single cell ATAC-seq data indicates that dual FLT3/LSD1 inhibition targets GMP-like leukemic blasts. Previous studies have established that leukemic blasts originate from a small number of leukemia stem cells (LSCs) (48). It has been suggested that to fully eradicate the bulk disease, new therapeutic strategies must not only target leukemic blasts, but also eliminate residual LSCs (49). Therefore, we wanted to understand the impact of combined FLT3/LSD1 inhibition on primary patient LSCs.
LSCs have been immunophenotypically characterized using cell surface markers. We utilized fluorescence-activated cell sorting (FACS) to enrich LSCs from three primary patient samples. Specifically, we isolated hematopoietic stem and progenitor cells (CD34+CD38-) expressing at least one previously identified LSC marker (IL1RAP, CD123, CD45RA; Supplementary Fig. 12A) (50–53). We sorted 40,000 enriched LSCs and performed colony forming assays in the presence of drug (Supplementary Fig. 12B). We observed a significant decrease in the number of colonies from LSCs treated with combined FLT3/LSD1 as compared to those exposed to vehicle (Supplementary Fig. 12C and D). This data suggests that combined FLT3/LSD1 disrupts primary patient clonogenicity.
DISCUSSION
Activating mutations in FLT3 are amongst the most common molecular events in AML (1). While FLT3 inhibitors are clinically available, they produce only modest improvements in survival (4,5). Here, we demonstrated that LSD1 inhibition potentiates the efficacy of FLT3 inhibition in FLT3-ITD AML cell lines and primary cell blasts. High-resolution transcriptomic and epigenetic profiling revealed that the mechanism of synergy is in part due to depletion of regulatory transcription factor binding, STAT5 and GFI1, at the MYC BENC. Moreover, we identified additional evidence that dual FLT3/LSD1 inhibition results in the accumulation repressive H3K9me1 marks at MYC-controlled proliferation genes. These findings reveal how epigenetic therapies augment the activity of kinase inhibitors in FLT3-ITD AML.
A crucial component to the mechanism of FLT3/LSD1 inhibitor synergy was altering MYC expression through regulation of the MYC BENC (Fig. 4 and Fig. 7). While others have demonstrated that MYC transcription can by altered by inhibiting general chromatin regulators, disruption of MYC BENC activity by combined epigenetic modulatory drugs and kinase inhibitors is a novel approach to targeting this central oncogenic regulator (13–16). Our single cell ATAC-seq analysis revealed substantial variation in the pattern of MYC BENC module utilization between AML samples at baseline and in response to drug treatment. Indeed, other studies have suggested that each BENC module is bound by a distinct set of transcription factors and regulates MYC expression in specific blood cell lineages (11). Understanding MYC BENC module utilization between molecularly-defined AML subtypes and its impact drug responses is an important question for future studies.
Prior work on LSD1 inhibitors has largely implicated the pro-differentiation effects of these drugs as the central mechanism of cytotoxicity. Our work here shows that LSD1 inhibition activates enhancers that are associated with PU.1 (Fig. 2). Other groups have shown that suppression of SPI1 expression results in a block in LSD1-inhibitor-induced differentiation and decreased cytotoxicity (45). While our work confirmed the role of PU.1 as a putative mediator of LSD1-inhibitor responses, we found that SPI1 knockdown had little effect on the transcriptional or cytotoxic response to dual FLT3/LSD1 inhibition (Fig. 5). Moreover, combined FLT3/LSD1 inhibition did not seem to have a major effect on myeloid differentiation (Supplementary Fig. 8). It is unclear whether the PU.1-associated transcriptional effects observed in our study are important to the drug effect. Investigation of the pro-differentiation effects of dual FLT3/LSD1 inhibition will be an important question for future investigation.
Previous studies of LSD1 inhibitors have also demonstrated that drug efficacy is dependent on the interruption of LSD1 scaffolding activity rather than its demethylation activity (18,54). Our work confirmed that a critical component of LSD1 inhibitor activity is the disruption of LSD1 binding to GFI1/CoREST (Fig. 5). However, LSD1 inhibition also resulted in the accumulation of repressive H3K9me1 marks at the promoters of MYC target genes. While LSD1 canonically demethylates activating H3K4 marks, alternative LSD1 complexes remove repressive H3K9 methylation marks in cells from other tissues, resulting in transcriptional activation (46,55). In prostate cancer, LSD1 forms a chromatin-associated complex with androgen receptor that demethylates H3K9 and de-represses androgen receptor target genes. In neuronal cells, on the other hand, an LSD1 isoform, LSD1+8a, complexes with supervillain and demethylates H3K9me2 to regulate neuronal differentiation. Interestingly, the H3K9 demethylation activity of LSD1 may be slightly antagonized by FLT3 inhibition as H3K9me1 signal in cells treated with the drug combination was lower than in those only treated with LSD1 inhibition. Further work needs to be done to nominate binding factors with LSD1 or LSD1 isoforms that, as a complex, functions as a transcriptional activator by H3K9 demethylation and how these complexes are affected by FLT3 inhibitors.
Collectively, our work demonstrates that LSD1 inhibition enhances the activity of FLT3 inhibition in FLT3-ITD AML. The efficacy of the drug combination is dependent on the simultaneous disruption of STAT5 and GFI1 from the MYC blood super-enhancer complex, resulting in repressed MYC expression, as well as the accumulation of repressive H3K9me1 at MYC target genes.
Supplementary Material
IMPLICATIONS.
This work establishes the synergistic efficacy of combined FLT3 and LSD1 inhibition in FLT3-ITD AML by disrupting STAT5 and GFI1 binding at the MYC blood-specific super-enhancer complex.
ACKNOWLEDGEMENTS
We would like to thank all of the BeatAML patients for their precious time and donation of samples supporting this research. We appreciate the following OHSU core facilities for their assistance: Advanced Light Microscopy, Flow Cytometry Shared Resource, Massive Parallel Sequencing Shared Resource, ExaCloud Cluster Computational Resource, and the Advanced Computing Center. This research was funded by the National Cancer Institute (K08 CA245224 to T.P. Braun; U54 CA224019 to S.K. McWeeney, J.W. Tyner, B.J. Druker, and J.E. Maxson; 5 R01 CA065823-25 to B.J. Druker; 5 R01 CA247943-02 to J.E. Maxson), the National Institutes of Health (T32 GM109835 to W.M. Yashar), the American Society of Hematology (American Society of Hematology Fellow Scholar Award (no number assigned) to T.P. Braun; American Society of Hematology Research Restart Award (no number assigned) to T.P. Braun), the American Cancer Society (P30 CA016672-40 to T.P. Braun), and the Leukemia & Lymphoma Society (Leukemia & Lymphoma Society Scholar Award (no number assigned) to J.E. Maxson).
Financial Support
W.M. Yashar: National Institutes of Health T32 GM109835. S.K. McWeeney: National Cancer Institute U54 CA224019. J.W. Tyner: National Cancer Institute U54 CA224019. B.J. Druker: National Cancer Institute U54 CA224019, National Cancer Institute 5 R01 CA065823-25. J.E. Maxson: National Cancer Institute U54 CA224019, National Cancer Institute 5 R01 CA247943-02, Leukemia & Lymphoma Society Scholar Award. T.P. Braun: American Society of Hematology Research Restart Award, American Society of Hematology Fellow Scholar Award, National Cancer Institute K08 CA245224, American Cancer Society Research Scholar Award P30 CA016672-40. The Functional Proteomics RPPA Core is supported by MD Anderson Cancer Center Support Grant #5 P30 CA016672-40.
Footnotes
Conflict of Interest
W.M. Yashar is a former employee of Abreos Biosciences, Inc. and was compensated in part with common stock options. Pursuant to the merger and reorganization agreement between Abreos Biosciences, Inc. and Fimafeng, Inc., W.M.Y. surrendered all of his common stock options in 03/2021. J.E. Maxson discloses a collaboration with Ionis pharmaceuticals, research funding from Gilead Sciences, research funding from Kura Oncology and research funding from Blueprint Medicines. J.W. Tyner has received research support from Acerta, Agios, Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Kronos, Meryx, Petra, Schrodinger, Seattle Genetics, Syros, Takeda, and Tolero. B.J. Druker potential competing interests-- SAB: Adela Bio, Aileron Therapeutics (inactive), Therapy Architects/ALLCRON (inactive), Cepheid, DNA SEQ, Nemucore Medical Innovations, Novartis, RUNX1 Research Program; SAB & Stock: Aptose Biosciences, Blueprint Medicines, Enliven Therapeutics, Iterion Therapeutics, GRAIL, Recludix Pharma; Board of Directors & Stock: Amgen, Vincerx Pharma; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Advisory Committee: Multicancer Early Detection Consortium; Founder: VB Therapeutics; Sponsored Research Agreement: Enliven Therapeutics, Recludix Pharma; Clinical Trial Funding: Novartis, Astra-Zeneca; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license, one CytoImage, Inc. exclusive license, and one Sun Pharma Advanced Research Company non-exclusive license); US Patents 4326534, 6958335, 7416873, 7592142, 10473667, 10664967, 11049247. T.P. Braun has received research support from AstraZeneca, Blueprint Medicines as well as Gilead Sciences and is the institutional PI on the FRIDA trial sponsored by Oryzon Genomics. The authors certify that all compounds tested in this study were chosen without input from any of our industry partners. The other authors do not have competing interests, financial or otherwise.
REFERENCES
- 1.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy VE, Smith CC. FLT3 Mutations in Acute Myeloid Leukemia: Key Concepts and Emerging Controversies. Front Oncol [Internet]. 2020. [cited 2021 May 7];10. Available from: 10.3389/fonc.2020.612880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herold T, Rothenberg-Thurley M, Grunwald VV, Janke H, Goerlich D, Sauerland MC, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia [Internet]. Nature Publishing Group; 2020. [cited 2022 Apr 21];34:3161–72. Available from: https://www.nature.com/articles/s41375-020-0806-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. New England Journal of Medicine. Massachusetts Medical Society; 2017;377:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. New England Journal of Medicine. Massachusetts Medical Society; 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 6.Kim K-T, Baird K, Davis S, Piloto O, Levis M, Li L, et al. Constitutive Fms-like tyrosine kinase 3 activation results in specific changes in gene expression in myeloid leukaemic cells. British Journal of Haematology [Internet]. 2007. [cited 2022 Mar 21];138:603–15. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1365-2141.2007.06696.x [DOI] [PubMed] [Google Scholar]
- 7.Basit F, Andersson M, Hultquist A. The Myc/Max/Mxd Network Is a Target of Mutated Flt3 Signaling in Hematopoietic Stem Cells in Flt3-ITD-Induced Myeloproliferative Disease. Stem Cells International [Internet]. Hindawi; 2018. [cited 2022 Mar 27];2018:e3286949. Available from: https://www.hindawi.com/journals/sci/2018/3286949/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohanian M, Kantarjian HM, Rozovski U, Loghavi S, Huh Y, Abruzzo L, et al. Clinical significance of MYC expression in acute myeloid leukemia. JCO. Wolters Kluwer; 2014;32:7094–7094. [Google Scholar]
- 9.Park HJ, Gregory MA, Zaberezhnyy V, Goodspeed A, Jordan CT, Kieft JS, et al. Therapeutic resistance to FLT3 inhibition is driven by a novel ATM/mTOR pathway regulating oxidative phosphorylation [Internet]. bioRxiv; 2022. [cited 2022 Mar 27]. page 2022.02.14.480468. Available from: https://www.biorxiv.org/content/10.1101/2022.02.14.480468v1 [DOI] [PMC free article] [PubMed]
- 10.Wang C, Zhang J, Yin J, Gan Y, Xu S, Gu Y, et al. Alternative approaches to target Myc for cancer treatment. Sig Transduct Target Ther. 2021;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553:515–20. [DOI] [PubMed] [Google Scholar]
- 12.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe J-S, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev [Internet]. 2013. [cited 2021 Sep 13];27:2648–62. Available from: 10.1101/gad.232710.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roe J-S, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Molecular Cell. 2015;58:1028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature [Internet]. 2015. [cited 2021 Sep 13];526:273–6. Available from: 10.1038/nature14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhagwat AS, Lu B, Vakoc CR. Enhancer dysfunction in leukemia. Blood [Internet]. 2018. [cited 2021 Sep 13];131:1795–804. Available from: 10/gdf8rg [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Liu Q, Acharya P, Stengel KR, Sheng Q, Zhou X, et al. High-Resolution Mapping of RNA Polymerases Identifies Mechanisms of Sensitivity and Resistance to BET Inhibitors in t(8;21) AML. Cell Rep [Internet]. 2016;16:2003–16. Available from: 10/gnzb2b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–195. [DOI] [PubMed] [Google Scholar]
- 18.Fiskus W, Mill CP, Nabet B, Perera D, Birdwell C, Manshouri T, et al. Superior efficacy of co-targeting GFI1/KDM1A and BRD4 against AML and post-MPN secondary AML cells. Blood Cancer J [Internet]. 2021. [cited 2021 Oct 27];11:1–16. Available from: https://www.nature.com/articles/s41408-021-00487-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedicona F, Casado P, Hijazi M, Gribben JG, Rouault-Pierre K, Cutillas PR. Targeting the lysine-specific demethylase 1 rewires kinase networks and primes leukemia cells for kinase inhibitor treatment. Science Signaling [Internet]. American Association for the Advancement of Science; [cited 2022 Apr 21];15:eabl7989. Available from: https://www.science.org/doi/10.1126/scisignal.abl7989 [DOI] [PubMed] [Google Scholar]
- 20.Smith BM, VanCampen J, Kong GL, Yashar W, Tsang YH, Horton W, et al. PU.1 and MYC transcriptional network defines synergistic drug responses to KIT and LSD1 inhibition in acute myeloid leukemia [Internet]. bioRxiv. 2021. [cited 2022 Mar 31]. Available from: https://www.biorxiv.org/content/10.1101/2021.08.24.456354v2 [DOI] [PMC free article] [PubMed]
- 21.Shi Y-J, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. [DOI] [PubMed] [Google Scholar]
- 22.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, et al. Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase. Molecular Cell [Internet]. 2006. [cited 2022 Apr 21];23:377–87. Available from: https://www.sciencedirect.com/science/article/pii/S1097276506004928 [DOI] [PubMed] [Google Scholar]
- 23.Braun TP, Coblentz C, Smith BM, Coleman DJ, Schonrock Z, Carratt SA, et al. Combined inhibition of JAK/STAT pathway and lysine-specific demethylase 1 as a therapeutic strategy in CSF3R/CEBPA mutant acute myeloid leukemia. PNAS. National Academy of Sciences; 2020;117:13670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Research [Internet]. 2020. [cited 2022 Nov 1];48:W488–93. Available from: 10.1093/nar/gkaa216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current Protocols in Molecular Biology. 2015;109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nature Communications. 2019;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell. 2018;173:1535–1548.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Reinberg D, editor. eLife. eLife Sciences Publications, Ltd; 2017;6:e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell. Elsevier; 2018;173:430–442.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Reports [Internet]. 2016. [cited 2022 Nov 1];17:1193–205. Available from: https://www.sciencedirect.com/science/article/pii/S2211124716313353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep. 2018;22:3641–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A [Internet]. 2010;107:21931–6. Available from: 10/bwpgsr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shigeyasu K, Toden S, Ozawa T, Matsuyama T, Nagasaka T, Ishikawa T, et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol Cancer [Internet]. 2020;19:155. Available from: 10/gnz6rs [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Gu X, Baraoidan K, Ibanez V, Sharma A, Kadkol S, et al. RUNX1 regulates corepressor interactions of PU.1. Blood [Internet]. 2011. [cited 2021 May 11];117:6498–508. Available from: 10/ffn9fh [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet [Internet]. Nature Publishing Group; 2007. [cited 2022 Mar 24];39:311–8. Available from: https://www.nature.com/articles/ng1966 [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell [Internet]. 2004. [cited 2022 Jan 9];119:941–53. Available from: 10/fd45dw [DOI] [PubMed] [Google Scholar]
- 40.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–4. [DOI] [PubMed] [Google Scholar]
- 41.Price DH. Regulation of RNA Polymerase II Elongation by c-Myc. Cell Elsevier; 2010;141:399–400. [DOI] [PubMed] [Google Scholar]
- 42.Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, et al. Constitutive Activation of STAT5 by a Point Mutation in the SH2 Domain*. Journal of Biological Chemistry [Internet]. 2000. [cited 2023 Jan 9];275:24407–13. Available from: https://www.sciencedirect.com/science/article/pii/S002192581962185X [DOI] [PubMed] [Google Scholar]
- 43.Mallette FA, Gaumont-Leclerc M-F, Huot G, Ferbeyre G. Myc Down-regulation as a Mechanism to Activate the Rb Pathway in STAT5A-induced Senescence. Journal of Biological Chemistry [Internet]. 2007. [cited 2023 Jan 9];282:34938–44. Available from: https://www.sciencedirect.com/science/article/pii/S0021925820546309 [DOI] [PubMed] [Google Scholar]
- 44.Pinz S, Unser S, Rascle A. Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Molecular Biology. 2016;17:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cusan M, Cai SF, Mohammad HP, Krivtsov A, Chramiec A, Loizou E, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPα-dependent enhancers in AML. Blood. 2018;131:1730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. [DOI] [PubMed] [Google Scholar]
- 47.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proceedings of the National Academy of Sciences [Internet]. Proceedings of the National Academy of Sciences; 2000. [cited 2022 Aug 10];97:2229–34. Available from: https://www.pnas.org/doi/10.1073/pnas.050586197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129:1577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Kao Y-R, Sun D, Todorova TI, Reynolds D, Narayanagari S-R, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med [Internet]. 2019;25:103–10. Available from: 10/gfkwkf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5:31–42. [DOI] [PubMed] [Google Scholar]
- 52.Askmyr M, Ågerstam H, Hansen N, Gordon S, Arvanitakis A, Rissler M, et al. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 2013;121:3709–13. [DOI] [PubMed] [Google Scholar]
- 53.Ågerstam H, Karlsson C, Hansen N, Sandén C, Askmyr M, von Palffy S, et al. Antibodies targeting human IL1RAP (IL1R3) show therapeutic effects in xenograft models of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2015;112:10786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Reports [Internet]. 2018. [cited 2021 Mar 8];22:3641–59. Available from: https://www.sciencedirect.com/science/article/pii/S2211124718303413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell. 2015;57:957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE190785.
For more information, please see Supplementary Materials & Methods.