Abstract
The number of children diagnosed with autism spectrum disorder (ASD) has increased substantially over the past two decades. Current research suggests that both genetic and environmental risk factors are involved in the etiology of ASD. The goal of this paper is to examine how one specific environmental factor, early social experience, may be correlated with DNA methylation (DNAm) changes in genes associated with ASD. We present an innovative model which proposes that polygenic risk and changes in DNAm due to social experience may both contribute to the symptoms of ASD. Previous research on genetic and environmental factors implicated in the etiology of ASD will be reviewed, with an emphasis on the oxytocin receptor gene, which may be epigenetically altered by early social experience, and which plays a crucial role in social and cognitive development. Identifying an environmental risk factor for ASD (e.g., social experience) that could be modified via early intervention and which results in epigenetic (DNAm) changes, could transform our understanding of this condition, facilitate earlier identification of ASD, and guide early intervention efforts.
Keywords: Autism spectrum disorder, epigenetics, DNA methylation, oxytocin, oxytocin receptor gene, social experience
Graphical Abstract
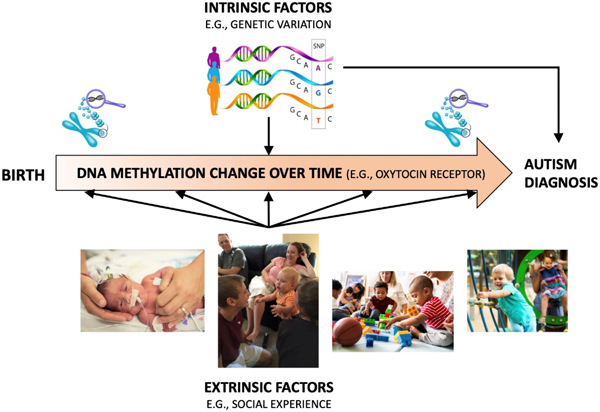
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disability that includes challenges in social communication and repetitive/restricted behaviors and interests (APA, 2013). Diagnosis of ASD can occur as early as 18 months to three years of age, when caregivers or healthcare professionals note developmental delays, particularly in social communication and social-emotional reciprocity. The developmental trajectory of individuals diagnosed with ASD varies based on symptom severity, response to intervention, and presence of comorbid disorders (e.g., intellectual impairment). Regardless, individuals diagnosed with ASD will most likely continue to experience the associated symptoms to some extent throughout their lifespan and require ongoing intervention and assistance in several contexts (e.g., education, vocation, family) (Buescher, Cidav, Knapp, & Mandell, 2014). For individuals with moderate to severe symptoms and who are diagnosed early in life, intervention focuses on intensive behavioral therapies and medications to treat comorbidities rather than core ASD symptoms. However, the ability to treat or prevent the core symptoms of ASD is currently inadequate, in large part because the etiological and biological mechanisms underlying ASD are not fully understood.
Notably, the number of children diagnosed with ASD in the United States has soared over the past two decades (Figure 1) (Baio et al., 2018; Christensen et al., 2016; Maenner et al., 2020; Maenner et al., 2021), with recent estimates of prevalence based on representative population samples indicating one in 36 children 3–17 years are diagnosed with ASD (Xu, Strathearn, Liu, & Bao, 2018). Potential explanations for the increased prevalence of ASD include improvements in diagnostic tools, expansion of diagnostic criteria, and increased awareness about ASD leading to more referrals for assessment. However, such explanations do not fully account for the increased prevalence (Weintraub, 2011), warranting investigation of other causal mechanisms. Currently, there are models which propose that genetic and environmental factors are involved in the etiology of ASD (Geschwind, 2011; Jiang et al., 2004). Specifically, genetic (or intrinsic) and environmental (or extrinsic) factors may independently alter the susceptibility for developing ASD; however, such factors may also interact, with environmental factors affecting activity of ASD-related genes via epigenetic changes (Figure 2).
Figure 1:
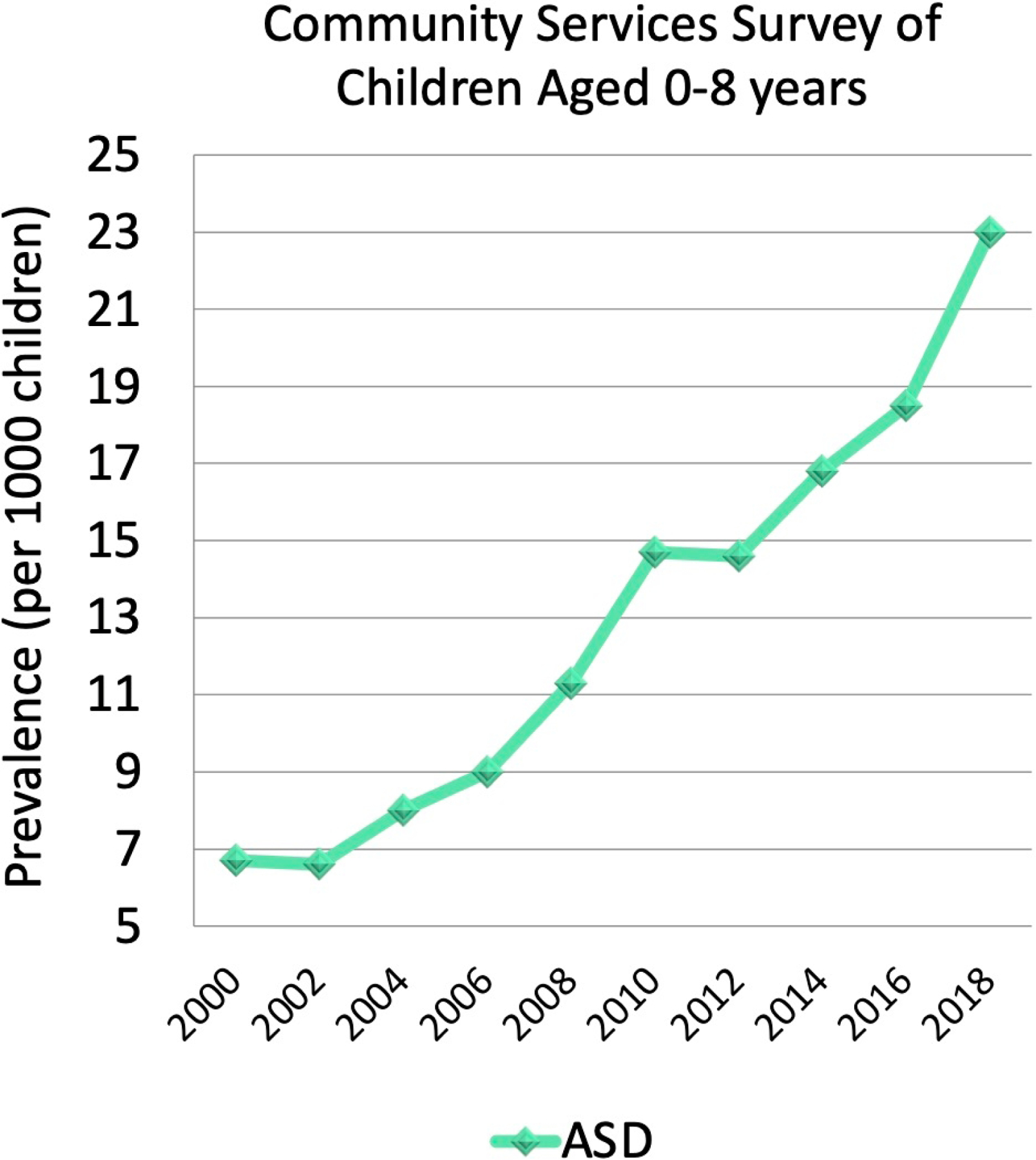
ASD prevalence in children 0–8 years from 2000 to 2018, based on the 11-site ASD and Developmental Disabilities Monitoring Network.
Figure 2:
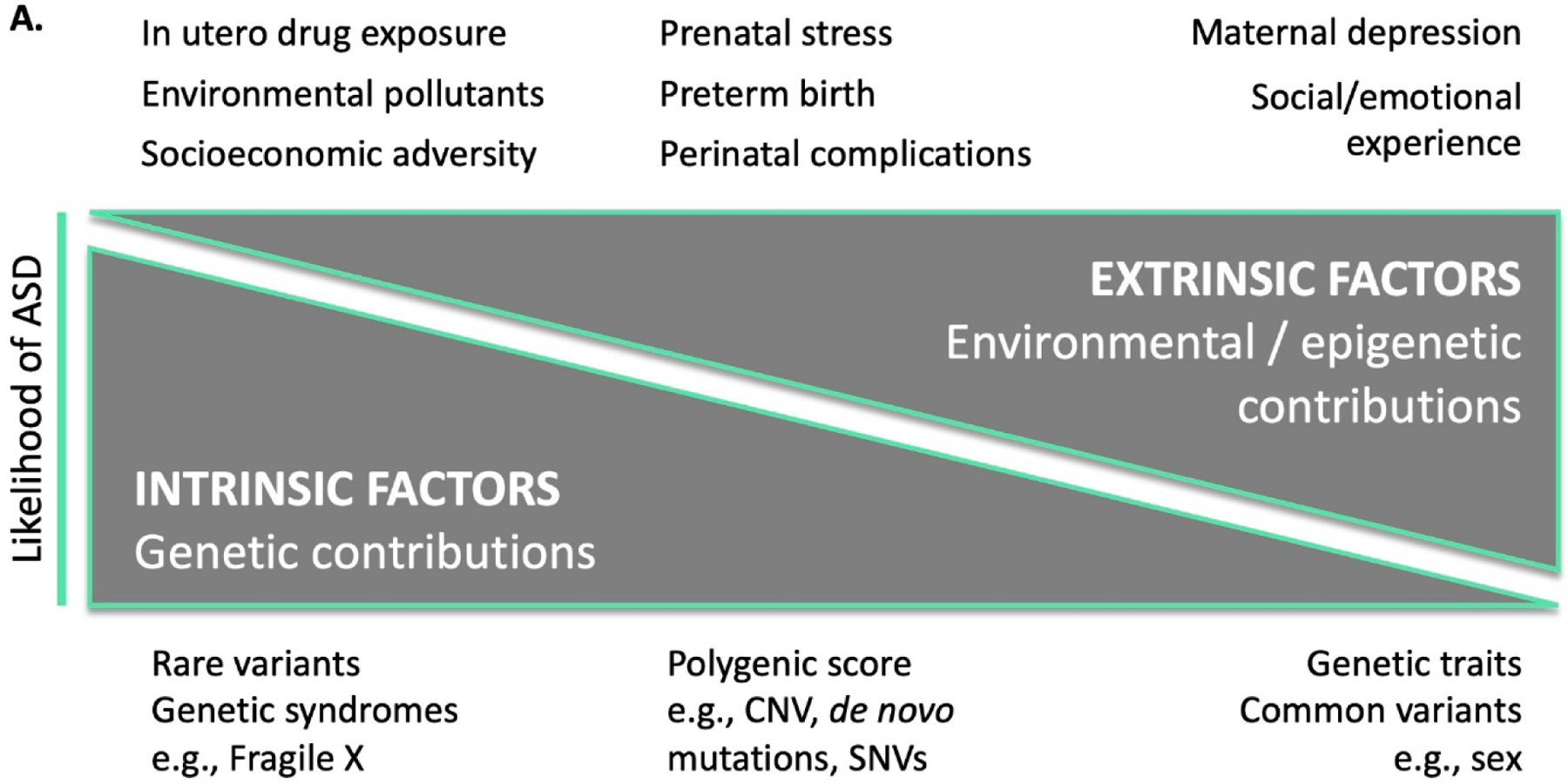
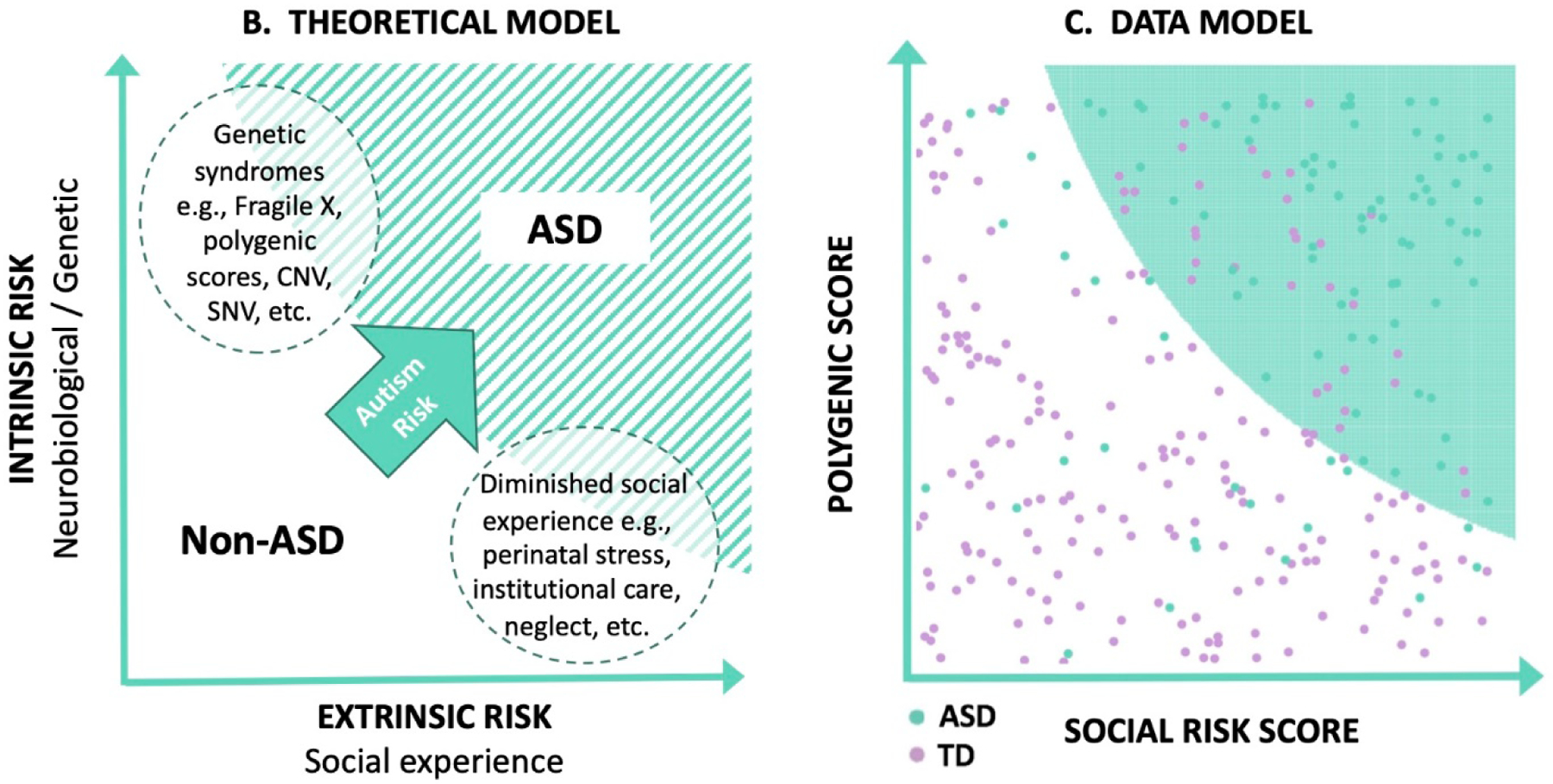
A. Model of intrinsic and extrinsic factors for increasing the likelihood of ASD. B. Theoretical model of how polygenic scores and social risk may predict the development of ASD, with examples given from each axis. C. Simulated model of intrinsic and extrinsic scores for ASD based on multiplicative interaction effects, along with unknown contributions to risk. For illustration purposes, half of the ASD liability derives from the combination of genetic and environmental factors, while unknown causes account for the other half. The shaded green area represents an estimate of the region where children are at high likelihood of ASD, based on an analysis of the simulated data.
CNV, copy number variation; SNV, single nucleotide variant.
Social experience is an environmental factor of particular interest given the well-established impact on social and cognitive development (Barendse et al., 2013), and the potential to modify social experience via intervention. Additionally, social experience has been shown in animal models to alter DNAm, which changes gene expression without altering the genomic sequence (Moore, Le, & Fan, 2013). For example, rodent studies have shown that early social experience, such as maternal licking and grooming of rat pups, alters DNAm of genes important for social development, such as the oxytocin receptor gene (Oxtr) (Beery, McEwen, MacIsaac, Francis, & Kobor, 2016). In this case, increased DNAm downregulated gene expression and appeared to affect behavioral development and stress response. Some translational evidence also exists in human infants (Krol, Moulder, Lillard, Grossmann, & Connelly, 2019).
We previously hypothesized that social experience may interact with genetic vulnerabilities to increase ASD risk via epigenetic mechanisms (Strathearn, 2009). The following sections of this paper will expand upon this to discuss, in relation to ASD, (1) genetic risk factors; (2) environmental factors; (3) the potential interplay of epigenetics and social experience; and (4) the intersection of the genome and epigenome.
2. Genetic Factors in ASD
Estimates of heritability for ASD range from 65 to 90%, with the most recent and largest international study estimating ~80% (Bai et al., 2019). While heritability estimates are helpful in quantifying the magnitude of genetic contribution to etiology, identification of specific variants that contribute to the observed phenotype is key to elucidating mechanisms and identifying biomarkers with potential risk prediction utility. These intrinsic risk factors range from rare variants (such as SHANK3 and SCN2A) (Antaki et al., 2022) and known genetic syndromes that have a relatively large effect size in predicting ASD, to relatively common genetic variants and trait markers, including sex. Research has revealed several hundred variants that are associated with ASD, including single nucleotide variants (SNVs; one nucleotide is substituted for another) and copy number variants (CNVs; duplication or deletion that changes the number of copies of a particular segment of DNA) (Ramaswami & Geschwind, 2018). While many common genetic variants are inherited, studies have shown an increase in rare de novo (new, non-inherited) genetic variants among individuals diagnosed with ASD (Waye & Cheng, 2018; Woodbury-Smith & Scherer, 2018). In fact, de novo CNVs occur four times as frequently in children diagnosed with ASD compared to their non-ASD siblings (Ramaswami & Geschwind, 2018). Several specific genetic syndromes have been strongly associated with ASD, such as mutations in TSC1 and TSC2 (which leads to Tuberous Sclerosis Complex, a disorder in which 61% of affected individuals also present with ASD) (Vignoli et al., 2015) and the FMR1 gene variant (the leading genetic association with ASD) found in Fragile X Syndrome (Varghese et al., 2017). In more recent years, common genetic variants for ASD have emerged (e.g., NEGR1, PTBP2, CADPS, KCNN2, KMT2E, and MACROD2) (Grove et al., 2019) through large genetic cohort studies such as the Psychiatric Genetics Consortium, Autism Workgroup (PGC-AUT), the Danish iPSYCH study (https://ipsych.dk/en/about-ipsych), and the Simons Powering Autism Research for Knowledge (or SPARK) study (pfeliciano@simonsfoundation.org & Consortium, 2018). The possible combined effect of common genetic variants has also been calculated as a “polygenic score” for ASD, or an aggregate genome-wide value indicating the number of ASD variants carried by an individual (Antaki et al., 2022; Guo et al., 2017) (Figure 2A). Although genetic factors clearly contribute to the development of ASD, this does not exclude the possibility of environmental effects. Although ASD has a high heritability estimate, the statistical models used in heritability studies often assume that genes do not interact with the environment, or with other genes, to influence phenotype, which is clearly not the case. Additionally, while an increasing number of genetic variants associated with ASD have been identified, the population level effect sizes are invariably small, and any individual variant is insufficient to explain the heritability estimates (Owen & Williams, 2021).
3. Environmental Factors in ASD
Just as numerous genetic variants have been linked to ASD, several environmental factors have also been identified. Given the early manifestation of symptoms in ASD, epidemiological research has focused on the pre- and peri-natal periods, identifying risk factors such as gestational hypertension and diabetes, fetal distress, birth injury or trauma, low birth weight, small for gestational age, and advanced parental age (Modabbernia, Velthorst, & Reichenberg, 2017; Wang, Geng, Liu, & Zhang, 2017). In utero exposure to valproate has likewise been associated with ASD-like symptoms in animals and human studies, potentially via oxidative stress-induced damage to the brain (Chaliha et al., 2020; Ornoy, 2009). A meta-analysis also found an increased risk of ASD in the children of mothers who used selective serotonin reuptake inhibitor (SSRI) drugs during pregnancy. However, when SSRI-exposed mothers were compared to non-SSRI-exposed mothers with psychiatric conditions, no significant difference in ASD risk was observed (Ames et al., 2021; Kobayashi, Matsuyama, Takeuchi, & Ito, 2016). Considering that maternal depression may itself be a risk factor for ASD (Rai et al., 2013; Wiggins et al., 2019) maternal psychiatric history may have been a confounding factor in these studies of anti-depressant use in pregnancy.
Early life exposure to pollutants has also been associated with ASD. However, a recent study by McGuinn et al. (2019) found this association to be confounded by environmental deprivation, defined by eight measures of socioeconomic status. Some studies have found higher median income to be associated with higher risk, but this is likely biased by increased access to healthcare services (Durkin et al., 2017; Thomas et al., 2011). When access biases are eliminated, such as in studies in countries with universal healthcare and routine developmental screenings, children of families with lower median incomes are at higher risk of ASD (Delobel-Ayoub et al., 2015; Rai et al., 2012).
Social experience is known to profoundly affect social and cognitive development, as demonstrated by decades of animal research (Caldji et al., 1998; F. Champagne & Meaney, 2001; F. A. Champagne & Meaney, 2007; Francis & Meaney, 1999; Ladd et al., 2000; Meaney, 2001; Weaver et al., 2004) and by human experimental and epidemiological studies (Bick & Nelson, 2017; Kuhl, 2004; Strathearn et al., 2020). Both Dawson (2008) and Schultz (2005) have hypothesized that basic deficits in social perception and experience may underlie many developmental and behavioral differences seen in ASD, and that a set of defining experiences early in life (or lack thereof) may adversely affect the development of multiple cascading neural pathways. Just as visual deprivation during a critical period of development may result in permanent disruption of the visual pathways and long-term visual impairment (Wiesel & Hubel, 1965), restricted social experience—either extrinsically or intrinsically derived—may lead to long-lasting impairment in social development. A child’s social experience may be affected by a variety of factors, ranging from perinatal stress and premature birth to socioeconomic adversity and parental psychopathology (Figure 2).
Prior studies have demonstrated associations between early social experience and the development of social communicative abilities in both neurotypical and neurodivergent populations. A prospective study of infant siblings of children with ASD showed links between language exposure at home during the first year of life and subsequent language development in toddlers later diagnosed with ASD (Swanson et al., 2019). Specific characteristics have also been noted in children at-risk of ASD, based on videotaped behavior and naturalistic recording of vocalizations and language exposure, including an impaired ability to respond to a caregiver’s social bids (Gangi, Ibanez, & Messinger, 2014; Swanson et al., 2018). Infants who are later diagnosed with ASD previously exhibited a reduced frequency of vocalizing with speech sounds (consonant-vowel syllables) (pfeliciano@simonsfoundation.org & Consortium, 2018; Plumb & Wetherby, 2013). Likewise, toddlers later diagnosed with ASD used fewer deictic gestures (show/give/point) and initiated joint attention at a lower rate than typically developing toddlers (Shumway & Wetherby, 2009). Each of these developmental characteristics may be associated with variation in early social experience.
Extreme examples of the impact of restricted social experience are found in multiple studies of children raised in Romanian orphanages. Exposed to severe physical and social deprivation before they were adopted into the U.K., over 10% these children were clinically indistinguishable from children with “typical” ASD at age 4, and almost 20% had autistic-like features (Sonuga-Barke et al., 2017). Furthermore, the children’s symptom severity was directly correlated with length of time spent in the institution. A more recent randomized intervention study confirmed that children raised in institutions, compared with non-institutional family-centered care, were at increased risk of ASD and deficits in social communication, whereas children randomly assigned to live in foster care showed an intermediate risk (Levin, Fox, Zeanah, & Nelson, 2015). That is, living in a home environment characterized with richer social experiences was a protective factor, and the earlier a child was placed into foster care, the less likely the child was to develop ASD-like symptoms. While most children do not experience these extreme social conditions, and a direct causal relationship between social deprivation and ASD is not implied, these studies provide insight into possible modifiable environmental factors that could be enriched, for example, to potentially ameliorate or reduce symptoms of ASD. Furthermore, differences in social experiences and perception may also underlie many other developmental and behavioral differences experienced by individuals diagnosed with ASD. Numerous studies have also shown that ASD interventions focused on providing parents with additional parenting tools and supports (e.g., training parents in Applied Behavior Analysis) are more likely to produce sustained improvements in child behavior and social development (Lindgren et al., 2020; Pickles et al., 2016; Steiner, Koegel, Koegel, & Ence, 2012; B. Tonge, Brereton, Kiomall, Mackinnon, & Rinehart, 2014; B. J. Tonge, Bull, Brereton, & Wilson, 2014). In fact, randomized trials of parent-mediated interventions for children at increased likelihood of developing ASD revealed significant reductions in symptoms which persisted up to three years post-intervention (Green et al., 2017; Whitehouse et al., 2021).
4. Epigenetics, Social Experience, and ASD
Given the evidence that gene and environment may both play a role in the etiology of ASD, the study of epigenetic mechanisms such as DNAm may be particularly relevant (Yoon, Choi, Lee, & Do, 2020). DNAm, or the addition of a methyl or hydroxymethyl group to the 5’ cytosine residue on DNA (Greenberg & Bourc’his, 2019), has been shown to vary across the lifespan, and involves both genetic and epigenetic contributions (Szyf & Bick, 2013). In general, DNAm regulates gene expression with this reversible modification, particularly when cytosine (C) is adjacent to guanine (G), separated by a phosphate bond (CpG). These CpG-rich regions often occur in gene regulatory regions, such as promoters. DNAm, the most studied epigenetic modification, has a critical role in human development and has the potential to be used as a biomarker for neurodevelopment and health conditions. DNAm has also been associated with environmental exposures and conditions, leading to investigations of its use as a marker of environmental influences. There is broad evidence supporting DNAm differences in ASD, utilizing both blood and brain tissue (Andrews et al., 2017; Ladd-Acosta et al., 2014). One study identified 20 loci in Danish newborn dried blood spots with differential methylation patterns seen in children later diagnosed with ASD (Table 1, Set A) (Hannon et al., 2018). A meta-analysis of post-diagnosis case-control blood samples from 796 ASD cases and 858 controls, aged 5–17 years, identified seven methylation differences, with suggestive ASD association p-values (Table 1, Set B) (Andrews et al., 2018). Several of these loci had a similar direction of hypo/hyper-methylation, and an even greater effect size, in brain samples. While some CpG sites remain unchanged between birth and later ASD diagnosis (Hannon et al., 2018), it is unknown whether these 27 ASD-related DNAm profiles change between birth and ASD diagnosis, and whether these changes are associated with social experience. Broadly, the genes identified in the latter study are involved in methylation machinery (Kundakovic et al., 2014; Siu & Weksberg, 2017), hippocampal plasticity (Henriquez et al., 2013), epigenetic modulation of social and motivational behavior (Johnstone et al., 2018), neural cell differentiation (Carelli et al., 2019), and glucocorticoid metabolism and stress response (Lester et al., 2018; Weaver et al., 2004).
Table 1:
Proposed DNA methylation loci associated with the pathogenesis of ASD. Set A involves differentially methylated loci at birth; Set B at time of ASD diagnosis; and Set C involving OXTR promoter region.
| Study | Nearest Gene | Chromosome | CpG Loci ID | Position |
|---|---|---|---|---|
| A. Hannon et al (2018) - Dried blood spots - N=1263 - Infants at birth, who developed ASD |
RALY | 20 | cg12699865 | 32583031 |
| 16 | cg03697766 | 54848022 | ||
| UNC84A | 7 | cg25203085 | 887678 | |
| 5 | cg20712043 | 16392700 | ||
| TG | 8 | cg04918350 | 134124199 | |
| 6 | cg21986027 | 169238138 | ||
| TRIM2 | 4 | cg14001992 | 154073813 | |
| RD3 | 1 | cg00692367 | 154073813 | |
| 1 | cg16254267 | 1073529 | ||
| C2orf85 | 2 | cg03270969 | 242813189 | |
| LHCGR | 2 | cg06995408 | 48977089 | |
| PAG1 | 8 | cg09973676 | 82006417 | |
| ZCCHC24 | 10 | cg25485956 | 81146099 | |
| 14 | cg23256480 | 93252030 | ||
| KLF8 | X | cg22829182 | 56258808 | |
| CCDC147 | 10 | cg02803139 | 106113391 | |
| LOC100128573 | 19 | cg03260991 | 7539710 | |
| 10 | cg04089434 | 94516971 | ||
| KDR | 4 | cg02723107 | 55987799 | |
| KCNJ10 | 1 | cg20064848 | 160037877 | |
| B. Andrews et al (2018) - Blood - N=1654 - Children/adolescents with ASD and controls |
CENPM | 22 | cg21151899 | 42337657 |
| FENDRR | 16 | cg03731974 | 86531598 | |
| SNRNP200 | 2 | cg09962502 | 96971189 | |
| PGLYRP4 | 1 | cg01798266 | 153347938 | |
| EZH1 | 17 | cg01716316 | 40897182 | |
| DIO3 | 14 | cg16234726 | 101632839 | |
| CCDC181 | 1 | cg09671955 | 169460734 | |
| C. Gregory et al (2009) - Blood (PBMC) - N=40 - ASD |
OXTR | 3 | CpG-924, Intron 1, MT2 | 8769047* |
| 8769121* | ||||
| 8769146* |
from Genome Reference Consortium Human Build 38 (hg38). All other position numbers from hg19.
The oxytocin receptor gene (OXTR in humans, Oxtr in rodents) is also of particular interest, given that its substrate, oxytocin, is a neuropeptide implicated in social salience, including eye gaze, empathy, and pair-bonding behavior (Mitre, Minder, Morina, Chao, & Froemke, 2018). Oxytocin and OXTR are programmed by early life experience, influence patterns of eye gaze and face perception, and play crucial roles in mammalian social development. Moreover, there are several DNAm sites in OXTR associated with ASD in humans. Oxtr expression in rodents appears to be programmed by early life experience, with decreased expression seen in the blood and brains of animals who receive lower levels of social experience in infancy (Beery et al., 2016; Francis, Champagne, & Meaney, 2000), and Oxtr knockout mice have impaired social memory and recognition (Lee, Caldwell, Macbeth, Tolu, & Young, 2008) and impaired mother-offspring interactions (Nishimori et al., 2008). In addition, OXTR hypermethylation has been associated with suppressed gene expression (Kusui et al., 2001), reduced circulating oxytocin (Dadds et al., 2014), and decreased OXTR expression in the temporal cortex of the brain of ASD vs. non-ASD controls (Elagoz Yuksel, Yuceturk, Karatas, Ozen, & Dogangun, 2016) (Table 1, Set C). More recently, a human longitudinal study showed that decreased social experience during infancy predicted increased methylation in a conserved regulatory site of the OXTR gene one year later, and that increased OXTR methylation at 18 months reflected differences in child behavior relevant to ASD (Krol et al., 2019). Thus, early life social experience may influence epigenetic levels, and ultimately, affect gene expression in the brain and social behaviors relevant to ASD.
5. The Intersection of Genome and Epigenome in ASD
Understanding cross-omics relationships, e.g. how genetic variation is related to gene expression, DNAm, or protein levels, can provide important biologic insights into how genetic risk variants manifest into phenotypes and health outcomes. With the emergence of cost-efficient genome-scale measurement tools, many studies have now measured multiple -omics from the same individuals and have shown that gene expression levels can be controlled by genetic variation, including in a tissue specific manner. Single nucleotide polymorphisms (SNPs) that control gene expression levels are called expression quantitative trait loci (eQTLs). Similarly, SNPs can also regulate DNAm levels; these are commonly referred to as meQTLs. These relationships can occur in -cis, i.e. in close genomic proximity, or in -trans, where the SNP and the gene whose expression or methylation that it regulates is very far away in linear distance or even on a different chromosome.
Multiple studies have identified SNPs that control blood DNAm levels at specific CpGs in the OXTR gene locus (Reiner et al., 2015; Rijlaarsdam et al., 2017; Smearman et al., 2016). For example, a recent analysis revealed methylation levels at 18 OXTR gene CpGs are controlled by nearby SNP genotypes (Smearman et al., 2016). Furthermore, one CpG methylation target associated with two SNPs modified the effect of social adversity (abuse) exposure on depression outcomes which suggests potential epigenetic mechanisms for gene-environment interactions at this locus. In addition to results reported in publications, a query of publicly available databases such as GTeX (https://www.gtexportal.org/home/gene/OXTR) the meQLT portals (http://www.mqtldb.orgfl) reveals dozens of SNPs control OXTR gene expression and DNAm levels in numerous brain regions. Targeted studies are needed to fully evaluate the relationship of these cross-omics data and in the context of social environments, ASD, and ASD-related quantitative traits. Genetic variants associated with ASD diagnosis are postulated to play an important role in gene regulation. Identification of ASD genetic variants that regulate gene expression or DNAm levels, and in which cell types, can provide information on biologic mechanisms that contribute to ASD etiology. This can provide candidate genes and/or biologic pathways that could be intervened upon to reduce disabilities associated with ASD (Pavlides et al., 2016; Zhu et al., 2014). Studies using eQTL and meQTL maps have shown ASD risk variants, discovered via GWAS, are enriched for being eQTLs (Cheng, Quinn, & Weiss, 2013; Davis et al., 2012) and meQTLs (Andrews et al., 2017) when compared to non-ASD SNPs with similar properties, particularly in neurodevelopmentally relevant tissue types such as fetal brain. Additionally, these studies have identified potential regulatory gene targets of ASD SNPs that would not have been otherwise identified when considering genomic location of the SNP alone. Additional studies are needed to specifically evaluate associations between ASD genetic variants and gene expression and methylation patterns at OXTR in relevant biospecimen types. Integration of genome and epigenome data has provided insights into ASD etiology and may inform future avenues of treatment research. It is also worth investigating whether DNAm and expression patterns at loci targeted by genetic variants are also susceptible to environmental exposures, which could implicate shared pathways and targets for different risk factors and/or potential mechanisms for gene-environment interactions. Another important line of future research will be to expand integration of genetic data with other omics measures including the proteome, metabolome, and microbiome, among others. In addition to providing insights into biologic mechanisms, the genome and epigenome can be used for predictive biomarker purposes. While the proportion of phenotypic variance in a population that can be explained by genetic variants, in aggregate, is often very small, there is evidence that including epigenetic measures in predictive models can improve these estimates (McCartney et al., 2018). Finally, genetic and epigenetic variation may be important contributors to ASD comorbidities and co-occurring conditions, which have substantial impacts on the health and quality of life for individuals diagnosed with ASD. Future studies on this topic are also needed to identify biologic processes and pathways that could be targeted to reduce their impact on individuals diagnosed with ASD.
6. Conclusion
Recognizing and understanding the significance of predictive markers for ASD has been a vexing challenge over the past two decades, especially in view of its dramatically increasing prevalence. While numerous genetic and environmental factors have been identified, the interaction between such factors is less well understood and warrants additional investigation. Of particular interest is the extent to which variability in social experiences may modify DNAm of genes that are known to be involved in social processes, such as OXTR, and thus confer risk for social deficits, a core feature of ASD. Understanding the intersection of genetic, epigenetic, and environmental factors in the etiology of ASD would transform our understanding of this disorder, and potentially identify modifiable pathways for intervention, which would likely have far-reaching impacts on individuals with ASD and their families.
Highlights.
There is evidence that both genes and environment contribute to symptoms in autism
Early social experience may be related to DNA methylation patterns in autism
DNA methylation in the OXTR gene may influence social and cognitive outcomes
Autism genetic risk variants may control DNA methylation and developmental outcomes
Acknowledgements
We acknowledge and thank Patrick Breheny, PhD, who prepared the theoretical model shown in Figure 2B and C.
Funding
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under Award Number P50 HD103556. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- ASD
autism spectrum disorder
- CNV
copy number variation
- CpG
Cytosine-phosphate-Guanine
- DNAm
DNA methylation
- eQTL
expression quantitative trait loci
- meQTL
methylation quantitative trait loci
- OXTR
oxytocin receptor gene
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interests
Dr. Ladd-Acosta is an Associate Professor in the Department of Epidemiology at the Johns Hopkins University Bloomberg School of Public Health and is engaged in this research as a private consultant or advisor and not in her capacity as a Johns Hopkins faculty member. She was compensated for these services by the University of Iowa in the form of income.
Credit Author Statement
Drs. Strathearn and Momany produced the first draft of the manuscript, with Dr. Ladd-Acosta contributing section 5. Emese Kovács and William Guiler assisted with researching subtopics and all authors reviewed and edited the final manuscript.
REFERENCES
- Ames JL, Ladd-Acosta C, Fallin MD, Qian Y, Schieve LA, DiGuiseppi C, … Croen LA (2021). Maternal Psychiatric Conditions, Treatment With Selective Serotonin Reuptake Inhibitors, and Neurodevelopmental Disorders. Biol Psychiatry, 90(4), 253–262. doi: 10.1016/j.biopsych.2021.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SV, Ellis SE, Bakulski KM, Sheppard B, Croen LA, Hertz-Picciotto I, … Fallin MD (2017). Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat Commun, 8(1), 1011. doi: 10.1038/s41467-017-00868-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SV, Sheppard B, Windham GC, Schieve LA, Schendel DE, Croen LA, … Ladd-Acosta C (2018). Case-control meta-analysis of blood DNA methylation and autism spectrum disorder. Mol Autism, 9, 40. doi: 10.1186/s13229-018-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antaki D, Guevara J, Maihofer AX, Klein M, Gujral M, Grove J, … Sebat J (2022). A phenotypic spectrum of autism is attributable to the combined effects of rare variants, polygenic risk and sex. Nat Genet, 54(9), 1284–1292. doi: 10.1038/s41588-022-01064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai D, Yip BHK, Windham GC, Sourander A, Francis R, Yoffe R, … Sandin S (2019). Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA Psychiatry doi: 10.1001/jamapsychiatry.2019.1411 [DOI] [PMC free article] [PubMed]
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, … Dowling NF (2018). Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ, 67(6), 1–23. doi: 10.15585/mmwr.ss6706a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse EM, Hendriks MP, Jansen JF, Backes WH, Hofman PA, Thoonen G, … Aldenkamp AP (2013). Working memory deficits in high-functioning adolescents with autism spectrum disorders: neuropsychological and neuroimaging correlates. J Neurodev Disord, 5(1), 14. doi: 10.1186/1866-1955-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery AK, McEwen LM, MacIsaac JL, Francis DD, & Kobor MS (2016). Natural variation in maternal care and cross-tissue patterns of oxytocin receptor gene methylation in rats. Horm Behav, 77, 42–52. doi: 10.1016/j.yhbeh.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J, & Nelson CA (2017). Early experience and brain development. Wiley Interdiscip Rev Cogn Sci, 8(1–2). doi: 10.1002/wcs.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher AV, Cidav Z, Knapp M, & Mandell DS (2014). Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr, 168(8), 721–728. doi: 10.1001/jamapediatrics.2014.210 [DOI] [PubMed] [Google Scholar]
- Caldji C, Tannenbaum B, Sharma S, Francis D, Plotsky PM, & Meaney MJ (1998). Maternal care during infancy regulates the development of neural systems mediating the expression of fearfulness in the rat. Proc Natl Acad Sci U S A, 95(9), 5335–5340. doi: 10.1073/pnas.95.9.5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli S, Giallongo T, Rey F, Latorre E, Bordoni M, Mazzucchelli S, … Di Giulio AM (2019). HuR interacts with lincBRN1a and lincBRN1b during neuronal stem cells differentiation. RNA Biol, 16(10), 1471–1485. doi: 10.1080/15476286.2019.1637698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaliha D, Albrecht M, Vaccarezza M, Takechi R, Lam V, Al-Salami H, & Mamo J (2020). A Systematic Review of the Valproic-Acid-Induced Rodent Model of Autism. Dev Neurosci, 42(1), 12–48. doi: 10.1159/000509109 [DOI] [PubMed] [Google Scholar]
- Champagne F, & Meaney MJ (2001). Like mother, like daughter: evidence for non-genomic transmission of parental behavior and stress responsivity. Prog Brain Res, 133, 287–302. doi: 10.1016/s0079-6123(01)33022-4 [DOI] [PubMed] [Google Scholar]
- Champagne FA, & Meaney MJ (2007). Transgenerational effects of social environment on variations in maternal care and behavioral response to novelty. Behav Neurosci, 121(6), 1353–1363. doi: 10.1037/0735-7044.121.6.1353 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Quinn JF, & Weiss LA (2013). An eQTL mapping approach reveals that rare variants in the SEMA5A regulatory network impact autism risk. Hum Mol Genet, 22(14), 2960–2972. doi: 10.1093/hmg/ddt150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, … Yeargin-Allsopp M (2016). Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years--Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ, 65(3), 1–23. doi: 10.15585/mmwr.ss6503a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadds MR, Moul C, Cauchi A, Dobson-Stone C, Hawes DJ, Brennan J, & Ebstein RE (2014). Methylation of the oxytocin receptor gene and oxytocin blood levels in the development of psychopathy. Dev Psychopathol, 26(1), 33–40. doi: 10.1017/S0954579413000497 [DOI] [PubMed] [Google Scholar]
- Davis LK, Gamazon ER, Kistner-Griffin E, Badner JA, Liu C, Cook EH, … Cox NJ (2012). Loci nominally associated with autism from genome-wide analysis show enrichment of brain expression quantitative trait loci but not lymphoblastoid cell line expression quantitative trait loci. Mol Autism, 3(1), 3. doi: 10.1186/2040-2392-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G (2008). Early behavioral intervention, brain plasticity, and the prevention of autism spectrum disorder. Dev Psychopathol, 20(3), 775–803. doi: 10.1017/S0954579408000370 [DOI] [PubMed] [Google Scholar]
- Delobel-Ayoub M, Ehlinger V, Klapouszczak D, Maffre T, Raynaud J-P, Delpierre C, & Arnaud C (2015). Socioeconomic Disparities and Prevalence of Autism Spectrum Disorders and Intellectual Disability. PLoS ONE, 10(11), e0141964. doi: 10.1371/journal.pone.0141964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin MS, Maenner MJ, Baio J, Christensen D, Daniels J, Fitzgerald R, … Yeargin-Allsopp M (2017). Autism Spectrum Disorder Among US Children (2002–2010): Socioeconomic, Racial, and Ethnic Disparities. American Journal of Public Health, 107(11), 1818–1826. doi: 10.2105/AJPH.2017.304032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagoz Yuksel M, Yuceturk B, Karatas OF, Ozen M, & Dogangun B (2016). The altered promoter methylation of oxytocin receptor gene in autism. J Neurogenet, 30(3–4), 280–284. doi: 10.1080/01677063.2016.1202951 [DOI] [PubMed] [Google Scholar]
- Francis DD, Champagne FC, & Meaney MJ (2000). Variations in maternal behaviour are associated with differences in oxytocin receptor levels in the rat. J Neuroendocrinol, 12(12), 1145–1148. doi: 10.1046/j.1365-2826.2000.00599.x [DOI] [PubMed] [Google Scholar]
- Francis DD, & Meaney MJ (1999). Maternal care and the development of stress responses. Curr Opin Neurobiol, 9(1), 128–134. doi: 10.1016/s0959-4388(99)80016-6 [DOI] [PubMed] [Google Scholar]
- Gangi DN, Ibanez LV, & Messinger DS (2014). Joint attention initiation with and without positive affect: risk group differences and associations with ASD symptoms. J Autism Dev Disord, 44(6), 1414–1424. doi: 10.1007/s10803-013-2002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH (2011). Genetics of autism spectrum disorders. Trends Cogn Sci, 15(9), 409–416. doi: 10.1016/j.tics.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Pickles A, Pasco G, Bedford R, Wan MW, Elsabbagh M, … British Autism Study of Infant Siblings, T. (2017). Randomised trial of a parent-mediated intervention for infants at high risk for autism: longitudinal outcomes to age 3 years. J Child Psychol Psychiatry, 58(12), 1330–1340. doi: 10.1111/jcpp.12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, & Bourc’his D (2019). The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol, 20(10), 590–607. doi: 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, … Borglum AD (2019). Identification of common genetic risk variants for autism spectrum disorder. Nat Genet, 51(3), 431–444. doi: 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Samuels JF, Wang Y, Cao H, Ritter M, Nestadt PS, … Shugart YY (2017). Polygenic risk score and heritability estimates reveals a genetic relationship between ASD and OCD. Eur Neuropsychopharmacol, 27(7), 657–666. doi: 10.1016/j.euroneuro.2017.03.011 [DOI] [PubMed] [Google Scholar]
- Hannon E, Schendel D, Ladd-Acosta C, Grove J, i, P.-B. A. S. D. G., Hansen CS, … Mill J (2018). Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Med, 10(1), 19. doi: 10.1186/s13073-018-0527-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, & van Zundert B (2013). Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Mol Cell Neurosci, 57, 130–143. doi: 10.1016/j.mcn.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Jiang YH, Sahoo T, Michaelis RC, Bercovich D, Bressler J, Kashork CD, … Beaudet AL (2004). A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am J Med Genet A, 131(1), 1–10. doi: 10.1002/ajmg.a.30297 [DOI] [PubMed] [Google Scholar]
- Johnstone AL, O’Reilly JJ, Patel AJ, Guo Z, Andrade NS, Magistri M, … Wahlestedt C (2018). EZH1 is an antipsychotic-sensitive epigenetic modulator of social and motivational behavior that is dysregulated in schizophrenia. Neurobiol Dis, 119, 149–58. doi: 10.1016/j.nbd.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Matsuyama T, Takeuchi M, & Ito S (2016). Autism spectrum disorder and prenatal exposure to selective serotonin reuptake inhibitors: A systematic review and meta-analysis. Reprod Toxicol, 65, 170–178. doi: 10.1016/j.reprotox.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Krol KM, Moulder RG, Lillard TS, Grossmann T, & Connelly JJ (2019). Epigenetic dynamics in infancy and the impact of maternal engagement. Sci Adv, 5(10), eaay0680. doi: 10.1126/sciadv.aay0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl PK (2004). Early language acquisition: cracking the speech code. Nat Rev Neurosci, 5(11), 831–843. doi: 10.1038/nrn1533 [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, & Champagne FA (2014). DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1408355111 [DOI] [PMC free article] [PubMed]
- Kusui C, Kimura T, Ogita K, Nakamura H, Matsumura Y, Koyama M, … Murata Y (2001). DNA methylation of the human oxytocin receptor gene promoter regulates tissue-specific gene suppression. Biochem Biophys Res Commun, 289(3), 681–686. doi: 10.1006/bbrc.2001.6024 [DOI] [PubMed] [Google Scholar]
- Ladd CO, Huot RL, Thrivikraman KV, Nemeroff CB, Meaney MJ, & Plotsky PM (2000). Long-term behavioral and neuroendocrine adaptations to adverse early experience. Prog Brain Res, 122, 81–103. doi: 10.1016/s0079-6123(08)62132-9 [DOI] [PubMed] [Google Scholar]
- Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, & Feinberg AP (2014). Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry, 19(8), 862–871. doi: 10.1038/mp.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Caldwell HK, Macbeth AH, Tolu SG, & Young WS 3rd. (2008). A conditional knockout mouse line of the oxytocin receptor. Endocrinology, 149(7), 3256–3263. doi: 10.1210/en.2007-1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, Conradt E, LaGasse LL, Tronick EZ, Padbury JF, & Marsit CJ (2018). Epigenetic Programming by Maternal Behavior in the Human Infant. Pediatrics, 142(4). doi: 10.1542/peds.2017-1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AR, Fox NA, Zeanah CH Jr., & Nelson CA (2015). Social communication difficulties and autism in previously institutionalized children. J Am Acad Child Adolesc Psychiatry, 54(2), 108–115 e101. doi: 10.1016/j.jaac.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren S, Wacker D, Schieltz K, Suess A, Pelzel K, Kopelman T, … O’Brien M (2020). A Randomized Controlled Trial of Functional Communication Training via Telehealth for Young Children with Autism Spectrum Disorder. J Autism Dev Disord, 50(12), 4449–4462. doi: 10.1007/s10803-020-04451-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Shaw KA, Baio J, Washington A, Patrick M, DiRienzo M, … Dietz PM (2020). Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill Summ, 69(4), 1–12. doi: 10.15585/mmwr.ss6904a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, … Cogswell, M. E. (2021). Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2018. MMWR Surveill Summ, 70(11), 1–16. doi: 10.15585/mmwr.ss7011a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Hillary RF, Stevenson AJ, Ritchie SJ, Walker RM, Zhang Q, … Marioni RE (2018). Epigenetic prediction of complex traits and death. Genome Biol, 19(1), 136. doi: 10.1186/s13059-018-1514-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinn LA, Windham GC, Messer LC, Di Q, Schwartz J, Croen LA, … Daniels JL (2019). Air pollution, neighborhood deprivation, and autism spectrum disorder in the Study to Explore Early Development. Environ Epidemiol, 3(5). doi: 10.1097/ee9.0000000000000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ (2001). Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci, 24(1), 1161–1192. doi: 10.1146/annurev.neuro.24.1.1161 [DOI] [PubMed] [Google Scholar]
- Mitre M, Minder J, Morina EX, Chao MV, & Froemke RC (2018). Oxytocin Modulation of Neural Circuits. Curr Top Behav Neurosci, 35, 31–53. doi: 10.1007/7854_2017_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modabbernia A, Velthorst E, & Reichenberg A (2017). Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol Autism, 8, 13. doi: 10.1186/s13229-017-0121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, & Fan G (2013). DNA methylation and its basic function. Neuropsychopharmacology, 38(1), 23–38. doi: 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimori K, Takayanagi Y, Yoshida M, Kasahara Y, Young LJ, & Kawamata M (2008). New aspects of oxytocin receptor function revealed by knockout mice: sociosexual behaviour and control of energy balance. Prog Brain Res, 170, 79–90. doi: 10.1016/s0079-6123(08)00408-1 [DOI] [PubMed] [Google Scholar]
- Ornoy A (2009). Valproic acid in pregnancy: how much are we endangering the embryo and fetus? Reprod Toxicol, 28(1), 1–10. doi: 10.1016/j.reprotox.2009.02.014 [DOI] [PubMed] [Google Scholar]
- Owen MJ, & Williams NM (2021). Explaining the missing heritability of psychiatric disorders. World Psychiatry, 20(2), 294–295. doi: 10.1002/wps.20870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlides JM, Zhu Z, Gratten J, McRae AF, Wray NR, & Yang J (2016). Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med, 8(1), 84. doi: 10.1186/s13073-016-0338-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- pfeliciano@simonsfoundation.org, S. C. E. a., & Consortium, S. (2018). SPARK: A US Cohort of 50,000 Families to Accelerate Autism Research. Neuron, 97(3), 488–493. doi: 10.1016/j.neuron.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles A, Le Couteur A, Leadbitter K, Salomone E, Cole-Fletcher R, Tobin H, … Green J (2016). Parent-mediated social communication therapy for young children with autism (PACT): long-term follow-up of a randomised controlled trial. Lancet, 388(10059), 2501–2509. doi: 10.1016/S0140-6736(16)31229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb AM, & Wetherby AM (2013). Vocalization Development in Toddlers With Autism Spectrum Disorder. Journal of Speech, Language, and Hearing Research, 56(2), 721–734. doi: 10.1044/1092-4388(2012/11-0104) [DOI] [PubMed] [Google Scholar]
- Rai D, Lee BK, Dalman C, Golding J, Lewis G, & Magnusson C (2013). Parental depression, maternal antidepressant use during pregnancy, and risk of autism spectrum disorders: population based case-control study. BMJ, 346, f2059. doi: 10.1136/bmj.f2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Lewis G, Lundberg M, Araya R, Svensson A, Dalman C, … Magnusson C (2012). Parental socioeconomic status and risk of offspring autism spectrum disorders in a Swedish population-based study. J Am Acad Child Adolesc Psychiatry, 51(5), 467–476 e466. doi: 10.1016/j.jaac.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Ramaswami G, & Geschwind DH (2018). Genetics of autism spectrum disorder. Handb Clin Neurol, 147, 321–329. doi: 10.1016/b978-0-444-63233-3.00021-x [DOI] [PubMed] [Google Scholar]
- Reiner I, Van IMH, Bakermans-Kranenburg MJ, Bleich S, Beutel M, & Frieling H (2015). Methylation of the oxytocin receptor gene in clinically depressed patients compared to controls: The role of OXTR rs53576 genotype. J Psychiatr Res, 65, 9–15. doi: 10.1016/j.jpsychires.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Rijlaarsdam J, van IMH, Verhulst FC, Jaddoe VW, Felix JF, Tiemeier H, & Bakermans-Kranenburg MJ (2017). Prenatal stress exposure, oxytocin receptor gene (OXTR) methylation, and child autistic traits: The moderating role of OXTR rs53576 genotype. Autism Res, 10(3), 430–438. doi: 10.1002/aur.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RT (2005). Developmental deficits in social perception in autism: the role of the amygdala and fusiform face area. Int J Dev Neurosci, 23(2–3), 125–141. doi: 10.1016/j.ijdevneu.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shumway S, & Wetherby AM (2009). Communicative acts of children with autism spectrum disorders in the second year of life. J Speech Lang Hear Res, 52(5), 1139–1156. doi: 10.1044/1092-4388(2009/07-0280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu MT, & Weksberg R (2017). Epigenetics of Autism Spectrum Disorder. Adv Exp Med Biol, 978, 63–90. doi: 10.1007/978-3-319-53889-1_4 [DOI] [PubMed] [Google Scholar]
- Smearman EL, Almli LM, Conneely KN, Brody GH, Sales JM, Bradley B, … Smith AK (2016). Oxytocin Receptor Genetic and Epigenetic Variations: Association With Child Abuse and Adult Psychiatric Symptoms. Child Dev, 87(1), 122–134. doi: 10.1111/cdev.12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonuga-Barke EJS, Kennedy M, Kumsta R, Knights N, Golm D, Rutter M, … Kreppner J (2017). Child-to-adult neurodevelopmental and mental health trajectories after early life deprivation: the young adult follow-up of the longitudinal English and Romanian Adoptees study. The Lancet, 389(10078), 1539–1548. doi: 10.1016/s0140-6736(17)30045-4 [DOI] [PubMed] [Google Scholar]
- Steiner AM, Koegel LK, Koegel RL, & Ence WA (2012). Issues and theoretical constructs regarding parent education for autism spectrum disorders. J Autism Dev Disord, 42(6), 1218–1227. doi: 10.1007/s10803-011-1194-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathearn L (2009). The elusive etiology of autism: nature and nurture? Front Behav Neurosci, 3(JUL), 11. doi: 10.3389/neuro.08.011.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathearn L, Giannotti M, Mills R, Kisely S, Najman J, & Abajobir A (2020). Long-term Cognitive, Psychological, and Health Outcomes Associated With Child Abuse and Neglect. Pediatrics, 146(4), e20200438. doi: 10.1542/peds.2020-0438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MR, Donovan K, Paterson S, Wolff JJ, Parish-Morris J, Meera SS, … Network I (2019). Early language exposure supports later language skills in infants with and without autism. Autism Res, 12(12), 1784–1795. doi: 10.1002/aur.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MR, Shen MD, Wolff JJ, Boyd B, Clements M, Rehg J, … Network I (2018). Naturalistic Language Recordings Reveal “Hypervocal” Infants at High Familial Risk for Autism. Child Dev, 89(2), e60–e73. doi: 10.1111/cdev.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, & Bick J (2013). DNA methylation: a mechanism for embedding early life experiences in the genome. Child Dev, 84(1), 49–57. doi: 10.1111/j.1467-8624.2012.01793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Zahorodny W, Peng B, Kim S, Jani N, Halperin W, & Brimacombe M (2011). The association of autism diagnosis with socioeconomic status. Autism, 16(2), 201–213. doi: 10.1177/1362361311413397 [DOI] [PubMed] [Google Scholar]
- Tonge B, Brereton A, Kiomall M, Mackinnon A, & Rinehart NJ (2014). A randomised group comparison controlled trial of ‘preschoolers with autism’: a parent education and skills training intervention for young children with autistic disorder. Autism, 18(2), 166–177. doi: 10.1177/1362361312458186 [DOI] [PubMed] [Google Scholar]
- Tonge BJ, Bull K, Brereton A, & Wilson R (2014). A review of evidence-based early intervention for behavioural problems in children with autism spectrum disorder: the core components of effective programs, child-focused interventions and comprehensive treatment models. Curr Opin Psychiatry, 27(2), 158–165. doi: 10.1097/yco.0000000000000043 [DOI] [PubMed] [Google Scholar]
- Varghese M, Keshav N, Jacot-Descombes S, Warda T, Wicinski B, Dickstein DL, … Hof PR (2017). Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol, 134(4), 537–566. doi: 10.1007/s00401-017-1736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli A, La Briola F, Peron A, Turner K, Vannicola C, Saccani M, … Canevini MP (2015). Autism spectrum disorder in tuberous sclerosis complex: searching for risk markers. Orphanet J Rare Dis, 10, 154. doi: 10.1186/s13023-015-0371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Geng H, Liu W, & Zhang G (2017). Prenatal, perinatal, and postnatal factors associated with autism: A meta-analysis. Medicine (Baltimore), 96(18), e6696. doi: 10.1097/MD.0000000000006696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye MMY, & Cheng HY (2018). Genetics and epigenetics of autism: A Review. Psychiatry Clin Neurosci, 72(4), 228–244. doi: 10.1111/pcn.12606 [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, … Meaney MJ (2004). Epigenetic programming by maternal behavior. Nat Neurosci, 7(8), 847–854. doi: 10.1038/nn1276 [DOI] [PubMed] [Google Scholar]
- Weintraub K (2011). The prevalence puzzle: Autism counts. Nature, 479(7371), 22–24. doi: 10.1038/479022a [DOI] [PubMed] [Google Scholar]
- Whitehouse AJO, Varcin KJ, Pillar S, Billingham W, Alvares GA, Barbaro J, … Hudry K (2021). Effect of Preemptive Intervention on Developmental Outcomes Among Infants Showing Early Signs of Autism: A Randomized Clinical Trial of Outcomes to Diagnosis. JAMA Pediatr, e213298. doi: 10.1001/jamapediatrics.2021.3298 [DOI] [PMC free article] [PubMed]
- Wiesel TN, & Hubel DH (1965). Comparison of the effects of unilateral and bilateral eye closure on cortical unit responses in kittens. J Neurophysiol, 28(6), 1029–1040. doi: 10.1152/jn.1965.28.6.1029 [DOI] [PubMed] [Google Scholar]
- Wiggins LD, Rubenstein E, Daniels J, DiGuiseppi C, Yeargin-Allsopp M, Schieve LA, … Levy SE (2019). A Phenotype of Childhood Autism Is Associated with Preexisting Maternal Anxiety and Depression. J Abnorm Child Psychol, 47(4), 731–740. doi: 10.1007/s10802-018-0469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury-Smith M, & Scherer SW (2018). Progress in the genetics of autism spectrum disorder. Dev Med Child Neurol, 60(5), 445–451. doi: 10.1111/dmcn.13717 [DOI] [PubMed] [Google Scholar]
- Xu G, Strathearn L, Liu B, & Bao W (2018). Prevalence of autism spectrum disorder among US children and adolescents, 2014–2016. JAMA, 319(1), 81–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Choi J, Lee WJ, & Do JT (2020). Genetic and Epigenetic Etiology Underlying Autism Spectrum Disorder. J Clin Med, 9(4). doi: 10.3390/jcm9040966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, … Jiang YH (2014). Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum Mol Genet, 23(6), 1563–1578. doi: 10.1093/hmg/ddt547 [DOI] [PMC free article] [PubMed] [Google Scholar]


