Abstract
Innate CD8 T cells correspond to a population of terminally differentiated effector T cells that phenotypically appear as antigen-experienced memory cells and functionally resemble proinflammatory CD8 T cells, expressing copious amounts of IFNγ. Innate CD8 T cells, however, are distinct from conventional effector-memory CD8 T cells as they acquire functional maturity during their generation in the thymus. Understanding the molecular mechanisms that drive their thymic development and differentiation is an intensely studied subject in T cell immunity, and here we identified the cytokine receptor γc as a critical mediator of innate CD8 T cell generation that promotes their selection even in the absence of classical MHC-I molecules. Consequently, overexpression of γc resulted in a dramatic increase of innate CD8 T cells in KbDb-deficient mice. We mapped its underlying mechanism to the expansion of IL-4-producing invariant NKT cells, so that it is the increased availability of intrathymic IL-4 which augments the selection of innate CD8 T cells. Collectively, these results unravel the selection of innate CD8 T cells being mediated by non-classical MHC-I molecules and being modulated by the abundance of the γc cytokine, IL-4.
Keywords: IFNγ, IL-4, iNKT cells, PLZF, thymus
1. Introduction
CD8 cytotoxic T cells are generated in the thymus upon a series of developmental selection processes that render them self-MHC/peptide reactive but also self-tolerant. The full maturation and acquisition of cytolytic effector function are thought to happen only after their export into and upon antigen encounter in peripheral tissues. Contrary to these conventional CD8 T cells, innate CD8 T cells acquire their effector function during their development in the thymus and prior to immune activation by foreign antigens [1]. Thus, innate CD8 T cells have been considered critical for establishing immediate-early immune response, but they also pose a risk of overt immune reaction and autoimmunity as they are already highly activated and proinflammatory under steady-state conditions. Consequently, the frequency and number of innate CD8 T cells are strictly controlled, and multiple mechanisms have been proposed to regulate and determine their abundance [1].
While their developmental pathway remains incompletely mapped, we previously documented a role of the γc cytokine receptor in promoting the generation of innate CD8 T cells [2]. Here, we expand on this observation, and we document that increased γc expression expands the pool of innate CD8 T cells through increased thymic selection and not by proliferation. Moreover, such γc-driven increase of innate CD8 T cell number was not restricted to classical MHC-I molecules, documenting a fundamental difference in their MHC-specificity and cytokine requirement with conventional CD8 T cells. These findings set the stage for further studies into assessing the distinct antigen reactivity and functional divergence of innate CD8 T cells.
2. Material and methods
2.1. Mice
C57BL/6 mice were purchased from the Charles River Laboratories. KbDb-deficient mice (JAX stock #019995) and CD1d-deficient mice (JAX stock #008881) were obtained from Jackson Laboratories. γc-transgenic mice were previously reported [3] and maintained on C57BL/6 background. Zbtb16–/– mice and Qa-1–/– mice were previously described and kindly provided by P.P. Pandolfi (Harvard Medical School) and Dorian McGavern (NINDS), respectively [4, 5]. KbDb–/–γcTg and KbDb–/–Zbtb16–/– mice were generated in-house by crossing KbDb–/– mice with γcTg and Zbtb16–/– mice, respectively. CD1d–/–γcTg mice were generated in-house by breeding CD1d–/– mice with γcTg mice. All animal experiments were approved by the NCI Animal Care and Use Committee. All mice were cared for in accordance with the NIH guidelines.
2.2. Cell isolation and purification
Thymocytes were isolated by gently teasing the thymus with forceps, then resuspending the processed tissues in ice-cold harvest media (10% FCS in RPMI-1640), before filtering the cell suspension through a 70 μm Nylon filter mesh (Millipore Sigma). Cells were washed once in harvest media by centrifugation for 7 min at 1,500 rpm, and resuspended in FACS buffer (0.5% BSA, 0.1 % sodium azide in HBSS) for staining. Liver mononuclear cells (MNC) were processed by pressing the liver tissues through a 70 μm cell strainer (BD Biosciences) into ice-cold PBS. The liver cell suspension was then briefly centrifuged at 700 rpm for 3 min to remove debris, and supernatants were collected, spun down, and the pellet was washed one more time with ice-cold PBS. The cell pellet was resuspended in 10 ml of 40% Percoll in PBS (GE Life Sciences) and layered on 10 ml of 70% Percoll in PBS. The Percoll density gradient was generated by centrifugation at room temperature for 25 min at 1,135 g. Lymphocytes at the interphase were harvested and then washed twice with harvest media (10% FCS in RPMI-1640) before further analysis. MNCs were identified by CD45 expression.
2.3. Flow cytometry
Fluorescent antibody-stained single-cell suspensions were analyzed using LSRFortessa or LSRII flow cytometers (BD Biosciences). Dead cells were excluded by adding propidium iodide. Alternatively, cells were stained with Ghost Dye Violet 510 (Tonbo) for exclusion of dead cells, followed by surface staining and fixation with Foxp3 fixation buffer for transcription factors (eBioscience) or intracellular fixation buffer for cytokines (eBioscience). Excess reagents from the staining were removed by extensive washing in FACS buffer before analyzing cells by flow cytometry.
2.4. Antibodies
Antibodies specific for the following antigens were used for staining: TCRβ (H57-597), CD4 (GK1.5), CD8α (53-6.7), CD8β (53-5.8), CXCR3 (CXCR3-173), IL-4Rα (M1), CD44 (IM7), CD45 (30-F11), CD24 (M1/69), CD69 (H1.2F3), NK1.1 (PK136), IFNγ (XMG1.2), IL-4 (11B11), Eomes (Dan11mag), T-bet (4B10), PLZF (9E12), and RORγt (Q31-378). PBS-57-loaded mouse CD1d tetramers were obtained from the NIH Tetramer Core Facility (Emory University, Atlanta, GA).
2.5. In vitro cytokine production assay
Thymocytes of the indicated mice were resuspended at 5×106 cells/mL. The cells were then stimulated in vitro with PMA (50 ng/mL) and ionomycin (1μM) in the presence of brefeldin A (3 μg/mL) for 4 hours. Following stimulation, cells were surface stained, fixed with IC Fixation Buffer (Invitrogen) and permeabilized (Permeabilization Buffer, Invitrogen) according to the manufacturer’s protocol. Fixed and permeabilized cells were then intracellularly stained with IFNγ (XMG1.2) and IL-4 (11B11) antibodies for one hour. Cells were washed and resuspended in FACS buffer prior to flow cytometry analysis.
2.6. EdU injection and incorporation assays
To assess in vivo cell proliferation, we employed EdU incorporation assays followed by Click-iT chemistry-based detection (Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit, Thermo Fisher). In brief, mice were i.p. injected with 1 mg of EdU diluted in 200 μL of PBS and analyzed 24 hours after injection for intranuclear EdU incorporation in thymocytes and thymic iNKT cells.
2.7. Statistics
Data are shown as the mean ± SEM. Two-tailed Student’s t-test was used to calculate P values. P values of less than 0.05 were considered significant, where *, P < 0.05; **, P < 0.01; and ***, P < 0.001; NS, not significant. Statistical data were analyzed using the GraphPad Prism 8 software.
3. Results
Cytotoxic CD8 T cells are generated from immature CD4, CD8 double-positive (DP) thymocytes upon positive selection by self-peptide/MHC-I molecules [6]. In mice, MHC-I molecules are primarily encoded in the H-2K and H-2D gene loci, and the genetic deficiency of MHC-I results in the impaired generation of CD8 T cells, as shown by C57BL/6 mice that are deficient for H-2 KbDb (KbDb–/–) (Fig. 1A and Suppl. Fig. 1A, 1B) [7]. Importantly, KbDb-deficiency selectively affects the generation of CD8 single-positive (SP) but not CD4SP thymocytes, resulting in a paucity of TCRβhi mature CD8SP cells without affecting CD4SP cells (Fig. 1A and Suppl. Fig. 1B). Altogether, the generation of CD8 T cells in the thymus critically depends on their TCR engagement with classical MHC-I molecules that correspond to H-2 KbDb in wild-type (WT) C57BL/6 mice.
Fig. 1: γc overexpression drives the generation of innate CD8 T cells in KbDb–/– mice.
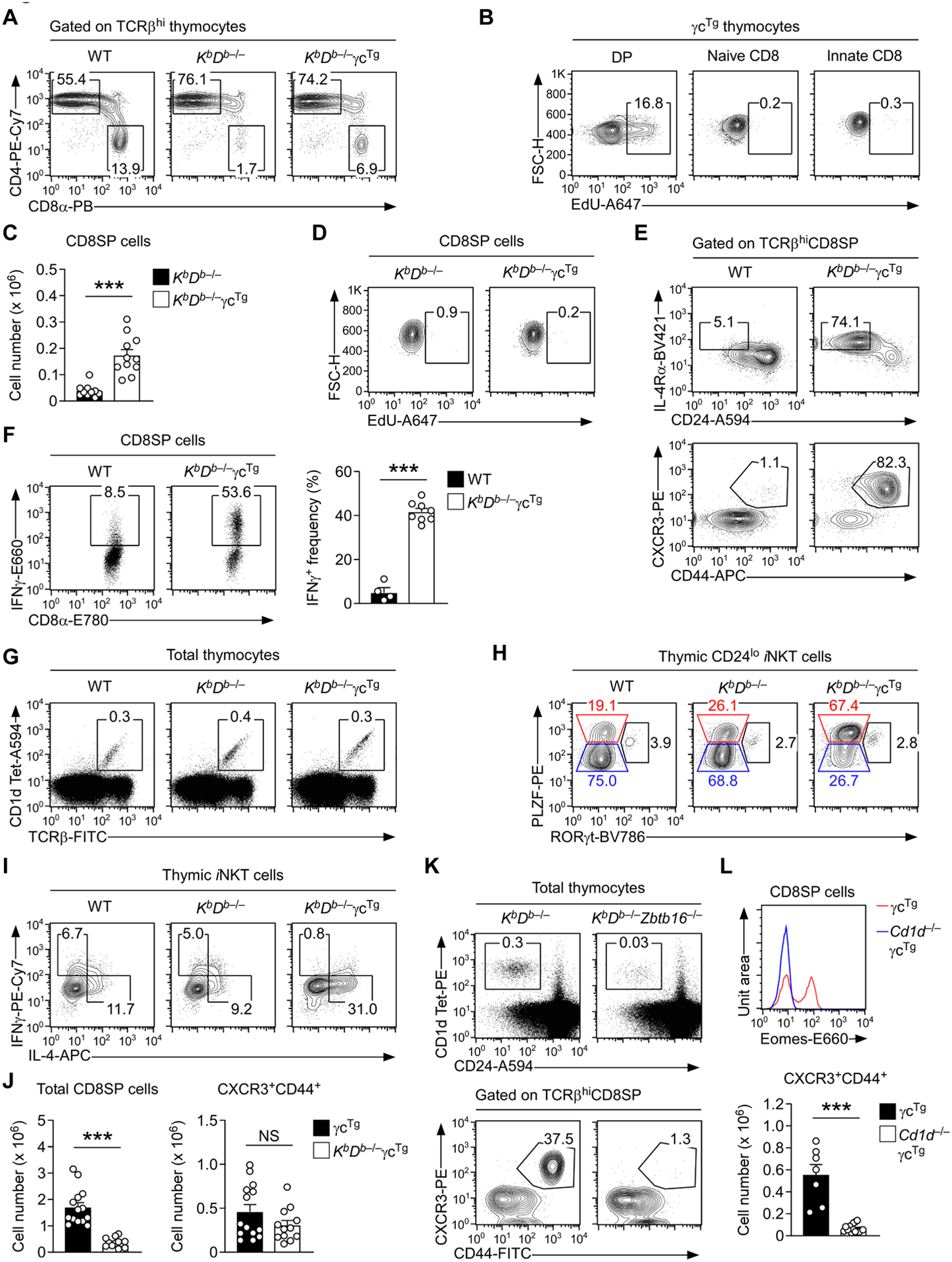
A. CD4 versus CD8 expression of TCRβhi mature thymocytes in WT, KbDb–/–, and KbDb–/–γcTg mice. The results are representative of at least eight independent experiments.
B. EdU incorporation in immature DP cells versus naïve and innate CD8 T cells of γcTg thymocytes. Mice were i.p. injected with 1 mg of EdU and analyzed 24 hours after EdU administration. The results are representative of two independent experiments.
C. Cell number of CD8SP thymocytes in KbDb–/– and KbDb–/–γcTg mice. The results are the summary of six independent experiments with a total of 10 KbDb–/– and 11 KbDb–/–γcTg mice.
D. EdU incorporation in mature CD8SP cells of KbDb–/– and KbDb–/–γcTg mice. Mice were i.p. injected with 1 mg of EdU and analyzed 24 hours after EdU administration. The results are representative of two independent experiments.
E. Identification of innate CD8 T cells by IL-4Rα versus CD24 and CXCR3 versus CD44 expression in thymic CD8SP cells of WT and KbDb–/–γcTg mice. The contour plots are representative of five independent experiments.
F. IFNγ expression in PMA- and ionomycin-stimulated CD8SP thymocytes of WT and KbDb–/– γcTg mice. The dot plots are representative (left) of five independent experiments, and the bar graph shows the summary of five independent experiments.
G. Dot plots identify and show the frequency of thymic iNKT cells in WT, KbDb–/–, and KbDb–/– γcTg mice. The results are representative of at least six independent experiments.
H. Subset composition of mature thymic iNKT cells in WT, KbDb–/–, and KbDb–/–γcTg mice as assessed by intranuclear staining for PLZF versus RORγt expression. Mature iNKT cells were identified by CD24 versus PBS-57-loaded CD1d-tetramer (CD1dTet) staining as CD24loCD1dTet+ cells. The results are representative of five independent experiments.
I. IL-4 and IFNγ production by thymic iNKT cells in WT, KbDb–/–, and KbDb–/–γcTg mice. Cytokine expression was assessed upon 4 hours of PMA and ionomycin stimulation in the presence of brefeldin A by intracellular staining, gating on mature iNKT cells. Results are representative of three independent experiments.
J. Cell numbers of total and CXCR3+CD44+ innate CD8 T cells in γcTg and KbDb–/–γcTg thymocytes. The bar graphs show the summary of six independent experiments with a total of 14 γcTg and 12 KbDb–/–γcTg mice.
K. Identification of mature iNKT cells by PBS-57-loaded CD1d-tetramer and CD24 staining (top) as well as innate CD8 T cells by CXCR3 versus CD44 expression (bottom) in thymic CD8SP cells of KbDb–/–, and KbDb–/–Zbtb16–/– mice. The contour plots are representative of five independent experiments.
L. Innate CD8 T cells in γcTg and CD1d–/–γcTg mice. Histogram (top) shows Eomes expression in TCRβhiCD8SP thymocytes of γcTg and CD1d–/–γcTg mice. Bar graph (bottom) shows the cell numbers of CXCR3+CD44+ innate CD8 T cells in γcTg and CD1d–/–γcTg thymocytes. Results show the summary of six independent experiments with a total of 7 γcTg and 16 CD1d–/–γcTg mice.
Lineage differentiation into CD8 cytotoxic T cells also depends on signaling by cytokines that are mainly transduced through cytokine receptors of the common γ-chain (γc) family [8]. Notably, the intrathymic abundance of the γc cytokine interleukin-4 (IL-4) plays a unique role in the differentiation of CD8SP thymocytes because increased levels of IL-4 induce the appearance of a distinct population of CD8 T cells with an innate-like phenotype and function that are commonly referred to as innate CD8 T cells [9]. Such IL-4-dependent innate CD8 T cells differ from unconventional “innate-like” CD8 T cells that are prominent in the liver [10], and which uniquely express the surface markers CD8αα, CD69, and NK1.1 (Suppl. Fig. 2), while depending on the transcription factor PLZF and the non-classical MHC-I molecule Qa-1 for their generation (Suppl. Fig. 3) [10]. IL-4-dependent innate CD8 T cells are also distinct from innate-like CD8αα T cells because they express the transcription factor Eomes that equips them with effector functions [11].
As such, innate CD8 T cells produce copious amounts of IFNγ, and they are marked by expressing large amounts of CD44, CXCR3, and IL-4Rα, and having downregulated the maturation marker CD24 [1]. Along these lines, we previously showed that γc overexpression promotes the generation of innate CD8 T cells by increasing the availability of intrathymic IL-4, so that γc-transgenic mice (γcTg) harbor large numbers of thymic innate CD8 T cells [2]. Thus, IL-4 is a critical factor in driving innate CD8 T cell differentiation. However, the involved mechanism remains unknown.
To address this issue, we hypothesized two distinct but not mutually exclusive mechanisms whereby either increased cell proliferation or increased selection may result in increased generation of innate CD8 T cells. To discriminate between these possibilities, we first assessed the rate of cell proliferation using EdU incorporation assays in conventional and innate CD8 T cells of γcTg mice [2]. In vivo EdU labeling for 24 hours showed strong EdU incorporation in immature DP thymocytes of γcTg mice but minimal EdU incorporation in post-selection mature CD8SP thymocytes (Fig. 1B), indicating that mature CD8 T cells were not actively proliferating. Moreover, the EdU incorporation did not differ between innate CD8 T cells and naïve CD8 T cells in the same mice (Fig. 1B). Collectively, these results indicated that IL-4 expands the size of the thymic innate CD8 T cell pool by mechanisms independent of increased cell proliferation.
We next hypothesized that IL-4 would increase the number of innate CD8 T cells by promoting their positive selection in the thymus. If such were the case, we considered it important to determine whether innate CD8 T cells are also selected by classical MHC-I molecules and thus would compete with conventional naïve CD8 T cells for MHC-I engagement. To this end, we generated γcTg mice that are KbDb-deficient (KbDb–/–γcTg) and assessed their CD8 T cell differentiation in the thymus. The generation of mature CD8SP thymocytes was severely impaired in KbDb–/– mice (Fig. 1A), but the forced expression of γc dramatically increased the overall frequency and number of CD8SP cells among KbDb–/– thymocytes (Fig. 1C and Suppl.Fig. 4A). Notably, γcTg did not increase CD8SP cell numbers by increasing cell proliferation because the EdU incorporation in CD8 T cells in KbDb–/–γcTg mice was minimal and did not differ from that in control KbDb–/– mice (Fig. 1D).
On the other hand, and in agreement with the effect of γc overexpression on CD8 T cell differentiation [2], we found that the CD8SP pool was highly enriched for innate CD8 T cells. Specifically, CD8SP thymocytes in KbDb–/–γcTg mice contained a large population of IL-4RαhiCD24lo and CXCR3+CD44+ innate phenotype cells (Fig. 1E, and Suppl. Fig. 4B) that produced copious amounts of the proinflammatory cytokine IFNγ (Fig. 1F). Together, these findings demonstrated that the forced expression of γc promotes the generation of innate CD8 T cells even in the absence of the classical MHC-I molecules, H-2 KbDb.
Innate CD8 T cell generation requires IL-4, and the intrathymic source of IL-4 has been identified as the NKT2 subset of invariant NKT (iNKT) cells [2, 9]. NKT2 cells are distinct from other thymic iNKT subsets as they express large amounts of the transcription factor PLZF but lack RORγt [12]. Overexpression of γc promotes the generation of NKT2 cells [2], and we previously demonstrated that the increased number of NKT2 cells is the cause of innate CD8 T cell generation in γcTg mice [2]. To confirm that the increase in NKT2 cells underpins innate CD8 T cell generation in KbDb–/–γcTg mice, we next assessed the thymic iNKT subset composition of KbDb–/– and KbDb–/–γcTg mice. Because iNKT cells are selected by the non-classical MHC-Ib molecule CD1d, the lack of classical KbDb molecules did not affect their overall development and maturation in the thymus (Fig. 1G and Suppl. Fig. 5A). Compared to WT control and KbDb–/– mice, however, the iNKT subsets of KbDb–/–γcTg thymocytes were dramatically skewed to NKT2 cells and conversely diminished in NKT1 cells (Fig. 1H and Suppl. Fig. 5B, 5C). Accordingly, IL-4 production was substantially increased among iNKT cells of KbDb–/–γcTg mice compared to control WT mice (Fig. 1I and Suppl. Fig. 5D). Collectively, these results suggested that classical MHC-I molecules are dispensable, but that IL-4 expression is critical for innate CD8 T cell generation in the thymus.
We next asked next whether innate CD8 T cell development in WT γcTg mice, where KbDb expression is intact, also depends on non-classical MHC-I molecules. If such were the case, we expected that KbDb-deficiency would only impair the generation of conventional naïve CD8 T cells but not of innate CD8 T cells. Enumeration of naïve and innate CD8 T cells in KbDb–/–γcTg and KbDb–/– thymocytes fully supported this hypothesis (Fig. 1J). While the number of total CD8SP thymocytes were significantly decreased in KbDb–/–γcTg mice, the number of thymic innate CD8 T cells remained unaltered between γcTg and KbDb–/–γcTg mice (Fig. 1J). Moreover, these CD8 T cells corresponded to Eomes-positive conventional innate T cells (Suppl. Fig. 6A) but not unconventional CD8αα innate-like T cells (Suppl. Fig. 6B). Collectively, these results suggested that innate CD8 T cells are generated through positive selection by non-classical MHC-I molecules and that IL-4 signaling is necessary for the lineage specification and acquisition of innate-like effector function and phenotype [8].
The developmental pathways of conventional versus innate CD8 T cells differ in their cytokine requirement because IL-4 is only required for innate but not for conventional CD8 T cells [8, 9]. Whether the MHC requirement also differs between these two distinct CD8 T cell populations is not clear. In this regard, it has been previously reported that Tec family kinase Itk-deficient mice (Itk–/–) generate a large number of innate-like CD8 T cells and that the genetic deletion of KbDb does not prevent the generation of these cells [13]. Thus, the positive selection of CD8 T cells by non-classical MHC-I molecules is also observed in other mouse models, suggesting that the MHC requirement of innate CD8 T cells is distinct from that of conventional naïve CD8 T cells [13]. In fact, the seminal notion that CD8 T cells selected by non-classical MHC-Ib molecules display features and function of innate-like cells [14] foreshadows the unique selection and maturation features of innate CD8 T cells that require MHC-Ib and IL-4, which differs from conventional CD8 T cells that mostly depend on classical MHC-I and IL-7 [6, 8].
We further confirmed such an IL-4/NKT2 cell requirement for MHC-Ib-dependent innate CD8 T cells by generating KbDb–/–mice that additionally lack PLZF, which is encoded by the Zbtb16 gene (KbDb–/–Zbtb16–/–) (Suppl. Fig. 7A); these mice failed to generate functional iNKT cells (Fig. 1K, top, and Suppl. Fig. 7B) [15] and thus did not generate IL-4-producing NKT2 cells. Moreover, we found that the few innate CD8 T cells that develop in KbDb–/–mice were completely absent in KbDb–/–Zbtb16–/– mice (Fig. 1K, bottom, and Suppl. Fig. 7C). The same lack of innate CD8 T cells was observed when deleting the non-classical MHC-I molecule CD1d, which is absolutely required for iNKT cell generation (Suppl. Fig. 8A) [16], and which also turned out to be indispensable for the generation of Eomes-expressing innate CD8 T cell (Fig. 1L and Supp. Fig. 8B). Altogether, these results affirm a central and non-redundant role of iNKT cells and NKT2-derived intra-thymic IL-4 as drivers of innate CD8 T cells.
4. Discussion
In this study, we document a distinct cytokine requirement, i.e., IL-4, in the generation of innate CD8 T cells, whereby IL-4 is necessary to promote the selection but not the proliferation/expansion of innate CD8 T cells in the thymus. Moreover, our data also show that IL-4 is not necessary because it drives the conversion of CD8 T cells that have been selected by classical MHC-I molecules into innate CD8 T cells. Instead, we found that IL-4 promotes the overall selection of innate CD8 T cells, establishing a pool of CD8 effector T cells that are restricted to recognize antigens in the context of non-classical MHC-I molecules. It remains unclear whether innate CD8 T cells are cross-reactive to classical MHC-I molecules and whether they are promiscuous in their TCR reactivity. Nonetheless, our present findings established that IL-4/NKT2-mediated generation of innate CD8 T cells is independent of classical MHC-I KbDb molecules and thus, distinct in their selection and differentiation processes from conventional CD8 T cells. Understanding the role and requirement of innate CD8 T cells in immune surveillance and the immediate-early immune response is still in its infancy. Our findings showing that thymic innate CD8 T cells are restricted to antigens in the context of non-classical MHC-I molecules provide a new perspective on their antigen reactivity. The fact that increased expression of the cytokine receptor γc prompts increased generation of innate CD8 T cells puts a twist into their developmental pathway which will impact our further assessment of this unique population of effector T cells.
Supplementary Material
Highlights.
Cytokine receptor γc promotes innate CD8 T cell generation through upregulating IL-4
Increased innate CD8 T cell generation by γc is independent of classical MHC-I
Classical MHC-I-deficiency reveals an iNKT/IL-4 axis of innate CD8 T cell selection
Funding
This work was supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research and by a National Research Foundation of Korea grant (NRF-2018R1A5A2024418) funded by the Korea Ministry of Science and ICT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflicts of interest.
References
- [1].Jameson SC, Lee YJ, Hogquist KA. Innate memory T cells. Adv Immunol, 2015;126:173–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Park JY, Won HY, DiPalma DT, Hong C, Park JH. Protein abundance of the cytokine receptor gammac controls the thymic generation of innate-like T cells. Cell Mol Life Sci, 2021;79:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hong C, Luckey MA, Ligons DL, Waickman AT, Park JY, Kim GY et al. Activated T cells secrete an alternatively spliced form of common gamma-chain that inhibits cytokine signaling and exacerbates inflammation. Immunity, 2014;40:910–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet, 2000;25:166–72. [DOI] [PubMed] [Google Scholar]
- [5].Hu D, Ikizawa K, Lu L, Sanchirico ME, Shinohara ML, Cantor H. Analysis of regulatory CD8 T cells in Qa-1-deficient mice. Nat Immunol, 2004;5:516–23. [DOI] [PubMed] [Google Scholar]
- [6].Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol, 2008;8:788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vugmeyster Y, Glas R, Perarnau B, Lemonnier FA, Eisen H, Ploegh H. Major histocompatibility complex (MHC) class I KbDb −/− deficient mice possess functional CD8+ T cells and natural killer cells. Proc Natl Acad Sci U S A, 1998;95:12492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Etzensperger R, Kadakia T, Tai X, Alag A, Guinter TI, Egawa T et al. Identification of lineage-specifying cytokines that signal all CD8(+)-cytotoxic-lineage-fate ‘decisions’ in the thymus. Nat Immunol, 2017;18:1218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol, 2013;14:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sheng H, Marrero I, Maricic I, Fanchiang SS, Zhang S, Sant’Angelo DB et al. Distinct PLZF(+)CD8alphaalpha(+) Unconventional T Cells Enriched in Liver Use a Cytotoxic Mechanism to Limit Autoimmunity. J Immunol, 2019;203:2150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity, 2009;31:122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Park JY, DiPalma DT, Kwon J, Fink J, Park JH. Quantitative Difference in PLZF Protein Expression Determines iNKT Lineage Fate and Controls Innate CD8 T Cell Generation. Cell Rep, 2019;27:2548–57 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity, 2006;25:93–104. [DOI] [PubMed] [Google Scholar]
- [14].Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol, 2002;3:772–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol, 2008;9:1055–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shissler SC, Webb TJ. The ins and outs of type I iNKT cell development. Mol Immunol, 2019;105:116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


