Abstract
Magnetic resonance fingerprinting (MRF) is an approach to quantitative magnetic resonance imaging that allows for efficient simultaneous measurements of multiple tissue properties, which are then used to create accurate and reproducible quantitative maps of these properties. As the technique has gained popularity, the extent of preclinical and clinical applications has vastly increased. The goal of this review is to provide an overview of currently investigated preclinical and clinical applications of MRF, as well as future directions. Topics covered include MRF in neuroimaging, neurovascular, prostate, liver, kidney, breast, abdominal quantitative imaging, cardiac, and musculoskeletal applications.
Keywords: MR fingerprinting, clinical applications, liver, kidneys, prostate, neuroimaging, musculoskeletal imaging, relaxometry, MRF
Magnetic resonance imaging (MRI) has become a mainstay of medical imaging over the past several decades. Magnetic resonance imaging is widely known for its excellent soft tissue contrast and is often a preferred imaging modality because it also does not use ionizing radiation. Currently, MRI heavily relies on subjective interpretation of signal characteristics on various MR sequences including T1-weighted, T2-weighted, proton density-weighted, diffusion-weighted, and postcontrast/perfusion sequences. These sequences are generated by systematic variation of numerous parameters such as echo time, repetition time (TR), and flip angle (FA).1 Radiologists generally evaluate signal using intensity descriptors such as “hyperintense” or “hypointense” and use the expected signal appearance of various tissues and associated pathologies to render a differential diagnosis. However, this subjective analysis of relatively “weighted” images is only a fraction of the tissue property information that could be provided by MRI techniques. The very concept of “weighting” can start to break down when more than 1 tissue is being compared.2,3 In addition, limitations of current MRI technologies include variable image acquisition across scanners, limited reproducibility of images, and known interreader variability in image interpretation. Quantitative MRI provides a unique and discrete analysis of tissue parameters and holds promise in addressing some of these limitations.4
To date, the most common quantitative MRI properties used in clinical practice are diffusion-weighted imaging, fat fraction (FF) mapping, and perfusion mapping. Quantification and mapping of T1 and T2 relaxation times, which affect all MR signals, are not commonly performed given the additional significant scan time that would be required in traditional serial MRI protocols. Magnetic resonance fingerprinting (MRF) is an approach to quantitative MRI that strays from traditional acquisitions for property mapping. Instead of an approach in which multiple images differently weighted by the property at hand are acquired and fit to a model, a pseudorandomized acquisition in a single sequence is used, such that any unique combination of these properties each generates unique signal evolutions or “fingerprints.” These can be matched to a predefined dictionary of possible signal evolutions to generate quantitative maps, allowing for measurement and analysis of multiple different MR parameters.5 The ideal MRF experiment yields rapid and accurate mapping of interesting properties, and the approach has been shown to be repeatable and reproducible in early applications.6-13 The MRF approach provides an opportunity for clinically feasible quantitative mapping of multiple tissue properties in disease diagnosis and follow-up. Multiple clinical applications of the technique have been attempted in recent years. The goal of this review is to provide a brief technical overview of MRF acquisition and processing, to review currently investigated clinical applications, to discuss challenges in MRF clinical application, and to look to the future of MRF in medical imaging.
TECHNICAL OVERVIEW
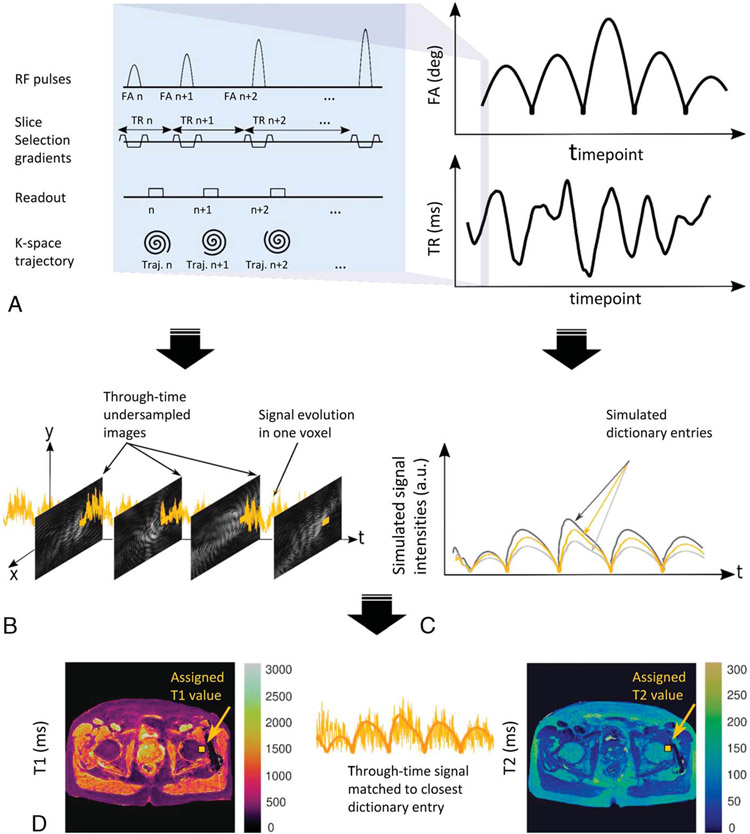
Magnetic resonance fingerprinting was proposed as a method to deliver simultaneously collected, accurate, and reproducible maps of multiple tissue properties in a single scan, while minimizing imaging time to facilitate clinical adoption. Magnetic resonance fingerprinting takes a different approach from conventional MRI for data acquisition and image reconstruction. Conventional MR uses repeated acquisition parameters in a particular sequence until all of k-space is filled, and then this is done serially for other sequences. Image contrast is created by reconstructing images that weight relaxation of protons in one effect; weighting toward optimization of longitudinal relaxation corresponds to “T1-weighting,” and weighting toward optimization of transverse relaxation corresponds to “T2-weighting.” In contrast, in MRF, the goal is to drive the signal away from a steady state by creating transient variabilities in signal that are highly sensitive to the tissue properties of interest. To do this, for data acquisition, parameters such as FA, TR, and echo time are varied in a pseudorandom manner (Fig. 1A) to generate a unique signal time-course for each tissue property of interest. The unique signal time-course can be simulated or calculated from MR scanner settings and basic physical principles. Each predicted signal evolution based on tissue properties can be stored within a comprehensive dictionary of potential time-courses (Fig. 1C).
FIGURE 1.
Representative example of MRF framework, with the typical steps shown: A, example of MR parameter variation for data acquisition; B, measured signal time-course, generated for each voxel; C, generation of simulated time-courses as dictionary entries; D, template matching algorithm used to match the voxel-specific signal time-course to the closest dictionary entry, repeated for each voxel to generate an MRF property map.
When the MRF sequence is run on the scanner, a measured signal time-course is generated for each voxel (Fig. 1B). Each voxel-specific signal evolution can then be differentiated into tissue type by pattern recognition. A template matching algorithm (Fig. 1D) is used to match the voxel-specific signal to the closest dictionary entry. Subsequently, the property values that went into calculating the dictionary entry are assigned to the voxel in representative MR property maps. The name “MR fingerprinting” references an analogy to human fingerprinting, in which the obtained signal (fingerprint) is compared with a dictionary of potential signals (or fingerprints), and the best match is selected to identify the property (or person) of interest. Repeating this process for every voxel yields MRF property maps.
Individual images obtained to generate the voxel time-courses are typically highly undersampled to minimize acquisition time and cannot be used diagnostically. However, the experiment is designed to vary acquisitions so that artifacts appear spatially and temporally incoherent, such that the eventual maps are not degraded and can be used diagnostically for spatial localization of pathology. This is an advantage of MRF data acquisition; although the rate-limiting step in conventional MRI is minimizing artifact to optimize diagnostic quality of each image, in MRF, this is not as important as the primary goal is to obtain usable quantitative data that can be accurately matched to its corresponding dictionary entry. It has been shown that, despite undersampling of underlying images, the maps are still quantitatively accurate, anatomy can be well characterized, and the measurements are reproducible and repeatable.5,14
The MRF framework offers great flexibility, since nearly any sequence structure and readout trajectory can be used if the signal can be sensitized sufficiently to the property that is to be mapped. Although the concept was initially introduced and demonstrated for mapping of T1 and T2 relaxation times, the approach can be used for any property of interest, and indeed even non-MR data could be analyzed in this manner. The example provided in Figure 1 illustrates a representative sequence structure and its corresponding TR and FA patterns. A Bloch equation simulation15 or extended phase graph16,17 has been used to calculate the dictionary. A notable caveat is that creation and use of the MRF dictionary vary based on what is being measured and the clinical question. For most clinical uses, the dictionary can be calculated once based on the acquisition parameters, and the same dictionary can be used across multiple patients. However, there are certain clinical uses for which a new dictionary needs to be generated for each patient. In MRF for cardiac imaging, a new dictionary is calculated for each patient to adjust for variable heart rates.18 Once an MRF application has been designed for a certain organ or pathology, a key step toward widespread clinical integration is assessing repeatability and reproducibility across multiple sites, scanners, and patients. This is central to the goals of objective and reliable differentiation of pathology or treatment response.4 Significant repeatability and reproducibility research has been performed for individual clinical scenarios; as appropriate, these studies are discussed in representative clinical sections below.
Since the introduction of MRF in 2013, substantial technical work has been performed to improve every step of the fingerprinting experiment, including image acquisition, sensitization to various parameters, dictionary generation, pattern matching, and quantitative analysis. These studies are extensive but beyond the scope of this review, which is focused primarily on clinical application of the technology. The reader is directed to excellent technical reviews of the subject.18-22
CURRENT CLINICAL APPLICATIONS
Neuroimaging
Magnetic resonance fingerprinting was initially applied largely in neuroimaging, which is unsurprising given the first published description of MRF itself was in the brain.5 Since then, the last decade has seen rapid technical developments and expansion of multiple clinical applications of MRF in neuroimaging. Various brain MRF techniques have been used for characterization of healthy brain tissue, neurological tumors, epilepsy imaging, neonatal neuroimaging, and neuro-aging. There is also a nascent but growing body of work focusing on neurovascular imaging applications.
From a technical perspective, the first clinically validated brain MRF sequence was a multislice 2-dimensional (2D) MRF sequence based on a balanced steady state free precession (bSSFP) acquisition, given known bSSFP high signal-to-noise ratio and T1/T2 contrast.5,23 This initial MRF sequence provided simultaneous T1, T2, and proton density (M0) mapping.5,24 Since then, brain MRF technology has progressed, including the development of isotropic 3-dimensional (3D) MRF, which can be obtained in a total of 4–10 minutes and provides further improved resolution with generation of anatomically-aligned T1 and T2 quantitative maps. The initial 3D MRF sequence described by Ma et al25 is based on the fast imaging with steady-state precession (FISP) acquisition instead of the bSSFP used for 2D MRF. Magnetic resonance fingerprinting–FISP was validated in neuroimaging after the initial use of bSSFP, with Jiang et al26 showing that MRF-FISP provides similar values to bSSFP but with additional benefit of eliminating the classic banding artifact seen in bSSFP from field inhomogeneities. As FISP on higher strength magnets remains at risk for transmit field (B1) inhomogeneity, a separate 3D B1 map can be acquired in conjunction with the 3D MRF acquisition to account for this problem. The B1 values are simulated into the MRF dictionary to correct for inaccurate FAs and field inhomogeneity resulting from the large volume brain coverage, to further improve accuracy of T1 and T2 values provided in 3D brain MRF. Another elegant way of correcting for B1 inhomogeneity is to incorporate B1 as an MRF parameter, as demonstrated by Buonincontri and Sawiak.27 Other 3D MRF sequences have used varying readout trajectories, parallel imaging, and deep learning to accelerate data sampling, accelerate dictionary matching, and to reduce artifacts.28,29
Brain MRF has also been modified for measurement of quantitative properties beyond T1 and T2. Examples include MRF techniques for measurement of combined T1, T2, and T2* mapping30-33; MRF with b-tensor encoding for combined T1, T2, and apparent diffusion coefficient (ADC) mapping34; and CEST-MRF for brain tumors,35 vascular MR fingerprinting,36 and MRF–arterial spin labeling (ASL) for cerebral perfusion.37 The test-retest repeatability as well as interscanner repeatability across multiple sites have been established for both 2D MRF and 3D MRF sequences.7,38-40
Magnetic resonance fingerprinting has been widely validated in brain tumors. T1 and T2 relaxation times obtained from both 2D MRF and 3D MRF have been used to differentiate low-grade gliomas, high-grade gliomas, and metastases.41,42 In a study of 31 adult patients with untreated brain gliomas and metastases, Badve et al41 demonstrated that mean T2 values could differentiate low-grade glioma solid tumor from metastasis (mean, 172 ± 53 milliseconds, and 105 ± 27 milliseconds, respectively; P = 0.004) and that the mean T1 of peritumoral white matter surrounding low-grade gliomas was significantly lower than peritumoral white matter around glioblastomas (mean 1066 ± 218 milliseconds, and 1578 ± 331 milliseconds, respectively; P = 0.004). In a similar study of 23 children and young adults by de Blank et al,42 high-grade gliomas had significantly higher T1 and T2 compared with low-grade gliomas (T1: 1863 ± 70 milliseconds vs 1355 ± 187 milliseconds, P = 0.007; T2: 90 ± 13 milliseconds vs 56 ± 19 milliseconds, P = 0.013), and peritumoral white matter around low-grade gliomas had significantly lower T1 than peritumoral white matter around high-grade gliomas (T1: 1154 ± 253 milliseconds vs 1581 ± 476 milliseconds, P = 0.039). In a study by Konar et al,43 both treated and untreated brain metastases showed higher T1 and T2 values than normal brain structures. Of note, in both the study by de Blank et al42 and Konar et al,43 there was no significant difference found between treated and untreated lesions, possibly due to small sample size, and this remains to be explored.42,43
Efforts have also been made to differentiate grade and subtypes of gliomas using MRF. Radiomic analysis of 2D MRF and 3D MRF has demonstrated features helpful for glioma grade differentiation,44-46 and MRF radiomics have also shown utility in prediction of IDH1 mutations.47 Haubold et al46 investigated the feasibility of multiparametric MRI (mpMRI), MRF, and 18F-fluoethyl-L-tyrosine (18F-FET) on a PET/MRI scanner to predict mutational status in 42 patients with primary brain gliomas. Magnetic resonance fingerprinting T1 and M0 were useful for prediction of 1p19q mutations (area under the curve [AUC], 0.978) and IDH1 mutation (AUC, 0.88). Similar results were obtained by Tippareddy et al,45 who found that radiomics features derived from precontrast 3D MRF T1 maps could predict IDH1 mutation status and also correlate with overall survival. These initial results are intriguing as the most useful radiomics features are derived from precontrast MRF T1 maps, whereas in current clinical practice, precontrast T1-weighted images have limited utility compared with postcontrast. Of note, although radiomics features derived from MRF are highly repeatable across various step sizes in a particular dictionary, they are limited by interdictionary variations.48 Since MRF-derived T1 and T2 may not require intensity normalization algorithms usually needed for qualitative MR images, MRF radiomics may be well suited for longitudinal studies across sites and scanners.49
Magnetic resonance fingerprinting has also been used to characterize nonglioma tumors such as meningiomas and pituitary macroadenomas.50-53 Zhang et al52 compared MRF-derived T1 and T2 values between pathologically confirmed subtypes of meningiomas and found that meningothelial meningiomas had significantly higher T1 and T2 values than transitional and fibrous meningiomas. Magnetic resonance fingerprinting T1 with MRF T2 showed ability to differentiate meningothelial meningiomas from other subtypes, whereas conventional weighted imaging and ADC values did not predict differentiation.52 In preoperative evaluation of pituitary macroadenomas, gonadotroph pituitary macroadenomas had significantly higher MRF-derived T1 and T2 values than nongonadotroph pituitary macroadenomas, and a combination of T1 and T2 could differentiate between the 2 subtypes. This distinction can have prognostic value, as cavernous sinus invasion is more likely with nongonadotroph pituitary macroadenomas.53
Because of flexibility in sequence designs and multiproperty mapping, MRF may be beneficial for radiation therapy treatment planning protocols for longitudinal assessment of changes in primary brain tumoral properties.54 The feasibility of MRF has been demonstrated on both high- and low-field strength hybrid MR-guided linear accelerators used for radiation therapy.55,56 Because MRF on MR-guided linear accelerator systems has been shown to be technically feasible, able to pick up subtle quantitative changes, and adds little scan time, there is potential for monitoring of posttherapy change and possible image-guided dose plan adaptation in the future.55,56 In addition, despite the large variations in B1 fields with the flexible MR coils used in radiation therapy, 3D MRF T1 maps show less B1-inhomogeneity induced intensity differences compared with qualitative T1-weighted images.10 Depending on the sequence structure selected, MRF can be made relatively tolerant to geometric distortion secondary to susceptibility artifact at air-tissue interfaces common in the skull (eg, skull base, sinonasal region, pituitary, and optic nerves) as compared with other functional sequences, especially those based on echo-planar imaging.10
The role of MRF in the brain has also expanded beyond tumor-related imaging. In patients with epilepsy, MRI can play an important role in detection of epileptogenic brain lesions.57 However, many pathologically proven epileptogenic lesions can be “MRI invisible” on visual assessment of conventional images, and quantitative analysis has the potential to improve detection.58 Magnetic resonance fingerprinting has been explored in both temporal lobe epilepsy (TLE)59 and extratemporal lobe epilepsy.60 When MRF was added to a clinical protocol in 33 patients with mesial TLE, Liao et al59 found that MRF increased correct diagnosis rate (correct diagnosis on MRF: 32/33 [96.9%] vs on conventional MRI: 23/33 [69.7%]) of TLE. The MRF T1 and T2 values were significantly higher in hippocampal sclerosis compared with normal hippocampus in healthy controls. Patients with hippocampal sclerosis have also shown increased T1 and T2 of ipsilateral temporal lobe white matter compared with the contralateral side as well as to healthy controls.61 In extratemporal lobe epilepsy, partial-volume analysis of MRF maps can generate gray matter and white matter maps to improve detection of focal cortical dysplasia and subtle epileptogenic lesions at the gray-white matter junction.62,63 In a study by Ma et al,62 3D MRF was added to the clinical protocol in 15 patients with epilepsy; in 4/15 (27%) patients, MRF maps detected additional findings that were not visible on conventional high-resolution T1-weighted T2-weighted, and fluid-attenuated inversion recovery images. Furthermore, 3D MRF-detected epileptogenic lesions were concordant with stereotactic electroencephalogram localization. In patients with multifocal lesions such as in periventricular nodular heterotopias, voxels within stereotactic electroencephalogram-localized seizure onset zones have shown higher T1 values compared with voxels outside these zones.60 In addition, in these patients, MRF T1 values have been shown to be associated with ictal-onset electroencephalogram waveforms, suggesting potential associations between tissue relaxometry and electrophysiological changes.60 In patients with medically intractable focal epilepsy, 3D MRF T1 and T2 maps combined with machine learning have shown potential in lateralizing seizure onset zones in patients with visually normal basal ganglia and thalami.64
Research has also been done in using brain MRF for assessment of pediatric brain myelination and evaluation of neurodevelopmental disorders.63,65,66 Magnetic resonance fingerprinting combined with partial volume analysis can be used to generate coregistered T1, T2, and myelin water fraction maps in neonates and small children within a clinically feasible time.63,65 Chen et al65 scanned 28 healthy children between 0 and 5 years old with multislice 2D MRF without sedation and demonstrated that myelin water fraction is undetectable at 0–6 months of age but increases after 6 months of age. Depending on brain regions, rapid increases in myelin water fraction were noted between 6 and 12 months, and between 6 and 18 months of age followed by a slower increase after 20 months of age. Age-related dependence was noted with earlier myelination in the corpus callosum, followed by parieto-occipital white matter, and lastly in frontotemporal white matter. Neonates demonstrated longer T1 and T2 in unmyelinated white matter compared with older children, with a marked decrease in T1 and T2 values until 20 months of age due to progressive myelination. These quantitative trends correlate with known age-dependent myelination patterns seen on conventional T1- and T2-weighted MRI.65 In a separate study, Yu et al67 demonstrated the feasibility of 3D MRF in 25 neonates with whole-brain coverage (scan time <5 minutes) and demonstrated regional differences in white matter T1 and T2.
Magnetic resonance fingerprinting–relaxometry has also been used to provide additional insights into neuroaging and neurodegenerative conditions,68,69 multiple sclerosis,70 and traumatic brain injury.71 Patients with multiple sclerosis have shown differences in splenium T1 and T2 and normal-appearing white matter compared with age-matched healthy controls, with negative correlation of splenium heterogeneity with increasing time since multiple sclerosis diagnosis.70 Whole-brain coverage with 3D MRF can be used for detection of demyelinating plaques and to quantify longitudinal changes.70,72 In patients with frontotemporal dementia, when compared with healthy controls, significantly longer T1 relaxation times have been found in cortical gray matter and global white matter, longer T1 and T2 relaxation times have been found in hippocampi, and T1 relaxation times in the amygdala and hippocampi are strongly correlated with duration of clinical symptoms and disease severity respectively.68 Finally, a small cohort study has shown promise for following traumatic brain injury using MRF. Gerhalter et al71 evaluated 22 patients with mild traumatic brain injury, approximately 1 month and 3 months after injury with conventional weighted imaging, MRF, and diffusion tensor imaging. Although there were no differences in baseline MRF-derived T1 and T2 between patients and healthy controls, a higher T1 at baseline with serially increasing T1 on follow-up correlated with poor recovery at 3 months. T1 showed better prospective identification of nonrecovered patients compared with T2, ADC, and fractional anisotropy from diffusion tensor imaging.71
To supplement research of diffuse neurological disease processes, brain atlases have been generated from 3D MRF in healthy volunteers, which provide highly repeatable and reproducible normative data for T1 and T2 maps, cortical thickness, and subcortical volumes.40,73,74 These 3D MRF-derived normative relaxometry and morphometry atlases can potentially be used as references to evaluate diseases with diffuse brain involvement (eg, multiple sclerosis, epilepsy, and dementia) to detect subtle changes, which may not be visible on qualitative assessment of images.
Magnetic resonance perfusion applications are also beginning to emerge. Magnetic resonance fingerprinting can be adapted to ASL framework (MRF-ASL) to provide noninvasive perfusion parameters of the whole brain. The evaluation of MRF-ASL in the ischemic stroke setting has shown ability to differentiate between stroke regions and contralateral normal brain, with stroke regions showing lower cerebral blood flow, longer bolus arrival time, and longer T1 (P < 0.001). In the same study, MRF-ASL parameters were shown to be predictive of National Institute of Health Stroke Scale score.75 Magnetic resonance fingerprinting–ASL has also been preliminarily explored in the setting of Moyamoya disease, with Su et al76 finding that bolus arrival time was prolonged (P < 0.001) and T1 values were longer (P < 0.05) in stenotic internal carotid artery territories compared with healthy controls. Perfusion parameters from MRF-ASL in Moyamoya disease were significantly correlated with parameters derived from conventional ASL perfusion and contrast-enhanced dynamic susceptibility contrast perfusion.76 Further similar techniques and direct clinical implications of such measurements are likely to emerge in the future.
Prostate Imaging
Men with known or suspected prostate cancer are increasingly evaluated with prostate mpMRI followed by transrectal ultrasound-mpMRI fusion-guided biopsy. This workflow has led to increased diagnosis of clinically significant prostate cancer and avoidance of unnecessary prostate biopsies.77 Prostate mpMRI uses T2-weighted images, diffusion-weighted images (high b-value sequences and ADC maps), and dynamic contrast-enhanced images. The evaluation of these sequences for cancer suspicious lesions is guided by recommendations provided in the Prostate Imaging and Reporting Data System (PI-RADS) 5-point scoring system, currently in version 2.1.78 Generally, lesions are more suspicious for cancer if they are T2 hypointense, have impeded water self-diffusivity, and show early postcontrast enhancement. The extent of suspicious features on T2-weighted images serves as the primary determinant of the final PI-RADS score in the transition zone (TZ) of the prostate, whereas diffusion-weighted image characteristics serve as the primary determinant of final PI-RADS score in the peripheral zone (PZ).79
Unfortunately, interpretation of prostate mpMRI suffers from several known limitations, many of which are related to the subjective nature of assessment and reporting. First, it is known that cancer detection is variable by reader experience, and that there is a learning curve to prostate mpMRI.80,81 In addition, prostate mpMRI acquisition can be variable across scanners, and the appearance of the prostate is heterogeneous across multiple glands, which further complicates the learning process for novice radiologists. Finally, despite the creation of PI-RADS, there remains known interreader disagreement, even among expert readers.82 Differences in appearances across images from different vendors are a known problem in prostate imaging.4 Given these limitations, there is interest in the community in investigating quantitative solutions. Moreover, reliable differentiation of lesions according to grade remains an important goal in addition to diagnosis, as this could decrease both needed biopsies and overtreatment. The ADC values have shown inverse correlation with prostate cancer pathology score, and studies have suggested that there may be benefit to using these values as an adjunct to mpMRI interpretation.83-87 T2 mapping has also been explored as a possible quantitative measure, and it has been shown that there is T2 relaxometry differentiation in PZ versus TZ as well as in differentiating prostatitis, benign prostatic hyperplasia, and prostate cancer.88-90 It has also been suggested that T2 mapping can be combined with ADC values to improve detection of prostate cancer.91-93 Unfortunately, conventional T2 mapping of the prostate takes up to 6 minutes, which has not yet been routinely incorporated into prostate MRI examinations.
Prostate MRF can provide reproducible rapid quantitative data on multiple tissue properties and help differentiate tissue changes in benign and malignant disease. A summary of early prostate MRF clinical validation is provided in Table 1.94-97
TABLE 1.
Published Values in Prostate MRF
| Study and Field Strength | Patient population, Mapped properties |
Reported Values | Study Notes |
|---|---|---|---|
| Peripheral zone (3 T)94 |
140 patients MRF-derived T1, T2 ADC mapping |
PCa: P < 0.0001 from NPZ T1: 1628 ± 344 ms T2: 73 ± 27 ms ADC: 0.773 × 10−3 mm2/s ± 0.331 Prostatitis: P < 0.0005 from NPZ T1: 1707 ± 377 ms T2: 79 ± 37 ms ADC: 0.911 × 10−3 mm2/s ± 0.239 NPZ: T1: 2247 ± 450 ms T2: 169 ± 61 ms ADC: 1.711 × 10−3 mm2/s ± 0.269 |
Notable AUCs (95% CI): PCa vs NPZ: 0.99 (0.98, 0.99) [ADC, T1, T2] High-grade vs low-grade PCa: 0.83 (0.71, 0.94) [ADC, T2] Prostatitis vs NPZ: 0.99 (0.97, 1.00) [ADC, T1] |
| Peripheral zone (3 T)95 |
85 patients MRF-derived T1, T2 ADC mapping |
PCa: (P < 0.05 from Prostatitis, NPZ) T1: 1660 ± 270 ms T2: 56 ± 20 ms ADC: 0.70 × 10−3 mm2/s ± 0.24 × 10−3 Prostatitis: T1: 1730 ± 350 ms T2: 77 ± 36 ms ADC: 1.00 × 10−3 mm2/s ± 0.30 × 10−3 NPZ: T1: 1810 ± 250 ms T2: 71 ± 37 ms ADC: 1.00 × 10−3 mm2/s ± 0.33 × 10−3 |
Notable AUCs: PCa vs NPZ: 0.83 [T1, ADC] PCa vs noncancer (NPZ + Prostatitis): 0.80 [T1, ADC]; 0.80 [ADC] High-grade vs low-grade PCa; 0.90 [ADC, T2] Proposed cutoffs (clinically significant cancer vs noncancer): T1: 1720 to 1730 ms T2: 52 to 60 ms ADC: 0.75 to 0.78 × 10−3 mm2/s |
| Transition zone (3 T)96 |
67 patients MRF-derived T1, T2 ADC mapping |
PCa: P < 0.001 from NTZ T1: 1450 ± 110 ms T2: 36 ± 11 ms ADC: 0.57 × 10−3 mm2/s ± 0.13 × 10−3 NTZ: T1: 1800 ± 150 ms T2: 65 ± 22 ms ADC: 1.13 × 10−3 mm2/s ± 0.19 × 10−3 Noncancer (NTZ + prostatitis): T1: 1620 ± 120 ms T2: 47 ± 16 ms ADC: 0.82 × 10−3 mm2/s ± 0.13 × 10−3 |
Notable AUCs (95% CI): PCa vs NTZ: 0.99 (0.98, 1) [ADC, T2] PCa vs noncancer: 0.94 (0.88, 0.98) [ADC, T1] Clinically significant PCa vs insignificant lesions (Gleason 6 + noncancers): 0.81 (0.69, 0.91) [ADC, T1] PI-RADS category 3 noncancers vs cancer: 0.79 (0.57, 0.96) [T1] |
| Pregadolinium and postgadolinium (3 T)97 |
14 patients MRF-derived T1, T2 measured pre- and post- gadolinium administration |
PZ lesions (pregadolinium): T1: 1640 ± 368.1 ms T2: 507.8 ± 292.7 ms* NPZ (pregadolinium): T1: 2521 ± 405.9 ms T2: 546.7 ± 294 ms* TZ lesions (pregadolinium): T1: 1696 ± 200.5 ms T2: 372.2 ± 209 ms* NTZ (pregadolinium): T1: 1753 ± 444.7 ms T2: 451 ± 228 ms* Postgadolinium T1 and T2 also provided, demonstrating shortening of both values. |
Conventional mapping comparisons - PZ lesions (pregadolinium): VFA T1: 1986 ± 629.5 ms MSE T2: 89.2 ± 21.8 ms NPZ (pregadolinium): VFA T1: 2188 ± 813.9 ms MSE T2: 139.4 ± 79.12 ms TZ lesions (pregadolinium): VFA T1: 2002 ± 413.7 ms MSE T2: 69.3 ± 9.8 ms NTZ (pregadolinium): VFA T1: 2118 ± 732.1 ms MSE T2: 88.56 ± 11.67 ms |
MRF-derived T1, T2 values are provided from various studies, along with ADC values if available. Values provided as mean ± standard deviation. Please refer to the referenced articles for further detail.
MRF T2 values were unreliable in this study.
MRF, magnetic resonance fingerprinting; ADC, apparent diffusion coefficient; PCa, prostate cancer; AUC, area under the curve; CI, confidence interval; NPZ, normal peripheral zone; NTZ, normal transition zone; PZ, peripheral zone; TZ, transition zone; VFA, variable flip angle; MSE, multiple spin echo.
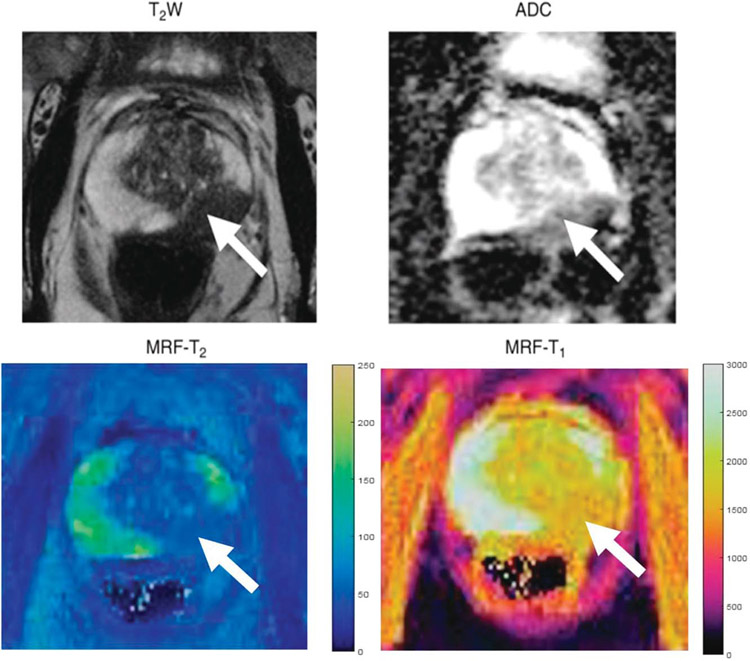
Initial prostate MRF was applied in the PZ, where a majority of prostate cancers are currently detected. An example of MRF-derived T1 and T2 mapping in the prostate PZ is provided in Figure 2. Yu et al94 showed that MRF T1 and T2 values were found to be significantly lower in PZ cancers when compared with normal PZ (P < 0.0001). Conventionally obtained ADC values were also found to be significantly lower in cancer than in normal PZ, and in combined models using ADC, T1, and T2, an AUC of 0.99 was achieved in differentiation of cancer from normal PZ. In addition, there was utility in combining ADC and T2 in distinguishing high-grade from low-grade prostate cancer. Using targeted biopsy of suspicious lesions, these findings were confirmed by Panda et al,95 with again significantly lower T1, T2, and ADC values in prostate cancer when compared with noncancer and high predictive of ability of T2 + ADC in distinguishing high-grade from low-grade prostate cancer (AUC, 0.90). In both studies, it was also demonstrated that MRF T1 mapping may help play a role in distinguishing prostate cancer from prostatitis in the PZ.
FIGURE 2.
Images in a 72-year-old man referred for elevated prostate-specific antigen level of 9.87 ng/mL with minimal urinary symptoms who underwent limited MR imaging and targeted biopsy of lesion in left mid prostate. Prostate adenocarcinoma with Gleason score 4 + 3 = 7 was diagnosed at cognitively targeted biopsy. T2-weighted image, ADC map, MRF T2 map, and MRF T1 map show corresponding hypointense lesion in mid prostate (arrow) and NPZ in right hemiprostate. Figure adapted with permission from Yu et al.94
Distinguishing cancer in the TZ comes with more challenges, as the TZ is heterogenous, and cancer detection can be confounded by both prostatitis and benign prostatic hyperplasia. Panda et al96 investigated the utility of MRF in detection of TZ cancer, and found that MRF-derived T1 and T2 values and conventional ADC values are significantly lower compared with normal TZ (P < 0.001). Apparent diffusion coefficient combined with MRF T2 had an AUC of 0.99 for distinguishing TZ cancer from normal TZ in cancer suspicious lesions, and ADC combined with MRF T1 had an AUC of 0.81 for distinguishing prostate cancer from noncancer. There is traditionally emphasis placed on the value of T2-weighted imaging in distinguishing prostate cancer in the TZ, including in PI-RADS, but this study suggests that there may be value to using T1 as a quantitative parameter in this assessment.
Although the above studies are validated with prostate biopsy, Shiradkar et al98 explored a histomorphometric basis for the different MRF-derived T1 and T2 values seen in prostate cancer and prostatitis using whole-mount prostate histopathology as the ground truth. On direct comparison with whole-mount prostate histopathology from 14 patients, it was found that MRF-derived T1 and T2 and ADC values in normal PZ are significantly higher than these values in prostatitis and prostate cancer (P < 0.05), similar to biopsy-based studies. Prostatitis had significantly higher T2 and ADC values than prostate cancer in the PZ. Furthermore, in analysis by tissue compartments within prostate cancer, there was a higher epithelium ratio (and decreasing lumen density and stromal content) with increasing prostate cancer grade in the PZ. This increasing epithelium ratio was found to be correlated with decreasing T1, T2, and ADC values in the PZ. In the TZ, T1 and ADC values (and not T2) were found to be negatively correlated with epithelium ratio, although comparison with normal TZ was not performed.98
The technique used in prostate MRF is FISP-based.94 Fast imaging with steady-state precession is used because it maintains scan efficiency while minimizing off-resonance effects.26 This is especially important when imaging the pelvis, where there is bowel gas and peristalsis resulting in artifact. The prostate MRF technique has been shown to have good intrascanner repeatability, although reproducibility across multiple different scanners has been shown to be slightly more variable but similar to other quantitative measures.9 T1 and T2 values have also been shown to be reproducible with both 1.5 T and 3 T acquisition.99 Continued assessment of reproducibility will enable creation of reliable normative quantitative data.
Prostate MRF is exciting because it seems feasible to easily use MRF-derived quantitative parameters as an adjunct to existing MRI interpretation, especially in conjunction with the already existing quantitative information provided by ADC. In addition, there is a known limitation of prostate MRI in detection of all cancers, so MRF may have the ability to detect subtle tissue changes.100 However, there remains work to be done in optimization of the prostate MRF protocol. Currently, a 2D acquisition is used, and each slice acquisition can take up to 50 seconds. Three-dimensional prostate MRF acquisition would accelerate acquisition and also produce higher-resolution images and is currently being investigated with initial promising results.101-103
Abdominal Imaging
Clinical uses of MRF in abdominal imaging have also been investigated. Abdominal MRI is increasingly used as a problem-solving tool for various organs, especially for clinical questions requiring superior soft tissue contrast and multiproperty characterization. Radiologists currently primarily use qualitative criteria described for each organ to evaluate for disease processes. Image acquisition variability and reader experience remain sources for inhomogeneity in the care received by patients in centers around the world. There is evidence that quantitative parameters can be measured for various imaging findings in the abdomen, can be used in characterization of liver disease, and can be used for therapeutic response assessment.13,104-115 Magnetic resonance fingerprinting provides a simultaneous multiparametric rapid quantitative analysis that could be highly valuable to abdominal organ assessment.
Imaging of the abdomen comes with unique challenges primarily because the included FOV is very large, making it more subject to field inhomogeneities and transmit field inhomogeneities. Abdominal MR is additionally limited by physiologic motion created by breathing and bowel peristalsis.116 The field inhomogeneities along with breath-hold requirements and the need to encode the large field of views make it difficult to achieve reliable mapping with the spatial resolution necessary to characterize smaller lesions. Multiple abdominal MRF techniques have been explored to overcome these limitations and to explore different clinical questions.
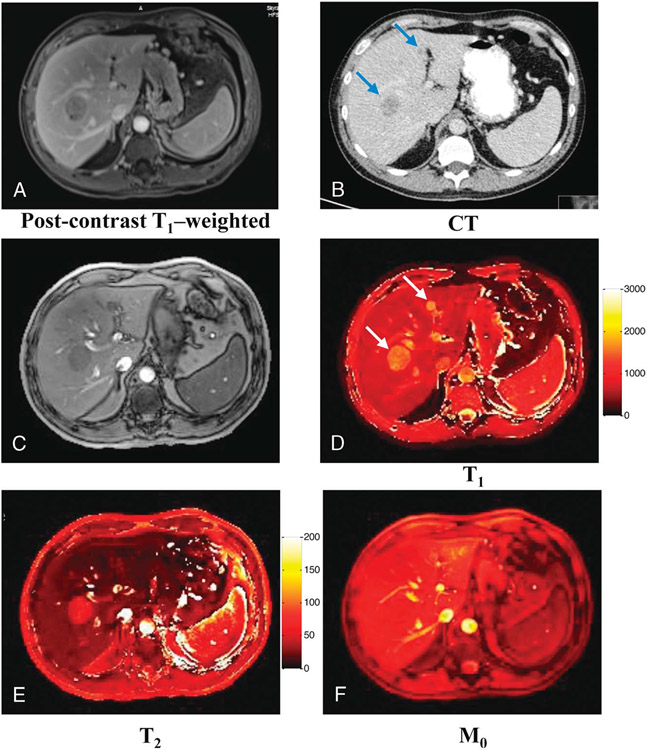
In work published by Chen et al,116 abdominal MRF was used for rapid quantitative abdominal imaging and for assessment of differentiation of hepatic metastatic lesions from hepatic parenchyma. Magnetic resonance fingerprinting was acquired with FISP in a single breath hold and with an interleaved spiral acquisition, with the goals of minimizing motion artifact and sampling the center of k-space multiple times to further reduce artifact. As transmit field (B1) inhomogeneities are of greater concern at higher fields, a B1 map was acquired in conjunction with the MRF acquisition, and the MRF output was provided both without and with B1 correction. This technique was shown to provide similar T1 and T2 relaxation times compared with the literature for normal liver tissue, renal medulla, renal cortex, spleen, skeletal muscle, and fat. Hepatic metastases also demonstrated significantly longer T1 and T2 relaxation times compared with hepatic parenchyma, showing promise for differentiation on MRF (Fig. 3).
FIGURE 3.
Quantitative maps acquired from a patient with metastatic lung adenocarcinoma. A, Conventional postcontrast T1-weighted image. B, CT image. C, Image summed up from all undersampled images. D–F, Quantitative T1, T2, and M0 maps. Two metastatic lesions with sizes of 32.9 and 9.8 mm were observed from both conventional images and quantitative maps. Figure originally printed in Chen et al.116
Diffuse liver disease is a physiological process, which is currently finally diagnosed invasively via random liver biopsy. However, liver disease is associated with various known imaging findings that can be easily quantified noninvasively. Specifically, T1, T2, T2*, and FF can be used for assessing hepatic inflammation, fibrosis, siderosis, and steatosis, respectively. Although each of these values can be quantified individually using conventional methods, MRF offers exciting potential to simultaneously measure these values for quick characterization of hepatic parenchyma. One of the earlier studies to do this was performed by Jaubert et al111; a gradient echo-based golden angle radial MRF approach was used to simultaneously measure hepatic parenchyma T1, T2, T2*, and FF within a single breath hold (~14 seconds). In this primarily phantom-based experiment, this MRF framework was found to have similar values relative to reference quantitative methods.111 This framework was later expanded to also include simultaneous T1rho mapping, given evidence that T1rho can play a role in liver fibrosis assessment.117,118 Velasco et al112 found that simultaneous mapping of T1, T2, T2*, T1rho, and FF again provided comparable measurements to conventional techniques in phantoms and was feasible in vivo with excellent output of high-quality reconstructed maps. More recently, Fujita et al13 validated MRF-derived simultaneous T1, T2, T2*, and FF maps by comparing to reference maps and to biopsy histopathology. High agreement was found with reference quantitative mapping with high repeatability (low coefficient of variation) of the measurements. In comparison with histopathology, MRF-derived values had the ability to differentiate moderate/severe inflammation from no/mild inflammation, steatosis, and siderosis. Certain parameters correlated with certain liver diseases. For example, MRF-derived high proton density fat fraction (PDFF) was associated with higher steatosis, higher T1 was associated with more inflammation, and lower T2* was associated with siderosis.13 Broader validation of hepatic disease characterization remains to be explored, but these results are promising because of the wide range of simultaneously and reproducibly measured coregistered properties, with broad clinical implications for multiple pathologies.
Other early uses of MRF in the abdomen include quantitative renal imaging and pancreatic imaging. In renal imaging, investigated by MacAskill et al,113 MRF has been performed using FISP, with output provided both with and without B1 correction. Output from this showed appropriate corticomedullary differentiation with comparable renal T1 and T2 values to conventional mapping. In addition, in this study, there was no effect of B1 on output, suggesting that the extra step to correct magnetic field inhomogeneities in abdominal MRF may not be needed for this application. Serrao et al119 explored use of MRF in pancreatic imaging, acquiring MRF at varying magnetic field strengths, in both the axial and coronal planes, in the presence of free-breathing motion, and without B1 correction. In the presence of free-breathing motion, it was found that coronal acquisition minimizes motion artifact when compared with axial. Quantitative measurements in the pancreas were found to be homogenous, enabling differentiation from pancreatic lesions. As a secondary outcome, abdominal organs could be differentiated using MRF-derived measurements at varying field strengths (both 1.5 and 3 T). This work is promising for the further development of more advanced pancreatic imaging in an especially sick population. It is also exciting from a technical standpoint as it indicates that we may be able to optimize abdominal MRF even with free-breathing and at varying field strengths.
Early feasibility of MRF has also been evaluated in ovarian malignancies and cervical carcinomas. A pilot study evaluating use of MRF on PET/MRI found that treated high-grade serous epithelial ovarian tumors had lower T1 and T2 values compared with untreated high-grade serous epithelial ovarian tumors.120 In another study, early application of MRF to cervical imaging in healthy volunteers was repeatable for both T1 and T2 measurements. In patients with cervical cancer, significantly higher T1 values were measured in tumor (1529 ± 112 milliseconds) compared with normal cervical mucosa (1430 ± 129 milliseconds, P = 0.031) and normal cervical stroma (1258 ± 101 milliseconds, P < 0.001). Conversely, cervical tumors showed significantly lower T2 values (69 ± 9 milliseconds) compared with normal cervical mucosa (88 ± 16 milliseconds, P < 0.001); however, not significantly different from normal cervical stroma (68 ± 10 milliseconds, P = 0.919).11
Breast Imaging
Magnetic resonance imaging has become a widely used modality for breast cancer screening, lesion characterization, and breast cancer staging, especially in high-risk patients. Currently, the Breast Imaging-Reporting and Data System (BI-RADS) provides MRI assessment guidance for assignment of a BI-RADS category, which is used to communicate suspicion for malignancy and further management recommendations.121 Magnetic resonance imaging has been shown to have higher sensitivity than mammography and ultrasound in detection of malignancy.122 However, BI-RADS MR guidance remains subjective and subject to interreader variation.123-125 In addition, while postcontrast perfusion assessment on MR is semiquantitative, it can be confounded by significant background breast enhancement.126 Therefore, truly quantitative parameters that can constitute a basis for intrinsic breast tissue differentiation are of great interest. Assessment and prediction of successful response to treatment remains another important goal in the field. Prior work has demonstrated utility of T1 and T2 relaxation times in characterization of normal fibroglandular tissue, benign tissue, and breast malignancy.127,128 T2 relaxation times have also been proven to be useful in monitoring of malignancy after initiation of therapy.129,130 Unfortunately, it is difficult to rely on these parameters, as reproducibility is variable across the multiple published mapping techniques and across different magnet strengths.131,132 Therefore, continued investigation of quantitative parameters is needed.
Breast MRF is currently in very early translational stages, with initial development published by Chen et al. 133 Initial breast MRF has been performed as a volumetric 3D FISP acquisition, rather than traditional 2D MRF applications. This is important in breast imaging, as 3D data are important in detection of multicentric and multifocal disease. Each partition is composed of segments that use their own preparatory pulses followed by data acquisition; total time for acquisition of 48 partitions in initial studies is approximately 6 minutes. The high fat content found in breast tissue also poses a unique challenge, as this results in higher magnetic field inhomogeneities. Magnetic resonance fingerprinting already compensates for heterogeneity with high artifact tolerance in the mapping process, and FISP is also less sensitive to field inhomogeneity as described previously. The T2 preparation module used for each segment of breast MRF uses 4 composite 180-degree pulses to further reduce acquisition sensitivity to inhomogeneities, and a spectral-selective fat-saturation module is used for fat suppression.
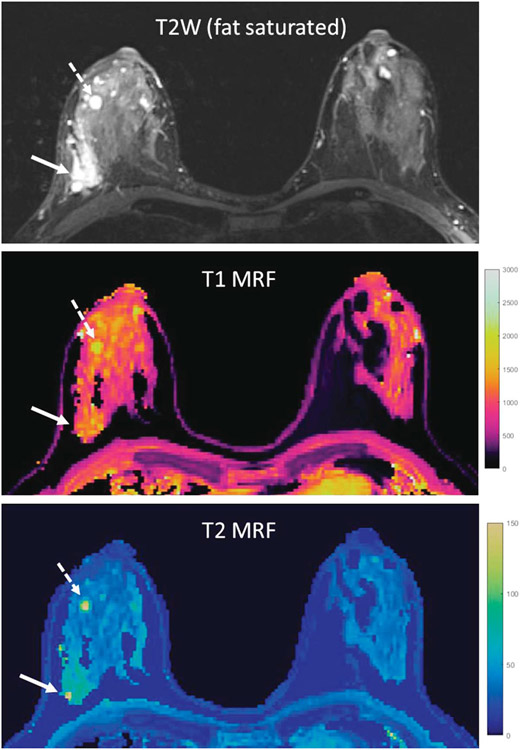
On initial phantom studies, this breast MRF technique showed good technical validation with minimal T1 and T2 variation in measurements across the partitions and strong correlation with conventionally acquired T1 and T2 values. In addition, images acquired in patient volunteers showed successful fat suppression with minimization of signal variation from field inhomogeneity. In a small series published by Panda et al6 using 3D breast MRF, the MRF-derived T1 and T2 values were reproducible across 2 different scanners and showed good test-retest repeatability within the same imaging visit, as well as in patients who were scanned twice in different phases of their menstrual cycle. In a small sample size of breast cancer patients, Chen et al found that longer T1 and T2 relaxation times were observed in tumor and peritumoral regions when compared with normal fibroglandular tissue, and this T2 relaxation time difference was significant (P < 0.001). They also found that T1 and T2 relaxation times were even longer in benign breast cysts.133 A representative example comparing cancer and cyst tissue to fibroglandular tissue is provided in Figure 4. These findings suggest tissue differentiation potential with 3D breast MRF. Future work remains to be done in further validating this technique and in establishing expected T1 and T2 values across age groups and breast densities.
FIGURE 4.
Example of breast MRF in a patient with right-sided breast cancer and multiple right-sided breast cysts. The breast tissue in this patient was classified as heterogeneously dense. Conventional T2W imaging and T1 and T2 MRF maps are provided. Values obtained in the breast cancer (solid arrow) and one breast cyst (dashed arrow) were higher than in normal breast tissue. Values obtained were breast cancer (solid arrow): T1 = 1171 milliseconds, T2 = 76 milliseconds; breast cyst (dashed arrow): T1 = 2079 milliseconds, T2 = 153 milliseconds; normal breast tissue (left breast): T1 = 954 milliseconds, T2 = 33 milliseconds.
Work has also been done in exploring the utility of an alternative 2D Dixon-based breast MRF acquisition. This technique was developed by Nolte et al134 and uses a 3-point Dixon water-fat separation and blurring correction to correct the fat blurring into adjacent voxels that occurs due to off-resonance effects in a spiral readout. Without correction, this fat blurring otherwise interferes with accurate T1 and T2 measurements and is a limitation of 2D spiral acquisition when imaging tissues containing both aqueous and fat components (such as the breast). It was found that this 2D MRF-Dixon technique produces clearer maps that show delineation between fibroglandular and fatty tissue and improved accuracy of T1 and T2 measurements. Zanderigo et al135 performed validation of this technique in a prospective study on 14 patients with both precontrast and postcontrast MRF maps. They found that using MRF-Dixon, T1 and T2 values of breast cancer were significantly higher than fibroglandular tissue on precontrast maps (T1: P = 0.007; T2: P = 0.03). On postcontrast maps, T1 in breast cancer was significantly higher than in fibroglandular tissue (P = 0.0005), but no significant difference was observed between T2 values of breast cancer versus fibroglandular tissue. On evaluation of the maps by 3 radiologists, at least 50% of the cancers were visually detectable on the precontrast T1 MRF maps, and this was improved on the postcontrast T1 MRF maps. Visual detection of malignancy on the T2 MRF maps was more variable. Overall, the precontrast findings were similar to those obtained by Chen et al133 and suggest potential use of MRF to supplement precontrast differentiation of breast tissue.133,135 This remains to be validated on a larger scale.
Cardiac Imaging
There has been considerable interest in the development and clinical translation of quantitative cardiac MRI parameter mapping for detection of cardiac disease. Several studies have previously described the accuracy and complimentary roles of mapping T1, T2, T2*, and extracellular volume fraction (ECV)—a property derived from native and postcontrast T1 measurements—for earlier detection of pathology compared with qualitative MRI. This has been shown in various infiltrative/inflammatory/ischemic cardiomyopathies, myocarditis, and postcardiac transplant imaging.136-139 Measurement of these parameters is now routinely recommended by the Society for Cardiovascular Magnetic Resonance and endorsed by the European Association of Cardiovascular Imaging in patients with amyloidosis, Anderson-Fabry disease, myocarditis, and iron overload.139 The consensus statement also recommends mapping in patients with heart failure to evaluate potential diffuse myocardial disease. Although T1, T2, T2*, and ECV are increasingly used in clinical cardiac MRI, other tissue properties may offer additional value. For example, PDFF mapping may have value in assessing intramyocardial fat infiltration and lipomas, and T1rho mapping could enable detection of focal and diffuse fibrosis without the need for exogenous contrast agent.140,141
Conventional cardiac MRI mapping techniques require a separate scan to measure each tissue property. Scans are typically 2D and breath held, and they often use electrocardiogram (ECG) triggering to limit the imaging window to diastole to minimize cardiac motion artifacts. A small number of T1- or T2-weighted images are collected using a single-shot readout with 1 image acquired per heartbeat. At each pixel, the signal intensities from the weighted images are fit to an exponential function to obtain a quantitative map. In clinical practice, T1 mapping is often performed using modified Look-Locker inversion recovery (MOLLI) or saturation recovery shingle-shot acquisition (SASHA). Commonly used T2 mapping sequences include T2-prepared bSSFP and gradient spin echo.142-145
Despite their widespread use, conventional mapping approaches have some limitations. A separate breath hold scan is needed to map each tissue property. If several properties are assessed, multiple breath holds are required, and the maps most likely will not be coregistered due to differences in breath hold position and cardiac motion. Many conventional techniques are affected by confounding factors that can alter the measured tissue property values. For example, MOLLI has been shown to be influenced by heart rate, T2 relaxation, off-resonance, B1 inhomogeneities, and magnetization transfer.146
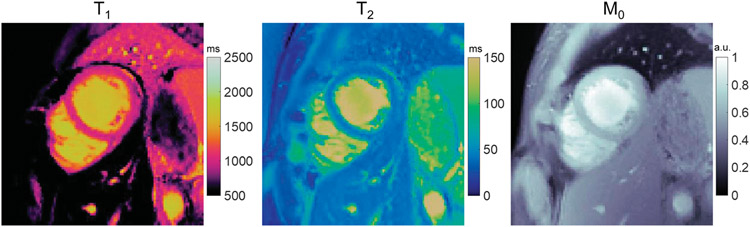
Cardiac MRF enables simultaneous mapping of multiple tissue properties in a single acquisition and yields coregistered maps, as shown in Figure 5.18,147 The first implementation of MRF for cardiac imaging was provided by Hamilton et al147 for joint T1, T2, and M0 mapping with diastolic ECG triggering and a breath hold duration of 15–16 heartbeats. There are several technical differences between cardiac MRF compared with other MRF implementations. Multiple inversion and T2 preparation pulses are applied throughout the scan to enhance the sensitivity of the sequence to T1 and T2, and small angles (below 25 degrees) are used to reduce the sensitivity to B1 effects. Because of the use of prospective ECG triggering, the subject’s heart rate influences the MRF sequence timings and the resulting fingerprints. Therefore, in cardiac MRF, a new dictionary is generated after every acquisition that incorporates the subject’s cardiac rhythm timings (obtained from the ECG signal) in the Bloch equation simulation. This step is essential to obtain accurate T1 and T2 measurements that are independent of heart rate. In contrast, most other MRF applications compute the dictionary once and reuse it for subsequent scans since the sequence timings are fixed.
FIGURE 5.
Simultaneous T1, T2, and M0 mapping in a healthy subject at 1.5 T using cardiac MRF. Data were acquired using a FISP-based MRF sequence during a 15-heartbeat breath hold and a 250 milliseconds ECG-triggered acquisition window. The sequence used variable flip angles of 4–25 degrees and multiple inversion and T2-preparation pulses. To generate the dictionary, a Bloch equation simulation was performed that incorporated the subject’s cardiac rhythm timings derived from the ECG signal.
The cardiac MRF framework has the potential to correct for confounding factors, which may improve the accuracy of tissue property measurements and reduce variability across scanners. For example, slice profile imperfections and signal relaxation during the inversion and T2 preparation pulses can be modeled in the MRF dictionary.148 Including these corrections was shown to reduce measurement variability among cardiac MRF sequences with different acquisition parameters. One disadvantage is additional computation time; for example, modeling these effects increases the dictionary calculation time from approximately 10 seconds to 5 minutes.
Artificial intelligence offers a promising solution for overcoming several computational bottlenecks in cardiac MRF.149 Artificial intelligence can be used to accelerate the dictionary generation process in MRF. One approach that has been investigated uses a neural network that receives the cardiac rhythm timings from the ECG signal as an input and outputs the dictionary in under 1 second, thus eliminating the need for a time-consuming Bloch equation simulation.150 Artificial intelligence methods for cardiac MRF have also been proposed that perform parameter estimation without a dictionary. One technique used a fully connected network to estimate T1, T2, and M0 values directly from the signal time-courses in undersampled cardiac MRF images in under 400 milliseconds. No differences in myocardial T1 and T2 values were observed compared with the standard dictionary-based reconstruction, which required nearly 5 minutes.151
Artificial intelligence has also been leveraged to improve the image quality in cardiac MRF. The MRF deep image prior uses a u-net to generate cardiac T1, T2, and M0 maps with reduced aliasing artifacts and noise compared with previous reconstruction methods. It also does not require any additional training data, as network training is performed using the undersampled MRF data from a single scan. This technique has been applied to shorten the cardiac MRF breath hold from 15 to 5 heartbeats and the diastolic acquisition window from 250 to 150 milliseconds to mitigate motion artifact.152
Although the original cardiac MRF technique focused on 2D T1, T2, and M0 mapping, several extensions have been proposed recently. Simultaneous multislice imaging has been combined with cardiac MRF to enable mapping from 3 slices during one breath hold.153 An approach for 3D MRF T1 and T2 mapping in 7 minutes of free-breathing has been proposed that combines the respiratory bellows with autofocus translational motion correction.154 Cine MRF techniques that use a continuous acquisition with retrospective ECG gating have been developed for cardiac phase-resolved (or cine) T1, T2, and M0 mapping.155,156 The maps can be used to generate contrast-weighted cine images to quantify ventricular volumes, mass, and ejection fraction. Thus, cine MRF enables tissue characterization and evaluation of cardiac function during a single acquisition. Finally, while many studies have focused on T1 and T2 mapping, additional tissue properties of interest in cardiac diseases have also been investigated. These include T1rho, T2*, postcontrast ECV mapping, and FF from either a water-fat DIXON sequence or rosette trajectory for water-fat separation.157-162
Table 2 summarizes several studies that have validated various cardiac MRF techniques against conventional cardiac mapping techniques.147,148,153-159,162,163 Generally, although absolute T1 and T2 values differ between MRF-derived mapping and conventional mapping, there is good correlation between MRF and conventional mapping sequences in all studies.
TABLE 2.
Published Values Using Various Cardiac MRF Techniques
| Study and Field Strength | Quantitative Properties Obtained Via Cardiac MRF Framework |
Cardiac MRF Values (Mean ± SD) | Conventional Mapping Sequences Values (Mean ± SD) |
|---|---|---|---|
| Original cardiac MRF (3 T)147 | Native myocardial T1, T2, M0 | T1: 1213 ± 75 ms T2: 35 ± 4 ms |
MOLLI T1: 1257 ± 61 ms T2 bSSFP: 42 ± 5 ms |
| MRF with corrected confounders (3 T)148 | Native myocardial T1, T2, M0 | T1: 1323 ± 47 ms T2: 37 ± 4 ms |
MOLLI T1: 1227 ± 30 ms T2 FLASH: 38 ± 3 ms |
| Simultaneous multislice MRF (3 T)153 | Native myocardial T1, T2 | T1: 1320 ± 52 ms T2: 32 ± 1 ms |
MOLLI T1: 1242 ± 32 ms T2 FLASH: 38 ± 2 ms |
| Water-fat Dixon MRF (1.5 T)159 | Native myocardial water-specific T1, T2, and FF | T1: 1045 ± 32 ms T2: 43 ± 3 ms FF in septum: 1.3% ± 3% |
MOLLI T1: 1026 ± 28 ms SASHAT1: 1129 ± 38 ms T2 GRASE: 52 ± 2 ms Good correlation with in vivo PDFF mapping: R2 > 0.98 |
| Cine MRF (3 T)155 | Native myocardial T1, T2, and LVEF | T1 diastole/systole: 1398/1391 ms T2 diastole/systole: 31/30 ms LVEF: mean bias −1.0% (95% LoA −3.4%, 1.3%) |
MOLLI T1 diastole/systole: 1234/1212 ms T2-bSSFP diastole/systole: 38/41 ms |
| Free-running (non-ECG triggered) MRF (1.5 T)156 | Native myocardial T1, T2, and LVEF | T1: 1160 ± 79 ms T2: 45 ± 4 ms LVEF: mean bias −0.83% (95% LoA −5.8%, 4.1%) |
MOLLI T1: 1043 ± 48 ms SASHA T1: 1150 ± 100 ms T2 GRASE: 52 ± 4 ms |
| 3D free-breathing MRF (1.5 T)154 | Native myocardial T1, T2 | T1: 1093 ± 24 ms T2: 45 ± 2 ms |
MOLLI T1: 1054 ± 19 ms SASHA T1: 1146 ± 20 ms T2 GRASE: 52 ± 2 ms |
| Rosette MRF for water-fat separation (1.5 T and 3 T)161 | Native myocardial T1, T2 | T1 at 3 T: 1329 ± 35 ms T1 at 1.5 T: 1044 ± 30 ms T2 at 3 T: 33 ± 2 ms* T2 at 1.5 T: 39 ± 1 ms* |
MOLLI T1 at 3 T: 1205 ± 31 ms MOLLI T1 at 1.5 T: 996 ± 18 ms T2 FLASH at 3 T: 39 ± 1 ms T2 bSSFP at 1.5 T: 47 ± 1 ms |
| Rosette MRF for water-fat separation and PDFF (1.5 T)162 | Native myocardial T1, T2, and PDFF | T1: 1081 ± 32 ms T2: 41 ± 1 ms PDFF: 0.4% (range, −4.5% to 5.7%) |
MOLLI T1: 1054 ± 19 ms T2 bSSFP: 46 ± 2 ms No comparison sequence for PDFF |
| T1rho MRF (1.5 T)157 | Native myocardial T1, T2, and T1rho | T1: 1133 ± 33 ms T2: 39 ± 4 ms T1rho: 52 ± 4 ms |
MOLLI T1: 1053 ± 47 ms T2-GRASE: 51 ± 4 ms T1rho turbo field echo: 56 ± 2 ms |
| Low-rank motion corrected MRF (1.5 T)158 | Native myocardial T1, T2, T2*, and FF | T1: 1148 ± 17 ms T2: 43 ± 1 ms T2*: 35 ± 2 ms FF: 0.8% ± 0.2% |
MOLLI T1: 1056 ± 11 ms T2 GRASE: 51 ± 1 ms T2* GRE: 40 ± 6 ms FF: 0.3% ± 0.9% |
| Shortened diastolic acquisition window MRF (1.5 T)163 | Native myocardial T1, T2, and postcontrast T1 9 healthy volunteers: both 250 ms diastolic window (MRF250ms) and 154 ms diastolic window (MRF154ms) acquired 61 patients referred for cardiac MRI: MRF154ms acquired only |
9 healthy volunteers: Native T1: MRF154ms: 1020 ± 45 ms; MRF250ms: 1026 ± 34 ms Native T2: MRF154ms: 42.4 ± 2.4 ms; MRF250ms: 38.5 ± 2.6 ms 61 patients: Native T1: MRF154ms: 1015 ± 61 ms Native T2: MRF154ms: 44.5 ± 5.5 ms; postcontrast T1: MRF154ms: 395 ± 50 ms; ECV: MRF154ms: 28.4% ± 4.8% |
9 healthy volunteers: MOLLI T1: 1005 ± 20 ms T2 bSSFP: 45.4 ± 2.0 ms 61 patients: MOLLI T1: 1029 ± 53 ms T2 bSSFP: 46.4 ± 3.7 ms Postcontrast MOLLI T1: 444 ± 53 ms ECV MOLLI: 26.0% ± 4.0% |
Comparison is provided to conventional mapping techniques.
Rosette MRF T2 values were higher than original spiral MRF T2 (P < 0.05, differences 3–5 ms).
MRF, magnetic resonance fingerprinting; MOLLI, modified Look-Locker inversion recovery; bSSFP, balanced steady state free precession; SASHA, saturation recovery shingle-shot acquisition; PDFF, proton density fat fraction; LVEF, left ventricle ejection fraction; ECG, electrocardiogram; FF, fat fraction; GRASE, gradient spin echo; FLASH, fast low angle shot.
For normative data assessment, cardiac MRF was evaluated in a cohort of 58 healthy subjects at 1.5 T and compared with conventional T1 and T2 mapping, presented in the first row of Table 3.12 In this study, MRF maps exhibited good test-retest repeatability (intraclass correlation coefficients ranging, 0.85–0.87). In addition, the MRF T1 and T2 maps were rated qualitatively higher than conventional T1 and T2 maps on Likert score analysis, based on sharpness of borders, absence of artifacts, and overall diagnostic confidence. When randomly presented with either MRF or conventional T1 and T2 maps in a blinded fashion, MRF maps were preferred over conventional maps in 80% of cases by all 3 radiologists.12 In a preliminary analysis performed by the same group, a small sample size (18 patients) across 2 sites showed good intersite reproducibility.170
TABLE 3.
Published Normative and Disease Cardiac MRF Values
| Normative Data and Clinical Studies | |||
|---|---|---|---|
| Study and Field Strength | Patient Population, Mapped Properties |
Cardiac MRF Values (Mean ± SD) | Conventional Mapping Sequences Values (Mean ± SD) |
| 2D spiral MRF (1.5 T)12 |
58 healthy volunteers Native myocardial T1, T2 |
T1: 1007 ± 97 ms (basal) 995 ± 102 ms (mid) 1001 ± 101 ms (apical) T2: 41 ± 7 ms (basal) 41 ± 6 ms (mid) 44 ± 7 ms (apical) |
MOLLI T1: 990 ± 45 ms (basal) 996 ± 60 ms (mid) 1007 ± 101 ms T2 bSSFP: 46 ± 4 ms (basal) 47 ± 4 ms (mid) 48 ± 4 ms (apical) |
| 2D Dixon MRF in patients with suspected cardiovascular disease (1.5 T)160 |
19 patients and 11 healthy volunteers Native myocardial T1, T2, and FF Post-contrast T1 ECV |
T1: 1062 ± 45 ms (volunteers) 1056 ± 33 ms (patients) T2: 43 ± 3 ms (volunteers) 44 ± 5 ms (patients) FF: 0.2% ± 2.5% (volunteers) −1.9% ± 2.4% (patients) Patients: Postcontrast T1: 452 ± 56 ms Synthetic ECV: 26% ± 2.2% |
MOLLI T1: 1025 ± 33 ms (volunteers) 995 ± 29 ms (patients) T2 GRASE: 52 ± 1 ms (volunteers) 52 ± 4 ms (patients) FF: 0.1% ± 1.4% (volunteers) −0.7% ± 1.7% (patients) Postcontrast conventional mapping sequences not performed |
| Nonischemic dilated cardiomyopathy (1.5 T)164 |
9 patients and 9 healthy controls Native myocardial T1, T2 ECV |
Patients: T1: 979 ± 50 ms* T2: 47 ± 4 ms ECV: 27% ± 4%* Controls: T1: 913 ± 68 ms* T2: 43 ± 4 ms ECV: 24% ± 3%* |
Patients: MOLLI T1: 1030 ± 45 ms* T2 bSSFP: 48 ± 3 ms ECV: 23% ± 3%* Controls: MOLLI T1: 993 ± 23 ms* T2 bSSFP: 47 ± 2 ms ECV: 19% ± 2%* |
| Cardiac amyloidosis (3 T)165 |
9 patients and 5 healthy controls Native myocardial T1, T2 |
Patients: T1: 1395 ± 121 ms* T2: 37 ± 3 ms* Controls: T1: 1240 ± 36 ms* T2: 32 ± 3 ms* |
Not performed |
| Inflammatory cardiomyopathy (1.5 T)166 |
24 patients Native myocardial T1, T2 |
T1: 1028 ± 64 ms T2: 53 ± 4 ms |
MOLLI T1: 1019 ± 53 ms T2 bSSFP: 49 ± 3 ms |
| Hypertrophic cardiomyopathy (1.5 T)167 |
6 patients and 12 healthy controls Native Myocardial T1, T2 Postcontrast T1 Postcontrast T2 ECV |
Patients: T1: 1017 ± 31 ms* T2: 45 ± 6 ms Postcontrast T1: 351 ± 43 ms Postcontrast T2: 36 ± 3 ms* ECV: 37% ± 4%* Controls: T1: 921 ± 65 ms* T2: 44 ± 4 ms Postcontrast T1: 369 ± 30 ms Postcontrast T2: 33 ± 3 ms* ECV: 25% ± 3%* |
Patients: T1: 1057 ± 76 ms* T2: 45 ± 4 ms Postcontrast T1: 336 ± 34 ms ECV: 32% ± 2%* Controls: T1: 997 ± 23 ms* T2: 44 ± 2 ms Postcontrast T1: 371 ± 53 ms ECV: 21% ± 2%* |
| Hypertrophic cardiomyopathy (3 T)168 |
23 patients Native myocardial T1, T2, postcontrast T1, ECV |
T1: 1397 ± 40 ms T2: 29 ± 3 ms Postcontrast T1: 521 ± 46 ms ECV: 28% ± 3%† |
T1: 1252 ± 25 ms T2 bSSFP: 40 ± 2 ms Postcontrast T1: 488 ± 36 ms ECV: 23% ± 2%† |
| Postcardiac transplant (3 T)169 |
13 patients and 5 healthy controls Native myocardial T1, T2 Postcontrast T1, T2 |
Patients: T1: 1259 ± 37 ms (basal) T1: 1252 ± 53 ms (mid) T2: 41 ± 4 ms (basal) T2: 42 ± 5 ms (mid) Postcontrast T1: 620 ± 81 ms (basal) Postcontrast T1: 630 ± 91 ms (mid) Controls: T1: 1206 ± 30 ms (basal) T1: 1219 ± 38 ms (mid) T2: 38 ± 4 ms (basal) T2: 38 ± 4 ms (mid) |
Patients: MOLLI T1: 1269 ± 35 ms (basal) MOLLI T1: 1261 ± 63 ms (mid) T2 bSSFP: 41 ± 2 ms (basal) T2 bSSFP: 41 ± 2 ms (mid) Controls: MOLLI T1: 1174 ± 38 ms (basal) MOLLI T1: 1171 ± 22 ms (mid) T2 bSSFP: 38 ± 2 ms (basal) T2 bSSFP: 37 ± 1 ms (mid) |
Comparison is provided to conventional mapping values if available.
Significant differences between controls and patients (P < 0.05).
Significant differences between cMRF-T1 derived ECV and MOLLI-T1 derived ECV (P < 0.05).
MRF, magnetic resonance fingerprinting; 2D, 2-dimensional; MOLLI, modified Look-Locker inversion recovery; bSSFP, balanced steady state free precession; ECV, extracellular volume; LVEF, left ventricle ejection fraction; FF, fat fraction.
Translational work has also been performed in certain diseases, with early MRF-based relaxometry studies published in suspected cardiovascular disease,160 cardiac amyloidosis,165 postcardiac transplant,169 nonischemic cardiomyopathy,164 hypertrophic cardiomyopathy,167,168 and inflammatory cardiomyopathy patients166 (remainder of Table 3). In many of these studies, MRF provides significant discriminatory information between healthy controls and patients with the cardiac disease of interest. So far, this has been shown in various cardiomyopathies and amyloidosis patients. A subset of this population often has implantable cardioverter-defibrillators, and in these patients, MRF has shown lower artifacts and higher image quality scores compared with conventional T1 and T2 maps.166 In patients who are postcardiac transplant, MRF provides rapid quantification to allow for longitudinal assessment and provides a baseline to monitor for future rejection.169 These early translational studies show great promise for the future of MRF in rapid characterization of cardiac disease.
Musculoskeletal Applications
Early work is starting to emerge in application of MRF techniques to the musculoskeletal system. For example, Sharafi et al171 reported low coefficients of variation for MRF measurements of T1, T2, and T1rho in lower leg musculature, suggesting feasibility of MRF for muscle characterization. Marty et al172-174 have applied simultaneous measurements of T1, T2, and FF on fat-infiltrated muscles, with implication for pathologies such as neuromuscular diseases. Work has also been done in using MRF to characterize multiple musculoskeletal structures, including cortical bone, which is usually barely detectable on conventional MRI due to its ultrashort T2 time (on the order of 110 milliseconds). Li et al175 have successfully been able to apply MRF on long, short, and ultrashort structures with successful differentiation of cortical bone and production of bone-enhanced MRF maps that mimic CT scans. The potential for MRF has also been explored in the setting of prostate cancer metastases; in a study comparing ADC values, unenhanced MRF T1 values, unenhanced MRF T2 values, postcontrast MRF T1 values, and postcontrast MRF T2 values of bone metastases and normal bone marrow in the pelvis, significant differences were found in the values between normal bone marrow and metastases across all quantitative parameters (P < 0.001). In addition, measurement of these parameters showed moderate to good interreader agreement for all parameters across treated lesions, untreated lesions, and normal bone marrow (intraclass correlation coefficients, 0.519–0.915).176
Preliminary clinical work in MRF on cartilage imaging has included using 3D MRF to quantify age-dependent differences in medial tibial and femoral cartilage between healthy patients and patients with knee osteoarthritis.177,178 Quantification of hip cartilage using MRF has also been explored, with Sharafi et al179 able to successfully characterize subregions of hip cartilage in both hips with excellent repeatability. Cloos et al180 were able to use MRF to enable semiautomatic hip cartilage segmentation and for T1/T2/proton density quantitative mapping, enabling an efficient and highly reproducible workflow for quantitative hip cartilage assessment. Finally, there are benefits of MRF in overcoming field heterogeneity created by prosthetic implants, with particular success using an MRF technique called plug-and-play MRF.181
CHALLENGES AND FUTURE
As can be seen, MRF has been extensively used to generate reproducible and repeatable tissue property maps of various parts of the body and for characterization of multiple diseases. However, there remain both technical and clinical barriers to overcome before widespread clinical adoption. Detailed discussion of specific remaining technical challenges is beyond the scope of this review, but advances continue at all steps of the pipeline of the MRF experiment. Some challenges for clinical utilization of the techniques being developed are discussed below, although these are by no means comprehensive.
One major obstacle to clinical adoption is that MRF maps must remain accurate, reproducible, and repeatable while being obtained in a fast enough acquisition such that patient table time is not greatly increased. If the resolution of these maps can be sufficiently high (to match conventional weighted images), the possibility exists to reconstruct weighted images or even to replace weighted images altogether, and there is significant ongoing work in this direction.28,40,182 As new technical advances arise in acquisition and reconstruction, it is also very important to make these advances directly available at the scanner so that clinical adoption can be facilitated. To do this, some advances (particularly in reconstruction and large dictionary generation) must often be coupled with appropriate implementation via graphic processing units and tensor processing units, which is not a trivial computing challenge.9,183 Finally, accuracy, reproducibility, and repeatability are central to all imaging advances and must be assessed. Unfortunately, careful testing of sequences using accepted phantoms (such as the ISMRM/NIST phantom) and on volunteers/patients is not often performed by authors purporting to perform quantitative imaging, which makes it difficult to compare quantitative data from MRF or other techniques.
In clinical application, as has been shown in this review, MRF has great potential. However, studies remain small and often are limited to one site. There is a need to generate sufficient normative data to be able compare pathologic tissue to normal tissue and to accurately differentiate different degrees of pathology. To accomplish this, large multicenter studies are needed for more widespread validation and adoption of new MRF techniques. In addition, it is hoped that MRF and other quantitative approaches can be used to assess response to treatment and ideally even identify early response to treatment. This would require these quantitative approaches to be validated via systematic laboratory or pathology-based testing, which is currently an unmet need in the rollout of MRF applications. When evaluating potential for widespread clinical use, most MRF sequences are currently developed on single vendor platforms. Execution of new MRF sequences across platforms requires collaboration with multiple vendors, which poses obvious practical problems that must be overcome as an MR community. With widespread clinical use, there would also be opportunity for computed or automated assessment of quantitative imaging data, which is a possibility that has not yet been deeply explored.184
We have aimed to provide a summary of many major developing clinical applications of MRF; however, given the excitement surrounding this topic and rapid evolution of the literature, we can by no means claim that this review is entirely comprehensive.
CONCLUSIONS
There has been a rising demand for personalized medicine, and objective, reliable, and quantitative diagnosis and treatment follow-up will be needed to provide this overarching goal in medicine. However, precision diagnosis and widespread quantitative imaging remain somewhat elusive goals in radiology. Rapid and reproducible quantitative MR would be a huge asset to this evolving landscape. Quantitative approaches could also provide another tool for dealing with ever-increasing volumes as quantitative data lend themselves well to computerized or semiautomated initial evaluation. Magnetic resonance fingerprinting is a tool for providing reproducible, simultaneously obtained (and thus perfectly registered) anatomic maps of multiple interesting properties, which can then be used for disease diagnosis, differentiation, and follow-up. Although initial application of MRF throughout the body has centered on T1 and T2 relaxation times, multiple additional properties have been explored and continue to be added to the possible maps that can be obtained in this manner.
ACKNOWLEDGMENTS
The authors acknowledge grant support from the National Institutes of Health: 1R01CA208236, 1R37CA263583, and 1R01HL163030.
Conflicts of interest and sources of funding:
none declared. S.G. is a recipient of royalties from the National Institutes of Health for licensed intellectual property. J.H. discloses support from Siemens Healthineers and NIH/NHLBI R01 HL163030. V.G. and Y.J. receive research support from Siemens Healthineers and have intellectual property that is licensed (royalties paid in the past) by Siemens Healthineers. V.G. consults for Cook Medical. We would like to acknowledge grant support from the National Institutes of Health: 1R01CA208236 and 1R37CA263583.
REFERENCES
- 1.Bitar R, Leung G, Perng R, et al. MR pulse sequences: what every radiologist wants to know but is afraid to ask. Radiographics. 2006;26:513–537. [DOI] [PubMed] [Google Scholar]
- 2.Yokoo T, Bae WC, Hamilton G, et al. A quantitative approach to sequence and image weighting. J Comput Assist Tomogr. 2010;34:317–331. [DOI] [PubMed] [Google Scholar]
- 3.Young IR, Szeverenyi NM, Du J, et al. Pulse sequences as tissue property filters (TP-filters): a way of understanding the signal, contrast and weighting of magnetic resonance images. Quant Imaging Med Surg. 2020;10:1080–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulani V, Seiberlich N. Quantitative MRI: Rationale and Challenges: Advances in Magnetic Resonance Technology and Applications. London, United Kingdom: Academic Press; 2020:xxxvii–li. [Google Scholar]
- 5.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda A, Chen Y, Ropella-Panagis K, et al. Repeatability and reproducibility of 3D MR fingerprinting relaxometry measurements in normal breast tissue. J Magn Reson Imaging. 2019;50:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buonincontri G, Biagi L, Retico A, et al. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0 T. Neuroimage. 2019;195:362–372. [DOI] [PubMed] [Google Scholar]
- 8.Konar AS, Qian E, Geethanath S, et al. Quantitative imaging metrics derived from magnetic resonance fingerprinting using ISMRM/NIST MRI system phantom: an international multicenter repeatability and reproducibility study. Med Phys. 2021;48:2438–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo WC, Bittencourt LK, Panda A, et al. Multicenter repeatability and reproducibility of MR fingerprinting in phantoms and in prostatic tissue. Magn Reson Med. 2022;88:1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Chen Y, Shen C, et al. Initial assessment of 3D magnetic resonance fingerprinting (MRF) towards quantitative brain imaging for radiation therapy. Med Phys. 2020;47:1199–1214. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Perucho JAU, Cao P, et al. Repeatability of MR fingerprinting in normal cervix and utility in cervical carcinoma. Quant Imaging Med Surg. 2021;11:3990–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton JI, Pahwa S, Adedigba J, et al. Simultaneous mapping of T(1) and T(2) using cardiac magnetic resonance fingerprinting in a cohort of healthy subjects at 1.5 T. J Magn Reson Imaging. 2020;52:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita S, Sano K, Cruz G, et al. MR fingerprinting for liver tissue characterization: a histopathologic correlation study. Radiology. 2023;306:150–159. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Ma D, Keenan KE, et al. Repeatability of magnetic resonance fingerprinting T(1) and T(2) estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med. 2017;78:1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloch F. The principle of nuclear induction. Science. 1953;118:425–430. [DOI] [PubMed] [Google Scholar]
- 16.Hennig J. Multiecho imaging sequences with low refocusing flip angles. J Magn Reson (1969). 1987;78:397–07. [Google Scholar]
- 17.Weigel M. Extended phase graphs: dephasing, RF pulses, and echoes—pure and simple. J Magn Reson Imaging. 2015;41:266–295. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Hamilton J, Rajagopalan S, et al. Cardiac magnetic resonance fingerprinting: technical overview and initial results. JACC Cardiovasc Imaging. 2018;11:1837–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asslander J. A perspective on MR fingerprinting. J Magn Reson Imaging. 2021;53:676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bipin Mehta B, Coppo S, Frances McGivney D, et al. Magnetic resonance fingerprinting: a technical review. Magn Reson Med. 2019;81:25–46. [DOI] [PubMed] [Google Scholar]
- 21.McGivney DF, Boyacioglu R, Jiang Y, et al. Magnetic resonance fingerprinting review part 2: technique and directions. J Magn Reson Imaging. 2020;51:993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D. Chapter 28—MR fingerprinting: concepts, implementation and applications. In: Choi I, Jezzard P, eds. Advanced Neuro MR Techniques and Applications. Cambridge, MA: Academic Press; 2021:435–339. [Google Scholar]
- 23.Scheffler K, Lehnhardt S. Principles and applications of balanced SSFP techniques. Eur Radiol. 2003;13:2409–2418. [DOI] [PubMed] [Google Scholar]
- 24.Badve C, Yu A, Rogers M, et al. Simultaneous T(1) and T(2) brain relaxometry in asymptomatic volunteers using magnetic resonance fingerprinting. Tomography. 2015;1:136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma D, Jiang Y, Chen Y, et al. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn Reson Med. 2018;79:2190–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, Ma D, Seiberlich N, et al. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74:1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buonincontri G, Sawiak SJ. MR fingerprinting with simultaneous B1 estimation. Magn Reson Med. 2016;76:1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Fang Z, Hung SC, et al. High-resolution 3D MR fingerprinting using parallel imaging and deep learning. Neuroimage. 2020;206:116329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao C, Bilgic B, Manhard MK, et al. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. Neuroimage. 2017;162:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang CY, Coppo S, Mehta BB, et al. Magnetic resonance fingerprinting with quadratic RF phase for measurement of T(2)(*) simultaneously with delta(f), T(1), and T(2). Magn Reson Med. 2019;81:1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyacioglu R, Wang C, Ma D, et al. 3D magnetic resonance fingerprinting with quadratic RF phase. Magn Reson Med. 2021;85:2084–2094. [DOI] [PubMed] [Google Scholar]
- 32.Rieger B, Akcakaya M, Pariente JC, et al. Time efficient whole-brain coverage with MR fingerprinting using slice-interleaved echo-planar-imaging. Sci Rep. 2018;8:6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong T, Han D, Kim DH. Simultaneous estimation of PD, T(1), T(2), T(2)(*), and ΔB(0) using magnetic resonance fingerprinting with background gradient compensation. Magn Reson Med. 2019;81:2614–2623. [DOI] [PubMed] [Google Scholar]
- 34.Afzali M, Mueller L, Sakaie K, et al. MR fingerprinting with b-tensor encoding for simultaneous quantification of relaxation and diffusion in a single scan. Magn Reson Med. 2022;88:2043–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen O, Yu VY, Tringale KR, et al. CEST MR fingerprinting (CEST-MRF) for brain tumor quantification using EPI readout and deep learning reconstruction. Magn Reson Med. 2023;89:233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemasson B, Pannetier N, Coquery N, et al. MR vascular fingerprinting in stroke and brain tumors models. Sci Rep. 2016;6:37071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su P, Mao D, Liu P, et al. Multiparametric estimation of brain hemodynamics with MR fingerprinting ASL. Magn Reson Med. 2017;78:1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korzdorfer G, Kirsch R, Liu K, et al. Reproducibility and repeatability of MR fingerprinting Relaxometry in the human brain. Radiology. 2019;292:429–437. [DOI] [PubMed] [Google Scholar]
- 39.Buonincontri G, Kurzawski JW, Kaggie JD, et al. Three dimensional MRF obtains highly repeatable and reproducible multi-parametric estimations in the healthy human brain at 1.5 T and 3 T. Neuroimage. 2021;226:117573. [DOI] [PubMed] [Google Scholar]
- 40.Fujita S, Cencini M, Buonincontri G, et al. Simultaneous relaxometry and morphometry of human brain structures with 3D magnetic resonance fingerprinting: a multicenter, multiplatform, multifield-strength study. Cereb Cortex. 2022;33:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badve C, Yu A, Dastmalchian S, et al. MR fingerprinting of adult brain tumors: initial experience. AJNR Am J Neuroradiol. 2017;38:492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Blank P, Badve C, Gold DR, et al. Magnetic resonance fingerprinting to characterize childhood and Young adult brain tumors. Pediatr Neurosurg. 2019;54:310–318. [DOI] [PubMed] [Google Scholar]
- 43.Konar AS, Shah AD, Paudyal R, et al. Quantitative relaxometry metrics for brain metastases compared to normal tissues: a pilot MR fingerprinting study. Cancers (Basel). 2022;14:5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dastmalchian S, Kilinc O, Onyewadume L, et al. Radiomic analysis of magnetic resonance fingerprinting in adult brain tumors. Eur J Nucl Med Mol Imaging. 2021;48:683–693. [DOI] [PubMed] [Google Scholar]
- 45.Tippareddy C, Onyewadume L, Sloan AE, et al. Novel 3D magnetic resonance fingerprinting radiomics in adult brain tumors: a feasibility study. Eur Radiol. 2022;33:836–844. [DOI] [PubMed] [Google Scholar]
- 46.Haubold J, Demircioglu A, Gratz M, et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric (18)F-FET PET-MRI and MR fingerprinting. Eur J Nucl Med Mol Imaging. 2020;47:1435–1445. [DOI] [PubMed] [Google Scholar]
- 47.Springer E, Cardoso PL, Strasser B, et al. MR fingerprinting—a radiogenomic marker for diffuse gliomas. Cancers (Basel). 2022;14:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita S, Hagiwara A, Yasaka K, et al. Radiomics with 3-dimensional magnetic resonance fingerprinting: influence of dictionary design on repeatability and reproducibility of radiomic features. Eur Radiol. 2022;32:4791–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu TLL, Chen Y, Lian J, et al. Quantitative evaluation of repeatability and reproducibility of MR fingerprinting and conventional contrast-weighted MRI for human brain radiomics. Joint American Association of Physicists in Medicine, Canadian Organization of Medical Physicists Annual Meeting, Virtual. 2020. [Google Scholar]
- 50.Mostardeiro TR, Panda A, Witte RJ, et al. Whole-brain 3D MR fingerprinting brain imaging: clinical validation and feasibility to patients with meningioma. MAGMA. 2021;34:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai Y, Zhang R, Zhang X, et al. Magnetic resonance fingerprinting for preoperative meningioma consistency prediction. Acad Radiol. 2022;29:e157–e165. [DOI] [PubMed] [Google Scholar]
- 52.Zhang R, Shen Y, Bai Y, et al. Application of magnetic resonance fingerprinting to differentiate grade I transitional and fibrous meningiomas from meningothelial meningiomas. Quant Imaging Med Surg. 2021;11:1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai Y, Shen Y, Chen R, et al. Magnetic resonance fingerprinting for preoperative differentiation between gonadotroph and non-gonadotroph pituitary macroadenomas. Eur Radiol. 2021;31:8420–8428. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Lu L, Zhu T, et al. Technical overview of magnetic resonance fingerprinting and its applications in radiation therapy. Med Phys. 2022;49:2 846–2860. [DOI] [PubMed] [Google Scholar]
- 55.Mickevicius NJ, Kim JP, Zhao J, et al. Toward magnetic resonance fingerprinting for low-field MR-guided radiation therapy. Med Phys. 2021;48:6930–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruijnen T, van der Heide O, Intven MPW, et al. Technical feasibility of magnetic resonance fingerprinting on a 1.5 T MRI-linac. Phys Med Biol. 2020;65:22NT01. [DOI] [PubMed] [Google Scholar]
- 57.Bernasconi A, Bernasconi N. The role of MRI in the treatment of drug-resistant focal epilepsy. Eur Neurol. 2022;85:333–341. [DOI] [PubMed] [Google Scholar]
- 58.Bernasconi N, Wang I. Emerging trends in neuroimaging of epilepsy. Epilepsy Curr. 2021;21:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao C, Wang K, Cao X, et al. Detection of lesions in mesial temporal lobe epilepsy by using MR fingerprinting. Radiology. 2018;288:804–812. [DOI] [PubMed] [Google Scholar]
- 60.Choi JY, Krishnan B, Hu S, et al. Using magnetic resonance fingerprinting to characterize periventricular nodular heterotopias in pharmacoresistant epilepsy. Epilepsia. 2022;63:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Cao X, Wu D, et al. Magnetic resonance fingerprinting of temporal lobe white matter in mesial temporal lobe epilepsy. Ann Clin Transl Neurol. 2019;6:1639–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma D, Jones SE, Deshmane A, et al. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. J Magn Reson Imaging. 2019;49:1333–1346. [DOI] [PubMed] [Google Scholar]
- 63.Deshmane A, McGivney DF, Ma D, et al. Partial volume mapping using magnetic resonance fingerprinting. NMR Biomed. 2019;32:e4082. [DOI] [PubMed] [Google Scholar]
- 64.Tang Y, Su TY, Choi JY, et al. Characterizing thalamic and basal ganglia nuclei in medically intractable focal epilepsy by MR fingerprinting. Epilepsia. 2022;63:1998–2010. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Chen MH, Baluyot KR, et al. MR fingerprinting enables quantitative measures of brain tissue relaxation times and myelin water fraction in the first five years of life. Neuroimage. 2019;186:782–793. [DOI] [PubMed] [Google Scholar]
- 66.Hung SC, Chen Y, Yap PT, et al. Magnetic resonance fingerprinting of the pediatric brain. Magn Reson Imaging Clin N Am. 2021;29:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu N, Kim JY, Han D, et al. Three-dimensional magnetic resonance fingerprinting in neonates: quantifying regional difference and maturation in the brain. Invest Radiol. 2022;57:44–51. [DOI] [PubMed] [Google Scholar]
- 68.Keil VC, Bakoeva SP, Jurcoane A, et al. MR fingerprinting as a diagnostic tool in patients with frontotemporal lobe degeneration: a pilot study. NMR Biomed. 2019;32:e4157. [DOI] [PubMed] [Google Scholar]
- 69.Naganawa S, Nakane T, Kawai H, et al. Detection of IV-gadolinium leakage from the cortical veins into the CSF using MR fingerprinting. Magn Reson Med Sci. 2020;19:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mostardeiro TR, Panda A, Campeau NG, et al. Whole brain 3D MR fingerprinting in multiple sclerosis: a pilot study. BMC Med Imaging. 2021;21:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerhalter T, Cloos M, Chen AM, et al. T(1) and T(2) quantification using magnetic resonance fingerprinting in mild traumatic brain injury. Eur Radiol. 2022;32:1308–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hermann I, Golla AK, Martinez-Heras E, et al. Lesion probability mapping in MS patients using a regression network on MR fingerprinting. BMC Med Imaging. 2021;21:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi JY, Hu S, Su TY, et al. Normative quantitative relaxation atlases for characterization of cortical regions using magnetic resonance fingerprinting. Cereb Cortex. 2022;bhac292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujita S, Buonincontri G, Cencini M, et al. Repeatability and reproducibility of human brain morphometry using three-dimensional magnetic resonance fingerprinting. Hum Brain Mapp. 2021;42:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan H, Su P, Lin DDM, et al. Simultaneous hemodynamic and structural imaging of ischemic stroke with magnetic resonance fingerprinting arterial spin labeling. Stroke. 2022;53:2016–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su P, Liu P, Pinho MC, et al. Non-contrast hemodynamic imaging of Moyamoya disease with MR fingerprinting ASL: a feasibility study. Magn Reson Imaging. 2022;88:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol. 2019;76:340–351. [DOI] [PubMed] [Google Scholar]
- 79.Dutruel SP, Jeph S, Margolis DJA, et al. PI-RADS: what is new and how to use it. Abdom Radiol (NY). 2020;45:3951–3960. [DOI] [PubMed] [Google Scholar]
- 80.Gaziev G, Wadhwa K, Barrett T, et al. Defining the learning curve for multiparametric magnetic resonance imaging (MRI) of the prostate using MRI-transrectal ultrasonography (TRUS) fusion-guided transperineal prostate biopsies as a validation tool. BJU Int. 2016;117:80–86. [DOI] [PubMed] [Google Scholar]
- 81.Salka BR, Shankar PR, Troost JP, et al. Effect of prostate MRI interpretation experience on PPV using PI-RADS version 2: a 6-year assessment among eight fellowship-trained radiologists. AJR Am J Roentgenol. 2022;219:453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosenkrantz AB, Ginocchio LA, Cornfeld D, et al. Interobserver reproducibility of the PI-RADS version 2 lexicon: a multicenter study of six experienced prostate radiologists. Radiology. 2016;280:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaur S, Harmon S, Rosenblum L, et al. Can apparent diffusion coefficient values assist PI-RADS version 2 DWI scoring? A correlation study using the PI-RADSv2 and International Society of Urological Pathology Systems. AJR Am J Roentgenol. 2018;211:W33–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hambrock T, Hoeks C. Hulsbergen-van de Kaa C, et al. Prospective assessment of prostate cancer aggressiveness using 3-T diffusion-weighted magnetic resonance imaging-guided biopsies versus a systematic 10-core transrectal ultrasound prostate biopsy cohort. Eur Urol. 2012;61:177–184. [DOI] [PubMed] [Google Scholar]
- 85.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259:453–461. [DOI] [PubMed] [Google Scholar]
- 86.Kitajima K, Takahashi S, Ueno Y, et al. Do apparent diffusion coefficient (ADC) values obtained using high b-values with a 3-T MRI correlate better than a transrectal ultrasound (TRUS)-guided biopsy with true Gleason scores obtained from radical prostatectomy specimens for patients with prostate cancer? Eur J Radiol. 2013;82:1219–1226. [DOI] [PubMed] [Google Scholar]
- 87.Boesen L, Chabanova E, Logager V, et al. Apparent diffusion coefficient ratio correlates significantly with prostate cancer Gleason score at final pathology. J Magn Reson Imaging. 2015;42:446–453. [DOI] [PubMed] [Google Scholar]
- 88.Jambor I, Pesola M, Merisaari H, et al. Relaxation along fictitious field, diffusion-weighted imaging, and T2 mapping of prostate cancer: prediction of cancer aggressiveness. Magn Reson Med. 2016;75:2130–2140. [DOI] [PubMed] [Google Scholar]
- 89.Yamauchi FI, Penzkofer T, Fedorov A, et al. Prostate cancer discrimination in the peripheral zone with a reduced field-of-view T(2)-mapping MRI sequence. Magn Reson Imaging. 2015;33:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W, Turkbey B, Senegas J, et al. Accelerated T2 mapping for characterization of prostate cancer. Magn Reson Med. 2011;65:1400–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chatterjee A, Devaraj A, Mathew M, et al. Performance of T2 maps in the detection of prostate cancer. Acad Radiol. 2019;26:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simpkin CJ, Morgan VA, Giles SL, et al. Relationship between T2 relaxation and apparent diffusion coefficient in malignant and non-malignant prostate regions and the effect of peripheral zone fractional volume. Br J Radiol. 2013;86:20120469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gibbs P, Liney GP, Pickles MD, et al. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 tesla. Invest Radiol. 2009;44:572–576. [DOI] [PubMed] [Google Scholar]
- 94.Yu AC, Badve C, Ponsky LE, et al. Development of a combined MR fingerprinting and diffusion examination for prostate cancer. Radiology. 2017;283:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panda A, O'Connor G, Lo WC, et al. Targeted biopsy validation of peripheral zone prostate cancer characterization with magnetic resonance fingerprinting and diffusion mapping. Invest Radiol. 2019;54:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Panda A, Obmann VC, Lo WC, et al. MR fingerprinting and ADC mapping for characterization of lesions in the transition zone of the prostate gland. Radiology. 2019;292:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sushentsev N, Kaggie JD, Buonincontri G, et al. The effect of gadolinium-based contrast agent administration on magnetic resonance fingerprinting-based T1 relaxometry in patients with prostate cancer. Sci Rep. 2020;10:20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shiradkar R, Panda A, Leo P, et al. T1 and T2 MR fingerprinting measurements of prostate cancer and prostatitis correlate with deep learning-derived estimates of epithelium, lumen, and stromal composition on corresponding whole mount histopathology. Eur Radiol. 2021;31:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sushentsev N, Kaggie JD, Slough RA, et al. Reproducibility of magnetic resonance fingerprinting-based T1 mapping of the healthy prostate at 1.5 and 3.0 T: a proof-of-concept study. PLoS One. 2021;16:e0245970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Borofsky S, George AK, Gaur S, et al. What are we missing? False-negative cancers at multiparametric MR imaging of the prostate. Radiology. 2018;286:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Han D, Choi MH, Lee YJ, et al. Feasibility of novel three-dimensional magnetic resonance fingerprinting of the prostate gland: phantom and clinical studies. Korean J Radiol. 2021;22:1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee YS, Choi MH, Lee YJ, et al. Magnetic resonance fingerprinting in prostate cancer before and after contrast enhancement. Br J Radiol. 2022;95:20210479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barbieri M, Lee PK, Brizi L, et al. Circumventing the curse of dimensionality in magnetic resonance fingerprinting through a deep learning approach. NMR Biomed. 2022;35:e4670. [DOI] [PubMed] [Google Scholar]
- 104.Goldberg MA, Hahn PF, Saini S, et al. Value of T1 and T2 relaxation times from echoplanar MR imaging in the characterization of focal hepatic lesions. AJR Am J Roentgenol. 1993;160:1011–1017. [DOI] [PubMed] [Google Scholar]
- 105.Jamin Y, Tucker ER, Poon E, et al. Evaluation of clinically translatable MR imaging biomarkers of therapeutic response in the TH-MYCN transgenic mouse model of neuroblastoma. Radiology. 2013;266:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weidensteiner C, Allegrini PR, Sticker-Jantscheff M, et al. Tumour T1 changes in vivo are highly predictive of response to chemotherapy and reflect the number of viable tumour cells—a preclinical MR study in mice. BMC Cancer. 2014;14:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raza SA, Funicelli L, Sohaib SA, et al. Assessment of colorectal hepatic metastases by quantitative T2 relaxation time. Eur J Radiol. 2012;81:e536–e540. [DOI] [PubMed] [Google Scholar]
- 108.Gambarota G, Veltien A, van Laarhoven H, et al. Measurements of T1 and T2 relaxation times of colon cancer metastases in rat liver at 7 T. MAGMA. 2004;17(3–6):281–287. [DOI] [PubMed] [Google Scholar]
- 109.Curtis WA, Fraum TJ, An H, et al. Quantitative MRI of diffuse liver disease: current applications and future directions. Radiology. 2019;290:23–30. [DOI] [PubMed] [Google Scholar]
- 110.Banerjee R, Pavlides M, Tunnicliffe EM, et al. Multiparametric magnetic resonance for the non-invasive diagnosis of liver disease. J Hepatol. 2014;60:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jaubert O, Arrieta C, Cruz G, et al. Multi-parametric liver tissue characterization using MR fingerprinting: simultaneous T1, T2, T2 *, and fat fraction mapping. Magn Reson Med. 2020;84:2625–2635. [DOI] [PubMed] [Google Scholar]
- 112.Velasco C, Cruz G, Jaubert O, et al. Simultaneous comprehensive liver T1, T2, T2 *, T1rho, and fat fraction characterization with MR fingerprinting. Magn Reson Med. 2022;87:1980–1991. [DOI] [PubMed] [Google Scholar]
- 113.MacAskill CJ, Markley M, Farr S, et al. Rapid B(1)-insensitive MR fingerprinting for quantitative kidney imaging. Radiology. 2021;300:380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Labranche R, Gilbert G, Cerny M, et al. Liver Iron quantification with MR imaging: a primer for radiologists. Radiographics. 2018;38:392–412. [DOI] [PubMed] [Google Scholar]
- 115.Zhang YN, Fowler KJ, Hamilton G, et al. Liver fat imaging—a clinical overview of ultrasound, CT, and MR imaging. Br J Radiol. 2018;91:20170959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Y, Jiang Y, Pahwa S, et al. MR fingerprinting for rapid quantitative abdominal imaging. Radiology. 2016;279:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh A, Reddy D, Haris M, et al. T1rho MRI of healthy and fibrotic human livers at 1.5 T. J Transl Med. 2015;13:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Allkemper T, Sagmeister F, Cicinnati V et al. Evaluation of fibrotic liver disease with whole-liver T1rho MR imaging: a feasibility study at 1.5 T. Radiology. 2014;271:408–415. [DOI] [PubMed] [Google Scholar]
- 119.Serrao EM, Kessler DA, Carmo B, et al. Magnetic resonance fingerprinting of the pancreas at 1.5 T and 3.0 T. Sci Rep. 2020;10:17563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaggie JD, Deen S, Kessler DA, et al. Feasibility of quantitative magnetic resonance fingerprinting in ovarian tumors for T(1) and T(2) mapping in a PET/MR setting. IEEE Trans Radiat Plasma Med Sci. 2019;3:509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.D'Orsi CJ, Sickles EA, Mendelson EB, et al. ACR BI-RADS ® Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology; 2013. [Google Scholar]
- 122.Patani N, Mokbel K. The utility of MRI for the screening and staging of breast cancer. Int J Clin Pract. 2008;62:450–453. [DOI] [PubMed] [Google Scholar]
- 123.Kriege M, Brekelmans CT, Boetes C, et al. Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition. N Engl J Med. 2004;351:427–437. [DOI] [PubMed] [Google Scholar]
- 124.Boyer B, Canale S, Arfi-Rouche J, et al. Variability and errors when applying the BIRADS mammography classification. Eur J Radiol. 2013;82:388–397. [DOI] [PubMed] [Google Scholar]
- 125.Berg WA, Campassi C, Langenberg P, et al. Breast imaging reporting and data system: inter- and intraobserver variability in feature analysis and final assessment. AJR Am J Roentgenol. 2000;174:1769–1777. [DOI] [PubMed] [Google Scholar]
- 126.Sippo DA, Rutledge GM, Mercaldo SF, et al. Impact of background parenchymal enhancement on diagnostic performance in screening breast MRI. Acad Radiol. 2020;27:663–671. [DOI] [PubMed] [Google Scholar]
- 127.McSweeney MB, Small WC, Cerny V et al. Magnetic resonance imaging in the diagnosis of breast disease: use of transverse relaxation times. Radiology. 1984;153:741–744. [DOI] [PubMed] [Google Scholar]
- 128.Merchant TE, Thelissen GR, de Graaf PW, et al. Application of a mixed imaging sequence for MR imaging characterization of human breast disease. Acta Radiol. 1993;34:356–361. [PubMed] [Google Scholar]
- 129.Tan PC, Pickles MD, Lowry M, et al. Lesion T(2) relaxation times and volumes predict the response of malignant breast lesions to neoadjuvant chemotherapy. Magn Reson Imaging. 2008;26:26–34. [DOI] [PubMed] [Google Scholar]
- 130.Manton DJ, Chaturvedi A, Hubbard A, et al. Neoadjuvant chemotherapy in breast cancer: early response prediction with quantitative MR imaging and spectroscopy. Br J Cancer. 2006;94:427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Edden RA, Smith SA, Barker PB. Longitudinal and multi-echo transverse relaxation times of normal breast tissue at 3 Tesla. J Magn Reson Imaging. 2010;32:982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rakow-Penner R, Daniel B, Yu H, et al. Relaxation times of breast tissue at 1. 5 T and 3 T measured using IDEAL. J Magn Reson Imaging. 2006;23:87–91. [DOI] [PubMed] [Google Scholar]
- 133.Chen Y, Panda A, Pahwa S, et al. Three-dimensional MR fingerprinting for quantitative breast imaging. Radiology. 2019;290:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nolte T, Gross-Weege N, Doneva M, et al. Spiral blurring correction with water-fat separation for magnetic resonance fingerprinting in the breast. Magn Reson Med. 2020;83:1192–1207. [DOI] [PubMed] [Google Scholar]
- 135.Zanderigo E, Huck L, Distelmaier M, et al. Feasibility study of 2D Dixon-magnetic resonance fingerprinting (MRF) of breast cancer. Eur J Radiol Open. 2022;9:100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tahir E, Sinn M, Bohnen S, et al. Acute versus chronic myocardial infarction: diagnostic accuracy of quantitative native T1 and T2 mapping versus assessment of edema on standard T2-weighted cardiovascular MR images for differentiation. Radiology. 2017;285:83–91. [DOI] [PubMed] [Google Scholar]
- 137.Bohnen S, Radunski UK, Lund GK, et al. Performance of T1 and T2 mapping cardiovascular magnetic resonance to detect active myocarditis in patients with recent-onset heart failure. Circ Cardiovasc Imaging. 2015;8:e003073. [DOI] [PubMed] [Google Scholar]
- 138.Puntmann VO, Nagel E. T1 and T2 mapping in nonischemic cardiomyopathies and agreement with endomyocardial biopsy. J Am Coll Cardiol. 2016;68:1923–1924. [DOI] [PubMed] [Google Scholar]
- 139.Messroghli DR, Moon JC, Ferreira VM, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson. 2017;19:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kellman P, Hernando D, Shah S, et al. Multiecho Dixon fat and water separation method for detecting fibrofatty infiltration in the myocardium. Magn Reson Med. 2009;61:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Witschey WR, Zsido GA, Koomalsingh K, et al. In vivo chronic myocardial infarction characterization by spin locked cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2012;14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Messroghli DR, Radjenovic A, Kozerke S, et al. Modified look-locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52:141–146. [DOI] [PubMed] [Google Scholar]
- 143.Chow K, Flewitt JA, Green JD, et al. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med. 2014;71:2082–2095. [DOI] [PubMed] [Google Scholar]
- 144.Giri S, Chung YC, Merchant A, et al. T2 quantification for improved detection of myocardial edema. J Cardiovasc Magn Reson. 2009;11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sprinkart AM, Luetkens JA, Traber F, et al. Gradient spin echo (GraSE) imaging for fast myocardial T2 mapping. J Cardiovasc Magn Reson. 2015;17:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hamilton JI, Jiang Y, Chen Y, et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med. 2017;77:1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hamilton JI, Jiang Y, Ma D, et al. Investigating and reducing the effects of confounding factors for robust T(1) and T(2) mapping with cardiac MR fingerprinting. Magn Reson Imaging. 2018;53:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Velasco C, Fletcher TJ, Botnar RM, et al. Artificial intelligence in cardiac magnetic resonance fingerprinting. Front CardiovascMed. 2022;9:1009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hamilton JI, Seiberlich N. Machine learning for rapid magnetic resonance fingerprinting tissue property quantification. Proc IEEE Inst Electr Electron Eng. 2020;108:69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hamilton JI, Currey D, Rajagopalan S, et al. Deep learning reconstruction for cardiac magnetic resonance fingerprinting T(1) and T(2) mapping. Magn Reson Med. 2021;85:2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hamilton JI. A self-supervised deep learning reconstruction for shortening the breathhold and acquisition window in cardiac magnetic resonance fingerprinting. Front Cardiovasc Med. 2022;9:928546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hamilton JI, Jiang Y, Ma D, et al. Simultaneous multislice cardiac magnetic resonance fingerprinting using low rank reconstruction. NMR Biomed. 2019;32:e4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cruz G, Jaubert O, Qi H, et al. 3D free-breathing cardiac magnetic resonance fingerprinting. NMR Biomed. 2020;33:e4370. [DOI] [PubMed] [Google Scholar]
- 155.Hamilton JI, Jiang Y, Eck B, et al. Cardiac cine magnetic resonance fingerprinting for combined ejection fraction, T(1) and T(2) quantification. NMR Biomed. 2020;33:e4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jaubert O, Cruz G, Bustin A, et al. Free-running cardiac magnetic resonance fingerprinting: joint T1/T2 map and cine imaging. Magn Reson Imaging. 2020;68:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Velasco C, Cruz G, Lavin B, et al. Simultaneous T(1), T(2), and T(1rho) cardiac magnetic resonance fingerprinting for contrast agent-free myocardial tissue characterization. Magn Reson Med. 2022;87:1992–2002. [DOI] [PubMed] [Google Scholar]
- 158.Lima da Cruz GJ, Velasco C, Lavin B, et al. Myocardial T1, T2, T2*, and fat fraction quantification via low-rank motion-corrected cardiac MR fingerprinting. Magn Reson Med. 2022;87:2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jaubert O, Cruz G, Bustin A, et al. Water-fat Dixon cardiac magnetic resonance fingerprinting. Magn Reson Med. 2020;83:2107–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jaubert O, Cruz G, Bustin A, et al. T1, T2, and fat fraction cardiac MR fingerprinting: preliminary clinical evaluation. J Magn Reson Imaging. 2021;53:1253–1265. [DOI] [PubMed] [Google Scholar]
- 161.Liu Y, Hamilton J, Eck B, et al. Myocardial T(1) and T(2) quantification and water-fat separation using cardiac MR fingerprinting with rosette trajectories at 3 T and 1.5 T. Magn Reson Med. 2021;85:103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Y, Hamilton J, Jiang Y, et al. Cardiac MRF using rosette trajectories for simultaneous myocardial T(1), T(2), and proton density fat fraction mapping. Front Cardiovasc Med. 2022;9:977603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rumac S, Pavon AG, Hamilton JI, et al. Cardiac MR fingerprinting with a short acquisition window in consecutive patients referred for clinical CMR and healthy volunteers. Sci Rep. 2022;12:18705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cavallo AU, Liu Y, Patterson A, et al. CMR fingerprinting for myocardial T1, T2, and ECV quantification in patients with nonischemic cardiomyopathy. JACC Cardiovasc Imaging. 2019;12(8 Pt 1):1584–1585. [DOI] [PubMed] [Google Scholar]
- 165.Eck BL, Seiberlich N, Flamm SD, et al. Characterization of cardiac amyloidosis using cardiac magnetic resonance fingerprinting. Int J Cardiol. 2022;351:107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vincenti G, Coristine AJ, Hamilton J, et al. Cardiac Magnetic Resonance Fingerprinting for the Investigation of Suspected Inflammatory Cardiomyopathy. Montreal, Quebec, Canada: International Society for Magnetic Resonance Medicine 27th Annual Meeting & Exhibition; 2019. [Google Scholar]
- 167.Patterson A, Li Y, Cavallo AU, et al. Initial Utility of Cardiac Magnetic Resonance Fingerprinting for Quantitative T1/T2 Parametric Mapping in Hypertrophic Cardiomyopathy. Montreal, Quebec, Canada: International Society for Magnetic Resonanace in Medicine 27th Annual Meeting & Exhibition; 2019. [Google Scholar]
- 168.Wintersperger B, Hamilton J, Houbois C, et al. Quantitative Multiparametric Myocardial Evaluation in Hypertrophic Cardiomyopathy Using Cardiac Magnetic Resonance Fingerprinting: Comparison to Conventional Cardiac Relaxometry. London, England: Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting; 2022. [Google Scholar]
- 169.Coristine AJ, Hamilton J, van Heeswijk RB, et al. Cardiac Magnetic Resonance Fingerprinting in Heart Transplant Recipients. Paris, France: Joint Annual Meeting ISMRM-ESMRMB; 2018. [Google Scholar]
- 170.Liu Y, Hopman LHGA, Hamilton J, et al. Inter-Site Reproducibility of Cardiac Magnetic Resonance Fingerprinting T1 and T2 Quantification in the ISMRM/NIST MRI System Phantom and Human Heart. Montreal, Quebec, Canada: International Society for Magnetic Resonance in Medicine 27th Annual Meeting & Exhibition; 2019. [Google Scholar]
- 171.Sharafi A, Medina K, Zibetti MWV, et al. Simultaneous T1, T2, and T1rho relaxation mapping of the lower leg muscle with MR fingerprinting. Magn Reson Med. 2021;86:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marty B, Lopez Kolkovsky AL, Araujo ECA, et al. Quantitative skeletal muscle imaging using 3D MR fingerprinting with water and fat separation. J Magn Reson Imaging. 2021;53:1529–1538. [DOI] [PubMed] [Google Scholar]
- 173.Marty B, Reyngoudt H, Boisserie JM, et al. Water-fat separation in MR fingerprinting for quantitative monitoring of the skeletal muscle in neuromuscular disorders. Radiology. 2021;300:652–660. [DOI] [PubMed] [Google Scholar]
- 174.Marty B, Carlier PG. MR fingerprinting for water T1 and fat fraction quantification in fat infiltrated skeletal muscles. Magn Reson Med. 2020;83:621–634. [DOI] [PubMed] [Google Scholar]
- 175.Li Q, Cao X, Ye H, et al. Ultrashort echo time magnetic resonance fingerprinting (UTE-MRF) for simultaneous quantification of long and ultrashort T(2) tissues. Magn Reson Med. 2019;82:1359–1372. [DOI] [PubMed] [Google Scholar]
- 176.Choi MH, Lee SW, Kim HG, et al. 3D MR fingerprinting (MRF) for simultaneous T1 and T2 quantification of the bone metastasis: initial validation in prostate cancer patients. Eur J Radiol. 2021;144:109990. [DOI] [PubMed] [Google Scholar]
- 177.Sharafi A, Zibetti MVW, Chang G, et al. MR fingerprinting for rapid simultaneous T1, T2, and T1 rho relaxation mapping of the human articular cartilage at 3 T. Magn Reson Med. 2020;84:2636–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kijowski R, Sharafi A, Zibetti MVW, et al. Age-dependent changes in knee cartilage T(1), T(2), and T(1p) simultaneously measured using MRI fingerprinting. J Magn Reson imaging. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sharafi A, Zibetti MVW, Chang G, et al. Simultaneous bilateral T(1), T(2), and T(1rho) relaxation mapping of the hip joint with magnetic resonance fingerprinting. NMR Biomed. 2022;35:e4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cloos MA, Asslander J, Abbas B, et al. Rapid radial T(1) and T(2) mapping of the hip articular cartilage with magnetic resonance fingerprinting. J Magn Reson Imaging. 2019;50:810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cloos MA, Knoll F, Zhao T, et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun. 2016;7:12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Konar AS, Shah AD, Paudyal R, et al. Quantitative synthetic magnetic resonance imaging for brain metastases: a feasibility study. Cancers (Basel). 2022;14:2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lu T, Marin T, Zhou Y, et al. Nonuniform fast Fourier transform on TPUs. IEEE 18th International Symposium on Biomedical Imaging (ISBI). 2021;783–787. [Google Scholar]
- 184.Hsieh JJL, Svalbe I. Magnetic resonance fingerprinting: from evolution to clinical applications. J Med Radiat Sci. 2020;67:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]