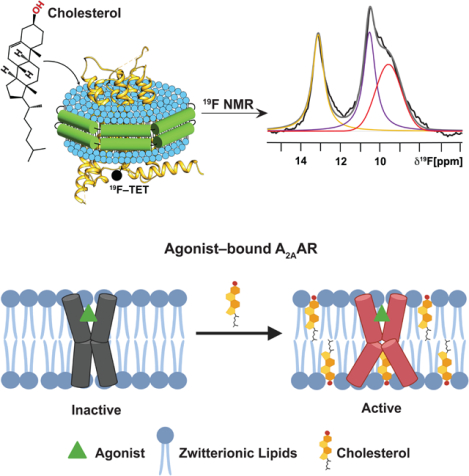
SUMMARY
Cholesterol is a critical component of mammalian cell membranes and an allosteric modulator of G protein-coupled receptors (GPCRs), but divergent views exist on the mechanisms by which cholesterol influences receptor functions. Leveraging the benefits of lipid nanodiscs, i.e., quantitative control of lipid composition, we observe distinct impacts of cholesterol in the presence and absence of anionic phospholipids on the function-related conformational dynamics of the human A2A adenosine receptor (A2AAR). Direct receptor-cholesterol interactions drive activation of agonist-bound A2AAR in membranes containing zwitterionic phospholipids. Intriguingly, the presence of anionic lipids attenuates cholesterol’s impact through direct interactions with the receptor, highlighting a more complex role for cholesterol that depends on membrane phospholipid composition. Targeted amino acid replacements at two frequently predicted cholesterol interaction sites showed distinct impacts of cholesterol at different receptor locations, demonstrating the ability to delineate different roles of cholesterol in modulating receptor signaling and maintaining receptor structural integrity.
Graphical Abstract
eTOC Blurb
Ray and Thakur et al. use 19F-NMR to observe how cholesterol affects the conformational equilibria of the A2A adenosine receptor. The impact of cholesterol varies depending on the type of phospholipids present and differs at different receptor locations, affecting either structural stability or receptor signaling activity.
INTRODUCTION
Cholesterol is a major component of mammalian cell membranes and an important regulator of the functions of membrane proteins, as documented extensively in the literature.1–9 Alterations in cholesterol distribution and metabolism, either through genetic or environmental causes, are highly correlated with numerous health disorders, including Alzheimer’s disease,10 heart disease,11 and diabetes.12 Cholesterol is also increasingly known for its role as an allosteric modulator of G protein-coupled receptors (GPCRs), integral membrane sensory proteins, and targets of over 35% of FDA-approved drugs.13
Literature data have reported the impacts of cholesterol on essentially every aspect of GPCR pharmacology and biology.14–19 Cholesterol has been shown experimentally to act as an allosteric modulator of GPCRs, altering the binding affinities of orthosteric ligands, as observed with, for example, the oxytocin receptor,20 CXCR4 chemokine receptor,21 serotonin1A receptor,22 cannabinoid CB1 receptor23 and CCR5 chemokine receptor.24 Structural and biochemical evidence also support the role of cholesterol as a direct orthosteric agonist for some GPCRs, including the smoothened receptor (SMO).25 In addition to cholesterol’s impacts on receptor pharmacology, experimental and computational studies have identified roles of cholesterol in GPCR oligomerization,26–29 receptor sorting and trafficking,30–33 and formation of signaling complexes.19,34,35
Three general mechanisms have been proposed to understand the influence of cholesterol on GPCR function: direct cholesterol-GPCR interactions, indirectly via modulation of bulk membrane properties, or a combination of both.36 Support for direct receptor-cholesterol interactions has come, in part, from the observation of cholesterol or cholesterol analogs closely associated with the transmembrane regions of more than 60 GPCRs in X-ray crystallography and cryo-electron microscopy structures, as reviewed.37 NMR studies of the β2-adrenergic receptor (β2AR) indicated that the receptor showed a specific preference to associate with cholesterol over ergosterol,38 and high-pressure NMR studies reported the cholesterol analog, cholesteryl hemisuccinate (CHS), specifically associated with the β1-adrenergic receptor (β1AR) to inhibit its activation.39 However, there is no consensus on the extent to which specific cholesterol-receptor interactions influence the function of GPCRs, and earlier studies of rhodopsin reported evidence for the influence of cholesterol on rhodopsin function both through direct interactions with rhodopsin40 and by modulating the properties of the lipid bilayer.41
We investigated the mechanism by which cholesterol modulates the function-related dynamics of the human A2A adenosine receptor (A2AAR), a class A GPCR. A2AAR is a model receptor for investigating mechanisms underlying GPCR signaling phenomena, owing to a large number of structures42–46 and the availability of spectroscopic data in different membrane mimetics.47–53 A2AAR is an important and validated drug target for Parkinson’s disease54 and cancers55 for which cholesterol may play a role. A growing body of literature has documented the impact of cholesterol on A2AAR signaling and pharmacology, though there is no consensus on the precise role of cholesterol or mechanisms by which cholesterol may influence A2AAR function. In HEK293 cells treated with methyl-β-cyclodextrin (MβCD) to deplete cholesterol, A2AAR signaling was observed to significantly decrease,56 indicating cholesterol was required to achieve a maximum signaling response to agonist stimulation. In a separate study, treatment of C6 glioma cells with MβCD showed an increase in the specific binding of an A2AAR antagonist, suggesting cholesterol inhibited antagonist binding.15 Canonical cholesterol recognition motifs and cholesterol-binding “hotspots” have been predicted for A2AAR as evidence for specific receptor-cholesterol interactions.57–59 In contrast to this view, 19F-NMR experiments with A2AAR labeled at position V229C on transmembrane helix 6 concluded cholesterol was a weak positive allosteric modulator and impacts of cholesterol were likely due more to cholesterol-mediated changes in membrane bilayer fluidity rather than direct interactions.60
To reconcile these divergent views, we integrated 19F-NMR data with correlative functional and pharmacological assays to investigate the impact of cholesterol on signaling-related A2AAR conformational dynamics. We leveraged the benefits of lipid nanodiscs to quantitatively control lipid composition in all samples. By systematically exploring a wider range of lipid compositions than previously studied, we observed evidence for both the direct and indirect influence of cholesterol. The mechanism of cholesterol influence depended both on the presence of cholesterol and the composition of phospholipids in the membrane. Using conditions where we observe specific cholesterol-receptor interactions, we explored variations in the cholesterol chemical scaffold to test the extent to which cholesterol-A2AAR interactions depended on the structural details of cholesterol molecules. The same conditions also allowed us to investigate two of the most frequently predicted cholesterol interaction sites.61–63 Our findings reveal a clear and significant impact of cholesterol on the activation of agonist-bound A2AAR in nanodiscs that contain zwitterionic lipids. This effect is observable even for samples containing only 1–2 molecules of cholesterol per nanodisc. An investigation of cholesterol analogs also yielded comparable results. Interestingly, this response is obscured when A2AAR is embedded in nanodiscs containing mixtures of zwitterionic and anionic lipids, suggesting a potential interplay between anionic lipids and cholesterol. These results are discussed in the context of integrating in vitro and in vivo data to provide a more comprehensive understanding of the role of cholesterol in GPCR activation mechanisms.
RESULTS
Cholesterol manifests different impacts on the pharmacology of A2AAR antagonists and agonists
For biophysical and functional experiments, a variant of human A2AAR containing a single cysteine replacement at position 289 located at the intracellular surface of helix 7 (Figure S1), A2AAR[A289C], was expressed in Pichia pastoris using previously described protocols.64 The location of the introduced cysteine was selected based on literature data from 19F NMR studies of A2AAR, which demonstrated NMR spectra of A2AAR labeled with 19F-2,2,2-trifluoroethanethiol at this position were sensitive to differences in the efficacies of bound ligands.51 This position was also shown to be sensitive to modulation of the A2AAR conformational ensemble by membrane phospholipids.64 C289 was the only cysteine available for chemical conjugation, as previously demonstrated.51
To investigate the role of cholesterol on the conformational equilibria of A2AAR, we prepared samples of A2AAR[A289C] in lipid nanodiscs containing binary mixtures of POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine) and cholesterol, or ternary mixtures of POPC and POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine) and cholesterol, or POPC and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol)) and cholesterol, where the amount of cholesterol was varied from 0 to 15 mol%. We observed homogenous preparations could be achieved for nanodiscs containing up to 15 mol% cholesterol and attempts to prepare nanodiscs with more resulted in heterogeneous, aggregated samples. This is consistent with analogous observations described previously, which reported higher concentrations of cholesterol inhibited nanodisc formation, possibly due to increased membrane rigidity.60,65,66 We therefore focused our study on membrane compositions containing ≤15 mol% cholesterol. While this is lower than what has been observed in some mammalian cell membranes,67 this amount still allowed us to investigate possible mechanisms of cholesterol influence on A2AAR.
To obtain homogeneous lipid compositions in nanodiscs, mixtures of lipids were quantitatively weighed and dissolved in chloroform, dried down to form a thin film, and then reconstituted in buffer (STAR Methods). Analytical size exclusion chromatography characterization of purified lipid nanodiscs containing A2AAR, phospholipids, and varying amounts of cholesterol showed monodispersed and homogeneous samples (Figure S2). The phospholipid composition of nanodisc samples containing A2AAR was verified by 31P-NMR, which showed the relative amounts of each phospholipid species in the nanodisc samples quantitatively agreed with the intended amounts (Figure S3). The amount of cholesterol in each nanodisc sample was verified using the Amplex Red Cholesterol Assay, a fluorometric method for the enzymatic detection of cholesterol68, and confirmed to be consistent with targeted amounts (Table S1). We also note that this assay has been used to quantify concentrations of other sterols including ergosterol69, which is produced by P. pastoris. Using the fluorometric assay we did not detect the presence of other sterols in nanodisc samples that were not prepared by purposely adding sterols. To confirm the receptor was folded in all employed lipid compositions, we recorded fluorescence thermal shift assays using a thiol-reactive dye, N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM), for A2AAR[A289C] in complex with the full agonist 5’-N-ethylcarboxamidoadenosine (NECA). As the nanodisc scaffold protein MSP1D1 contains no cysteines, the assay thus observed a direct response of the thermal unfolding of the receptor. A2AAR was folded in all studied lipid compositions across the full range of cholesterol concentrations (Figure S4). We observed at most a minimal impact of cholesterol on the measured melting temperature of A2AAR[A289C] for nanodisc samples prepared either with POPC and cholesterol or ternary mixtures of POPC, POPS, and cholesterol (Figure S4). The more significant impact was the presence or absence of POPS, consistent with earlier studies.64
To investigate the impact of cholesterol on the pharmacological activity of A2AAR, we measured affinities for the antagonist ZM241385 and agonist NECA with radioligand competition binding experiments in lipid nanodiscs containing variable amounts of cholesterol. For all samples studied, we observed clear evidence of ligand binding, indicating A2AAR[A289C] retained pharmacological activity across the range of cholesterol concentrations used in subsequent NMR experiments (Figure 1). For experiments with the antagonist ZM241385, we measured at most a factor of ~3 difference in binding affinities in samples containing 0 to 10 mol% cholesterol, with no clear relationship between the amount of cholesterol and determined KD value for the antagonist (Figure 1A and Table S2), suggesting these relatively smaller differences were likely unrelated to the amount of cholesterol present. For the agonist NECA, we observed a difference of ~4.5 in measured binding affinities between samples containing 0 mol% cholesterol and 10 mol% cholesterol, with a linear decrease in the agonist dissociation constant with increasing amounts of cholesterol (Figure 1B and Table S2). This suggested a potential correlation between the presence of cholesterol and affinity for agonists, though less than an order of magnitude difference in agonist affinities between samples containing 0 to 10 mol% cholesterol.
Figure 1. Pharmacological activity of A2AAR[A289C] in lipid nanodiscs containing varying amounts of cholesterol.
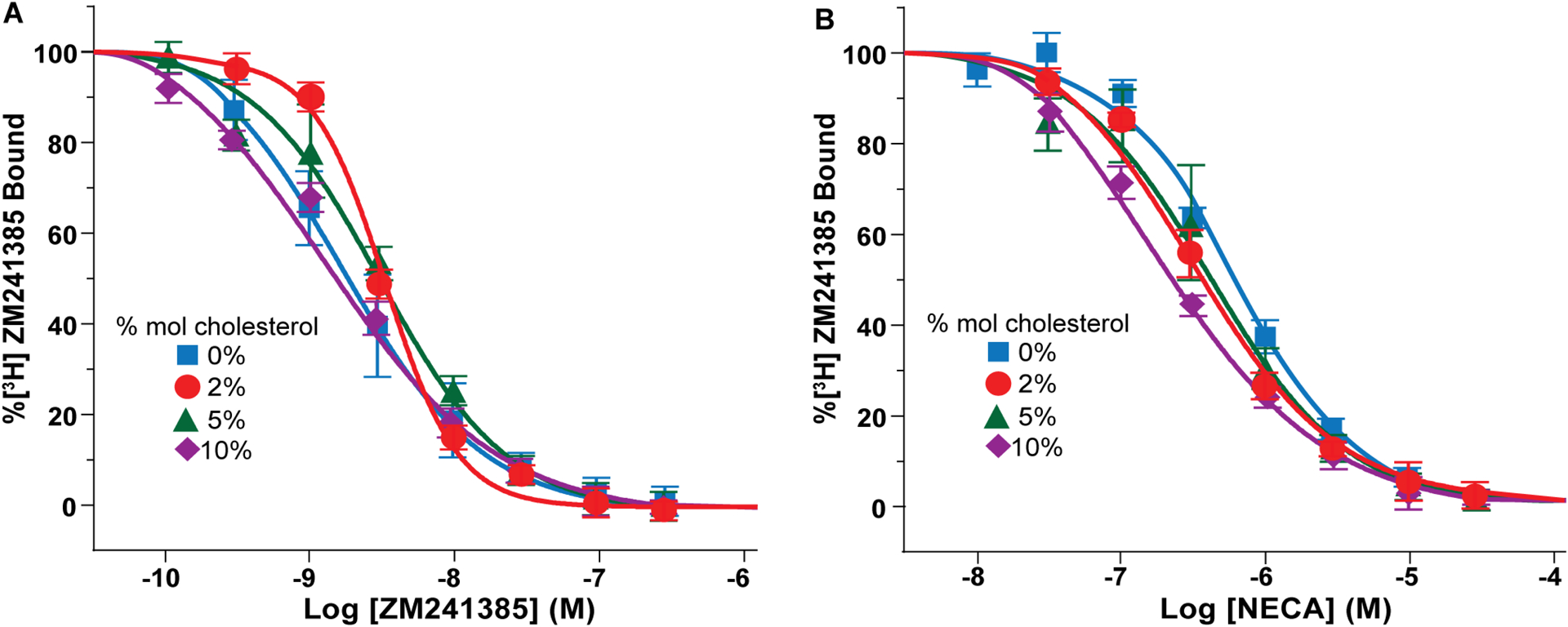
(A) Homologous competition binding experiments with the antagonist ZM241385. Data were measured for A2AAR[A289C] prepared in nanodiscs containing POPC and varying amounts of cholesterol, as indicated. Error bars indicate the s.e.m for 3 independent trials done in triplicate. The determined KD values are listed in Table S2.
(B) Competition binding experiments with the agonist NECA for A2AAR[A289C] prepared in nanodiscs containing POPC and varying amounts of cholesterol. Same presentation details as (A). The determined KI values are listed in Table S2. See also Figure S1, Figure S3 and Table S1.
Cholesterol is required to populate an active A2AAR ensemble in zwitterionic phospholipid membranes
We investigated the impact of cholesterol on the function-related conformational dynamics of A2AAR by NMR using a 19F-NMR reporter group introduced at position C289, located in transmembrane helix 7, with 2,2,2-trifluoroethanethiol (TET) covalently attached via an in-membrane chemical modification approach,70 yielding A2AAR[A289CTET]. Earlier studies demonstrated no other cysteines were available for 19F-labeling.51 The 19F-NMR reporter at position C289 was shown to be highly sensitive to function-related changes in the efficacies of bound drugs51 and changes in membrane phospholipid composition,64 providing ‘fingerprints’ for the corresponding functional states.
We recorded 19F-NMR spectra of A2AAR[A289CTET] in complex with the antagonist ZM241385 in lipid nanodiscs containing POPC and either without cholesterol or with 5 mol% cholesterol (Figure 2A). For both samples, highly similar spectra were observed containing two components, labeled P1 and P3, with P3 being the dominant component in the spectrum. In earlier studies, 19F-NMR spectra of antagonist-bound A2AAR[A289CTET] in DDM/CHS micelles,51 P3 had been assigned to an inactive conformation of the receptor and P1 had been observed for both antagonist-bound and agonist-bound A2AAR[A289CTET].51 The spectra of antagonist-bound A2AAR[A289CTET] shown in Figure 2A are also highly similar to spectra reported of antagonist-bound A2AAR[A289CTET] in lipid nanodiscs of binary mixtures of phospholipids that did not contain cholesterol.64 The chemical shifts for components P1 and P3 appear to be highly similar for preparations of antagonist-bound A2AAR[A289CTET] either in DDM/CHS micelles51 or lipid nanodiscs without cholesterol.64 Further, only relatively smaller variations in the relative intensities of components P1 and P3 were observed in the absence and presence of cholesterol for antagonist-bound A2AAR (Figure 2A), compared with more striking changes observed for agonist-bound A2AAR[A289CTET] upon addition of cholesterol (see below). Specifically, for antagonist-bound A2AAR[A289CTET] we observed a ~15% decrease in the relative population for state P3 and a ~30% increase in the population of state P1 upon addition of cholesterol (Table S4). The dominant component in the spectrum was still clearly P3, indicating the inactive conformational ensemble remained similar in the absence and the presence of cholesterol.
Figure 2. NMR-observed conformational states of A2AAR[A289CTET] in nanodiscs with POPC and varying amounts of cholesterol.
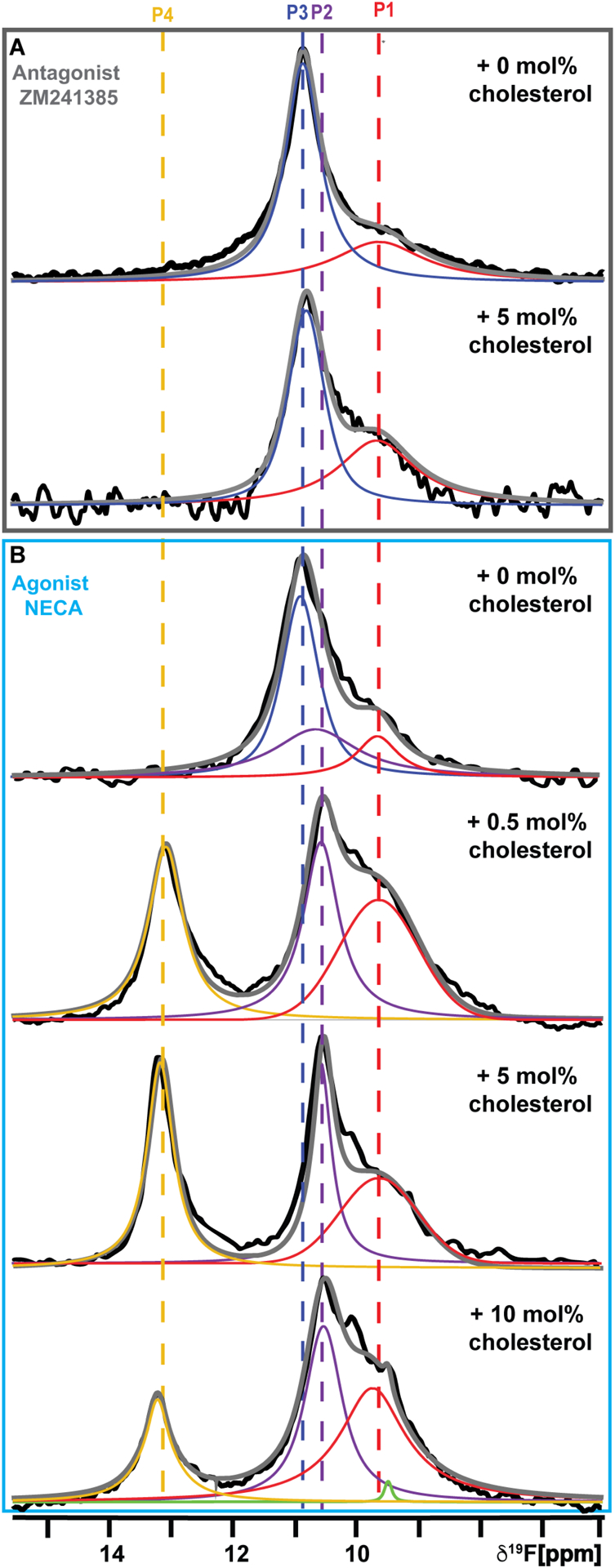
(A) The 1-dimensional 19F-NMR spectra of A2AAR[A289CTET] in complex with the antagonist ZM241385 in nanodiscs containing POPC and without cholesterol or with 5 mol% cholesterol.
(B) The 1-dimensional 19F-NMR spectra of A2AAR[A289CTET] in complex with the agonist NECA in nanodiscs containing POPC and varying amounts of cholesterol, as indicated. The NMR spectra shown are interpreted using Lorentzian deconvolutions with the minimal number of components required to provide a good fit, labeled P1 to P4. The grey lines superimposed on the spectra are the total sums of the individual deconvolutions. The green line superimposed on the bottom spectrum is a Lorentzian deconvolution of a minor contribution from free TET present in this sample. The chemical shifts of P1 to P4 are indicated by the colored dashed vertical lines. See also Figures S3 – S6, Figure S11, Table S3, and Table S4.
In contrast to our observations with antagonist-bound A2AAR, 19F-NMR spectra of agonist-bound A2AAR[A289CTET] showed a striking dependence on the presence of cholesterol. 19F-NMR spectra of A2AAR[A289CTET] in complex with the agonist NECA in lipid nanodiscs containing POPC were highly similar to spectra of A2AAR[A289CTET] in complex with the antagonist ZM241385 (Figure 2B). This indicated we observed an inactive A2AAR conformational ensemble even with a saturating amount of agonist present, consistent with earlier studies in nanodiscs containing only zwitterionic phospholipids.64 From the radioligand binding data in Figure 1, we concluded the pharmacological activity of A2AAR was preserved in the employed lipid composition, and thus the observation of the inactive conformational ensemble was not due to impaired ligand binding.
Compared to the results in nanodiscs containing only POPC, we observed a clear-cut response to cholesterol for agonist-bound A2AAR[A289CTET] in nanodiscs containing POPC. 19F-NMR spectra of agonist-bound A2AAR in nanodiscs containing POPC and 0.5 mol% cholesterol showed the hallmarks of an active ensemble, noted by the disappearance of peak P3 and the emergence of two new peaks P2 and P4 (Figure 2B), which had highly similar chemical shifts to signals identified for agonist-bound A2AAR in DDM/CHS micelles.51 Previously, we assigned P4 to the fully active A2AAR conformation by carefully examining experimental conditions in which we observed a population of P4 only upon complex formation of A2AAR[A289CTET] with mini-GS.64 The composition of 0.5 mol% cholesterol corresponds on average to 1 to 2 molecules of cholesterol per nanodisc (see Discussion), which was verified in fluorometric assays as described above. At this relatively low concentration of cholesterol, we did not anticipate cholesterol would have a significant impact on membrane fluidity, which may also potentially impact A2AAR. To verify this, we recorded fluorescence spectra of nanodiscs containing POPC, varying amounts of cholesterol, and the lipophilic fluorescent probe Laurdan, which is a sensitive probe of membrane fluidity.71 We observed that nanodiscs containing 0.5 mol% cholesterol showed almost identical membrane phase behavior compared with nanodiscs containing only POPC (Figure S5). Based on these collective observations, we concluded the effects of cholesterol observed on populating the active state ensemble of agonist-bound A2AAR must be due to a direct influence on the receptor rather than to the modulation of the bulk properties of the membranes in the nanodiscs.
We recorded additional 19F-NMR spectra of agonist-bound A2AAR[A289CTET] in nanodiscs containing POPC and 5 mol% cholesterol or 10 mol% cholesterol. The fits for the data for all spectra were evaluated by calculating the residual difference between the raw data and the sum of the individually fit components (Figure S6), from which we conclude that no new signals were observed from earlier studies of A2AAR in DDM/CHS51 or in lipid nanodiscs.64 The 19F-NMR spectrum of agonist-bound A2AAR[A289CTET] in nanodiscs containing POPC and 5 mol% cholesterol qualitatively resembled the spectrum for A2AAR in nanodiscs containing 0.5 mol% cholesterol with marginally reduced linewidths observed for states P4 and P2 (Figure 2B), possibly indicating reduced conformational plasticity with a higher concentration of cholesterol. While the 19F-NMR spectrum A2AAR[A289CTET] in nanodiscs containing POPC and 10 mol% cholesterol qualitatively resembled spectra with lower amounts of cholesterol, we observed a slightly diminished intensity for state P4 (Figure 2B). In the membrane fluidity measurements with Laurdan, we observed a reduction in membrane fluidity with increasing amounts of cholesterol. Samples prepared with 10 mol% cholesterol showed the largest increase in order (Figure S5). This observation may correlate with the slightly diminished intensity for state P4 observed with increasing cholesterol concentration. We also recorded 19F-NMR spectra at a higher temperature of 300 K of A2AAR[A289CTET] in complex with NECA in nanodiscs containing POPC and either 0.5 mol% cholesterol or 5 mol% cholesterol (Figure S7). At 300 K, we observed a ~50% increase in the popultion of state P4 with respect to the total integrated spectrum (Table S4). We anticipate that at even higher temperatures we would likely observe additional marginal increases in the population of P4.
Cholesterol analogs also significantly influence the conformational equilibria of agonist-bound A2AAR
Having established experimental conditions where we observed a clear impact of cholesterol on populating the active ensemble of agonist-bound A2AAR, we investigated whether this impact was specific to cholesterol or could be observed also with other sterols. Cholesterol-protein interactions are thought to be nuanced, and it has been proposed that details of the chemical structure of cholesterol facilitate its interactions with membrane proteins. MD simulations have predicted that specific structural features of cholesterol molecules form unique interactions with membrane proteins at cholesterol interaction sites.56–59,72 We recorded 19F-NMR spectra of A2AAR[A289CTET] in complex with the agonist NECA in lipid nanodiscs containing POPC and 0.5% mol fraction of one of three different sterols: epicholesterol, ergosterol, or digoxigenin (Figure 3). The three sterols were chosen to explore the extent to which the influence of cholesterol on the conformational ensemble of A2AAR depended on specific features of the cholesterol chemical structure, as epicholesterol is an enantiomer and ergosterol is a diastereomer of cholesterol. 19F-NMR spectra of agonist-bound A2AAR[A289CTET] in the presence of POPC and 0.5 mol% of epi-cholesterol, ergosterol, or digoxigenin were similar to spectra recorded in the presence of 0.5 mol% cholesterol, showing an active state ensemble (Figure 3 and Figure S8). The relative population of state P4 varied among the spectra recorded with different sterols, with the sample prepared with digoxigenin showing the lowest relative population of P4 and the sample prepared with epicholesterol showing approximately twice the relative population (Table S4). Despite these variations, there was a distinct difference in the overall spectral fingerprint of all samples prepared with 0.5 mol% sterol compared to agonist-bound receptor in the absence of cholesterol. These observations suggest the influence of cholesterol on the A2AAR conformational ensemble does not depend on specific details of the cholesterol chemical scaffold.
Figure 3. Sterols with different chemical structures shift the conformational equilibria of the A2AAR-agonist complex toward an active ensemble.
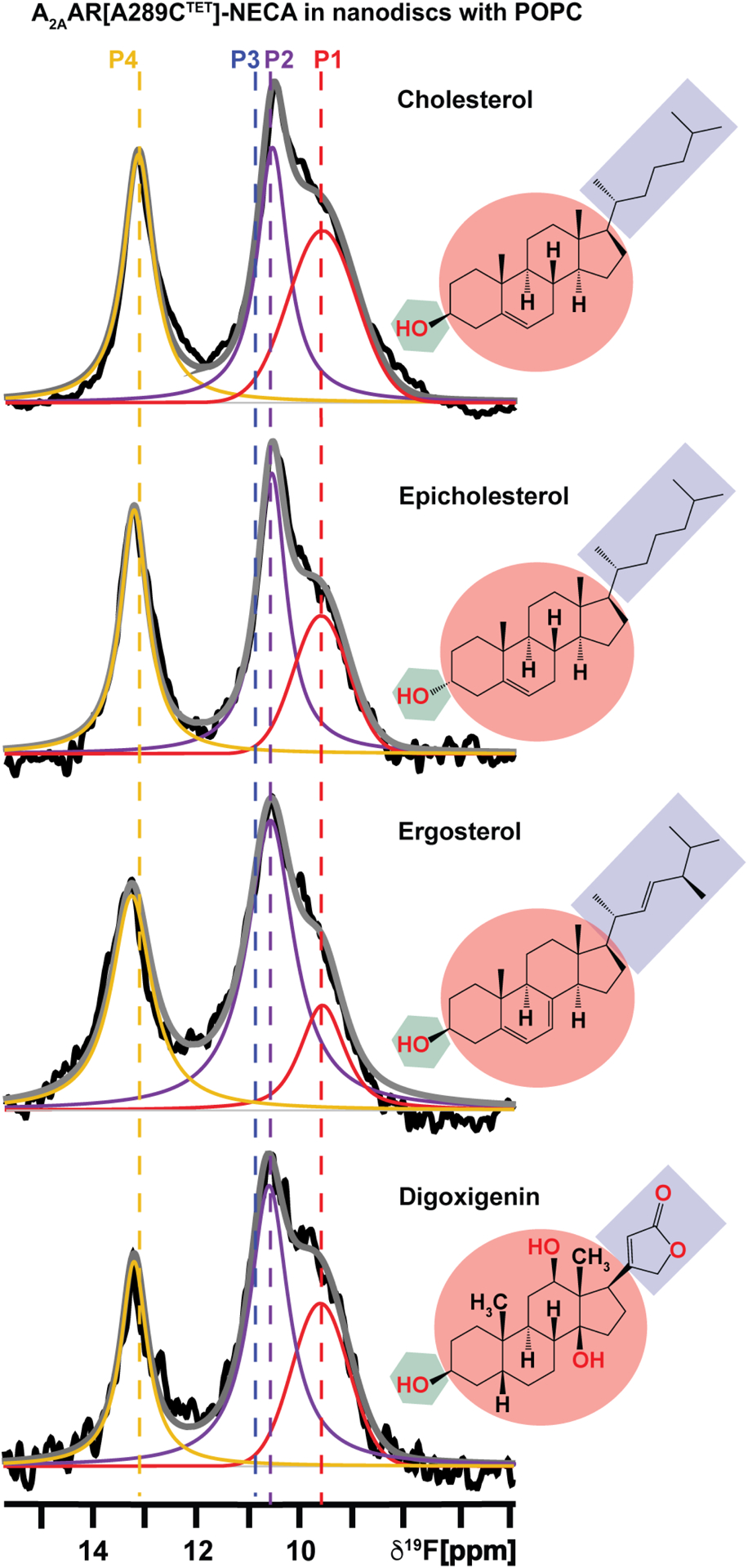
1-dimesional 19F-NMR spectra of A2AAR in complex with the agonist NECA in nanodiscs containing POPC and 0.5 mol% of different sterols, as indicated. The chemical structures of each sterol are colored according to the different chemical groups. The green hexagon highlights the flexible hydroxyl head group, the maroon circle highlights the fused tetracyclic ring, and the purple box highlights the flexible iso-octyl chain of cholesterol, and the corresponding sterol analogs. Other presentation details are the same as Figure 2. See also Table S4.
Anionic phospholipids modulate the impact of cholesterol on the A2AAR conformational equilibria
In a previous study, we showed that the presence of anionic lipids, including POPS and POPG, modified the conformational equilibrium of agonist-bound A2AAR.64 Our findings revealed that 19F-NMR spectra exhibited an inactive conformational ensemble for agonist-bound A2AAR[A289CTET] in nanodiscs containing zwitterionic lipids. Upon the introduction of anionic phospholipids, an active conformational ensemble was observed.64 It was therefore of interest to investigate how the A2AAR conformational equilibria were affected by ternary mixtures of zwitterionic phospholipids, anionic phospholipids and cholesterol. We prepared samples of A2AAR reconstituted into nanodiscs containing POPC, 5 mol% cholesterol, and either POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine) or POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol)). 19F-NMR spectra of the A2AAR[A289CTET] complex with the antagonist ZM241385 were highly similar in the presence and absence of 5 mol% cholesterol, showing slightly narrower lines in the presence of cholesterol with no significant changes in the relative populations of observed signals (Figure 4A, Figure S9, and Table S4). For agonist-bound A2AAR, in contrast to our observations with nanodiscs containing POPC and cholesterol, we observed at most subtle dependence on the presence of cholesterol in 19F-NMR spectra of A2AAR[A289CTET] in nanodiscs containing ternary mixtures of POPC and POPS and cholesterol or POPC and POPG (Figures 4B and 4C). In all cases, we observed the signature of an active ensemble with only subtle changes in the relative populations of P4, P2, and P1 (Figures 4B and 4C, Table S4). 19F-NMR spectra of A2AAR[A289CTET] in nanodiscs containing ternary mixtures of POPC and POPS exhibited a ~20% decrease in the relative population of state P2 upon addition of 5 mol% cholesterol whereas the population of state P1 remained nearly unchanged (Table S4). 19F-NMR spectra of A2AAR[A289CTET] in nanodiscs containing ternary mixtures of POPC and POPG exhibited a ~50% decrease in the population of state P2 upon addition of 5 mol% cholesterol and the population of P1 increased by ~65% (Table S4). 19F-NMR spectra of agonist-bound A2AAR[A289CTET] in nanodiscs containing a smaller amount of anionic lipids (POPC:POPG, 93:7 molar ratio) also show a diminished effect of the addition of cholesterol (Figure S10) as compared to spectra measured with cholesterol in nanodiscs containing only POPC. We observed an increase by ~20% in the population of state P4, a ~20% reduction in the population of state P2, and the relative population of state P1 remained nearly the same (Table S4).
Figure 4. The presence of anionic lipids diminishes the observed impact of cholesterol on the A2AAR conformational equilibria.
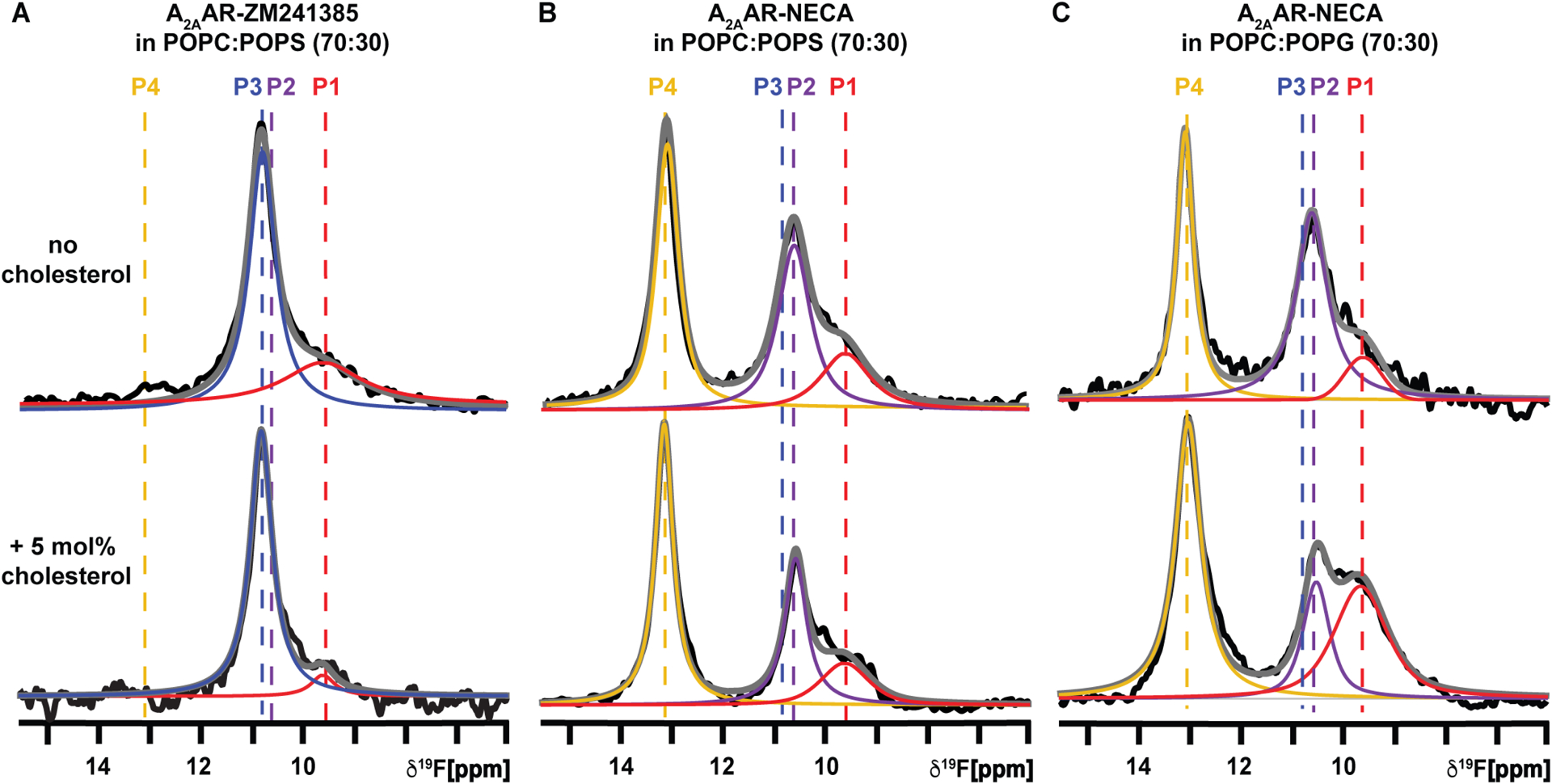
(A and B) The 1-dimensional 19F-NMR spectra of A2AAR[A289CTET] in lipid nanodiscs containing POPC and POPS (70:30 molar ratio), without cholesterol or containing 5 mol% cholesterol, for complexes with (A) the antagonist ZM241385 and (B) the agonist NECA. (C) A2AAR[A289CTET] in lipid nanodiscs containing POPC and POPG (70:30 molar ratio), without cholesterol or with 5 mol% cholesterol, in complex with the agonist NECA. Other presentation details are the same as Figure 2. See also Figures S2, S3, S4, S10, S11, Table S3 and Table S4.
We further explored the impact of cholesterol on A2AAR function in binary and ternary mixtures of phospholipids by observing the impact of cholesterol and anionic lipids on A2AAR-catalyzed nucleotide exchange using a GTP hydrolysis assay73 (Figure S11 and Table S3). This assay monitors the exchange of GTP for GDP bound to the stimulatory G protein GαS upon complex formation with A2AAR[A289C]. In lipid nanodiscs containing POPC, we observed a significant increase in GTPase activity for agonist-bound A2AAR over antagonist-bound A2AAR, as expected (Figure S11). We observed a statistically significant increase in GTPase activity for agonist-bound A2AAR in nanodiscs containing 0.5 mol% cholesterol (Figure S11). A further increase in GTPase activity was observed for A2AAR in nanodiscs containing 5 mol% cholesterol. In nanodiscs containing POPC and 10 mol% cholesterol, activity was still present but appeared to be decreased compared with nanodiscs containing 0.5 and 5 mol% cholesterol. This observation appears consistent with our NMR data showing an active-state ensemble with a lower P4 population for nanodiscs containing 10 mol% cholesterol, as discussed above (Figure 2 and Table S4), which may reflect an impact of the decrease in membrane fluidity on the A2AAR conformational ensemble. In nanodiscs containing mixtures of POPC and POPS, the addition of anionic lipids correlated with a significant increase in GTPase activity for agonist-bound A2AAR (Figure S11), consistent with our earlier observations of the impact of anionic phospholipids on populating an A2AAR active state ensemble.64 For binary phospholipid compositions, the presence of cholesterol showed a positive impact on GTPase activity; however, the impact of cholesterol was significantly muted in the presence of anionic lipids compared with the GTPase activity of A2AAR in nanodiscs containing only zwitterionic lipids, which correlated with the corresponding 19F-NMR spectra (Figure 4, Table S4).
Distinct roles for different cholesterol interaction sites influence signaling and receptor stability
Sequence analysis and structure-based computational modeling have identified several putative sites for cholesterol interactions with A2AAR, described as cholesterol recognition amino acid consensus (CRAC) sequences or cholesterol consensus motifs (CCM). One CCM site predicted for A2AAR from all-atom simulation data74 involved residues Y432.41, S472.45, K1224.44, I1254.46 and W1294.50 (superscripts indicate the Ballesteros-Weinstein nomenclature75). This location has also been proposed to be a cholesterol interaction site in other class A GPCRs, including the β2AR.76 Amino acid replacement of these residues in A2AAR resulted in reduced ligand specific binding and reduced cAMP signaling in HEK293 cells.61 Additional cholesterol interaction sites were observed in molecular dynamics simulations, including a location at the intracellular surface between TMVI and TMVII62 involving R2917.56 (Figure 5A). From our earlier results showing the involvement of R291 in facilitating A2AAR interactions with anionic lipids,64 we sought to test if this residue was also important for mediating the effects of cholesterol on the A2AAR active-state conformational ensemble.
Figure 5. Impact of cholesterol interaction sites on the conformational dynamics of A2AAR.
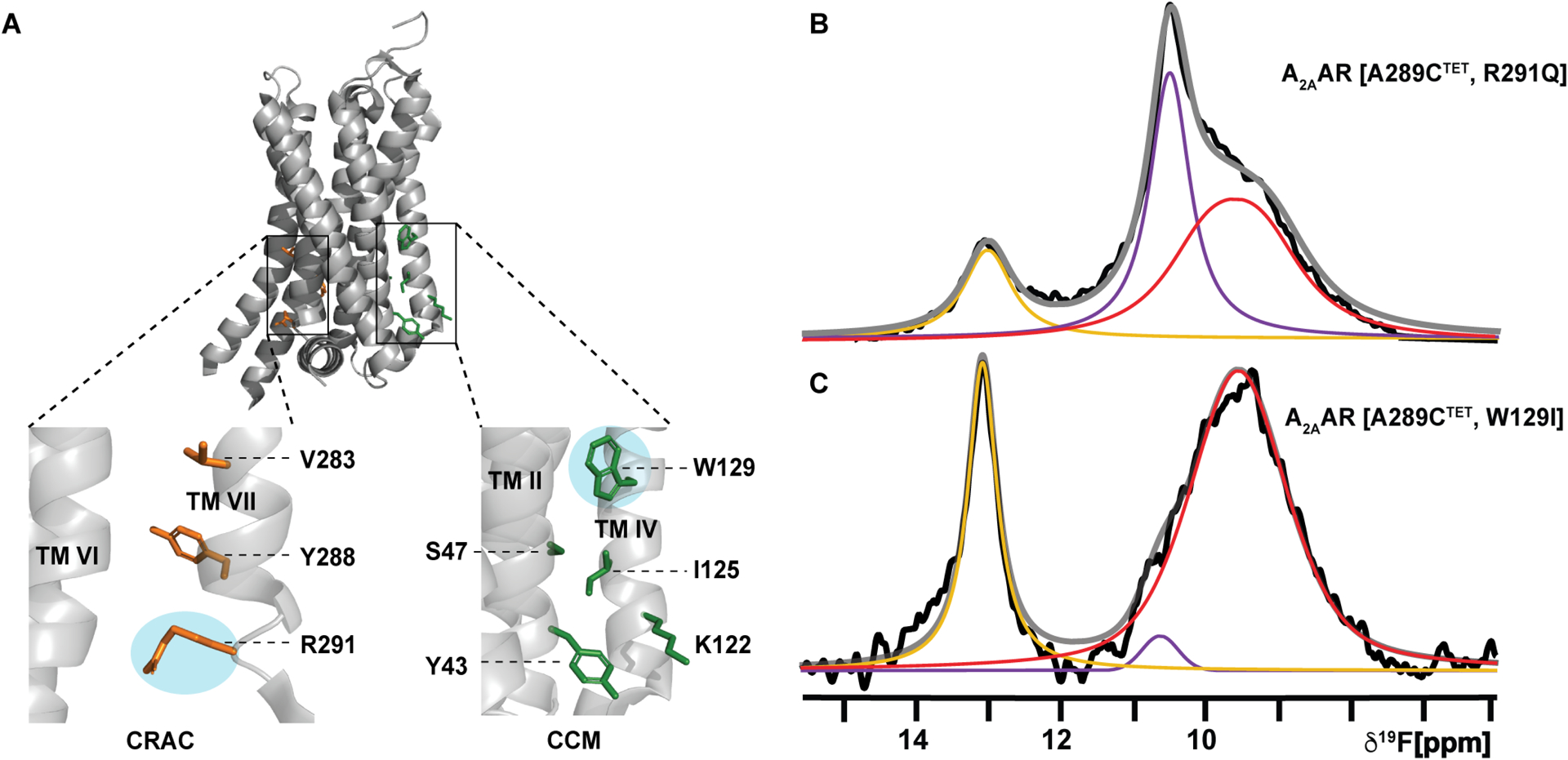
(A) Crystal structure of A2AAR (PDB: 5G53) showing the predicted CRAC site62 (orange) and CCM site74 (green). Expanded insets show the individual residues of each motif predicted to interact with cholesterol. Residues shown in cyan ellipses were mutated for 19F NMR analysis.
(B and C) 1-dimesional 19F NMR spectra of A2AAR variants in complexes with the agonist NECA and in nanodiscs containing POPC and 0.5 mol% cholesterol for (B) A2AAR[A289CTET, R291Q], and (C) A2AAR[A289CTET, W129I]. Other presentation details are the same as in Figure 2. See also Table S4.
We selected W129 and R291 for amino acid replacement to test if the variants A2AAR[A289C,R291Q] and A2AAR[A289C,W129I] showed evidence for disrupting cholesterol-A2AAR interactions. W129 was replaced with isoleucine to disrupt potential ring stacking between cholesterol and A2AAR, and R291 was replaced with the relatively shorter, uncharged amino acid glutamine to potentially alter interactions with the cholesterol 3β-hydroxyl group, which orients cholesterol to the membrane surface. 19F-NMR spectra of agonist complexes with the A2AAR variants A2AAR[A289CTET,R291Q] and A2AAR[A289CTET,W129I] were recorded using conditions we had previously identified where a clear response to cholesterol was observed for agonist-bound A2AAR in nanodiscs containing POPC and 0.5 mol% cholesterol (1–2 cholesterol molecules per nanodisc) (Figure 2). 19F-NMR spectra of agonist-bound A2AAR[A289CTET,R291Q] in nanodiscs containing POPC and 0.5 mol% cholesterol showed the spectral signature of an active-state ensemble but with a significant decrease in the population of the active-state P4 (Figure 5B). We had previously shown that pharmacological activity was preserved for this variant and 19F-NMR spectra of agonist-bound A2AAR[A289CTET,R291Q] in lipid nanodiscs containing only POPC resembled an inactive conformational ensemble.64 We had also shown that the R291Q amino acid replacement increased the basal activity of the receptor, manifesting in the 19F-NMR spectra of A2AAR[A289CTET,R291Q] as an increased population of the state P4.64 The presence of 0.5 mol% cholesterol did populate an active ensemble but with a reduced impact, which suggests that this site may contribute toward the impact of cholesterol in addition to one or more other locations. Thus, in the present study it is remarkable that the relative population of P4 is observed to be decreased for A2AAR[A289CTET,R291Q] in conditions where we observe a much larger population for P4 for A2AAR[A289CTET]. 19F-NMR spectra of agonist bound A2AAR[A289CTET,W129I] in nanodiscs containing POPC and 0.5 mol% cholesterol also showed an altered conformational equilibria with changes in the relative populations of states P1 and P2 (Figure 5C), with an increase in the population of P1 by ~60% and a decrease in the population of P2 by a factor of 5–6 (Table S4). However, we observed a significant population for state P4 similar to the observations with A2AAR[A289CTET], indicating the presence of a substantial active state population. The distinct 19F-NMR spectra of the agonist complexes with A2AAR[A289CTET,R291Q] and A2AAR[A289CTET,W129I] in the presence of 0.5 mol% cholesterol suggest potentially different roles for cholesterol interactions in the two locations.
DISCUSSION
The pharmacological data in Figure 1 show no correlation between measured binding affinities for antagonists and cholesterol and a potential positive correlation between binding affinities of agonists and cholesterol concentrations in lipid nanodiscs, albeit less than a factor of 10 over the range of cholesterol concentrations studied. Previous studies reported an increase in specific binding of the antagonist ZM241385 to A2AAR upon using MβCD to remove cholesterol from C6 glioma cells.15 As our pharmacological data were recorded for A2AAR in lipid nanodiscs, it suggests the observations in glioma cells may reflect a more complicated interplay between cholesterol and additional membrane lipids in the cellular environment that could impact A2AAR pharmacology.
In nanodiscs containing POPC, 19F-NMR spectra of agonist-bound A2AAR showed a clear response to the presence of just 1–2 molecules cholesterol per nanodisc (Figure 2). There was no observable impact of this relatively lower amount of cholesterol on the fluidity of the membranes in nanodiscs (Figure S5). We concluded the impact of cholesterol on the A2AAR conformational ensemble was due to direct interactions with the receptor. The estimation of 0.5 mol% cholesterol corresponding to 1 to 2 molecules of cholesterol, on average, was based on both experimental data presented in this study and earlier computational studies that estimated the number of lipid molecules in each nanodisc. In the 19F-NMR spectra of agonist-bound A2AAR in nanodiscs containing POPC and 0.5 mol% cholesterol, there is no observable population of state P3 (Figure 2B), whereas in the absence of any cholesterol, the signal for P3 is clearly observed as the most intense signal in the spectrum (Figure 2A). These results indicate that for samples prepared with 0.5 mol% cholesterol, nearly all nanodiscs must contain at least 1 molecule of cholesterol, and the number of nanodiscs that do not contain cholesterol is below the detection limit of the experiment. Therefore, the NMR data provide a lower limit for the number of molecules of cholesterol per nanodisc. Previous attempts to estimate the number of lipid molecules in nanodiscs relied mainly on computational approaches, including those by Denisov and coworkers77, which estimated the number of lipid molecules in nanodiscs made from the MSP1D1 scaffold protein to range between 112 and 160 in multiple simulations.77 However, these simulations were acknowledged to have limitations, including the lack of an experimental structure of MSP1D1 in its nanodisc state and challenges in accurately modeling the flexibility and deformations of the scaffold protein that are anticipated to be observed for real samples. Additionally, simulations have considered systems containing a single lipid component and did not consider the role of cholesterol or compositions of multiple lipid types.77 Based on our experimental data, we estimate that the number of lipid molecules per nanodisc is likely closer to the upper end of the previously estimated range, and may even somewhat exceed this range in the specific conditions employed in our experiments.
The NMR observations from Figure 2 aligned with our GTP hydrolysis assay results, which showed a positive correlation of nucleotide exchange for nanodiscs containing cholesterol (Figure S11). These observations also appear qualitatively consistent with A2AAR cell signaling experiments, which showed a positive correlation of cAMP production and the presence of cholesterol in membranes.56 The 19F-NMR data of Figure 2 showed a very different response of A2AAR to cholesterol than what has been reported in previous 19F-NMR studies of A2AAR with an NMR probe in helix VI in nanodiscs containing a mixture of POPC and POPG (3:2 molar ratio).60 In this earlier study, much more subtle changes were observed in the 19F-NMR spectra between samples in nanodiscs without and with cholesterol, which, combined with additional data from the same study, supported the view that the impact of cholesterol on A2AAR was more likely through modulation of membrane fluidity rather than direct interactions with A2AAR.60 The more subtle changes in the 19F-NMR spectra with and without cholesterol from this earlier NMR study do appear more in line with our observations of A2AAR in nanodiscs containing ternary mixtures of POPC, cholesterol, and either POPS or POPG, where we could not distinguish the impact of cholesterol from that of anionic lipids (Figure 4). Indeed, in the presence of just 7 mol% POPG, one could no longer distinguish potential effects of cholesterol from those of the presence of anionic lipids on the agonist-bound A2AAR conformational ensemble (Figure S10). Taken together with earlier spectroscopic and signaling studies, this suggests the mechanism of cholesterol’s influence on A2AAR depends on the phospholipid composition of the membrane system used in experiments. A corollary from this conclusion is that for in vivo experiments, a full understanding of the impact of cholesterol on receptor function may also require a more complete inventory of the phospholipid composition of cellular membranes, especially if the profile of phospholipids changes upon the addition or depletion of cholesterol.
The presented data interpreted within the context of previous studies suggest cholesterol and anionic lipids may have interchangeable roles in populating an active conformational ensemble for agonist-bound A2AAR (Figure 6). Anionic lipids and cholesterol may also compete for similar locations of interaction with A2AAR. Molecular dynamics simulations observed the displacement of cholesterol by the anionic lipid phosphatidylinositol 4,5-bisphosphate (PIP2) from the A2AAR intracellular surface between helices VI and VII involving interactions with R291.63 19F-NMR spectra of agonist-bound A2AAR[A289CTET,R291Q] in nanodiscs containing POPC and 0.5 mol% cholesterol showed a significantly diminished impact of cholesterol on the active state conformational ensemble (Figure 5). We had previously shown this residue was also involved in facilitating receptor interactions with anionic lipids.64
Figure 6. Visualization of the role of cholesterol on A2AAR function in binary and ternary lipid mixtures.
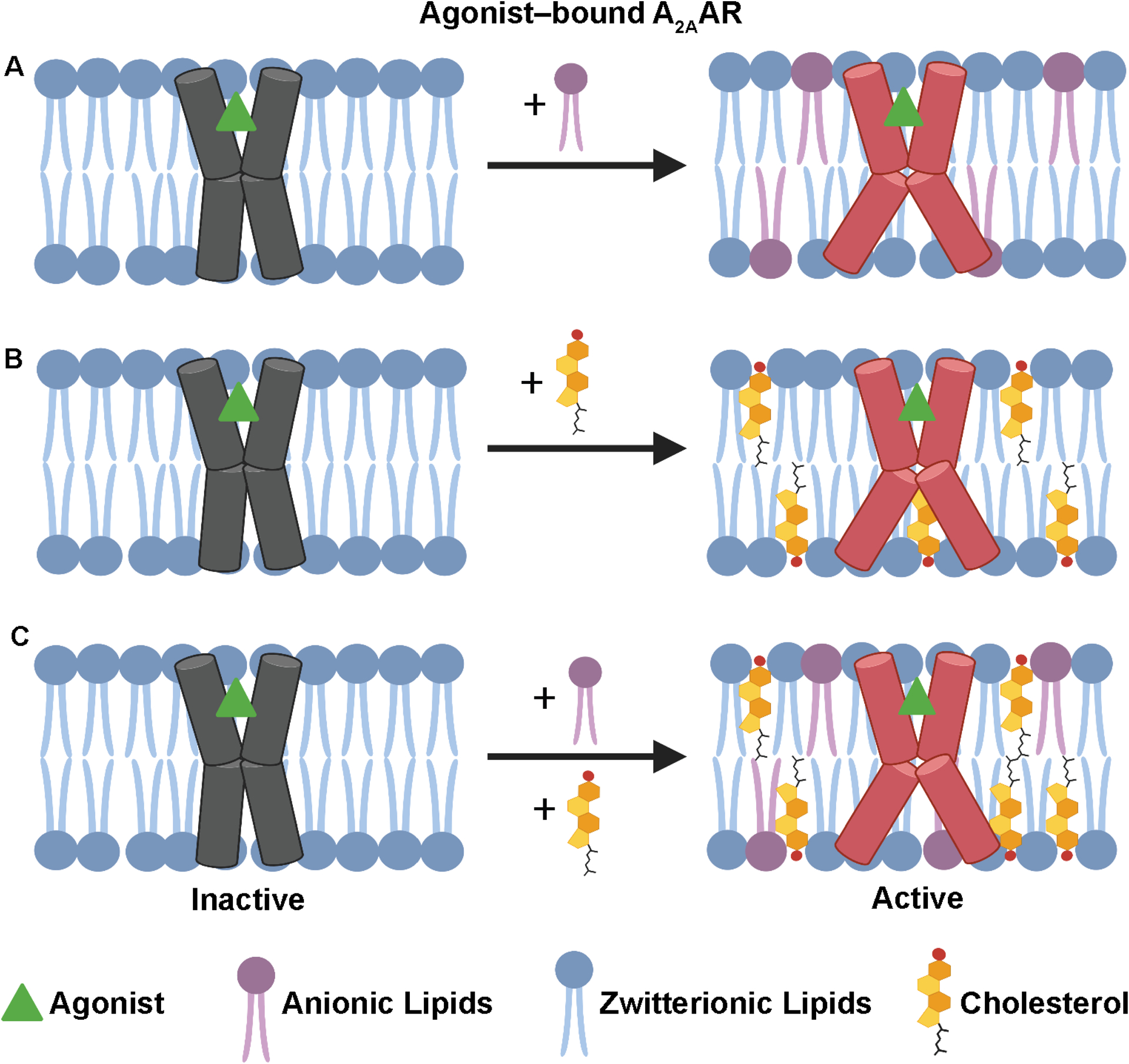
(A-C) Schematic side-views of agonist-bound A2AAR in phospholipid membranes illustrating the impact of adding (A) anionic lipids, (B) cholesterol, or (C) both anionic lipids and cholesterol to membranes containing zwitterionic lipids. This figure was prepared with Biorender.com.
Cholesterol is observed to associate with receptors at multiple locations in the majority of GPRC crystal and cryo-EM structures,14,72 and it remains an open question whether potential cholesterol interaction sites can be assigned to different roles. In A2AAR, while mutation of R291Q resulted in diminished impact of cholesterol on the active state conformational ensemble, mutation of a second predicted cholesterol interaction site, W129, resulted in changes to the relative populations of populations P1 and P2 but did not significantly perturb the active-state population P4 (Figure 5 and Table S4). Earlier in vivo studies demonstrated the W129 mutation significantly decreased agonist-stimulated cAMP signaling and the number of functional A2AAR molecules on the cell surface.61 This suggests interactions of cholesterol near position W129 may be more important for maintaining the structural integrity of the receptor rather than signaling. This interpretation is consistent with receptor engineering efforts that introduced tryptophan at the same position in other class A GPCRs when it was not present endogenously to improve receptor expression, stability, and facilitate crystallization.76,78–83 The results from Figure 5 also suggest cholesterol may play different roles at different interaction sites: cholesterol interaction at W129 may be more important to facilitate receptor stability whereas interactions near R291 may be more involved directly in modulating signaling. The approach described here provides a means for systematically evaluating the potentially different roles of cholesterol at distinct receptor locations.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Lead Contact: Dr. Matthew Eddy (matthew.eddy@chem.ufl.edu). Further information and requests for resources or reagents should be directed to and will be fulfilled by the Lead Contact.
Materials Availability
All unique reagents generated in this study are available from the Lead Contact by request and with a completed Materials Transfer Agreement.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbes
XL-10 E. coli cells were cultured in LB media, BL21(DE3) and BL21(DE3)-RIL cells were cultured in TB media, and the BG12 strain of P. pastoris was cultured in BMGY and BMMY media.
Cell lines
All cell lines used in this study were authenticated by the suppliers and were chosen to remain consistent with previous studies.
METHOD DETAILS
Construct Design
The gene encoding human A2AAR (1-316) was cloned into a pPIC9K vector (Invitrogen) at the BamHI and NotI restriction sites and is consistent with a previously characterized construct.84 64,85 The gene contained a single amino acid replacement, N154Q, to remove the only glycosylation site in the receptor, an N-terminal FLAG tag, a 10 X C-terminal His tag, and amino acid replacement in helix VII, A2897.54C, to generate A2AAR[A289C]. This plasmid was used as a template to perform PCR-based site-directed mutagenesis using the Accuprime Pfx SuperMix (ThermoFisher Scientific) to generate the A2AAR variants R2917.56Q, and W1294.50I.
A2AAR Production
Plasmids were transformed into competent cells of the Pichia pastoris BG12 strain (Biogrammatics). Selection of high-expressing clones was carried out following previously reported protocols,85 and glycerol stocks of highly expressing clones were prepared and stored for future use.
A2AAR[A289C] and A2AAR[A289C,R291Q] were expressed using the protocol described by Thakur et al..85 Glycerol stocks of highly expressing colonies were used to inoculate 4 mL cultures in buffered minimal glycerol (BMGY) media, grown at 30 °C for 48 h and used to inoculate 50 mL BMGY medium and grown at 30 °C for 60 h. Each 50 mL culture was used to inoculate 500 mL BMGY medium and grown at 30 °C for 48 h. The cells were then harvested by centrifugation and then resuspended in 500 mL of buffered minimal methanol (BMMY) media without methanol. Cultures were starved for 6 h at 28 °C to remove remaining glycerol before induction of protein expression by methanol to a final concentration of 0.5% w/v. Two further aliquots of 0.5% w/v methanol were added to the cultures at 12 h intervals after induction for a total expression time of 36 h. A2AAR[A289C,W129I] was expressed with modified conditions; specifically, expression was induced by the addition of methanol to a final concentration of 0.5% w/v at 20 °C in the presence of 1% (v/v) DMSO and 1 mM theophylline. Four further aliquots of 0.5% w/v methanol were added to the cell cultures at 12 h intervals after induction for a total expression time of 64 h. All cells were harvested by centrifugation after the end of the expression period and stored at −80 °C for purification.
A2AAR Purification and 19F-Labeling
Purification and 19F-labeling of A2AAR[A289C] and all A2AAR variants were carried out as described previously.85 86 Cells expressing A2AAR were resuspended in lysis buffer (50 mM sodium phosphate pH 7.0, 100 mM NaCl, 5% glycerol (w/v), and in-house prepared protease inhibitor solution) and lysed using a cell disruptor (Pressure Biosciences) at 40k PSI. The membranes were isolated by ultracentrifugation at 200,000 * g, homogenized in buffer (10 mM HEPES pH 7.0, 1 M NaCl, 10 mM KCl, and 20 mM MgCl2) and incubated for 1 h at 4 °C with 1 mM of 4,4’-dithiodipyridine (aldrithiol-4) and protease inhibitor cocktail solution (prepared in-house). The suspended membranes were pelleted using ultracentrifugation, homogenized in the same buffer without aldrithiol-4 and incubated with 1 mM of 2,2,2-trifluoroethaethiol (TET) for 1 h at 4°C. The suspended membranes were pelleted again using ultracentrifugation to remove excess TET. The resulting pellet was homogenized in the same buffer without TET and incubated with 1 mM theophylline and protease inhibitor solution for 30 min at 4 °C. Subsequently, protein was extracted from the membrane in a solubilization buffer (50 mM HEPES pH 7.0, 500 mM NaCl, 0.5% (w/v) n-Dodecyl-β-D-Maltopyranoside (DDM), and 0.05% cholesteryl hemisuccinate (CHS)) for 6 hours at 4 °C. Then insolubilized material was separated by ultracentrifugation at 200,000 × g for 30 min, and the supernatant was incubated overnight with Co2+-charged affinity resin (Talon) and 30 mM imidazole at 4 °C. The resin was collected and washed with 20 CV of wash buffer 1 (50 mM HEPES pH 7.0, 500 mM NaCl, 10 mM MgCl2, 30 mM imidazole, 8 mM ATP, 0.05% DDM, and 0.005% CHS), and 20 CV of wash buffer 2 (25 mM HEPES pH 7.0, 250 mM NaCl, 5% glycerol, 30 mM imidazole, 0.05% DDM, 0.005% CHS, and ligand) twice. A2AAR was eluted with an elution buffer (50 mM HEPES pH 7.0, 250 mM NaCl, 5% glycerol, 300 mM imidazole, 0.05% DDM, 0.005% CHS, and ligand) and the protein was finally exchanged into a final buffer (25 mM HEPES pH 7.0, 75 mM NaCl, 0.05% DDM, 0.005% CHS, and ligand), using a PD-10 desalting column (Cytiva) for use in all further experiments. All buffers were prepared with a saturating concentration of the required ligand.
Expression and Purification of MSP1D1
MSP1D1 was expressed and purified following previously described protocols.85 87 A single colony of BL21(DE3) cells transformed with the MSP1D1 plasmid was used to inoculate 5 mL of LB broth (supplemented with 50 μg/mL of kanamycin) and grown at 37 °C overnight. These 5 mL cultures were consequently used to inoculate 1 L of terrific broth (TB) media and allowed to grow at 37 °C till the OD600nm reached 0.6–0.8. At this point, protein expression was induced by addition of 1 mM isopropyl β-D-1thiogalactopyranoside (IPTG) and expression was continued for 4 h at 30 °C. The cells were then harvested, and cell pellets were stored at −80 °C for purification. Cell pellets were thawed and resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 1% triton X-100, and in-house protease inhibitor solution). The cells were lysed using a cell disruptor (Pressure Biosciences) at 20 kPSI and the cell lysate was spun down at 20000 * g for 45 min at 4 °C and the resulting supernatant was incubated with Ni-NTA resin (pre-equilibrated with wash buffer 1 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 1% (w/v) Triton X-100)) for 2 h at 4°C. The resin was then collected and washed with 5 CVs of wash buffer 1, 5 CVs of wash buffer 2 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 50 mM cholate), 5 CVs of wash buffer 3 (50 mM Tris-HCl, pH 8.0, and 500 mM NaCl) and finally 5 CVs of wash buffer 4 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 20 mM imidazole). The MSP1D1 was eluted using an elution buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 500 mM imidazole). The eluted MSP1D1 was dialyzed against a dialysis buffer (50 mM Tris-HCl, pH 8.0, 20 mM NaCl, and 0.5 mM EDTA) in dialysis tubing with 10 kDa MWCO. Consequently, MSP1D1 was incubated with TEV protease at a ratio of 1:100 (TEV: MSP1D1) (w/w) overnight at 4 °C. Following this, MSP1D1 was isolated by incubating the mixture with Ni-NTA resin and collecting the flow-through. The MSP1D1 was dialyzed in a storage buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 0.5 mM EDTA) for 4 h at 4 °C. The purified MSP1D1 was then concentrated to 1 mM, aliquoted and flash frozen for future use.
Nanodisc Assembly
Lipid stocks of POPC, POPC:POPS (70:30, molar ratio), and POPC:POPG (70:30, molar ratio) in chloroform were co-dried with varying amounts of cholesterol to make a lipid film and then vacuum dried for 16 h. The dried lipid film was resuspended in cholate buffer (25 mM Tris-HCl, pH 8.0, 200 mM sodium cholate and 150 mM NaCl) to a final phospholipid concentration of 100 mM.
Nanodiscs samples containing A2AAR were assembled using an earlier described protocol.85 27 μM of purified A2AAR was mixed with purified MSP1D1 and detergent-solubilized lipids in a molar ratio of 1:5:250, respectively. After incubation for 2 h at 4 °C, the mixture was incubated overnight with pre-washed bio-beads at 4 °C. Consequently, the bio-beads were removed, and the resulting mixture was incubated with Ni-NTA resin for 24 h at 4 °C. After incubation, the Ni-NTA resin was washed with 2 CV of a wash buffer (50 mM HEPES, pH 7.0, 150 mM NaCl, and 10 mM imidazole), and A2AAR-containing nanodiscs were eluted with an elution buffer (50 mM HEPES, pH 7.0, 150 mM NaCl, 300 mM imidazole and ligand). Using a PD-10 desalting column, all nanodisc samples were exchanged into a final buffer (25 mM HEPES pH 7.0, 75 mM NaCl, 100 μM TFA and ligand) to use for further experiments. All ligand-containing buffers were prepared with a saturating concentration of ligand. All nanodisc samples were characterized using analytical size exclusion chromatography on an Agilent HPLC equipped with a Sepax Nanofilm SEC-200 column operating at a flow rate of 0.5 mL/min. The observed elution time of the nanodisc samples corresponded to particles with an estimated size of approximately 160 KDa, which is consistent with previously reported studies.88
Radioligand Binding Assays
Competition binding assays with nanodiscs containing A2AAR were recorded as previously described.64,89 ZM241385 and NECA binding affinities (KD or KI) were determined using competition binding experiments. Increasing concentrations of cold ligands (ZM241385 and NECA) were incubated with 0.0625 μg of A2AAR in nanodiscs in buffer (25 mM HEPES, pH 7.0, and 75 mM NaCl) with 0.5 nM of [3H]ZM241385 for 60 min at 25 °C. Following termination of binding reactions by filtration through Microbeta filtermat-96 cell harvester, radioactivity was measured with a Microbeta2 microplate scintillation counter.
NMR Experiments
NMR samples containing A2AAR in nanodiscs were concentrated to ~200 μM in 280 μL using a Vivaspin-6 concentrator with a 30,000 MWCO. 20 μL of 99.8% D2O was added85 and gently mixed with the sample. 19F NMR and 31P NMR experiments were recorded on a Bruker Avance III HD spectrometer operating at 600 MHz 1H nutation frequency using TopSpin 3.6.2 and equipped with a Bruker 5-mm BBFO probe. 19F-NMR spectra were measured at 280 K for direct comparison to earlier results.64,84 31P-NMR experiments were measured at 300 K. Temperatures were calibrated from a standard sample of 4% methanol in D4-MeOH.
The 1-dimensional 19F data were recorded with a data size of 32k complex points, an acquisition period of 360 ms, 16k scans, 120 μs dwell time, and 0.3 s recycle delay for a total experimental time of about 3 hours per experiment. Each final data set averaged six to eight experiments of 16k scans each for total acquisition times between ~18 to ~24 hours. All 31P NMR experiments were acquired with an acquisition time of 900 ms, 2k scans, and 0.3 s recycle delay for a total experiment time of 42 min per experiment.
Quantification of Cholesterol in Nanodisc Samples
The amount of cholesterol in nanodiscs was quantified using the Amplex® Red Cholesterol Assay Kit (Invitrogen) following the manufacturer’s protocol. Nanodiscs containing cholesterol were first incubated with 0.2% (v/v) of Triton X-100 for 10 min and then mixed in a 1:1 (v/v) ratio with 1X reaction buffer (300 μM Amplex® Red reagent, 2 U/mL horseradish peroxidase (HRP), 2 U/mL cholesterol oxidase, 0.2 U/mL cholesterol esterase) and incubated in the dark for 30 min at 37 °C. After incubation, fluorescence was detected in a CLARIOstar (BMG Labtech) plate-reader with an excitation range of 530 – 560 nm and emission at 590 nm. For each measurement, the background fluorescence was subtracted, and the resulting fluorescence readout was used to quantify the amount of cholesterol in each nanodisc sample (Table S1). All values reported are mean ± s.e.m for n≥3 independent experiments.
Fluorescence Thermal Shift Assays
Fluorescent thermal shift experiments were carried out following a protocol adapted from earlier publications.90 91 10 μg of nanodisc samples was incubated with 10 μM of N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM; Invitrogen) in buffer (50 mM HEPES pH 7.0, 150 mM NaCl) for 30 min on ice in the dark. Fluorescent thermal shift was measured using a Cary Eclipse spectrofluorometer over a temperature range of 20 °C to 90 °C with a fixed heating rate of 2 °C /min. The excitation and emission wavelength for the CPM dye is 387 nm and 463 nm respectively.
GαS Expression and Purification
Human GαS containing an N-terminal 6X polyhistidine tag and an N-terminal TEV cleavage site was expressed and purified following a protocol adapted from earlier publications.92 Glycerol stocks of BL21(DE3)-RIL cells transformed with the GαS plasmid was were grown in 4 mL LB media (supplemented with 0.2% glucose, 34 μg/mL chloramphenicol and 100 μg/mL carbenicillin) at 37 °C for 8 h. This culture was used to inoculate 75 mL of the same media and continued to grow at 30 °C overnight. After ~16 hours, the cells were harvested and used to inoculate 1 L of M9 media and grown at 30 °C. Protein expression was induced with 50 μM of IPTG when the OD600nm reached 0.8, and expression continued for 16 h at 25 °C. The cells were harvested by centrifugation and stored for future use.
Cells containing GαS were resuspended in lysis buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM MgCl2, 5 μM GDP, in-house protease inhibitor cocktail85) and lysed using a cell disruptor at 22 kPSI. The cell lysate was centrifuged at 35,000 × g for 35 min at 4 °C. The lysate was then incubated with Ni-NTA resin overnight at 4 °C. After incubation, the resin was washed with 20 CVs of wash buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM MgCl2, 5 mM imidazole, 5 μM GDP, and 2 mg/mL iodoacetamide) and the protein was eluted using an elution buffer (25 mM Tris-HCl pH 8.0, 250 mM NaCl, 1 mM MgCl2, 250 mM imidazole, 10% (v/v) glycerol, and 5 μM GDP). The eluted protein was exchanged into buffer (25 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM MgCl2, 10% (v/v) glycerol, 5 μM GDP) and incubated with TEV protease at a ratio of 1:50 (TEV: GαS) (w/w) overnight at 4 °C. Following this, GαS was isolated by incubating the mixture with Ni-NTA resin and collecting the flow-through. GαS was further purified by gel-filtration with buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% (v/v) glycerol, and 5 μM GDP), concentrated to 50 μM and either immediately used or frozen and stored for future use.
Membrane Fluidity Measurements
Nanodiscs containing POPC and various amounts of cholesterol were prepared and mixed with Laurdan (Tocris Bioscience) at a molar ratio of 500:1 (total lipids:Laurdan), and the mixture was incubated in the dark at 25 °C for 1 hour. Following this, excess Laurdan was removed via buffer exchange using a PD MiniTrap G-25 column (Cytiva) equilibrated with buffer (25 mM HEPES pH 7.0 and 75 mM NaCl). Fluorescence emission curves for Laurdan embedded in nanodiscs were measured in a Cary Eclipse spectrofluorometer, using an excitation wavelength of 366 nm and observing emission between 400 nm and 600 nm.
GTP Hydrolysis Assay
GTP turnover for A2AAR in nanodiscs was measured using the GTPase-Glo™ assay (Promega) following manufacturer’s protocols. Purified nanodiscs containing A2AAR were mixed with the Gαs protein in a molar ratio of 4:1 in the presence of 5 μM GTP in buffer (25 mM HEPES pH 7.0, 75 mM NaCl) and incubated for 1 hour. Subsequently, GTP turnover was measured using a luminescence readout on a CLARIOstar (BMG Labtech) plate-reader.
QUANTIFICATION AND STATISTICAL ANALYSIS
Radioligand Binding Data Analysis
All radioligand competition experiments were conducted with 3 or more replicates. IC50 values were determined using a nonlinear, least-square regression analysis in Prism 8 (GraphPad Software, Inc.). The KI values were calculated from the IC50 using the Cheng−Prusoff equation.93 Error bars for each sample were calculated as the standard error of mean (s.e.m) for 3 independent experiments done in triplicate.
Fluorescence Thermal Shift Data Analysis
The raw data were fit according to previously reported procedures90 and analyzed using Origin (OriginLab Corporation). The raw data was fit to a Boltzmann sigmoidal curve to determine the melting temperature (Tm). The error bars are reported as standard error of mean (s.e.m) for 3 or more independent experiments.
Membrane Fluidity Data Analysis
The emission spectra were normalized to the corresponding emission maxima. Following the protocol from Yu et al.,71 the generalized polarization (GP) of Laurdan was calculated using the formula:
where I440 and I490 are the fluorescence emission intensities at 440 nm and 490 nm, respectively. All GP values reported are the mean ± s.e.m. for 3 independent measurements.
GTP Hydrolysis Assay Analysis
The percentage of GTP hydrolyzed was calculated by subtracting background luminescence. All values reported in Table 3 are the mean ± s.e.m. for 3 or more independent experiments. To determine the statistical significance of reported differences in GTP hydrolysis, statistical analysis was conducted using a two-tailed unpaired t-test. P values less than 0.05 were considered to be statistically significant. Statistically significant values are illustrated as *P<0.05, **P<0.005, and ***P<0.0005, respectively.
NMR Data Analysis
All NMR data were processed and analyzed in Topspin 4.0.8 (Bruker Biospin). Prior to Fourier transformation, all 1-dimensional 19F-NMR data were zero-filled to 64k points and multiplied by an exponential window function with 40 Hz line broadening. All 19F spectra were referenced to a TFA signal at −75.8 ppm, which was set to 0 ppm. Deconvolution of the overlapping 19F-NMR data were done in accordance to previously published procedures64,84 with MestreNova version 14.1.1–24571 (MestreLab Research S.L). The 19F-NMR spectra were fit to double- or triple-Lorentzian functions, and the quality of the fits was assessed from the residual difference of the experimental data and the sum of the computed components. The relative populations of A2AAR conformational states were calculated as a ratio of the integrated area of each deconvoluted peak with respect to the total integral of all signals from 7.5 ppm to 14.5 ppm.
All 31P NMR data were processed identically. Prior to Fourier transformation, 31P spectra were zero-filled to 64k points and multiplied by an exponential window function with 50 Hz line broadening.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG® M2-Alkaline Phosphatase antibody produced in mouse | Sigma-Aldrich | Cat#A9469 |
| Bacterial and virus strains | ||
| XL10-Gold ultracompetent cells | Agilent | Cat#200314 |
| BL21 (DE3) cells | Agilent | Cat#200132 |
| BL21(DE3)-RIL cells | Agilent | Cat#230245 |
| Chemicals, peptides, and recombinant proteins | ||
| n-Dodecyl-b-D-Maltopyranoside (DDM) | Anatrace | Cat#D310 |
| Cholesteryl hemisuccinate (CHS) | Sigma-Aldrich | Cat#C6512 |
| Adenosine 5’-triphosphate disodium salt hydrate (ATP) | Sigma-Aldrich | Cat#34369078 |
| 4-(2-[7-Amino-2-(2-furyl)[1,2,4]triazolo[2,3-a][1,3,5]triazin-5-ylamino]ethyl)phenol (ZM241385) | Tocris | Cat#1036 |
| 5’-N-Ethylcarboxamidoadenosine (NECA) | Tocris | Cat#1691 |
| Theophylline | Sigma-Aldrich | Cat#T1633 |
| [3H]4-[2-[7-amino-2-(2-furyl)-1,2,4-triazolo[1,5-a][1,3,5]triazin-5-yl-amino]ethylphenol ([3H]ZM241385) | American Radiolabeled Chemicals | Cat#ART0884-50 |
| TALON Metal Affinity Resin | Gold bio | Cat#H-310-5 |
| Ni-NTA Metal Affinity Resin | Sigma-Aldrich | Product#70666 |
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti | SKU#850457 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (POPS) | Avanti | SKU#840034 |
| 1 -palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’- rac-glycerol) (sodium salt) (POPG) | Avanti | SKU#840457 |
| Cholic Acid, Sodium Salt | Anatrace | Cat#S1010S |
| Isopropyl β- d-1-thiogalactopyranoside (IPTG) | Goldbio | Cat#I2481C |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| SnakeSkin™ Dialysis Tubing, 10K MWCO, 22 mm | ThermoFischer Scientific | Cat#68100 |
| Bio-beads SM-2 Adsorbents | BioRad | Cat#1523920 |
| N-(2-Hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid), 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | Sigma-Aldrich | Cas#7365-45-9 |
| TRIS Hydrochloride (Tris-HCl) | Sigma-Aldrich | Cas#1185-53-1 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cas#60-00-4 |
| Cholesterol | Sigma-Aldrich | Cas#57-88-5 |
| Ergosterol | Cayman Chemicals | Cas#57-87-4 |
| Epi-cholesteryl methyl ether | Sigma-Aldrich | Cas#2867-93-8 |
| Digoxigenin | Sigma-Aldrich | Cas#1672-46-4 |
| Biotin | Sigma-Aldrich | Cas#58-85-5 |
| Imidazole | Sigma-Aldrich | Cas#288-32-4 |
| Glycerol | Sigma-Aldrich | Cas#56-81-5 |
| Sodium phosphate monobasic anhydrous | Sigma-Aldrich | Cas#7558-80-7 |
| Sodium phosphate dibasic anhydrous | Sigma-Aldrich | Cas#7558-79-4 |
| Magnesium Chloride | Sigma-Aldrich | Cas#7791-18-6 |
| Methanol | Sigma-Aldrich | Cas#67-56-1 |
| DMSO | Fisher Scientific | Cas#67-68-5 |
| Sodium Chloride | Sigma-Aldrich | Cas#7647-14-5 |
| Potassium Chloride | Sigma-Aldrich | Cas#7447-40-7 |
| Aldrithiol-4 | Sigma-Aldrich | Cas#2645-22-9 |
| 2,2,2-Trifluoroethanethiol (TET) | Sigma-Aldrich | Cas#1544-53-2 |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich | Cas#76051 |
| N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) | Invitrogen | Cat#D346 |
| Guanosine-5’-diphosphate (disodium salt) (GDP) | ThermoFisher Scientific | Cas#7415-69-2 |
| 2-Iodoacetamide | TCI Chemicals | Cas#144-48-9 |
| Laurdan | Tocris | Cas#74515-25-6 |
| Deposited data | ||
| A2AAR in complex with ZM241385 | Jaakola et al., 200846 | PDB: 3EML |
| A2AAR in complex with NECA bound to an engineered G protein | Carpenter et al., 201644 | PDB:5G53 |
| Experimental models: Organisms/strains | ||
| P. pastoris: Bg12 | BioGrammatics | Cat#PS004-01 |
| Oligonucleotides | ||
| A2AAR[A289C,W129I] Forward: [5’AAGGGCATCATTGCCATCTGCATTGTGCTGTCGIIIGCCATCGGCCTGAC3’] | This study | n/a |
| A2AAR[A289C,W129I] Reverse: [5’GTCAGGCCGATGGCAAACGACAGCACAATGCAGATGGCAATGATGCCCTT3’] | This study | n/a |
| Recombinant DNA | ||
| Plasmid: human A2AAR (1-316) in pPIC9K | Eddy et al., 201850 | n/a |
| Plasmid: human A2AAR[A289C] | Thakur et al., 202364 | n/a |
| Plasmid: human A2AAR[A289C,R291Q] | Thakur et al., 202364 | n/a |
| Software and algorithms | ||
| GraphPad prism | GraphPad Software Inc. | https://www.graphpad.com |
| MestreNova 14.1.1-24571 | MestreLab Research S.L | https://mestrelab.com/download/mnova/ |
| Topspin 4.0.8 | Bruker Biospin | https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html |
| UCSF Chimera | UCSF | https://www.cgLucsf.edu/chimera/ |
| Other | ||
| PD-10 column | Cytiva | Cat#17085101 |
| Vivaspin® 6 ultrafiltration spin columns MWCO 30 kDa | Cytiva | Cat#28932361 |
| Vivaspin® 6 ultrafiltration spin columns MWCO 10 kDa | Sigma-Aldrich | SKU#GE28932296 |
| AccuPrime Pfx SuperMix | ThermoFischer Scientific | Cat#12344040 |
| SnakeSkin™ Dialysis Tubing, 10K MWCO, 22 mm | ThermoFischer Scientific | Cat#68100 |
| Amplex Red Cholesterol Assay Kit | ThermoFischer Scientific | Cat#A12216 |
| GTPase-Glo Assay | Promega | Cat#V7681 |
Highlights.
Specific cholesterol-A2AAR interactions activate receptor in POPC lipid bilayers
Influence of sterols on agonist-bound A2AAR observed with cholesterol analogs
Cholesterol and anionic lipids share complementary roles in A2AAR activation
Different cholesterol interaction sites distinctly impact structure and function
ACKNOWLEDGEMENTS
This work is supported by the National Institutes of Health, NIGMS MIRA grant R35GM138291 (M.T.E., N.T., A.R. and N.G.P). A portion of this work was supported by the McKnight Brain Institute at the National High Magnetic Field Laboratory’s AMRIS Facility, which is funded by National Science Foundation Cooperative Agreement No. DMR-1644779 and the State of Florida
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References:
- 1.Yeagle PL (2014). Non-covalent binding of membrane lipids to membrane proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes 1838, 1548–1559. 10.1016/J.BBAMEM.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Yeagle PL (1991). Modulation of membrane function by cholesterol. Biochimie 73, 1303–1310. 10.1016/0300-9084(91)90093-G. [DOI] [PubMed] [Google Scholar]
- 3.Elkins MR, Williams JK, Gelenter MD, Dai P, Kwon B, Sergeyev IV, Pentelute BL, and Hong M (2017). Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci 114, 12946–12951. 10.1073/pnas.1715127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fantini J, and Barrantes FJ (2013). How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol 4. 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epand RM (2006). Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res 45, 279–294. 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Grouleff J, Irudayam SJ, Skeby KK, and Schiøtt B (2015). The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochimica et Biophysica Acta (BBA) - Biomembranes 1848, 1783–1795. 10.1016/J.BBAMEM.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Carruthers A, and Melchior DL (1986). How bilayer lipids affect membrane protein activity. Trends Biochem. Sci 11, 331–335. 10.1016/0968-0004(86)90292-6. [DOI] [Google Scholar]
- 8.Song Y, Kenworthy AK, and Sanders CR (2014). Cholesterol as a co-solvent and a ligand for membrane proteins: Cholesterol-Protein Interactions. Protein Sci. 23, 1–22. 10.1002/pro.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastiaanse E (1997). The effect of membrane cholesterol content on ion transport processes in plasma membranes. Cardiovasc. Res 33, 272–283. 10.1016/S0008-6363(96)00193-9. [DOI] [PubMed] [Google Scholar]
- 10.Di Paolo G, and Kim T-W (2011). Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci 12, 284–296. 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein Joseph L., and Brown Michael S. (2015). A Century of Cholesterol and Coronaries: From Plaques to Genes to Statins. Cell 161, 161–172. 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perego C, Da Dalt L, Pirillo A, Galli A, Catapano AL, and Norata GD (2019). Cholesterol metabolism, pancreatic β-cell function and diabetes. Biochim. Biophys. Acta Mol. Basis Dis 1865, 2149–2156. 10.1016/j.bbadis.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, and Gloriam DE (2017). Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Disc 16, 829–842. 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan AL, Song W, and Sansom MSP (2020). Lipid-Dependent Regulation of Ion Channels and G Protein–Coupled Receptors: Insights from Structures and Simulations. Annual Review of Pharmacology and Toxicology 60, 31–50. 10.1146/ANNUREV-PHARMTOX-010919-023411. [DOI] [PubMed] [Google Scholar]
- 15.Guixà-González R, Albasanz JL, Rodriguez-Espigares I, Pastor M, Sanz F, Martí-Solano M, Manna M, Martinez-Seara H, Hildebrand PW, Martín M, and Selent J (2017). Membrane cholesterol access into a G-protein-coupled receptor. Nat. Comm 8, 1–12. 10.1038/ncomms14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedger G, Koldsø H, Chavent M, Siebold C, Rohatgi R, and Sansom MSP (2019). Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened. Structure 27, 549–559.e542. 10.1016/j.str.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, Latorraca NR, Faust B, Dror RO, Beachy PA, et al. (2019). Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284–288. 10.1038/s41586-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena R, and Chattopadhyay A (2012). Membrane cholesterol stabilizes the human serotonin1A receptor. Biochimica et Biophysica Acta (BBA) - Biomembranes 1818, 2936–2942. 10.1016/J.BBAMEM.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Lu M, Zhao W, Han S, Lin X, Xu T, Tan Q, Wang M, Yi C, Chu X, Yang W, et al. (2022). Activation of the human chemokine receptor CX3CR1 regulated by cholesterol. Science Advances 8, 8048–8048. 10.1126/SCIADV.ABN8048/SUPPL_FILE/SCIADV.ABN8048_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimpl G, Klein U, Reilaender H, and Fahrenholz F (1995). Expression of the Human Oxytocin Receptor in Baculovirus-Infected Insect Cells: High-Affinity Binding Is Induced by a Cholesterol-Cyclodextrin Complex. Biochemistry 34, 13794–13801. 10.1021/bi00042a010. [DOI] [PubMed] [Google Scholar]
- 21.Babcock GJ, Farzan M, and Sodroski J (2003). Ligand-independent Dimerization of CXCR4, a Principal HIV-1 Coreceptor. J. Biol. Chem 278, 3378–3385. 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- 22.Chattopadhyay A, Jafurulla M, Kalipatnapu S, Pucadyil TJ, and Harikumar KG (2005). Role of cholesterol in ligand binding and G-protein coupling of serotonin1A receptors solubilized from bovine hippocampus. Biochem. Biophys. Res. Commun 327, 1036–1041. 10.1016/j.bbrc.2004.12.102. [DOI] [PubMed] [Google Scholar]
- 23.Ruthirakuhan M, Herrmann N, Andreazza AC, Verhoeff NPLG, Gallagher D, Black SE, Kiss A, and Lanctôt KL (2019). 24S-Hydroxycholesterol Is Associated with Agitation Severity in Patients with Moderate-to-Severe Alzheimer’s Disease: Analyses from a Clinical Trial with Nabilone. J. Alzheimers Dis 71, 21–31. 10.3233/JAD-190202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calmet P, Cullin C, Cortès S, Vang M, Caudy N, Baccouch R, Dessolin J, Maamar NT, Lecomte S, Tillier B, and Alves ID (2020). Cholesterol impacts chemokine CCR5 receptor ligand-binding activity. FEBS J. 287, 2367–2385. 10.1111/febs.15145. [DOI] [PubMed] [Google Scholar]
- 25.Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, and Rohatgi R (2016). Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife 5, e20304. 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluhackova K, Gahbauer S, Kranz F, Wassenaar TA, and Böckmann RA (2016). Dynamic Cholesterol-Conditioned Dimerization of the G Protein Coupled Chemokine Receptor Type 4. PLoS Comput. Biol. 12, e1005169. 10.1371/journal.pcbi.1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sengupta D, Kumar GA, and Chattopadhyay A (2017). Interaction of Membrane Cholesterol with GPCRs: Implications in Receptor Oligomerization. In G-Protein-Coupled Receptor Dimers, Herrick-Davis K, Milligan G, and Di Giovanni G, eds. (Springer International Publishing; ), pp. 415–429. [Google Scholar]
- 28.Prasanna X, Sengupta D, and Chattopadhyay A (2016). Cholesterol-dependent Conformational Plasticity in GPCR Dimers. Sci. Rep 6, 31858. 10.1038/srep31858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty H, and Chattopadhyay A (2015). Excitements and Challenges in GPCR Oligomerization: Molecular Insight from FRET. ACS Chem. Neurosci 6, 199–206. 10.1021/cn500231d. [DOI] [PubMed] [Google Scholar]
- 30.Kumar GA, Sarkar P, Stepniewski TM, Jafurulla M, Singh SP, Selent J, and Chattopadhyay A (2021). A molecular sensor for cholesterol in the human serotonin 1A receptor. Sci. Adv 7, eabh2922. 10.1126/sciadv.abh2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar GA, and Chattopadhyay A (2020). Statin-Induced Chronic Cholesterol Depletion Switches GPCR Endocytosis and Trafficking: Insights from the Serotonin 1A Receptor. ACS Chem. Neurosci 11, 453–465. 10.1021/acschemneuro.9b00659. [DOI] [PubMed] [Google Scholar]
- 32.Maza N, Wang D, Kowalski C, Stoveken HM, Dao M, Sial OK, Giles AC, Grill B, and Martemyanov KA (2022). Ptchd1 mediates opioid tolerance via cholesterol-dependent effects on μ-opioid receptor trafficking. Nat. Neurosci 25, 1179–1190. 10.1038/s41593-022-01135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunselman JM, Lott J, and Puthenveedu MA (2021). Mechanisms of selective G protein–coupled receptor localization and trafficking. Curr. Opin. Cell Biol 71, 158–165. 10.1016/j.ceb.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeliseev A, Iyer MR, Joseph TT, Coffey NJ, Cinar R, Zoubak L, Kunos G, and Gawrisch K (2021). Cholesterol as a modulator of cannabinoid receptor CB2 signaling. Sci. Rep 11, 3706. 10.1038/s41598-021-83245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyerowitz JG, Robertson MJ, Barros-Álvarez X, Panova O, Nwokonko RM, Gao Y, and Skiniotis G (2022). The oxytocin signaling complex reveals a molecular switch for cation dependence. Nat. Struct. Mol. Biol 29, 274–281. 10.1038/s41594-022-00728-4. [DOI] [PubMed] [Google Scholar]
- 36.Oates J, and Watts A (2011). Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol 21, 802–807. 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar P, and Chattopadhyay A (2020). Cholesterol interaction motifs in G protein-coupled receptors: Slippery hot spots? WIREs Syst. Biol. Med 12. 10.1002/wsbm.1481. [DOI] [PubMed] [Google Scholar]
- 38.Gater Deborah L., Saurel O, Iordanov I, Liu W, Cherezov V, and Milon A (2014). Two Classes of Cholesterol Binding Sites for the β 2 AR Revealed by Thermostability and NMR. Biophys. J 107, 2305–2312. 10.1016/j.bpj.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abiko LA, Dias Teixeira R, Engilberge S, Grahl A, Mühlethaler T, Sharpe T, and Grzesiek S (2022). Filling of a water-free void explains the allosteric regulation of the β1-adrenergic receptor by cholesterol. Nat. Chem 14, 1133–1141. 10.1038/s41557-022-01009-9. [DOI] [PubMed] [Google Scholar]
- 40.Albert AD, Young JE, and Yeagle PL (1996). Rhodopsin-cholesterol interactions in bovine rod outer segment disk membranes. Biochim. Biophys. Acta Biomembr 1285, 47–55. 10.1016/S0005-2736(96)00145-9. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell DC, Straume M, Miller JL, and Litman BJ (1990). Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry 29, 9143–9149. 10.1021/bi00491a007. [DOI] [PubMed] [Google Scholar]
- 42.Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AGW, and Tate CG (2011). Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 2011 474:7352 474, 521–525. 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García-Nafría J, Lee Y, Bai X, Carpenter B, and Tate CG (2018). Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7. 10.7554/eLife.35946.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter B, Nehmé R, Warne T, Leslie AGW, and Tate CG (2016). Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536, 104–107. 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, et al. (2012). Structural Basis for Allosteric Regulation of GPCRs by Sodium Ions. Science 337, 232–236. 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, and Stevens RC (2008). The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217. 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang SK, Pandey A, Tran DP, Villanueva NL, Kitao A, Sunahara RK, Sljoka A, and Prosser RS (2021). Delineating the conformational landscape of the adenosine A2A receptor during G protein coupling. Cell 184, 1884–1894. 10.1016/j.cell.2021.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye L, Van Eps N, Zimmer M, Ernst OP, and Scott Prosser R (2016). Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268. 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 49.Eddy MT, Martin BT, and Wüthrich K (2021). A2A adenosine receptor partial agonism related to structural rearrangements in an activation microswitch. Structure 29, 170–176. 10.1016/j.str.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddy MT, Lee M-Y, Gao Z-G, White KL, Didenko T, Horst R, Audet M, Stanczak P, McClary KM, Han GW, et al. (2018). Allosteric coupling of drug binding and intracellular signaling in the A2A adenosine receptor. Cell 172, 68–80. 10.1016/j.cell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sušac L, Eddy MT, Didenko T, Stevens RC, and Wüthrich K (2018). A2A adenosine receptor functional states characterized by 19F-NMR. Proc. Natl. Acad. Sci 115, 12733–12738. 10.1073/pnas.1813649115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eddy MT, Gao Z-G, Mannes P, Patel N, Jacobson KA, Katritch V, Stevens RC, and Wüthrich K (2018). Extrinsic tryptophans as NMR probes of allosteric coupling in membrane proteins: application to the A2A adenosine receptor. J. Am. Chem. Soc 140, 8228–8235. 10.1021/jacs.8b03805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizumura T, Kondo K, Kurita M, Kofuku Y, Natsume M, Imai S, Shiraishi Y, Ueda T, and Shimada I (2020). Activation of adenosine A2A receptor by lipids from docosahexaenoic acid revealed by NMR. Science Advances 6, eaay8544. 10.1126/sciadv.aay8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bara-Jimenez W, Sherzai A, Dimitrova T, Favit A, Bibbiani F, Gillespie M, Morris MJ, Mouradian MM, and Chase TN (2003). Adenosine A2A receptor antagonist treatment of Parkinson’s disease. Neurology 61, 293–296. 10.1212/01.WNL.0000073136.00548.D4. [DOI] [PubMed] [Google Scholar]
- 55.Young A, Ngiow SF, Barkauskas DS, Sult E, Hay C, Blake SJ, Huang Q, Liu J, Takeda K, Teng MWL, et al. (2016). Co-inhibition of CD73 and A2AR Adenosine Signaling Improves Anti-tumor Immune Responses. Cancer Cell 30, 391–403. 10.1016/J.CCELL.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 56.McGraw C, Yang L, Levental I, Lyman E, and Robinson AS (2019). Membrane cholesterol depletion reduces downstream signaling activity of the adenosine A2A receptor. Biochim. Biophys. Acta Biomembr 1861, 760–767. 10.1016/j.bbamem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouviere E, Arnarez C, Yang L, and Lyman E (2017). Identification of Two New Cholesterol Interaction Sites on the A2A Adenosine Receptor. Biophys. J 113, 2415–2424. 10.1016/j.bpj.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song W, Yen HY, Robinson CV, and Sansom MSP (2019). State-dependent Lipid Interactions with the A2a Receptor Revealed by MD Simulations Using In Vivo-Mimetic Membranes. Structure 27, 392–403.e393. 10.1016/j.str.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lovera S, Cuzzolin A, Kelm S, De Fabritiis G, and Sands ZA (2019). Reconstruction of apo A2A receptor activation pathways reveal ligand-competent intermediates and state-dependent cholesterol hotspots. Sci. Rep 9, 14199. 10.1038/s41598-019-50752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang SK, Almurad O, Pejana RJ, Morrison ZA, Pandey A, Picard L-P, Nitz M, Sljoka A, and Prosser RS (2022). Allosteric modulation of the adenosine A2A receptor by cholesterol. eLife 11, e73901. 10.7554/eLife.73901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGraw C, Koretz KS, Oseid D, Lyman E, and Robinson AS (2022). Cholesterol Dependent Activity of the Adenosine A2A Receptor Is Modulated via the Cholesterol Consensus Motif. Molecules 27, 3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taghon GJ, Rowe JB, Kapolka NJ, and Isom DG (2021). Predictable cholesterol binding sites in GPCRs lack consensus motifs. Structure 29, 499–506.e493. 10.1016/j.str.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song W, Yen H-Y, Robinson CV, and Sansom MSP (2019). State-dependent lipid interactions with the A2a receptor revealed by MD simulations using in vivo-mimetic membranes. Structure 27, 392–403. 10.1016/j.str.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thakur N, Ray AP, Sharp L, Jin B, Duong A, Pour NG, Obeng S, Wijesekara AV, Gao Z-G, McCurdy CR, et al. (2023). Anionic phospholipids control mechanisms of GPCR-G protein recognition. Nat. Comm 14, 794. 10.1038/s41467-023-36425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Aalst E, and Wylie BJ (2021). Cholesterol Is a Dose-Dependent Positive Allosteric Modulator of CCR3 Ligand Affinity and G Protein Coupling. Front. Mol. Biosci 8. 10.3389/fmolb.2021.724603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheidelaar S, Koorengevel Martijn C., Pardo Juan D., Meeldijk Johannes D., Breukink E, and Killian JA (2015). Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophys. J 108, 279–290. 10.1016/j.bpj.2014.11.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harayama T, and Riezman H (2018). Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol 19, 281–296. 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 68.Amundson DM, and Zhou M (1999). Fluorometric method for the enzymatic determination of cholesterol. Journal of Biochemical and Biophysical Methods 38, 43–52. 10.1016/S0165-022X(98)00036-0. [DOI] [PubMed] [Google Scholar]
- 69.Oliveira FFM, Paes HC, Peconick LDF, Fonseca FL, Marina CLF, Bocca AL, Homem-de-Mello M, Rodrigues ML, Albuquerque P, Nicola AM, et al. (2020). Erg6 affects membrane composition and virulence of the human fungal pathogen Cryptococcus neoformans. Fungal Genet. Biol 140, 103368. 10.1016/j.fgb.2020.103368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sušac L, O’Connor C, Stevens RC, and Wüthrich K (2015). In-membrane chemical modification (IMCM) for site-specific chromophore labeling of GPCRs. Angew. Chem 127, 15461–15464. 10.1002/ange.201508506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu W, So PT, French T, and Gratton E (1996). Fluorescence generalized polarization of cell membranes: a two-photon scanning microscopy approach. Biophys. J 70, 626–636. 10.1016/S0006-3495(96)79646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakubík J, and El-Fakahany EE (2021). Allosteric Modulation of GPCRs of Class A by Cholesterol. Int. J. Mol. Sci 22, 1953–1953. 10.3390/IJMS22041953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mondal S, Hsiao K, and Goueli SA (2015). A Homogenous Bioluminescent System for Measuring GTPase, GTPase Activating Protein, and Guanine Nucleotide Exchange Factor Activities. ASSAY and Drug Development Technologies 13, 444–455. 10.1089/adt.2015.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JY, Patel R, and Lyman E (2013). Ligand-dependent cholesterol interactions with the human A2A adenosine receptor. Chemistry and Physics of Lipids 169, 39–45. 10.1016/j.chemphyslip.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ballesteros JA, and Weinstein H (1995). Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In Methods in neurosciences, (Elsevier; ), pp. 366–428. [Google Scholar]
- 76.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, Velasquez J, Kuhn P, and Stevens RC (2008). A Specific Cholesterol Binding Site Is Established by the 2.8 Å Structure of the Human β2-Adrenergic Receptor. Structure 16, 897–905. 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denisov IG, and Sligar SG (2017). Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 117, 4669–4713. 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roth CB, Hanson MA, and Stevens RC (2008). Stabilization of the Human β2-Adrenergic Receptor TM4–TM3–TM5 Helix Interface by Mutagenesis of Glu1223.41, A Critical Residue in GPCR Structure. J. Mol. Biol 376, 1305–1319. 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, and Stevens RC (2007). High-Resolution Crystal Structure of an Engineered Human β2-Adrenergic G Protein–Coupled Receptor. Science 318, 1258–1265. 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johansson LC, Stauch B, McCorvy JD, Han GW, Patel N, Huang X-P, Batyuk A, Gati C, Slocum ST, Li C, et al. (2019). XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 569, 289–292. 10.1038/s41586-019-1144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stauch B, Johansson LC, McCorvy JD, Patel N, Han GW, Huang X-P, Gati C, Batyuk A, Slocum ST, Ishchenko A, et al. (2019). Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288. 10.1038/s41586-019-1141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wacker D, Wang C, Katritch V, Han GW, Huang X-P, Vardy E, McCorvy JD, Jiang Y, Chu M, Siu FY, et al. (2013). Structural Features for Functional Selectivity at Serotonin Receptors. Science 340, 615–619. 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, and Stevens RC (2010). Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science 330, 1091–1095. 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sušac L, Eddy MT, Didenko T, Stevens RC, and Wüthrich K (2018). A2A adenosine receptor functional states characterized by 19F-NMR. Proceedings of the National Academy of Sciences of the United States of America 115, 12733–12738. 10.1073/pnas.1813649115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thakur N, Wei S, Ray AP, Lamichhane R, and Eddy MT (2022). Production of human A2AAR in lipid nanodiscs for 19F-NMR and single-molecule fluorescence spectroscopy. STAR Protocols 3, 101535–101535. 10.1016/J.XPRO.2022.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sušac L, O’Connor C, Stevens RC, and Wüthrich K (2015). In-Membrane Chemical Modification (IMCM) for Site-Specific Chromophore Labeling of GPCRs. Angew Chem Int Ed Engl 54, 15246–15249. 10.1002/anie.201508506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hagn F, Nasr ML, and Wagner G (2018). Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat Protoc 13, 79–98. 10.1038/nprot.2017.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thakur N, Wei S, Ray AP, Lamichhane R, and Eddy MT (2022). Production of human A2AAR in lipid nanodiscs for 19F-NMR and single-molecule fluorescence spectroscopy. STAR Protocols 3, 101535. 10.1016/j.xpro.2022.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei S, Thakur N, Ray AP, Jin B, Obeng S, McCurdy CR, McMahon LR, Gutiérrez-de-Terán H, Eddy MT, and Lamichhane R (2022). Slow conformational dynamics of the human A2A adenosine receptor are temporally ordered. Structure 30, 329–337.e325. 10.1016/J.STR.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, and Stevens RC (2008). Microscale Fluorescent Thermal Stability Assay for Membrane Proteins. Structure 16, 351–359. 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 91.White KL, Eddy MT, Gao ZG, Han GW, Lian T, Deary A, Patel N, Jacobson KA, Katritch V, and Stevens RC (2018). Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure 26, 259–269.e255. 10.1016/j.str.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu Q, and Shokat KM (2018). Disease-causing mutations in the G protein Gαs subvert the roles of GDP and GTP. Cell 173, 1254–1264. e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng Y, and Prusoff WH (1973). Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


