Abstract
We report a refractory and relapsed visceral leishmaniasis case in a child male patient followed from 2016 to 2020, whose clinical isolates from multiple relapses were analyzed at genome level. To the best of our knowledge, it is the first report that both visceral leishmaniasis and non-ulcerated cutaneous leishmaniasis have concomitantly manifested in the same patient. Importantly, sequence analysis revealed that the patient was co-infected with Leishmania infantum and a Crithidia-related parasite, which was previously found in a fatal case of visceral leishmaniasis from the same endemic region.
Keywords: Leishmania infantum, Crithidia sp, trypanosomatid co-infection, visceral leishmaniasis (VL), non-ulcerated cutaneous leishmaniasis (NUCL), whole-genome sequencing analysis
Graphical Abstract
In the Americas, Brazil is the most affected country by Visceral Leishmaniasis (VL), the most serious form of Leishmaniases, which can be lethal if left untreated or misdiagnosed [1]. Leishmania infantum is the causative species, however amplicon-based next-generation sequencing has revealed that VL patients can be co-infected with multiple Leishmania species and even with Trypanosoma [2]. Since 1980’s there have been occasional reports of monoxenous-trypanosomatid infection in humans, which have typically been associated with immunosuppression. Recently, there has been an increase in reported cases of such infections, both in immunocompromised and some immunocompetent individuals, involving co-infection of Leishmania with supposedly monoxenous trypanosomatids [3]. In Brazil, in 2019, Crithidia-related parasites were reported for the first time in humans, with specimens being isolated from a fatal case of visceral leishmaniasis in Sergipe (SE) [4]. This discovery thereby provides evidence to indicate that these trypanosomatid species may be associated with severe VL. Here, we describe isolation of the same Crithidia-related species from a further human patient with severe VL in the same area. The patient presented with multiple relapses, clinical complications, and a rare cutaneous manifestation concomitant with the visceral disease. In this article, we present a case study of a male child who suffered from refractory and relapsed VL from 2016 to 2020, with an emphasis on the genome sequencing-based analysis of clinical isolates.
In 2016, a 9-year-old male from Aracaju, SE, was admitted to the University Hospital of the Federal University of Sergipe (HUUFS-SE) with severe symptoms indicative of VL. Figure 1A depicts a timeline of the key events in this case (details of which are presented in the Supplemental Material). By 2020, the patient had been hospitalized five times with VL, during which time he undergone treatment and met the criteria for hospital discharge. During his third and fourth admissions, parasite strains were isolated from his bone marrow (LVH117) and spleen (LVH120), respectively, and during the fourth period of hospitalization, he underwent splenectomy to resolve the hypersplenism (Supplementary Figure S1) and removal of a probable parasite reservoir. In 2020, in addition to the VL symptoms, he developed non-ulcerative papular skin lesions (Figure S2). LVH161 and LVH161a parasite strains were isolated from his bone marrow and skin biopsies, respectively. Four clinical isolates obtained during his several relapses were cultivated to obtain cultured isolates, and his tissue samples were cryopreserved. These cultures and samples were subsequently used for nucleic acid isolation, and all available samples were sequenced for detection of the small subunit ribosomal ribonucleic acid (SSU rRNA) taxonomic marker. Phylogenetic analyses indicated that both polyclonal and clonal cultures of parasites isolated in 2017 (LVH117 and LVH120) were more closely related to monoxenous Leishmaniinae members, such as Crithidia, than to the L. donovani complex. These parasites were similar to the clinical isolates (LVH60/LVH60a; hereafter referred to as Crithidia-like parasites) obtained from a fatal case involving a patient from the same geographical region [4] (Figure S3). However, SSU sequences detected in samples from patient bone marrow collected in 2017 (BMVL), LVH161 (bone marrow, 2020), and LVH161a (skin, 2020) isolate cultures closely clustered with L. infantum.
Figure 1.
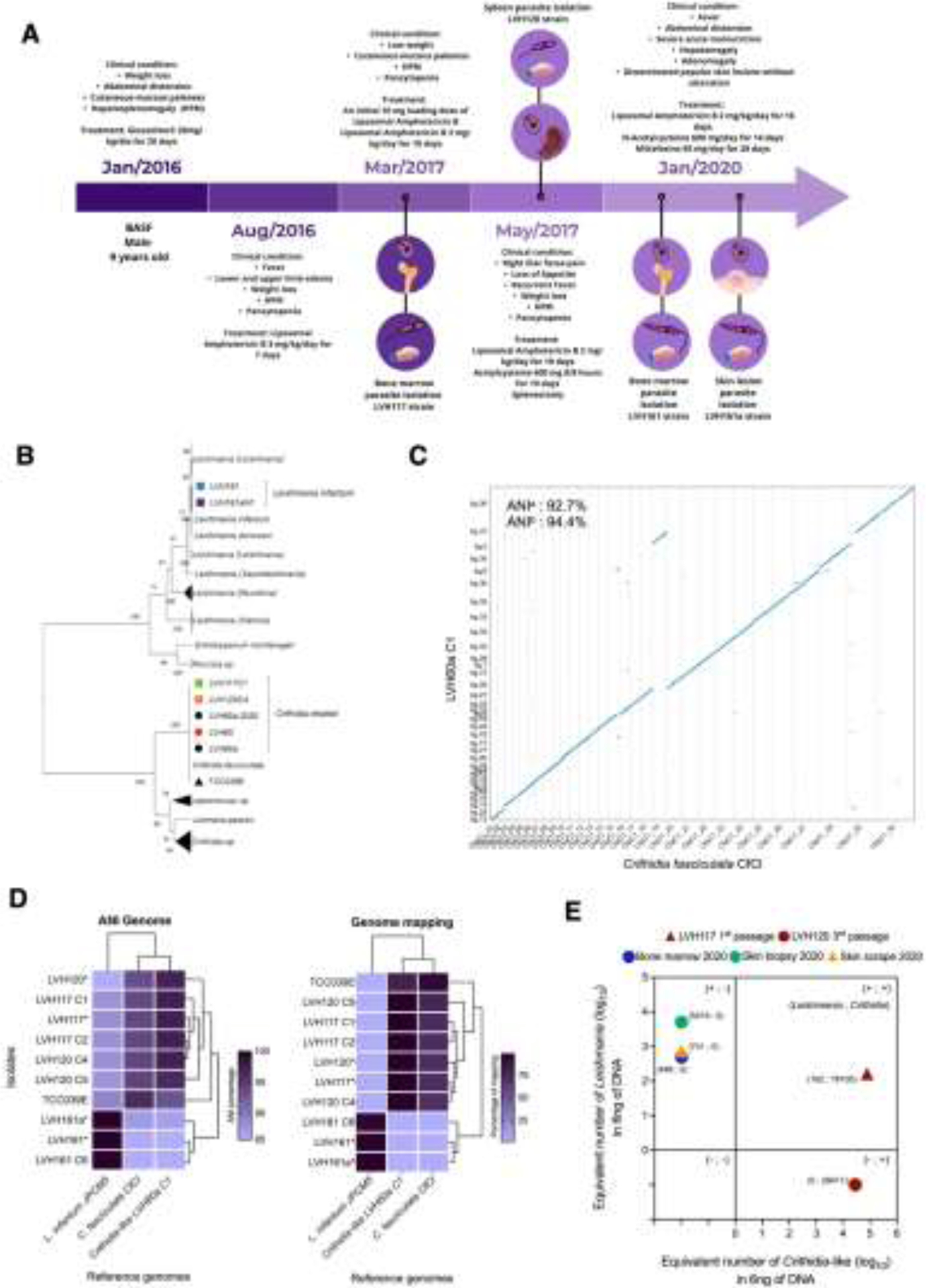
VL case report analysis. (A) Timeline of the patient’s admissions depicting parasite isolations. (B) Phylogenomic analysis of Leishmaniinae parasites. Clinical isolates are highlighted with coloured icons. (C) Nucmer dot plot displaying the whole-genome alignment of chromosome between LVH60a-C1 Crithidia-like and C. fasciculata-CfCl. ANI: average nucleotide identity. (D) Clustered heatmaps showing the percentage of ANI and read mapping values for the genomes of clinical isolates against reference genomes. (E) qPCR-based detection of parasites in parasite cultures and patient tissue samples.
Genomic DNAs obtained from cultured isolates, their clones, and C. fasciculata (TCC039E strain) were sequenced using an Illumina platform, and phylogenomic analysis was performed using the TOMM method [5]. The findings using this phylogenomic approach were found to be consistent with those obtained based on SSU marker analysis, with LVH117-C1 (clone 1, from bone marrow/2017) and LVH120-C4 (clone 4, from spleen/2017) cultured isolates being clustered with LVH60, LVH60a, and C. fasciculata, whereas the LVH161 and LVH161a isolates were placed in the same cluster with L. infantum and L. donovani (Figure 1B).
To enhance the previously obtained genome assembly of Crithidia-like species [4], Oxford Nanopore technology (ONT) was used to perform whole-genome sequencing of the LVH60a-C1 clone. Genome assembly of the LVH60a-C1 parasite strain yielded 38 contigs with a predicted genome size of ~34.4 Mb. This assembly was used as a reference genome for the Crithidia-like species, enabling further genome analysis. Whole-genome pairwise alignment between LVH60a-C1 and C. fasciculata revealed high linearity, with an average nucleotide identity (ANI) of ~93.5%, as calculated using MUMmer [6] (Figure 1C).
Although there is no threshold ANI value on which to base classification of trypanosomatids at the species levels, the observed values of ANI within cultured isolates of the L. donovani complex are over 99% [7]. Compared with the reference genomes, dermotropic and viscerotropic species of Leishmania have ANI values of ~94% (Table S8), whereas Crithidia-like isolates have ANI values ranging between 85.8% and 97.3%. ANI values within isolates of the same species are >96.43%, whereas that between Crithidia and Leishmania was ~87% (Figure 1D; Tables S8 and S9). These findings accordingly provide evidence to indicate the potential existence of a novel species within the genus Crithidia, although further population genomics analysis is needed to determine interbreeding and separate species.
Short read mapping to reference genomes confirmed these findings, revealing that LVH117, LVH120, and their respective clones have a percentage alignment >98% compared with that of the Crithidia-like LVH60a-C1 reference genome, which was reduced to 73.7% when mapped to the C. fasciculata reference genome (Figure 1D; Table S10). LVH161 and LVH161a-N1 isolates showed mapping percentages >95% with L. infantum-JPCM5, thereby confirming that isolates obtained during the final relapse period are those of Leishmania infantum, which is endemic to Brazil.
To detect and quantify L. infantum and Crithidia-like parasites in cultured isolates and patient samples, we used an in-house quantitative PCR (qPCR) assay using species-specific primers (Supplemental Material). Using this assay, L. infantum was detected in bone marrow and skin biopsy samples collected during the fifth relapse (2020). Although no tissue samples were collected during the first and second hospital admissions (2016), the earliest passage cultures from the third and fourth relapses (2017) tested positive for Crithidia-like species. The LVH117 clinical isolate cultured from the first passage (Figure 1E) contained both parasites. On the basis of these findings and those obtained from SSU-rRNA marker analysis, we concluded that the patient was co-infected in 2017 with Crithidia-like and L. infantum parasites. In the LVH120 isolate obtained from the spleen (culture at the third passage), only Crithidia-like parasites were detected. Nevertheless, given that Crithidia-like parasites might potentially outgrow the more fastidious Leishmania, we should not overlook the possibility of co-infection of the spleen with both parasites.
Leishmania infantum is occasionally associated with papular, nodular, or ulcerated lesions [8]. Although non-ulcerated cutaneous leishmaniasis is observed in patients after VL treatment (cured patients) or without previous VL symptoms, the case reported herein is considered unique, in that the patient experienced both VL and non-ulcerated cutaneous leishmaniasis during his fifth remission. The patient was also co-infected with a Crithidia-related parasite, which appears to be an isolate of the same species involved in a fatal case of VL [4]. This raises concerns regarding the emergence of a novel cryptic parasite species within Crithidia genus. Although the transmission routes or possible vectors for Crithidia-like parasites are unknown, previous studies have detected Crithidia sp. in sandflies, which are known vectors of a range of pathogens [9], and Crithidia and Leishmania have been co-detected in non-phlebotomine vectors [10].
Supplementary Material
Highlights.
Severe Visceral Leishmaniasis (VL) with non-ulcerated cutaneous leishmaniasis.
A Crithidia-related parasite was isolated from bone marrow aspirate and spleen.
Whole-genome sequencing of clinical isolates suggested a novel parasite species.
Clinical implications of this trypanosomatid co-infection remain unknown in VL.
Acknowledgements
We thank Lucile Floeter-Winter and the Trypanosomatid Culture Collection of the USP for kindly providing the C. fasciculata TCC039E, Felipe Teixeira for his generous and continuing support and Sally James and the Genomics Laboratory at the University of York for their assistance with Oxford Nanopore sequencing workflow. This work utilized the Viking cluster at the University of York.
Funding
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2016/20258–0 and 2017/16328–6) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 309776/2018–0) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, code 001). Scholarships from FAPESP were awarded to LAR (2020/14011–8), NTT (2021/12464–8), and TYT (2019/03095–9, 2019/12142–0).
Footnotes
Disclosure statement
The authors declare that they have no potential conflicts of interest.
Ethical approval statement
Sample collection was performed in accordance with the Brazilian Human Research Ethics Evaluation System (CEP/CONEP) with approval from the local ethics committee (Federal University of Sergipe, protocol CAAE 04587312.2.0000.0058).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The whole-genome sequencing data and genome assembly of Crithidia-like LVH60a-C1 parasite have been deposited in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA940846 (Reviewer link for peer review: https://dataview.ncbi.nlm.nih.gov/object/PRJNA940846?reviewer=1afbteko9ahl5bmih3lq9tb tkd). The SSU rRNA amplicons have been deposited in GenBank/NCBI under accession numbers OQ581228-OQ581240.
References
- [1].Serafim TD, Iniguez E, Oliveira F. Leishmania infantum. Trends Parasitol 2020; 36: 80–81. doi: 10.1016/j.pt.2019.10.006. [DOI] [PubMed] [Google Scholar]
- [2].Castillo-Castañeda A, Patiño LH, Muñoz M et al. Amplicon-based next-generation sequencing reveals the co-existence of multiple Leishmania species in patients with visceral leishmaniasis. Int J Infect Dis 2022; 115: 35–38. doi: 10.1016/j.ijid.2021.11.029. [DOI] [PubMed] [Google Scholar]
- [3].Boucinha C, Andrade-Neto VV, Ennes-Vidal V et al. A Stroll Through the History of Monoxenous Trypanosomatids Infection in Vertebrate Hosts. Front Cell Infect Microbiol 2022; 12. doi: 10.3389/fcimb.2022.804707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Maruyama SR, Santana AKM de, Takamiya NT et al. Non-Leishmania Parasite in Fatal Visceral Leishmaniasis–Like Disease, Brazil. Emerg Infect Dis 2019; 25: 2088–2092. doi: 10.3201/eid2511.181548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maruyama SR, Rogerio LA, Freitas PD, Teixeira MMG, Ribeiro JMC. Total Ortholog Median Matrix as an alternative unsupervised approach for phylogenomics based on evolutionary distance between protein coding genes. Sci Rep 2021; 11: 3791. doi: 10.1038/s41598-021-81926-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLOS Comput Biol 2018; 14: e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fernández-Arévalo A, El Baidouri F, Ravel C et al. The Leishmania donovani species complex: A new insight into taxonomy☆. Int J Parasitol 2020; 50: 1079–1088. doi: 10.1016/j.ijpara.2020.06.013. [DOI] [PubMed] [Google Scholar]
- [8].Sandoval C, Araujo G, Sosa W et al. In situ cellular immune response in non-ulcerated skin lesions due to Leishmania (L.) infantum chagasi infection. J Venom Anim Toxins Incl Trop Dis 2021; 27: e20200149. doi: 10.1590/1678-9199-JVATITD-2020-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang J, Gou Q, Luo G, Hou X, Liang G, Shi M. Total RNA sequencing of Phlebotomus chinensis sandflies in China revealed viral, bacterial, and eukaryotic microbes potentially pathogenic to humans. Emerg Microbes Infect 2022; 11: 2080–2092. doi: 10.1080/22221751.2022.2109516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Songumpai N, Promrangsee C, Noopetch P, Siriyasatien P, Preativatanyou K. First Evidence of Co-Circulation of Emerging Leishmania martiniquensis, Leishmania orientalis, and Crithidia sp. in Culicoides Biting Midges (Diptera: Ceratopogonidae), the Putative Vectors for Autochthonous Transmission in Southern Thailand. Trop Med Infect Dis 2022; 7: 379. doi: 10.3390/tropicalmed7110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequencing data and genome assembly of Crithidia-like LVH60a-C1 parasite have been deposited in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA940846 (Reviewer link for peer review: https://dataview.ncbi.nlm.nih.gov/object/PRJNA940846?reviewer=1afbteko9ahl5bmih3lq9tb tkd). The SSU rRNA amplicons have been deposited in GenBank/NCBI under accession numbers OQ581228-OQ581240.


