Abstract
Many bacteria use protein-based organelles known as bacterial microcompartments (BMCs) to organize and sequester sequential enzymatic reactions. Regardless of their specialized metabolic function, all BMCs are delimited by a shell made of multiple structurally redundant, yet functionally diverse, hexameric (BMC-H), pseudohexameric/trimeric (BMC-T), or pentameric (BMC-P) shell protein paralogs. When expressed without their native cargo, shell proteins have been shown to self-assemble into 2D sheets, open-ended nanotubes and closed shells of ~40 nm diameter that are being developed as scaffolds and nanocontainers for applications in biotechnology. Here by leveraging a strategy for affinity-based purification we demonstrate that a wide range of empty synthetic shells, many differing in end cap structures, can be derived from a glycyl radical enzyme-associated microcompartment (GRM). The range of pleomorphic shells observed, which span ~2 orders of magnitude in size from ~25 nm to ~1.8 μm, reveals the remarkable plasticity of BMC-based biomaterials. In addition, we observe new capped nanotube and nanocone morphologies that are consistent with a multicomponent geometric model in which architectural principles are shared among asymmetric carbon, viral protein, and BMC-based structures.
Keywords: bacterial microcompartments, self-assembly, nanotubes, nanocones, fullerenes, synthetic biology
Graphical Abstract

Self-assembling shell proteins derived from bacterial microcompartments are a promising new biomaterial with applications in drug delivery, metabolic engineering, cell-free synthesis, and bioelectronics. A synthetic platform produces pleomorphic shells spanning the nano- to microscale that are rapidly purified. Geometric models of new morphologies suggest shared design principles with asymmetric carbon and viral protein analogs.
1. Introduction
Bacterial microcompartments (BMCs) are self-assembling subcellular organelles consisting of a core of catabolic (metabolosomes) or anabolic (carboxysomes) enzymes surrounded by a shell composed of hexameric (BMC-H), trimeric (BMC-T), and pentameric (BMC-P) proteins 1. When expressed in the absence of their native enzymatic cargo, BMC shell proteins and their engineered derivatives self-assemble into an array of higher-order structures, including 2D hexagonal sheets 2,3, small prolates 4,5, open-ended nanotubes 2,6-8, dodecahedra 9, icosahedra with diameters up to 40nm 4,10-14, and irregular polyhedra 8,11,15. The building blocks of these genetically tractable BMC-based biomaterials are structurally redundant, forming either hexagons (BMC-H and BMC-T) 16,17 or pentagons (BMC-P) 18. Nonetheless, shell protein paralogs are functionally diverse, with surfaces, pores, and interfaces that reflect the permeability, encapsulation, and assembly of their respective native organelles.
The propensity for synthetic BMC architectures to self-assemble in the absence of their native cargos in vivo and in vitro 19, as well as their plasticity for accommodating structural modifications 20-23, make them attractive targets for synthetic biologists and bioengineers aiming to confine and coordinate chemistries with precise spatial control 24. However, only a few examples of synthetic metabolosome shell systems have been characterized to date, with isolation procedures typically involving differential centrifugation. These include shells derived from B12-dependent 1,2-propanediol utilizing (PDU) 8 or ethanolamine utilizing (EUT) BMCs 15, a BMC of unknown function from Haliangium ochraceum (HO) 14, and, more recently, a choline-metabolizing glycyl radical enzyme-associated microcompartment (GRM2) from Klebsiella pneumoniae 11. This limited complement of characterized systems stands in stark contrast to our growing appreciation of the natural abundance and diversity of BMCs, with 68 subtypes now identified in ~20% of bacterial genomes spanning 45 phyla 25. However, this dichotomy between experimentally characterized and bioinformatically-identified BMCs represents an opportunity to expand our portfolio of building blocks, as well as advance our understanding of the molecular mechanisms governing their self-assembly to inform the design of sustainable bio-based architectures for drug delivery, cell-free synthesis, metabolic engineering, and bioelectronics.
In this work, we set out to expand the repertoire of characterized synthetic metabolosome BMC shell chassis by applying the complementation-based affinity purification (CAP) method 20 to rapidly isolate heterologously assembled shells derived from the B12-independent 1,2-propanediol metabolizing GRM3C organelle from the purple non-sulfur bacterium Rhodopseudomonas palustris BisB18 25,26. The building block set of this shell system is composed of four distinct types of hexamers, a trimer, and a pentamer. The synthetic GRM3C architectures produced from a self-assembling mixture of these 6 types of proteins are morphologically diverse, some evoking parallels to single walled carbon nanotubes and mature HIV-1 capsids. These structures span the nano- to microscale, and serve as an exemplar for the remarkable structural plasticity of BMC-based biomaterials. Moreover, we propose that pleomorphic capped nanotubes and nanocones that we observed reflect an underlying geometric organization of hexagon and pentagon shell proteins analogous to the 6- and 5-member carbon rings of fullerenes, in which the asymmetrical distribution of the pentamers influences morphology. Our results suggest the existence of shared architectural principles among asymmetric carbon, viral protein, and BMC-based architectures.
2. Results and Discussion
2.1. Operon design
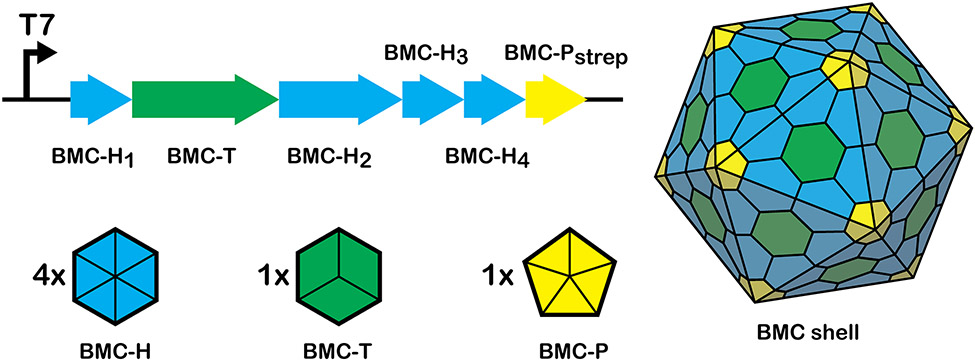
The native GRM3C locus in R. palustris BisB18 encodes a total of six shell proteins: four hexameric BMC-H paralogs (pfam00936), one pseudo-hexameric trimer BMC-T (2xpfam00936), and one pentameric BMC-P (pfam03319). Our synthetic GRM3C operon encodes all six of these shell proteins under the control of a T7 promoter (Figure 1).
Figure 1. GRM3C shell proteins and design of the synthetic operon.
All six genes encoding shell proteins in the GRM3C locus from R. palustris BisB18 are expressed in E. coli under the control of a T7 promoter. The Ribosome binding site (RBS) preceding BMC-H1 is derived from pET29b 27. Native RBS sequences were used for BMC-T, BMC-H2, and BMC-H3 by including the intergenic sequences upstream of each gene in the R. palustris BisB18 genome. A synthetic RBS 28 and intergenic region from Synechocystis sp. PCC 6803 were used preceding BMC-H4 and BMC-Pstrep, respectively. Due to a ~100 amino acid C-terminal extension, BMC-H2 is almost twice the length as BMC-H1-3. We modified the native BMC-P sequence to include a C-terminal Strep Tag II sequence for purification (BMC-Pstrep). The shell proteins can be further classified into clades relative to all identified BMC shell protein sequences: BMC-H1: H_robinEggBlue, BMC-H2: H_pumpkin, BMC-H3: H_tan, BMC-H4: Hp_GrpU_ickyGreen, BMC-T: Tsp_turquoiseBlue, BMC-P: P_stone according to the classification of Sutter et al. 25 and Melnicki et al. 29.
BMC-P shell proteins are defining elements of BMC loci 25,30 and despite their low numerical abundance relative to hexagonal shell protein building blocks (BMC-H and BMC-T), they play an important structural role by sealing the vertices of a polyhedral BMC shell 18,31,32. The ubiquity of BMC-P across functionally diverse BMC types, coupled with reports demonstrating that incorporation of a Strep Tag II modified BMC-P homolog enabled rapid purification of both empty synthetic metabolosome 20 and β-carboxysome shells 4, led us to hypothesize that this affinity purification strategy could be used to isolate synthetic GRM3C shells assembled heterologously in Escherichia coli. To that end, we added a (Gly-Ser)2 linker and the 8-amino acid Strep Tag II sequence to the C-terminus of the only BMC-P shell protein encoded in our synthetic operon as a handle to isolate empty GRM3C shells using complementation-based affinity purification (CAP) 20.
2.2. Purification of structurally diverse synthetic GRM3C shells
To identify the shell proteins in our purified GRM3C sample, concentrated eluate from a StrepTrap affinity column was analyzed by SDS-PAGE and mass spectrometry (Figure 2A). Bands 1 and 2 both contain a mixture of BMC-H1 (9.6 kDa), BMC-H3 (9.4 kDa), and BMC-H4 (10.7 kDa). As expected, we also observed BMC-Pstrep (10.2 kDa) in Band 2. Band 3 contained the remaining two shell proteins, BMC-T (27.4 kDa) and BMC-H2 (18.7 kDa). Two additional gel fragments spanning the 15-20 kDa range (where we expected BMC-H2 to migrate based on its predicted molecular weight) were also excised for mass spectrometry analysis and provided evidence of low amounts of BMC-H2, BMC-T, as well as some possible BMC-H1, BMC-H3, and BMC-H4 dimers not completely disassembled in the reducing SDS sample buffer (Table S2). Next, we measured the relative abundance of the six shell proteins in concentrated eluates from independent purifications using mass spectrometry. The amount of each shell protein from three biological replicates was averaged and normalized relative to the BMC-Pstrep content. The ratio of GRM3C shell proteins in our StrepTrap purified samples was 1.2 ± 0.2 (BMC-H1) : 0.4 ± 0.1 (BMC-H2) : 1.1 ± 0.2 (BMC-H3) : 0.5 ± 0.1 (BMC-H4) : 0.9 ± 0.1 (BMC-T) : 1.0 ± 0.2 (BMC-Pstrep), or when all four BMC-H proteins are combined, 3.2 ± 0.3 (BMC-H1-4) : 0.9 ± 0.1 (BMC-T) : 1.0 ± 0.2 (BMC-Pstrep) (Table S3). The successful identification of all six shell proteins in the StrepTrap eluate, coupled with the abundance of untagged hexagonal building blocks (BMC-H and BMC-T) relative to BMC-Pstrep, suggests higher-order interactions between shell proteins and is consistent with the self-assembly of synthetic shells. Due to the use of the BMC-Pstrep as an affinity purification handle, the abundance of this protein is higher than expected from the observed structures since it includes some protein that is not incorporated into assemblies. At the same time, any structures formed that do not include a BMC-P subunit, such as previously observed 2D hexagonal sheets 2,3 or open-ended nanotubes 2,6-8, would not be captured.
Figure 2. Composition, size, and morphology of synthetic GRM3C shells.
A. Coomassie blue-stained SDS-polyacrylamide gel of concentrated StrepTrap eluate containing GRM3C shell proteins expressed in E. coli BL21(DE3). Numbered bands were analyzed by mass spectrometry. B. Hydrodynamic diameter of purified GRM3C particles measured by Dynamic Light Scattering (DLS). Data represent the average and standard deviation of three measurements. C/D. Negative-stained (C) and cryo electron micrographs (D) of GRM3C shells. Arrows indicate examples of different shell morphologies: small spheres (black), irregular polyhedra (yellow), nanotubes (cyan), ovoids (magenta), and nested shells (white). Scale bars are 200 nm.
To confirm the assembly of GRM3C shells, we examined the StrepTrap eluate containing all six shell proteins using dynamic light scattering (DLS) and negative-stained electron microscopy (EM). The DLS profile of our GRM3C sample shows a broad distribution of particle sizes ranging from ~30 – 200 nm (Figure 2B). Electron micrographs collected after sample dehydration and staining with uranyl acetate confirmed the presence of shells and reveal a remarkably diverse array of morphologies (Figure 2C). In addition to small ~25-30 nm symmetric particles (Figure 2C, black arrow) similar in appearance to synthetic icosahedral shells from other systems 4,10,11, we also observed larger, irregular polyhedra ~100-200 nm in diameter reminiscent of native BMCs (Figure 2C, yellow arrow) 33,34. Furthermore, GRM3C shells also appeared to form both capped nanotubes (Figure 2C, cyan arrow) as well as ovoid structures with regions of sharp curvature (Figure 2C, magenta arrow). Interestingly, we also observed examples of nested shells, with one or more smaller shells encapsulated within a larger polyhedron (Figure 2C, white arrow), similar to those reported previously 5. These results demonstrate that the CAP method can successfully isolate shells of drastically different sizes and shapes, including the unique capped nanotubes and ovoid morphologies that had not been characterized previously.
We undertook Cryo-EM analyses of GRM3C synthetic shell samples to substantiate the pleomorphic distribution of shell assemblies observed with negative-stained EM, including populations of small spherical particles (Figure 2D, black arrow), large irregular polyhedra (Figure 2D, yellow arrow), ovoids (Figure 2D, magenta arrow), and capped nanotubes (Figure 2D, cyan arrow). Furthermore, a range of different cap morphologies are also observed in our cryo-electron micrographs (Figure 2D), confirming the diversity seen with negative-stained EM.
2.3. Structural characterization and modeling of GRM3C icosahedra
Because of our purification method, all GRM3C shells observed, including the icosahedral shells, are expected to incorporate BMC-Pstrep, the only BMC-P shell protein encoded within our synthetic operon. On the other hand, the presence and relative abundance of the five hexagonal shell proteins (BMC-H1-4 and BMC-T) within each of the different shell morphologies remains an open question. In the cryo-EM structures of the two icosahedral GRM3C shells we obtained (Figure S1, Supplemental Text 1), the pentamers are, as expected, in the vertex positions. However, we could not distinguish the different hexagonal BMC-H and BMC-T building blocks of the facets. The cryo-EM maps of T=4 and T=7 shells are best fit by BMC-H1, a member of the basal hexamer shell protein clade, which are typically the major components of functionally diverse shell systems 25,29. However, the resolution of our data does not allow us to confidently distinguish between BMC-H1, BMC-H2, and BMC-H3. Therefore, shells likely contain a mixture of BMC-H components with varying positions within the shell. Additionally, BMC-H3 has 77% sequence identity to BMC-H1 and, like BMC-H2 (53% identical to BMC-H1 when comparing their BMC domains), shares similar interface and pore motifs with BMC-H1 (Figure S2). We did not detect any evidence for the ~100 amino acid C-terminal extension of unknown function of BMC-H2 in our shell structures, in agreement with a PONDR® 35 analysis predicting this sequence to be disordered. BMC-H1 and BMC-H3, the most abundant shell proteins in our purified GRM3C shell samples, are present in roughly equal amounts, indicating that, in addition to their incorporation into these smaller icosahedra, they likely also contribute to the formation of the larger GRM3C shells.
2.4. Structural characterization and modeling of capped GRM3C nanotubes
2.4.1. Comparison of capped GRM3C to open-ended BMC-H based nanotubes
When expressed heterologously in the absence of other shell proteins, some BMC-H homologs self-assemble into single-component nanotubes ~18-20 nm in diameter that can exceed 1 μm in length 2,6,7. A BMC-Tsp-type 25 trimer, PduB, has also been reported to assemble into similar, albeit wider, nanotubes with diameters of ~63 nm 6. However, none of the previously reported BMC-based nanotubes form closed shells. Instead, they have been conceptualized as the product of rolling a 2-D hexagonal array of either BMC-H and/or BMC-T into an open-ended 3-D cylinder in a manner analogous to the rolling of a sheet of graphene into a single-walled carbon nanotube (SWCNT) 6,7,36. In this model, depending on how the hexagonal lattice is rolled up, the orientation of individual hexagons relative to the longitudinal axis of the resulting tube changes will result in either an armchair, zig-zag, or chiral arrangement (Figure S3). A similar tiling of BMC-H and BMC-T likely also explains the cylindrical bodies of GRM3C nanotubes. Notably, the GRM3C BMC-T, and BMC-H1 and BMC-H3 paralogs are closely related to the known open-ended nanotube-forming shell proteins PduB, PduA and PduJ, respectively 29,37 (Table S1).
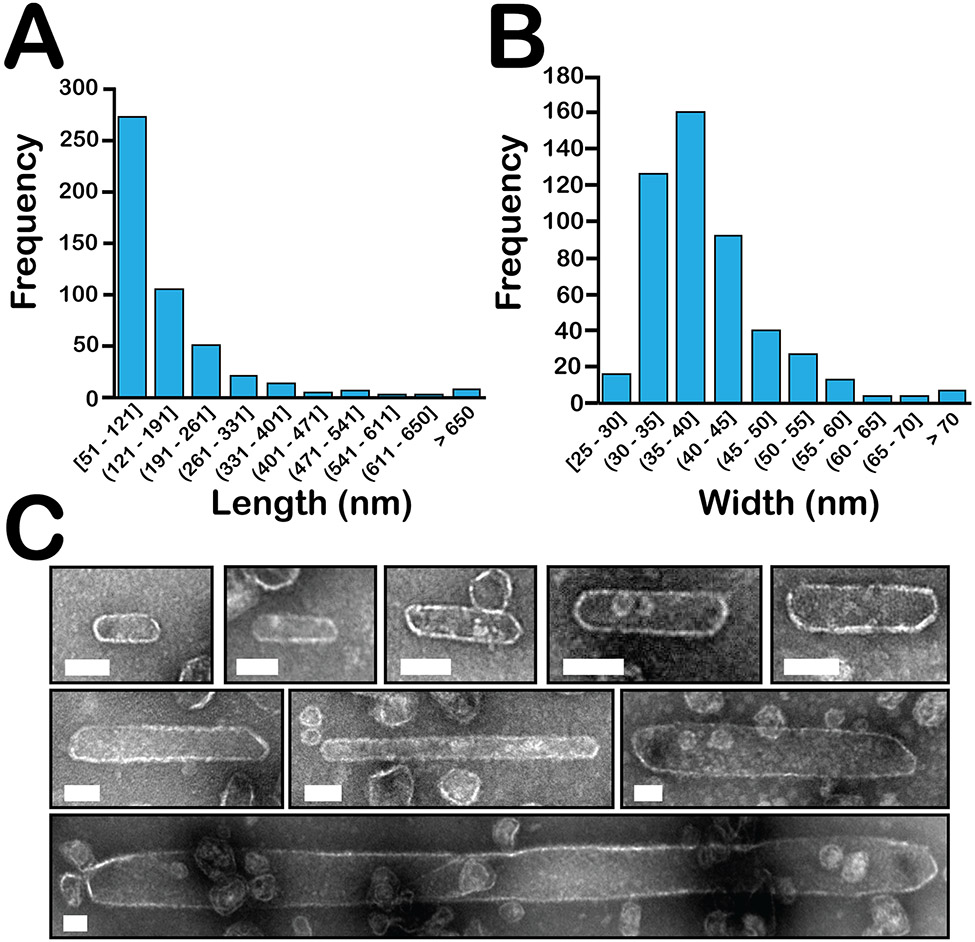
Unlike the previously described BMC-based nanotube forming systems, the nanotubes observed in GRM3C shell samples appear to be closed shells, with curved end caps terminating their cylindrical bodies. These unique capped nanotubes span more than an order of magnitude in length, with 56% (272/489) of nanotubes between 51-121 nm long, and the remaining 44% (217/489) ranging from 121 nm up to ~1.8 μm (Figure 3A). Moreover, the frequency of long tubes may be underestimated, given that even the mild centrifugation (15,000 x g for 5 min) and filtration (0.45 μm) steps used during the CAP purification could feasibly remove some of these extremely large species prior to the affinity chromatography step. In contrast to the broad distribution of observed lengths, the vast majority (89%, 439/489) of nanotubes have diameters between 25-50 nm (Figure 3B). Preparation of the negatively stained samples leads to flattening or collapse of the nanotubes and can affect their apparent diameter. However, our cryo-EM images (Figure 4) show that nanotube diameter ranges that are very similar to the negative stained samples.
Figure 3. Negative-stained EM characterization of capped GRM3C nanotubes.
A. Frequency of observed nanotube lengths and B. widths measured from negative-stained EM images; n = 489. Widths represent the average of 2-5 measurements per nanotube. Round brackets are exclusive and square brackets are inclusive. C. Negative-stained EM images showing GRM3C nanotube diversity. Scale bars are 50 nm.
Figure 4. Cryo-EM characterization and geometric model for GRM3C nanotubes.
A. Representative cryo-electron micrograph of purified synthetic GRM3C shells. Arrows indicate examples of different shell morphologies: small spheres (black), irregular polyhedra (yellow), nanotubes (cyan), and ovoids (magenta). Scale bar is 100 nm. B. Examples of nanotube heterogeneity observed by cryo-EM; the top panel, an enlargement from A, shows a nanotube ~135 nm long, with a diameter of ~36 nm. Scale bars are 50 nm. C. Four class averages of cylindrical nanotube cross sections 27 nm, 31 nm, 33 nm, and 36 nm in diameter. For additional class averages see Figure S4. D. End-on view of computational models of two example fullerene caps. The distribution of their six respective pentagons (yellow, numbered 1-6 and 7-12) are defined according to the nomenclature of Lair et al. 41: 5-2d, 3a (Cap 1, cyan) and 6-0a, 2e (Cap 2, magenta). E. Two orientations of a fullerene-based model of a GRM3C nanotube incorporating the two caps in panel D. The zig-zag (16,0) nanotube cylinder is 16 hexagons in circumference and 20 hexagons long, corresponding to a diameter of ~36 nm and length of ~140 nm if constructed from hexagonal BMC-H shell proteins with a side length of 4 nm.
2.3.2. Structural diversity of nanotube end caps
Although the geometry of the cylindrical body of a GRM3C nanotube can be explained by the rolling-up of a 2-D hexagonal array of shell proteins like open-ended BMC-H or BMC-T nanotubes, the observation of end-capped nanotubes in our electron micrographs (Figure 3C), and the involvement of BMC-Pstrep (selected for by our affinity-based purification method), points to important differences in their structures and underlying assembly principles. Reports of smaller, ~20-40 nm long, prolate BMC shells derived from β-carboxysome 4 and GRM2 5,11 model shells provide some insight into how the end caps can be formed by analogy to those described for carbon nanotubes 38. For example, BMC-H and BMC-P can close a cylindrical tube made of hexagonal shell proteins by splitting an icosahedron into two equal halves, with the circumference of the icosahedron dictating the nanotube diameter, and a variable number of hexamer belts inserted between the caps, dictating the nanotube length. For the β-carboxysome, there is one hexamer belt between icosahedral hemispheres 4, whereas GRM2 shells have been observed with 1 to 4 belts of BMC-H 5,11. Despite their substantially shorter lengths compared to the GRM3C nanotubes, the end caps sealing these short prolates can theoretically be used to close a tube of any length. This geometric model has also been used as a framework for interpreting the structures of a number of short prolate viral capsids 39, as well as elongated (~ 50-100 nm) alfalfa mosaic virus (AMV) nanotubes 40. However, the GRM3C BMC proteins are capable of forming end capped BMC nanotubes of up to 1 μm in length, vastly longer than those seen in any previous studies. Furthermore, in stark contrast to end caps derived from bisected icosahedra, GRM3C end caps appear to vary dramatically in their apparent symmetry, and different caps are often observed at opposite ends of the same tube (Figure 3C, Figure 4A and B) suggesting a not yet observed plasticity in BMC end cap formation.
Consistent with the heterogeneity observed in GRM3C nanotube caps, we did not identify a subpopulation of nanotubes amenable to structural determination from our cryo-EM data. However, our analyses generated 11 discrete class averages of ~70 nm-long segments of cylindrical nanotube bodies (Figure 4C, Figure S4). In some classes, a repeating unit of ~3.2 nm in length is observed that roughly corresponds to half the diameter of the hexagonal shell building blocks (i.e. hexameric BMC-H and trimeric BMC-T). Four representative class averages for nanotubes of 27, 31, 33, and 36 nm in diameter are shown in Figure 4C (see Figure S4 for additional class averages). These averages show that the walls of GRM3C nanotubes are ~3.9 nm thick, and therefore likely constructed, as expected, from a single layer of shell proteins (a single BMC-H or BMC-T shell protein is ~3 nm thick 12).
2.3.3. Geometric models for capped BMC nanotubes
According to Euler’s theorem, additional hexagons and exactly 12 pentagons (six in each cap) are required to close a SWCNT 41. One way to satisfy these conditions is to generate end caps by bisecting an icosahedron, as is the case for the synthetic prolate β-carboxysome and GRM2 shells, viral capsids, or the elongated AMV particles discussed above. Such icosahedron-derived caps could theoretically explain some of the more rounded, symmetric ends of the GRM3C nanotubes, and would be in line with the propensity for BMC shell proteins to form icosahedral shapes when expressed heterologously 4,11,12. As noted above, GRM3C shell proteins also assemble into icosahedra with diameters of ~25 nm (T=4) and ~31 nm (T=7) (Figure S1). Short prolates, albeit at much lower abundance (183 particles vs a total of 4012 for spherical shells), are also formed (Figure S5A, top right corner). However, a model where caps are derived only from icosahedral hemispheres fails to fully explain the observed morphological diversity in our sample, and why, for example, in the case of GRM3C nanotubes with a diameter matching the ~31 nm circumference of our T=7 icosahedron (Figure 4C), we did not identify a homogenous subpopulation of symmetric caps, further strengthening the concept of end cap plasticity.
If we allow for more flexibility in the spatial distribution of the six pentamers per end cap, the potential to create a wide variety of end cap isomers capable of closing a nanotube with a given diameter increases dramatically and allows for the formation of asymmetric end cap shapes reminiscent of those observed in our GRM3C nanotubes. These fullerene end caps have been extensively characterized in the context of SWCNTs 41,42, and depending on the arrangement of pentagons, can seal armchair, zig-zag, or chiral hexagonal cylinders (Figure S3). Following the systematic classification of Lair et al. 41, SWCNT end caps can be constructed from the perspective of a base shape at the pole of the cap, either a pentagon (in our case, BMC-P) or hexagon (in our case, BMC-H or BMC-T), with the relative positions of the remaining 5 (or 6) pentagons determining the final shape and circumference of the nanotube. The ability of GRM3C nanotubes to incorporate diverse, asymmetric fullerene-based end caps could therefore explain their underlying structural heterogeneity, especially when we consider that two different end caps, with unique distributions of pentagons and relative orientations, can seal opposite ends of the same tube.
This geometric model can recapitulate the observed heterogeneity of GRM3C nanotubes. We constructed a fullerene nanotube with two different end caps in silico, one with a pentagon (Figure 4D, Cap 1) and the other a hexagon base plate (Figure 4D, Cap 2). A nanotube model incorporating these two example caps (Figure 4E), viewed perpendicular to its longitudinal axis to imitate the orientation of GRM3C nanotubes in our EM images, closely resembles our empirically observed morphologies (see, for example, Figure 4B, top). Moreover, rotating this model by 90 degrees highlights the angular dependence of the apparent asymmetry of the end caps (in particular, Cap 1). How a nanotube is deposited on a negative-stained EM grid, or oriented in vitreous ice for cryo-EM, could therefore potentially obscure characterization of structural heterogeneity during visual inspection. Importantly, our observations of elongated capped nanotubes also bear striking resemblance to a sub-population of elongated BMC architectures observed in a recent computational study 43. Although these simulations contained only hexagonal shell components, and explicitly account for the presence of cargo molecules, the simulated architecture contains 12 pentameric defects consistent with both our geometric model and biochemical evidence for incorporation of BMC-Pstrep within the GRM3C nanotubes.
2.4. Structural characterization and modeling of GRM3C nanocones
Our proposed geometric model for capped BMC nanotubes suggests plasticity in the interactions between BMC-H, BMC-T, and BMC-P, consistent with their ability to assemble into irregular polyhedra. Moreover, our model can explain the formation of other regular fullerene-based architectures such as nanocones, that also have an asymmetric distribution of pentagon subunits. At its apex, a fullerene cone is like the nanotube end cap, except less than 6 pentagons (P) are incorporated before a helical propagation of hexagons extends towards its base. In accordance with Euler’s theorem, the base of the cone can also be closed with a multitude of structurally unique end caps containing (12-P) pentagons. The ovoid-shaped GRM3C shells (Figure 2C, 4A, magenta arrows) are consistent with this fullerene-based nanocone model. These GRM3C nanocones, observed in both negative-stained and cryo-EM images (Figure 5A), range in length between 58-280 nm, and 98% of particles are between 58-175 nm (211/216) (Figure 5B). Apex angles measured from negative-stained EM images further support a conical geometry, and follow a bimodal distribution with modes at 29° and 57° degrees, consistent with the narrowest two theoretical angles of a cone constructed from a hexagonal array (i.e. 30°, 60°) (Figure S6) 44. While the negative stain process can slightly affect the morphology due to flattening or collapse of the walls we see the same morphologies in our cryo-EM images (Figure 5A) so a substantial deviation of the measured angles can be excluded. Rare observations of hybrid nanocone-nanotube structures, where it appears the hexagonal extension of a cone’s apex takes on a constant curvature resulting in the formation of a nanotube, further supports a common helical arrangement of BMC-H/-T shell proteins in both morphologies (Figure S7).
Figure 5. EM characterization and geometric model of GRM3C nanocones.
A. Examples of nanocones from purified GRM3C samples imaged by negative-stained (top) or cryo-EM (bottom). Scale bars are 50 nm. B. Distribution of nanocone lengths measured from negative-stained EM images (n = 216). Round brackets are exclusive and square brackets are inclusive. C. A geometric model of a fullerene cone derived from the atomic-level structure of a mature HIV-1 capsid (PDB ID: 3J3Q) 45 comprised of 216 hexagons and 12 pentagons (yellow, numbered). When built with BMC shell proteins, this model corresponds to a nanocone of about 75 nm in length.
The GRM3C nanocones observed in our analyses evoke EM images of mature HIV-1 capsids 44,45. Functionally and evolutionarily unrelated to BMC proteins, the HIV-1 capsid is assembled from a single two domain subunit that can oligomerize into either hexamers or pentamers 46. In its mature form, these quasi-equivalent building blocks assemble into fullerene cones ~100 nm in length 44,47. When assembled in vitro, HIV-1 capsid proteins can also readily form nanotubes 44. By leveraging the decades of detailed biophysical studies of the mature HIV-1 capsid, culminating in atomic-level structures outlining the arrangement of hexagonal and pentagonal subunits 45, we propose an analogous geometric model to interpret the structures of GRM3C nanocones (Figure 5C). Consistent with the observed morphological diversity of both the HIV-1 and GRM3C architectures, the proposed conical model, like other fullerenes, can vary dramatically in both size (depending on the number of hexagons) and shape (depending on the distribution of the 12 pentagons at the apices and end caps). In contrast to the HIV-1 capsid, which is formed by a single protein protomer that can assemble into hexagons and pentagons, and even switch between the two confirmations 45,48,49, BMCs have dedicated proteins forming pentagons (BMC-P) or hexagons (BMC-H/-T). This difference in the architecture of these proteinaceous nanocones likely has consequences of material properties, like rigidity and strain resistance, which must contribute to their distinct functions in biology.
3. Conclusions
Although a comprehensive understanding of the determinants of BMC shell assembly remains enigmatic, recent computational studies have successfully reproduced many of the complex irregular polyhedral shapes observed empirically 43,50-52. These reports have highlighted how the relative abundance, absolute concentration, interaction strength, and flexibility of shell proteins, as well as the presence of cargo, can impact the size and shape of a BMC shell. The insights from these computational studies, coupled with the conservation of common interface motifs between BMC-H and BMC-P shell proteins, suggest a molecular explanation for the pleomorphic diversity of GRM3C shells that likely involves multiple factors, including intrinsic properties of the individual subunits, their abundance, as well as their spatial distribution within the shell. Given the multiple distinct hexamers and the pseudohexamer present in our assembly mixture, it is reasonable to assume that the different morphologies observed result from different hexamer lattice compositions, and that these may vary in flexibility to accommodate local curvature introduced by the pentamers/end caps; this would alter the frequency with which pentamers are incorporated into the hexagonal lattice. To test this hypothesis, systematically characterizing the structural diversity formed by specific combinations of shell proteins, is a fundamental next step in developing BMC shell proteins into a platform technology for predictive building of nanocompartments and microcompartments for a range of applications in biomedicine and bioengineering. Indeed, being able to predictably construct nanocontainers of defined size, shape and charge is not only important for compartmentalizing metabolic pathways, but is critical for other potential applications of shells, including those involving uptake by cellular targets and cargo delivery. In this context, icosahedral particles previously characterized are of limited size (20-40 nm). The larger compartments described here may be better suited for cargo delivery applications; for example, packaging DNA or RNA molecules into the capsid-like nanocone shell. Despite recent advances in BMC shell protein engineering, the number of synthetic metabolosome shell systems that have been experimentally characterized remains limited. The rapid isolation of pleomorphic shells from a new synthetic GRM3C system that span almost two-orders of magnitude in size from 25 nm to ~1.8 μm shows that we have only begun to explore the functional and structural potential of this remarkable self-assembling and genetically tractable biomaterial. In addition to shapes observed previously, such as small symmetric icosahedra and large irregular polyhedra, GRM3C shell proteins also form regular, yet asymmetric, capped nanotube and nanocone architectures that incorporate BMC-P. The heterogeneity of these new shapes is consistent with a model based on the hexagon and pentagon composition of fullerenes, and suggests that common geometric principles underly the assembly of asymmetric carbon, viral protein, and BMC-based architectures.
4. Materials and Methods
Synthetic operon design:
The six genes encoding shell proteins in the GRM3C locus from Rhodopseudomonas palustris BisB18, referred to in this manuscript as BMC-H1 (Locus tag: RPC_1165; Uniprot accession: ABD86728), BMC-T (RPC_1166; ABD86729), BMC-H2 (RPC_1167; ABD86730), BMC-H3 (RPC_1168; ABD86731), BMC-H4 (RPC_1175; ABD86738), and BMC-P (RPC_1172; ABD86735), were PCR amplified from R. palustris BisB18 genomic DNA (see Table S1 for amino acid sequences of all proteins). The intergenic region between cpcB and cpcA was PCR amplified from the Synechocystis sp. PCC 6803 genome. The PCR products were cloned into pBbE2K27 by Gibson assembly53 to form a synthetic operon, in which the cpcB-cpcA intergenic region served as the intergenic region between BMC-H4 and BMC-Pstrep (See Sequence S1 for complete plasmid DNA sequence) and a synthetic ribosome binding site28 was used in front of the BMC-H4 gene. A flexible (GlySer)2 linker and Strep Tag II were added on the C-terminus of BMC-P using Gibson assembly. The amino acid sequences of all shell proteins encoded in the synthetic GRM3C operon are listed in Table S1.
Growth, expression, and purification of synthetic GRM3C shells from E. coli:
Lysogeny broth starter cultures supplemented with 50 μg ml−1 kanamycin were inoculated with Escherichia coli BL21(DE3) cells carrying the pBbE2K plasmid containing the synthetic GRM3C operon from R. palustris BisB18 and grown overnight at 37 °C while shaking at 200 RPM. 10 ml of the starter culture was used to inoculate flasks containing 1 L of lysogeny broth with 50 μg ml−1 kanamycin and the culture shaken at 37 °C and 200 RPM until they reached an OD600 of ~0.6-0.8, at which point protein expression was induced by adding IPTG to a final concentration of 250 μM. After adding IPTG, the temperature was lowered to 18 °C and protein expression allowed to continue overnight while shaking at 200 RPM. Cells were harvested by centrifugation at 8,000xg for 10 min at 25 °C and resuspended using B-PER detergent lysis reagent (ThermoFischer) supplemented with 1XSigmaFast protease inhibitor (Sigma Aldrich), 2 μl of 50 mg ml−1 lysozyme per ml lysis reagent, and 1 mg DNaseI, at a ratio of 4 ml g−1 of cells according to the manufacturers’ guidelines. Unbroken cells were removed by centrifugation at 15,000xg for 5 min at 25 °C and the resulting lysate filtered through a 0.45 μm syringe filter before loading onto a 5 mL StrepTrap column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0 with 300 mM NaCl (Buffer A) at 5 ml min−1 using an Akta Pure FPLC at 10 °C; for GRM3C sample preparations for Cryo-EM, Buffer A was composed of 20 mM HEPES pH 7.4 and 50 mM NaCl. After loading the cell lysate, the column was washed with 5 column volumes of Buffer A before eluting shells with Buffer A containing 2.5 mM desthiobiotin. Pooled StrepTrap elution fractions were concentrated with a 100 kDa MWCO centrifugal filter at 4 °C at 4,500xg to a final volume of ~ 0.5-1 ml. For SDS-PAGE, 15 μl of the concentrated shell sample was mixed with 5 μl of 4X reducing SDS sample buffer and boiled for 10 min before loading onto an SDS-polyacrylamide gel (4% stacking/18% separating).
Mass Spectrometry sample preparation, data collection, and analysis
Proteolytic digestion
Gel bands excised from the 18% acrylamide separating gel of purified GRM3C shells were digested in-gel according to Shevchenko, et. al.54 with modifications. Briefly, gel bands were dehydrated using 100% acetonitrile and incubated with 10 mM dithiothreitol in 100 mM ammonium bicarbonate, pH ~ 8, at 56 °C for 45 min, dehydrated again and incubated in the dark with 50 mM iodoacetamide in 100 mM ammonium bicarbonate for 20 min. Gel bands were then washed with ammonium bicarbonate and dehydrated again. Sequencing grade modified trypsin was prepared to 0.01μg μl−1 in 50 mM ammonium bicarbonate and ~100 μl of this was added to each gel band so that the gel was completely submerged. Bands were then incubated at 37 °C overnight. Peptides were extracted from the gel by water bath sonication in a solution of 60% Acetonitrile (ACN)/1% Trifluoroacetic acid (TFA) and vacuum dried to ~2 μl. Dried peptides were then re-suspended in 2% ACN/0.1% TFA to 20 μl prior to injection.
LC/MS/MS analysis
An injection of 5 μl was automatically made using a Thermo (www.thermo.com) EASYnLC 1000 onto a Thermo Acclaim PepMap RSLC 0.1 mm x 20 mm C18 trapping column and washed for ~5 min with buffer A. Bound peptides were then eluted over 35 min onto a Thermo Acclaim PepMap RSLC 0.075mm x 250mm resolving column with a gradient of 5%B to 40%B in 24 min, ramping to 90%B at 25 min and held at 90%B for the duration of the run (Buffer A = 99.9% Water/0.1% Formic Acid, Buffer B = 80% Acetonitrile/0.1% Formic Acid/19.9% Water) at a constant flow rate of 300 nl min−1. Column temperature was maintained at a constant temperature of 50 °C using and integrated column oven (PRSO-V1, Sonation GmbH, Biberach, Germany).
Eluted peptides were sprayed into a ThermoScientific Q-Exactive mass spectrometer (www.thermo.com) using a FlexSpray spray ion source. Survey scans were taken in the Orbi trap (35000 resolution, determined at m/z 200) and the top 15 ions in each survey scan are then subjected to automatic higher energy collision induced dissociation (HCD) with fragment spectra acquired at 17,500 resolution. The resulting MS/MS spectra are converted to peak lists using Mascot Distiller, v2.8.0.1 (www.matrixscience.com) and searched against R. palustris BisB18 shell protein sequences and all E. coli protein sequences (downloaded 2020-11-19, www.uniprot.org) appended with common laboratory contaminants (downloaded from www.thegpm.org, cRAP project) using the Mascot searching algorithm, v 2.7. The Mascot output was then analyzed using Scaffold, v5.0.0 (www.proteomesoftware.com) to probabilistically validate protein identifications. Assignments validated using the Scaffold 2% FDR confidence filter and containing 4 unique peptides are considered true. The resulting percent sequence coverage for each GRM3C shell protein is listed in Table S2. Mascot parameters for all databases were as follows: (i) allow up to 2 missed tryptic sites, (ii) fixed modification of Carbamidomethyl Cysteine, (iii) variable modification of Oxidation of Methionine, (iv) peptide tolerance of ± 10ppm, (v) MS/MS tolerance of 0.02 Da, (vi) FDR calculated using randomized database search.
Mass spectrometry quantification of relative shell protein abundances
To calculate the relative abundance of each shell protein, we analyzed biological replicates of purified shells from three independent 1 L cultures. Total protein content of reducing SDS-PAGE samples prepared from concentrated StrepTrap eluates (see Materials and Methods: Growth, expression, and purification of synthetic GRM3C shells from E. coli) were normalized to an A280 of 0.513 and equal volumes loaded onto a polyacrylamide gel with a 4% acrylamide stacking layer. The gel was run at room temperature for 20 min at 50 V to concentrate the shell samples into a single band within the 4% acrylamide stacking layer. The gel was stained with Coomassie-blue and the concentrated band representing the total protein content of each eluate was excised and placed into separate tubes with 100 μl of 5% acetic acid. Samples were proteolyzed and analyzed as described above. The normalized average total ion current (TIC) for each shell protein was quantified using Scaffold, v5.0.0 (www.proteomesoftware.com) as described above, and used to calculate the abundance of each BMC-H and BMC-T shell protein relative to the amount of BMC-P (Table S3).
Negative-stained transmission electron microscopy
Uranyl acetate-stained GRM3C shells were imaged using a JEM-1400Flash microscope (JOEL) with a bottom-mounted Matataki Flash sCMOS camera (JOEL). Grids were prepared as described in Ferlez et al. 21 by floating a carbon-coated copper grid on top of 5 μl of StrepTrap eluate containing GRM3C shells for 30 seconds, wicking the excess solution away using Whatman paper, followed by floating the grid on top of 5 μl of 1% uranyl acetate for 15 seconds before removing excess stain with Whatman paper.
Dynamic light scattering
Dynamic light scattering data were collected using a Wyatt DynaPro Nanostar. 5 μl of the pooled StrepTrap elution fractions were loaded into a 1 μl cuvette and three scans, each consisting of 20 five second acquisitions, were used to measure the size distribution of GRM3C shells in solution. The percent scattered light intensity for the three scans were averaged and plotted along with their standard deviation.
Cryo-EM specimen preparation and data collection
A thin film of continuous carbon (prepared in-house) was floated onto R1.2/1.3 holey carbon grids or UltrAufoil R1.2/1.3 gold foil grids (Quantifoil Microtools). After drying (at least overnight), the grids were plasma cleaned using a Tergeo plasma cleaner (PIE Scientific). 4 μl of GRM3C shell sample at a concentration of 1.5 mg ml−1 were applied to the grid and incubated for approximately 30 sec in the humidity chamber of a Vitrobot Mk IV (Thermo Fisher Scientific) to allow the particles to adsorb to the carbon film. Subsequently, the liquid was blotted away for 5-7 sec and the specimen was vitrified by plunging it into a mixture of liquid ethane and propane at liquid nitrogen temperature.55
The grids were first screened using a Tecnai F20 cryo-transmission electron microscope (FEI) operated at 120 kV acceleration voltage and equipped with a US4000 CCD camera (Gatan). Selected grids were then loaded into a Talos Arctica cryo-transmission electron microscope (Thermo Fisher Scientific) operated at 200 kV acceleration voltage. Data were acquired at 0.5-1.5 μm underfocus using a K3 direct electron detector (Gatan) operated in super-resolution counting mode, with electron micrograph movies fractionated into 50 frames at a total exposure of 25 electrons Å−2, and with the microscope set to 35,638 x magnification, resulting in a pixel size of 1.403 Å on the object scale (super-resolution pixel size 0.7015 Å). The data collection was automated using SerialEM56 and accelerated by using image shift to acquire data in 9 holes per stage position. A total of 1,102 movies were recorded, of which 378 were acquired on one holey carbon grid and 724 on one holey gold grid (both of them carbon-coated, as described above).
Cryo-EM data processing
All data processing was performed in RELION 3.0 57,58, unless stated otherwise. Beam-induced motion in the electron micrograph movies was corrected using the CPU-based implementation of motion correction within RELION (movies subdivided into 3 x 3 patches), followed by CTF parameter fitting using CTFFIND 4 59 from within RELION. Based on the quality of the CTF parameter fits, 858 aligned and summed movies were retained for further processing.
For the analysis of tubular shell structures from cryo-electron micrographs, all tubular assemblies were picked manually from 858 selected micrographs. Particles were extracted at a pixel size of 1.05 Å and a step size of 70 Å along the tube axis. The extracted dataset was subjected to repeated 2D classification to remove tube caps or aberrant particles, such that tubes of different diameters could be resolved in a final round of 2D classification (shown in Figure 3C and S4). Nanotube wall thickness and overall diameter of GRM3C nanotube class averages were measured with Fiji using a pixel size of 1.05 Å (see Figure S4 for example measurements).
For reconstruction of symmetrical shells (Figure S1, S5), particles with roughly spherical appearance (i.e. putative icosahedral symmetry), were manually picked from a subset of micrographs because of the extreme heterogeneity of the specimen, and 2D classification was used to generate picking references, notably including shells of different diameters, for subsequent automated selection of a dataset of particles with putative icosahedral symmetry. A total of 90,863 particle coordinates were identified, which included a diverse array of particles, comprising round (i.e. symmetrical or quasi-symmetrical) particles and tips of elongated or tubular structures. The auto-picked particles were subjected to two rounds of 2D classification, initially selecting 6,436 spherical, putatively icosahedral, particles of different sizes, and then identifying large and small diameter shell classes (1,154 large particles with hexagonal appearance, 1,518 large particles with round appearance, 1,340 small particles) within those initially-selected particles. Additionally, 183 bullet-shaped, or prolate particles were identified, similar to those observed in other microcompartment systems 4,5. However, this subset could not be refined reliably, likely owing to the small number of particles and reduced symmetry. All other classes represented irregular or tubular structures. Refinement of the 1,154 large and 1,340 small shell particles revealed that they correspond to T=7 (large) and T=4 (small) icosahedral shells. These two particle classes were subsequently processed independently.
For the T=7 shell, the particle subsets with hexagonal and round appearance in 2D classes were initially processed separately. Particles belonging to the classes with hexagonal appearance, which appeared more sharply defined than the round particles of the same diameter, were extracted at their original 1.403 Å pixel size, followed by 3D auto-refinement and CTF refinement (magnification anisotropy, per-particle defocus, beam tilt), 3D auto-refinement, and 3D classification. The best 3D class was again refined and subjected to CTF refinement, resulting in a 3.9 Å-resolution map from 870 particles. The particles belonging to the 2D classes of similar size to the T=7 shell, but with round appearance, were of lower apparent quality (as judged from the resolution of 3D auto-refinements) and were therefore initially 2D-subclassified, 3D auto-refined, and 3D-subclassified using the binned 4.209 Å/pixel particle images. The 431 remaining particles were extracted at the original pixel size (1.403 Å), and subsequently 3D auto-refined, subjected to two rounds of CTF refinement (anisotropic magnification, per-particle defocus, beam tilt), and further refined, resulting in a reconstruction at 4.7 Å resolution. Further 3D classification identified 362 higher-quality particles, which were added to the previously identified 870-particle subset of T=7 shells.
The joined dataset of 1,232 T=7 shell particles was refined and CTF refined (per-particle defocus and astigmatism, beam tilt). The dataset was then symmetry expanded with icosahedral (I2) symmetry (resulting in 73,920 particles), which appeared to aid with subsequent CTF refinement (which included per-particle defocus, per-micrograph astigmatism, and beam tilt, followed by beam tilt, trefoil and 4th order aberrations). 11,400 particles originating from micrographs with poor ice quality (as judged by manual micrograph inspection) were excluded, resulting in a 3.6 Å resolution reconstruction of the T=7 shell from 62,520 particles (Figure S5A). For the T=4 shell, 1,340 particles were selected and refined using the binned 4.209 Å/pixel data, followed by extraction of unbinned particle images with improved centering quality. The unbinned particles were 3D auto-refined to 4.4 Å resolution. After CTF refinement (magnification anisotropy followed by per-particle defocus and beam tilt), 3D auto-refinement and 3D classification, 1,190 high-quality particles were selected. This subset was subjected to two rounds of CTF refinement (per-particle defocus and astigmatism followed by 3D-refinement and refinement of per-particle defocus, per-particle astigmatism, and beam tilt) and 3D-auto refined to 3.5 Å resolution. The dataset was symmetry expanded (resulting in 71,400 particles) and subjected to CTF-refinement (per-particle defocus, per-micrograph astigmatism, beam tilt) and refined. Of the 71,400 particles, 57,840 were selected based on micrograph ice quality (as judged by manual inspection, as above) and refined, resulting in a 3.4 Å resolution reconstruction of the T=4 shell (Figure S5B). The cryo-EM maps for the icosahedral structures were deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under accessions EMD-16401 for T=4 and EMD-16402 for T=7.
Nanotube and nanocone models
The nanotube model in Figure 3E was built using carbon atoms with optimized molecular geometry in Avogadro 1.2.0 60. Two different end caps compatible with a zig-zag nanotube were selected to close a (16,0) graphene nanotube 20 hexagons in length. The distribution of pentagons in each of the two end caps is described following the nomenclature of Lair et al.;41 Cap 1 is 5-2d,3a and Cap 2 is 6-0a,2e. PyMOL (The PyMOL molecular Graphics System, Version 2.3.4, Schrodinger LLC) was used for visualization and to generate the final image; pentamers were colored using Adobe Illustrator.
The nanocone model in Figure 5C was built with PyMOL by extracting the positions of Ala204 of the HIV-1 proteins from the pdb file 3J3Q 45, which is located at the corner position between three subunits, and using those coordinates to draw lines in between them.
Statistical Analysis
Sample sizes for size measurements from electron microscopy images are indicated in the Figure legends. A Fourier Shell Correlation (FSC) curve was used to assess the resolution of the icosahedral T=4 and T=7 structures. Statistical analysis methods for analysis of mass-spectrometry data are is detailed in the LC/MS/MS analysis section of the methods.
Supplementary Material
Acknowledgements
We would like to thank Carrie Harwood for providing the culture of R. palustris BisB18 and the MSU Metabolomics Center, in particular Douglas Whitten, for both collecting the mass spectrometry data and providing valuable assistance with data analysis. EN is a Howard Hughes Medical Institute Investigator. This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-06. Research by CAK and MS is supported as part of the Center for Catalysis in Biomimetic Confinement, an Energy Frontier Research Center funded by the U.S. Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES), under award DE-SC0023395.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Bryan H. Ferlez, MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA; Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
Henning Kirst, Environmental Genomics and Systems Biology and Molecular Biophysics and Integrative Bioimaging Divisions, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA; MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA.
Basil J. Greber, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, California 94720, USA; Molecular Biophysics and Integrative Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA
Eva Nogales, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, California 94720, USA; Molecular Biophysics and Integrative Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA; Howard Hughes Medical Institute, University of California, Berkeley, California 94720, USA; Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Markus Sutter, Environmental Genomics and Systems Biology and Molecular Biophysics and Integrative Bioimaging Divisions, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA; MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA.
Cheryl A. Kerfeld, Environmental Genomics and Systems Biology and Molecular Biophysics and Integrative Bioimaging Divisions, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA; MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA; Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
References
- 1.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F & Sutter M Bacterial microcompartments. Nature Reviews Microbiology 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pang A, Frank S, Brown I, Warren MJ & Pickersgill RW Structural Insights into Higher Order Assembly and Function of the Bacterial Microcompartment Protein PduA *. 289, 22377–22384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutter M. et al. Visualization of Bacterial Microcompartment Facet Assembly Using High-Speed Atomic Force Microscopy. Nano Letters 16, 1590–1595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutter M. et al. Structure of a synthetic beta-carboxysome shell. Plant Physiology pp.00885.2019 (2019) doi: 10.1104/pp.19.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesle EE, Filimonenko A, Tars K & Kalnins G Variety of size and form of GRM2 bacterial microcompartment particles. Protein Science: A Publication of the Protein Society (2021) doi: 10.1002/pro.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uddin I, Frank S, Warren MJ & Pickersgill RW A Generic Self-Assembly Process in Microcompartments and Synthetic Protein Nanotubes. Small (Weinheim an Der Bergstrasse, Germany) 14, e1704020 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Noël CR, Cai F & Kerfeld CA Purification and Characterization of Protein Nanotubes Assembled from a Single Bacterial Microcompartment Shell Subunit. Advanced Materials Interfaces 3, 1500295 (2016). [Google Scholar]
- 8.Parsons JB et al. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Molecular cell 38, 305–15 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Jorda J, Leibly DJ, Thompson MC & Yeates TO Structure of a novel 13 nm dodecahedral nanocage assembled from a redesigned bacterial microcompartment shell protein. Chemical Communications (Cambridge, England) 52, 5041–5044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai F, Bernstein SL, Wilson SC & Kerfeld CA Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiology 170, 1868–1877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalnins G. et al. Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nature communications 11, 388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutter M, Greber B, Aussignargues C & Kerfeld CA Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutter M, McGuire S, Ferlez B & Kerfeld CA Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies. ACS Synthetic Biology 8, 668–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lassila JK, Bernstein SL, Kinney JN, Axen SD & Kerfeld C. a. Assembly of Robust Bacterial Microcompartment Shells Using Building Blocks from an Organelle of Unknown Function. Journal of Molecular Biology 426, 2217–2228 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Held M. et al. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Scientific Reports 6, 24359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerfeld CA et al. Protein Structures Forming the Shell of Primitive Bacterial Organelles. 1–12. [DOI] [PubMed] [Google Scholar]
- 17.Klein MG et al. Identification and Structural Analysis of a Novel Carboxysome Shell Protein with Implications for Metabolite Transport. Journal of Molecular Biology 392, 319–333 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Tanaka S. et al. Atomic-Level Models of the Bacterial Carboxysome Shell. Science 319, 1083 LP – 1086 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Hagen AR et al. In Vitro Assembly of Diverse Bacterial Microcompartment Shell Architectures. Nano Letters 18, 7030–7037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen A, Sutter M, Sloan N & Kerfeld CA Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nature Communications 9, 2881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferlez B, Sutter M & Kerfeld CA A designed bacterial microcompartment shell with tunable composition and precision cargo loading. Metabolic Engineering 54, 286–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MJ et al. De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments. Nature Communications 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirst H. et al. Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate. Proceedings of the National Academy of Sciences 119, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerfeld CA & Sutter M Engineered bacterial microcompartments: apps for programming metabolism. Current Opinion in Biotechnology 65, 225–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter M, Melnicki MR, Schulz F, Woyke T & Kerfeld CA A catalog of the diversity and ubiquity of bacterial microcompartments. Nature Communications 12, 3809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferlez B, Sutter M & Kerfeld CA Glycyl Radical Enzyme-Associated Microcompartments: Redox-Replete Bacterial Organelles. mBio 10, e02327–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TS et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. Journal of Biological Engineering 5, 12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin-Karp A. et al. Quantifying Translational Coupling in E. coli Synthetic Operons Using RBS Modulation and Fluorescent Reporters. ACS Synthetic Biology 2, 327–336 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Melnicki MR, Sutter M & Kerfeld CA Evolutionary relationships among shell proteins of carboxysomes and metabolosomes. Current Opinion in Microbiology 63, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axen SD, Erbilgin O & Kerfeld CA A Taxonomy of Bacterial Microcompartment Loci Constructed by a Novel Scoring Method. PLOS Computational Biology 10, e1003898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai F. et al. The Pentameric Vertex Proteins Are Necessary for the Icosahedral Carboxysome Shell to Function as a CO2 Leakage Barrier. PLoS ONE 4, e7521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheatley M, Gidaniyan SD, Liu Y, Cascio D & Yeates TO Bacterial Microcompartment Shells of Diverse Functional Types Possess Pentameric Vertex Proteins. (2013) doi: 10.1002/pro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy NW et al. Apparent size and morphology of bacterial microcompartments varies with technique. PLOS ONE 15, e0226395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, Cheng S, Fan C & Bobik TA The PduM Protein Is a Structural Component of the Microcompartments Involved in Coenzyme B12-Dependent 1,2-Propanediol Degradation by Salmonella enterica. Journal of Bacteriology 194, 1912–1918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero P, Obradovic Z, Kissinger C, Villafranca JE & Dunker AK Identifying disordered regions in proteins from amino acid sequence. in Proceedings of International Conference on Neural Networks (ICNN’97) vol. 1 90–95 vol.1 (1997). [Google Scholar]
- 36.Dresselhaus MS, Dresselhaus G & Saito R Physics of carbon nanotubes. Carbon 33, 883–891 (1995). [Google Scholar]
- 37.Uddin I, Frank S, Warren MJ & Pickersgill RW A Generic Self-Assembly Process in Microcompartments and Synthetic Protein Nanotubes. Small (Weinheim an Der Bergstrasse, Germany) 14, e1704020 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Lair SL, Herndon WC, Murr LE & Quinones SA End cap nucleation of carbon nanotubes. Carbon 44, 447–455 (2006). [Google Scholar]
- 39.Luque A, Zandi R & Reguera D Optimal architectures of elongated viruses. Proceedings of the National Academy of Sciences 107, 5323–5328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hull R, Rees MW & Short MN Studies on alfalfa mosaic virus. I. The protein and nucleic acid. Virology 37, 404–415 (1969). [DOI] [PubMed] [Google Scholar]
- 41.Lair SL, Herndon WC, Murr LE & Quinones SA End cap nucleation of carbon nanotubes. Carbon 44, 447–455 (2006). [Google Scholar]
- 42.Melle-Franco M, Brinkmann G & Zerbetto F Modeling Nanotube Caps: The Relationship Between Fullerenes and Caps. The Journal of Physical Chemistry A 119, 12839–12844 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Mohajerani F, Sayer E, Neil C, Inlow K & Hagan MF Mechanisms of Scaffold-Mediated Microcompartment Assembly and Size Control. ACS Nano 15, 4197–4212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganser BK, Li S, Klishko VY, Finch JT & Sundquist WI Assembly and Analysis of Conical Models for the HIV-1 Core. Science 283, 80–83 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Zhao G. et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Hill CP, Sundquist WI & Finch JT Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Gelderblom HR, Hausmann EH, Ozel M, Pauli G & Koch MA Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology 156, 171–176 (1987). [DOI] [PubMed] [Google Scholar]
- 48.Byeon I-JL et al. Motions on the Millisecond Time Scale and Multiple Conformations of HIV-1 Capsid Protein: Implications for Structural Polymorphism of CA Assemblies. Journal of the American Chemical Society 134, 6455–6466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinn CM et al. Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations. Proceedings of the National Academy of Sciences 115, 11519–11524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y. et al. Computational and Experimental Approaches to Controlling Bacterial Microcompartment Assembly. ACS Central Science 7, 658–670 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vernizzi G, Sknepnek R & de la Cruz MO Platonic and Archimedean geometries in multicomponent elastic membranes. Proceedings of the National Academy of Sciences 108, 4292–4296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohajerani F & Hagan MF The role of the encapsulated cargo in microcompartment assembly. 1–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Shevchenko A, Wilm M, Vorm O & Mann M Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Analytical Chemistry 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Tivol WF, Briegel A & Jensen GJ An improved cryogen for plunge freezing. Microscopy and Microanalysis: The Official Journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada 14, 375–379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schorb M, Haberbosch I, Hagen WJH, Schwab Y & Mastronarde DN Software tools for automated transmission electron microscopy. Nature Methods 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zivanov J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zivanov J, Nakane T & Scheres SHW Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanwell MD et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics 4, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.