Abstract
Background:
Cardiac fibrosis represents a key element in the pathophysiology of heart failure with preserved ejection fraction (HFpEF), a condition highly prevalent amongst geriatric patients, especially if diabetic. The microRNA 181c (miR-181c) has been shown to be associated with the response to exercise training in HFpEF patients and has been also linked to diabetic cardiovascular complications. However, the underlying mechanisms have not been fully elucidated.
Objective:
To measure circulating miR-181c in elderly patients with HFpEF and diabetes mellitus (DM) and identify gene targets pathophysiologically relevant in HFpEF.
Methods:
We quantified circulating miR-181c in frail older adults with a confirmed diagnosis of HFpEF and DM, and, as control, we enrolled age-matched subjects without HFpEF and without DM. We validated in human cardiac fibroblasts the molecular mechanisms linking miR-181c to a pro-fibrotic response.
Results:
51 frail patients were included :34 patients with DM and HFpEF and 17 age-matched controls. We observed that miR-181c was significantly upregulated (p < 0.0001) in HFpEF patients vs controls. We confirmed in vitro that miR-181c is targeting PRKN and SMAD7.
Conclusions:
We demonstrate that miR-181c levels are significantly increased in frail elderly adults with DM and HFpEF and that miR-181c targets PRKN and SMAD7 in human cardiac fibroblasts.
Keywords: Aging, Diabetes, Fibrosis, HFpEF, MiRNA, Parkin, SMAD7
1. Introduction
Heart failure (HF) is one of the leading causes of mortality and hospitalization worldwide. According to the 2022 guidelines (Santulli et al., 2022) of the American Heart Association (AHA), the American College of Cardiology (ACC), and the Heart Failure Society of America (HFSA), HF is classified according to the left ventricular ejection fraction (EF), identifying four classes, namely HF with reduced EF (HFrEF), HF with improved EF (HFimpEF), HF with mildly reduced EF (HFmrEF), and HF with preserved EF (HFpEF). Of these forms, HFpEF is the most common in older adults, especially in patients with diabetes mellitus (DM), and is increasing in prevalence as the population ages, representing a major global public health problem in the elderly population (Abudureyimu et al., 2022; Sugita et al., 2023; Omote et al., 2022; Shi et al., 2021; Meng et al., 2023).
The pathobiology of HFpEF is complex and encompasses a number of mechanisms including endothelial dysfunction, inflammation, oxidative stress, and fibrosis (Besler et al., 2021; Fusco-Allison et al., 2021; Gu et al., 2016; Mone et al., 2023; Mordi et al., 2020; Tsujimoto and Kajio, 2018). Furthermore, diabetic cardiomyopathy has many points in common with HFpEF, including diastolic dysfunction and fibrosis (Aroor et al., 2021; Meagher et al., 2018).
microRNAs (miRNAs) are small (~21 nucleotides), single-stranded noncoding RNA molecules involved in the regulation of gene expression through translational repression (Avvisato et al., 2023; Bielska et al., 2022; Caravia et al., 2017; Fu et al., 2023; Ilieva et al., 2022; Izzo et al., 2023; Jusic et al., 2022; Laggerbauer and Engelhardt, 2022; Leitao and Enguita, 2022; Macvanin et al., 2023; Menezes Junior et al., 2023; Mone et al., 2021b; Olivieri et al., 2012; Rozhkov et al., 2022; Shimizu et al., 2020; Wronska, 2023). In the Optimizing Exercise Training in Prevention and Treatment of Diastolic Heart Failure (OptimEx) clinical trial, Andreas Gevaert and collaborators observed that miR-181c was associated with the response to exercise training in HF patients (Gevaert et al., 2021).
The same miR has been later linked also to diabetic cardiovascular complications (Cao et al., 2022; Morrison et al., 2021; Parker et al., 2022; Shu and Xiang, 2022; Solly et al., 2021). Notwithstanding, the exact underlying molecular mechanisms have not been fully investigated.
The aim of the current study is to measure circulating miR-181c in elderly patients with HFpEF and DM and identify gene targets pathophysiologically relevant in HFpEF.
2. Methods
2.1. Study design
We evaluated consecutive frail older adults with a confirmed diagnosis of HFpEF and DM who presented from October 2021 to March 2022 at the Sant’Angelo dei Lombardi Hospital, ASL (local health unit of the Italian Ministry of Health) Avellino, Italy. Inclusion criteria were: age > 65 years, a previous diagnosis of DM, frailty, and HFpEF. Patients who had experienced previous stroke, acute myocardial infarction, or cardiac revascularization and subjects with evidence of valvular, or pulmonary disease, or documented intracardiac shunting, were excluded. We also excluded patients with an implantable pacemaker and/or cardioverter–defibrillator, primary hypertrophic or restrictive cardiomyopathy, or any systemic illness associated with infiltrative heart disease (e.g. cardiac amyloidosis). Patients with HF with reduced EF (HFrEF), ischemic heart disease, myocarditis, HIV disease, drug/alcohol abuse, cancer and/or a history of chemotherapy, were also excluded, as well as patients with a history of HF and recovered EF. As a control population, we enrolled age-matched non-diabetic subjects with no evidence of HFpEF.
All patients underwent clinical evaluation and transthoracic echocardiography. A diagnosis of frailty was made when at least three of the following five, previously published (Fried et al., 2001; Mone et al., 2021a; Mone et al., 2022), criteria, were present: exhaustion (poor endurance and energy); slowness (walking speed under the lowest quintile adjusted for sex and height); weight loss (defined as unintentional loss ≥4.5 kg in the past twelve months); low physical activity (lowest quintile of kilocalories of physical activity during the previous seven days); weakness (handgrip strength in the lowest 20% quintile at baseline, adjusted for sex and body mass index).
Every patient (or a legally authorized representative) signed a written informed consent. The study was approved by the local Institutional Review Board Ethical Committee (Campania Nord) and was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
2.2. Blood collection and quantification of circulating miR-181c
Peripheral blood was collected from patients in EDTA-tubes and plasma was obtained by centrifugation as previously reported (Gambardella et al., 2023). We extracted microRNAs using the miRVana miRNA isolation kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol; the quality of miRNA was determined using the Agilent Small RNA Kit (Kansakar et al., 2022). Measurements of miR-181c levels were performed by reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR); the standard curve method was adopted for absolute quantification of the amplification products and specificity was determined by performing a melting curve analysis. U6 expression was utilized as a normalization standard and relative quantification of the amplification products was performed using the comparative Ct (2−ΔΔCt) method. We performed standard curves for miR-181c and U6 and then calculate the ratio. All samples were run in triplicate and Ct values were averaged for the replicates.
2.3. Cell culture and in vitro assays
Adult human cardiac fibroblasts were obtained from Merck (Darmstadt, Germany; catalog number #306–05A). Cells were cultured in a standard humidified atmosphere (37 °C) containing 5% CO2, according to the manufacturer’s instructions.
2.4. Identification and Validation of miR-181c as a regulator of PRKN and SMAD7
To identify pathophysiologically relevant genes among the potential targets of miRNAs we used TargetScanHuman 8.0, as we previously described (Wang et al., 2020b). We decided to validate two genes, namely PRKN (encoding Parkin), and “Mothers Against Decapentaplegic Homolog 7” (SMAD7), which are known from the literature to be involved in the regulation of two fundamental processes underlying the pathobiology of HFpEF, namely mitochondrial dysfunction and fibrosis (He et al., 2011; Humeres et al., 2022; Kubli et al., 2015; Kumar et al., 2019; Sweeney et al., 2020). To assess the effects of miR-181c on PRKN and SMAD7 gene transcription, we engineered luciferase reporters containing the 3′-UTR of the predicted miRNA interaction sites (Jin et al., 2013), both wild-type and mutated, in human cardiac fibroblasts. The mutant constructs of PRKN 3′-UTR (PRKN MUT) and SMAD7 3′-UTR (SMAD7 MUT), harboring nucleotide substitutions within the predicted miR-181c binding sites of their 3′-UTR were obtained using the NEBa-seChanger and Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA) as we described (Matarese et al., 2020). The luciferase complexes were constructed by ligating oligonucleotides containing the wild-type or mutated putative target sites of PRKN and SMAD7 3′ UTR) into the multicloning site of the pMirTarget luciferase reporter vector (Origene, Rockville, MD, USA), following the manufacturer’s instructions.
We transfected human cardiac fibroblasts with the 3′-UTR reporter plasmids (0.05 μg) and miR-181c mimic (ThermoFisher Scientific, Waltham MA, USA) or miR-181c inhibitor, as well as a non-targeting negative control (scramble), all employed at a final concentration of 50 nMol/L, using Lipofectamine RNAiMAX (ThermoFisher Scientific) (Kansakar et al., 2022). Firefly and Renilla luciferase activities were measured 48 h after transfection, using Luciferase Reporter Assay System (Promega, Madison, WI, USA), normalizing Firefly luciferase to Renilla luciferase activity (Santulli et al., 2014). The cellular expression of SMAD7 was determined using RT-qPCR as we previously described (Tang et al., 2021; Yuan et al., 2014), normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The sequences of oligonucleotide primers (Merck) are reported in Table 1.
Table 1.
Oligonucleotide primer sequences.
| Gene | Primer | Sequence (5′–3′) | Amplicon (bp) |
|---|---|---|---|
| PRKN | Forward | GAG TGT ACA CAG GGC CTT CT | 94 |
| Reverse | CCT GCT CGC TCT GAA TTG TC | ||
| SMAD7 | Forward | TTC AGA ACA CCC TCC TCC AC | 107 |
| Reverse | CCA GGG AAG GAA GGA CAC AT | ||
| GAPDH | Forward | GGC TCC CTT GGG TAT ATG GT | 94 |
| Reverse | TTG ATT TTG GAG GGA TCT CG |
Abbreviations: bp, base pairs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
2.5. Immunoblotting
Immunoblotting assays were performed as previously described and validated by our group (Ciccarelli et al., 2011; Santulli et al., 2015). Briefly, samples were lysed in ice-cold RIPA/SDS buffer [150 mM NaCl, 2 mM Na3VO4, 50 mM Tris-HCl (pH 7.4), 0.01 g/L NP-40, 0.0025 g/L deoxycholate, 0.2 g/L sodium dodecylsulphate, and cOmplete™ Protease Inhibtor cocktail (Merck)]. Protein concentration was determined using BCA assay kit (ThermoFisher Scientific). Equal amounts of total cellular extracts or immunocomplexes were electrophoresed on 4–12% SDS-PAGE gel and transferred to nitrocellulose filters (Merck). The membranes were blocked in Tris buffered saline containing 0.002 g/L Tween 20 (TBST) and 0.05 g/L nonfat dry milk After blocking, the membranes were washed three times in TBST and then incubated overnight at 4 °C in TBST containing 5% BSA with primary specific antibodies. The antibody for Parkin was purchased from abcam (Cambridge, UK; catalog number #ab77924); the antibody for SMAD7 was purchased from Santa Cruz (Dallas, TX; catalog number #SC-101152); the antibody for GAPDH was purchased from Novus Biologicals (Englewood, CO; catalog number #NB-300–221). The blots were then washed three times in TBST, incubated with appropriate HRP-conjugated secondary antibodies (1:2000, Santa Cruz), dissolved in TBST containing 5% nonfat dry milk and incubated for 1 h at room temperature (Cipolletta et al., 2015). After 3 additional washes with TBST, immunoreactive bands were visualized by enhanced chemiluminescence using the Pierce™ ECL-plus detection kit (ThermoFisher Scientific).
2.6. Statistical Analysis
All data were analyzed using the GraphPad software (Version 9.0; Dotmatics, San Diego, CA, USA). Data are expressed as means ± SD or numbers and percentages. Normal distribution was tested using the Shapiro-Wilk normality test. Differences in miRNA levels between groups were analyzed using two-tailed t-test.
3. Results
3.1. Patient characteristics
A total of 34 elderly patients with DM and HFpEF and 17 age-matched controls (frail patients without DM and without HFpEF) were successfully enrolled. The main characteristics of these subjects are shown in Table 2. No main differences between these groups were noted, except of course in terms of glycemia and left ventricular EF (Table 2).
Table 2.
Main characteristics of the patients.
| HFpEF | Control | |
|---|---|---|
| N | 34 | 17 |
| Age (years) | 81.6 ± 6.8 | 79.6 ± 8.7 |
| Female Sex, n (%) | 18 (55.8) | 9 (52.9) |
| BMI (kg/m2) | 27.2 ± 1.6 | 26.9 ± 1.9 |
| SBP (mmHg) | 118.7 ± 7.9 | 116.8 ± 7.5 |
| DBP (mmHg) | 79.1 ± 7.1 | 76.5 ± 8.7 |
| Heart rate (bpm) | 86 ± 8 | 78 ± 12 |
| LVEF (%) | 55.2 ± 5.1 * | 66.5 ± 7.1 |
| Comorbidities | ||
| Hypertension, n (%) | 15 (44.1) | 7 (41.2) |
| Dyslipidemia, n (%) | 23 (67.6) | 12 (70.6) |
| COPD, n (%) | 13 (38.23) | 7 (41.2) |
| CKD, n (%) | 12 (35.3) | 6 (35.3) |
| Laboratory parameters | ||
| Plasma glucose (mg/dl) | 160.5 ± 38.8 * | 105.1 ± 28.9 |
| Cholesterol (mg/dl) | 205.5 ± 19.8 | 202.2 ± 20.1 |
| LDL-cholesterol (mg/dl) | 133.1 ± 19.6 | 130.6 ± 15.7 |
| HDL-cholesterol (mg/dl) | 36.7 ± 3.3 | 34.6 ± 3.4 |
| HbA1c (mMol/Mol) | 56.5 ± 6.3 | - |
| BNP (pg/mL) | 445.1 ± 24.2 | - |
Data are means ± SD or n (%). “Control” refers to subjects who did not have any evidence of HFpEF or DM. BMI: body mass index; BNP: brain natriuretic peptide; DBP: diastolic blood pressure; CKD: chronic kidney disease; COPD: chronic obstructive pulmonary disease; HbA1c: glycated hemoglobin; HDL: high-density lipoprotein; LDL: low-density lipoprotein; LVEF: left ventricular ejection fraction; SBP: systolic blood pressure
: p < 0.05.
3.2. Circulating miR-181c in HFpEF patients and controls
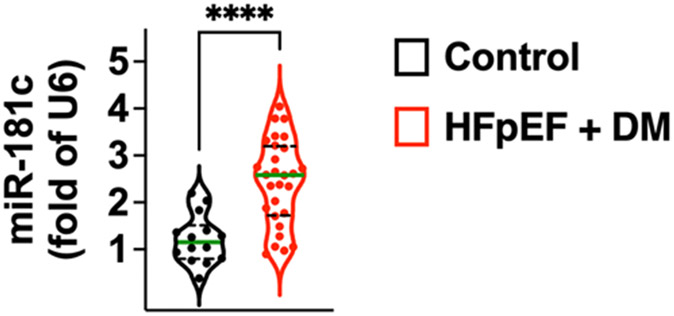
Our quantification of circulating miR-181c level revealed a significant upregulation in HFpEF patients vs controls, as shown in Fig. 1.
Fig. 1.
Quantification of circulating miR-181c in diabetic HFpEF patients vs control subjects (age-matched subjects without DM and without HFpEF). Values are normalized to U6. The violin plots indicate median (solid green line) and inter-quartile range (dashed lines); ****: p < 0.0001.
3.3. Molecular targets of miR-181c in HFpEF
We then sought to determine the main targets of this miRNA that could be biologically and clinically relevant in the pathophysiology of HFpEF. Previous studies in this sense, focusing on potential targets in cardiomyocytes, had yielded conflicting results. Indeed, whereas miR-181c overexpression was suggested to have protective actions in HF by impeding cardiomyocyte apoptosis through the PI3K/Akt pathway (Li et al., 2020b), on the other hand, strategies inhibiting miR-181c in cardiomyocytes could preserve cardiac function during obesity by improving mitochondrial function, most likely targeting MICU1 and Sp1 (George et al., 2021).
On these grounds, considering the pivotal role of interstitial cardiac fibrosis in HFpEF (Paulus and Zile, 2021; Sweeney et al., 2020), we focused our attention on signaling pathways involved in the regulation of myofibroblasts and the ensuing fibrotic response with the goal to validate some of the candidate target genes in human cardiac fibroblasts.
3.4. miR-181c targets PRKN and SMAD7 in different species
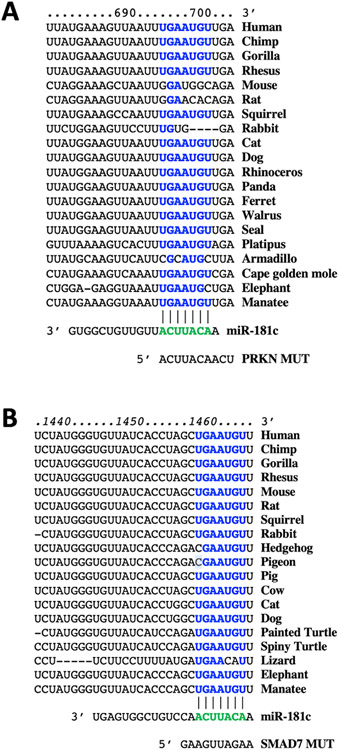
By combining bioinformatic analyses and functional assays, we identified hsa-miR-181c as a specific and highly conserved regulator of PRKN and SMAD7 (Fig. 2 and Fig. 3).
Fig. 2.
Identification of miR-181c as a specific modulator of PRKN and SMAD7. The complementary nucleotides between hsa-miR-181c and the target regions of PRKN 3′-UTR (A) and SMAD7 3′-UTR (B) are highly conserved across different species.
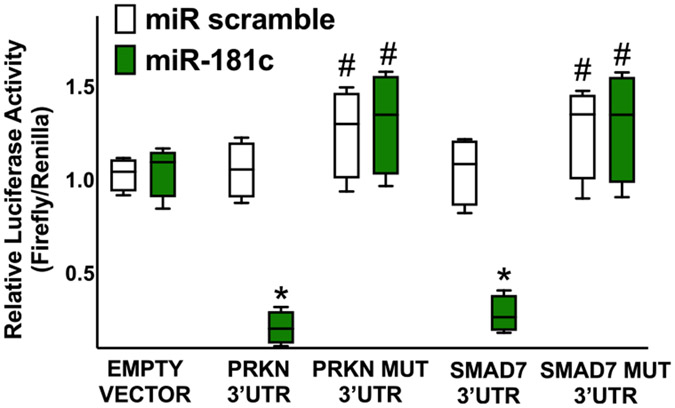
Fig. 3.
Validation of PARKN and SMAD7 targeting by miR-181c. Luciferase activity was measured in human cardiac fibroblasts 48 h after transfection, using empty vector, the vectors containing the wild-type forms of PRKN 3’UTR or SMAD7 3′-UTR, and the vectors containing the mutated forms PRKN MUT and SMAD7 MUT; a non-targeting miRNA (miR scramble) was employed as a further control. All experiments were performed at least in triplicate; the box-and-whiskers graph indicates medians and 5th–95th percentiles; * : p < 0.01 vs. miR scramble, #: p < 0.05 vs. wild type 3’UTR.
The PRKN gene (also known as PARK2) encodes Parkin, a 465-amino acid residue E3 ubiquitin ligase that is known to be a master regulator of the processes leading to the clearance of damaged mitochondria (Huo et al., 2022; Rahman and Kim, 2020). PARK has been shown to be involved in the activation of myofibroblast differentiation and proliferation, in the setting of pulmonary fibrosis (Kobayashi et al., 2016). On the other hand, SMAD7 is a well-established inhibitor of the TGF-β1-mediated fibrosis, deterring the activation of cardiac myofibroblasts (Humeres et al., 2022; Wang et al., 2020b).
Thus, we generated mutant constructs of PRKN 3′ untranslated region (UTR) and (PRKN MUT), SMAD7 3′-UTR (SMAD7 MUT), harboring nucleotide substitutions within the predicted miR-181c binding sites of their 3′-UTR; the sequences of SMAD7 3’UTR and PRKN 3’UTR are shown in Fig. 2 (human), alongside their mutated forms.
3.5. Validation in human cardiac fibroblasts of the transcriptional regulation of PRKN and SMAD7 by miR-181c
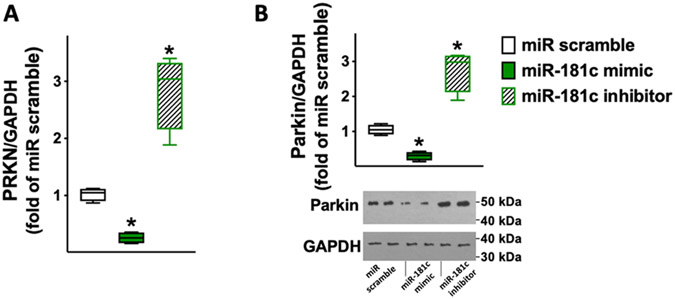
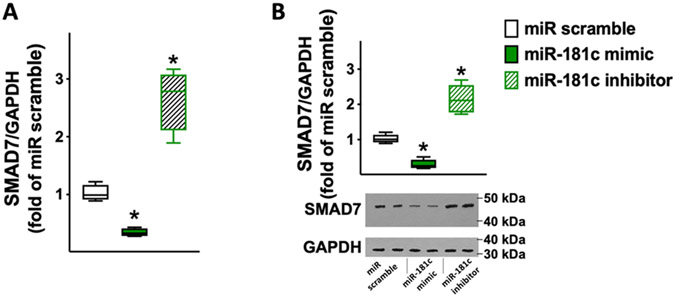
We validated, for the first time to the best of our knowledge, miR-181c as a functional modulator of PRKN and SMAD7 in human cardiac fibroblasts. We demonstrated via luciferase assays (Fig. 3) that PRKN and SMAD7 are specific molecular targets of miR-181c. Corroborating these findings, we also observed that miR-181c was able to reduce both mRNA and protein levels of PRKN (Fig. 4) and SMAD7 (Fig. 5).
Fig. 4.
PRKN expression in human cardiac fibroblasts is reduced by miR-181c and increased by miR-181c inhibitor. PRKN mRNA levels were measured using RT-qPCR (A) in human cardiac fibroblasts transfected with miR-181c mimic, inhibitor, or scramble (negative control) for 48 h, normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH); sequences of oligonucleotide primers are reported in Table 1. The results obtained via RT-qPCR were confirmed in terms of protein levels as shown by the representative immunoblots and their quantification (B). All experiments were performed at least in triplicate; the box-and-whiskers graphs indicate medians and 5th–95th percentiles; * : p < 0.01 vs. miR scramble.
Fig. 5.
SMAD7 expression in human cardiac fibroblasts is reduced by miR-181c and increased by miR-181c inhibitor. SMAD7 mRNA levels were measured using RT-qPCR (A) in human cardiac fibroblasts transfected with miR-181c mimic, inhibitor, or scramble (negative control) for 48 h, normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH); sequences of oligonucleotide primers are reported in Table 1. The results obtained via RT-qPCR were confirmed in terms of protein levels as shown by the representative immunoblots and their quantification (B). All experiments were performed at least in triplicate; the box-and-whiskers graphs indicate medians and 5th–95th percentiles; * : p < 0.01 vs. miR scramble.
4. Discussion
In this study, we have demonstrated that miR-181c is differentially expressed in frail elderly patients with DM and HFpEF compared to controls.
Several investigators have assessed the expression of miRNAs in HF patients, for instance when attempting to distinguish HFpEF from HFrEF (Florijn et al., 2018; Chen et al., 2019; Meng et al., 2023; Vilella-Figuerola, 2022). Our group has also identified a miRNA signature of endothelial dysfunction, a crucial feature of HFpEF, in patients with this disease (Mone et al., 2023). In the present study, we focused on miR-181c and we were able to identify two molecular targets essential in the pro-fibrotic process.
In line with our data, previous studies had indicated the importance of miR-181c in cardiovascular disorders (Morrison et al., 2021; Solly et al., 2021; Wang et al., 2015; Wang et al., 2020a). Chronic cardiac overexpression of miR-181c in rats using cationic nanovectors led to an impaired mitochondrial metabolism (Das et al., 2014), triggering mitochondrial Ca2+ overload and subsequent increased generation of reactive oxygen species (ROS). Equally important, the double-stranded RNA-dependent protein kinase activating protein (PACT), an RNA-binding protein that is part of the RNA-induced silencing complex, was recently shown to represent a post-transcriptional brake on mitochondrial biogenesis by promoting the maturation of miR-181c (Dogan et al., 2022). Nonetheless, the direct actions of miR-181c on cardiac fibroblasts had not been explored hitherto.
The importance of miR-181c in DM-related cardiovascular complications attributed to hyperglycemia is substantiated by the observation that this miR had been shown to promote high-glucose-induced dysfunction in human umbilical vein endothelial cells (Shen et al., 2018). Our results linking miR-181c with SMAD7 are consistent with a previous report showing that miR-181c regulates the formation of insulin-producing cells from human induced pluripotent stem cells (Li et al., 2020a). Similarly, the association of miR-181c and PRKN confirms the observation that miR-181a suppresses mitophagy in the human neuroblastoma cell line SH-SY5Y (Cheng et al., 2016).
We validated SMAD7 as a molecular target of miR-181c in human cardiac fibroblasts, providing a potential new target to combat fibrosis. The main molecular mechanism by which SMAD7 attenuates cardiac myofibroblast activation and reduces synthesis of structural and matricellular extracellular matrix proteins have been recently demonstrated by Humeres and colleagues (Humeres et al., 2022). Indeed, miR-181c relieves the pro-fibrotic process from the SMAD7-mediated inhibition, hence its antagonism would eventually reduce fibrosis.
4.1. Strengths and limitations
The major strength of this study is the combination of clinical investigations, in a homogeneous real-world population of elderly patients with HFpEF and DM, and basic research performed in vitro in human cells. This report is not exempt from limitations, including the lack of a comparison with a population of patients with HFrEF or HFmEF. Besides, our observations were limited to a population of elderly patients with DM. Further prospective investigations are needed to validate the actual relevance of miR-181c as a diagnostic and/or prognostic biomarker of HFpEF in the general population.
5. Conclusion
Taken together, our data demonstrate that circulating miR-181c levels are significantly augmented in frail elderly patients with DM and HFpEF compared to control subjects and that miR-181c targets PRKN and SMAD7 in human cardiac fibroblasts.
Acknowledgements
The authors thank Drs. Wilson and Wang for helpful discussion and technical assistance.
Funding
The Santulli’s Lab is currently supported in part by the National Institutes of Health (NIH): National Heart, Lung, and Blood Institute (NHLBI: R01-HL164772, R01-HL159062, R01-HL146691, T32-HL144456), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK: R01-DK123259, R01-DK033823), National Center for Advancing Translational Sciences (NCATS: UL1-TR002556–06, UM1-TR004400) to G.S., by the Diabetes Action Research and Education Foundation (to G.S.), and by the Monique Weill-Caulier and Irma T. Hirschl Trusts (to G.S.). S.S.J. is supported in part by a postdoctoral fellowship of the American Heart Association (AHA-21POST836407). F. V. is supported in part by a postdoctoral fellowship of the American Heart Association (AHA-22POST915561). U.K. is supported in part by a postdoctoral fellowship of the American Heart Association (AHA-23POST1026190). J.G. is supported by a postdoctoral fellowship of the American Heart Association (AHA-20POST35211151).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
CRediT authorship contribution statement
SSJ, PM, RA, FV, SDG, UK, GM, JG and MDM, were responsible for investigation, manuscript preparation, writing, analysis, tables, and figures under the supervision of GS. PM, LS, AC, SF, JG, TT, and GS gave a substantial contribution to the methodology, critical discussion, and preparation of the manuscript. The final manuscript was revised and approved by all authors.
Data Availability
Data will be made available upon reasonable request.
References
- Abudureyimu M, Luo X, Wang X, Sowers JR, Wang W, Ge J, Ren J, Zhang Y, 2022. Heart failure with preserved ejection fraction (HFpEF) in type 2 diabetes mellitus: from pathophysiology to therapeutics. J. Mol. Cell Biol 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroor AR, Mummidi S, Lopez-Alvarenga JC, Das N, Habibi J, Jia G, Lastra G, Chandrasekar B, DeMarco VG, 2021. Sacubitril/valsartan inhibits obesity-associated diastolic dysfunction through suppression of ventricular-vascular stiffness. Cardiovasc Diabetol. 20, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvisato R, Mone P, Jankauskas SS, Varzideh F, Kansakar U, Gambardella J, De Luca A, Matarese A, Santulli G, 2023. miR-4432 targets FGFBP1 in human endothelial cells. Biology (Basel) 12, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besler C, Rommel KP, Kresoja KP, Morbitz J, Kirsten H, Scholz M, Klingel K, Thiery J, Burkhardt R, Buttner P, Adams V, Thiele H, Lurz P, 2021. Evaluation of phosphodiesterase 9A as a novel biomarker in heart failure with preserved ejection fraction. ESC Heart Fail 8, 1861–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska A, Niemira M, Bauer W, Sidorkiewicz I, Szalkowska A, Skwarska A, Raczkowska J, Ostrowski D, Gugala K, Dobrzycki S, Kretowski A, 2022. Serum miRNA profile in diabetic patients with ischemic heart disease as a promising non-invasive biomarker. Front Endocrinol. (Lausanne) 13, 888948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Zhao C, Gong L, Cheng X, Yang J, Zhu M, Lv X, 2022. MiR-181 enhances proliferative and migratory potentials of retinal endothelial cells in diabetic retinopathy by targeting KLF6. Curr. Eye Res 47, 882–888. [DOI] [PubMed] [Google Scholar]
- Caravia XM, Roiz-Valle D, Moran-Alvarez A, Lopez-Otin C, 2017. Functional relevance of miRNAs in premature ageing. Mech. Ageing Dev 168, 10–19. [DOI] [PubMed] [Google Scholar]
- Cheng M, Liu L, Lao Y, Liao W, Liao M, Luo X, Wu J, Xie W, Zhang Y, Xu N, 2016. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 7, 42274–42287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli M, Sorriento D, Cipolletta E, Santulli G, Fusco A, Zhou RH, Eckhart AD, Peppel K, Koch WJ, Trimarco B, Iaccarino G, 2011. Impaired neoangiogenesis in beta(2)-adrenoceptor gene-deficient mice: restoration by intravascular human beta(2)-adrenoceptor gene transfer and role of NFkappaB and CREB transcription factors. Br. J. Pharm 162, 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta E, Rusciano MR, Maione AS, Santulli G, Sorriento D, Del Giudice C, Ciccarelli M, Franco A, Crola C, Campiglia P, Sala M, Gomez-Monterrey I, De Luca N, Trimarco B, Iaccarino G, Illario M, 2015. Targeting the CaMKII/ERK interaction in the heart prevents cardiac hypertrophy. PLoS One 10, e0130477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C, 2014. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One 9, e96820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan AE, Hamid SM, Yildirim AD, Yildirim Z, Sen G, Riera CE, Gottlieb RA, Erbay E, 2022. PACT establishes a posttranscriptional brake on mitochondrial biogenesis by promoting the maturation of miR-181c. J. Biol. Chem 298, 102050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, Seeman T, Tracy R, Kop WJ, Burke G, McBurnie MA, Cardiovascular Health Study Collaborative Research, G., 2001. Frailty in older adults: evidence for a phenotype. J. Gerontol. A Biol. Sci. Med Sci 56, M146–M156. [DOI] [PubMed] [Google Scholar]
- Fu X, Mishra R, Chen L, Arfat MY, Sharma S, Kingsbury T, Gunasekaran M, Saha P, Hong C, Yang P, Li D, Kaushal S, 2023. Exosomes mediated fibrogenesis in dilated cardiomyopathy through a MicroRNA pathway. iScience 26, 105963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco-Allison G, Li DK, Hunter B, Jackson D, Bannon PG, Lal S, O’Sullivan JF, 2021. Optimizing the discovery and assessment of therapeutic targets in heart failure with preserved ejection fraction. ESC Heart Fail 8, 3643–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella J, Fiordelisi A, Sorriento D, Cerasuolo F, Buonaiuto A, Avvisato R, Pisani A, Varzideh F, Riccio E, Santulli G, Iaccarino G, 2023. Mitochondrial microRNAs are dysregulated in patients with Fabry disease. J. Pharm. Exp. Ther 384, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MN, Leavens KF, Gadue P, 2021. Genome editing human pluripotent stem cells to model beta-cell disease and unmask novel genetic modifiers. Front Endocrinol. (Lausanne) 12, 682625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert AB, Witvrouwen I, Van Craenenbroeck AH, Van Laere SJ, Boen JRA, Van de Heyning CM, Belyavskiy E, Mueller S, Winzer E, Duvinage A, Edelmann F, Beckers PJ, Heidbuchel H, Wisloff U, Pieske B, Adams V, Halle M, Van Craenenbroeck EM, OptimEx-Clin Study G, 2021. miR-181c level predicts response to exercise training in patients with heart failure and preserved ejection fraction: an analysis of the OptimEx-Clin trial. Eur. J. Prev. Cardiol 28, 1722–1733. [DOI] [PubMed] [Google Scholar]
- Gu J, Fan YQ, Bian L, Zhang HL, Xu ZJ, Zhang Y, Chen QZ, Yin ZF, Xie YS, Wang CQ, 2016. Long-term prescription of beta-blocker delays the progression of heart failure with preserved ejection fraction in patients with hypertension: a retrospective observational cohort study. Eur. J. Prev. Cardiol 23, 1421–1428. [DOI] [PubMed] [Google Scholar]
- He X, Gao X, Peng L, Wang S, Zhu Y, Ma H, Lin J, Duan DD, 2011. Atrial fibrillation induces myocardial fibrosis through angiotensin II type 1 receptor-specific Arkadia-mediated downregulation of Smad7. Circ. Res 108, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeres C, Shinde AV, Hanna A, Alex L, Hernandez SC, Li R, Chen B, Conway SJ, Frangogiannis NG, 2022. Smad7 effects on TGF-beta and ErbB2 restrain myofibroblast activation and protect from postinfarction heart failure. J. Clin. Invest 132 e146926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Lu W, Tian Y, Hou Q, Man HY, 2022. Prkn knockout mice show autistic-like behaviors and aberrant synapse formation. iScience 25, 104573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva M, Panella R, Uchida S, 2022. MicroRNAs in cancer and cardiovascular disease. Cells 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo C, Visco V, Gambardella J, Ferruzzi GJ, Rispoli A, Rusciano MR, Toni AL, Virtuoso N, Carrizzo A, Di Pietro P, Iaccarino G, Vecchione C, Ciccarelli M, 2023. Cardiovascular Implications of microRNAs in Coronavirus Disease 2019. J. Pharm. Exp. Ther 384, 102–108. [DOI] [PubMed] [Google Scholar]
- Jin Y, Chen Z, Liu X, Zhou X, 2013. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol. Biol 936, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusic A, Thomas PB, Wettinger SB, Dogan S, Farrugia R, Gaetano C, Tuna BG, Pinet F, Robinson EL, Tual-Chalot S, Stellos K, Devaux Y, 2022. Noncoding RNAs in age-related cardiovascular diseases. Ageing Res Rev. 77, 101610. [DOI] [PubMed] [Google Scholar]
- Kansakar U, Gambardella J, Varzideh F, Avvisato R, Jankauskas SS, Mone P, Matarese A, Santulli G, 2022. miR-142 Targets TIM-1 in human endothelial cells: potential implications for stroke, COVID-19, Zika, Ebola, Dengue, and other viral infections. Int J. Mol. Sci 23, 10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Araya J, Minagawa S, Hara H, Saito N, Kadota T, Sato N, Yoshida M, Tsubouchi K, Kurita Y, Ito S, Fujita Y, Takasaka N, Utsumi H, Yanagisawa H, Hashimoto M, Wakui H, Kojima J, Shimizu K, Numata T, Kawaishi M, Kaneko Y, Asano H, Yamashita M, Odaka M, Morikawa T, Nakayama K, Kuwano K, 2016. Involvement of PARK2-mediated mitophagy in idiopathic pulmonary fibrosis pathogenesis. J. Immunol 197, 504–516. [DOI] [PubMed] [Google Scholar]
- Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson AB, 2015. PINK1 is dispensable for mitochondrial recruitment of parkin and activation of mitophagy in cardiac myocytes. PLoS One 10 e0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y, Ito K, Yoshioka Y, Sakai S, 2023. Association of complication of type 2 diabetes mellitus with hemodynamics and exercise capacity in patients with heart failure with preserved ejection fraction: a case-control study in individuals aged 65-80 years. Cardiovasc. Diabetol 22, 97 e0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AA, Kelly DP, Chirinos JA, 2019. Mitochondrial dysfunction in heart failure with preserved ejection fraction. Circulation 139, 1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Engelhardt S, 2022. MicroRNAs as therapeutic targets in cardiovascular disease. J. Clin. Invest 132 e159179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitao AL, Enguita FJ, 2022. A Structural View of miRNA Biogenesis and Function. Noncoding. RNA 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Jiang D, He Q, He F, Li Y, Deng C, Li F, 2020a. microRNA-181c-5p promotes the formation of insulin-producing cells from human induced pluripotent stem cells by targeting smad7 and TGIF2. Cell Death Dis. 11, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhong J, Zeng Z, Wang H, Li J, Liu X, Yang X, 2020b. MiR-181c protects cardiomyocyte injury by preventing cell apoptosis through PI3K/Akt signaling pathway. Cardiovasc Diagn. Ther 10, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florijn BW, Bijkerk R, van der Veer EP, van Zonneveld AJ, 2018. Gender and cardiovascular disease: are sex-biased microRNA networks a driving force behind heart failure with preserved ejection fraction in women? Cardiovasc Res. 114, 210–225. [DOI] [PubMed] [Google Scholar]
- Macvanin MT, Gluvic Z, Radovanovic J, Essack M, Gao X, Isenovic ER, 2023. Diabetic cardiomyopathy: The role of microRNAs and long non-coding RNAs. Front Endocrinol. (Lausanne) 14, 1124613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese A, Gambardella J, Lombardi A, Wang X, Santulli G, 2020. miR-7 Regulates GLP-1-mediated insulin release by targeting beta-arrestin 1. Cells 9, 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher P, Adam M, Civitarese R, Bugyei-Twum A, Connelly KA, 2018. Heart failure with preserved ejection fraction in diabetes: mechanisms and management. Can. J. Cardiol 34, 632–643. [DOI] [PubMed] [Google Scholar]
- Menezes Junior ADS, Ferreira LC, Barbosa LJV, Silva DME, Saddi VA, Silva A, 2023. Circulating MicroRNAs as specific biomarkers in atrial fibrillation: a meta-analysis. Noncoding RNA 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone P, Gambardella J, Pansini A, de Donato A, Martinelli G, Boccalone E, Matarese A, Frullone S, Santulli G, 2021a. Cognitive impairment in frail hypertensive elderly patients: role of hyperglycemia. Cells 10, 2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone P, Gambardella J, Wang X, Jankauskas SS, Matarese A, Santulli G, 2021b. miR-24 targets the transmembrane glycoprotein neuropilin-1 in human brain microvascular endothelial cells. NonCoding RNA 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone P, Lombardi A, Gambardella J, Pansini A, Macina G, Morgante M, Frullone S, Santulli G, 2022. Empagliflozin improves cognitive impairment in frail older adults with type 2 diabetes and heart failure with preserved ejection fraction. Diabetes Care 45, 1247–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone P, Lombardi A, Kansakar U, Varzideh F, Jankauskas SS, Pansini A, Marzocco S, De Gennaro S, Famiglietti M, Macina G, Frullone S, Santulli G, 2023. Empagliflozin improves the MicroRNA signature of endothelial dysfunction in patients with heart failure with preserved ejection fraction and diabetes. J. Pharm. Exp. Ther 384, 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordi IR, Tee A, Palmer CN, McCrimmon RJ, Doney ASF, Lang CC, 2020. Microvascular disease and heart failure with reduced and preserved ejection fraction in type 2 diabetes. ESC Heart Fail 7, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison KR, Solly EL, Shemesh T, Psaltis PJ, Nicholls SJ, Brown A, Bursill CA, Tan JTM, 2021. Elevated HDL-bound miR-181c-5p level is associated with diabetic vascular complications in Australian Aboriginal people. Diabetologia 64, 1402–1411. [DOI] [PubMed] [Google Scholar]
- Chen YT, Wong LL, Liew OW, Richards AM, 2019. Heart Failure with Reduced Ejection Fraction (HFrEF) and Preserved Ejection Fraction (HFpEF): The Diagnostic Value of Circulating MicroRNAs. Cells 16, 1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, Galeazzi R, Abbatecola AM, Marcheselli F, Monti D, Ostan R, Cevenini E, Antonicelli R, Franceschi C, Procopio AD, 2012. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech. Ageing Dev 133, 675–685. [DOI] [PubMed] [Google Scholar]
- Omote K, Verbrugge FH, Borlaug BA, 2022. Heart failure with preserved ejection fraction: mechanisms and treatment strategies. Annu Rev. Med 73, 321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DC, Wan M, Lohman K, Hou L, Nguyen AT, Ding J, Bertoni A, Shea S, Burke GL, Jacobs DR, Post W, Corcoran D, Hoeschele I, Parks JS, Liu Y, 2022. Monocyte miRNAs are associated with type 2 diabetes. Diabetes 71, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus WJ, Zile MR, 2021. From systemic inflammation to myocardial fibrosis: the heart failure with preserved ejection fraction paradigm revisited. Circ. Res 128, 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Kim YS, 2020. PINK1-PRKN mitophagy suppression by mangiferin promotes a brown-fat-phenotype via PKA-p38 MAPK signalling in murine C3H10T1/2 mesenchymal stem cells. Metabolism 107, 154228. [DOI] [PubMed] [Google Scholar]
- Rozhkov AN, Shchekochikhin DY, Ashikhmin YI, Mitina YO, Evgrafova VV, Zhelankin AV, Gognieva DG, Akselrod AS, Kopylov PY, 2022. The profile of circulating blood microRNAs in outpatients with vulnerable and stable atherosclerotic plaques: associations with cardiovascular risks. noncoding. RNA 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Wang X, Mone P, 2022. Updated ACC/AHA/HFSA 2022 guidelines on heart failure: what is new? From epidemiology to clinical management. Eur Heart J Cardiovasc Pharmacother. 11, e23–e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Wronska A, Uryu K, Diacovo TG, Gao M, Marx SO, Kitajewski J, Chilton JM, Akat KM, Tuschl T, Marks AR, Totary-Jain H, 2014. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J. Clin. Invest 124, 4102–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Pagano G, Sardu C, Xie W, Reiken S, D’Ascia SL, Cannone M, Marziliano N, Trimarco B, Guise TA, Lacampagne A, Marks AR, 2015. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J. Clin. Invest 125, 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Li Y, Sun G, Guo D, Bai X, 2018. miR-181c-3p and -5p promotes high-glucose-induced dysfunction in human umbilical vein endothelial cells by regulating leukemia inhibitory factor. Int J. Biol. Macromol 115, 509–517. [DOI] [PubMed] [Google Scholar]
- Shi K, Yang MX, Huang S, Yan WF, Qian WL, Li Y, Guo YK, Yang ZG, 2021. Effect of diabetes mellitus on the development of left ventricular contractile dysfunction in women with heart failure and preserved ejection fraction. Cardiovasc Diabetol. 20, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Taguchi A, Higashijima Y, Takubo N, Kanki Y, Urade Y, Wada Y, 2020. PERK-mediated suppression of microRNAs by sildenafil improves mitochondrial dysfunction in heart failure. iScience 23, 101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu M, Xiang C, 2022. Relationship between peripheral Blood miR-181c, miR-101, and cognitive impairment in patients with diabetes mellitus complicated with acute stroke. Emerg. Med Int 2022, 5777106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Solly EL, Psaltis PJ, Bursill CA, Tan JTM, 2021. The role of miR-181c in mechanisms of diabetes-impaired angiogenesis: an emerging therapeutic target for diabetic vascular complications. Front Pharm. 12, 718679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M, Corden B, Cook SA, 2020. Targeting cardiac fibrosis in heart failure with preserved ejection fraction: mirage or miracle? EMBO Mol. Med 12, e10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XH, Gambardella J, Jankauskas S, Wang X, Santulli G, Gudas LJ, Levi R, 2021. A retinoic acid receptor beta 2 agonist improves cardiac function in a heart failure model. J. Pharm. Exp. Ther 379, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto T, Kajio H, 2018. Efficacy of renin-angiotensin system inhibitors for patients with heart failure with preserved ejection fraction and mild to moderate chronic kidney disease. Eur. J. Prev. Cardiol 25, 1268–1277. [DOI] [PubMed] [Google Scholar]
- Meng C, Chai K, Li YY, Luo Y, Wang H, Yang JF, 2023. Prevalence and prognosis of frailty in older patients with stage B heart failure with preserved ejection fraction. ESC Heart Fail 10, 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li J, Chi H, Zhang F, Zhu X, Cai J, Yang X, 2015. MicroRNA-181c targets Bcl-2 and regulates mitochondrial morphology in myocardial cells. J. Cell Mol. Med 19, 2084–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ge L, Zhang D, Wang L, Liu H, Ye X, Liang W, Li J, Ma H, Cai Y, Xia Z, 2020a. MiR-181c-5p promotes inflammatory response during hypoxia/reoxygenation injury by downregulating protein tyrosine phosphatase nonreceptor type 4 in H9C2 cardiomyocytes. Oxid. Med Cell Longev 2020, 7913418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Morelli MB, Matarese A, Sardu C, Santulli G, 2020b. Cardiomyocyte-derived exosomal microRNA-92a mediates post-ischemic myofibroblast activation both in vitro and ex vivo. ESC Heart Fail 7, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech M, Barandiarán AA, van Empel V, van Bilsen M, Schroen B, 2018. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Cardiovasc Res. 114, 782–793.29462282 [Google Scholar]
- Vilella-Figuerola A, Gallinat A, Escate R, Mirabet S, Padró T, Badimon L, 2022. Systems Biology in Chronic Heart Failure-Identification of Potential miRNA Regulators. Int J Mol Sci. 23, 15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wronska A, 2023. The Role of microRNA in the development, diagnosis, and treatment of cardiovascular disease: recent developments. J. Pharm. Exp. Ther 384, 123–132. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Chen Z, Santulli G, Gu L, Yang ZG, Yuan ZQ, Zhao YT, Xin HB, Deng KY, Wang SQ, Ji G, 2014. Functional role of Calstabin2 in age-related cardiac alterations. Sci. Rep 4, 7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available upon reasonable request.