Abstract
Multicyclic peptides are appealing candidates for peptide-based drug discovery. While various methods are developed for peptide cyclization, few allow multi-cyclization of native peptides. Herein we report a novel crosslinker DCA-RMR1, which elicits facile bicyclization of native peptides via N-terminus-Cys-Cys crosslinking. The bicyclization is fast, affords quantitative conversion and tolerates various side chain functionalities. Importantly, the resulting diazaborine linkage, while stable at neutral pH, can readily reverse upon mild acidification to give pH-responsive peptides.
Graphical Abstarct

Multicyclic peptides have shown promise in peptide-based drug discovery and as a toolkit in chemical biology.1 The greater conformational rigidity and metabolic stability make multicyclic peptides appealing candidates for peptide-based drug development.2 An increasing number of multicyclic peptides are found to show antibody-like affinity and specificity for challenging protein targets such as protein-protein interactions.3 Importantly, selected multicyclic peptides have shown remarkable oral availability, which presents an enormous advantage over antibody-based therapeutics.4 Although there are various methods developed for peptide cyclization, few methods exist for multi-cyclization of native peptides.5 Furthermore, facile multi-cyclization of native peptides is highly desirable for constructing genetically encoded peptide libraries to streamline molecular discovery.6
Cyclization of native peptides calls for efficient and chemoselective conjugation of protein nucleophiles.7 Not surprisingly, known examples of native peptide cyclization largely rely on Cys-Cys crosslinking due to the superior nucleophilicity of the thiol group (Figure 1a).8 Biocompatible and chemoselective reactions for other protein nucleophiles remain scarce.9 We recently reported a novel lysine conjugation, in which a synthetic warhead, RMR1, exclusively reacts with a primary amine to give a diazaborine hydrate.10 Furthermore, RMR1 was found to react with an unhindered amine such as a lysine side chain over the alpha-amine of the N-terminus (Figure 1b). However, we report herein that RMR1 can react with N-terminal amine efficiently in an intramolecular setting to give cyclic peptides. In particular, when conjugated with a Cys-Cys crosslinker, RMR1 elicits N-terminus-Cys-Cys crosslinking to render bicyclization of native peptides with quantitative yields (Figure 1c).
Figure 1:

Native peptide bicyclization. (a) Cysteine mediated bicyclization of native peptide in aqueous conditions (b) Diazaborine chemistry for targeting amines (c) Present work on diazaborine mediated bicyclization on native peptides.
RMR1 conjugates with primary amines to give diazaborines via an imine intermediate. This conjugation mechanism affords exclusive selectivity to amines and full autonomous reversibility, with the conjugate dissociating over the time scale of hours.10 In contrast, salicylaldehyde and 2-formyl/acetyl-phenylboronic acid (2-FPBA/2-APBA), which are also known to reversibly conjugate with amines, give imine conjugates of much lower kinetic stability. Consistent with the kinetic stability of the diazaborines, RMR1 binds amines with at least 10 times greater thermodynamic propensity in comparison to salicylaldehyde or APBA.10,11 These attributes of the diazaborine chemistry motivated us to investigate its use for peptide cyclization. To this end, we designed a bifunctional molecule DCA-RMR1 (Figure 1c), which derivatizes RMR1 with 1,3-dichloroacetone (DCA), a moiety commonly used for Cys-Cys crosslinking.12 Note that DCA reacts with Cys with a rate of 0.1-1 M−1s−1 (reference 13), while the RMR1-amine conjugation is >10 times slower (k2: ~10−2 M−1s−1, reference 10). We envisioned that this hierarchical reactivity will ensure quick Cys-Cys crosslinking followed by RMR1 conjugation to proximal amines.
The synthesis of DCA-RMR1 is summarized in Scheme 1 with more details given in the ESI (Scheme S1–S4). With DCA-RMR1 in hand, we first investigated its potential to elicit peptide bicyclization using a CX6C peptide (CPFPSAWC) revealed from our earlier sortase inhibition studies.12 A Gly residue was installed onto the N-terminus of this peptide (Figure 2a) as the glycine amine is known to conjugate with RMR1 efficiently.10 The resulting peptide P1 was incubated with DCA-RMR1 in PBS (pH ~8) and the bicyclization reaction was monitored via LC-MS. Satisfyingly, clean conjugation and efficient bicyclization was observed after 3 hr incubation, giving 90% of the bicyclic product (Figure 2b, S2). It is important to note that the 10% peptide remaining was found to be disulfide-cyclized. In that sense, the diazaborine-mediated bicyclization is quantitative as no monocyclic peptide was observed. We further synthesized the peptide P2, which carries an N-terminal Ala. P2 also underwent quantitative bicyclization despite the reduced propensity of RMR1 to bind hindered amines (Figure 2c, S3–4). In contrast, the N-acetylated P2 just afforded a Cys-Cys crosslinked monocyclic peptide (Figure S5), consistent with the N-terminus mediated bicyclization of P2. The kinetic profiles of bicyclization were recorded by monitoring the UV-vis absorption of the diazaborine conjugate, which fits nicely to a single exponential equation (Figure S6). The curve fitting gives a t1/2 of 21.7 and 27.6 min for P1 and P2 respectively.
Scheme 1:

Synthesis of DCA-RMR1
Figure 2:
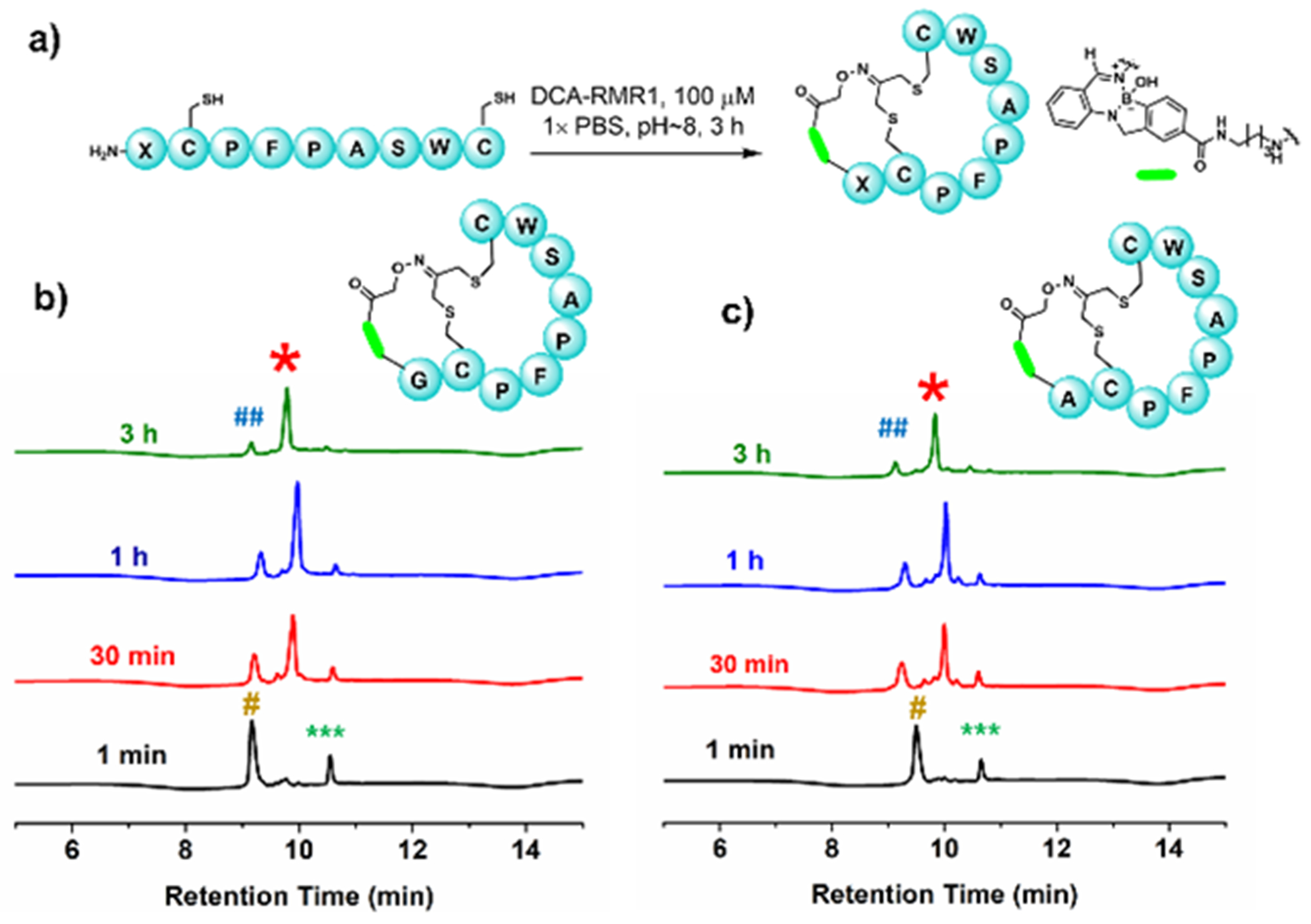
DCA-RMR1 elicited peptide bicyclization. (a) Bicyclization reaction using DCA-RMR1 on P1 (X = G) and P2 (X = A). (b) and (c) LC traces of P1 and P2 bicyclization over time (recorded with absorption at 280 nm). # and *** denotes linear peptide and DCA-RMR1, respectively. * represents the desired bicyclic product and ## denotes the disulfide-cyclized peptide.
Motivated by the promise of bicyclic peptide libraries for inhibitor discovery,14 We investigated the applicability of our DCA-RMR1 mediated cyclization to diverse peptide sequences. Specifically, we synthesized a series of peptides (Figure 3), which harbor a varied N-terminal residue as well as varied ring sizes of cyclization. All peptides were incubated with DCA-RMR1 at 100 μM concentration (PBS, pH ~8). The efficiency and kinetics of bicyclization were respectively measured using LC-MS and UV-vis absorption. The LC-MS results (Figure 3a–c, S8–12) revealed efficient bicyclization for all peptides with an N-terminal Gly or Ala (P3, P4-G/A, and P5-G/A), regardless of the resulting ring sizes or the exact peptide sequences. Furthermore, all these peptides exhibited comparable kinetics of bicyclization, although the P4-A and P5-A appeared to be slighter slower (by a factor of 2) than P4/5-G. We then expanded our investigation of the N-terminal residue effect on bicyclization using additional P4/5-X peptides. Interestingly, while most N-terminal residues elicited efficient bicyclization (Figure 3g), an N-terminal Thr caused some erosion of bicyclization efficiency, particularly within the P5 peptide series. It is important to note that, in all reactions, the only side products observed are the monocyclic peptides with unreacted RMR1 and the disulfide-cyclized peptides at low percentages (Figure 3d–e, S13–14). This less favorable bicyclization of P5-T is presumably due to the steric hindrance of the β-branched side chain of Thr as well as the expanded ring of the RMR1-N-terminus cyclization.
Figure 3:

Bicyclization of diverse peptide sequences. (a-c) LC traces (absorption at 280 nm) of the bicyclization reaction of P3, P4-G, and P5-A respectively. (d) and (e) LC traces (absorption at 280 nm) of selected bicyclized P4 and P5 peptides. * Represents the desired bicyclized peptides. Whereas # and *** denote the linear peptide and DCA-RMR1 (starting materials) respectively. ## and ** denote a corresponding disulfide-cyclized peptide monocyclic (RMR1 unreacted) peptide, respectively. (f) Kinetic parameters of the bicyclization reaction on various peptides. (g) N-terminal residue effect on bicyclization. All the bicyclization reactions were carried out at 100 μM conc. of both peptide and DCA-RMR1 in 1× PBS, pH 8.
The RMR1-amine conjugation is fully and autonomously reversible with the diazaborine dissociation happening on the time scale of hours.10 To assess the implications of this reversibility in biological milieu, we first tested the stability of diazaborine-cyclized peptides in the presence of lysine as potential competitors. Specifically, we mixed the bicyclized P2 with lysine at 10 mM and 100 mM respectively. The lysine-triggered peptide uncyclization was recorded using LC-MS analysis, which revealed negligible uncyclization induced by 10 mM lysine over 27 h incubation. Even at 100 mM, lysine only resulted in ~20% uncyclization with ~80 % bicyclic peptide remaining (Figure S16). These results suggest the diazaborine-cyclized peptides are quite resistant to the attack of endogenous amines. This robust stability is perhaps not surprising given the intramolecular nature of the diazaborine-mediated cyclization.
Although resistant to lysine competition, we found that the diazaborine-mediated cyclization could be readily reversed upon acidification. To this end, we first examined the pH dependence of RMR1-lysine conjugation using 1H-NMR. Specifically, the 1H-NMR signature of a 10 mM RMR1-lysine mixture was recorded under varied pH’s (Figure S17). The results clearly show that lower pH triggered dissociation of the diazaborine conjugate. Such pH tunable properties have found wide use in the development of smart peptides for drug delivery and nanotechnological applicatioins.15,16 This prospect motivated us to further explore the pH dependence of the DCA-RMR1 mediated peptide bicyclization (Figure 4a). While the Cys-Cys crosslinking by DCA is irreversible, the diazaborine-bicyclized peptides can be reversed back to its monocyclic forms. To study this monocycle-bicycle transition, we incubated the bicyclized P2 in buffers of varied pH and determined the extent of bicyclic peptide remaining using LC-MS. Consistent with the pH dependence of the RMR1-lysine conjugation, the diazaborine-bicyclized peptide appeared as the dominant species in neutral and basic pH, while the monocyclic peptide was found to be the major product under acidic conditions (Figure 4b, S18). Quantitative analysis of the monocycle-bicycle conversion revealed a mid-point of transition around pH 6, which is quite relevant to the local environment of endosomes, lysosomes as well as selected tumor tissues. The kinetics of the uncyclization at acidic pH was measured by dissolving the bicyclic P2 in a pH 3 buffer and monitoring the UV-Vis absorbance at 425 nm over time. Fitting the kinetic profile to a single exponential equation yielded a t1/2 of 81.9 ± 1.0 min, which is an order of magnitude faster than the diazaborine dissociation at neutral conditions (Figure 4c).10
Figure 4:

pH dependent bicyclization of DCA-RMR1 cyclized P2. (a) A schematic illustration. (b) pH dependent bicyclization efficiency determined via LC-MS analysis. (c) Kinetic profile of bicycle to monocycle transformation upon acidification (1× PBS, pH 3) recorded by monitoring the diazaborine absorbance (425 nm). All the experiments were set up with 100 μM of peptide.
In conclusion, we report herein a facile peptide bicyclization strategy, in which a trifunctional crosslinker DCA-RMR1 elicits efficient N-terminus-Cys-Cys crosslinking under physiological conditions. Note that having lysines in the peptide sequence can complicate the bicyclization due to their competition against the N-terminus. This method furnishes near quantitative bicyclization for diverse peptide sequences and tolerates various side chain functionalities on native peptides. The peptide bicycles feature a novel and rigid diazaborine crosslink, which results from the reversible conjugation of RMR1 with the N-terminal amine of the peptides. Although reversible, the diazaborine linkage exhibits robust stability under neutral and basic conditions, showing strong resistance to the attack of endogenous amines. However, the diazaborine-mediated cyclization can be readily reversed upon mild acidification, giving rise to a new strategy of designing pH-responsive peptides for biological applications.
Supplementary Material
ACKNOWLEDGMENT
We thank National Institutes of Health (GM124231), National Science Foundation (CHE-2204078), and the Ono Pharma Foundation for the funding to support this work.
Footnotes
Supporting Information Statement
The Supporting Information is available free of charge on the ACS Publications website. Typical experimental procedures, peptide bicyclization procedures, LC-MS trace of the peptides and characterization for all compounds (PDF)
Data Availability Statement
The data underlying this study are available in the published article and its Supporting Information.
REFERENCES
- 1.(a) Vinogradov AA; Yin Y; Suga H Macrocyclic peptides as drug candidates: recent progress and remaining challenges. J. Am. Chem. Soc 2019, 141, 4167–4181. [DOI] [PubMed] [Google Scholar]; (b) Muttenthaler M; King GF; Adams DJ; Alewood PF Trends in peptide drug discovery. Nat. Rev. Drug Discov 2021, 20, 309–325. [DOI] [PubMed] [Google Scholar]
- 2.(a) Baeriswyl V; Heinis C Polycyclic peptide therapeutics. ChemMedChem 2013, 8, 377–384. [DOI] [PubMed] [Google Scholar]; (b) Zorzi A; Deyle K; Heinis C Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol 2017, 38, 24–29. [DOI] [PubMed] [Google Scholar]; (c) Wang CK; Craik DJ Cyclic peptide oral bioavailability: Lessons from the past. Biopolymers 2016, 106, 901–909. [DOI] [PubMed] [Google Scholar]
- 3.(a) Milroy LG; Grossmann TN; Hennig S; Brunsveld L; Ottmann C Modulators of protein-protein interactions. Chem. Rev 2014, 114, 4695–4748. [DOI] [PubMed] [Google Scholar]; (b) Passioura T; Katoh T; Goto Y; Suga H Selection-based discovery of druglike macrocyclic peptides. Annu. Rev. Biochem 2014, 83, 727–752. [DOI] [PubMed] [Google Scholar]
- 4.(a) Owens B Faster, deeper, smaller-the rise of antibody-like scaffolds. Nat. Biotechnol 2017, 35, 602–603. [DOI] [PubMed] [Google Scholar]; (b) Morrison C Constrained peptides’ time to shine? Nat. Rev. Drug Discov 2018, 17, 531–533. [DOI] [PubMed] [Google Scholar]
- 5.(a) Reguera L; Rivera DG Multicomponent reaction toolbox for peptide macrocyclization and stapling. Chem. Rev 2019, 119, 9836–9860. [DOI] [PubMed] [Google Scholar]; (b) White CJ; Yudin AK Contemporary strategies for peptide macrocyclization. Nat. Chem 2011, 3, 509–524. [DOI] [PubMed] [Google Scholar]
- 6.(a) Heinis C; Rutherford T; Freund S; Winter G Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol 2009, 5, 502–507. [DOI] [PubMed] [Google Scholar]; (b) Heinis C; Winter G Encoded libraries of chemically modified peptides. Curr. Opin. Chem. Biol 2015, 26, 89–98. [DOI] [PubMed] [Google Scholar]; (c) Wong JY-K; Mukherjee R; Miao J; Bilyk O; Triana V; Miskolzie M; Henninot A; Dwyer JJ; Kharchenko S; Iampolska A; Volochnyuk DM; Lin Y-S; Postovit L-M; Derda R Genetically-encoded discovery of proteolytically stable bicyclic inhibitors for morphogen NODAL. Chem. Sci 2021, 12, 9694–9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Yudin AK Macrocycles: lessons from the distant past, recent developments, and future directions. Chem. Sci 2015, 6, 30–49. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Derda R; Jafari MR Synthetic cross-linking of peptides: molecular linchpins for peptide cyclization. Protein Pept. Lett 2018, 25, 1051–1075. [DOI] [PubMed] [Google Scholar]; (c) Zhang Y; Zhang Q; Wong CTT; Li X Chemoselective peptide cyclization and bicyclization directly on unprotected peptides. J. Am. Chem. Soc 2019, 141, 12274–12279. [DOI] [PubMed] [Google Scholar]
- 8.(a) Bechtler C; Lamers C Macrocyclization strategies for cyclic peptides and peptidomimetics. RSC. Med. Chem 2021, 12, 1325– 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Oppewal TR; Jansen ID; Hekelaar J; Mayer C A strategy to select macrocyclic peptides featuring asymmetric molecular scaffolds as cyclization units by phage display. J. Am. Chem. Soc 2022, 144, 3644–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chen F-J.; Zheng M; Nobile V; Gao J. Chem. -Eur. J 2022, 28, e2022000. [Google Scholar]
- 9.Tamura T; Hamachi I Chemistry for covalent modification of endogenous/native proteins: from test tubes to complex biological systems. J. Am. Chem. Soc 2019, 141, 2782–2799. [DOI] [PubMed] [Google Scholar]
- 10.Reja RM; Wang W; Lyu Y; Haeffner F; Gao J Lysine targeting reversible covalent inhibitors with long residence time. J. Am. Chem. Soc 2022, 144, 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cambray S; Gao J Versatile conjugation chemistries of ortho-boronyl aryl ketones and aldehydes. Acc. Chem. Res 2018, 51, 2198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Zheng M; Chen F-J; Li K; Reja RM; Haeffner F; Gao J Lysine-targeted reversible covalent ligand discovery for proteins via phagedisplay. J. Am. Chem. Soc 2022, 144, 15885–15893. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen S; Lovell S; Lee S; Fellner M; Mace PD; Bogyo M Identification of highly selective covalent inhibitors by phage display. Nat. Biotechnol 2021, 39, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ng S; Derda R Phage-displayed macrocyclic glycopeptide libraries. Org. Biomol. Chem 2016, 14, 5539–5545. [DOI] [PubMed] [Google Scholar]
- 13.Chen F-J.; Gao J. Fast cysteine bioconjugation chemistry. Chem. Eur. J 2022, 28, e202201843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diderich P; Heinis C Directed evolution of bicyclic peptides for therapeutic application. CHIMIA 2013, 67, 910–915. [DOI] [PubMed] [Google Scholar]
- 15.Branco MC; Sigano DM; Schneider JP Materials from peptide assembly: towards the treatment of cancer and transmittable disease. Curr. Opin. Chem. Biol 2011, 15, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Zhou J; Xu B Enzyme-instructed self-assembly: a multistep process for potential cancer therapy. Bioconjugate Chem. 2015, 26, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bandyopadhyay A; Gao J Iminoboronate-based peptide cyclization that responds to pH, oxidation, and small molecule modulators. J. Am. Chem. Soc 2016, 138, 2098–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this study are available in the published article and its Supporting Information.


