Abstract
Objectives:
To characterize the oral microbiota among middle-aged men and identify differences between men with a prevalent oral high-risk (oncogenic) HPV infection and those without.
Materials and Methods:
This was a case-control study nested within a prospective screening study for HPV-related cancers among middle-aged men. 16S rRNA sequencing was used to characterize the oral microbiota and the cobas HPV Test was used to detect presence of oral high-risk HPV types. We determined the overall composition of the oral microbiota and assessed differences in relative abundance of bacterial taxa as well as alpha and beta diversity among men with a prevalent oral high-risk HPV infection compared to men who were HPV-negative.
Results:
Among 13 high-risk HPV-positive and 30 HPV-negative men, we found significant differences in beta diversity but not alpha diversity. Fretibacterium, F0058, Kingella, Treponema, and Prevotella were more abundant among the high-risk HPV-positive men while Neisseria and Lactobacillus were more abundant among the HPV-negative men.
Conclusion:
This study adds to the evidence that the oral microbiota varies according to oral HPV infection status and may be associated with the natural history of oral HPV infection.
Keywords: 16S rRNA sequencing, HPV, human papillomavirus infection, oral microbiome, oral microbiota, oropharynx, case-control studies
Introduction
Human papillomavirus (HPV) is a common sexually transmitted infection that causes cancers of the anogenital region and oropharynx. There has been a substantial increase in the incidence of oropharyngeal cancer (OPC) over the last several decades in the United States and other high-income countries, attributed to an increase in the proportion of HPV-positive OPC [1–3].
It has been established that a persistent infection with high-risk (oncogenic) HPV is required for malignant transformation [4]. Although an incident HPV infection is normally cleared within several months, inability of the immune system to clear an infection will lead to persistence that in some individuals can progress to cancer. The natural history of HPV is influenced by the microbial environment, and dysregulation of the local microbiome with a shift to a pro-inflammatory state may increase the risk for cancer [5, 6].
The “oral microbiota,” the full complement of microorganisms that live in the oral cavity and oropharynx, is among the most diverse in the human body [7]. Influenced by genetics, diet, and environment, it has long been known to affect both local and systemic health states, and dysregulation may predispose an individual to disease, including cancer [8, 9]. As with genital HPV infection and progression to cervical cancer, we hypothesize that microbial dysbiosis also plays a significant role in oral HPV infection and progression to OPC. Therefore, understanding how the oral microbiota affects the natural history of oral HPV infection and predisposition to HPV-related OPC will help inform future prevention efforts. The goal of this study was to characterize the oral microbiota among a cohort of middle-aged men and to identify variations according to prevalent oral high-risk HPV infection status.
Materials and Methods
Study design and population
This was a case-control study nested within the HPV-related Oropharyngeal and Uncommon Cancers Screening Trial of Men (HOUSTON), a prospective screening trial for HPV-related cancers among middle-aged men (NCT02897427) [10, 11]. In brief, 553 men aged 50–64 years without a history of HPV-related cancer were enrolled between April 2017 and December 2019. At the initial study visit, study subjects completed a self-administered questionnaire pertaining to demographic and socioeconomic characteristics, alcohol and tobacco exposure, sexual behaviors, and history of sexually transmitted diseases, including HPV-related disease in partners and themselves. They also provided blood and oral rinse samples for biologic testing. Because the collection of samples for microbiome analysis was started later, only samples from subjects enrolled after June 2018 were included in this study. The study was approved by the institutional review board and all subjects provided written informed consent.
Laboratory methods
Oral rinse collection and HPV DNA detection:
Participants were asked to swish and gargle 10 ml of Scope mouthwash (Procter & Gamble, Cincinnati, OH) for 15 seconds each. Samples were immediately placed at 4°C and were processed within the same day. For microbiome analysis, a 1 ml aliquot was saved at −80°C prior to processing. The remaining sample was washed with 10 ml of phosphate buffered saline (PBS) and resuspended in 1 ml of PBS. The cobas HPV Test (Roche Diagnostics, Indianapolis, IN) was used to detect high-risk HPV types. The assay provides individual results for HPV16 and HPV18 and pooled results for 12 other high-risk types (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68).
Microbial DNA extraction and bacterial 16S rRNA gene sequencing:
One milliliter of oral rinse was centrifuged at 3000 g for 10 minutes. The cell pellet was processed in 200 μl cell lysis buffer (20 mM Tris-Cl [pH 8], 2 mM EDTA, 1.2% Triton X-100) supplemented with lysozyme and mutanolysin at 37°C for 30 minutes, followed by addition of proteinase K and Buffer AL and incubation at 56°C for 30 minutes. Bacterial genomic DNA was extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany). A 400 bp fragment of the16S rDNA gene was amplified by PCR from purified genomic DNA. PCR was performed with 16S rRNA primers (515 forward primer: 5’-ATGATACGGCGACCACCGAGATCTACACGCTXXXXXXXXXXXXTATGGTAATTGTGTGYCAGCMGCCGCGGTAA-3’, where XXXXXXXXXXXX is an index sequence for multiplexing libraries. 806 reverse primer: 5’- CAAGCAGAAGACGGCATACGAGATAGTCAGCCAGCCGGACTACNVGGGTWTCTAAT-3’). PCR conditions were as follows: 98°C for 3 minutes, then 27 cycles of 98°C for 50 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final cycle of 72°C for 5 minutes. Libraries were purified using Zymo I-96 columns and analyzed on an Agilent 4200 Tapestation system (Agilent, Santa Clara, CA). The barcoded amplicons were pooled in equal concentrations, quantified by Qubit fluorometer, and molarity was calculated based on the size of the amplicon. The sequencing run was performed using 2×250 bp paired-end on the Illumina MiSeq platform using custom primers (Read1 seq primer: 5’-TATGGTAATTGTGTGYCAGCMGCCGCGGTAA -3’; Read2 seq primer: 5’-AGTCAGCCAGCCGGACTACNVGGGTWTCTAAT-3’; and index primer: AATGATACGGCGACCACCGAGATCTACACGCT) and included positive and negative controls.
Bioinformatic analysis pipeline for 16S rRNA gene sequencing.
After sequencing, the reads were de-multiplexed using QIIME. Subsequently, the reads were merged and dereplicated to remove chimeras using VSEARCH. To denoise the reads, the UNOISE 3 algorithm command was employed. The resulting sequences were then assigned to operational taxonomic units (OTUs) using the Mothur method and the SILVA database version 138. An OTU table was generated using USEARCH. Alpha and beta diversity were measured using QIIME, with sample sequences being rarefied to a level below that of the sample with the least sequences.
Statistical analysis.
The primary outcome was prevalent high-risk HPV DNA. We included all oral high-risk HPV-positive participants and a random subset of high-risk HPV-negative participants in a 1:2 ratio. Other variables included in the analysis were age, race, smoking and alcohol use, and sexual behavior history (lifetime number of any [vaginal, anal, and oral] sex partners and oral sex partners). Ever smoking and ever drinking were defined as regular current or former smoking or alcohol drinking, respectively. Sexual behaviors were dichotomized at the median for comparison between the groups. Frequencies, means, and medians were calculated for the major variables of interest. We assessed differences between groups with respect to alpha diversity (richness and evenness), beta diversity (in-between sample differences), and taxa abundances. We used weighted and unweighted UniFrac to calculate the distances (beta diversity) between samples, where weighted takes into account the relative abundance of each organism and unweighted uses only presence/absence data to determine distance. Principal coordinate analysis (PCoA) plots under metrics suitable for microbiome data (Bray Curtis [12], unweighted UniFrac [13], and weighted UniFrac [14]) were used to project high dimensional OTU information into two dimensions to visually examine the association between the oral microbiota and high-risk HPV positivity as well as demographic factors, smoking, alcohol use, and sexual behaviors. Permutation multivariate analysis of variance (PERMANOVA) was adopted to evaluate the association [15]. P-values were adjusted for multiple comparisons using the Benjamin-Hochberg method. All the statistical analyses were implemented in R. OUT counts and abundance tables are provided as supplementary materials.
Results
Thirteen of 15 available oral high-risk HPV DNA-positive samples were adequate for evaluation and were included in the analysis along with 30 oral high-risk HPV DNA-negative samples. The demographic and behavioral characteristics of the study population are shown in Table 1. There were no significant differences with respect to age, race, or history of smoking, alcohol use, or sexual behaviors between the two groups.
Table 1.
Characteristics of the 13 high-risk HPV-positive and 30 HPV-negative men.
| HPV (−) N = 30 |
High-risk HPV (+) N = 13 |
p-value | |
|---|---|---|---|
| Median (IQR) | Median (IQR) | ||
| Age, years | 57.5 (55–60) | 56 (54–59) | 1.0a |
| N (%) | N (%) | ||
| Race | 1.0 | ||
| Non-Hispanic white | 28 (93.3) | 12 (92.3) | |
| Hispanic/Latino | 2 (6.7) | 1 (7.7) | |
| Black | 0 | 0 | |
| Smoking status | 0.736 | ||
| Never | 19 (63.3) | 7 (53.9) | |
| Ever | 11 (36.7) | 6 (46.2) | |
| Alcohol status | 1.0 | ||
| Never | 2 (6.7) | 0 | |
| Ever | 28 (93.3) | 13 (100.0) | |
| Lifetime number of sex partners | 0.181 | ||
| 1 – 8 | 16(57.1) | 4 (30.8) | |
| > 8 | 12 (42.9) | 9 (69.2) | |
| Lifetime number of oral sex partners | 0.172 | ||
| 1 – 5 | 17 (65.4) | 5 (38.5) | |
| > 5 | 9 (34.6) | 8 (61.5) |
Median regression
Abbreviations: HPV: human papillomavirus; /IQR: interquartile range
Taxonomic composition of the oral microbiota
Figure 1 shows the relative abundance of the most common taxa by oral high-risk HPV status at the phylum (Figure 1A) and genus level (Figure 1B). Overall, the most abundant phyla were Proteobacteria, Cyanobacteria, Synergistota, Spirochaetota, Campilobacteroda, Fusobacterota, Actinobacteria, Firmicutes, and Bacteroidota with interindividual variation observed between the samples. To determine whether microbial composition varied significantly according to HPV status, we next evaluated the alpha and beta diversity of the samples.
Figure 1.
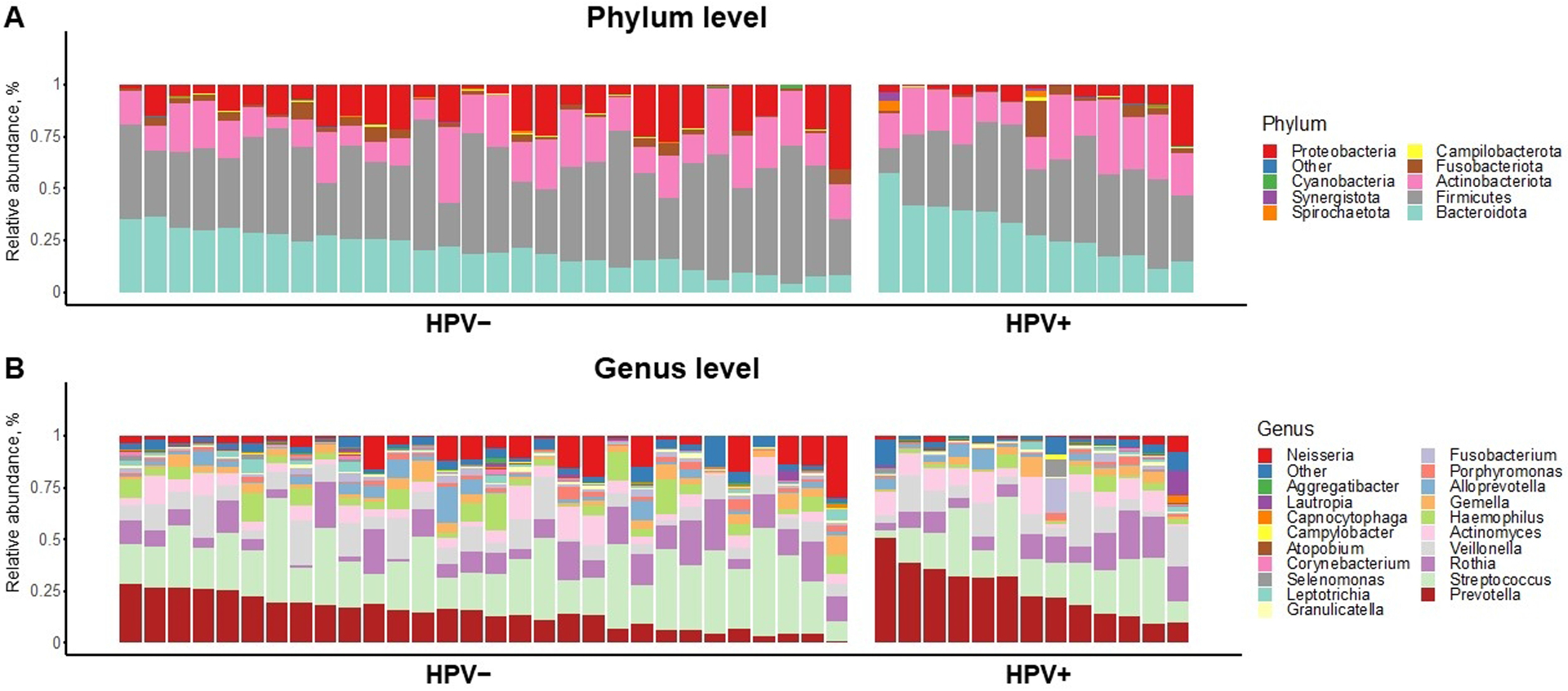
Taxonomic profiles of oral rinse samples at A) genus level and B) species level. Stacked bar chart of relative abundances of common bacterial taxa by prevalent oral high-risk human papillomavirus (HPV) status (left: HPV-negative, n = 30 and right, high-risk HPV-positive, n = 13).
Alpha and beta diversity of the oral microbiota by oral high-risk HPV status
We found no difference in alpha diversity between oral high-risk HPV-positive and -negative samples (Figure 2). The two groups were similar with respect to observed richness (p = 0.5) and diversity (Shannon index, p = 0.2 and Simpson reciprocal, p = 0.3). To better understand the microbiota diversity and its connection with HPV status, smoking status, lifetime number of sex partners, and lifetime number of oral sex partners, beta diversity was evaluated using weighted-UniFrac distances, and a principal coordinate analysis (PCoA) was conducted. The results demonstrated a clear differentiation in clustering between HPV-positive and HPV-negative samples (p = 0.001; Figure 3); however, we did not see notable differential clustering for microbial beta diversity on smoking status, lifetime number of sex partners, and lifetime number of oral sex partners (data not shown).
Figure 2.
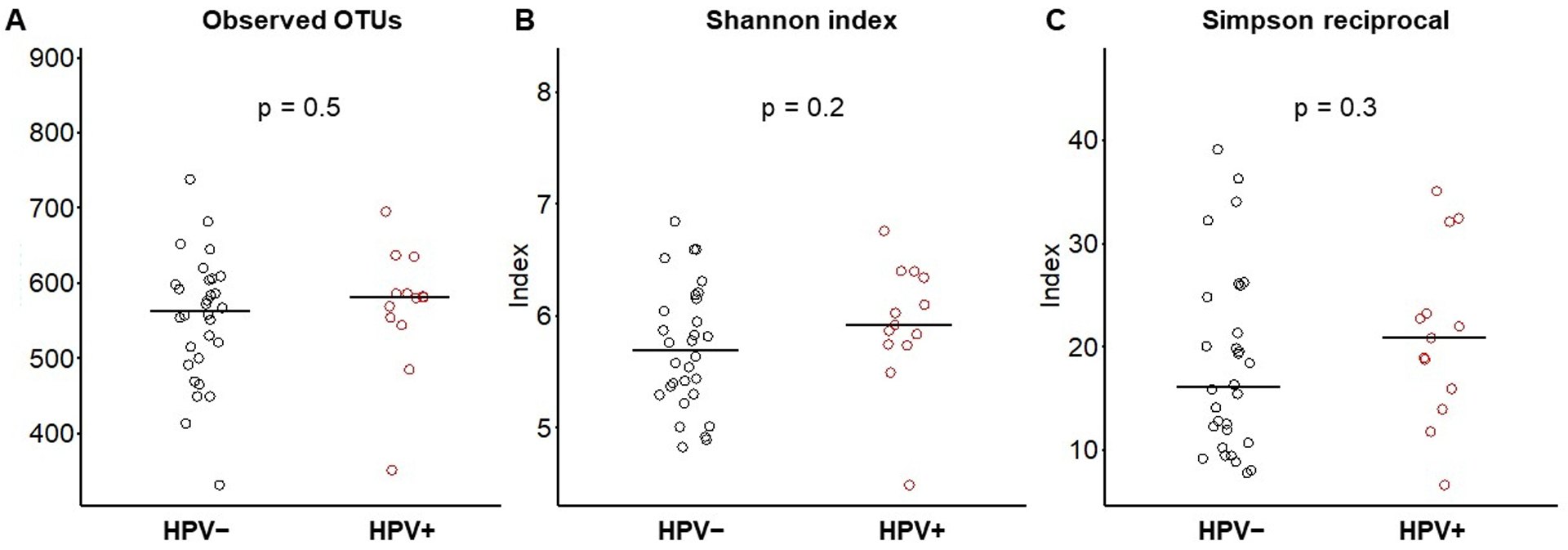
Boxplot showing alpha diversity indices with p-values for each by prevalent oral high-risk human papillomavirus (HPV) status. A) Observed species (richness), p = 0.5; B) Shannon index (diversity), p = 0.25; and C) Simpson reciprocal index (dominance), p = 0.3.
Figure 3.
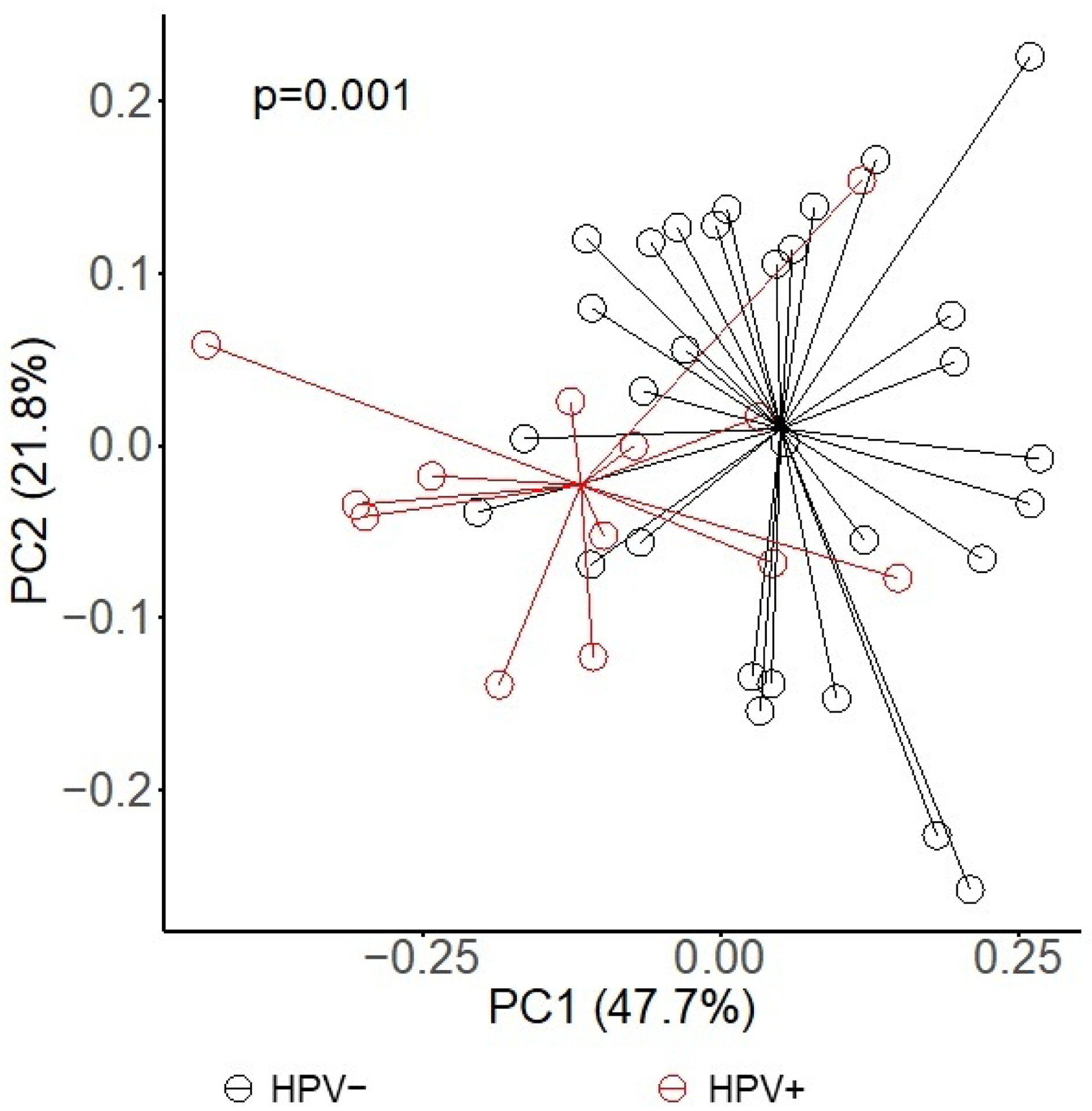
Principal coordinate analysis (PCoA) of beta-diversity with weighted-UniFrac distances by prevalent oral high-risk human papillomavirus status, p = 0.001.
Differential abundance of the oral microbiota by oral high-risk HPV status
To determine any statistically significant differences, we next performed permutation analysis with 5,000 permutations to determine which microbial taxa differed in abundance with false discovery rate (FDR) < 0.05 among the two groups. At the phylum level, relative abundance of Synergistota, Bacteroidota, and Spirochaetota was increased in HPV-positive samples and Proteobacteria was decreased. At the family level, we observed increased relative abundance of Synergistaceae and Prevotellaceae and decreased abundance of Neisseriaceae and Lactobacillaceae in the HPV-positive samples. At the genus level, we found seven taxa that were differentially abundant (Figure 4). Specifically, genus Fretibacterium, F0058, Kingella, Treponema, and Prevotella were more abundant in the HPV-positive samples while Neisseria and Lactobacillus were more abundant in the HPV-negative samples. Relative abundance is shown in a volcano plot with observations where abundance was significantly increased shaded blue for HPV-negative samples and red for HPV-positive samples (Figure 4A). Additionally, relative abundance of genera that varied significantly between groups is shown for each individual sample in a heatmap along with their p-values (Figure 4B). As shown, Fretibacterium and Kingella were more abundant in HPV-positive samples at p < 0.001 while F0058, Treponema, and Prevotella were more abundant at p < 0.01. For HPV-negative samples, both Neisseria and Lactobacillus were significantly more abundant at p < 0.01.
Figure 4.
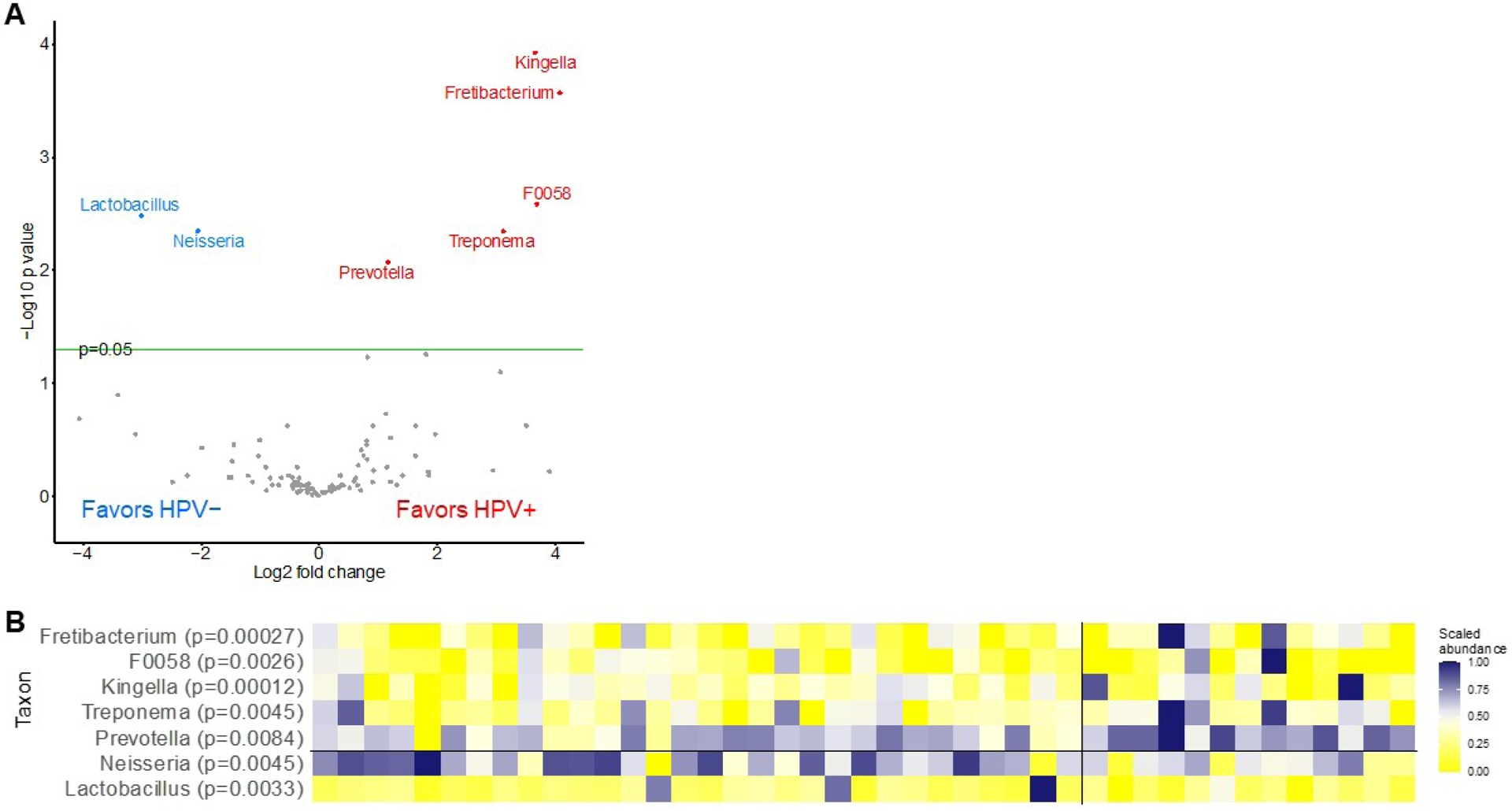
Differential abundance of microbial genera by oral high-risk HPV status. A) Volcano plot depicting the permutation-based test to identify differentially abundant microbial taxa in human papillomavirus (HPV)-positive and HPV-negative men. Differentially abundant taxa at false discovery rate (FDR) < 0.05 are indicated by colored circles where blue favors HPV-negative (left) and red favors HPV-positive (right) and B) Heatmap showing the differential abundance of microbial genera at false discovery rate (FDR) <0.05 for individual samples with HPV-negative shown on the left and high-risk HPV-positive on the right. Level of relative abundance is indicated by intensity of color (blue = increasing abundance and yellow = decreasing abundance).
Discussion
Using 16S rRNA sequencing, we characterized the oral microbiota of 13 oral high-risk HPV-positive and 30 HPV-negative middle-aged men. We did not find a difference in alpha diversity (within-sample) between HPV-positive and -negative participants. However, beta diversity (between-sample) varied significantly according to HPV status. This study further helps clarify the natural history of oral HPV and suggests a possible association between the oral microbiota and prevalent oral HPV infection status.
Microbial species diversity has been established as a risk factor for HPV infection, persistence, and progression of precancer to cancer [5, 16, 17]. To our knowledge, there are few studies investigating the association between the oral microbiota and oral HPV infection. Although the association between the oral microbiota and head and neck cancer has been established in multiple studies, they are typically not stratified by HPV status and/or include non-HPV-related head and neck sites [18–21].
The largest study to date investigating the association between oral HPV and the microbiota included 495 participants, 68 with prevalent oral HPV DNA detected [22]. In agreement with our findings, alpha diversity was not significantly different between oral HPV-positive and -negative individuals while beta diversity varied significantly between the two groups. The authors found that the families Prevotellaceae, Actinomycetaceae, Veillonellaceae, and Campylobacteraceae, and Bacteroidetes were more abundant among individuals who were oral HPV-positive while Gemellaceae was less abundant. Although results were not reported at the genus level, this is in general agreement with our finding of greater relative abundance of genus Prevotella (within family Prevotellaceae), although we did not confirm the finding of significant variation between groups for family Gemellaceae.
Genus Prevotella is associated with a pro-inflammatory response and may promote chronic inflammation [23]. This is consistent with earlier studies showing that prior oral inflammatory disease is a risk factor for head and neck cancer, in particular OPC, and that elevated salivary Prevotella melaninogenica is positively associated with oral squamous cell carcinoma [24, 25]. A small study that investigated differences in the oral microbiota between women with and without a prevalent oral HPV infection (n = 13 and n = 26, respectively) found that increased richness was associated with oral HPV infection [26], which was in contrast to our findings. That study also found that the relative abundance of members of phylum TM7 was increased in HPV-positive samples as was the members of phylum Firmicutes, TM73, and species Selenomonas and Megasphaera, while species Haemophilus (phylum Proteobacteria) was higher among those who were HPV-negative [26].
For the association between head and neck cancer and the oral microbiota, a recent study that included 13 cases of OPC, of which eight were HPV-positive, found differential clustering in the taxonomic profiles of the oral microbiota according to HPV status. Specifically, the genera Treponema, Neisseria, and Veillonella were associated with HPV status; conversely, no association between the oral microbiota and smoking status was observed [27]. In a large population-based nested case-control study, Hayes et al. determined that an abundance of Corynebacterium and Kingella were associated with a decreased risk for head and neck cancer, consistent with the carcinogen metabolism capacity of these genera [18]. However, in the same study overall composition of the oral microbiota was not associated with risk for head and neck cancer. In one study investigating the association between the oral microbiota, HPV, and head and neck cancer, Lactobacillus spp. were significantly associated with HPV-positive tumors compared with HPV-negative tumors [19]. Consistent with studies of the cervical microbiota, several species of acid tolerant anaerobic bacteria, specifically Veillonella, Megasphaera, and Anaerolineae, were identified as markers for HPV-positive tumors. A low pH hypoxic microenvironment is typical of solid tumors, favoring the growth of this taxa [28]. In another study, the same group determined that Lactobacillus gasseri/johnsonii and Lactobacillus vaginalis, commensal vaginal flora species, were more highly abundant in saliva from patients with OPC compared with healthy controls and was hypothesized to be transmitted during oral sex [20]. It remains unknown what role these species play in OPC oncogenesis, but taken together, the results from these studies show a key role for the microbiota in oral HPV infection and OPC, with a potential for interventions to prevent infection or increase clearance rates of oral HPV [29].
Although not directly comparable with oral HPV infection and risk for OPC, vaginal bacterial communities have been shown to protect against sexually transmitted infections including HPV [29]. Cross-sectional studies of women with cervical lesions, including cervical cancer, have found that increased species diversity and reduced abundance of Lactobacillus spp. are associated with increasing severity of cervical disease in a dose-dependent manner [5, 30]. Additionally, a pro-inflammatory response induced by some bacteria leads to tissue damage that promotes oncogenesis by HPV [31]. It is plausible that similar mechanisms might also promote HPV infection, persistence, and oncogenesis in OPC.
This pilot study had a few limitations: 1) First, the small sample size may have resulted in masking statistically significant associations. This also precluded performing multivariable analysis. Second, we had only oral rinse samples available for analysis. Although oral rinse is well-established for oral HPV detection, the oral microbiota varies according to sampling site and specimen type and our results may have differed had we used another sampling strategy. Third, our study population was homogenous with respect to age, sex, and race/ethnicity. In future studies, we will aim to include a more diverse population of men and women of all ages and races/ethnicities to be more representative of the population as a whole. Differences between our findings and others could be due to sampling site, methodological differences with respect to 16S rRNA sequencing or bioinformatics pipeline, and choice of study population. However, our main results, greater oral microbial betadiversity among those with an HPV infection, agree with previously published studies by other groups.
Conclusions
We found that microbial composition, but not richness, were associated with prevalent oral high-risk HPV status in oral rinse samples from middle-aged men. Larger studies are needed to understand the influence of the oral microbiota on the natural history, including persistence and clearance, of oral HPV. We are currently planning a larger study with longitudinal follow-up to gain further insight into the natural history of oral HPV infection, including its interaction with the oral microbiota.
Supplementary Material
Highlights.
There was an association between the oral microbiota and prevalent oral oncogenic HPV infection status
Clear differential clustering was observed when stratifying by prevalent oral oncogenic HPV status but not smoking or sexual behavior status on principal coordinate analysis (PCoA)
Microbial beta-diversity (between-sample diversity) but not alpha-diversity (within-sample diversity) differed between oral oncogenic HPV-positive and -negative middle-aged men
Neisseria spp. were significantly more abundant among HPV-negative men and Prevotella spp. were more abundant among high-risk HPV-positive men
Acknowledgements:
We used the MD Anderson Cancer Center Microbiome Core Facility.
Funding:
This work was supported by The University of Texas MD Anderson Cancer Center [internal research grant]; Cancer Prevention & Research Institute of Texas [grant number RP200025], the Stiefel Oropharyngeal Research Fund; the Christopher and Susan Damico Chair in Viral Associated Malignancies; the MD Anderson Cancer Center Microbiome Core Facility is supported by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant [grant number P30CA016672]. Roche Diagnostics provided laboratory reagents for HPV DNA testing of oral rinse specimens.
Role of the Funding Source:
The funding source had no role in study design, data collection, analyses, interpretation of the data, or decision to submit results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Dr. Dahlstrom: Roche Diagnostics provided laboratory reagents for HPV DNA testing of oral rinse specimens.
Dr. Sturgis: Roche Diagnostics provided laboratory reagents for HPV DNA testing of oral rinse specimens and mucosal swabs, 2021 Board Chair for The Immunization Partnership non-profit organization, and Medical Advisory Board Member of The Prevent Cancer Foundation non-profit organization.
No other authors have any interests to declare, including financial interests or relationships or affiliations that are relevant to the subject of this manuscript.
References
- [1].Viens LJ, Henley SJ, Watson M, et al. Human papillomavirus-associated cancers - United States, 2008–2012. MMWR Morb Mortal Wkly Rep. 2016;65(26):661–666. Published 2016 Jul 8. doi: 10.15585/mmwr.mm6526a1 [DOI] [PubMed] [Google Scholar]
- [2].Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33(29):3235–3242. doi: 10.1200/JCO.2015.61.6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Doorbar J, Zheng K, Aiyenuro A, et al. Principles of epithelial homeostasis control during persistent human papillomavirus infection and its deregulation at the cervical transformation zone. Curr Opin Virol. 2021;51:96–105. doi: 10.1016/j.coviro.2021.09.014 [DOI] [PubMed] [Google Scholar]
- [5].Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. Published 2015 Nov 17. doi: 10.1038/srep16865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Łaniewski P, Cui H, Roe DJ, et al. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci Rep. 2019;9(1):7333. Published 2019 May 14. doi: 10.1038/s41598-019-43849-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. Published 2012 Jun 13. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Le Bars P, Matamoros S, Montassier E, et al. The oral cavity microbiota: between health, oral disease, and cancers of the aerodigestive tract. Can J Microbiol. 2017;63(6):475–492. doi: 10.1139/cjm-2016-0603 [DOI] [PubMed] [Google Scholar]
- [9].Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15(2):109–128. doi: 10.1038/nrmicro.2016.171 [DOI] [PubMed] [Google Scholar]
- [10].Dahlstrom KR, Anderson KS, Guo M, et al. Screening for HPV-related oropharyngeal, anal, and penile cancers in middle-aged men: Initial report from the HOUSTON clinical trial. Oral Oncol. 2021;120:105397. doi: 10.1016/j.oraloncology.2021.105397 [DOI] [PubMed] [Google Scholar]
- [11].TRINITY. Clinicaltrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US); Throat and Other HPV-Related Cancers in Men: Identifying Them Early (TRINITY), Identifier NCT02897427 . [updated 2022 Feb 11; cited 2022 May 25]. Available from: https://clinicaltrials.gov/ct2/show/NCT02897427.
- [12].Bray JR, Curtis JT. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27(4):325–349. doi: 10.2307/1942268 [DOI] [Google Scholar]
- [13].Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anderson MJ. Permutational multivariate analysis of variance(PERMANOVA). In: Balakrishnan N, Colton T, Everitt B, Piegorsch W, Ruggeri F, Teugels JL, editors. Wiley StatsRef: Statistics Reference Online. New York: John Wiley & Sons; 2017. doi: 10.1002/9781118445112.stat07841 [DOI] [Google Scholar]
- [16].Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila). 2016;9(5):357–366. doi: 10.1158/1940-6207.CAPR-15-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reimers LL, Mehta SD, Massad LS, et al. The cervicovaginal microbiota and its associations with human papillomavirus detection in HIV-infected and HIV-uninfected women. J Infect Dis. 2016;214(9):1361–1369. doi: 10.1093/infdis/jiw374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hayes RB, Ahn J, Fan X, et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018;4(3):358–365. doi: 10.1001/jamaoncol.2017.4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7(32):51320–51334. doi: 10.18632/oncotarget.9710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guerrero-Preston R, White JR, Godoy-Vitorino F, et al. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget. 2017;8(67):110931–110948. Published 2017 Sep 7. doi: 10.18632/oncotarget.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pushalkar S, Mane SP, Ji X, et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol. 2011;61(3):269–277. doi: 10.1111/j.1574-695X.2010.00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang Y, D’Souza G, Fakhry C, et al. Oral HPV associated with differences in oral microbiota beta diversity and microbiota abundance [published online ahead of print, 2022 Jan 17]. J Infect Dis. 2022;jiac010. doi: 10.1093/infdis/jiac010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grønhøj Larsen C, Gyldenløve M, Jensen DH, et al. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Br J Cancer. 2014;110(6):1587–1594. doi: 10.1038/bjc.2014.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tezal M,A Sullivan M, R. Marshall J. The role of chronic periodontitis in prevention and treatment of head and neck cancers. Cur Cancer Ther Rev. 2010;6(4):323–333. doi: 10.2174/157339410793358057 [DOI] [Google Scholar]
- [25].Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. Published 2005 Jul 7. doi: 10.1186/1479-5876-3-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tuominen H, Rautava S, Syrjänen S, Collado MC, Rautava J. HPV infection and bacterial microbiota in the placenta, uterine cervix and oral mucosa. Sci Rep. 2018;8(1):9787. Published 2018 Jun 28. doi: 10.1038/s41598-018-27980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zakrzewski M, Gannon OM, Panizza BJ, Saunders NA, Antonsson A. Human papillomavirus infection and tumor microenvironment are associated with the microbiota in patients with oropharyngeal cancers-pilot study. Head Neck. 2021;43(11):3324–3330. doi: 10.1002/hed.26821 [DOI] [PubMed] [Google Scholar]
- [28].Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975. doi: 10.1038/nrc2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brotman RM. Vaginal microbiome and sexually transmitted infections: An epidemiologic perspective. J Clin Invest. 2011;121(12):4610–4617. doi: 10.1172/JCI57172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PLoS One. 2016;11(4):e0153274. Published 2016 Apr 26. doi: 10.1371/journal.pone.0153274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Anahtar MN, Byrne EH, Doherty KE, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42(5):965–976. doi: 10.1016/j.immuni.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


