Abstract
Heptamethine cyanine dyes enable deep tissue fluorescence imaging in the near infrared (NIR) window. Small molecule conjugates of the benchmark dye ZW800-1 have been tested in humans. However, long-term imaging protocols using ZW800-1 conjugates are limited by their instability, primarily because the chemically labile C4’-O-aryl linker is susceptible to cleavage by biological nucleophiles. Here, we report a modular synthetic method that produces novel doubly strapped zwitterionic heptamethine cyanine dyes, including a structural analogue of ZW800-1, with greatly enhanced dye stability. NIR-I and NIR-II versions of these doubly strapped dyes can be conjugated to proteins, including monoclonal antibodies, without causing undesired fluorophore degradation or dye stacking on the protein surface. The fluorescent antibody conjugates show excellent tumor-targeting specificity in a xenograft mouse tumor model. The enhanced stability provided by doubly strapped molecular design will enable new classes of in vivo NIR fluorescence imaging experiments with possible translation to humans.
Keywords: antibodies, cyanines, dyes/pigments, fluorescent probes, imaging agents
Graphical Abstract
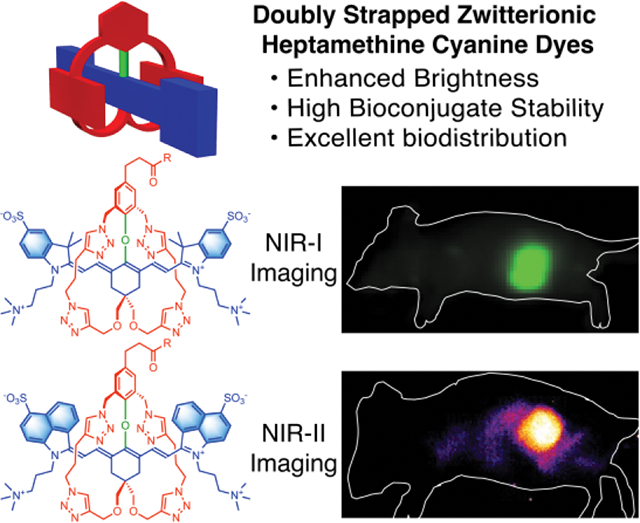
A versatile and efficient synthetic method produces conjugatable sterically shielded heptamethine cyanine dyes for in vivo NIR-I or NIR-II fluorescence imaging. The fluorochromes are enclosed by two flanking straps which enhance fluorescence brightness and chemical stability, prevent dye stacking on the surface of an antibody, and enable high performance fluorescence imaging of a mouse tumor.
Introduction
Fluorescent near-infrared (NIR) fluorophores with absorption and emission wavelengths in the NIR-I (700–1000 nm) or NIR-II (1000–1700 nm) windows are very attractive for biomedical imaging since there is relatively deep penetration of the NIR light through skin and tissue.[1–3] For many decades the clinical NIR heptamethine cyanine dye, Indocyanine Green (ICG), with absorption/emission wavelengths around 800 nm has been used for optical imaging and diagnostics (Scheme 1). [4,5] While ICG is broadly applied, its ultimate potential is limited because it cannot be conjugated to targeting biomolecules. This deficiency has led to a methodical exploration of conjugatable heptamethine cyanine dyes that have a desirable mixture of chemical, physical and spectral properties.[6–8] A notable commercial NIR-I dye is IRdye800CW which has a carboxyl group for bioconjugation and multiple sulfonates to ensure good water solubility. Many bioconjugates have been prepared and evaluated for in vivo imaging performance. [9,10] In some cases, concerns have been noted about undesired dye or fluorescent bioconjugate accumulation in off-target organs tissue,[11–15] which led to the development of NIR-I dye structures with a geometric balance of opposing charges (so-called zwitterionic dyes) that diminishes dye association with biological surfaces. [16,17] A leading example is ZW800-1 which has been used for more than ten years for different types of bioconjugation.[17,18] The dye itself, or small-molecule conjugates, are very hydrophilic and they quickly clear from the bloodstream through the kidney.[18,19] One particularly promising conjugate, with an appended cyclic-RGD peptide motif that targets cancerous tissue, has progressed to clinical trials.[20,21] There are also a growing number of reports that use ZW800-1 or close structural analogues to label macromolecules for in vivo imaging and histology applications, [17,22–24] or biomaterials such as cell surfaces, hydrogels, nanoparticles, 3D printed biocomposites, or extracellular matrix biopolymers for tissue engineering applications.[25–32] A necessary dye performance property for most of these applications is long-term chemical stability of the fluorescent bioconjugate and this requirement highlights a potential chemical reactivity problem with ZW800-1; namely, linker cleavage due to nucleophilic substitution of the dye’s 4’-phenoxy group by biological nucleophiles (illustrated by the pink arrow in Scheme 1).[33] Studies that have incubated samples of antibodies or nanoparticles labeled with ZW800-1 in serum for 24 hours have observed substantial loss of fluorescent signal.[33,34] Under these near-physiological conditions there are a multitude of possibilities for intramolecular and intermolecular attack of the dye by various biological nucleophiles.[35,36] The loss of fluorescence signal due to dye degradation introduces uncertainty into longitudinal experiments that aim to measure long-term degradation or trafficking of implanted biomaterials that have been labeled with the dye.[37]
Scheme 1.
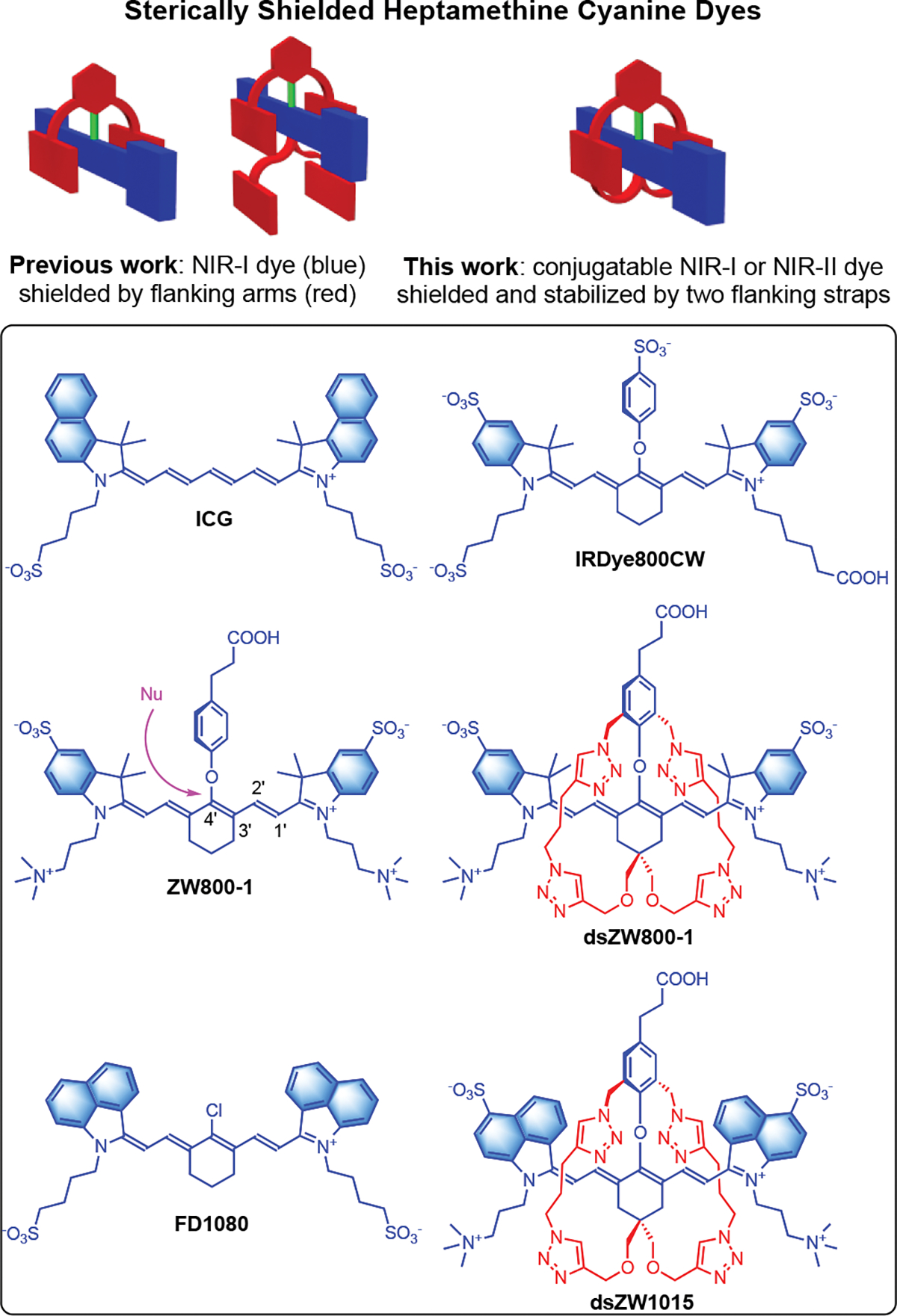
(top) Underlying molecular concepts. (bottom) Chemical structures of NIR-I and NIR-II dyes. Nu: Nucleophile.
As summarized in the top of Scheme 1, we have recently developed synthetic methods to produce NIR-I cyanine dyes that are sterically shielded by two or four flanking, linear arms that project over each face of the planar dye. [7,38–40] Also shown in Scheme 1 is the specific focus of this current report; that is, a new class of doubly strapped and conjugatable NIR-I and NIR-II heptamethine cyanine dyes with greatly enhanced performance properties. The literature on doubly strapped dyes includes several organic-soluble examples [41–46] and one water-soluble molecule.[47] Most do not emit in the NIR region and none have been designed for bioconjugation in water. As described below, we have solved the ZW800-1 degradation problem by making a stabilized, doubly strapped version that we call dsZW800-1. Moreover, the versatile synthetic methodology provided access to a doubly strapped NIR-II analogue called dsZW1015 with absorption/emission maxima >1000 nm and we generated a dsZW1015-labeled antibody conjugate for successful in vivo NIR-II fluorescence imaging of a mouse tumor.
Results and Discussion
Synthesis of dsZW800-1
The synthesis of dsZW800-1 is summarized in Scheme 2. The key synthetic intermediate is unstrapped dye 3 which was assembled under a mild condition by substituting the 4’-chloro atom in precursor 1 with phenol 2 whose structure bears two appended arms containing terminal azido groups. A subsequent saponification reaction converted 3 into unstrapped dye 4, which was followed by a Cu-catalyzed azide-alkyne cycloaddition reaction that covalently connected the two flanking straps to produce dsZW800-1 in 62 % yield.
Scheme 2.
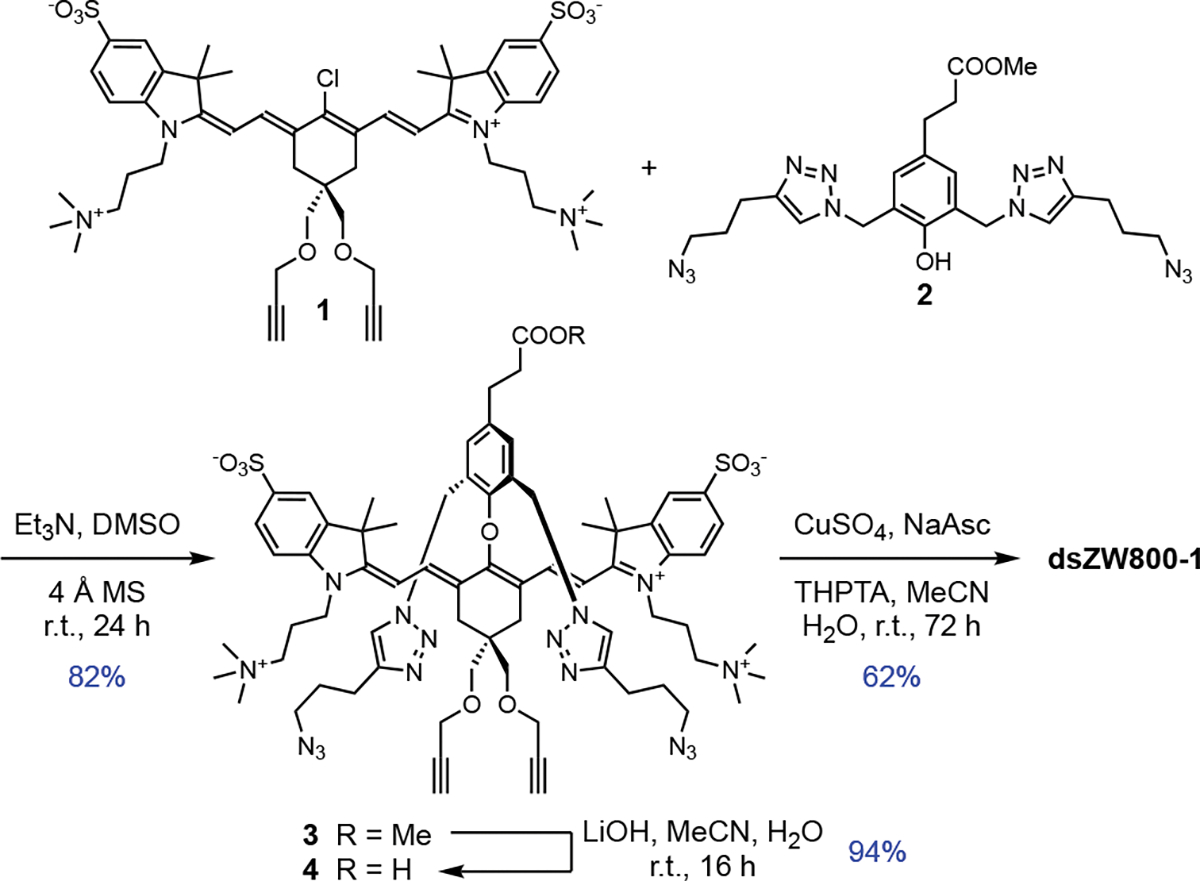
Synthesis of intermediate 4 and dsZW800-1
Molecular Structure of dsZW800-1
The molecular structures of all compounds were characterized by standard spectrometric methods. Figure S1 contains a comparison of 1H NMR spectra in DMSO-d6 for ZW800-1, unstrapped dye 4, and doubly strapped dsZW800-1. The chemical shifts for ZW800-1 and unstrapped dye 4 closely match, suggesting similar molecular conformations. In contrast, the spectrum for dsZW800-1 includes specific upfield chemical shifts for the polymethine protons, indicating through space magnetic shielding by the triazole rings that are part of the two flanking straps. Support for this intramolecular shielding effect was gained when an X-ray crystal structure of dsZW800-1 was obtained (Figure 1).[48] The crystal structure of dsZW800-1 revealed several other structural features that are consistent with the NMR data. Notably, the embedded heptamethine fluorochrome adopts the expected low-energy, all-trans conformation with relatively small variation in the C-C bond lengths of the polymethine units (i.e., little bond length alternation, see Figure S26) reflecting a high level of π-electron delocalization throughout the polyene. A comparison with literature crystal structures of unstrapped heptamethine dyes that have 4’-phenoxy substituents reveals two unique structural features caused by the two flanking straps in dsZW800-1. One feature is the orientation of the 4’-phenoxy ring relative to the heptamethine fluorochrome. Normally the longitudinal axis of the 4’-phenoxy ring points away from the vertical plane of the heptamethine,[49] but in the case of dsZW800-1 the 4’-phenoxy ring is forced by the flanking straps to align with the vertical heptamethine plane and rotate its face towards one end of the dye. The other unique feature of crystalline dsZW800-1 is the intramolecular lattice packing distance. Normally, heptamethine cyanine dyes exhibit slipped cofacial stacking of the polymethine fluorochrome and a typical intermolecular distance of ~3.5–5.0 Å. [49,50] But in the case of dsZW800-1, the flanking straps enforce spatial isolation of each molecule, and the intermolecular solid-state distance between adjacent fluorochromes is ~9 Å. Thus, even if multiple copies of dsZW800-1 are forced together under self-aggregation conditions, the flanking straps ensure enough spatial separation to prevent strong coupling of the dye transition dipoles.
Figure 1.
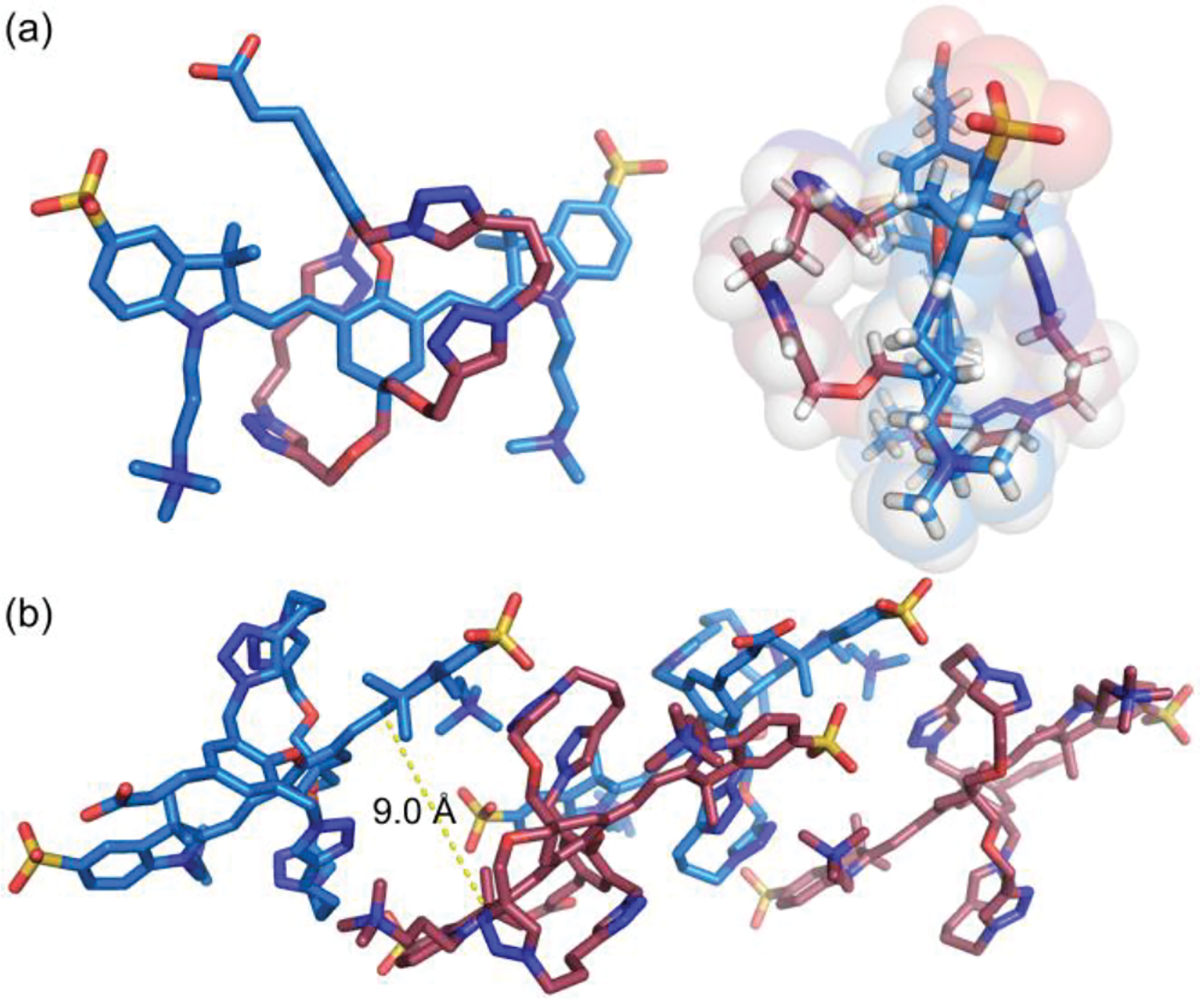
(a) Side and end views of X-ray crystal structure of dsZW800-1. (b) Lattice packing of four copies of dsZW800-1; the distance between adjacent heptamethine fluorochromes is ~9 Å. CCDC No.: 2236818.
Spectral Properties and Stability of dsZW800-1
A comparison of the dye photophysical properties in Table 1 indicates that the two flanking straps in dsZW800-1 do not significantly change the maximum absorption and emission wavelengths or the fluorescence peak width compared to ZW800-1 (Figure S4). Interestingly, the fluorescence quantum yield in PBS for dsZW800-1 (11.0%) is measurably higher than the value for unstrapped 4 (8.1%) which is formally a structural isomer. This difference suggests that the constrained flanking straps in dsZW800-1 are more effective at inhibiting non-radiative energy transfer from the dye excited state to the surrounding hydration shell which is usually a major energy relaxation pathway for highly conjugated NIR dyes in water.[51–53]
Table 1.
Photophysical properties at room temperature.
| Dye | Solvent[a] | λabsmax (nm) | λemmax (nm) | ε (105 M−1cm−1) | ΦF (%)[b] | Brightness[c] |
|---|---|---|---|---|---|---|
|
| ||||||
| ZW800–1 | PBS | 768 | 785 | 1.98 | 9.1 | 18,000 |
| 4 | PBS | 769 | 788 | 1.91 | 8.1 | 16,000 |
| dsZW800–1 | PBS | 769 | 788 | 2.10 | 11.0 | 23,000 |
| 5 | PBS | 1040 (m) 845 (a)[d] | -[e] | 0.27 | -[e] | - |
| 5 | DMSO | 1050 | 1081 | 1.02 | 0.029 | 29 |
| 7 | PBS | 985 (m) 875(a)[d] | 1015 | 1.35 | 0.006 | 8 |
| 7 | DMSO | 1015 | 1056 | 1.38 | 0.11 | 150 |
| dsZW1015 | PBS | 980 | 1015 | 1.37 | 0.014 | 19 |
| dsZW1015 | DMSO | 1030 | 1056 | 1.62 | 0.10 | 160 |
PBS = pH 7.4 phosphate buffered saline; DMSO = dimethylsulfoxide. Dye concentration range is 0 – 5 μM. All measurements conducted at room temperature.
For ZW800–1, 4, and dsZW800–1, absolute fluorescence quantum yield was measured directly by photon counting, error is ±5%. For dyes 5, 7, and dsZW1015, the measurements are relative to IR26 in DCM (0.05%).[54]
Brightness = ε × ΦF, error is ±15%.
m = monomer; a = aggregate.
Not measurable by our instrument.
The photostabilities of ZW800-1, 4, and dsZW800-1, were quantified by conducting simple photobleaching experiments that irradiated separate dye solutions in PBS using a xenon lamp with 620 nm long-pass filter and fitting the bleaching curves to a one-phase exponential decay (Figure 2). The measured order of photostability was 4 > dsZW800-1 > ZW800-1. The major photobleaching pathway for heptamethine cyanine dyes is bimolecular reaction of photogenerated singlet oxygen with the polymethine fluorophore, followed by bond cleavage and formation of non-fluorescent carbonyl fragments.[55] The relatively poor photostability of ZW800-1 is consistent with the high reactivity of electrophilic singlet oxygen with heptamethines that have electron donating 4’phenoxy substituents.[7] The relatively enhanced photostabilities of 4 and dsZW800-1 are attributed to steric shielding provided by the proximal linear arms or straps, which may decrease the efficiency of oxygen photosensitization and/or inhibit the subsequent bimolecular reaction with singlet oxygen.[56]
Figure 2.
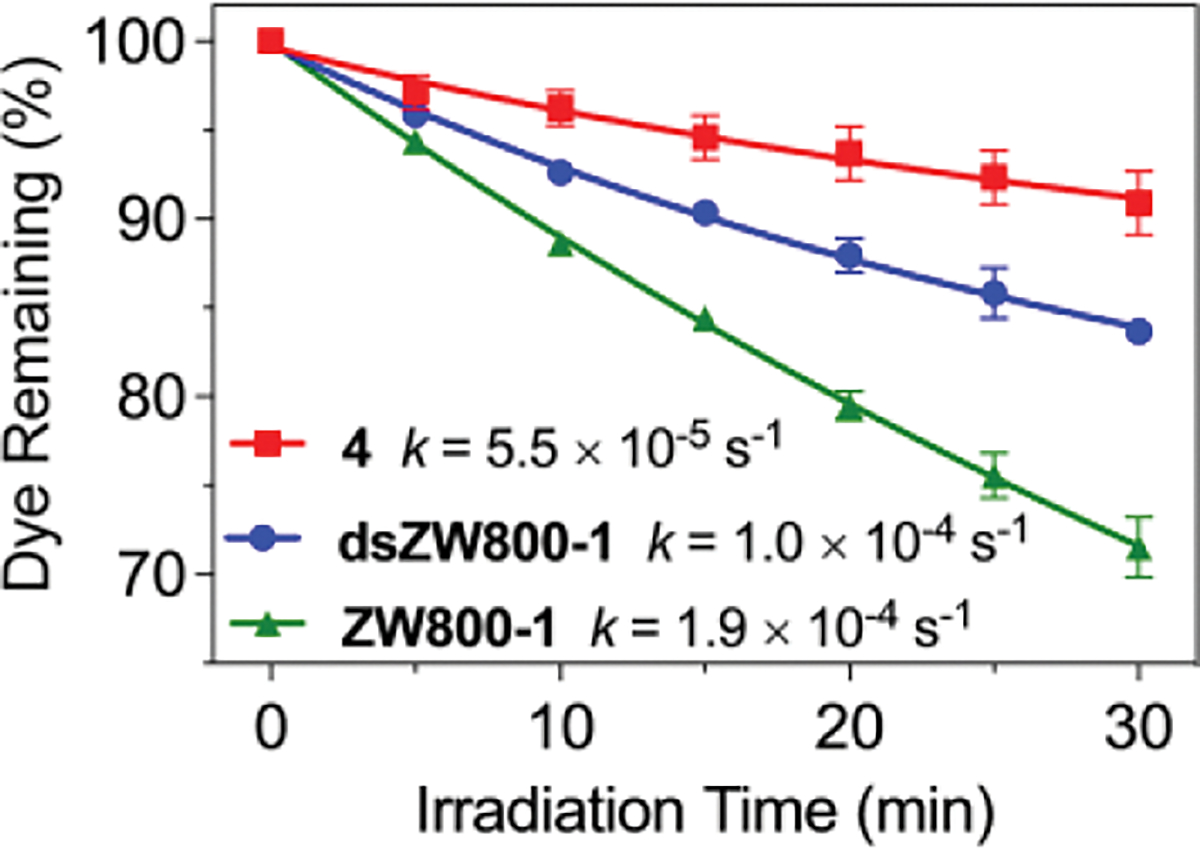
Lamp irradiation: Three separate cuvettes, each containing 2 μM dye in PBS buffer, pH 7.4, were irradiated by a 150 W Xenon lamp with a 620 nm long-pass filter (0.5 mW/cm2). The errors bars indicate standard deviation for the triplicate measurements. Each plot was fit to a one-phase exponential decay and the rate constants were obtained by fitting to a linear pseudo-first order reaction model.
The chemical stability of each dye was evaluated by a set of spectroscopic experiments that incubated separate solutions of the dyes mixed with 1 mM glutathione (GSH) in pH 7.4 PBS buffer. Substitution of the heptamethine 4’phenoxy group by the nucleophilic thiol in GSH is easily tracked since the reaction produces a noticeable red-shift in the heptamethine absorbance (Figure S8).[40,57,58] Shown in Figure 3a is the chemical product of GSH reaction with ZW800-1 as proved by NMR and mass spectrometry (Figure S9). The plot in Figure 3b compares the stability profiles of the three dyes in pH 7.4 PBS buffer with 1 mM GSH. The unstrapped dyes, ZW800-1 and 4, were rapidly and completely consumed with half-lives (t1/2) of 7.1 and 27 minutes, respectively. In dramatic contrast, incubation of dsZW800-1 with 1 mM GSH for 10 hours produced no change in the absorption spectrum indicating no reaction (confirmed by mass spectrometry). Since glutathione levels can reach 10 mM inside certain cells,[59] a second sample of dsZW800-1 was incubated with 10 mM GSH for 10 hours with still no evidence of any dsZW800-1 degradation (Figure S8). The protection from GSH attack is a compelling demonstration of the effectiveness of the two flanking straps in dsZW800-1 to completely stop substitution of the labile heptamethine 4’phenoxy group by biological nucleophiles.
Figure 3.
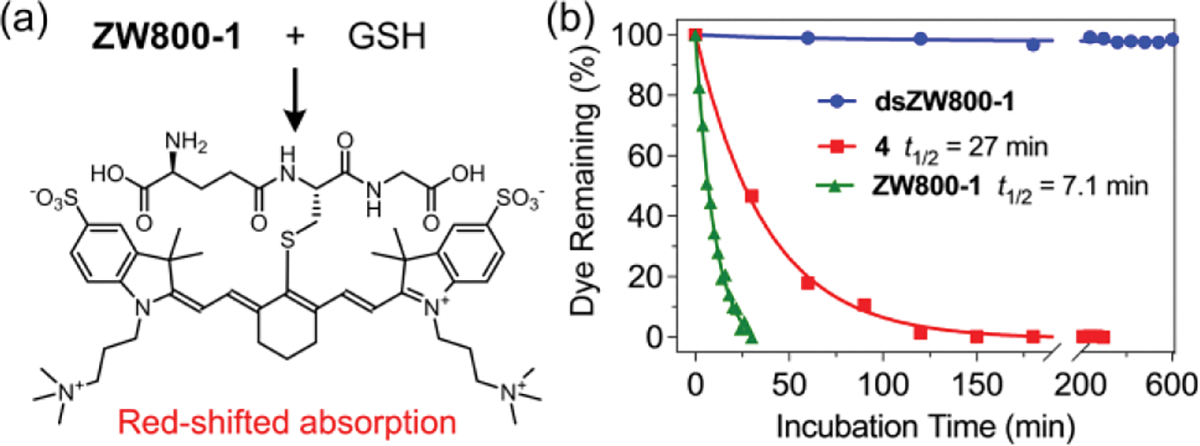
(a) Reaction of ZW800-1 with glutathione (GSH) to give product with red-shifted absorption maxima band. (b) Comparison of dye stabilities in pH 7.4 PBS buffer with 1 mM GSH, room temperature. Each plot was fit to a one-phase exponential decay and the half-lives were obtained by fitting to a linear pseudo-first order reaction model.
Antibody Labeling using dsZW800-1
The dye degradation caused by GSH suggests that unstrapped ZW800-1 and 4 are not good choices for protein labeling because the labile heptamethine 4’phenoxy linker can be cleaved over time by nucleophilic side-chains on the protein surface, such as the ε-amino groups of surface lysine residues. This prediction was borne out by experiments that assessed the suitability of ZW800-1 and dsZW800-1 for protein conjugation. Standard amide bond formation chemistry was used to attach ZW800-1 or dsZW800-1 to goat IgG antibody or bovine serum albumin (BSA) (Scheme S1 and Figure S10). Shown in Figures S11 and S12 are the absorption and fluorescence spectra for the purified bioconjugates. In short, protein conjugation with dsZW800-1 produced a higher Degree of Labeling (DOL) compared to protein conjugation with ZW800-1 and the fluorescence intensities of the proteins labeled with dsZW800-1 were 3–4 times higher. Focusing on the stability of the IgG conjugates, the NIR signal for IgG-dsZW800-1 was observed to be substantially more stable than IgG-ZW800-1, as proved by experiments that tracked the loss of dye absorption over time for samples in PBS or serum (Figure S13). The known chemical degradation of IgG-ZW800-1 is a significant technical limitation,[33] and the greatly enhanced stability of IgG-dsZW800-1 is very welcome. dsZW800-1 will be especially useful as a stable NIR fluorescent label for use within longitudinal experiments that measure degradation or trafficking of implanted dye-labeled cells or biomaterials.[25–28,30,32,60]
In Vivo Imaging using dsZW800-1
The in vivo imaging performance of a dsZW800-1-labeled antibody was assessed by attaching dsZW800-1 to Panitumumab (Pan), a clinical monoclonal antibody that targets EGFR-positive tumors. Fluorescent versions of Pan are highly desired as cancer imaging agents for pathology and clinical procedures such as fluorescence guided surgery; however, it is known that in vivo imaging performance of a Pan-dye conjugate depends greatly on the structure of the appended dye.[13] In this regard, it is notable that the absorption spectrum of Pan-dsZW800-1 (DOL = 3.1) in Figure 4a exhibits a relatively narrow peak at 772 nm whose excitation produces the same fluorescence spectrum as free dye. There is no evidence of a blue-shifted absorption peak corresponding to H-stacking of adjacent appended dsZW800-1 dyes on the surface of the antibody which would be undesired because dye H-stacking quenches dye fluorescence and can potentially alter antibody biodistribution. Furthermore, the absorption spectrum of Pan-dsZW800-1 was unchanged after 15 days at 4 °C (Figure S15a) suggesting that it has a very similar storage lifetime as the reconstituted Pan that is used clinically. The in vivo imaging performance was tested by intravenous injection of Pan-dsZW800-1 into EGFR+ JIMT-1 (triple-negative breast cancer) tumor-bearing mice (Figure S16). Specific and high tumor uptake was observed using a commercial In Vivo Imaging Station (IVIS) with NIR-I fluorescence imaging settings, and the average tumor-to-background ratio (TBR) reached ~ 8 at 72 h post-injection with negligible liver signal (Figure 4b, 4c, S17, S18). The mice were sacrificed at 72 h post-injection and NIR-I fluorescence imaging of the excised organs confirmed the very high tumor specificity of Pan-dsZW800-1 (Figure 4d, 4e, S19 and S20).
Figure 4.
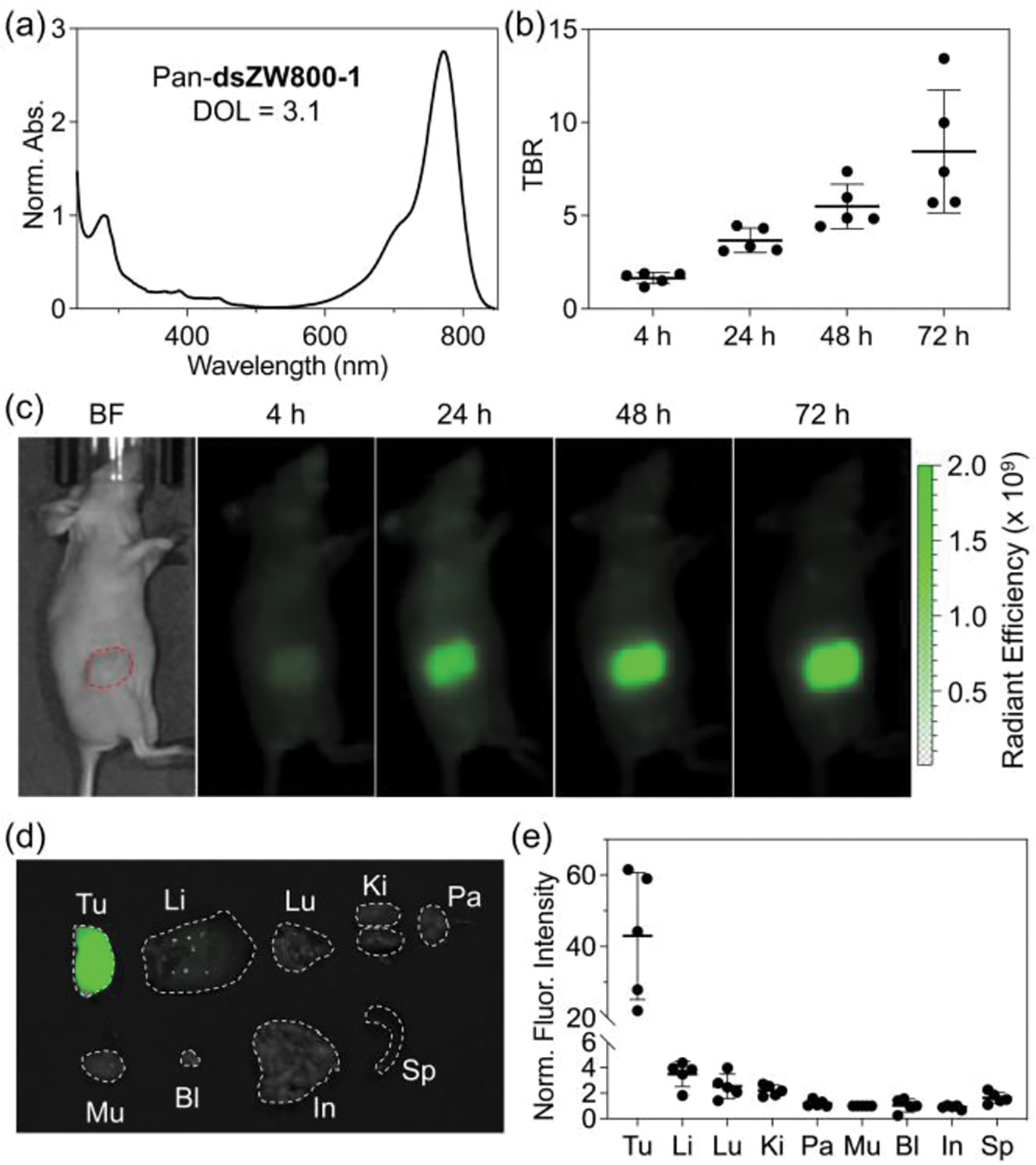
(a) Normalized absorption spectrum of Pan-dsZW800-1 (Degree of labeling, DOL = 3.1) in pH 7.2 PBS at room temperature. (b) Tumor-to-background ratio (TBR) from in vivo NIR-I imaging (ex: 745 ± 15 nm, em: 800 ± 10 nm) of JIMT-1 tumor-bearing mice (N = 5) after intravenous injection of Pan-dsZW800-1 (50 μg dose) and imaging at 4, 24, 48, and 72 h post-injection. (c) Representative in vivo NIR-I fluorescence images of the JIMT-1 tumor-bearing mice. (d) Representative overlaid brightfield and NIR-I fluorescence images of excised organs, and (e) Biodistribution of Pan-dsZW800-1 determined by quantifying the NIR-I fluorescence of excised organs (N = 5). Tu: tumor, Li: liver, Lu: lung, Ki: kidneys, Pa: pancreas, Mu: muscle, Bl: bladder, In: small intestines, Sp: spleen.
Synthesis of dsZW1015
The next step in the project was to expand the spectral scope of doubly strapped heptamethine dyes to fluorochromes that absorb and emit in the NIR-II region (1000–1700 nm) which improves in vivo imaging.[61] The recent availability of commercial InGaAs cameras has greatly facilitated fluorescent NIR-II imaging, but a current limitation to further advancement is the lack of relatively small (MW < 2000 Da) hydrophilic and zwitterionic NIR-II dyes that can be attached to targeting biomolecules, such as an antibody or affibody, without forming non-fluorescent dye H-stacks or altering the targeting specificity of the biomacromolecule. [62–65] The extensive hydrophobic surface area of NIR-II dyes makes it very challenging to produce water-soluble bioconjugates for targeted bioimaging.[62] A well-known NIR-II cyanine dye is FD-1080 (Scheme 1) whose structure includes hydrophobic benzo[c,d]indole heterocycles as the terminal units.[66,67] In PBS solution, FD-1080 (and close structural analogues)[68] forms non-fluorescent H-aggregates and thus it has to be formulated as a non-covalent complex with serum blood proteins, or packaged inside self-assembled micelles or liposomes for in vivo imaging studies.[50,69–71] A second functional drawback with FD-1080 is the lack of a suitable protein conjugation site.[72] To address both concerns, we employed the modular synthetic sequence shown in Scheme 3 to prepare the doubly strapped, charged balanced NIR-II dye dsZW1015. The key synthetic precursor was heptamethine dye 5 which was prepared in a four-step sequence (see Supporting Information) that started with commercial benzo[c,d]indol-2(1H)-one. Two subsequent steps introduced the conjugatable carboxyl group and produced unstrapped heptamethine 7, which was converted to the desired doubly strapped version, dsZW1015.[73] It is worth noting that the non-planar structures of all the compounds listed in Scheme 3 greatly facilitated purification by column chromatography.
Scheme 3.
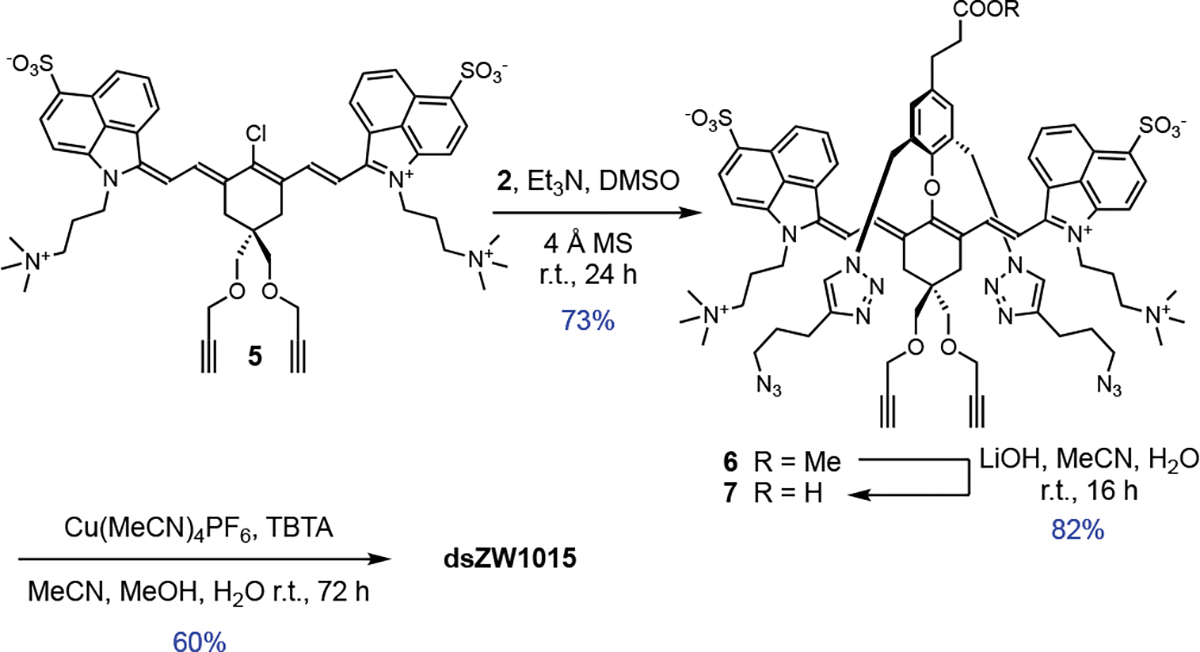
Synthesis of doubly strapped, conjugatable NIR-II dye dsZW1015.
Spectral Properties of dsZW1015
NIR-II dyes have inherently low fluorescence quantum yields and the measured values in DMSO of 0.10 % for dsZW1015 and 0.11 % for unstrapped isomer 7 (Table 1) are comparable to literature NIR-II dyes in nonpolar organic solvents.[74] In water, NIR-II fluorescence quantum yields are even lower, [51–53] but we were pleased to find that the fluorescence quantum yield in PBS for dsZW1015-1 (0.014%) is more than double the value for unstrapped isomer 7 (0.006%). There are two likely reasons for the increased brightness. One is enhanced steric shielding of the NIR-II fluorochrome from the surrounding hydration sphere (analogous to the above comparison of ZW800-1 and 4), and the second is a noticeable difference in dye self-aggregation. Evidence for the latter factor was gained by inspecting the absorption spectra in Figure S6 which show that unstrapped 7 in PBS exhibits a second, blue-shifted and non-fluorescent, H-aggregate absorption band at ~850 nm, whereas doubly strapped dsZW1015 only exists as an emissive monomer band at 980 nm.
In Vivo Imaging using dsZW1015
The NHS ester of dsZW1015 was used to produce Pan-dsZW1015 conjugate with DOL = 1.2. The absorption spectrum for Pan-dsZW1015 only exhibits an emissive monomeric dye peak at 1000 nm (Figure 5a) with no evidence for H-stacking of multiple appended dyes and no change in spectral properties after 15 days at 4 °C (Figure S15b). To the best of our knowledge dsZW1015 is the first zwitterionic heptamethine NIR-II dye to be conjugated to a biomacromolecule.[61,75] The imaging performance of Pan-dsZW1015 was evaluated in the JIMT-1 mouse tumor model. The in vivo whole-body NIR-II fluorescent images (Figure S22 and S23) showed a strong initial hepatic signal (4 h and 24 h) attributed to the influence of the appended hydrophobic benzo[c,d]indole-based dye. Nonetheless, clear tumor targeting was observed at 72 h post-injection with very low off-target signal (Figure 5b and 5c). The imaging results demonstrate that dsZW1015 with its favorable structural combination of steric shielding and balanced charge enables the production of a labeled antibody with specific targeting properties for effective NIR-II imaging of living subjects.
Figure 5.
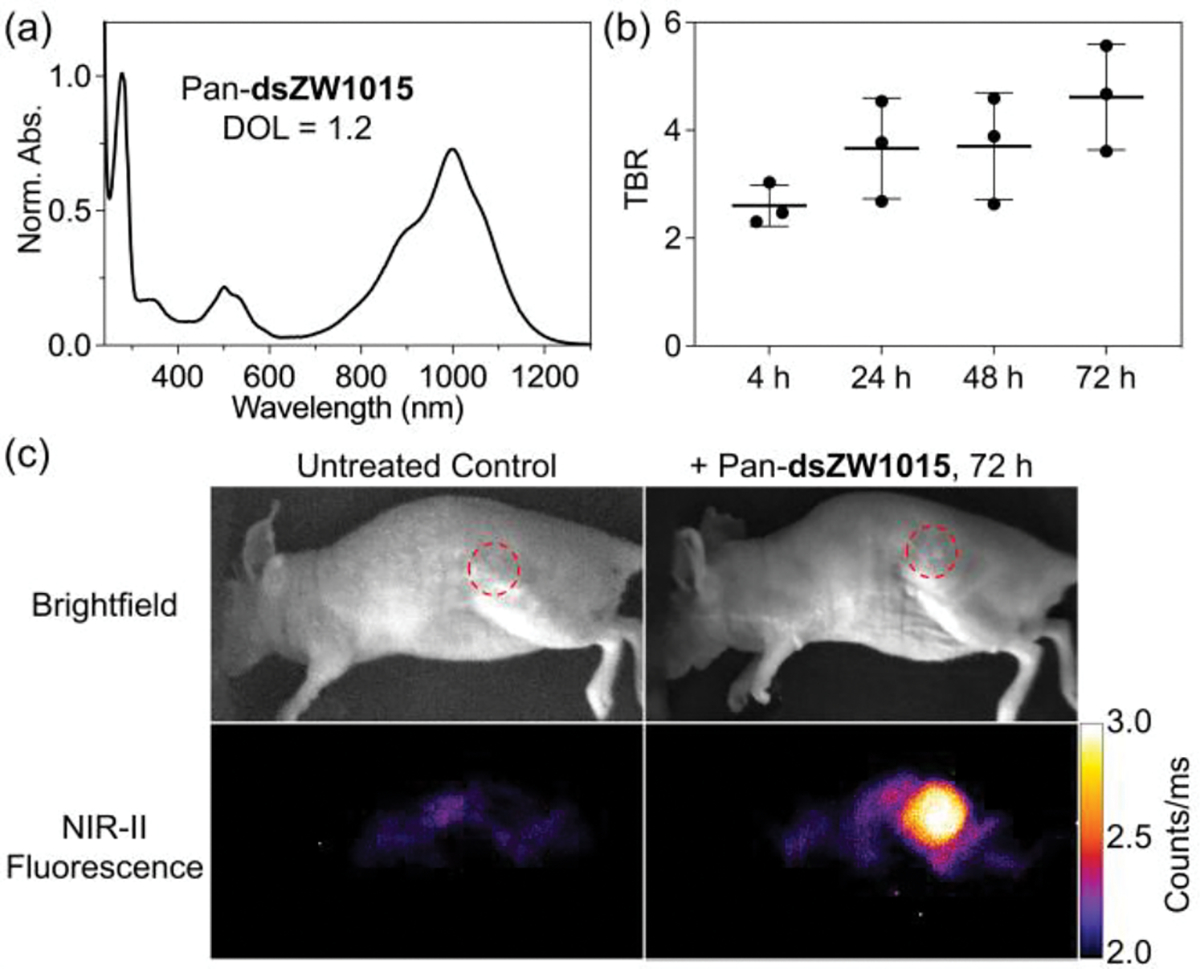
(a) Normalized absorption spectrum of Pan-dsZW1015 (DOL = 1.2) in pH 7.2 PBS at room temperature. (b) Tumor-to-background ratio (TBR) from in vivo NIR-II imaging of JIMT-1 tumor-bearing mice (N = 3) after intravenous injection of Pan-dsZW1015 (250 μg dose) and imaging at 4, 24, 48, and 72 h post-injection. (c) Representative in vivo brightfield and NIR-II fluorescence images of a Pan-dsZW1015 treated or untreated JIMT-1 tumor-bearing mouse (ex: 890 nm, em: >1150 nm LP filters).
Conclusion
In summary, we have validated doubly strapped heptamethine cyanine dyes as a new class of high performance, conjugatable NIR fluorophores. Compared to the benchmark NIR-I dye ZW800-1, the doubly strapped analogue dsZW800-1 exhibits slightly higher fluorescence brightness, substantially greater chemical stability and twice the photostability. Notably, dsZW800-1 is superior to ZW800-1 for fluorescence imaging using a dye-labeled antibody and will be much better suited for longitudinal experiments that measure long-term degradation or trafficking of implanted dye-labeled biomaterials. It is worth noting that there is a growing family of fluorescent probes that utilize NIR-I heptamethine cyanine dyes with structures that are closely related to ZW800-1 and each is likely to have stability limitations due to the presence of a reactive 4’-phenoxy group. [35,36,76–80] In each case, it should be straightforward to prepare doubly strapped versions of these NIR-I dyes with an expectation of improved stability and imaging performance. An attractive feature of the modular synthetic method is the straightforward manner that the terminal heterocycles can be varied, as highlighted by the preparation of the doubly strapped heptamethine cyanine dye, dsZW1015, with terminal benzo[c,d]indole heterocycles that extend the absorption/emission wavelengths into the NIR-II region.[73] The structural combination of steric shielding and balanced charge counters the enhanced hydrophobicity of the fluorochrome within dsZW1015 and produces dye-labeled antibodies for effective NIR-II fluorescence imaging of tumor in living mice. Future design iterations can incorporate suitable functional groups within the terminal heterocycles to produce bioresponsive versions of these doubly strapped cyanine dyes for bioconjugation and deployment as next-generation “turn on” or “ratiometric” near-infrared fluorescent probes.[81,82]
Supplementary Material
Acknowledgements
We are grateful for funding by the NIH (R35GM136212). This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), NCI-CCR. We acknowledge Lai Thang (Small Animal Imaging Program), and Chelsea Sanders and Dr. Simone Difilippantonio (Laboratory Animal Sciences Program) for expert assistance with the in vivo study.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Wu J, Shi Z, Zhu L, Li J, Han X, Xu M, Hao S, Fan Y, Shao T, Bai H, Peng B, Hu W, Liu X, Yao C, Li L, Huang W, Adv. Opt. Mater. 2022, 10, 2102514. [Google Scholar]
- [2].Shi C, Wu JB, Pan D, J. Biomed. Opt. 2016, 21, 050901. [DOI] [PubMed] [Google Scholar]
- [3].Du Y, Liu X, Zhu S, Front. Chem. 2021, 9, 718709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Reinhart MB, Huntington CR, Blair LJ, Heniford BT, Augenstein VA, Surg. Innov. 2016, 23, 166–175. [DOI] [PubMed] [Google Scholar]
- [5].Papayan G, Akopov A, Photodiagnosis Photodyn. Ther. 2018, 24, 292–299. [DOI] [PubMed] [Google Scholar]
- [6].Li Y, Zhou Y, Yue X, Dai Z, Adv. Healthc. Mater. 2020, 9, 2001327. [DOI] [PubMed] [Google Scholar]
- [7].Li D, Schreiber CL, Smith BD, Angew. Chem. Int. Ed. 2020, 59, 12154–12161; Angew. Chem. 2020, 132, 12252–12259; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Luciano MP, Crooke SN, Nourian S, Dingle I, Nani RR, Kline G, Patel NL, Robinson CM, Difilippantonio S, Kalen JD, Finn MG, Schnermann MJ, ACS Chem. Biol. 2019, 14, 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Korb ML, Hartman YE, Kovar J, Zinn KR, Bland KI, Rosenthal EL, J. Surg. Res. 2014, 188, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schouw HM, Huisman LA, Janssen YF, Slart RHJA, Borra RJH, Willemsen ATM, Brouwers AH, van Dijl JM, Dierckx RA, van Dam GM, Szymanski W, Boersma HH, Kruijff S, Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4272–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Debie P, Van Quathem J, Hansen I, Bala G, Massa S, Devoogdt N, Xavier C, Hernot S, Mol. Pharm. 2017, 14, 1145–1153. [DOI] [PubMed] [Google Scholar]
- [12].Buckle T, van Willigen DM, Spa SJ, Hensbergen AW, van der Wal S, de Korne CM, Welling MM, van der Poel HG, Hardwick JCH, van Leeuwen FWB, J. Nucl. Med. 2018, 59, 986–992. [DOI] [PubMed] [Google Scholar]
- [13].Usama SM, Thapaliya ER, Luciano MP, Schnermann MJ, Curr. Opin. Chem. Biol. 2021, 63, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pauli J, Pochstein M, Haase A, Napp J, Luch A, Resch-Genger U, ChemBioChem 2017, 18, 101–110. [DOI] [PubMed] [Google Scholar]
- [15].García de Jalón E, Kleinmanns K, Fosse V, Davidson B, Bjørge L, Haug BE, McCormack E, Mol. Imaging Biol. 2023, 25, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Erfani A, Seaberg J, Aichele CP, Ramsey JD, Biomacromolecules 2020, 21, 2557–2573. [DOI] [PubMed] [Google Scholar]
- [17].Choi HS, Gibbs SL, Lee JH, Kim SH, Ashitate Y, Liu F, Hyun H, Park G, Xie Y, Bae S, Henary M, V Frangioni J, Nat. Biotechnol. 2013, 31, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Choi HS, Nasr K, Alyabyev S, Feith D, Lee JH, Kim SH, Ashitate Y, Hyun H, Patonay G, Strekowski L, Henary M, Frangioni JV, Angew. Chem. Int. Ed. 2011, 50, 6258–6263; Angew. Chem. 2011, 123, 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].de Valk KS, Handgraaf HJ, Deken MM, Sibinga Mulder BG, Valentijn AR, Terwisscha van Scheltinga AG, Kuil J, van Esdonk MJ, Vuijk J, Bevers RF, Peeters KC, Holman FA, Frangioni JV, Burggraaf J, Vahrmeijer AL, Nat. Commun. 2019, 10, 3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].de Valk KS, Deken MM, Handgraaf HJM, Bhairosingh SS, Bijlstra OD, van Esdonk MJ, Terwisscha van Scheltinga AGT, Valentijn ARPM, March TL, Vuijk J, Peeters KCMJ, Holman FA, Hilling DE, Mieog JSD, Frangioni JV, Burggraaf J, Vahrmeijer AL, Clin. Cancer Res. 2020, 26, 3990–3998. [DOI] [PubMed] [Google Scholar]
- [21].Handgraaf HJM, Boonstra MC, Prevoo HAJM, Kuil J, Bordo MW, Boogerd LSF, Sibinga Mulder BG, Sier CFM, Vinkenburg-van Slooten ML, Valentijn ARPM, Burggraaf J, van de Velde CJH, Frangioni JV, Vahrmeijer AL, Oncotarget 2017, 8, 21054–21066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee S, Lim W, Jung JS, Jo D, Jo G, Park MH, Hyun H, Macromol. Res. 2018, 26, 1251–1256. [Google Scholar]
- [23].Gibbs SL, Genega E, Salemi J, Kianzad V, Goodwill HL, Xie Y, Oketokoun R, Khurd P, Kamen A, Frangioni JV, Mol. Imaging 2015, 14, DOI 10.2310/7290.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Inoue K, Gibbs SL, Liu F, Lee JH, Xie Y, Ashitate Y, Fujii H, Frangioni JV, Choi HS, J. Nucl. Med. 2014, 55, 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim SH, Lee JH, Hyun H, Ashitate Y, Park G, Robichaud K, Lunsford E, Lee SJ, Khang G, Choi HS, Sci. Rep. 2013, 3, 1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim SH, Kwon JS, Cho JG, Park KG, Lim TH, Kim MS, Choi HS, Park CH, Lee SJ, Bioeng. Transl. Med. 2021, 6, 1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Suh YJ, Lim TH, Choi HS, Kim MS, Lee SJ, Kim SH, Park CH, Materials 2020, 13, 4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park GK, Kim SH, Kim K, Das P, Kim BG, Kashiwagi S, Choi HS, Hwang NS, Theranostics 2019, 9, 4255–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee S, Jo G, Jung JS, Yang DH, Hyun H, Artif. Cells, Nanomedicine Biotechnol. 2020, 48, 1144–1152. [DOI] [PubMed] [Google Scholar]
- [30].Kim SH, Park JH, Kwon JS, Cho JG, Park KG, Park CH, Yoo JJ, Atala A, Choi HS, Kim MS, Lee SJ, Biomaterials 2020, 258, 120267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Garifo S, Stanicki D, Boutry S, Larbanoix L, Ternad I, Muller RN, Laurent S, Nanoscale 2021, 13, 16509–16524. [DOI] [PubMed] [Google Scholar]
- [32].Kang H, Kang M-W, Kashiwagi S, Choi HS, J. Immunother. Cancer 2022, 10, e004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hyun H, Owens EA, Narayana L, Wada H, Gravier J, Bao K, Frangioni JV, Choi HS, Henary M, RSC Adv. 2014, 4, 58762–58768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Deng H, Konopka CJ, Cross TL, Swanson KS, Dobrucki LW, Smith AM, ACS Nano 2020, 14, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zaheer A, Wheat TE, Frangioni JV, Mol. Imaging 2002, 1, 354–364. [DOI] [PubMed] [Google Scholar]
- [36].Mahalingam SM, Kularatne SA, Myers CH, Gagare P, Norshi M, Liu X, Singhal S, Low PS, J. Med. Chem. 2018, 61, 9637–9646. [DOI] [PubMed] [Google Scholar]
- [37].Wang G, Zannikou M, Lofchy L, Li Y, Gaikwad H, Balyasnikova IV, Simberg D, ACS Nano 2021, 15, 11880–11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schreiber CL, Li D-H, Smith BD, Anal. Chem. 2021, 93, 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gamage RS, Li D-H, Schreiber CL, Smith BD, ACS Omega 2021, 6, 30130–30139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li D-H, Gamage RS, Smith BD, J. Org. Chem. 2022, 87, 11593–11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ahrens L, Tverskoy O, Weigold S, Ganschow M, Rominger F, Freudenberg J, Bunz UHF, Angew. Chem. Int. Ed. 2021, 60, 9270–9273; Angew. Chem. 2021, 133, 9356–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Royakkers J, Minotto A, Congrave DG, Zeng W, Patel A, Bond AD, Bučar DK, Cacialli F, Bronstein H, J. Org. Chem. 2020, 85, 207–214. [DOI] [PubMed] [Google Scholar]
- [43].Zeng K, Lu Y, Tang W, Zhao S, Liu Q, Zhu W, Tian H, Xie Y, Chem. Sci. 2019, 10, 2186–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sugiyasu K, Ogi S, Takeuchi M, Polym. J. 2014, 46, 674–681. [Google Scholar]
- [45].Royakkers J, Guo K, Toolan DTW, Feng LW, Minotto A, Congrave DG, Danowska M, Zeng W, Bond AD, Al-Hashimi M, Marks TJ, Facchetti A, Cacialli F, Bronstein H, Angew. Chem. Int. Ed. 2021, 60, 25005–25012; Angew. Chem. 2021, 133, 25209–25216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Singh Mehra K, Jha S, Bhandary S, Mandal D, Mishra R, Sankar J, Angew. Chem. Int. Ed. 2022, 61, e202205600; Angew. Chem. 2022, 134, e202205600. [DOI] [PubMed] [Google Scholar]
- [47].Yang F, Li R, Wei W, Ding X, Xu Z, Wang P, Wang G, Xu Y, Fu H, Zhao Y, Angew. Chem. Int. Ed. 2022, 61, e202202491; Angew. Chem. 2022, 134, e202202491. [DOI] [PubMed] [Google Scholar]
- [48].Deposition number 2236818 contains the supplementary crystallographic data for dsZW800-1. The data is provided free of charge by the joint Cambridge Crystallographic Data Centre and Fachinformationszentrum Karlsruhe Access Structures service. [Google Scholar]
- [49].Xu X, Strongin RM, Fronczek FR, CSD Commun. 2015, CCDC 1404767 and 1404768. [Google Scholar]
- [50].Sun C, Li B, Zhao M, Wang S, Lei Z, Lu L, Zhang H, Feng L, Dou C, Yin D, Xu H, Cheng Y, Zhang F, J. Am. Chem. Soc. 2019, 141, 19221–19225. [DOI] [PubMed] [Google Scholar]
- [51].Yang Q, Ma H, Liang Y, Dai H, Acc. Mater. Res. 2021, 2, 170–183. [Google Scholar]
- [52].Matikonda SS, Hammersley G, Kumari N, Grabenhorst L, Glembockyte V, Tinnefeld P, Ivanic J, Levitus M, Schnermann MJ, J. Org. Chem. 2020, 85, 5907–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Maillard J, Klehs K, Rumble C, Vauthey E, Heilemann M, Fuerstenberg A, Chem. Sci. 2021, 12, 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Semonin OE, Johnson JC, Luther JM, Midgett AG, Nozik AJ, Beard MC, J. Phys. Chem. Lett. 2010, 1, 2445–2450. [Google Scholar]
- [55].Nani RR, Kelley JA, Ivanic J, Schnermann MJ, Chem. Sci. 2015, 6, 6556–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hirakawa K, Hirano T, Nishimura Y, Arai T, Nosaka Y, J. Phys. Chem. B 2012, 116, 3037–3044. [DOI] [PubMed] [Google Scholar]
- [57].Exner RM, Cortezon-Tamarit F, Pascu SI, Angew. Chem. Int. Ed. 2021, 60, 6230–6241; Angew. Chem. 2021, 133, 6295–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lin C-M, Usama SM, Burgess K, Molecules 2018, 23, 2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Forman HJ, Zhang H, Rinna A, Mol. Aspects Med. 2009, 30, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gurumurthy B, Tucci MA, Fan LW, Benghuzzi HA, Pal P, Bidwell GL, Salazar Marocho SM, Cason Z, Gordy D, Janorkar AV, Adv. Healthc. Mater. 2020, 9, 1901385. [DOI] [PubMed] [Google Scholar]
- [61].Mu J, Xiao M, Shi Y, Geng X, Li H, Yin Y, Chen X, Angew. Chem. Int. Ed. 2022, 61, e202114722; Angew. Chem. 2022, 134, e202114722. [DOI] [PubMed] [Google Scholar]
- [62].Lei Z, Zhang F, Angew. Chem. Int. Ed. 2021, 60, 16294–16308; Angew. Chem. 2021, 133, 16430–16444. [DOI] [PubMed] [Google Scholar]
- [63].Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Zhang X, Yaghi OK, Alamparambil ZR, Hong X, Cheng Z, Dai H, Nat. Mater. 2016, 15, 235–242. [DOI] [PubMed] [Google Scholar]
- [64].Zhu S, Yang Q, Antaris AL, Yue J, Ma Z, Wang H, Huang W, Wan H, Wang J, Diao S, Zhang B, Li X, Zhong Y, Yu K, Hong G, Luo J, Liang Y, Dai H, Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bandi VG, Luciano MP, Saccomano M, Patel NL, Bischof TS, Lingg JGP, Tsrunchev PT, Nix MN, Ruehle B, Sanders C, Riffle L, Robinson CM, Difilippantonio S, Kalen JD, Resch-Genger U, Ivanic J, Bruns OT, Schnermann MJ, Nat. Methods 2022, 19, 353–358. [DOI] [PubMed] [Google Scholar]
- [66].Li B, Lu L, Zhao M, Lei Z, Zhang F, Angew. Chem. Int. Ed. 2018, 57, 7483–7487; Angew. Chem. 2018, 130, 7605–7609. [DOI] [PubMed] [Google Scholar]
- [67].Henary M, Mojzych M, Say M, Strekowski L, J. Heterocycl. Chem. 2009, 46, 84–87. [Google Scholar]
- [68].Ma X, Huang Y, Abedi SAA, Kim H, Davin TTB, Liu X, Yang WC, Sun Y, Liu SH, Yin J, Yoon J, Yang GF, CCS Chem. 2022, 4, 1961–1976. [Google Scholar]
- [69].Gao D, Luo Z, He Y, Yang L, Hu D, Liang Y, Zheng H, Liu X, Sheng Z, Small, 2023, 19, 2206544. [DOI] [PubMed] [Google Scholar]
- [70].Qian H, Cheng Q, Tian Y, Dang H, Teng C, Yan L, J. Mater. Chem. B 2021, 9, 2688–2696. [DOI] [PubMed] [Google Scholar]
- [71].Zhao M, Ding J, Mao Q, Zhang Y, Gao Y, Ye S, Qin H, Shi H, Nanoscale 2020, 12, 6953–6958. [DOI] [PubMed] [Google Scholar]
- [72].US patent application US2019/0100654 A1 describes a dye called D2 that is a derivative of FD1080 with a conjugatable carboxyl residue and five sulfonates. Absorption spectra indicate that D2 is dispersed a monomer in water and that it can be conjugated to mouse IgG. A homologous dye called D3, has only two sulfonates, and forms H-aggregates in water. The patent does not mention any dye homologue with a charge balanced molecular structure. [Google Scholar]
- [73].Success making dsZW1015 is a welcome synthetic advance since we cannot synthesize analogous shielded NIR-II dyes in useful yields using the literature Zincke method described in reference 7.
- [74].Pan H-M, Wu C-C, Lin C-Y, Hsu C-S, Tsai Y-C, Chowdhury P, Wang C-H, Chang K-H, Yang C-H, Liu M-H, Chen Y-C, Su S-P, Lee Y-J, Chiang HK, Chan Y-H, Chou P-T, J. Am. Chem. Soc. 2023, 145, 516–526. [DOI] [PubMed] [Google Scholar]
- [75].Liu C, Ma H, Hu Z, Tian R, Ma R, Xu Y, Wang X, Zhu X, Yu P, Zhu S, Sun H, Liang Y, Front. Chem. 2021, 9, 739802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mahalingam SM, Dudkin VY, Goldberg S, Klein D, Yi F, Singhal S, O’Neil KT, Low PS, Bioconjug. Chem. 2017, 28, 2865–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Roy J, Kaake M, Low PS, Oncotarget 2019, 10, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mahalingam SM, Chu H, Lin X, Leamon CP, Low PS, Bioconjug. Chem. 2018, 29, 3320–3331. [DOI] [PubMed] [Google Scholar]
- [79].Tung CH, Han MS, Shen Z, Gray BD, Pak KY, Wang J, ACS Sensors 2021, 6, 3657–3666. [DOI] [PubMed] [Google Scholar]
- [80].Kularatne SA, Thomas M, Myers CH, Gagare P, Kanduluru AK, Crian CJ, Cichocki BN, Clin. Cancer Res. 2019, 25, 177–187. [DOI] [PubMed] [Google Scholar]
- [81].Lan Q, Yu P, Yan K, Li X, Zhang F, Lei Z, J. Am. Chem. Soc. 2022, 144, 21010–21015. [DOI] [PubMed] [Google Scholar]
- [82].Chen S, Xiao L, Li Y, Qiu M, Yuan Y, Zhou R, Li C, Zhang L, Jiang Z-X, Liu M, Zhou X, Angew. Chem. Int. Ed. 2022, 61, e202213495; Angew. Chem. 2022, 134, e202213495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


