Abstract
The persistent environmental aryl hydrocarbon receptor agonist and hepatotoxin TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) induces hepatic lipid accumulation (steatosis), inflammation (steatohepatitis) and fibrosis. Thousands of liver-expressed, nuclear-localized lncRNAs with regulatory potential have been identified; however, their roles in TCDD-induced hepatoxicity and liver disease are unknown. We analyzed single nucleus (sn)RNA-seq data from control and subchronic (4 wk) TCDD-exposed mouse liver to determine liver cell-type specificity, zonation and differential expression profiles for thousands of IncRNAs. TCDD dysregulated >4,000 of these lncRNAs in one or more liver cell types, including 684 lncRNAs specifically dysregulated in liver non-parenchymal cells. Trajectory inference analysis revealed major disruption by TCDD of hepatocyte zonation, affecting >800 genes, including 121 IncRNAs, with strong enrichment for lipid metabolism genes. TCDD also dysregulated expression of >200 transcription factors, including 19 Nuclear Receptors, most notably in hepatocytes and Kupffer cells. TCDD-induced changes in cell–cell communication patterns included marked decreases in EGF signaling from hepatocytes to non-parenchymal cells and increases in extracellular matrix-receptor interactions central to liver fibrosis. Gene regulatory networks constructed from the snRNA-seq data identified TCDD-exposed liver network-essential lncRNA regulators linked to functions such as fatty acid metabolic process, peroxisome and xenobiotic metabolism. Networks were validated by the striking enrichments that predicted regulatory IncRNAs showed for specific biological pathways. These findings highlight the power of snRNA-seq to discover functional roles for many xenobiotic-responsive lncRNAs in both hepatocytes and liver non-parenchymal cells and to elucidate novel aspects of foreign chemical-induced hepatotoxicity and liver disease, including dysregulation of intercellular communication within the liver lobule.
Keywords: Ah receptor, single cell gene regulatory networks, xenobiotic-responsive lncRNAs, liver nuclear receptors, hepatic intercellular communication, TCDD-responsive nuclear receptors
Introduction
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a persistent hepatotoxic environmental chemical that directly binds to aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor that dysregulates hundreds of genes (Kulkarni et al., 2008; Bock, 2020). TCDD-activated AHR translocates to the nucleus, where it heterodimerizes with ARNT and then binds to chromatin at thousands of regulatory sequences controlling transcription of linked target genes (Cholico et al., 2022). TCDD also induces AHR-dependent nongenomic signaling via changes in intracellular Ca2+ and activation of the tyrosine kinase SRC and downstream kinases (Larigot et al., 2018; Patrizi and Siciliani de Cumis, 2018). TCDD exposure leads to a broad range of hepatotoxic and other effects in both mice and humans (Pelclova et al., 2006; Girer et al., 2020), including immune suppression, hepatic lipid accumulation and progression to non-alcoholic steatohepatitis (NASH) with fibrosis (Pierre et al., 2014; Nault et al., 2016; Nault et al., 2017b). TCDD also has the potential to induce epigenetic alterations that may be passed along trans-generationally (Viluksela and Pohjanvirta, 2019). Many metabolic and other pathways dysregulated by TCDD have been identified (Fader and Zacharewski, 2017), but the precise mechanisms of TCDD action and in particular the role of AHR downstream genes that mediate the effects of subchronic TCDD exposure on liver metabolic diseases, including NASH and liver fibrosis, remains an area of active investigation.
The liver carries out a wide array of physiological processes, including energy homeostasis, gluconeogenesis, serum protein synthesis and xenobiotic metabolism. These diverse processes are enabled by the liver’s complex architecture, where hepatocytes and non-parenchymal cells (NPCs) are organized within local repeating structures known as the liver lobule. Hepatocytes constitute about 70% of the liver mass while the remaining 30% consists of NPCs, primarily endothelial cells, hepatic stellate cells, and various immune cell populations, including Kupffer cells (liver resident macrophages) (Kegel et al., 2016). Single-cell transcriptomics has the power to elucidate cell-specific responses and spatial zonation (Halpern et al., 2017; Ben-Moshe et al., 2019) and has transformed our understanding of the roles these diverse cells play in liver homeostasis (MacParland et al., 2018) and pathophysiology (Dobie et al., 2019; Xiong et al., 2019), including following subchronic TCDD exposure (Nault et al., 2021).
Long non-coding RNAs (lncRNAs) are RNAs longer than 200 bp with little or no protein-coding potential. Thousands of liver-expressed IncRNAs are enriched in the nucleus where many bind tightly to liver chromatin (Goldfarb and Waxman, 2021), enabling them to play essential regulatory roles in modulating chromatin function and gene expression (Statello et al., 2021). Several hundred lncRNAs show significant differences in expression between male and female mouse liver (Melia and Waxman, 2019; Goldfarb and Waxman, 2021; Goldfarb et al., 2022) and/or respond to drug and foreign chemical exposure (Lodato et al., 2017; Dempsey and Cui, 2019; Karri and Waxman, 2020; Goldfarb and Waxman, 2021). Individual lncRNAs have been linked to hepatotoxicity induced by foreign chemical exposures (Zeng et al., 2019; Zhang et al., 2020), however, a global understanding of the roles lncRNAs play in foreign chemical-induced hepatotoxicity is lacking. Prior studies identified specific, individual lncRNAs with roles in certain extrahepatic effects of TCDD exposure (Garcia et al., 2018; Lee et al., 2020; He et al., 2021) but did not investigate effects in liver, a major target of TCDD action. Recently, Nault et al used snRNA-seq to characterize liver cell type-specific transcriptional changes in a subchronic (28 day) TCDD exposure model and reported widespread dysregulation of gene expression in multiple liver cell types, as well as marked expansion of the liver Kupffer cell/macrophage population and extensive TCDD-induced changes in hepatic zonation (Nault et al., 2021; Nault et al., 2023). However, the effects of TCDD on global lncRNA expression, including the many thousands of novel liver-expressed lncRNAs recently identified (Karri and Waxman, 2023), were not considered.
Here, we utilized a single cell transcriptome reference gene set comprised of 76,011 mouse genes, including 48,261 mouse liver-expressed lncRNAs whose gene structures and isoform patterns we recently characterized (Karri and Waxman, 2023), to discover lncRNAs whose expression and zonation across the liver lobule is dysregulated in either hepatocytes or liver NPCs following subchronic TCDD exposure. We also characterize the liver cell type-dependent effects of TCDD on expression of more than 1,500 transcription factors, including Ahr itself and many members of the nuclear receptor (NR) superfamily. Furthermore, we map the global landscape of intercellular signaling between liver cell types and explore TCDD-induced changes in intrahepatic cell-cell communication, which plays a critical role in tissue homeostasis and injury response (Jin et al., 2021) and may contribute to TCDD hepatotoxicity. Finally, we construct gene regulatory networks to link individual TCDD-responsive lncRNAs to biological functions, and using network centrality metrics, we identify network-essential regulatory nodes (Madhamshettiwar et al., 2012; Iacono et al., 2019) that give systems-level mechanistic insight into the regulatory functions of both lncRNAs and protein-coding genes contributing to the widespread hepatic effects of TCDD exposure.
Methods
Data processing of snRNA-seq samples –
Single nucleus-based RNA-seq raw sequencing data (Fastq files) for control male C57BL/6 mice, or male mice gavaged with 30 μg/kg TCDD every 4 d for 28 d (subchronic exposure) (Nault et al., 2021) were downloaded from GEO (accession #GSE148339; https://www.ncbi.nlm.nih.gov/geo/), processed using CellRanger (v3.1.0) (Zheng et al., 2017) and aligned to the mouse mm10 reference genome (Table S1A). Mapped snRNA-seq reads were assigned to individual genes using a custom, tab-delimited GTF (Gene Transfer Format) file, which provides detailed gene structure information and is used to count sequence reads mapping to the genomic regions specified for each gene (see below). The custom GTF file used here includes entries that span the full gene body of each gene (rather than exonic regions only, as is typically carried out when counting single cell-RNA-seq reads) to enable counting of intronic reads, which comprise a substantial fraction of the snRNA-seq reads. For gene pairs that overlap each other on the same strand (whose gene counts would be excluded by the 10X Genomics CellRanger’s counting algorithm across the entire region of overlap), we modified the GTF file entries to include all exons of both overlapping genes, plus all non-exonic gene regions that were unique to each gene, as described elsewhere (Goldfarb et al., 2022). Thus, sequence reads that map to an exon of one gene but overlap an intronic region of the other, overlapping gene were uniquely counted for the first gene by excluding the overlapping intronic region of the second gene from the GTF file annotation for that gene.
The custom GTF file used for counting, GTFB_FullGeneBody_MouseLiver_snRNAseq.gtf, is available at https://tinyurl.com/GTF-MouseLiver48kLncRNAs and includes a total of 76,011 mouse mm10 genes, plus 91 ERCC spike-in control sequences: 1) 20,973 RefSeq protein-coding genes including 13 mitochondrial genes; 2) 2,077 RefSeq non-coding genes (i.e., RefSeq genes assigned NR accession numbers that do not overlap the set of 48,261 lncRNAs described immediately below); and 3) a total of a total of 52,961 lncRNA genes, comprised of 48,261 liver-expressed lncRNAs (Karri and Waxman, 2023) plus 4,700 other lncRNA genes, namely, 4,697 Ensembl noncoding lncRNAs that do not overlap the RefSeq NR dataset or the set of 48,261 liver-expressed lncRNAs and 3 other lncRNAs (lnc-LFAR1, LeXis, Lnclgr) that were absent from the above lists. (Of note, the set of 4,697 Ensembl genes is comprised of 3,176 genes designated ‘lincRNA’ and 1,521 genes designated ‘anti-sense’ with respect to a protein-coding gene in the Supplemental tables). Human and rat lncRNA sequences orthologous to the above sets of mouse lncRNAs were identified using methods described previously (Karri and Waxman, 2020) and are included in Table S1B. A large majority of the set of 48,261 liver-expressed lncRNAs are novel; only 5,057 are represented in the Gencode reference database, which includes many well characterized lncRNAs; these 5,057 lncRNAs are identified by their NR gene names in Table S8A of (Karri and Waxman, 2023), which presents extensive other characterization data for the full set of lncRNAs. The 48,261 lncRNAs account for 10.7-17.8% of all snRNA-seq UMI counts across the various liver cell types (Table S1C), with the ubiquitous lncRNA Malat1 (Inc31752*) comprising 26-46% of all lncRNA UMI counts.
Integration and clustering –
Feature-barcode matrices were generated using the 10x Genomics Cell Ranger pipeline (v3.1.0) (Zheng et al., 2017). snRNA-seq data from the control and TCDD-exposed liver samples were combined using the CellRanger aggr command. The resulting aggregated matrix was processed using Seurat package (v3.0) (Stuart et al., 2019) in R (v.3.6.0). Cells with fewer than 200 genes detected, less than 400 UMI per cell, or >5% mitochondrial contamination were excluded. Doublets and multiplets were identified and removed from the single-cell sequencing data using scDblFinder (Germain et al., 2021), a doublet detection algorithm that uses several methods, including artificial doublet generation, parameter optimization and thresholding (with default parameters). The final count matrix used for clustering included 15,573 nuclei (9,597 nuclei from control liver; 5,976 nuclei from TCDD-exposed liver).
The integrated count matrix data from Cellranger was normalized by dividing the UMI count per gene by the total UMI count in the corresponding cell and then log-transforming the data. Highly variable genes were identified using the FindVariableGenes function of Seurat (v3) with default parameters. Variable genes were projected onto a low-dimensional space using principal component (PC) analysis, with the number of PCs set to 10 based on an inspection of elbow plots of the variance explained for each dataset. Batch correction was performed using Harmony (Korsunsky et al., 2019) using the Seurat object as input, with default parameters to remove from the embedding the influence of dataset-of-origin factors. We input to Harmony normalized gene matrix files saved as a Seurat object along with pre-calculated PC analysis embedding based on 10 PCs using default parameters. Graph-based clustering of the cells based on their gene expression profiles was implemented using the FindClusters function in Seurat. The clusters obtained were visualized in Seurat v3 using the UMAP function (resolution: 0.2 and PC=10). Cell identities were assigned to each cluster based on the expression of established marker genes (Fig. S1B).
Differential expression analysis -
Differential gene expression between cell clusters under a given set of biological conditions (e.g., periportal vs pericentral hepatocytes from control liver), or between biological conditions for a given cell cluster (e.g., control endothelial cells vs TCDD-exposed endothelial cells) was performed using the Locally Distinguishing function of the 10x Genomics Loupe Browser (v.6.0) (see Loupe Browser file at https://doi.org/10.6084/m9.figshare.22661596.v1). This method implements the negative binomial test based on the sSeq method (Yu et al., 2013) with Benjamini-Hochberg correction for multiple tests; it uses log-normalized average expression values and the distribution of UMIs across the two specific cell populations being compared to obtain fold-change and FDR values for each pair-wise gene expression comparison. Genes were identified as showing significant differential expression between two cell populations if they met the threshold of |fold-change| >4 at FDR <0.05. This stringent, 4-fold cutoff was adopted to exclude from the list of regulated genes low expressed lncRNAs that show moderate differences in expression. The total number of genes expressed at any level in our dataset was determined by global differential expression analysis between each cell type cluster analyzed across all liver samples (Globally Distinguishing feature option within Loupe Browser); overall, 32,758 lncRNAs from the full list of 48,261 liver lncRNAs were detected in one or more of the snRNA-seq samples (Table S1C).
Functional enrichment analysis -
Enrichment analysis of protein-coding gene lists from various outputs was performed using DAVID (Sherman et al., 2022) with default parameters, except that GO FAT terms were used in place of GO DIRECT terms to include a broader range of enrichment terms than the default GO DIRECT option. We used a custom script prepared by Dr. Alan Downey-Wall of this laboratory (https://github.com/adowneywall/Davidaggregator) to aggregate and reformat DAVID output files from multiple gene lists, with each row presenting top enriched annotation clusters and individual columns presenting enrichment score, p-value, FDR and other such data. Top terms from each DAVID annotation cluster having a cluster enrichment score > 3 and top FDR < 0.05 were used in downstream analysis.
TCDD-elicited perturbations in hepatocyte zonation -
Monocle2 (Trapnell et al., 2014) was used to infer spatial trajectories for hepatocyte nuclei from control and TCDD-exposed liver. Genes showing significant zonation in control hepatocytes at q-value <0.001 were identified using generalized linear models via the differentialGeneTest function in Monocle. Genes that were differentially zonated between control liver and TCDD-exposed liver were identified by deriving a common pseudotime trajectory using Monocle2. We used the conditionTest function implemented in the package tradeSeq in Bioconductor (Van den Berge et al., 2020) to compare the two conditions (control and TCDD exposure) along a common trajectory to detect gene expression changes indicative of differential progression. The conditionTest function tests the null hypothesis that genes have identical expression patterns in each condition across a pseudotime. We extracted perturbed genes that were significant at FDR < 0.001 and created matched heatmaps for both control and TCDD-exposed liver using the pheatmap function of Monocle2. Heatmaps were further clustered individually for each condition to assign the zonation labels shown in Table S2C.
Transcription factor (TF) expression analysis -
The Dotplot function in Seurat (v3) was used to present expression profiles for each control mouse liver cell cluster for all 49 mouse Nr genes and for Ahr. Differential expression analysis between control and TCDD-exposed liver was carried out for those 50 TF genes, and also for a much larger listing of 1,533 mouse TFs (Table S3B), which were downloaded from AnimalTFDB3.0 (http://bioinfo.life.hust.edu.cn/AnimalTFDB/) along with their classifications into 72 families based on DNA-binding domains (Hu et al., 2019), by using the Locally Distinguishing function of the 10x Genomics Loupe Browser (v.6.0) (Table S3C), as described above. Fisher’s exact test was used to identify those TF families whose gene members were significantly enriched (at p-value <0.05) in the set of differentially expressed TFs from TCDD-exposed liver (Table S3D).
Cell-cell communication analysis -
CellChat (Jin et al., 2021) inputs gene expression data, assigns cell labels as input, and then models cell-cell communication probabilities by integrating gene expression with prior knowledge of the interactions between signaling ligands, receptors and their cofactors. We input to CellChat processed transcriptomic data from the 15,573 nuclei from control liver, and from TCDD-exposed liver, together with cell labels extracted from the Seurat object. Next, we inferred cell-cell communication probabilities using the computeCommunProb function, which infers biologically significant interactions by assigning to each interaction a probability value and then performing a permutation test. We ran CellChat analysis on the control liver and on the TCDD-exposed liver datasets separately and then merged the CellChat objects from each analysis using the mergeCellChat function. This yielded a total of 1,087 interactions (225 unique interactions) in control and TCDD-exposed liver (Table S4A). These interactions were grouped into 52 signaling pathways, as defined by CellChatDB (Fig. 5B). Pathways that were either conserved between biological conditions or were specific to TCDD-exposed liver were identified by comparing the information flow for each signaling pathway, which is defined by the sum of communication probabilities among all pairs of cell groups in the inferred network (i.e., the total weights in the network). Cell-cell interaction networks were visualized using the circle plot layout in CellChat, with edge colors consistent with the cell sources, and with edge weights proportional to the interaction strength (i.e., using a thicker edge to indicate a stronger signal).
Fig. 5. TCDD-induced changes in intercellular communication within mouse liver.
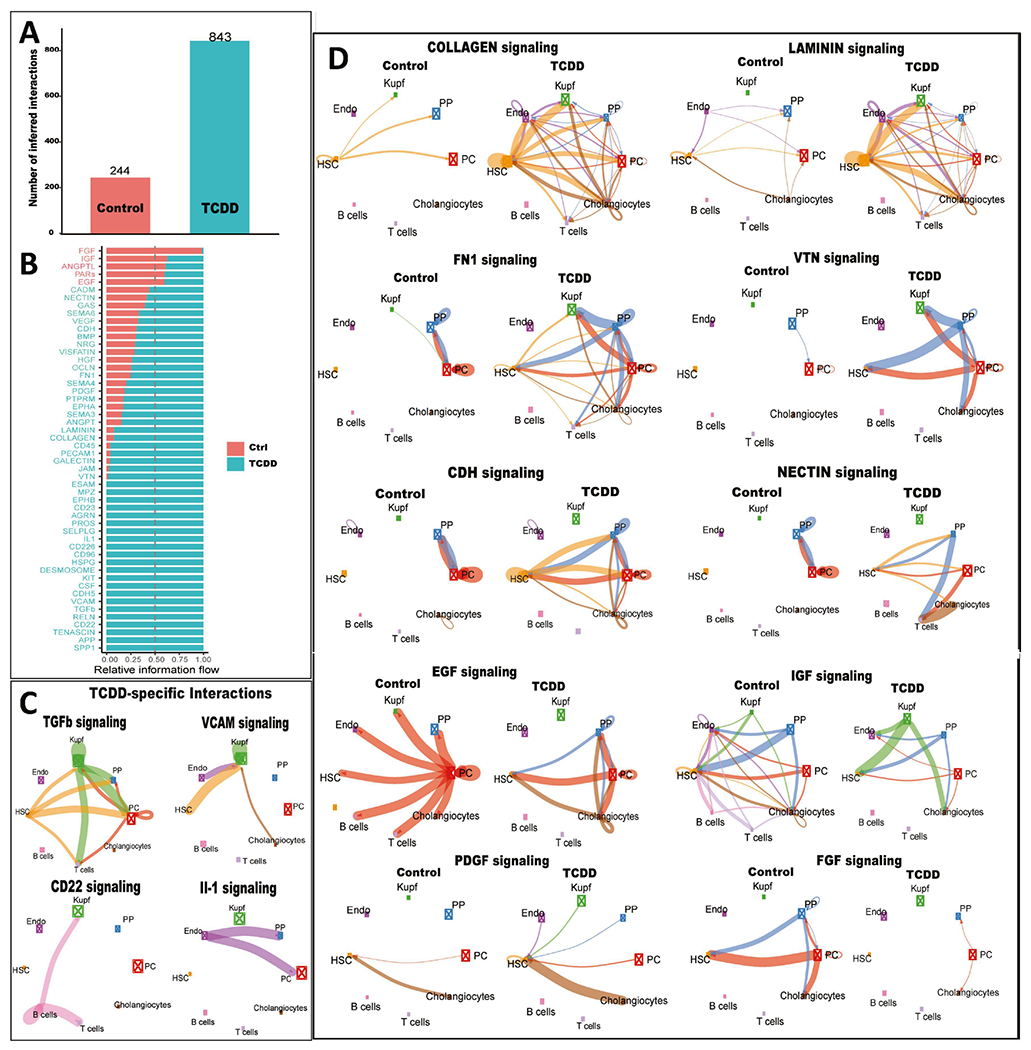
A. Bar plot showing number of ligand-receptor interactions identified by CellChat in snRNA cell clusters from control and TCDD-exposed liver. B. Ligand-receptor interactions identified in each dataset are grouped into 52 signaling pathways. X-axis shows the relative information flow of each signaling pathway along the Y-axis, indicating intercellular signaling pathways whose activity increases or decreases between control and TCDD-exposed liver. C. Circle plots showing examples of TCDD-specific intercellular communication patterns. Edge weights are proportional to the interaction strength: a thicker edge line indicates a stronger signal, while sizes of the boxes with X in the center are proportional to the number of cells in each cell type. D. Circle plots showing examples of differential intercellular communication patterns between control and TCDD-exposed liver. Also see Table S4.
Gene regulatory network analysis:
Gene regulatory networks for control liver and for TCDD-exposed liver were inferred separately by analysis of the snRNA-seq data using bigSCale2 (Iacono et al., 2019). Nuclei from control liver and from TCDD-exposed liver were split into two Seurat objects and passed to bigSCale2 (Iacono et al., 2019), where each network was constructed using the normal (default) clustering parameter, with granularity set to the highest setting. bigSCale2 uses recursive clustering to cluster the single cells at the highest feasible granularity (i.e., it generates as many truly distinct cell clusters and subclusters as possible), then runs an iterative differential expression analysis between all pairs of clusters, the results of which are used to generate a Z-score for each gene representing the likelihood of an expression change between two clusters. Correlations between gene pairs are then computed using Z-scores instead of UMI-based expression values, which circumvents the serious limitation of using standard coefficients to assess gene-gene expression correlations from noisy and highly sparse scRNA-seq data, with its many 0s and 1s. Strong Z-score-based gene correlations are obtained when two genes follow similar patterns of differential expression between cell sub-types (Iacono et al., 2019). We applied an edge cutoff corresponding to the top 99.9 quantile for correlation coefficient when constructing regulatory networks. bigSCale2 retains edges that are expected to represent actual regulatory links by removing genes that do not have a direct edge with a known GO regulator (GO sub-setting step). This was achieved by using a list of GO regulators consisting of the union of the genes in GO:0006355 “regulation of transcription, DNA-templated” (N=2,776) and GO:000370 “DNA-binding transcription factor activity” (N=1,001). We also included our list of 52,961 lncRNAs (see above) as potential regulators in the GO sub-setting step. Network specifications for the networks generated are shown in Table S5G. The networks obtained were converted into json files for visualization by Cytoscape (Shannon et al., 2003) using the toCytoscape function of the package iGraph (v1.3.5) (https://igraph.org/r) in R. Specifically, the network json file was imported in Cytoscape using “Import Network from File”. Next, we used “forced-directed” layout in Cytoscape, where each network’s layout was derived from 10,000 iterations of the Fruchterman-Reingold algorithm (Gajdoš et al., 2016) and using the Nogrid parameter (seed=7). The resultant networks and derived subnetworks shown in the various figures are available are available at Ndexbio.org at https://tinyurl.com/snTCDDliverNetworksKarriWaxman. Gene modules in each network were discovered using the Glay community cluster algorithm (Su et al., 2010) in Cytoscape (Shannon et al., 2003), whereby the overall network topology was used to subdivide the overall networks generated by bigSCale2 into functional modules. Genes in each module were input to DAVID (Sherman et al., 2022) for gene ontology enrichment analysis. Node rank was calculated for each of four key bigSCale2 network metrics (Betweenness, Closeness, Degree, PAGERANK). Protein-coding genes that ranked within the top 100 nodes for any one of the above four network metrics were deemed to be network-essential nodes that identify regulatory protein-coding genes; and lncRNAs that ranked within the top 50 nodes for any one of the above four network metrics were deemed to be network-essential nodes corresponding to regulatory lncRNAs (Table S5B)
Master regulators –
To identify master regulators, we extracted a subnetwork compromised of the network-essential genes, i.e., the top 100 ranked nodes for protein-coding genes, and the top 50 ranked nodes for lncRNAs, considered separately for each of the 4 network metrics, and then recalculated network metrics. The critical nodes in these subnetworks were identified using five network metrics: Stress, Degree, Betweenness, Closeness and Neighborhood Connectivity, and were calculated using the “Analyze Network” option in Cytoscape. We then designated as master regulators, i.e., critical nodes regulating the regulators, the top 5 ranked nodes, considering each of the 5 metrics in the ranking independently (Table S5B).
IPA Analysis –
We used Ingenuity Pathway Analysis software (http://www.ingenuity.com) to validate the regulatory role of protein-coding gene master regulators from the control liver and from the TCDD-exposed liver networks. Putative gene targets of protein-coding gene master regulators were provided as input to IPA. IPA core analysis of these gene lists yielded information on the upstream regulators, top enriched canonical pathways, diseases, and toxicological functions. The resulting data output by IPA for all protein-coding gene master regulators is included in Table S5E.
Enrichment heatmap –
Protein-coding genes that make direct connections to a regulatory lncRNAs in a bigSCale2 network were input to Metascape (Zhou et al., 2019). We then performed multi-list comparative analysis within and across the networks to identify pathways that are shared between or are specific to a lncRNA’s gene targets. The clustered heatmaps obtained depict −log10 P-value of top enriched pathways across multiple lncRNAs target lists.
Results
Cell-type specific responses of liver lncRNAs to TCDD exposure -
We integrated and clustered snRNA-seq datasets representing a total of 15,573 nuclei extracted from livers of mice exposed to TCDD subchronically (28 days), or vehicle control (Nault et al., 2021) (Table S1A). Eight major cell clusters representing 8 distinct liver cell subpopulations were identified using established liver cell type-specific marker genes (Fig. S1). Major treatment-dependent changes in the proportions of key liver cell types were found, including large decreases in the proportion of periportal hepatocytes and hepatic stellate cells, and increases in Kupffer cells (liver macrophages), endothelial cells, B cells and T cells (Table S1A), consistent with (Nault et al., 2021). The snRNA-seq data was of high quality and sufficient sequencing depth for us to detect 32,758 liver-expressed lncRNA genes (Table S1C) using a custom GTF file encompassing a total of 76,011 genes, including 52,961 lncRNA genes (see Methods).
Differential expression analysis applied to each liver cell cluster identified a total of 7,821 genes, including 4,016 lncRNA genes, that were dysregulated following TCDD exposure in at least one liver cell type at an expression |fold-change| > 4 and FDR <0.05 (Table S1B). The largest number of TCDD gene responses was seen in pericentral hepatocytes, where Ahr, the receptor for TCDD, is most highly expressed. Fewer gene responses were seen in periportal hepatocytes followed by Kupffer cells/liver macrophages (Fig. 1, Table S1A), where Ahr expression is much lower (see below). In hepatocytes, many more genes were induced by TCDD than were repressed, both for protein-coding genes and for lncRNAs, whereas in the NPC clusters gene down regulation was generally more extensive than gene up regulation (Fig. 1, Fig. S2). We observed large differences in the cell type-specificity of TCDD-induced gene responses for lncRNAs as compared to protein-coding genes. Thus, 75% of TCDD-responsive lncRNAs, but only 30% of TCDD-responsive protein-coding genes, showed a response to TCDD in hepatocytes but not in NPCs; whereas 54% of TCDD-responsive protein-coding genes, but only 17% of TCDD-responsive lncRNAs, responded to TCDD in one or more NPCs, but not in hepatocytes (Fig. S2D).
Fig. 1. TCDD-elicited gene responses in individual liver cell populations.
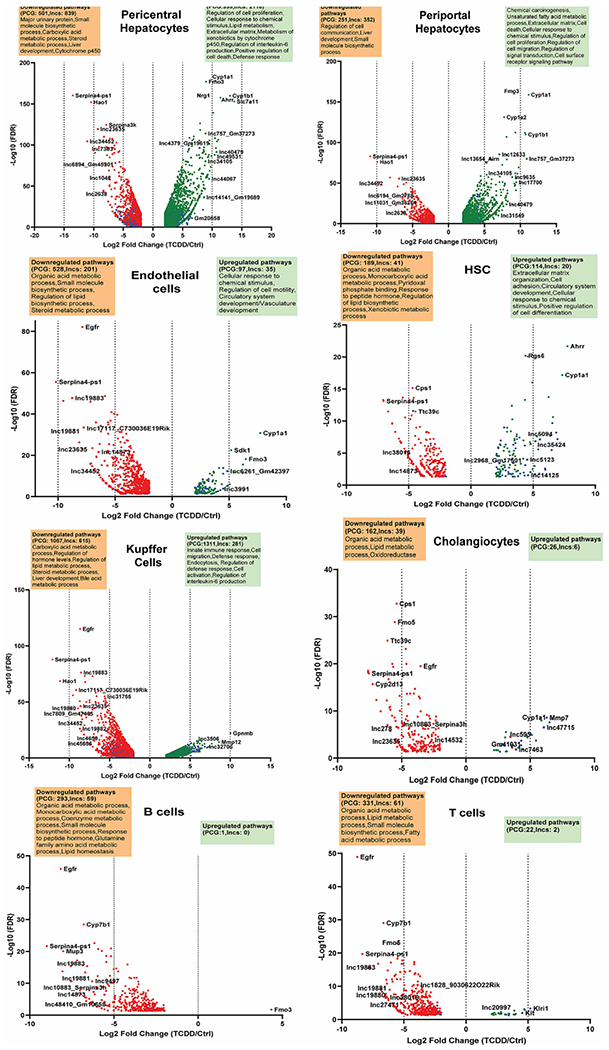
Volcano plots showing protein-coding genes and lncRNA genes whose expression is dysregulated by TCDD at |fold-change| >4 and FDR <0.05 in each of the 8 indicated liver cell clusters. See Table S1B for full gene listing.
Top TCDD-responding protein-coding genes include classic AHR target genes, such as Cyp1a1 and Cyp1a2 (Fig. S3A). In hepatocytes, the most highly inducible lncRNAs included lnc17700* and Gm2968 (lnc19317) (Fig. S3B), with fold-change values > 400 (Table S1B). Well-characterized lncRNAs induced by TCDD in both pericentral and periportal hepatocytes include Snhg15 (lnc9442*), whose increased expression has been linked to liver metastasis and poor overall survival in colorectal cancer (Huang et al., 2019), and Lrrc75-as1 (lnc9790*), which inhibits cell proliferation and migration in colorectal cancer (Chen et al., 2019). In contrast, Lhx1os (lnc9944), which is anti-sense to the transcription factor gene Lhx1, was induced 100-fold by TCDD specifically in pericentral hepatocytes, while 4933401D09Rik (lnc13688) was induced 45-fold specifically in periportal hepatocytes (Table S1B). TCDD responses that were highly specific for individual NPC populations include the >30-fold up regulation of two adjacent novel lncRNA genes (lnc47715, lnc47717) in TCDD-exposed cholangiocytes (Fig. S3A), and the >30-fold up regulation of a novel lncRNA (lnc5094) in hepatic stellate cells. Neat1 (lnc14746*), a positive regulator of liver fibrosis (Yu et al., 2017), was down regulated 18-fold in TCDD-exposed Kupffer cells. Thirteen lncRNAs, including lnc10883 and lnc17117, were significantly repressed by TCDD across all liver cell types (Table S1D, Fig. S3C).
Functional enrichment analysis of the set of TCDD-responsive protein-coding genes identified overlapping groups of pathways that were down regulated in multiple NPC cell types. For example, genes associated with lipid homeostasis and related terms were down regulated in endothelial cells, hepatic stellate cells, cholangiocytes and immune cells (B cells, T cells, Kupffer cells). Pathways related to xenobiotic metabolism, cell proliferation and cytokine production were strongly up regulated in hepatocytes. In contrast, extracellular matrix was most strongly induced in hepatic stellate cells, while immune response pathways showed very strong increases in Kupffer cells (Fig. 1, Table S1E).
Zonated expression of lncRNAs in hepatocytes -
Hepatocyte nuclei from healthy (control) mouse liver were resolved using the spatial inference algorithm Monocle2 (Trapnell et al., 2014) to identify 204 lncRNAs and 1,744 protein-coding genes (Table S2A) that showed significant zone-dependent expression in hepatocytes across the liver lobule (Table S2A). The functional enrichments obtained were consistent with prior findings using experimentally validated hepatocyte zonation marker RNAs (Halpern et al., 2017), such as Gulo (pericentral) and Cyp2f2 (periportal) (Fig. 2A, Table S2B). Examples of strong pericentral hepatocyte zonation bias include lnc34787 (Snhg11), which functions as a tumor promoter in hepatocellular carcinoma (Huang et al., 2020) and lnc32377 (Cyp2c53-ps). Strong pericentral zonation was found for lnc2311 (Platr4), which ameliorates steatohepatitis in mouse liver (Lin et al., 2021), and for lnc14025 (Dreh), which promotes cell proliferation in hepatitis B virus-associated hepatocellular carcinoma (Lv et al., 2017). Comparing zonal distributions between control and TCDD-exposed hepatocyte nuclei revealed major disruption of hepatocyte zonation following TCDD exposure, with 810 genes, including 121 lncRNAs, showing differential expression along the spatial trajectory between the two conditions (Table S2C). Fig. 2B shows examples of disrupted zonation of protein-coding genes (Car8, Etnk1) and lncRNAs (Cyp2c53-ps (lnc32377), lnc39934) in TCDD-exposed hepatocytes. Top enrichments for the zone-dysregulated protein-coding genes included carboxylic acid metabolic process, lipid metabolic process, apoptotic process and peroxisome (Table S2D).
Fig. 2. Dysregulation of hepatocyte zonation across the liver lobule in TCDD-exposed liver.
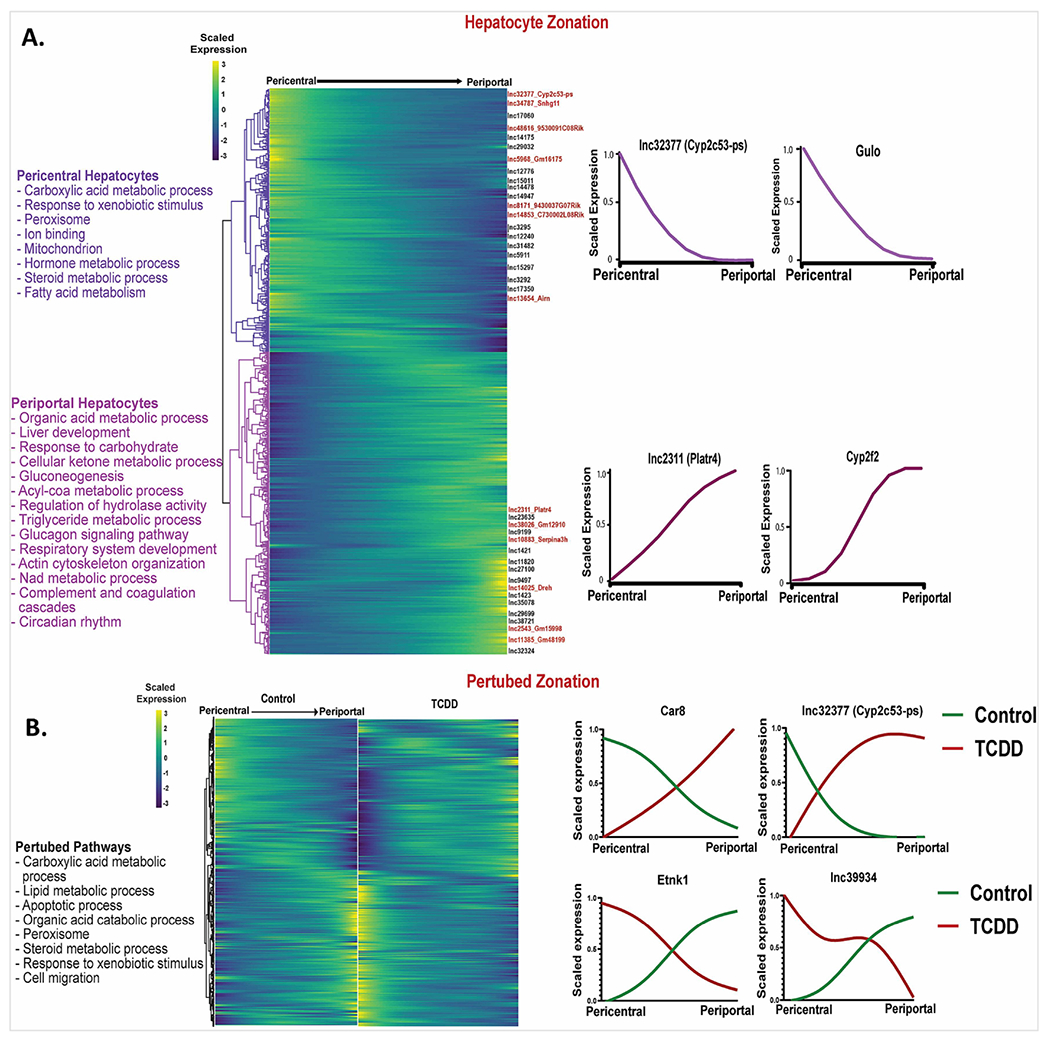
A. Heatmap showing relative expression patterns for 1,744 genes that show zonated expression in hepatocytes from healthy (control) mouse liver. Select zonated lncRNAs are marked along the right Y-axis. Zonation profiles for select genes are shown at the far right and top enriched terms for each hepatocyte zone are listed at the left. B. Matched heatmaps for 810 genes that are differentially zonated between control and TCDD-exposed hepatocytes at FDR <0.001. The data are scaled independently for the control and for the TCDD groups. See Table S2 for full datasets.
Impact of TCDD on expression profiles for Nr genes and Ahr –
While primary gene responses to TCDD, including lncRNA gene responses, are mediated by AHR, the subchronic TCDD exposure datasets analyzed here are expected to include many secondary gene responses, which may be mediated by other TFs whose expression or activity is altered by TCDD exposure. TFs from the Nr gene superfamily are of particular interest, given their activation by foreign chemicals that dysregulate key liver functions such as glucose and cholesterol metabolism, bile homeostasis and xenobiotic metabolism (Cave et al., 2016; Rudraiah et al., 2016). Here, we examined the impact of TCDD exposure on the expression of these Nr genes, with their potential to alter physiological processes and pathophysiological responses in many liver cell types. Specifically, we analyzed liver cell type-dependent expression profiles of all 49 mouse Nr genes (Zhao et al., 2015), as well as that of Ahr, under both basal conditions and following 4 wk TCDD exposure. The 49 Nr genes were grouped into five separate clusters based on their physiological functions (Gonzalez-Sanchez et al., 2015), as marked in Fig. 3A. Nr genes in the bile acid and xenobiotic metabolism cluster and in the lipid metabolism and energy homeostasis cluster generally showed the highest expression (Fig. 3A). Expression levels were similar between periportal and pericentral hepatocytes, with the exception of three receptors with a strong pericentral bias: Nr1i3 (CAR), an important regulator of hepatic drug and xenobiotic metabolism and energy metabolism (Cai et al., 2021); Esrrg, a key regulator of hepatic gluconeogenesis (Kim et al., 2012); and Ahr itself. Ppara, which along with Ppard and Pparg plays a critical role as a lipid sensor and regulator of lipid metabolism (Berger and Moller, 2002; Hernandez-Quiles et al., 2021), showed higher expression in both hepatocyte clusters than in the NPC clusters. Nr1h4 (FXRα), a bile acid sensor that regulates hepatic bile acid metabolism (Chiang and Ferrell, 2022), showed a liver cell type specificity similar to Ppara, except that its expression was high in hepatic stellate cells, which have a major role in the deposition of extracellular matrix during liver fibrosis. Of note, Nr1h4 (FXRα) suppresses liver fibrosis development, and its deficiency is associated with fatty liver and increased susceptibility to NASH in high fat diet-fed mice (Armstrong and Guo, 2017). In contrast, expression of Nr1h5 (FXRβ), which is activated by lanosterol but not bile acids (Otte et al., 2003), was largely restricted to hepatic stellate cells. Rarb, which also showed high specificity for hepatic stellate cell expression, inhibits hepatic stellate cell activation in NAFLD (Trasino et al., 2016). Finally, Rora, which decreases oxidative stress and has the ability to attenuate NAFLD progression (Han et al., 2014; Chai et al., 2020), was expressed in most liver cell types, with highest expression seen in cholangiocytes.
Fig. 3. snRNA-seq expression profiles for Ahr and members of the Nr superfamily.
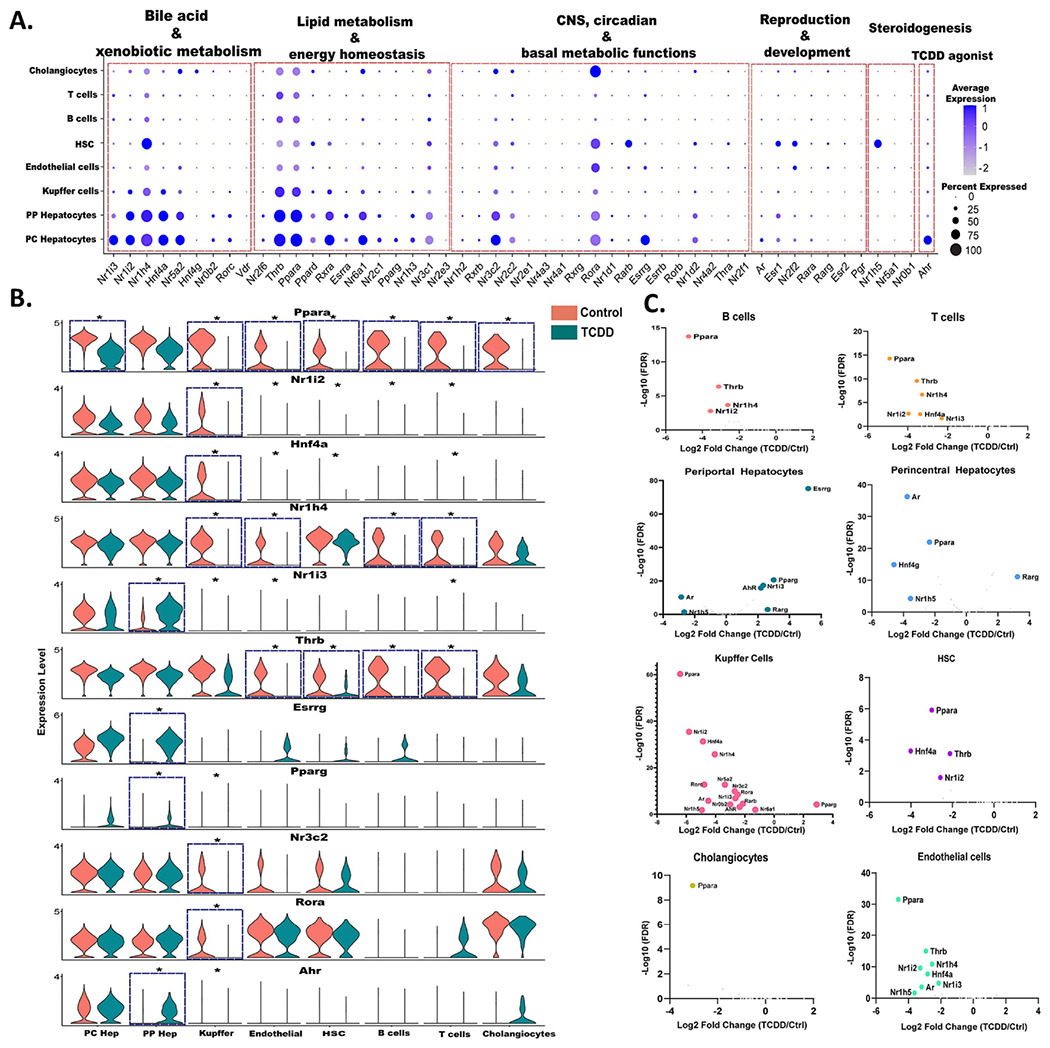
A. Dot plot expression profiles for all 49 mouse Nr genes, and for Ahr, in each of 8 mouse liver cell types from control mouse liver. B. Violin plots showing changes in expression of select Nr genes, and Ahr, between control and TCDD-exposed mouse liver in the 8 liver cell types. Asterisk indicates significant difference in expression at FDR <0.05. C. Volcano plots depicting control versus TCDD-exposed liver for differentially expressed Nr genes in each liver cell type. X-axis, log2 fold-change values for differential expression for control vs. TCDD within each cell cluster; Y-axis, - log10 (FDR) value to mark the significance of differential expression. Also see Table S3A.
Overall, TCDD altered the expression of Ahr and 19 out of the 49 Nr genes in at least one liver cell type. Ahr and four Nr genes (Nr1i3 (CAR), Esrrg, Pparg, Rarg) were up regulated by TCDD in periportal hepatocytes. The increase in Nr1i3 (CAR) is consistent with a prior report in bulk liver analysis (Lee et al., 2015) and leads to a reversal of Nr1i3 (CAR) zonation (Table S2C). Similarly, the periportal increases in Esrrg and Ahr largely abolish the pericentral zonation of those receptors seen in control hepatocytes (Fig. 3B, Table S2A). In addition, TCDD induced large decreases in expression of Ppara in all major liver cell types except periportal hepatocytes, which may contribute to TCDD disruption of PPARA-regulated lipid metabolism (Shaban et al., 2004). In Kupffer cells, TCDD stimulated widespread decreases in expression of many Nr genes, including Ppara, Nr1i2 (PXR), Nr1h4 (FXRα) and Hnf4a. The down regulation of Hnf4a may contribute to TCDD-induced liver injury, insofar as Hnf4a repression occurs in several mouse models of acute liver damage, including sepsis and liver injury following high dose exposure to acetaminophen or carbon tetrachloride (Joo et al., 2019; Dubois et al., 2020). Thrb was down regulated by TCDD in both endothelial cells and hepatic stellate cells, as well as in B cells and T cells (Fig. 3B, Fig. 3C). Hepatic thyroid hormone signaling is crucial in the development and progression of NASH, and reduced expression of Thrb has been linked to NASH in human liver (Krause et al., 2018).
Global evaluation of TF dysregulation in TCDD-exposed liver –
Given the major changes in Nr expression seen following TCDD exposure, above, we investigated the effects of TCDD on the expression of 1,533 TFs, which were grouped into 72 TF families (Table S3B). TCDD induced or repressed the expression of 239 of these TFs (including 19 Nr genes discussed above) by > 4-fold in at least one liver cell type (Table S3C). Five of the 72 TF families showed significant enrichment (p <0.05) for TFs showing dysregulated expression (Fig. S4).
Dysregulation of expression was most widespread in hepatocytes and Kupffer cells (Fig. 4A). Nine TFs, including the Nr genes Ppara and Nr1i2, were dysregulated by TCDD in five or more cell types; all but one of these TFs (Ahrr) showed down regulation in multiple cell types (Fig. 4B). Examples include: Onecut1, a liver expressed TF implicated in regulation of hepatic sex differences (Conforto et al., 2015), consistent with the loss of liver sex differences following TCDD exposure (Nault et al., 2017a); Mafb, which promotes macrophage polarization into an anti-inflammatory M2 state (Kim, 2017); and the lipogenic TF Mlxipl (CHREBP) (Filali-Mouncef et al., 2022), which was strongly down regulated in all six NPC populations, but not in hepatocytes.
Fig. 4. TCDD dysregulates TF expression across multiple liver cell types.
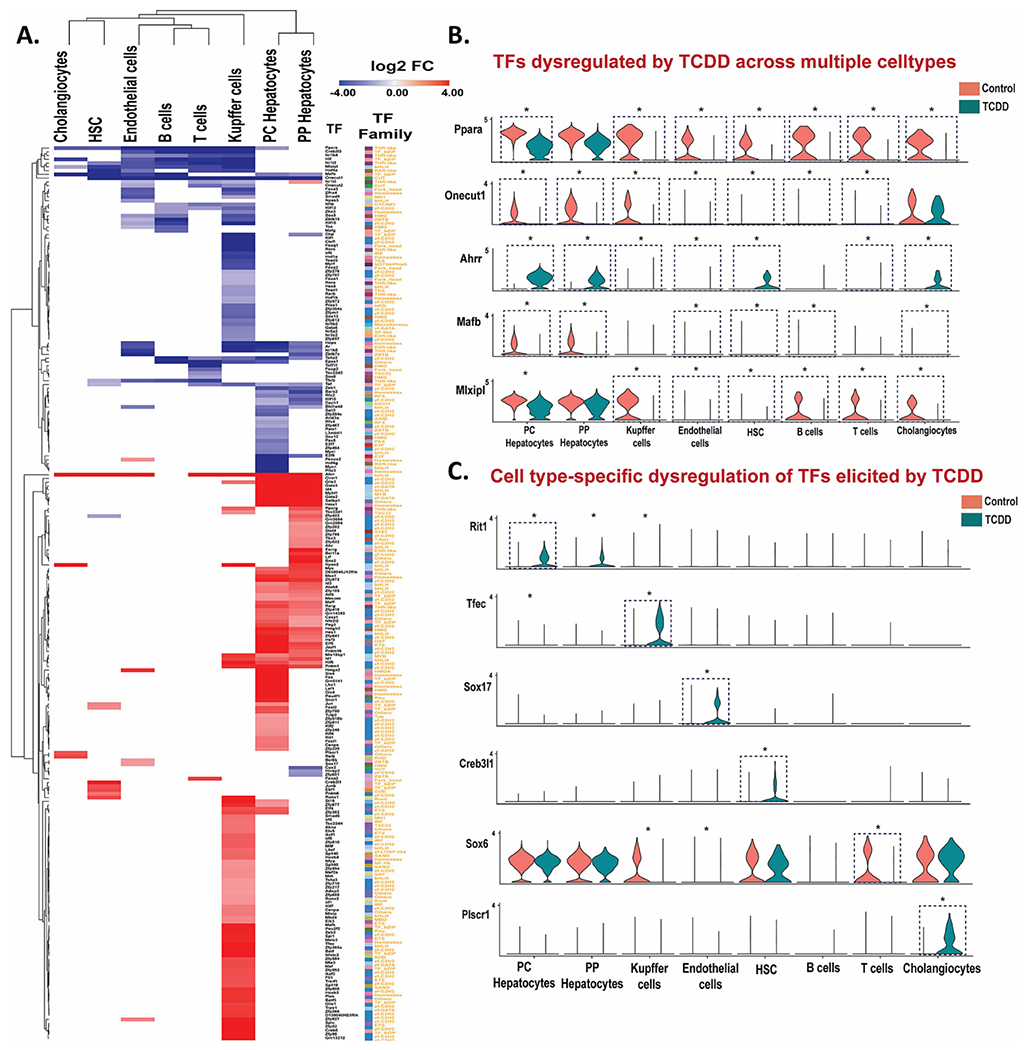
A. Heat maps of differentially expressed TFs in the indicated liver cell clusters. The colored cells in the heat map (red or blue) show log2 fold-change (FC) values for TFs whose expression is significantly (FDR <0.05) increased (red) or decreased (purple) by TCDD exposure. B, C. Violin plots showing expression data for TFs significantly dysregulated by TCDD (*) in multiple liver cell types (B) or in a cell type-specific manner (C). Blue dashed box with * marks TFs differentially expressed at |fold-change| >4 and FDR <0.05. Also see Table S3C.
TCDD increased expression of the AHR repressor Ahrr in all liver cell types, except B cells, with strongest increase seen in hepatocytes (log2 fold-change: 10-11). AHRR binds to the AHR heterodimerization partner ARNT to initiate a negative feedback loop regulating AHR function and TCDD hepatotoxicity (Vogel et al., 2019). Five other TF RNAs were strongly up regulated in both hepatocyte populations and in Kupffer cells, including Id1, which promotes HCC proliferation (Yin et al., 2017), and Klf6, a regulator of lipid homeostasis that is up regulated in models of liver injury and activates autophagy in hepatocytes (Sydor et al., 2017) (Table S3C). TF RNAs whose expression was dysregulated by TCDD in primarily one liver cell population include Tfec, a macrophage-specific TF (Rehli et al., 1999) that was induced 16-fold in Kupffer cells, and Sox17, a regulator of liver lipid metabolism with a role in endothelial regeneration during vasculature injury (Rommelaere et al., 2014; Liu et al., 2019), which was specifically induced in endothelial cells by 4-fold. Creb3l1 (OASIS), a key regulatory TF for liver fibrosis-related genes (Wang et al., 2021), was specifically induced, by 15-fold, in hepatic stellate cells, which may contribute mechanistically to TCDD-induced liver fibrosis. Finally, in Kupffer cells, TCDD induced Irf1, Irf5 and Irf8, all members of the Irf (Interferon regulatory factor) family that regulates innate and adaptive immune responses (Negishi et al., 2018), while a fourth family member, Irf6, was repressed (Table S3C).
TCDD-elicited alterations in cell-cell communications –
We investigated the effects of TCDD on intrahepatic cell–cell communication patterns, as judged by changes in expression of ligands and their cognate receptors in each liver cell cluster. These analyses were implemented using CellChat and its manually curated list of mouse ligand–receptor interactions (Jin et al., 2021). Strikingly, we observed a large increase in the number of inferred ligand receptor interactions following TCDD exposure (Fig. 5A). These interactions were grouped into a total of 52 signaling pathways, 47 of which have higher overall communication probability (greater relative information flow) in TCDD-exposed liver (Fig. 5B, Table S4). By comparing the overall communication probability between control and TCDD liver, we identified 22 signaling pathways that were specifically active in TCDD-exposed liver (Fig. 5B, Fig. 5C, Table S4). Important signaling pathways altered by subchronic TCDD exposure include: increased TGFβ signaling, which promotes the development of NASH in hepatocytes and mediates hepatic stellate cell activation, resulting in a wound-healing response and extracellular matrix deposition (Dooley and ten Dijke, 2012; Yang et al., 2014); increased signaling via the adhesion molecule VCAM1 to Kupffer cells from several NPCs, including cholangiocytes, which contributes to the persistence of liver inflammation (Afford et al., 2014) and promotes NASH (Furuta et al., 2021); B cell signaling by CD22, a regulator of B cell proliferation (Matsubara et al., 2018); and signaling from endothelial cells by the pro-inflammatory cytokine IL-1, which contributes to the hepatotoxic and tumor promoting effects of TCDD (Kennedy et al., 2014).
Thirty of the 52 intercellular communication pathways examined were shared between control and TCDD-exposed liver (Fig. 5B), but often with large differences between the two conditions (Fig. 5D). Thus, major increases in extracellular matrix-receptor interactions were seen in TCDD-exposed liver cells, including signaling via multiple collagens and laminins, and by fibronectin (FN1) and vitronectin (VTN). Increases in intercellular communication involving adhesion proteins, such as nectins and cadherins (CDH), was also found, consistent with AhR regulation of cell adhesion and matrix remodeling (Kung et al., 2009). PDGF, which promotes hepatic stellate cell proliferation and characterizes the fibrotic niche in human liver (Ramachandran et al., 2019; Roehlen et al., 2020), showed increased intercellular connectivity to hepatic stellate cells in TCDD-exposed liver. In contrast, intercellular connections were decreased by TCDD for several growth factor signaling pathways, including EGF, IGF and FGF, which may contribute to the pathological effects of TCDD (Croutch et al., 2005; Campion et al., 2016).
Network-essential regulatory lncRNAs in control and TCDD-exposed liver –
We utilized the rich transcriptomic profiles extracted by these snRNAseq datasets to construct gene regulatory networks, which we then used to identify lncRNAs whose expression is closely linked to specific biological functions. The algorithm we employed, bigSCale2 (Iacono et al., 2019), integrates expression data from all cell types in a tissue sample to construct gene regulatory networks, utilizing the diversity of expression data across all cell types to develop the robust Z-score-based correlations that underly the networks (see Methods). We obtained separate networks for control liver and for TCDD-exposed liver, each comprised of 5 functional gene modules. Each of the gene modules was enriched for distinct sets of pathways and biological functions (Fig. 6, Table S5A). For each network, we identified network-essential genes (nodes marked with gene names in Fig. 6) based on four key network centrality metrics, namely, Betweenness, PageRank centrality, Closeness, and Degree (Iacono et al., 2019), each of which serves as a proxy for a gene’s influence on the network (Table S5B). Strikingly, 41 lncRNAs occupied network-essential nodes in the control liver gene regulatory network and 66 lncRNAs in the TCDD-exposed liver network. Six of these lncRNA genes were identified as essential genes in both networks: Hnf4aos (lnc1966*), Gm36251 (lnc8349), Gm32063 (lnc23914*), AC118710.3 (lnc12772), 0610005C13Rik (lnc6166*), and Loc102632463 (lnc979*) (Fig. S5; Table S5B). Interestingly, 21 of the 41 control liver network-essential lncRNAs were dysregulated by TCDD, as were 44 of the 66 TCDD network-essential lncRNAs, many of which were down regulated by TCDD in Kupffer cells and/or in endothelial cells (Table S5B, column AI). Five TCDD network-essential lncRNAs (3 with human orthologs) showed robust (>30-fold) induction by TCDD, specifically in periportal and pericentral hepatocytes: lnc12630*, lnc12633, lnc17289, lnc26884* and lnc40479*. Importantly, the overall liver regulatory networks were validated by the striking enrichments that the gene targets of the network-essential IncRNAs showed for specific biological pathways. Thus, individual liver network-essential lncRNA regulators were linked to gene ontology terms such as cell adhesion, cell migration and extracellular matrix in the control liver network, and to terms such as fatty acid metabolic process, peroxisome and xenobiotic metabolic process in the TCDD-exposed liver network (Table S5C, Table S5D).
Fig. 6. Gene regulatory networks for control and TCDD-exposed mouse liver.
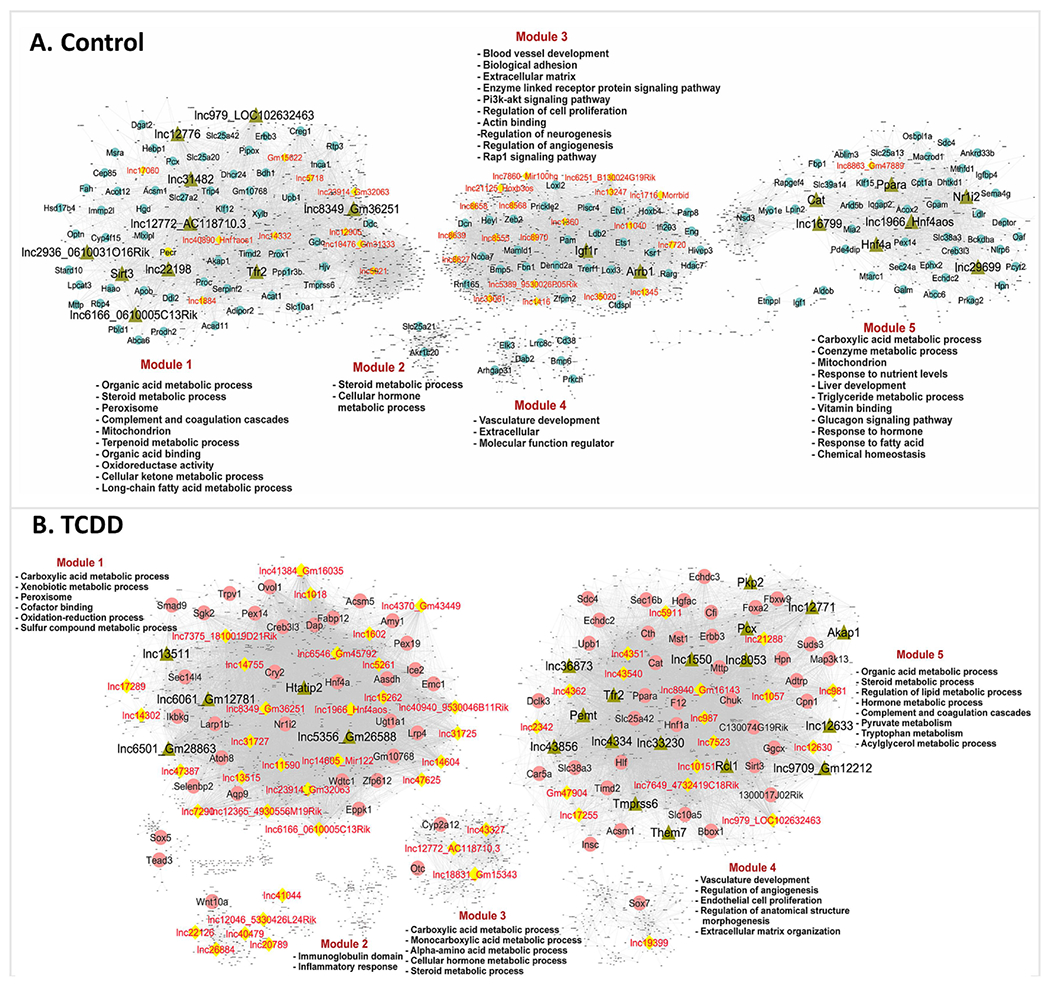
Shown are bigSCale2 networks for control (A) and TCDD-exposed (B) livers, where protein-coding genes or lncRNAs are nodes and the edges between them are correlations based on an adaptive threshold. All nodes with gene names displayed represent network-essential genes predicted based on top network metrics. Circular nodes represent protein-coding genes; yellow diamonds indicate lncRNAs; and green triangles identify master regulators, defined as nodes (genes) that have high network centrality metrics calculated in subnetworks extracted from all top 100 ranked protein-coding gene nodes or top 50 ranked lncRNA nodes. Each liver network is subdivided into gene modules that are enriched for the biological functions shown. See Table S5 and https://tinyurl.com/snTCDDliverNetworksKarriWaxman for complete datasets.
Next, we recalculated network metrics using a subnetwork comprised of all top network-essential genes to identify putative master regulators of each network (green triangular nodes in Fig. 6). We identified 19 putative master regulator genes (including 11 lncRNAs) in the control liver network and 22 genes (including 13 lncRNAs) in the TCDD-exposed liver network (Table S5B, Table S5E). Interestingly, one of the lncRNA master regulators identified in the control liver network, lnc12772, is anti-sense to the Cyp2d13 gene and showed strong down regulation in all 8 liver cell types in TCDD liver (Table S5B). Importantly, the gene targets of several of the protein-coding gene master regulators showed strong functional enrichment for the known functions of those genes, validating our approach for discovery of network regulatory genes. Examples include the master regulators Nr1i2 (PXR), Sirt3, Ppara and Hnf4a in control liver, and Tmprss6 and Htatip2 in TCDD-exposed liver (Table S5E). Further validation was obtained by IPA Upstream Regulator Analysis, which revealed that 3 of the protein-coding gene master regulators from the control liver network (Nr1i2, Ppara, Hnf4a) are significant Upstream Regulators of their network-determined gene targets (Table S5E).
Functional clustering of regulatory network-essential lncRNAs:
Protein-coding genes that were targets of the network-essential lncRNAs from the control and TCDD-exposed liver networks were input to Metascape (Zhou et al., 2019) to facilitate cross-comparison analysis to identify lncRNAs whose network-predicted gene targets share common biological pathways. Network-essential lncRNAs from the control liver network clustered into three major functional groups (Fig. S6), one of which was enriched for metabolic processes and lipid homeostasis, while the other two were variously enriched for terms related to collagen biosynthesis, extracellular matrix, cell-cell adhesion and other non-metabolic processes. Similarly, we identified three clusters of TCDD network-essential lncRNAs, which were distinguished by the functional enrichment terms of their gene targets (Fig. S7). Two of the lncRNA clusters were enriched for both metabolic and non-metabolic processes (angiogenesis, complement cascade), while the third cluster showed strong enrichment for carbon metabolism, xenobiotic metabolism, peroxisome processes and PPAR signaling. Pathways that were either common or specific to the gene targets of the regulatory lncRNAs from each network are shown in Fig. S8.
Discussion
Thousands of liver-expressed lncRNAs have been identified, a subset of which are responsive to foreign chemical exposures (Lodato et al., 2017; Dempsey and Cui, 2019; Karri and Waxman, 2020; Goldfarb and Waxman, 2021). Individual lncRNAs have been shown to promote liver pathologies, including liver diseases associated with dietary or xenobiotic exposures (Jin et al., 2020; Massart et al., 2022; Rajak et al., 2022), however, a global assessment of their roles in foreign chemical-induced hepatotoxicity, such as subchronic toxicity induced by TCDD exposure, has been lacking. Here, we addressed this issue by introducing an innovative, two step approach to elucidate the roles of liver-expressed lncRNAs in TCDD hepatotoxicity. First, we employed recent high-quality public snRNA-seq datasets of subchronic TCDD exposure (Nault et al., 2021) to characterize liver cell type-specific expression patterns for thousands of liver-expressed lncRNAs; and second, we utilized the thousands of resulting single nucleus-based transcriptomes to construct gene regulatory networks that were then used to identify network-essential regulatory lncRNAs. The overall strategy was made possible by the strong nuclear enrichment seen for a majority of liver-expressed lncRNAs, many of which are tightly bound to the chromatin fraction (Goldfarb and Waxman, 2021); this, in turn, increases the sensitivity of snRNA-seq for lncRNA detection as compared to scRNA-seq analysis, both in liver and in other tissues (Zeng et al., 2016; Goldfarb et al., 2022). The global reach of our analysis was increased by including more than 48,000 liver-expressed lncRNAs in the custom GTF file used to analyze the snRNA-seq datasets, including many thousands of novel lncRNA genes and isoform structures discovered by de novo transcriptome assembly from > 2,000 diverse mouse liver RNA-seq samples (Karri and Waxman, 2023). Remarkably, 32,758 of the 48,000 lncRNAs were detected in one of more liver cell types, of which 4,016 were dysregulated by TCDD in at least one cell type. Importantly, the use of snRNA-seq for single cell-based transcriptomics has been validated for both mouse and human liver, and offers important advantages over single cell-RNA-seq; these include the ability to analyze flash frozen liver tissue and to capture nuclei representing all cells present in bulk liver tissue, while minimizing the bias with respect to recovery or viability of individual cell types (Nault et al., 2021; Andrews et al., 2022; Goldfarb et al., 2022; Oh et al., 2022) that is seen when performing scRNA-seq using single cells dissociated from fresh liver tissue (Su et al., 2021).
As noted, TCDD dysregulated more than 4,000 lncRNAs in one or more liver cell types, including 684 lncRNAs specifically deregulated in NPCs. We also identified 121 lncRNAs, and 689 protein-coding genes, whose zonated expression in hepatocytes across the liver lobule is disrupted by TCDD, the latter finding being consistent with the major zonal toxicity recently reported for TCDD (Nault et al., 2023). To tackle the major challenge of elucidating lncRNA function on a global scale, we used the snRNA-seq expression datasets to construct gene regulatory networks, which enabled us to identify network-essential regulatory lncRNAs showing strong enrichments for specific functional gene ontology terms. Key examples include cell adhesion, cell migration and extracellular matrix in control (healthy) liver, and fatty acid metabolic process, peroxisome, and xenobiotic metabolic process in TCDD-exposed liver. These findings illustrate how public snRNA-seq datasets for a toxicologically relevant environmental chemical exposure model (Pierre et al., 2014; Nault et al., 2016; Nault et al., 2017b) can be used to discover large numbers of previously unknown and uncharacterized regulatory lncRNAs, characterize their cell type-specific expression patterns, and then link them to specific biological functions impacted by foreign chemical exposure.
The snRNA-seq data used for this study (Nault et al., 2021) was obtained from mice persistently exposed to the AhR agonist ligand TCDD over a 4-wk period, which results in hepatic lipid accumulation and promotes progression to steatohepatitis with liver fibrosis (Nault et al., 2017b). We observed major changes in lncRNA expression patterns in pericentral hepatocytes, where Ahr, which encodes the receptor for TCDD, is most highly expressed. Extensive responses were also seen in periportal hepatocytes, where basal expression of Ahr is very low prior to TCDD exposure, but which respond to TCDD with a 5-fold increase in Ahr expression. The latter increase undoubtedly contributes to the large number of TCDD-induced gene responses, including lncRNA responses, that we found in the periportal hepatocyte subpopulation. Two TFs from the Nr superfamily, Nr1i3 (CAR) and Esrrg, showed the same general pattern of low basal expression in periportal hepatocytes combined with strong periportal induction following TCDD-exposure, which abolished the pericentral bias in their zonation seen in healthy liver. Nr1i3 (CAR) and Esrrg are both important regulators of liver metabolic homeostasis and responses to liver injury (Misra et al., 2017; Cai et al., 2021; Jung et al., 2021) and their induction in periportal hepatocytes may be part of the injury response to TCDD. Widespread gene responses to TCDD were also seen in various NPCs, consistent with prior results (Nault et al., 2021) and despite the low expression of Ahr in those NPCs even after TCDD exposure. Many of the latter gene expression changes are likely to be indirect, downstream effects of TCDD associated with the widespread liver pathology that TCDD induces in this exposure model (Nault et al., 2017b) and perhaps involve the actions of one of more TCDD-responsive TFs or regulatory lncRNAs that we identified.
Genes active in xenobiotic metabolism and other metabolic processes are often regulated by TFs from the Nr superfamily, most notably those whose proteins act as xenosensors (Waxman, 1999; Klotz and Steinbrenner, 2017). These NRs crosstalk with each other and with other TFs that regulate processes such as bile acid synthesis, lipid metabolism and liver fibrosis and inflammation (Puengel et al., 2022). We mined the liver snRNA-seq datasets to characterize both basal and TCDD dysregulated transcriptomic profiles of all 49 mouse Nr genes across all 8 liver cell types. Expression of 18 of the 49 Nr genes was undetectable, which in some cases could be due to intrinsic sensitivity limitations of the single cell sequencing technology. In contrast, Nr genes with established functions linked to lipid metabolic processes and liver disease, most notably Ppara, Nr1h4 (FXRα), Thrb and Rora, were expressed at high levels in both hepatocytes and various NPCs, including hepatic stellate cells, endothelial cells and immune cells. Strikingly, TCDD suppressed the expression of three of these Nr genes (Ppara, Nr1h4 (FXRα), Thrb) in several NPC populations, including Kupffer cells/liver macrophages, which undergo extensive changes following TCDD exposure (Nault et al., 2021) and more generally during NASH pathogenesis (Kazankov et al., 2019). The strong repression of Ppara in TCDD-exposed liver across all major liver cell types is consistent with an earlier report (Shaban et al., 2004) and with the finding that reduced Ppara expression promotes steatosis and NASH development (Liss and Finck, 2017). These observations, together with our finding that TCDD dysregulates large numbers of other liver-expressed TFs (Fig. 4), give important insights into underlying mechanisms for the transcriptional crosstalk between AHR and other TFs, including NRs, and may help elucidate some of the underlying complexities of TCDD hepatotoxicity.
We investigated the impact of TCDD exposure on cell-cell communication patterns and signaling crosstalk, as deduced from changes in the expression of ligands and their cognate receptors in each liver cell population (Jin et al., 2021). Ligand-receptor interactions within and between liver cell clusters were grouped into signaling pathways, enabling us to capture changes in intra-hepatic cell-cell communication patterns induced by TCDD. Key features of the responses to TCDD include an increase in extracellular matrix and collagen-related signaling pathways and increased PDGF signaling, strong indications of hepatic stellate cell activation leading to liver fibrosis (Roehlen et al., 2020), a hallmark of subchronic exposure to TCDD (Fling et al., 2020; Li et al., 2020). Changes in cell-cell signaling patterns linked to fibrosis and other TCDD-induced hepatotoxic responses include increased TGFβ signaling, which promotes the development of NASH (Dooley and ten Dijke, 2012; Yang et al., 2014), increases in signaling by multiple collagens and laminins, which contribute to liver fibrosis, and decreases in intercellular connections for several growth factor signaling pathways associated with pathological effects of TCDD, including EGF, IGF and FGF (Croutch et al., 2005; Campion et al., 2016). A potential limitation of this approach is that transcriptomic changes in the expression of ligand-receptor pairs do not always correlate with proteomic data (Buccitelli and Selbach, 2020; Armingol et al., 2021). Increased confidence for such predictions may be obtained by integrating information from proteomics-based technologies that combine scRNA-seq with intracellular protein measurements to profile signaling activity (Katzenelenbogen et al., 2020) or that use mass spectrometry to detect ligand–receptor interactions and assay changes across conditions (Gault et al., 2020).
Single-cell expression data are especially well suited for computing gene regulatory networks, as they do not average out biological signals, unlike bulk transcriptomic data. We used the control and TCDD-exposed snRNA-seq datasets to construct gene regulatory networks, which enabled us to associate individual lncRNAs with functional network modules. Further, we used network centrality metrics (Iacono et al., 2019) to identify network-essential genes (putative regulatory genes) that drive various biological processes in the liver. We observed only partial overlap between the functional modules identified in the healthy liver (control) and the TCDD-exposed liver networks, consistent with the extensive changes in gene expression and gene regulation that accompany TCDD hepatotoxicity and disease development. The overall network density was higher in the TCDD network (Edges/Nodes=18.1) as compared to the control liver network (Edges/Nodes=8.5) (Table S5G), indicating increased gene regulatory interactions. The network-essential protein-coding gene RNAs identified included Nr1i2, Ppara, and Hnf4a, all three being major, well-established transcriptional regulators in the liver. Furthermore, in an entirely independent approach, we verified by Ingenuity Pathway Analysis that these three TFs are significant Upstream Regulators of their network-determined gene targets. Taken together, our findings establish the utility of our gene regulatory networks and validate the overall approach described here for discovery of lncRNA genes with high regulatory potential, and likely high relevance to environmental chemical exposure-induced tissue pathologies and hepatic disease.
Study limitations -
First, while snRNA-seq does have increased sensitivity compared to scRNA-seq for detection of the very large majority of liver-expressed lncRNAs showing strong nuclear enrichment in the chromatin-bound fraction (Goldfarb and Waxman, 2021), transcript dropout, which is intrinsic to all current single cell-based RNA-seq methods, still limits the sensitivity for detection of lowly expressed lncRNAs. Furthermore, we can anticipate a decrease in detection sensitivity for those genes whose RNA transcripts are enriched in the cytoplasm as compared to the nucleus (Goldfarb and Waxman, 2021). Another limitation is that our analysis was limited to the specific set of 48,261 liver-expressed lncRNAs that we recently identified (Karri and Waxman, 2023), in addition to 4,700 other lncRNA genes that we collected from established databases. Our finding that TCDD dysregulates more lncRNAs than protein-coding genes in hepatocytes, whereas the opposite is true for each of the NPC cell types (Fig. S2), suggests a bias favoring the discovery of TCDD-responsive lncRNAs in hepatocytes as compared to NPCs. There may be a bias in our set of 48,261 liver-expressed lncRNAs in favor of hepatocyte-expressed lncRNAs, which are very likely to dominate the set of lncRNA sequence reads present in the bulk liver RNA-seq samples used for transcriptome assembly and lncRNA gene and isoform discovery (Karri and Waxman, 2023). Another factor contributing to the apparent lower detection sensitivity of NPC-expressed lncRNAs may be the lower cell numbers in the NPC cell clusters. A further limitation of this study is our use of correlation-based computational methods, such as the pseudo-time trajectories used to infer TCDD-induced changes in lncRNA zonation patterns across the liver lobule, and the disruption of intercellular signaling pathways, determined by CellChat analysis, both of which will require experimental validation. The inferred nature of the gene regulatory networks, their disruption by TCDD-exposure, and our discovery of key network-essential lncRNAs will also require experimental validation, e.g., using lncRNA knockout or knockdown mouse models to further elucidate regulatory functions and molecular mechanisms of action.
Supplementary Material
Highlights:
Subchronic TCDD exposure dysregulates >4,000 mouse liver-expressed lncRNAs
snRNA-seq-based gene regulatory networks identify TCDD-responsive regulatory lncRNAs
TCDD dysregulates >200 transcription factors in multiple liver cell types
TCDD disrupts intercellular communication within the liver lobule
Acknowledgements:
The authors thank Dr. Alan Downey-Wall for sharing scripts used for analysis of DAVID output files.
Grant funding:
Supported in part by NIH grant ES024421 (to DJW).
Abbreviations:
- AhR
aryl hydrocarbon receptor
- DEG
differentially expressed gene
- GTF
Gene Transfer Format
- lncRNA
long non-coding RNA
- lnc
followed by a number: numbering system for set of 48,261 mouse liver expressed lncRNAs, where an asterisk (*) marks mouse liver lncRNAs with a human ortholog
- NPC
non-parenchymal cell
- PC
principal components
- scRNA-seq
single cell RNA sequencing
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TF
transcription factor
- TGI
transcript-genome identity
- TTI
transcript-transcript identity
- UMAP
Uniformed Manifold Approximation and Projection
- UMI
unique molecular identifier
- VSMC
vascular smooth muscle cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability:
Supplementary figures and tables are available online at the journal’s website. A Loupe Browser file, Liver_Control_TCDD_GSE148339, is available at https://doi.org/10.6084/m9.figshare.22661596.v1 and can be used to query cell clusters in the UMAP shown in Fig. S1 for individual genes, including lncRNA genes, and to perform further downstream data analysis.
References
- Afford SC, Humphreys EH, Reid DT, Russell CL, Banz VM, Oo Y, Vo T, Jenne C, Adams DH, Eksteen B, 2014. Vascular cell adhesion molecule 1 expression by biliary epithelium promotes persistence of inflammation by inhibiting effector T-cell apoptosis. Hepatology 59, 1932–1943. [DOI] [PubMed] [Google Scholar]
- Andrews TS, Atif J, Liu JC, Perciani CT, Ma XZ, Thoeni C, Slyper M, Eraslan G, Segerstolpe A, Manuel J, Chung S, Winter E, Cirlan I, Khuu N, Fischer S, Rozenblatt-Rosen O, Regev A, McGilvray ID, Bader GD, MacParland SA, 2022. Single-Cell, Single-Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol Commun 6, 821–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armingol E, Officer A, Harismendy O, Lewis NE, 2021. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet 22, 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong LE, Guo GL, 2017. Role of FXR in Liver Inflammation during Nonalcoholic Steatohepatitis. Curr Pharmacol Rep 3, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, Itzkovitz S, 2019. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab 1, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Moller DE, 2002. The mechanisms of action of PPARs. Annu Rev Med 53, 409–435. [DOI] [PubMed] [Google Scholar]
- Bock KW, 2020. Aryl hydrocarbon receptor (AHR)-mediated inflammation and resolution: Non-genomic and genomic signaling. Biochem Pharmacol 182, 114220. [DOI] [PubMed] [Google Scholar]
- Buccitelli C, Selbach M, 2020. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21, 630–644. [DOI] [PubMed] [Google Scholar]
- Cai X, Young GM, Xie W, 2021. The xenobiotic receptors PXR and CAR in liver physiology, an update. Biochim Biophys Acta Mol Basis Dis 1867, 166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion CM, Leon Carrion S, Mamidanna G, Sutter CH, Sutter TR, Cole JA, 2016. Role of EGF receptor ligands in TCDD-induced EGFR down-regulation and cellular proliferation. Chem Biol Interact 253, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave MC, Clair HB, Hardesty JE, Falkner KC, Feng W, Clark BJ, Sidey J, Shi H, Aqel BA, McClain CJ, Prough RA, 2016. Nuclear receptors and nonalcoholic fatty liver disease. Biochim Biophys Acta 1859, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C, Cox B, Yaish D, Gross D, Rosenberg N, Amblard F, Shemuelian Z, Gefen M, Korach A, Tirosh O, Lanton T, Link H, Tam J, Permyakova A, Ozhan G, Citrin J, Liao H, Tannous M, Hahn M, Axelrod J, Arretxe E, Alonso C, Martinez-Arranz I, Betes PO, Safadi R, Salhab A, Amer J, Tber Z, Mengshetti S, Giladi H, Schinazi RF, Galun E, 2020. Agonist of RORA Attenuates Nonalcoholic Fatty Liver Progression in Mice via Up-regulation of MicroRNA 122. Gastroenterology 159, 999–1014 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lan J, Ye Z, Duan S, Hu Y, Zou Y, Zhou J, 2019. Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma. Exp Biol Med (Maywood) 244, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JYL, Ferrell JM, 2022. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol Cell Endocrinol 548, 111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholico GN, Nault R, Zacharewski TR, 2022. Genome-Wide ChIPseq Analysis of AhR, COUP-TF, and HNF4 Enrichment in TCDD-Treated Mouse Liver. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Steinhardt G.F.t., Waxman DJ, 2015. Cross Talk Between GH-Regulated Transcription Factors HNF6 and CUX2 in Adult Mouse Liver. Mol Endocrinol 29, 1286–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croutch CR, Lebofsky M, Schramm KW, Terranova PF, Rozman KK, 2005. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and 1,2,3,4,7,8-hexachlorodibenzo-p-dioxin (HxCDD) alter body weight by decreasing insulin-like growth factor I (IGF-I) signaling. Toxicol Sci 85, 560–571. [DOI] [PubMed] [Google Scholar]
- Dempsey JL, Cui JY, 2019. Regulation of Hepatic Long Noncoding RNAs by Pregnane X Receptor and Constitutive Androstane Receptor Agonists in Mouse Liver. Drug Metab Dispos 47, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagorska A, Pendem SV, Hudson TE, Wu MM, Budas GR, Breckenridge DG, Harrison EM, Mole DJ, Wigmore SJ, Ramachandran P, Ponting CP, Teichmann SA, Marioni JC, Henderson NC, 2019. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep 29, 1832–1847 e1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley S, ten Dijke P, 2012. TGF-beta in progression of liver disease. Cell Tissue Res 347, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V, Gheeraert C, Vankrunkelsven W, Dubois-Chevalier J, Dehondt H, Bobowski-Gerard M, Vinod M, Zummo FP, Guiza F, Ploton M, Dorchies E, Pineau L, Boulinguiez A, Vallez E, Woitrain E, Bauge E, Lalloyer F, Duhem C, Rabhi N, van Kesteren RE, Chiang CM, Lancel S, Duez H, Annicotte JS, Paumelle R, Vanhorebeek I, Van den Berghe G, Staels B, Lefebvre P, Eeckhoute J, 2020. Endoplasmic reticulum stress actively suppresses hepatic molecular identity in damaged liver. Mol Syst Biol 16, e9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader KA, Zacharewski TR, 2017. Beyond the Aryl Hydrocarbon Receptor: Pathway Interactions in the Hepatotoxicity of 2,3,7,8-Tetrachlorodibenzo-p-dioxin and Related Compounds. Curr Opin Toxicol 2, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali-Mouncef Y, Hunter C, Roccio F, Zagkou S, Dupont N, Primard C, Proikas-Cezanne T, Reggiori F, 2022. The menage a trois of autophagy, lipid droplets and liver disease. Autophagy 18, 50–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling RR, Doskey CM, Fader KA, Nault R, Zacharewski TR, 2020. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) dysregulates hepatic one carbon metabolism during the progression of steatosis to steatohepatitis with fibrosis in mice. Sci Rep 10, 14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Guo Q, Pavelko KD, Lee JH, Robertson KD, Nakao Y, Melek J, Shah VH, Hirsova P, Ibrahim SH, 2021. Lipid-induced endothelial vascular cell adhesion molecule 1 promotes nonalcoholic steatohepatitis pathogenesis. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdoš P, Ježowicz T, Uher V, Dohnálek P, 2016. A parallel Fruchterman–Reingold algorithm optimized for fast visualization of large graphs and swarms of data. Swarm and evolutionary computation 26, 56–63. [Google Scholar]
- Garcia GR, Shankar P, Dunham CL, Garcia A, La Du JK, Truong L, Tilton SC, Tanguay RL, 2018. Signaling Events Downstream of AHR Activation That Contribute to Toxic Responses: The Functional Role of an AHR-Dependent Long Noncoding RNA (slincR) Using the Zebrafish Model. Environ Health Perspect 126, 117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault J, Liko I, Landreh M, Shutin D, Bolla JR, Jefferies D, Agasid M, Yen HY, Ladds M, Lane DP, Khalid S, Mullen C, Remes PM, Huguet R, McAlister G, Goodwin M, Viner R, Syka JEP, Robinson CV, 2020. Combining native and ‘omics’ mass spectrometry to identify endogenous ligands bound to membrane proteins. Nat Methods 17, 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain PL, Lun A, Garcia Meixide C, Macnair W, Robinson MD, 2021. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girer NG, Tomlinson CR, Elferink CJ, 2020. The Aryl Hydrocarbon Receptor in Energy Balance: The Road from Dioxin-Induced Wasting Syndrome to Combating Obesity with Ahr Ligands. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb CN, Karri K, Pyatkov M, Waxman DJ, 2022. Interplay Between GH-regulated, Sex-biased Liver Transcriptome and Hepatic Zonation Revealed by Single-Nucleus RNA Sequencing. Endocrinology 163, bqac059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb CN, Waxman DJ, 2021. Global analysis of expression, maturation and subcellular localization of mouse liver transcriptome identifies novel sex-biased and TCPOBOP-responsive long non-coding RNAs. BMC Genomics 22, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sanchez E, Firrincieli D, Housset C, Chignard N, 2015. Nuclear receptors in acute and chronic cholestasis. Dig Dis 33, 357–366. [DOI] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S, 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YH, Kim HJ, Kim EJ, Kim KS, Hong S, Park HG, Lee MO, 2014. RORalpha decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid Redox Signal 21, 2083–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Liu X, Liu X, Cui L, Yuan Y, Zhang H, Chen Y, Tao Y, Yu Z, 2021. The role of MEG3 in the proliferation of palatal mesenchymal cells is related to the TGFβ/Smad pathway in TCDD inducing cleft palate. Toxicol Appl Pharmacol 419, 115517. [DOI] [PubMed] [Google Scholar]
- Hernandez-Quiles M, Broekema MF, Kalkhoven E, 2021. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front Endocrinol (Lausanne) 12, 624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY, 2019. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 47, D33–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lin H, Kang L, Huang P, Huang J, Cai J, Xian Z, Zhu P, Huang M, Wang L, Xian CJ, Wang J, Dong J, 2019. Aberrant expression of long noncoding RNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer. J Cell Physiol 234, 7032–7039. [DOI] [PubMed] [Google Scholar]
- Huang W, Huang F, Lei Z, Luo H, 2020. LncRNA SNHG11 Promotes Proliferation, Migration, Apoptosis, and Autophagy by Regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther 13, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono G, Massoni-Badosa R, Heyn H, 2019. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol 20, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wahlang B, Shi H, Hardesty JE, Falkner KC, Head KZ, Srivastava S, Merchant ML, Rai SN, Cave MC, Prough RA, 2020. Dioxin-like and non-dioxin-like PCBs differentially regulate the hepatic proteome and modify diet-induced nonalcoholic fatty liver disease severity. Med Chem Res 29, 1247–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q, 2021. Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo MS, Koo JH, Kim TH, Kim YS, Kim SG, 2019. LRH1-driven transcription factor circuitry for hepatocyte identity: Super-enhancer cistromic analysis. EBioMedicine 40, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Kim YH, Radhakrishnan K, Kim J, Lee IK, Cho SJ, Kim DK, Dooley S, Lee CH, Choi HS, 2021. Orphan nuclear receptor ERRgamma regulates hepatic TGF-beta2 expression and fibrogenic response in CCl(4)-induced acute liver injury. Arch Toxicol 95, 3071–3084. [DOI] [PubMed] [Google Scholar]
- Karri K, Waxman DJ, 2020. Widespread Dysregulation of Long Noncoding Genes Associated With Fatty Acid Metabolism, Cell Division, and Immune Response Gene Networks in Xenobiotic-exposed Rat Liver. Toxicol Sci 174, 291–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karri K, Waxman DJ, 2023. Dysregulation of murine long non-coding single cell transcriptome in non-alcoholic steatohepatitis and liver fibrosis. RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, Bornstein C, Moshe A, Keren-Shaul H, Cohen M, Wang SY, Li B, David E, Salame TM, Weiner A, Amit I, 2020. Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 182, 872–885 e819. [DOI] [PubMed] [Google Scholar]
- Kazankov K, Jorgensen SMD, Thomsen KL, Moller HJ, Vilstrup H, George J, Schuppan D, Gronbaek H, 2019. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol 16, 145–159. [DOI] [PubMed] [Google Scholar]
- Kegel V, Deharde D, Pfeiffer E, Zeilinger K, Seehofer D, Damm G, 2016. Protocol for Isolation of Primary Human Hepatocytes and Corresponding Major Populations of Non-parenchymal Liver Cells. J Vis Exp, e53069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy GD, Nukaya M, Moran SM, Glover E, Weinberg S, Balbo S, Hecht SS, Pitot HC, Drinkwater NR, Bradfield CA, 2014. Liver tumor promotion by 2,3,7,8-tetrachlorodibenzo-p-dioxin is dependent on the aryl hydrocarbon receptor and TNF/IL-1 receptors. Toxicol Sci 140, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-K, Ryu D, Koh M, Lee M-W, Lim D, Kim M-J, Kim Y-H, Cho W-J, Lee C-H, Park SB, 2012. Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. Journal of Biological Chemistry 287, 21628–21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, 2017. The transcription factor MafB promotes anti-inflammatory M2 polarization and cholesterol efflux in macrophages. Sci Rep 7, 7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz LO, Steinbrenner H, 2017. Cellular adaptation to xenobiotics: Interplay between xenosensors, reactive oxygen species and FOXO transcription factors. Redox Biol 13, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S, 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause C, Grohs M, El Gammal AT, Wolter S, Lehnert H, Mann O, Mittag J, Kirchner H, 2018. Reduced expression of thyroid hormone receptor beta in human nonalcoholic steatohepatitis. Endocr Connect 7, 1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni PS, Crespo JG, Afonso CA, 2008. Dioxins sources and current remediation technologies--a review. Environ Int 34, 139–153. [DOI] [PubMed] [Google Scholar]
- Kung T, Murphy KA, White LA, 2009. The aryl hydrocarbon receptor (AhR) pathway as a regulatory pathway for cell adhesion and matrix metabolism. Biochem Pharmacol 77, 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larigot L, Juricek L, Dairou J, Coumoul X, 2018. AhR signaling pathways and regulatory functions. Biochim Open 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Prokopec SD, Watson JD, Sun RX, Pohjanvirta R, Boutros PC, 2015. Male and female mice show significant differences in hepatic transcriptomic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics 16, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cho SG, Ko SG, Ahrmad SA, Puga A, Kim K, 2020. Regulation of a long noncoding RNA MALAT1 by aryl hydrocarbon receptor in pancreatic cancer cells and tissues. Biochem Biophys Res Commun 532, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu Y, Dong Z, Xu M, Gao M, Cong M, Liu S, 2020. TCDD promotes liver fibrosis through disordering systemic and hepatic iron homeostasis. J Hazard Mater 395, 122588. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wang S, Gao L, Zhou Z, Yang Z, Lin J, Ren S, Xing H, Wu B, 2021. Oscillating lncRNA Platr4 regulates NLRP3 inflammasome to ameliorate nonalcoholic steatohepatitis in mice. Theranostics 11, 426–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss KH, Finck BN, 2017. PPARs and nonalcoholic fatty liver disease. Biochimie 136, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang L, Marsboom G, Jambusaria A, Xiong S, Toth PT, Benevolenskaya EV, Rehman J, Malik AB, 2019. Sox17 is required for endothelial regeneration following inflammation-induced vascular injury. Nat Commun 10, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato NJ, Melia T, Rampersaud A, Waxman DJ, 2017. Sex-Differential Responses of Tumor Promotion-Associated Genes and Dysregulation of Novel Long Noncoding RNAs in Constitutive Androstane Receptor-Activated Mouse Liver. Toxicol Sci 159, 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D, Wang Y, Zhang Y, Cui P, Xu Y, 2017. Downregulated long non-coding RNA DREH promotes cell proliferation in hepatitis B virus-associated hepatocellular carcinoma. Oncol Lett 14, 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, Cheng ML, Liu LY, Camat D, Chung SW, Seliga RK, Shao Z, Lee E, Ogawa S, Ogawa M, Wilson MD, Fish JE, Selzner M, Ghanekar A, Grant D, Greig P, Sapisochin G, Selzner N, Winegarden N, Adeyi O, Keller G, Bader GD, McGilvray ID, 2018. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhamshettiwar PB, Maetschke SR, Davis MJ, Reverter A, Ragan MA, 2012. Gene regulatory network inference: evaluation and application to ovarian cancer allows the prioritization of drug targets. Genome Med 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart J, Begriche K, Corlu A, Fromenty B, 2022. Xenobiotic-Induced Aggravation of Metabolic-Associated Fatty Liver Disease. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara N, Imamura A, Yonemizu T, Akatsu C, Yang H, Ueki A, Watanabe N, Abdu-Allah H, Numoto N, Takematsu H, Kitazume S, Tedder TF, Marth JD, Ito N, Ando H, Ishida H, Kiso M, Tsubata T, 2018. CD22-Binding Synthetic Sialosides Regulate B Lymphocyte Proliferation Through CD22 Ligand-Dependent and Independent Pathways, and Enhance Antibody Production in Mice. Front Immunol 9, 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T, Waxman DJ, 2019. Sex-Biased lncRNAs Inversely Correlate With Sex-Opposite Gene Coexpression Networks in Diversity Outbred Mouse Liver. Endocrinology 160, 989–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J, Kim DK, Choi HS, 2017. ERRgamma: a Junior Orphan with a Senior Role in Metabolism. Trends Endocrinol Metab 28, 261–272. [DOI] [PubMed] [Google Scholar]
- Nault R, Fader KA, Ammendolia DA, Dornbos P, Potter D, Sharratt B, Kumagai K, Harkema JR, Lunt SY, Matthews J, Zacharewski T, 2016. Dose-Dependent Metabolic Reprogramming and Differential Gene Expression in TCDD-Elicited Hepatic Fibrosis. Toxicol Sci 154, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R, Fader KA, Bhattacharya S, Zacharewski TR, 2021. Single-Nuclei RNA Sequencing Assessment of the Hepatic Effects of 2,3,7,8-Tetrachlorodibenzo-p-dioxin. Cell Mol Gastroenterol Hepatol 11, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R, Fader KA, Harkema JR, Zacharewski T, 2017a. Loss of liver-specific and sexually dimorphic gene expression by aryl hydrocarbon receptor activation in C57BL/6 mice. PLoS One 12, e0184842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R, Fader KA, Lydic TA, Zacharewski TR, 2017b. Lipidomic Evaluation of Aryl Hydrocarbon Receptor-Mediated Hepatic Steatosis in Male and Female Mice Elicited by 2,3,7,8-Tetrachlorodibenzo-p-dioxin. Chem Res Toxicol 30, 1060–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R, Saha S, Bhattacharya S, Sinha S, Maiti T, Zacharewski T, 2023. Single-cell transcriptomics shows dose-dependent disruption of hepatic zonation by TCDD in mice. Toxicol Sci 191, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H, Taniguchi T, Yanai H, 2018. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JM, An M, Son DS, Choi J, Cho YB, Yoo CE, Park WY, 2022. Comparison of cell type distribution between single-cell and single-nucleus RNA sequencing: enrichment of adherent cell types in single-nucleus RNA sequencing. Exp Mol Med 54, 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, 2003. Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Molecular and cellular biology 23, 864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrizi B, Siciliani de Cumis M, 2018. TCDD Toxicity Mediated by Epigenetic Mechanisms. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelclova D, Urban P, Preiss J, Lukas E, Fenclova Z, Navratil T, Dubska Z, Senholdova Z, 2006. Adverse health effects in humans exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Rev Environ Health 21, 119–138. [DOI] [PubMed] [Google Scholar]
- Pierre S, Chevallier A, Teixeira-Clerc F, Ambolet-Camoit A, Bui LC, Bats AS, Fournet JC, Fernandez-Salguero P, Aggerbeck M, Lotersztajn S, Barouki R, Coumoul X, 2014. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol Sci 137, 114–124. [DOI] [PubMed] [Google Scholar]
- Puengel T, Liu H, Guillot A, Heymann F, Tacke F, Peiseler M, 2022. Nuclear Receptors Linking Metabolism, Inflammation, and Fibrosis in Nonalcoholic Fatty Liver Disease. Int J Mol Sci 23, 2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajak S, Raza S, Tewari A, Sinha RA, 2022. Environmental Toxicants and NAFLD: A Neglected yet Significant Relationship. Dig Dis Sci 67, 3497–3507. [DOI] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, Taylor RS, Efremova M, Vento-Tormo R, Carragher NO, Kendall TJ, Fallowfield JA, Harrison EM, Mole DJ, Wigmore SJ, Newsome PN, Weston CJ, Iredale JP, Tacke F, Pollard JW, Ponting CP, Marioni JC, Teichmann SA, Henderson NC, 2019. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehli M, Lichanska A, Cassady AI, Ostrowski MC, Hume DA, 1999. TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J Immunol 162, 1559–1565. [PubMed] [Google Scholar]
- Roehlen N, Crouchet E, Baumert TF, 2020. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere S, Millet V, Vu Manh TP, Gensollen T, Andreoletti P, Cherkaoui-Malki M, Bourges C, Escaliere B, Du X, Xia Y, Imbert J, Beutler B, Kanai Y, Malissen B, Malissen M, Tailleux A, Staels B, Galland F, Naquet P, 2014. Sox17 regulates liver lipid metabolism and adaptation to fasting. PLoS One 9, e104925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudraiah S, Zhang X, Wang L, 2016. Nuclear Receptors as Therapeutic Targets in Liver Disease: Are We There Yet? Annu Rev Pharmacol Toxicol 56, 605–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban Z, El-Shazly S, Abdelhady S, Fattouh I, Muzandu K, Ishizuka M, Kimura K, Kazusaka A, Fujita S, 2004. Down regulation of hepatic PPARalpha function by AhR ligand. J Vet Med Sci 66, 1377–1386. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W, 2022. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50, W216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M, 2021. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, 2019. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G, Kuchinsky A, Morris JH, States DJ, Meng F, 2010. GLay: community structure analysis of biological networks. Bioinformatics 26, 3135–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Kim SY, Adewale F, Zhou Y, Aldler C, Ni M, Wei Y, Burczynski ME, Atwal GS, Sleeman MW, Murphy AJ, Xin Y, Cheng X, 2021. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver. iScience 24, 103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydor S, Manka P, Best J, Jafoui S, Sowa JP, Zoubek ME, Hernandez-Gea V, Cubero FJ, Kalsch J, Vetter D, Fiel MI, Hoshida Y, Bian CB, Nelson LJ, Moshage H, Faber KN, Paul A, Baba HA, Gerken G, Friedman SL, Canbay A, Bechmann LP, 2017. Kruppel-like factor 6 is a transcriptional activator of autophagy in acute liver injury. Sci Rep 7, 8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasino SE, Tang XH, Jessurun J, Gudas LJ, 2016. A retinoic acid receptor beta2 agonist reduces hepatic stellate cell activation in nonalcoholic fatty liver disease. J Mol Med (Berl) 94, 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K, Roux de Bezieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L, 2020. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 11, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viluksela M, Pohjanvirta R, 2019. Multigenerational and Transgenerational Effects of Dioxins. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CFA, Ishihara Y, Campbell CE, Kado SY, Nguyen-Chi A, Sweeney C, Pollet M, Haarmann-Stemmann T, Tuscano JM, 2019. A Protective Role of Aryl Hydrocarbon Receptor Repressor in Inflammation and Tumor Growth. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Keogh A, Waldt A, Cuttat R, Neri M, Zhu S, Schuierer S, Ruchti A, Crochemore C, Knehr J, Bastien J, Ksiazek I, Sanchez-Taltavull D, Ge H, Wu J, Roma G, Helliwell SB, Stroka D, Nigsch F, 2021. Single-cell and bulk transcriptomics of the liver reveals potential targets of NASH with fibrosis. Sci Rep 11, 19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, 1999. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys 369, 11–23. [DOI] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, Zhou L, Chen Z, Leon-Mimila P, Chung MT, Kurabayashi K, Opp J, Campos-Perez F, Villamil-Ramirez H, Canizales-Quinteros S, Lyons R, Lumeng CN, Zhou B, Qi L, Huertas-Vazquez A, Lusis AJ, Xu XZS, Li S, Yu Y, Li JZ, Lin JD, 2019. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 75, 644–660 e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Roh YS, Song J, Zhang B, Liu C, Loomba R, Seki E, 2014. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology 59, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Tang B, Li JH, Wang Y, Zhang L, Xie XY, Zhang BH, Qiu SJ, Wu WZ, Ren ZG, 2017. ID1 promotes hepatocellular carcinoma proliferation and confers chemoresistance to oxaliplatin by activating pentose phosphate pathway. J Exp Clin Cancer Res 36, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Huber W, Vitek O, 2013. Shrinkage estimation of dispersion in Negative Binomial models for RNA-seq experiments with small sample size. Bioinformatics 29, 1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Jiang Z, Chen B, Dong P, Zheng J, 2017. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl) 95, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Zeng N, Zhang Z, Jiang H, Li R, Chang C, Wang F, Xu D, Fan Q, Wang T, Xiao Y, Chen W, Shan Z, Huang Z, Wang Q, 2019. LncRNA-241 inhibits 1,2-Dichloroethane-induced hepatic apoptosis. Toxicol In Vitro 61, 104650. [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang S, Kong X, El-Ali N, Ball AR Jr., Ma CI, Hashimoto N, Yokomori K, Mortazavi A, 2016. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res 44, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang T, Yang Y, Li R, Chen Y, Li R, Jiang X, Wang L, 2020. Tanshinone IIA attenuates acetaminophen-induced hepatotoxicity through HOTAIR-Nrf2-MRP2/4 signaling pathway. Biomed Pharmacother 130, 110547. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang K, Giesy JP, Hu J, 2015. Families of nuclear receptors in vertebrate models: characteristic and comparative toxicological perspective. Sci Rep 5, 8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary figures and tables are available online at the journal’s website. A Loupe Browser file, Liver_Control_TCDD_GSE148339, is available at https://doi.org/10.6084/m9.figshare.22661596.v1 and can be used to query cell clusters in the UMAP shown in Fig. S1 for individual genes, including lncRNA genes, and to perform further downstream data analysis.


