Abstract
Objectives.
Matching treatment intensity to tumor biology is critical to precision oncology for head and neck squamous cell carcinoma (HNSCC) patients. We sought to identify biological features of tumor cell multinucleation, previously shown by us to correlate with survival in oropharyngeal (OP) SCC using a machine learning approach.
Materials and Methods.
Hematoxylin and eosin images from an institutional OPSCC cohort formed the training set (DTr). TCGA HNSCC patients (oral cavity, oropharynx and larynx/hypopharynx) formed the validation set (DV). Deep learning models were trained in DTr to calculate a multinucleation index (MuNI) score. Gene set enrichment analysis (GSEA) was then used to explore correlations between MuNI and tumor biology.
Results.
MuNI correlated with overall survival. A multivariable nomogram that included MuNI, age, race, sex, T/N stage, and smoking status yielded a C-index of 0.65, and MuNI was prognostic of overall survival (2.25, 1.07-4.71, 0.03), independent of the other variables. High MuNI scores correlated with depletion of effector immunocyte subsets across all HNSCC sites independent of HPV and TP53 mutational status although the correlations were strongest in wild-type TP53 tumors potentially due to aberrant mitotic events and activation of DNA-repair mechanisms.
Conclusion.
MuNI is associated with survival in HNSCC across subsites. This may be driven by an association between high levels of multinucleation and a suppressive (potentially exhausted) tumor immune microenvironment. Mechanistic studies examining the link between multinucleation and tumor immunity will be required to characterize biological drivers of multinucleation and their impact on treatment response and outcomes.
Keywords: oral cavity cancer, multinucleation, tumor infiltrating lymphocytes
Introduction
Head and neck squamous cell carcinoma (HNSCC) is often refractory to conventional treatment, and recurrent HNSCC is nearly uniformly fatal.1–3 Although survival for patients with human papillomavirus (HPV) - associated HNSCC is excellent, for patients with HPV-independent disease, it has not improved in 3 decades.3–5 The dichotomization between “low-risk” HNSCC with good treatment response and survival and “high-risk” HNSCC with limited response and high disease specific mortality generates an urgent need for better risk stratification algorithms that can match treatment intensity to disease biology. To be useful, the tests or mechanisms for better risk stratification must be easy to apply and widely available.
Tumor cell multinucleation (high power field with 3 tumor cells with 3 or more nuclei in the same cell) is detectable in HNSCC specimens in concert with tumor cell anaplasia (high power field with 3 or more tumor cells with a nuclear diameter ≥ the diameter of five resting lymphocyte nuclei).6 Multinucleation is strongly correlated with reduced survival in multiple solid tumors including SCC of the oropharynx site (OPSCC), both in the setting of HPV-associated and HPV-independent disease.6–8 A large scale analysis demonstrated that multinucleation, quantified using state-of-the-art machine learning algorithms, can risk stratify OPSCC patients already stratified by HPV/p16 status.7 In parallel, a smaller single institution analysis demonstrated that high levels of multinucleation accompany “high-risk” HPV-associated OPSCC, particularly in patients with an extensive tobacco exposure history and tumors depleted of effector immunocytes.8 Although multinucleation appears to correlate with survival in the setting of OPSCC, and seems to be more common in HPV-independent disease, the biological mechanisms which give rise to this pathologic feature, or alternatively permit it to exist, remain unclear. Our previous work showed that tumors with high levels of multinucleation demonstrated increased expression of genes associated with suppressed immunity.8 These data led us to hypothesize that multinucleation represents a phenotype correlated with exhausted or absent anti-tumor immunity. We sought to address this hypothesis using data from The Cancer Genome Atlas (TCGA) which allowed us to conduct in depth genomic and transcriptomic analyses of HNSCC tumors as a function of multinucleation based on the MuNI machine learning algorithm.7–9
Materials and Methods
Data set preparation:
Following approval of the Institutional Review Board of the Southern California Permanente Medical Group in Woodland Hills, California, USA, data from 171 patients with HPV-associated OPSCC (defined using p16 immunohistochemistry) were utilized as the training set, DTR, for model development and risk cutoff definition.7 The slides were digitized at ×40 resolution (0.25 μ/pixel resolution) using a Ventana iScan HT slide scanner at Case Western Reserve University. Whole slide image (WSI) quality was checked using HistoQC,10 an automatic quality control tool for computational pathology. Images annotated to the TCGA genomic and transcriptomic data were obtained from a public repository11 and analyzed in a similar manner.12
MuNI calculation:
For MuNI calculations, the present study utilized the previously established models.7 MuNI quantifies the frequency of multinucleated tumor cells within epithelial regions with the help of two generative adversarial networks.7 The first model, MMN, was established to localize multinucleated cells, as well as other type of cells, and the second one, MEP, to designate epithelial regions to identify epithelial cells and to remove falsely detected multinucleated cells, which were outside of epithelial regions. MMN and MEP were trained on 0.25μ/pixel and 1.00μ/pixel resolutions, respectively. Once the multinucleated and epithelial cells were segmented from a WSI, the ratio of the total number multinucleated cells to the total number of epithelial cells were defined as MuNI for this WSI (Eq1). For a WSI, SMN and SEP are segmented multinucleated and epithelial cells in the WSI, respectively. MuNI is then defined as follows:
| Eq1 |
MuNI is a continuous variable and its correlation with OS was analyzed with a Cox proportional hazards regression model using GraphPad Prism v9 software, while its Pearson correlation with other continuous variables was analyzed with JMP13 (SAS) statistical software. For some analyses, samples were dichotomized into high and low MuNI groups using either the previously published cut-off threshold of 0.000380 representing the median of an independent HPV-associated OPSCC cohort13, or a biological cut-off of 0.0004670 below which 90% of HPV-associated OPSCC samples in this study fell (i.e., sample mean plus 0.842 x std).
Genomic and transcriptomic analysis.
Upper quartile normalization of RNA-seq FPKM data from the TCGA were derived as previously described14 and used in all transcriptomic analyses including single sample gene set enrichment analyses (ssGSEA). The list of top 30 significantly mutated cancer drivers in HNSCC were taken from the MutSig analysis available through the Broad Firehose portal and the mutational status of cancer driver genes for each OCSCC sample was extracted from the TCGA HNSCC mutation annotation file, also available through the data portal.15 For each driver, the OCSCC cohort samples with MuNI scores available were partitioned into wild type or mutant groups and multiple T-tests performed using the Benjamini Hochberg correction to control the FDR.
Statistical and immune analysis.
Associations between variables were determined by two-sided Fisher’s exact tests. Survival was analyzed using the Kaplan-Meier method coupled to log-rank statistics. Multivariate analysis was performed using Cox regression. Statistical calculations were performed with SPSS (IBM SPSS Statistics version 25) or JMP v13. P-values were considered to be significant if below a threshold of 0.05 (two-sided) or false discovery rate (FDR) <0.1 as indicated. The tumor immune microenvironment was analyzed from bulk RNA-seq data using published and vetted gene lists specific to each leukocyte subtype to run single sample gene set enrichment analysis (ssGSEA), a module available from the Broad Institute’s Gene Pattern portal, as we previously described.14–16 ssGSEA values are continuous variables that can be negative or positive, with larger values corresponding to greater enrichment.
Results
Multinucleation varies across sites and genomic backgrounds.
A total of 436 patients from the TCGA HNSCC cohort underwent quantitative analysis of multinucleation (Supplementary Table 1).7 In HPV-independent tumor samples (Figure 1A), average MuNI scores were similar among SCC from the larynx/hypopharynx (LHSCC), oral cavity (OCSCC), and oropharynx (OPSCC). HPV-associated tumors had lower MuNI scores in all subsites and HPV-associated OPSCC had the lowest MuNI scores, nearly twice as low as HPV-independent OPSCC (p <0.0001). Among HPV-independent OCSCC and LHSCC tumors, African American patients had higher MuNI (Supplementary Figure 1) compared to non-African American patients (0.00075 vs 0.00063; p=0.0051) which correlates with our recent report.9
Figure 1. Multinucleation varies as a function of disease site and genomic background.
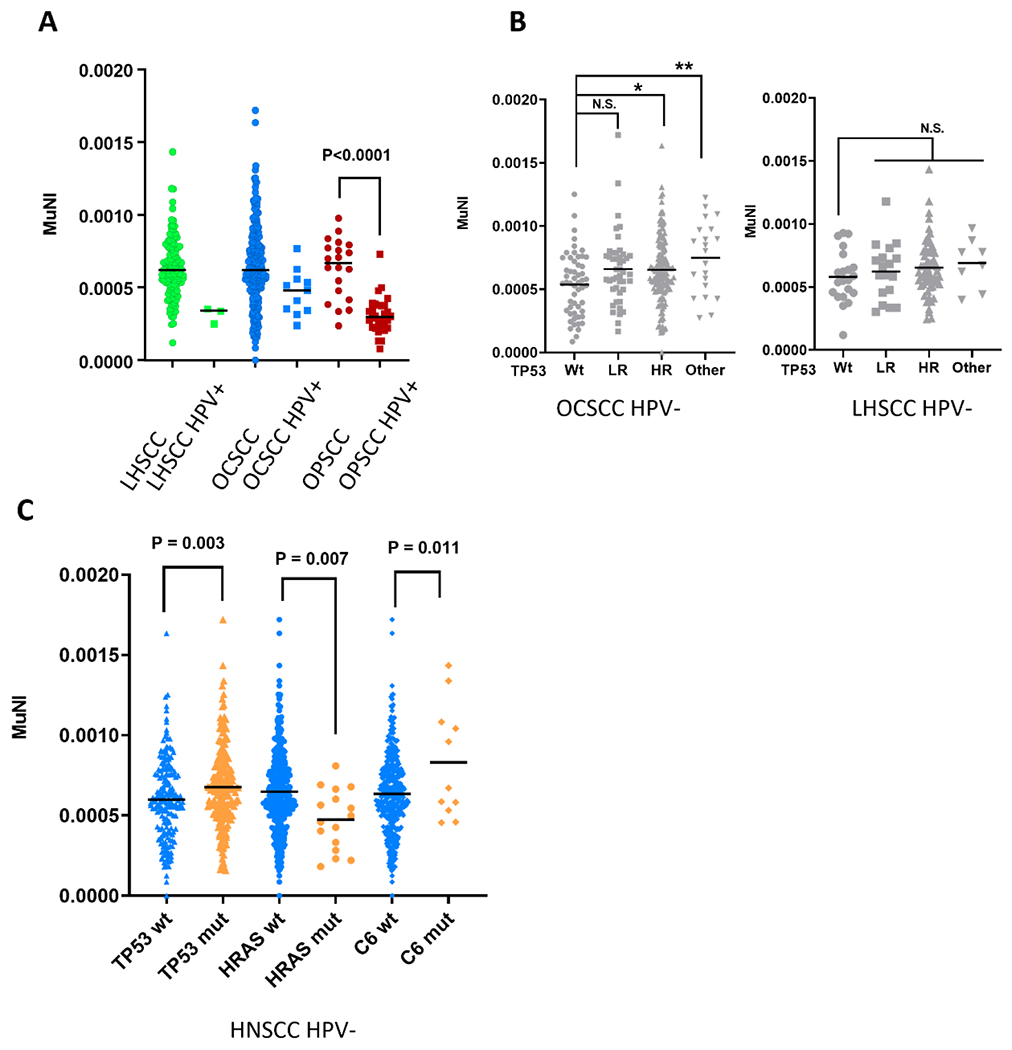
MuNI as a function of disease site (A), TP53 mutational status (B) and genomic background (C). wt= wild type; mut= mutant; HR= high risk; HPV+ = HPV-associated; HPV- = HPV-independent
TP53 is considered to be the “guardian” of the genome, but is mutated in roughly 80% of HPV-independent HNSCC.11 We examined if TP53 mutations, categorized as low or high risk using the evolutionary action scoring algorithm, impacted MuNI scores. 17,18 In HPV-independent OCSCC samples, there was a trend towards increased MuNI score across all classes of TP53 mutation, but only the high-risk or undefined risk TP53 mutations (splice or inframe) reached significance (Figure 1B). Although the same trend existed in LHSCC tumors, differences did not reach statistical significance (Figure 1B). To globally examine whether MuNI was associated with a specific genomic HNSCC background, we compared MuNI values among 387 TCGA HPV-independent HNSCC samples with available exome sequencing data that were either wild type or mutant for the top 30 cancer drivers (Supplementary Table 2).11 MuNI values were higher in samples with a mutated TP53 or Complement 6 (C6) gene and lower when an HRAS mutation was present (Figure 1C and Supplementary Table 2).
Multinucleation correlates with survival in HNSCC.
We previously published that high MuNI scores were associated with poor patient overall survival (OS) in a large cohort of HPV-associated OPSCC, using a MuNI cutoff score of 0.000380 derived from the median value of our training set from that study.7 As OCSCC and LHSCC are biologically distinct subtypes of HNSCC not generally driven by HPV, we investigated the relationship between MuNI and OS in these disease subsites as well. Initially, MuNI was treated as a continuous variable in a Cox proportional hazard model and found to be highly correlated with OS in the combined cohort of OCSCC and LHSCC patients (Figure 2A, p=0.00036). Expected cumulative survival was modeled for the minimum, lower quartile, middle quartile, upper quartile, and maximum observed MuNI values and a clear trend of decreasing survival (e.g., median OS and 5-year OS) was observed with increasing MuNI (Figure 2A). A multivariable nomogram that included MuNI, age, race, gender, T/N stage, and smoking status yielded a C-index of 0.65, where MuNI was prognostic of OS (2.25, 1.07-4.71, 0.03), independent from other clinical variables (Supplementary Table 3). High risk HPV is detected infrequently in OCSCC (e.g., 5%) and LHSCC (e.g., 3%)11 and has not been associated with improved survival in these disease subsites. When HPV-associated tumors were excluded from the TCGA OCSCC/LHSCC cohort, MuNI was still highly associated with OS in a Cox proportional hazards regression model (p=0.0065), consistent with the observation that the presence of HPV had no detectable impact among OCSCC patients (Figure 2B), which accounted for 80% of HPV cases (i.e., 11/14) outside of OPSCC.
Figure 2. Multinucleation correlates with overall survival.
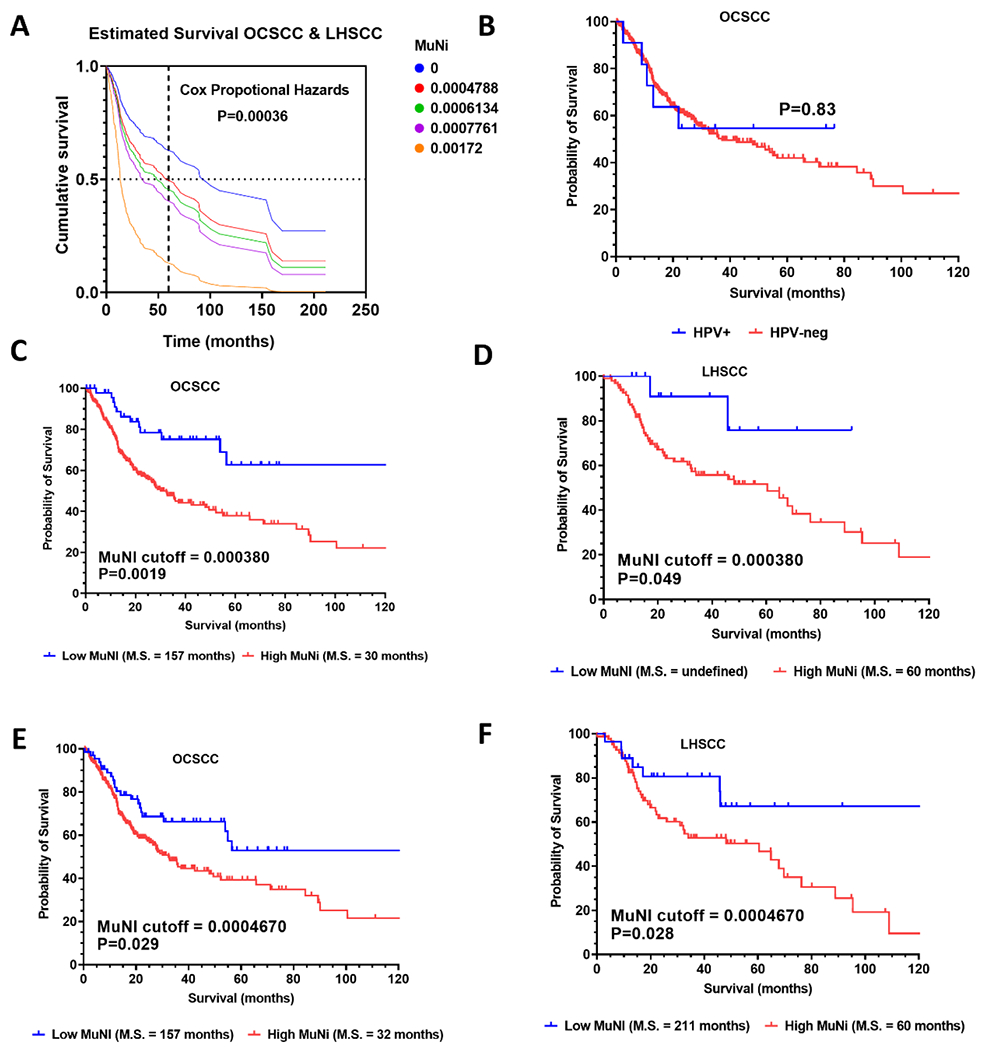
A significant relationship between MuNI and overall survival was found in the OCSCC/LHSCC TCGA cohort (A) using a Cox proportional hazards regression model (p=0.00036, Log-likelihood ratio), which showed decreases in estimated median and 5-year survival for MuNI values for the cohort minimum, lower quartile, median, upper quartile, and maximum. In the OCSCC cohort, the presence of HPV was not associated with survival differences (B). The MuNI cut-off (0.00038) derived from the median of HPV-associated OPSCC samples in a prior study was validated for survival stratification of TCGA OCSCC and LHSCC samples (C and D). The biological cut-off (0.0004670) derived from distribution of HPV-associated OPSCC samples performed well for survival stratification when applied to OCSCC and LHSCC (E, F). Survival is represented by Kaplan-Meier curves (C-F) evaluated for significance by a log-rank test.
Next, we examined whether our previously published MuNI cutoff (e.g., 0.000380) derived from a large independent cohort of HPV-associated OPSCC was also prognostic in OCSCC and LHSCC, which are largely HPV-independent. This cutoff threshold significantly separated both OCSCC (p=0.0019) and LHSCC patients (p=0.049) into good and poor prognosis groups (Figure 2C, D) with hazard ratios of 2.33 (95% confidence interval 1.54-3.52) and 3.71 (95% confidence interval = 1.64-8.38), respectively. Because our original MuNI cut-off employed the median approach (often used apriori for dichotomization) we investigated whether a cut-off based on biology might perform similar or better. For biological context, we took advantage of the finding that a subset of HPV-independent OCSCC and LHSCC had MuNI values in the range of HPV-associated OPSCC that on average have a better prognosis. We utilized the mean MuNI value and standard deviation of the HPV-associated OPSCC samples in the current study (e.g., 0.000308 ± 0.000126) to derive a new cut-off of 0.00046, below which 90% of HPV-associated OPSCC specimens were expected. When this biological cut-off was applied to OCSCC and LHSCC (Figure 2E, F), patients in the high MuNI groups from both subsites had significantly worse OS (p=0.029 and p=0.028, respectively) with very similar median survival times regardless of whether the new or previous cutoff was used. As the biological cut-off was more robust when both disease subsites were considered collectively and had a better underlying rationale, we applied it for all subsequent analyses. Lastly, a multivariable nomogram that included MuNI, age, race, gender, T/N stage, and smoking status yielded a C-index of 0.65, where MuNI was prognostic of OS (2.25, 1.07-4.71, 0.03), independent from other clinical variables (Supplementary Table 3).
Higher multinucleation correlates with suppressed anti-tumor immunity.
We previously showed that patients whose tumors had higher levels of infiltration by effector immunocytes had longer survival compared to their counterparts across multiple solid tumors.14,15 Using a published immune clustering approach based on single sample gene set enrichment analysis (ssGSEA) of immunocyte transcriptomic data14, we clustered OCSCC samples by relative leukocyte infiltration and examined the relationship between tumor immune status and MuNI levels (Figure 3A; Supplementary Table 4) after dichotomizing the latter into high or low MuNI based on our biological cut-off of 0.00046 (as shown above). First, we compared the distributions of high and low MuNI tumor samples among the immune clusters (Figure 3C). Immune cluster 1, which contained the highest degree of overall immune infiltration (very hot) was enriched for tumors with low MuNI status, in contrast to immune cluster 7 (cold) that was enriched for high MuNI samples (p=0.010). To analyze whether this effect is potentially driven by immune editing of multinucleated cells, we compared absolute levels of MuNI among just high MuNI tumors in either hot immune clusters or the cold immune cluster and found MuNI scores to be significantly higher in tumors with a cold immune microenviroment (Figure 3B).
Figure 3. MuNI correlation with anti-tumor immunity in OCSCC.
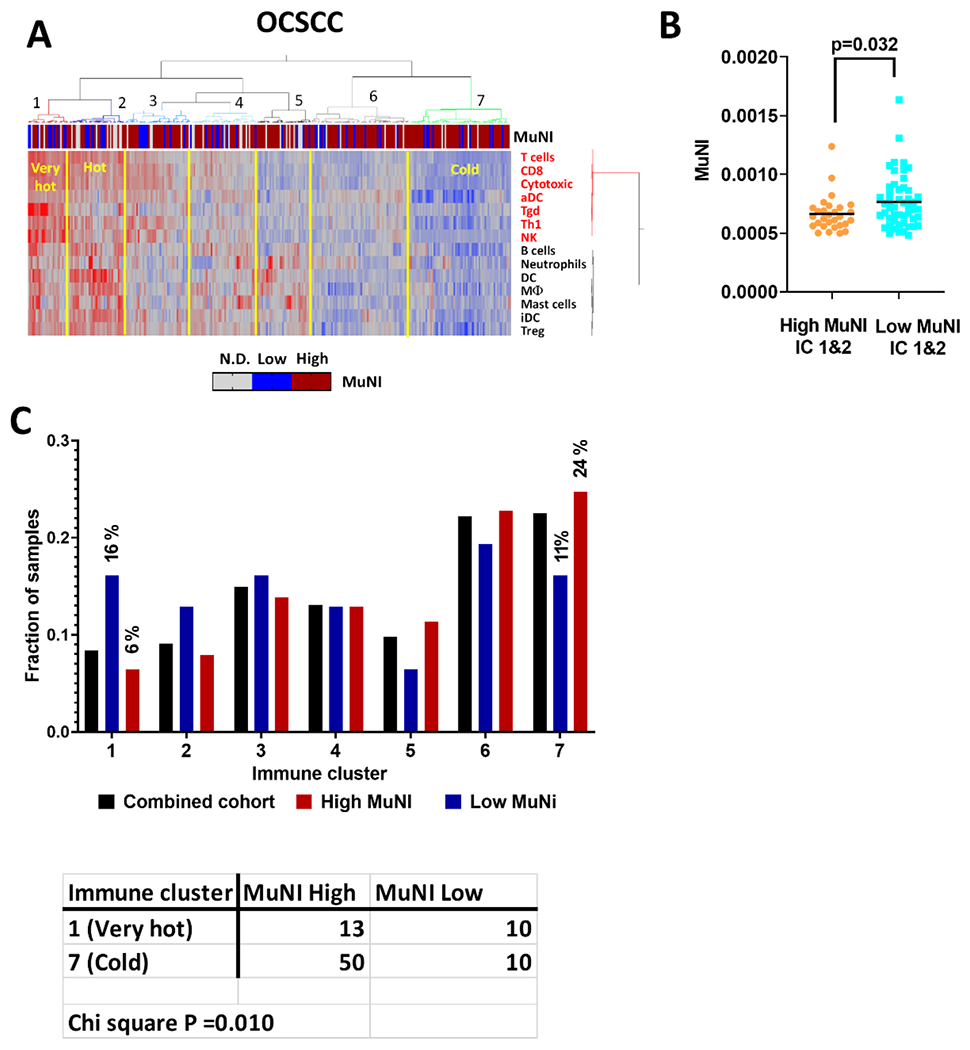
A) ssGSEA scores from 14 different immune subsets derived from gene expression data were used with consensus hierarchical clustering to identify subgroups of OCSCC patients (N=305) with varying degree of leukocyte infiltration. Cluster 1 and 2 were immunologically “very hot” and “hot” based on relative ssGSEA scores for T cells, CD8, cytotoxic and NK cells, compared to cluster 7 defined as immunologically cold. Tumors classified as high or low MuNI using a threshold of 0.00296 (see methods) are annotated across the top. B) High MuNI samples from the very hot/hot immune clusters had significantly lower MuNI scores than their counterparts in the cold immune cluster 7, which suggests immune editing. C) The very hot immune cluster 1 is enriched in samples from the low MuNI group while the cold immune cluster is enriched for samples with high MuNI. Bars represent the fraction (% in parenthesis) of each MuNI group, or the entire cohort represented in the immune cluster.
We used individual leukocyte subtype ssGSEA scores (Supplementary Table 4) to identify correlations with MuNI values among all HPV-independent OCSCC and LHSCC, as well as HPV-independent and HPV-associated OPSCC samples (Supplementary Table 5). In OCSCC, the majority of leukocyte subtypes were anti-correlated with MuNI scores, albeit the correlation values themselves were low (−0.14 to −0.21). A number of immune subsets (B cells, CD8+ cells, T cells) were modestly anti-correlated with MuNI values in HPV-associated OPSCC, consistent with our previously published results.8 However, little correlation with immune subsets was found in HPV-independent LHSCC. To further dissect relationships between MuNI and immune response, we stratified OCSCC samples by TP53 mutational status and discovered that wild type TP53 tumors showed the greatest degree of anti-correlation with MuNI values for multiple immune subsets (Supplementary Table 6), including T cells (ρ = −0.35, FDR =0.044), CD8+ cells (ρ = −0.32, FDR =0.044), cytotoxic cells (ρ = −0.31, FDR =0.044), and NK cells (ρ = −0.40, FDR =0.039). In OCSCC tumors with mutant TP53, these anti-correlations were much lower and unadjusted p-values frequently failed to be significant (Supplementary Table 6; Supplementary Figure 2). To better understand why immune correlations with MuNI had greater magnitude among tumors with wild type TP53, we stratified HPV-independent OCSCC by TP53 status alone and compared their ssGSEA scores. Tumors with wild type TP53 status had significantly greater infiltration of aDC, CD8+ cells, Cytotoxic cells, NK, T cells, Tgd, and Th1 compared to those with a high-risk or splice/frameshift mutation (Supplementary Table 7; Supplementary Figure 3A). In contrast, no significant differences in immune ssGSEA scores were found between tumors with wild type or mutant TP53 status among HPV-independent LHSCC samples (Supplementary Figure 3B) which could explain reduced impact of TP53 status on MuNI levels in LHSCC. Possibly, higher levels of cytotoxic and dendritic cells found in OCSCC tumors with wild type TP53, compared to those harboring HR TP53 mutations, are able to clear MuNI tumor cells though immune editing and significantly lower MuNI values (e.g. Figure 1B). Lack of increased leukocyte subsets in the LHSCC TP53 wt tumors could explain why such differences in MuNI were not found in this subsite. Although alternative explanations cannot be ruled out, the connection between TP53 and MuNI appears unrelated to genomic instability, as the latter would have impacted OCSCC and LHSCC similarly.
To further validate the anti-correlations between MuNI and tumor immune microenvironment components, we examined correlations between expression of the 116 individual immune-related genes defining the leukocyte ssGSEA scores and MuNI values among TP53 wild type (wt) and TP53 mutant (mut) HPV-independent OSCC tumors. In the TP53 wt tumors, 44 of the immune-related genes were significantly anti-correlated (p<0.05) and none were positively correlated (Supplementary Table 8; Figure 4). In TP53 mut (high-risk and other), however, only two immune-related genes (IDO/aDC, and FCRL2/B-cells) were negatively correlated with MuNI and again none were positively correlated. Among top ranked genes anti-correlating with MuNI in the TP53 wt tumors were those defining T cells (e.g., TRAT1, CD3G), CD8+ cells (e.g., CD8B, Peforin/PRF1), Cytotoxic cells (Granzyme A/GZMA) and NK cells (e.g., KIR3DL2).
Figure 4. Individual immune-gene correlates with MuNI.
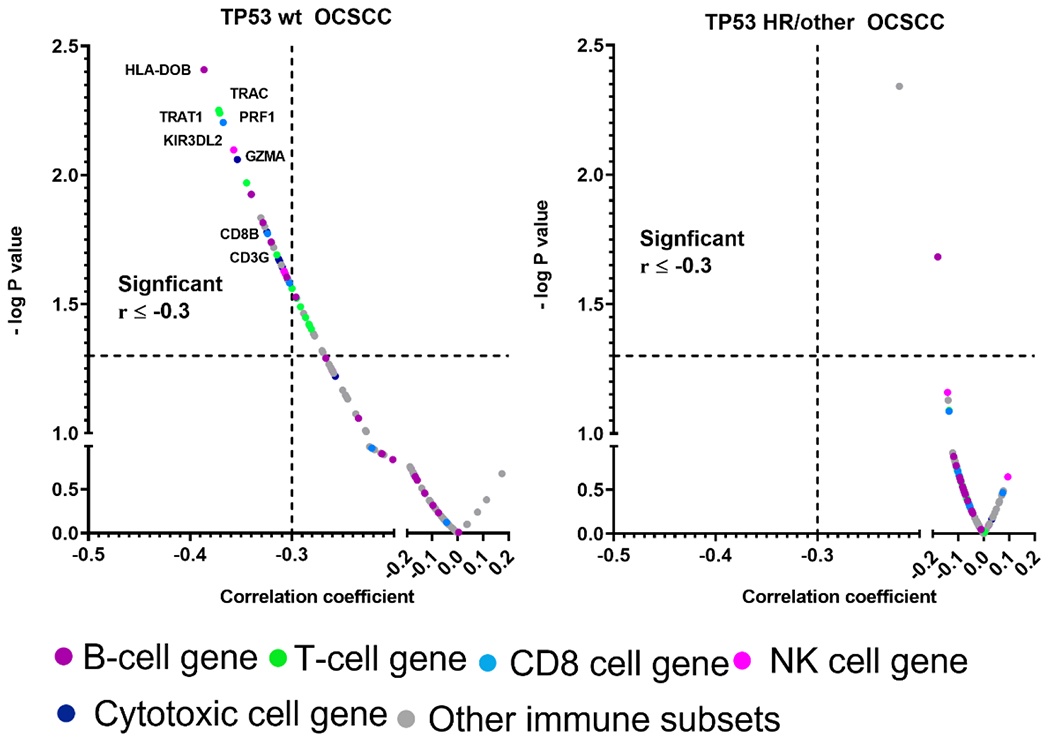
Correlation between individual genes (colors designate whether individual genes are included in ssGSEA sets for specific immunocytes) and MuNI in wt TP53 and mut TP53 OCSCC tumors. Dashed vertical and horizontal lines denote cut-off for significance (i.e., raw P value <0.05) or anti-correlation magnitude > 0.3, respectively.
Global analysis of genes associated with MuNI.
To identify genes which may pre-dispose tumors to have increased or decreased MuNI, independent of the immune microenvironment, we analyzed whole transcriptomic RNA-seq expression data to look for correlations with MuNI in TP53 wt and TP53 mut (high-risk/other) HPV-independent OCSCC. Genes that had significant positive or negative correlations (p<0.05) with MuNI at a magnitude ≥ 0.25 (Supplementary Table 9) were filtered by subtracting genes significantly correlated or anti-correlated with the presence of T-cells using matching ssGSEA scores (Figure 5). In OCSCC, the expression of more than one hundred genes meeting these criteria were either positively or negatively correlated with MuNI when OCSCC tumors were TP53 wt, and those numbers dropped precipitously to just a handful of genes correlating with MuNI in tumors harboring high risk/other TP53 mutations (Supplementary Table 9; Figure 5). A parallel analysis identified 18 genes positively correlated in TP53 wt OCSCC that were similarly correlated with MuNI in LHSCC, sometimes in the TP53 wt LHSCC group, sometimes in the TP53 mut LHSCC group, and sometimes regardless of TP53 status. Among the genes positively correlating with MuNI in both OCSCC and LHSCC were genes coding for transporters, BCAP31 which can regulate apoptosis, and death-associated protein kinase 3 (DAPK3) which has previously been linked to regulating cytokinesis and increased multi-nucleation.19 There were nine genes negatively correlated with MuNI in both TP53 wt OCSCC and LHSCC that were either TP53 wt or mutant, and these included genes regulating mitotic spindles, DNA replication, DNA damage, and apoptosis (Supplementary Table 9; Figure 5).
Figure 5. Individual gene correlates with MuNI.
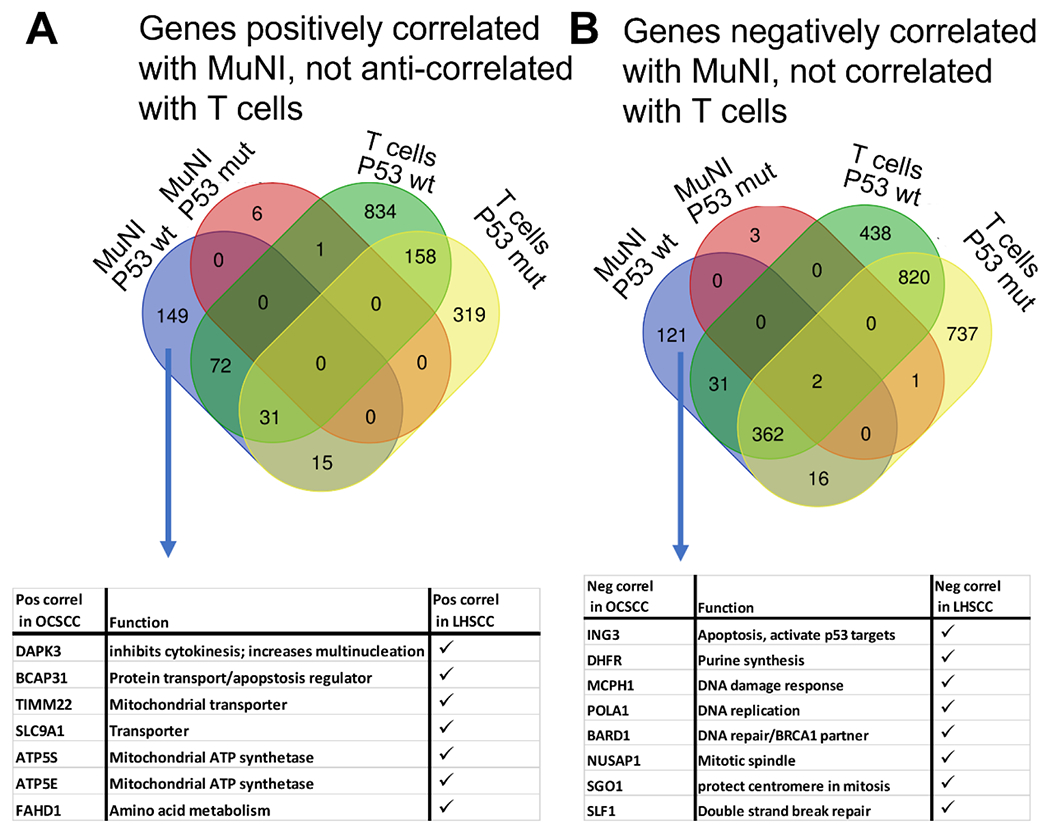
Venn diagrams showing relationship between genes significantly correlated with MuNI for TP53 wt or TP53 mutant (i.e., HR or other) OCSCC HPV-independent tumors and genes also correlating with T cell ssGSEA scores to identify those genes independent of the TIME which may be related instead to tumor biology. In (A) genes anti-correlated with T cell scores were used for analysis while in (B) genes that positively correlated with T cell scores were used (e.g., green and yellow ovals) because MuNI itself was found to inversely correlated with immune infiltration (see text). Tables in the bottom panels indicate specific genes of interest, independent of immune correlation, that were also found to significantly correlate with MuNI in the HPV-independent LHSCC cohort.
Because elevated expression of certain genes linked to DNA fidelity were associated with lower levels of MuNI in both OCSCC and LHSCC, we performed an expanded analysis to look for possible ties to the DNA damage response. We previously developed a 64 gene expression signature of the DNA damage response (DDR), which was validated in tumors harboring deletion of known DNA repair genes (manuscript under review). Therefore, we examined whether MuNI scores correlated with DDR scores of HPV-independent OCSCC and LHSCC, when stratified by TP53 mutational status. A strong anti-correlation was observed between DDR score and MuNI in the TP53 mut (high-risk/other) LHSCC (r =−0.4; p=0.0006, Figure 6) but for not for the other disease sites or mutational groups.
Figure 6. Correlation between MuNI and DNA damage repair activation (DDR).
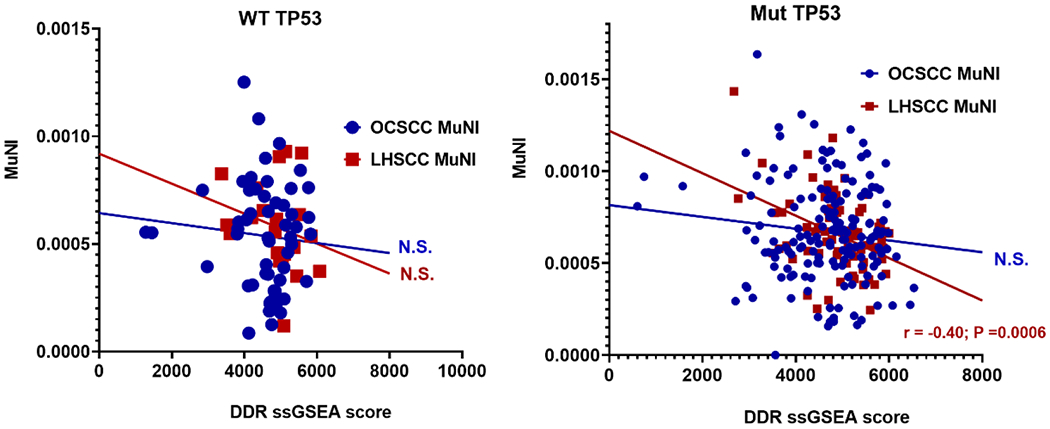
Using data from OCSCC and LHSCC tumors, dichotomized by TP53 mutational status we correlated activation of DDR (ssGSEA) with MuNI.
Discussion
Predicting response to treatment remains an unsolved problem in HNSCC. To date, T-N-M classification remains the gold standard for risk prediction and decisions regarding HNSCC treatment intensity (e.g. addition of adjuvant radiation post surgically). Unfortunately, T-N-M classification is primarily driven by tumor biology as it relates to rapid proliferation and metastatic potential, biological characteristics which have only indirect links with relative responsiveness to chemotherapy, radiation and immune checkpoint inhibitors (ICIs). This is demonstrated in the setting of HPV-associated HNSCC which overwhelmingly displays nodal metastasis at initial presentation yet at the same time retains an excellent prognosis due to a robust response to chemo-radiation. This very fact led to the de-emphasis of nodal metastasis in the 8th edition of the AJCC Staging Manual for HPV-associated OPSCC.20
Because HPV-associated OPSCCs demonstrate superior chemo-radiation response compared to HPV-independent HNSCC, HPV status is now used as a secondary prognostic classifier in HNSCC. However, it alone remains an insufficient predictor to identify patients who would safely benefit from a reduction in radiation dose, elimination of conventional chemotherapy or its replacement with targeted agents.21–23 We and others have identified additional risk classifiers, even in the context of HPV-based risk stratification, that can significantly segregate patients based on survival and potentially improve clinical algorithms aimed at de-escalation.4,5,7,8,24 Unfortunately, many of the new risk classifiers remain biological “black boxes” - statistical classifiers without a clear putative mechanism of action. This limits their potential in the clinical setting where we need to understand how to predict response to disparate treatment regimens. Simply put, black box classifiers are prognostic of survival, but without biological/mechanistic data they cannot become predictors that which can inform treatment selection and intensity.
We previously evaluated multinucleation using machine learning (MuNI) in multiple cohorts of surgically and non-surgically treated OPSCC patients and identified an excellent prognostic potential.7,8 A limited analysis of OPSCC specimens demonstrated a strong correlation between MuNI and cytotoxic immunocyte infiltration of tumors (pre-treatment) as well as expression of immune related genes.4,5,7,8,24 In the current study, we expanded the depth of genomic and transcriptomic analyses and breadth of the HNSCC disease sites interrogated to gain further insight into biological factors that may be influencing MuNI. It now appears that the frequency of multinucleated cells may be affected by multiple biological processes that interact including the tumor immune microenvironment (TIME), the presence of HPV, mutational status of known oncogenic drivers such as TP53 and expression of genes involved in DNA replication and repair. The degree to which each factor contributes seems to depend on the context of the other factors. HPV-associated tumors clearly demonstrated diminished frequency of multinucleated cells. Three biological features normally standout for HPV-associated HNSCC tumors that may contribute to this phenotype: 1) higher levels of pre-treatment infiltrating leukocytes such as T cells and cytotoxic cells.8,25 2) higher expression of DDR genes, which translates into higher DDR scores and 3) TP53 wild-type status. Along these lines, we found lower MuNI levels in wild-type TP53 HPV-independent OCSCC were associated with significantly higher infiltration of T cells, CD8+ cells, cytotoxic cells, and NK cells within this genomic subtype. Collectively, the evidence suggests immune editing of multinucleated cells could be taking place. In other words, low MuNI may be a good surrogate for a more favorable TIME whereas high MuNI may mean the opposite.
In mutant TP53 LHSCC, we measured a significant anti-correlation between expression of DDR genes and MuNI levels, which is reminiscent of HPV-associated tumors which also have higher DDR scores and lower MuNI. Possibly, tumor cells with enhanced machinery to divert replication stress or DNA damage are inherently less prone to mitotic catastrophe and/or MuNI events.26,27 More specifically, when we globally examined correlations between gene expression and MuNI we found a handful of the same genes involved in DNA replication and DNA damage to be inversely correlated in wild type TP53 OCSCC and in LHSCC, where the TP53 status seemed less important for the latter. The picture that emerges from our analysis is that there are likely intrinsic tumor biology mechanisms pertaining to mutational drivers, cell division and DNA replication/repair that can alter the background rate at which multinucleation appears, but the simultaneous presence or absence of functional cytotoxic lymphocytes in the TIME may impact how many of the multinucleated cells get eliminated. In retrospective analyses it is difficult to distinguish cause and effect. An alternative explanation could be that the presence of multinucleated cells suppresses a robust immune response directly. The extremely low frequency of multinucleated cells inside HNSCC tumors makes this a fairly unlikely scenario. Nevertheless, we are currently deploying spatial transcriptomic approaches to directly answer this question. If our hypothesis is correct, and high MuNI scores reflect a functionally defective TIME, this could have substantial implications for pre-treatment evaluations of HNSCC TIME and therapeutic algorithms predicated on active anti-tumor immunity. In contrast to genomic or transcriptomic functional TIME signatures, quantification of MuNI from H&E slides is cheap, fast and highly reproducible across clinical grade tissue samples.28 We have now utilized a discovery institutional OPSCC dataset to correlate MuNI to anti-tumor immunity and have validated our findings across the entire TCGA HNSCC cohort. We are deploying preclinical immune-competent models of HNSCC to study the mechanistic links between MuNI, DNA damage generation and repair and TIME. Although our results are promising, there are caveats to using survival data from the TCGA, including lack of uniform treatment and follow-up. Efforts are underway to further validate these correlations in prospective clinical datasets from HNSCC patients treated with definitive chemo-radiation or ICIs in order to address the inherent limitations of our correlative studies and develop MuNI into a predictive rather than simply prognostic biomarker. In conclusion, we have shown that MuNI represents not only a robust marker of survival in HNSCC, but one that seems to denote a functionally exhausted TIME and which may be a functional and phenotypic link between intrinsic biological events within HNSCC tumor cells and activation of effector immunocytes.
Supplementary Material
Highlights.
Multinucleation levels vary as a function of disease site and intrinsic tumor biology (e.g. HPV, TP53 mutational status).
Multinucleation correlates strongly with survival across head and neck disease sites.
Multinucleation correlates with anti-tumor immunity independent of HPV and/or TP53 mutational status and may be linked to an immunosuppressed tumor microenvironment.
Funding:
This work was supported by a Career Development Award to VCS from the Veterans Administration Clinical Science Research and Development division (1IK2CX001953). Dr Madabhushi’s work is supported from grants supported by the National Cancer Institute under award numbers 1U24CA199374-01, R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01, National Heart, Lung and Blood Institute 1R01HL15127701A1, R01HL15807101A1, National Institute of Biomedical Imaging and Bioengineering 1R43EB028736-01, National Center for Research Resources under award number 1 C06 RR12463-01 VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404, W81XWH-21-1-0345), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Mayo Clinic Breast Cancer SPORE grant P50 CA116201 from the National Institutes of Health, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. Sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, and Astrazeneca. KY was supported by the Computational Genomic Epidemiology of Cancer (CoGEC) Program at Case Comprehensive Cancer Center (T32CA094186), Young Investigator Award from ASCO Conquer Cancer Foundation, RSNA Research Resident Grant, and Cleveland Clinic VeloSano Impact Award.
Conflicts of interest:
There are no competing interests or conflicts of interest directly related to this work. Dr. Madabhushi is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently he serves on the advisory board of Picture Health, Aiforia Inc, and SimBioSys. He also currently consults for Roche, Biohme, and Castle Biosciences. He also has sponsored research agreements with AstraZeneca, Boehringer-Ingelheim, Eli-Lilly and Bristol Myers-Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. He is also involved in 3 different R01 grants with Inspirata Inc. Dr. Koyfman serves on the advisory boards of Merck and Regeneron and has received funding from Merck, BMS and Castle Biosciences as well as honoraria from UptoDate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bauml JM, Vinnakota R, Anna Park YH, et al. Cisplatin versus cetuximab with definitive concurrent radiotherapy for head and neck squamous cell carcinoma: An analysis of Veterans Health Affairs data. Cancer. Feb 1 2019;125(3):406–415. doi: 10.1002/cncr.31816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard P, Baujat B, Holostenco V, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): a comprehensive analysis by tumour site. Radiother Oncol. Jul 2011;100(1):33–40. doi: 10.1016/j.radonc.2011.05.036 [DOI] [PubMed] [Google Scholar]
- 3.Sandulache VC, Wilde DC, Sturgis EM, Chiao EY, Sikora AG. A Hidden Epidemic of “Intermediate Risk” Oropharynx Cancer. Laryngoscope Investig Otolaryngol. Dec 2019;4(6):617–623. doi: 10.1002/lio2.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandulache VC, Hamblin J, Lai S, et al. Oropharyngeal squamous cell carcinoma in the veteran population: Association with traditional carcinogen exposure and poor clinical outcomes. Head Neck. Sep 2015;37(9):1246–53. doi: 10.1002/hed.23740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elhalawani H, Mohamed ASR, Elgohari B, et al. Tobacco exposure as a major modifier of oncologic outcomes in human papillomavirus (HPV) associated oropharyngeal squamous cell carcinoma. BMC Cancer. Sep 23 2020;20(1):912. doi: 10.1186/s12885-020-07427-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis JS Jr., Scantlebury JB, Luo J, Thorstad WL. Tumor cell anaplasia and multinucleation are predictors of disease recurrence in oropharyngeal squamous cell carcinoma, including among just the human papillomavirus-related cancers. Am J Surg Pathol. Jul 2012;36(7):1036–46. doi: 10.1097/PAS.0b013e3182583678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyuncu CF, Lu C, Bera K, et al. Computerized tumor multinucleation index (MuNI) is prognostic in p16+ oropharyngeal carcinoma: A multi-site validation study. J Clin Invest. Mar 2 2021;doi: 10.1172/JCI145488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilde DC, Castro PD, Bera K, et al. Oropharyngeal cancer outcomes correlate with p16 status, multinucleation and immune infiltration. Mod Pathol. Feb 18 2022;doi: 10.1038/s41379-022-01024-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koyuncu CF, Nag R, Lu C, et al. Image analysis reveals differences in tumor multinucleations in Black and White patients with human papillomavirus-associated oropharyngeal squamous cell carcinoma. Cancer. Nov 1 2022;128(21):3831–3842. doi: 10.1002/cncr.34446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides. JCO Clin Cancer Inform. Apr 2019;3:1–7. doi: 10.1200/CCI.18.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. Jan 29 2015;517(7536):576–82. doi: 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68–77. doi: 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyuncu CF, Lu C, Bera K, et al. Computerized tumor multinucleation index (MuNI) is prognostic in p16+ oropharyngeal carcinoma. J Clin Invest. Apr 15 2021;131(8)doi: 10.1172/JCI145488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazi MA, Veeramachaneni R, Deng D, et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. Journal for ImmunoTherapy of Cancer. 2022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frederick M, Skinner HD, Kazi SA, Sikora AG, Sandulache VC. High expression of oxidative phosphorylation genes predicts improved survival in squamous cell carcinomas of the head and neck and lung. Sci Rep. Apr 14 2020;10(1):6380. doi: 10.1038/s41598-020-63448-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Chen Y, Putluri N, et al. Evolution of cisplatin resistance through coordinated metabolic reprogramming of the cellular reductive state. Br J Cancer. Apr 3 2023;doi: 10.1038/s41416-023-02253-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neskey DM, Osman AA, Ow TJ, et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res. Apr 1 2015;75(7):1527–36. doi: 10.1158/0008-5472.CAN-14-2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman AA, Neskey DM, Katsonis P, et al. Evolutionary Action Score of TP53 Coding Variants Is Predictive of Platinum Response in Head and Neck Cancer Patients. Cancer Res. Apr 1 2015;75(7):1205–15. doi: 10.1158/0008-5472.CAN-14-2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono T, Terada F, Okumura M, Chihara T, Hamao K. Impairment of cytokinesis by cancer-associated DAPK3 mutations. Biochem Biophys Res Commun. Dec 17 2020;533(4):1095–1101. doi: 10.1016/j.bbrc.2020.09.078 [DOI] [PubMed] [Google Scholar]
- 20.O’Sullivan B, Huang SH, Su J, et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol. Apr 2016;17(4):440–451. doi: 10.1016/S1470-2045(15)00560-4 [DOI] [PubMed] [Google Scholar]
- 21.Yom SS, Torres-Saavedra P, Caudell JJ, et al. Reduced-Dose Radiation Therapy for HPV-Associated Oropharyngeal Carcinoma (NRG Oncology HN002). J Clin Oncol. Mar 20 2021;39(9):956–965. doi: 10.1200/JCO.20.03128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehanna H, Robinson M, Hartley A, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet. Jan 5 2019;393(10166):51–60. doi: 10.1016/S0140-6736(18)32752-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillison ML, Trotti AM, Harris J, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet. Jan 5 2019;393(10166):40–50. doi: 10.1016/S0140-6736(18)32779-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corredor G, Toro P, Koyuncu C, et al. An Imaging Biomarker of Tumor-Infiltrating Lymphocytes to Risk-Stratify Patients with HPV-Associated Oropharyngeal Cancer. J Natl Cancer Inst. Nov 29 2021;doi: 10.1093/jnci/djab215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemnade JO, Elhalawani H, Castro P, et al. CD8 infiltration is associated with disease control and tobacco exposure in intermediate-risk oropharyngeal cancer. Sci Rep. Jan 14 2020;10(1):243. doi: 10.1038/s41598-019-57111-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha D, Duijf PHG, Khanna KK. Mitotic slippage: an old tale with a new twist. Cell Cycle. Jan 2019;18(1):7–15. doi: 10.1080/15384101.2018.1559557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranjan A, Iwakuma T. Non-Canonical Cell Death Induced by p53. Int J Mol Sci. Dec 9 2016;17(12)doi: 10.3390/ijms17122068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damotte D, Warren S, Arrondeau J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J Transl Med. Nov 4 2019;17(1):357. doi: 10.1186/s12967-019-2100-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


