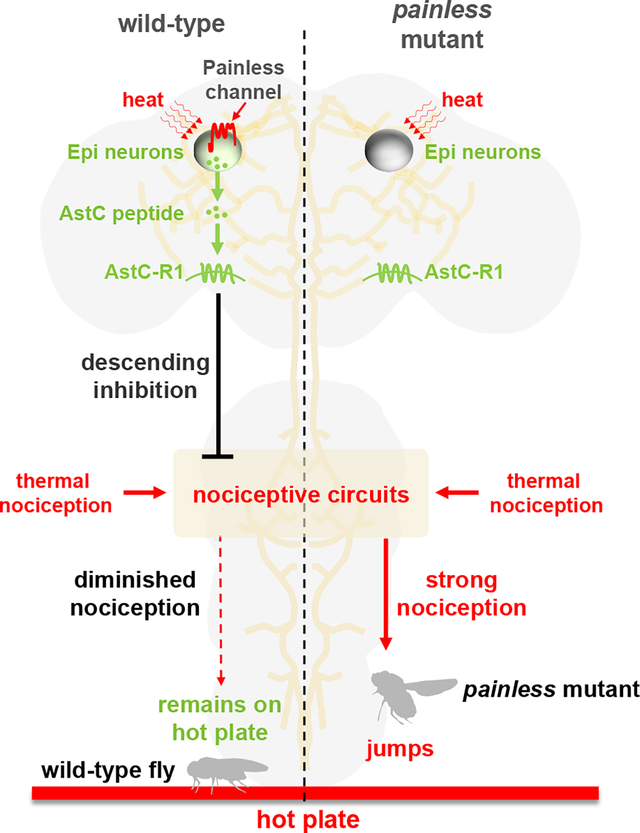
SUMMARY
Acute avoidance of dangerous temperatures is critical for animals to prevent or minimize injury. Therefore, surface receptors have evolved to endow neurons with the capacity to detect noxious heat, so that animals can initiate escape behaviors. Animals including humans have evolved intrinsic pain-suppressing systems to attenuate nociception under some circumstances. Here, using Drosophila melanogaster, we uncovered a new mechanism through which thermal nociception is suppressed. We identified a single descending neuron in each brain hemisphere, which is the center for suppression of thermal nociception. These Epi neurons, for Epione—the goddess of soothing of pain, express a nociception-suppressing neuropeptide Allatostatin C (AstC), which is related to a mammalian anti-nociceptive peptide, somatostatin. Epi neurons are direct sensors for noxious heat, and when activated they release AstC, which diminishes nociception. We found that Epi neurons also express the heat-activated TRP channel, Painless (Pain), and thermal activation of Epi neurons, and the subsequent suppression of thermal nociception, depends on Pain. Thus, while TRP channels are well known to sense noxious temperatures to promote avoidance behavior, this work reveals the first role for a TRP channel for detecting noxious temperatures for the purpose of suppressing rather than enhancing nociception behavior in response to hot thermal stimuli.
Keywords: Drosophila, nociception, pain, pain suppression, neuromodulation, neuropeptide, Allatostatin C, TRP channel, Painless, Epione neurons, temperature, thermal, hot
eTOC blurb
Sensing noxious stimuli is critical for survival. Animals are also endowed with mechanisms to suppress nociception. Using Drosophila, Liu et al. identify a pair of neurons in the brain (Epi neurons) critical for suppressing thermal nociception. Epi neurons sense heat via a TRP channel, and then release a neuropeptide that suppresses nociception.
Graphical Abstract
INTRODUCTION
Endogenous pain inhibitory systems can temporarily provide relief. Millions of people suffer from chronic and debilitating pain, some of which might be induced by abnormalities in the descending pain modulatory system.1–12 In mammals, neurotransmitters and neuromodulators, including endogenous opioids (β-endorphin, encephalin and dynorphin) and endogenous cannabinoids play important roles in nociception inhibition.13,14 Brain imaging and electrophysiological studies indicate that the pain-suppressing descending modulatory circuit receives input from multiple brain regions including the rostral anterior cingulate cortex, the periaqueductal gray region and the rostral ventromedial medulla.12,13 However, the key neurons that are activated in the inhibitory pathway, and the target neurons that are silenced have not been clearly delineated.
A pain inhibitory system has also been documented in worms. In C. elegans avoidance responses that are mediated through the polymodal ASH neurons,15,16 are suppressed by complex signaling pathways initiated by octopamine and neuropeptides.17 Drosophila has also been employed to study the inhibition of nociception,18 in addition to the far more extensive studies focusing on the mechanisms for detecting noxious stimuli, such as excessive heat, to initiate escape responses.19,20 Tracey et al. revealed that a Drosophila channel, Painless (Pain),21 which is related to the TRP channel in the fly’s compound eye,22 is critical for sensing noxious heat. This work, which followed the seminal discovery of TRPV1 as a heat sensor in mammals,23 and the finding that a related TRPV channel (Osm-9) contributes to several other sensory modalities in C. elegans,33 contributed significantly to the notion that TRP channels are evolutionarily conserved polymodal sensors.24 In addition to Pain, two other Drosophila TRP channels also function in sensing high temperatures to promote escape behavior: Pyrexia (Pyx) and TRPA1.25–28 However, it is unclear if any TRP channel serves to detect noxious heat for the purpose of alleviating thermally-induced nociception.
In this work, we used the fruit fly, Drosophila melanogaster, to investigate an intrinsic system for suppression of thermal nociception. We identified a pair of bilaterally-symmetrical neurons in the brain that is required for decreasing the nociceptive response to hot temperatures. These Epi neurons respond directly to heat and release a neuropeptide, Allatostatin C (AstC), which is required for suppression of nociception. The ability of Epi neurons to sense noxious heat depends on Pain, demonstrating a role for a thermo-TRP in suppressing rather than enhancing the nociceptive response to high temperatures.
RESULTS
Epi neurons function in suppression of thermal nociception
To identify neurons that may play roles in the suppression of thermal nociception, we devised a thermal nociception assay (Figures 1A, S1A and S1B). We placed flies on a hot plate, and assayed the percentage that jumped within 10 seconds. To prevent the insects from flying away during the assay, we first amputated their wings and allowed them to recover for 24 hours.
Figure 1. Identification of a pair of nociception-inhibitory neurons in the central brain.
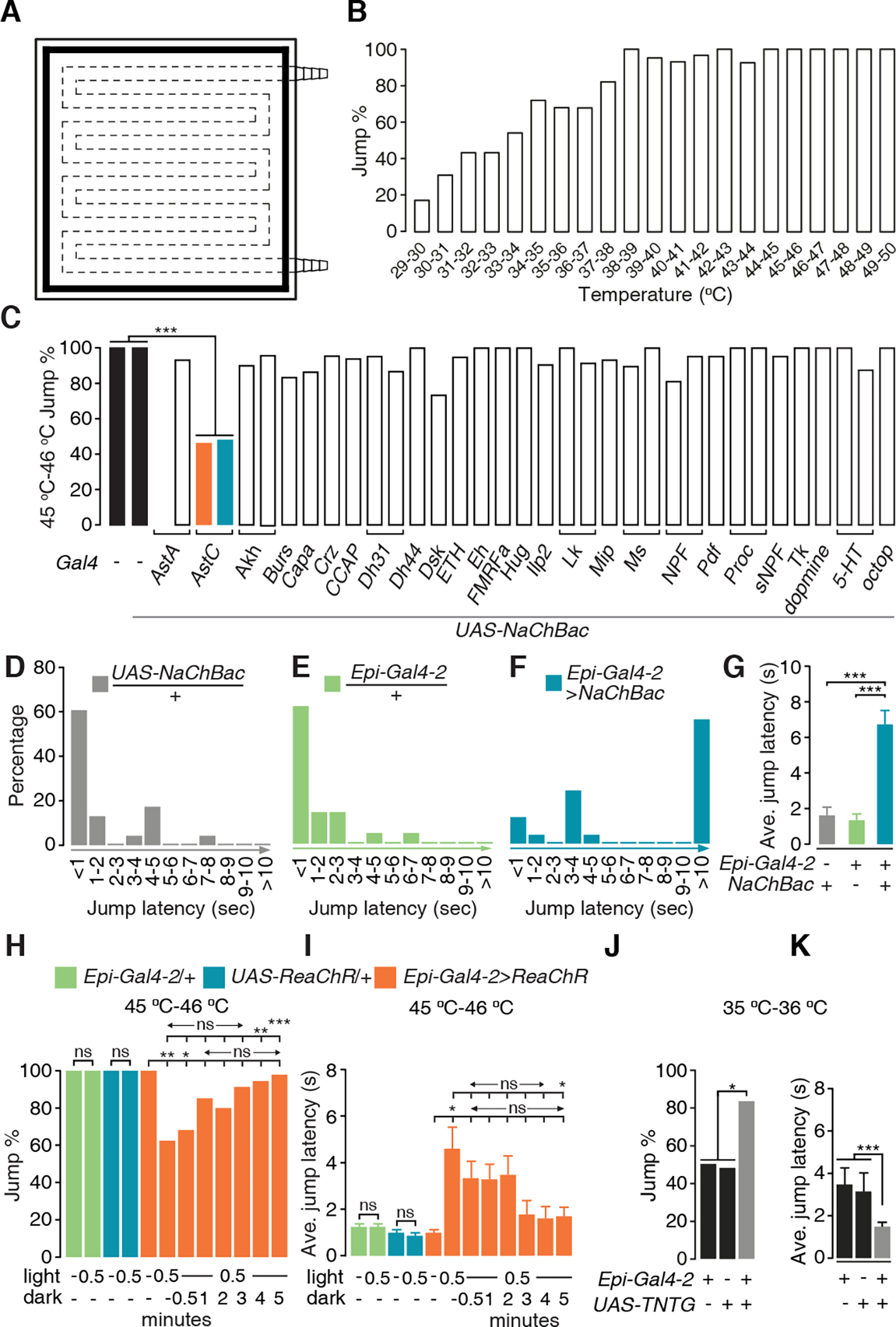
(A) Design of the hot plate for the jump assay. The dashed lines indicate water tunnels inside the plate. The black shading around the periphery represents the moat.
(B) Jump percentages of the control line (w1118) at different temperatures. n ≥ 20.
(C) Jump percentages (45 °C—46 °C) exhibited by flies expressing peptidergic and aminergic Gal4 lines driving expression NaChBac. The control lines (black bars) are w1118 (left bar) and UAS-NaChBac /+ (second bar from the left). n ≥ 20. Fisher’s exact test. *** P < 0.001.
(D-F) Percentages of flies showing the indicated jump latencies (sec) on a 45 °C—46 °C hot plate. (D) UAS-NaChBac only control. (E) Epi-Gal4-2 only control. (F) Epi-Gal4-2>NaChBac.
(G) Average values of jump latencies on a 45 °C—46 °C hot plate. n ≥ 20. Error bars indicate S.E.M.s. Mann-Whitney test. ***P < 0.001.
(H and I) Jump percentages (H) and average jump latencies (I) of flies on a 45 °C—46 °C hot plate, when Epi neurons were optogenetically activated for 0.5 min and then allowed to recover in the dark for 0.5—5 min. To stimulate the Epi neurons, the flies expressed UAS-ReaChR under control of the Epi-Gal4-2 and were stimulated with red lights. n ≥ 20. Error bars indicate S.E.M.s. Fisher’s exact test (H). Mann-Whitney test (I). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
(J and K) Jump percentages (J) and average jump latencies (K) on a 35 °C—36 °C hot plate using flies in which the Epi neurons were blocked with TNTG. n ≥ 20. Error bars indicate S.E.M.s. Fisher’s exact test (J). Mann-Whitney test (K). * P < 0.05, ***P < 0.001.
To establish the relationship between temperature and the jump response, we exposed control flies (w1118) to a series of temperatures ranging from 29 °C to 50 °C. At 29 °C—30 °C, few flies exhibited jump responses (17.2%), while at temperatures between 33 °C—34 °C more than half (54.2%) of the flies jumped (Figure 1B). The percentage of flies that responded continued to increase with temperature. Between 38 °C—44 °C, nearly all the files jumped (92.6%—100%; Figure 1B). Once the temperature exceeded 44 °C, all of the flies reacted to the noxious stimuli (Figure 1B).
To interrogate candidate neurons that might contribute to suppression of thermal nociception, we tested the effects of activating peptidergic and aminergic neurons. We chronically activated different subsets of neurons by expressing the bacterial Na+ channel NaChBac (UAS-NaChBac) under the control of 31 peptidergic and 4 aminergic Gal4 drivers, and monitored the percentage of flies that jumped at 45 °C—46 °C. Expression of UAS-NaChBac under the control of most Gal4 lines had no significant effect (Figure 1C). In contrast, two distinct drivers corresponding to the Allatostatin C (AstC) gene, AstC-Gal4-1 (BL39283)29 and AstC-Gal4-2 (BL52017) significantly diminished the percentage of flies that jumped (Figure 1C). We refer to these lines as Epi-Gal4-1 and Epi-Gal4-2, respectively. We also tested whether activation of the neurons labeled by the Epi-Gal4 lines impacted on jump latency. When we placed control flies on a 45 °C—46 °C surface, most jumped in ≤1 second (Figures 1D, 1E and S2A; UAS-NaChBac/+, 60.9%. Epi-Gal4-1/+, 71.4%. Epi-Gal4-2/+, 61.9%). In addition, the average jump latencies were only ~1.0—1.6 seconds (Figure 1G; UAS-NaChBac/+, 1.63 ±0.46 s. Epi-Gal4-1/+, 0.95 ±0.28 s. Epi-Gal4-2/+, 1.35 ±0.34 s; see Methods for calculation of ave. jump latencies). In contrast, activation of the Epi-Gal4-positive neurons dramatically increased the latency before jumping. The majority of Epi>NaChBac flies required ≥10 second (Figures 1F and S2B; Epi-Gal4-1>NaChBac, 57.5%. Epi-Gal4-2>NaChBac, 56.0%), and the average jump latencies were also greatly increased (Figures 1G and S2C; Epi-Gal4-1>NaChBac, 7.52±0.57 s. Epi-Gal4-2>NaChBac, 6.72±0.78 sec). At higher noxious temperatures (48 °C—50 °C), Epi>NaChBac flies reacted similarly to control flies (Figures S2D—S2H), indicating the limits of Epi>NaChBac neuronal activation in suppressing the jump reaction. These data also show that the reduced jump reactions at 45 °C—46 °C were not due to impaired jump ability.
To address whether acute activation of Epi-Gal4 positive neurons is sufficient to alleviate nociception, we used an optogenetic approach. We expressed the red-shifted channelrhodopsin, ReaChR (UAS-ReaChR) under control of the Epi-Gal4-1 or Epi-Gal4-2, exposed the animals to red lights (655 nm peak) for 30 seconds, and assayed their jump responses when placed on a 45 °C—46 °C surface. We found that acute activation of the Epi-Gal4 neurons significantly reduced the percentages of flies that jumped (Figures 1H and S2I) and increased the jump latency (Figures 1I and S2J). To determine how long the nociception-alleviation effects continued following acute activation of the Epi-Gal4 neurons, we exposed the UAS-ReaChR flies to light for 30 seconds and then maintained them in the dark for 0.5—5 minutes before testing their jump responses. We found that the alleviation of nociception gradually diminished over time. After only 30 to 60 seconds in the dark, the jump percentages and the average jump latencies were not statistically different from flies that did not have their Epi neurons light activated (Figures 1H, 1I, S2I, and S2J). However, ~3 minutes was required before the decreased jump percentages and latencies were significantly different from flies immediately after activation (Figures 1I, 1J, S2I and S2J). Because several minutes were required for full restoration of the nociceptive response, a neuromodulator such as AstC, which is longer lasting than a neurotransmitter, might underlie this relatively slow recovery.
To confirm the role of Epi-Gal4 neurons in suppressing nociception, we tested the effects of inhibiting synaptic transmission from these neurons and on the reaction to a 35 °C—36 °C heat stimulus. We expressed tetanus toxin (UAS-TNTG)30 under control of the Epi-Gal4-2 driver, and found that disrupting signaling from these neurons significantly increased the jump percentage (Figure 1J) and decreased the average jump latency (Figure 1K).
To identify the neurons expressing the Epi-Gal4-1 and the Epi-Gal4-2, we used these lines to drive expression of UAS-mCD8::GFP and performed staining with anti-GFP. The Epi-Gal4-1 reporter stained multiple brain regions, including neurons in a region near the optic lobe (OL), neurons in the primary taste center (the subesophageal zone; SEZ), and a cluster of neurons projecting to the ellipsoid body (EB) of the central complex, which functions in multisensory integration (Figures 2A and 2B).31–33 The Epi-Gal4-2 displayed a much more restricted pattern—labeling one pair of neurons in the brain with large cell bodies (Figures 2E and 2F). We expressed a nuclear GFP (UAS-Stinger 2), which confirmed that there were only two cells labeled in the brain using the Epi-Gal4-2 (Figure S3A). We did not detect Epi-Gal4-2 expression in other organs, including the legs, wings, antenna, proboscis and the digestion system (Figures S3B—S3H). The Epi-Gal4-2-positive neurons arborized a portion of the dorsal region in the brain (Figures 2E and 2F), and projected to multiple segments in the ventral nerve cord (VNC; Figures 2G and 2H). The arborization was more extensive with the Epi-Gal4-1 (Figures 2A—2D). Based on the position and arborization pattern, the neurons labeled by the Epi-Gal4-2 (arrows, Figures 2E and 2F) also appeared to be labeled by the Epi-Gal4-1 (arrows, Figures 2A and 2B). We refer to this pair of neurons as Epione (Epi) neurons after the Greek goddess of soothing of pain,34 since chronic or acute activation of these neurons is sufficient to alleviate thermal nociception.
Figure 2. Spatial localization of Epi neurons.
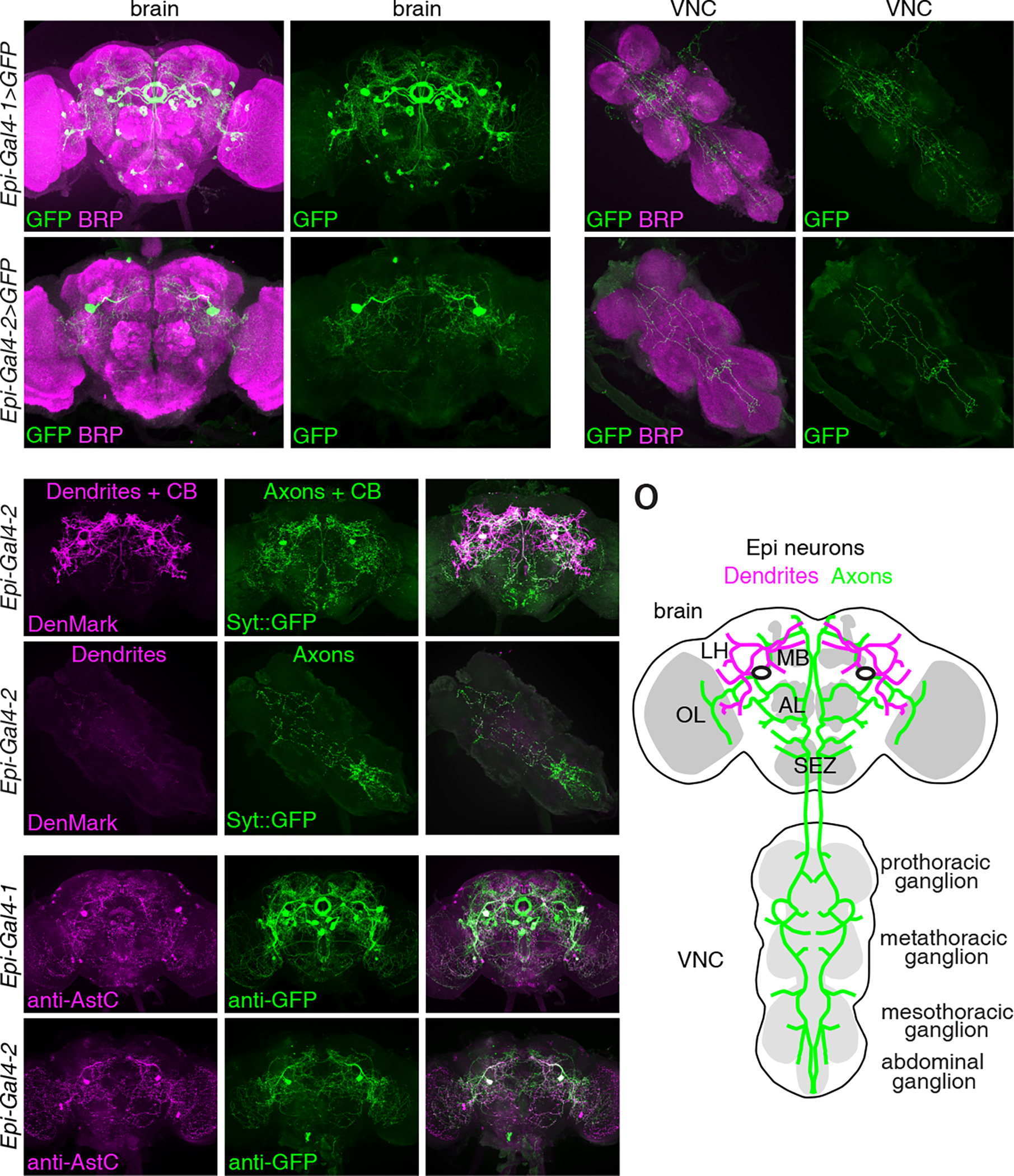
(A-H) Expression patterns of Epi-Gal4-1 (A-D) and Epi-Gal4-2 (E-H) in the brain and VNC. Gal4-driven expression of UAS-mCD8::GFP is indicated in green (anti-GFP) and the neuropil marker BRP is labeled in magenta (anti-BRP). Scale bars indicate 50 μm. (I-N) Axons (green) and dendrites (magenta) of Epi-Gal4-2 positive neurons labeled with Syt::eGFP and DenMark, respectively. (I-K) Brain. (L-N) VNC. Scale bars indicate 50 μm. CB, cell body. OL, optic lobe. LH, lateral horn. MB, mushroom body. AL, antenna lobe. SEZ, subesophageal zone.
(O) Schematic illustration of the dendrites (magenta), axons (green) and cell bodies (two black ovals) of Epi neurons in the brain and VNC.
(P-U) Adult brains stained with anti-AstC (magenta) and anti-GFP (green). (P-R) Epi-Gal4-1-driven expression of UAS-mCD8::GFP. (S-U) Epi-Gal4-2-driven expression of UAS-mCD8::GFP. Scale bars indicate 50 μm.
See also Figure S3.
To better visualize the distribution of the dendrites and axons of the Epi neurons, we used the Epi-Ga4-2 to drive expression of DenMark and synaptotagmin::GFP (Syt::GFP), which label dendrites and axons, respectively.35,36 Both DenMark and Syt::GFP also label the cell bodies. We found that the dendrites of the Epi neurons innervated multiple regions in the brain, including the optic lobe (OL), the lateral horn (LH), and areas close to the mushroom body (MB; Figures 2I, 2K and 2O). However, there was very limited DenMark staining in the VNC (Figure 2L). The axonal signals, which were marked by Syt::GFP, branched extensively in the brain, including regions in the optic lobe (OL), mushroom bodies (MB) and the subesophageal zone (SEZ) (Figures 2J, 2K and 2O). The axons also projected to multiple segments in the VNC, including the prothoracic, metathoracic, mesothoracic and abdominal ganglion (Figures 2M—2O).
AstC is required in Epi neurons to control nociception
AstC receptors has been proposed to be expressed in nociceptive neurons,37 but the neurons secreting AstC to inhibit nociception have not been identified. To determine if Epi neurons express the AstC neuropeptide, we performed double labeling. We found that the two most prominent neurons that were labeled with anti-AstC also expressed the Epi-Gal4-1 and the Epi-Gal4-2, which drove the expression of UAS-mCD8::GFP (Figures 2P—U). Thus, Epi neurons express AstC.
AstC is related to the mammalian neuropeptide, somatostatin, which plays a role in suppressing thermally-induced pain.38 Expression of AstC in Epi neurons raised the possibility that it is a nociceptive-suppressing neuromodulator produced by Epi neurons. To address this idea, we used the Epi-Gal4-2 to knock down AstC (UAS-AstC RNAi). This approach was effective since the anti-AstC staining was virtually eliminated in the Epi neurons (Figures S4A and S4B). To determine whether the reduction in AstC increased the nociceptive response, we tested whether the flies exhibited hypersensitivity to 35 °C—36 °C. We found that knockdown of AstC in Epi neurons (Epi-Gal4-2 >UAS-AstC RNAi) greatly increased the percentage of flies that jumped (Figure 3A) and decreased the jump latency (Figure 3B).
Figure 3. AstC contributes to inhibition of nociception.
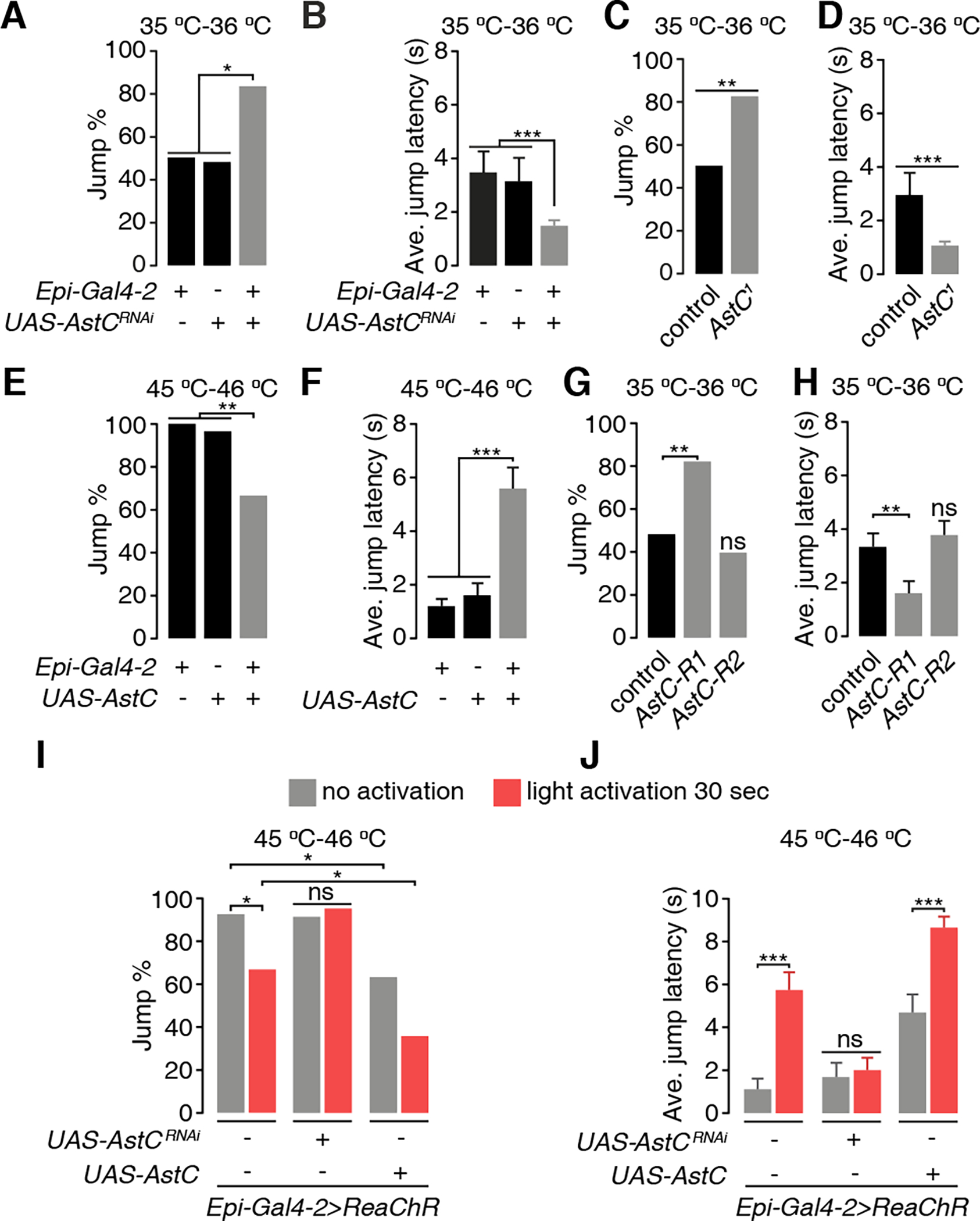
(A and B) Effects of UAS-AstCRNAi knock down in Epi neurons (Epi-Gal4-2) on the jump percentages (A) and the average jump latencies (B) on a 35 °C—36 °C hot-plate. (C and D) Jump percentage (D) and average jump latency (E) of AstC1 flies on a 35 °—36 °C hot-plate.
(E and F) Jump percentages (F) and average jump latencies (G) of flies overexpressing AstC in Epi neurons.
(G and H) Jump percentages (G) and average jump latencies (H) of AstC-R1 and AstC-R2 mutants.
(I and J) Effects of optogenetic stimulation of Epi neurons (Epi-Gal4-2 and UAS-ReaChR) on the jump percentages (H) and average jump latencies (I) to 45 °C—46 °C. The Epi neurons were activated by red lights for 30 sec. The flies tested either had AstC knocked down (UAS-AstCRNAi) or AstC overexpressed (UAS-AstC) in Epi neurons. n ≥ 20.
Error bars indicate S.E.M.s. Fisher’s exact test (A, D, F, H). Mann-Whitney test (B, E, G, I). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
See also Figure S4.
To confirm a role for AstC as a nociception modulator, we created a mutant that deleted most of the region coding for the 122 amino acid protein that is the precursor for the 15 residue AstC peptide. However, the line was lethal. Therefore, we generated an allele (AstC1), which changed the C-terminal two residues from cysteine and phenylalanine to leucine and lysine (Figure S4D). The mutation reduced expression of the AstC peptide below the level of detection and was therefore a strong allele (Figure S4C). However, AstC1 was not a null since it did not cause lethality. Consistent with the RNAi knockdown phenotype, AstC1 flies showed an increased propensity to jump upon contacting a 35 °C—36 °C surface and a decreased jump latency (Figures 3C and 3D). Because knockdown of AstC in Epi neurons increased nociception, we tested the idea that increased expression of AstC would render flies less sensitive to nociception. Therefore, we expressed UAS-AstC under control of the Epi-Gal4-2, and tested the response to 45 °C—46 °C. We found that the percentage of flies that jumped declined, while the jump latency increased (Figures 3E and 3F).
We then tested the idea that artificially activating Epi neurons would decrease their sensitivity to nociception. We expressed ReaChR in Epi neurons to acutely activate them with red lights (655 nm), and tested the response of the flies to 45 °C—46 °C. Again, the percentage of flies that jumped declined and the jump latency increased (Figures 3I and 3J). The reduction in thermal nociception (45 °C—46 °C) induced by optogenetic activation of Epi neurons was suppressed by RNAi knockdown of AstC (Figures 3I and 3J), while overexpression of AstC in combination with optogenetic stimulation of these neurons resulted in a greater alleviation of thermal nociception (Figures 3I and 3J).
Drosophila encodes two AstC receptors, AstC-R1 and AstC-R2.39,40 To address which of these two receptors might function in the suppression of thermal nociception, we examined the jump reactions of AstC-R1MI04794 and AstC-R2f01336 mutants in response to a 35 °C—36 °C surface. We found that mutation of AstC-R1 increased the percentage of flies that jumped and decreased the jump latency, while disruption of AstC-R2 had no effect (Figures 3G and 3H).
Epi neurons are sensors in the brain for noxious heat
To test whether Epi neurons respond to noxious heat, we expressed the genetically-encoded Ca2+ sensor, GCaMP6f41 in Epi neurons. We dissected out the brains, and monitored fluorescence changes (ΔF/F0) in the cell bodies as we applied an 18 °C—44 °C temperature ramp, and then decreased the temperature back to 18 °C (Figure 4A). We found the signals in Epi neurons were not increased by the lower temperatures. Between ~18 °C—40 °C, the ΔF/F0 dipped slightly (Figure 4D). Once the temperature reached ~39 °C—40 °C, the Ca2+ signals increased (Figures 4D and 4G; quantification was limited to the cells bodies, indicated by the dashed circles). Then during the 44 °C to 18 °C ramp, the fluorescence declined (Figures 4D and 4G). When we limited the temperature rise to 32 °C (Figure 4B), there was no increase in Ca2+ (Figures 4E and 4H).
Figure 4. Epi neurons are direct sensors for nociception.
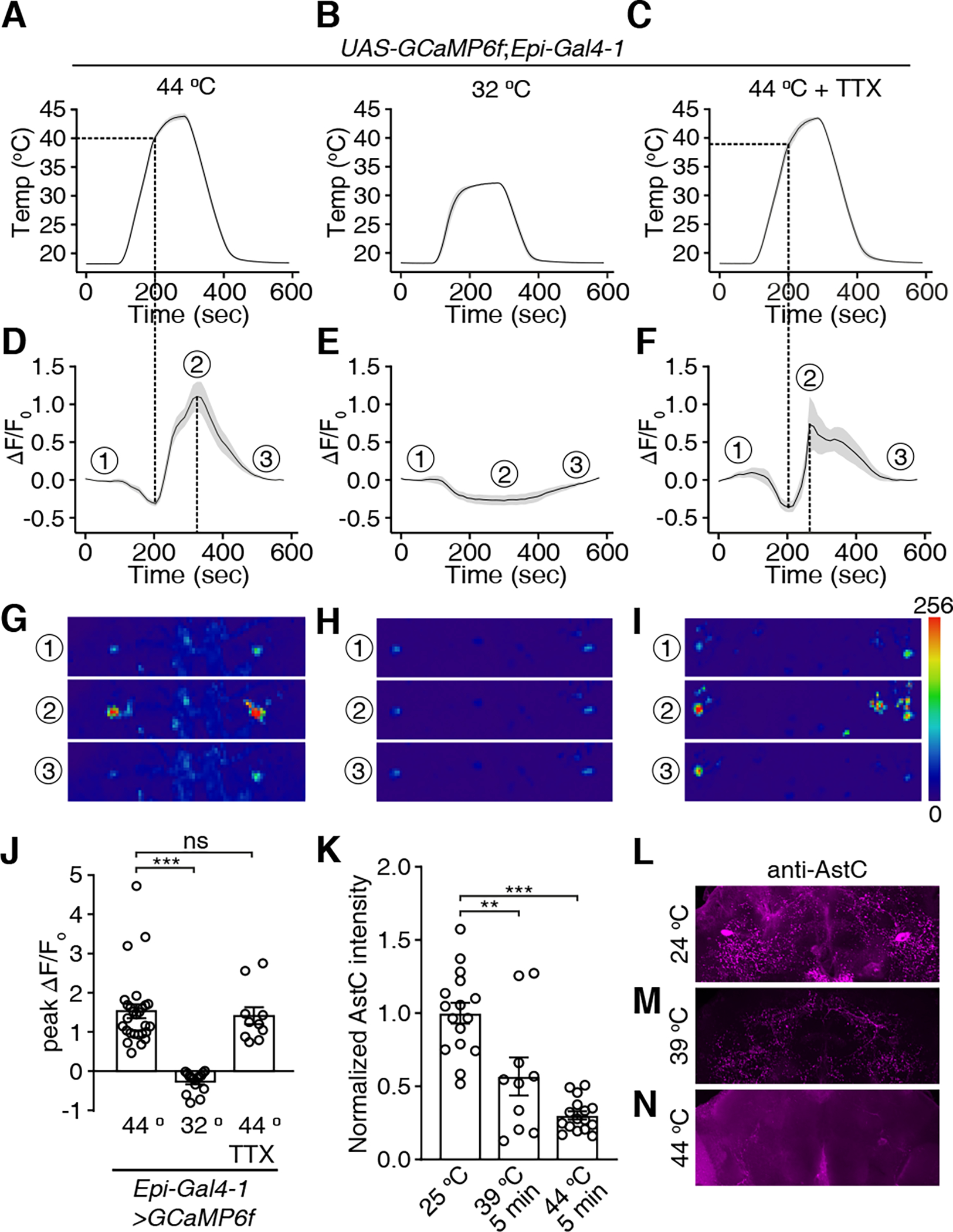
Ca2+ signals of Epi neurons during temperature changes. UAS-GCaMP6f was expressed under control of the Epi-Gal4-1. Brains were dissected and GCaMP6f signals were monitored in response to temperature ramps in the absence or presence of tetrodotoxin (TTX).
(A-C) Temperature ramps with the maximum temperatures indicated. TTX was added to the bath in (C).
(D) Changes in GCaMP6f signals (ΔF/F0) in response to the temperature ramp in (A). The ①, ②, and ③ indicate the time points for the sample images in (G-I).
(E) Changes in GCaMP6f signals (ΔF/F0) in response to the temperature ramp in (B).
(F) Changes in GCaMP6f signals (ΔF/F0) in response to the temperature ramp in (C) in the presence of TTX.
(G-I) Images of the GCaMP6f signals in Epi neurons (the dashed circles indicate the cell bodies) at three time points during the Ca2+ imaging as indicated in (D-F), respectively. Scale bars indicate 50 μm.
(J) Average peak ΔF/F0 exhibited by Epi neurons in (A-C). n = 10–27 neurons from ≥6 dissected brains. Error bars indicate S.E.M.s. Mann-Whitney test. ***P < 0.001, ns, not significant.
(K) Normalized anti-AstC signals in Epi neurons. The brains were incubated at room temperature (~ 24 °C), at 39 °C, or at 44 °C for 5 minutes. n = 10–16 neurons from ≥5 dissected brains. Error bars indicate S.E.M.s. Mann-Whitney test. **P < 0.01. (L-N) Sample images of anti-AstC signals at 25 °C (L), 39 °C (M) and 44 °C (N).
See also Figure S5.
To address whether the Epi neurons directly sense noxious heat, we applied the voltage-gated Na+ channel blocker tetrodotoxin (TTX) to the brain to inhibit action potential firing. Epi neurons responded to noxious heat even in the presence of TTX (Figures 4C, 4F, 4I and 4J; dashed circles indicate cell bodies), supporting the conclusion that Epi neurons are direct internal sensors for noxious heat. In some samples, we were able to detect GCaMP6f signals in arborizations from the Epi neurons (Figure 4I, panel 2). However, for consistency these Ca2+ signals were not included in the quantification. The absence of effect of TTX on the responses was not due to technical difficulties in applying the TTX to the brain since TTX inhibited GCaMP signals induced by ATP in neurons expression the ATP-gated P2X2 cation channel (Figures S5A—S5E).
We also compared AstC signals within Epi neurons before and after a 5-minute heat shock. We examined the anti-AstC signals from flies at 25 °C and after heating to 39 °C (near the threshold for detecting changes in GCaMP6f fluorescence) or to 44 °C. We found that either the 39 °C or 44 °C treatment significantly decreased the anti-AstC signals in Epi neurons (Figures 4K—4N), suggesting that Epi neurons that are exposed to heat release AstC.
Painless required in Epi neurons to sense noxious heat
The TRP channel, Painless (Pain), is a prime candidate temperature sensor in Epi neurons since it has an activation threshold in vivo in the mid 30 °C range,21 and is widely expressed in the brain.42 To test whether pain is expressed in Epi neurons, we used a Gal4 reporter to drive membrane GFP (UAS-mCD8::GFP) and performed double-labeling using anti-AstC, and anti-GPF. The pain reporter was widely expressed in the brain, including AstC-positive Epi neurons (Figures 5A—5C).
Figure 5. painless functions in Epi neurons,.
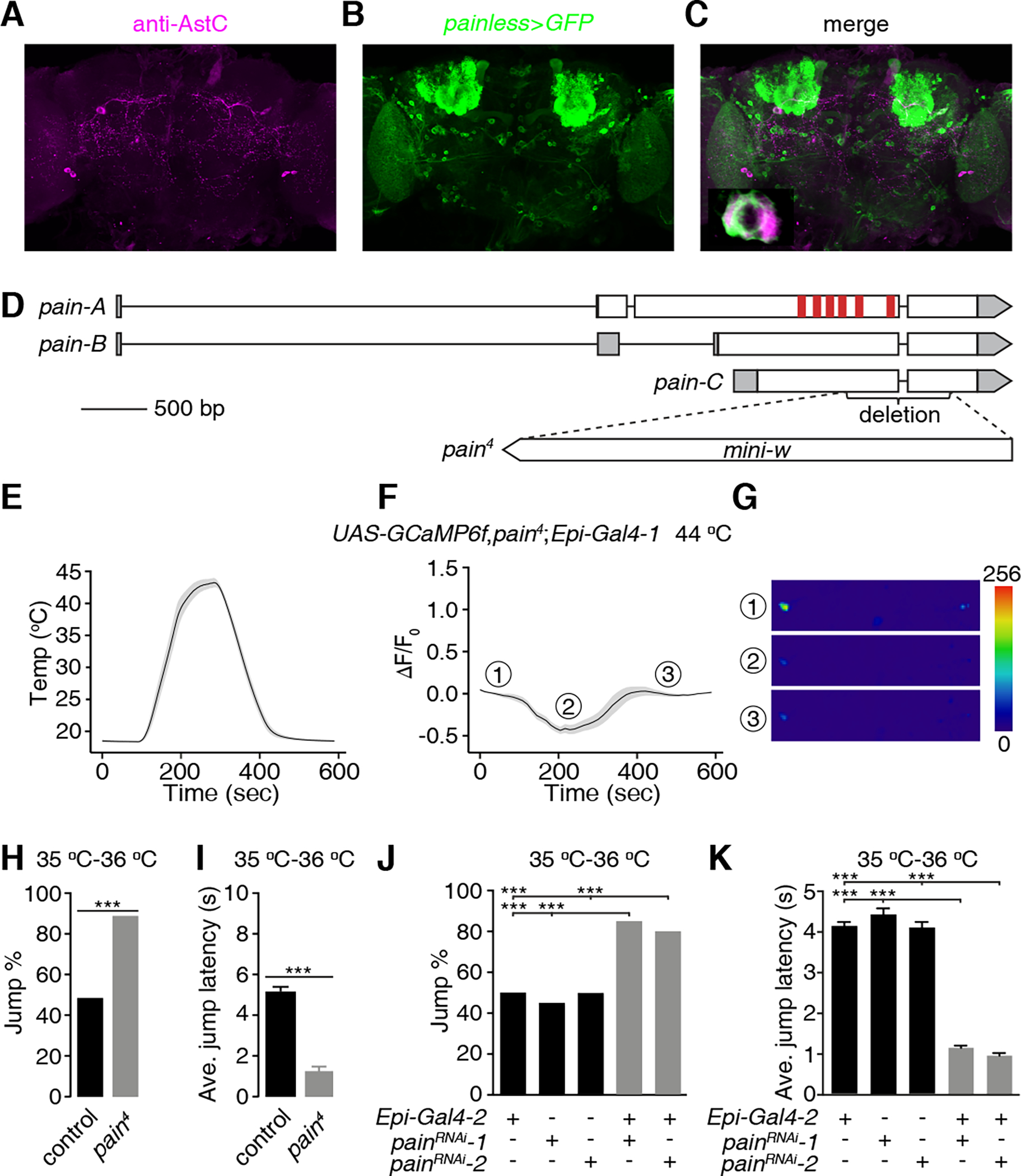
(A-C) Testing for co-localization of anti-AstC staining with the pain reporter. A brain from a fly expressing UAS-mCD8::GFP under control of the pain-Gal4 (painless>GFP) was stained with anti-AstC and anti-GFP (green, B). (A) Rabbit anti-AstC (magenta). Scale bar indicates 50 μm. (B) Chicken anti-GFP. (C) Merge of (A and B). Bottom-left panel is an enlarged image of the framed area. Scale bar indicates 5 μm.
(D) Schematic of the gene structure of the wild-type pain gene and the pain4 mutant. The red vertical bars represent the six transmembrane domains.
(E-G) GCaMP6f responses of Epi neurons from pain4 brains during a temperature ramp (maximum 44 °C). (E) Temperature ramp. (F) Changes in GcaMP6f signals (ΔF/F0) in response to the temperature ramp in (E). The ①, ②, and ③ indicate the time points for the sample images in (G). (G) Sample images of GCaMP6f signals displayed by Epi neurons (indicated by the dashed circles) at three time points during the Ca2+ imaging in (F). Scale bar indicates 50 μm. n = 23 neurons from 15 dissected brains.
(H and I) Jump percentages (H) and average jump latencies (I) of control and pain4 flies on a 35 °C—36 °C hot-plate. n ≥ 20. Error bars indicate S.E.M.s. Fisher’s exact test (H). Mann-Whitney test (I). ***P < 0.001.
(J and K) Effect of RNAi knockdown of pain in Epi neurons on jump percentages (J) and average jump latencies (K) using flies on a 35 °C—36 °C hot-plate. n ≥ 20. Error bars indicate S.E.M.s. Fisher’s exact test (J). Mann-Whitney test (K). ***P < 0.001. See also Figure S5.
To test whether the heat responsiveness of Epi neurons depends on the pain gene, we generated a pain null allele by inserting mini-white in the coding region, thereby creating a deletion that removed +1877—2660 base pairs, including part of transmembrane domain 4, and all of the transmembrane domains 5 and 6 (pain4; Figure 5D). The pain4 mutation abolished the response of the Epi neurons to temperatures ≥40 °C (Figures 5E, 5F and 5G). The small, initial dip in ΔF/F0 during the early phase of the temperature ramp still occurred (Figure 5F), demonstrating that this phase is pain independent.
To determine whether mutation of pain increases thermal nociception we employed the jump assay. We tested 35 °C—36 °C since 45 °C—46 °C causes 100% of control flies to jump (Figure 1B). In response to 35 °C—36 °C, 49.6% of control flies jump, and they do so with a latency of 5.1 ±0.2 seconds (Figures 5H and 5I). We found that null pain4 mutants exhibited a large increase in the percentage of flies that jumped (Figure 5H; 88.4%) and a 4.1-fold decrease in the jump latency (Figure 5I control, 5.1 ±0.2 sec; pain4, 1.2 ±0.3 sec). Another Drosophila TRP channel, Pyrexia (Pyx), is heat activated with a threshold of ~40 °C28, which is in a similar range as Pain.21,43 Opposite to the pain mutant phenotype, we found that pyxex44 mutants exhibited lower jump percentages and increased jump latencies at both 35 °C—36 °C and 45 °C—46 °C (Figures S5F—S5I). These results indicate that Pyx contributes to the thermal nociceptive response, rather than suppressing the response.
To address whether the pain mutant phenotype reflects a requirement for pain in Epi neurons, we used two effective RNAi lines,45 to knockdown pain under control of the Epi-Gal4-2. We found that knockdown of pain specifically in Epi neurons increased the jump percentage (Figure 5J) and reduced the average jump latency (Figure 5K). Thus, Pain is required in Epi neurons for suppressing the nociceptive response to heat.
To address whether expression of Pain affects expression of AstC in Epi neurons, we stained both control and pain4 mutant brains with anti-AstC. We found that pain is also required for expression of AstC in Epi neurons (Figures S4E and S4F). However, AstC expression was still detected in other neurons in the pain4 brain, such as the pair of small neurons proximal to the optic lobes (Figures S4E and S4F; arrowheads).
DISCUSSION
We found that a single pair of bilaterally-symmetrical Epi neurons in the fly brain is critical for suppressing thermal nociception. The importance of Epi neurons is underscored by the observation that artificial activation of these neurons is sufficient to suppress the aversive jump response to hot temperatures, and that inhibition of signaling from these neurons increases the jump responses to moderate heat. The profound effect of a single pair of neurons in reducing thermal nociception is surprising given that multiple brain regions appear to function in pain suppression in mammals.46
The dendrites of Epi neurons arborize to multiple regions of the brain, such as the optic lobes, the lateral horn and to a region near the mushroom bodies, indicating that Epi neurons receive multiple signal inputs. The lateral horn is a higher order processing center that receives input from the antennal (olfactory) lobes, and then sends relays to other brain regions such as the mushroom bodies.47 Therefore, it is intriguing to speculate that the Epi neurons may be activated by noxious odorants, and aversive visual cues, which attenuate the avoidance behavioral responses to these stimuli. Epi neurons might also receive input from attractive olfactory and visual cues, which in turn diminish the escape responses to noxious stimuli such as high temperatures. In addition, the axons of Epi neurons project to the ventral nerve cord, consistent with a role in descending control of motor output.
A key question is the mechanism through which Epi neurons respond to hot temperatures and alleviate thermal nociception. We found that Epi neurons are directly activated by hot temperatures, and do so through activation of the thermo-TRP channel, Pain,21 which is expressed in Epi neurons. The Pain channel is critical for suppressing nociception since mutation of the pain gene causes an increase in thermal pain sensitivity (hyperalgesia). While Epi neurons respond directly to heat and are anti-nociceptors, other neurons in the fly brain, the so-called anterior cell neurons, respond directly to suboptimal warm temperatures.48 In contrast to the anti-nociceptive Epi neurons, the AC neurons function in thermal avoidance, which is mediated through thermal activation of TRPA1.48
The next question is the mechanism through which activation of Epi neurons suppresses thermal pain. Epi neurons express a neuropeptide, AstC, which binds to receptors that have sequence homology (39.0% identity for AstC-R1, 38.5% identity for AstC-R2) to human opioid receptors,49 which function in the suppression of nociception in mammals.50,51 Moreover, mutation of AstC or knockdown of AstC in Epi neurons causes thermal hyperalgesia, and mutation of AstC-R1 elicits a similar phenotype. Heat stimulation diminishes the level of AstC in Epi neurons, indicating that activation of these neurons promotes release of AstC. We conclude that Epi neurons alleviate thermal nociception through a mechanism that depends on heat sensing by the Pain channel, leading to release of AstC.
Surprisingly, mutation of pain also reduced expression of AstC in Epi neurons below the level of detection. This effect was not due to elimination of Epi neurons since pain mutant brains express UAS-GCaMP6f under control of the Epi-Gal4. Expression of neuropeptides have been linked to neuronal activity.52 Moreover, there is an example in which a thermosensory TRPV channel, affects expression of a neuropeptide receptor.53,54 Pain is activated by thermal heat, with the most pronounced activation in the noxious heat range.21,43 However, even at temperatures significantly below the flex point in which a given temperature rapidly opens the gate of a thermosensory TRP, such as Pain, there is some channel activity. We suggest that low levels of Pain and Epi neuron activities are necessary for expression of AstC, while high levels of activities that are induced by noxious heat are required for release of the AstC.
A feature of activation of Epi neurons is that the pain suppression due to an acute 30-second activation of Epi neurons is sustained for several minutes. We suggest that the slow termination of the pain suppression following stimulation of these neurons is mediated by release of the neuromodulator AstC, which persists for several minutes. Epi neurons appear to be non-adapting, as chronic activation of these neurons with the NaChBac channel leads to similar levels of pain suppression as acute stimulation with channelrhodopsin. This non-adapting feature of Epi neurons may be beneficial because it allows for pain suppression under conditions in which the aversive response to heat needs to be suppressed sufficiently long enough to allow activities that promote survival. Given that fruit flies are poikilothermic, and their body temperature equilibrates with the environment, direct activation of Epi neurons would allow the flies to suppress nociception and enter excessively warm environments to feed or avoid predators. In conclusion, this study unveils a molecular and cellular basis for pain suppression in Drosophila. The observation that Pain is essential for suppressing nociception is surprising given that all other thermal-TRP channels function in avoidance of suboptimal or noxious temperatures. Mutation of pain in fly larvae eliminates the sensitivity to hot temperatures (hypoalgesia).21 Thus, it is remarkable that the same TRP channel has opposite functions in nociception and anti-nociception in larvae and adults.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
The lead contact is Craig Montell (cmontell@ucsb.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction. The fly stocks generated in this study will be deposited with the Bloomington Stock Center for public distribution (http://flystocks.bio.indiana.edu/). Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact.
Data and Code Availability
This study did not generate any standardized datatypes.
This study did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly stocks and husbandry
All experiments were performed with the indicated strains of adult male and female Drosophila melanogaster. Flies were raised at 25 °C and 60% relative humidity on standard cornmeal fly food under a 12 hr light:12 hr dark cycles. The control flies were w1118, unless indicated otherwise. The following fly lines were from the Bloomington Stock Center (Indiana University): w1118 (BL5905), AstA (Allatostatin A)-Gal4 (BL51978, BL51979), Epi-Gal4-1 (AstC-Gal4-1; BL39283), Epi-Gal4-2 (AstC-Gal4-2; BL52017), Akh (Adipokinetic hormone)-Gal4 (BL25683, BL25684), Burs (Bursicon)-Gal4 (BL51980), Capa (Capability)-Gal4 (BL51970), Crz (Corazonin)-Gal4 (BL51977), CCAP (Crustacean cardioactive peptide)-Gal4 (BL25685), Dh31 (Diuretic hormone 31)-Gal4 (BL51988, BL51989), Dh44 (Diuretic hormone 44)-Gal4 (BL51987), Dsk (Drosulfakinin)-Gal4 (BL51981), ETH (Ecdysis triggering hormone)-Gal4 (BL51982), Eh (Eclosion hormone)-Gal4 (BL6301), FMRFa (FMRFamide)-Gal4 (BL51990), Hug (Hugin)-Gal4 (BL58769), Ilp2 (Insulin-like peptide 2)-Gal4 (BL37516), Lk (Leucokinin)-Gal4 (BL51992, BL51993), Mip (Myoinhibiting peptide precursor)-Gal4 (BL51984), Ms (Myosuppressin)-Gal4 (BL51985, BL51986), NPF (neuropeptide F)-Gal4 (BL25681, BL25682), Pdf (Pigment-dispersing factor)-Gal4 (BL6899), Proc (Proctolin)-Gal4 (BL51971, BL51972), sNPF (short neuropeptide F precursor)-Gal4 (BL51991), Tk (Tachiykinin)-Gal4 (BL51975), TH (tyrosine hydroxylase)-Gal4 (BL8848), Trh (trachealess)-Gal4 (BL38388, BL38389), Tdc2 (Tyrosine decarboxylase 2)-Gal4 (BL9313), UAS-NaChBac (BL9469), UAS-mCD8::GFP (BL5137), UAS-DenMark,UAS-syt.eGFP (BL33064), UAS-ReaChR (BL53741), UAS-AstCRNAi (BL25868), UAS-GCaMP6f (BL42747), UAS-Stinger 2 (BL84277), pain (painless)-Gal4 (BL27894), 70FLP,70I-SceI (BL6935), UAS-TNTG (BL28838), AstC-R1MI04794 (BL59307), AstC-R2f01336 (BL18622), AstA-R1-Gal4 (BL49032), and AstA-LexA (BL53625). The following fly lines were from the Vienna Drosophila Resource Center: UAS-painRNAi-1 (v39477), UAS-painRNAi-2 (v39478). We described pyxex previously.44 UAS-GCaMP3;LexAop-P2X2 was from Dr. Orie Shafer (University of Michigan).55 The following flies were outcrossed to w1118 (BL5905) for 6 generations: Epi-Gal4-1 (AstC-Gal4-1; BL39283), Epi-Gal4-2 (AstC-Gal4-2; BL52017), UAS-ReaChR (BL53741), UAS-AstCRNAi (BL25868), UAS-NaChBac (BL9469), UAS-painRNAi-1 (v39477), and UAS-painRNAi-2 (v39478).
METHOD DETAILS
Generation of UAS-AstC
To generate UAS-AstC, we first cloned the AstC cDNA. We extracted mRNA from w1118 heads, performed reverse transcription PCR (RT-PCR), and inserted the cDNA into the pUAST vector between the EcoRI and XhoI sites. We added the Drosophila Kozak consensus sequence (CAAA)56 immediately 5’ to the start codon. The primers for cloning the AstC cDNA were: forward: AGGGAATTGGGAATTCAAAATGATGAAATTCGTGCAGATATTATTGTGC; reverse: TAGAGGTACCCTCGAGTTACTTCCTAAAGCAGGAGATGGGATT. We injected the pUAST-AstC construct into embryos from phiC31 transgenic flies that contained the attP2 docking site (BestGene).
Generation of the AstC1 mutant using CRISPR-Cas9
The primary translation product of AstC is 122 residues, and the 15 amino acid AstC peptide is encoded by residues 104–115. We used CRISPR/Cas957–60 to generate the AstC1 mutant by changing the last two residues of AstC (amino acids 118—119 of the primary translation product) from cysteine and phenylalanine to leucine and lysine. We designed the CRISPR targets with the CRISPR Optimal Target Finder (http://targetfinder.flycrispr.neuro.brown.edu/). We inserted the 20 base pair guide sequence (GAACTTACTTCCTAAAGC) targeting the region coding for the AstC peptide (Figure S4C) into pU6-BbsI-chiRNA.58 We then injected this plasmid into embryos of Cas9 transgenic flies (BestGene). To screen for the AstC1 allele, we performed PCR to amplify the flanking regions and genotyped by DNA sequencing. The primers for DNA sequencing of AstC1 and AstC2 were: forward: TTTTTGCGTGAACTCGCCTC; reverse: CCATTATGCTAATCTTTGTGTTGTGG.
Generation of pain4 mutant by ends-out homologous recombination
We generated painless knock-out flies (pain4) by ends-out homologous recombination.61 The targeting construct deleted a 782-bp region encompassing part of exons 3 and 4 (encoding amino acid residues 606 to 844), which removes part of transmembrane domain 4, and the entirety of transmembrane domains 5 and 6 (Fig. 5D). To generate the construct for the ends-out recombination, we inserted two 3 kb genomic fragments into the NotI site and the BamHI sites of pw35,61 respectively. Transgenic flies carrying the targeting construct on the third chromosome were crossed to 70FLP,70I-SceI flies, and the progeny were screened for gene targeting by DNA sequencing. The primers used for DNA sequencing were: forward: AGTGAGCGACACCCAAGT; reverse: TAAGTAGTTCGGGTAGATGTT. These primers amplified pain from wild-type and pain4/+ flies, but not from the pain4 homozygous mutant
Construction of the hot plate apparatus for the thermal jump assays
The apparatus for the assay (Figure S1) was fabricated at the UCSB Physics Machine Shop (https://www.physics.ucsb.edu/resources/machineshops/shop) and was a copper plate (19 mm thick, 190 mm wide and long) with 8 internal 10 mm water tunnels. The hot plate contained a 3 mm deep moat near the perimeter, which was filled with water so that the flies with clipped wings would not be able to escape from the hot plate surface. The temperature of the plate was controlled by water circulating through the tunnels in the plate from a water bath (PolyScience 9106, Refrigerated/Heated 6L Circulating Bath). We used silicone tubing (1/4” ID x 3/8” OD x 1/16” wall) to connect the outlets of the water bath to the two connectors on the plate. After the water bath was turned on, we allowed the temperature of the plate to equilibrate for 30 min. We used a thermometer (Fluke 51II) with a thermocouple (Fluke K type) to measure the temperature of the surface of the plate.
Hot plate jump assays
To perform the jump assays on a hot plate, we amputated the wings from 2—3 day-old flies using Vannas Spring scissors (No. 15000–04, Fine Science Tools), and allowed them to recover for 24 hrs on standard corn meal/molasses fly food in vials (AS-516, Fisherbrand). We tapped down individual flies from the vials to transfer them to the hot plate and determined whether the flies jumped. We recorded the behavior of the flies using a webcam (Logitech C615 HD Webcam) and manually analyzed the percentage of flies that jumped as well as the jump latencies (sec), by examining the individual frames on a VLC media player for 10 sec. The jump responses were straightforward to score since the assessments of flies that left the surface of the hot plate were unambiguous. We displayed the results in 1 sec bins. All flies that did not jump within the 10 sec window were placed in the >10 sec bin. The average jump latencies were calculated by averaging the individual jump latencies. For flies that jumped within 10 sec, we used the exact number. For flies that did not jump within the first 10 sec, we used a value of 10 sec to determine the jump latencies. Therefore, the average jump latencies are an underestimate, especially for experiments in which the majority of flies jumped in >10 sec.
Immunostaining
We performed the dissections and immunostaining as we described previously.62 Briefly, we dissected out the brains and fixed them in 4% paraformaldehyde in PBST [0.3% Triton X-100 (Sigma) in 1X PBS (diluted from 10X PBS; AAJ62036K2, Fisher Scientific)] at room temperature for 30 min. We washed the fixed brains 3x briefly with PBST and blocked them with 5% normal goat serum (Fisher Scientific, ICN19135680) in PBST at room temperature for 1 hr. We then incubated the brains with primary antibodies overnight at 4 °C. After three washes (15 min each) in PBST, we incubated the brains with secondary antibodies overnight at 4 °C. We imaged the samples using a Zeiss LSM700 Confocal Laser Scanning Microscope using a 20x/0.8 Plan-Apochromat DIC objective and Zen software. We used the following primary antibodies: chicken anti-GFP (1:500; A10262, Invitrogen), rabbit anti-DsRed (1:500; 632496, Clontech), mouse anti-BRP (nc82, Developmental Studies Hybridoma Bank) and rabbit anti-AstC (a generous gift from Dr. Jan Veenstra).63 We used the following secondary antibodies: Alexa Fluor 488 goat-anti-chicken IgG (1:1000; A11039, Thermo Fisher Scientific), Alexa Fluor 555 donkey-anti-rabbit IgG (1:1000; A31572, Thermo Fisher Scientific) and Alexa Fluor 633 goat-anti-mouse IgG (1:1000; A21050, Thermo Fisher Scientific).
Optogenetic stimulation of Epi neurons for thermal jump assays
To stimulate Epi neurons with light, we expressed UAS-ReaChR under control of the Epi-Gal4-2. For some experiments, we knocked down or overexpressed AstC in Epi neurons by introducing either UAS-AstCRNAi or UAS-AstC, respectively. Prior to initiating the experiments, we amputated the wings from 2—3 day-old flies using Vannas Spring scissors (No. 15000–04, Fine Science Tools). We allowed them to recover for 24 hrs in vials (AS-516, Fisherbrand) with ~10 mL of standard corn meal/molasses fly food in which we added 50 μL of 100 mM all-trans-retinal (R2500-1G, Sigma) to the food surface.
To carry out the optogenetic experiments, we collected the flies in transparent vials (AS-516, Fisherbrand) and stimulated the animals for 30 sec with red lights using an AmScope CF-4 Color Filter with an AmScope HL250YA 150W Fiber Optic Dual-Gooseneck Stereo Microscope Light Illuminator. The light intensity was 5.2 mW/cm2. We then transferred the flies to a hot plate quickly (< 5 sec) or after a 0.5—5 min delay, and assayed their jump responses to temperatures ranging from 45 °C to 50 °C. We recorded the behavior of the flies using a webcam (Logitech C615 HD Webcam) and manually analyzed the jump latencies by examining individual frames using a VLC media player.
Ex vivo GCaMP imaging with temperature ramps
To perform the Ca2+ imaging assays with temperature ramps, we used 3—5 day-old flies that expressed UAS-GCaMP6f under control of the Epi-Gal4-1, which drove higher UAS-GCaMP6f expression than Epi-Gal4-2. To enhance the expression level of GCaMP6f, we used a fly stock with two copies of Epi-Gal4-1 and UAS-GCaMP6f. We controlled the temperature as previously described.64 Briefly, we used a QE-1HC quick exchange heating/cooling platform with a CL-100 bipolar temperature controller (Warner Instruments). We dissected the brains from adult flies in Drosophila imaging saline (108 mM NaCl, 5 mM KCl, 8.2 mM MgCl2, 2 mM CaCl2, NaHCO3 4 mM, NaH2PO4 1 mM, trehalose 5 mM, sucrose 10 mM, HEPES 5 Mm, pH 7.5), and quickly transferred the brains to the center of a copper chamber (1–5/8” in diameter, 1/16” thick and a 1/4” glass window on the bottom) filled with the saline solution, which fits the QE-1HC. For experiments with TTX, which suppresses voltage-gated Na+ channels and synaptic transmission, we pre-dissolved 1 μM TTX in Drosophila imaging saline before transferring the brains to a copper chamber. We placed a temperature probe (IT-18, Physitemp) next to the brain to detect the ambient temperatures that the brain was exposed to. We recorded from each brain only once, and collected images of the GCaMP6f fluorescence using an upright Zeiss LSM700 Confocal Laser Scanning Microscope, and a 488-nm laser at a resolution of 256 × 256 pixels using a 20x/1.0 Plan-Apochromat water immersion objective. We recorded from each brain for 10 min in total and at a rate of 400 ms/frame. ~15 Z axial sections were imaged in one time-series cycle. The section interval was ~6 μm. The time intervals between each cycle were ~8 sec. The images were analyzed using ImageJ/Fiji. The increases in Ca2+ levels in each neuron were indicated by ΔF/F0. F0 was the average baseline fluorescence of GCaMP6f during the first 90 seconds of the experiment before the temperature was increased, and ΔF equals F-F0. We rectified the photobleaching effect according to the following equation: . To obtain the photobleaching factor k, we measured the fluorescent signals before and after the temperature stimuli, and assumed that they were the same without photobleaching. Then, we used the factor k to compensate for photo-bleaching: .
To perform the Ca2+ imaging assays with AstA-R1 neurons, we dissected 3—5 day-old flies and transferred individual brains to 35 mm plastic Petri dishes (35 3001, Falcon) filled with 2 mL Drosophila imaging saline, and immobilized the brain with a metal harp (SHD-26GH/10, Warner Instruments). We imaged the basal GCaMP3 signals for 15 cycles, and then added 200 μl 50 mM ATP (pH adjusted to 7.0, A2383-5G, Sigma) to the Petri dishes, resulting in a final ATP concentration of 5 mM. For experiments with TTX, we pre-dissolved 1 μM TTX in Drosophila imaging saline before transferring the brains to the 35 mm plastic Petri dishes. We collected images of the GCaMP3 fluorescence using an upright Zeiss LSM700 Confocal Laser Scanning Microscopy and a 488-nm laser at a resolution of 256 × 256 pixels using a 20x/1.0 Plan-Apochromat water immersion objective. ~10 Z axial sections were imaged in one time-series cycle. The section intervals were ~1 μm. The time intervals between each cycle were ~2 sec. The images were analyzed using ImageJ/Fiji. The increases in Ca2+ levels in each neuron were indicated by ΔF/F0. F0 was the average baseline fluorescence of GCaMP for 5 cycles immediately before ATP application, and ΔF equals F-F0.
Quantification and statistical analysis
Descriptions, results, and sample sizes of each test are provided in the figure legends. All replicates were biological replicates using different flies. Data for all quantitative experiments were collected on at least three different days. For the hot plate jump assays, each “n” represents an individual fly. Based on our experience and common practices in this field, we used a sample size of n ≥ 20 for each genotype or treatment for the hot plate jump assays. Each “n” for the Ca2+ imaging experiments represents a single neuron from ≥6 independent flies. Each “n” for quantification of the AstC staining intensities represents a single neuron from ≥5 independent flies. GraphPad Prism 9 software or MS Excel were used for statistical tests. We used Fisher’s exact test (in MS Excel) and Mann-Whitney test for non-parametric tests (in GraphPad Prism 9). Sample sizes were determined based on previous publications and are cited in the figure legends. In all graphs, error bars represent the standard error of the mean (S.E.M.). We set the significance level, α = 0.05. Asterisks indicate statistical significance: *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP (chicken) | Invitrogen | Cat # A-10262 RRID: AB_2534023 |
| anti-DsRed (rabbit) | Clontech Laboratories, Inc. | Cat # 632496 RRID: AB_10013483 |
| anti-BRP (mouse) | Developmental Studies Hybridoma Bank | Cat # nc82 RRID: AB_2314866 |
| Goat anti-chicken, Alexa Fluor 488 | Thermo Fisher Scientific | Cat # A-11039 RRID: AB_2534096 |
| Donkey anti-rabbit, Alexa Fluor 555 | Thermo Fisher Scientific | Cat # A-11036 RRID: AB_162543 |
| Goat anti-mouse, Alexa Fluor 633 | Thermo Fisher Scientific | Cat # A-21050 RRID: AB_2535718 |
| anti-AstC (rabbit) | From Dr. Jan Veenstra | NA |
| Chemicals | ||
| Paraformaldehyde | Electron Microscopy Sciences | Cat # 15710 |
| PBS | Fisher Scientific | Cat # AAJ62036K2 |
| Triton X-100 | Sigma | Cat # X100 |
| goat serum | Fisher Scientific | Cat # ICN19135680 |
| VECTASHIELD anti-fade mounting media | Vector Labs | Cat # H-1200 |
| all-trans-retinal | Sigma | Cat # R2500-1G |
| NaCl | Fisher Scientific | Cat # S271-500 |
| KCl | Sigma | Cat # P9541 |
| MgCl2 | Sigma | Cat # M2670-500G |
| CaCl2 | Sigma | Cat # C3881-500G |
| NaHCO3 | Sigma | Cat # S6014-500G |
| trehalose | Sigma | Cat # T0167-100G |
| HEPES | Fisher Scientific | Cat # 15630080 |
| Experimental Models: Organisms/Strains | ||
| Drosophila: w1118 | Bloomington Drosophila Stock Center | Cat # BL5905 |
| Drosophila: AstC1 | In this paper | NA |
| Drosophila: pain4 | In this paper | NA |
| Drosophila: UAS-AstC | In this paper | NA |
| Drosophila: Epi-Gal4-1 | Bloomington Drosophila Stock Center | Cat # BL39283 |
| Drosophila: Epi-Gal4-2 | Bloomington Drosophila Stock Center | Cat # BL52017 |
| Drosophila: AstC-R1MI04794 | Bloomington Drosophila Stock Center | Cat # BL59307 |
| Drosophila: AstC-R2f01336 | Bloomington Drosophila Stock Center | Cat # BL18622 |
| Drosophila: AstA-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51978 |
| Drosophila: AstA-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51979 |
| Drosophila: Akh-Gal4 | Bloomington Drosophila Stock Center | Cat # BL25683 |
| Drosophila: Akh-Gal4 | Bloomington Drosophila Stock Center | Cat # BL25684 |
| Drosophila: Burs-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51980 |
| Drosophila: Capa-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51970 |
| Drosophila: Crz-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51977 |
| Drosophila: CCAP-Gal4 | Bloomington Drosophila Stock Center | Cat # BL25685 |
| Drosophila: Dh31-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51988 |
| Drosophila: Dh31-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51989 |
| Drosophila: Dh44-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51987 |
| Drosophila: Dsk-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51981 |
| Drosophila: ETH-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51982 |
| Drosophila: EH-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51990 |
| Drosophila: FMRFa-Gal4 | Bloomington Drosophila Stock Center | Cat # BL58769 |
| Drosophila: Hug-Gal4 | ^Bloomington Drosophila Stock Center | Cat # BL58769 |
| Drosophila: Ilp2-Gal4 | Bloomington Drosophila Stock Center | Cat # BL37516 |
| Drosophila: Lk-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51992 |
| Drosophila: Lk-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51993 |
| Drosophila: Mip-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51984 |
| Drosophila: Ms-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51985 |
| Drosophila: Ms-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51986 |
| Drosophila: NPF-Gal4 | Bloomington Drosophila Stock Center | Cat # BL25681 |
| Drosophila: NPF-Gal4 | Bloomington Drosophila Stock Center | Cat # BL25682 |
| Drosophila: Pdf-Gal4 | Bloomington Drosophila Stock Center | Cat # BL6899 |
| Drosophila: Proc-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51971 |
| Drosophila: Proc-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51972 |
| Drosophila: sNPF-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51991 |
| Drosophila: Tk-Gal4 | Bloomington Drosophila Stock Center | Cat # BL51975 |
| Drosophila: TH-Gal4 | Bloomington Drosophila Stock Center | Cat # BL8848 |
| Drosophila: Trh-Gal4 | Bloomington Drosophila Stock Center | Cat # BL38388 |
| Drosophila: Trh-Gal4 | Bloomington Drosophila Stock Center | Cat # BL38389 |
| Drosophila: Tdc2-Gal4 | Bloomington Drosophila Stock Center | Cat # BL9313 |
| Drosophila: UAS-NaChBac | Bloomington Drosophila Stock Center | Cat # BL9469 |
| Drosophila: UAS-mCD8::GFP | Bloomington Drosophila Stock Center | Cat # BL5137 |
| Drosophila: UAS-DenMark, UAS-syt.eGFP | Bloomington Drosophila Stock Center | Cat # BL33064 |
| Drosophila: UAS-ReaChR | Bloomington Drosophila Stock Center | Cat # BL53741 |
| Drosophila: UAS-AstCRNAi | Bloomington Drosophila Stock Center | Cat # BL25868 |
| Drosophila: UAS-GCaMP6f | Bloomington Drosophila Stock Center | Cat # BL42747 |
| Drosophila: UAS-Stinger 2 | Bloomington Drosophila Stock Center | Cat # BL84277 |
| Drosophila: pain (painless)-Gal4 | Bloomington Drosophila Stock Center | Cat # BL27894 |
| Drosophila: 70FLP, 70I-SceI | Bloomington Drosophila Stock Center | Cat # BL6935 |
| Drosophila: UAS-TNTG | ^Bloomington Drosophila Stock Center | Cat # BL28838 |
| Drosophila: AstAR1-Gal4 | Bloomington Drosophila Stock Center | Cat # BL49032 |
| Drosophila: AstA-LexA | Bloomington Drosophila Stock Center | Cat # BL53625 |
| Drosophila: UAS-painRNAi-1 | Vienna Drosophila Resource Center | Cat # v39477 |
| Drosophila: UAS-painRNAi-2 | Vienna Drosophila Resource Center | Cat # v39478 |
| Drosophila: pyxex | From Dr. Craig Montell | NA |
| Drosophila: UAS-GCaMP3, LexAop-P2X2 | From Dr. Orie Shafer | NA |
| Oligonucleotides | ||
| For cloning AstC cDNA. Forward primer: AGGGAATTGGGAATTCAAAATGATGAAATTCGTGCAGATATTATTGTGC | This paper | NA |
| For cloning AstC cDNA.. Reverse primer: TAGAGGTACCCTCGAGTTACTTCCTAAAGCAGGAGATGGGATT | This paper | NA |
| For DNA sequencing of AstC1. Forward primer: TTTTTGCGTGAACTCGCCTC | This paper | NA |
| For DNA sequencing of AstC1. Reverse primer: CCATTATGCTAATCTTTGTGTTGTGG | This paper | NA |
| For DNA sequencing of pain4. Forward primer: AGTGAGCGACACCCAAGT | This paper | NA |
| For DNA sequencing of pain4. Reverse primer: TAAGTAGTTCGGGTAGATGTT | This paper | NA |
| Recombinant DNA | ||
| pUAST | cDNA vector66 | |
| pU6-BbsI-chiRNA | gRNA vector | Addgene Plasmid #45946 |
| pw35 | [59] Donor vector | NA |
| Software and Algorithms | ||
| Prism9 | Software |
https://www.graphpad.com/scientific-software/prism/ RRID:SCR_002798 |
| Fiji | Software |
https://imagej.net/Fiji RRID:SCR_002285 |
| Other | ||
| Hot plate in jump assay | UCSB Physics Machine Shop | https://www.physics.ucsb.edu/resources/machineshops/shop |
| Refrigerated/Heated 6L Circulating Bath | PolyScience | Cat # 9106 |
Highlights.
Epi neurons are descending neurons in the brain that suppress thermal nociception
A TRP channel (Painless) is required in Epi neurons to suppress thermal nociception
Activation of Epi neurons by noxious heat depends on Painless
Allatostatin C is a neuropeptide released by Epi neurons that reduces nociception
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute on Deafness and other Communication Disorders (DC007864) and National Eye Institute (EY008117) to C.M. We would like to thank Dr. Zhefeng Gong (Zhejiang University, China), who generously allowed J.L. to carry out experiments in his laboratory during 2020 when he was restricted from returning to the USA due to the pandemic. We thank Dr. Jan Veenstra (Université de Bordeaux, France) for sharing the AstC antibodies, and Drs. Junjie Luo, Hsiang-Chin Chen, and Yijin Wang for discussions and technical advice. We also thank the Physics Machine Shop (UC Santa Barbara) for fabricating the behavioral plates.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lutz J, Jager L, de Quervain D, Krauseneck T, Padberg F, Wichnalek M, Beyer A, Stahl R, Zirngibl B, Morhard D, et al. (2008). White and gray matter abnormalities in the brain of patients with fibromyalgia: a diffusion-tensor and volumetric imaging study. Arthritis Rheum. 58, 3960–3969. [DOI] [PubMed] [Google Scholar]
- 2.Geha PY, Baliki MN, Harden RN, Bauer WR, Parrish TB, and Apkarian AV (2008). The brain in chronic CRPS pain: abnormal gray-white matter interactions in emotional and autonomic regions. Neuron 60, 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt-Wilcke T, Leinisch E, Ganssbauer S, Draganski B, Bogdahn U, Altmeppen J, and May A (2006). Affective components and intensity of pain correlate with structural differences in gray matter in chronic back pain patients. Pain 125, 89–97. [DOI] [PubMed] [Google Scholar]
- 4.Apkarian AV, Sosa Y, Sonty S, Levy RM, Harden RN, Parrish TB, and Gitelman DR (2004). Chronic back pain is associated with decreased prefrontal and thalamic gray matter density. J. Neurosci. 24, 10410–10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porreca F, Ossipov MH, and Gebhart GF (2002). Chronic pain and medullary descending facilitation. Trends Neurosci. 25, 319–325. [DOI] [PubMed] [Google Scholar]
- 6.Jensen KB, Kosek E, Petzke F, Carville S, Fransson P, Marcus H, Williams SC, Choy E, Giesecke T, Mainguy Y, et al. (2009). Evidence of dysfunctional pain inhibition in Fibromyalgia reflected in rACC during provoked pain. Pain 144, 95–100. [DOI] [PubMed] [Google Scholar]
- 7.Gerstner G, Ichesco E, Quintero A, and Schmidt-Wilcke T (2011). Changes in regional gray and white matter volume in patients with myofascial-type temporomandibular disorders: a voxel-based morphometry study. J. Orofac. Pain 25, 99–106. [PubMed] [Google Scholar]
- 8.Lewis GN, Rice DA, and McNair PJ (2012). Conditioned pain modulation in populations with chronic pain: a systematic review and meta-analysis. J. Pain 13, 936–944. [DOI] [PubMed] [Google Scholar]
- 9.Moayedi M, Weissman-Fogel I, Salomons TV, Crawley AP, Goldberg MB, Freeman BV, Tenenbaum HC, and Davis KD (2012). White matter brain and trigeminal nerve abnormalities in temporomandibular disorder. Pain 153, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 10.Staud R (2012). Abnormal endogenous pain modulation is a shared characteristic of many chronic pain conditions. Expert Rev. Neurother. 12, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabó N, Kincses ZT, Párdutz A, Tajti J, Szok D, Tuka B, Király A, Babos M, Vörös E, Bomboi G, et al. (2012). White matter microstructural alterations in migraine: a diffusion-weighted MRI study. Pain 153, 651–656. [DOI] [PubMed] [Google Scholar]
- 12.Bushnell MC, Ceko M, and Low LA (2013). Cognitive and emotional control of pain and its disruption in chronic pain. Nat. Rev. Neurosci. 14, 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ossipov MH, Dussor GO, and Porreca F (2010). Central modulation of pain. J. Clin. Invest. 120, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millan MJ (2002). Descending control of pain. Prog. Neurobiol. 66, 355–474. [DOI] [PubMed] [Google Scholar]
- 15.Bargmann CI, Thomas JH, and Horvitz HR (1990). Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 55, 529–538. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan JM, and Horvitz HR (1993). A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90, 2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills H, Wragg R, Hapiak V, Castelletto M, Zahratka J, Harris G, Summers P, Korchnak A, Law W, Bamber B, et al. (2012). Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J. 31, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi H, and Sakai T (2018). Leucokinin signaling regulates hunger-driven reduction of behavioral responses to noxious heat in Drosophila. Biochem. Biophys. Res. Commun. 499, 221–226. [DOI] [PubMed] [Google Scholar]
- 19.Barbagallo B, and Garrity PA (2015). Temperature sensation in Drosophila. Curr. Opin. Neurobiol. 34C, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montell C (2021). Drosophila sensory receptors—a set of molecular Swiss Army Knives. Genetics 217, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tracey WD, Wilson RI, Laurent G, and Benzer S (2003). painless, a Drosophila gene essential for nociception. Cell 113, 261–273. [DOI] [PubMed] [Google Scholar]
- 22.Montell C, and Rubin GM (1989). Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron 2, 1313–1323. [DOI] [PubMed] [Google Scholar]
- 23.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, and Julius D (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824. [DOI] [PubMed] [Google Scholar]
- 24.Montell C (2005). The TRP superfamily of cation channels. Sci. STKE 2005(272), re3. [DOI] [PubMed] [Google Scholar]
- 25.Neely GG, Keene AC, Duchek P, Chang EC, Wang QP, Aksoy YA, Rosenzweig M, Costigan M, Woolf CJ, Garrity PA, et al. (2011). TrpA1 regulates thermal nociception in Drosophila. PLoS One 6, e24343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong L, Bellemer A, Yan H, Honjo K, Robertson J, Hwang RY, Pitt GS, and Tracey WD (2012). Thermosensory and non-thermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat sensor domains of a thermoTRP channel. Cell Rep. 1, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu P, Gong J, Shang Y, Wang F, Ruppell KT, Ma Z, Sheehan AE, Freeman MR, and Xiang Y (2019). Polymodal nociception in Drosophila requires alternative splicing of TrpA1. Curr. Biol. 29, 3961–3973 e3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Lee Y, Lee J, Bang S, Hyun S, Kang J, Hong ST, Bae E, Kaang BK, and Kim J (2005). Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat. Genet. 37, 305–310. [DOI] [PubMed] [Google Scholar]
- 29.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney ST, Broadie K, Keane J, Niemann H, and O’Kane CJ (1995). Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14, 341–351. [DOI] [PubMed] [Google Scholar]
- 31.Turner-Evans DB, and Jayaraman V (2016). The insect central complex. Curr. Biol. 26, R453–457. [DOI] [PubMed] [Google Scholar]
- 32.Pfeiffer K, and Homberg U (2014). Organization and functional roles of the central complex in the insect brain. Annu. Rev. Entomol. 59, 165–184. [DOI] [PubMed] [Google Scholar]
- 33.Strauss R (2002). The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 12, 633–638. [DOI] [PubMed] [Google Scholar]
- 34.Mylonopoulos I (2013). Epione. In The Encyclopedia of Ancient History, Bagnall RS, Brodersen K, Champion CB, Erskine A and Huebner SR, eds. (John Wiley & Sons, Ltd; ). [Google Scholar]
- 35.Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, and Hassan BA (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc. Natl. Acad. Sci. USA 107, 20553–20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YQ, Rodesch CK, and Broadie K (2002). Living synaptic vesicle marker: synaptotagmin-GFP. Genesis 34, 142–145. [DOI] [PubMed] [Google Scholar]
- 37.Bachtel ND, Hovsepian GA, Nixon DF, and Eleftherianos I (2018). Allatostatin C modulates nociception and immunity in Drosophila. Sci. Rep. 8, 7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Polgar E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, et al. (2018). Circuit dissection of the role of somatostatin in itch and pain. Nat. Neurosci. 21, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenz C, Williamson M, and Grimmelikhuijzen CJ (2000). Molecular cloning and genomic organization of a second probable allatostatin receptor from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 273, 571–577. [DOI] [PubMed] [Google Scholar]
- 40.Birgül N, Weise C, Kreienkamp HJ, and Richter D (1999). Reverse physiology in Drosophila: identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. EMBO J. 18, 5892–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu SY, Cang CL, Liu XF, Peng YQ, Ye YZ, Zhao ZQ, and Guo AK (2006). Thermal nociception in adult Drosophila: behavioral characterization and the role of the painless gene. Genes Brain Behav. 5, 602–613. [DOI] [PubMed] [Google Scholar]
- 43.Sokabe T, Tsujiuchi S, Kadowaki T, and Tominaga M (2008). Drosophila Painless is a Ca2+-requiring channel activated by noxious heat. J. Neurosci. 28, 9929–9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SH, Lee Y, Akitake B, Woodward OM, Guggino WB, and Montell C (2010). Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc. Natl. Acad. Sci. USA 107, 8440–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Guo Y, Wang F, and Wang Z (2011). Drosophila TRPA channel painless inhibits male-male courtship behavior through modulating olfactory sensation. PLoS One 6, e25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamotová A (2019). Endogenous antinociceptive system and potential ways to influence It. Physiol. Res. 68, S195–S205. [DOI] [PubMed] [Google Scholar]
- 47.Schultzhaus JN, Saleem S, Iftikhar H, and Carney GE (2017). The role of the Drosophila lateral horn in olfactory information processing and behavioral response. J. Insect Physiol. 98, 29–37. [DOI] [PubMed] [Google Scholar]
- 48.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. (2010). A genome-wide Drosophila screen for heat nociception identifies ⍺2δ3 as an evolutionarily conserved pain gene. Cell 143, 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson EC, Bohn LM, Barak LS, Birse RT, Nassel DR, Caron MG, and Taghert PH (2003). Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-β-arrestin2 interactions. J. Biol. Chem. 278, 52172–52178. [DOI] [PubMed] [Google Scholar]
- 51.Kreienkamp HJ, Larusson HJ, Witte I, Roeder T, Birgül N, Hönck HH, Harder S, Ellinghausen G, Buck F, and Richter D (2002). Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, from Drosophila melanogaster and their ligands by reverse pharmacology. J. Biol. Chem. 277, 39937–39943. [DOI] [PubMed] [Google Scholar]
- 52.Uhl GR, and Nishimori T (1990). Neuropeptide gene expression and neural activity: assessing a working hypothesis in nucleus caudalis and dorsal horn neurons expressing preproenkephalin and preprodynorphin. Cell. Mol. Neurobiol. 10, 73–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gruner M, Nelson D, Winbush A, Hintz R, Ryu L, Chung SH, Kim K, Gabel CV, and van der Linden AM (2014). Feeding state, insulin and NPR-1 modulate chemoreceptor gene expression via integration of sensory and circuit inputs. PLoS Genet. 10, e1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glauser DA, Chen WC, Agin R, Macinnis BL, Hellman AB, Garrity PA, Tan MW, and Goodman MB (2011). Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics 188, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao Z, Macara AM, Lelito KR, Minosyan TY, and Shafer OT (2012). Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol. 108, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cavener DR (1987). Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 15, 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassett AR, Tibbit C, Ponting CP, and Liu JL (2013). Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O’Connor-Giles KM (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo S, and Ueda R (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, and Gao G (2013). Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gong WJ, and Golic KG (2003). Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y, and Montell C (2013). Drosophila TRPA1 functions in temperature control of circadian rhythm in pacemaker neurons. J. Neurosci. 33, 6716–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veenstra JA, Agricola HJ, and Sellami A (2008). Regulatory peptides in fruit fly midgut. Cell Tissue Res. 334, 499–516. [DOI] [PubMed] [Google Scholar]
- 64.Luo J, Shen WL, and Montell C (2017). TRPA1 mediates sensation of the rate of temperature change in Drosophila larvae. Nat. Neurosci. 20, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veenstra JA (2009). Allatostatin C and its paralog allatostatin double C: the arthropod somatostatins. Insect Biochem. Mol. Biol. 39, 161–170. [DOI] [PubMed] [Google Scholar]
- 66.Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any standardized datatypes.
This study did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


