Abstract
Apolipoprotein (apo) E functions in lipoprotein metabolism as a low density lipoprotein receptor ligand. ApoE is comprised of two structural domains, a 22 kDa N-terminal (NT) domain that adopts a helix bundle conformation and a 10 kDa C-terminal domain with strong lipid binding affinity. The NT domain is capable of transforming aqueous phospholipid dispersions into discoidal reconstituted high density lipoprotein (rHDL) particles. Given the utility of apoE-NT as a structural component of rHDL, expression studies were conducted. A plasmid construct encoding a pelB leader sequence fused to the N-terminus of human apoE4 (residues 1–183) was transformed into Escherichia coli. Upon expression, the fusion protein is directed to the periplasmic space where leader peptidase cleaves the pelB sequence, generating mature apoE4-NT. In shaker flask expression cultures, apoE4-NT escapes the bacteria and accumulates in the medium. In a bioreactor setting, however, apoE4-NT was found to combine with gas and liquid components in the culture medium to generate large quantities of foam. When this foam was collected in an external vessel and collapsed into a liquid foamate, analysis revealed that apoE4-NT was the sole major protein present. The product protein was further isolated by heparin affinity chromatography (60 – 80 mg/liter bacterial culture), shown to be active in rHDL formulation, and documented to serve as an acceptor of effluxed cellular cholesterol. Thus, foam fractionation provides a streamlined process to produce recombinant apoE4-NT for biotechnology applications.
Keywords: Apolipoprotein E, reconstituted high density lipoprotein, bioreactor, foam fractionation, nanodisc, nanodisk
Introduction
Human apolipoprotein (apo) E is a 299 amino acid amphipathic exchangeable apolipoprotein. In solution, apoE is a two domain protein, including a 22 kDa N-terminal (NT) four helix bundle that is linked to a 10 kDa C-terminal domain with strong lipid binding affinity [1]. NT domain helical segments are amphipathic and, in the absence of lipid, align such that each helix projects their hydrophilic face toward the aqueous environment while their opposing hydrophobic faces interact with one another in the bundle interior. The C-terminal domain is attached to the NT domain via a largely unstructured hinge region. Three major isoforms of human apoE exist that differ from one another at residues 112 and 158 in the NT domain. ApoE2 contains Cys at both sites, apoE3 has Cys at 112 and Arg at 158, while apoE4 contains Arg at both sites [2]. In vivo, apoE binds to circulating plasma lipoproteins and serves as a ligand for the low-density lipoprotein (LDL) receptor in the liver and the LDL receptor related protein 1 in the brain [3,4].
ApoE has also gained notoriety for its association with Alzheimer’s disease (AD), a neurodegenerative disorder characterized by severe memory loss [5]. AD is associated with the presence of extracellular plaque deposits of amyloid β (Aβ) peptide and intracellular neurofibrillary tangles comprised of hyper-phosphorylated Tau protein [6]. Individuals homozygous for the ε4 allele experience an increased risk of AD and a lower age of onset while individuals homozygous for the ε2 allele show reduced rates of disease prevalence, as compared to those homozygous for the ε3 allele [7,8]. The mechanism underlying apoE isoform-specific differences in AD risk remains enigmatic [9], although apoE is a known component of Aβ plaque deposits [10,11]. ApoE4 is not only associated with increased AD risk but also with elevated plasma cholesterol levels, as compared to patients with apoE3. This effect has been ascribed to an increased propensity of apoE4 to associate with very low density lipoproteins [12], a property related to isoform-specific interactions between the N and C terminal domains of apoE4.
Reconstituted high density lipoproteins (rHDL) are a class of nanoparticles comprised of a disk-shaped lipid bilayer circumscribed by an amphipathic “scaffold protein”. These particles constitute a mimic of nascent HDL and are often prepared using phosphatidylcholine as the bilayer forming lipid and apoA-I as the scaffold protein [13]. These rHDL have the ability to promote removal of cholesterol from plaque deposits on arterial walls and have been investigated in human clinical trials as a treatment for atherosclerosis [14]. Moreover, rHDL lipid and protein components are interchangeable, providing enhanced versatility in terms of their structure - function properties [13,15]. rHDL containing apoE4-NT have been formulated and characterized previously [16,17,18]. rHDL prepared with apoE-NT, or mimetic peptides, have been used to investigate topics including hydrophobic bioactive agent transport [19], targeted delivery via the low-density lipoprotein receptor [20] and as a miniature membrane to analyze SNARE-induced fusion pores [21]. In a similar manner, rHDL have been used as a transport vehicle for imaging agents in theranostic applications [22], a model membrane for ligand binding studies [23] and as a miniature membrane for transmembrane protein incorporation and characterization [24].
Despite the importance of apoE in lipid transport, metabolism, and disease, production of this protein at scale suffers from poor yield and high cost, thereby hampering its utility as an rHDL scaffold. Herein, a facile method for production of untagged apoE4-NT in Escherichia coli is described that employs a bioreactor and takes advantage of the intrinsic biosurfactant-like properties of this protein. The method described utilizes a plasmid construct that encodes a pelB leader sequence fused to the coding sequence of apoE4-NT. The leader sequence directs the nascent fusion protein to the periplasmic space where the pelB sequence is cleaved by leader peptidase, generating mature apoE4-NT which then escapes from the bacterium into the culture medium. Upon interacting with gas and liquid components of the medium, apoE4-NT induces formation of a thick foam that contains apoE4-NT as the sole major protein component, simplifying downstream processing and isolation of this protein.
Materials and Methods
Plasmid construct design / pelB leader sequence.
A pET22b+ plasmid vector containing the coding sequence for residues 1 – 183 of mature human apoE4 (apoE4-NT) was generated in silico and synthesized by Genscript Biotech (Piscataway, NJ). The human apoE4-NT coding sequence was optimized for bacterial expression by changing 22 Arg codons from CGG to CGC. Although both encode Arg, the latter is strongly favored by E. coli. The construct encodes a pelB leader sequence fused at the N-terminus of apoE4-NT. The plasmid was then transformed into BL21 competent cells and maintained on streaked agar plates or as a frozen glycerol stock.
Shaker flask expression.
A 500 ml shaker flask containing 125 ml NZCYM media and 100 μg/ml ampicillin was inoculated with a single E. coli bacterial colony harboring the pelB - apoE4-NT plasmid construct. The bacterial culture was grown at 37 °C until the optical density (OD) of the medium at 600 nm reached ~0.7. PelB-apoE4-NT fusion protein expression was induced with isopropyl β-D-thiogalactopyranoside (IPTG; 1 mM), followed by culturing the bacteria for 5 h at 37 °C. The bacteria were then pelleted by centrifugation at 11,200 × g for 10 min at 4 °C and the supernatant recovered [25]. Isolated supernatant (125 ml) was vacuum filtered (0.45 μm) and applied to a 5 ml HiTrap Heparin HP column at a rate of 5 ml/min. Subsequently, the column was washed with 10 column volumes of binding buffer (10 mM sodium phosphate, pH 7.0) followed by 3 column volumes of elution buffer (10 mM sodium phosphate, pH 7.0, 1.5 M NaCl). Protein eluting under these conditions was recovered, dialyzed against phosphate buffered saline (PBS, 100 mM sodium phosphate, pH 7.0, 150 mM NaCl) for 16 h and stored at 4 °C until further use. Sample protein content was measured using the bicinchoninic acid assay, following manufacturer instructions (Thermo Scientific, San Jose, CA).
Bioreactor-based expression of apoE4-NT.
A Distek Model BiOne 1250 bioprocessor (5 L capacity) was employed for expression studies. The bioreactor control tower was calibrated for specific running parameters including dissolved oxygen, pH, temperature, and impeller rotation rate. Dissolved oxygen and pH were monitored using an OxyFerm FDA VP 325 dissolved oxygen sensor and an EasyFerm Plus PHI VP 325 Pt100 pH probe (Hamilton Co., Reno, NV). For standard apoE4-NT expression experiments, the bioreactor chamber contained 3 L NZCYM medium supplemented with 100 μg/ml ampicillin and 30 g/L D-glucose. The bioreactor chamber was seeded with a saturated overnight culture of E. coli (50 ml/L culture volume). Following a growth period of 5 h at 37 °C, when the OD600 reached 7.0 – 9.0, IPTG was added (1 mM final concentration). Approximately 1 h post induction, foam production became continuous and culture pH was observed to decline over the next 2 h. At this point, 2 M NaOH was titrated into the culture to maintain the pH between 6.5 – 6.8. Air flow was monitored and controlled by the dissolved oxygen cascade, which is established using the proportional integral derivative value determined as a function of the current dissolved oxygen percentage within the bioreactor chamber. Flow of compressed air and/or O2 was maintained at 5–15 psi using an internal sparge unit. Sterility was maintained by flowing the gases through 0.22 μm sterile filters prior to entering the sparge unit.
Foam Collection.
Foam was reliably generated in cultures expressing the pelB – apoE4-NT fusion protein at ~1 h post induction. Foam production continued for a period of 5 – 7 h. Foam diversion into a collection vessel relied on the internal pressure of the bioreactor chamber. Once the chamber filled with foam, pressure within the chamber forced foam into flexible tubing attached to a port in the bioreactor headplate, where it flowed into the collection vessel (a 30 L carboy placed on ice). When foam production slowed, or the collection vessel filled to capacity, it was transferred to a 4 °C cold room for 12 – 14 h, allowing the foam to collapse into a liquid foamate. The foamate was then centrifuged at 11,200 × g for 20 min at 4 °C. The resulting supernatant was collected, vacuum filtered through a 0.45 μm membrane and subjected to heparin affinity chromatography as described above.
SDS-PAGE Analysis.
SDS-PAGE was performed on 4–20% Bio-Rad Mini-PROTEAN TGX Precast Gels. Gels were electrophoresed at 200 V for 30 min and stained with a solution (80% methanol and 20% glacial acetic acid) containing Naphthol Blue Black (20 μg/ml, Sigma) for 1 h. Gels were de-stained in a solution containing 40% deionized water / 40% methanol / 20% glacial acetic acid for 3 h. Stained gels were imaged using a Bio-Rad ChemiDoc instrument.
Liquid chromatography / mass spectrometry analysis.
Protein content of individual samples was estimated using EZQ Protein Quantitation Kit (Thermo Scientific Cat R33200). One hundred μg protein was reduced, alkylated with iodoacetamide, and digested with trypsin/Lys-C protease mixture using Thermo Scientific EasyPep Mini MS Sample prep kit (Cat #A40006). Proteases were added in a 1:10 (enzyme to protein) ratio for digestion and samples were cleaned using the column provided with the kit. Samples were analyzed using an UltiMate 3000 RSLCnano system (Thermo Scientific). Peptides were trapped prior to separation on a 300 μm i.d. × 5 mm C18 PepMap 100 trap (Thermo Scientific) for 5 min at 10 μL/min. Separation was conducted on a 50 cm uPAC C18 nanoLC column (PharmaFluidics, Ghent, Belgium) with a 20 μm fused silica emitter and a Nanospray Flex ion source (Thermo Scientific). Separation was performed at 350 nL/min using a gradient from 1% - 45% (Solvent A 0.1% Formic Acid, Solvent B Acetonitrile, 0.1% Formic Acid). Data-dependent analysis (DDA) mass spectrometry was performed using an Orbitrap Eclipse mass spectrometer (Thermo Scientific). The MS1 precursor selection range was from 375–1500 m/z at a resolution of 120K with a normalized automatic gain control (AGC) target of 250% and an automatic maximum injection time. Quadrupole isolation was 0.7 Th for MS2 isolation and CID fragmentation in the linear ion trap, with a collision energy of 35% and a 10 ms activation time. The MS2 AGC was in standard mode with a 35 ms maximum injection time. The instrument was operated in data-dependent mode with a 3 s cycle time, the most intense precursor priority and the dynamic exclusion set to 60 s with a 10 ppm tolerance. Data analysis was performed using Sequest (Thermo Scientific, version v.27, rev. 11) and Scaffold Q+ (Proteome Software, Portland, OR).
Mass Spectrometry – Direct Infusion.
Direct Infusion was performed on 1 mg/ml solutions of apE4-NT at a flow rate of 5 μl/min using electrospray ionization (ESI) on an Orbitrap Eclipse mass spectrometer (Thermo Scientific) set to intact protein mode. ESI spray voltage was set to 3030 V with an ion transfer tube temperature of 310 °C. The MS1 was acquired within the orbitrap mass analyzer with a precursor selection range from 6002000 m/z at a resolution of 120K with a normalized AGC target of 250% and an automatic maximum injection time. Quadrupole isolation was 0.7 Th for MS2 isolation and CID fragmentation in the linear ion trap with a collision energy of 35% had a 10 ms activation time. The MS2 AGC was in standard mode with a 35 ms maximum injection time. The instrument was operated in data-dependent mode with a 3 second cycle time with the most intense precursor priority and the dynamic exclusion set to an exclusion duration of 60 s with a 10 ppm tolerance. Deconvolution was performed using the Xcalibur Xtract algorithm (Thermo Scientific, version 3.0.63). The data was then analyzed using Sequest (Thermo Scientific, version v.27, rev. 11) and Scaffold Q+ (Proteome Software, Portland, OR).
ApoE4-NT rHDL formation.
Five mg dimyristoylphosphatidylcholine (DMPC, Avanti Polar Lipids) was dissolved in chloroform/methanol (3:1 v/v) and the solvent evaporated under a gentle stream of N2 gas, leaving a thin film of phospholipid on the vessel wall. Eight hundred μL of 10 mM sodium phosphate, pH 3.0, was added and the sample vortexed to form an aqueous dispersion of DMPC. Two mg isolated apoE4-NT (in 200 μL 10 mM sodium phosphate, pH 3.0) was then added and the sample bath sonicated at 24 °C until the solution clarified (<15 min).
FPLC gel filtration chromatography.
ApoE4-NT rHDL particles (200 μL) were analyzed on a GE AKTA Pure FPLC system fitted with a Superdex 200 Increase 10/300 GL column equilibrated in 10 mM sodium phosphate, pH 3.4. Isolated apoE4-NT and apoE4-NT rHDL were applied to the column, respectively, and eluted at a rate of 0.75 ml/min. Absorbance was continuously monitored at 280 nm with collection of 1.5 mL fractions.
Cholesterol efflux assay.
Human THP-1 monocytic cells were obtained from American Type Culture Collection (ATCC TIB-202, Cedarlane, Ontario). Phorbol 12-myristate-12 acetate (PMA) was obtained from Sigma-Aldrich (Oakville, Ontario, Canada) while cholesterol, [1,2-3H(N)] was purchased from Perkin Elmer (Norwalk, CT). Human oxidized low-density lipoprotein (ox-LDL) was from Kalen Biomedical. Cells were cultured in T175 flasks in bicarbonate-buffered RMPI medium containing 10% heat-inactivated FBS, 50 mmol/L β-mercaptoethanol, and 100 U/ml penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. Cells were used between passage 2 and 5. Experiments were carried out in 24-well tissue culture plates at a cell density of 1.0 × 106/mL in the presence of 10% FBS and 200 nM PMA for 48 h. Subsequently, the cells were labeled with 2 μCi/ml [3H]-cholesterol and ox-LDL (50 μg/ml) in RPMI media / 1% FBS / 200 nM PMA for 48 h. Cells were washed twice with RPMI media prior to efflux assays [26]. Efflux assays were performed in triplicate by incubating cells for 6 h at 37 °C with 40 μg/ml of specified cholesterol acceptors (as protein). Following incubation, the culture medium was collected, and cells were washed twice with PBS. The medium was centrifuged at 3000 × g for 5 min to remove cell debris and 0.2 mL of the supernatant fraction was mixed with scintillation liquid and counted for radioactivity. The cells were lysed by overnight incubation with 0.5 mL 0.1 N NaOH, with shaking. A 0.1 mL aliquot of each cell lysate was mixed with scintillation liquid and counted for radioactivity. Cholesterol efflux capacity was calculated as percent cholesterol efflux = [3H] cpm medium / ([3H] cpm medium + [3H] cpm cells) × 100%. Diffusional efflux (control condition) was subtracted from total efflux to determine net efflux.
Statistical Analysis.
Statistical analysis was performed by one-way ANOVA followed by Tukey multiple comparison test. Statistical tests were performed using Graphpad Prism version 9.5.1 for Windows (Graphpad Software, San Diego, CA).
Results
Bioreactor production of recombinant apoE4-NT.
Herein, the ability of recombinant apoE4-NT to escape E. coli when expressed as a fusion protein with an N-terminal pelB leader sequence extension has been exploited to streamline isolation of this protein. Following expression of the pelB – apoE4-NT fusion, apoE4-NT is directed to the bacterial periplasmic space where the pelB leader sequence is cleaved and ultimately mature apoE4-NT transits to the extracellular space [25, 27]. In a bioreactor setting, apoE4-NT expression coincides with production of copious quantities of dense, viscous foam (Figure 1). Given that foam was not produced when pelB - apoE4-NT was expressed in shaker flask cultures, foam generation in the bioreactor chamber was hypothesized to result from apoE4-NT interaction with gas / liquid bubbles that deliver air and O2 to the culture media.
Figure 1. Bioreactor-dependent apoE4-NT-induced foam production.

Photograph obtained 5 h post-induction of apoE4-NT expression depicting the bioreactor control panel (right), culture chamber (middle) and foam collection vessel (left). As foam fills the bioreactor chamber, it is forced out through flexible tubing attached to a headplate port and into a 30 L carboy collection vessel. The foam deposited in the carboy is dense and piles up on itself to occupy a large volume of the collection vessel.
Foam analysis.
To examine foam generated in the bioreactor chamber during pelB – apoE4-NT fusion protein expression, it was diverted to a collection vessel and condensed to a liquid foamate. To investigate the potential correlation between foam production and apoE4-NT expression, SDS-PAGE was performed on supernatant fractions obtained from aliquots of culture medium and foamate (Figure 2). A band corresponding to apoE4-NT appears in both the culture medium and foamate supernatants, indicating apoE4-NT is associated with foam produced in the bioreactor chamber during expression.
Figure 2. SDS-PAGE analysis of fractions from bioreactor-based expression of pelB-apoE4-NT.
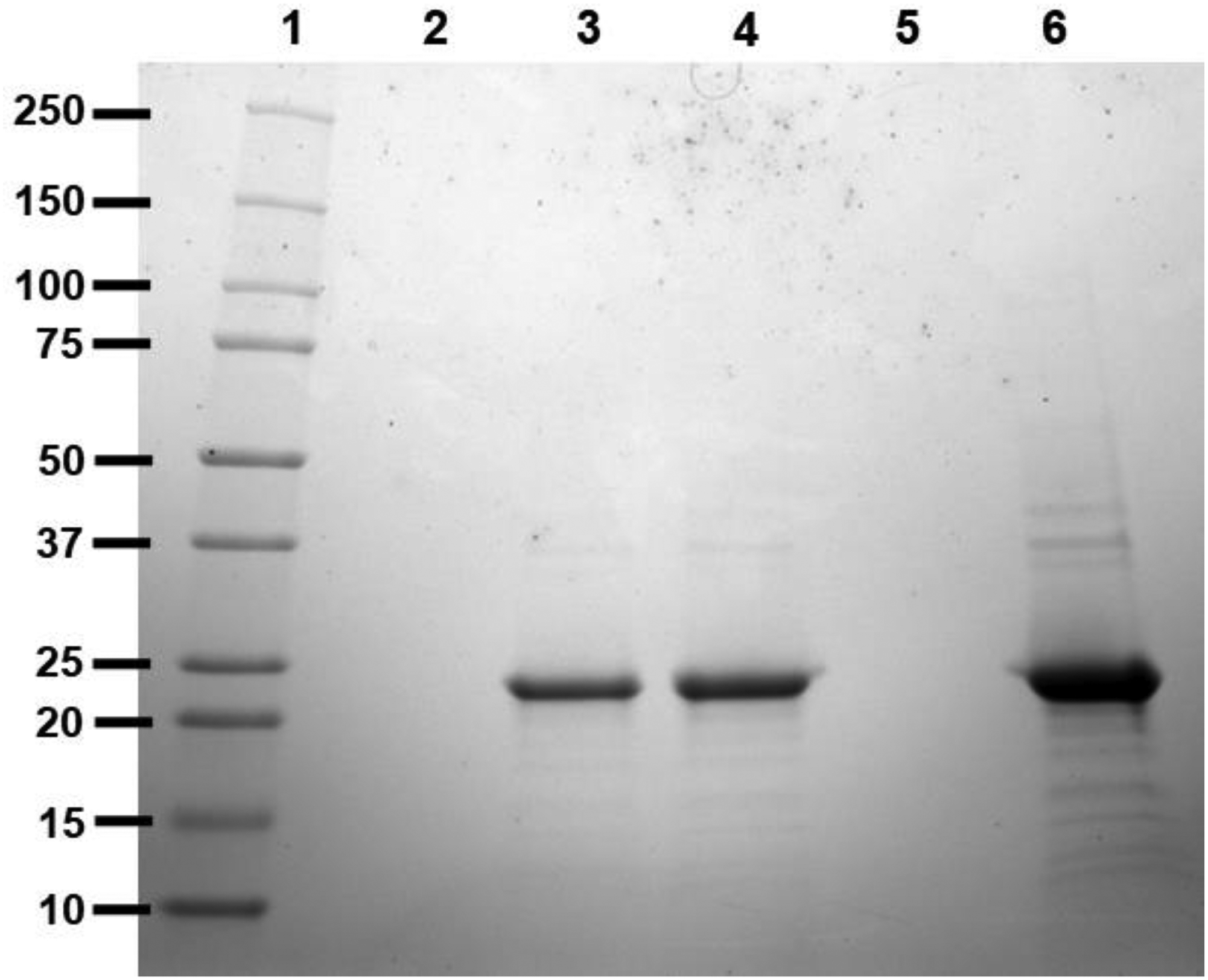
E. coli transformed with a pET22b+ plasmid harboring the pelB-apoE4-NT fusion construct were used to seed a bioreactor expression culture as described in Materials and Methods. Aliquots were collected at various stages of the expression / isolation procedure and subjected to SDS-PAGE analysis. Lane 1) Molecular weight standards; Lane 2) uninduced bioreactor culture medium supernatant; Lane 3) Induced bioreactor culture medium supernatant; Lane 4) foamate fraction. Lane 5) flow through collected following application of foamate onto a HiTrap Heparin HP column; Lane 6) eluate obtained following application of high salt buffer to the HiTrap Heparin HP column.
Foam processing and apoE4-NT isolation.
Approximately 5 h after induction of pelB - apoE4-NT expression, foam production slowed or stopped. At this point, the carboy containing collected foam was stored at 4 °C to promote foam collapse into a liquid foamate. From a 3 L bioreactor culture, ~1.3 L of liquid foamate was recovered. Following centrifugation and filtration of the supernatant, the sample was subjected to heparin affinity chromatography. ApoE4-NT bound to the column in low salt buffer and eluted upon application of buffer containing 1.5 M NaCl. The eluted protein was dialyzed against low salt buffer and used in structure-function characterization studies. The yield of apoE4-NT ranged from 60 to 80 mg per liter of bioreactor culture medium.
Characterization of apoE4-NT.
Given the nature of foam fractionation method, it is conceivable that recombinant apoE4-NT was modified or damaged during this process. To investigate this, mass spectrometry analysis was performed on shaker flask- and bioreactor-derived apoE4-NT samples. The proteins were digested with trypsin/Lys-C prior to analysis by liquid chromatography / mass spectrometry (Table 1). The peptides observed in both samples were identical and covered ~80% of the apoE4-NT sequence. Although evidence of methionine oxidation was obtained, no differences were observed between shaker flask- and bioreactor-derived, suggesting methionine oxidation is not specific to foam fractionated apoE4-NT samples. In other experiments, intact apoE4-NT from shaker flask- and bioreactor-based expressions were subjected to direct infusion mass spectrometry. The results revealed a single major protein population (MW = 21,461 Da) is present in both samples, in good agreement with the calculated mass of apoE4-NT (MW = 21,245 Da).
Table 1.
Liquid chromatography – mass spectrometry analysis of apoE4-NTa
| Bioreactor Sample | ||||||
|---|---|---|---|---|---|---|
| ApoE4-NT Fragment List | Modifications | Avg mass (observed) | Avg mass (calculated) | Theoretical mass | Start Residue Stop Residue | |
| VEQAVETEPEPELR | None | 813.4 | 1,624.80 | 1,624.80 | 2 | 15 |
| QQTEWQSGQR | None | 624.29 | 1,246.57 | 1,246.57 | 16 | 25 |
| WELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELR | None | 1,109.82 | 4,435.25 | 4,432.26 | 26 | 61 |
| ALMDETMK | Oxidation | 477.72 | 953.42 | 937.42 | 62 | 69 |
| AYKSELEEQLTPVAEETR | None | 1,047.02 | 2,092.03 | 2,092.03 | 73 | 90 |
| LGADMEDVR | Oxidation | 511.24 | 1,020.45 | 1,004.46 | 104 | 112 |
| LVQYRGEVQAMLGQSTEELR | Oxidation | 775.06 | 2,322.16 | 2,306.17 | 115 | 134 |
| LLRDADDLQK | None | 396.22 | 1,185.64 | 1,185.64 | 148 | 157 |
| LAVYQAGAR | None | 474.77 | 947.52 | 947.52 | 159 | 167 |
| Shaker Flask Sample | ||||||
| ApoE4-NT Fragment List | Modifications | Avg mass (observed) | Avg mass (calculated) | Theoretical mass | Start Residue Stop Residue | |
| VEQAVETEPEPELR | None | 813.4 | 1624.79 | 1624.79 | 2 | 15 |
| QQTEWQSGQR | None | 624.29 | 1246.57 | 1246.57 | 16 | 25 |
| WELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELR | None | 1109.82 | 4435.27 | 4435.26 | 26 | 61 |
| ALMDETMK | Oxidation | 477.72 | 953.42 | 937.42 | 62 | 69 |
| AYKSELEEQLTPVAEETR | None | 1047.02 | 2092.02 | 2092.03 | 73 | 90 |
| LGADMEDVR | Oxidation | 511.24 | 1020.45 | 1004.46 | 104 | 112 |
| LVQYRGEVQAMLGQSTEELR | Oxidation | 775.06 | 2322.16 | 2306.17 | 115 | 134 |
| LLRDADDLQK | None | 396.22 | 1185.64 | 1185.64 | 148 | 157 |
| LAVYQAGAR | None | 474.77 | 947.52 | 947.52 | 159 | 167 |
Bioreactor-derived (foam fractionated) and shaker flask-derived apoE4-NT were digested with trypsin/Lys-C prior to analysis
Biological activity of foam fractionated apoE4-NT.
The ability of apoE4-NT to transform phospholipid vesicles into rHDL was evaluated as described previously [28]. Briefly, an aqueous dispersion of DMPC was incubated with isolated recombinant apoE4-NT. Mild bath sonication induced transformation of the sample from an opaque lipid dispersion into a clear solution, indicative of rHDL formation. FPLC gel filtration chromatography was then performed to assess rHDL particle size and homogeneity. When foam-derived, lipid-free apoE4-NT was examined (Figure 3A), a single major absorbance peak at ~17.0 mL was observed, consistent with the monomeric molecular weight of apoE4-NT (~22 kDa). By contrast, apoE4-NT rHDL eluted as a single absorbance peak at ~11.5 ml (Figure 3B). This peak corresponds to a MW in the range of 100–150 kDa, consistent with the expected size of apoE4-NT rHDL. Thus, apoE4-NT isolated from bioreactor foam retains the ability to form rHDL.
Figure 3. FPLC analysis of foam-fractionated lipid-free apoE4-NT and apoE4-NT rHDL.
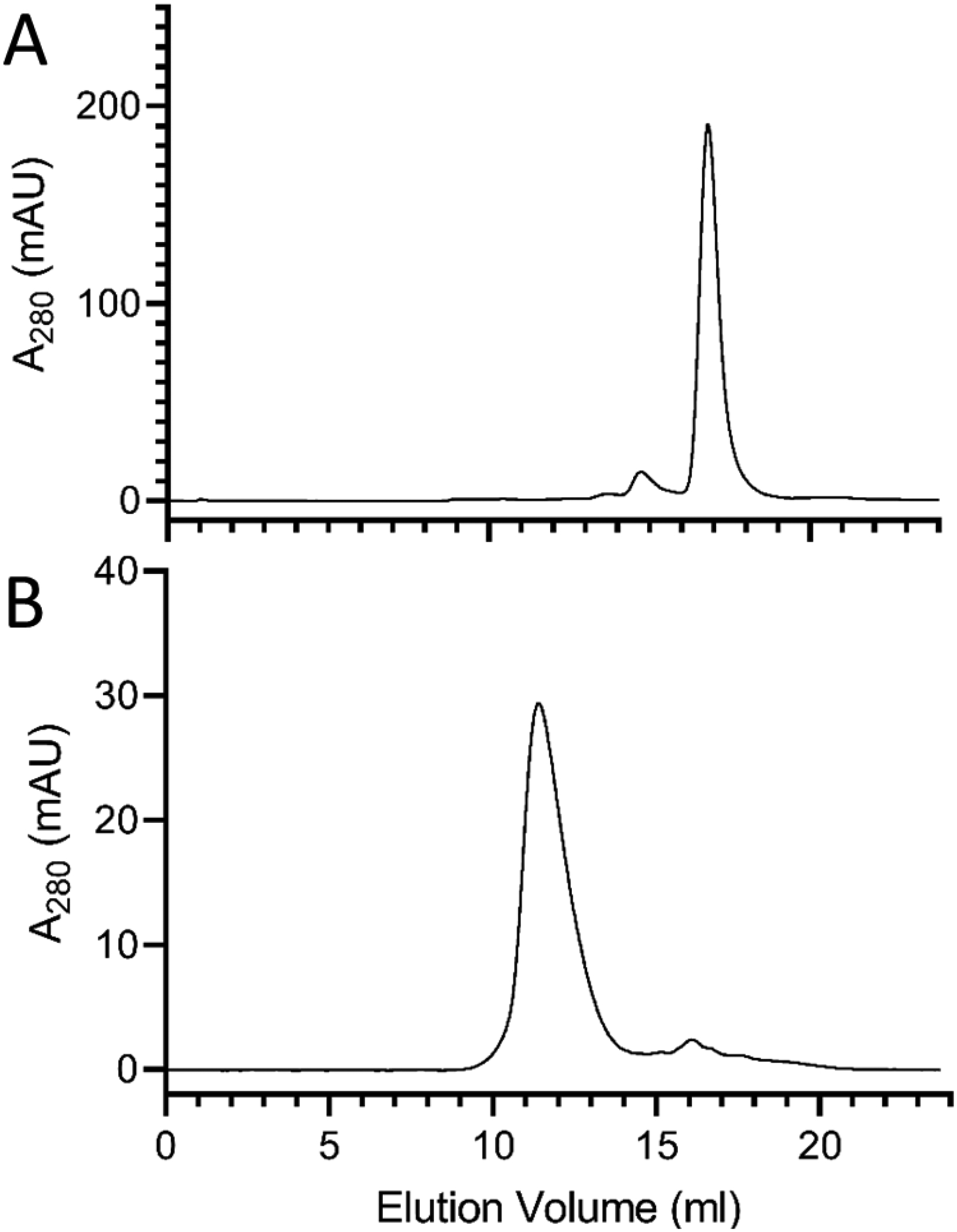
Gel filtration chromatography of a 300 μL aliquot of the eluate obtained following HiTrap Heparin HP chromatography of foamate (Panel A) and a 300 μL aliquot of foam-derived apoE4-NT-rHDL (Panel B). Samples were applied to a Superdex 200 Increase 10/300 column and eluted with 10 mM sodium phosphate, pH 3.4, at 0.75 mL/min with continuous monitoring of absorbance at 280 nm.
Cholesterol efflux capacity of foam fractionated apoE4-NT.
To test the functionality of apoE4-NT, cholesterol efflux assays were performed. This assay examined the ability of lipid-free apolipoproteins, or rHDL, to serve as acceptors of cholesterol effluxed from cultured cells by the ATP binding cassette (ABC) transporters (ABCA1 and/or ABCG1). ABCA1 specifically interacts with lipid-free apolipoprotein to promote formation of phospholipid/cholesterol/apoA-I complexes, while ABCG1 preferentially interacts with lipid-bound apolipoprotein and promotes efflux of cholesterol to HDL, LDL, and phospholipid vesicles [29]. Differentiated THP-1 monocytes were cultured, loaded with a mass of cholesterol, and labeled with [3H]-cholesterol as a tracer of cholesterol mobilization. Compared to lipid-free apoA-I (positive control), apoE4-NT from shaker flask expressions and apoE4-NT derived from bioreactor foamate, were slightly less effective in promoting cholesterol efflux (Figure 4). Compared to apoA-I rHDL, apoE4-NT rHDL derived from shaker flask expressions was marginally less effective, while apoE4-NT rHDL, formulated from bioreactor-derived foamate, was equally effective at promoting cholesterol efflux. These results demonstrate that the apoE4-NT isolated from bioreactor foamate is functionally competent as a cholesterol efflux acceptor.
Figure 4. Characterization of apoE4-NT cholesterol efflux activity.
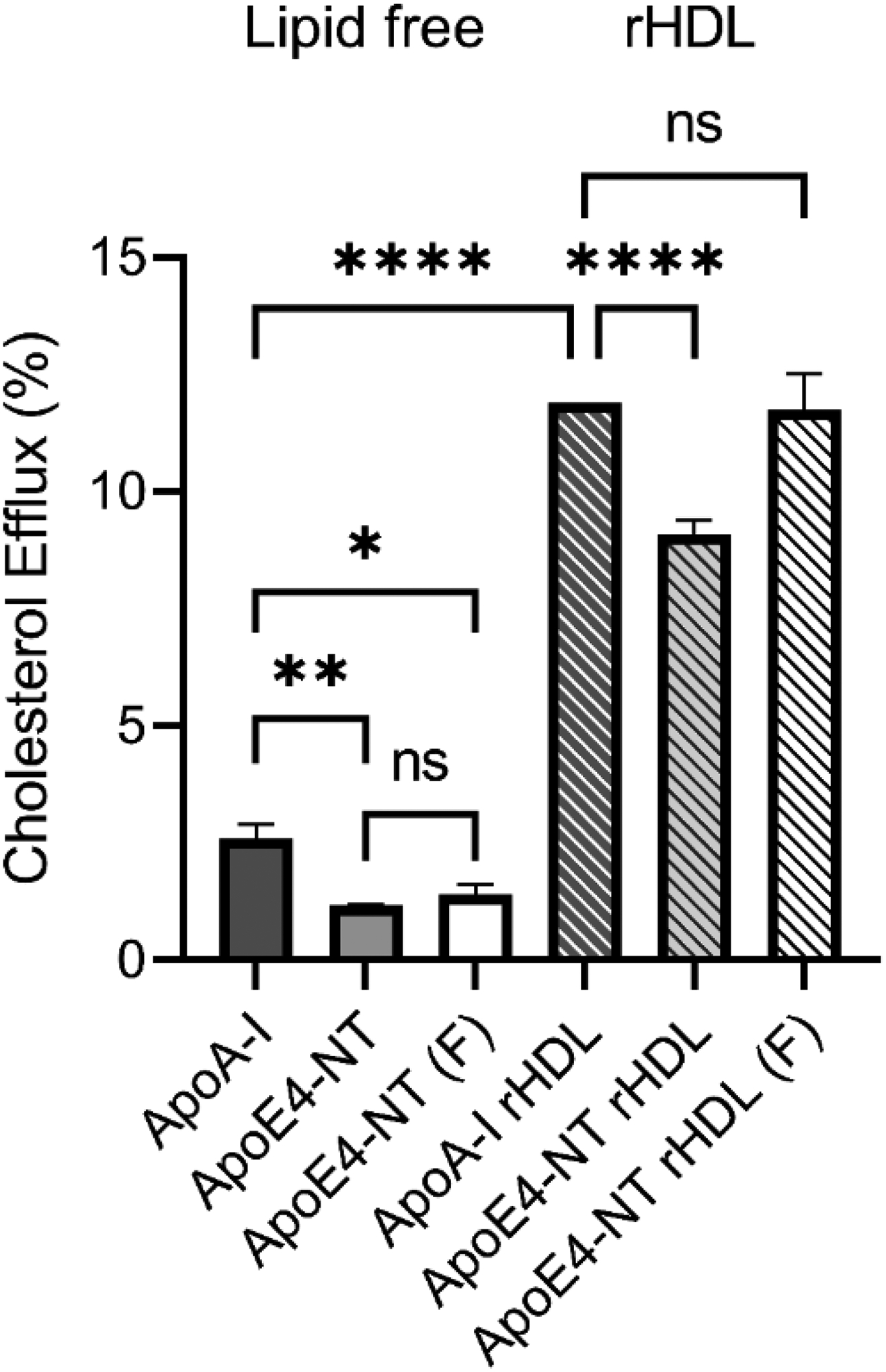
Cholesterol efflux assays were performed using cholesterol-loaded and radiolabeled THP-1 monocyte / macrophages as described in Materials and Methods. Lipid-free apoA-I, lipid-free apoE4-NT (isolated from a shaker flask expression), and lipid-free, foamate-derived, apoE4-NT (F) were incubated with cells for 6 h at 37 °C. In a similar manner, efflux experiments were performed using apoA-I rHDL, shaker flask-derived apoE4-NT rHDL, and foamate derived apoE4-NT rHDL (F). Experiments were performed in triplicate, and control nonspecific efflux was subtracted from each condition to calculate apolipoprotein or rHDL specific efflux. Statistical significance was determined using one-way ANOVA multiple comparison with a Tukey’s post hoc test. Values reported are the mean ± standard deviation (n=3) *, p<0.05; **, p<0.001; ****, p<0.0001; ns, not significant.
Discussion
Foam fractionation has previously been utilized for production of numerous biosurfactant molecules secreted by bacteria [30,31]. These biosurfactants are generally low molecular weight, non-proteinaceous compounds such as glycolipids, biopolymers, and lipopeptides [32]. Microbial biosurfactant production using foam fractionation technology is well established and these biosurfactants are widely used as emulsification agents in the food, petro-chemical, antimicrobial, biomedical, and pharmaceutical industries [33–37]. Extension of this methodology to recombinant biosurfactant-like apolipoproteins was recently demonstrated for insect apolipophorin-III [38].
It is evident that a required step in foam formation is apoE4-NT transit from the bacterial cell into the culture medium. A key step in this process involves pelB leader sequence targeting of apoE4-NT to the periplasmic space. A less well understood process is the subsequent movement of mature apoE4-NT from the periplasm to the culture medium. Given that this behavior is not common, it likely relates to the intrinsic structural properties of this protein. E. coli outer cell membranes are comprised of approximately 20% negatively charged phospholipid (i.e. phosphatidylglycerol) [39]. ApoE4-NT possesses a natural propensity to bind negatively charged phospholipids [28], whereupon it undergoes a conformational change [40]. In aqueous solutions, apoE4-NT exists as a bundle of four elongated amphipathic α-helices. Each α-helix segment possesses distinct hydrophobic and hydrophilic faces. The hydrophobic faces of apoE4-NT α-helix segments are directed toward the interior of the bundle where they contact one another via hydrophobic interactions, thereby stabilizing the bundle conformation [41]. Upon exposure to an appropriate lipid surface, however, the molecule undergoes a conformational change in which the protein adopts an “open” conformation that exposes the hydrophobic faces of its amphipathic helix segments, promoting their interaction with lipid surfaces [40]. Thus, it is possible that, as apoE4-NT gains access to the periplasmic space, it interacts with lipids in the outer membrane of E. coli and undergoes a conformational change that facilitates its transit into the culture media.
In a similar fashion, when utilized as a component in rHDL particles, apoE4-NT circumscribes the perimeter of a nanoscale disk-shaped phospholipid bilayer, wherein the hydrophobic faces of its amphipathic α-helices contact phospholipid fatty acyl chains at the edge of the disk. Indeed, this process is analogous to apoE’s function in lipoprotein binding and is required for LDL receptor binding activity [42]. In the present study we posit that, just as it interacts with lipid surfaces, when apoE4-NT interacts with gas/liquid bubble interfaces in the culture media, it generates foam.
Given that foam fractionation represents a novel approach to recombinant apolipoprotein production and isolation, it was important to assess if this method alters, or otherwise damages, apoE4-NT structure or function. Analysis of bioreactor-derived, foam fractionated, apoE4-NT by mass spectrometry indicated that, while methionine oxidation was detected, no differences were observed between shaker flask-derived and foam fractionation-derived expression cultures. The molecular weight increment observed for intact apoE4-NT samples versus the value calculated based on amino acid sequence may have arisen from the electro-spray ionization methodology [43]. A related concern is whether recombinant apoE4-NT retains its functional properties following isolation by foam fractionation. To examine this, the ability of apoE4-NT to generate rHDL was examined. Both shaker flask-derived [28] and foam fractionation-derived apoE-NT (this study) possess the ability for transform dispersed phospholipid vesicles into fully aqueous soluble rHDL. Moreover, apoE4-NT produced in shaker flask cultures and from bioreactor-dependent foam fractionation retained the ability to promote cholesterol efflux from cultured cells. Based on these results, it may be concluded that apoE4-NT obtained from bioreactor-generated foam is functional.
An advantage of foam fractionation methodology for production of recombinant apoE4-NT relates to downstream processing following protein production at bioreactor scale, an important consideration for industrial and academic applications. ApoE-NT has previously been shown to function as a scaffold protein for a specialized type of rHDL, termed nanodisks, that represent an advancement in drug delivery technology [13, 44]. In general, rHDL technology has been exploited in a wide range of applications, including cholesterol efflux enhancement, use as a bioactive agent delivery vehicle and receptor targeting (LDL receptor or N-acetyl-d-galactosamine / asialoglycoprotein receptor) [13,15,45]. Efforts to commercialize these, and other, applications of rHDL technology will be facilitated by the availability of a cost effective, high yield recombinant scaffold protein production methodology. Towards that end, the method described herein represents an attractive approach to meet the growing demand for apolipoproteins.
The findings reported in this study also suggest that other members of the class of amphipathic exchangeable apolipoproteins may be isolated using foam fractionation methods. For example, it is conceivable that apoA-I and/or full-length apoE (299 amino acids) will be amenable to foam fractionation methods when expressed in a bioreactor as pelB – apolipoprotein chimeras. Furthermore, investigation into the structural properties of recombinant apolipoproteins, as it pertains to their biosurfactant-like behavior and extracellular secretion, may provide insight into specialized fields such as lipopeptide biosurfactants.
Highlights.
A pelB leader sequence:apolipoprotein (apo) E4 N-terminal (NT) fusion was generated
Expression of recombinant fusion protein in a bioreactor led to foam production
When the foam was collapsed into a liquid foamate it was found to possess apoE4-NT
Foam-derived apoE4-NT was active in reconstituted high density lipoprotein formation
Lipid-free and lipid-associated apoE4-NT were active in cholesterol efflux assays
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R37 HL-64159) and a Nevada Center for Proteomics INBRE Award. The authors acknowledge support from the Alice and Fred Ottoboni Endowed Chair in Diet and Disease Prevention (ROR).
Abbreviations:
- Apo
apolipoprotein
- NT
N-terminal
- rHDL
reconstituted high density lipoprotein
- LDL
low density lipoprotein
- DMPC
dimyristoylphosphatidylcholine
- ABC
ATP binding cassette
- IPTG
isopropyl β-D-thiogalactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- 1.Phillips MC (2014). Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life, 66, 616–623. [DOI] [PubMed] [Google Scholar]
- 2.Weisgraber KH (1994) Apolipoprotein E: structure-function relationships. Adv. Protein Chem 45, 249–302. [DOI] [PubMed] [Google Scholar]
- 3.Huebbe P, Rimbach G (2017). Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res. Rev 37, 146–161. [DOI] [PubMed] [Google Scholar]
- 4.Amaro AB, Curco AS, Garcia E, Julve J, Rives J, Benitez S, Cortes VL (2021) Apolipoprotein and LRP1-based peptides as new therapeutic tools in atherosclerosis. J Clin Med 10(16), 3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser PS, Ryan RO (2013) Impact of apolipoprotein E on Alzheimer’s disease. Curr Alzheimer Res 10, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser PS, Narayanaswami V, Ryan RO (2011) Apolipoprotein E: from lipid transport to neurobiology. Prog. Lipid Res 50, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu CC, Kanekiyo T, Xu H, Bu G (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms, and therapy. Nat Rev Neurol, 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumsden AL, Mulugeta A, Zhou A, Hypponen E (2020). Apolipoprotein E (APOE) genotype-associated disease risks: a phenome-wide, registry-based, casecontrol study utilizing the UK Biobank. Ebiomed 59, 102954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao N, Liu CC, Qiao W, Bu G. (2018) Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol. Psychiatry, 83, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD (1993) Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA 90, 9649–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K (1991). Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res 541:163–6. [DOI] [PubMed] [Google Scholar]
- 12.Dong LM, Weisgraber KH (1996) Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem 271, 19053–19057. [DOI] [PubMed] [Google Scholar]
- 13.Ryan RO (2010) Nanobiotechnology applications of reconstituted high density lipoprotein. J. Nanobiotechnology 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tricoci P, D’Andrea DM, Gurbel PA, Yao Z, Cuchel M, Winston B (2015) Infusion of reconstituted high-density lipoprotein, CSL112, in patients with atherosclerosis: safety and pharmacokinetic results from a phase 2a randomized clinical trial. J. Am. Heart Assoc 4, e002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox CA, Moschetti A, Ryan RO (2021). Reconstituted HDL as a therapeutic delivery device. BBA Mol. Cell Biol. Lipids 1866, 159025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, Coleman MA, Henderson PT, Hinz AK, Kuhn EA, Pesavento JB, Segelke BW, Sulchek TA, Tarasow T, Walsworth VL, Hoeprich PD (2007) Different apolipoproteins impact nanolipoprotein particle formation. J. Am. Chem. Soc 129, 14348–54. [DOI] [PubMed] [Google Scholar]
- 17.Blanchette CD, Law R, Benner WH, Pesavento JB, Cappuccio JA, Walsworth V, Kuhn EA, Corzett M, Chromy BA, Segelke BW, Coleman MA, Bench G, Hoeprich PD, Sulchek TA (2008) Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. J Lipid Res 49, 1420–1430. [DOI] [PubMed] [Google Scholar]
- 18.Katzen F, Fletcher JE, Yang JP, Kang D, Peterson TC, Cappuccio JA, Blanchette CD, Sulchek T, Chromy BA, Hoeprich PD, Coleman MA, Kudlicki W (2008) Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. J Proteome Res 7, 3535–3542. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh M, Ryan RO (2014) ApoE enhances nanodisk-mediated curcumin delivery to glioblastoma multiforme cells. Nanomed. (Lond) 9:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, Hasegawa M, Yoshimoto N, Hoshikawa K, Mukai T (2019) Preparation of lipid nanodisks containing apolipoprotein E-derived synthetic peptides for biocompatible delivery vehicles targeting low-density lipoprotein receptor. Biol. Pharm. Bull 42, 1376–1383. [DOI] [PubMed] [Google Scholar]
- 21.Bello OD, Auclair SM, Rothman JE, Krishnakumar SS (2016) Using apoE nanolipoprotein particles to analyze SNARE-induced fusion pores. Langmuir 32, 3015–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang ST, Cruz S, Narayanaswami V (2020) Reconfiguring nature’s cholesterol accepting lipoproteins as nanoparticle platforms for transport and delivery of therapeutic and imaging agents. Nanomaterials (Basel) 10, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox CA, Ryan RO (2022). Studies of the cardiolipin interactome. Prog. Lipid Res 88, 101195. [DOI] [PubMed] [Google Scholar]
- 24.Hagn F, Etzkorn M, Raschle T, Wagner G (2013) Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc 135, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher CA, Wang J, Francis GA, Sykes BD, Kay CM, Ryan RO (1997). Bacterial overexpression, isotope enrichment, and NMR analysis of the N-terminal domain of human apolipoprotein E. Biochem. Cell Biol 75, 45–53. [PubMed] [Google Scholar]
- 26.Kritharides L, Christian A, Stoudt G, Morel D, Rothblat GH (1998) Cholesterol metabolism and efflux in human THP-1 macrophages. ATV 18(10), 1589–1599. [DOI] [PubMed] [Google Scholar]
- 27.Ryan RO, Schieve D, Wientzek M, Narayanaswami V, Oikawa K, Kay CM, Agellon LB. (1995). Bacterial expression and site-directed mutagenesis of a functional recombinant apolipoprotein. J Lipid Res, 36, 1066–1072. [PubMed] [Google Scholar]
- 28.Weers PM, Narayanaswami V, Ryan RO (2001) Modulation of the lipid binding properties of the N-terminal domain of human apolipoprotein E3. Eur J Biochem 268, 3728–3735. [DOI] [PubMed] [Google Scholar]
- 29.Yvan-Charvet L, Wang N, Tall AR (2010) Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol 30, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai JD, Banat IM, (1997) Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev 61, 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakur P, Saini NK, Thakur VK, Gupta VK, Saini RV, Saini AK (2021) Rhamnolipid the glycolipid biosurfactant: emerging trends and promising strategies in the field of biotechnology and biomedicine. Microb Cell Fact 20, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mnif I, Ghribi D (2015) Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers 104(3), 129–47. [DOI] [PubMed] [Google Scholar]
- 33.Castro GR, Panilaitis B, Kaplan DL (2008) Emulsan, a tailorable biopolymer for controlled release. Bioresour. Technol 99, 4566–4571. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Lu Y, Shan M, Zhao H, Lu Z (2022) A mini-review: mechanism of antimicrobial action and application of surfactin. Wold J Microbiol Biotechnol 38, 143. [DOI] [PubMed] [Google Scholar]
- 35.Cho WY, Ng JF, Yap WH, Goh BH (2022) Sophorolipids-bio-based antimicrobial formulating agents for applications in food and health. Molecules 27, 5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christopher FC, Ponnusamy SK, Ganesan JJ, Ramamurthy R (2019) Investigating the prospects of bacterial biosurfactants for metal nanoparticle synthesis- a comprehensive review. IET Nanobiotechnol 13, 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handa S, Aggarwal Y, Puri S, Chatterjee M (2022) Pharmaceutical prospects of biosurfactants produced from fungal species. J. Basic Microbiol 62, 1307–1318. [DOI] [PubMed] [Google Scholar]
- 38.Lethcoe K, Fox CA, Ryan RO (2022). Foam fractionation of a recombinant biosurfactant apolipoprotein. J Biotechnol 343, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundstedt E, Kahne D, Ruiz N (2021). Assembly and maintenance of lipids at the bacterial outer membrane. Chem Rev, 121(9), 5098–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher CA, Ryan RO (1999) Lipid binding-induced conformational changes in the N-terminal domain of human apolipoprotein E. J. Lipid Res, 40, 93–99. [PubMed] [Google Scholar]
- 41.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA (1991) Threedimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252, 1817–1822. [DOI] [PubMed] [Google Scholar]
- 42.Fisher CA, Narayanaswami V, Ryan RO (2000) The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E. J. Biol. Chem 275, 33601–33606. [DOI] [PubMed] [Google Scholar]
- 43.Chen M, Cook KD (2007) Oxidation artifacts in the electrospray mass spectrometry of Abeta Peptide. Anal Chem 79, 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox CA, Moschetti A, Ryan RO (2021) Reconstituted HDL as a therapeutic delivery devise. BBA Mol. Cell Biol. Lipids 1866, 159025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato Y, Kinami Y, Hashiba K, Harashima H (2020) Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J Control Release, 322, 217–226. [DOI] [PubMed] [Google Scholar]


