Abstract
With the exponential discovery of new inborn errors of immunity (IEI), it is becoming increasing difficult to differentiate between a number of the more recently defined disorders. This is compounded by the fact that while IEI primarily present with immunodeficiency, the spectrum of disease is broad and often extends to features typical of autoimmunity, autoinflammation, atopic disease, and/or malignancy. Here we use case studies to discuss the laboratory and genetic tests utilized that ultimately led to the specific diagnoses.
Keywords: diagnosis of inborn errors of immunity, functional testing, genetic testing, flow cytometry, laboratory immunology
Introduction
In this manuscript, brief histories of individual patients currently followed (by AFF) are presented and linked to a discussion of the most appropriate laboratory tests based on the clinical phenotype in order to develop the appropriate diagnosis. Owing to the fact that there are now over 485 genetic defects linked to inborn errors of immunity (IEI) (1) (IEI, also referred to as primary immunodeficiencies/immune dysregulation syndromes), it is obviously not possible to cover all known diseases. Through select cases, we will focus primarily on disorders impacting antibody production, T cell function, T cell interaction with monocytes/macrophages, and neutrophil function to provide general guidance for an immunologic evaluation in the setting of different recurrent/chronic infections and/or immune dysregulation/autoimmunity. We will present assays that are generally available in medical centers with clinical immunologists caring for patients with IEIs as well as major commercial laboratories (2, 3). The focus is on tests that evaluate specific aspects of immune function, but reference to genetic testing is necessary particularly in the context of the specific cases presented.
Cases presenting with recurrent sinopulmonary infections:
Case 1.
A 43-year-old man presented for evaluation of persistent cellulitis affecting the lower extremity. Starting at 7 months of age, he developed recurrent sinopulmonary infections affecting the respiratory tract and middle ear. The ear infections led to recurrent mastoiditis requiring several surgeries. He started immune globulin replacement therapy (IgRT) in early childhood resulting in decreased sinopulmonary infections and improved overall health. Blood cultures at the time of the most recent presentation were positive for Campylobacter jejuni, a bacterial species that can be associated with chronic cellulitis in patients with congenital agammaglobulinemia, the diagnosis he had carried since childhood.
Case 2.
A 70-year-old woman presented for bronchiectasis related to recurrent pulmonary infections. In childhood, the patient had repeated diagnoses of viral respiratory tract infections, that did not require hospitalizations. In her 40s, she developed frequent sinusitis and pneumonias that responded to antibiotic therapy but were also often associated with recurrence. In her mid-50s, she underwent an immune evaluation that showed low total serum immunoglobulins with undetectable antibodies to diphtheria toxoid, tetanus toxoid and pneumococcal polysaccharide antigens. Furthermore, vaccination with diphtheria and tetanus toxoid as well as the polyvalent pneumococcal polysaccharide vaccine induced no response. Chest CT imaging at that time showed bronchiectasis and pulmonary function tests demonstrated decreased FEV1 and DLCO. She was started on IgRT resulting in a decrease in the frequency of infections.
In both cases, the clinical story strongly suggests a problem with antibody production manifesting as increased susceptibility to high grade encapsulated bacteria (e.g. Haemophilus influenzae, Streptococcus pneumoniae) involving primarily the sinopulmonary tract. The history typically provides evidence that these infections are responsive to antibiotics, but successful treatment often requires a more protracted course and/or parenteral administration of the antibiotic. It is important to distinguish those patients with recurrent Infections in infancy (typically starting after six months of age when maternally transferred IgG is waning) versus those presenting later in childhood or adulthood.
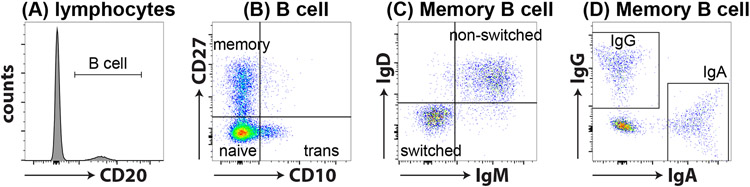
The first step in evaluating either case presented should focus on a basic screening test by obtaining serum immunoglobulin levels (i.e. IgG, IgA, IgM, IgE). This revealed agammaglobulinemia in the first case and as noted, low serum immunoglobulins (IgG 117 mg/dL, IgA 22 mg/dL, IgM <13 mg/dL, and IgE <2 IU/ml) in the second case. It is critical that these results are compared to the age-based control values (generally provided by the performing lab) since immunoglobulin production undergoes maturation throughout childhood with adult levels not achieved until adolescence. The finding of low circulating immunoglobulins in either case could be evaluated for possible protein loss as the explanation that would be associated with a markedly decreased serum albumin level. Importantly, a convincing clinical infection history in the setting of decreased immunoglobulins should be further evaluated by assessing in vivo antibody responses. This should include response to both protein antigen vaccines and carbohydrate antigen vaccines by assessing pre- and 4-week post-vaccine antibody levels looking for protective levels and/or a 4-fold increase (2-fold increase for carbohydrate antigens) in levels following immunization (4). In the first case, vaccine challenge would not be necessary due to the early clinical presentation along with the essential absence of serum immunoglobulins while in the second case vaccine responses were evaluated and found to be absent in response to both protein antigens (tetanus and diphtheria toxoid) and carbohydrate antigens (polyvalent pneumococcal vaccine [Pneumovax]). At this stage in the evaluation there was clear evidence for a humoral immune defect and the next step should focus on characterizing circulating B cells and B cell subsets by flow cytometry. The initial objective is to establish whether or not B cells are present in order to distinguish between congenital defects that block normal B cell development (e.g. X-lined agammaglobulinemia [XLA]) (5, 6) and functional B cell defects that impact on the capacity of B cells to make an antigen specific response (e.g. common variable immune deficiency [CVID]) (7). In the first case, the finding that circulating CD19+ and/or CD20+ B cells were absent associated with agammaglobulinemia along with the clinical history suggests congenital agammaglobulinemia with the most common cause being XLA, a diagnosis established in this patient via genetic evaluation of the gene encoding BTK (c.1559 G>A). In the second case, the B cells were present but low and this finding was followed by evaluating additional B cell subset focusing on memory B cells to include non-switched (CD20+/CD27−/sIgM+/sIgD+) and switched memory B cells (CD20+/CD27+/sIgM−/sIgD−) (Figure 1; Table 1) (8). These studies revealed that this patient had very low switched memory B cells, a finding when combined with the clinical history and the other immunologic studies is consistent with common variable immune deficiency (CVID). These results must be compared to age matched control data since memory B cells change as the immune system matures and is exposed to antigenic challenge (4). There are additional B cell subsets (i.e. transitional B cells, CD21low atypical B cells) that can be evaluated (Figure 1; Table 1). Finally, there are phenotypic classifications of CVID into subtypes based on the Freiburg (9) and EUROClass (10) criteria, but the interpretation of these results generally would be best served by consulting directly with a clinical immunologist.
Figure 1: B cell subsets.
B cell subsets can be easily identified through flow cytometry. (A) Within lymphocytes, B cells are identified as CD20+ cells. (B) CD20+ B cells can be further divided into transitional (CD10+CD27−; trans), naïve (CD10−CD27−) and memory (CD10−CD27+) subsets using CD10 and CD27. (C) Memory B cells can be further subdivided as Ig switched (IgD−IgM−) or non-switched (IgD+IgM+) B cells (D) a proportion of IgD−IgM− Ig switched cells express IgG or IgA.
Table 1:
Lymphocyte subsets
| Cell type | Phenotypic marker(s) |
|---|---|
| B cells | |
| Total B cells | CD20+ |
| Transitional B cells | CD20+CD10+CD27− |
| Naïve B cells | CD20+CD10−CD27− |
| Memory B cells | CD20+CD10−CD27+ |
| Unswitched B cells | CD20+IgD+IgM+ |
| Switched B cells | CD20+IgD−IgM− |
| Switched IgG+ B cells | CD20+IgD−IgM−IgG+ |
| Switched IgA+ B cells | CD20+IgD−IgM−IgA+ |
| Atypical/CD21low B cells | CD20+CD9hiCD21low |
| T cells | |
| Total T cells | CD3+ |
| CD4+ helper T (Th) cell | CD3+CD4+CD8− |
| CD8+ cytotoxic T cell | CD3+CD4−CD8+ |
| Naïve T cell | CCR7+CD45RA+ |
| Central memory T cell | CCR7+CD45RA− |
| Effector memory T cell | CCR7−CD45RA− |
| Revertant memory T cell | CCR7−CD45RA+ |
| Regulatory T cells | CD3+CD4+CD25highCD127low |
| T follicular helper cell | CD3+CD4+ CD45RA−CXCR5+ |
| Th1 | CD3+CD4+ CD45RA−CXCR3+CCR6− |
| Th17 | CD3+CD4+ CD45RA−CXCR3−CCR6+ |
| Unconventional T cells | |
| NK T cells | CD3+TCRVα24JαQ+ |
| γδ T cells | CD3+TCRVαβ−TCRVγδ+ |
| MAIT cells | CD3+CD161+TCRVα7.2+ |
| Other lymphocytes | |
| NK cells | CD3−CD56+ |
CVID patients with recurrent infections plus other clinical problems/complications have significantly higher morbidity and mortality compared to CVID patients who only have recurrent infections (11). Furthermore, genetic testing has become more common for patients with antibody defects, as the presence of monogenetic immune defects has been demonstrated in 30-35% of CVID patients (12). Genetic testing can be done via immunodeficiency panels often consisting of >200 genes known to be associated with IEIs, whole exome sequencing (WES) or whole genome sequencing (WGS), the latter of which is necessary to reliably identify mutations in intronic and non-coding regions (13). Genetic evaluation may also include samples from other affected family members as well as samples from the patient’s parents. But even without inclusion of family members, a defect may be found; interestingly, more than 20% of more recently identified genes identified to cause IEI have been from single-patient studies (14). Identification of defined genetic defects associated with a CVID-like presentation has continued to increase and allowed identification of novel targeted therapies that can provide additional clinical benefit for some of these patients.
Monogenic disorders presenting with a CVID-like disorder
Case 3.
A 25-year-old man presented for evaluation of immune deficiency in the setting of arthritis and persistent diarrhea. The patient developed diarrhea in early childhood with watery stools from 3-12 times daily. He has had repeated intestinal Campylobacter infection with poor response to antibiotic therapy and malabsorption that led to significant weight loss. His medical history also included eczema staring early in childhood, one hospitalization for pneumonia, recurrent cold sores, a persistent wart, hypothyroidism, and pernicious anemia. Family history was significant for his mother dying in her 20s with pernicious anemia, hypothyroidism, diarrhea, and cough. Laboratory findings were significant for low serum IgG, IgA, and IgM (250 mg/dL, 15 mg/dL, 7 mg/dL respectively) as well as T and B lymphopenia. The finding of hematologic disease, endocrinopathy and GI disease together with low immunoglobulins suggested that genetic testing was indicated, and results revealed a pathogenic heterozygous missense variant in CTLA4 (15).
Although routine lab tests are not available that enable a reliable diagnosis of CTLA4 haploinsufficiency, flow cytometry has proven useful as supporting evidence for this genetic diagnosis. Specifically, CTLA4 haploinsufficiency is typically associated with low CD4+ T cells, in particular naïve (CD45RA+CD62L+ and/or CD45RA+CCR7+) CD4+ T cells (Figure 2; Table 1) and increased expression of the exhaustion marker PD-1 (Figure 2; Table 1), indicative of hyperproliferation (15, 16). T regulatory cells (Tregs) in healthy individuals are CD4+CD25highCD127low (Figure 3, Table 1), but a large proportion of Tregs in CTLA4 haploinsufficiency patients are CD25− and functionally defective in suppressing the proliferation of cocultured activated autologous or allogenic T cells (15). With respect to the B cell compartment, CTLA4 haploinsufficiency results in a decrease in CD27+ switched memory B cells (Figure 1; Table 1) and a significant elevation in CD21low B cells (Table 1), which often increase over time.
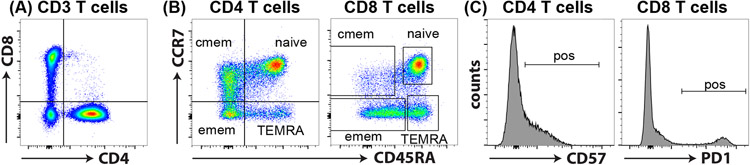
Figure 2: T cell subsets.
(A) CD3+ T cells can be divided into CD4+ helper and CD8+ cytotoxic T cells. (B) CD45RA and CCR7 in combination are used to identify naïve (CD45RA+CCR7+), central memory (CD45RA−CCR7+; cmem), effector memory (CD45RA−CCR7−; emem) and revertant memory (CD45RA+CCR7−; TEMRA) subsets. (C) These subsets represent different stages of T cell differentiation and express varying levels of activation markers such as CD57 and PD1. In general, these markers are absent on naïve T cells and upregulated in response to T cell activation.
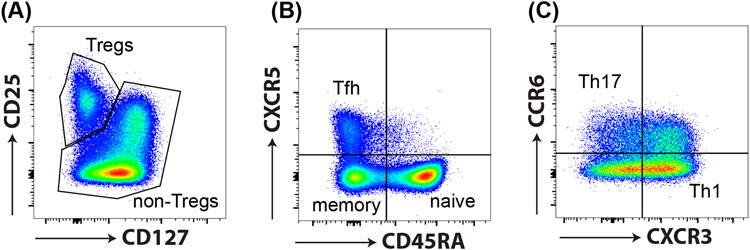
Figure 3: CD4+ T helper cell subsets.
CD4+ T helper cells can be identified by various cell surface markers and chemokine receptors. (A) Tregs have a CD25hiCD127low phenotype, and (B) T follicular helper cells (CXCR5+CD45RA−; Tfh), (C) Th1 cells (CXCR3+CCR6−), and Th17 (CXCR3−CCR6+) can be identified via expression of the chemokine receptors CXCR5, CXCR3 and CCR6, respectively.
Case 4.
A 36-year-old man reported recurrent sinopulmonary and ear infections starting in early childhood. At 13 years of age, he developed Evan’s syndrome (autoimmune anemia and thrombocytopenia) ultimately requiring splenectomy. Hypogammaglobulinemia was found leading to initiation of IgRT. In early adulthood he had recurrent respiratory tract infections and was diagnosed with interstitial lung disease. Chronic diarrhea developed with autoimmune enteropathy diagnosed based on villous blunting and intraepithelial lymphocytes found on biopsy plus chronic norovirus infection was also present. Lymphocyte phenotyping revealed low naïve CD4+ and CD8+ T cells however these findings did not yield a specific diagnosis and genetic testing ordered due to the findings of autoimmune cytopenia and enteropathy revealed a pathogenic variant in STAT3 causing STAT3 gain of function (GOF) disease (17-20).
Unless the genetic finding is a previously published STAT3 variant known to be associated with GOF or a hyper IgE (dominant negative [DN]) phenotype), the mutation alone does not establish the diagnosis. However, the clinical histories are distinct between GOF and DN disease, and further immunologic testing can point towards the functional impact of the variant. Interestingly, none of the reported STAT3 GOF mutations result in increased expression of STAT3 unlike STAT1 GOF where this is the characteristic finding. Instead, STAT3 GOF results in enhanced basal STAT3 activity, increased cytokine dependent STAT3 activation, stability of activated STAT3 homodimers, increased affinity of STAT3 to DNA binding targets or impaired/delayed STAT3 dephosphorylation (17-20). In terms of cellular phenotype detected by flow cytometry, the majority of STAT3 GOF patients have an increase in T cell receptor (TCR) αβ+ CD4−CD8− (double negative) T cells, a finding reminiscent of the αβ+ CD4−CD8− double negative T cells observed in patients with autoimmune lymphoproliferative syndrome (ALPS, due to defects in the FAS pathway) (21, 22). A proportion of STAT3 GOF patients have increased circulating CXCR5+ T follicular helper (Tfh) like cells and ~40% have a decrease in Tregs, in particular CD25 expression (17-20, 23) (Figure 3, Table 1). Patients usually display CD4 T cell and B cell lymphopenia, and hypogammaglobulinemia (19). Within the B cell compartment, STAT3 GOF patients typically have an expansion in CD21low B cells (18, 24).
Monogenic Disorders with Infection Susceptibility and Atopy
Patients may present with an initial infectious history involving recurrent sinopulmonary infections, but also demonstrate markedly elevated IgE, eosinophilia, and atopy as well as other infectious and/or somatic findings as exemplified by the following case vignettes (25, 26).
Case 5.
15-year-old boy presented at 11 days of age with a rash, followed by recurrent pneumonias starting in infancy. A pneumatocele formed in the right upper lobe, which became infected and ultimately required resection with a post-op course complicated by a prolonged requirement for a chest tube due to an air leak. Additional history revealed significant eczema early in childhood, at least 4 episodes of cutaneous abscesses requiring antibiotic therapy and the development of recurrent thrush. In addition, the patient had a spontaneous bowel perforation requiring surgical repair. Other identified medical issues included hyperextensible joints, three minimal trauma bone fractures, scoliosis requiring rod placement, and retention of primary teeth. Laboratory findings were significant for an elevated serum IgE of 7651 IU/ml, eosinophilia in the 1000s, and poor antibody response to vaccination. The clinical history was not consistent with CVID and the additional clinical findings made the clinical picture strongly suggestive of Job’s Syndrome (autosomal dominant hyper IgE syndrome resulting from a DN defect in STAT3) (27, 28). Genetic testing revealed a previously demonstrated pathogenic heterozygous defect in STAT3 (1144>T) establishing the suspected diagnosis.
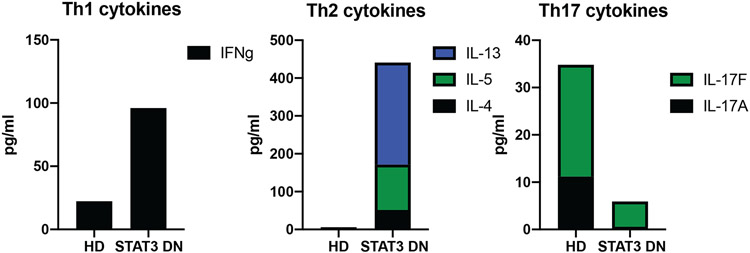
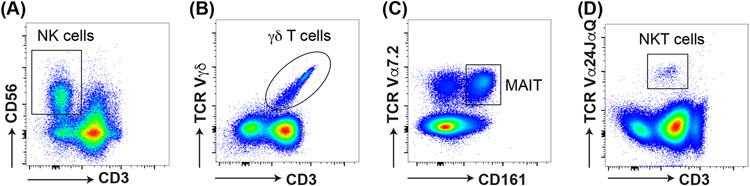
Flow cytometry and other laboratory tests can be informative in clarifying STAT3 DN disease. While CD4+ and CD8+ T cells are usually present in normal frequencies, there is skewing towards CD45RA+CCR7+ naïve T cells at the expense of CD45RA− memory T cells. Further investigation into the CD4+ T cell compartment for T helper cell subsets, often reveals a decrease in Th17 (CD4+CCR6+) cells (Figure 3; Table 1)(29, 30). This is supported by in vitro culture of memory CD4+ T cells from STAT3 DN patients compared to normal healthy donors, revealing decrease in Th17 cytokines (IL7A, IL-17F, IL-22) (Figure 4). In addition, there is often a decrease in CD4+CXCR5+ Tfh cells with the production of the Tfh cytokine, IL-21, as well as an increase in Th1 (IFNγ) and increase in Th2 (IL-4, IL-5, IL-13) cytokines both by intracellular flow or secretion into culture supernatants (29-32) (Figure 4). In terms of the B cell compartment, again STAT3 DN individuals have normal B cell frequencies, but these B cells tend to be more immature as highlighted by an increase in transitional/naive and concurrent reduction in memory B cells (33). Interestingly, despite the severe decrease in memory B cells in STAT3 DN patients, the memory B cells that are present have undergone normal Ig isotype switching albeit with a slight preference towards IgG and away from IgA (33). STAT3 DN defects also affect innate and innate-like lymphocytes with normal NK and γδ T cells but decreased NKT and mucosal-associated invariant T (MAIT) cells (34) (Figure 5; Table 1). Taken together these features provide a relatively distinct lymphocyte phenotype for STAT3 DN patients compared to healthy controls. This lymphocyte signature can aid in the identification of potential STAT3 DN patients and provides valuable insights into disease susceptibility and a cellular/molecular explanation for the infectious and atopic history of STAT3 deficiency. For instance, CD4+CCR6+ Th17 cells (Figure 3; Table 1) have been implicated in protective immunity against bacterial and fungal infections and a severe reduction in these cells in STAT3 DN patients may explain their extreme susceptibility to Candida albicans and Staphylococcus aureus (20, 29). Furthermore, the significant reductions in memory B cells and Tfh cells in the absence of intact STAT3 signalling, are likely to account for the humoral defects presented by STAT3 DN patients. Nevertheless, genetic testing is key and absolutely required for a STAT3 DN diagnosis, as provided in this case.
Figure 4: Cytokine secretion by memory CD4+ T cells.
CD4+ helper T cells from healthy controls secrete cytokines characteristic of different T helper cell subsets. This can be examined in vitro by activating memory CD4+ T cells with stimuli such as anti-CD3, anti-CD28 and anti-CD2 for 5 days and looking at cytokine secretion into the culture supernatant by cytometric bead arrays (BD Biosciences) or ELISA. Compared to normal healthy donors (HD), memory CD4+ T cells from STAT3 DN patients have an increase in Th1 cytokines (IFNγ), increase in TH2 cytokines (IL-4, IL-5, IL-13) and decrease in Th17 cytokines (IL-17A, IL-17F).
Figure 5: identification of Innate-like lymphocytes.
Flow cytometry can be used to identify innate-like lymphocytes in the blood. This includes (A) CD3−CD56+ NK cells, (B) CD3+Vγδ+ γδ T cells, (C) CD3+CD161+Vα7.2+ mucosal associated invariant T (MAIT) cells, and (D) CD3+Vα24JαQ+ NKT cells.
Case 6.
A 9-year-old girl presented with persistent widespread molluscum lesions. She had a history of severe eczema as a toddler that persisted and was complicated by occasional Staphylococcal infections. At age 7 years she developed molluscum lesions that rapidly became widespread and recalcitrant to therapy. In addition, she had recurrent ear and sinus infections requiring frequent antibiotics as well as mild asthma. Immunologic evaluation demonstrated an IgE of > 10,000 IU/ml, eosinophilia (>1000 cells/mm3), poor antibody responses despite a normal IgG, low serum IgM (17 mg/dL), mild T lymphopenia and low memory B cells. The combination of recurrent infections, elevated IgE, eosinophilia, low IgM, severe cutaneous viral infections, and atopy strongly suggested a defect in DOCK8.
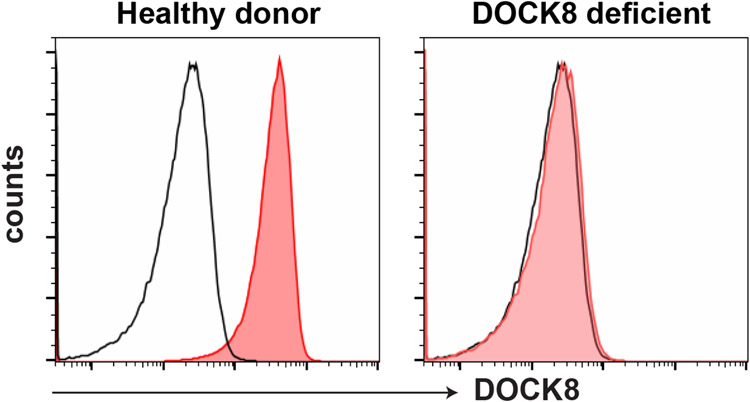
While there is not a standard laboratory test to establish the diagnosis of DOCK8 deficiency, several laboratories have established protocols for evaluating DOCK8 expression in human PBMCs (35-40) (Figure 6). Furthermore, the lymphocyte compartment of DOCK8-deficent patients display a distinct phenotype characterized by decreased CD4, but typically normal CD8 T cell frequencies, resulting in an inverted CD4:CD8 T cell ratio compared to healthy controls. In addition to this, there is a decrease in naïve and increase in effector and CD45RA+ revertant memory CD4 and CD8 T cells (Figure 2). The memory T cell compartment displays exhaustion as indicated by decrease in CD27, CD28 and CD127 and increase in CD57, CD95 and PD1 expression (36, 39-41) (Figure 2; Table 1). The B cell compartment in DOCK8-deficient patients contain normal frequencies of B cells but an overall increase in naïve and decrease in memory B cells. NKT and MAIT cells are also decreased, NK cells are normal, while γδ T cell frequencies are increased (36, 40) (Figure 5; Table 1). Functionally, DOCK8 deficiency impairs the activation and proliferation of T cells, which has been replicated by in vitro proliferation assays (41-43). In addition, DOCK8-deficient memory CD4+ T cells are skewed towards Th2 cytokine producing cells (IL-4, IL-13, IL-5) at the expense of both Th1 and Th17 cells (36, 39, 40, 44). Nevertheless, despite this characteristic phenotype revealed by flow cytometry and in vitro functional assays, the gold standard in diagnosis is still genetic testing which in this case revealed compound heterozygous deletions in the gene encoding DOCK8.
Figure 6: DOCK8 expression.
Intracellular DOCK8 expression is routinely performed in research labs around the world. PBMCs are fixed and then strained with fluorochrome conjugated DOCK8 antibody or an unconjugated DOCK8 antibody followed by a secondary fluorochrome conjugated antibody in the presence of a permeabilization agent such as saponin. Lymphocytes from DOCK8-deficient patients do not express DOCK8 protein. Red histogram: DOCK8 antibody; black histogram: isotype control antibody.
Combined Immune Deficiencies
Case 7.
A 21-year-old male patient presented as a toddler with recurrent ear and sinus infections. He was diagnosed with multiple infections by the age of 2 years. At age 6 years he developed disseminated Histoplasmosis, which recurred after stopping antifungal therapy. Other infections included two episodes of clinically diagnosed Varicella and a cutaneous abscess requiring intravenous antibiotics. His maternal half-brother had similar recurrent bacterial infections starting in infancy. The story of this patient clearly suggests a defective antibody response but the history is also consistent with clinically significant T cell dysfunction (i.e. disseminated histoplasma) and a likely X-linked recessive pattern of inheritance, such that the evaluation of this patient should also include studies focused on the T cell compartment.
When patients develop recurrent or chronic infections with viruses, fungi or other opportunistic microbes (e.g. P. jirovecci), it strongly suggests an underlying defect affecting T cell immunity. The most severe form of T cell deficiency is associated with severe combined immunodeficiency (SCID) (45). In countries such as the United States, the UK and Australia this category of IEI is routinely screened for using the T cell receptor excision circle (TREC) assay that is performed on the standard newborn blood spot (i.e. Guthrie card) obtained during the immediate newborn period. Consequently, an abnormal TREC screen is followed up by an evaluation using standard testing approaches in order to establish this diagnosis during infancy enableing early prophylactic measures as well as moving directly to curative immune reconstitution prior to the development of serious infectious problems and failure to thrive. An abnormal TREC screening assay also identifies a small number of infants who do not have SCID, but is associated with complete DiGeorge syndrome, ataxia telangiectasis, marked prematurity, delayed T cell maturation, as well as certain other congenital disorders (46).
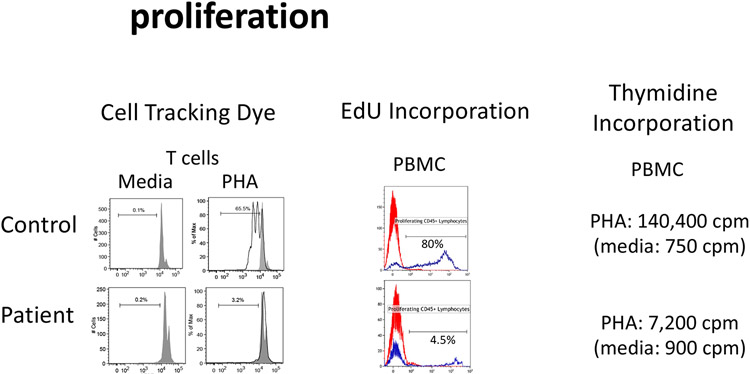
TREC screening fails to identify most disorders that do not fulfill the criteria for SCID leaving a number of monogenic defects impacting cellular immunity to be evaluated following their clinical presentation. The approach to evaluating this group of patients typically includes evaluating an absolute lymphocyte count as T cells represent 65-80% of all circulating lymphocytes such that any defect that impacts T cell development or increases T cell destruction/loss will likely present with lymphopenia. As with most immune tests it is important that these results are compared to age specific control values. Following (or simultaneous to) the lymphocyte count, a general assessment of circulating T cells including evaluating CD3+/CD4+ and CD3+/CD8+ T cells enables assessing for the possibility that a major T cell subset is absent (Table 1), a finding that can define specific disorders (e.g. defects in ZAP70, MHC class I, MHC class II) (47-49). The presence or absence of naïve and memory CD3+/CD4+ and CD3+/CD8+ T cells (Figure 2; Table 1) can provide further insight into disorders associated with T cell dysfunction. In addition, evaluation for the expression of surface and/or intracellular proteins can be performed to screen for specific defects suggested by the clinical history. The assessment of specific T cell subsets and specific cell proteins is usually followed by assessing T cell function based on assays of in vitro T cell proliferation. This typically focuses on T cell response to non-specific stimuli using mitogens (e.g. phytohemagglutinin [PHA], concanavalin A [Con A]) and/or monoclonal antibodies that bind to the TCR (e.g. anti-CD3) with or without a costimulatory signal (e.g. anti-CD28) with or without IL-2. An alternative approach that involves direct T cell stimulation not involving TCR engagement can be performed using a calcium ionophore (ionomycin) and phorbol myristate acetate (PMA). Each of these various assays involve culturing lymphocytes and assessing proliferation by uptake of tritiated thymidine or evaluation by flow cytometry using either a cell tracking dye (such as carboxyfluorescein succinimidyl ester [CFSE], cell trace violet) (50) or a flow evaluation that is analogous to tritiated thymidine using 5-ethynyl-2'-deoxyuridine (EdU) (Figure 7). Lymphocyte function can also be evaluated using recall antigens (e.g tetanus toxoid) that requires a longer culture period (6-7 days) before assessing proliferation. The difficulty with the latter approach is not knowing the status of prior exposure to the specific antigen since proliferation only occurs as a recall response. Additional assays involve evaluating cytokine production (i.e. intracellular cytokine evaluation) or cytokine secretion into the culture medium following stimulation (29, 31, 32, 51, 52) with the various agents discussed above. Another functional approach to evaluating T cells depends on response to cytokines as measured by the rapid induction of phosphorylated STAT proteins in response to specific cytokines as noted in the STAT3 GOF discussion (Figure 8) (52).
Figure 7: Cell proliferation.
The ability of lymphocytes to undergo proliferation can be analyzed in vitro. Cells are labelled with cell tracking dyes such as CFSE or cell trace violet which are evenly distributed to daughter cells following each round of division (evidenced by a decrease in fluorescence). Alternatively, cells are activated in vitro followed by evaluating the uptake of tritiated thymidine or analogue of tritiated thymidine, ethynyl deoxyuridine (EdU).
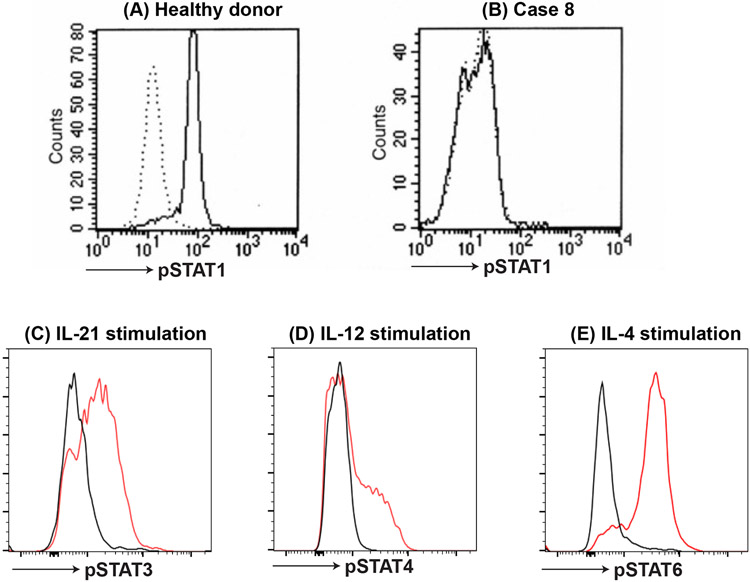
Figure 8: phosphor STAT staining.
STATs become phosphorylated following activation by specific cytokines and this can be used to evaluate different cytokine/STAT pathways. For instance, (A) pSTAT1 is phosphorylated in monocytes from healthy controls following stimulation with IFNγ. However, (B) phosphorylation of STAT1 is defective in IFNGR1 or IFNGR2 deficient patients (Case 8). Similarly, (C) pSTAT3, (D) pSTAT4 and (E) pSTAT6 can be detected following stimulation with IL-21, IL-12 or IL-4, respectively (red histogram; control without cytokine: black histogram). Intracellular pSTATs are detected by flow cytometry using monoclonal antibodies to pSTAT specific residue following fixation and permeabilization of the cells.
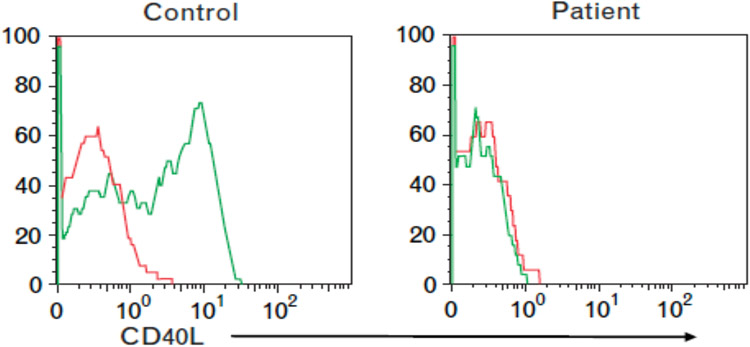
In case 7, the key finding was based on the initial measurement of serum immunoglobulins that revealed a normal serum IgM but low serum IgA and IgG. This is consistent with an abnormality in class switching associated with a defect in the expression of either CD40ligand (CD40L/CD154) or CD40 (53-55). In either of these monogenic disorders the normal interaction between CD40 expressed on B cells and CD40L expressed on activated CD4 T cells is disrupted preventing class switching from IgM to IgG, IgA, and IgE. This leads to recurrent bacterial sinopulmonary infections as well as increased susceptibility to certain intracellular pathogens and opportunists. Either genetic defect is associated with normal B cell numbers as was found in this patient but decreased numbers of class switched B cells. The easiest screening test for these defects involves evaluating for constitutive CD40 expression on B cells and CD40L (CD154) expression on activated T cells (Figure 9) (52). The latter test requires pre-activation of T cells focusing on CD4 T cells before testing for CD40L expression using a standard surface expression assay (monoclonal antibody to CD40L). In addition, because in some patients with a monogenic defect in CD40L the protein is expressed but is functionally defective, it is optimal to also perform a functional test of CD40L using fluorochrome labelled CD40-Ig (55). This paradigm is true for other proteins affected in IEIs, namely absence of protein expression is diagnostic but detection using a protein specific monoclonal antibody to identify protein expression, does not distinguish a dysfunctional from a function protein. Thus, in the former circumstance a study directed at evaluating the function of the protein would be required. In this case, genetic studies confirmed a pathogenic variant in CD40L (c.289-15 T>A) in the patient as well as in his maternal half-brother.
Figure 9: CD40L expression.
CD40L is expressed on T cells from healthy controls following activation but not by CD40L deficient patient. Green histogram: CD40L monoclonal antibody; red histogram: isotype control antibody.
Cases demonstrating defects in the interaction between monocytes/macrophages and T cells
Case 8.
A 4year-old Mennonite boy presented with disseminated Mycobacterium avium complex (MAC). The patient was well until about 15 months of age when he was hospitalized for RSV bronchiolitis, followed by repeated episodes of fever and wheezing leading to the diagnosis of pulmonary MAC with airway granulomas causing narrowing of the airways. He had initial improvement on combination antibiotics, but then had worsening fevers and abdominal distension. CT imaging showed left sided perihilar consolidation, bulky retroperitoneal adenopathy, hepatosplenomegaly, and ascites. Blood cultures were positive for Mycobacterium avium.
Case 9.
A 3 ½ year-old boy from Peru presented at 2 years of age with an eyelid lesion that spread to the periorbital space and was associated with fevers. The lesions progressed despite appropriate antibiotics with the formation of a scalp lesion which upon brain imaging showed multiple osteolytic lesions involving multiple sites of the scalp and the orbit. Biopsy of skull lesions showed AFB on microscopy and cultures positive for Mycobacteria tuberculosis complex, later confirmed as M. bovis BCG strain. Further imaging revealed multi-focal osteomyelitis with lesions of the skull, mandible, ribs, and right femur.
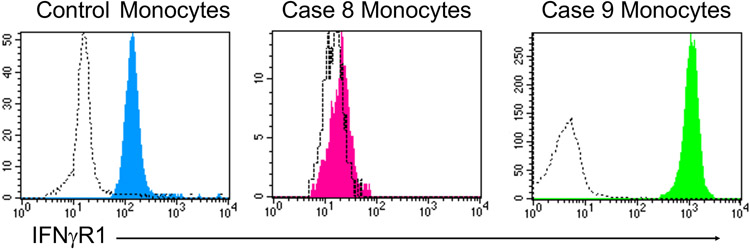
The finding of chronic and often severe non-tuberculous mycobacterial disease (environmental mycobacteria or BCG) is referred to as Mendelian susceptibility to mycobacterial disease (MSMD) and the associated defects can also result in infections due to Salmonella species, fungi (e.g. histoplasmosis, toxoplasmosis) and viruses (56). The susceptibility to disseminated BCG clarifies that immunization with this live organism vaccine is absolutely contraindicated in any patient suspected to be at risk for MSMD. The common feature of the defects in MSMD is their impact on the normal inflammatory response involving activated T cell (also NK cell) production of IFNγ with induction of IL-12 (and IL-23) production by macrophages. A critical feature of this circuit is the resultant killing of intracellular pathogens by the activated macrophages. The general approach to evaluating these patients involves screening for defective surface receptor expression on the surface of monocytes (IFNγR1, IFNγR2) or on the surface of activated T cells (IL-12Rp40, part of the IL-12 and IL-23 receptor complexes) as well as evaluation of STAT1 phosphorylation by monocytes following exposure to IFNγ ex vivo (Figure 8). Generally, this would be complemented by genetic testing focused on evaluating the multiple genes linked to MSMD (56, 57). Using this approach revealed the following findings in the two cases. Case 8 had absent IFNγR1 on the surface of circulating monocytes establishing that this patient has a defect affecting both alleles of the gene encoding IFNγR1 (homozygous defect c.672G>A) (Figure 10). This finding is associated with severe disease that often proves to be fatal without immune reconstitution via hematopoietic stem cell transplantation despite aggressive antibiotic therapy. The same study focused on IFNγR1 expression applied to case 9 demonstrated excessive expression of IFNγR1 (Figure 10) that defines the autosomal dominant defect of this protein in which the abnormal allele codes for a protein that does not combine with its partner protein (i.e. IFNγR2) resulting in the mutant protein binding IFNγ but not signaling. In addition, the mutant protein is not recycled properly resulting in increased surface expression and a decreased response to IFNγ with the typical clinical presentation of osteomyelitis involving environmental Mycobacteria. As expected, case 9 had a heterozygous pathogenic IFNγR1 variant (c.819_822delTAAT) that led to the overexpression of the mutant protein. Interestingly, phenotypically there is not much difference between the T and B cell compartment of patients with IFNGR defects compared to healthy controls with respect to naïve and memory CD4+ and CD8+ T cells and B cells (57). Furthermore, PBMCs evaluated for phospho (p)STAT1 in response to IFNγ stimulation is defective (effectively absent in case 8 and a dose shift in case 9) compared to healthy controls (Figure 8).
Figure 10: IFNGR1 expression.
IFNγR1 expression on the surface of circulating monocytes is evaluated in healthy control as well as 2 IFNGR1 deficient patients. In Case 8, the patient had defective expression due to homozygous defect c.672G>A variant. In contrast, monocytes from Case 9 had excessive expression of IFNγR1 due to the autosomal dominant defect with a pathogenic IFNγR1 variant (c.819_822delTAAT), where the abnormal allele codes for a protein that is expressed but does not combine with its partner protein IFNγR2 and is not recycled properly.
Cases of neutrophil dysfunction
Case 10.
A 39-year-old man presented in early childhood with recurrent pneumonias and lymphadenitis. At 12 years of age, he developed a Staphylococcus aureus liver abscess, which led to the diagnosis of CGD. Over the years, he continued to have intermittent bacterial and mold pneumonias. He also suffered from inflammatory bowel disease with fistula formation, requiring immune suppression. He had two affected brothers who had similar complications and died as a result of their disease.
Case 11.
A 28-year-old woman presented in infancy with peri-anal cellulitis and skin infections with ulcerations and cellulitis leading to the diagnosis of leukocyte adhesion deficiency type 1 (LAD1). Since that time, she has had repeated Gram-positive and Gram-negative bacterial skin infections, sinusitis, lymphadenitis and one pneumonia, all with minimal evidence of purulence. She had severe periodontitis/gingivitis leading to full dental extractions by 12 years of age. Her baseline white blood cells range from 10-15,000 with neutrophil predominance and the neutrophilia increased further during episodes of infection.
A history of recurrent bacterial and/or fungal infections involving the skin and deep organs (e.g. lungs, liver, bone) suggests either a numerical deficiency or a functional defect in neutrophils. Clearly, the starting point for an evaluation under this clinical scenario should involve an absolute neutrophil count to rule out neutropenia. This should be repeated at least 3-4 times each separated by 4-5 days to rule out cyclic neutropenia. If there is no evidence for neutropenia, the next step should focus on two monogenic disorders that impact neutrophil function or migration to sites of infection. These can be evaluated using flow cytometry assays that establish the diagnosis.
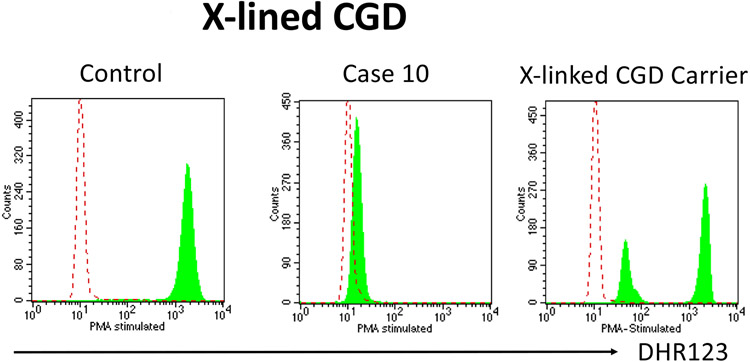
Using a flow cytometric assay that assesses oxidative burst in neutrophils (DHR flow assay) one can rule in or rule out the four most common genetic causes of chronic granulomatous disease (CGD) (58). This disorder typically involves skin, lung, and other deep-seated infections primarily involving five organisms: S. aureus, S. marcecsens, Burkholderia species, Nocardia species, Aspergillus species although there are also less common infections with other organisms observed in these patients. In the DHR flow assay, density gradient separated neutrophils are loaded with the non-fluorescent dye dihydrorhodamine 123 (DHR123) and stimulated with PMA resulting in the rapid generation of reactive oxygen species that render the dye fluorescent. Hence neutrophils from controls show approximately a two-log increase in fluorescence following PMA stimulation while neutrophils from CGD patients show no or very little increase in fluorescence (Figure 11). The actual degree of the defect in reactive oxygen species generation via the DHR assay can predict patient outcome (59). In addition, this assay can identify carriers of X-linked CGD as evidenced by the finding of two populations of neutrophils, one with normal increase in fluorescence and the second with markedly diminished fluorescence (Figure 11). This finding is the result of somatic cells in females having only one active X chromosome and this occurs randomly in early embryogenesis (a process referred to as lyonization). Recently, the proportion of normal neutrophils via the DHR assay also identifies carriers at risk for CGD-like infections in the setting where normal neutrophils were <20% (60). The clinical history of case 10 is strongly suggestive for CGD and the family history of two affected brothers is consistent with X-linked CGD based on a defect in gp91phox (CYBB). The DHR flow cytometry assay confirmed this diagnosis and demonstrated essentially no evidence of superoxide formation (Figure 11). Genetic testing confirmed a pathogenic mutation in the CYBB gene. DHR flow testing of the mother demonstrated two populations of neutrophils, one with relatively normal increase in fluorescence following PMA stimulation (active wild type X chromosome) and the other virtually no fluorescence (active mutant X chromosome) (Figure 11).
Figure 11: DHR123 expression.
Neutrophils are loaded with the non-fluorescent dye dihydrorhodamine 123 (DHR123) and stimulated with PMA. Neutrophils from normal healthy controls rapidly generate reactive oxygen species that induces a change in the dye rendering it fluorescent. In contrast neutrophils from the X-linked CGD patient do not show an increase in fluorescence and neutrophils from a X-linked CGD carrier shows two populations of neutrophils, one with normal increase in fluorescence (active wild-type X chromosome) and the second with markedly diminished fluorescence (active mutant X chromosome).
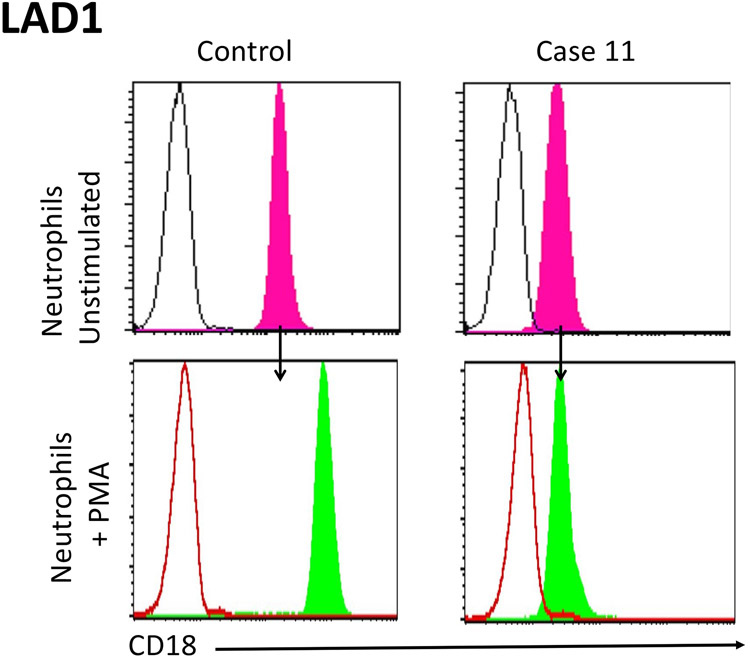
The second neutrophil defect that can routinely be established via standard laboratory studies is linked to a defect in the β2 integrins producing a defect in the normal migration of neutrophils to sites of infection (i.e. moving from the intravascular space to the infected tissues). This disorder is leukocyte adhesion deficiency type 1 (LAD1) and has unique clinical findings that include necrotic skin lesions and periodontitis. Because of the migration defect, these patients typically have a neutrophilia that increases further during times of infection meaning that the absolute neutrophil counts obtained to rule out neutropenia likely will reveal a baseline neutrophilia. The specific laboratory evaluation for LAD1 focuses on expression of CD18 on the surface of neutrophils (61). This protein normally combines with CD11a, 11b or 11c to form three different β2 integrins, absence or decreased expression of CD18 interferes with the expression of all three heterodimers. In most LAD1 patients the expression of CD18 is markedly decreased while in a minority of these patients it is absent (Figure 12). The latter individuals tend to have more severe infections early in life and immune reconstitution is typically required in their management. An additional laboratory feature that characterizes LAD1 neutrophils is failure to upregulate CD18 following neutrophil activation using PMA, a phenomenon that occurs within minutes following activation of control neutrophils (Figure 12).
Figure 12: CD18 expression.
CD18 expression is assessed on the surface of neutrophils from normal healthy control and Case 11 (LAD1-deficent patient). Testing neutrophils from the normal healthy control, CD18 expression is detected, and increases within minutes following stimulation with PMA. In contrast, CD18 expression is markedly decreased on neutrophils from Case 11 and there is no upregulation in CD18 expression following stimulation with PMA.
Case 11 was studied using flow cytometry to evaluate CD18 expression on neutrophils that was markedly decreased (Figue 12). In addition, there was no upregulation of protein expression following neutrophil activation with PMA (Figure 12). This result is sufficient to establish the diagnosis of LAD1 but further studies involving a genetic evaluation revealed a homozygous deletion in ITGB2 gene encoding integrin subunit beta 2 (CD18) supporting the diagnosis of LAD1.
Summary
The capacity to effectively screen for an IEI has become readily available via routine laboratory testing. This process is very direct in evaluating the humoral immune system based on measuring serum immunoglobulin levels followed by assessing the capacity to produce specific antibodies following immunization. These latter studies should involve the response to both protein and carbohydrate antigens. This in vivo functional evaluation should clearly identify any disorder that results in a defective antibody response and this can be followed by methods such as flow cytometry to assess for the presence or absence of B cells (i.e. the cells that differentiate into antibody producing cells), as well as CD4+ T cells such as Tfh cells which are required for intact humoral immunity (2, 3). In fact, in cases where humoral defects are present, but B cell numbers are intact, CD4+ T function should be investigated (2, 3). Defects in cellular immunity do not lend themselves to in vivo studies, hence the approach turns to ex vivo testing focused on cell identification using methods similar to those used in evaluating B cells plus a variety of ex vivo T cell functional tests. Again, this evaluation should provide clear evidence of a T cell defect if an IEI involving cellular immunity is present. The capacity to evaluate components of the innate system follow the same paradigm as noted in the cases presented including defects in the IFNγ-IL12/23 circuit associated with MSMD as well as specific neutrophil defects linked to recurrent bacterial and fungal infections. The latter disorders can be definitively diagnosed with specific tests that are routinely available through most clinical immunology laboratories. Other innate defects not covered including those involving TLR signaling and complement function also lend themselves to screening assays.
However, with the ever-increasing number of monogenic defects resulting in IEI involving the merging of immune deficiency with immune dysregulation/autoimmunity, it has become quite clear that in addition to the types of assays discussed in this paper, genetic testing has evolved as a critical approach not only as a vehicle for discovering new genetic defects but also for differentiating between disorders with overlapping clinical phenotypes. This approach will likely result in the identification of more monogenic defects resulting in IEIs and importantly, also open the door to new targeted forms of therapy. This issue requires a close connection between more basic laboratory studies to clarify the potential impact of a specific genetic defect on the function of the immune system linked to the clinical findings associated with identified and currently unidentified genetic defects resulting in IEIs.
Acknowledgements
CSM is supported by grants and fellowships from the National Health and Medical Research Council (NHMRC) of Australia (grant ID 2017463), Allergy and Immunology Foundation of Australasia (ASCIA) and American Association of Immunologists (AAI). AFF is supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (NIAID).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
References
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne K, Li W, Salomon R, Ma CS. OMIP-063: 28-Color Flow Cytometry Panel for Broad Human Immunophenotyping. Cytometry A. 2020;97(8):777–81. [DOI] [PubMed] [Google Scholar]
- 3.Ma CS, Tangye SG. Flow Cytometric-Based Analysis of Defects in Lymphocyte Differentiation and Function Due to Inborn Errors of Immunity. Front Immunol. 2019;10:2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh RA, Orange JS. Antibody deficiency testing for primary immunodeficiency: A practical review for the clinician. Ann Allergy Asthma Immunol. 2019;123(5):444–53. [DOI] [PubMed] [Google Scholar]
- 5.Cardenas-Morales M, Hernandez-Trujillo VP. Agammaglobulinemia: from X-linked to Autosomal Forms of Disease. Clin Rev Allergy Immunol. 2022;63(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conley ME, Dobbs AK, Farmer DM, Kilic S, Paris K, Grigoriadou S, et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92(1):34–48. [DOI] [PubMed] [Google Scholar]
- 8.Tangye SG, Liu YJ, Aversa G, Phillips JH, de Vries JE. Identification of functional human splenic memory B cells by expression of CD148 and CD27. J Exp Med. 1998;188(9):1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warnatz K, Denz A, Drager R, Braun M, Groth C, Wolff-Vorbeck G, et al. Severe deficiency of switched memory B cells (CD27(+)IgM(−)IgD(−)) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood. 2002;99(5):1544–51. [DOI] [PubMed] [Google Scholar]
- 10.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111(1):77–85. [DOI] [PubMed] [Google Scholar]
- 11.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119(7):1650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abolhassani H, Hammarstrom L, Cunningham-Rundles C. Current genetic landscape in common variable immune deficiency. Blood. 2020;135(9):656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucciol G, Van Nieuwenhove E, Moens L, Itan Y, Meyts I. Whole exome sequencing in inborn errors of immunity: use the power but mind the limits. Curr Opin Allergy Clin Immunol. 2017;17(6):421–30. [DOI] [PubMed] [Google Scholar]
- 14.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehn HS, Ouyang W, Lo B, Deenick EK, Niemela JE, Avery DT, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345(6204):1623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap JY, Gloss B, Batten M, Hsu P, Berglund L, Cai F, et al. Everolimus-Induced Remission of Classic Kaposi's Sarcoma Secondary to Cryptic Splicing Mediated CTLA4 Haploinsufficiency. J Clin Immunol. 2020;40(5):774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46(8):812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haapaniemi EM, Kaustio M, Rajala HL, van Adrichem AJ, Kainulainen L, Glumoff V, et al. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood. 2015;125(4):639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leiding JW, Vogel TP, Santarlas VGJ, Mhaskar R, Smith MR, Carisey A, et al. Monogenic early-onset lymphoproliferation and autoimmunity: Natural history of STAT3 gain-of-function syndrome. J Allergy Clin Immunol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125(4):591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marlies A, Udo G, Juergen B, Bernd S, Herrmann M, Haas JP. The expanded double negative T cell populations of a patient with ALPS are not clonally related to CD4+ or to CD8+ T cells. Autoimmunity. 2007;40(4):299–301. [DOI] [PubMed] [Google Scholar]
- 22.Rieux-Laucat F, Magerus-Chatinet A. Autoimmune lymphoproliferative syndrome: a multifactorial disorder. Haematologica. 2010;95(11):1805–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagle S, Heeg M, Grun S, Rensing-Ehl A, Maccari ME, Klemann C, et al. Distinct molecular response patterns of activating STAT3 mutations associate with penetrance of lymphoproliferation and autoimmunity. Clin Immunol. 2020;210:108316. [DOI] [PubMed] [Google Scholar]
- 24.Keller B, Strohmeier V, Harder I, Unger S, Payne KJ, Andrieux G, et al. The expansion of human T-bet(high)CD21(low) B cells is T cell dependent. Sci Immunol. 2021;6(64):eabh0891. [DOI] [PubMed] [Google Scholar]
- 25.Nelson RW, Geha RS, McDonald DR. Inborn Errors of the Immune System Associated With Atopy. Front Immunol. 2022;13:860821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaseghi-Shanjani M, Smith KL, Sara RJ, Modi BP, Branch A, Sharma M, et al. Inborn errors of immunity manifesting as atopic disorders. J Allergy Clin Immunol. 2021;148(5):1130–9. [DOI] [PubMed] [Google Scholar]
- 27.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–19. [DOI] [PubMed] [Google Scholar]
- 28.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–62. [DOI] [PubMed] [Google Scholar]
- 29.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119(17):3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma CS, Wong N, Rao G, Avery DT, Torpy J, Hambridge T, et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J Allergy Clin Immunol. 2015;136(4):993–1006 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207(1):155–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson RP, Ives ML, Rao G, Lau A, Payne K, Kobayashi M, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. 2015;212(6):855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pai SY, de Boer H, Massaad MJ, Chatila TA, Keles S, Jabara HH, et al. Flow cytometry diagnosis of dedicator of cytokinesis 8 (DOCK8) deficiency. J Allergy Clin Immunol. 2014;134(1):221–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillay BA, Avery DT, Smart JM, Cole T, Choo S, Chan D, et al. Hematopoietic stem cell transplant effectively rescues lymphocyte differentiation and function in DOCK8-deficient patients. JCI Insight. 2019;5(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su HC, Jing H, Angelus P, Freeman AF. Insights into immunity from clinical and basic science studies of DOCK8 immunodeficiency syndrome. Immunol Rev. 2019;287(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tangye SG, Gray PE, Pillay BA, Yap JY, Figgett WA, Reeves J, et al. Hyper-IgE Syndrome due to an Elusive Novel Intronic Homozygous Variant in DOCK8. J Clin Immunol. 2022;42(1):119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tangye SG, Pillay B, Randall KL, Avery DT, Phan TG, Gray P, et al. Dedicator of cytokinesis 8-deficient CD4(+) T cells are biased to a T(H)2 effector fate at the expense of T(H)1 and T(H)17 cells. J Allergy Clin Immunol. 2017;139(3):933–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pillay BA, Fusaro M, Gray PE, Statham AL, Burnett L, Bezrodnik L, et al. Somatic reversion of pathogenic DOCK8 variants alters lymphocyte differentiation and function to effectively cure DOCK8 deficiency. J Clin Invest. 2021;131(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randall KL, Chan SS, Ma CS, Fung I, Mei Y, Yabas M, et al. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J Exp Med. 2011;208(11):2305–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124(6):1289–302 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361(21):2046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambe T, Crawford G, Johnson AL, Crockford TL, Bouriez-Jones T, Smyth AM, et al. DOCK8 is essential for T-cell survival and the maintenance of CD8+ T-cell memory. Eur J Immunol. 2011;41(12):3423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dvorak CC, Haddad E, Heimall J, Dunn E, Buckley RH, Kohn DB, et al. The diagnosis of severe combined immunodeficiency (SCID): The Primary Immune Deficiency Treatment Consortium (PIDTC) 2022 Definitions. J Allergy Clin Immunol. 2023;151(2):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puck JM. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia. Immunol Rev. 2019;287(1):241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanna S, Etzioni A. MHC class I and II deficiencies. J Allergy Clin Immunol. 2014;134(2):269–75. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Almeida JR, Darko S, van der Burg M, DeRavin SS, Malech H, et al. Human syndromes of immunodeficiency and dysregulation are characterized by distinct defects in T-cell receptor repertoire development. J Allergy Clin Immunol. 2014;133(4):1109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharifinejad N, Jamee M, Zaki-Dizaji M, Lo B, Shaghaghi M, Mohammadi H, et al. Clinical, Immunological, and Genetic Features in 49 Patients With ZAP-70 Deficiency: A Systematic Review. Front Immunol. 2020;11:831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tempany JC, Zhou JH, Hodgkin PD, Bryant VL. Superior properties of CellTrace Yellow as a division tracking dye for human and murine lymphocytes. Immunol Cell Biol. 2018;96(2):149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, et al. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest. 2005;115(4):1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma CS, Wong N, Rao G, Nguyen A, Avery DT, Payne K, et al. Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets. J Exp Med. 2016;213(8):1589–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen RC, Armitage RJ, Conley ME, Rosenblatt H, Jenkins NA, Copeland NG, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259(5097):990–3. [DOI] [PubMed] [Google Scholar]
- 54.Ferrari S, Giliani S, Insalaco A, Al-Ghonaium A, Soresina AR, Loubser M, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci U S A. 2001;98(22):12614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrari S, Plebani A. Cross-talk between CD40 and CD40L: lessons from primary immune deficiencies. Curr Opin Allergy Clin Immunol. 2002;2(6):489–94. [DOI] [PubMed] [Google Scholar]
- 56.Bustamante J. Mendelian susceptibility to mycobacterial disease: recent discoveries. Hum Genet. 2020;139(6-7):993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol. 2014;26(6):454–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jirapongsananuruk O, Malech HL, Kuhns DB, Niemela JE, Brown MR, Anderson-Cohen M, et al. Diagnostic paradigm for evaluation of male patients with chronic granulomatous disease, based on the dihydrorhodamine 123 assay. J Allergy Clin Immunol. 2003;111(2):374–9. [DOI] [PubMed] [Google Scholar]
- 59.Kuhns DB, Alvord WG, Heller T, Feld JJ, Pike KM, Marciano BE, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363(27):2600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marciano BE, Zerbe CS, Falcone EL, Ding L, DeRavin SS, Daub J, et al. X-linked carriers of chronic granulomatous disease: Illness, lyonization, and stability. J Allergy Clin Immunol. 2018;141(1):365–71. [DOI] [PubMed] [Google Scholar]
- 61.Uzel G, Tng E, Rosenzweig SD, Hsu AP, Shaw JM, Horwitz ME, et al. Reversion mutations in patients with leukocyte adhesion deficiency type-1 (LAD-1). Blood. 2008;111(1):209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]