Summary
Background
Only around 20–30% of patients with non-small-cell lung cancer (NCSLC) have durable benefit from immune-checkpoint inhibitors. Although tissue-based biomarkers (eg, PD-L1) are limited by suboptimal performance, tissue availability, and tumour heterogeneity, radiographic images might holistically capture the underlying cancer biology. We aimed to investigate the application of deep learning on chest CT scans to derive an imaging signature of response to immune checkpoint inhibitors and evaluate its added value in the clinical context.
Methods
In this retrospective modelling study, 976 patients with metastatic, EGFR/ALK wild-type NSCLC treated with immune checkpoint inhibitors at MD Anderson and Stanford were enrolled from Jan 1, 2014, to Feb 29, 2020. We built and tested an ensemble deep learning model on pretreatment CTs (Deep-CT) to predict overall survival and progression-free survival after treatment with immune checkpoint inhibitors. We also evaluated the added predictive value of the Deep-CT model in the context of existing clinicopathological and radiological metrics.
Findings
Our Deep-CT model demonstrated robust stratification of patient survival of the MD Anderson testing set, which was validated in the external Stanford set. The performance of the Deep-CT model remained significant on subgroup analyses stratified by PD-L1, histology, age, sex, and race. In univariate analysis, Deep-CT outperformed the conventional risk factors, including histology, smoking status, and PD-L1 expression, and remained an independent predictor after multivariate adjustment. Integrating the Deep-CT model with conventional risk factors demonstrated significantly improved prediction performance, with overall survival C-index increases from 0·70 (clinical model) to 0·75 (composite model) during testing. On the other hand, the deep learning risk scores correlated with some radiomics features, but radiomics alone could not reach the performance level of deep learning, indicating that the deep learning model effectively captured additional imaging patterns beyond known radiomics features.
Interpretation
This proof-of-concept study shows that automated profiling of radiographic scans through deep learning can provide orthogonal information independent of existing clinicopathological biomarkers, bringing the goal of precision immunotherapy for patients with NSCLC closer.
Introduction
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer-related death in the USA and worldwide.1 Immune checkpoint inhibitors targeting PD-1 or PD-L1 have become the mainstay of therapy for patients without targeted treatment options. However, durable response remains limited to a minority of patients (20–30%).2 Robust predictive biomarkers might help identify patients who are likely to benefit from immune checkpoint inhibitor-based therapies versus those who might benefit from alternative therapeutic strategies.
Although many molecular predictors of immune checkpoint inhibition response have been proposed,3 and the US Food and Drug Administration have approved PD-L1 and tumour mutational burden as biomarkers to guide treatment selection in patients with NSCLC,4,5 none of these features fully capture the variability in outcomes, suggesting that benefit from immune checkpoint inhibition is orchestrated by the interaction of the tumour, the microenvironment, and the host.6 These tissue-based tests are further limited by inadequate biopsy specimens,7,8 intratumour heterogeneity,9,10 and high cost. By contrast, radiological images are already routinely obtained for cancer staging and therapeutic response assessment and can capture the entire tumour as well as its background parenchyma non-invasively. Moreover, the distinct biology of the tumour and its interaction with the tumour immune microenvironment give rise to distinct imaging phenotypes that, with machine learning and deep learning algorithms, can be mined to predict clinical outcomes and genomic features, known as radiomics and radiogenomics.11–15 Consequently, radiological images coupled with deep learning might provide a non-invasive means of identifying the biology relevant to immune checkpoint inhibition response, and developing predictive biomarkers to inform immune checkpoint inhibitor treatment selection.
Previous studies have shown the feasibility of developing radiomic or deep learning markers to predict immunotherapy outcomes.16–19 However, these analyses have used under-powered patient cohorts mixed with biologically heterogeneous cancer phenotypes. Furthermore, they aimed to use radiogenomics to develop imaging surrogates for already imperfect molecular signatures (PD-L1, EGFR mutation status, and CD8+ T-cell infiltrate), rather than determining whether radiographic features can independently improve outcome prediction.
In this study, we aimed to assess whether machine learning analysis could extract clinically relevant features from routine CT scans that are predictive of immune checkpoint inhibition outcomes and are complementary to known clinicopathological and radiological risk factors. We launched a multi-institutional, multidisciplinary collaboration to develop and validate a deep learning framework to comprehensively characterise individual patients’ imaging patterns and predict their clinical benefit from immune checkpoint inhibition.
Methods
Study design and participants
In this retrospective modelling study, clinicopathological data, radiographic reports, and patient prognostic information were retrieved from Epic electronic medical record software and baseline CT images were curated from the IntelliSpace PACS system (Philips). These data were used for model development and testing. To mitigate the heterogeneous protocols in image acquisition, we applied a series of image processing algorithms to harmonise the CT data and facilitate robust feature extraction (appendix p 27).
We queried the MD Anderson GEMINI database (a database containing detailed clinicopathological, radiological, and survival information, as well as molecular data for patients with lung disease) for patients with metastatic NSCLC enrolled from Jan 1, 2014, to Feb 29, 2020. Patients were included if they met the following criteria: (1) histologically confirmed NSCLC; (2) metastatic disease at the time of immune checkpoint inhibitor initiation; (3) treatment with anti-PD-1 or anti-PDL1 inhibitors for at least two cycles, either as monotherapy or combined with chemotherapy; (4) EGFR and ALK wild-type; (5) available follow-up data for progression-free survival and overall survival analysis; and (6) available high quality CT images that were obtained within 3 months before the start of immune checkpoint inhibition. Through balancing the distribution of demographics, radiological factors, and histopathological factors, we divided the MD Anderson cohort into training, validation, and testing cohorts at a ratio of 6:1:3 for use in model development and validation.
The same inclusion and exclusion criteria were applied to a previously published cohort from Stanford University of patients with metastatic lung cancer treated with immunotherapy20 for use as an external validation cohort with overall survival as the primary endpoint. Progression-free survival data were not available for this cohort.
This study was granted ethical approval by the institutional review board of The University of Texas MD Anderson Cancer Center and Stanford University, and was performed in accordance with the ethical standards of the 1964 Declaration of Helsinki. Informed consent was waived due to the retrospective nature of the study.
Model construction
Our overall approach is summarised in figure 1A. Briefly, we performed patient and imaging curation, then trained, internally validated, and tested a CT-derived deep learning signature using an ensemble learning scheme to stratify a patient’s progression-free survival and overall survival after immune checkpoint inhibitor therapy. We then tested this model on the external Stanford cohort, and interpreted it through correlating to hand-crafted radiomic features as well as blood and genomic biomarkers. In parallel, we derived a benchmark clinical model by using informative clinicopathological and radiological variables to fit into a random survival forest model. After verifying that our newly proposed CT-based deep learning model complemented the benchmark clinical model, we integrated these two approaches into a composite model and further evaluated its clinical performance. These steps are described in more detail in the following sections.
Figure 1: Overall study design.
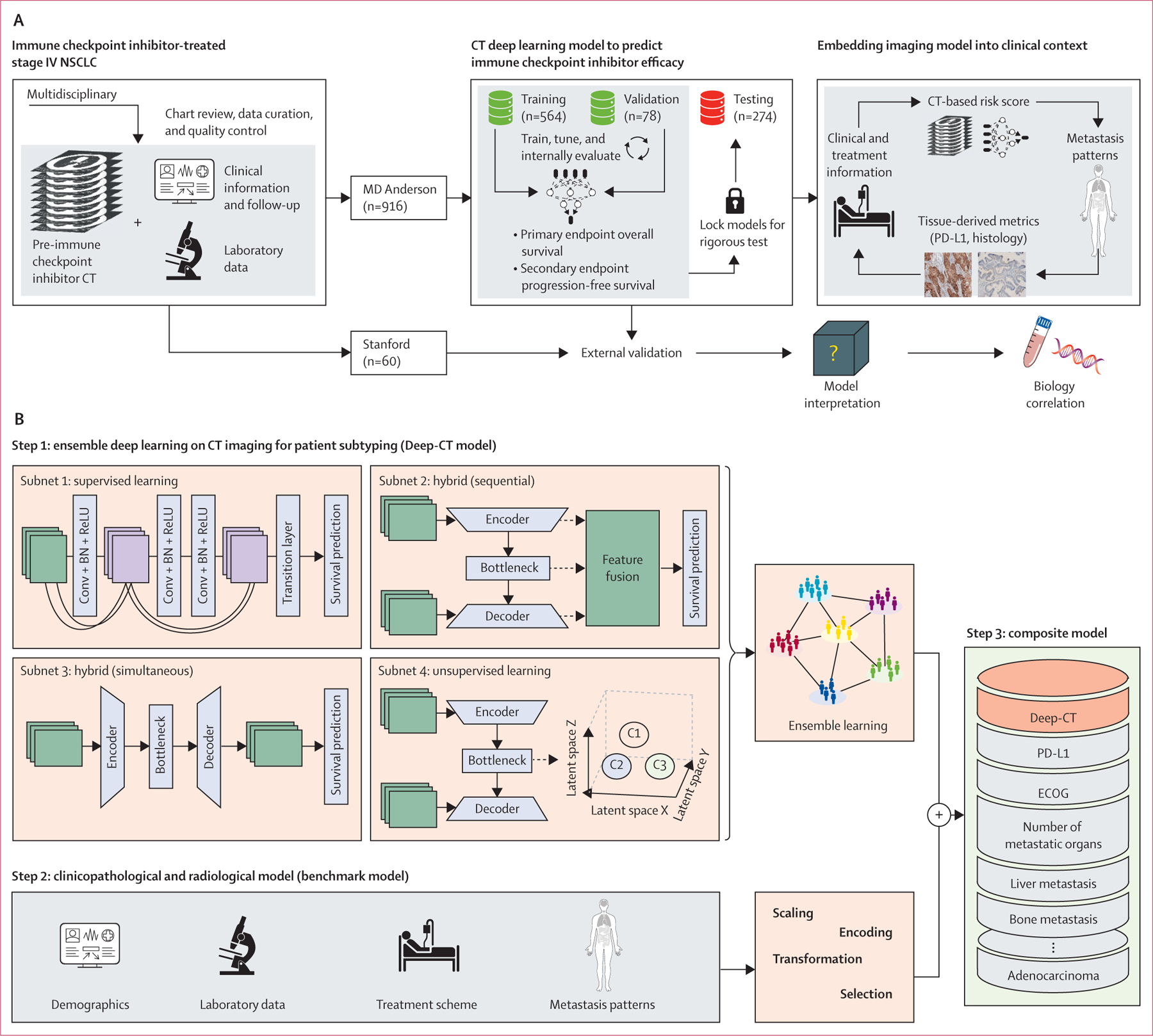
(A) Schematic plot for developing the CT deep learning model and evaluating it in a clinical context. (B) The details of different model development and integration. Parts of figure created with BioRender (biorender.com). Conv + BN + ReLu=Convolution plus batch normalisation plus rectified linear unit. ECOG=Eastern Cooperative Oncology group. NSCLC=non-small-cell lung cancer.
For step 1, ensembled CT deep learning model for patient subtyping (hereafter referred to as Deep-CT), we aimed to develop a deep learning-powered prognostic framework applied to baseline CT images to automate the quantification of patients’ risk of progression or death on immune checkpoint inhibitors. Given the clinical observation that the metastatic patterns as manifested on CT can vary substantially between patients with stage IV lung cancer, we focused on lung parenchymal and tumour lesions. To mitigate the uncertainty inevitable in one particular type of network model, we adopted an ensemble learning strategy to integrate fundamentally different but potentially complementary convolutional neural network architectures to increase the model’s generalisability. The ensemble framework consisted of four three-dimensional convolutional neural network models (figure 1B), including a supervised learning network (subnetwork 1), two hybrid networks that merged supervised and unsupervised learning differently (subnetworks 2 and 3), and an unsupervised learning network (subnetwork 4). The key difference between subnetwork 2 and subnetwork 3 was the way they integrated supervised and unsupervised modules: in subnetwork 2, a sequential scheme was used to optimise the unsupervised module before tuning the supervised one, whereas in subnetwork 3, both modules were optimised simultaneously. The individual models are described in detail in the appendix (pp 4–5, 30).
For the four subnetwork model training and validation in step 1, we adopted a state-of-the-art deep learning training strategy to develop a model on the MD Anderson cohort, where our training set was used for hyper-parameter tuning and the validation set was assessed to select the model with optimal performance. To mitigate overfitting, we used data augmentation (including rotation, flipping, scale, and contrast adjustment), dropout, and L1 regularisation for network parameters. We used the Adam algorithm for optimisation. The epoch was set as 300; batch size to 40; learning rate was set to 0·001; and learning rate decay and early stop were used. All subnetworks were developed using open source PyTorch version 1.4.0 and trained independently on NVIDIA DGX A100 station.
Step 1 also included patient subtyping by ensemble learning of predictions from the four subnetwork models. Using the individual risk stratification from the four subnetwork models, we built an integrated model using different ensemble strategies, including voting-based, attention-based, clustering-based, and tree-based algorithms. The aim was to capture common and complimentary signals (strong similarities) present across different networks while reducing individual models’ bias and variance by aggregating across multiple model predictions. The optimal ensemble algorithm was locked, then tested on the discovery cohort for validation.
For comparison purposes, in step 2 we trained a benchmark model from existing conventional clinicopathological factors and radiological factors for predicting progression-free survival and overall survival by implementing a two-step strategy on the training cohort. First, by fitting a univariate Cox proportional hazards model we evaluated the clinical value of individual features: (1) demographic features, including age, sex, race, BMI, and smoking status; (2) radiological features, including stage, metastasis patterns (liver, adrenal, bone, and brain), and number of metastatic organs; (3) pathological features, including histology and PD-L1 tumour proportion score; (4) treatment features, including line of treatment, therapy regimen, and previous local therapy; and (5) Eastern Cooperative Oncology Group performance score. To build a benchmark clinicopathological predictive model, four different machine learning models were trained, including LASSO, Elastic Net with Cox model, random survival forest, and gradient boosting, and the model with optimal performance on the discovery cohort was locked for validation on the testing cohort.
In step 3, we assessed the added clinical value of Deep-CT in addition to the benchmark model, and integrated them to form a composite model. Given patient stratification by either Deep-CT or the benchmark model, we first evaluated the effect of their joint stratification on the prediction of specified clinical endpoints. In particular, we assessed the progression-free survival or overall survival differences when both models agreed on predicting as high-risk versus when only one model predicted as high-risk. Second, we built a composite model that integrated patient stratification by Deep-CT (high risk versus low risk) and risk score from the benchmark model in random survival forests.
Clinical value assessment
We evaluated the prognostic value of the developed models for predicting progression-free survival and overall survival. The ensemble Deep-CT model was trained on the training cohort and internally validated on the validation cohort from the MD Anderson set. To be consistent, the benchmark model and composite model were also trained and validated on the training and validation sets, respectively. To prevent information leakage, the Deep-CT model was locked in order to be rigorously evaluated on the hold-out discovery cohort from the MD Anderson cohort as well as the external Stanford cohort. To assess whether Deep-CT was predictive for immune checkpoint inhibition rather than prognostic, we also tested it on a set of radiotherapy-treated patients with NSCLC (n=240). Then, we correlated Deep-CT stratification with clinicopathological and radiological risk factors. Furthermore, to evaluate any potential complementary effects among Deep-CT and existing clinicopathological variables, we modelled their relation to survival time in a multivariate Cox model and reported their effects (as measured by hazard ratio) in a forest plot analysis.
We further evaluated the prognostic significance of the proposed signatures (Deep-CT model and composite model) in clinically relevant subgroups, as separately stratified by (1) demographic information, including race (White vs non-White), sex, age (≥65 years vs <65 years); (2) tissue-derived metrics, including histology (adenocarcinoma vs squamous cell carcinoma vs other types), PD-L1 tumour proportion score (high [≥50%] vs intermediate [1–49%] vs low [<1%]); (3) metastasis pattern and stage, including stage at immune checkpoint inhibitor start (IVA vs IVB), liver metastasis status, and bone metastasis status; (4) treatment regimen, including line of therapy (first line vs second line vs other lines) and therapeutic regimens (immune checkpoint inhibitor-monotherapy vs immune checkpoint inhibitor plus chemotherapy); (5) previous local treatment (with surgery or radiotherapy vs without surgery or radiotherapy); and (6) CT modality (with contrast vs without contrast).
Radiological characteristics associated with patient subtyping
To overcome the challenge of pinpointing the areas or appearances on original CT scans that contributed to the inferred machine-learning output, we correlated Deep-CT stratification with interpretable radiomics metrics, including the disease burden measurements from radiologists’ manual annotation as well as classic radiomic metrics21 extracted from primary tumour or lung region measuring intensity and texture (the full list of 54 features is shown in appendix pp 16–17). Then, we linked these metrics with patient risk predictions and stratifications by the deep learning models. To understand the difference between radiomics and the Deep-CT model, we built a radiomics model and compared it with a deep-learning-based model (appendix pp 5–6).
Biological characteristics associated with patient subtyping
We evaluated the gene mutation patterns associated with Deep-CT model risk stratifications using genomic data obtained as part of the MD Anderson database through routine clinical-panel-based sequencing from tissue or blood. Furthermore, the imaging model was correlated to lung immune prognostic index, based on the ratio of derived neutrophils to leukocytes minus neutrophils and lactate dehydrogenase concentration within 30 days before immune checkpoint inhibitor treatment.22
Statistical analysis
The primary endpoint was overall survival and the secondary endpoint was progression-free survival. Overall survival was defined as the time from immune checkpoint inhibitor start until death from any cause. Patients who were alive at the last follow-up were censored for the overall survival analysis. Progression-free survival was defined as the time from immune checkpoint inhibitor initiation until progression or death. Progression was identified on the basis of imaging reports of tumour growth or new disease sites and the assessment by the treating physician. Patients who were alive without disease progression were censored at their last image assessment. Clinical data collection was locked for outcome analysis on Sept 10, 2020. Given the retrospective nature of this study, we chose not to perform a power calculation.
A Cox proportional hazard regression model was used to adjust for relevant clinicopathological variables in multivariable analysis. Kaplan-Meier analysis and log-rank tests were used to evaluate statistical significance of patient stratification by the proposed signatures. The cutoff value of a continuous risk score was optimised by the log-rank test based on the training and validation cohorts, which was locked for testing. Antolini’s concordance index (C-index) was used to measure the goodness of fit between models predicted by risk scores and progression-free survival or overall survival time. The net reclassification index was used to quantify the incremental value of the new model compared with the baseline model. The Wilcoxon signed-rank test and χ² test were used to test the differences for continuous and categorical variables, respectively. To adjust for multiple statistical testing, the Benjamini-Hochberg method was used to control the false discovery rate. All statistical tests were two-sided, with a p value less than 0·05 considered statistically significant. All statistical analyses were performed in R version 3.6.1.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
We identified 3428 patients with metastatic NSCLC in the MD Anderson GEMINI database, of whom 916 (27%) were EGFR/ALK wild-type, had received immune checkpoint inhibitor treatment, and met the other inclusion criteria (appendix p 28). Detailed information on demographic and clinical characteristics are shown in the table, and the change in line-of-therapy and immune checkpoint inhibitor regimen over time is shown in the appendix (p 29). Patients were stratified into training (n=564), validation (n=78), and testing cohorts (n=274) for model discovery, validation, and independent evaluation, respectively, at a ratio of 6:1:3. We conducted a balanced split across demographic variables, with no statistically significant differences in training, validation, and testing cohorts (table), and imaging scanner parameters and protocols were similar across cohorts (appendix p 7). The overall mean age at immune checkpoint inhibitor start was 65·2 years (SD 9·9), 498 (54%) patients were male, and 418 (46%) were female. There were 152 (17%) patients who had never smoked, and 202 (22%) patients with high PD-L1 expression (tumour proportion score ≥50%). An external cohort with the same inclusion criteria from Stanford (n=60) was used for external validation (appendix p 8).
Table:
Demographics and clinical characteristics of patients in the MD Anderson cohort
| Training Cohort |
Validation Cohort |
p value* | Testing Cohort |
p value† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients alive at data cutoff (n=294) | Patients who died (n=270) | p value‡ | Patients alive at data cutoff (n=40) | Patients who died (n=38) | p value‡ | Patients alive at data cutoff (n=140) | Patients who died (n=134) | p value‡ | |||
| Mean age, years | 64·9 (10·8) | 65·3 (9·6) | 0·46 | 62·9 (10·5) | 65·4 (10·3) | 0·17 | 0·61 | 65·8 (9·5) | 65·9 (8·9) | 0·50 | 0·51 |
| Sex | ·· | ·· | 1·0 | ·· | ·· | 0·17 | 0·78 | ·· | ·· | 0·44 | 0·92 |
| Female | 133 (45%) | 125 (46%) | ·· | 22 (55%) | 15 (39%) | ·· | ·· | 66 (47%) | 57 (43%) | ·· | ·· |
| Male | 161 (55%) | 145 (54%) | ·· | 18 (45%) | 23 (61%) | ·· | ·· | 74 (53%) | 77 (57%) | ·· | ·· |
| Histology | ·· | ·· | 0·25 | ·· | ·· | 0·098 | 0·78 | ·· | ·· | 0·31 | 0·47 |
| Adenocarcinoma | 242 (82%) | 209 (77%) | ·· | 36 (90%) | 29 (76%) | ·· | ·· | 108 (77%) | 100 (75%) | ·· | ·· |
| Squamous cell carcinoma | 41 (14%) | 44 (16%) | ·· | 2 (5%) | 8 (21%) | ·· | ·· | 25 (18%) | 21 (16%) | ·· | ·· |
| Other | 11 (4%) | 17 (6%) | ·· | 2 (5%) | 1 (3%) | ·· | ·· | 7 (5%) | 13 (10%) | ·· | ·· |
| Smoking history | ·· | ·· | 0·32 | ·· | ·· | 0·80 | 0·25 | ·· | ·· | 0·023 | 0·33 |
| Current | 44 (15%) | 43 (16%) | ·· | 7 (18%) | 7 (18%) | ·· | ·· | 36 (26%) | 17 (13%) | ·· | ·· |
| Former | 191 (65%) | 186 (69%) | ·· | 28 (70%) | 28 (74%) | ·· | ·· | 84 (60%) | 93 (69%) | ·· | ·· |
| Never | 59 (20%) | 41 (15%) | ·· | 5 (13%) | 3 (8%) | ·· | ·· | 20 (14%) | 24 (18%) | ·· | ·· |
| Stage immune checkpoint inhibitors started | ·· | ·· | <0·0001 | ·· | ·· | 0·014 | 0·79 | ·· | ·· | 0·034 | 0·96 |
| IVA | 130 (44%) | 74 (27%) | ·· | 19 (48%) | 8 (21%) | ·· | ·· | 59 (42%) | 40 (30%) | ·· | ·· |
| IVB | 164 (56%) | 196 (73%) | ·· | 21 (53%) | 30 (79%) | ·· | ·· | 81 (58%) | 94 (70%) | ·· | ·· |
| Liver metastasis | ·· | ·· | 0·021 | ·· | ·· | 0·31 | 0·76 | ·· | ·· | 0·15 | 0·39 |
| No | 259 (88%) | 219 (81%) | ·· | 35 (88%) | 30 (79%) | ·· | ·· | 127 (91%) | 114 (85%) | ·· | ·· |
| Yes | 35 (12%) | 51 (19%) | ·· | 5 (13%) | 8 (21%) | ·· | ·· | 13 (9%) | 20 (15%) | ·· | ·· |
| Adrenal metastasis | ·· | ·· | 0·071 | ·· | ·· | 0·92 | 0·84 | ·· | ·· | 0·34 | 0·94 |
| No | 252 (86%) | 216 (80%) | ·· | 33 (83%) | 31 (82%) | ·· | ·· | 118 (84%) | 107 (80%) | ·· | ·· |
| Yes | 42 (14%) | 54 (20%) | ·· | 7 (18%) | 7 (18%) | ·· | ·· | 22 (16%) | 27 (20%) | ·· | ·· |
| Bone metastasis | ·· | ·· | 0·0021 | ·· | ·· | 0·013 | 0·68 | ·· | ·· | 0·023 | 0·63 |
| No | 191 (65%) | 141 (52%) | ·· | 28 (70%) | 16 (42%) | ·· | ·· | 87 (62%) | 65 (49%) | ·· | ·· |
| Yes | 103 (35%) | 129 (48%) | ·· | 12 (30%) | 22 (58%) | ·· | ·· | 53 (38%) | 69 (51%) | ·· | ·· |
| Brain metastasis | ·· | ·· | 0·40 | ·· | ·· | 0·48 | 0·18 | ·· | ·· | 0·12 | 0·39 |
| No | 204 (69%) | 196 (73%) | ·· | 30 (75%) | 31 (82%) | ·· | ·· | 105 (75%) | 89 (66%) | ·· | ·· |
| Yes | 90 (31%) | 74 (27%) | ·· | 10 (25%) | 7 (18%) | ·· | ·· | 35 (25%) | 45 (34%) | ·· | ·· |
| Line of immune checkpoint inhibitor therapy | ·· | ·· | <0·0001 | ·· | ·· | 0·27 | 0·73 | ·· | ·· | 0·0049 | 0·89 |
| 1 | 212 (72%) | 138 (51%) | ·· | 28 (70%) | 22 (58%) | ·· | ·· | 100 (71%) | 74 (55%) | ·· | ·· |
| ≥2 | 82 (28%) | 132 (49%) | ·· | 12 (30%) | 16 (42%) | ·· | ·· | 40 (29%) | 60 (45%) | ·· | ·· |
| Therapy regimen | ·· | ·· | <0·0001 | ·· | ·· | 0·15 | 0·61 | ·· | ·· | 0·14 | 0·87 |
| Anti-PD-1 immune checkpoint inhibitor monotherapy | 119 (40%) | 154 (57%) | ·· | 17 (43%) | 18 (47%) | ·· | ·· | 68 (49%) | 66 (49%) | ·· | ·· |
| Anti-PD-L1 immune checkpoint inhibitor monotherapy | 14 (5%) | 17 (6%) | ·· | 0 | 5 (13%) | ·· | ·· | 4 (3%) | 11 (8%) | ·· | ·· |
| <1 immune checkpoint inhibitor combination | 39 (13%) | 23 (9%) | ·· | 3 (8%) | 2 (5%) | ·· | ·· | 20 (14%) | 12 (9%) | ·· | ·· |
| Immune checkpoint inhibitor plus chemotherapy | 119 (40%) | 69 (26%) | ·· | 19 (48%) | 12 (32%) | ·· | ·· | 46 (33%) | 40 (30%) | ·· | ·· |
| Immune checkpoint inhibitor plus other therapy | 3 (1%) | 7 (3%) | ·· | 1 (3%) | 1 (3%) | ·· | ·· | 2 (1%) | 5 (4%) | ·· | ·· |
| PD-L1 expression (tumour proportion score) | ·· | ·· | <0·0001 | ·· | ·· | 0·43 | 0·49 | ·· | ·· | <0·0001 | 0·51 |
| Low (<1%) | 75 (26%) | 62 (23%) | ·· | 10 (25%) | 11 (29%) | ·· | ·· | 27 (19%) | 32 (24%) | ·· | ·· |
| Intermediate (1–49%) | 88 (30%) | 61 (23%) | ·· | 10 (25%) | 8 (21%) | ·· | ·· | 34 (24%) | 29 (22%) | ·· | ·· |
| High (50–100%) | 74 (25%) | 51 (19%) | ·· | 9 (23%) | 4 (11%) | ·· | ·· | 46 (33%) | 18 (13%) | ·· | ·· |
| Unknown | 57 (19%) | 96 (36%) | ·· | 11 (28%) | 15 (39%) | ·· | ·· | 33 (24%) | 55 (41%) | ·· | ·· |
| Imaging type | ·· | ·· | 0·91 | ·· | ·· | 0·51 | 0·76 | ·· | ·· | 0·96 | 0·74 |
| CT | 222 (76%) | 203 (75%) | ·· | 32 (80%) | 28 (74%) | ·· | ·· | 109 (78%) | 104 (78%) | ·· | ·· |
| PET–CT | 72 (24%) | 67 (25%) | ·· | 8 (20%) | 10 (26%) | ·· | ·· | 31 (22%) | 30 (22%) | ·· | ·· |
| Image resources | ·· | ·· | 0·0010 | ·· | ·· | 0·42 | 0·23 | ·· | ·· | 0·37 | 0·23 |
| MD Anderson cohort | 156 (53%) | 180 (67%) | ·· | 25 (63%) | 27 (71%) | ·· | ·· | 76 (54%) | 80 (60%) | ·· | ·· |
| External | 138 (47%) | 90 (33%) | ·· | 15 (38%) | 11 (29%) | ·· | ·· | 64 (46%) | 54 (40%) | ·· | ·· |
| Median progression-free survival, months | 8·4 (3·5–20·6) | 4·1 (2·0–8·7) | ·· | 15·7 (6·2–30·5) | 4·2 (2·0–7·7) | ·· | 0·24 | 11·6 (4·4–23·2) | 6·4 (2·5–16·0) | ·· | 0·22 |
| Median overall survival, months | 18·1 (9·4–28·9) | 11·4 (6·3–18·7) | ·· | 21·1 (9·7–36·3) | 9·7 (5·0–14·8) | ·· | 0·87 | 19·8 (10·7–31·8) | 10·1 (6·3–18·2) | ·· | 0·86 |
Data are mean (SD), n (%), or median (IQR).
Comparison between training and validation cohorts.
Comparison between training, validation, and testing cohorts.
Derived from the comparison of patients alive at data cutoff and patients who died within a cohort.
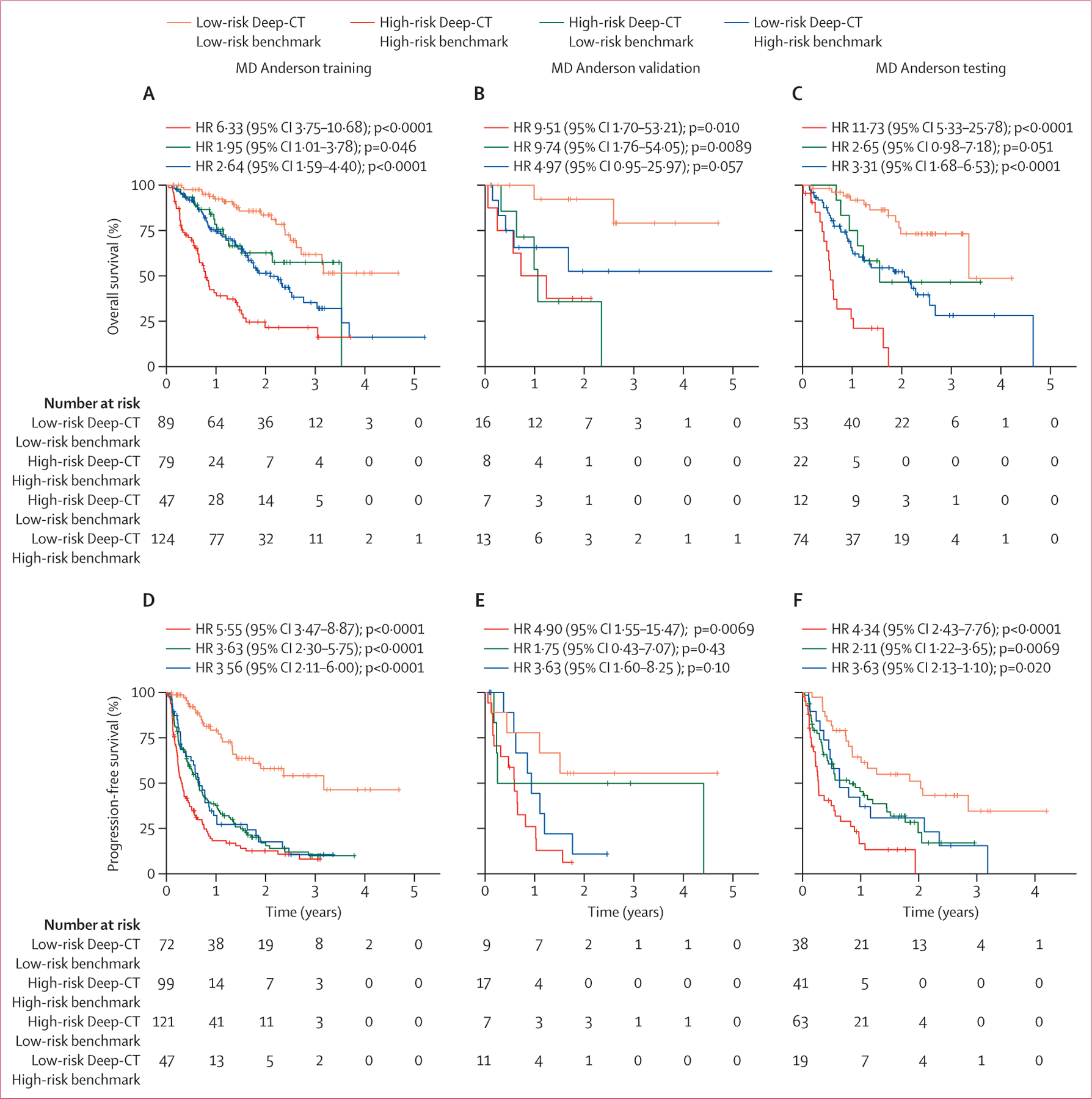
We developed an ensemble-based deep learning model to predict patients’ risk of progression or death after immune checkpoint inhibition from their baseline lung CT images (figure 1). After image preprocessing (appendix p 27), we applied distinct convolutional neural network architectures to build four individual deep learning subnetworks (appendix p 30), thereby mitigating the uncertainty inevitable to one particular type of network model. Each subnetwork significantly correlated to overall survival and progression-free survival (appendix pp 31–32), with similar performance across different models. The detailed ablation analysis to optimise subnetworks 1 and 2 is shown in the appendix (pp 9–10). Among several integration algorithms investigated to ensemble the individual subnetworks (four standalone deep learning models), the tree-based ensemble strategy achieved the optimal performance (appendix p 11), and was thus used to build an ensemble deep learning model (Deep-CT model, figure 1B). The ablation test showed that use of all four models was needed to achieve the optimal performance (appendix p 12). The optimised Deep-CT model clustered patients into two groups and showed a more stable C-index and a more consistently significant p value across training, validation, and testing sets than any of the individual deep learning models (appendix pp 13–14), suggesting superior robustness and reduced overfitting from the ensemble approach. Furthermore, the ensemble Deep-CT model achieved higher net reclassification index value than any individual model (appendix p 15), suggesting its superior added value.
The Deep-CT model stratified patients with significant differences in overall survival (figure 2A–C) and progression-free survival (figure 2D–F). Compared with the high-risk groups, in the low-risk groups median overall survival was 5·4–6·5 months longer and median progression-free survival was 15·5–17·3 months longer. When tested on the Stanford external validation dataset, the locked Deep-CT model stratified patients into high-risk and low-risk groups (figure 2G). The ensemble Deep-CT model again had superior performance compared with the individual deep learning models on overall stratification (appendix p 33). Because the Stanford cohort was small, we ran additional simulations to compare the Deep-CT model performance between external validation (on the Stanford set) and internal testing (on the hold-out MD Anderson set). We observed that the simulated C-index distributions and widths from the randomly sampled MD Anderson set were similar to the Stanford set testing (appendix p 34), adding confidence for the validation results on Stanford data to warrant its further validation on a larger set.
Figure 2: Prognostic performance of the ensemble deep learning CT model (Deep-CT).
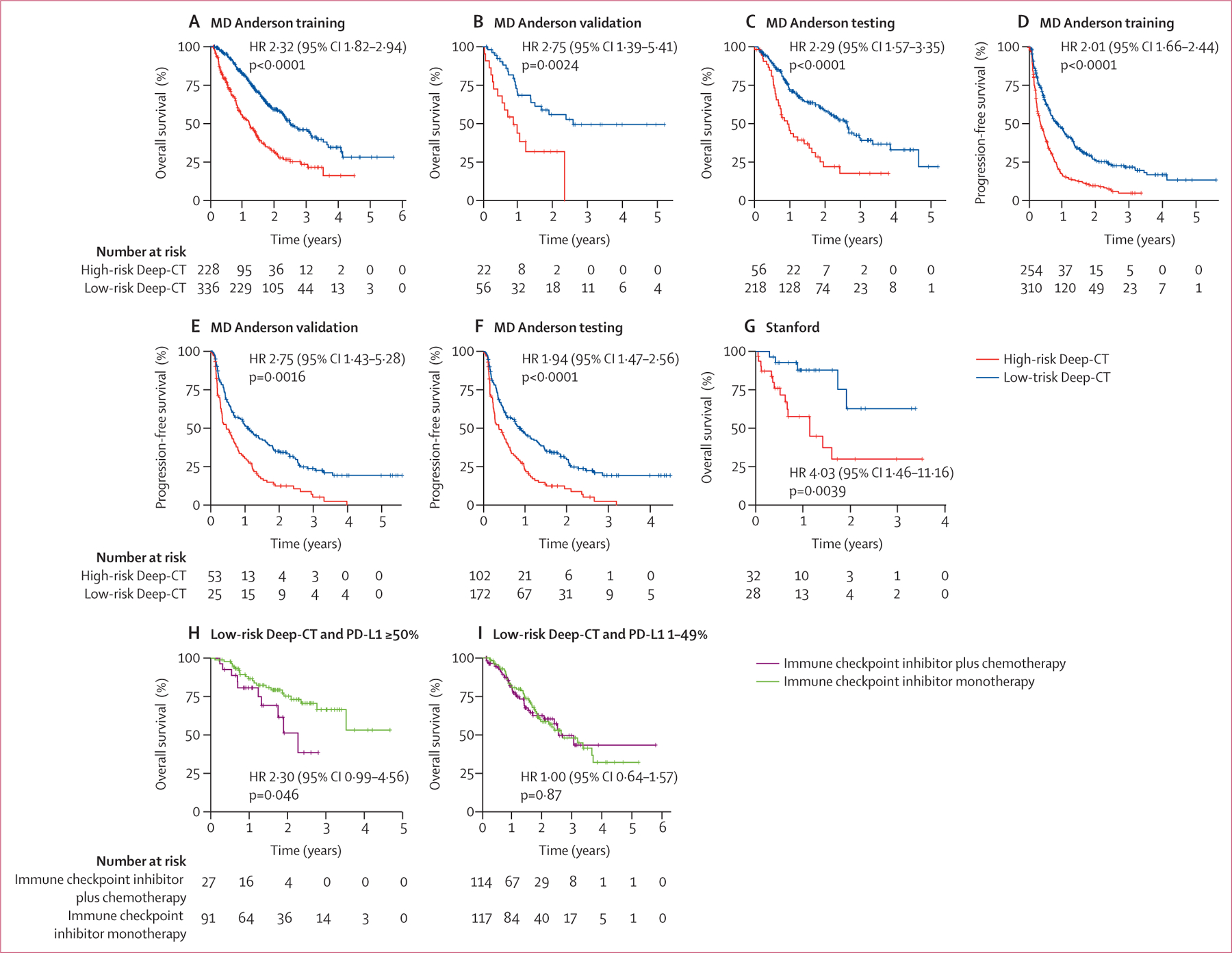
Kaplan-Meier survival curves for the MD Anderson cohort for (A) overall survival for training, (B) overall survival for validation, (C) overall survival for testing, (D) progression-free survival for training, (E) progression-free survival for validation, and (F) progression-free survival for testing. (G) Kaplan-Meier survival curve for external validation on the Stanford set. Joint stratification by Deep-CT and PD-L1 expression levels in patients treated with (H) immune checkpoint inhibitor monotherapy and (I) combination immune checkpoint inhibitor plus chemotherapy. HR=hazard ratio.
When testing the Deep-CT model on a stage III NSCLC radiotherapy set (n=240), there was no statistically significant difference for overall survival between the predicted high-risk versus low-risk groups in the overall population or in the stage IIIa or stage IIIb subgroups (appendix p 34). When mixing the radiotherapy cohort with the immune checkpoint inhibitor-treated MD Anderson cohort, we tested the interaction between therapeutic regimens (radiotherapy versus immune checkpoint inhibitor) and risk stratification from the deep learning model and observed a strong interaction effect (p=0·0008).
To understand the attention mechanism of the deep learning model, we plotted out the attention map (appendix p 35) and observed that the model weighted more on the tumour regions. To better understand the imaging characteristics underlying the predictive power of the CT models (including the four subnetwork models and their final ensemble model), we correlated their risk score stratifications with hand-crafted radiomics features, including those that measure disease burden, intensity, or texture heterogeneity from tumour or lung regions defined by a radiologist’s manual annotation (see feature list in appendix pp 16–17). Some of these radiomics features were significantly associated with the deep learning predictions from the individual subnetworks and the ensemble model (appendix p 36). 13 features positively correlated with all deep learning models, and an additional six features negatively correlated with all deep learning models (see detailed lists in appendix pp 18–20). Inspection of these features showed that they measured intratumoural and background lung texture heterogeneity (figure 3A). To illustrate these differences, we focused on two pairs of patients who had similar primary tumour volume and total disease burden but significantly different overall survival and progression-free survival outcomes and risk score predictions from the deep learning model (figure 3B). We observed that the textural heterogeneity from lung and tumour regions captured by Deep-CT separated these pairs, indicating its added value over disease burden alone for predicting a patient’s benefit from immune checkpoint inhibition. However, the radiomics model based solely on these hand-crafted features fluctuated substantially during model training, validation, and testing, and did not reach the robust performance level of Deep-CT, indicating there are additional imaging patterns learned by Deep-CT beyond the conventional radiomics domain.
Figure 3: Radiological interpretation of the CT deep learning models.
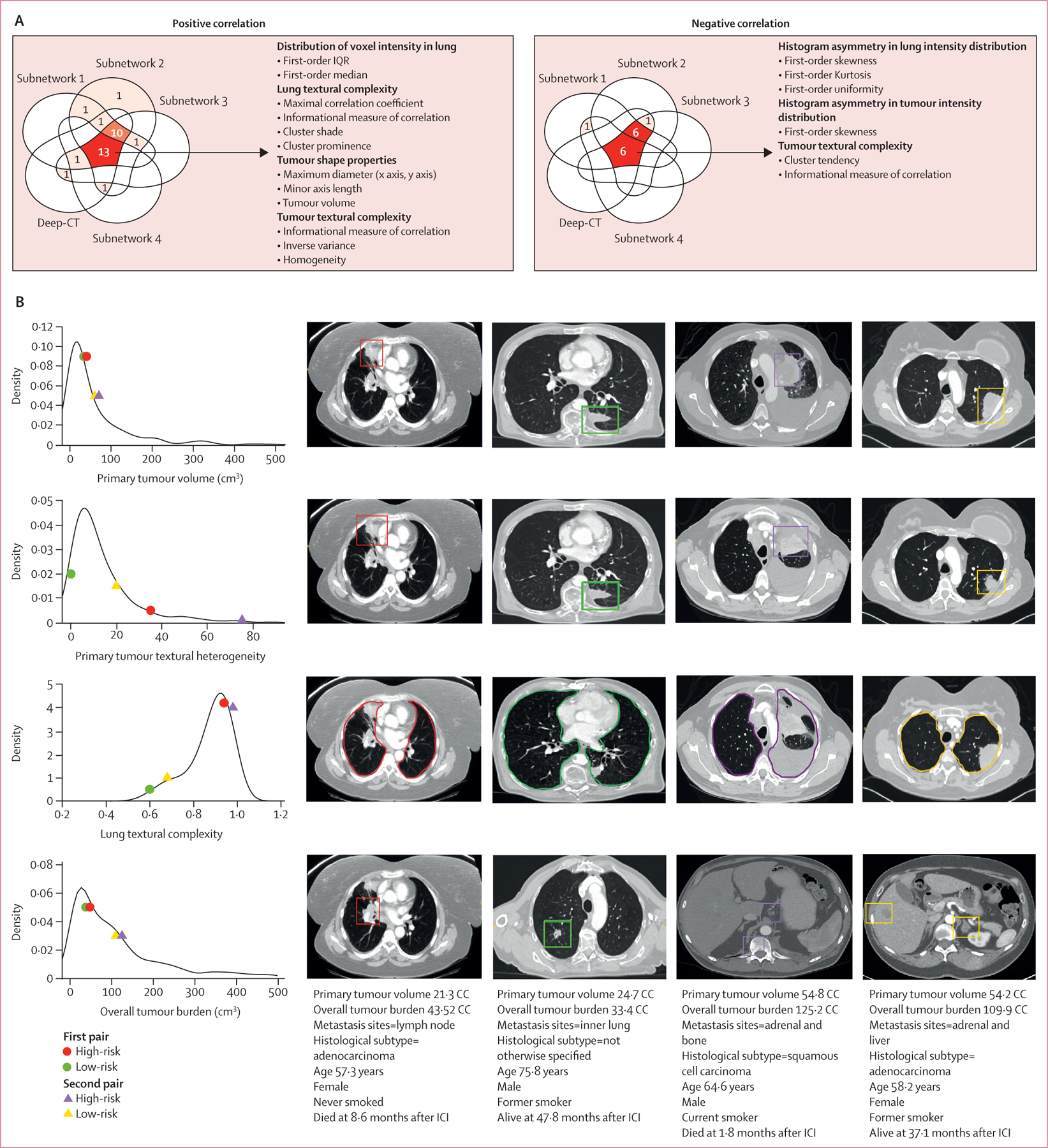
(A) Venn diagrams representing the number of features that showed significant correlation with the four individual subnetwork models and their ensemble model, with positive correlation and negative correlation. The red colour denotes the features agreed to be significant by more than one model. (B) Visualisation of different prognostic information captured by ensemble Deep-CT on selected four patients. The charts show radiomic feature score distribution. The first and second columns of scans show the first pair of patients (high-risk vs low-risk predicted by Deep-CT), and the last two columns of scans show another set of patients (high-risk vs low-risk). Each row shows how a feature score corresponds to radiological characteristic as seen in CT images. Each pair of patients were reported with roughly similar size of primary tumour volume and overall tumour burden, but with significant difference in overall survival and progression-free survival. The Deep-CT model captured non-conventional prognostic information including textural complexity and heterogeneity in primary tumour and background lung parenchyma beyond disease burden. ICI=immune checkpoint inhibitor.
To understand how the high-risk and low-risk groups from Deep-CT mapped onto known clinicopathological and radiographical factors, we assessed whether there were any correlations between the risk groups and these factors; we did not observe any correlations (appendix pp 21–23). When we compared the predictive power of the Deep-CT signature versus clinicopathological features, the deep learning signature showed a significant association with overall survival in the discovery and testing cohorts, although stage, bone metastasis, Eastern Cooperative Oncology Group (ECOG) performance status, and number of metastatic organs also showed consistent associations (figure 4A). After multivariate adjustment, Deep-CT risk remained the most significant predictor of overall survival. Similar patterns were also observed in univariate and multivariate analysis for progression-free survival (figure 4B). Consistent with these findings, when we analysed subgroups stratified by clinical variables including histology, PD-L1 expression, stage, liver or bone metastasis status, and age, we observed that the Deep-CT stratification demonstrated significant predictive value even within these subgroups (appendix pp 37–40). Notably, for patients with a PD-L1 tumour proportion score of 50% or more and Deep-CT low risk, immune checkpoint inhibitor monotherapy tended to be correlated with better prognosis than immune checkpoint inhibition plus chemotherapy (figure 2H). On the other hand, for a PD-L1 tumour proportion score of 1–49% and Deep-CT low-risk patients, immune checkpoint inhibitor monotherapy appeared to work equally well as immune checkpoint inhibition plus chemotherapy (figure 2I). When stratifying patients by previous local treatment (with or without surgery or radiotherapy) as well as by input CT modality (with or without contrast, routine CT vs PET–CT), the Deep-CT model performed consistently well (appendix pp 41–43).
Figure 4: Evaluation of Deep-CT in clinical context.
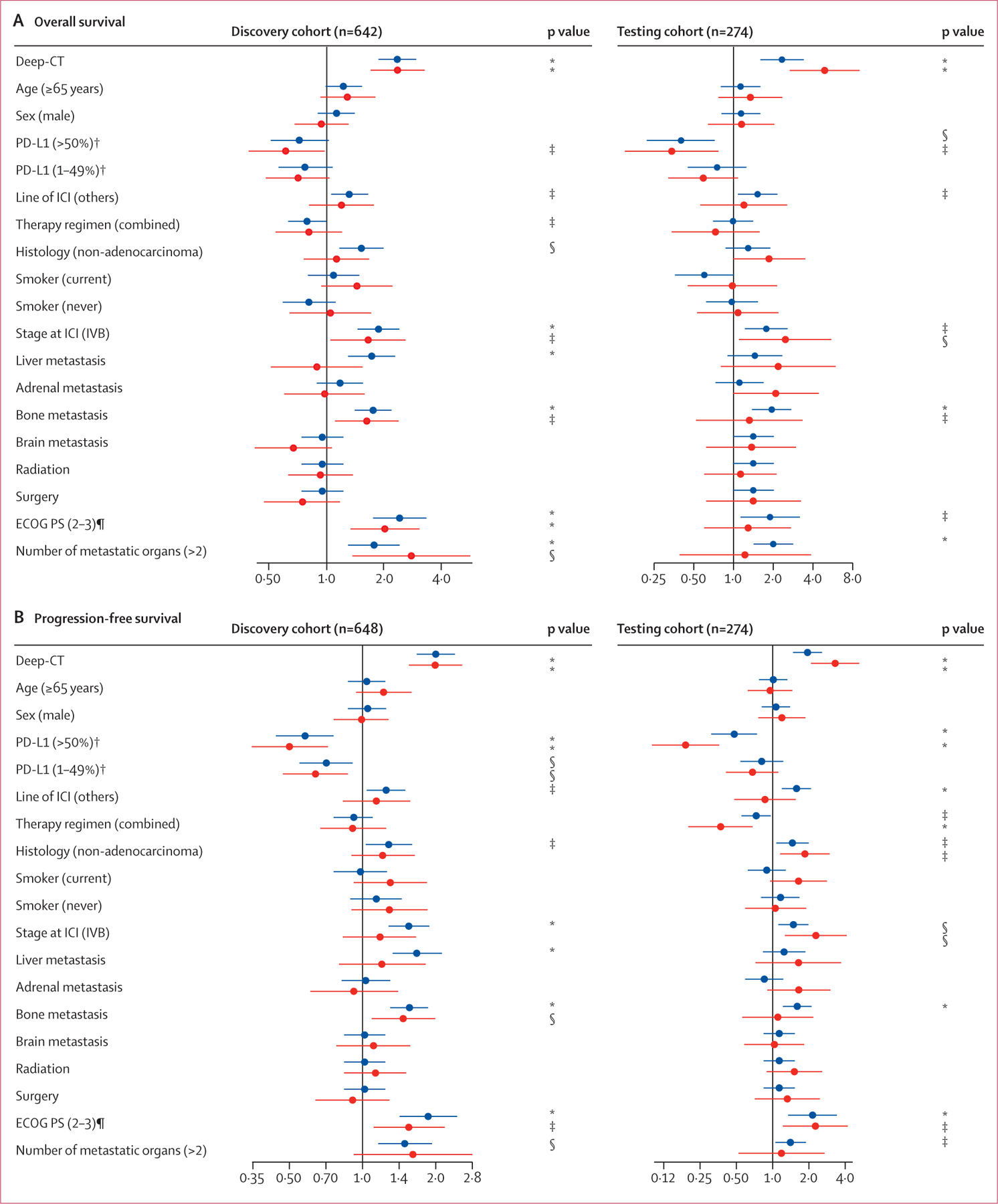
Forest plots summarise the univariate and multivariate analyses for the CT-deep learning model. (A) Overall survival. (B) Progression-free survival. Blue data points are from univariate analysis. Red datapoints are from multivariate analysis. Multivariate analysis of progression-free survival used smaller cohort sizes in the discovery cohort (n=383) and the testing cohort (n=161) due to missing values. The exact p values are shown in the appendix (pp 25–26). ECOG PS=Eastern Cooperative Oncology Group Performance Status. ICI=immune checkpoint inhibitor. *p<0·001. †For overall survival, PD-L1 expression used smaller cohort sizes in the discovery cohort (n=463) and testing cohort (n=186) due to missing values. ‡p<0·05. §p<0·01. ¶For overall survival, ECOG PS used smaller cohort sizes in the discovery cohort (n=512) and testing cohort (n=221) due to missing values.
To benchmark our Deep-CT model, we built an optimised model for predicting progression-free survival and overall survival based on conventional clinicopathological and radiological features using a random survival forest, as this model architecture achieved the best performance (see benchmark models in appendix p 24). The discriminative ability of the benchmark model alone in predicting progression-free survival and overall survival was also assessed, with C-index ranging between 0·67 and 0·72 for overall survival and 0·62 and 0·68 for progression-free survival in training, validation, and testing cohorts. Significant differences were also observed in the Kaplan-Meier plots for overall survival and progression-free survival (appendix p 44). Notably, the Deep-CT model that automatically abstracts radiographic characteristics has achieved an equivalent patient risk stratification compared with the benchmark models of multi-faceted clinical variables, including PD-L1 and metastasis patterns from radiologist reports.
To assess the interaction between the Deep-CT and benchmark models, we directly integrated their stratifications into four subgroups as follows: harmonised prediction, where both models predicted patients as high risk or low risk, and discrepant prediction, where the models disagreed with each other (high risk in Deep-CT and low risk in benchmark, or low risk in Deep-CT and high risk in benchmark). As expected, the subgroup that was low risk in both models consistently correlated with the best outcomes, and the subgroup that was high risk in both models was associated with the worst outcomes. The discrepant groups were found to have an intermediate prognosis (figure 5A–F). These observations suggest that the benchmark model and the Deep-CT model capture orthogonal prognostic features and therefore that there might be potential synergies between our proposed deep learning and conventional benchmark models.
Figure 5: Added prognostic value of the Deep-CT model on the clinical benchmark model.
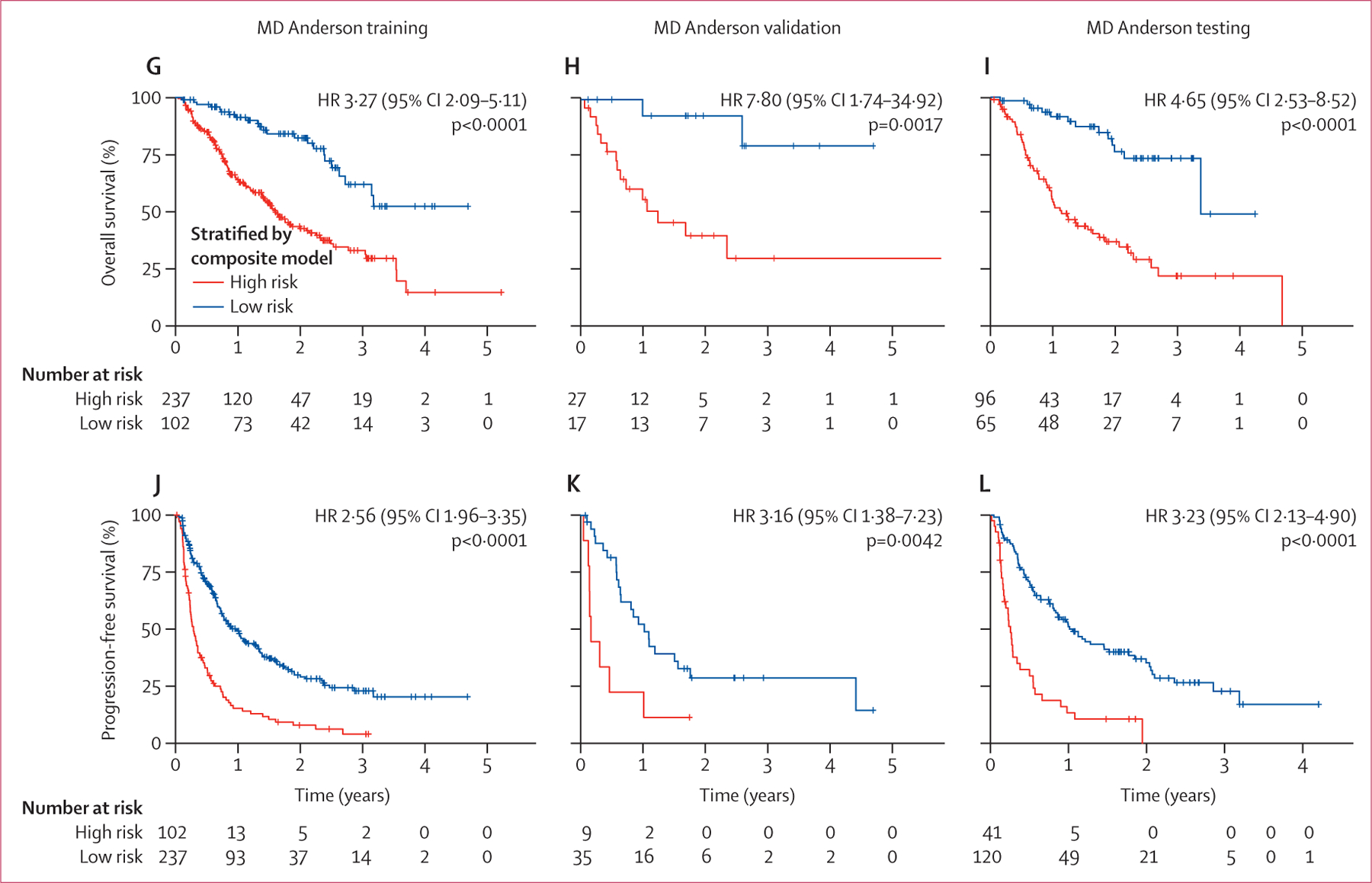
The Kaplan-Meier survival curves jointly stratified by the Deep-CT model and the benchmark model for (A) overall survival training, (B) overall survival validation, (C) overall survival testing, (D) progression-free survival training, (E) progression-free survival validation, and (F) progression-free survival testing. The composite model integrating the Deep-CT model and the benchmark model for (G) overall survival training, (H) overall survival validation, (I) overall survival testing, (J) progression-free survival training, (K) progression-free survival validation, and (L) progression-free survival testing.
When correlating Deep-CT stratification with the serum-based lung immune prognostic index biomarker, we observed that the proportion of patients predicted to be high risk by Deep-CT gradually increased when the lung immune prognostic index category changed from good to intermediate to poor (appendix p 45). Through radiogenomic analsyis (appendix p 45), some gene mutation patterns were correlated to Deep-CT stratification. For instance, ERBB2 and CDH1 genes were frequently mutated in the high-risk group, and by contrast, ATM, BAP1, and NTRK3 were frequently mutated in the Deep-CT low-risk group.
We built a composite model by refitting the Deep-CT and benchmark predictors using random survival forest. This composite model achieved the best prediction during discovery, validation, and testing, outperforming Deep-CT, the benchmark model, or conventional metrics including PD-L1 tumour proportion score, disease burden, and radiomics features alone, based on a subset of patients with complete information (figure 6). Moreover, the composite model was well calibarated (appendix pp 46–47). Patient stratifications using the composite signature were also significant (figure 5G–L). Notably, when compared with the benchmark model, the composite model significantly improved prediction accuracy, and reclassified 196 (56%) of 348 high-risk patients into low risk for progression-free survival. Furthermore, the composite model showed robust performance in subgroup analysis (appendix pp 48–51). When testing the composite model in the PD-L1 high group, we did not observe its predictive value to stratify response to immune checkpoint inhibitor monotherapy immune checkpoint inhibitor plus chemotherapy (appendix p 52).
Figure 6: C-index comparison of six different prediction models for overall survival and progression-free survival.
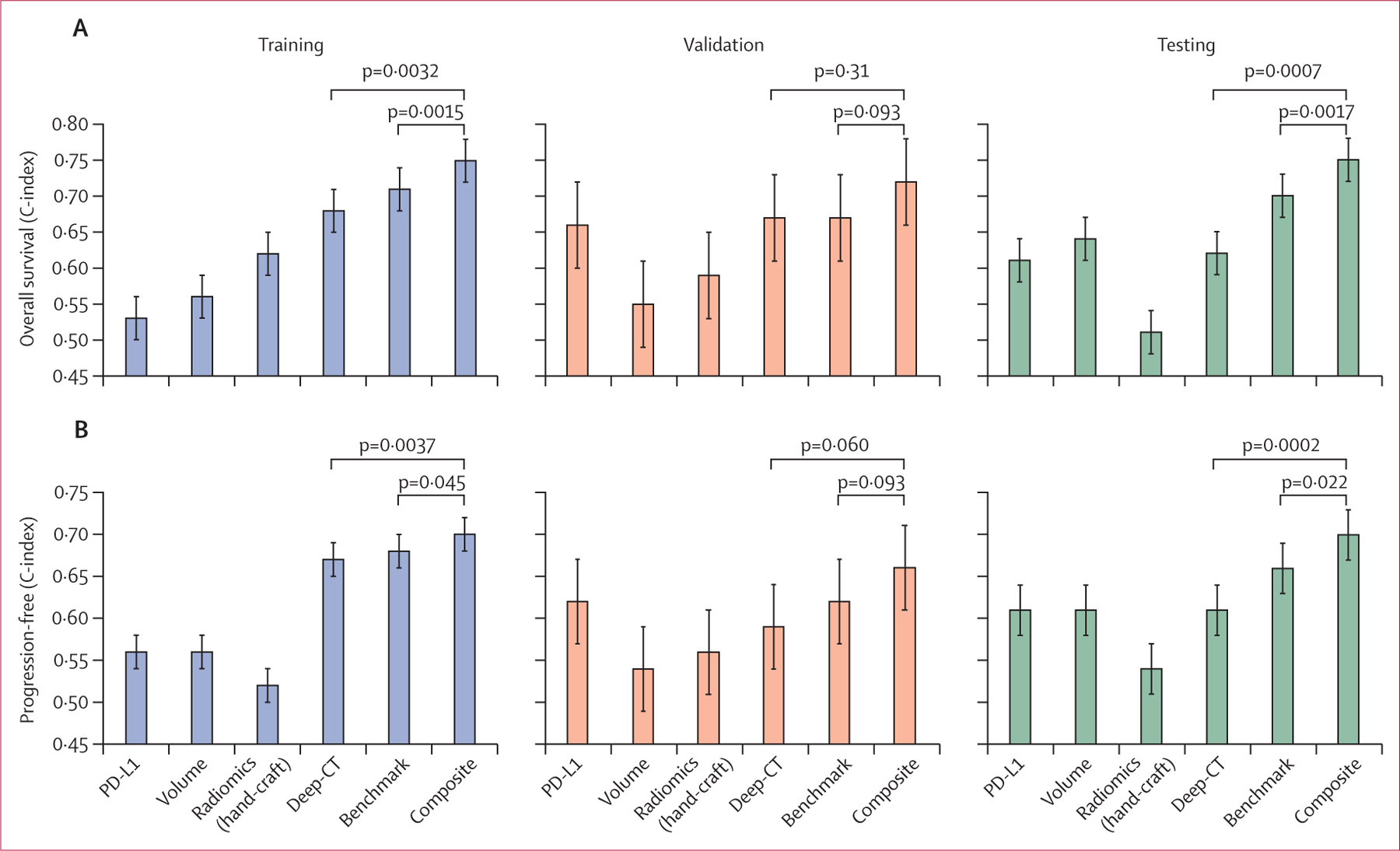
C-index comparison of six different prediction models (PD-L1, overall disease volume, radiomics model, Deep-CT, benchmark model, and the composite model that integrates Deep-CT and benchmark models) for (A) overall survival and (B) progression-free survival. The comparison was made on a smaller cohort size that contains the overlapped population with PD-L1 expression.
Discussion
In this study, we developed a deep learning CT signature that successfully predicted survival in patients with NSCLC treated with immune checkpoint inhibitors using large real-world data from two cancer centres. The proposed imaging signature shows robust stratification in clinically meaningful subgroups as defined by PD-L1 level, histology, age, sex, and race. We observed a synergy between this deep learning signature and existing clinicopathological and radiological risk factors, and their integration into a joint model achieved the best prediction. Taken together, our proof-of-concept study shows that automated profiling of routine radiographic scans through deep learning can add orthogonal information to existing clinicopathological biomarkers, bringing the goal of precision immunotherapy in NSCLC closer.
Previous radiomic analyses to predict benefit of immune checkpoint inhibition consisted of smaller pilot studies that focused on establishing imaging surrogates for specific molecular biomarkers, including CD8+ T-cell infiltrate,19 EGFR mutation,17,23 or PD-L1 expression.17,24 However, although these studies established proof-of-principle that radiomics can capture relevant biology, these efforts were limited by the fundamental fact that these biological features are themselves only limited predictors of immune checkpoint inhibitor response that capture only a small portion of the complex and heterogenous molecular features underlying responsiveness. Consequently, the imaging surrogates of these molecular markers are not likely to exceed the performance of the markers themselves, nor is imaging positioned to provide any complementary value to augment the prediction. To overcome this limitation, we aimed to harness the full power of imaging by leveraging an artificial intelligence framework to directly predict outcomes.
In contrast to conventional radiomics studies that built models based on hand-crafted features of tumour regions, we derived the deep learning signature of whole three-dimensional lung regions on CT scans under the hypothesis that both tumour and background lung parenchyma contain phenotypical patterns contributing to patient outcome. Existing deep learning studies in the medical imaging field have typically been built on a single network structure,25 and as such are prone to uncertainty from stochastic processes and model overfitting, which remains a general challenge for model deployment. To address this, we took advantage of fundamentally distinct and potentially complementary neural network architectures to comprehensively profile individual patients from their baseline CT scans. Combining these networks under an ensemble learning framework can mitigate the uncertainty in modelling and achieve enhanced prediction, as evidenced by our ensemble model’s robust performance during external validation on the Stanford dataset.
An additional strength of our study is the strict selection of patients with patients with stage IV NSCLC without EGFR or ALK alterations, in line with current treatment guidelines.26–28 Our cohort is, to the best of our knowledge, the first radiomics study on this clinically relevant population for predicting response to immune checkpoint inhibitors. Our large sample size provides sufficient statistical power for developing the model and accounting for potential confounders, without mixing heterogeneous cancer types as was done previously. Additionally, although most patients were from the MD Anderson cohort, 41% of CT scans from MD Anderson patients were performed at other hospitals with different scanners and imaging protocols, mimicking diverse data in a multi-institutional study, and adding confidence as to the generalisability of our machine learning models, which was further supported by the consistent performance in the Stanford cohort. Additionally, the availability of complete and high quality metadata (clinical, radiological, pathological, treatment, and follow-up information) enabled us to evaluate the proposed deep learning model in clinically meaningful subgroups. In contrast to the reduced performance of artificial intelligence models in historically under-served populations,29 we observed consistent performance of the proposed imaging model in subgroups as stratified by race or ethnicity, age, or sex.
Moreover, we observed that the newly obtained imaging signature exhibited similar or superior prediction power compared with known prognostic factors, including PD-L1 expression, metastatic patterns, histology, ECOG performance status, age, and stage. To quantify the clinical oncologist’s predictive assessment, a clinical benchmark model was built based on these clinically important factors. The integration of the deep learning model and benchmark clinical model into a composite model resulted in significant improvement over individual known risk factors, suggesting that clinicopathological features and deep learning capture orthogonal predictive information that together provide the best predictive power. Notably, patients with high PD-L1 expression (tumour proportion score ≥50%) and low-risk imaging score might benefit the most from immune checkpoint inhibitor monotherapy, which is associated with fewer toxicities and a trend towards improved long-term outcomes. Similarly, patients with a PD-L1 tumour proportion score of 1–49% and a low-risk Deep-CT score derived equal benefit from treatment with immune checkpoint inhibitor plus chemotherapy and immune checkpoint inhibitor monotherapy, suggesting that monotherapy might be also an option for this patient population. If further validated, this provides guidance for frontline treatment selection by serving as a predictive marker to select between immune checkpoint inhibition as monotherapy and in combination with chemotherapy, a choice not currently informed by markers in routine clinical use. Conversely, this approach might also more reliably identify those patients at high risk of progressing on immune checkpoint inhibitor therapy, which might enable providers to escalate therapy or perform earlier response assessment, and might also help support important research efforts by reproducibly identifying an immune checkpoint inhibitor resistant population to target in clinical trials aimed at augmenting antitumour immune responses.
To augment the interpretability of our deep-learning model, we developed an approach of mapping the model features back to interpretable radiographic patterns, including disease burden, primary tumour texture, and background lung parenchymal heterogeneity. We found the deep learning risk scores had high statistical correlations with several of these conventional radiomics features, including ones that measure tumour and lung parenchymal heterogeneity. This is in line with previous radiomics studies, which reported that intratumoural heterogeneity is associated with response to immune checkpoint inhibitors.16,30 However, the deep learning approach improves on classical radiomics by automatically learning radiographic patterns without requiring manual tumour segmentation or preprocessing, thereby minimising errors in reproducibility. It also demonstrated substantially improved performance compared with the classical radiomics models, suggesting that deep learning uses informative patterns beyond these known entities to predict immunotherapy outcomes.
Our study has several important limitations. First, as a retrospective, real-world study of metastatic disease, it might be biased by data heterogeneity in image protocol, therapy regimen, previous local treatment, line-of-treatment, outcome, and response metrics. The robust validation of our putative model on pilot Stanford data was intriguing, yet it warrants further validation on large prospective cohorts with homogeneous patient populations, treatments, and image modalities. Second, although the deep learning model shows prognostic value for patients treated with immune checkpoint inhibitors but not with radiotherapy, the specificity of the predictive value of this model to immune checkpoint inhibitors should be further validated by comparing with cohorts of patients with chemotherapy-treated metastatic NSCLC. Third, tailoring the treatment decision between immune checkpoint inhibitor monotherapy and immune checkpoint inhibitor plus chemotherapy is the most important unmet need in clinical thoracic oncology. Unfortunately, our cohort was not powered to address this important question. Herein, we are reporting the current model as a proof-of-concept that a deep learning model can extract important, potentially predictive information from thoracic CT scans to guide immune checkpoint inhibitor selection. Deep-CT models built and validated on patients with metastatic lung cancer treated with immune checkpoint inhibitor monotherapy versus immune checkpoint inhibitor plus chemotherapy are needed to facilitate decision making in the first-line setting. Fourth, we chose to focus on lung and chest regions to mitigate the heterogeneity among patients with stage IV NSCLC with different metastatic patterns in other organs. However, it is known that sites of metastasis play key roles in determining patient outcomes, and we did observe that the number of involved metastatic organs correlates to patient survival. Future efforts are needed to profile all measurable lesions to provide a holistic view of each patient’s disease, as well as assess its relationship with intrathoracic disease. Finally, more work is needed to improve the interpretability of our deep learning model. As hypothesis generation, we did observe correlations between the deep learning model and hand-crafted radiomics features as well as gene mutations. Future studies to understand the mechanisms underlying the differences between these models and their performances will help establish causal radio-immunogenomic relationships to uncover the biology driving deep learning prediction.
In conclusion, we have developed and validated an ensemble deep-learning-based CT signature to predict benefit of immune checkpoint inhibition for patients with metastatic, EGFR/ALK wild-type NSCLC. The Deep-CT signature has been validated comprehensively on multicentre data and demonstrated additional prognostic value beyond known risk factors. Moreover, the Deep-CT signature showed the potential to help identify patients who might benefit from immune checkpoint inhibition alone rather than combined with chemotherapy. These results warrant further verification in future large prospective trials to refine such findings and test clinical utility of our proposed imaging-based biomarker to guide individualised therapeutic selection.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for peer-reviewed, English-language journal and conference articles, published up to Nov 1, 2022, using the terms “lung cancer” AND (“EGFR wild” OR “driver negative”) AND (“immunotherapy” OR “immune checkpoint inhibitor”) AND (“prognosis” OR “survival”) AND (“deep learning” OR “convolutional neural network”). We also examined the reference lists of relevant publications. Our search did not identify any previous studies on the use of deep learning analysis of radiological images for predicting immnunotherapy related prognosis in patients with EGFR/ALK wild-type non-small-cell lung cancer (NSCLC).
Added value of this study
To our knowledge, this retrospective study is the first to develop an imaging-based model to predict benefit from immune checkpoint inhibitor therapy in EGFR/ALK wild-type NSCLC, in large multi-institution cohorts (n=976). An ensemble deep learning signature was developed and externally validated for the accurate prediction of progression-free survival and overall survival from baseline CT images, independent of known clinicopathological variables including PD-L1. When integrating the deep learning model with a clinical benchmark model that included PD-L1 and metastasis distribution, the composite model achieved the highest predictive performance. Additionally, the deep learning model complemented PD-L1 in identifying patients who were most likely to benefit from immune checkpoint inhibitor monotherapy versus combination immune checkpoint inhibitor plus chemotherapy.
Implications of all the available evidence
A deep learning-based radiomic model might complement molecular and clinical biomarkers to improve prediction of benefit from immune checkpoint inhibitors. Our proof-of-concept study warrants further optimisation and validation in larger cohorts prospectively. If validated, such efforts might contribute to more personalised use of immune checkpoint inhibitors in patients with NSCLC and identify patients with resistant disease who might benefit from novel or enhanced treatment approaches.
Acknowledgments
This work was supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Moon Shot Program and the MD Anderson Cancer Center Support Grant P30 CA016672. This research was partially supported by the National Institutes of Health grants R00 CA218667, and R01 CA262425. This work was supported by the Tumor Measurement Initiative through the MD Anderson Strategic Initiative Development Program (STRIDE). NIV is supported by the Mark Foundation Damon Runyon Foundation Physician Scientist Award, Conquer Cancer Foundation YIA, and SITC-Genentech Women in Cancer Research Fellowship. This work was supported by generous philanthropic contributions from Andrea Mugnaini and Edward L C Smith. This work was supported by the Rexanna’s Foundation for Fighting Lung Cancer.
Funding
National Institutes of Health, Mark Foundation Damon Runyon Foundation Physician Scientist Award, MD Anderson Strategic Initiative Development Program, MD Anderson Lung Moon Shot Program, Andrea Mugnaini, and Edward L C Smith.
Footnotes
Declaration of interests
NIV receives consulting fees from Sanofi, Regeneron, Oncocyte, and Eli Lilly; and research funding from Mirati, outside the submitted work. SHL reports research funding from STCube Pharmaceuticals, Beyond Spring Pharmaceuticals, and Nektar Therapeutics; being on an advisory board for AstraZeneca and Creatv Microtech; and receiving consultation fees from XRAD Therapeutics, all outside the submitted work. PPL reports personal fees from Viewray and AstraZeneca; personal fees and non-financial support from Varian; and personal fees from Genentech, outside the submitted work. SG reports research support from AstraZeneca, BMS, and Millenium Pharmaceuticals, all outside the submitted work. JYC reports research funding from BMS-MDACC and consultation fees from Legion Healthcare Partners. MFG reports research support from Varian Medical Systems and RefleXion Medical. HAW reports research funding from ACEA Biosciences, Arrys Therapeutics, AstraZeneca/Medimmune, BMS, Clovis Oncology, Genentech/Roche, Merck, Novartis, SeaGen, Xcovery, and Helsinn; being on an advisory board for AstraZeneca, Blueprint, Mirati, Merck, and Genentech/Roche; and has leadership roles with the International Association for the Study of Lung Cancer, and ECOG-ACRIN. JWN reports honoraria from CME Matters, Clinical Care Options Continuing Medical Education (CME), Research to Practice CME, Medscape CME, Biomedical Learning Institute CME, MLI Peerview CME, Prime Oncology CME, Projects in Knowledge CME, Rockpointe CME, MJH Life Sciences CME, Medical Educator Consortium, and HMP Education; consulting or advisory roles for AstraZeneca, Genentech/Roche, Exelixis, Jounce Therapeutics, Takeda Pharmaceuticals, Eli Lilly, Calithera Biosciences, Amgen, Iovance Biotherapeutics, Blueprint Pharmaceuticals, Regeneron Pharmaceuticals, Natera, Sanofi/Regeneron, D2G Oncology, Surface Oncology, Turning Point Therapeutics, Mirati Therapeutics, Gilead Sciences, and AbbVie; and research funding from Genentech/Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, Nektar Therapeutics, Takeda Pharmaceuticals, Adaptimmune, GSK, Janssen, and AbbVie. H-SL reports research funding from Samyang Biopharmaceutical USA. VV reports consulting fees from BMS, Merck, Novartis, Amgen, Foundation Medicine, and AstraZeneca. YL reports research funding from Merck, MacroGenics, Tolero Pharmaceuticals, AstraZeneca, Vaccinex, Blueprint Medicines, Harpoon Therapeutics, Sun Pharma Advanced Research, Bristol Myers Squibb, Kyowa Pharmaceuticals, Tesaro, Bayer HealthCare, Mirati Therapeutics, and Daiichi Sankyo; has been on scientific advisory boards for AstraZeneca Pharmaceuticals, Janssen Pharmaceutical, Lilly Oncology, and Turning Point Therapeutics; has received consultation fees from AstraZeneca; and has received honoraria from Clarion Health Care. MP reports research funding from Novartis Institutes for Biomedical Research. XLe reports research funding from Eli Lilly, EMD Serono, Regeneron, and Boehringer Ingelheim; and consultant fees from EMD Serono (Merck KGaA), AstraZeneca, Spectrum Pharmaceutics, Novartis, Eli Lilly, Boehringer Ingelheim, Hengrui Therapeutics, Janssen, Blueprint Medicines, Sensei Biotherapeutics, and AbbVie, outside the submitted work. YYE discloses research support from AstraZeneca, Takeda, Eli Lilly, Xcovery, Tuning Point Therapeutics, Blueprint, Elevation Oncology, Spectrum, and Nuvalent; having advisory roles for AstraZeneca, Eli Lilly, Takeda, Specturm, Bristol Myers Squibb, and Turning Point Therapeutics; and accommodation expenses from Eli Lilly. MVN has been on scientific advisory boards for Mirati, Merck/MSD, and Genentech; and has received research funding from Mirati, Novartis, Checkmate, Alaunos/Ziopharm, AstraZeneca, Pfizer, and Genentech. FS reports consulting fees and advisory roles from Amgen, AstraZeneca Pharmaceuticals, Novartis, BeiGene, Tango Therapeutics, Calithera Biosciences, Navire Pharma, Medscape, Intellisphere, Guardant Health, and BergenBio; speaker fees from BMS, RV Mais Promoção e Eventos, Visiting Speakers Programme in Oncology at McGill University and the Université de Montréal, AIM Group International, and ESMO; fees for travel, food, and beverages from Tango Therapeutics, AstraZeneca Pharmaceuticals, Amgen, Guardant Health, and Dava Oncology; stock or stock options in BioNTech and Moderna; research grants (to institution) from Amgen, Mirati Therapeutics, Boehringer Ingelheim, Merck & Co, and Novartis; study chair funds (to institution) from Pfizer; and research grants (spouse, to institution) from Almmune. CMG reports fees for advisory committees from AstraZeneca, Bristol Myers Squibb, Jazz Pharmaceuticals, and Monte Rosa Therapeutics; research support from AstraZeneca; and speaker’s fees from AstraZeneca and Beigene. TC reports speaker fees and honoraria from The Society for Immunotherapy of Cancer, Bristol Myers Squibb, Roche, Medscape, and PeerView; having an advisory role or receiving consulting fees from AstraZeneca, Bristol Myers Squibb, EMD Serono, Merck & Co, Genentech, and Arrowhead Pharmaceuticals; and institutional research funding from AstraZeneca, Bristol Myers Squibb, and EMD Serono. IIW reports grants and personal fees from Genentech/Roche, Bayer, Bristol Myers Squibb, AstraZeneca, Pfizer, HTG Molecular, Merck, Guardant Health, Novartis, and Amgen; personal fees from GSK, Flame, Sanofi, Daiichi Sankyo, Oncocyte, Janssen, MSD, and Platform Health; and grants from Adaptimmune, Adaptive, 4D, EMD Serono, Takeda, Karus, Iovance, Johnson & Johnson, and Akoya outside the submitted work. JDH is on the Scientific Advisory Board of Imagion Biosystems. DLG reports honoraria for scientific advisory boards from AstraZeneca, Sanofi, Alethia Biotherapeutics, Menarini, Eli Lilly, 4D Pharma, and Onconova; and research support from Janssen, Takeda, Astellas, Ribon Therapeutics, NGM Biopharmaceuticals, Boehringer Ingelheim, Mirati Therapeutics, and AstraZeneca. JVH reports being on scientific advisory boards for AstraZeneca, Boehringer Ingelheim, Genentech, GlaxoSmithKline, Eli Lilly, Novartis, Spectrum, EMD Serono, Sanofi, Takeda, Mirati Therapeutics, BMS, and Janssen Global Services; receiving research support from AstraZeneca, Takeda, Boehringer Ingelheim, and Spectrum; and receiving licensing fees from Spectrum. JZ reports research funding from Merck, Johnson & Johnson, and Novartis; and consultant fees from BMS, Johnson & Johnson, AstraZeneca, Geneplus, OrigMed, Novartis, and Innovent, outside the submitted work. CCW reports research support from Medical Imaging and Data Resource Center from NIBIB/University of Chicago and royalties from Elsevier. All other authors declare no competing interests.
Data sharing
The data that support the findings of this study are available through data access agreement from the corresponding authors. De-identified clinical data will be provided on reasonable request. The image data are not publicly available because they contain sensitive information that could compromise patient privacy. The source code for the deep learning model is available at https://github.com/WuLabMDA/Deep_CT-prognostic-biomarker.
For Epic electronic medical record software see https://www.epic.com/
See Online for appendix
Contributor Information
Maliazurina B Saad, Department of Imaging Physics.
Lingzhi Hong, Department of Imaging Physics, Department of Thoracic/Head and Neck Medical Oncology.
Muhammad Aminu, Department of Imaging Physics.
Natalie I Vokes, Department of Thoracic/Head and Neck Medical Oncology, Department of Genomic Medicine.
Pingjun Chen, Department of Imaging Physics.
Morteza Salehjahromi, Department of Imaging Physics.
Kang Qin, Department of Thoracic/Head and Neck Medical Oncology.
Sheeba J Sujit, Department of Imaging Physics.
Xuetao Lu, Department of Biostatistics.
Elliana Young, Department of Enterprise Data Engineering and Analytics.
Qasem Al-Tashi, Department of Imaging Physics.
Rizwan Qureshi, Department of Imaging Physics.
Carol C Wu, Department of Thoracic Imaging.
Brett W Carter, Department of Thoracic Imaging.
Steven H Lin, Department of Radiation Oncology.
Percy P Lee, Department of Radiation Oncology, Department of Radiation Oncology, City of Hope National Medical Center, Los Angeles, CA, USA.
Saumil Gandhi, Department of Radiation Oncology.
Joe Y Chang, Department of Radiation Oncology.
Ruijiang Li, The University of Texas MD Anderson Cancer Center, Houston, TX, USA; Department of Radiation Oncology, Stanford University School of Medicine, Palo Alto, CA, USA.
Michael F Gensheimer, The University of Texas MD Anderson Cancer Center, Houston, TX, USA; Department of Radiation Oncology, Stanford University School of Medicine, Palo Alto, CA, USA.
Heather A Wakelee, Department of Medicine, Division of Oncology, Stanford University School of Medicine, Stanford, CA, USA, Stanford Cancer Institute, Stanford, CA, USA.
Joel W Neal, Department of Medicine, Division of Oncology, Stanford University School of Medicine, Stanford, CA, USA, Stanford Cancer Institute, Stanford, CA, USA.
Hyun-Sung Lee, Systems Onco-Immunology Laboratory, David J Sugarbaker Division of Thoracic Surgery, Michael E DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX, USA.
Chao Cheng, Institute for Clinical and Translational Research, Baylor College of Medicine, Houston, TX, USA.
Vamsidhar Velcheti, Department of Hematology and Oncology, New York University Langone Health, New York, NY, USA.
Yanyan Lou, Division of Hematology and Oncology, Mayo Clinic, Jacksonville, FL, USA.
Milena Petranovic, Department of Radiology, Massachusetts General Hospital, Boston, MA, USA.
Waree Rinsurongkawong, Department of Biostatistics.
Xiuning Le, Department of Thoracic/Head and Neck Medical Oncology.
Vadeerat Rinsurongkawong, Department of Biostatistics.
Amy Spelman, Department of Thoracic/Head and Neck Medical Oncology.
Yasir Y Elamin, Department of Thoracic/Head and Neck Medical Oncology.
Marcelo V Negrao, Department of Thoracic/Head and Neck Medical Oncology.
Ferdinandos Skoulidis, Department of Thoracic/Head and Neck Medical Oncology.
Carl M Gay, Department of Thoracic/Head and Neck Medical Oncology.
Tina Cascone, Department of Thoracic/Head and Neck Medical Oncology.
Mara B Antonoff, Department of Thoracic and Cardiovascular Surgery.
Boris Sepesi, Department of Thoracic and Cardiovascular Surgery.
Jeff Lewis, Department of Biostatistics.
Ignacio I Wistuba, Department of Translational Molecular Pathology.
John D Hazle, Department of Imaging Physics.
Caroline Chung, Department of Radiation Oncology, Department of Neuroradiology.
David Jaffray, Department of Imaging Physics, Department of Radiation Physics.
Don L Gibbons, Department of Thoracic/Head and Neck Medical Oncology.
Ara Vaporciyan, Department of Thoracic and Cardiovascular Surgery.
J Jack Lee, Department of Biostatistics.
John V Heymach, Department of Thoracic/Head and Neck Medical Oncology.
Jianjun Zhang, Department of Thoracic/Head and Neck Medical Oncology, Department of Genomic Medicine.
Jia Wu, Department of Imaging Physics, Department of Thoracic/Head and Neck Medical Oncology.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022; 72: 7–33. [DOI] [PubMed] [Google Scholar]
- 2.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018; 359: 1350–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019; 16: 341–55. [DOI] [PubMed] [Google Scholar]
- 4.Doroshow DB, Bhalla S, Beasley MB, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 2021; 18: 345–62. [DOI] [PubMed] [Google Scholar]
- 5.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018; 378: 2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dejima H, Hu X, Chen R, et al. Immune evolution from preneoplasia to invasive lung adenocarcinomas and underlying molecular features. Nat Commun 2021; 12: 2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green MR, Willey J, Buettner A, Lankford M, Neely DB, Ramalingam SS. Molecular testing prior to first-line therapy in patients with stage IV nonsquamous non-small cell lung cancer (NSCLC): a survey of U.S. medical oncologists. J Clin Oncol 2014; 32 (suppl): 8097. [Google Scholar]
- 8.Aggarwal C, Rolfo CD, Oxnard GR, Gray JE, Sholl LM, Gandara DR. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol 2021; 18: 56–62. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014; 346: 256–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuben A, Zhang J, Chiou S-H, et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun 2020; 11: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–62. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Mayer AT, Li R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. Semin Cancer Biol 2020; 84: 310–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin 2019; 69: 127–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol 2022; 19: 132–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019; 30: 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu W, Jiang L, Shi Y, et al. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer 2021; 9: e002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khorrami M, Prasanna P, Gupta A, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol Res 2020; 8: 108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018; 19: 1180–91. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Li C, Gensheimer M, et al. Radiological tumor classification across imaging modality and histology. Nat Mach Intell 2021; 3: 787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mezquita L, Auclin E, Ferrara R, et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol 2018; 4: 351–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mu W, Jiang L, Zhang J, et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun 2020; 11: 5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian P, He B, Mu W, et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics 2021; 11: 2098–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou SK, Greenspan H, Davatzikos C, et al. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc IEEE 2021; 109: 820–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016; 22: 4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negrao MV, Skoulidis F, Montesion M, et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J Immunother Cancer 2021; 9: e002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019; 30: 1321–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med 2021; 27: 2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ligero M, Garcia-Ruiz A, Viaplana C, et al. A CT-based radiomics signature is associated with response to immune checkpoint inhibitors in advanced solid tumors. Radiology 2021; 299: 109–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


