Abstract
Ethylene glycol oligomers have been studied systematically as non-nucleotide loop replacements in short hairpin oligoribonucleotides. Structural optimization concerns the length of the linkers and is based on the thermodynamic stabilities of the corresponding duplexes. The optimum linker is derived from heptakis (ethylene glycol) provided that the duplex end to be bridged comprises solely the terminal base pair; the optimum linker is derived from hexakis(ethylene glycol) if a dangling unpaired nucleotide is incorporated into the loop. Moreover, these linkers have been compared to other commonly used linker types which consist of repeating units of tris- or tetrakis(ethylene glycol) phosphate, or of 3-hydroxypropane-1-phosphate. In all cases, the correlation between linker length and duplex stability is independent of the kind of counter ions used (Na+, Na+/Mg2+, K+ or Li+). Furthermore, all duplexes with non-nucleotide loop replacements are less stable than those with the corresponding standard nucleotide loop. The results corroborate that the linkers are solvent-exposed and do not specifically interfere with the terminal nucleotides at the bridged duplex end.
INTRODUCTION
In the last decade, tethered oligonucleotides have found broad attention mainly for three reasons. First, they represent synthetically altered nucleic acids and are therefore potential targets in therapeutics and diagnostics. Non-natural modifications promise higher resistance in biological media, improved ability for cellular uptake and improved hybridization proporties (1). Second, the architecture of large, structurally defined nucleic acids can be reduced to the functionally important domains under the challenge of retaining their biological activity (2,3). The shortened synthesis is a welcomed circumstance. Third, functionalization by linking small synthetic molecules to oligonucleotides offers specific manipulation of nucleic acid properties (4–7).
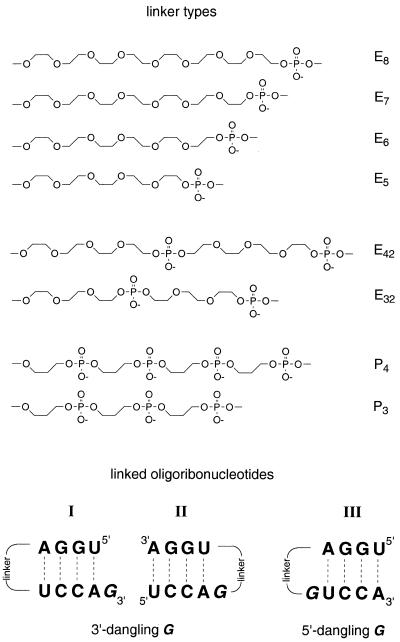
Our interest in tethered oligonucleotides, in particular for non-nucleotide loop replacements in short hairpin oligoribonucleotides, arises from the recent development of a codon–anticodon pairing model (8,9). Therefore, we cyclize RNA duplexes consisting of three base pairs via flexible linkers on both helix ends. As we intend to study stacking interactions within the double helix it is required that the linkers do not interfere with the base pairs themselves. Recently introduced linker types that contain aromatic carbocycles stack on the terminal base pair and are therefore not suitable (10,11). However, ethylene glycol based (Ex) or ethylene glycol derived (Exy, Px) linkers should meet these requirements as they have the advantage of good water solubility and high conformational flexibility (Fig. 1) (2,3,12–14). Despite the numerous reported applications for linkers of this kind, a detailed structural optimization in terms of linker length and stability of the corresponding duplexes is documented only in a DNA series (15). Here we report the structural optimization of various flexible non-nucleotide linkers as loop replacements in short double helical RNAs under the aspects of duplex stability, linker length and dangling unpaired nucleotides.
Figure 1.
Constitution of linkers and base sequences of corresponding hairpin oligoribonucleotides.
MATERIALS AND METHODS
RNA synthesis and purification
5′-O-DMT-2′-O-TOM-protected nucleoside phosphoramidites were obtained from Xeragon AG (Switzerland). The syntheses of linker phosphoramidites were based on procedures described elsewhere (12,15).
All oligoribonucleotides were synthesized on CPG supports on a Pharmacia Gene Assembler Plus following slightly modified DNA standard methods; detritylation: dichloroacetic acid/1,2-dichloroethane (4/96, 2 min); coupling: phosphoramidites/acetonitrile (0.1 M × 120 µl, 2.5 min) were activated by benzyl thiotetrazole/acetonitrile (0.35 M × 360 µl); capping: A: Ac2O/sym-collidine/THF (10/10/80), B: N-methylimidazole/THF (16/84); oxidation, I2/H2O/pyridine/THF (3/2/20/75, 45 s). Amidite solutions, tetrazole solutions and acetonitrile were dried over activated molecular sieves overnight. All sequences were synthesized trityl-off.
Deprotection and cleavage of the oligonucleotides from solid support were achieved with MeNH2 in EtOH (8 M, 0.75 ml) and MeNH2 in water (40%, 0.75 ml) for 3–6 h at 33°C; the solution was then evaporated to dryness. Removal of the 2′-O-silyl acetyls was afforded by treatment with tetrabutylammonium fluoride trihydrate (TBAF.3H2O) in THF (1 M, 0.90 ml) for at least 12 h at room temperature. The reaction was quenched by the addition of Tris–HCl (1 M, pH 7.4, 0.95 ml). The volume of the solution was reduced to 1 ml and directly applied on a Sephadex G 10 column (30 × 1.5 cm) controlled by UV-detection at 260 nm. The product was eluted with water and evaporated to dryness.
All oligoribonucleotides were purified by ion-exchange chromatography on a MONO Q HR 5/5 column. Flow rate: 1 ml/min; eluant A: 10 mM Na2HPO4 in H2O (pH 10.5); eluant B: 10 mM Na2HPO4/1 M NaCl in H2O (pH 10.5); detection at 260 nm; gradient I (for checking the purity of crude products after deprotection): 30 min 0–100% B in A; gradient II (for purification): Δ30% B in A within 30 min. Fractions containing the purified oligonucleotide were desalted by loading onto a C18 SepPak cartridge (Waters/Millipore), followed by elution with 0.1–0.2 M (Et3NH)HCO3, water, and then H2O/CH3CN (6/4). Combined fractions containing the oligonucleotide were lyophilized to dryness.
The masses expected have been confirmed for all oligoribonucleotides by MALDI-TOF mass spectrometry.
Thermal denaturation studies
Absorbance versus temperature profiles were recorded at 265 nm on a Cary-100 spectrophotometer equipped with a multiple cell holder and a Peltier temperature control device. Sample preparation: oligonucleotides were lyophilized to dryness, dissolved in the corresponding buffer from stock solutions, and subsequently degassed by treatment with ultrasonics. A layer of silicon oil was placed on the surface of the solution. Concentrations were 4 µM. Data were collected after a complete cooling and heating cycle at a rate of 0.5°C/min. Melting transitions were reversible for all sequences. Hyperchromicities of melting profiles were in a range of 16–19% for all tethered hairpin sequences and of 11–14% for the reference sequences with nucleotidic loops. Concentration independence of the melting temperature was demonstrated for the hairpin sequences rUGGA-E7-UCCAG and rUCCAG-E6-UGGA.
Values of ΔH° and ΔS° were derived from a two-state van’t Hoff analysis by fitting the shape of the individual absorbance versus temperature curve according to Breslauer (16) and Turner (17,18).
Circular dichroism
CD spectra were recorded at 20°C on a Jasco-710 spectropolarimeter equipped with a Peltier temperature control device; samples were prepared as described for UV samples.
RESULTS AND DISCUSSION
The requirement for minimum interference of the synthetic loop replacements with the nucleobases implies that the most reasonable bridging spans from one terminal ribose phosphate to the phosphate group of the opposite strand. That offers a simple synthetic strategy: commonly available diols are derivatized to the corresponding α-tritylated ω-phosphoramidite linkers which are then incorporated into oligoribonucleotides following standard procedures (19).
Sequence design
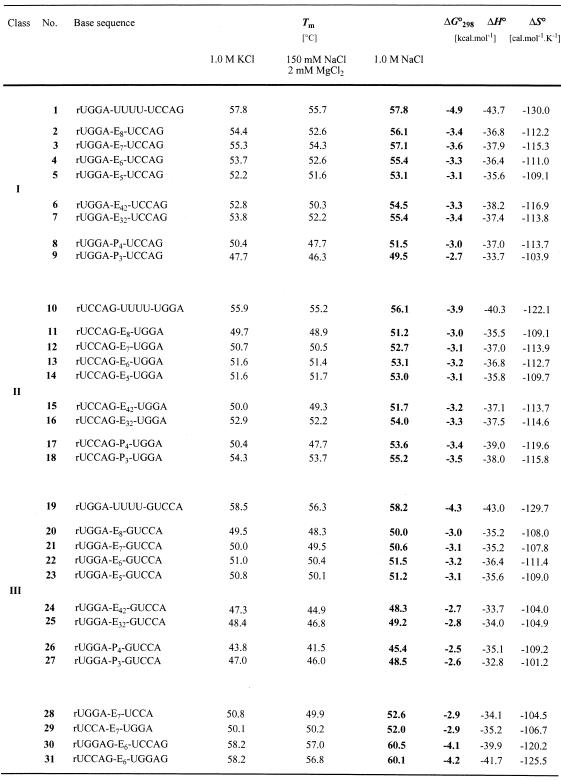
All 31 hairpin oligoribonucleotides listed in Table 1 possess the same base sequence as core duplex, namely 5′-rUGGA pairing with 5′-rUCCA. The sequences are subdivided into three different classes (I, II and III; Fig. 1, Table 1). Two of them, class I (2–9) and class II (11–18) oligoribonucleotides possess a dangling guanosine attached to the 3′-end of rUCCA. However, the distinction is made by the position of their loops. On the one hand, such a design enables the optimization of linkers for a simple blunt-end duplex (I) as well as for a duplex end with an unpaired nucleotide incorporated into the loop (II). On the other hand, as these oligoribonucleotides are of the same base sequence, differences in their thermal stabilities are to be attributed to specific linker-based properties and not to sequence-dependent ones. In oligoribonucleotides of class III (20–27) an unpaired guanosine is also incorporated into the loop; however, this dangling nucleotide is attached to the 5′-end of rUCCA. The oligoribonucleotides 1 (I), 10 (II) and 19 (III) are reference sequences with a tetrauridine nucleotide loop; the oligoribonucleotides 28–31 are reference sequences in order to understand stability differences of the same core duplex arising from a combination of influences that are dangling bases, different linker positions and solvatization effects of double helix ends.
Table 1. Thermal melting temperatures and thermodynamic data of hairpin oligoribonucleotides with non-nucleotide loop replacements and corresponding reference sequences measured in aqueous HPO42– buffer solutions with different kinds of cations at pH 7.0.
Linker constitution, linker length and effects on double helix stability
Linker constitution. Three types of flexible synthetic linkers are investigated in the present study. They are derived from oligo(ethylene glycols) (Ex), oligo(ethylene glycol)-phosphates (Exy) and 3-hydroxypropane-1-phosphates (Px). Their structural formulas are depicted in Figure 1 in the order of decreasing hydrophobicity. Linkers derived from 3-hydroxypropane-1-phosphates directly mimic the charged P-O3′-C3′-C4′-C5′-O5′-P nucleotide backbone. They are more demanding with regard to stereoelectronic and conformational aspects than ethylene glycol-based linkers.
Linker length. For A-RNA double-helical structures, a distance analysis shows that the phosphor–phosphor bridging distance is 18 Å between a nucleotide and its pairing partner (20). This distance is ~1–2 Å more for the same nucleotide to a nucleotide attached at the 5′ end of its pairing partner but ~1–2 Å less to a corresponding nucleotide at its 3′-end. In the DNA series, the length of the optimum ethylene glycol-based linker (E7) in extended conformation (27 Å) exceeds the straight-line distance to be spanned (17 Å) (15). This can be reasoned as the starting vectors of bridging and the preference for a helical conformation of oligo(ethylene glycols) in solution (21) has to be taken into account. Furthermore, as there is evidence that linkers of single tris- or tetrakis(ethylene glycol) units distort the geometry of the terminal base pair, linkers of these lengths are not included in the present RNA study (22).
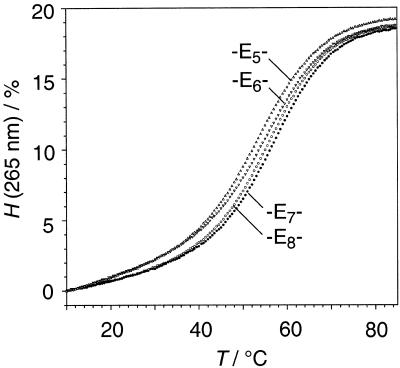
Effects on RNA double helix stability. With respect to the three oligoribonucleotide classes I, II and III, the thermal and thermodynamic data listed in Table 1 show small but distinct differences caused by structure and length of the linkers (see also Fig. 2). Concerning the family of oligo(ethylene glycol) tethered oligoribonucleotides, linker length was increased stepwise by single ethylene glycol units from pentakis- to octakis(ethylene glycol) (E5–E8). The optimum linker is derived from heptakis(ethylene glycol) provided that the duplex end to be bridged comprises solely the terminal base pair (3); the optimum linker is derived from hexakis(ethylene glycol) if a dangling unpaired nucleotide is incorporated into the loop (13, 22).
Figure 2.
Absorbance versus temperature profiles of rUGGA-Ex-UCCAG, (x = 5–8), 2–5; 4 µM, 1 M NaCl, 10 mM Na2HPO4, pH 7.0; H = hyperchromicity.
For class I and class III oligoribonucleotides, linkers based on tris- and tetrakis(ethylene glycol)-phosphate or 3-hydroxypropane-1-phosphate are inferior to their heptakis- and hexakis(ethylene glycol) counterparts, respectively (Tm: 3 > 6–9; Tm: 22 > 24–27). However, if an unpaired nucleotide is incorporated into the loop at the 3′-end (II), these linker types lead to slightly higher stabilities of the corresponding hairpin structures (Tm: 13 < 16–18). The reason for this reversed behavior is not yet clear, however, a detailed NMR analysis of a hexakis(ethylene glycol) linked class II hairpin is currently in progress in the laboratory of B. Jaun (ETH Zürich) and should lead to deeper insight.
Comparison to nucleotide loops. All duplexes in the present study with non-nucleotide loop replacements are less stable than their corresponding reference sequence comprising a nucleotide loop. The difference in thermal stability between the optimum heptakis(ethylene glycol) linked sequence and the one containing a tetrauridine loop is <1°C (Tm: 1 > 3), however, it is reflected clearly in a ΔΔG°298 value of 1.3 kcal/mol. The differences are of the same order for the replacement of a linker with tetrauridine in class II and class III oligoribonucleotide hairpins (Tm: 10 > 13; 19 > 22). Thereby, formation of an additional U–G base pair within the pentanucleotidic loop structure is likely (10; 19).
Effects of cations
In order to investigate whether the correlation between double helix stability and structure of the linkers is influenced by the kind of cations in aqueous buffer solutions, we performed Tm determinations in three different buffer systems (10 mM HPO42–, pH 7.0): 1.0 M NaCl; 0.15 M NaCl, 2 mM MgCl2; and 1.0 M KCl (Table 1). Moreover, a 1.0 M LiCl buffer (10 mM HPO42–, pH 7.0) has been tested additionally for the sequences numbered 2–5. The data verify that in all buffer systems the same dependence of duplex stability on linker length and structure is observed. Therefore, no specific chelating of cations within the synthetic loop replacements is ascertainable (Table 1).
Linker position, dangling bases and solvatization of helix ends
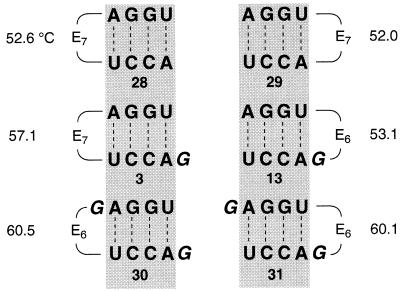
Figure 3 depicts a selection of hairpin oligoribonucleotides containing the optimized oligo(ethylene glycol) linkers. All six sequences are of the same core double helix rUGGA/rUCCA, however, the position of loop replacements and the number of dangling unpaired bases is varied. The well-known feature that dangling bases at the 3′-end enhance the stability of oligoribonucleotide duplexes (17) is correctly reflected in the melting temperatures Tm of the six sequences (Tm: 28 < 3 < 30; 29 < 13 < 31), even if the unpaired guanidine becomes part of the loop (Fig. 3). Interestingly, sequence 3 melts 4°C higher than sequence 13, both sequences showing identical CD spectra. We do not mainly attribute this distinction to the different linker positions (Tm: 28 ≈ 29; 30 ≈ 31) or to the different linker lengths (Tm: 3 ≈ 4; 12 ≈ 13), we rather render solvatization at the unbridged helix ends responsible. Solvatization of a helix end that comprises a dangling unpaired base is energetically more favorable than solvatization of a blunt-end terminal base pair. In the latter case the hydrophobic π-electron system is fully exposed to the aqueous solution, whereas an unpaired dangling base shields this plane and provides a better transition to the hydrophilic environment by formation of hydrogen bonds between the dangling base and water molecules (23,24). These considerations are further supported by the observation that the differences in stability between class I and class II oligo(ethylene glycol) linked hairpins are more pronounced if their core duplexes consist of a low number of base pairs compared to the ones consisting of a higher number.
Figure 3.
Selection of hairpin oligoribonucleotides (3, 13, 28–31) containing the optimized oligo(ethylene glycol) linkers: influence of different linker positions, 3′-dangling bases and solvatization effects on double helix stablity.
CONCLUSIONS
In the present study oligo(ethylene glycol) linkers have been systematically investigated as loop-replacements in short hairpin oligoribonucleotides. According to the criterion of highest duplex stability the optimum linker is the one derived from heptakis(ethylene glycol), provided that the duplex end to be bridged comprises solely the terminal base pair; it is derived from hexakis(ethylene glycol) if a dangling unpaired nucleotide is incorporated into the loop. In general, ethylene glycol-based linkers are superior to other commonly used flexible linkers. Only if a dangling unpaired nucleotide at the 3′-end is part of the loop, linkers based on tris(ethylene glycol)-phosphate or 3-hydroxypropane-1-phosphate result in hairpins of slightly higher stability. Importantly, the correlation between linker constitution and stability of the corresponding oligoribonucleotide is independent of the kind of counter ions present in the aqueous buffer.
We consider the results to assist in the selection of a proper linker system for the various applications encountered in both fundamental RNA research and pharmaceutical development (2,3,8,12,14,25).
Acknowledgments
ACKNOWLEDGEMENTS
R.M. thanks Prof. Eschenmoser (Zürich) and Prof. Falk (Linz) for generous hospitality in their laboratories. We thank Dr Krishnamurthy (Scripps, CA) for mass analyses and Prof. Grubmayr (Linz) for critical comments on the manuscript. R.M. acknowledges the Austrian Academy of Science for an APART fellowship (Austrian Programme for Advanced Research and Technology). This work was supported by the Austrian Science Fund (P13216-CHE), Vienna.
REFERENCES
- 1.Seitz O. (1999) Angew. Chem. Int. Ed. Engl., 111, 3466–3469. [DOI] [PubMed] [Google Scholar]
- 2.Benseler F., Fu,D.-j., Ludwig,J. and McLaughlin,L.W. (1993) J. Am. Chem. Soc., 115, 8483–8484. [Google Scholar]
- 3.Thomson J.B., Tuschl,T. and Eckstein,F. (1993) Nucleic Acids Res., 21, 5600–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X. and Pitsch,S. (1998) Nucleic Acids Res., 26, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempcy R.O., Kutyavin,I.V., Mills,A.G., Lukhtanov,E.A. and Meyer R.B. (1999) Nucleic Acids Res., 27, 2931–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiederholt K. and McLaughlin,L.W. (1999) Nucleic Acids Res., 27, 2487–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajur S.B., Robles,J., Wiederholt,K., Kuimelis,R.G. and McLaughlin,L.W. (1997) J. Org. Chem., 62, 523–529. [DOI] [PubMed] [Google Scholar]
- 8.Micura R., Pils,W. and Grubmayr,K. (2000) Angew. Chem. Int. Ed. Engl., 112, 922–925. [DOI] [PubMed] [Google Scholar]
- 9.Micura R. (1999) Chem. Eur. J., 5, 2077–2082. [Google Scholar]
- 10.Lewis F.D., Liu,X., Wu,Y., Miller,S.E., Wasielewski, Letsinger,R.L., Sanishvili,R., Joachimiak,A., Tereshko,V. and Egli,M. (1999) J. Am. Chem. Soc., 121, 9905–9906. [Google Scholar]
- 11.Bevers S., O’Dea,T.P. and McLaughlin,L.W. (1998) J. Am. Chem. Soc., 120, 11004–11005. [Google Scholar]
- 12.Ma M.Y.-X., Reid,L.S., Climie,S.C., Lin,W.C., Kuperman,R., Sumner-Smith,M. and Barnett,R.W. (1993) Biochemistry, 32, 1751–1758. [DOI] [PubMed] [Google Scholar]
- 13.Durand M., Chevrie,K., Chassignol,M., Thuong,N.T. and Maurizot,J.C. (1990) Nucleic Acids Res., 18, 6353–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moses A.C. and Schepartz,A. (1997) J. Am. Chem. Soc., 119, 11591–11597. [Google Scholar]
- 15.Rumney S. and Kool,E.T. (1995) J. Am. Chem. Soc., 117, 5635–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marky L. and Breslauer,K. (1987) Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- 17.Xia T., Mathews,D.H. and Turner,D.H. (1999) In Söll,D., Nishimura,S. and Moore,P. (eds), Comprehensive Natural Product Chemistry, Vol. 8. Elsevier, Oxford, UK, S. 21–47.
- 18.Petersheim M. and Turner,D.H. (1983) Biochemistry, 22, 256–263. [DOI] [PubMed] [Google Scholar]
- 19.Beaucage S. and Iyer,R. (1992) Tetrahedron, 48, 2223–2311. [Google Scholar]
- 20.Haasnoot C.A.G., Hilbers,C.W., van der Marel,G.A., van Boom,J.H., Singh,U.C., Pattabiraman,N. and Kollman,P.A. (1986) J. Biomol. Struct. Dyn., 3, 843–857. [DOI] [PubMed] [Google Scholar]
- 21.Abe A. and Mark,J.E. (1976) J. Am. Chem. Soc., 98, 6468–6476. [Google Scholar]
- 22.Gao H., Chidambaram,N., Chen,B.C., Pelham,D.E., Patel,R., Yang,M., Zhou,L., Cook,A. and Cohen,J.S. (1994) Bioconjug. Chem., 5, 445–453. [DOI] [PubMed] [Google Scholar]
- 23.Williams D.H. (1991) Aldrichimica Acta, 24, 71–80. [Google Scholar]
- 24.Auffinger P. and Westhof,E. (1998) J. Biomol. Struct. Dyn., 16, 693–707. [DOI] [PubMed] [Google Scholar]
- 25.Ma M.Y.-X., McCallum,K., Climie,S.C., Kuperman,R., Lin,W.C., Sumner-Smith,M. and Barnett,R.W. (1993) Nucleic Acids Res., 21, 2585–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]