Abstract
The complement system is an important part of innate immunity. Through complement-dependent cytotoxicity (CDC), it plays an important role in the clearance of invading pathogens but also cancerous host cells. Therapy with anti-CD20 monoclonal antibodies (mAbs), for example, rituximab and ofatumumab, is a well-established treatment for lymphoid malignancies, and CDC is one of the main mechanisms underlying their anti-cancer activity. However, there are still some issues with the clinical application of anti-CD20 antibodies. On the one hand, anti-CD20 can cause some clinical side effects; on the other hand, anti-CD20 has low potency in some patients, and increasing the dosage does not enhance its effectiveness in these patients. Previous studies have reported that a gain-of-function in a certain complement component can boost the cytolytic activity of anti-CD20 mAbs. Through reviewing the literature on complement system control and anti-CD20 mAbs, this article aims to provide a thorough understanding of the potential of targeting complement components in lymphoma therapy.
Keywords: Innate immunity, complement system, anti-CD20, anti-cancer, monoclonal antibody therapy
Introduction
Lymphoma is a group of cancers that start in the lymph system. Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL) are two main types of lymphoma, yet there also exist additional1,2 subclasses of lymphoma based on lymphoid lineage cells that expand and undergo malignant transformation.3,4 Lymphoma can occur in all age groups, yet HL is more common among people aged 15–39 years and 75 years or older than other age groups while the rates of NHL grow higher as people get old. 4 In 2020, HL and NHL accounted for 23,376 and 259,793 deaths globally, respectively.5,6 Though HL is relatively rare, it is the most common type of cancer among teens aged 15–19 years. 6
Thanks to the development of antibody therapy, the survival rate of lymphoma has increased. Monoclonal antibodies (mAbs) have been developed into a mature strategy for cancer treatment for the past 20 years; however, there is still much space for improvement, in particular anti-CD20 mAbs, which have been reported to have various detrimental symptoms and/or low response in some patients.7–10 Manipulating complement cascade can serve as an effective strategy to increase the anti-cancer efficacy of anti-CD20 mAbs.1,2,11–14
In innate immunity, the complement system is a key component. The activation of the complement cascade is strictly restricted by different regulators in order to protect host tissues from being attacked, though, it can be detected on the surface of cancer cells and the fluids of cancer patients.15,16 Previous studies have found that blocking negative regulators of the complement system or introducing gain-of-function complement protein mutants can enhance the lymphocytolysis efficacy of anti-CD20 mAbs.
The activation and regulation of complement system
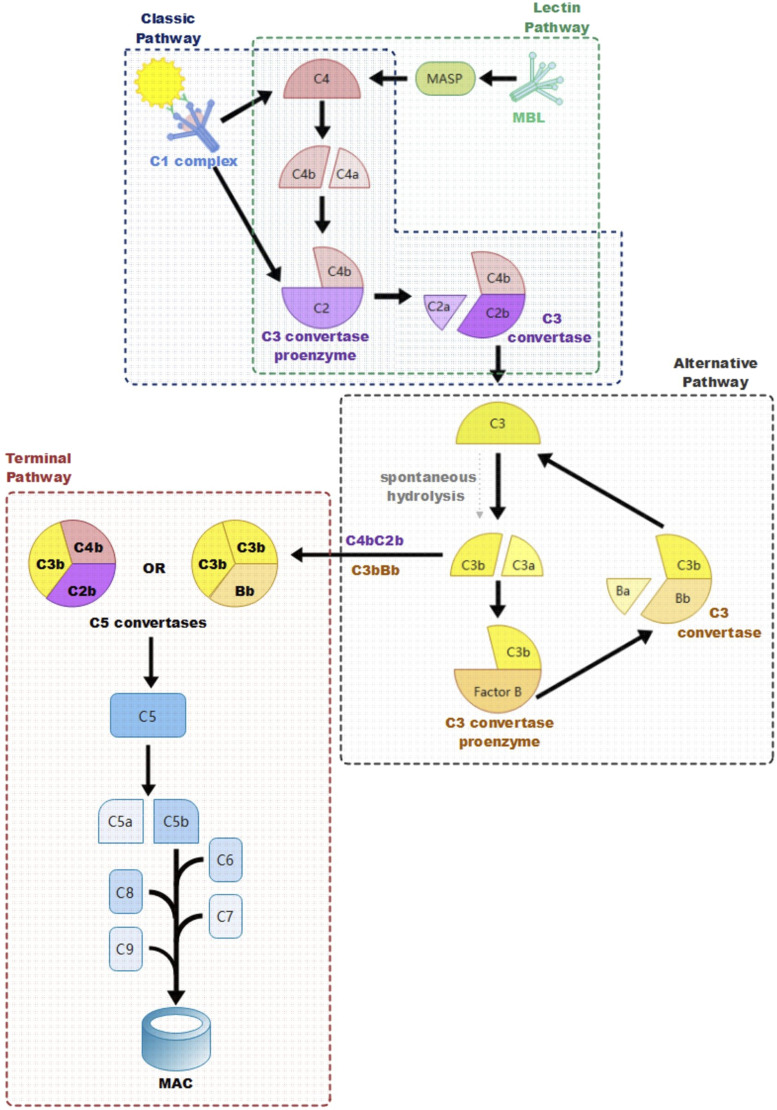
The complement system is ubiquitous in innate immune defense; However, it is also involved in acquired immunity, for example, when recruited by antigen-antibody complexes. Studies from recent years have shown components of the complement cascade are also involved in adaptive immune responses. For instance, intracellular protease-activated complement cascade component C3 inside CD4+ T cells participates in the induction of Th1 and Th17 responses; And regulators of complement activation CD46 and CD55 have been proven to affect T cell activation.17–19 There are three pathways to initiate the complement cascade: the classical pathway (CP), lectin pathway (LP), and alternative pathway (AP). All those pathways converge in the assembly of C3 convertase and C5 convertase and end by forming the same terminal product named membrane attack complex (MAC).17,20,21
CP is activated by antigen-antibody immune complexes binding to C1. 22 The C1 complex (C1qC1r2C1s2) is a pentamer composed of a characteristic bouquet-shaped module C1q and a heterotetramer C1r2C1s2.22–24 C1q serves as a scaffold for the heterotetramer, its collagen-like stem can bind with the fragment crystallizable (Fc) of antibodies IgG and IgM after their binding with cognate antigens.23,24 As a result of the interaction between C1q and Fc, serine proteases on the C1 complex, C1r and C1s, are activated. 22 Next, complement C4 is cleaved into a small inactive fragment (C4a) and a large active fragment (C4b) by C1s. Then, complement C2 binds to C4b, forming the C3 convertase proenzyme. 22 Afterward, C2 is cleaved into C2a and C2b by C1s, the larger fragment containing the catalytic site, C2b, stays bound with C4b, forming the activated C3 convertase C4bC2b, whilst the small inactive fragment C2a is released.17,22,24,25Figure 1
Figure 1.
Activation of Three Complement Pathways. The three complement activation pathways: classic pathway, lectin pathway, and alternative pathway converge on cleaving complement component C3 into the principal effector molecule C3 and forming C5 convertase.
LP is similar to CP in how it also activates serine proteases and leads to cleavage of C4 and C2; The main difference between these pathways is that LP is initiated by pattern recognition molecules: collectins, ficolins, and mannose-binding lectins (MBL).22,26–29 These pattern recognition molecules initiate LP by recognizing and binding to sugar residues on microbial surfaces; As a result, MBL serine proteases (MASP-1, MASP-2, and MASP-3) are activated, leading to cleavage of C4 and C2 and assembly and activation of C3 convertase as in CP.17,21 C3 convertase acts on complement protein C3 which is the pivotal point of all three complement initiation pathways; The activated C3 convertase cleaves C3 into a smaller fragment C3a and a larger fragment C3b.17,30 The released C3a part acts as an anaphylatoxin, decreasing in concentration the further away from infection; The larger C3b deposits on the microbial surface covalently and acts as an opsonin and aids in phagocytosis.31,32 In the complement cascade, C3b is the principal effector molecule, after C3 is cleaved by C3 convertase into C3a and C3b, C3b further participates in the generation of C5 convertase and the assembly of MAC Figure 1. 30
In AP, C3b is produced by the spontaneous hydrolysis of complement C3 or downstream reaction of the complement cascade, leading to a positive feedback amplification loop.21,33 C3b recruits complement protein factor B, which is the homologue of C2 in AP and binds to it, forming the C3 convertase proenzyme which is called C3bB. 21 Then complement factor D cleaves factor B into smaller leaving fragment Ba and catalytic fragment Bb, causing the transformation of the proenzyme into its active form: C3bBb. 21 Irrespective of the initially activated pathways, AP serves as an amplification loop to generate and increase C3b and C3 convertase Figure 1.17,34
All three pathways converge on forming C5 convertases; Once C3b is formed, it binds with C3 convertase and generates C5 convertases (C4bC2bC3b in CP and LP or C3bBbC3b in AP).17,21 C5 convertases cleave complement C5, releasing a C5a fragment that acts as an anaphylatoxin, creating a chemical gradient decreasing in concentration the further away from the infection site and producing C5b which initiates the Terminal Lytic Pathway, which is identical in all three pathways.17,21 C5b first binds to complement component C6, forming a stable complex C5b6 which further binds to C7; the newly generated complex C5b67 anchors to the membrane surface. Afterward, complement protein C8 binds to it, forming C5b678 and leading to the first membrane penetration; subsequently, multiple repeats of complement C9 are arranged on complex C5b678, enlarging the transmembrane channel and leading to the formation of the terminal complement component, MAC.17,20–22,26 As a consequence of the complement cascade, a pore on the cell membrane is caused by MAC, resulting in cell lysis through osmotic flux.34–36
To protect host tissues from attack from the complement system, the activation of the complement cascade is strictly controlled by soluble and membrane-bound complement regulators and inhibitory proteins. Except for properdin, the only known positive regulator in the regulation of the complement cascade, all complement regulators and inhibitory proteins down-regulate the activity of the complement cascade.21,37–39 There are four ways for complement regulators and inhibitory proteins to down-regulate the complement system: inhibition of complement cascade activators; inhibition of MAC formation, proteolytic inactivation of active complement components (C3b and/or C4b); and decay-accelerating C3/C5 convertase. C1 esterase inhibitor (C1INH) is a soluble complement regulating factor which is an important inhibitor in CP and LP and was also found to have down-regulating activity on AP. 40 C1INH prevents the initiation of the complement cascade in CP by binding to C1s and C1r and thus inhibiting the activation of C1q and inactivating the C1 complex; in LP by binding MASP-1 and MASP-2 and inhibiting activation of MBL.41–43 Unlike in CP and LP, C1INH down-regulation in AP might lie in its possible decay-accelerating activity, as it can compete with factor B to reversibly bind with C3. 40 CD59, vitronectin, and clusterin regulate complement cascade by preventing the formation and/or the cytolytic activity of MAC. CD59, also known as homologous restriction factor 20 (HRF20) and protectin, can prohibit the formation of MAC through inhibition of C9 binding to C5b678.44–46 Vitronectin, which is also called S protein and epibolin, binds at the membrane binding site of C5b67 and blocks its membrane insertion activity; in addition, vitronectin can also inhibit the polymerization of C9 on C5b678.47,48 Clusterin, alias apoprotein J, can competitively form stable complexes with C5b67, C5b678, and MAC to prevent their cytolytic activity.49,50 Proteolytic inactivation of C3b and/or C4b is dependent on factor I (CFI) in both membrane-bound and fluid phases; nonetheless, CFI alone cannot accomplish this cleavage, and it must be aided with cofactors as factor H (CFH), C4b-binding protein (C4BP), CD35 (complement receptor type 1, CR1), and CD46 (membrane cofactor protein, MCP).37,38,51 In the presence of CFI and appropriate cofactors, C3b and C4b are degraded into inactive fragments iC3b and iC4b. iC3b then participates in other immune activities, including down-regulating inflammasome activation, enhancing B cell-mediated response, and targeting pathogens for phagocytosis. 52 However, no immunological function has been found in iC4b so far. 53 CD55, namely, decay-accelerating factor (DAF), down-modulates complement cascades by promoting the disassembly of key enzymes C3 convertase and C5 convertase in all three pathways. 54 Notably, CFI cofactors CFH, C4BP, and CD35 also have similar decay-accelerating activity in complement regulation; as CFH and C4BP dissociate C3/C5 convertases C3bBb/C3bBbC3b and C4bC2a/C4bC2aC3b in AP and CP/LP, respectively, while C4BP degrades C3/C5 convertases in all three pathways.37,38 In contrast to decay-accelerating activity, properdin up-regulates AP of complement cascade by stabilizing C3 and C5 convertases. 39
Complement system in cancer treatment
The activation of the complement system is limited to foreign tissues, yet its components can be detected on the surface of cancerous host cells and in the biological fluids of cancer patients.15,16 An elevated level of complement activation has been reported in patients diagnosed with solid cancer such as breast, ovarian, lung, digestive tract, brain cancers, and blood cell tumors.55–64 Complement activation can have both anti and pro-cancer effects. It triggers the clearance of cancerous cells by CDC, whereas its products (like C3a and C5a) are involved in inflammation as well as in angiogenesis in cancer progression.65,66 Despite the controversial role of some complement components in cancer progression, CDC itself is detrimental to cancerous cells. In order to escape from CDC, cancer cells overexpress negative complement regulators and inhibitory proteins including both membrane (CD55, CD59, CD46) and soluble (CFH, CFI, C4BP) ones.55,56,65 The over-expression of negative complement regulators and inhibitory proteins, together with other immune escape strategies of cancer cells, leads to a lack of specificity and potency for the response of an innate immune system, including the complement system, to cancer cells.67,68Figure 2
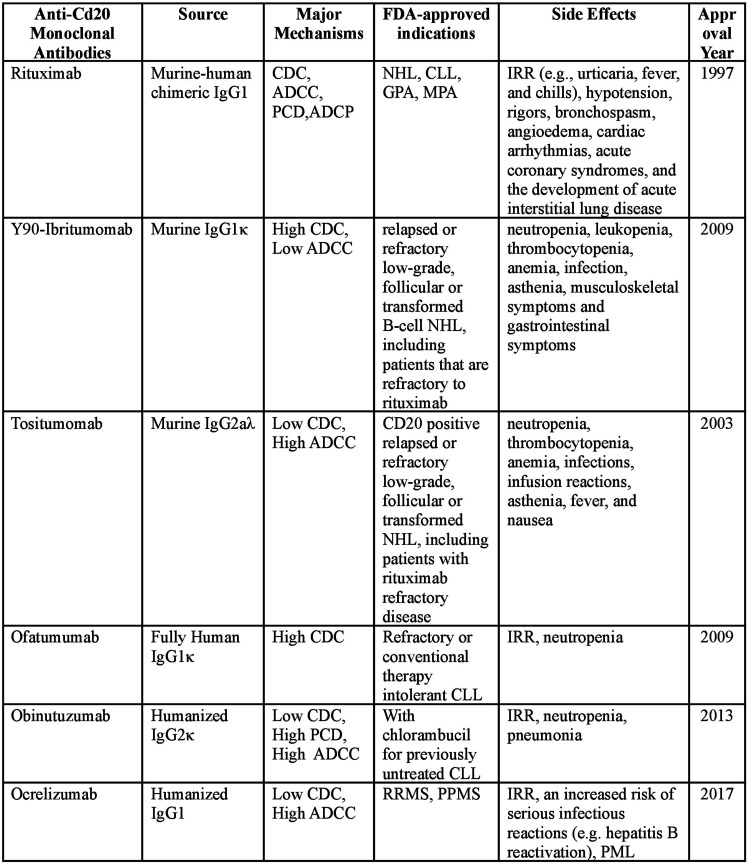
Figure 2.
FDA-approved anti-CD20 mAbs in clinical treatment.69–78 Abbreviation: CDC, complement-dependent lysis; ADCC, antibody-dependent cell-mediated cytotoxicity; PCD, programmed cell death; ADCP, Antibody-dependent cellular phagocytosis; NHL, non-Hodgkin’s lymphoma; CLL, chronic lymphocytic leukemia; RA, Rheumatoid arthritis; GPA, granulomatosis with polyangiitis; MPA, microscopic polyangiitis; IRR, Infusion-related reactions; RRMS, relapsing-remitting multiple sclerosis; PPMS, pemphigus multiple sclerosis; PML, progressive multifocal leukoencephalopathy.
Therapeutic mAbs are widely applied in cancer treatment by redirecting the immune system to attack and/or suppress cancer cells, and the complement system plays a critical role in this process, in particular, in the elimination of lymphoma B cells by anti-CD20 mAbs. 68 CD20 is a non-glycosylated, four-transmembrane phosphoprotein expressed during B cell differentiation from the pro-B cell to the plasma cell. As a general marker of B cells, CD20 has been the best-studied biomarker and mAb drug target in lymphoma treatment in the past two decades. Indeed, CD-20 targeting mAb rituximab, a murine-human chimeric mAb developed for NHL treatment, is the first FDA (U.S. Food and Drug Administration) approved therapeutic mAb. 79 Second generation anti-CD20 mAbs, including ofatumumab, ocrelizumab, and veltuzumab are Fc-engineered to be more humanized compared to rituximab, especially ofatumumab, which is an FDA-approved fully human anti-CD20 mAbs.7–9 Obinutuzumab and ocaratuzumab are third generation anti-CD20 mAbs that are fully humanized and engineered. 69 However, patients might suffer various detrimental symptoms in anti-CD20 mAbs treatment. As anti-CD20 mAbs cause B-cell deletion and some degree of immunodeficiency, infectious complications are not uncommon in patients; infusion-related reactions (IRR) (e.g., urticaria, fever, and chills) are also common adverse events in anti-CD20 therapy and some high-grade adverse events can even be fatal.7–9 Monoclonal antibodies binding to CD20 antigens can elicit cytolytic responses by multiple parallel effector pathways, including CDC, antibody-dependent cell-mediated cytotoxicity (ADCC), and direct induction of apoptosis. 80 CDC and ADCC interact in a complementary manner and ADCC is synchronously promoted once CDC is activated. 81 Previous in vitro studies have suggested that CDC and ADCC together enhance mAb-dependent NK cell-mediated cytotoxicity. 81 This review would mainly discuss the CDC potency of anti-CD20 mAbs.
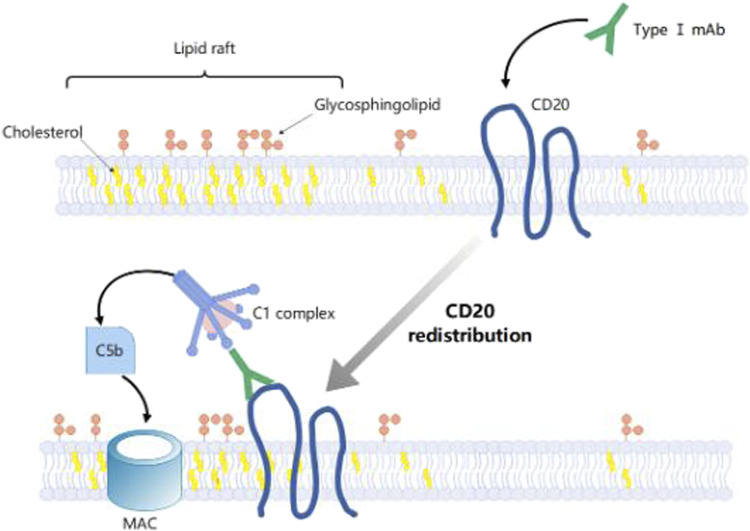
According to their different effector mechanisms, anti-CD20 mAbs can be classified into two types. Type I mAbs, including rituximab, ocrelizumab, ofatumumab, veltuzumab, and ocaratuzumab, are potent in inducing CDC, but less effective in activating cell death; By contrast, type II mAbs like obinutuzumab is an effective activator of apoptotic or non-apoptotic cell death with no significant effect in complement activation. 80 Both type I and type II mAbs can recruit complement, though, type I mAb has a semi-saturation value 10 to 1000 times lower than type II mAbs when recruiting complement to a similar level, and some type II mAbs hardly activate any CDC. 82 Due to its hydrophobic properties, CD20 has a rather low affinity to lipid rafts, causing difficulties for MAC deposition around mAb:CD20 binding area on the cell surface. Lipid rafts are cholesterols and glycosphingolipids enriched microdomains on the cell surface facilitating bioactivities of the bilayer.80,83,84 Cholesterol enrichment on the cell surface provides a favorable environment for MAC deposition, further elevating CDC.80,83,84 Previous research on human erythrocytes has shown that in cholesterol-rich cells with antisera and complement, a 30%–50% hemolysis could be observed, whereas cholesterol-depleted cells under the same incubation conditions remained nearly intact. 85 Only type I mAbs can redistribute CD20 molecules to lipid rafts on the cell surface, and thus type I anti-CD20 mAbs rather than type II are effective in activating CDC Figure 3.80,83 Besides, the different binding properties of type I and type II mAbs to CD20 can result in their different cytotoxic effects. The study of Diebolder et al. revealed that complement is activated by IgG hexamers assembled at the surface of the target cell. 86 In the hexameric IgG-C1 binding model, the hexameric platform in the center was arranged by six Fc fragments, one Fab arm of each antibody was positioned at the height of the platform while the other Fab arm was placed vertically to the platform and bound the membrane-associated antigen, and the collagen-like stem of C1q bound to Fc fragments at the hexameric platform. 86 Kumar et al. found that when the Fab fragments of type I mAbs rituximab and ofatumumab bound to CD 20 with the Fab arms oriented in opposite directions, which was similar to the conformation of Fab fragments in the hexameric IgG-C1 binding model; and in contrast to type I, Fab fragments of type II mAb obinutuzumab bound to CD 20 nearly vertically to the membrane plane. 87 On the other hand, type I mAbs bind twice as many type II mAbs to a certain B cell type. Type II forms 1:2 (mAb: CD20) “terminal” complexes that preclude binding of additional mAbs; While type I forms 1:2 or 2:1 (mAb: CD20) “seeding” complexes that enable subsequent concatenation of mAb or CD20 molecules, leading to an increased local concentration of Fc fragments and an enhancement of their oligomerization for C1q recruitment. 87
Figure 3.
Type I anti-CD20 mAbs binding redistributes CD20 to Raft Shaft Domain on Cell Surface.
The off-rate of antibody may also account for the different CDC potency of anti-CD20 mAbs, however, there are contradictory results about the contribution of anti-CD20 mAbs’ off-rate to their CDC potency in previous studies. 88 Li et al. found that in rituximab and its modulated mutants, CDC potency was independent of the off-rate. 89 Goldenberg et al. also suggested that the off-rate difference between mAbs is not related to an enhanced CDC, as though they observed a significantly slower off-rate in veltuzumab compared to rituximab in all three cell lines used in their studies (Daudi, Raji, and Ramos), only in Daudi cells veltuzumab showed an enhanced CDC. 90 By contrast, Teeling et al. reported that using a non-complement activating F (ab’)2 anti-human κ reagent to mitigate rituximab’s off-rate can enhance its CDC potency; 91 However, in their follow-on research, they generated a new anti-CD20 mAb 2C6 (IgG1) and found that 2C6 which had a faster off-rate than rituximab showed a significantly higher potency than rituximab in activating CDC. 92
Manipulations of complement cascade as anti-cancer strategies
Even though rituximab functions successfully in treating most lymphoma cases, its effectiveness can be limited due to inter-individual variability, resulting in rituximab resistance in a subset of patients. 10 Early phase II trials have shown that the response rate of rituximab for follicular or low-grade lymphomas is only about 50% and that for intermediate- to high-grade lymphomas is even lower.10,93 A dose-escalation trial on CLL patients has demonstrated that a higher rituximab dose could lead to a higher response rate. 94 On the other hand, a higher level of rituximab can also cause worse adverse reactions in patients with little difference in their survival rate. 95 Some previous studies have suggested that the dosage of rituximab should be tailored precisely to patients’ conditions, or much simpler, increased overall. 79 However, in order to increase therapeutic efficacy, and preserve safety in the clinic, it is more rational to reduce rituximab dosage and increase anti-cancer efficacy via combination therapies. For example, in CLL treatment, apoptosis-inducing agents, such as fludarabine, cyclophosphamide, and bendamustine combined with rituximab have been the most commonly used chemoimmunotherapy regimens for patients. 11 Venetoclax, a mimetic of the anti-apoptotic B-cell lymphoma-2 (Bcl-2) protein that results in programmed cell death of CLL cells, is also used in combination with rituximab. 12 However, all these compounds targeting the apoptosis pathway can also cause modest to severe side effects, especially for CLL patients who are elderly and/or have comorbidities. 13
CDC pathway can be a promising target for combination therapy development. Previous studies have found that manipulating the complement cascade can enhance the CDC of type I anti-CD20 mAbs in lymphoma cells in vitro. Some recent research has reported that by introducing gain-of-function complement protein mutants, an enhancement effect on lymphocytolysis mediated by type I anti-CD20 mAbs can be observed in vitro. Felberg A. et al. have found that under a restricted complement concentration, adding quadruple hyperactive factor B mutants D279 G, F286 L, K323 E, Y363 A, or selected single mutants could lead to a significantly increased cytolysis in ofatumumab-resistant lymphoma cells (Namalwa and SU-DHL-8 cells), as well as complete lysis of moderately sensitive lymphoma cells (Raji and WSU-NHL cells). 1 The increased rituximab-mediated CDC might result from hyperactive convertases formed by those mutants, which can be observed to have a more intense cytolysis effect at the same time and a slower decay compared with wild-type convertases in the hemolytic assay. 1 Notably, in 2020, Urban A. et al. further reported a complement C2 gain-of-function mutant, S250 C, in an atypical hemolytic uremic syndrome (aHUS) patient. They found that this natural mutant, S250 C, together with the positive control in their study, a C2 mutant designed in silico, Y347 A (equivalent to Y363 A mutation in factor B), has a significantly higher cytolysis activity in lymphoma Raji and Ramos cell lines compared with wildtype C2 and other mutants found in aHUS patients 2. In addition, S250 C and Y347 A are also examined to be able to form hyperactive and more stable C3/C5 convertases in the presence of ofatumumab. 2 Their latest research published in 2022 suggested that optimized C2 variants had a universal property of enhancing anti-CD20 mAbs combination therapy, not limited to type I mAbs. 96 For type I mAbs, they reported that supplementation of patients’ sera with gain-of-function C2 mutants can reduce the effective rituximab dose; Besides, gain-of-function C2 mutants can also enhance CDC activated by type I anti-CD20 mAbs toward resistant CLL cells. 96 As for type II mAbs, they observed a significantly increased cytocidal effect in Raji cells that were sensitized with type II anti-CD20 mAb obinutuzumab and supplemented with gain-of-function C2 variants; However, the mechanism behind the enhanced CDC still needs more experimental evidence to discover. 96
Manipulation of complement cascade by down-regulating negative complement regulators and inhibitory proteins can also serve as a rational strategy for increasing the anti-cancer activity of anti-CD20 mAbs. Ge et al. also demonstrated that in rituximab-resistant Raji 32 and LY8 cells, herbal products curcumin and perillyl alcohol can suppress the expression of only inducible CD59 but not CD20 and consequently sensitize those rituximab-resistant B lymphoma cells to CDC effect. 97 Besides, in a series of in vitro studies using blood samples from prolymphocytic leukemia (PLL) and B-cell chronic leukemia (B-CLL) patients, Golay J. and his colleagues also found that in the presence of antibodies blocking negative complement regulators and inhibitory proteins, the lymphocytolysis efficacy of rituximab could be significantly improved. 98 They reported a lymphocytolysis at 2- to 3-fold increase with adding a single blocking antibody and up to 10-fold increase with adding both anti-CD55 and anti-CD59 in samples from patients with low rituximab-mediated CDC (less than 10%); However, in high responders group (more than 50% rituximab-mediated CDC), lymphocytolysis was complete in the presence of single blocking antibodies. 98 Golay J et al. also reported that the expression of CD55 and CD59 shows high individual variability; this was observed as an up to 100-fold (CD55) or 10-fold (CD59) difference in the mean fluorescence intensity respectively. However, it was also demonstrated that there is no correlation between lymphocytolysis and the expression level of CD55 and CD59, and that the complement cascade could be efficiently blocked even at low levels of CD55 and/or CD59. 98 Hence, we might expect that the stability of C3/C5 convertases, rather than the expression level of their inhibitors, plays a more important role in alleviating decay-accelerating and further enhancing the efficacy of type I anti- CD20 mAbs. However, studies on the role of inhibitors and the gain-of-function components of complement convertases in CDC mediated by type I anti-CD20 mAbs are quite limited, and the hypothesis that improving the stability of complement convertases could enhance the efficacy of type I anti-CD20 mAbs still needs more experimental evidence to back up. Besides, as we mentioned before, the products of C3/C5 convertases: C3a and C5a, play a controversial role in cancer progression. However, as these anaphylatoxins participate in the promotion of angiogenesis and the recruitment of myeloid-derived suppressor cells, this is more the case in solid tumors such as ovarian cancer, their role in lymphoma and immunotherapy may have limited importance.31,99 The development of a rational clinical strategy for anti-CD20 mAbs application based on complement cascade manipulation, especially in blocking the decay-accelerating of complement convertases, still has a long way to go through.
Discussion
This review discussed the role of the complement system in anti-CD20 therapy for lymphoma. The complement system is an important part of innate immune defense against invading pathogens as well as cancerous host cells. To protect host tissues from CDC, the complement cascade is restricted by complement regulators and inhibitory proteins. Overexpressing negative complement regulators and inhibitory proteins to block CDC is an important immune escape strategy for cancerous cells. Anti-CD20 mAbs are well-established therapies for B cell lymphomas that can induce cytolytic responses including CDC, ADCC, and direct induction of apoptosis. Manipulation of the complement cascade is a promising strategy to improve therapeutic efficacy and preserve the safety of anti-CD20 mAs. Anti-CD20 mAbs can be classified into two types by their different effector mechanisms. Type I mAbs like rituximab and ofatumumab are potent CDC activators but are less effective in activating cell death; In contrast to type I, type II mAbs like obinutuzumab are potent apoptotic or non-apoptotic cell death activators with no significant effect in CDC activation.
Previous in vitro studies suggested that CDC can be enhanced in the presence of optimized complement proteins. Felberg A. et al. found that gain-of-function factor B mutants could lead to increase cytolysis in ofatumumab-resistant lymphoma cells as well as complete lysis of moderately sensitive lymphoma cells. 1 Urban A. et al. also reported that in the presence of gain-of-function C2 mutants, the cytolysis activity in lymphoma Raji and Ramos cell lines significantly increased compared with wildtype and other C2 mutants 2 ; their follow research further revealed that gain-of-function C2 mutants could not only enhance CDC activated by type I mAbs but also increase the cytocidal effect of by type II mAbs in lymphoma cell lines. 96 Another strategy to manipulate the complement cascade is the down-regulation of negative complement regulators and inhibitory proteins. The study of Ge et al. indicated that herbal products curcumin and perillyl alcohol could activate rituximab-mediated CDC in rituximab-resistant B lymphoma cell lines by the inhibition of CD59 expression. 97 In blood samples from lymphoma patients with less than 10% rituximab-mediated CDC, Golay J et al. observed a 2- to 3-fold increase of lymphocytolysis by adding a single anti-CD55 or anti-CD59 blocking antibody and an up to 10-fold increase by adding both antibodies. 98
There are still limitations in this review. On the one hand, as this review focused on the complement system in anti-CD20 mAbs therapies, there is a lack of in-depth expansion on the effects of anti-CD20 mAbs on ADCP and apoptosis. On the other hand, though models of anti-CD20 mAbs binding to C1q and CD20 were discussed in this review, a systematic sorting-out of the research results about the protein structural study of complement components has not been included. Besides, due to the lack of publication about the in vivo experimental evidence of optimized complement proteins or inhibitors of negative complement regulators and inhibitory proteins in the use of boosting the efficacy of anti-CD20 monoclonal antibodies, this review can only provide a theoretical suggestion of the clinical prospect of manipulating the complement system in lymphoma treatment. In a word, this review is based on the author’s own analysis and summary of publications about the complement system and anti-CD20 mAbs. Although the analysis process is as objective as possible, it still has subjectivity and limitation to some extent.
Conclusion
Anti-CD20 mAbs therapy is a mutual strategy for lymphoma treatment, and the activation of CDC is an important mechanism of anti-CD20 mAbs eliminating cancerous cells. Previous in vitro studies have suggested that manipulation of the complement cascade can boost the cytolysis activity of anti-CD20 mAbs in lymphoma cell lines. There are two manipulation strategies that are reported to be able to result in an increased mAbs-mediated CDC: adding gain-of-function complement C2/factor B mutants and inhibiting negative complement regulators and inhibitory proteins. However, the in vivo evidence for the potency of complement system manipulation in treating lymphoma is still lacking. Therefore, the development of a practical clinical strategy for anti-CD20 mAbs application based on complement cascade manipulation still has a long way to go through.
Footnotes
Authors’ contributions: Yiwen Gao contributed to the conception of the study, performed the literature analysis and review and wrote the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD
Yiwen Gao https://orcid.org/0000-0002-2942-6676
References
- 1.Felberg A, Urban A, Borowska A, et al. (2019) Mutations resulting in the formation of hyperactive complement convertases support cytocidal effect of anti-cd20 immunotherapeutics. Cancer Immunology, Immunotherapy 68: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urban A, Volokhina E, Felberg A, et al. (2020) Gain-of-function mutation in complement C2 protein identified in a patient with aHUS. The Journal of Allergy and Clinical Immunology 146: 916–919. [DOI] [PubMed] [Google Scholar]
- 3.Neelapu S, Adkins S, Brody J, et al. (2020) Society for immunotherapy of cancer (sitc) clinical practice guideline on immunotherapy for the treatment of lymphoma. Journal for ImmunoTherapy of Cancer 8: e001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Lymphoma. https://www.cdc.gov/cancer/lymphoma/index.htm. (Accessed 10 November 2022).
- 5.Diseases G, Collaborators I. (2020) Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of disease study 2019. Lancet 396: 1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Pang W, Lok V, et al. (2022) Incidence, mortality, risk factors, and trends for hodgkin lymphoma: a global data analysis. Journal of Hematology and Oncology 15: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cragg M, Walshe C, Ivanov A, et al. (2005) The Biology of CD20 and its Potential as a Target for mAb Therapy. Tnf Pathophysiology: Molecular and Cellular Mechanisms 8: 140–174. [DOI] [PubMed] [Google Scholar]
- 8.Marshall M, Stopforth R, Cragg M. (2017) Therapeutic antibodies: what have we learnt from targeting cd20 and where are we going? Frontiers in Immunology 8: 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du F, Mills E, Mao-Draayer Y. (2017) Next-generation anti-CD20 monoclonal antibodies in autoimmune disease treatment. Auto- Immunity Highlights 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman C, Sehn L. (2018) A tale of two antibodies: obinutuzumab versus rituximab. British Journal of Haematology 182: 29–45. [DOI] [PubMed] [Google Scholar]
- 11.Schwa¨nen C, Hecker T, Hu¨binger G, et al. (2002) In vitro evaluation of bendamustine induced apoptosis in b-chronic lymphocytic leukemia. Leukemia 16: 2096–2105. [DOI] [PubMed] [Google Scholar]
- 12.Roberts A, Huang D. (2017) Targeting BCL2 with BH3 mimetics: basic science and clinical application of venetoclax in chronic lymphocytic leukemia and related B cell malignancies. Clinical Pharmacology and Therapeutics 101: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallek M, Shanafelt T, Eichhorst B. (2018) Chronic lymphocytic leukaemia. Lancet 391: 1524–1537. 10.1016/S0140-6736(18)30422-7 [DOI] [PubMed] [Google Scholar]
- 14.Golay J, Zaffaroni L, Vaccari T, et al. (2000) Biologic Response of B Lymphoma Cells to Anti-CD20 Monoclonal Antibody Rituximab in Vitro: CD55 and CD59 Regulate Complement- Mediated Cell Lysis. Blood 15: 3900–3980. [PubMed] [Google Scholar]
- 15.Woo S, Corrales L, Gajewski T. (2015) Innate immune recognition of cancer. Annual Review of Immunology 33: 445–474. [DOI] [PubMed] [Google Scholar]
- 16.Fishelson Z, Kirschfink M. (2019) Complement C5b-9 and cancer: mechanisms of cell damage, cancer counteractions, and approaches for intervention. Frontiers in Immunology 10: 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy K, Weaver C. (2017) Janeway’s Immunobiology. Ninth edition. New York City: Garland Science. [Google Scholar]
- 18.Liszewski M, Elvington M, Kulkarni H, et al. (2017) Complement’s hidden arsenal: new insights and novel functions inside the cell. Molecular Immunology 84: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killick J, Morisse G, Sieger D, et al. (2018) Complement as a regulator of adaptive immunity. Seminars in Immunopathology 40: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peitsch M, Tschopp J. (1991) Assembly of macromolecular pores by immune defense systems. Current Opinion in Cell Biology 3: 710–716. [DOI] [PubMed] [Google Scholar]
- 21.Walport M. (2001) Complement. First of two parts. The New England Journal of Medicine 344: 1058–1066. [DOI] [PubMed] [Google Scholar]
- 22.Wallis R, Mitchell D, Schmid R, et al. (2010) Paths reunited: initiation of the classical and lectin pathways of complement activation. Immunobiology 215: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortensen S, Sander B, Jensen J, et al. (2017) Structure and activation of C1, the complex initiating the classical pathway of the complement cascade. Proceedings of The National Academy of Sciences of The United States of America 114: 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, Kishore U. (2017) C1 Complex: An adaptable proteolytic module for complement and non-complement functions. Frontiers in immunology 8: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortensen S, Jensen J, Andersen G. (2016) Solution structures of complement C2 and Its C4 complexes propose pathway- specific mechanisms for control and activation of the complement proconvertases. Journal of Biological Chemistry 291: 16494–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CJ P. (1992) Regulation of complement by membrane proteins: an overview. Current Topics In Microbiology and Immunology 178: 1–6. [DOI] [PubMed] [Google Scholar]
- 27.Jacquet M, Cioci G, Jacquet G, et al. (2018) C1q and mannose- binding lectin interact with CR1 in the same region on CCP24-25 modules. Frontiers in immunology 9: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira V, Cortes C. (2022) The complement system. Encyclopedia of Infection and Immunity 1: 144–169. DOI: 10.1016/B978-0-12-818731-9.00056-2 [DOI] [Google Scholar]
- 29.Zewde N, Hsu RV, Morikis D, Palermo G. (2021) Systems biology modeling of the complement system under immune susceptible pathogens. Frontiers in Physics 9: 9. DOI: 10.3389/fphy.2021.603704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schramm E, Roumenina L, Rybkine T, et al. (2015) Mapping interactions between complement C3 and regulators using mutations in atypical hemolytic uremic syndrome. Blood 125: 2359–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ajona D, Ortiz-Espinosa S, Pio R. (2019) Complement anaphylatoxins C3a and C5a: emerging roles in cancer progression and treatment. Seminars in Cell and Developmental Biology 85: 153–163. [DOI] [PubMed] [Google Scholar]
- 32.Juul-Madsen H, Viertlboeck B, Hartle S, et al. (2014) Innate immune responses. In Editors: Schat K, Kaspers B and Kaiser P (eds.), Avian Immunology. Academic Press: Cambridge, Massachusetts, USA. [Google Scholar]
- 33.Parker C. (1992) Regulation of complement by membrane proteins: an overview. Current Topics In Microbiology and Immunology 178: 1–6. [DOI] [PubMed] [Google Scholar]
- 34.Merle N, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. (2015) Complement system part II: role in immunity. Frontiers in Immunology 6: 257. DOI: 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayly-Jones C, Bubeck D, Dunstone M. (2017) The mystery behind membrane insertion: a review of the complement membrane attack complex. Philosophical Transactions of The Royal Society Of London Series B: Biological Sciences 372: 20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnen A, Henneke P. (2014) Structural biology of the membrane attack complex. Subcellular Biochemistry 80: 83–116. DOI: 10.1007/978-94-017-8881-6-6 [DOI] [PubMed] [Google Scholar]
- 37.Noris M, Remuzzi G. (2013) Overview of complement activation and regulation. Seminars in Nephrology 33: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipfel P, Skerka C. (2009) Complement regulators and inhibitory proteins. Nature Reviews Immunology 9: 729–740. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Cortes C, Ferreira V. (2018) Properdin: a multifaceted molecule involved in inflammation and diseases.Molecular Immunology 102: 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, Wagner E, Zhang H, Frank MM. (2001) Complement 1 inhibitor is a regulator of the alternative complement pathway. The Journal of Experimental Medicine 194: 1606–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis A, Lu F, Mejia P. (2010) C1 inhibitor, a multi-functional serine protease inhibitor. Thrombosis and Haemostasis 104: 886–893. [DOI] [PubMed] [Google Scholar]
- 42.Gigli I, Ruddy S, Austen K. (1968) The stoichiometric measurement of the serum inhibition of the first component of complement by the inhibition of immune hemolysis. Journal of Immunology 100: 1154–1164. [PubMed] [Google Scholar]
- 43.Harpel P, Cooper N. (1975) Studies on human plasma C1 inactivator-enzyme interactions. I. Mechanisms of interaction with c1s, plasmin, and trypsin. Journal of Clinical Investigation 55: 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiso T, Mizuno M, Nasu J, et al. (2002) Enhanced expression of decay-accelerating factor and CD59/homologous restric- tion factor 20 in intestinal metaplasia, gastric adenomas and intestinal-type gastric carcinomas but not in diffuse-type carcinomas. Histopathology 40: 339–347. [DOI] [PubMed] [Google Scholar]
- 45.Okada N, Harada R, Fujita T, et al. (1989) A novel membrane glycoprotein capable of inhibiting membrane attack by homologous complement. International Immunology 1: 205–208. [DOI] [PubMed] [Google Scholar]
- 46.Sugita Y, Masuho Y. (1995) CD59: its role in complement regulation and potential for therapeutic use. Immunotechnology 1: 157–168. [DOI] [PubMed] [Google Scholar]
- 47.Milis L, Morris C, Sheehan M, et al. (1993) Vitronectin-mediated inhibition of complement: evidence for different binding sites for C5b-7 and C9. Clinical and Experimental Immunology 92: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schvartz I, Seger D, Shaltiel S. (1999) Vitronectin. International Journal of Biochemistry and Cell Biology 31: 539-544. [DOI] [PubMed] [Google Scholar]
- 49.Tschopp J, Chonn A, Hertig S, et al. (1993) Clusterin, the human apolipoprotein and complement inhibitor, binds to complement C7, C8 beta, and the b domain of C9. Journal of Immunology 151: 2159–2165. [PubMed] [Google Scholar]
- 50.Schwarz M, Spath L, Lux C, et al. (2008) Potential protective role of apoprotein j (clusterin) in atherogenesis: binding to enzymatically modified low-density lipoprotein reduces fatty acid-mediated cytotoxicity. Thrombosis and Haemostasis 100: 110–118. [DOI] [PubMed] [Google Scholar]
- 51.Masaki T, Matsumoto M, Nakanishi I, et al. (1992) Factor I- dependent inactivation of human complement C4b of the classical pathway by C3b/C4b receptor (CR1, CD35) and membrane cofactor protein (MCP, CD46). Journal of Biochemistry 111: 573–578. [DOI] [PubMed] [Google Scholar]
- 52.Alcorlo M, Mart´ınez-Barricarte R, Ferna´ndez F, et al. (2011) Unique structure of iC3b resolved at a resolution of 24 A by 3D- electron microscopy. Proceedings of The National Academy of Sciences of The United States of America 108: 13236–13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hair P, Wagner S, Friederich P, et al. (2012) Complement regulator C4BP binds to staphylococcus aureus and decreases opsonization. Molecular Immunology 50: 253–261. [DOI] [PubMed] [Google Scholar]
- 54.Spendlove I, Ramage J, Bradley R, et al. (2006) Complement decay accelerating factor (DAF)/CD55 in cancer. Cancer Immunology Immunotherapy 55: 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fishelson Z, Donin N, Zell S, et al. (2003) Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Molecular Immunology 40: 109–123. [DOI] [PubMed] [Google Scholar]
- 56.Okroj M, Hsu YF, Ajona D, et al. (2008) Non-small cell lung cancer cells produce a functional set of complement factor I and its soluble cofactors. Molecular Immunology 45:169–179. [DOI] [PubMed] [Google Scholar]
- 57.Niculescu F, Rus H, Retegan M, et al. (1992) Persistent complement activation on tumor cells in breast cancer. American Journal of Pathology 140: 1039–1043. [PMC free article] [PubMed] [Google Scholar]
- 58.Bjørge L, Hakulinen J, Vintermyr O, et al. (2005) Ascitic complement system in ovarian cancer. British Journal of Cancer 92: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ajona D, Pajares M, Corrales L, et al. (2013) Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. Journal of The National Cancer Institute 105: 1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corrales L, Ajona D, Rafail S, et al. (2012) Anaphylatoxin C5acreates a favorable microenvironment for lung cancer progression. Journal of Immunology 189: 4674–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gminski J, Mykala-Ciesla J, Machalski M, et al. (1992) Immunoglobulins and complement components levels in patients with lung cancer. Romanian Journal of Internal Medicine 30: 39–44. [PubMed] [Google Scholar]
- 62.Ytting H, Christensen I, Thiel S, et al. (2005) Serum mannan-binding lectin-associated serine protease 2 levels in colorectal cancer: relation to recurrence and mortality. Clinical Cancer Research 11: 1441–1446. [DOI] [PubMed] [Google Scholar]
- 63.Maness P, Orengo A. (1977) Serum complement levels in patients with digestive tract carcinomas and other neoplastic diseases.Oncology 34: 87–89. [DOI] [PubMed] [Google Scholar]
- 64.Matsutani M, Suzuki T, Hori T, et al. (1984) Cellular immunity and complement levels in hosts with brain tumours. Neurosurgical Review 7: 29–35. [DOI] [PubMed] [Google Scholar]
- 65.Afshar-Kharghan V. (2017) The role of the complement system in cancer. Journal of Clinical Investigation 127: 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeon H, Han S, Lee S, et al. (2018) Activation of the complement system in an osteosarcoma cell line promotes angiogenesis through enhanced production of growth factors. Scientific Reports 8: 5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beatty G, Gladney W. (2015) Immune escape mechanisms as a guide for cancer immunotherapy. Clinical Cancer Research 21: 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melis J, Strumane K, Ruuls S, et al. (2015) Complement in therapy and disease: regulating the complement system with antibody-based therapeutics. Molecular Immunology 67: 117–130. [DOI] [PubMed] [Google Scholar]
- 69.Luo C, Wu G, Huang X, et al. (2021) Efficacy and safety of new anti-CD20 monoclonal antibodies versus rituximab for induction therapy of CD20+ B-cell non-Hodgkin lymphomas: a systematic review and meta-analysis. Scientific Reports 11: 3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD. (2018) Complement in cancer: untangling an intricate relationship. Nature Reviews Immunology 18(1): 5–18. DOI: 10.1038/nri.2017.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maloney DG. (2012) Anti-CD20 antibody therapy for B-cell lymphomas. New England Journal of Medicine 366(21): 2008–2016. DOI: 10.1056/NEJMct1114348 [DOI] [PubMed] [Google Scholar]
- 72.Plosker GL, Figgitt DP. (2003) Rituximab. Drugs 63(8): 803–843. DOI: 10.2165/00003495-200363080-00005 [DOI] [PubMed] [Google Scholar]
- 73.Hadjinicolaou AV, Nisar MK, Parfrey H, Chilvers ER, Ostor AJK. (2012) Non-infectious pulmonary toxicity of rituximab: a systematic review. Rheumatology 51(4): 653–662. DOI: 10.1093/rheumatology/ker290 [DOI] [PubMed] [Google Scholar]
- 74.Harigai M, Tanaka Y, Maisawa S, et al. (2012) Safety and efficacy of various dosages of ocrelizumab in Japanese patients with rheumatoid arthritis with an inadequate response to methotrexate therapy: A placebo-controlled double-blind parallel-group study. The Journal of Rheumatology 39(3): 486–495. DOI: 10.3899/jrheum.110994 [DOI] [PubMed] [Google Scholar]
- 75.Marcus R, Davies A, Ando K, Klapper W, Opat S, Owen C, et al. (2017) Obinutuzumab for the first-line treatment of follicular lymphoma. New England Journal of Medicine 377(14): 1331–1344. DOI: 10.1056/NEJMoa1614598 [DOI] [PubMed] [Google Scholar]
- 76.Boross P, Leusen JHW. (2012) Mechanisms of action of CD20 antibodies. American Journal of Cancer Research 2(6): 676–690. [PMC free article] [PubMed] [Google Scholar]
- 77.Iodine-131 Tositumomab (2003) Iodine-131 Tositumomab. BioDrugs 17(4): 290–295. DOI: 10.2165/00063030-200317040-00009 [DOI] [PubMed] [Google Scholar]
- 78.McKinney M, Beaven A. (2014) Yttrium-90 ibritumomab tiuxetan in the treatment of non-hodgkin lymphoma. Blood Lymphat Cancer 2014: 45–59. [Google Scholar]
- 79.Pierpont T, Limper C, Richards K. (2018) Past, present, and future of rituximab-the world’s first oncology monoclonal antibody therapy. Frontiers in Oncology 8: 163 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winiarska M, Glodkowska-Mrowka E, Bil J, et al. (2011) Molecular mechanisms of the antitumor effects of anti-CD20 antibodies. Frontiers in Bioscience (Landmark Edition) 16: 277–306. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Huang K, Liu L, et al. (2019) Effects of complement and serum IgG on rituximab-dependent natural killer cell-mediated cytotoxicity against Raji cells. Oncology Letters 17: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herter S, Frank H, Mundigl O, et al. (2013) Preclinical activity of the type II CD20 antibody GA101 (Obinutuzumab) compared with rituximab and ofatumumab in vitro and in xenograft models. Molecular Cancer Therapeutics 12: 2031–2042. [DOI] [PubMed] [Google Scholar]
- 83.Semac I, Palomba C, Kulangara K, et al. (2003) Anti-CD20 therapeutic antibody rituximab modiies the functional organization of rafts/microdomains of b lymphoma cells. Cancer Research 63: 534–540. [PubMed] [Google Scholar]
- 84.Lingwood D, Simons K. (2010) Lipid rafts as a membrane- organizing principle.Science 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 85.Cohen A, Shinitzky M. (1982) Modulation of complement lysis of human erythrocytes by the membrane lipid viscosity. Vox Sanguinis 43: 23–27. [DOI] [PubMed] [Google Scholar]
- 86.Diebolder CA, Beurskens FJ, de Jong RN, et al. (2014) Complement is activated by IgG hexamers assembled at the cell surface. Science 343: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar A, Planchais C, Fronzes R, et al. (2020) Binding mechanisms of therapeutic antibodies to human CD20. Science 369: 793–799. [DOI] [PubMed] [Google Scholar]
- 88.Meyer S, Evers M, Jansen JHM, et al. (2018) New insights in Type I and II CD20 antibody mechanisms-of-action with a panel of novel CD20 antibodies. British Journal of Haematology 180: 808–820. [DOI] [PubMed] [Google Scholar]
- 89.Li B, Zhao L, Guo H, et al. (2009) Characterization of a rituximab variant with potent antitumor activity against rituximab-resistant b-cell lymphoma. Blood 114: 5007–5015. [DOI] [PubMed] [Google Scholar]
- 90.Goldenberg DM, Rossi EA, Stein R, Cardillo TM, Czuczman MS, Hernandez-Ilizaliturri FZ, et al. (2009) Properties and structure-function relationships of veltuzumab (hA20), a humanized anti-CD20 monoclonal antibody. Blood 113(21):1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teeling JL, Mackus WJ, Wiegman LJ, et al. (2004) Characterization of new human CD20 monoclonal antibodies with potent cytolyticactivity against non-hodgkin lymphomas. Blood 104: 1793–1800. [DOI] [PubMed] [Google Scholar]
- 92.Teeling JL, Mackus WJ, Wiegman LJ, et al. (2006) The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. Journal of Immunology 177: 362–371. [DOI] [PubMed] [Google Scholar]
- 93.Rezvani A, Maloney D. (2011) Rituximab resistance. Best Practice and Research Clinical Haematology 24: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Brien S, Kantarjian H, Thomas D, et al. (2001) Rituximab dose- escalation trial in chronic lymphocytic leukemia. Journal of Clinical Oncology 19: 2165–2170. [DOI] [PubMed] [Google Scholar]
- 95.Pfreundschuh M, Murawski N, Zeynalova S, et al. (2017) Optimiza- tion of rituximab for the treatment of DLBCL: increasing the dose for elderly male patients. British Journal of Haematology 179: 410–420. [DOI] [PubMed] [Google Scholar]
- 96.Urban A, Majeranowski A, Stasiłojć G, et al. (2022) In silico designed gain-of-function variants of complement C2 support cytocidal activity of anticancer monoclonal antibodies. Cancers (Basel) 14: 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ge X, Du Y, Chen J, et al. (2021) Herbal NF-κB inhibitors sensitize rituximab-resistant b lymphoma cells to complement-mediated cytolysis. Frontiers in Oncology 11: 751904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Golay J, Lazzari M, Facchinetti V, et al. (2001) CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood 98: 3383–3389. [DOI] [PubMed] [Google Scholar]
- 99.Markiewski MM, DeAngelis RA, Benencia F, et al. (2008) Modulation of the antitumor immune response by complement. Nature Immunology 9: 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]